Abstract
Background
Plexins are large transmembrane receptors for the semaphorin family of signalling proteins. Semaphorin-plexin signalling controls cellular interactions that are critical during development as well as in adult life stages. Nine plexin genes have been identified in humans, but despite the apparent importance of plexins in development, only biallelic PLXND1 and PLXNA1 variants have so far been associated with Mendelian genetic disease.
Methods
Eight individuals from six families presented with a recessively inherited variable clinical condition, with core features of amelogenesis imperfecta (AI) and sensorineural hearing loss (SNHL), with variable intellectual disability. Probands were investigated by exome or genome sequencing. Common variants and those unlikely to affect function were excluded. Variants consistent with autosomal recessive inheritance were prioritised. Variant segregation analysis was performed by Sanger sequencing. RNA expression analysis was conducted in C57Bl6 mice.
Results
Rare biallelic pathogenic variants in plexin B2 (PLXNB2), a large transmembrane semaphorin receptor protein, were found to segregate with disease in all six families. The variants identified include missense, nonsense, splicing changes and a multiexon deletion. Plxnb2 expression was detected in differentiating ameloblasts.
Conclusion
We identify rare biallelic pathogenic variants in PLXNB2 as a cause of a new autosomal recessive, phenotypically diverse syndrome with AI and SNHL as core features. Intellectual disability, ocular disease, ear developmental abnormalities and lymphoedema were also present in multiple cases. The variable syndromic human phenotype overlaps with that seen in Plxnb2 knockout mice, and, together with the rarity of human PLXNB2 variants, may explain why pathogenic variants in PLXNB2 have not been reported previously.
Keywords: Dentistry; Dentistry; Genetic Research; Genetics; Genetics, Medical
WHAT IS ALREADY KNOWN ON THIS TOPIC
Plexins are large transmembrane proteins that act as receptors for the semaphorin family of signalling proteins. Semaphorin-plexin signalling controls cellular interactions that are critical during development as well as in adult life stages. Nine plexin genes have been identified in humans, but despite the apparent importance of plexins in development, only biallelic PLXND1 and PLXNA1 variants have so far been associated with Mendelian genetic disease.
WHAT THIS STUDY ADDS
We identify rare biallelic pathogenic variants in PLXNB2 as a cause of a new autosomal recessive, phenotypically diverse syndrome with amelogenesis imperfecta and sensorineural hearing loss as core features. Intellectual disability, ocular disease, ear developmental abnormalities and lymphoedema were also present in multiple cases. The variable syndromic human phenotype overlaps with that seen in Plxnb2 knockout mice, and, together with the rarity of human PLXNB2 variants, may explain why pathogenic variants in PLXNB2 have not been reported previously.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Individuals presenting with amelogenesis imperfecta and sensorineural hearing loss should be screened for mutations in PLXNB2 and tested for other features of the syndrome. PLXNB2 should be added to amelogenesis imperfecta, deafness and intellectual disability gene panels to improve mutation detection rates. Affected families should receive appropriate genetic counselling.
Introduction
Development is a cascade of highly dynamic, time-critical, complex processes involving many signalling molecules and receptors that act to regulate cell proliferation, migration, adhesion and differentiation. This results in the formation of complex tissues, which further organise to effect organogenesis. Plexins are large transmembrane proteins that act as receptors for the semaphorin family of signalling proteins. Semaphorin-plexin signalling controls cellular interactions that are critical during development as well as in adult life stages (reviewed by Perälä et al).1 Semaphorin signalling modulates changes to both actin and microtubule organisation and therefore to the overall cytoskeleton, cell morphology, cell adhesion and cell motility (reviewed in Alto and Terman2).
The plexin gene family was originally identified in humans, and members were grouped according to the domain structure of the encoded proteins.3 4 Nine genes have been identified in both humans and mice, with class A plexins consisting of four genes (A1–A4), class B of three (B1–B3), and class C (C1) and class D (D1) of one each.1 4 Plexin family members share a common structure, with extracellular and intracellular portions. The extracellular portions contain a sema domain that binds with semaphorin ligands to activate signalling, as well as two or three PSI (plexin, semaphorin and integrin) domains and two or three glycine-proline rich IPT (immunoglobulin, plexin and transcription factor) domains. The intracellular portions are highly conserved5 and contain two R-Ras GAP motifs and one set of Plexin Rho-GTPase Association Motifs.
Class B plexins have an additional intracellular C-terminal PSD95, DLG1 and ZO1 (PDZ) interaction domain6 and an extracellular cleavage site for proprotein convertases.1 Plexin B2 (PLXNB2) participates in axonal guidance and cell migration.7 It is expressed widely but it also demonstrates a specific temporospatial pattern of expression throughout development, and its expression is distinct from that of other plexins, suggesting non-redundancy.8 RNA transcripts are detectable in mice from early fetal stages to adulthood within the brain.9 In situ hybridisation in E14 mouse embryos revealed high expression within several regions of the central nervous system, including many regions of the brain and retina.8 Expression was also high in the developing tooth bud, oral epithelium and in cartilage, with lower expression also detected in the cochlea, lung, kidney, epidermis and intestine.8 The exact role of PLXNB2 in tooth development is currently unknown, but semaphorins and plexin B1 have been found to be important for innervation of tooth buds10 and for dental stem cell migration ex vivo.11
Plxnb2 knockout mice (Plxnb2 -/-) vary in phenotype, depending on their genetic background. Plxnb2 -/- mice produced on inbred backgrounds did not survive gestation.12 13 The majority developed exencephaly, reflecting the importance of PLXNB2 activity for neural tube closure and potentially also its influence on the actin cytoskeleton. Other defects noted included abnormal development of the dentate gyrus, defects in cerebellar foliation and lamination, retarded development of the olfactory bulb and impaired neuronal proliferation. In contrast, when the same pathogenic variant was introduced into outbred CD1 mice, neural tube closure defects were less common, and after four generations, around 30% of Plxnb2 -/- mice were viable and fertile. Despite the knockout mice having no obvious behavioural or motor defects, their cerebella were smaller and major brain foliation defects were still present.12 Heterozygous Plxnb2 +/- mice had no apparent abnormalities.
Human PLXNB2 variants (MIM*604293) and aberrant PLXNB2 expression have been associated with lung cancer,14 15 acute myeloid leukaemia,16 amyotrophic lateral sclerosis,17 glioblastoma,18 autism spectrum disorders with regression,19 psoriasis20 and first trimester euploid miscarriage.21 In contrast, despite the apparent importance of plexins in development, only biallelic PLXND1 (MIM*620282) and PLXNA1 (MIM*601055) variants have so far been associated with Mendelian genetic disease in humans. PLXND1 variants cause multiple types of congenital heart defects (MIM#620294).22 PLXNA1 variants cause Dworschak-Punetha neurodevelopmental syndrome which includes speech regression, autistic features and hyperactivity, variable sensorineural hearing loss (SNHL), and ocular, brain, facial and skin abnormalities (MIM#619955).23 The same authors also suggested that pathogenic variants in other plexins may be embryonic lethal or may cause a range of phenotypes that have not yet been recognised as part of one syndrome.23
Here we describe six families with probands carrying rare biallelic PLXNB2 variants. Affected individuals manifest a complex, variable syndromic phenotype, the core features of which appear to be SNHL and amelogenesis imperfecta (AI), with intellectual disability also present in most cases.
