Abstract
Up to 80% of Parkinson's disease patients develop dementia, but time to dementia varies widely from motor symptom onset. Dementia with Lewy bodies presents with clinical features similar to Parkinson’s disease dementia, but cognitive impairment precedes or coincides with motor onset. It remains controversial whether dementia with Lewy bodies and Parkinson's disease dementia are distinct conditions or represent part of a disease spectrum. The biological mechanisms underlying disease heterogeneity, in particular the development of dementia, remain poorly understood, but will likely be the key to understanding disease pathways and, ultimately, therapy development. Previous genome-wide association studies in Parkinson's disease and dementia with Lewy bodies/Parkinson's disease dementia have identified risk loci differentiating patients from controls. We collated data for 7804 patients of European ancestry from Tracking Parkinson’s, The Oxford Discovery Cohort, and Accelerating Medicine Partnership—Parkinson's Disease Initiative. We conducted a discrete phenotype genome-wide association study comparing Lewy body diseases with and without dementia to decode disease heterogeneity by investigating the genetic drivers of dementia in Lewy body diseases. We found that risk allele rs429358 tagging APOEe4 increases the odds of developing dementia, and that rs7668531 near the MMRN1 and SNCA-AS1 genes and an intronic variant rs17442721 tagging LRRK2 G2019S on chromosome 12 are protective against dementia. These results should be validated in autopsy-confirmed cases in future studies.
Keywords: Lewy body diseases, dementia, genome-wide association studies, APOE
Lewy body diseases are heterogeneous and could present with or without dementia. Wu et al. conducted a case–case genome-wide association study in 7804 patients and identified variant tagging APOE e4 to increase odds of developing dementia and variants near SNCA-AS1 and tagging LRRK2 to be protective.
Graphical Abstract
Graphical Abstract.
Introduction
Parkinson's disease, Parkinson’s disease dementia and dementia with Lewy bodies, which we describe here jointly as Lewy body diseases, are characterized pathologically by alpha-synuclein aggregates forming Lewy bodies and Lewy neurites.1 Parkinson’s disease is a common degenerative movement disorder presenting with tremor, rigidity and bradykinesia. Non-motor features, including cognitive impairment and dementia, develop with disease progression in Parkinson’s disease. Approximately 24% of Parkinson’s disease patients have mild cognitive impairment at the time of diagnosis,2 and up to 80% of Parkinson’s disease patients eventually progress to dementia (Parkinson’s disease dementia),3 which is associated with worse functioning, poorer quality of life, care home admission and significant morbidity.4 However, the time to dementia from motor symptom onset varies widely between patients. Dementia with Lewy bodies is a synucleinopathy presenting with symptoms similar to Parkinson’s disease dementia, including dementia, cognitive fluctuations, visual hallucinations and REM sleep behaviour disorder in conjunction with existing or latent parkinsonism.5 Clinically, Parkinson’s disease dementia and dementia with Lewy bodies are distinguished by the ‘1-year rule’, where Parkinson’s disease dementia is diagnosed when dementia develops in the context of well-established Parkinson’s disease more than 1 year after motor symptom onset, while a diagnosis of dementia with Lewy bodies is given when cognitive impairment precedes or coincides with motor impairment. Parkinson’s disease dementia is distinguished from dementia with Lewy bodies by the temporal sequence of symptoms.
Neuropathologically, Parkinson’s disease usually differs from Parkinson’s disease dementia/dementia with Lewy bodies in the extent of Lewy body pathology in the brain, as inclusions are limited to the limbic system or brainstem in Parkinson’s disease without dementia. However, the pathological delineation of Parkinson’s disease dementia from dementia with Lewy bodies is extremely difficult. Both are characterized by Lewy bodies in cortical areas and a high frequency of Alzheimer’s disease co-pathology. Indeed, about 50% of Parkinson’s disease dementia patients have beta-amyloid plaques and neurofibrillary tangles at postmortem, which may be a better predictor of dementia than the extent of cortical alpha-synuclein pathology.6 The majority of dementia with Lewy bodies brains also fulfil criteria for a secondary diagnosis of Alzheimer’s disease.7 While a recent pathological study examining 110 Parkinson’s disease dementia and 78 dementia with Lewy bodies postmortem brains showed more severe synuclein cortical load, Alzheimer’s disease-related pathological changes and cerebral amyloid angiopathy in the dementia with Lewy bodies brains,8 it is generally agreed that there are no clear hallmark features distinguishing the two diseases.9 The separation of Parkinson’s disease dementia and dementia with Lewy bodies as discrete clinical and pathological entities is controversial.
Lewy body diseases are primarily sporadic. Case–control genome-wide association studies (GWAS) in the past decade have identified 90 common variant risk loci associated with Parkinson’s disease10 and 5 risk loci associated with dementia with Lewy bodies.11 Variation in several genes, including GBA1, TMEM175 and SNCA, confers risk for both diseases, suggesting overlapping pathogenesis and underlying biological dysfunction. Strikingly, TMEM175 and SNCA also modulate age at onset in Parkinson’s disease.12 On the other hand, there are distinct loci for dementia with Lewy bodies compared with Parkinson’s disease encompassing different genes (e.g. APOE and BIN1 for dementia with Lewy bodies), and, in some cases, distinct association signals at the same locus. In a study using targeted high-throughput sequencing, two distinct regions of the SNCA gene at the 3′ and 5′ ends were found to be differentially associated with Parkinson’s disease and dementia with Lewy bodies risk, respectively.13 While the consequences of these distinct signals remain to be clarified, it has been hypothesized that these distinct association signals could relate to the control of gene expression in different brain regions, leading to different phenotypes.14 Genome-wide survival analysis of Parkinson’s disease identified RIMS215 and LRP1B16 as common risk loci for progression from Parkinson’s disease to Parkinson’s disease dementia; however, they do not seem to be relevant to dementia with Lewy bodies.
Heterozygous mutations in GBA1 are among the strongest genetic risk factors for Parkinson’s disease and dementia with Lewy bodies.17,18 GBA1 encodes glucocerebrosidase, a lysosomal enzyme involved in the metabolism of glycosphingolipid. A meta-analysis of Parkinson’s disease patients showed that GBA1 mutations are associated with a 2.4-fold increase in the incidence of cognitive impairment.19 Moreover, mutation carriers tend to have earlier disease onset12 and shorter survival.20 In a large multicentre study of GBA1 mutation carriers, GBA1 was also found to be strongly associated with Parkinson’s disease dementia as well as dementia with Lewy bodies, providing evidence that GBA1 mutations lead to impaired cognition in synucleinopathies.21 However, as is the case for SNCA, the specific variants associated with Parkinson’s disease and dementia with Lewy bodies differ.22 Although the role of these GBA1 variants in pathogenesis remains unclear, studies in postmortem tissue showed that reduced lysosomal GCase is associated with alpha-synuclein aggregation, inflammation and cellular damage,22 suggesting an important role for GCase in the propagation of the alpha-synuclein pathology. This could explain the spread of Lewy bodies to limbic and neocortical areas of Parkinson’s disease patients with GBA1 mutations.
The apolipoprotein E (APOE) ε4 allele, a well-known risk locus for Alzheimer’s disease, has also been identified as a strong genetic risk factor for developing Parkinson’s disease dementia/dementia with Lewy bodies.11 APOE4 promotes amyloid-beta oligomerization and its pathological accumulation.23 The role of APOE4 in dementia with Lewy bodies pathogenesis is still unclear. It has been suggested that APOE4 might be a driver of amyloid-beta deposition, which presents as a co-pathology in the majority of dementia with Lewy bodies brains.7 However, there is some evidence showing that APOE may contribute to cognitive decline independently of amyloid. In autopsy studies, APOE4 was associated with dementia and diffuse LB pathology in ‘pure’ dementia with Lewy bodies patients (i.e. with absent or low levels of amyloid) as well as Parkinson’s disease dementia.24,25 Mouse models of synucleinopathy have also demonstrated that APOE4 exacerbated alpha-synuclein pathology in the absence of amyloid.26,27
Parkinson’s disease, Parkinson’s disease dementia and dementia with Lewy bodies share common risk genes. However, specific risk loci within these genes may vary across diseases, potentially leading to different phenotypes, which ultimately relate to the involvement of different cell types. Previous GWAS for Parkinson’s disease and dementia with Lewy bodies have compared Parkinson’s disease and dementia with Lewy bodies cases with controls. Here, in a study of almost 8000 cases, we aim to define the genetic determinants of dementia in Lewy body diseases by taking a different approach. We have taken a disease classification agnostic approach by comparing all Lewy body diseases with dementia (LBD-D), including both Parkinson’s disease dementia and dementia with Lewy bodies, to Parkinson's disease cases without dementia (LBD-ND). This ‘case–case’ approach should help identify specific variants that are associated with more extensive LB and Alzheimer’s disease pathology that contribute to cognitive impairment, rather than variants that are related to the initiation of the LB pathology as compared with unaffected controls.
