Abstract
Infectious hematopoietic necrosis virus (IHNV) infection in tissue culture cells has previously been shown to result in the shutdown of host protein synthesis, cell rounding, and cell death. We report here an investigation of the cytopathogenicity of the viral phosphoprotein (P or M1), matrix (M or M2), and nonvirion (NV) proteins in cultured fish cells. The expression of M alone potently inhibited reporter gene expression from a viral and an interferon (IFN)-inducible promoter, whereas P and NV did not produce a similar effect. Northern blot analysis further revealed a reduction in the steady-state level of reporter mRNA when the M gene was cotransfected into cells; conversely, M mRNA was not drastically reduced in the same cells. By immunofluorescence confocal microscopy, fragmented nuclei were found in some cells expressing M protein but not in cells expressing P, NV, or β-galactosidase protein. Electron microscopy revealed the morphological changes associated with apoptosis in the M-transfected cells. Furthermore, IHNV infection was shown to produce DNA “laddering” in cultured cells. Taken together, these data suggested at least two functions for M protein in an IHNV infection: down regulation of host transcription and the induction of programmed cell death. In the course of these experiments, we also discovered that NV expression was associated with cell rounding, the first biological effect on cells to be attributed to the NV gene.
The rhabdovirus matrix (M) protein has many different functions in virus replication, the most obvious one being the initiation of virion assembly by forming a bridge between the host plasma membrane and the ribonucleocapsid core (6, 12, 13). For vesicular stomatitis virus (VSV), the M protein has been shown to be solely responsible for the cytopathic effect typically seen as rounding of polygonal cells in culture (11). VSV M protein is also a potent inhibitor of host-directed transcription in mammalian cells when expressed in the absence of other viral components (1, 8, 9, 17, 31, 35). It was first shown by double transient-transfection experiments that VSV M protein could inhibit the transcription of a cotransfected plasmid, pSVCAT (simian virus 40 early-promoter-controlled chloramphenicol acetyltransferase [CAT]), while it stimulated the translation of the CAT mRNA (8, 9). The combined effect was a greater-than-20-fold inhibition of the reporter CAT activity in the M- and CAT-cotransfected cells (9). VSV M protein also inhibited other viral as well as cellular promoters including the human beta interferon (IFN-β) promoter (17, 35). Most recently, Ahmed and Lyles (1) have shown that VSV M protein is capable of suppressing the transcription directed by each of the three RNA polymerases (RNAP): RNAPI, RNAPII, and RNAPIII. We sought to determine whether the M proteins of a rhabdovirus from an entirely different genus could function in the same manner.
We examined the effect of M protein expression on fish cells for a fish rhabdovirus, infectious hematopoietic necrosis virus (IHNV). This virus is a member of the new genus Novirhabdovirus of the family Rhabdoviridae (35a) and is characterized by a sixth gene, encoding a nonvirion protein, located between the glycoprotein and polymerase genes (5, 28, 37). The six genes of IHNV are mapped on the genome in the following order: 3′-N-P(M1)-M(M2)-G-NV-L-5′, where N is the nucleocapsid protein, P or M1 is the phosphoprotein, M or M2 is the matrix protein, G is the glycoprotein, NV is the nonvirion protein, and L is the polymerase protein (27). The virus kills young salmonid fish (2), and most survivors become carriers (14, 25). In tissue culture cells, IHNV infection causes the shutdown of host protein synthesis (23, 30) and cytopathology characterized by cell rounding and cell death. Persistent infection has also been established in fish cells infected with IHNV (15). Thus, the virus produces a cytolytic response in fish cells much like that observed for VSV in mammalian cells.
We report here a study on the role of the IHNV P, M, and NV proteins in viral cytopathogenesis. Using double transient transfections, we demonstrate that expression of the M gene can inhibit reporter gene expression from the human cytomegalovirus (CMV) immediate-early promoter (IEP) and from the cellular IFN- and double-stranded RNA-inducible 561 gene promoter (4). Northern blot analysis demonstrated a reduction in the level of reporter mRNA in the transfected cells. Further studies of M in transfected cells by immunofluorescence confocal microscopy and electron microscopy revealed the nuclear fragmentation characteristic of apoptosis. Therefore, it appears that M acts in IHNV infection by shutting down host transcription and triggering programmed cell death. In the course of these studies, we also found that NV expression was associated with cell rounding, a cytopathic characteristic found in IHNV-infected cells in culture. This observation is the first biological phenomenon attributed to the NV gene.
[This article reports a portion of the work encompassed by a thesis submitted to Department of Microbiology, Oregon State University, in partial fulfillment of the requirements for the Ph.D. degree for P. P. Chiou.
Plasmids pP(+), pP(−), and pM(+) were constructed by Patty A. Ormonde as part of her work toward a Master's degree at Oregon State University (34).]
MATERIALS AND METHODS
Cells and viruses.
The chinook salmon embryonic cell line (CHSE-214) (20) and the epithelioma papulosum cyprini cell line (EPC) (18) were grown at 17°C in minimum essential medium (MEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) (Intergen), 100 IU of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine. The cells were maintained in an incubator culture chamber (C.B.S. Scientific) perfused with a blood-gas mixture composed of 9.9% (mol/mol) CO2, 10.2% (mol/mol) O2, and 79.9% (mol/mol) N2. Two IHNV isolates were used in the DNA fragmentation assay: RB1, a type 1 isolate, taken from an adult steelhead trout (Oncorhynchus mykiss) at the Round Butte Hatchery in central Oregon in 1975, and RA, a type 2 isolate, obtained in 1983 from dead rainbow trout fry at the International Aquaculture Research Center (Rangen Research), Hagerman, Idaho (23).
Plasmid DNA constructs and DNA transfection.
The P, M, and NV genes from the IHNV RB1 isolate (27, 34) were subcloned into the eukaryotic expression vector pcDNA3 (Invitrogen). The viral genes were placed downstream of the CMV IEP to generate plasmids pcDNA3-P(+), pcDNA3-P(−), pcDNA3-M(+), pcDNA3-M(−), pcDNA3-NV(+), and pcDNA3-NV(−). We shortened the plasmid nomenclature to pP(+), pP(−), pM(+), pM(−), pNV(+), and pNV(−), respectively. The construction of pP(+), pP(−), and pM(+) was reported previously (34) and is described briefly here. Plasmids pP(+) and pP(−) contain the entire P gene in the protein-encoding and noncoding orientations, respectively. Plasmid pM(+) contains the complete M gene in the protein-encoding orientation. A fragment containing the entire open reading frame and the 3′ nontranslated region of the M gene was generated by PCR from pM(+) and was subcloned in the noncoding orientation into the EcoRI-EcoRV site of pcDNA3 to generate plasmid pM(−). A fragment containing the complete open reading frame of the NV gene was generated by PCR from plasmid pNV137 (27) and was used to generate plasmids pNV(+), containing the PCR-amplified fragment inserted into the BamHI-EcoRV site of pcDNA3 in the protein-encoding orientation, and pNV(−), containing the amplified fragment inserted into the HindIII-BamHI site in the noncoding orientation.
