SUMMARY
Post-acute sequelae of COVID-19 (PASC, “Long COVID”) pose a significant global health challenge. The pathophysiology is unknown, and no effective treatments have been found to date. Several hypotheses have been formulated to explain the etiology of PASC, including viral persistence, chronic inflammation, hypercoagulability, and autonomic dysfunction. Here, we propose a mechanism that links all four hypotheses in a single pathway and provides actionable insights for therapeutic interventions. We find that PASC are associated with serotonin reduction. Viral infection and type I interferon-driven inflammation reduce serotonin through three mechanisms: diminished intestinal absorption of the serotonin precursor tryptophan; platelet hyperactivation and thrombocytopenia, which impacts serotonin storage; and enhanced MAO-mediated serotonin turnover. Peripheral serotonin reduction, in turn, impedes the activity of the vagus nerve and thereby impairs hippocampal responses and memory. These findings provide a possible explanation for neurocognitive symptoms associated with viral persistence in Long COVID, which may extend to other post-viral syndromes.
In brief
Post-viral syndromes are associated with serotonin reduction, which may contribute to the neurological and cognitive symptoms seen in individuals with Long COVID.
Graphical Abstract
INTRODUCTION
Post-viral syndromes arise in a subset of individuals and can persist for months to years after disease onset.1 The accompanying symptoms are diverse and often include fatigue, post-exertional malaise, memory loss, and other neurocognitive impairments.2 A major post-viral syndrome is “Long COVID,” manifesting as post-acute sequelae of COVID-19 (PASC), which are experienced by a subset of individuals after SARS-CoV-2 infection.3 The molecular etiology of most post-viral syndromes, including Long COVID, remains unclear. Several hypotheses have been proposed to explain the persistence of symptoms, including the presence of a viral reservoir that is not cleared after the initial infection,4 chronic inflammation, auto-antibody development, and tissue damage as a result of non-resolving anti-viral responses.1 Another common feature that has been associated with post-viral syndromes is platelet dysfunction and hypercoagulability.5 Finally, Long COVID and other post-viral syndromes have been linked to autonomic nervous system dysfunction.6 A deeper understanding of whether these mechanisms occur in different subsets of patients or jointly drive disease persistence is urgently needed.
In this study, we perform a metabolomics investigation and find that serotonin levels are a possible discriminator between recovered individuals and Long COVID patients. Using a combination of human cohort studies, animal models of viral infection, and organoid cultures, we determine that the presence of viral RNA and downstream interferon responses cause a decrease in serotonin. Several mechanisms account for this phenomenon, including diminished uptake of the serotonin precursor tryptophan in the gastrointestinal tract, reduced storage in platelets due to thrombocytopenia, and enhanced turnover by serotonin-metabolizing enzymes. One important consequence of peripheral serotonin deficiency is reduced activity of the vagus nerve, which in turn is associated with hippocampal dysfunction and memory loss. Our findings suggest that many of the current hypotheses for the pathophysiology of PASC might be interconnected and offer actionable therapeutic insights.
RESULTS
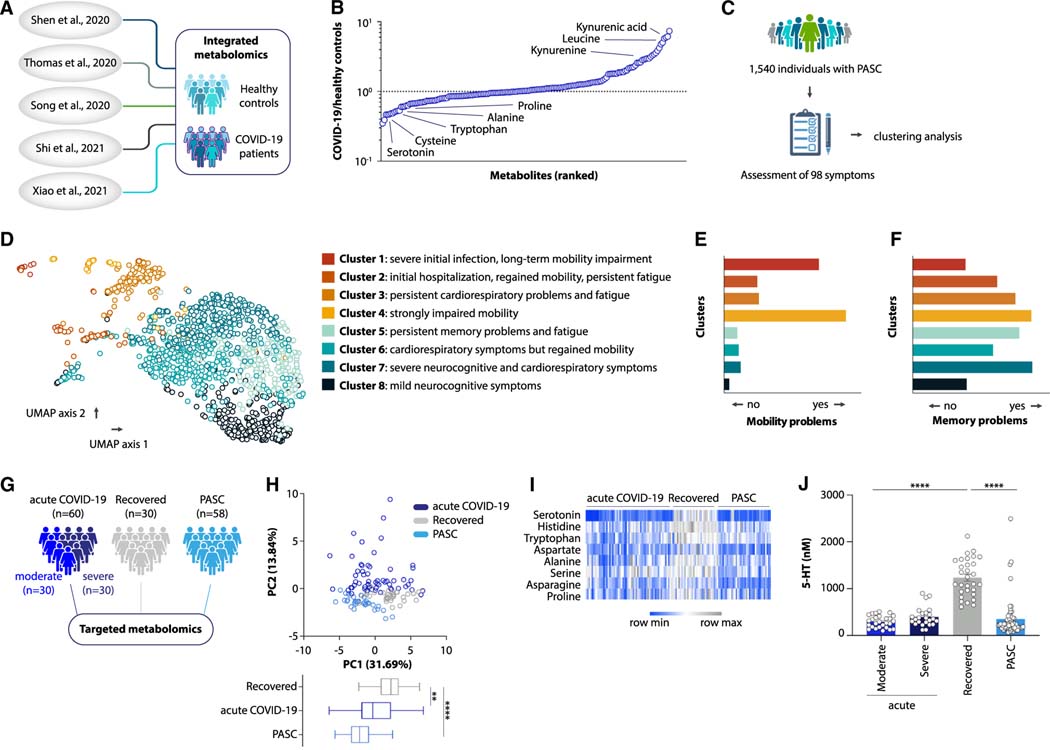
PASC can be characterized by serotonin reduction
We began our explorations by defining a consensus metabolomics signature of acute COVID-19. We integrated previously published metabolomics datasets across different cohort studies7–11 and ranked the metabolites detected in COVID-19 patients by their degree of deviation from the healthy state (Figures 1A and 1B). Among the metabolites most strongly altered during acute COVID-19 were amino acids and their derivatives (Figure 1B). We thus focused on possible roles for these metabolites in Long COVID. We followed a cohort of 1,540 individuals with PASC at Penn Medicine and performed a systematic symptom analysis based on questionnaire surveys and chart review (Figures 1C and S1A–S1C; Table S1). Dimensionality reduction analysis defined eight subtypes of PASC based on symptom similarity (Figure 1D), categorized by different degrees of initial hospitalization for acute infection, mobility impairment, visceral malaise, cardiorespiratory problems, and neurocognitive symptoms (Figures 1D–1F and S1D–S1S). We then performed targeted plasma metabolomics on 58 Long COVID patients who were representative of different clusters (Figure S1T) and experienced persistent symptoms 3 to 22 months after acute infection (Figure S2A). We compared them to 60 individuals with acute COVID-19 and 30 individuals with symptom-free recovery from COVID-19 (Figures 1G and S2B–S2D; Table S2). Notably, the metabolite profile of Long COVID patients was distinct from individuals who recovered to a symptom-free state after SARS-CoV-2 infection (Figure 1H). To determine those molecules that drive the altered metabolomics state in Long COVID, we correlated the abundance of each amino acid metabolite with the presence of symptoms (Figure S2E). We identified a set of molecules whose levels were depleted in both acute and post-acute COVID-19 (Figure 1I), the most significant of which was serotonin (5-hydroxytryptamine, 5-HT) (Figures 1J and S2E). In the post-acute state of infection, serotonin levels were predictive of whether a patient fully recovered or developed long-term sequelae (Figure S2F). Several other amino acids and their derivatives were either unaffected during acute COVID-19 or returned to normal levels in both recovered individuals and Long COVID patients (Figures S2G–S2I).
Figure 1. Serotonin deficiency in PASC.
(A and B) Study schematic (A) and differential abundance ranking (B) of metabolomics data from COVID-19 patients vs. healthy controls.
(C and D) Study schematic (C) and uniform manifold approximation and projection (UMAP) clusters (D) of symptom presentation in UPenn PASC cohort.
(E and F) Symptom distribution in PASC cohort clusters.
(G–J) Study schematic (G), principal component analysis (PCA) plot and PC1 values from targeted metabolomics data (H), heatmap of metabolites decreasing in acute COVID-19 and not recovering in PASC (I), and plasma serotonin (J) in acute COVID-19, recovered, and PASC patients.
Plotted are means ± SEM. **p < 0.01, ****p < 0.0001. See also Figures S1 and S2.
We sought to verify this finding in other cohorts. In a metabolomics study of Long COVID patients and healthy controls (from Cork, Ireland12), serotonin was among the metabolites whose abundance was most strongly depleted in individuals with PASC (Figures S2J–S2L). In contrast, no serotonin reduction was observed in participants of the UNCOVR cohort13 (Figures S2M and S2N; Table S3). In this study, patients were enrolled during acute COVID-19 and then longitudinally provided follow-up blood samples and symptom questionnaires.13 Conversely, the participants at Penn Medicine were enrolled after seeking treatment at a post-COVID clinic. We thus speculated that the severity of PASC might be greater in a cohort that presents for treatment than in a longitudinal recovery cohort. Indeed, the average number of symptoms was higher in the Penn Medicine cohort compared to UNCOVR (Figure S2O). To corroborate whether different levels of circulating serotonin can be explained by differences in PASC symptoms, we measured plasma serotonin levels in a separate longitudinal study (UCSF LIINC cohort14), which includes individuals with a wide range of symptoms (Figure S2P; Table S4). Indeed, in this cohort, serotonin levels negatively correlated with the number of symptoms that participants reported three to four months after acute infection (Figure S2Q). Serotonin levels during the acute phase of SARS-CoV-2 infection were not predictive of the development of PASC (Figures S2R and S2S). Taken together, these investigations reveal that serotonin levels are diminished during acute COVID-19 and remain reduced in severe cases of PASC.
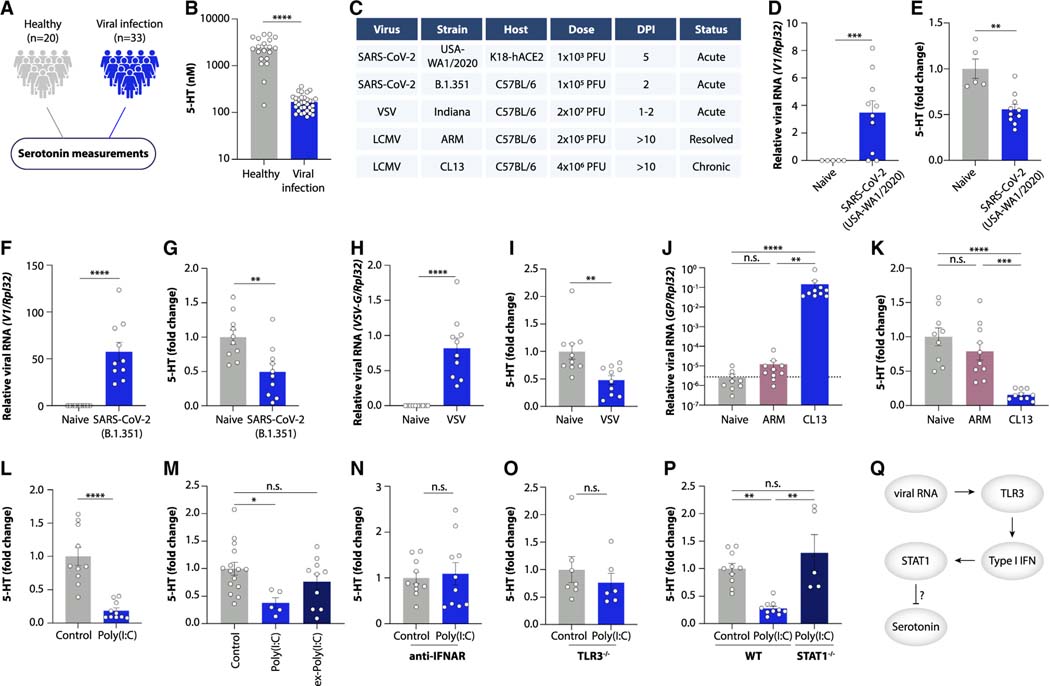
Viral inflammation decreases plasma serotonin levels
Given the centrality of serotonin in regulating a large array of physiological processes,15 we investigated the mechanisms underlying its decrease during acute infection and Long COVID. We first explored whether serotonin depletion was unique to COVID-19 or whether other acute viral infections led to a similar decrease. To this end, we measured serotonin levels in the plasma of 33 individuals with non-SARS-CoV-2 systemic viral infections and compared them to 20 healthy controls (Figures 2A and S3A–S3D; Tables S5 and S6). As in acute COVID-19, serotonin levels were strongly decreased by other viral infections (Figure 2B), suggesting that this might be a more general characteristic of systemic viral infection.
Figure 2. Viral inflammation drives serotonin deficiency.
(A and B) Study schematic (A) and plasma serotonin (B) in viremia patients vs. healthy controls.
(C) Overview of viral infections in mice.
(D and E) Viral RNA load in lungs (D) and plasma serotonin levels (E) in K18-hACE2 mice infected with SARS-CoV-2 (USA-WA 1/2020).
(F and G) Viral RNA load in lungs (F) and plasma serotonin levels (G) in mice infected with SARS-CoV-2 (B.1.351).
(H and I) Viral RNA load in spleen (H) and plasma serotonin levels (I) in mice infected with VSV.
(J and K) Viral RNA load in ileum (J) and plasma serotonin levels (K) in mice infected with LCMV Armstrong (ARM) or Clone 13 (CL13) for 15 days.
(L–P) Plasma serotonin levels in control and poly(I:C)-treated mice (L and M), ex-poly(I:C) mice (M), anti-IFNAR-treated mice (N), TLR3−/− mice (O), and STAT1−/− mice (P).
(Q) Schematic of serotonin reduction by viral RNA.
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S3.
To investigate the mechanisms underlying this association, we used mouse models of viral infection. We first infected mice expressing human ACE2 (K18-ACE2) with the ancestral strain of SARS-CoV-2 (Figures 2C and 2D). Notably, SARS-CoV-2 infection of K18-ACE2 mice led to a reduction in circulating serotonin (Figure 2E). We also observed reduced serotonin in wild-type mice infected with the beta variant of SARS-CoV-216 (Figures 2C, 2F, and 2G). Consistent with our human cohorts, this was not a unique property of SARS-CoV-2, since infection of mice with vesicular stomatitis virus (VSV) similarly decreased plasma serotonin levels (Figures 2C, 2H, and 2I).
Several studies indicate that viral persistence might be a characteristic feature of PASC.17–21 We addressed this question using the lymphocytic choriomeningitis virus (LCMV) mouse model of persistent viral infection (Figure 2C). While serotonin levels returned to baseline after clearance of an acute infection (LCMV Armstrong), chronic viral infection sustained serotonin reduction (LCMV clone 13) (Figures 2J, 2K, and S3E–S3H). We thus speculated that reduced serotonin levels in Long COVID might be a consequence of unresolved inflammation induced by viral products. To test this, we recreated viral-induced inflammation in the absence of a replicating pathogen by repeatedly injecting mice with the synthetic double-stranded RNA polyinosinic:polycytidylic acid (poly(I:C)), which mimics viral replication intermediates. Notably, poly(I:C) treatment was sufficient to diminish serotonin levels (Figures 2L and S3I) both in total plasma and in isolated platelets, which are the major reservoir of circulating serotonin (Figures S3J and S3K). This effect was reversible since normal serotonin levels were restored within a week of poly(I:C) cessation (Figure 2M).
Both viral infection and poly(I:C) treatment induce type I interferon (IFN) signaling. Indeed, exposure to SARS-CoV-2, infection with VSV, persistence of LCMV, or injection of poly(I:C) all strongly upregulated the levels of interferon-stimulated genes (ISGs; Figures S3L–S3O). Importantly, sustained elevation of type I interferons has been observed in Long COVID patients.22 We therefore asked whether the interferon response caused serotonin reduction. Inhibiting interferon signaling through the interferon alpha receptor (IFNAR) prevented poly(I:C)-induced serotonin reduction (Figure 2N). Moreover, mice with genetic deficiency in either the poly(I:C) receptor TLR3 or in the ISG-inducing transcription factor STAT1 (Figures S3P and S3Q) were resistant to the effects of poly(I:C) on serotonin levels (Figures 2O and 2P). Serotonin depletion did not appear to contribute to host defense, since pharmacological inhibition of the serotonin-synthesizing enzyme TPH1 enhanced viral loads and pathogenesis during VSV infection and had no effect on SARS-CoV-2 replication (Figures S3R–S3V). Collectively, these findings suggest that the canonical pathway of viral RNA sensing and type I interferon induction by TLR3 leads to serotonin depletion (Figure 2Q).