Materials and methods
Cohort description and study design
We analysed three large independent cohorts: Tracking Parkinson's (TPD, www.parkinsons.org.uk/),28 Oxford Parkinson's Disease Centre Discovery (OPDC, www.dpag.ox.ac.uk/opdc/)29 and Accelerating Medicine Partnership—Parkinson's Disease Initiative (AMP-PD v2.5, https://www.amp-pd.org/) (Table 1, Supplementary Table 1). The AMP-PD data set is enriched for patients with LRRK2 p.G2019S. Participants were included in the present study based on their most recent clinical diagnosis or final pathological diagnosis of Parkinson’s disease, Parkinson’s disease dementia or dementia with Lewy bodies. A status of ‘case’ for LBD-D was defined if the patient had a clinical diagnosis of dementia with Lewy bodies5 or met the Movement Disorder Society task force Parkinson’s disease dementia diagnostic criteria.30 In detail, for Parkinson’s disease dementia, the criteria included (i) scoring below the threshold for dementia on the Montreal Cognitive Assessment (MoCA score < 21/30); (ii) having cognitive deficits that are severe enough to interfere with activities of daily living (MDS-Unified Parkinson's disease Rating Scale (UPDRS) part I 1.1 ≥ 2 score) and (iii) and the absence of severe depression defined using the MDS-UPDRS (MDS-UPDRS part I 1.3 < 4). LBD-ND was given a status of ‘control’. These patients did not have dementia based on the available clinical data. Patients with a change of diagnosis to a non-Lewy body disorder during the follow-up period were removed from analyses. AMP-PD is a unified cohort consisting of eight longitudinal studies with similar sample collection protocols. All studies were approved by local and multicentre ethics committees and are in compliance with the Declaration of Helsinki. Appropriate data use agreements were approved.
Table 1.
Cohort demographics
| Cohort | TPD | OPDC | AMP-PD | Total | ||||
|---|---|---|---|---|---|---|---|---|
| LBD-D | LBD-ND | LBD-D | LBD-ND | LBD-D | LBD-ND | LBD-D | LBD-ND | |
| N | 159 | 1377 | 93 | 737 | 2656 | 2782 | 2908 | 4896 |
| N male (%) | 128 (81) | 863 (63) | 72 (77) | 460 (62) | 1694 (64) | 1695 (61) | 1894 (65)a | 3017 (61) |
| Age diagnosis, years | 69.0 (7.9) | 64.3 (9.9) | 68.2 (9.0) | 64.2 (9.6) | 76.5 (8.8) | 60.4 (10.5) | 75.8 (9.1)b | 62.1 (10.4) |
| Array/sequencing | Illumina HumanCore Exome array | Illumina HumanCore Exome-12 v1.1 or Illumina InfiniumCoreExome-24 v1.1 | Illumina HiSeq X Ten | |||||
Means (SD) are shown unless otherwise indicated. Data shown are only in individuals who had both clinical and genetic data after quality control filters have been applied within each cohort.
aThere are significantly more males in the LBD-ND group (P = 2.31e−16).
bLBD-D is significantly older than LBD-BD (P = 2e−16).
TPD, Tracking Parkinson's; OPDC, Oxford Parkinson's Disease Centre Discovery, Accelerating Medicine Partnership—Parkinson's Disease Initiative; LBD-D: Lewy body disease with dementia; and LBD-ND: Lewy body disease without dementia.
Genotyping and quality control
DNA was extracted from whole blood or brain tissue as detailed in the protocols of each study. TPD used the Illumina HumanCoreExome array for genotyping. OPDC generated genotype data using the Illumina HumanCoreExome-12 v1.1 and Illumina Infinium HumanCoreExome-24 v1.1 SNP arrays. Whole genome sequencing for AMP-PD samples was performed using Illumina HiSeq X Ten sequencer, and data were processed against Human Genome Reference Build 38 (https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/GRCh38_reference_genome/). Data cleaning was performed using PLINK v1.9 (RRID:SCR_001757; https://www.cog-genomics.org/plink/1.9/)31 and PLINK v2.0 (https://www.cog-genomics.org/plink/2.0/). For quality control at the sample level, we excluded individuals from analysis if they had a low genotyping call rate (≤95%), excessive heterozygosity rates (>±0.15 F-statistic) or a mismatch between clinically reported and genetically determined sex by the X chromosome. We also excluded duplicate or related samples (kinship coefficient > 0.088). We removed individuals that were not of European ancestry by performing a principal component analysis from pruned genetic data of each cohort included in the analysis. We used Hapmap3 as the reference panel to derive ancestry groups. Individuals that deviated by more than 3 standard deviations from the mean of the first two principal components of the HapMap3 CEU group were removed from the analysis.
For quality control at the variant level, we removed SNPs from analysis if they had a low genotyping rate (<0.99%), deviated significantly from the Hardy–Weinberg equilibrium (P < 1E−8), had a minor allele frequency <1% and were non-autosomal (X, Y, mitochondrial chromosomes). After quality control, genetic data for TPD and OPDC were imputed separately against the TOPMed (https://imputation.biodatacatalyst.nhlbi.nih.gov/#!)32 r2 panel with Eagle v2.4 phasing on the TOPMed Imputation Server using Minimac4.33,34 We used the Rsq info measure of imputation accuracy to exclude variants that were not confidently imputed. We filtered out variants with an Rsq lower than 0.8. We also removed SNPs if missingness was >5%, and minor allele frequency was <1%. The two data sets were then merged, with only shared variants retained.
Statistical analysis for single-variant associations
Clinical data were cleaned and analysed using R v4.1.3 (RRID:SCR_001905; R Project for Statistical Computing, version 4.1.3; https://www.R-project.org/). We used logistic regression in PLINK to perform two separate genome-wide association studies for LBD-D (dementia with Lewy bodies and Parkinson’s disease dementia) compared with LBD-ND (Parkinson’s disease cases without dementia) in AMP-PD and in the merged TPD/OPDC data sets, respectively. The following covariates were incorporated in our model: age at onset (for TPD/OPDC cohorts) or age at diagnosis (for AMP-PD), sex, study and the first five genetic principal components. We meta-analysed the summary results for TPD/OPDC and AMP-PD using METAL (RRID:SCR_002013; http://csg.sph.umich.edu//abecasis/Metal/)35 under a random effects model using genomic control correction. We only included variants present in all cohorts and with a minor allele frequency variability below 15% across studies. We used Cochran’s Q to test for heterogeneity in the meta-analysis and excluded variants with P-value < 0.05 and I2 statistic≤ 80%. We considered P-values below 5 × 10−8 to be genome-wide significant and nominally significant below 5 × 10−6. We used LocusZoom to generate the Manhattan plot and the regional association plots (RRID:SCR_021374; http://locuszoom.org/).36
Conditional analysis
In order to determine whether there were single or multiple independent signals at each genome-wide significant locus, we carried out a conditional and joint multiple-SNP analysis (COJO) on the GWAS summary statistics. We used the AMP-PD cohort as the reference panel to estimate the LD between the SNPs and apply corrections to the models as it is the largest participating cohort in the meta-analysis. COJO was performed using GCTA (v1.93.0, GCTA | Yang Lab).37
Colocalization analysis
We performed a colocalization analysis to investigate whether there is a shared causal variant between the risk of dementia in Lewy body disease cases and expression quantitative trait loci (eQTLs). We used the coloc R package (version 5.1.0; https://cran.rproject.org/web/packages/colocr/index.html)38 and colochelpR as a wrapper (version 0.99.0).39 coloc is based on a Bayesian statistical approach to compute a posterior probability (PP) for the following hypotheses: there is no association with either trait (H0); there is an association with the Lewy body dementia trait, but not the eQTL trait (H1); there is an association with the eQTL trait, but not the Lewy body dementia trait (H2); there is an association with a Lewy body dementia and an eQTL variant, but the causal variants are independent (H3); and there is a shared causal variant associated with Lewy body dementia and eQTL within the analysed region (H4). coloc was run using default per SNP priors p1 = 10–4, p2 = 10–4 and p12 = 10–5. A PPH4 > 0.80 was considered a statistically significant support for colocalization. We used Cis-eQTL data from eQTLGen, which include 31 684 individuals (https://www.eqtlgen.org/cis-eqtls.html) and compare genetic variation with blood RNA and PsychEncode. PsychEncode includes 1387 individuals (http://resource.psychencode.org/) and compares genetic variation with brain RNA. We extracted all the genes from ±1 Mb of the significant hits from the GWAS and performed a colocalization analysis on each gene. Since the cis-eQTL and the GWAS summary statistics were in different builds, we converted the summary statistics of the meta-analysis from hg38 to hg19 using the LiftOver tool (RRID:SCR_018160; https://genome.sph.umich.edu/wiki/LiftOver).
Polygenic risk score
To assess the genetic overlap between LBD-D and Parkinson’s disease, dementia with Lewy bodies and Alzheimer’s disease risk profile, we computed a polygenic risk score (PRS) on all the LBD-D cases and LBD-ND for comparison. We used previously published Parkinson’s disease, dementia with Lewy bodies and Alzheimer’s disease GWAS10,11,40 as the reference data. After performing QC on summary statistics of the base data sets, we used PRSice-2 (version 2.3.5; RRID:SCR_017057; https://choishingwan.github.io/PRSice/)41 to calculate the PRS with the C + T method, which involves clumping SNPs and performing P-value thresholding. After clumping, 1 284 510 SNPs were included to generate the Parkinson’s disease PRS, 380 274 SNPs for the dementia with Lewy bodies PRS and 11 931 for the Alzheimer’s disease PRS. We then conducted general linear regression adjusted for age at onset, sex and PC1–PC5 to test if the PRS predicted the development of dementia. Results from the regression were meta-analysed in R with the meta package (RRID:SCR_019055; https://cran.r-project.org/web/packages/meta/index.html).
Results
After QC, a total of 7804 individuals were selected, including 2908 LBD-D (2552 dementia with Lewy bodies, 357 Parkinson’s disease dementia) and 4896 LBD-ND. Case selection is summarized in Supplementary Fig. 1. Demographic characteristics are summarized in Table 1. LBD-D patients were significantly older than LBD-ND at diagnosis (Kruskal–Wallis Chi-squared = 2627, df = 1, P-value < 2.2e−16). There are more men than women in our study, and they are more likely to have a dementia phenotype (P-value = 3.09e−06). We determined that with the sample size we had, we were very well-powered (100% power) to detect genetic variants associated with dementia, assuming an odds ratio of 1.4 and a minor allele frequency of 0.15 under an additive model (see Supplementary Fig. 2).