Materials and methods
Patients
Affected individuals and family members were recruited in accordance with the principles outlined by the Declaration of Helsinki, with local ethical approval. Clinical evaluation captured disease features as part of routine patient care. Genomic DNA was obtained from venous blood samples using a salt-based extraction protocol, or from saliva using Oragene DNA Sample Collection Kits (DNA Genotek, Ottawa, Ontario, Canada), as detailed in the manufacturer’s instructions.
Sequencing and analysis
Individuals were recruited and genomic DNA was subjected to SNP genotyping, exome or genome sequencing at different institutions. Sequencing and analysis methods for each family can be found in the online supplemental materials and methods. In summary, variants identified in next generation short-read sequencing data were filtered to exclude all changes other than missense, frameshift or stop variants, exonic insertion/deletions or variants located at splice consensus sites (up to 8 bp within introns or 3 bp within exons away from splice junctions). Synonymous variants outside of the splice region were discarded. Variants in the Genome Aggregation Database (gnomAD) (v2.2.1)24 were excluded if present at a global minor allele frequency of 1% or higher. Variants were also filtered based on the mode of inheritance. In families known to be consanguineous, homozygous variants were prioritised. Population-specific high-frequency variants and platform artefacts were excluded by removing variants also present in exomes of individuals of the same ethnicity without dental disease that had been sequenced using the same reagents and platform. Splicing prediction analysis was carried out using NetGene2 (v2.4.2)25 and Splice AI.26 CADD v1.6,27 REVEL,28 Polyphen-2 (HumVar model)29 and SIFT30 were used to assess each variant’s potential to be disease causing. CNVs were identified using ExomeDepth (v1.0.0).31 Variants were confirmed and segregation analysis was performed for all available family members by Sanger sequencing. Primer sequences used are shown in online supplemental table 1. Genomic coordinates are based on the GRCh37 human reference genome, the reference gene sequence used for PLXNB2 is MANE Select transcript NM_012401.4 (ENST00000359337.9) and protein variant nomenclature for PLXNB2 is based on RefSeq protein NP_036533.2 (ENSP00000352288.4). The corresponding references used for CRYBB3 are MANE Select transcript NM_004076.5 (ENST00000215855.7) and for CRYBB3 RefSeq protein NP_004067.1 (ENSP00000215855.2). All variants identified as part of this study were uploaded to ClinVar: SCV002822954–SCV002822961. In silico modelling of the effect of the variants on the PLXNB2 protein tertiary structure was completed using I-TASSER-MTD32 using the default parameters. The protein structures were visualised with UCSF Chimera.33
jmg-2023-109728supp001.pdf (18.3MB, pdf)
Mouse tissue preparation
All animals were maintained in accordance with the French Ministry of Agriculture guidelines for the use of laboratory animals under study (SC67-218-37-IGBMC and APAFIS 3957-2016020516359388v1) and in accordance with the National Institutes of Health guidelines provided in the Guide for the Care and Use of Laboratory Animals. All methods and experimental procedures were reviewed and approved by an institutional safety committee.
Mouse embryos/fetuses were collected at E14.5, E16.5, E19.5 or on the day of birth and analysed as detailed in the online supplemental materials and methods.
Results
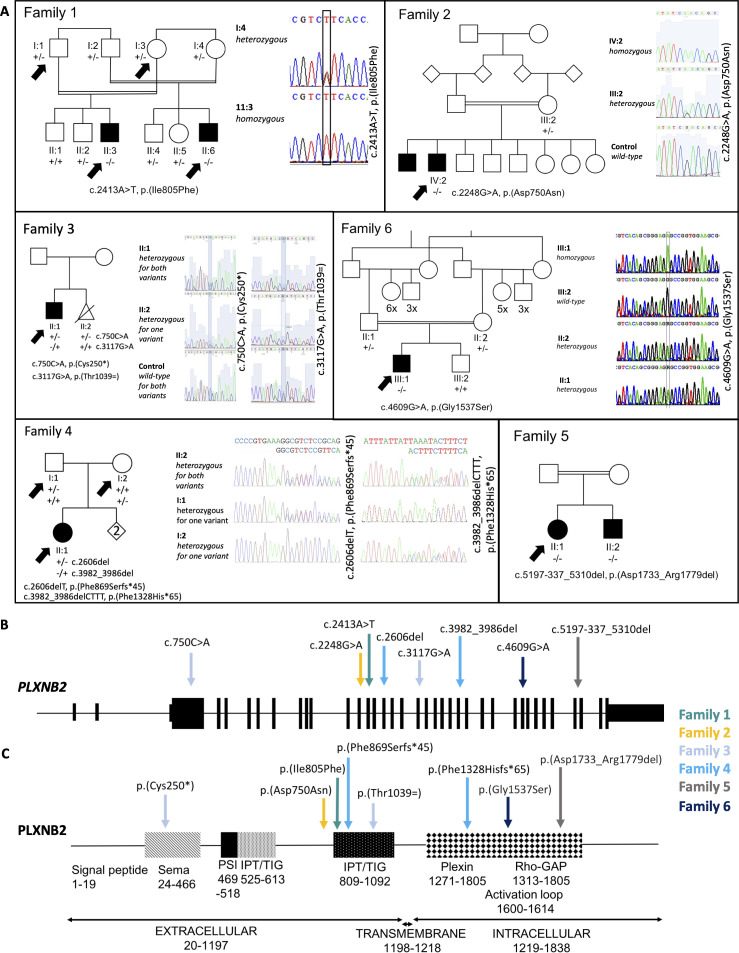
Initially, we recruited two consanguineous families, one Turkish (Family 1) and one Omani (Family 2) (figure 1). In Family 1, two male double first cousins have bilateral SNHL, intellectual disability, AI and severe myopia (online supplemental figure 1). In addition to the shared phenotype, one (II:3) also has bilateral cataracts and the other (II:6) has unilateral renal agenesis and pyloric stenosis. Analysis of GeneChip Human Mapping 250K Nsp SNP data from affected individuals II:3 and II:6 showed two homozygous regions encompassing chr7:34,080,354-41,760,000 and chr22:49,714,781-51,1756,26. Exome sequencing of II:3 and II:6 revealed a shared homozygous variant in PLXNB2 within the region on chromosome 22, c.2413A>T, p.(Ile805Phe) (table 1) which segregates with disease in the family (online supplemental figure 2). It affects the extracellular portion of the protein, specifically the first cell surface receptor IPT domain, and is predicted to be damaging by all pathogenicity prediction software tested (online supplemental table 2). The residue affected is highly conserved (online supplemental figure 3) and the variant is absent from gnomAD. Individual II:3 was also found to carry variant c.388G>A, p.(Glu130Lys) in crystallin beta 3 (CRYBB3; MIM*123630) (online supplemental figure 4), which is likely to explain the bilateral cataracts observed in him (MIM#609741).34 Variants that passed population and pathogenicity prediction filters but were not investigated further are detailed in online supplemental table 3 for each family.