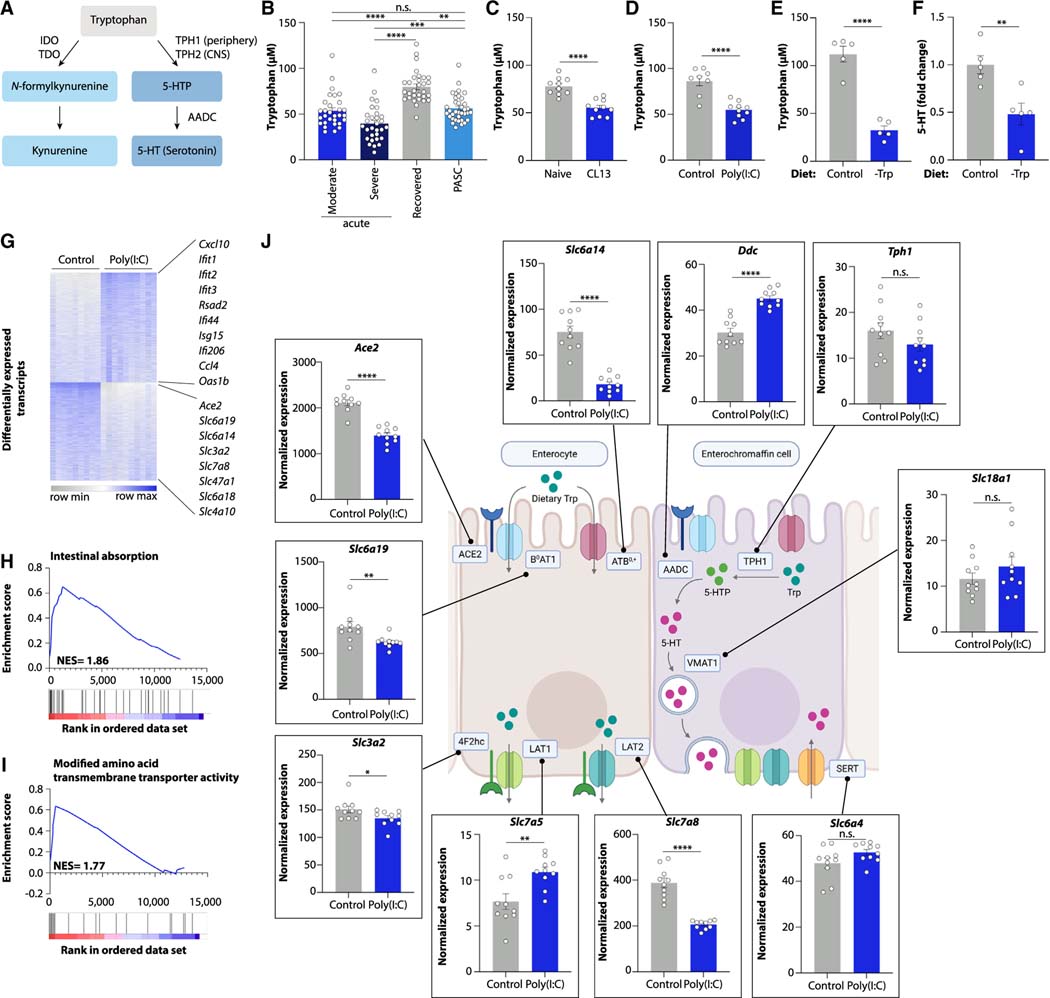
Viral inflammation blocks intestinal tryptophan uptake
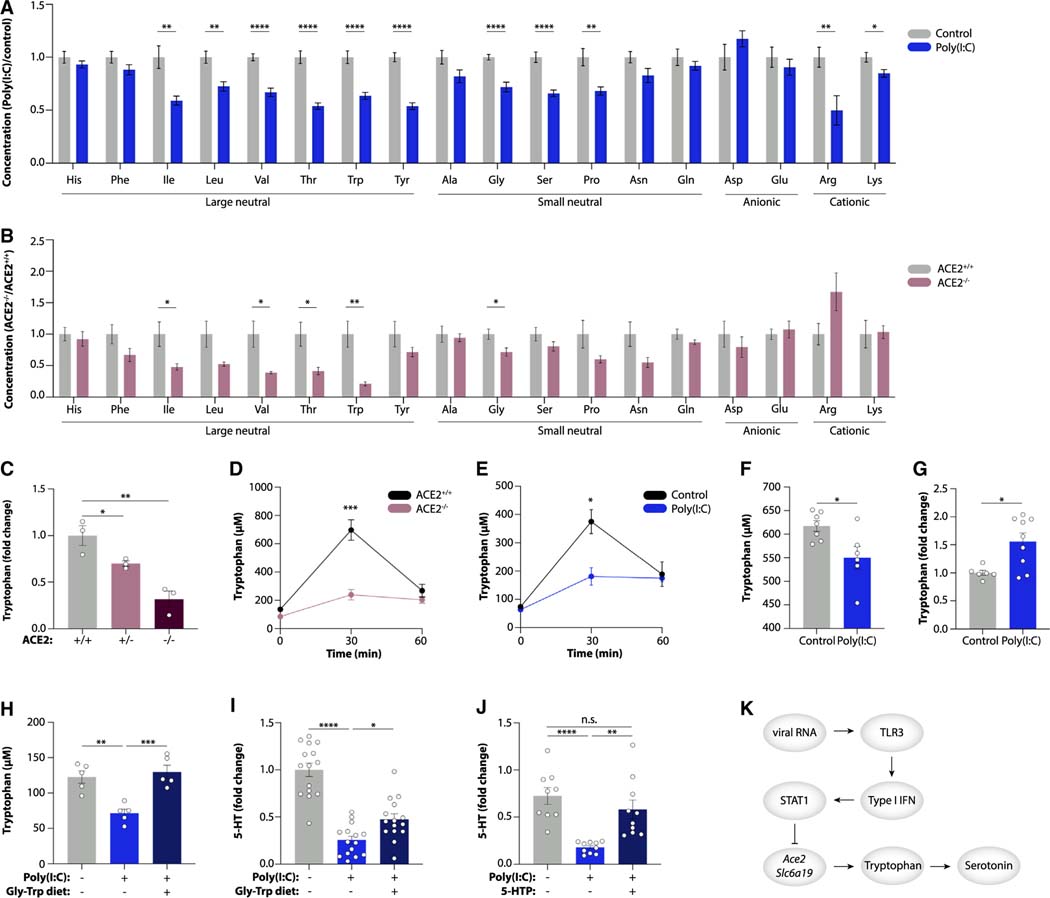
We next investigated the mechanisms by which viral-induced inflammation reduces serotonin levels. The large majority of circulating serotonin is produced in the gastrointestinal tract, where it is synthesized from dietary tryptophan in enterochromaffin cells23 (Figure 3A). We thus investigated whether serotonin production during viral infection might be limited by reduced tryptophan availability. Indeed, individuals with acute COVID-19 showed reduced plasma tryptophan levels (Figures 1B and 3B).24–26 Moreover, tryptophan levels were decreased in Long COVID patients (Figures 1B and 3B). A similar decrease in tryptophan levels was observed in the UCSF LIINC cohort and in another independent Long COVID study (Rush University)14,27 (Figures S4A and S4B; Table S4). Plasma tryptophan concentrations were likewise reduced during chronic LCMV infection and after poly(I:C) treatment of mice (Figures 3C and 3D), suggesting that lower tryptophan availability may cause serotonin reduction by substrate limitation. Consistently, feeding a tryptophan-deficient diet to mice phenocopied the effect of poly(I:C) treatment on plasma serotonin levels in mice (Figures 3E and 3F).
Figure 3. Viral inflammation suppresses genes involved in intestinal amino acid absorption.
(A) Schematic of kynurenine and serotonin biosynthesis.
(B–F) Plasma levels of tryptophan (B–E) and serotonin (F) in acute COVID-19, recovered, and PASC patients (B), mice infected with LCMV CL13 for 30 days (C), poly(I:C)-treated mice (D), and mice fed a tryptophan-deficient diet (E and F).
(G) Differentially expressed genes in ileum of poly(I:C)-treated mice vs. controls.
(H and I) Gene set enrichment analysis plots of ileal genes downregulated by poly(I:C) treatment.
(J) Ileal expression of genes involved in tryptophan uptake and serotonin biosynthesis.
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S4.
Generally, tryptophan deficiency can be caused by either reduced intestinal absorption or by enhanced conversion into tryptophan derivatives such as kynurenine (Figure 3A). Kynurenine levels are elevated during viral infection, and numerous reports have highlighted kynurenine as a metabolite strongly induced by SARS-CoV-2 infection7–11 (Figure 1B). Indeed, kynurenine levels were increased during acute COVID-19 in our cohort (Figure S4C) and likewise elevated by poly(I:C) treatment of mice (Figure S4D). We therefore hypothesized that serotonin reduction was a consequence of tryptophan depletion due to increased kynurenine production. However, the increase in kynurenine levels did not persist in individuals with PASC (Figure S4C). Furthermore, mice lacking the kynurenine-producing enzyme IDO1, which are deficient in kynurenine production, still presented with reduced serotonin upon poly(I:C) treatment (Figures S4E and S4F). Similarly, pharmacological inhibition of the alternative kynurenine-producing enzyme TDO2 did not restore serotonin levels (Figures S4G and S4H). These findings make it unlikely that kynurenine production is the major cause for serotonin depletion during viral inflammation.
We therefore explored intestinal amino acid uptake as a possible cause of tryptophan deficiency and serotonin depletion. Since poly(I:C) treatment reduces food intake (Figure S4I),28,29 we speculated that tryptophan deficiency may result from diminished consumption of this essential amino acid. However, the poly(I:C)-induced tryptophan and serotonin reduction was seen even after an extended fast, in paired feeding experiments, and in experiments in which we supplemented food to poly(I:C)-injected mice (Figures S4J–S4N). The number of serotonin-producing enterochromaffin cells was unaltered by poly(I:C) treatment, ruling out enzymatic synthesis of serotonin as the critical bottleneck (Figures S4O and S4P). We thus used an unbiased approach to explore the impact of viral inflammation on intestinal nutrient absorption. We performed RNA-sequencing of small intestinal tissue of poly(I:C)-treated mice and controls, which revealed strong alterations in intestinal gene expression (Figure S4Q). Expectedly, most upregulated genes belonged to viral recognition and inflammation pathways (Figures 3G and S4R). Remarkably, the gene functions most significantly diminished by poly(I:C) treatment were involved in nutrient metabolism, including amino acid absorption (Figures 3G–3I, S4R, and S4S). For example, the expression of the apical global amino acid transporter ATB0,+ (Slc6a14), the neutral amino acid transporter B0AT1 (Slc6a19), and the B0AT1 chaperone ACE2 were all strongly decreased in poly(I:C)-treated mice (Figures 3G and 3J). The expression of transporters on the basolateral side, such as LAT2 (Slc7a8), were likewise reduced (Figures 3G and 3J). In contrast, the biosynthetic pathway converting tryptophan into serotonin, including the rate-limiting enzyme TPH1, was not affected (Figure 3J). These data highlight transcriptional downregulation of key amino acid absorption genes during viral inflammation, which we verified by qPCR of intestinal tissue from poly(I:C)-treated mice (Figures S5A–S5K).
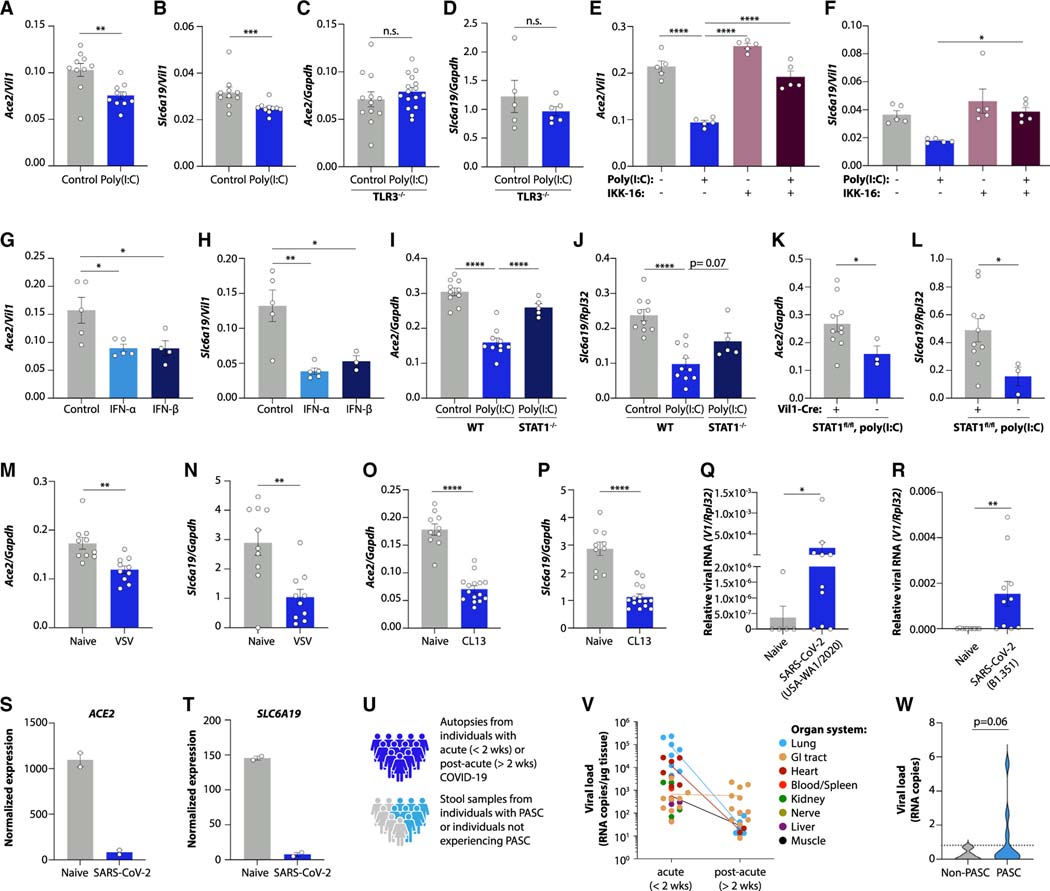
We next used both mice and intestinal organoids to reconstruct the poly(I:C)-induced signaling pathway leading to transcriptional alteration in tryptophan uptake genes (Figures S5A and S5L). As in intestinal tissue, small intestinal organoids responded to poly(I:C) with downregulation of Ace2 and Slc6a19 (Figures 4A, 4B, and S5M). TLR3 deletion prevented the downregulation of these genes after poly(I:C) injection (Figures 4C and 4D). Inhibition of the transcription factor NF-κB, which signals downstream to TLR3, blunted the induction of an interferon response and the downregulation of Ace2 and Slc6a19 in organoids (Figures 4E, 4F, S5N, and S5O). Notably, exposure to type I interferons was sufficient to reduce the expression of genes involved in tryptophan absorption (Figures 4G, 4H, and S5P). The interferon receptor signals via STAT1, and we verified marked STAT1 phosphorylation in response to poly(I:C) treatment in both organoids and intestinal epithelial cells (Figures S5Q and S5R). STAT1 was required for the transcriptional inhibition of Ace2 and Slc6a19 (Figures 4I and 4J) in an epithelial-intrinsic manner (Figures 4K and 4L).
Figure 4. Mechanisms of viral inflammation-induced intestinal gene expression changes.
(A–P) Ace2 and Slc6a19 expression in poly(I:C)-treated small intestinal organoids (A and B), poly(I:C)-treated TLR3−/− mice (C and D), poly(I:C) and IKK-16-treated small intestinal organoids (E and F), IFN-α- and IFN-β-treated small intestinal organoids (G and H), poly(I:C)-treated STAT1−/− mice (I and J), poly(I:C)-treated VillinCre–ERT/+ STAT1flox/flox mice (K and L), ileum of VSV-infected mice (M and N), and ileum of LCMV CL13-infected mice 27 days post-infection (O and P).
(Q and R) Intestinal viral RNA after infection with the indicated strains of SARS-CoV-2.
(S and T) Normalized expression of Ace2 (S) and Slc6a19 (T) in SARS-CoV-2-infected human small intestinal organoids.30
(U–W) Study schematic (U), SARS-CoV-2 RNA detected in tissues obtained from autopsies during the acute or post-acute phase after infection (V), and SARS-CoV-2 RNA detected in stool obtained from individuals with PASC and a control group of individuals with prior SARS-CoV-2 infection but no persistent symptoms (W).
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S5.
To explore the connection between viral persistence in the gut and transcriptional regulation of tryptophan uptake genes, we examined gastrointestinal samples from both mice and humans after viral infection. Indeed, we observed downregulation of Ace2 and Slc6a19 in both acute (VSV) and chronic (LCMV clone 13) settings of viral infection (Figures 4M–4P). Acute SARS-CoV-2 infection in mice also resulted in detectable viral RNA in intestinal tissue (Figures 4Q and 4R), and data from SARS-CoV-2-infected human intestinal organoids30 revealed strong transcriptional inhibition of ACE2 and SLC6A19 (Figures 4S and 4T). Numerous reports have suggested that SARS-CoV-2 can replicate in the human gastrointestinal tract and remain detected long after the acute infection.17,30,31 We confirmed these findings in tissue samples obtained from autopsies during the acute (<2 weeks) and post-acute (>2 weeks) phase after SARS-CoV-2 infection (Figure 4U). While viral RNA could be amplified from several organs during the acute phase (Figure 4V), the gastrointestinal tract stayed viral-RNA-positive in samples obtained from the post-acute phase (Figures 4V and S5S). To determine whether viral persistence in the gastrointestinal tract was associated with the development of PASC, we collected stool samples from individuals with PASC as well as a control group of individuals with prior SARS-CoV-2 infection but no persistent symptoms (Figure 4U). Viral RNA was indeed detected in the stool of a subset of individuals with PASC (Figure 4W), highlighting a possible connection between the presence of viral components in the gastrointestinal tract and the persistence of long-term symptoms in certain individuals.
We next assessed the consequences of reduced epithelial expression of amino acid uptake genes during viral inflammation. In addition to tryptophan, we noted a pronounced reduction in the plasma concentrations of several amino acids in mice injected with poly(I:C), particularly in neutral amino acids (Figure 5A). This amino acid profile resembled the one in mice lacking ACE2 (Figure 5B), which together with B0AT1 is required for the transport of neutral amino acids across the apical membrane of intestinal epithelial cells.32 We confirmed that the successive loss of functional Ace2 alleles in heterozygous and homozygous Ace2-deficient mice led to a stepwise reduction in tryptophan levels (Figure 5C). Mice lacking ACE2 were also unable to absorb an oral bolus of tryptophan (Figure 5D), in line with previous findings.32 Notably, the same phenomenon was observed with poly(I:C) treatment of heterozygous Ace2-deficient mice (Figure 5E), indicating that transcriptional downregulation of Ace2 in these mice phenocopied the homozygous Ace2-deficient state. While the systemic levels of tryptophan were reduced, ileal tryptophan accumulated after poly(I:C) injection (Figures 5F and 5G). Isotope tracing confirmed that circulating tryptophan is derived from the orally supplemented source (Figure S6A), highlighting that poly(I:C) treatment prevented tryptophan absorption.
Figure 5. Viral inflammation inhibits intestinal amino acid absorption.
(A and B) Targeted plasma metabolomics in poly(I:C)-treated mice vs. controls (A) and ACE2−/− vs. ACE2+/+ mice (B).
(C) Plasma tryptophan in ACE2+/+, ACE2+/−, and ACE2−/− mice.
(D and E) Plasma tryptophan in ACE2+/+ vs. ACE2−/− mice (D) and poly(I:C)-treated mice vs. controls (E) after tryptophan gavage.
(F and G) Tryptophan levels in sera (F) and ileal content (G) of poly(I:C)-treated mice 30 min following tryptophan gavage.
(H–J) Plasma tryptophan (H) and serotonin (I and J) in poly(I:C)-treated mice fed a Gly-Trp dipeptide diet (H and I) or given the serotonin precursor 5-HTP (J).
(K) Schematic of serotonin reduction by viral RNA via reduced tryptophan uptake.
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S6.
If tryptophan uptake was abrogated by poly(I:C) treatment, tryptophan supplementation should elevate serotonin levels even during viral inflammation. To corroborate this, we used a diet containing a glycine-tryptophan dipeptide, which bypasses the need for B0AT1 and enables tryptophan uptake via dipeptide transporters.33 This diet compensated for impaired uptake in poly(I:C)-treated mice and led to an increase in both tryptophan and serotonin levels in systemic circulation (Figures 5H and 5I). Similarly, supplementation with the serotonin precursor 5-hydroxytryptophan (5-HTP), which bypasses the requirement for tryptophan, rescued serotonin levels in poly(I:C)-injected mice (Figure 5J). Collectively, these data demonstrate that viral-RNA-induced inflammation impairs intestinal tryptophan uptake, which causes systemic serotonin depletion (Figure 5K).