Identification of risk loci for dementia in Lewy body diseases
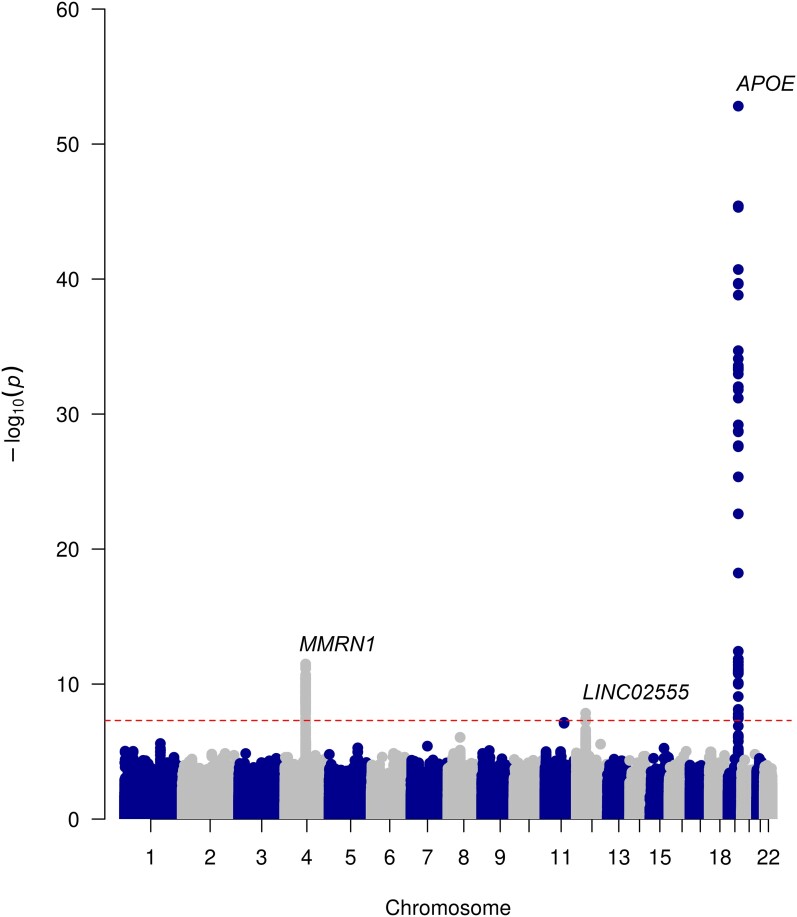
Using a case–case GWAS approach comparing patients with LBD-D and LBD-ND, we analysed 6 226 081 SNPs and identified three genome-wide significant loci (Fig. 1, Table 2).
Figure 1.
Manhattan plot of LBD-D versus LBD-ND. A Manhattan plot representing the results of the case–case genome-wide association study results (n = 2908 Lewy body disorders with dementia and n = 4896 Lewy body disorders without dementia), where 6 877 765 variants have been analysed under a logistic regression model. The plot highlights genome-wide significant single nuclear variants on chromosome 4 (rs7668531, P = 3.25e−12), 12 (rs17442721, P = 1.44e−08) and 19 (rs429358, P = 3.25e−57). Negative logarithm P-value is represented on the y-axis, while chromosome position is represented on the x-axis. The dotted line indicates genome-wide significant threshold (5 × 10−8).
Table 2.
Top SNPs from meta-analysis
| CHR | BP | SNP ID | Effect allele | Nearest gene | Effect allele frequency | OR | CI | P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tracking Parkinson's and Oxford Discovery | AMP-PD | gnomAD | ||||||||||
| LBD-D | LBD-ND | LBD-D | LBD-ND | HC | ||||||||
| 19 | 44908684 | rs429358 | C | APOE | 0.2 | 0.13 | 0.27 | 0.13 | 0.15 | 2.606 | 2.307–2.943 | 3.25E−57 |
| 4 | 89870668 | rs7668531 | G | MMRN1 | 0.43 | 0.47 | 0.43 | 0.49 | 0.52 | 0.719 | 0.656–0.789 | 3.25E−12 |
| 12 | 40141971 | rs17442721 | G | LINC02555 | 0.01 | 0.02 | 0.02 | 0.07 | 0.02 | 0.427 | 0.318–0.573 | 1.44E−08 |
| 11 | 82697450 | rs11233271 | G | MIR4300HG | 0.1 | 0.12 | 0.11 | 0.13 | 0.12 | 1.482 | 1.284–1.709 | 6.78E−08 |
Independent lead SNPs identified by LocusZoom. Genome coordinates are in build GRCh38. Allele frequency in European (non-Finnish) general population extracted from the gnomAD database (https://gnomad.broadinstitute.org/).
BP, base pair; chr, chromosome; CI, confidence interval; HC, healthy controls; OR, odds ratio; SNP, single nucleotide polymorphism.
The lead SNP was rs429358 in the APOE gene on chromosome 19 (OR = 2.606, 95% CI = 2.307–2.943, P = 3.25e−57; Supplementary Fig. 3A). APOE encodes apolipoprotein E, a known genetic factor for Alzheimer’s disease and dementia with Lewy bodies. It has also been identified as a risk factor for dementia in Parkinson’s disease.16,42 Conditional analysis on the lead SNP detected a secondary independent signal at the APOE locus at 19:32848205.
The second genome-wide significant SNP was rs7668531, an intergenic SNP between the MMRN1 gene and the SNCA-AS1 gene (OR = 0.719, 95% CI = 0.656–0.789, P = 3.25e−12; Supplementary Fig. 3B) located 170 323 kb downstream of the SNCA gene. This SNP is close to and in linkage disequilibrium with rs7680557 (D′ = 0.9959, r2 = 0.9196), which is associated with dementia as identified in the most recent Lewy body disease case–control GWAS.11 The rs7668531 signal is no longer genome-wide significant when we condition on the top rs7680557 in our data set, which suggests that rs7668531 is not independent and most likely tags SNCA-AS1.
The third genome-wide significant SNP was rs17442721 in the noncoding RNA LINC02555, which was protective against developing dementia (OR = 0.427, 95% CI = 0.318–0.573, P = 1.44e−08; Supplementary Fig. 3C). LINC02555 is potentially a regulatory locus for LRRK2 expression in specific cell types43 and may mediate PSP survival.44 However, this SNP is in LD with LRRK2 G2019S (rs34637584, r2 = 0.54, D′ = 0.97). To confirm whether rs17442721 is independent of LRRK2 G2019S, we performed a conditional analysis. Results show that rs17442721 is no longer genome-wide significant after conditioning on the G2019S variant, confirming that it tags LRRK2 G2019S, and there is no difference in dementia related to this SNP when the data are stratified by G2019S status (Supplementary Table 2). In this data set, the rate of dementia in LRRK2 G2019S carriers is 5% as compared with 39% in the total data set (Supplementary Table 3). rs17442721 was not in a linkage disequilibrium with PSP progression variant rs2242367 (r2 < 0.05).
Rs11233271 on chromosome 11 near the MIR4300HG gene approached genome-wide significance (OR = 1.48, 95% CI = 1.28–1.71, P = 6.78e−08), although this will need further evaluation in future work.
Common variant GBA E326K was nominally, but not genome-wide significant (OR = 2.01, 95% CI = 1.44–2.83, P = 2.517e−06). The Parkinson’s disease case–control GWAS LRRK2 rs76904798 variant was also not genome-wide significant (OR = 1.02, 95% CI = 0.88–1.18, P = 0.7759).
Colocalization
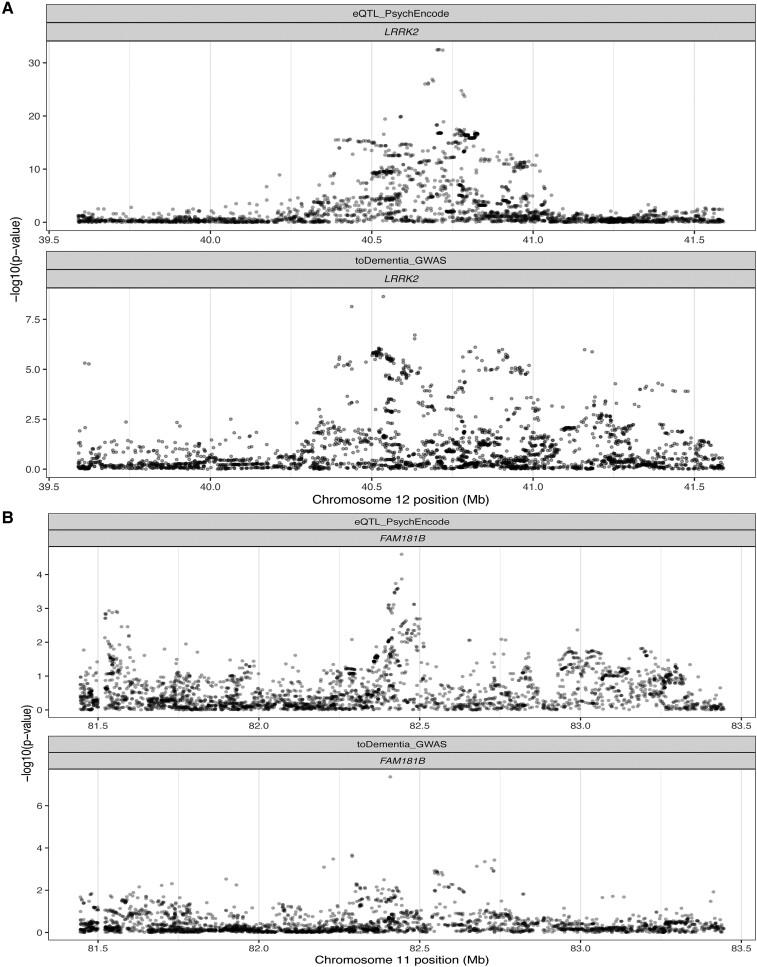
We performed a colocalization analysis to assess the probability of a shared causal signal between dementia status and genetically determined gene expression regulation. eQTLs were obtained from eQTLGen and PsychENCODE. eQTLGen comprises gene expression derived from blood, and psychENCODE data set comprises gene expression from bulk RNA sequencing from the frontal cortex. We found a suggestive colocalization between the genome-wide significant signal on chromosome 12 and cis-eQTL data from eQTLGen (PPH4 = 0.7154) for LRRK2, and rs11233271 on chromosome 11 suggestively colocalized with FAM181B (PPH4 = 0.7009), a protein-coding gene that is expressed in the brain (Fig. 2).