Figure 1.
Pedigrees, Sanger sequencing and schematic diagram of the PLXNB2 protein. (A) DNA was available for all labelled individuals on each pedigree. Arrows indicate the individuals whose DNA was exome or genome sequenced. Affected status is as reported by the families for individuals for which DNA was not available. Sanger sequencing traces showing the segregation of each variant with disease in each family, except for Family 5, for which this is shown in online supplemental figure 9. The schematic diagram shows (B) the PLXNB2 transcript (ENST00000359337.9, NM_012401.4; 6409 bp) and (C) the PLXNB2 protein (ENSP00000352288.4, NP_036533.2; 1838 amino acids), with the positions marked for the pathogenic variants identified in this study. IPT, immunoglobulin, plexin and transcription factor; PSI, plexin, semaphorin and integrin; TIG, transcription factor immunoglobin.
Table 1.
The variants in PLXNB2 identified in six families and the disease features observed
| Patient (sex/age range) | Zygosity and variants (NM_012401.4, NP_036533.2) | Phenotype | ||||
| Auditory | Dental | Developmental/neurological | Vision | Other | ||
| Family 1 II:3 (M/12–18) |
Homozygous c.2413A>T: p.(Ile805Phe); c.2413A>T: p.(Ile805Phe) | SNHL | AI, conical permanent incisors | Global developmental delay, moderate intellectual disability | Myopia, horizontal nystagmus (congenital cataract due to CRYBB3 variant) | Ear lobe skin blind-ended tracts |
| Family 1 II:6 (M/19–21) |
Homozygous c.2413A>T: p.(Ile805Phe); c.2413A>T: p.(Ile805Phe) | SNHL; labyrinthine malformation | AI, conical permanent incisors | Global developmental delay, epilepsy, moderate intellectual disability | Severe myopia with scattered papillae, horizontal nystagmus, macular atrophy | Ear lobe skin blind-ended tracts, unilateral renal agenesis, pyloric stenosis, asthma, recurrent bronchitis, intrauterine growth retardation, finger pads, watch glass toenails, overweight |
| Family 2 IV:2 (M/12–18) | Homozygous c.2248G>A: p.(Asp750Asn); c.2248G>A: p.(Asp750Asn) | SNHL | AI | Intellectual disability | No obvious abnormality, not examined | |
| Family 3 II:1 (M/6–11) |
Compound heterozygous c.750C>A: p.(Cys250*); c.3117G>A: p.(Thr1039=) | SNHL (mild) 500–4000 Hz | AI with hypoplasia | Normal | Developmental macular abnormality with pale fundus, attenuated blood vessels, high myopia, nystagmus, microcornea | Ear lobe skin blind-ended tracts, cleft palate, hypertelorism, keratopathy |
| Family 4 II:1 (F/19–21) |
Compound heterozygous c.2606del: p.(Phe869Serfs*45); c.3982_3986del: p.(Phe1328His*65) | SNHL | AI; missing upper permanent lateral incisors | Normal | No obvious abnormality, not examined | Ear lobe skin blind-ended tracts, bilateral primary lower limb lymphoedema (onset aged 3), nevus, cellulitis |
| Family 5 II:1 (F/50–59) |
Homozygous c.5197-337_5310del: p.(Asp1733_Arg1779del); c.5197-337_5310del: p.(Asp1733_Arg1779del) | SNHL | AI | Mild/moderate intellectual disability | No obvious abnormality, not examined | Bilateral primary lower limb lymphoedema |
| Family 5 II:2 (M/50–59) |
Homozygous c.5197-337_5310del: p.(Asp1733_Met1770del); c.5197-337_5310del: p.(Asp1733_Met1770del) | SNHL | AI | Intellectual disability | No obvious abnormality, not examined | Unilateral lymphoedema of one foot |
| Family 6 III:1 (M/2–5) |
Homozygous c.4609G>A: p.(Gly1537Ser); c.4609G>A: p.(Gly1537Ser) | Could not be assessed; no current indication of hearing loss | Clinical tooth failure (cause unclear); could not be assessed further | Profound intellectual disability, non-verbal, autistic features, hyperactive behaviour | Strabismus, no other obvious abnormality, could not be assessed | Mild generalised muscular hypotonia |
Splicing prediction tools suggest that exon 35 is entirely skipped leading to p.(Asp1733_Arg1779del) instead of p.(Asp1733_Met1770del) as would be predicted from the proportion of the gene deleted. Age ranges are shown for individuals to maintain anonymity. Age of onset was from birth unless otherwise stated. Variants are based on genome build GRCh37 and PLXNB2 transcript ENST00000359337.9, NM_012401.4 and PLXNB2 protein ENSP00000352288.4, NP_036533.2.
AI, amelogenesis imperfecta; SNHL, sensorineural hearing loss.
In Family 2, two Omani brothers born of a first cousin union were found to have bilateral SNHL, intellectual disability and AI (table 1, figure 1 and online supplemental figure 5). Exome sequencing of one affected brother revealed a homozygous missense variant in PLXNB2, c.2248G>A, p.(Asp750Asn). This replaces a charged residue with an uncharged one, affecting a highly conserved residue in the extracellular portion of the protein (online supplemental figure 3). SIFT predicts the variant to be deleterious and it was not identified in gnomAD (online supplemental table 2).
Family 3 was identified independently via the UK Inherited Retinal Dystrophy Consortium. They are a white British family with an affected male child born with facial clefting, who also presented with nystagmus shortly after birth. In middle childhood, he was diagnosed with retinal dystrophy, high myopia, microcorneas and mild keratopathy. He also has mild bilateral SNHL for sounds ranging from 500 to 4000 Hz, and AI (table 1, figures 1 and 2 and online supplemental figure 6). Exome sequencing revealed biallelic compound heterozygous variants in PLXNB2, one a nonsense variant, c.750C>A, p.(Cys250*), and one synonymous variant altering the final base of the splice donor site of exon 19, predicted to affect splicing, c.3117G>A, p.(Thr1039=). Analysis with splice prediction tools Splice AI26 and NetGene225 predicted loss of the donor site, suggesting that some of intron 19 may be retained in the mature transcript (online supplemental table 4, online supplemental figure 7). Neither variant was identified in gnomAD (online supplemental table 2). A second fetus, who presented with clefting, was electively aborted. Genotyping showed that the fetus was heterozygous for the nonsense PLXNB2 variant only.
Figure 2.