Three reporter plasmids were employed in the study, pCMV4EAL (pLuc), p561-Luc, and pcDNA3-βgal [p(gal)]. pLuc contains the firefly luciferase gene under control of the CMV IEP in plasmid pCMV4 as described by Anderson et al. (3). p561-Luc (4) harbors the luciferase gene under control of the IFN- and double-stranded RNA (dsRNA)-inducible 561 gene promoter, which consists of nucleotides −134 to +1 of the 561 gene and the IFN-stimulated response element within the region (4). This plasmid was kindly provided by G. T. Leonard, Jr. (Case Western Reserve University). The third construct, p(gal), contains the HindIII-BamHI β-galactosidase fragment from pSV-β-Galactosidase plasmid (Promega) inserted downstream of the CMV IEP at the HindIII-BamHI site of the pcDNA3 vector.
Unless otherwise stated, transfections of the cells were performed in six-well plates (Corning). For CHSE-214 cells, monolayers were incubated for 6 h at 17°C with a solution of 1 ml of Opti-MEM (Gibco-BRL) per ml containing 18 μg of Lipofectamine (Gibco-BRL) and 2 μg of total DNA. Transfected cells were then washed twice with Opti-MEM, resupplemented with 2 ml of MEM containing 10% FBS, and maintained in an incubator culture chamber perfused with a blood-gas mixture. For EPC cells cotransfected with p561-Luc, monolayers were incubated with 1 ml of Opti-MEM containing 12 μg of Lipofectamine and 2 μg of total DNA. Transfected cells were incubated for 6 h at 20 to 22°C, washed, and then incubated with MEM without FBS.
Luciferase assays and β-galactosidase assays.
Luciferase assays were performed with the enhanced luciferase detection system as specified by the manufacturer (Analytical Luminescence Laboratory). Luciferase activity was measured and integrated over a 30-s period in a Beckman LS 8000 liquid scintillation counter on single-photon mode. The resultant counts per minute (cpm) was converted into the total cpm by using the dilution factor of each sample.
The β-galactosidase activity was determined in situ as described by Fischer et al. (19). Briefly, transfected cells were fixed and incubated for 5 h at 37°C in a solution containing 0.2% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 10 mM sodium phosphate buffer (pH 7.0), 150 mM NaCl, 1 mM MgCl2, 3.3 mM K4Fe(CN)6 · 3H2O, and 3.3 mM K3Fe(CN)6, and the blue cells were identified under a light microscope.
Southern and Northern blot analysis.
CHSE-214 cells (107) were cotransfected with either pM(+)/pLuc plasmid DNAs or pM(−)/pLuc plasmid DNAs at a 1:1 ratio. At 12 h posttransfection, the cells were washed with phosphate-buffered saline (PBS) and subjected to DNase I treatment. The cells were harvested and lysed for 30 min at 4°C in ice-cold lysis buffer (10 mM Tris, 0.5% Triton X-100 [pH 7.5]) containing proteinase K (0.6 μg/ml). Total DNA was extracted with phenol-chloroform, digested with SmaI, and electrophoresed on a 1% agarose gel. The separated DNA bands were transferred onto a Nytran membrane (Schleicher & Schuell), and the luciferase gene was detected by a luciferase-specific dsDNA probe, which was labeled with [α-32P]dCTP (Amersham) and generated by a random priming reaction using the complete luciferase gene from pLuc as template.
At 24 and 72 h posttransfection, total RNA was extracted from the above transfected cells with RNAzol (Tel-Test, Inc.) as recommended by the manufacturer. Polyadenylated RNA was separated from total RNA with Oligotex-dT (Qiagen). The RNA samples were electrophoresed on 1% agarose gels after denaturation with glyoxal and dimethyl sulfoxide and then transferred to a Nytran membrane. To detect the M transcripts, a dsDNA probe labeled with [α-32P]dCTP was generated by a random priming reaction using the full-length M gene as template. The relative intensity of the specific signal was obtained by scanning the blot in a PhosphorImager scanner (Molecular Dynamics).
Immunofluorescence and confocal microscopy analysis.
CHSE-214 cells were grown in eight-well chamber slides (Fisher) to 70 to 80% confluency and were transfected with pP(+), pM(+), pNV(+), or p(gal). At 48 h posttransfection, the transfected cells were washed twice with PBS and fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. For permeabilization, the cells were incubated with 0.5% Triton X-100 in PBS for 5 min at room temperature. A solution of 1% bovine serum albumin–0.1% Tween 20 in PBS was used as a blocking agent and incubated with the cells for 30 min at room temperature. At this point, the blocking agent was used as the diluent for the primary- and secondary-antibody preparations. The samples were incubated with a 1:200 dilution of rabbit anti-P (16), rabbit anti-M (16), or rabbit anti-NV (unpublished data) serum for 1 h. The cells were washed and subsequently incubated with goat anti-rabbit immunoglobulin conjugated with Texas Red (10 μg/ml; Molecular Probes, Inc.) for 1 h. After the washing step, the nucleic acid-staining dye DAPI (4′,6-diamidino-2-phenylindole) at 250 nM was added for 5 min. The cells were washed and allowed to air dry, and the slides were mounted in Cytoseal (Stephens Scientific). Confocal images were captured using a Leica TCS 4D confocal microscope and combined using Photoshop software (Adobe, Mountain View, Calif.).
Electron microscopy.
Mock-transfected and pM(+)-transfected cells were collected, washed in PBS, and centrifuged to form a pellet. The pelleted cells were fixed in a mixture of glutaraldehyde and osmium tetroxide and processed by standard procedures. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Philips CM12 transmission electron microscope.
DNA fragmentation assay.
CHSE-214 cells in a 150 cm2 plate (2 × 107 cells) were infected with virus at multiplicity of infection (MOI) of 10. The low-molecular-weight DNA was extracted at 6, 12, and 24 h postinfection as described by Takizawa et al. (39). Briefly, the cells were washed twice in PBS, trypsinized, resuspended in 1.5 ml of PBS, and transferred to a 2-ml microcentrifuge tube. The cells were centrifuged for 15 s at 13,000 × g at 4°C and resuspended in 2 ml of PBS. These cells were centrifuged once more, and the pelleted cells were lysed in 800 ml of ice-cold lysis buffer and incubated on ice for 30 min. After centrifugation of the lysates for 10 min at 13,000 × g at 4°C, the supernatant fluids were extracted with buffered phenol followed by buffered phenol-chloroform-isoamyl alcohol. DNA was then precipitated with ethanol and treated with RNase A to a final concentration of 1.0 mg/ml for 30 min at 37°C. Aliquots of the DNA sample were electrophoresed through a 2% agarose gel and stained with ethidium bromide.
RESULTS
IHNV M inhibits host-directed expression of plasmid-encoded target genes.