Figure 2.
Regional association plot for eQTL and GWAS signals. Results from colocalization analysis presented via regional association plot for expression quantitative trait loci and (A) genome-wide association signals in the region close to LRRK2 (posterior probability H4 = 0.72, 4026 variants analysed) and (B) in the region close to FAM181B (posterior probability H4 = 0.70, 4357 variants analysed). Negative logarithm P-value is represented on the y-axis, while chromosome position is represented on the x-axis.
Polygenic risk score
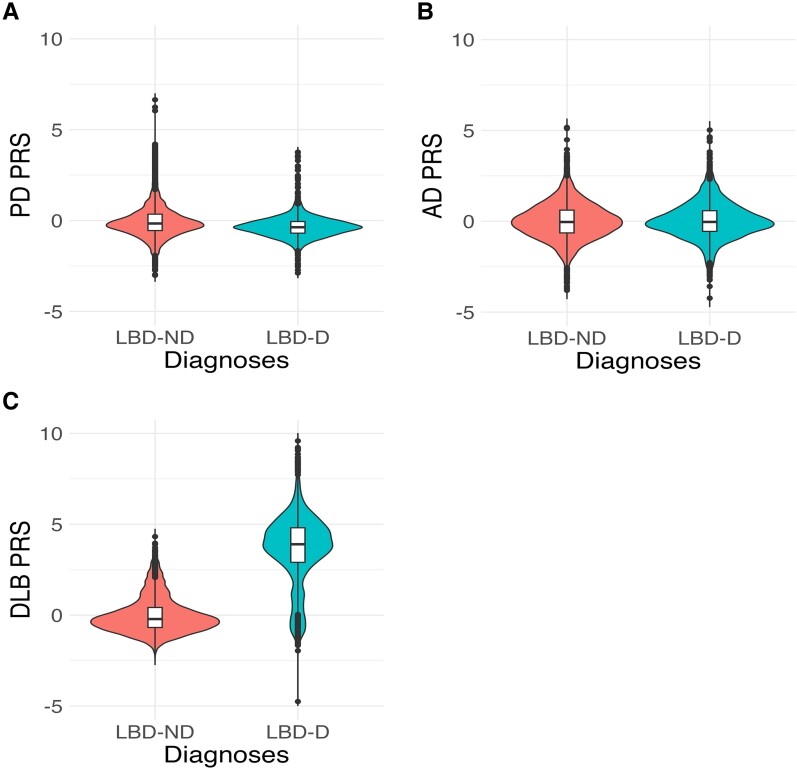
We applied a Parkinson’s disease, Alzheimer’s disease and dementia with Lewy bodies PRS derived from the most recent GWAS to the LBD-D patients identified in each of our cohorts, as well as to LBD-ND patients for comparison. We used a general linear regression model to assess if the normalized PRS predicted dementia and meta-analysed the regressions using a random effects model. Optimized P-value PRS based on genome-wide significant and sub-genome significant SNPs indicated that patients with a higher Parkinson’s disease PRS score (based on 1 284 510 SNPs) were less likely to develop dementia (OR = 0.74, 95% CI = 0.56–0.98, P = 0.03), and the Alzheimer’s disease risk profile (based on 31 000 SNPs) was not significantly different between the two groups (OR = 0.99, 95% CI = 0.82–1.20, P = 0.93). LBD-D was significantly associated with higher (pure) dementia with Lewy bodies PRS (OR = 2.69, 95% CI = 0.69–10.42, P = 0.01); however, this needs to be interpreted with caution as the confidence interval is very large (Fig. 3).
Figure 3.
Polygenic risk score from Parkinson’s disease, Alzheimer’s disease and dementia with Lewy bodies GWAS. Violin plot comparing z-transformed (A) Parkinson's disease, (B) Alzheimer's disease and (C) dementia with Lewy body polygenic risk score (PRS) distributions in Lewy body disease with dementia (LBD-D, n = 2908) with those without (LBD-ND, n = 4896). The centreline of the box plot represents the median, and the box limits are the interquartile range. Dots correspond to outliers. A general linear regression model was applied to assess the odds PRS-predicted dementia. High Parkinson’s disease PRS predicts lower odds of developing dementia (OR = 0.74, 95% CI = 0.56–0.98, P = 0.03), while high dementia with Lewy bodies PRS predicts increased odds of developing dementia (OR = 2.69, 95% CI = 0.69–10.42, P = 0.01).
Discussion
We have conducted a large-scale genome-wide case–case analysis to understand the genetic drivers of dementia in Lewy body diseases by comparing Lewy body diseases with and without dementia and identified three independent genome-wide significant signals in a novel case–case analysis by comparing LB cases with dementia with cases unaffected by dementia.
In line with previous studies, we showed that APOE e4 is the strongest risk factor for dementia in Lewy body diseases. Given the role of APOE e4 in Alzheimer’s disease, this may modulate the risk of dementia via Alzheimer’s disease pathology in at least a subset of the LBD-D cases; however, previous work has been inconsistent. A substantial proportion (30–40%) of patients with Parkinson’s disease and 50–80% of patients with dementia with Lewy bodies have co-occurring Alzheimer’s disease pathology.45 However, it is unclear whether APOE e4 drives dementia via Alzheimer’s disease pathology or independently. Our results indicate that the Alzheimer’s disease PRS does not drive dementia in Lewy body diseases, suggesting that APOE e4 may drive dementia in these cases by an Alzheimer’s disease pathology-independent mechanism. Consistent with our findings, postmortem studies have found that APOE e4 was associated with dementia in Lewy body diseases in both ‘pure’ Lewy body diseases and those with Alzheimer’s disease co-pathology.25 It is also possible that APOE e4 mediates neurodegenerative processes via neuroinflammation independently of amyloid and tau pathology.46,47 In fact, inflammation markers are apparent before protein aggregation.47 For instance, a longitudinal study showed that blood–brain barrier dysfunction at baseline predicted future cognitive decline in APOE4 carriers, but not in non-carriers.48 Further research is needed to clarify the role of APOE e4 in the Lewy body disease pathology.
We also found a SNP between MMRN1 and the 5′ end of SNCA, but not at the 3′ end to be significantly associated with lower odds of developing dementia, consistent with previous candidate gene studies13 and GWAS.11 Postmortem studies have found that alpha-synuclein in cortical areas is a predictor of dementia in Lewy body diseases. The finding that SNCA-AS1 is specific to LBD-D makes it an interesting potential therapeutic target. Indeed, LBD-D tends to have a much more aggressive disease course with faster progression to mortality. Targeting SNCA-AS1 could therefore be a potential solution to reducing the alpha-synuclein pathology in the cortex and the progression to dementia in Lewy body diseases. Our study has separated the 3′ signal in SNCA, which is associated with Parkinson’s disease risk, from the 5′ signal associated with dementia in Lewy body diseases. We hypothesize that the 3′ signal is important for the level of SNCA expression and the initiation of the Parkinson’s disease process, particularly in subcortical areas, whereas the 5′ SNP is associated with the expression of SNCA in the cortex.49 Indirectly, this suggests that local SNCA expression is important, distinct from cell to cell spread from subcortical areas.
The third genome-wide significant signal was located near LINC02555, which is potentially a regulator of LRRK2. However, we confirmed that this SNP is tagging LRRK2 G2019S. In the present study, we did not exclude LRRK2 mutation carriers from the main analysis. As previously described, we have shown in this study that LRRK2 G2019S carriers are less likely to develop dementia.50 Moreover, LRRK2 likely does not play a major role in dementia with Lewy bodies.51 Our results confirm that in Lewy body diseases, the LRRK2 G2019S mutation status is associated with decreased odds of progression towards dementia.
Rs11233271 on chromosome 11 close to MIR4300HG was nominally significant in our GWAS. This SNP may regulate the expression of FAM181B, a protein-coding gene involved in the development of the nervous system.52 FAM181B was also associated with working memory in a gene-based study on cognitive measures in adolescence.53 Furthermore, this locus has been associated with variation in the microbiome. Further studies are needed to investigate the role of this locus in Lewy body dementia.
Interestingly, GBA1, BIN1 and TMEM175, which are associated with case–control Lewy body disease GWAS11, did not appear significant when comparing LBD-D with LBD-ND. Since GBA1 is a known risk gene for both Parkinson’s disease and dementia with Lewy bodies, our analysis shows that variation in GBA1 does not distinguish between LBD-D and LBD-ND within a study of this size. Similarly, TMEM175 is a risk factor in both Lewy body diseases with and without dementia. Therefore, it is not surprising that the signal disappears when we make a head-to-head comparison. BIN1 encodes bridging integrator 1 and is the second strongest signal associated with Alzheimer’s disease, but was not genome-wide significantly associated with LBD-D in this study (P = 2.276e−05).54 Increased BIN1 expression is associated with a higher load of tau in the Alzheimer’s disease brain, but not amyloid.55 While some studies found the tau load to be a correlate of dementia in Parkinson’s disease and dementia with Lewy bodies, other studies have not. Autopsy studies have found tau to colocalize with alpha-synuclein in Lewy bodies in both Parkinson’s disease and dementia with Lewy bodies.56 A small autopsy study in LRRK2 carriers found that 100% of the brains had tau pathology.57 Therefore, it is possible that the Lewy body disease risk genes associated with tau pathology are not good candidates to distinguish LBD-D from LBD-ND. RIMS2 was identified as a progression locus in a genome-wide survival study of Parkinson’s disease dementia15; however, this was not genome-wide significant in the present study (P = 0.016).