Viral inflammation impairs serotonin storage
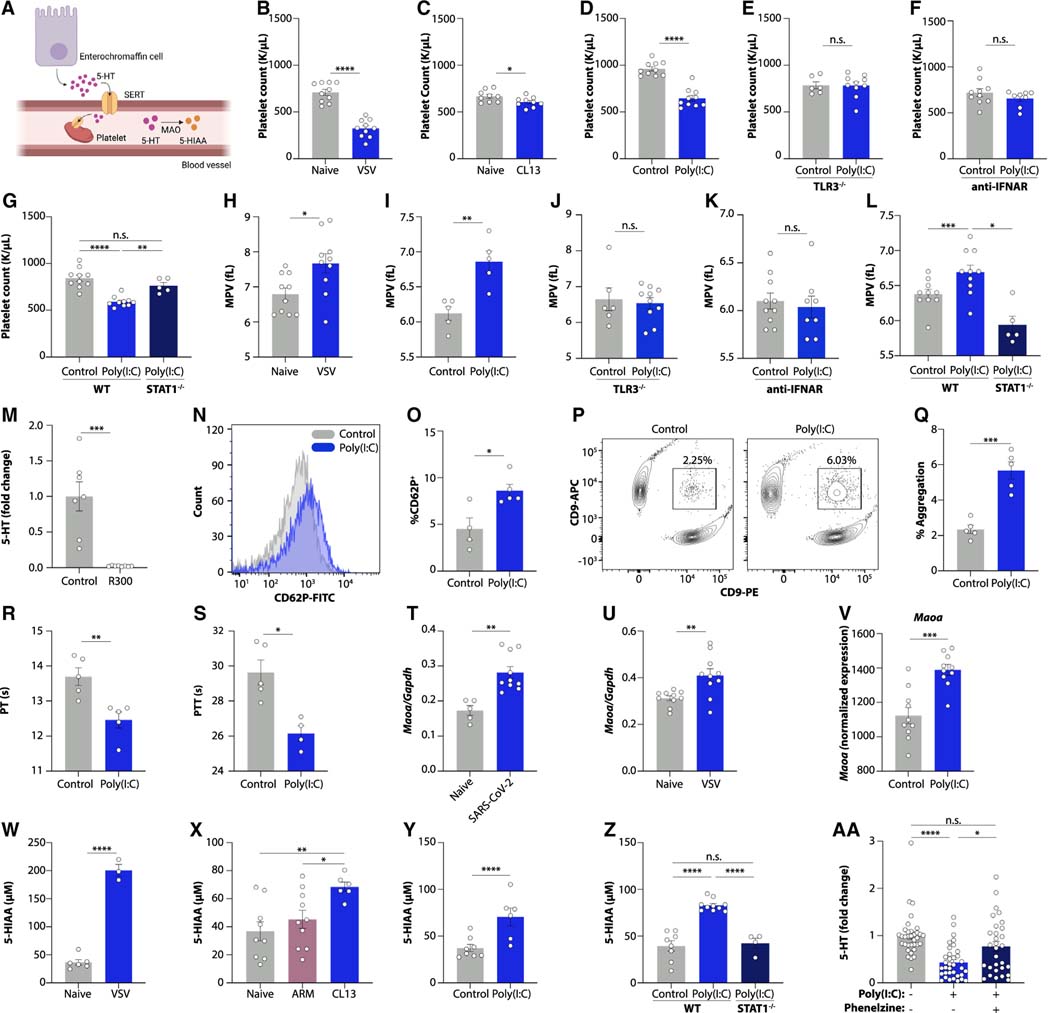
Upon synthesis in enterochromaffin cells, circulating serotonin is transported inside platelets, while free serotonin is rapidly degraded by monoamine oxidase (MAO) enzymes (Figure 6A).34 We noted that platelet counts were strongly decreased after acute VSV infection, chronic LCMV infection, and poly(I:C) injection,35 providing a possible explanation for reduced circulating serotonin levels (Figures 6B–6D). Poly(I:C)-induced thrombocytopenia was dependent on the TLR3-IFN-STAT1 signaling pathway (Figures 6E–6G). The overall white blood cell count was unchanged by poly(I:C) treatment (Figure S6B). Erythrocyte, hemoglobin, and hematocrit counts were reduced (Figures S6C–S6E), while mean corpuscular volume and mean corpuscular hemoglobin were not affected (Figures S6F and S6G). Increased mean platelet volumes (Figures 6H and 6I) were indicative of increased destruction of platelets,36–40 which was likewise dependent on TLR3, type I interferon signaling, and STAT1 (Figures 6J–6L). Tryptophan supplementation was unable to restore platelet counts (Figure S6H), indicating that reduced intestinal amino acid uptake and platelet depletion were independent effects of poly(I:C) injection. Consistently, platelet depletion41 abolished circulating serotonin levels (Figures 6M and S6I) without affecting intestinal tryptophan uptake genes (Figures S6J and S6K).
Figure 6. Viral inflammation drives thrombocytopenia and serotonin turnover.
(A) Cartoon of serotonin transport and degradation.
(B–G) Platelet counts in naive or VSV-infected mice (B); LCMV CL13-infected mice at day 15 post-infection (C); and poly(I:C)-treated wild-type (D), TLR3−/− (E), anti-IFNAR-receiving (F), and STAT1−/− mice.
(H–L) Mean platelet volume of VSV-infected mice (H) and poly(I:C)-treated wild-type (I), TLR3−/− (J), anti-IFNAR-receiving (K), and STAT1−/− mice (L).
(M) Plasma serotonin in mice treated with a platelet-depleting antibody.
(N–Q) Representative FACS plot (N and P) and quantification (O and Q) of platelet CD62P expression (N and O) and platelet aggregation (P and Q) in poly(I:C)-treated mice.
(R and S) Prothrombin (R) and partial thromboplastin (S) time in poly(I:C)-treated mice.
(T–V) Ileal Maoa expression in mice treated with SARS-CoV-2 (USA-WA 1/2020) (T), VSV (U), or poly(I:C) (V).
(W–Z) 5-HIAA levels in urine from mice infected with VSV (W) and LCMV ARM or CL13 at day 15 post-infection (X), as well as poly(I:C)-treated wild-type (Y) and STAT1−/− mice (Z).
(AA) Platelet serotonin levels of poly(I:C)-treated mice receiving the MAO inhibitor phenelzine.
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S6.
We next investigated the causes for thrombocytopenia during viral inflammation. The number and size of megakaryocytes in the bone marrow was increased in poly(I:C)-treated mice (Figures S6L–S6N), while thrombopoietin levels were unchanged (Figures S6O and S6P). We noted that the baseline activation status of platelets was increased by poly(I:C) treatment (Figures 6N and 6O). Consistently, platelet aggregation was markedly enhanced (Figures 6P and 6Q). Prothrombin time (PT) and partial thromboplastin time (PTT) were reduced (Figures 6R and 6S), further indicative of hypercoagulability.42 We ruled out changes in the concentrations of fibrinogen, tissue factor, or TAT complexes as alternative explanations (Figures S6Q–S6S). Collectively, these results indicate that viral inflammation drives platelet hyperactivation, resulting in hypercoagulability and thrombocytopenia in an interferon-dependent manner. Consequently, platelet-mediated systemic serotonin transport is impaired.
Since free serotonin is the target of rapid degradation,34 we next focused on MAO-mediated serotonin turnover. We found that intestinal transcript levels of Maoa were increased in SARS-CoV-2-infected, VSV-infected, and poly(I:C)-treated mice in a TLR3-dependent manner (Figures 6T–6V and S6T). Consistently, the levels of the serotonin degradation product 5-hydroxyindoleacetic acid (5-HIAA) were increased in the urine of virally infected mice and in mice injected with poly(I:C) (Figures 6W–6Y and S6U). STAT1-deficient mice were protected from the accumulation of 5-HIAA (Figure 6Z). Notably, pharmacological inhibition of MAO prevented the accumulation of 5-HIAA and restored serotonin levels in poly(I:C)-treated mice (Figures 6AA and S6V). These findings indicate that serotonin turnover is enhanced during viral inflammation.
Serotonin reduction impairs vagal signaling and memory function
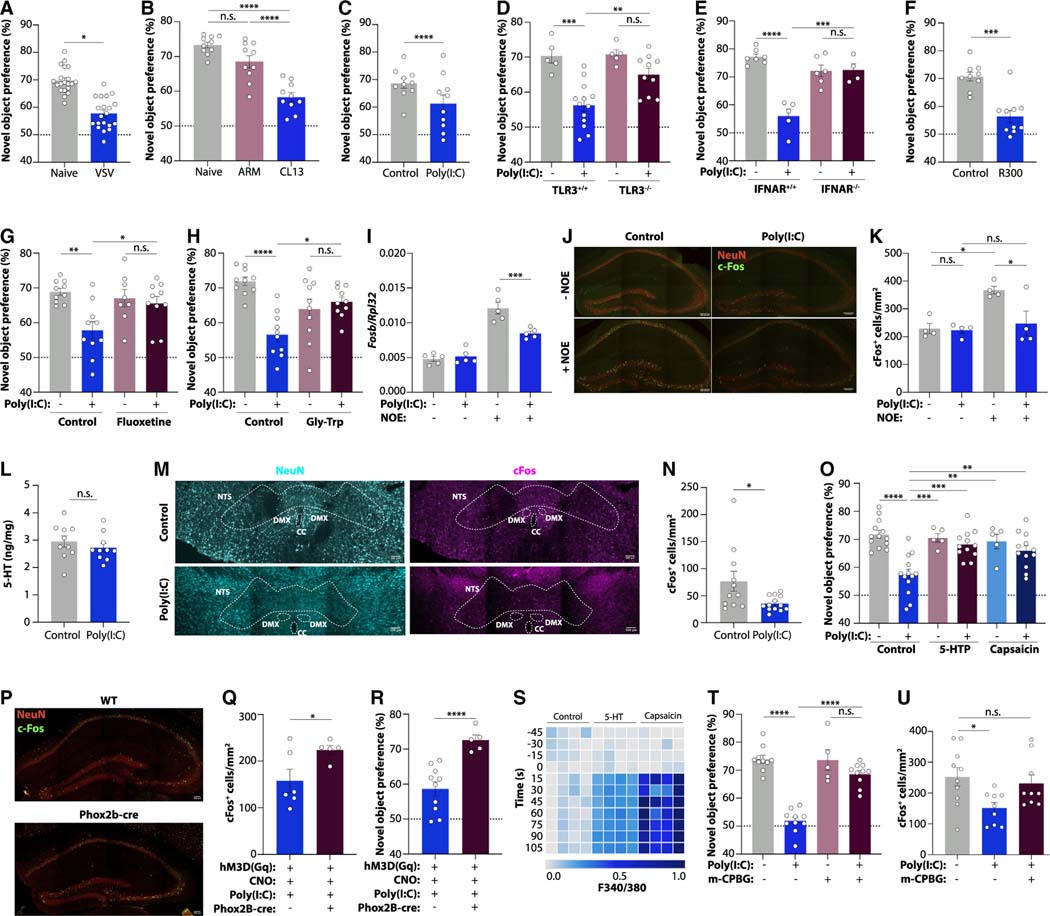
Finally, we explored the consequences of peripheral serotonin depletion on individuals experiencing PASC. In a symptom questionnaire administered at the time of blood draw, the majority of patients in our cohort reported fatigue, cognitive difficulties, headaches, loss of endurance, problems with sleep, anxiety, and memory loss (Figure S7A). To investigate possible mechanisms underlying the association between serotonin reduction and prevalent neurocognitive manifestations, we again turned to mouse models. We observed cognitive impairment in the setting of acute VSV infection, chronic LCMV persistence, and in poly(I:C)-treated mice as assessed by the novel object recognition paradigm43 (Figures 7A–7C). This was dependent on TLR3 and type I interferon signaling44,45 (Figures 7D and 7E). Platelet depletion similarly impaired memory function (Figure 7F). We therefore hypothesized that serotonin reduction may be responsible for poor cognitive performance after poly(I:C) injection. Indeed, treatment of mice with the selective serotonin reuptake inhibitor (SSRI) fluoxetine restored novel object recognition (Figure 7G), and rescue of tryptophan levels by glycine-tryptophan supplementation reinstated normal cognitive performance in poly(I:C)-treated mice (Figure 7H). Differences in explorative behavior did not affect the results across all of these experiments (Figures S7B–S7H).
Figure 7. Serotonin deficiency drives cognitive dysfunction via vagal signaling.
(A–H) Novel object preference in mice infected with VSV (A) or LCMV ARM or CL13 at day 14 post-infection (B); poly(I:C)-treated wild-type (C), TLR3−/− (D), and IFNAR−/− mice (E); platelet-depleted mice (F); poly(I:C)-treated mice receiving the SSRI fluoxetine (G); and poly(I:C)-treated mice fed a Gly-Trp dipeptide diet (H).
(I) Fosb expression in the hippocampus of poly(I:C)-treated mice with or without novel object exposure (NOE).
(J and K) Representative images (J) and quantification (K) of cFos+ cells in the dentate gyrus of poly(I:C)-treated mice with or without NOE. Scale bars, 100 μm.
(L) Serotonin concentrations in the brains of poly(I:C)-treated mice.
(M and N) Representative images (M) and quantification (N) of cFos+ cells in the nucleus tractus solitarii (NTS) of poly(I:C)-treated mice. Scale bars, 100 μm. Outlined are NTS, dorsal motor nucleus (DMX), and central canal (CC).
(O) Novel object preference in mice receiving poly(I:C), 5-HTP, or capsaicin.
(P–R) Representative images (P) and quantification of cFos+ cells in the dentate gyrus following NOE (Q) and novel object preference (R) of Phox2b-cre mice injected with AAV-hM3Dq, CNO, and poly(I:C). Scale bars, 100 μm.
(S) Calcium signaling of cultured vagal neurons exposed to capsaicin or serotonin.
(T and U) Novel object preference (T) and quantification of cFos+ cells in the dentate gyrus (U) of mice treated with poly(I:C) and the 5-HT3 receptor agonist m-CPBG.
Plotted are means ± SEM. n.s. p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S7.
The acquisition of short-term memories is driven by the hippocampus,46 and studies have described reduced hippocampal activity in COVID-19 patients.47,48 We found that hippocampal activation in response to novel object exposure was blunted in poly(I:C)-treated mice (Figures 7I–7K, S7I, and S7J). This was not accounted for by changes in hippocampal neurogenesis (Figures S7K–S7N). Since serotonin plays an important role in hippocampal function,49–51 we hypothesized that serotonin reduction directly impaired the generation of hippocampus-dependent memories. However, serotonin levels in the brain were unaffected by viral inflammation (Figure 7L), suggesting that the peripheral reduction of serotonin was responsible for cognitive impairment.
Circulating serotonin does not cross the blood-brain barrier15 but can influence the brain via afferent sensory neurons.52 To explore the impact of peripheral serotonin on sensory neurons, we measured neuronal activation in sensory terminals of the nucleus tractus solitarii (NTS) in the brainstem. Novelty exposure led to an increase in cFos+ cells in the NTS, but this response was abrogated upon poly(I:C) treatment (Figures 7M and 7N), suggesting that serotonin depletion causes cognitive impairment through reduced sensory neuron activity. Consistently, restoration of peripheral serotonin levels using 5-HTP rescued cognition in poly(I:C)-treated mice (Figures 7O and S7O), and so did the TRPV1 agonist capsaicin, a strong stimulant of sensory neurons (Figure 7O). Of note, capsaicin treatment did not affect peripheral serotonin levels (Figure S7O), and neither capsaicin nor 5-HTP treatment ameliorated poly(I:C)-induced ISG responses in the brain (Figure S7P), highlighting that restoration of sensory input from the periphery is able to rescue cognition despite serotonin deficiency or ongoing neuroinflammation. Peripheral serotonin reduction alone, as in the case of platelet depletion, did not trigger inflammation in the brain (Figure S7Q).
TRPV1+ sensory neurons can be broadly categorized as vagal and spinal cord afferents. To distinguish between both possibilities, we chemogenetically activated Phox2b-expressing neurons, which are restricted to the vagus nerve. Indeed, stimulation of Phox2b neurons during poly(I:C) treatment restored activation of hippocampal neurons and the formation of short-term memories (Figures 7P–7R and S7R). To determine the mechanism by which serotonin influences the activity of vagal neurons, we used an in vitro system in which we cultured neurons from nodose ganglia and exposed them to serotonin. Vagal neurons robustly responded to serotonin treatment, as evidenced by rapid calcium influx (Figure 7S), suggesting a possible direct effect of peripheral serotonin on the vagus nerve. Single-cell transcriptomics data52 showed high and selective expression of the serotonin receptor 5-HT3 on vagal neurons (Figure S7S). To determine whether serotonin signaling via 5-HT3 receptors was sufficient to restore cognition during viral inflammation, we used the pharmacological 5-HT3 receptor agonist meta-Chlorophenylbiguanide (m-CPBG). Indeed, m-CPBG treatment normalized both novelty responses of hippocampal neurons and performance in the novel object recognition paradigm (Figures 7T and 7U). Taken together, these findings suggest that serotonin reduction dampens vagal signaling and thereby impairs cognitive function.
DISCUSSION
The emergence of PASC poses a global health challenge. The pathophysiology of post-viral syndromes remains poorly understood,1,3 leaving medical systems across the world unprepared for the large number of individuals developing cardiorespiratory, neurocognitive, gastrointestinal, and musculoskeletal symptoms in the months and years53 following acute COVID-19. While vaccination may reduce the risk of developing PASC,54,55 instances of Long COVID after breakthrough infections continue to occur.56 A deeper understanding of the molecular and cellular etiopathology of PASC is thus urgently needed.
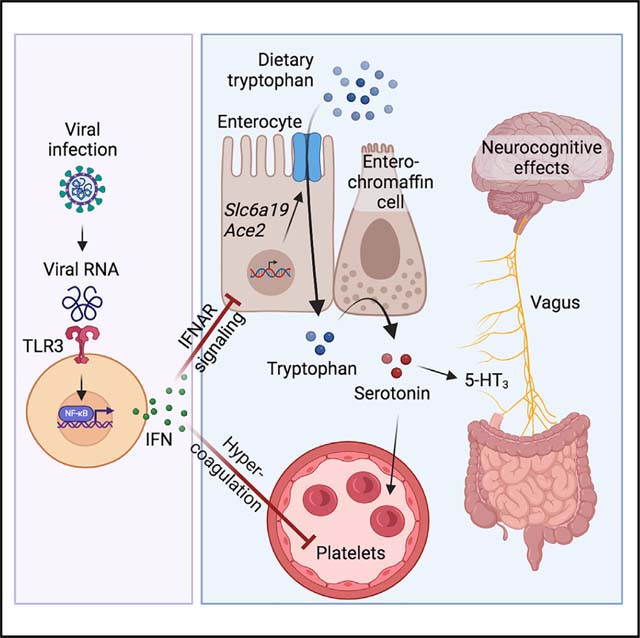
In this study, we have investigated metabolite signatures associated with Long COVID. We have focused on metabolites whose concentrations are perturbed both in acute COVID-19 and in patients with PASC. Among the metabolites we measured, the molecule most significantly associated with PASC was serotonin. We show that viral inflammation-driven serotonin depletion can be caused by reduction of tryptophan absorption, thrombocytopenia, and increased MAO expression. This response is TLR3-, IFNAR-, and STAT1-dependent and results in decreased vagal and hippocampal activation as well as cognitive impairment.