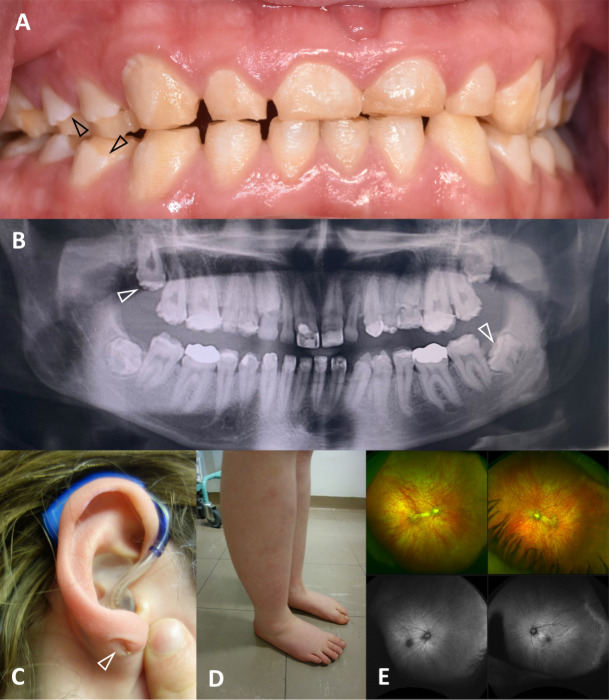
Clinical images that are illustrative of the shared and variable clinical features for affected individuals. (A) The anterior clinical photograph showing primary teeth, the posterior teeth have changes consistent with loss of enamel (arrowheads) due to fracturing, whereas the enamel of the anterior teeth is developmentally thin and optically abnormal (Family 3 II:1). (B) Panoramic radiograph of the adult permanent dentition illustrates that the enamel is more radiodense than the supporting dentine, but with a variably reduced enamel volume and irregular occlusal cusp morphology (arrowhead) consistent with amelogenesis imperfecta (AI) characterised by enamel that is hypomineralised with variable hypoplasia (Family 4 II:2). (C) Sensorineural hearing loss (SNHL) was typical, with provision of hearing aids in childhood (Family 4 II:2). Blind-end skin tracts involving the skin of the ears or adjacent tissues (arrowhead) were observed in at least 3/6 families. (D) Lower limb lymphoedema was observed in two families (Families 4 and 5, with Family 4 illustrated). (E) Fundus autofluorescence images (Family 3 II:1) illustrate a developmental macular abnormality with pale fundus and attenuated blood vessels. The individual also has high myopia, nystagmus and microcornea.
Next, we searched the UK 100,000 Genomes dataset35 for patients with biallelic variants in PLXNB2 and a similar phenotype. We identified one affected white British female (Family 4) with SNHL, AI, lower limb lymphoedema and cellulitis (table 1, figures 1 and 2, online supplemental figure 8). She carries biallelic compound heterozygous frameshift variants c.2606delT, p.(Phe869Serfs*45) and c.3982_3986delCTTT, p.(Phe1328Hisfs*65) in PLXNB2 (online supplemental figure 9), both predicted to produce transcripts that are subject to nonsense mediated decay.36 Variant c.2606delT, p.(Phe869Serfs*45) has previously been identified in gnomAD as a heterozygous variant in one individual, suggesting an allele frequency of 4.024×10–6. Variant c.3982_3986delCTTT, p.(Phe1328Hisfs*65) was not present in gnomAD.
With increased understanding of the clinical presentation associated with biallelic PLXNB2 variants, we identified Family 5 through further collaboration. Two affected siblings of Pakistani origin, born of a consanguineous union (figure 1), presented with deafness, AI, intellectual disability and lower limb lymphoedema (figure 2, table 1, online supplemental figures 10 and 11). Exome sequencing of II:1 and subsequent ExomeDepth analysis revealed a homozygous deletion spanning exons 34 and 35 (Reads ratio 0.0291, Bayes factor 18.3; online supplemental table 2). The breakpoints predicted in the exome sequence (online supplemental figure 12), chr22:50,715,085 and chr22:50,715,672, were confirmed by PCR, which also confirmed the deletion was present in II:2. This deletion is in-frame and is predicted to delete at least 38 amino acids (aa) (p.(Asp1733_Met1770del)) from the 1838aa protein, including all of exon 34 and part of exon 35. Splice prediction tool NetGene2 (v2.4.2) predicts that exon 35 will be skipped entirely (online supplemental table 5), resulting in an in-frame deletion of 47aa, p.(Asp1733_Arg1779del), suggesting that an abnormal PLXNB2 protein may be produced. The deleted region is part of the Rho-GAP catalytic domain critical to the protein’s function, but the deletion would leave the catalytic arginine residues at 1395, 1396 and 1691 intact.
Using GeneMatcher,37 we identified one further patient with biallelic PLXNB2 variants and an overlapping disease phenotype. Family 6 is of Iraqi origin. One affected male was born of first cousin consanguineous parents (figure 1). In early childhood, he has severe developmental delay and autistic features, and his tooth enamel shows evidence of severe damage from a limited visual inspection (table 1, online supplemental figure 13). However, it was not possible to carry out a detailed dental examination or to obtain dental radiographs. The presence of AI, as opposed to severe caries, could therefore not be confirmed, and testing for SNHL and eye disease was not possible. On exome sequencing, the affected individual was found to carry a homozygous PLXNB2 variant c.4609G>A, p.(Gly1537Ser), which affects the highly conserved Rho-GAP domain that lies within the intracellular portion of PLXNB2 and changes the residue from a non-polar to a polar residue (table 1, online supplemental table 2, online supplemental figure 3). The residue is conserved in all species examined and this variant was not present in gnomAD.
We next used I-TASSER-MTD to try to assess the effect of each of the variants on the overall predicted structure of PLXNB2 (online supplemental figure 14). In silico predictions of WT PLXNB2 structure (panel A) and of these variants are likely to be of limited use due to the small percentage of PLXNB2 covered by known crystal structures and the lack of appropriate homologous protein structures on which to base the WT structure. The local structural changes for the missense variants (Asp750Asn, Ile805Phe and Gly1537Ser) are shown in panels B, C and D. There were minor changes in the solvent accessibility for Asp750Asn and Ile805Phe. For Ile805Phe, the mutant structure is characterised as undefined, in comparison to the WT strand structure. The model for the deletion Asp1733_Arg1779del shows extensive structural differences to the WT protein.
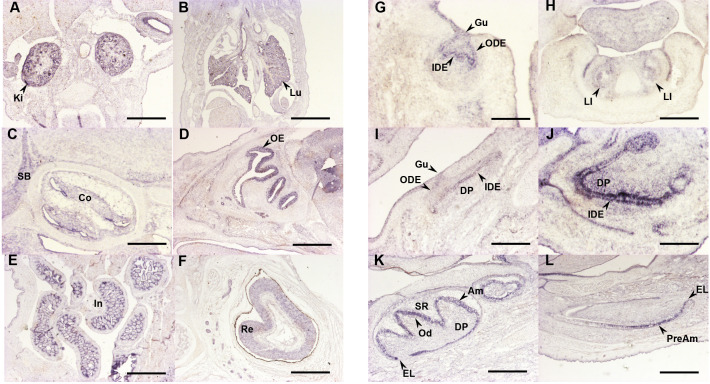
To gain insight into Plxnb2 expression in mice, we performed in situ hybridisation using a digoxigenin-labelled antisense riboprobe generated from the same DNA template as previously used by the EURExpress consortium (www.eurexpress.org) (online supplemental figure 15). Mouse embryos were analysed at E14.5, E16.5 and E19.5. Plxnb2 expression was detected in the kidney and lung (figure 3A,B). Discrete expression was observed in the developing inner ear (cochlea: figure 3C). Expression was also detected in olfactory epithelium and retina (figure 3D,F) and in the small and large intestines (figure 3E).