We acknowledge several limitations of our study. First, the analysis only included patients of European ancestry and is therefore not generalizable to other populations. As in previous studies, there are more men than women in our study.58,59 Our main results apply to men and women, but we have not carried out a sex stratified analysis to look for sex-specific loci associated with dementia. In addition, it is possible that some patients were censored as non-demented based on the clinical data available, but who might have developed dementia if followed-up for a longer period of time. We grouped patients who developed dementia at any time point together in the design of our study. However, it is possible that genetic risk factors and associated neuropathology leading to dementia at onset are different from those associated with dementia later in the disease course. We hypothesize that Parkinson’s disease patients developing dementia early in the disease course will be genetically closer to dementia with Lewy bodies, while those developing dementia much later on will present with a different genetic profile. Future studies should aim to identify risk factors leading to a more aggressive disease course in Lewy body diseases to improve prognosis and care.
In conclusion, in a pooled analysis of dementia with Lewy bodies, Parkinson’s disease and Parkinson’s disease dementia, we have shown that APOE e4 is the major determinant of Lewy body diseases with dementia. We have also shown that variation at the 5′ end of the SNCA gene and variant tagging LRRK2 G2019S are associated with a significantly reduced risk of dementia. Although APOE is associated with dementia, other Alzheimer’s disease risk loci defined by PRS analysis were not associated with LBD-dementia. Increasing sample sizes in collaborative international studies will help resolve the disease pathogenesis, the nosological overlap between Parkinson’s disease dementia and dementia with Lewy bodies, and ultimately help define new treatments.
Supplementary Material
Acknowledgements
Data used in the preparation of this article were obtained from the AMP-PD Knowledge Platform (https://www.amp-pd.org). AMP-PD is a public–private partnership managed by the FNIH and funded by Celgene, GSK, Michael J. Fox Foundation for Parkinson's Research, the National Institute of Neurological Disorders and Stroke (NINDS), Pfizer and Verily. TPD and OPDC cohorts are primarily funded and supported by Parkinson's UK (https://www.parkinsons.org.uk/) and supported by the National Institute for Health and Care Research (NIHR) Clinical Research Network (CRN). The TPD study is also supported by NHS Greater Glasgow and Clyde. The OPDC cohort is also supported by the NIHR Oxford Biomedical Research Centre based at the Oxford University Hospitals NHS Trust and the University of Oxford. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging and National Institute of Neurological Disorders and Stroke.
Appendix
International LBD Genomics Consortium (iLBDGC)
United States
Yevgeniya Abramzon, B.Sc. (Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Sarah Ahmed, B.Sc. (Neurodegenerative Diseases Research Unit, Laboratory of Neurogenetics, National Institute of Neurological Disorders and Stroke, Bethesda, MD, 20892, USA)
Camille Alba, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Marilyn S. Albert, Ph.D. (Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD, 21287, USA)
Dagmar Bacikova, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Matthew J. Barrett, M.D. (Department of Neurology, University of Virginia School of Medicine, Charlottesville, VA, 22903, USA)
Thomas G. Beach, M.D., Ph.D. (Civin Laboratory for Neuropathology, Banner Sun Health Research Institute, Sun City, AZ, 85006, USA)
David A. Bennett, M.D. (Rush Alzheimer's Disease Center, Rush University, Chicago, IL, 60612, USA)
Lilah M. Besser, Ph.D., M.Ph. (Institute for Human Health and Disease Intervention, Florida Atlantic University, Boca Raton, FL, 33431, USA)
Eileen H. Bigio, M.D. (Mesulam Center for Cognitive Neurology and Alzheimer's Disease, Northwestern University Feinberg School of Medicine, Chicago, IL, 60611, USA)
Bradley F. Boeve, M.D. (Center for Sleep Medicine, Mayo Clinic, Rochester, MN, 55905, USA)
Ryan C. Bohannan, B.Sc. (Department of Neurobiology and Behavior, University of California Irvine, Irvine, CA, 92697, USA)
Chad A. Caraway, Ph.D. (Institute for Memory Impairments and Neurological Disorders, University of California Irvine, Irvine, CA, 92697, USA)
Jose-Alberto Palma, M.D., Ph.D. (Department of Neurology, New York University School of Medicine, New York, NY, 10016, USA)
Ruth Chia, Ph.D. (Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Clifton L. Dalgard, Ph.D. (Department of Anatomy, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA; The American Genome Center, Collaborative Health Initiative Research Program, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Dennis Dickson, M.D. (Department of Neuroscience, Mayo Clinic, Jacksonville, FL, 32224, USA)
Jinhui Ding, Ph.D. (Computational Biology Core, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Kelley Faber, M.Sc. (Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, 46202, USA)
Tanis Ferman, Ph.D. (Department of Psychiatry and Psychology, Mayo Clinic, Jacksonville, FL, 32224, USA)
Luigi Ferrucci, M.D., Ph.D. (Longitudinal Studies Section, National Institute on Aging, Baltimore, MD, 21224, USA)
Margaret E. Flanagan, M.D. (Northwestern University Feinberg School of Medicine, Chicago, IL, 60611, USA)
Tatiana M. Foroud, M.D. (Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, 46202, USA)
Bernardino Ghetti, M.D. (Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN, 46202, USA)
J. Raphael Gibbs, Ph.D. (Computational Biology Core, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Alison Goate, Ph.D. (Nash Family Department of Neuroscience, Department of Genetics and Genomic Sciences, and Department of Pathology, Icahn School of Medicine at Mount Sinai, New York, NY, 10029, USA)
David Goldstein, M.D. (Clinical Neurocardiology Section, National Institute of Neurological Disorders and Stroke, Bethesda, MD, 20892, USA)
Neill R. Graff-Radford, M.D. (Department of Neurology, Mayo Clinic Florida, 4500 San Pablo Road South, Jacksonville, FL, 32224, USA)
Heng-Chen Hu, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Daniel Hupalo, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Scott M. Kaiser, M.B.A. (Department of Neuropathology, Indiana University School of Medicine, Indianapolis, IN, 46202, USA)
Horacio Kaufmann, M.D. (Department of Neurology, New York University School of Medicine, New York, NY, 10016, USA)
Ronald C. Kim, M.D. (Department of Neuropathology, School of Medicine, University of California Irvine, Irvine, CA, 92697, USA)
Gregory Klein (Rush Alzheimer's Disease Center, Rush University, Chicago, IL, 60612, USA)
Walter Kukull, Ph.D. (National Alzheimer's Coordinating Center (NACC), University of Washington, Seattle, WA, 98195, USA)
Amanda Kuzma, M.Sc. (Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA)
James Leverenz, M.D. (Cleveland Lou Ruvo Center for Brain Health, Neurological Institute, Cleveland Clinic, Cleveland, OH, 44195, USA)
Grisel Lopez, M.D. (Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MD, 20892, USA)
Qinwen Mao, M.D., Ph.D. (Northwestern University Feinberg School of Medicine, Chicago, IL, 60611, USA)
Elisa Martinez-McGrath, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Eliezer Masliah, M.D. (Molecular Neuropathology Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Ed Monuki, M.D., Ph.D. (Department of Pathology & Laboratory Medicine, School of Medicine, University of California Irvine, Irvine, CA, 92697, USA)
Kathy L. Newell, M.D. (Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS, 66160, USA)
Lucy Norcliffe-Kaufmann, Ph.D. (Department of Neurology, New York University School of Medicine, New York, NY, 10016, USA)
Matthew Perkins, B.Sc. (Michigan Brain Bank, University of Michigan Medical School, Ann Arbor, MI, 48109, USA)
Olga Pletnikova, M.D. (Department of Pathology [Neuropathology], Johns Hopkins University Medical Center, Baltimore, MD, 21287, USA)
Alan E. Renton, Ph.D. (Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, 10029, USA)
Susan M. Resnick, M.D. (Laboratory of Behavioral Neuroscience, National Institute on Aging, Baltimore, MD, 21224, USA)
Owen A. Ross, Ph.D. (Department of Neuroscience & Department of Clinical Genomics, Mayo Clinic Florida, 4500 San Pablo Road South, Jacksonville, FL, 32224, USA)
Marya S. Sabir, B.Sc. (Neurodegenerative Diseases Research Unit, Laboratory of Neurogenetics, National Institute of Neurological Disorders and Stroke, Bethesda, MD, 20892, USA)
Clemens R. Scherzer, M.D. (Precision Neurology Program, Brigham & Women's Hospital, Harvard Medical School, Boston, MA, 02115, USA)
Sonja W. Scholz, M.D., Ph.D. (Neurodegenerative Diseases Research Unit, Laboratory of Neurogenetics, National Institute of Neurological Disorders and Stroke, Bethesda, MD, 20892, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD, 21287, USA)
Geidy Serrano, Ph.D. (Civin Laboratory for Neuropathology, Banner Sun Health Research Institute, Sun City, AZ, 85006, USA)
Vikram Shakkotai, M.D., Ph.D. (Department of Neurology, University of Michigan Medical School, Ann Arbor, MI, 48109, USA)
Ellen Sidransky, M.D. (Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MD, 20892, USA)
Andrew B. Singleton, Ph.D. (Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA)
Toshiko Tanaka, Ph.D. (Longitudinal Studies Section, National Institute on Aging, Baltimore, MD, 21224, USA)
Nahid Tayebi, Ph.D. (Medical Genetics Branch, National Human Genome Research Institute, Bethesda, MD, 20892, USA)
Bryan J. Traynor, M.D., Ph.D. (Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, 20892, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD, 21287, USA)
Juan C. Troncoso, M.D. (Department of Pathology [Neuropathology], Johns Hopkins University Medical Center, Baltimore, MD, 21287, USA)