These findings have several important implications. First, they highlight the profound consequences that persistent viral reservoirs can have. Numerous studies have provided evidence for the presence of viral components17–19 and persistently high levels of type I interferons in the blood 8 months after infection.22 Our data indicate that the presence of viral components and resultant interferon response might be a causative factor in the development of PASC-associated symptoms.
Second, our study highlights a mechanism by which viral infection can alter amino acid uptake. Deviations from homeostatic concentrations of amino acids can exert profound effects on tissue function.57 While we focused on serotonin in this study, tryptophan serves as the precursor for many other important metabolites, including niacin, NAD, and melatonin.23,58 The evolutionary teleology of reduced intestinal amino acid absorption during viral inflammation remains unclear, but it is possible that acute downregulation of genes involved in amino acid uptake is part of a cellular response to interferon stimulation aimed at abrupt cessation of cellular metabolism during viral infection. In the case of non-resolving viral inflammation, this response may persist and result in nutrient deficiency.
Third, a common feature of both acute and post-acute SARS-CoV-2 infection is the formation of microthrombi as a result of hypercoagulability.59–61 Our findings imply that thrombocytopenia may diminish the carrying capacity of the systemic circulation for serotonin. Reduced serotonin storage, coupled with the induction of MAO enzymes, may enhance the turnover of serotonin and excretion of its degradation products. Thus, hypercoagulability in acute COVID-19 and Long COVID may have implications beyond its cardiovascular effects.
Fourth, our study indicates a role for the vagus nerve in mediating the impact of serotonin reduction on the brain.62 Neurological symptoms are widespread in patients with both acute and post-acute COVID-19.63,64 Since unequivocal evidence for SARS-CoV-2 replication in the brain is lacking, recent studies have focused on the cognitive consequences of peripheral immune activation as well as neuroinflammation.63,65 Based on our data, we suggest that afferent sensory neurons may play a critical role in the neurocognitive manifestations of both acute and post-acute viral infections. The vagus nerve is an important mediator of sickness behavior,66 responds to peripheral serotonin levels,67 and has been implicated in the pathophysiology of chronic fatigue syndrome.68 While the precise circuit by which the vagus nerve is involved in the development of PASC remains unclear, sensory neurons may emerge as an important element in relaying the effect of peripheral viral inflammation to the brain.
Finally, our findings indicate possible targets for clinical interventions aimed at the prevention and treatment of PASC. Our animal models demonstrate that serotonin levels can be restored and memory impairment reversed by precursor supplementation or SSRI treatment. While the effectiveness of SSRIs in acute COVID-19 has been a subject of debate,69–74 no systematic exploration of SSRIs in individuals with PASC has been performed to date. Our study, together with recent findings linking depression with cognitive impairment in Long COVID75 and the effect of SSRIs on vagus nerve activity,67 call for the assessment of targeting serotonin signaling for the prevention or treatment of neurocognitive manifestations.
Given the dual role of ACE2 as both a mediator of intestinal tryptophan absorption32 and a receptor for SARS-CoV-2,76 it is possible that virus-induced receptor internalization augments the effect of interferons on ACE2 downregulation and serotonin reduction. In principle, however, none of the mechanisms described in this study are unique to SARS-CoV-2 infection. Indeed, reduced serotonin levels have been reported in other settings of viral inflammation, such as dengue virus infection,77 which is the trigger of another post-viral syndrome.78 The connection between serotonin reduction and vagus nerve dysfunction may thus be relevant beyond Long COVID. The fact that low serotonin levels are also found in non-viral conditions characterized by elevated interferon levels, such as systemic lupus erythematosus or multiple sclerosis,79–81 suggests that the pathway described in this study may even apply beyond viral infections.
Limitations of this study
The degree of serotonin reduction is variable across the four cohorts of individuals with PASC that we have examined in this study. While modes of recruitment, number of symptoms, and degree of disease severity might provide possible sources of this variability, there are likely further differences that we have not accounted for. The manifestations of Long COVID are highly heterogeneous,82 and the subtypes of PASC that are studied in individual cohorts are likely different. Our results indicate that serotonin reduction is not specific to any particular subset of PASC, but much larger numbers of longitudinal samples are required to comprehensively characterize serum metabolite levels across the different endotypes of Long COVID.
In addition, while we provide evidence for serotonin reduction in acute COVID-19, individuals with PASC, and acutely and chronically infected mice, mouse models for Long COVID are still lacking, and thus our study does not establish a direct causal connection between post-acute SARS-CoV-2 infection, tryptophan uptake, thrombocytopenia, and serotonin levels. The chronic LCMV and poly(I:C) models used in this study recapitulate important features of SARS-CoV-2 infection but have clear limitations. For instance, when administered systemically, poly(I:C) may not accurately mimic the tissue-level inflammatory processes induced by persistent viral reservoirs. Furthermore, while the persistent presence of circulating spike protein may be a useful marker for PASC,18 it remains unclear whether remnants of SARS-CoV-2 nucleic acid play any functional role in Long COVID.
Finally, our assessment of viral persistence in the gastrointestinal tract of individuals with PASC is based on a limited number of participants. Similarly, we have not demonstrated a direct connection between intestinal viral persistence and chronically elevated levels of type I interferons in humans, which would require collecting a large number of intestinal biopsies from Long COVID patients. Our results thus call for the large-scale investigation of the causal connection between the presence of a viral reservoir in the gastrointestinal tract, sustained inflammatory responses, and manifestations of Long COVID.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maayan Levy (maayanle@pennmedicine.upenn.edu).
Materials availability
Animal strains used in this study are available from The Jackson Laboratory, Taconic Biosciences, or were provided by the indicated investigators.
Data and code availability
All data and code to understand and assess the conclusions of this research are available in the main text and supplementary materials. RNA-seq data have been deposited and are publicly available. Accession numbers are listed in the key resources table.
This paper does not contain original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Anti-doublecortin antibody | Abcam | ab18723; RRID:AB_732011 |
| Anti-NeuN Antibody, clone A60 | Millipore Sigma | MAB377; RRID:AB_2298772 |
| Anti-mouse GPIbα | Emfret | R300; RRID:AB_2721041 |
| Anti-mouse IFNAR-1 antibody | Bio X Cell | BE0241; RRID:AB_2687723 |
| APC anti-mouse CD9 Antibody | Biolegend | 124811; RRID:AB_2783070 |
| APC Rat Anti-Mouse CD41 | Biolegend | 133913; RRID:AB_11126751 |
| c-Fos (9F6) Rabbit mAb | Cell Signaling Technology | 2250; RRID:AB_2247211 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Thermo Fisher Scientific | A-21202; RRID:AB_141607 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 | Thermo Fisher Scientific | A-31573; RRID:AB_2536183 |
| FITC Rat Anti-Mouse CD62P | BD | 561923; RRID:AB_10896149 |
| GAPDH (D16H11) XP® Rabbit mAb | Cell Signaling Technology | 5174S; RRID:AB_10622025 |
| Goat anti-Human IgM-HRP | SouthernBiotech | 2020–05; RRID:AB_2795603 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | A-11008; RRID:AB_143165 |
| Ki-67 Monoclonal Antibody (SolA15) | eBioscience | 14–5698-82; RRID:AB_10854564 |
| Non-immune rat immunoglobulins (IgG) | Emfret | C301; RRID:AB_2734715 |
| PE anti-mouse CD9 Antibody | Biolegend | 124805; RRID:AB_1279327 |
| Peroxidase AffiniPure Goat Anti-Human IgG (H + L) | Jackson ImmunoResearch Laboratories | 109–035-088; RRID:AB_2337584 |
| Phospho-Stat1 (Tyr701) (58D6) Rabbit mAb | Cell Signaling Technology | 9167S; RRID:AB_561284 |
| Rabbit anti-chromogranin A antibody | Novus Biologicals | NB120–15160 |
| Stat1 (D1K9Y) Rabbit mAb | Cell Signaling Technology | 14994S; RRID:AB_2737027 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology | sc-47778; RRID:AB_2714189 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pAAV-hSyn-DIO-hM3Dq-mCherry | Addgene | 44361 |
| LCMV (Armstrong strain) | John Wherry, University of Pennsylvania | N/A |
| LCMV (Clone 13 strain) | John Wherry, University of Pennsylvania | N/A |
| SARS-CoV-2, Isolate B.1.351 | Andy Pekosz, Johns Hopkins University | N/A |
| SARS-CoV-2, Isolate USA-WA1/2020 | BEI Resources | NR-52281 |
| Vesicular stomatitis virus (Indiana strain) | Sara Cherry, University of Pennsylvania | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Acute and recovered COVID-19 cohort plasma samples | Mathew et al.83 | N/A |
| Healthy and PASC stool samples | This study | N/A |
| Healthy cohort plasma samples | Una O’Doherty, University of Pennsylvania | N/A |
| Human autopsy tissues | This study | N/A |
| RUSH PASC cohort plasma samples | Giron et al.27 | N/A |
| UCSF LIINC cohort plasma samples | Peluso et al.14 | N/A |
| UNCOVR cohort plasma samples | Su et al.13 | N/A |
| UPenn PASC cohort plasma samples | This study | N/A |
| Viremia cohort plasma samples | Reilly et al.84 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| L-tryptophan (13C11, 99%) | Cambridge Isotope Laboratories | CLM-4290-H-0.1 |
| 1-(3-Chlorophenyl)biguanide hydrochloride | Tocris | 440 |
| 4-Chloro-DL-phenylalanine methyl ester hydrochloride | Sigma-Aldrich | C3635 |
| 5-HT | Millipore Sigma | 14927 |
| 5-hydroxy-L-tryptophan | Cayman Chemical Company | 20539 |
| 680C91 | Selleck Chemicals | S8997 |
| Capsaicin | Sigma-Aldrich | M2028 |
| Clozapine N-oxide hydrochloride | Sigma-Aldrich | SML2304 |
| Collagenase 1A | Gibco | 17100017 |
| Dithiothreitol (DTT) | Sigma-Aldrich | 1.02E+10 |
| Fluoxetine oral solution, USP | Aurobindo | NDC 65862–306-12 |
| Fura- 2-AM | Thermo Fisher Scientific | F-1221 |
| Gly-Pro-Arg-Pro | Sigma-Aldrich | G1895 |
| Glycyl-L-tryptophan hydrate | VWR | 100276–390 |
| HEPES | Thermo Fisher Scientific | 5–630-080 |
| HMW poly(I:C) | InvivoGen | tlrl-pic |
| IKK-16 | Selleck Chemicals | No.S2882 |
| L-tryptophan | Sigma-Aldrich | T8941 |
| LMW poly(I:C) | InvivoGen | tlrl-picw |
| Mouse NGF 7S Subunit protein | Gibco | 549 13–290-010 |
| N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide | Sigma-Aldrich | 394882 |
| Nickel–nitrilotriacetic acid (Ni-NTA) resin | Qiagen | 30210 |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15140122 |
| Phenelzine sulfate salt | Sigma-Aldrich | P6777 |
| Poly-L-lysine | Sigma Aldrich | P4707 |
| Poly(I:C) | Sigma-Aldrich | P1530 |
| Recombinant Mouse IFN-α | BioLegend | 752802 |
| Recombinant Mouse IFN-β1 | BioLegend | 581302 |
| Sodium pyruvate | Corning | MT25000CI |
| SureBlue 3,3′,5,5′-tetramethylbenzidine substrate | KPL | 5120–0075 |
| Thrombin | Sigma-Aldrich | T4648 |
|
| ||
| Critical commercial assays | ||
|
| ||
| 5-HIAA ELISA kits | Abnova | KA1881 |
| Fibrinogen ELISA kits | Abcam | ab213478 |
| High-Capacity cDNA Reverse Transcription kits | Thermo Fisher Scientific | 43–688-13 |
| Kynurenine ELISA kits | Abnova | KA6140 |
| LUNA Universal PCR kit | New England Biolabs | M3003E |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| QuantiFast SYBR Green PCR kit | Qiagen | 204056 |
| RNAeasy mini kits | Qiagen | 74104 |
| Serotonin ELISA kits | Novus biologicals | KA1894 |
| Thrombin-Antithrombin Complexes ELISA kits | Abcam | ab137994 |
| Tissue factor ELISA kits | Abcam | ab214091 |
| Tryptophan ELISA kits | Novus biologicals | KA11916 |
| Waters AccQTag Ultra derivatization kit | Waters corporation | 86003836 |
| Viral RNA Mini Kit | Qiagen | 52906 |
|
| ||
| Deposited data | ||
|
| ||
| Human intestinal organoid RNA-sequencing | Lamers et al.30 | GSE149312 |
| Metabolomics of COVID-19 patients | Shen et al., Shi et al., Song et al., Thomas et al., and Xiao et al.7–11 | N/A |
| Metabolomics of patients with PASC | Sadlier et al.12 | N/A |
| Single-cell RNA-seq of vagal neurons | Kupari et al.52 | GSE124312 |
| RNA-seq of ileum from poly(I:C)-treated mice | This study | PRJNA1007416 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| TLR3−/− | The Jackson Laboratory | 005217 |
| ACE2−/− | Taconic Biosciences | 18180 |
| IDO1−/− | The Jackson Laboratory | 005867 |
| IFNAR1−/− | The Jackson Laboratory | 028288 |
| K18-HuACE2 | The Jackson Laboratory | 034860 |
| Phox2b-Cre | The Jackson Laboratory | 016223 |
| STAT1−/− | The Jackson Laboratory | 012606 |
| STAT1flox/flox | Klover et al., Neoplasia, 201085 | Klover et al.85 |
| Villin-creERT2 | The Jackson Laboratory | 020282 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Forward primer for 18S qPCR: 5′- AACCCGTTGAACCCCATT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for 18S qPCR: 5′- CCATCCAATCGGTAGTAGCG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ddc qPCR: 5′-TAGCTGACTATCTGGATGGCAT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ddc qPCR: 5′-GTCCTCGTATGTTTCTGGCTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit1 qPCR: 5′-CAGAAGCACACATTGAAGAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit1 qPCR: 5′-TGTAAGTAGCCAGAGGAAGG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit2 qPCR: 5′-GGGAAAGCAGAGGAAATCAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit2 qPCR: 5′-TGAAAGTTGCCATACAGAAG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit3 qPCR: 5′-GCCGTTACAGGGAAATACTGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit3 qPCR: 5′-CCTCAACATCGGGGCTCT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Mx1 qPCR: 5′-GACTACCACTGAGATGACCCAGC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Mx1 qPCR: 5′-ATTTCCTCCCCAAATGTTTTCA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Nsp14 qPCR: 5′-TGGGGYTTTACRGGTAACCT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Nsp14 qPCR: 5′-AACRCGCTTAACAAAGCACTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Oas1b qPCR: 5′-TTCTACGCCAATCTCATCAGTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Oas1b qPCR: 5′-GGTCCCCCAGCTTCTCCTTAC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Rpl32 qPCR: 5′-TTCCTGGTCCACAATGTCAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Rpl32 qPCR: 5′-GGCTTTTCGGTTCTTAGAGGA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc18a1 qPCR: 5′-GTCCCGGAAGCTGGTGTTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc18a1 qPCR: 5′-ACAGTGAGCAGCATATTGTCC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc3a2 qPCR: 5′-ACGGTGTGGATGGTTTCCAAT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc3a2 qPCR: 5′-TCCCTGCAATCAAAAGCCTGT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a14 qPCR: 5′-GACAGCTTCATCCGAGAACTTC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a14 qPCR: 5′-ATTGCCCAATCCCACTGCAT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a19 qPCR: 5′-AACGCTCATGTATAGCATCTGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a19 qPCR: 5′-CAGCCACAGTGACCACAAC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a4 qPCR: 5′-GACAGGGGTGTGGGTTGATGC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a4 qPCR: 5′-TCAGCCATGTAGCCAAGCACC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc7a5 qPCR: 5′-CTACGCCTACATGCTGGAGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc7a5 qPCR: 5′-GAGGGCCGAATGATGAGCAG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc7a8 qPCR: 5′-TCAGCGCCTGTGGTATCATTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc7a8 qPCR: 5′-TGATGCCTGTCACGATCCAGA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Tph1 qPCR: 5′-AACAAAGACCATTCCTCCGAAAG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Tph1 qPCR: 5′-TGTAACAGGCTCACATGATTCTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for VI qPCR (for detection of SARS-CoV-2 viral RNA): 5′-ATGCTGCAATCGTGCTACAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for VI qPCR (for detection of SARS-CoV-2 viral RNA): 5′-CCTCTGCTCCCTTCTGCGTA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Vil1 qPCR: 5′-TCAAAGGCTCTCTCAACATCAC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Vil1 qPCR: 5′-AGCAGTCACCATCGAAGAAGC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for VSV-G qPCR: 5′-CAAGTCAAAATGCCCAAGAGTCACA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for VSV-G qPCR: 5′-TTTCCTTGCATTGTTCTACAGATGG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for GP qPCR (for detection of LCMV viral RNA): 5′- GCAACTGCTGTGTTCCCGAAAC-3′ |
Integrated DNA technologies | Peluso et al.86 |
| Forward primer for GP qPCR (for detection of LCMV viral RNA): 5′- CATTCACCTGGACTTTGTCAGACTC-3′ |
Integrated DNA technologies | Peluso et al.86 |
| Ace2 Taqman assay | Thermo Scientific | Mm01159006_m1 |
| Doublecortin Taqman assay | Thermo Scientific | Mm00438400_m1 |
| Gapdh Taqman assay | Thermo Scientific | Mm99999915_g1 |
| Maoa Taqman assay | Thermo Scientific | Mm00558004_m1 |
| Vil1 Taqman assay | Thermo Scientific | Mm00494146_m1 |
|
| ||
| Software and algorithms | ||
|
| ||
| Agilent software | Agilent | N/A |
| Bioconductor v.3.8 | Bioconductor | https://www.bioconductor.org/ |
| Biorender | Biorender | https://biorender.com/ |
| Flowjo v10.6.2 | BD | https://www.flowjo.com/ |
| GSEA | Broad institute | https://www.gsea-msigdb.org/ |
| ImageJ v2.1.0/1.53c | NIH | https://imagej.nih.gov/ij/ |
| Kallisto v.0.46.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| Olympic cellSens imaging software | Olympus LS | https://www.olympus-lifescience.com/ |
| Prism v9.3.0 | Graphpad | https://graphpad.com |
| RStudio v.1.2.5019 | The R foundation | https://www.r-project.org/ |
|
| ||
| Other | ||
|
| ||
| 1% Glycyl-L-tryptophan Diet | Envigo | TD.210749 |
| 2″ binder clips | Amazon | ASIN B07C94YCR5 |
| Advanced DMEM | Corning | MT10013CV |
| Amino acid control diet | Envigo | TD.01084 |
| B27 supplement | Gibco | 17504044 |
| BioDAQ cages | Research Diets, Inc. | N/A |
| DAPI mounting media | Electron Microscopy Sciences | 17985–50 |
| DNA/RNA Shield Fecal Collection Tubes | Zymo Research | R1137 |
| Elmer’s 0.77 oz glue sticks | Amazon | #E517 |
| FBS | Corning | MT35–010-CV |
| Immulon 4 HBX ELISA plates | Thermo Fisher Scientific | 3855 |
| Matrigel (GFR) | BD Biosciences | BD356231 |
| M-MLV Reverse Transcriptase (200 U/μL) | Invitrogen | 28025013 |
| Neurobasal-A medium | Thermo Fisher Scientific | 10888022 |
| Quantitative Synthetic SARA-CoV-2 RNA: ORF, E, N | ATTC | VR-3276SD |
| Random primers | Invitrogen | 48190011 |
| Serum gel tubes | Sarstedt | 41.1500.005 |
| Syringe-driven filter units, 0.22 μm low protein binding durapore membrane | Millipore | SLGVR33RS |
| Taqman Fast Advanced Master Mix | Thermo Scientific | 4444557 |
| TrypLE Express | Thermo Fisher Scientific | 12604013 |
| Tryptophan-deficient diet | Envigo | TD.130674 |
| Vacutainer EDTA tubes | BD | 365974 |
METHOD DETAILS
Mice
C57BL/6J (000664), TLR3−/− (005217), IFNAR1−/− (028288), K18-HuACE2 (034860), STAT1−/− (012606), Phox2b-Cre (016223), and IDO1−/− (005867) mice were purchased from The Jackson Laboratory. ACE2−/− (18180) mice were purchased from Taconic Biosciences. STAT1fl/fl Villin-creERT2 mice were obtained by crossing Villin-creERT2 mice (The Jackson Laboratory 020282) with STAT1fl/fl mice.85 At the beginning of each experiment, mice were randomly allocated into experimental groups. In all experiments, age- and sex-matched mice were used. In cases where littermates were not used, mice were cohoused to ensure consistency of common microbiota and genetic background. Mice were 5–12 weeks of age at the beginning of experiments. Both male and female mice were used for experiments, but within each experiment, they were sex matched. Mice were housed at 22.2°C and 52.1% humidity. Mice were given access to food and water ad libitum and were maintained under a 12 h light–dark cycle. All mice were maintained in filter-topped cages and given autoclaved food and water at the University of Pennsylvania University Laboratory Animal Resources (Penn ULAR) facility. All experiments were performed in accordance with the guidelines of the respective facilities and were approved by the regulations of the local institutional animal care and use committee (IACUC). No methods were used to predetermine sample size; rather, sample sizes were determined by pilot experiments to assess effect sizes and variability.