Figure 3.
Analysis of mouse Plxnb2 transcript distribution by in situ hybridisation. Selected sections illustrating Plxnb2 expression features in the developing skull bone, sensorial organs and viscera are shown in the left side panels (A–F), whereas right side panels focus on incisor (G, I and K) and molar (H, J and L) tooth development. Developmental stages and section planes are: E14.5 frontal (A, B, G and H), E16.5 sagittal (C, D, E, I and J); E19.5 sagittal (F, K and L) sections. Scale bars: 25 µm (J); 40 µm (H and I); 50 µm (C and K); 60 µm (F and H), 80 µm (A, D, E and L); 150 µm (B and G). Am, ameloblasts; Co, cochlea; DP, dental papilla; EL, epithelial loop; Gu, gubernaculum; IDE, inner dental epithelium; In, intestine; Ki, kidney; LI, lower incisor; Lu, lung; Od, odontoblasts; ODE, outer dental epithelium; OE, olfactory epithelium; Re, retina; SB, skull bone; SR, stellate reticulum.
We also interrogated the GTEx portal38 (https://gtexportal.org/) to determine the expression of PLXNB2 in human tissues. This showed that there was relatively high expression of PLXNB2 in the cerebellum and cerebellar hemisphere compared with other regions of the brain. There was also high expression in the kidney, salivary gland, ovary, prostate, spleen and thyroid, among other tissues (online supplemental figure 16).
Since all individuals studied had AI or were suspected to have AI, we investigated the expression of Plxnb2 in the mouse during various stages of tooth development (figure 3G,I,K lower molar and figure 3H,J,L lower incisor). Plxnb2 transcripts were observed at E14.5 in the epithelial tissues of the developing teeth, at E16.5 in the epithelial and mesenchymal compartments of the incisor and in the epithelial part of the molar. At E19.5, labelling was observed in differentiating ameloblasts.
Discussion
Here we describe biallelic pathogenic variants in PLXNB2 in six families of diverse ethnicity with individuals affected by a variable syndromic phenotype. Four of these are consanguineous families with affected individuals homozygous for an extremely rare variant that has almost certainly been passed down both branches of the family, and one includes affected cousins, demonstrating significant cosegregation of the disease with biallelic PLXNB2 variants. This syndrome has AI and SNHL as core symptoms, and intellectual disability, lower limb lymphoedema, ocular abnormalities and a variety of other conditions are also seen in some, but not all, cases. Given the varied and complex roles of PLXNB2 in development,1 it seems unsurprising that biallelic variants cause a syndromic disease phenotype. The variants all have pathogenicity scores indicating that they are predicted to be deleterious (online supplemental table 2). The case series we have accumulated, together with published evidence of an overlapping condition in Plxnb2 knockout mice, provides compelling evidence for biallelic variants in PLXNB2 as the cause of this recessively inherited condition.
All variants implicated are extremely rare, with all but one absent in gnomAD (v2.1.1), which contains approximately 124 000 individuals with good quality sequence across PLXNB2. The rarity of these variants, together with the constraint metrics available in gnomAD (o/e=0.19, pLI=0.99),39 suggests that PLXNB2 loss-of-function variants are not well tolerated and affect viability. However, the identification of an individual who may entirely lack PLXNB2 (Family 4 II:1) seems to contradict this. This is consistent with observations in Plxnb2 knockout mice, where homozygosity for the knockout allele was lethal on one genetic background but viable on another.12 13 The PLXNB2 variants identified herein include missense, frameshift and splice variants, a premature termination codon and a deletion spanning two exons, which are all observed to be protein-damaging variants.
One possible interpretation of these findings is that all the observed human variants have the effect of being functional knockouts, and that, as observed in mouse models, the viability of such embryos is determined by the genetic background. Genetic background might also explain the highly variable phenotypes observed in different cases, with only AI and SNHL as consistent features. Pathogenic variants in PLXNA1 cause an overlapping and similarly varying range of phenotypes to pathogenic variants in PLXNB2.23 The impact of genetic background on variation in disease phenotype, severity and survival has been noted for one Plxnb2 -/- mouse model,12 which suggests that other cosegregating variants may affect disease range and severity. This may suggest that, in spite of the distinct patterns of plexin expression,8 other plexins can sometimes partially compensate for loss of PLXNB2 or PLXNA1 to allow developmental processes vital to life to proceed. However, to prove such an effect would require the study of a large cohort of cases, and as yet no specific variants have been implicated in phenotype variation in either of these syndromes. Given that this cohort consists of only eight individuals with six different biallelic genotypes, the power to detect any genotype-phenotype correlation in this study is very limited.
Alternatively, the PLXNB2 variants and/or genotypes in the families described herein may each have unique effects on PLXNB2 function. Variants could cause partial loss of function through hypomorphic alleles, with expression of the normal transcript reduced but not abolished,40 or may act as ‘gain-of-function’ alleles, creating a protein with altered or enhanced function or inappropriate persistence within the cell. Consistent with this hypothesis, the affected cousins in Family 1 and affected siblings in Family 5, each having the same genotype, have remarkably similar phenotypes (table 1).
The missense variants identified in this study affect both the extracellular (p.(Ile805Phe) and p.(Asp750Asn)) and intracellular portions of the protein (p.(Gly1537Ser)), with no particular region or domain specifically affected by the variants identified in this study. In order to assess the effects of the variants on protein structure, we attempted to model the changes using I-TASSER-MTD.32 However, due to the lack of homologous structures available for the majority of the PLXNB2 protein, including the regions affected by the variants, the accuracy of the modelling is likely to be low. The variants identified herein could be altering the binding of PLXNB2 to semaphorins via the sema domain, the subsequent homodimerisation of PLXNB2 on binding semaphorin, catalytic activity via the GAP domain and interaction with GTPases, or the ability to interact with other proteins and to effect other types of downstream signalling.41 Further investigation will be required to better understand the effects of specific variants and the basis of variation on phenotype in the condition caused by PLXNB2 variants.