Coralie Viollet, Ph.D. (Department of Anatomy, Physiology and Genetics, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, 20814, USA)
Ronald L. Walton, B.Sc. (Department of Neuroscience, Mayo Clinic Florida, Jacksonville, FL, 32224, USA)
Randy Woltjer, M.D., Ph.D. (Department of Neurology, Oregon Health & Sciences University, Portland, OR, 97239, USA)
Zbigniew K. Wszolek, M.D. (Department of Neurology, Mayo Clinic Florida, 4500 San Pablo Road South, Jacksonville, FL, 32224, USA)
Canada
Sandra E. Black, M.D. (Institute of Medical Science, Faculty of Medicine, University of Toronto, 1 King's College Circle, Room 2374, Toronto, ON, M5S 1A8, Canada; Division of Neurology, Department of Medicine, University of Toronto, 27 King's College Circle, Toronto, ON, M5S 1A1, Canada; Heart and Stroke Foundation Canadian Partnership for Stroke Recovery, Sunnybrook Health Sciences Centre, University of Toronto, 1 King's College Circle, Room 2374, Toronto, ON, M5S 1A8, Canada; Hurvitz Brain Sciences Research Program, Sunnybrook Research Institute, University of Toronto, 2075 Bayview Avenue, Toronto, ON, M4N 3M5, Canada; LC Campbell Cognitive Neurology Research Unit, Sunnybrook Research Institute, University of Toronto, 2075 Bayview Avenue, Toronto, ON, M4N 3M5, Canada)
Ziv Gan-Or, M.D., Ph.D. (Montreal Neurological Institute and Hospital, Department of Neurology & Neurosurgery, McGill University, 3801 University Street, Montreal, QC, H3A 2B4, Canada)
Julia Keith, M.D. (Department of Anatomical Pathology, Sunnybrook Health Sciences Centre, University of Toronto, 1 King's College Circle, Room 2374, Toronto, ON, M5S 1A8, Canada)
Mario Masellis, M.D., Ph.D. (Cognitive & Movement Disorders Clinic, Sunnybrook Health Sciences Centre, University of Toronto, 1 King's College Circle, Room 2374, Toronto, ON, M5S 1A8, Canada; Department of Medicine, Division of Neurology, University of Toronto, Toronto, ON, M5S 1A8, Canada; Hurvitz Brain Sciences Research Program, Sunnybrook Research Institute, University of Toronto, Toronto, ON, M5S 1A8, Canada; LC Campbell Cognitive Neurology Research Unit, Sunnybrook Research Institute, University of Toronto, Toronto, ON, M5S 1A8, Canada)
Ekaterina Rogaeva, Ph.D. (Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, 1 King's College Circle, Room 2111, Toronto, ON, M5S 1A8, Canada)
United Kingdom
Dag Aarsland, M.D. (Department of Old Age Psychiatry, Institute of Psychiatry, Psychology and Neuroscience [IoPPN], King's College London, DeCrespigny Park, London, SE5 8AF, UK)
Safa Al-Sarraj, M.D., Ph.D. (Department of Clinical Neuropathology and London Neurodegenerative Diseases Brain Bank, Institute of Psychiatry, Psychology and Neuroscience [IoPPN], King's College Hospital and King's College London, DeCrespigny Park, London, SE5 8AF, UK)
Johannes Attems, M.D. (Mental Health, Dementia and Neurodegeneration, Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, NE4 5PL, UK)
Raffaele Ferrari, Ph.D. (Department of Molecular Neuroscience, Institute of Neurology, University College London, London, WC1B 5EH, UK)
Steve Gentleman, M.D. (Neuropathology Unit, Division of Brain Sciences, Department of Medicine, Imperial College London, London, W12 0NN, UK)
John A. Hardy, Ph.D. (Department of Neurodegenerative Disease, Reta Lila Weston Laboratories, Queen Square Genomics, UCL Dementia Research Institute, London WC1E 6BT, UK)
Angela K. Hodges, Ph.D. (Department of Old Age Psychiatry, Institute of Psychiatry, Psychology and Neuroscience [IoPPN], King's College London, Maurice Wohl Clinical Neuroscience Institute, London, SE5 9NU, UK)
Seth Love, M.D. (Dementia Research Group, School of Clinical Sciences, University of Bristol, Bristol, BS10 5NB, UK)
Ian McKeith, M.D. (Mental Health, Dementia and Neurodegeneration, Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, NE4 5PL, UK)
Christopher M. Morris, Ph.D. (Mental Health, Dementia and Neurodegeneration, Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, NE4 5PL, UK)
Huw R. Morris, M.D., Ph.D. (Department of Molecular Neuroscience, Institute of Neurology, University College London, London, WC1B 5EH, UK; Department of Clinical and Movement Neuroscience, Royal Free Campus UCL Institute of Neurology, University College London, London, NW3 2PF, UK)
Laura Palmer, Ph.D. (South West Dementia Brain Bank, Bristol Medical School, University of Bristol, Bristol, BS10 5NB, UK)
Stuart Pickering-Brown, Ph.D. (Division of Neuroscience and Experimental Psychology, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, M13 9PT, UK)
Regina H. Reynolds, M.Sc. (NIHR Great Ormond Street Hospital Biomedical Research Centre, University College London, London, UK; Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, University College London, London WC1E 6BT, UK; Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, University College London, London, UK)
Mina Ryten, M.D., Ph.D. (NIHR Great Ormond Street Hospital Biomedical Research Centre, University College London, London, UK; Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, University College London, London WC1E 6BT, UK)
Alan J. Thomas, M.B.B.S., Ph.D. (Mental Health, Dementia and Neurodegeneration, Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, NE4 5PL, UK)
Bension S. Tilley, M.B.B.S., Ph.D. (Neuropathology Unit, Division of Brain Sciences, Department of Medicine, Imperial College London, London, W12 0NN, UK)
Claire Troakes, Ph.D. (Department of Old Age Psychiatry, Department of Basic and Clinical Neuroscience, King's College London, DeCrespigny Park, London, SE5 8AF, UK)
Republic of Ireland
Francesca Brett, M.D. (Dublin Brain Bank, Neuropathology Department, Beaumont Hospital, Dublin, D2, Ireland)
France
Alexis Brice, M.D. (Paris Brain Institute, Paris Brain Institute, Sorbonne Universites, 47, boulevard de l'Hôpital, Paris, CS 21 414 – 75646, France)
Charles Duyckaerts, M.D. (Paris Brain Institute, Paris Brain Institute, Sorbonne Universites, 47, boulevard de l'Hôpital, Paris, CS 21 414 – 75646, France)
Suzanne Lesage, Ph.D. (Paris Brain Institute, ICM, Sorbonne Universites, 47, boulevard de l'Hôpital, Paris, CS 21 414 – 75646, France)
Italy
Maura Brunetti, M.Sc. (Rita Levi Montalcini Department of Neuroscience, University of Turin, Turin, 10126, Italy)
Andrea Calvo, M.D. (Rita Levi Montalcini Department of Neuroscience, University of Turin, Turin, 10126, Italy)
Antonio Canosa, M.D. (Rita Levi Montalcini Department of Neuroscience, University of Turin, Turin, 10126, Italy)
Adriano Chiò, M.D. (Rita Levi Montalcini Department of Neuroscience, University of Turin, Turin, 10126, Italy; Institute of Cognitive Sciences and Technologies, C.N.R., Via S. Martino della Battaglia, 44, Rome, 00185, Italy, Azienda Ospedaliero Universitaria Città della Salute e della Scienza, Corso Bramante, 88, Turin, 10126, Italy)
Gianluca Floris, M.D. (Department of Neurology, University Hospital of Cagliari, Via Ospedale, 54, Cagliari, 09124, Italy)
Giancarlo Logroscino, M.D., Ph.D. (Department of Basic Medical Sciences, Neurosciences and Sense Organs, University of Bari “Aldo Moro”, Bari, 70121, Italy)
Chiara Zecca, B.Sc. (Department of Basic Medical Sciences, Neurosciences and Sense Organs, University of Bari “Aldo Moro”, Bari, 70121, Italy)
Jordi Clarimon, Ph.D. (Universitat Autònoma de Barcelona, Carrer de Sant Quintí, 77-79, Barcelona, 08041, Spain, and Movement Disorders Units, Department of Neurology, University Hospital Mutua de Terrassa, Barcelona, 08221, Spain)
Monica Diez-Fairen, M.Sc. (Memory and Movement Disorders Units, Department of Neurology, University Hospital Mutua de Terrassa, 08221 Barcelona, Spain)
Juan Fortea, M.D., Ph.D. (Universitat Autònoma de Barcelona, Carrer de Sant Quintí, 77-79, Barcelona, 08041, Spain, and Movement Disorders Units, Department of Neurology, University Hospital Mutua de Terrassa, Barcelona, 08221, Spain)
Isabel González-Aramburu, M.D., Ph.D. (Neurology Service, University Hospital Marqués de Valdecilla-IDIVAL-UC, Santander, 39011, Spain)
Jon Infante, M.D., Ph.D. (Neurology Service, University Hospital Marqués de Valdecilla-IDIVAL-UC, Santander, 39011, Spain)
Carmen Lage, M.D. (Neurology Service, University Hospital Marqués de Valdecilla-IDIVAL-UC, Santander, 39011, Spain)
Alberto Lleó, M.D., Ph.D. (Universitat Autònoma de Barcelona, Carrer de Sant Quintí, 77-79, Barcelona, 08041, Spain, and Movement Disorders Units, Department of Neurology, University Hospital Mutua de Terrassa, Barcelona, 08221, Spain)
Pau Pastor, M.D., Ph.D. (Memory and Movement Disorders Units, Department of Neurology, University Hospital Mutua de Terrassa, 08221 Barcelona, Spain)
Laura Porcel-Molina, M.D., Ph.D. (Neurological Tissue Bank, Biobanc-Hospital Clinic - IDIBAPS, C/ Centre Esther Koplowitz, Barcelona, 08036, Spain)
Eloy Rodríguez-Rodríguez, M.D., Ph.D. (Neurology Service, University Hospital Marqués de Valdecilla-IDIVAL-UC, Santander, 39011, Spain)
Pascual Sanchez-Juan, M.D., Ph.D. (Neurology Service, University Hospital Marqués de Valdecilla-IDIVAL-UC, Santander, 39011, Spain)
Luxembourg
Rejko Krüger, M.D. (Luxembourg Center for Systems Biomedicine, University of Luxembourg, Esch-sur-Alzette, Luxembourg, L-4362, Luxembourg; Luxembourg Institute of Health (LIH), University of Luxembourg, Esch-sur-Alzette, Luxembourg, L-4362, Luxembourg; Centre Hospitalier de Luxembourg (CHL), University of Luxembourg, Esch-sur-Alzette, Luxembourg, L-4362, Luxembourg)
Patrick May, Ph.D. (Luxembourg Center for Systems Biomedicine, University of Luxembourg, Esch-sur-Alzette, Luxembourg, L-4362, Luxembourg)
Greece
Georgia Xiromerisiou, M.D., Ph.D. (Department of Neurology, University Hospital of Larissa, University of Thessalia, Mezourlo, Larissa, 41110, Greece)
Contributor Information
Lesley Yue Wu, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Raquel Real, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Alejandro Martinez-Carrasco, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
Ruth Chia, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20814, USA.