Poly(I:C) treatment
Except where noted, mice were intraperitoneally injected with 200 μg of low molecular weight (LMW) poly(I:C) (InvivoGen) once a day for 5 consecutive days, with the last injection occurring 3 h before sacrifice. High molecular weight (HMW) (InvivoGen) poly(I:C) and poly(I:C) from Sigma were also used where noted.
Novel object recognition test
Mice were allowed to acclimate in a rat cage with bedding for 1 h before testing. Following acclimation, mice were allowed to explore an object (glue sticks and 2″ binder clips) for 10 min. One hour after exposure to the familiar object, mice were allowed to explore the familiar object and the novel object for 10 min. Novel and familiar objects were randomized between mice. Objects were tested previously to ensure no inherent preference by mice. Interaction was defined as sniffing or direct contact with paws (excluding climbing and chewing behavior). Interaction time with each object was recorded until 30 s of total interaction time was reached. Mice taking longer than 10 min to reach 30 s of total interaction time between the two objects were excluded. The researcher was blinded to treatment groups during testing.
For novel object stimulation prior to sacrifice for hippocampus and NTS analysis, mice were allowed to explore an object (custom made 100 mL glass bottle filled with purple-colored water) for 10 min. One hour after exposure, mice were sacrificed, and brains were fixed for imaging or hippocampus was dissected out and snap frozen in liquid nitrogen and stored at −80°C for downstream analysis.
Radiolabeled tryptophan measurements
Tryptophan extraction and derivatization
Mice were fasted for 36 h before an oral administration of 13C11 L-tryptophan (200 mg/kg body weight). 30 min later, blood was collected via cardiac puncture and serum was snap frozen for downstream analysis. Ileal content was also collected, snap frozen, and weighed. Tryptophan was quantified via ELISA. Percent enrichment of labeled tryptophan was determined as follows: 10 μL of 100 μM norvaline was added to 20 μL of serum. 300 μL of 100% ice-cold acetone was then added to the serum and norvaline mixture and centrifuged at 10,000 x g for 10 min at 4°C. The supernatant containing metabolites was then dried by SpeedVac. The pellet was resuspended in 100 μL N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide and heated at 70°C for 1.5 h to derivatize. After derivatization, samples were centrifuged at 10,000 x g for 5 min at room temperature and the supernatant was transferred to a GC-MS vial with a volume reducing insert for analysis.
GC-MS protocol and tracing analysis
1 μL of sample was injected on splitless mode with an initial temperature of 60°C held for 1 min. The temperature increased at 10°C per minute up to 320°C. Analysis was performed on an Agilent 7890A series GC using a DB-5MS column coupled to a 5975C MSD. Isotopologue abundance was calculated using fluxfix and unlabeled samples from matched tissue. For tryptophan, the 244 m/z and 489 m/z ions of the 3TBDMS derivative were used for total carbon enrichment. Samples were analyzed up to m+11 (489 m/z and 244 m/z) to account for natural abundance.
In vivo treatments
5-HTP
5-hydroxy-L-tryptophan was administered in drinking water at 1.5 mg/mL for 5 days.
Capsaicin
Capsaicin was dissolved at 25 mg/mL in 10% Tween-80, 10% ethanol, and 80% PBS. 200 μL of capsaicin was injected intraperitoneally at 2 μM once daily for 5 days.
Phenelzine
Phenelzine sulfate salt was injected intraperitoneally at 50 mg/kg body weight once daily for 5 days.
680C91
680C91 was dissolved in DMSO and administered via oral gavage at 7.5 mg/kg body weight once daily for 5 days.
Fluoxetine
Fluoxetine oral solution was administered in drinking water at 160 mg/L for 5 weeks.
PCPA
pCPA methyl ester hydrochloride was dissolved in PBS and administered intraperitoneally at 300 mg/kg body weight once daily for 5 days.
m-CPBG
1-(3-Chlorophenyl)biguanide hydrochloride was dissolved in PBS and administered intraperitoneally 10 mg/kg body weight once daily for 5 days.
CNO
Clozapine N-oxide hydrochloride was administered intraperitoneally at 2 mg/kg body weight once daily for 5 days.
Tamoxifen
Tamoxifen was dissolved in corn oil and 1 mg of tamoxifen was administered to STAT1fl/fl Villin-creERT2 mice via oral gavage once daily for 4 consecutive days. Poly(I:C) or vehicle control injections were started one week after the last tamoxifen injection.
Anti-IFNAR1
500 μg of IFNAR1 blocking antibody was administered intraperitoneally one day before the first poly(I:C) injection (day −1) and at the start of poly(I:C) injections (day 0). 250 μg of IFNAR1 blocking antibody was then administered on day 1 and day 3 of poly(I:C) injections.
AAV injections
pAAV-hSyn-DIO-hM3Dq-mCherry (Addgene) was administered intravenously at 1011 PFU/mouse to Phox2B-Cre−/− or Phox2B-Cre+/− mice. 2 weeks later, mice were injected with CNO and poly(I:C) once a day for 5 consecutive days.
Patients, participants, and clinical data collection
Acute and recovered cohort
Plasma samples were obtained from a patient cohort previously described.83 Briefly, plasma samples were collected from patients admitted to the Hospital of the University of Pennsylvania with a positive SARS-CoV-2 PCR test between March and May 2020. Recovered donors self-reported a prior SARS-CoV-2 positive PCR test and met the definition of recovered as defined by the Centers for Disease Control and Prevention. Patients with acute SARS-CoV-2 infection were categorized as having moderate or severe disease based on admittance to the intensive care unit (ICU) (moderate patients were not admitted to the ICU, severe patients were hospitalized and admitted to the ICU). This sample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 808542. For metabolomics analysis, samples were heat-inactivated at 56°C for 1 h. For serotonin ELISA measurements, samples were not heat-inactivated. Participants provided written informed consent before inclusion in the study.
UPenn PASC cohort
Plasma samples were obtained from 58 patients with PASC seen at the Hospital of the University of Pennsylvania and Presbyterian Hospital. This biosample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 849140. Briefly, a blood sample was obtained from each patient and a questionnaire was obtained within 24 h of blood sample collection. For metabolomics analysis, samples were heat-inactivated at 56°C for 1 h. For serotonin ELISA measurements, samples were not heat-inactivated. Participants provided written informed consent before inclusion in the study. Participants were not offered any monetary compensation for participation. For symptom clustering analysis, questionnaire data from 1,540 individuals was used, UMAP coordinates were calculated, and average symptom levels per cluster were determined.
Viremia cohort
Plasma samples were obtained from a patient cohort previously described.84 Briefly, plasma samples were obtained from subjects admitted to the intensive care unit (ICU) with sepsis within 24 h of ICU admission. Source of sepsis was adjudicated by critical care physician investigators. This biosample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 808542. Participants provided written informed consent before inclusion in the study.
Healthy cohort
Plasma samples were obtained from healthcare workers in the apheresis unit at the Hospital of the University of Pennsylvania. This biosample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 843812. Participants provided written informed consent before inclusion in the study.
UCSF LIINC cohort
Plasma samples were obtained from a patient cohort previously described.14 Briefly, plasma samples were collected 90–160 days after the first positive SARS-CoV-2 quantitative PCR result. SARS-CoV-2 was not detected in the saliva of these patients at the time of sampling.86 Patients were divided into two groups based on symptom assessment at the time of sampling: patients with no COVID-19 attributed symptoms (recovered) and patients with two or more COVID-19 attributed symptoms (PASC). Individuals reporting one COVID-19 attributed symptom were not included.
RUSH PASC cohort
Plasma samples were obtained from a patient cohort previously described.27 Briefly, plasma samples were obtained from individuals with COVID-19 experiencing PASC symptoms 3–4 months after acute COVID-19.
UNCOVR cohort
Plasma samples were obtained from a patient cohort previously described.13 Briefly, plasma samples were obtained from individuals who had previously experienced acute COVID-19 at various time points (2–3, 6, 12, 18, and 24 months post-acute infection). Patients with 2 or more symptoms at the time of sample collection were defined as having PASC at that time point. Patients with 0 symptoms at the time of sample collection were defined as recovered. In order to compare the number of PASC symptoms experienced by patients with PASC in the Penn cohort and the UNCOVR cohort, questionnaires were compared and only questions that appeared on both questionnaires were taken into consideration. This totaled to be 26 questions from each questionnaire.
Human tissues
Material from 6 autopsies of patients who died of COVID-19 were obtained from family-consented research-only autopsies performed by the Department of Pathology and Laboratory Medicine at the Hospital of the University of Pennsylvania. Tissues were collected and placed in Trizol and processed for qPCR analysis of total RNA in BSL3 as approved by EHRS. Individuals were then categorized as having died during the acute phase of COVID-19 (within two weeks of the infection) or the post-acute phase of the infection (greater than two weeks after the infection).
Stool samples
Samples were processed as previously described.17 Healthy donor biosample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 833761. Long COVID biosample collection study was approved by the University of Pennsylvania Institutional Review Board, protocol number 849140. Briefly, stool samples were collected in tubes with DNA/RNA shield. Fecal samples were processed within 24 h of receipt by the lab. Upon receipt, samples were homogenized by vortexing for 30 s. Each sample was then aliquoted into cryovials, labeled with the patient ID, and then frozen at −80°C. Samples were thawed and centrifuged at 4000 x g for 10 min at 4°C and the supernatant was sterile filtered through 0.22 μm low protein binding durapore membranes. 140 μL of the filtered supernatant was transferred to a fresh Eppendorf tube for RNA extraction using the QiaAMP Viral RNA Mini kit. RNA extraction was performed as per manufacturer’s protocol and eluted in 60 μL of the elution buffer EB from the kit. Extracted RNA was stored at −80°C until further analysis.
Quantification of SARS-CoV-2 viral copies in stool samples
For cDNA synthesis, reverse transcription was performed with random primers and Moloney murine leukemia virus (M-MLV) reverse transcriptase. Synthesized SARS-CoV-2 RNA was used as a standard. Gene-specific primers to SARS-CoV-2 (Wuhan v1, NSP14) and SYBR green master mix were used to amplify viral RNA, and 18S rRNA primers were used to amplify cellular RNA using the QuantStudio 6 Flex RT–PCR system. Copy numbers of viral RNA were calculated using the absolute standard curve method.
Organoids
Tissues from the small intestine were washed with ice-cold PBS, opened longitudinally, and cut into 2 mm pieces. Intestinal pieces were then pipetted up and down three times in ice-cold PBS, and the PBS was then removed. This process was repeated 15–20 times. Crypts were then mechanically separated by shaking in HBSS-EDTA (10 mM) for 15 min and were then filtered through a 70 μm strainer into a 50 mL conical tube. Isolated crypts were embedded in Matrigel. Organoids were grown in a modified form of establishment media (described previously)87,88 for 3 days and then cultured in differentiation media (described previously)89 for two days. Organoids were maintained through passaging by adding ice-cold PBS to the Matrigel plug and digesting with TrypLE Express at 37°C for 2 min. Organoids were treated on day 5 of culture with 20 μg/mL LMW poly(I:C) for 4 h. Organoids were incubated with 1 ng/mL of recombinant mouse IFN-β1 or IFN-α for 4 h. To investigate the effects of NF-κB inhibition, organoids were treated with 1.5 μM IKK-16 or vehicle control for 2 h prior to poly(I:C) treatment.
Paired food intake and food gavage
Paired food intake
To ensure control and poly(I:C)-treated mice consumed the same amount of food daily, one cage containing 5 mice was given 3 g of food each day. Researcher confirmed that 100% of the food was eaten each day. To avoid competition between cage mates, only female mice were used in paired feeding experiments.
Food gavage
14 g of food (5010 rodent diet) was crushed using a mortar and pestle, passed through a metal sieve, and dissolved in 40 mL of sterile water. The mixture was then filtered through a 70 μm filter. Mice were given 300 μL of the food mixture or water via oral gavage, twice a day (morning and evening).
BioDAQ cages
Food intake was measured using BioDAQ food and water monitoring system cages (Research Diets, Inc.). Mice were allowed to acclimate for 3 days before daily injections of poly(I:C).
Platelet depletion
Platelets were depleted using a mixture of purified rat monoclonal antibodies directed against mouse GPIbα (CD42b). Control mice were injected with a mixture of non-immune rat antibodies (IgG). Mice were injected with 12.5 μg of antibody intravenously. 24 h post injection, mice were evaluated for serotonin depletion and novel object recognition.
Platelet aggregation FACS
Platelet aggregation was measured as previously described.90 Briefly, blood was collected via cardiac puncture into EDTA-coated tubes. Blood was diluted with 2X volume HEPES medium and PRP was collected after a 15-min spin at 50 x g. Platelets were counted and adjusted to equal concentrations between experimental conditions. PRP was divided into two equal portions. One portion was stained with PE-CD9 (1:100) and one portion was stained with APC-CD9 (1:100) for 15 min. Samples were then spun at 2250 x g for 5 min and resuspended in HEPES medium. The two singly stained portions for each sample were mixed 1:1 (volume:volume) before analysis using an LSR flow cytometer. Platelet aggregates were defined as APC+ PE+ cells.
Western blots
Intestinal epithelial cells were harvested by incubating ileal sections in 3 μM EDTA and 1.5 μM DTT on ice for 20 min. Ileal sections were then removed and incubated at 37°C for 10 min in 3 μM EDTA. Tubes were shaken for 30 s to release epithelium from basement membrane. Remnant tissue was removed, and the epithelial cells were pelleted by centrifugation at 800 x g for 5 min at 4°C. The IEC pellet was resuspended in RIPA buffer [0.1% SDS, 150 mM NaCl, 50 mM tris-HCl (pH 8.0), 0.5% sodium deoxycholate, and 1% NP-40] supplemented with a protease inhibitor cocktail (Sigma-Aldrich). Organoids were harvested by removing media and dissociating the Matrigel plug with ice-cold RIPA buffer. The cells were then centrifugated at 13,000 rpm for 15 min at 4°C, and cell lysates were used to measure protein concentration using the BCA kit. A protein concentration of 20 μg was then incubated with 5X sample buffer at 100°C for 10 min. Proteins were then separated on 4–15% SDS-polyacrylamide gel electrophoresis gels (Bio-Rad) and transferred to Immobilon-P polyvinylidene difluoride (PVDF) transfer membranes (Millipore). The membranes were blocked with 5% milk in trisbuffered saline with 0.1% Tween 20 (TBST) for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. After three 5-min washes with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:1000) for 1 h at room temperature. Membranes were then developed with ECL Western blotting reagents (Amersham). The signals were visualized using Amersham Imager 680 (Amersham).
Nodose ganglion extraction, culture, and calcium imaging
Nodose ganglia were extracted and cultured as previously described.91,92 Nodose ganglia were collected into Neurobasal-A medium and dissociated in 1 mg/mL collagenase 1A for 1 h at 37°C in serum-free media containing Advanced DMEM, sodium pyruvate solution, and HEPES buffered saline. Nodose ganglion were washed and triturated with glass Pasteur pipettes 60 times and centrifuged 500 x g at 4°C for 5 min. The cell pellet was resuspended into culture media (10% FBS, Neurobasal-A medium, B27 supplement, 50 ng/mL nerve growth factor [NGF], and penicillin/streptomycin) and subsequently seeded onto poly-L-lysine coated (at least 2 h at 37°C, 5% CO2) 96 well plates and cultured overnight at 37°C, 5% CO2 incubator. For calcium imaging, the nodose ganglion were washed with fresh serum-free culture media and loaded with 1 μM Fura-2-AM for 1 h at 37°C. The cells were then washed into modified extracellular Ringer’s solution containing 145 mM NaCl, 4.7 mM KCl, 3.4 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4,1 mM MgCl2, 10 mM glucose, and 10 mM HEPES. Baseline was measured prior to addition of 50 μL of either 2 μM capsaicin or 1 μM 5-HT, which were directly applied onto neurons. Imaging was conducted immediately in a plate reader at room temperature with wavelengths set to 340, 380, and 510 nm.