Heterozygous carriers of the variants identified in these families appear in general to be unaffected by disease, although one fetus (Family 3 II:2) did have facial clefting and carried a heterozygous nonsense PLXNB2 variant (c.750C>A, p.(Cys250*)). It is unknown whether this fetus would have developed other clinical features similar to their sibling. It is possible that another variant could be partially or entirely responsible for this particular phenotype rather than the PLXNB2 variant. In individuals with PLXNA1 pathogenic variants, disease has been observed with both biallelic and particular de novo heterozygous variants.23
The core phenotypes observed for the individuals carrying pathogenic PLXNB2 variants reflect negative impacts on the development or function of the cochlea and ameloblasts. This led us to consider whether other plexins are expressed in the cochlea and inner enamel epithelium at a similar time to PLXNB2, or whether PLXNB2 is expressed alone in these tissues, excluding the possibility of partial compensation by a closely related protein. Our investigation of the expression of Plxnb2 transcripts within the dental tissues of murine embryos gave similar results to the expression patterns previously detailed by Perälä et al.8 Their analysis of the expression of plexins in murine embryos using in situ hybridisation revealed that Plxnb2 transcripts are present in the brain, retina, cochlea and tooth bud at E14.8 Plxnb2 is the only plexin to be expressed at this timepoint in all four tissues, although the expression of Plxnd1 was not examined in this study and the expression of other plexins, most notably Plxnb1, does overlap that of Plxnb2 in many tissues. Plxnb2, Plxna2 and Plxna3 transcripts are all expressed in the cochlea, although the relative levels of expression of each were not determined. Similarly, Plxnb1 and Plxnb2 transcripts were detected at relatively high levels at E14 in the oral epithelium and tooth bud, but other plexins were also detected at lower levels of expression. Plxnb2 was shown to be expressed at relatively high levels in the inner enamel epithelium at E15, with expression sustained until at least E16. These findings suggest that PLXNA2, PLXNA3 and PLXNB1 are coexpressed in affected tissues at relevant timepoints, but are unlikely to be able to fully compensate for the loss of PLXNB2.
The variable phenotype and extremely low population frequency of the variants reported in this study may be the reasons why pathogenic variants in PLXNB2 have not previously been reported as causing syndromic disease in humans. We suggest that the tooth enamel phenotype, AI, is a consistent, but potentially easily missed feature that flags this genetic diagnosis. AI is a heterogeneous group of genetic conditions characterised by a deficit in enamel quantity and/or quality affecting all teeth of both dentitions.42–44 It can present as an isolated disease or can be part of more complex and diverse syndromes affecting other tissues and organs. Once formed and following tooth eruption, enamel has no capacity for cellular repair. Accordingly, AI provides a clear and persistent marker of abnormal development that is recognisable at an early age. However, due to the presence of neuronal deficits such as hearing loss and intellectual disability, AI might easily be overlooked or dismissed as dental caries due to suboptimal diet and/or poor dental hygiene. A differential diagnosis of AI as opposed to fluorosis or molar incisor hypomineralisation is also a possible confounding issue. Diagnosis of AI may therefore require a specialist paediatric dental professional. The disconnection between dental and general healthcare also presents barriers to diagnosis and has been flagged as problematic previously in the differential diagnosis of Usher and Heimler syndromes.45
The families presented have AI characterised by variable abnormalities of enamel volume (hypoplasia) and mineralisation (hypomineralised). This is consistent with the understanding of PLXNB2 function. Future laboratory investigation of enamel from affected individuals will give insight into the characteristics of the disruption to enamel rod morphology and mineralisation. It is unclear if the other dental morphological changes reported, including conical lateral incisors, missing lateral incisors, flattened occlusal surfaces and mild taurodontism, are consequent to PLXNB2 functional disruption or are coincidence in these families. As further families are described, the core features of the dental phenotype will become clearer and will be an important clinical indicator to consider PLXNB2 further.
In conclusion, we identify biallelic pathogenic variants in PLXNB2 as a cause of a new autosomal recessively inherited, phenotypically diverse syndrome including AI and SNHL as core symptoms, with intellectual disability, ocular disease, ear developmental abnormalities and lymphoedema also present in multiple cases.
Acknowledgments
We thank all of the families involved in this study, as well as the patient support group Amélogenèse France: https://amelogenesefrance.wixsite.com/amelogenese. We thank C Stoetzel and M Prasad, University of Strasbourg; J Clayton-Smith, University of Manchester; and S Al-Bahlani, Al Nadha Hospital, Muscat, Oman, for their help in the first steps of this project. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. The data used for the analyses described in this manuscript were obtained from https://gtexportal.org/home/gene/PLXNB2 on the GTEx portal on 21 July 2023.
Footnotes
@jamesapoulter, @ChrisM_Watson
CELS, VL-H, AM and AB-Z contributed equally.
Correction notice: This article has been corrected since it was published online. Reference 5 has been corrected.
Collaborators: The UK Inherited Retinal Dystrophy Consortium (UKIRDC) includes, in addition to listed authors CELS, RLT, GCB and CFI, Martin McKibbin, Manir Ali, Carmel Toomes, Stuart Ingram, Forbes Manson, Panagiotis Sergouniotis, Gavin Arno, Alison J Hardcastle, Andrew Webster, Nikolas Pontikos, Michael Cheetham, Alessia Fiorentino, Susan Downes, Jing Yu, Stephanie Halford, Suzanne Broadgate and Veronica van Heyningen. The Genomes England Research Consortium (GERC) includes Ambrose JC (Genomics England, London, UK), Arumugam P (Genomics England), Bevers R (Genomics England), Bleda M (Genomics England), Boardman-Pretty F (Genomics England; William Harvey Research Institute, Queen Mary University of London, UK), Boustred CR (Genomics England), Brittain H (Genomics England), Brown MA (Genomics England), Caulfield MJ (Genomics England; William Harvey Research Institute, Queen Mary University of London), Chan GC (Genomics England), Giess A (Genomics England), Griffin JN (Genomics England), Hamblin A (Genomics England), Henderson S (Genomics England; William Harvey Research Institute, Queen Mary University of London), Hubbard TJP (Genomics England), Jackson R (Genomics England), Jones LJ (Genomics England; William Harvey Research Institute, Queen Mary University of London), Kasperaviciute D (Genomics England; William Harvey Research Institute, Queen Mary University of London), Kayikci M (Genomics England), Kousathanas A (Genomics England), Lahnstein L (Genomics England), Lakey A (Genomics England), Leigh SEA (Genomics England), Leong IUS (Genomics England), Lopez FJ (Genomics England), Maleady-Crowe F (Genomics England), McEntagart M (Genomics England), Minneci F (Genomics England), Mitchell J (Genomics England), Moutsianas L (Genomics England; William Harvey Research Institute, Queen Mary University of London), Mueller M (Genomics England; William Harvey Research Institute, Queen Mary University of London), Murugaesu N (Genomics England), Need AC (Genomics England; William Harvey Research Institute, Queen Mary University of London), O‘Donovan P (Genomics England), Odhams CA (Genomics England), Patch C (Genomics England; William Harvey Research Institute, Queen Mary University of London), Perez-Gil D (Genomics England), Pereira MB (Genomics England), Pullinger J (Genomics England), Rahim T (Genomics England), Rendon A (Genomics England), Rogers T (Genomics England), Savage K (Genomics England), Sawant K (Genomics England), Scott RH (Genomics England), Siddiq A (Genomics England), Sieghart A (Genomics England), Smith SC (Genomics England), Sosinsky A (Genomics England; William Harvey Research Institute, Queen Mary University of London), Stuckey A (Genomics England), Tanguy M (Genomics England), Taylor Tavares AL (Genomics England), Thomas ERA (Genomics England; William Harvey Research Institute, Queen Mary University of London), Thompson SR (Genomics England), Tucci A (Genomics England; William Harvey Research Institute, Queen Mary University of London), Welland MJ (Genomics England), Williams E (Genomics England), Witkowska K (Genomics England; William Harvey Research Institute, Queen Mary University of London), Wood SM (Genomics England; William Harvey Research Institute, Queen Mary University of London) and Zarowiecki M (Genomics England).