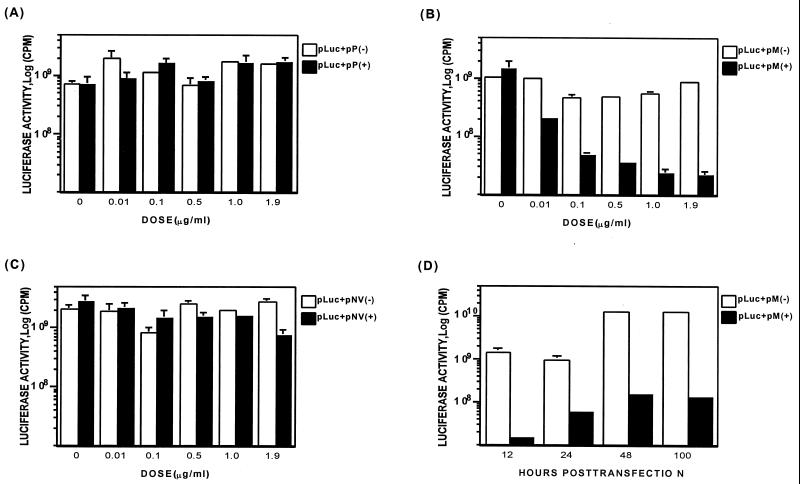
The effect of IHNV M, P, or NV gene expression on luciferase synthesis by a cotransfected plasmid, pLuc containing the firefly luciferase gene controlled by the CMV IEP, was examined in CHSE-214 cells. The experiment also included as controls three plasmids expressing the viral gene in antisense orientation, pP(−), pM(−), and pNV(−). CHSE-214 cells were cotransfected with the viral gene-encoding plasmid and a constant 0.1 μg of pLuc plasmid at DNA concentration ratios of 0:1, 0.1:1, 1:1, 5:1, 10:1, and 19:1 (Fig. 1). The total amount of plasmid DNA used in each transfection was kept at 2 μg per ml by the addition of pcDNA3, the parent plasmid of the viral gene-containing plasmids. At 24 h posttransfection, as shown in Fig. 1B, expression of M protein drastically inhibited the luciferase activity in a gene dosage-dependent manner. P and NV gene expression had no significant effect on target gene expression (Fig. 1A and C). The luciferase activity decreased 10-fold when the pM(+) plasmid was cotransfected at a 1:1 ratio with the pLuc plasmid and reached maximum inhibition (40-fold) at a ratio of 19:1. Even at a ratio as low as 0.1:1, there was a fivefold reduction in the luciferase activity. These results show that M expression alone inhibits the expression of the plasmid-located reporter gene.
FIG. 1.
Analysis of the effect of P, M, and NV genes on plasmid-encoded luciferase gene expression. (A to C) Monolayers of CHSE-214 cells were cotransfected with a viral gene-encoded plasmid and a constant 0.1 μg of pLuc plasmid at ratios of 0:1, 0.1:1, 1:1, 5:1, 10:1, and 19:1, respectively (see Materials and Methods for procedures). (A) Cells cotransfected with pP(+) or pP(−); (B) cells cotransfected with pM(+) or pM(−); (C) cells cotransfected with pNV(+) or pNV(−). Luciferase activity was assayed at 24 h posttransfection. The dose of viral gene-encoded plasmid in each transcription is shown on the x axis. (D) Time course of M-induced inhibition of the luciferase gene expression. CHSE-214 cells were cotransfected with pM(+) or pM(−) and a constant amount of pLuc at a ratio of 1.9:0.1 as described above. The cells were then lysed and analyzed at 12, 24, 48, and 100 h posttransfection.
Figure 1D depicts the data from a time course study of M-induced inhibition of luciferase gene expression. In the experiment, CHSE-214 cells were cotransfected with pM(+) or pM(−) and a constant amount of pLuc at a ratio of 1.9:0.1. At 12, 24, 48, and 100 h posttransfection, the transfected cells were harvested and analyzed for luciferase activity. Inhibition of luciferase activity was observed as early as 12 h posttransfection, and maximal inhibition was reached at about 48 h posttransfection, with a 100-fold reduction. In the assay, the medium was replaced with fresh medium daily to ensure a consistent supply of nutrients to the cells, and, as shown in Fig. 1D, the inhibition of luciferase activity persisted for at least 100 h.
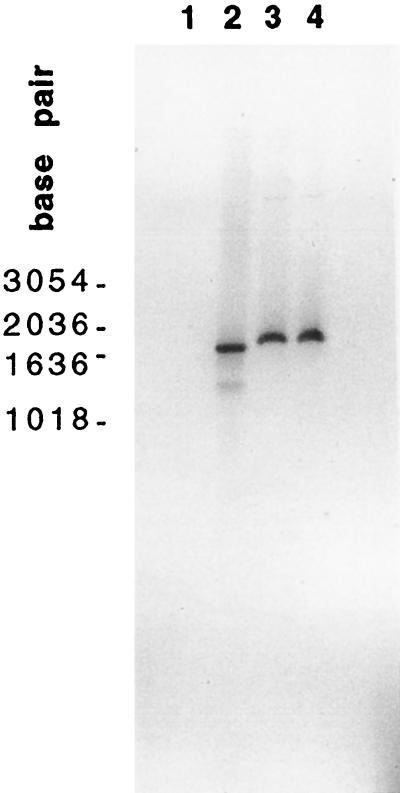
Southern blot analyses of the transfected cells were performed to determine whether the differences in luciferase expression might be due to differences in transfection efficiency. In this experiment, CHSE-214 cells were cotransfected at a 1:1 ratio with either pM(+) plus pLuc or pM(−) plus pLuc. At 12 h posttransfection, the cells were washed with PBS, subjected to DNase I treatment, and then lysed with proteinase K. Treatment with DNase I eliminated the possibility that the DNA may have been bound to the plasma membrane of the cells rather than internalized by the cells. Total DNA was extracted with phenol-chloroform and digested with SmaI, which cleaves the pLuc plasmid into two fragments of 1.9 kb (the luciferase insert) and 4.9 kb (the vector). The DNA samples were analyzed by Southern hybridization with a 32P-labeled dsDNA probe specific for the luciferase gene. Each lane was loaded with the same amount of DNA. As shown in Fig. 2, it appears that equal amounts of the 1.9-kb luciferase DNA were present in the cells cotransfected with pM(+) or pM(−) (lanes 3 and 4, respectively). The results indicate that the differences in luciferase activity in the M(+) and M(−) cell populations were not due to large differences in the transfection efficiency of the two populations.
FIG. 2.
Southern blot analysis of DNA from transfected cells. CHSE-214 cells (107) were cotransfected with either pM(+) plus pLuc or pM(−) plus pLuc at a 1:1 ratio. At 24 h posttransfection, the total cell DNA was extracted and digested with SmaI for Southern blots as described in Materials and Methods. Lanes: 1, the M fragment from pM(+); 2, a fragment of luciferase gene from the pCMV-Luc DNA by PCR amplification; 3, DNA from pM(−)-pLuc-cotransfected cells; 4, DNA from pM(+)-pLuc-cotransfected cells. SmaI cleaves the pLuc plasmid into two fragments of 1.9 kb (the luciferase insert) and 4.9 kb (the vector). Molecular sizes in base pairs are indicated at the left.