Intestinal immunofluorescence
Intestinal sections were fixed overnight in 10% formalin and stored in 70% ethanol before sectioning. Sections were deparaffinized and re-hydrated using serial 5-min incubations in xylene and ethanol gradient (100%–70%). The slides were then washed in PBS and antigen retrieval was performed in citrate buffer (10 mM citrate, pH 6) at 95°C for 1 h. The slides were then washed again in PBS and blocked in 20% normal goat serum and 0.05% Triton X-100 in PBS for 30 min. Sections were incubated with anti-chromogranin A primary antibody at 1:200 overnight at 4°C. Sections were washed with PBS and incubated with goat anti-rabbit AF488 at 1:400 for 2 h at room temperature. Finally, sections were washed and mounted with DAPI mounting media. Images were acquired using a Nikon fluorescence microscope. To quantify chromogranin A positive cells, the number of positive cells were counted in 5, 10x fields and averaged.
Hippocampus and nucleus tractus solitarius (NTS) immunofluorescence
Mice were terminally anesthetized with 2,2,2-Tribromoethanol (Avertin) and perfused with ice-cold PBS and 4% paraformaldehyde (PFA) before decapitation and brain dissection. Brains were then fixed in 4% paraformaldehyde for 4 h at 4°C. For free floating IF sections, brains were cut coronally at 70 μm using a Leica 1000S Vibratome. Free-floating sections were stained overnight at 4°C in primary antibody in PBS with 0.1% Triton and 1% bovine serum albumin (BSA). The primary antibodies used were cFos (1:1500), Ki67 (1:500), doublecortin (1:500), NeuN (1:500). Sections were washed three times in PBS and incubated with secondary antibodies (1:500) for 1 h at 37°C. After three washes in PBS, the sections were mounted onto charged glass slides and covered with coverslips with Vectashield antifade DAPI aqueous mounting medium. Sections were imaged on a Zeiss LSM 710 confocal microscope with a 1030.45 NA objective. The entire thickness of the section was imaged at 5 μm intervals and maximum intensity projections were used for analysis. cFos and Ki67 positive cells were quantified using ImageJ. Each data point is a single mouse.
To quantify doublecortin positive cells, we utilized an artificial neural network (ANN)-based approach. In brief, the network was loosely based on a yolo3 architecture with a total of 120 layers. It contained 79 convolutional layers built with consecutive 3×3 and 1×1 filters followed by a skip connection to help activations propagate through deeper layers without gradients diminishing. The network was entrained by manually providing cell counts of several sets of microscopic images of neurons as a ground truth and accuracy was further improved by giving manual feedback on the ANN-outputs on nontrained datasets.
Metabolomics
For targeted LC/MS metabolomics of amino acids in plasma, 100 μL aliquots of plasma on ice were spiked with isotopically-labelled amino acid internal standards and extracted with ice-cold methanol according to validated, optimized protocols in a previously published study.93 Extracted amino acids were derivatized using a Waters AccQTag Ultra derivatization kit. Separation and quantitation of derivatized amino acids was achieved using multiple reaction monitoring of calibration solutions and study samples on an Agilent 1290 Infinity UHPLC/6495 triple quadrupole mass spectrometer.93,94 Raw data were processed using Mass Hunter quantitative analysis software (Agilent). Calibration curves (R2 = 0.99 or greater) were either fitted with a linear or a quadratic curve with a 1/X or 1/X2 weighting.
For the integrative analysis of metabolomics datasets in acute COVID-19, publicly available data were integrated by calculating fold change rates between COVID-19 patients and healthy controls across five studies.7–11 Only metabolites detected in at least two studies were considered. Metabolites were than ranked by their average fold change from all five studies.
VSV infections
Vesicular stomatitis virus (Indiana strain) was intravenously administered 2 × 107 PFU. Since VSV does not produce severe infection in C57BL/6 mice, 500 μg anti-mouse IFNAR-1 antibody was injected intraperitoneally to control and VSV-infected mice, one day before the infection and on the day of the infection. Additionally, 250 μg of anti-IFNAR-1 was administered 1 day post infection. Mice were sacrificed 24–48 h post infection.
LCMV infections
LCMV Armstrong and LCMV clone 13 were grown in BHK cells (ATCC) and titers calculated by plaque assay as previously described.95 For acute infection, mice were injected intraperitoneally with 2 × 105 plaque-forming units of LCMV Armstrong in RPMI supplemented with 1% Fetal Bovine Serum. For chronic infection, mice were injected intravenously with 4 × 106 PFU of LCMV clone 13 in RPMI supplemented with 1% FBS.
Diets
Tryptophan-deficient diet and control amino acid diet were custom ordered from Envigo. Diets were matched for their source of macro- and micronutrient content and differed only in their tryptophan content. Glycine-tryptophan dipeptide diet was custom ordered from Envigo. Glycyl-L-tryptophan hydrate was supplemented in the diet at 10 mg dipeptide/1 g diet on the 5015 diet background (Envigo). Un-supplemented 5015 diet was used as the control diet.
SARS-CoV-2 mouse infections
Virus
SARS-CoV-2 (Isolate USA-WA1/2020) was obtained from BEI Resources (NR-52281) and infectious stocks were grown in Vero-E6 cells (ATCC) and stored at −80°C. SARS-CoV-2 (Isolate B.1.351) for mouse studies was obtained from Andy Pekosz. Infectious stocks were grown in Vero-Ace2-Tmprss2 cells (BEI Resources) and stored at −80°C. All work with infectious virus was performed in a biosafety level 3 laboratory and approved by the Institutional Biosafety Committee and Environmental Health and Safety.
Mouse infections
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania (protocol number 807017). Virus inoculations were performed under anesthesia which was induced and maintained with ketamine hydrochloride and xylazine. Animals were housed in groups and fed standard chow diets. Mice of different ages and both sexes (age and sex-matched within experiments) were administered 1 × 103 or 1 × 105 plaque-forming units (PFU) of SARS-CoV-2 isolate USA-WA1/2020 or isolate B.1.351, respectively, via intranasal administration.
Tryptophan gavage
Mice were fasted for 36 h and then given 200 mg/kg body weight L-tryptophan (dissolved in water at pH 3 and heated at 95°C until complete dissolution) via oral gavage. For analysis of tryptophan levels across time, plasma was collected via cheek bleed before gavage, and 30 and 60 min after.
PTT and aPTT
Blood was collected via cardiac puncture into tubes containing 3.2% trisodium citrate. The final concentration was 9 parts blood and 1 part sodium citrate. Samples were stored at room temperature until centrifugation and were centrifuged at 2000 x g for 10 min at room temperature within 30 min of collection. Plasma was collected and stored at −80°C until analysis. Coagulation testing was done at the IDEXX BioAnalytics N. Grafton, MA Laboratory on 3.2% sodium citrate samples using an STAGO STA Compact Max automated coagulation analyzer.
Flow cytometric assessment of platelet activation
Platelet activation and whole-blood cytometry was performed as described previously,96 with a few modifications. Briefly, 200 μL of whole blood was collected via cardiac puncture into tubes containing 25 μL of 3.8% trisodium citrate and 0.4 mM Gly-Pro-Arg-Pro (GPRP). Samples were centrifuged at 350 x g for 10 min at room temperature and 30 μL of the top layer (platelet-rich plasma) was carefully collected with a wide-bore pipette tip and transferred to a separate tube. 120 μL of room temperature PBS−/− was added and samples were centrifuged at 4500 x g for 10 min at room temperature. The supernatant was pipetted off and the platelet pellet was carefully resuspended in FACS buffer containing 0.4 mM GPRP. Half of each sample was stimulated with 0.1 U/mL thrombin at room temperature for 30 min and stained with a 1:100 dilution of rat mAb against mouse P-selectin, labeled with FITC, and a 1:100 dilution of rat mAb against mouse CD41, labeled with APC. Samples were washed with 1 mL of FACs buffer and fixed with 1% PFA on ice for 2 h before analysis using a FACSCanto II flow cytometer.
Hematology analysis
Initial hematology testing for control and poly(I:C)-treated mice was done at the IDEXX BioAnalytics N. Grafton, MA Laboratory. 150 μL of blood was collected via cardiac puncture into tubes containing 15 μL 0.5 M EDTA. Blood samples were kept at room temperature and CBCs were performed within 24 h of collection. Whole blood samples were analyzed using a Sysmex XT-iV automated hematology analyzer with the platelet count also checked by manual blood smear for clumping to ensure sample quality by technicians trained in murine hematology analysis. Subsequent CBCs were performed using The VetScan HM5 (Abaxis). Blood samples were kept at room temperature and CBCs were performed the same day as collection.
Megakaryocyte analysis
Femurs were fixed in 10% formalin for 48 h and then transferred to 20% EDTA (pH 7.4) and kept at 4°C with gentle agitation for 4 weeks. The EDTA solution was changed every 2 weeks. Femurs were then transferred to 70% ethanol, paraffin embedded, sectioned, and stained with hematoxylin and eosin. Evaluation was initially performed blinded to experimental group, with unblinding after evaluation to assist with interpretation of group differences. Megakaryocytes were counted by the number of individual megakaryocytes per single 400x high power field, with sum totals and average given for ten consecutive fields. Whenever possible, evaluation was done within the diaphysis, starting at the most proximal aspect, and extending distally. Using Olympic cellSens imaging software, the glass slides were examined for expedited photomicrographs as well as manual measurement of megakaryocyte diameter. Cells were measured at their widest points.
Platelet and platelet-rich-plasma isolation
Blood was collected via cardiac puncture into Vacutainer EDTA tubes and maintained at room temperature until centrifugation. Samples were centrifuged at 200 x g for 10 min at room temperature. 30 μL of the upper layer of platelet-rich-plasma was added to 120 μL of PBS−/−. Samples were then centrifuged at 4500 x g at 4°C for 10 min and the upper 120 μL was discarded without disturbing the platelet pellet. Samples were stored at −80°C for downstream analysis.
Metabolite and soluble factor measurements
Samples within experiments were run at the same time, on the same lot number of kits for all ELISA measurements. In cases where less than the required volume was available for individual mice, samples from 2 or more mice from the same experimental group were pooled.
Serotonin
Plasma, isolated platelets, platelet-rich-plasma, and perfused brains were prepared according to manufacturer instructions and serotonin was quantified with Serotonin ELISA kits. Brains were snap frozen in liquid nitrogen and stored at −80°C for downstream analysis. Whole brains were homogenized in PBS−/− and serotonin levels were normalized to total BCA protein.
Tryptophan
Plasma, platelet-rich-plasma, serum, and ileum content were prepared according to manufacturer instructions and tryptophan was measured with Tryptophan ELISA kits. Ileum content was normalized to total weight of the sample.
5-HIAA
Urine was prepared according to the manufacturer instructions and 5-HIAA was measured using 5-HIAA ELISA kits.
Kynurenine
Plasma and livers were prepared according to the manufacturer instructions and kynurenine was measured with Kynurenine ELISA kits. Liver samples were homogenized in PBS−/− and kynurenine levels were normalized to total BCA protein.
Tissue factor
Plasma was prepared according to the manufacturer instructions and tissue factor was measured using Tissue factor ELISA kits.
Fibrinogen
Plasma was prepared according to the manufacturer instructions and fibrinogen was measured using Tissue factor ELISA kits.
TAT complexes
Plasma was prepared according to the manufacturer instructions and Thrombin-antithrombin complexes were measured using Thrombin-Antithrombin Complexes ELISA kits.
BCA protein quantification
Brains and livers were homogenized in PBS−/− and total protein levels were quantified using the Pierce BCA Protein Assay Kit.
RNA sequencing analysis of human organoids
Relative expression of ACE2 and SLC6A19 in human small intestinal organoids infected with SARS-CoV-2 was obtained from publicly available RNA sequencing data.30 Briefly, data was analyzed from human intestinal organoids, which were grown in differentiation media and infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 1. Organoids were incubated for 24 h and transcripts from uninfected organoids were compared to infected.
Transcriptional profiling by bulk-RNA sequencing
Libraries were prepared using the Illumina TruSeq stranded mRNA kit with IDT for Illumina TruSeq Unique Dual indexes according to the manufacturer’s instructions. Quality and quantity control of RNA and libraries were performed using Agilent 4200 TapeStation and Qubit 4, respectively. Libraries were sequenced on an Illumina NextSeq 550 to produce 75-base pair single-end reads with an average sequencing depth of 7 million reads per sample. Raw reads were mapped to the mouse reference transcriptome (Ensembl; Mus musculus version 67) using Kallisto version 0.46.0. Subsequent analysis was carried out using the statistical computing environment R version 3.6.1 in RStudio version 1.2.5019 and Bioconductor version 3.8. Briefly, transcript quantification data were summarized to genes using the tximport package and normalized using the trimmed mean of M values (TMM) method in edgeR. Genes with <1 CPM in n+1 of the samples, where n is the size of the smallest group of replicates, were filtered out. Differentially expressed genes were identified with linear modeling using limma (FDR ≤0.05; absolute logFC ≥1) after correcting for multiple testing using Benjamini-Hochberg.
GSEA analysis
Differentially expressed transcripts from RNA-seq on ilea from control or poly(I:C) treated mice were compared to curated gene sets using GSEA software (Broad Institute) with the following parameters; Gene sets database: c5.all.v7.5.1.symbols.gmt [Gene ontology], Number of permutations: 1000, Permutation type: phenotype, Chip platform: Mouse_Gene_Symbol_Remapping_Human_Orthologs_MSigDB.v.7.5.1.chip.97,98
Single-cell RNA-sequencing analysis
Single cell datasets of mouse nodose ganglion was obtained from GEO (Accession number: GSE124312).52 Data was analyzed with Seurat v4.99 Data was normalized using SCTransform,100 clustered, and visualized using UMAP. Clusters were annotated and reproduced as described.52 Nodose ganglion neurons were subsetted, and expression levels for each gene across all nodose ganglion neurons was visualized using the DotPlot() function in Seurat.
Quantitative real-time PCR
Total RNA from tissues and organoids was extracted using TRIzol and RNAeasy mini kits, respectively. RNA was reversed transcribed using High-Capacity cDNA Reverse Transcription kits. RT-qPCR was performed using QuantiFast SYBR Green PCR kit, New England Biolabs LUNA Universal PCR kit, or Taqman Fast Advanced Master Mix. RT-qPCR was performed on an Applied Biosystems CFX96 machine.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as means ± SEM. Replicates represent biologically independent samples. In the figures, asterisks denote statistical significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) as assessed by two-tailed Mann-Whitney U test; unpaired two-tailed or one-tailed t test; one-way ANOVA with Tukey’s multiple comparisons test, Dunn’s multiple comparisons test, Šidák’s multiple comparisons test, or Dunnett’s multiple comparisons test; Kruskal-Wallis test with Tukey’s multiple comparisons test or Dunn’s multiple comparisons test; one-tailed linear regression, or hypergeometric test where appropriate. To determine the classification errors of a binary classifier, we calculated the Receiver Operating Characteristics (ROC) and determined the area under the sensitivity/specificity tradeoff curve. UMAP clustering of 1,540 individuals with PASC was performed in R using a matrix of patient records. Statistical analysis was performed in GraphPad PRISM 9 and Microsoft Excel. Graphics were generated in BioRender and Adobe Illustrator.
Supplementary Material
Highlights.
Long COVID is associated with reduced circulating serotonin levels
Serotonin depletion is driven by viral RNA-induced type I interferons (IFNs)
IFNs reduce serotonin through diminished tryptophan uptake and hypercoagulability
Peripheral serotonin deficiency impairs cognition via reduced vagal signaling
ACKNOWLEDGMENTS
We thank all study participants and acknowledge James Heath (ISB) for providing samples from the INCOV/UNCOVR study. We also acknowledge the following individuals and cores at UPenn for their contributions to this study: Eline Luning Prak (Human Immunology Core), MESSI Cohort Study (supported by grants HL137006 and HL137915), COVID Processing Unit, PennMedicine Biobank, M. Risman, J. Weaver, M. Livingstone, Post-COVID Assessment and Recovery Clinic, Center for Human Phenomic Science, Molecular Pathology and Imaging Core, CDB Microscopy Core Facility, Bates laboratory, Hensley laboratory, Animal Biosafety Level 3 Core, Matthew Lanza (Vet Comparative Pathology Core), Collman laboratory, J. Graham-Wooten, Muzykantov laboratory, Lee laboratory, and R. Hess. The Vet Comparative Pathology Core is supported by the Abramson Cancer Center (P30 CA016520). The Human Immunology Core is supported by the NIH (P30-CA016520 and P30-AI045008), the Department of Pathology and Laboratory Medicine, and the Penn Center for Precision Medicine. We thank E. Krause, K. Scott, and A. de Kloet (University of Florida) for providing mice. We acknowledge grants T32-AI-055400-19 (A.C.W.); F31HL160065 and T32-AI-141393 (P.L.); Boehringer Ingelheim Fonds MD fellowship (Jihee Kim); Canadian Institutes for Health Research and the Fonds de Recherche du Québec - Santé (R.D.P.); Hartwell Foundation (M.J.L.); T32-AI-055400 (B.M.M.); Cancer Research Institute Irvington Postdoctoral Fellowship (S.L.P.); Cancer Immunotherapy Bridge fellowship (J.B.); Australia NHMRC C.J. Martin Fellowship GNT1111469 and Mark Foundation Momentum Fellowship (S.N.); NIH R01 DK123733 (M.A.-M., A.L., and A.K.); and NIH R01AA029859 (A.K. and M.A.-M.). E.J.W. is supported by The Parker Institute for Cancer Immunotherapy; NIH grants AI155577, AI149680, AI108545, AI082630 and CA210944; and the Polybio Research Foundation. M.P is supported by NIH/NIAID 3R01AI141003-03S1 and NIH/NIAID R01AI158013. C.A.T. is a Pew biomedical scholar and a Kathryn W. Davis Aging Brain scholar and is supported by the NIH Director’s New Innovator Award (DP2-AG-067492), the Agilent Early Career Professor Award, and grants from the Edward Mallinckrodt, Jr. Foundation, the IDSA Foundation, the Thyssen Foundation, Polybio Research Foundation, the Kenneth Rainin Foundation, the PennCHOP Microbiome Program, Penn Institute for Immunology, Penn Center for Molecular Studies in Digestive and Liver Diseases (P30-DK-050306), Penn Skin Biology and Diseases Resource-based Center (P30-AR-069589), Penn Diabetes Research Center (P30-DK-019525), Penn Institute on Aging, the Penn Institute for Infectious & Zoonotic Diseases, the University Research Foundation, the Dean’s Innovation Fund of the University of Pennsylvania, and a Borrelli Family Lynch Syndrome grant. M. Levy is supported by the NIH Director’s New Innovator Award (DP2-AG-067511), an American Cancer Society Scholar Award, The Pew Scholar Award, the Searle Scholars Program, the Edward Mallinckrodt, Jr. Foundation, the W. W. Smith Charitable Trust, the Prevent Cancer Foundation, Polybio Research Foundation, V Foundation, The Burroughs Wellcome Fund, and grants from the Abramson Cancer Center (P30-CA-016420), the PennCHOP microbiome program, Penn Center for Nutritional Science and Medicine, Penn Coronavirus Center, Penn Institute for Immunology, Penn Center for Molecular Studies in Digestive and Liver Diseases (P30-DK-050306), Penn Center for Precision Medicine, Penn Institute on Aging, Penn Center of Excellence in Environmental Toxicology (P30-ES-013508), and the Borrelli Family Pilot Grant in Lynch Syndrome.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community.