Contributors: Conceptualisation: CFI, AJM, AB-Z. Data curation: CELS, VL-H, UKIRDC, GERC, RGF, GN, AR. Formal analysis: CELS, VL-H, UH, SB, RLT, RGF, JAM, CMW. Funding acquisition: CFI, AJM, AB-Z. Investigation: CELS, VL-H, UH, SB, RLT, JAP, SBW, RGF, JAM, SAB, GCB, CMW, SM, AJM, AB-Z, UKIRDC, GERC. Methodology: CELS, VL-H, UH, SB, RLT, JAP, UKIRDC, GERC, SBW, RGF, JAM, CFI, AJM, AB-Z. Project administration: CELS, CFI, AJM, AB-Z. Resources: GERC. Supervision: CELS, JAP, CMW, CFI, AJM, AB-Z. Validation: CELS, VL-H, UH, RLT, RGF, AR. Visualisation: CELS, VL-H, UH, SB, GN, AR, SBW, RGF, SM, AJM, AB-Z. Writing—original draft: CELS, VL-H. Writing—review and editing: CELS, CMW, CFI, AJM, AB-Z. Guarantor: CELS. All authors reviewed and commented critically on the final draft.
Funding: This work was supported by Rosetrees Trust Grant PGS19-2/10111, Retina UK and Fight for Sight UK (RP Genome Project GR586), the Wellcome Trust (grant number: 093113), the French Ministry of Health (National Program for Clinical Research, PHRC 2008 No 4266 Amelogenesis Imperfecta), the University Hospital of Strasbourg (HUS, API, 2009–2012, ‘Development of the oral cavity: from gene to clinical phenotype in Human’) and the grant ANR-10-LABX-0030-INRT, a French state fund managed by the Agence Nationale de la Recherche under the frame programme Investissements d’Avenir labelled ANR-10-IDEX-0002-02. It was also financed by and contributed to the actions of the Project No 1.7 ‘RARENET: a tri-national network for education, research and management of complex and rare disorders in the Upper Rhine’, cofinanced by the European Regional Development Fund (ERDF) of the European Union in the framework of the INTERREG V and previously INTERREG IV Upper Rhine programme as well as to the European Reference Network (ERN) CRANIO initiative. AB-Z is a USIAS 2015 Fellow of the Institute of Advanced Studies (Institut d’Etudes Avancées) de l’Université de Strasbourg, France. UH is the recipient of a Leeds Doctoral Scholarship (LDS). This work is part of the Projet E-GENODENT financed by the Fonds d’Intervention Régionale (FIR) of the Agence Régionale de Santé Grand Est (2019–2022). We are grateful for the funding provided by Filière TETECOU and ‘Pierre Henri et ses amis’ patient support groups.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
Collaborators: The UK Inherited Retinal Disease Consortium, Genomics England Research Consortium, Martin McKibbin, Manir Ali, Carmel Toomes, Stuart Ingram, Forbes Manson, Panagiotis Sergouniotis, Gavin Arno, Alison J Hardcastle, Andrew Webster, Nikolas Pontikos, Michael Cheetham, Alessia Fiorentino, Susan Downes, Jing Yu, Stephanie Halford, Suzanne Broadgate, Veronica van Heyningen, John C Ambrose, Prahbu Arumugam, Roel Bevers, Marta Bleda, Freya Boardman-Pretty, Chris R Boustred, Helen K Brittain, Matthew A Brown, Mark J Caulfield, Georgia C Chan, Adam Giess, John N Griffin, Angela Hamblin, Shirley Henderson, Tim JP Hubbard, Rob Jackson, Louise J Jones, Dalia Kasperaviciute, Melis Kayikci, Athanasios Kousathanas, Lea Lahnstein, Anna Louise Lakey, Sarah EA Leigh, Ivonne US Leong, Javier F Lopez, Fiona Maleady-Crowe, Meriel McEntagart, Federico Minneci, J Mitchell, Loukas Moutsianas, Michael Mueller, Nirupa Murugaesu, Anna C Need, Peter O‘Donovan, Chris A Odhams, Christine Patch, Daniel Perez-Gil, Marianna Buongermino Pereira, John Pullinger, Tahrima Rahim, Augusto Rendon, Tim Rogers, Kevin Savage, Kushmita Sawant, Richard H Scott, Afshan Siddiq, Alexander Sieghart, Samuel C Smith, Alona Sosinsky, Alexander Stuckey, Mélanie Tanguy, Ana Lisa Taylor Tavares, Ellen RA Thomas, Simon R Thompson, Arianna Tucci, Matthew J Welland, Eleanor Williams, Katarzyna Witkowska, Suzanne M Wood, and Magdalena Zarowiecki
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. Partial (anonymised) information can be provided on request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study underwent ethical review by the National Research Ethics Service Committee for Yorkshire & the Humber–South Yorkshire REC (ref: 13/YH/0028, IRAS project ID: 82448) and underwent local ethical approval at other centres involved in this study. The 100,000 Genomes Project is covered by REC reference approval 14/EE/1112. Participants gave informed consent to participate in the study before taking part.