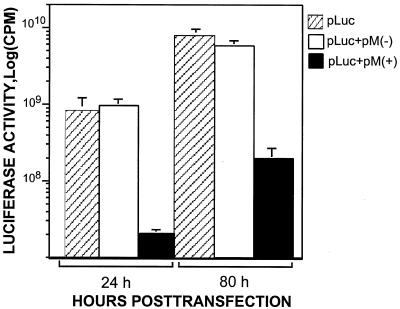
Extracellular luciferase activity was also measured to rule out the possibility that the differences in luciferase activity between the M(+) and M(−) populations were due to leakage of intracellular luciferase molecules through any disruption to the plasma membrane. In this experiment, the culture medium was not replaced daily, and so the accumulation of luciferase activity in the medium could be monitored. The culture medium was collected at 24 and 80 h posttransfection and then centrifuged to remove any cellular debris. As shown in Fig. 3, at 24 h posttransfection, approximately the same activity level of luciferase was detected in the culture medium of both the pM(−)-plus pLuc-cotransfected cells and the pLuc-transfected cells. The luciferase activity, however, was inhibited (40-fold) in the medium of the pM(+)-plus pLuc-cotransfected cells. This result correlated with the luciferase activity measured for the intracellular luciferase. Even at 80 h posttransfection, the extracellular luciferase activity of the pM(+)- plus pLuc-cotransfected cells was consistently lower (30- to 40-fold). This result demonstrated that the inhibition of luciferase expression in the pM(+)-transfected cells was not due to leakage of cytoplasmic components or detachment of the transfected cells.
FIG. 3.
Analysis of the luciferase activity in the culture medium. CHSE-214 cells were cotransfected with pM(+) or pM(−) and a constant amount of pLuc at the ratio of 1.9:0. In this assay, the culture medium was collected at 24 and 80 h posttransfection and then harvested by centrifugation to remove any cellular debris. The culture medium was not replaced daily, so that the accumulation of luciferase activity in the medium could be monitored.
The IHNV M-induced inhibitory effect is not gene specific.
We examined the possibility that the M-induced inhibitory effect could be a gene-specific event. In this experiment, the β-galactosidase gene under control of the same CMV promoter was used as a reporter. CHSE-214 cells were cotransfected with pM(+) and p(gal) or with pM(−) and p(gal) at ratios of 0:1 and 1:1. On day 5 after transfection, the cells were fixed in glutaraldehyde and incubated with X-Gal. The cells expressing β-galactosidase were then identified by their blue color under a light microscope, and the results are shown in Table 1. In the cells cotransfected with pM(+), there was a 96.5% reduction in the number of cells expressing β-galactosidase, while the reduction in β-galactosidase-expressing cells in the pM(−)-cotransfected population was far less, at 25%. These data provide in situ evidence that IHNV M inhibits reporter gene expression.
TABLE 1.
Analysis of the inhibitory effect of IHNV M gene on host-directed expression of reporter β-galactosidasea
| Plasmid used for transfection | No. of positive cells | Reduction (%)b |
|---|---|---|
| p(gal) (1 μg) | 5,544 | |
| pM(+) + p(gal) (1 μg/1 μg) | 192 | 96.5 |
| pM(−) + p(gal) (1 μg/1 μg) | 4,236 | 23.6 |
On day 5 after transfection, cells were fixed and stained with 0.2% X-Gal as described in the Materials and Methods. Cells expressing β-galactosidase were identified by the blue color under a light microscope.
Reduction is expressed by [(number of β-galactosidase-expressing cells in cells cotransfected with M-encoded plasmid − number of β-galactosidase-expressing cells in cells transfected with p(gal) alone)/the number of β-galactosidase-expressing cells in cells transfected with p(gal)alone] × 100.
IHNV M suppresses the expression of target gene mRNA.
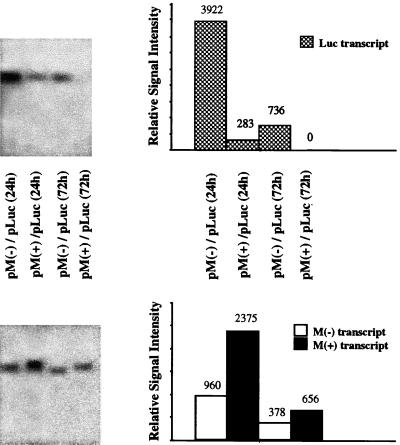
The quantity of luciferase mRNA produced in the presence or absence of M expression was compared by Northern analysis. In this assay, CHSE-214 cells were cotransfected with either pM(+) plus pLuc or pM(−) plus pLuc at a 1:1 ratio, and at different time points polyadenylated RNA was isolated from the transfected cells and analyzed in Northern blots probed with a 32P-labeled DNA probe specific to the luciferase gene. As shown in Fig. 4, the amount of luciferase mRNA was reduced in cells cotransfected with pM(+) in comparison to that in cells cotransfected with pM(−) at 24 h and 72 h posttransfection. The relative signal intensity of luciferase mRNA was measured in a PhosphorImager scanner and found to be decreased by 15-fold at 24 h and gone by 72 h in pM(+)- plus pLuc-cotransfected cells. The relative level of luciferase mRNA in cells cotransfected with pM(−) was decreased fivefold from 24 to 72 h posttransfection. Thus, the expression of the M gene resulted in a reduction in the concentration of luciferase mRNA.
FIG. 4.
Northern blot analysis of the luciferase and M transcripts. At 24 and 72 h posttransfection, polyadenylated RNA was isolated from the transfected cells in Fig. 2 and subjected to Northern blot analysis with a 32P-labeled dsDNA probe specific to the luciferase or M genes. Gels in both panels contain equal amounts of the same RNA samples.
The quantity of polyadenylated M transcripts from the pM(+) and pM(−) plasmids was determined with a 32P-labeled dsDNA probe capable of distinguishing between the positive- and negative-sense transcripts of the M gene based on their different sizes (Fig. 4). A comparison of the relative signal intensity revealed that the amount of the M(+) polyadenylated RNA was about twofold higher in the pM(+)-cotransfected cells than the amount of the M(−) polyadenylated RNA in the pM(−)-cotransfected cells at 24 and 72 h posttransfection. Thus, IHNV M did not suppress its own transcription as drastically as it suppressed that of the luciferase gene. Although there are a number of explanations for these results, such as differences in hybridization efficiency, differences in the specific activity of the M(+) and the M(−) probes, or the different stabilities of the M(+) and M(−) transcripts, the simplest explanation is that M does not suppress its own transcription.
IHNV M inhibits the expression of a target gene controlled by a cellular IFN/dsRNA-inducible promoter.
The effect of IHNV M on the expression from a cellular promoter was examined with a reporter gene under control of an IFN- and dsRNA-inducible 561 gene promoter. The 561 gene in human cells is strongly induced by type I IFNs (IFN-α and IFN-β) and by dsRNA or virus infection that produces dsRNA (41), and it is significantly induced by poly(I-C) dsRNA in some fish cell lines (Marc Johnson, Oregon State University, personal communication). The 561 gene promoter contains a single ISRE, which is a crucial cis-acting element for both IFN and dsRNA signaling (4). Although the 561 gene can be induced by both IFN and dsRNA, the induction by poly(I-C) in mammalian cells does not rely on the intermediate synthesis of IFN and seems to be mediated by a different pathway from the conventional IFN signaling pathway, i.e., the JAK-STAT pathway (4).