Michael A Lawton, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol BS8 2PS, UK.
Maryam Shoai, Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Neurodegenerative Diseases, UCL Queen Square Institute of Neurology, University College London, London, WC1N 3BG, UK; UK Dementia Research Institute, University College London, London WC1E 6BT, UK.
Catherine Bresner, Institute of Psychological Medicine and Clinical Neurosciences, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff CF24 4HQ, UK.
Cornelis Blauwendraat, Integrative Neurogenomics Unit, National Institute on Aging, Bethesda, MD 20814, USA; Center for Alzheimer’s and Related Dementias, National Institute on Aging, Bethesda, MD 20892, USA.
Andrew B Singleton, Center for Alzheimer’s and Related Dementias, National Institute on Aging, Bethesda, MD 20892, USA.
Mina Ryten, Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Genetics and Genomic Medicine, UCL Great Ormond Street Institute of Child Health, University College London, London WC1N 1EH, UK; Genetics and Genomic Medicine, NIHR Great Ormond Street Hospital Biomedical Research Centre, University College London, London WC1N 1EH, UK; UK Dementia Research Institute at The University of Cambridge, Cambridge, UK; Department of Clinical Neurosciences, School of Clinical Medicine, The University of Cambridge, Cambridge, UK.
Sonja W Scholz, Neurodegenerative Diseases Research Section, National Institute of Neurological Disorders and Stroke, Bethesda, MD 20892, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD 21287, USA.
Bryan J Traynor, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20814, USA; Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD 21287, USA; Reta Lila Weston Institute, UCL Queen Square Institute of Neurology, London WC1N 1PJ, UK.
Nigel M Williams, Institute of Psychological Medicine and Clinical Neurosciences, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff CF24 4HQ, UK.
Michele T M Hu, Nuffield Department of Clinical Neurosciences, Division of Clinical Neurology, University of Oxford, Oxford OX3 9DU, UK; Oxford Parkinson’s Disease Centre, University of Oxford, Oxford OX1 3QU, UK.
Yoav Ben-Shlomo, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol BS8 2PS, UK.
Donald G Grosset, School of Neuroscience and Psychology, University of Glasgow, Glasgow G12 8QQ, UK.
John Hardy, Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA; Department of Neurodegenerative Diseases, UCL Queen Square Institute of Neurology, University College London, London, WC1N 3BG, UK; UK Dementia Research Institute, University College London, London WC1E 6BT, UK; Reta Lila Weston Institute, UCL Queen Square Institute of Neurology, London WC1N 1PJ, UK; National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre, London W1T 7DN, UK; Institute for Advanced Study, The Hong Kong University of Science and Technology, Hong Kong SAR, China.
Huw R Morris, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK; UCL Movement Disorders Centre, University College London, London WC1N 3BG, UK; Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network, Chevy Chase, MD 20815, USA.
International Lewy Body Dementia Genomics Consortium:
Yevgeniya Abramzon, Sarah Ahmed, Camille Alba, Marilyn S Albert, Dagmar Bacikova, Matthew J Barrett, Thomas G Beach, David A Bennett, Lilah M Besser, Eileen H Bigio, Bradley F Boeve, Ryan C Bohannan, Chad A Caraway, Jose-Alberto Palma, Ruth Chia, Clifton L Dalgard, Dennis Dickson, Jinhui Ding, Kelley Faber, Tanis Ferman, Luigi Ferrucci, Margaret E Flanagan, Tatiana M Foroud, Bernardino Ghetti, J Raphael Gibbs, Alison Goate, David Goldstein, Neill R Graff-Radford, Heng-Chen Hu, Daniel Hupalo, Scott M Kaiser, Horacio Kaufmann, Ronald C Kim, Gregory Klein, Walter Kukull, Amanda Kuzma, James Leverenz, Grisel Lopez, Qinwen Mao, Elisa Martinez-McGrath, Eliezer Masliah, Ed Monuki, Kathy L Newell, Lucy Norcliffe-Kaufmann, Matthew Perkins, Olga Pletnikova, Alan E Renton, Susan M Resnick, Owen A Ross, Marya S Sabir, Clemens R Scherzer, Sonja W Scholz, Geidy Serrano, Vikram Shakkotai, Ellen Sidransky, Andrew B Singleton, Toshiko Tanaka, Nahid Tayebi, Bryan J Traynor, Juan C Troncoso, Coralie Viollet, Ronald L Walton, Randy Woltjer, Zbigniew K Wszolek, Sandra E Black, Ziv Gan-Or, Julia Keith, Mario Masellis, Ekaterina Rogaeva, Dag Aarsland, Safa Al-Sarraj, Johannes Attems, Raffaele Ferrari, Steve Gentleman, John A Hardy, Angela K Hodges, Seth Love, Ian McKeith, Christopher M Morris, Huw R Morris, Laura Palmer, Stuart Pickering-Brown, Regina H Reynolds, Mina Ryten, Alan J Thomas, Bension S Tilley, Claire Troakes, Francesca Brett, Alexis Brice, Charles Duyckaerts, Suzanne Lesage, Maura Brunetti, Andrea Calvo, Antonio Canosa, Adriano Chiò, Gianluca Floris, Giancarlo Logroscino, Chiara Zecca, Jordi Clarimon, Monica Diez-Fairen, Juan Fortea, Isabel González-Aramburu, Jon Infante, Carmen Lage, Alberto Lleó, Pau Pastor, Laura Porcel-Molina, Eloy Rodríguez-Rodríguez, Pascual Sanchez-Juan, Rejko Krüger, Patrick May, and Georgia Xiromerisiou
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
This research was funded in whole or in part by Aligning Science Across Parkinson's (ASAP 000478) through the Michael J. Fox Foundation for Parkinson's Research (MJFF) and Movement Disorders through integrated analysis of genetics and neuroPathology (MD-GAP) through the Medical Research Council (MRC). For the purpose of open access, the author has applied a CC BY public copyright licence to all author accepted manuscripts arising from this submission. This research was supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. The University College London Movement Disorders Centre is supported by the Edmond J. Safra Philanthropic Foundation.
Competing interests
H.R.M. is employed by UCL. In the last 12 months, he reports paid consultancy from Roche, Aprinoia, AI Therapeutics and Amylyx; lecture fees/honoraria from BMJ, Kyowa Kirin and Movement Disorders Society; and research grants from Parkinson's UK, Cure Parkinson's Trust, PSP Association, Medical Research Council and Michael J. Fox Foundation. H.R.M. is a co-applicant on a patent application related to C9ORF72—Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140).
Data availability
TPD data are available upon access request from https://www.dpag.ox.ac.uk/opdc/team/proband-tracking-parkinsons. AMP-PD data are available upon registration at https://www.amp-pd.org/. OPDC data are available upon request from the Dementias Platform UK (https://portal.dementiasplatform.uk/Apply). HapMap phase 3 data (HapMap3) are available for download at https://www.broadinstitute.org/medical-and-population-genetics/hapmap-3. Cis-QTL eQTLGen data were downloaded from (https://www.eqtlgen.org/cis-eqtls.html). eQTL data from eQTL catalogue can be ftp-accessed (https://www.ebi.ac.uk/eqtl/Data_access/). Summary statistics from the Parkinson’s disease GWAS (Nalls et al.)10 used to perform the PRS analysis are available from https://pdgenetics.org/resources. The source code is available on GitHub (https://github.com/huw-morris-lab/LBD-case-case-GWAS; https://doi.org/10.5281/zenodo.8335404).