DECLARATION OF INTERESTS
E.J.W. is an advisor for Danger Bio, Janssen, New Limit, Marengo, Pluto Immunotherapeutics Related Sciences, Rubius Therapeutics, Santa Ana Bio, Synthekine, and Surface Oncology. E.J.W. is a founder of and holds stock in Surface Oncology, Danger Bio, and Arsenal Biosciences. N.J.M. reports consulting fees from Endpoint Health Inc and AstraZeneca and receives funding from Quantum Leap Healthcare Collaborative outside of the published work.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2023.09.013.
REFERENCES
- 1.Choutka J, Jansari V, Hornig M, and Iwasaki A. (2022). Unexplained post-acute infection syndromes. Nat. Med. 28, 911–923. 10.1038/s41591-022-01810-6. [DOI] [PubMed] [Google Scholar]
- 2.Al-Aly Z, Xie Y, and Bowe B. (2021). High-dimensional characterization of post-acute sequelae of COVID-19. Nature 594, 259–264. 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 3.Davis HE, McCorkell L, Vogel JM, and Topol EJ (2023). Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146. 10.1038/s41579-022-00846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merad M, Blish CA, Sallusto F, and Iwasaki A. (2022). The immunology and immunopathology of COVID-19. Science 375, 1122–1127. 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 5.Pretorius E, Vlok M, Venter C, Bezuidenhout JA, Laubscher GJ, Steenkamp J, and Kell DB (2021). Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 20, 172. 10.1186/s12933-021-01359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dani M, Dirksen A, Taraborrelli P, Torocastro M, Panagopoulos D, Sutton R, and Lim PB (2021). Autonomic dysfunction in ‘long COVID’: rationale, physiology and management strategies. Clin. Med. 21, e63–e67. 10.7861/clinmed.2020-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L, et al. (2020). Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 182, 59–72.e15. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi D, Yan R, Lv L, Jiang H, Lu Y, Sheng J, Xie J, Wu W, Xia J,Xu K, et al. (2021). The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism 118, 154739. 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song JW, Lam SM, Fan X, Cao WJ, Wang SY, Tian H, Chua GH, Zhang C, Meng FP, Xu Z, et al. (2020). Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 32, 188–202.e5. 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas T, Stefanoni D, Reisz JA, Nemkov T, Bertolone L, Francis RO, Hudson KE, Zimring JC, Hansen KC, Hod EA, et al. (2020). COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 5, e140327. 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao N, Nie M, Pang H, Wang B, Hu J, Meng X, Li K, Ran X, Long Q, Deng H, et al. (2021). Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 12, 1618. 10.1038/s41467-021-21907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadlier C, Albrich WC, Neogi U, Lunjani N, Horgan M, O’Toole PW, and O’Mahony L. (2022). Metabolic rewiring and serotonin depletion in patients with postacute sequelae of COVID-19. Allergy 77, 1623–1625. 10.1111/all.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, Li S, Hong S, Zhang R, Xie J, et al. (2022). Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185, 881–895.e20. 10.1016/j.cell.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peluso MJ, Kelly JD, Lu S, Goldberg SA, Davidson MC, Mathur S, Durstenfeld MS, Spinelli MA, Hoh R, Tai V, et al. (2022). Persistence, Magnitude, and Patterns of Postacute Symptoms and Quality of Life Following Onset of SARS-CoV-2 Infection: Cohort Description and Approaches for Measurement. Open Forum Infect. Dis. 9, ofab640. 10.1093/ofid/ofab640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger M, Gray JA, and Roth BL (2009). The expanded biology of serotonin. Annu. Rev. Med. 60, 355–366. 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasui F, Matsumoto Y, Yamamoto N, Sanada T, Honda T, Munakata T, Itoh Y, and Kohara M. (2022). Infection with the SARS-CoV-2 B.1.351 variant is lethal in aged BALB/c mice. Sci. Rep. 12, 4150. 10.1038/s41598-022-08104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan A, Zlitni S, Brooks EF, Vance SE, Dahlen A, Hedlin H, Park RM, Han A, Schmidtke DT, Verma R, et al. (2022). Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med (N Y) 3, 371–387.e9. 10.1016/j.medj.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swank Z, Senussi Y, Manickas-Hill Z, Yu XG, Li JZ, Alter G, and Walt DR (2022). Persistent circulating SARS-CoV-2 spike is associated with post-acute COVID-19 sequelae. Clin. Infect. Dis. 10.1093/cid/ciac722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zollner A, Koch R, Jukic A, Pfister A, Meyer M, Rössler A, Kimpel J, Adolph TE, and Tilg H. (2022). Postacute COVID-19 is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 163, 495–506.e8. 10.1053/j.gastro.2022.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peluso MJ, Ryder D, Flavell R, Wang Y, Levi J, LaFranchi BH, Deveau TM, Buck AM, Munter SE, Asare KA, et al. (2023). Multimodal Molecular Imaging Reveals Tissue-Based T Cell Activation and Viral RNA Persistence for Up to 2 Years Following COVID-19. Preprint at medRxiv.. 10.1101/2023.07.27.23293177. [DOI] [Google Scholar]
- 21.Goh D, Lim JCT, Fernaíndez SB, Joseph CR, Edwards SG, Neo ZW, Lee JN, Caballero SG, Lau MC, and Yeong JPS (2022). Case report: Persistence of residual antigen and RNA of the SARS-CoV-2 virus in tissues of two patients with long COVID. Front. Immunol. 13, 939–989. 10.3389/fimmu.2022.939989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phetsouphanh C, Darley DR, Wilson DB, Howe A, Munier CML, Patel SK, Juno JA, Burrell LM, Kent SJ, Dore GJ, et al. (2022). Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 23, 210–216. 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 23.Roager HM, and Licht TR (2018). Microbial tryptophan catabolites in health and disease. Nat. Commun. 9, 3294. 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danlos FX, Grajeda-Iglesias C, Durand S, Sauvat A, Roumier M, Cantin D, Colomba E, Rohmer J, Pommeret F, Baciarello G, et al. (2021). Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 12, 258. 10.1038/s41419-021-03540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdás A, Moreno LO, Rello SR, Orduña A, Bernardo D, and Cifuentes A. (2022). Metabolomics study of COVID-19 patients in four different clinical stages. Sci. Rep. 12, 1650. 10.1038/s41598-022-05667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, Gorman N, Palmer CS, Tang HY, Shaikh MW, et al. (2021). Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front. Immunol. 12, 686240. 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giron LB, Peluso MJ, Ding J, Kenny G, Zilberstein NF, Koshy J, Hong KY, Rasmussen H, Miller GE, Bishehsari F, et al. (2022). Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-kappaB signaling. JCI Insight 7, e160989. 10.1172/jci.insight.160989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fortier ME, Kent S, Ashdown H, Poole S, Boksa P, and Luheshi GN (2004). The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287, R759–R766. 10.1152/ajpregu.00293.2004. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X, Levasseur PR, Michaelis KA, Burfeind KG, and Marks DL (2016). A distinct brain pathway links viral RNA exposure to sickness behavior. Sci. Rep. 6, 29885. 10.1038/srep29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, et al. (2021). Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644. 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer D, Camargo SMR, Ramadan T, Schäfer M, Mariotta L, Herzog B, Huggel K, Wolfer D, Werner S, Penninger JM, and Verrey F. (2012). Defective intestinal amino acid absorption in Ace2 null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G686–G695. 10.1152/ajpgi.00140.2012. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. (2012). ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481. 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf K, Braun A, Haining EJ, Tseng YL, Kraft P, Schuhmann MK, Gotru SK, Chen W, Hermanns HM, Stoll G, et al. (2016). Partially Defective Store Operated Calcium Entry and Hem(ITAM) Signaling in Platelets of Serotonin Transporter Deficient Mice. PLoS One 11, e0147664. 10.1371/journal.pone.0147664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivadeneyra L, Pozner RG, Meiss R, Fondevila C, Gómez RM, and Schattner M. (2015). Poly (I:C) downregulates platelet production and function through type I interferon. Thromb. Haemost. 114, 982–993. 10.1160/TH14-11-0951. [DOI] [PubMed] [Google Scholar]
- 36.Lee E, Kim M, Jeon K, Lee J, Lee JS, Kim HS, Kang HJ, and Lee YK (2019). Mean Platelet Volume, Platelet Distribution Width, and Platelet Count, in Connection with Immune Thrombocytopenic Purpura and Essential Thrombocytopenia. Lab. Med. 50, 279–285. 10.1093/labmed/lmy082. [DOI] [PubMed] [Google Scholar]
- 37.Norrasethada L, Khumpoo W, Rattarittamrong E, Rattanathammethee T, Chai-Adisaksopha C, and Tantiworawit A. (2019). The use of mean platelet volume for distinguishing the causes of thrombocytopenia in adult patients. Hematol. Rep. 11, 7732. 10.4081/hr.2019.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmoeller D, Picarelli MM, Paz Munhoz T, Poli de Figueiredo CE, and Staub HL (2017). Mean Platelet Volume and Immature Platelet Fraction in Autoimmune Disorders. Front. Med. 4, 146. 10.3389/fmed.2017.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anabel AS, Eduardo PC, Pedro Antonio HC, Carlos SM, Juana NM, Honorio TA, Nicolás VS, and Sergio Roberto AR (2014). Human platelets express Toll-like receptor 3 and respond to poly I:C. Hum. Immunol. 75, 1244–1251. 10.1016/j.humimm.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 40.D’Atri LP, Etulain J, Rivadeneyra L, Lapponi MJ, Centurion M, Cheng K, Yin H, and Schattner M. (2015). Expression and functionality of Toll-like receptor 3 in the megakaryocytic lineage. J. Thromb. Haemost. 13, 839–850. 10.1111/jth.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morodomi Y, Kanaji S, Won E, Ruggeri ZM, and Kanaji T. (2020). Mechanisms of anti-GPIbalpha antibody-induced thrombocytopenia in mice. Blood 135, 2292–2301. 10.1182/blood.2019003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamal AH, Tefferi A, and Pruthi RK (2007). How to interpret and pursue an abnormal prothrombin time, activated partial thromboplastin time, and bleeding time in adults. Mayo Clin. Proc. 82, 864–873. 10.4065/82.7.864. [DOI] [PubMed] [Google Scholar]
- 43.Antunes M, and Biala G. (2012). The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn. Process. 13, 93–110. 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costello DA, and Lynch MA (2013). Toll-like receptor 3 activation modulates hippocampal network excitability, via glial production of interferon-beta. Hippocampus 23, 696–707. 10.1002/hipo.22129. [DOI] [PubMed] [Google Scholar]
- 45.Chen C, Gao R, Li M, Wang Q, Chen H, Zhang S, Mao X, Behensky A, Zhang Z, Gan L, et al. (2019). Extracellular RNAs-TLR3 signaling contributes to cognitive decline in a mouse model of postoperative cognitive dysfunction. Brain Behav. Immun. 80, 439–451. 10.1016/j.bbi.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Clark RE, Zola SM, and Squire LR (2000). Impaired recognition memory in rats after damage to the hippocampus. J. Neurosci. 20, 8853–8860. 10.1523/JNEUROSCI.20-23-08853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douaud G, Lee S, Alfaro-Almagro F, Arthofer C, Wang C, McCarthy P, Lange F, Andersson JLR, Griffanti L, Duff E, et al. (2022). SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature 604, 697–707. 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soung AL, Vanderheiden A, Nordvig AS, Sissoko CA, Canoll P, Mariani MB, Jiang X, Bricker T, Rosoklija GB, Arango V, et al. (2022). COVID-19 induces CNS cytokine expression and loss of hippocampal neurogenesis. Brain 145, 4193–4201. 10.1093/brain/awac270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alenina N, and Klempin F. (2015). The role of serotonin in adult hippocampal neurogenesis. Behav. Brain Res. 277, 49–57. 10.1016/j.bbr.2014.07.038. [DOI] [PubMed] [Google Scholar]
- 50.Richter-Levin G, and Segal M. (1996). Serotonin, aging and cognitive functions of the hippocampus. Rev. Neurosci. 7, 103–113. 10.1515/revneuro.1996.7.2.103. [DOI] [PubMed] [Google Scholar]
- 51.Winterer J, Stempel AV, Dugladze T, Földy C, Maziashvili N, Zivkovic AR, Priller J, Soltesz I, Gloveli T, and Schmitz D. (2011). Cell-type-specific modulation of feedback inhibition by serotonin in the hippocampus. J. Neurosci. 31, 8464–8475. 10.1523/JNEUROSCI.6382-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kupari J, Häring M, Agirre E, Castelo-Branco G, and Ernfors P. (2019). An Atlas of Vagal Sensory Neurons and Their Molecular Specialization. Cell Rep. 27, 2508–2523.e4. 10.1016/j.celrep.2019.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowe B, Xie Y, and Al-Aly Z. (2023). Postacute sequelae of COVID-19 at 2 years. Nat. Med. 29, 2347–2357. 10.1038/s41591-023-02521-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azzolini E, Levi R, Sarti R, Pozzi C, Mollura M, Mantovani A, and Rescigno M. (2022). Association Between BNT162b2 Vaccination and Long COVID After Infections Not Requiring Hospitalization in Health Care Workers. JAMA 328, 676–678. 10.1001/jama.2022.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Y, Bowe B, and Al-Aly Z. (2021). Burdens of post-acute sequelae of COVID-19 by severity of acute infection, demographics and health status. Nat. Commun. 12, 6571. 10.1038/s41467-021-26513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Aly Z, Bowe B, and Xie Y. (2022). Long COVID after breakthrough SARS-CoV-2 infection. Nat. Med. 28, 1461–1467. 10.1038/s41591-022-01840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palm W, and Thompson CB (2017). Nutrient acquisition strategies of mammalian cells. Nature 546, 234–242. 10.1038/nature22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Platten M, Nollen EAA, Röhrig UF, Fallarino F, and Opitz CA (2019). Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 18, 379–401. 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 59.Ghebrehiwet B, and Peerschke EI (2020). Complement and coagulation: key triggers of COVID-19-induced multiorgan pathology. J. Clin. Invest. 130, 5674–5676. 10.1172/JCI142780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasini E, Corsetti G, Romano C, Scarabelli TM, Chen-Scarabelli C, Saravolatz L, and Dioguardi FS (2021). Serum Metabolic Profile in Patients With Long-Covid (PASC) Syndrome: Clinical Implications. Front. Med. 8, 714426. 10.3389/fmed.2021.714426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spyropoulos AC, and Bonaca MP (2022). Studying the coagulopathy of COVID-19. Lancet 399, 118–119. 10.1016/S0140-6736(21)01906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Proal AD, and VanElzakker MB (2021). Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 12, 698169. 10.3389/fmicb.2021.698169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aschman T, Mothes R, Heppner FL, and Radbruch H. (2022). What SARS-CoV-2 does to our brains. Immunity 55, 1159–1172. 10.1016/j.immuni.2022.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frontera JA, and Simon NM (2022). Bridging Knowledge Gaps in the Diagnosis and Management of Neuropsychiatric Sequelae of COVID-19. JAMA Psychiatr. 79, 811–817. 10.1001/jamapsychiatry.2022.1616. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez-Castaneda A, Lu P, Geraghty AC, Song E, Lee MH, Wood J, O’Dea MR, Dutton S, Shamardani K, Nwangwu K, et al. (2022). Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell 185, 2452–2468.e2416. 10.1016/j.cell.2022.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bin NR, Prescott SL, Horio N, Wang Y, Chiu IM, and Liberles SD (2023). An airway-to-brain sensory pathway mediates influenza-induced sickness. Nature 615, 660–667. 10.1038/s41586-023-05796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McVey Neufeld KA, Bienenstock J, Bharwani A, Champagne-Jorgensen K, Mao Y, West C, Liu Y, Surette MG, Kunze W, and Forsythe P. (2019). Oral selective serotonin reuptake inhibitors activate vagus nerve dependent gut-brain signalling. Sci. Rep. 9, 14290. 10.1038/s41598-019-50807-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.VanElzakker MB (2013). Chronic fatigue syndrome from vagus nerve infection: a psychoneuroimmunological hypothesis. Med. Hypotheses 81, 414–423. 10.1016/j.mehy.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 69.Bonnet U, and Juckel G. (2022). COVID-19 Outcomes: Does the Use of Psychotropic Drugs Make a Difference? Accumulating Evidence of a Beneficial Effect of Antidepressants-A Scoping Review. J. Clin. Psychopharmacol. 42, 284–292. 10.1097/JCP.0000000000001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee TC, Vigod S, Bortolussi-Courval É, Hanula R, Boulware DR, Lenze EJ, Reiersen AM, and McDonald EG (2022). Fluvoxamine for Outpatient Management of COVID-19 to Prevent Hospitalization: A Systematic Review and Meta-analysis. JAMA Netw. Open 5, e226269. 10.1001/jamanetworkopen.2022.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahdi M, Hermán L, Réthelyi JM, and Bálint BL (2022). Potential Role of the Antidepressants Fluoxetine and Fluvoxamine in the Treatment of COVID-19. Int. J. Mol. Sci. 23, 3812. 10.3390/ijms23073812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reis G, Dos Santos Moreira-Silva EA, Silva DCM, Thabane L, Milagres AC, Ferreira TS, Dos Santos CVQ, de Souza Campos VH, Nogueira AMR, de Almeida APFG, et al. (2022). Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the TOGETHER randomised, platform clinical trial. Lancet. Glob. Health 10, e42–e51. 10.1016/S2214-109X(21)00448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen W, Chen C, Tang J, Wang C, Zhou M, Cheng Y, Zhou X, Wu Q, Zhang X, Feng Z, et al. (2022). Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: a meta-analysis. Ann. Med. 54, 516–523. 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng W, Sun HL, Cai H, Zhang Q, Ng CH, and Xiang YT (2022). Antidepressants for COVID-19: A systematic review. J. Affect. Disord. 307, 108–114. 10.1016/j.jad.2022.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown LA, Ballentine E, Zhu Y, McGinley EL, Pezzin L, and Abramoff B. (2022). The unique contribution of depression to cognitive impairment in Post-Acute Sequelae of SARS-CoV-2 infection. Brain Behav. Immun. Health 22, 100460. 10.1016/j.bbih.2022.100460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui L, Lee YH, Thein TL, Fang J, Pang J, Ooi EE, Leo YS, Ong CN, and Tannenbaum SR (2016). Serum Metabolomics Reveals Serotonin as a Predictor of Severe Dengue in the Early Phase of Dengue Fever. PLoS Negl. Trop. Dis. 10, e0004607. 10.1371/journal.pntd.0004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seet RCS, Quek AML, and Lim ECH (2007). Post-infectious fatigue syndrome in dengue infection. J. Clin. Virol. 38, 1–6. 10.1016/j.jcv.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 79.Lood C, Tydén H, Gullstrand B, Klint C, Wenglén C, Nielsen CT, Heegaard NHH, Jönsen A, Kahn R, and Bengtsson AA (2015). Type I interferon-mediated skewing of the serotonin synthesis is associated with severe disease in systemic lupus erythematosus. PLoS One 10, e0125109. 10.1371/journal.pone.0125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.San Hernandez AM, Singh C, Valero DJ, Nisar J, Trujillo Ramirez JI, Kothari KK, Isola S, and Gordon DK (2020). Multiple Sclerosis and Serotonin: Potential Therapeutic Applications. Cureus 12, e11293. 10.7759/cureus.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyerhoff J, and Dorsch CA (1981). Decreased platelet serotonin levels in systemic lupus erythematosus. Arthritis Rheum. 24, 1495–1500. 10.1002/art.1780241207. [DOI] [PubMed] [Google Scholar]
- 82.Thompson RC, Simons NW, Wilkins L, Cheng E, Del Valle DM, Hoffman GE, Cervia C, Fennessy B, Mouskas K, Francoeur NJ, et al. (2023). Molecular states during acute COVID-19 reveal distinct etiologies of long-term sequelae. Nat. Med. 29, 236–246. 10.1038/s41591-022-02107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, et al. (2020). Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511. 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reilly JP, Anderson BJ, Hudock KM, Dunn TG, Kazi A, Tommasini A, Charles D, Shashaty MGS, Mikkelsen ME, Christie JD, and Meyer NJ (2016). Neutropenic sepsis is associated with distinct clinical and biological characteristics: a cohort study of severe sepsis. Crit. Care 20, 222. 10.1186/s13054-016-1398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, and Hennighausen L. (2010). Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia 12, 899–905. 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peluso MJ, Deitchman AN, Torres L, Iyer NS, Munter SE, Nixon CC, Donatelli J, Thanh C, Takahashi S, Hakim J, et al. (2021). Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 36, 109518. 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miyoshi H, and Stappenbeck TS (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482. 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, and Clevers H. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 89.Mizutani T, and Clevers H. (2020). Primary Intestinal Epithelial Organoid Culture. Methods Mol. Biol. 2171, 185–200. 10.1007/978-1-0716-0747-3_11. [DOI] [PubMed] [Google Scholar]
- 90.De Cuyper IM, Meinders M, van de Vijver E, de Korte D, Porcelijn L, de Haas M, Eble JA, Seeger K, Rutella S, Pagliara D, et al. (2013). A novel flow cytometry-based platelet aggregation assay. Blood 121, e70–e80. 10.1182/blood-2012-06-437723. [DOI] [PubMed] [Google Scholar]
- 91.Han W, and de Araujo IE (2021). Dissection and surgical approaches to the mouse jugular-nodose ganglia. STAR Protoc. 2, 100474. 10.1016/j.xpro.2021.100474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin YT, and Chen JC (2018). Dorsal Root Ganglia Isolation and Primary Culture to Study Neurotransmitter Release. J. Vis. Exp. 10.3791/57569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lanfear DE, Gibbs JJ, Li J, She R, Petucci C, Culver JA, Tang WHW, Pinto YM, Williams LK, Sabbah HN, and Gardell SJ (2017). Targeted Metabolomic Profiling of Plasma and Survival in Heart Failure Patients. JACC. Heart Fail. 5, 823–832. 10.1016/j.jchf.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gardell SJ, Zhang X, Kapoor N, Petucci C, and Coen PM (2019). Metabolomics Analyses of Muscle Atrophy Induced by Hind Limb Unloading. Methods Mol. Biol. 1996, 297–309. 10.1007/978-1-4939-9488-5_22. [DOI] [PubMed] [Google Scholar]
- 95.Odorizzi PM, Pauken KE, Paley MA, Sharpe A, and Wherry EJ (2015). Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137. 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burzynski LC, Pugh N, and Clarke MCH (2019). Platelet Isolation and Activation Assays. Bio. Protoc. 9, e3405. 10.21769/BioProtoc.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 98.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hafemeister C, and Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code to understand and assess the conclusions of this research are available in the main text and supplementary materials. RNA-seq data have been deposited and are publicly available. Accession numbers are listed in the key resources table.