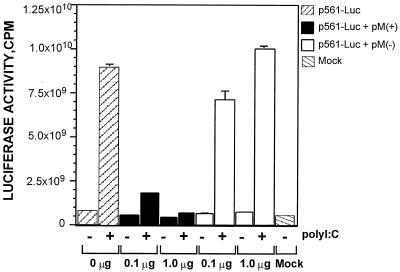
In this assay, a carp cell line, EPC, was cotransfected with pM(+) and p561-Luc or with pM(−) and p561-Luc at ratios of 1:1, 0.1:1, and 0:1 (Fig. 5). At 12 h posttransfection, the transfected cells were treated with medium without serum (MEM-0) or 100 μg of poly(I-C) dsRNA per ml in MEM-0. After exposure to poly I:C for 12 h, the cells were harvested and analyzed for luciferase activity. As shown in Fig. 5, the expression of M abolished the poly(I-C)-induced luciferase activity in a dosage-dependent fashion similar to that observed in Fig. 1B. The luciferase activity decreased fivefold when the M(+) plasmid was cotransfected at a 0.1:1 ratio with the p561-Luc plasmid and decreased 15-fold at a 1:1 ratio. In fact, the luciferase activity of pM(+)-cotransfected cells was abolished almost completely at a 1:1 ratio, as shown in Fig. 5. The result demonstrated that the IHNV M-induced inhibition of host directed gene expression is not a host-specific event and that gene expression from both a viral and a cellular promoter can be inhibited by M.
FIG. 5.
Analysis of the effect of IHNV M on the expression of luciferase under the control of the IFN- and dsRNA-inducible 561 gene promoter. EPC cells were cotransfected with pM(+) p561-Luc or pM(−) plus p561-Luc at ratios of 0:1, 0.1:1 and 1:1. Transfected cells were mock induced or induced with poly I:C (100 μg/ml) at 12 h posttransfection. Luciferase activity was analyzed at 12 h after poly(I-C) induction.
Transfection of IHNV M gene and IHNV infection causes morphological changes of apoptosis.
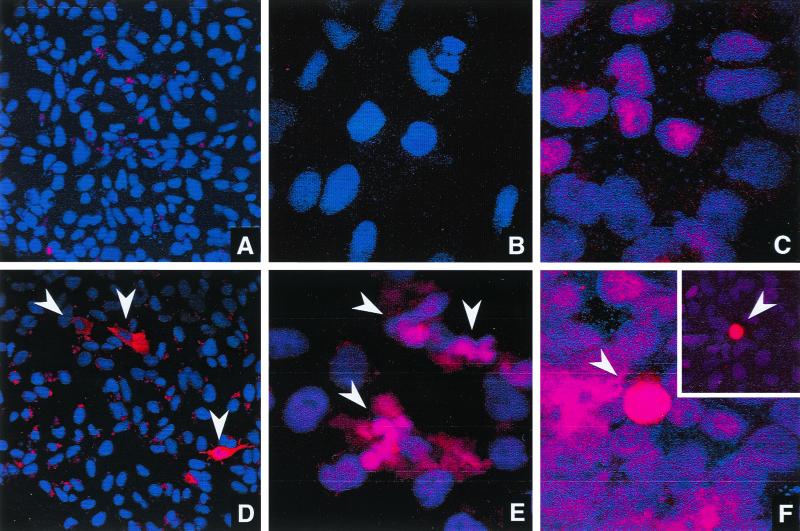
The presence of M protein in the transfected cells and its cytopathic effect were further examined by immunofluorescence confocal microscopy. CHSE-214 cells transfected with pP(+), pM(+), or pNV(+) were fixed and incubated with antiserum specific to P, M, or NV protein, respectively. Cells transfected with p(gal) were included as a control. A UV-excited blue-emitting fluorophore, DAPI, was used to fluorescently label the nuclei of these transfected cells. Confocal microscopy was conducted by capturing single optical sections through the nucleus of the cell. In IHNV-infected cells, M and NV proteins were expressed in both the nucleus and cytoplasm while the presence of P protein was confined to the cytoplasm (unpublished data). As shown in Fig. 6, P protein was expressed in the cytoplasm and NV protein was found in both the nucleus and the cytoplasm of transfected cells. Interestingly, NV protein was frequently identified in cells exhibiting cell rounding, a typical cytopathic effect caused by IHNV infection. M protein was detected in both the nucleus and the cytoplasm of transfected and IHNV-infected cells. Fragmented nuclei were found in approximately 10% of the cells expressing M protein (Fig. 6E). The nuclear fragmentation was observed only in the pM(+)-transfected cells, an observation made consistently in three separate sets of experiments.
FIG. 6.
Immunofluorescence analysis of the cellular localization of transgenic proteins. In panels D, E, and F, CHSE-214 cells were transfected with pP(+), pM(+) and pNV(+), respectively. Cells transfected with p(gal) (A to C) were included as control. At 48 h posttransfection, cells were fixed, labeled with antiserum specific to P (A and D), M (B and E), or NV (C and F), incubated with goat anti-rabbit immunoglobulin G conjugated to Texas Red (10 mg/ml), and counterstained with 250 nM DAPI. Confocal images were captured using a Leica TCS4 confocal microscope. Magnification: ×400 (A and D) and ×1,000 (to show more details in the M- and NV-transfected cells) (B, C, E, and F). All positive transfected cells in panels D to F are indicated by arrowheads. The inset in panel F was taken from a different field under the microscope due to the low frequency of NV-transfected cells.
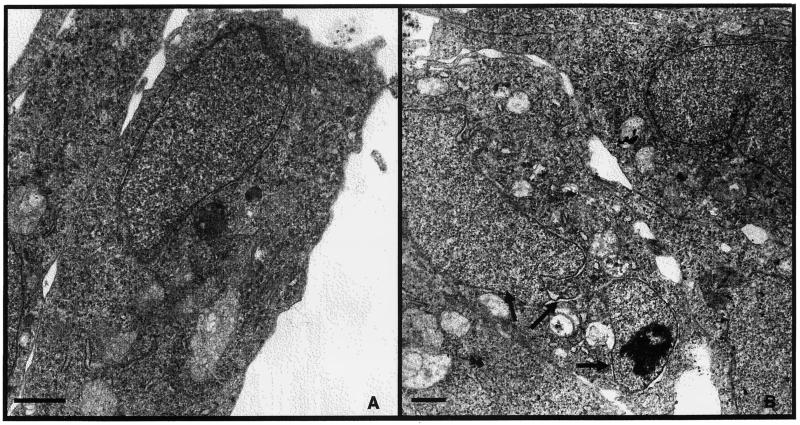
Fragmented nuclei are characteristic of cells undergoing programmed cell death (apoptosis). Other morphological features of apoptotic cells include cell shrinkage, plasma membrane blebbing, nuclear condensation and fragmentation, and, ultimately, the appearance of apoptotic bodies. Figure 7B shows the ultrastructure of a pM(+)-transfected CHSE-214 cell, revealing more morphological changes typical of apoptosis. In this cell, the nucleus is divided into three fragments (indicated by arrows) and cell shrinkage is apparent from the increased ratio of nucleus to cytoplasm. A mild degree of chromatin DNA condensation was also observed in one fragment. This observation provided another illustration of the nuclear fragmentation associated with M protein expression. Further studies that employ immunogold labeling of cells transfected with the M gene are needed to confirm the relationship between M protein expression and nuclear fragmentation in the electron micrographs.