References
- 1. Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9(1):13–24. [DOI] [PubMed] [Google Scholar]
- 2. Muslimovic D, Post B, Speelman JD, Schmand B. Cognitive profile of patients with newly diagnosed Parkinson disease. Neurology. 2005;65(8):1239–1245. [DOI] [PubMed] [Google Scholar]
- 3. Hely MA, Reid WGJ, Adena MA, Halliday GM, Morris JGL. The Sydney multicenter study of Parkinson’s disease: The inevitability of dementia at 20 years. Mov Disord. 2008;23(6):837–844. [DOI] [PubMed] [Google Scholar]
- 4. Lo RY, Tanner CM, Albers KB, et al. Clinical features in early Parkinson disease and survival. Arch Neurol. 2009;66(11):1353–1358. [DOI] [PubMed] [Google Scholar]
- 5. McKeith IG, Boeve BF, Dickson DW, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89(1):88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Compta Y, Parkkinen L, O’Sullivan SS, et al. Lewy- and Alzheimer-type pathologies in Parkinson's disease dementia: Which is more important? Brain. 2011;134(Pt 5):1493–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson JL, Lee EB, Xie SX, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141(7):2181–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jellinger KA. Are there morphological differences between Parkinson's disease–dementia and dementia with Lewy bodies? Parkinsonism Relat Disord. 2022;100:24–32. [DOI] [PubMed] [Google Scholar]
- 9. Weil RS, Lashley TL, Bras J, Schrag AE, Schott JM. Current concepts and controversies in the pathogenesis of Parkinson's disease dementia and dementia with Lewy bodies. F1000Res. 2017;6:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chia R, Sabir MS, Bandres-Ciga S, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53(3):294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blauwendraat C, Heilbron K, Vallerga CL, et al. Parkinson's disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord. 2019;34(6):866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guella I, Evans DM, Szu-Tu C, et al. α-Synuclein genetic variability: A biomarker for dementia in Parkinson disease. Ann Neurol. 2016;79(6):991–999. [DOI] [PubMed] [Google Scholar]
- 14. Menšíková K, Matěj R, Colosimo C, et al. Lewy body disease or diseases with Lewy bodies? NPJ Parkinsons Dis. 2022;8(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu G, Peng J, Liao Z, et al. Genome-wide survival study identifies a novel synaptic locus and polygenic score for cognitive progression in Parkinson's disease. Nat Genet. 2021;53(6):787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Real R, Martinez-Carrasco A, Reynolds RH, et al. Association between the LRP1B and APOE loci in the development of Parkinson's disease dementia. Brain. 2023;146(5):1873-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malek N, Weil RS, Bresner C, et al. Features of GBA-associated Parkinson's disease at presentation in the UK Tracking Parkinson's study. J Neurol Neurosurg Psychiatry. 2018;89(7):702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vergouw LJM, van Steenoven I, van de Berg WDJ, et al. An update on the genetics of dementia with Lewy bodies. Parkinsonism Relat Disord. 2017;43:1–8. [DOI] [PubMed] [Google Scholar]
- 19. Creese B, Bell E, Johar I, Francis P, Ballard C, Aarsland D. Glucocerebrosidase mutations and neuropsychiatric phenotypes in Parkinson's disease and Lewy body dementias: Review and meta-analyses. Am J Med Genet B Neuropsychiatr Genet. 2018;177(2):232–241. [DOI] [PubMed] [Google Scholar]
- 20. Stoker TB, Camacho M, Winder-Rhodes S, et al. Impact of GBA1 variants on long-term clinical progression and mortality in incident Parkinson's disease. J Neurol Neurosurg Psychiatry. 2020;91(7):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nalls MA, Duran R, Lopez G, et al. A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA Neurol. 2013;70(6):727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chouliaras L, Kumar GS, Thomas AJ, Lunnon K, Chinnery PF, O’Brien JT. Epigenetic regulation in the pathophysiology of Lewy body dementia. Prog Neurobiol. 2020;192:101822. [DOI] [PubMed] [Google Scholar]
- 23. Wisniewski T, Drummond E. APOE–amyloid interaction: Therapeutic targets. Neurobiol Dis. 2020;138:104784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dickson DW. Neuropathology of Parkinson disease. Parkinsonism Relat Disord. 2018;46(Suppl 1):S30–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsuang D, Leverenz JB, Lopez OL, et al. APOE ε4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70(2):223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davis AA, Inman CE, Wargel ZM, et al. APOE genotype regulates pathology and disease progression in synucleinopathy. Sci Transl Med. 2020;12(529):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao N, Attrebi ON, Ren Y, et al. APOE4 exacerbates α-synuclein pathology and related toxicity independent of amyloid. Sci Transl Med. 2020;12(529):1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Malek N, Swallow DMA, Grosset KA, et al. Tracking Parkinson's: Study design and baseline patient data. J Parkinsons Dis. 2015;5(4):947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szewczyk-Krolikowski K, Tomlinson P, Nithi K, et al. The influence of age and gender on motor and non-motor features of early Parkinson's disease: Initial findings from the Oxford Parkinson Disease Center (OPDC) discovery cohort. Parkinsonism Relat Disord. 2014;20(1):99–105. [DOI] [PubMed] [Google Scholar]
- 30. Dubois B, Burn D, Goetz C, et al. Diagnostic procedures for Parkinson's disease dementia: Recommendations from the Movement Disorder Society task force. Mov Disord. 2007;22(16):2314–2324. [DOI] [PubMed] [Google Scholar]
- 31. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(7):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loh PR, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuchsberger C, Abecasis GR, Hinds DA. minimac2: Faster genotype imputation. Bioinformatics. 2015;31(5):782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boughton AP, Welch RP, Flickinger M, et al. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics. 2021;37(18):3017–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reynolds RH. RHReynolds/colochelpR: v0.99.1 (0.99.1). Zenodo. 2021. doi: 10.5281/zenodo.5011869. [DOI]
- 40. Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi SW, O’Reilly PF. PRSice-2: Polygenic risk score software for biobank-scale data. Gigascience. 2019;8(7):1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jo S, Kim SO, Park KW, Lee SH, Hwang YS, Chung SJ. The role of APOE in cognitive trajectories and motor decline in Parkinson's disease. Sci Rep. 2021;11(1):7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herbst S, Lewis PA, Morris HR. The emerging role of LRRK2 in tauopathies. Clin Sci. 2022;136(13):1071–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jabbari E, Koga S, Valentino RR, et al. Genetic determinants of survival in progressive supranuclear palsy: A genome-wide association study. Lancet Neurol. 2021;20(2):107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jellinger KA, Wenning GK, Seppi K. Predictors of survival in dementia with Lewy bodies and Parkinson dementia. Neurodegener Dis. 2007;4(6):428–430. [DOI] [PubMed] [Google Scholar]
- 46. Ferrari-Souza JP, Lussier FZ, Leffa DT, et al. APOEε4 associates with microglial activation independently of Aβ plaques and tau tangles. Sci Adv. 2023;9(14):eade1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang W, Xiao D, Mao Q, Xia H. Role of neuroinflammation in neurodegeneration development. Signal Transduct Target Ther. 2023;8(1):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montagne A, Nation DA, Sagare AP, et al. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature. 2020;581(7806):71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krohn L, Wu RYJ, Heilbron K, et al. Fine-mapping of SNCA in rapid eye movement sleep behavior disorder and overt synucleinopathies. Ann Neurol. 2020;87(4):584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Srivatsal S, Cholerton B, Leverenz JB, et al. Cognitive profile of LRRK2-related Parkinson's disease. Mov Disord. 2015;30(5):728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heckman MG, Soto-Ortolaza AI, Contreras MYS, et al. LRRK2 variation and dementia with Lewy bodies. Parkinsonism Relat Disord. 2016;31:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bokhovchuk F, Mesrouze Y, Delaunay C, et al. Identification of FAM181A and FAM181B as new interactors with the TEAD transcription factors. Protein Sci. 2020;29(2):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Donati G, Dumontheil I, Meaburn EL. Genome-wide association study of latent cognitive measures in adolescence: Genetic overlap with intelligence and education. Mind Brain Educ. 2019;13(3):224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wightman DP, Jansen IE, Savage JE, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat Genet. 2021;53(9):1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Crotti A, Sait HR, McAvoy KM, et al. BIN1 favors the spreading of tau via extracellular vesicles. Sci Rep. 2019;9(1):9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arima K, Hirai S, Sunohara N, et al. Cellular co-localization of phosphorylated tau- and NACP/alpha-synuclein-epitopes in Lewy bodies in sporadic Parkinson's disease and in dementia with Lewy bodies. Brain Res. 1999;843(1-2):53–61. [DOI] [PubMed] [Google Scholar]
- 57. Henderson MX, Sengupta M, Trojanowski JQ, Lee VMY. Alzheimer’s disease tau is a prominent pathology in LRRK2 Parkinson's disease. Acta Neuropathol Commun. 2019;7(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cerri S, Mus L, Blandini F. Parkinson's disease in women and men: What’s the difference? J Parkinsons Dis. 2019;9(3):501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chiu SY, Wyman-Chick KA, Ferman TJ, et al. Sex differences in dementia with Lewy bodies: Focused review of available evidence and future directions. Parkinsonism Relat Disord. 2023;107:105285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Reynolds RH. RHReynolds/colochelpR: v0.99.1 (0.99.1). Zenodo. 2021. doi: 10.5281/zenodo.5011869. [DOI]
Supplementary Materials
Data Availability Statement
TPD data are available upon access request from https://www.dpag.ox.ac.uk/opdc/team/proband-tracking-parkinsons. AMP-PD data are available upon registration at https://www.amp-pd.org/. OPDC data are available upon request from the Dementias Platform UK (https://portal.dementiasplatform.uk/Apply). HapMap phase 3 data (HapMap3) are available for download at https://www.broadinstitute.org/medical-and-population-genetics/hapmap-3. Cis-QTL eQTLGen data were downloaded from (https://www.eqtlgen.org/cis-eqtls.html). eQTL data from eQTL catalogue can be ftp-accessed (https://www.ebi.ac.uk/eqtl/Data_access/). Summary statistics from the Parkinson’s disease GWAS (Nalls et al.)10 used to perform the PRS analysis are available from https://pdgenetics.org/resources. The source code is available on GitHub (https://github.com/huw-morris-lab/LBD-case-case-GWAS; https://doi.org/10.5281/zenodo.8335404).