References
- 1. Perälä N, Sariola H, Immonen T. More than nervous: the emerging roles of Plexins. Differentiation 2012;83:77–91. 10.1016/j.diff.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 2. Alto LT, Terman JR. Semaphorins and their signaling mechanisms. Methods Mol Biol 2017;1493:1–25. 10.1007/978-1-4939-6448-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maestrini E, Tamagnone L, Longati P, et al. A family of transmembrane proteins with homology to the MET-hepatocyte growth factor receptor. Proc Natl Acad Sci U S A 1996;93:674–8. 10.1073/pnas.93.2.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tamagnone L, Artigiani S, Chen H, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999;99:71–80. 10.1016/s0092-8674(00)80063-x [DOI] [PubMed] [Google Scholar]
- 5. Alves CJ, Yokoto K, Zou H, et al. Origin and evolution of Plexins, Semaphorins, and met receptor tyrosine Kinases. Scientific Reports 2019;9:1970. 10.1038/s41598-019-38512-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swiercz JM, Kuner R, Behrens J, et al. Plexin-B1 directly interacts with PDZ-Rhogef/LARG to regulate Rhoa and growth cone morphology. Neuron 2002;35:51–63. 10.1016/s0896-6273(02)00750-x [DOI] [PubMed] [Google Scholar]
- 7. Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the Guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-Rhogef. J Biol Chem 2002;277:43115–20. 10.1074/jbc.M206005200 [DOI] [PubMed] [Google Scholar]
- 8. Perälä NM, Immonen T, Sariola H. The expression of plexins during mouse embryogenesis. Gene Expr Patterns 2005;5:355–62. 10.1016/j.modgep.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 9. Worzfeld T, Püschel AW, Offermanns S, et al. Plexin-B family members demonstrate non-redundant expression patterns in the developing mouse nervous system: an anatomical basis for morphogenetic effects of sema4D during development. Eur J Neurosci 2004;19:2622–32. 10.1111/j.0953-816X.2004.03401.x [DOI] [PubMed] [Google Scholar]
- 10. Luukko K, Kettunen P. Integration of tooth morphogenesis and innervation by local tissue interactions, signaling networks, and semaphorin 3A. Cell Adh Migr 2016;10:618–26. 10.1080/19336918.2016.1216746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L, Han Y, Chen Q, et al. Sema4D-Plexin-B1 signaling in recruiting dental stem cells for vascular stabilization on a microfluidic platform. Lab Chip 2022;22:4632–44. 10.1039/d2lc00632d [DOI] [PubMed] [Google Scholar]
- 12. Friedel RH, Kerjan G, Rayburn H, et al. Plexin-B2 controls the development of cerebellar granule cells. J Neurosci 2007;27:3921–32. 10.1523/JNEUROSCI.4710-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng S, Hirschberg A, Worzfeld T, et al. Plexin-B2, but not Plexin-B1, critically modulates neuronal migration and Patterning of the developing nervous system in vivo. J Neurosci 2007;27:6333–47. 10.1523/JNEUROSCI.5381-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao N, Ruan M, Koestler DC, et al. Epigenome-wide scan identifies differentially methylated regions for lung cancer using pre-diagnostic peripheral blood. Epigenetics 2022;17:460–72. 10.1080/15592294.2021.1923615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H, Zhao H. Prognosis related miRNAs, DNA methylation, and epigenetic interactions in lung adenocarcinoma. Neoplasma 2019;66:487–93. 10.4149/neo_2018_181029N805 [DOI] [PubMed] [Google Scholar]
- 16. Lin L, Wang Y, Bian S, et al. A circular RNA derived from Plxnb2 as a valuable predictor of the prognosis of patients with acute myeloid leukaemia. J Transl Med 2021;19:123. 10.1186/s12967-021-02793-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saez-Atienzar S, Bandres-Ciga S, Langston RG, et al. Genetic analysis of amyotrophic lateral sclerosis identifies contributing pathways and cell types. Sci Adv 2021;7:eabd9036. 10.1126/sciadv.abd9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y, Tejero R, Lee VK, et al. Plexin-B2 facilitates glioblastoma infiltration by modulating cell biomechanics. Commun Biol 2021;4:145. 10.1038/s42003-021-01667-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yin J, Chun C-A, Zavadenko NN, et al. Next generation sequencing of 134 children with autism spectrum disorder and regression. Genes (Basel) 2020;11:853. 10.3390/genes11080853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hemida AS, Mareae AH, Elbasiony ASA, et al. Plexin-B2 in psoriasis; a clinical and immunohistochemical study. J Immunoassay Immunochem 2020;41:718–28. 10.1080/15321819.2020.1741385 [DOI] [PubMed] [Google Scholar]
- 21. Wang X, Shi W, Zhao S, et al. Whole exome sequencing in unexplained recurrent miscarriage families identified novel pathogenic genetic causes of euploid miscarriage. Hum Reprod 2023;38:1003–18. 10.1093/humrep/dead039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guimier A, de Pontual L, Braddock SR, et al. Biallelic alterations in Plxnd1 cause common arterial trunk and other cardiac malformations in humans. Hum Mol Genet 2023;32:353–6. 10.1093/hmg/ddac084 [DOI] [PubMed] [Google Scholar]
- 23. Dworschak GC, Punetha J, Kalanithy JC, et al. Biallelic and monoallelic variants in Plxna1 are implicated in a novel neurodevelopmental disorder with variable cerebral and eye anomalies. Genet Med 2021;23:1715–25. 10.1038/s41436-021-01196-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hebsgaard SM, Korning PG, Tolstrup N, et al. Splice site prediction in arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 1996;24:3439–52. 10.1093/nar/24.17.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell 2019;176:535–48. 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- 27. Rentzsch P, Schubach M, Shendure J, et al. CADD-splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med 2021;13:31. 10.1186/s13073-021-00835-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 2016;99:877–85. 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–81. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 31. Plagnol V, Curtis J, Epstein M, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012;28:2747–54. 10.1093/bioinformatics/bts526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou X, Zheng W, Li Y, et al. I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat Protoc 2022;17:2326–53. 10.1038/s41596-022-00728-0 [DOI] [PubMed] [Google Scholar]
- 33. Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 34. Reis LM, Tyler RC, Muheisen S, et al. Whole exome sequencing in dominant cataract identifies a new causative factor, Cryba2, and a variety of novel Alleles in known genes. Hum Genet 2013;132:761–70. 10.1007/s00439-013-1289-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. 100,000 Genomes Project Pilot Investigators, Smedley D, Smith KR, et al. 100,000 Genomes pilot on rare-disease diagnosis in health care. N Engl J Med 2021;385:1868–80. 10.1056/NEJMoa2035790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007;76:51–74. 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- 37. Sobreira N, Schiettecatte F, Valle D, et al. Genematcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015;36:928–30. 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melé M, Ferreira PG, Reverter F, et al. Human genomics. the human transcriptome across tissues and individuals. Science 2015;348:660–5. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bauwens M, Garanto A, Sangermano R, et al. Abca4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet Med 2019;21:1761–71. 10.1038/s41436-018-0420-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pascoe HG, Wang Y, Zhang X. Structural mechanisms of plexin signaling. Prog Biophys Mol Biol 2015;118:161–8. 10.1016/j.pbiomolbio.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morr T. Amelogenesis imperfecta: more than just an enamel problem. J Esthet Restor Dent 2023;35:745–57. 10.1111/jerd.13063 [DOI] [PubMed] [Google Scholar]
- 43. Dong J, Ruan W, Duan X. Molecular-based phenotype variations in amelogenesis imperfecta. Oral Dis 2023;29:2334–65. 10.1111/odi.14599 [DOI] [PubMed] [Google Scholar]
- 44. Smith CEL, Poulter JA, Antanaviciute A, et al. Amelogenesis imperfecta; genes, proteins, and pathways. Front Physiol 2017;8:435. 10.3389/fphys.2017.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith CEL, Poulter JA, Levin AV, et al. Spectrum of Pex1 and Pex6 variants in heimler syndrome. Eur J Hum Genet 2016;24:1565–71. 10.1038/ejhg.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmg-2023-109728supp001.pdf (18.3MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information. Partial (anonymised) information can be provided on request.