FIG. 7.
Electron micrographs of mock- and pM(+)-transfected CHSE-214 cells. Mock-transfected (A) or pM(+)-transfected (B) CHSE-214 cells were harvested and fixed at 24 h posttransfection. Ultrathin sections were stained and examined in a Philips CM2 transmission electron microscope. Note the fragmented nucleus (indicated by arrows), chromatin condensation, and cell shrinkage. Magnification: ×8,000 (A) and ×6,300 (B). Bar, 100 μm.
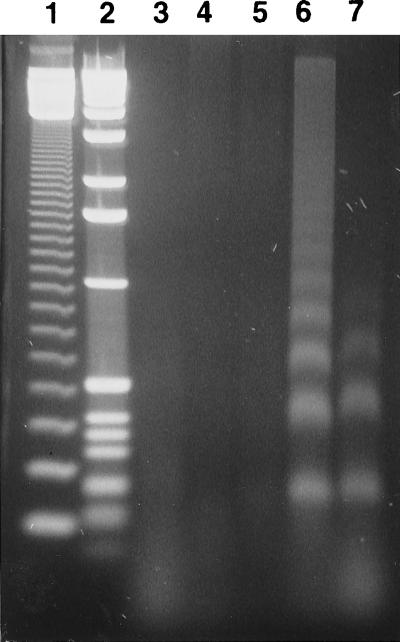
It was possible that M induction of apoptosis was a consequence of M overexpression rather than a typical consequence of IHNV infection. The effect of IHNV infection on the chromosomal DNA of CHSE-214 cells was evaluated in DNA-laddering gels. One hallmark of apoptosis is the activation of a calcium-dependent endonuclease that fragments chromosomal DNA into oligomeric multimers of 180 to 200 bp. In this assay, CHSE-214 cells were mock infected or infected with an IHNV RB1 or RA isolate at a MOI of 10. Cells were harvested, and the low-molecular-weight DNA was isolated at 6, 12, and 24 h postinfection and then analyzed by electrophoresis on a 2% agarose gel. As shown in Fig. 8, at 24 h postinfection, DNA from both RB1- and RA-infected CHSE-214 cells exhibited the apoptotic pattern of DNA laddering (lanes 6 and 7). This laddering was absent in DNA from mock-infected cells at 24 h (lane 3) and DNA from RB1-infected cells at 6 and 12 h postinfection (lanes 4 and 5). The result demonstrated that nuclear fragmentation can be triggered by IHNV infection and suggested that expression of M protein during IHNV infection may initiate programmed cell death in infected cells.
FIG. 8.
DNA fragmentation analysis of IHNV-infected CHSE-214 cells. CHSE-214 cells (2 × 107) were infected with virus at MOI of 10, and the low-molecular-weight DNA was obtained at 6, 12, and 24 h postinfection. DNA was treated with RNase A and then electrophoresed through 2% agarose. Lanes: 1, molecular size marker, 123-bp DNA ladder (Gibco BRL); 2, 1-kb DNA ladder (Gibco BRL); 3, DNA isolated from mock-infected cells at 24 h postinfection; 4, IHNV RB1-infected cells, 6 h; 5, IHNV RB1-infected cells, 12 h; 6, IHNV RB1-infected cells, 24 h; 7, IHNV RA-infected cells, 24 h.
DISCUSSION
Down regulation of protein synthesis is one effect of IHNV infection on the host cell. In this report, we show that the expression of IHNV M protein alone may be responsible for this shutdown. M expression results in the inhibition of plasmid DNA-directed transcription, leading to the inhibition of plasmid-directed protein synthesis. Our hypothesis is that the same mechanism is involved in the shutdown of host cell protein synthesis. Prior studies on the VSV M protein have shown that this protein is a potent inhibitor of host-directed transcription (1, 8, 9, 17, 31, 34). Our results with IHNV extend these observations and indicate that inhibition of host transcription is a common function of the matrix proteins of the Rhabdoviridae family.
Multiple mechanisms may contribute to the shutdown of host protein synthesis by rhabdovirus matrix proteins. It has been demonstrated recently that VSV M protein inhibits the transport of certain species of DNAs and proteins from the cytoplasm to the nucleus by interfering with the Ran-dependent nuclear transport system (22). VSV M also potently suppresses the transcription directed by the host RNA polymerases RNAPI, RNAPII, and RNAPIII (1). Furthermore, VSV infection leads to the inhibition of host RNAPII-dependent transcription as a result of inactivation of transcription factor IID, while the purified recombinant TATA-binding protein is able to reconstitute the transcription activity (46). IHNV M did not seem to suppress its own transcription as drastically as it suppressed that of the reporter gene driven by the same promoter, CMV IEP. Thus, the regulatory element(s) distinguishing IHNV-encoded genes from “other” genes in the transcriptional process may be present in the 5′ or 3′ noncoding region of the M gene. Such promoter-dependent inhibition has also been found in the VSV M-induced inhibition of transcription by RNAPIII.
Experiments showing that the VSV M protein suppresses gene expression from the human IFN-β promoter supports the theory that M protein plays a central role in regulating the host response to viral infection by down regulating IFN production (17). Ferran and Lucas-Lenard showed that the M gene from a VSV-Indiana mutant, which had been characterized as a good inducer of IFN, was defective in inhibiting transcription from the IFN-β promoter whereas wild-type VSV-Indiana M protein induced IFN poorly and potently suppressed expression from the IFN-β promoter. These results are very different from those of Marcus et al. (33), who reported that the predicted M-protein amino acid sequence of a wild-type VSV-Indiana field isolate that strongly induced IFN in chicken embryo cells was identical to the M sequence of both a poorly inducing wild-type field isolate and a noninducing wild-type laboratory strain. For IHNV, the M gene alone suppresses transcription from the IFN-inducible promoter for the mammalian 561 gene. Since this promoter contains only one ISRE and no other binding site for known transcriptional factors, the suppression presumably involves this ISRE site. The inhibition of p561-Luc by the IHNV M in fish cells provides further evidence that EPC cells contain the same or similar signal transduction systems for IFN induction as do mammalian cells. The trout IFN regulatory factor 1, STAT1, STAT3, STAT4, and STAT5 (Marc Johnson, personal communication), and the trout IFN-responsive genes, RbTMx1, RbTMx2, and RbTMx3 (42–44), are all highly conserved genes. Thus, the suppression of a mammalian IFN-inducible promoter in fish cells was not surprising.