This paper does not contain original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Anti-doublecortin antibody | Abcam | ab18723; RRID:AB_732011 |
| Anti-NeuN Antibody, clone A60 | Millipore Sigma | MAB377; RRID:AB_2298772 |
| Anti-mouse GPIbα | Emfret | R300; RRID:AB_2721041 |
| Anti-mouse IFNAR-1 antibody | Bio X Cell | BE0241; RRID:AB_2687723 |
| APC anti-mouse CD9 Antibody | Biolegend | 124811; RRID:AB_2783070 |
| APC Rat Anti-Mouse CD41 | Biolegend | 133913; RRID:AB_11126751 |
| c-Fos (9F6) Rabbit mAb | Cell Signaling Technology | 2250; RRID:AB_2247211 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Thermo Fisher Scientific | A-21202; RRID:AB_141607 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 | Thermo Fisher Scientific | A-31573; RRID:AB_2536183 |
| FITC Rat Anti-Mouse CD62P | BD | 561923; RRID:AB_10896149 |
| GAPDH (D16H11) XP® Rabbit mAb | Cell Signaling Technology | 5174S; RRID:AB_10622025 |
| Goat anti-Human IgM-HRP | SouthernBiotech | 2020–05; RRID:AB_2795603 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | A-11008; RRID:AB_143165 |
| Ki-67 Monoclonal Antibody (SolA15) | eBioscience | 14–5698-82; RRID:AB_10854564 |
| Non-immune rat immunoglobulins (IgG) | Emfret | C301; RRID:AB_2734715 |
| PE anti-mouse CD9 Antibody | Biolegend | 124805; RRID:AB_1279327 |
| Peroxidase AffiniPure Goat Anti-Human IgG (H + L) | Jackson ImmunoResearch Laboratories | 109–035-088; RRID:AB_2337584 |
| Phospho-Stat1 (Tyr701) (58D6) Rabbit mAb | Cell Signaling Technology | 9167S; RRID:AB_561284 |
| Rabbit anti-chromogranin A antibody | Novus Biologicals | NB120–15160 |
| Stat1 (D1K9Y) Rabbit mAb | Cell Signaling Technology | 14994S; RRID:AB_2737027 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology | sc-47778; RRID:AB_2714189 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pAAV-hSyn-DIO-hM3Dq-mCherry | Addgene | 44361 |
| LCMV (Armstrong strain) | John Wherry, University of Pennsylvania | N/A |
| LCMV (Clone 13 strain) | John Wherry, University of Pennsylvania | N/A |
| SARS-CoV-2, Isolate B.1.351 | Andy Pekosz, Johns Hopkins University | N/A |
| SARS-CoV-2, Isolate USA-WA1/2020 | BEI Resources | NR-52281 |
| Vesicular stomatitis virus (Indiana strain) | Sara Cherry, University of Pennsylvania | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Acute and recovered COVID-19 cohort plasma samples | Mathew et al.83 | N/A |
| Healthy and PASC stool samples | This study | N/A |
| Healthy cohort plasma samples | Una O’Doherty, University of Pennsylvania | N/A |
| Human autopsy tissues | This study | N/A |
| RUSH PASC cohort plasma samples | Giron et al.27 | N/A |
| UCSF LIINC cohort plasma samples | Peluso et al.14 | N/A |
| UNCOVR cohort plasma samples | Su et al.13 | N/A |
| UPenn PASC cohort plasma samples | This study | N/A |
| Viremia cohort plasma samples | Reilly et al.84 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| L-tryptophan (13C11, 99%) | Cambridge Isotope Laboratories | CLM-4290-H-0.1 |
| 1-(3-Chlorophenyl)biguanide hydrochloride | Tocris | 440 |
| 4-Chloro-DL-phenylalanine methyl ester hydrochloride | Sigma-Aldrich | C3635 |
| 5-HT | Millipore Sigma | 14927 |
| 5-hydroxy-L-tryptophan | Cayman Chemical Company | 20539 |
| 680C91 | Selleck Chemicals | S8997 |
| Capsaicin | Sigma-Aldrich | M2028 |
| Clozapine N-oxide hydrochloride | Sigma-Aldrich | SML2304 |
| Collagenase 1A | Gibco | 17100017 |
| Dithiothreitol (DTT) | Sigma-Aldrich | 1.02E+10 |
| Fluoxetine oral solution, USP | Aurobindo | NDC 65862–306-12 |
| Fura- 2-AM | Thermo Fisher Scientific | F-1221 |
| Gly-Pro-Arg-Pro | Sigma-Aldrich | G1895 |
| Glycyl-L-tryptophan hydrate | VWR | 100276–390 |
| HEPES | Thermo Fisher Scientific | 5–630-080 |
| HMW poly(I:C) | InvivoGen | tlrl-pic |
| IKK-16 | Selleck Chemicals | No.S2882 |
| L-tryptophan | Sigma-Aldrich | T8941 |
| LMW poly(I:C) | InvivoGen | tlrl-picw |
| Mouse NGF 7S Subunit protein | Gibco | 549 13–290-010 |
| N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide | Sigma-Aldrich | 394882 |
| Nickel–nitrilotriacetic acid (Ni-NTA) resin | Qiagen | 30210 |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15140122 |
| Phenelzine sulfate salt | Sigma-Aldrich | P6777 |
| Poly-L-lysine | Sigma Aldrich | P4707 |
| Poly(I:C) | Sigma-Aldrich | P1530 |
| Recombinant Mouse IFN-α | BioLegend | 752802 |
| Recombinant Mouse IFN-β1 | BioLegend | 581302 |
| Sodium pyruvate | Corning | MT25000CI |
| SureBlue 3,3′,5,5′-tetramethylbenzidine substrate | KPL | 5120–0075 |
| Thrombin | Sigma-Aldrich | T4648 |
|
| ||
| Critical commercial assays | ||
|
| ||
| 5-HIAA ELISA kits | Abnova | KA1881 |
| Fibrinogen ELISA kits | Abcam | ab213478 |
| High-Capacity cDNA Reverse Transcription kits | Thermo Fisher Scientific | 43–688-13 |
| Kynurenine ELISA kits | Abnova | KA6140 |
| LUNA Universal PCR kit | New England Biolabs | M3003E |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| QuantiFast SYBR Green PCR kit | Qiagen | 204056 |
| RNAeasy mini kits | Qiagen | 74104 |
| Serotonin ELISA kits | Novus biologicals | KA1894 |
| Thrombin-Antithrombin Complexes ELISA kits | Abcam | ab137994 |
| Tissue factor ELISA kits | Abcam | ab214091 |
| Tryptophan ELISA kits | Novus biologicals | KA11916 |
| Waters AccQTag Ultra derivatization kit | Waters corporation | 86003836 |
| Viral RNA Mini Kit | Qiagen | 52906 |
|
| ||
| Deposited data | ||
|
| ||
| Human intestinal organoid RNA-sequencing | Lamers et al.30 | GSE149312 |
| Metabolomics of COVID-19 patients | Shen et al., Shi et al., Song et al., Thomas et al., and Xiao et al.7–11 | N/A |
| Metabolomics of patients with PASC | Sadlier et al.12 | N/A |
| Single-cell RNA-seq of vagal neurons | Kupari et al.52 | GSE124312 |
| RNA-seq of ileum from poly(I:C)-treated mice | This study | PRJNA1007416 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| TLR3−/− | The Jackson Laboratory | 005217 |
| ACE2−/− | Taconic Biosciences | 18180 |
| IDO1−/− | The Jackson Laboratory | 005867 |
| IFNAR1−/− | The Jackson Laboratory | 028288 |
| K18-HuACE2 | The Jackson Laboratory | 034860 |
| Phox2b-Cre | The Jackson Laboratory | 016223 |
| STAT1−/− | The Jackson Laboratory | 012606 |
| STAT1flox/flox | Klover et al., Neoplasia, 201085 | Klover et al.85 |
| Villin-creERT2 | The Jackson Laboratory | 020282 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Forward primer for 18S qPCR: 5′- AACCCGTTGAACCCCATT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for 18S qPCR: 5′- CCATCCAATCGGTAGTAGCG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ddc qPCR: 5′-TAGCTGACTATCTGGATGGCAT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ddc qPCR: 5′-GTCCTCGTATGTTTCTGGCTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit1 qPCR: 5′-CAGAAGCACACATTGAAGAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit1 qPCR: 5′-TGTAAGTAGCCAGAGGAAGG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit2 qPCR: 5′-GGGAAAGCAGAGGAAATCAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit2 qPCR: 5′-TGAAAGTTGCCATACAGAAG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Ifit3 qPCR: 5′-GCCGTTACAGGGAAATACTGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Ifit3 qPCR: 5′-CCTCAACATCGGGGCTCT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Mx1 qPCR: 5′-GACTACCACTGAGATGACCCAGC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Mx1 qPCR: 5′-ATTTCCTCCCCAAATGTTTTCA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Nsp14 qPCR: 5′-TGGGGYTTTACRGGTAACCT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Nsp14 qPCR: 5′-AACRCGCTTAACAAAGCACTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Oas1b qPCR: 5′-TTCTACGCCAATCTCATCAGTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Oas1b qPCR: 5′-GGTCCCCCAGCTTCTCCTTAC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Rpl32 qPCR: 5′-TTCCTGGTCCACAATGTCAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Rpl32 qPCR: 5′-GGCTTTTCGGTTCTTAGAGGA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc18a1 qPCR: 5′-GTCCCGGAAGCTGGTGTTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc18a1 qPCR: 5′-ACAGTGAGCAGCATATTGTCC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc3a2 qPCR: 5′-ACGGTGTGGATGGTTTCCAAT-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc3a2 qPCR: 5′-TCCCTGCAATCAAAAGCCTGT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a14 qPCR: 5′-GACAGCTTCATCCGAGAACTTC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a14 qPCR: 5′-ATTGCCCAATCCCACTGCAT-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a19 qPCR: 5′-AACGCTCATGTATAGCATCTGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a19 qPCR: 5′-CAGCCACAGTGACCACAAC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc6a4 qPCR: 5′-GACAGGGGTGTGGGTTGATGC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc6a4 qPCR: 5′-TCAGCCATGTAGCCAAGCACC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc7a5 qPCR: 5′-CTACGCCTACATGCTGGAGG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc7a5 qPCR: 5′-GAGGGCCGAATGATGAGCAG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Slc7a8 qPCR: 5′-TCAGCGCCTGTGGTATCATTG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Slc7a8 qPCR: 5′-TGATGCCTGTCACGATCCAGA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Tph1 qPCR: 5′-AACAAAGACCATTCCTCCGAAAG-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Tph1 qPCR: 5′-TGTAACAGGCTCACATGATTCTC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for VI qPCR (for detection of SARS-CoV-2 viral RNA): 5′-ATGCTGCAATCGTGCTACAA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for VI qPCR (for detection of SARS-CoV-2 viral RNA): 5′-CCTCTGCTCCCTTCTGCGTA-3′ |
Integrated DNA technologies | N/A |
| Forward primer for Vil1 qPCR: 5′-TCAAAGGCTCTCTCAACATCAC-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for Vil1 qPCR: 5′-AGCAGTCACCATCGAAGAAGC-3′ |
Integrated DNA technologies | N/A |
| Forward primer for VSV-G qPCR: 5′-CAAGTCAAAATGCCCAAGAGTCACA-3′ |
Integrated DNA technologies | N/A |
| Reverse primer for VSV-G qPCR: 5′-TTTCCTTGCATTGTTCTACAGATGG-3′ |
Integrated DNA technologies | N/A |
| Forward primer for GP qPCR (for detection of LCMV viral RNA): 5′- GCAACTGCTGTGTTCCCGAAAC-3′ |
Integrated DNA technologies | Peluso et al.86 |
| Forward primer for GP qPCR (for detection of LCMV viral RNA): 5′- CATTCACCTGGACTTTGTCAGACTC-3′ |
Integrated DNA technologies | Peluso et al.86 |
| Ace2 Taqman assay | Thermo Scientific | Mm01159006_m1 |
| Doublecortin Taqman assay | Thermo Scientific | Mm00438400_m1 |
| Gapdh Taqman assay | Thermo Scientific | Mm99999915_g1 |
| Maoa Taqman assay | Thermo Scientific | Mm00558004_m1 |
| Vil1 Taqman assay | Thermo Scientific | Mm00494146_m1 |
|
| ||
| Software and algorithms | ||
|
| ||
| Agilent software | Agilent | N/A |
| Bioconductor v.3.8 | Bioconductor | https://www.bioconductor.org/ |
| Biorender | Biorender | https://biorender.com/ |
| Flowjo v10.6.2 | BD | https://www.flowjo.com/ |
| GSEA | Broad institute | https://www.gsea-msigdb.org/ |
| ImageJ v2.1.0/1.53c | NIH | https://imagej.nih.gov/ij/ |
| Kallisto v.0.46.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| Olympic cellSens imaging software | Olympus LS | https://www.olympus-lifescience.com/ |
| Prism v9.3.0 | Graphpad | https://graphpad.com |
| RStudio v.1.2.5019 | The R foundation | https://www.r-project.org/ |
|
| ||
| Other | ||
|
| ||
| 1% Glycyl-L-tryptophan Diet | Envigo | TD.210749 |
| 2″ binder clips | Amazon | ASIN B07C94YCR5 |
| Advanced DMEM | Corning | MT10013CV |
| Amino acid control diet | Envigo | TD.01084 |
| B27 supplement | Gibco | 17504044 |
| BioDAQ cages | Research Diets, Inc. | N/A |
| DAPI mounting media | Electron Microscopy Sciences | 17985–50 |
| DNA/RNA Shield Fecal Collection Tubes | Zymo Research | R1137 |
| Elmer’s 0.77 oz glue sticks | Amazon | #E517 |
| FBS | Corning | MT35–010-CV |
| Immulon 4 HBX ELISA plates | Thermo Fisher Scientific | 3855 |
| Matrigel (GFR) | BD Biosciences | BD356231 |
| M-MLV Reverse Transcriptase (200 U/μL) | Invitrogen | 28025013 |
| Neurobasal-A medium | Thermo Fisher Scientific | 10888022 |
| Quantitative Synthetic SARA-CoV-2 RNA: ORF, E, N | ATTC | VR-3276SD |
| Random primers | Invitrogen | 48190011 |
| Serum gel tubes | Sarstedt | 41.1500.005 |
| Syringe-driven filter units, 0.22 μm low protein binding durapore membrane | Millipore | SLGVR33RS |
| Taqman Fast Advanced Master Mix | Thermo Scientific | 4444557 |
| TrypLE Express | Thermo Fisher Scientific | 12604013 |
| Tryptophan-deficient diet | Envigo | TD.130674 |
| Vacutainer EDTA tubes | BD | 365974 |