The association of the IHNV M protein and nuclear fragmentation was consistently observed in this study despite the low frequency of cells expressing the transfected viral protein. Low transfection frequencies with plasmid DNA have always been a problem with the study of gene expression in fish cells. We routinely observe transfection frequencies ranging from 0.5 to 2% with an occasional transfection rate of 10% in CHSE-214 and EPC cells. In contrast, the transfection frequency in RTG-2 cells (rainbow trout gonad cells) is virtually negligible. This makes the characterization of cellular changes induced by single genes in fish cells difficult. Nevertheless, our observations were confirmed in repeated experiments involving the scanning of dozens of microscopic fields. Nuclear fragmentation was observed only in M-transfected cells, and cell rounding was observed only in NV-transfected cells. These observations were consistently made in at least three different experiments for each transfected gene.
Other markers for apoptosis, e.g., lamin A cleavage (38) and poly(ADP-ribose) polymerase-cleaving activity (29), were not examined in this study for several reasons. There are no available monoclonal antibody reagents for trout poly(ADP-ribose) polymerase, and a typical interleukin-1 cleavage enzyme does not appear to cleave trout interleukin-1β in the same position as in mammalian pro-interleukin-1β (47). Characterization of the reagents necessary to carry out these assays in fish cells would, in itself, be useful and should provide sufficient information for a separate paper. Because we were unable to carry out the lamin A cleavage and PARP assays, this report only demonstrates that the IHNV M protein induces the “morphological changes consistent with apoptosis” in cultured fish cells.
Cell death in rhabdovirus infections had long been considered a consequence of necrosis until recent reports showed that apoptotic cell death occurred in VSV-infected HeLa cells (26), in rabies virus-infected mouse thymocytes (32) and brains (24), and in EPC cells infected by the fish-pathogenic rhabdovirus spring viremia of carp virus (7). In this report, we have also demonstrated that IHNV infection can trigger apoptotic cell death as evidenced by the oligonucleosomal DNA fragmentation. Taking these results together, apoptosis is very probably a common outcome of rhabdovirus infection. There is mounting evidence that the induction of apoptosis contributes directly to the pathogenesis of a number of viruses. There is, for example, in vitro and in vivo evidence that strongly supports an important pathogenic role of apoptosis in producing the neurological disease in rabies (24). Although we have investigated evidence for IHNV-induced apoptosis only in cultured cells, it is possible that the programmed cell death plays a significant role in IHNV infection in fish. It would be interesting in future studies to determine what types of tissues are killed by IHNV-induced apoptosis in infected fish.
An interesting but unexpected finding in the study was the observation that overexpression of NV resulted in cell rounding. Cells transiently transfected with a plasmid expressing the NV gene, pNV(+), were found to undergo cell rounding. This is the first biological function we have been able to assign to NV. Cell rounding is a common and obvious cytopathic effect observed in rhabdovirus-infected cells. This rounding has been assigned to the VSV M protein, which causes dissociation of the cytoskeleton (11). We have not observed cell rounding in cells transfected with the IHNV M gene. Further studies of the NV function should include verification of the interaction between NV protein and the cytoskeleton in IHNV-infected or NV gene-transfected cells.
In summary, this report provides evidence that the inhibition of host-directed transcription may be a common function for the matrix proteins of the Rhabdoviridae family. We have also shown that overexpression of the IHNV M gene leads to apoptosis in tissue culture cells, a finding that adds IHNV to the list of DNA and RNA viruses with genes that induce apoptosis (for reviews, see references 21 and 40). For example, the adenovirus type 12 E1A protein blocks transcription of major histocompatibility complex class I (43) and induces apoptosis in infected cells (36, 45). A relationship between IHNV M inhibition of transcription and M-induced apoptosis has not been determined in this study. However, mutation analysis of the VSV M protein has shown that inhibition of host-directed gene expression is genetically separable from its function in virus assembly (10) but is correlated with the virus-induced cell rounding (30). Mutational analysis of the IHNV M will be the subject of another report.
ACKNOWLEDGMENTS
We thank Grant Trobridge for his helpful comments on the manuscript and G. T. Leonard, Jr., (Case Western Reserve University) for generously providing the p561-Luc plasmid.
This research was supported by the U.S. Department of Agriculture grant to the Western Regional Aquaculture Consortium under grant 92-38500-7195, project 92080441; an Oregon Sea Grant with funds from the National Oceanic and Atmospheric Administration, Office of Sea Grant, Department of Commerce, under grant NA89AA-D-SG108, project R/FSD-16, grant NA36RG451, projects F/FSD-23 and Amend. No. 5; and a grant from the National Oceanic and Atmospheric Administration (Saltonstall-Kennedy funds), NA46FD0490.
Footnotes
Oregon Agricultural Experiment Station technical paper 11,687.
REFERENCES
- 1.Ahmed M, Lyles D S. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J Virol. 1998;72:8413–8419. doi: 10.1128/jvi.72.10.8413-8419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amend D F, Yasutake W T, Mead R W. A hematopoietic virus disease of rainbow trout and sockeye salmon. Trans Am Fish Soc. 1969;98:796–804. [Google Scholar]
- 3.Anderson E D, Mourich D V, Leong J C. Gene expression in rainbow trout (Onchorhynchus mykiss) following intramuscular injection of DNA. Mol Marine Biol Biotechnol. 1996;5:105–113. [PubMed] [Google Scholar]
- 4.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated responses elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 5.Basurco B, Benmansour A. Distant strains of the fish rhabdovirus VHSV maintain a sixth functional cistron which codes for a nonstructural protein of unknown function. Virology. 1995;212:741–745. doi: 10.1006/viro.1995.1534. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann J E, Fusco P J. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol. 1988;107:1707–1715. doi: 10.1083/jcb.107.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorklund H V, Johansson T R, Rinne A. Rhabdovirus-induced apoptosis in a fish cell line is inhibited by a human endogenous acid cysteine proteinase inhibitor. J Virol. 1997;71:5658–5662. doi: 10.1128/jvi.71.7.5658-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black B L, Brewer G, Lyles D S. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J Virol. 1994;68:555–560. doi: 10.1128/jvi.68.1.555-560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black B L, Rhodes R B, McKenzie M, Lyles D S. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol. 1993;67:4814–4821. doi: 10.1128/jvi.67.8.4814-4821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blondel D, Harmison G G, Schubert M. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J Virol. 1990;64:1716–1725. doi: 10.1128/jvi.64.4.1716-1725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong L D, Rose J K. Interaction of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulon P, Deutsch V, Lafay F, Martinet-Edelist C, Wyers F, Herman R C, Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- 14.Drolet B, Chiou P P, Leong J C. Detection of truncated virus particles in a persistent RNA virus infection in vivo. J Virol. 1995;69:2140–2147. doi: 10.1128/jvi.69.4.2140-2147.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelking H M, Leong J C. IHNV persistently infects chinook salmon embryo cells. Virology. 1981;109:47–58. doi: 10.1016/0042-6822(81)90470-0. [DOI] [PubMed] [Google Scholar]
- 16.Engelking H M, Leong J C. The glycoprotein of infectious hematopoietic necrosis virus elicits neutralizing antibody and protective responses. Virus Res. 1989;13:213–230. doi: 10.1016/0168-1702(89)90017-8. [DOI] [PubMed] [Google Scholar]
- 17.Ferran M C, Lucas-Lenard J M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fijan N, Sulimovic D, Bearsotti M, Musinie D, Zwillenger L D, Chilmonczyk S, Vantherot J F, de Kinkelin P. Some properties of the Epithelioma papulosum cyprini (EPC) cell line form carp Cyprinus carpio. Ann Virol. 1983;134:207–220. [Google Scholar]
- 19.Fischer J A, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 20.Fryer J L, Yusha A, Pilcher K S. The in vitro cultivation of tissue and cells of Pacific salmon and steelhead trout. Ann NY Acad Sci. 1965;126:566–586. doi: 10.1111/j.1749-6632.1965.tb14303.x. [DOI] [PubMed] [Google Scholar]
- 21.Hale A J, Smith C A, Sutherland L C, Stoneman V E, Longthorne V L, Culhane A C, Williams G T. Apoptosis: molecular regulation of cell death. Eur J Biochem. 1996;236:1–26. doi: 10.1111/j.1432-1033.1996.00001.x. [DOI] [PubMed] [Google Scholar]
- 22.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 23.Hsu Y L, Engelking H M, Leong J C. Occurrence of different types of infectious hematopoietic necrosis virus in fish. Appl Environ Microbiol. 1986;52:1353–1361. doi: 10.1128/aem.52.6.1353-1361.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson A C, Rossiter J P. Apoptosis plays an important role in experimental rabies virus infection. J Virol. 1997;71:5603–5607. doi: 10.1128/jvi.71.7.5603-5607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim C H, Dummer D M, Chiou P P, Leong J A. Truncated particles produced in fish surviving infectious hematopoietic necrosis virus infection: mediators of persistence? J Virol. 1999;73:843–849. doi: 10.1128/jvi.73.1.843-849.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koyama A H. Induction of apoptotic DNA fragmentation by the infection of vesicular stomatitis virus. Virus Res. 1995;37:285–290. doi: 10.1016/0168-1702(95)00026-m. [DOI] [PubMed] [Google Scholar]
- 27.Kurath G, Ahern K G, Pearson G D, Leong J C. Molecular cloning of the six mRNA species of infectious hematopoietic necrosis, a fish rhabdovirus, and gene order determination by R-loop mapping. J Virol. 1985;53:462–468. doi: 10.1128/jvi.53.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurath G, Higman K H, Bjorklund H V. Distribution and variation of NV genes in fish rhabdoviruses. J Gen Virol. 1997;78:113–117. doi: 10.1099/0022-1317-78-1-113. [DOI] [PubMed] [Google Scholar]
- 29.Lazebnik Y A, Cole S, Cooke C A, Nelson W G, Earnshaw W C. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong J C, Hsu Y L, Engelking H M. Synthesis of the structural proteins of infectious hematopoietic necrosis virus. In: Crosa J, editor. Bacterial and viral diseases of fish. Molecular studies. Seattle: University of Washington Press; 1983. pp. 61–70. [Google Scholar]
- 31.Lyles D S, McKenzie M O. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- 32.Marcovistz R, Bertho A L, Matos D C S. Relationship between apoptosis and thymocyte depletion in rabies-infected mice. Braz J Med Biol Res. 1994;27:1599–1603. [PubMed] [Google Scholar]
- 33.Marcus P I, Sekellick M J, Spiropoulou C F, Nichol S T. Interferon induction by viruses. XXII. Vesicular stomatitis virus—Indiana: M-protein and leader RNA do not regulate interferon induction in chicken embryo cells. J Interferon Res. 1993;13:413–418. doi: 10.1089/jir.1993.13.413. [DOI] [PubMed] [Google Scholar]
- 34.Ormonde P A. Characterization of the matrix proteins of the fish rhabdovirus, infectious hematopoietic necrosis virus. M.S. thesis. Corvallis: Oregon State University; 1995. [Google Scholar]
- 35.Paik S-Y, Banerjea A C, Harmison G G, Chen C-J, Schubert M. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J Virol. 1995;69:3529–3537. doi: 10.1128/jvi.69.6.3529-3537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Pringle C R. Virus taxonomy—1999. The universal system of virus taxonomy, updated to include the new proposals ratified by the International Committee on Taxonomy of Viruses during 1998. Arch Virol. 1999;144:421–429. doi: 10.1007/s007050050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao L, Debbas M, Sabbatini P, Hockenberg D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis which is inhibited by the E1B 19k-Da and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schutze H, Enzmann P-J, Kuchling R, Mundt E, Niemann H, Mettenleiter T C. Complete genomic sequence of the fish rhabdovirus infectious hematopoietic necrosis virus. J Gen Virol. 1995;76:2519–2527. doi: 10.1099/0022-1317-76-10-2519. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi A, Alnemri E S, Lazebnik Y A, Fernandez-Alnemri T, Litwack G, Moir R D, Goldman R D, Poirier G G, Kaufmann S H, Earnshaw W C. Cleavage of lamin A by Mch2a but not CPP32-multiple interleukin 1b converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- 40.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 41.Tiwari R K, Kusari J, Kumar R, Sen G C. Gene induction by interferons and double-stranded RNA: selective inhibition by 2-aminopurine. Mol Cell Biol. 1988;8:4289–4294. doi: 10.1128/mcb.8.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trobridge G D, Leong J C. Characterization of a rainbow trout Mx gene. J Interferon Cytokine Res. 1995;15:691–702. doi: 10.1089/jir.1995.15.691. [DOI] [PubMed] [Google Scholar]
- 43.Trobridge G D, Chiou P P, Leong J C. Cloning of the rainbow trout (Oncorhynchus mykiss) Mx2 and Mx3 cDNAs and characterization of trout Mx protein expression in salmon cells. J Virol. 1997;71:5304–5311. doi: 10.1128/jvi.71.7.5304-5311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trobridge G D, Chiou P P, Kim C H, Leong J C. Induction of the Mx protein of rainbow trout Oncorhynchus mykiss in vitro and in vivo with poly I:C dsRNA and infectious hematopoietic necrosis virus. Dis Aquat Org. 1997;30:91–98. [Google Scholar]
- 45.Vasavada R, Eager K B, Barbanti-Brodano G, Caputo A, Ricciardi R P. Adenovirus type 12 early region 1A proteins repress class I HLA expression in transformed human cells. Proc Natl Acad Sci USA. 1986;83:5257–5261. doi: 10.1073/pnas.83.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan H, Yoza B K, Lyles D S. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology. 1998;251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]
- 47.Zou J, Grabowski P S, Cunningham C, Secombes C J. Molecular cloning of interleukin 1beta from rainbow trout Oncorhynchus mykiss reveals no evidence of an ice cut site. Cytokine. 1999;11:552–560. doi: 10.1006/cyto.1998.0470. [DOI] [PubMed] [Google Scholar]