Significance
Ryanodine receptors (RyRs) are Ca2+ release channels on the sarcoplasmic reticulum (SR) that serves as an internal Ca2+ storage organelle surrounding the contractile apparatus in striated muscles. SR Ca2+ release by RyR channels plays a crucial role in mediating excitation–contraction coupling, a process in which electrical signaling is converted to mechanical muscle contraction. The small Ca2+-binding protein S100A1 modulates SR Ca2+ transients and contractile performance by binding to regulatory target proteins including RyR channels. We present here cryoelectron microscopy (cryo-EM) structures of skeletal muscle type-1 RyR in complex with S100A1 under Ca2+-free and Ca2+-saturated conditions revealing Ca2+-dependent differences in the S100A1 binding poses and providing structural insights into the regulation of RyR channels by S100A1.
Keywords: calcium, S100A1, ryanodine receptor, cryo-EM
Abstract
S100A1, a small homodimeric EF-hand Ca2+-binding protein (~21 kDa), plays an important regulatory role in Ca2+ signaling pathways involved in various biological functions including Ca2+ cycling and contractile performance in skeletal and cardiac myocytes. One key target of the S100A1 interactome is the ryanodine receptor (RyR), a huge homotetrameric Ca2+ release channel (~2.3 MDa) of the sarcoplasmic reticulum. Here, we report cryoelectron microscopy structures of S100A1 bound to RyR1, the skeletal muscle isoform, in absence and presence of Ca2+. Ca2+-free apo-S100A1 binds beneath the bridging solenoid (BSol) and forms contacts with the junctional solenoid and the shell-core linker of RyR1. Upon Ca2+-binding, S100A1 undergoes a conformational change resulting in the exposure of the hydrophobic pocket known to serve as a major interaction site of S100A1. Through interactions of the hydrophobic pocket with RyR1, Ca2+-bound S100A1 intrudes deeper into the RyR1 structure beneath BSol than the apo-form and induces sideways motions of the C-terminal BSol region toward the adjacent RyR1 protomer resulting in tighter interprotomer contacts. Interestingly, the second hydrophobic pocket of the S100A1-dimer is largely exposed at the hydrophilic surface making it prone to interactions with the local environment, suggesting that S100A1 could be involved in forming larger heterocomplexes of RyRs with other protein partners. Since S100A1 interactions stabilizing BSol are implicated in the regulation of RyR-mediated Ca2+ release, the characterization of the S100A1 binding site conserved between RyR isoforms may provide the structural basis for the development of therapeutic strategies regarding treatments of RyR-related disorders.
S100 proteins are small Ca2+-binding proteins that are involved in multiple intracellular and extracellular signaling pathways regulating a wide range of biological functions including cell division, differentiation, cytoskeleton organization, and cell motility (1, 2). A prominent member of the S100 protein family is the S100A1 isoform. S100A1 is expressed in the heart, skeletal muscle, and brain (3–5). Subcellularly, S100A1 localizes with higher densities at the endoplasmic/sarcoplasmic reticulum (ER/SR) and Z-lines (6, 7). At the SR of cardiac and skeletal myocytes, S100A1 modulates Ca2+ cycling that determines the sarcomere contractile function (7). Animal models overexpressing cardiac S100A1 are characterized by enhanced SR Ca2+ release and augmented SR Ca2+ reuptake resulting in larger Ca2+ transients and improved myocardial performance (8, 9). In contrast, S100A1 knockout (SKO) mice are asymptomatic at resting conditions exhibiting normal baseline cardiac function and heart rate. During exercise or stress, the responsiveness to β-adrenergic stimulation is abnormally deficient in SKO mice displaying reduced responses in contraction and relaxation rates that may reflect impaired Ca2+ sensitivity in the cardiac reserve due to SKO (10). Skeletal muscle fibers of SKO mice show abnormal action potential-evoked Ca2+ transients with decreased peak amplitudes and prolonged rising phases, resulting in an impaired maximal isometric force and a faster onset of muscle fatigue upon repetitive stimulation (11, 12). Interestingly, cardiac S100A1 levels are decreased in heart failure animal models and patients (13, 14). Cardiac adenoviral S100A1 gene transfer in pigs with postischemic heart failure has been shown to restore Ca2+ homeostasis, reverse maladaptive remodeling, and stabilize cardiac function (15). S100A1 is considered an attractive candidate for gene therapy clinical trials (16).
One key regulatory target of the S100A1 interactome is the ryanodine receptor (RyR), a huge homotetrameric Ca2+ release channel (~2.3 MDa) on the SR localized to triad junctions in skeletal myocytes (RyR1) or to dyadic junctions in cardiomyocytes (RyR2). The SR Ca2+ release by RyR channels regulates many cellular processes including excitation–contraction (E-C) coupling that converts electrical signaling (action potentials) to mechanical muscle contraction. In hearts, dysfunctional RyR2 channels can cause diastolic SR Ca2+ leak associated with cardiac arrhythmias and heart failure (17, 18). Similarly, RyR1-mediated SR Ca2+ leak can impair skeletal muscle function. Accordingly, several RyR1-related myopathies have been reported including malignant hyperthermia (MH), central core disease, and centronuclear myopathy (18, 19). Targeting the impaired regulation of dysfunctional RyR channels represents a promising therapeutic strategy for RyR-related disorders (19, 20).
Calmodulin (CaM) is another small Ca2+-binding protein of the EF-hand superfamily involved in regulation of E-C coupling. CaM and S100A1 share similar but not identical target recognition specificities in binding preferentially to amphipathic α-helices upon activation by Ca2+ (1, 12, 21). Both proteins can modulate SR Ca2+ release by direct binding to RyR channels (22–24) or indirectly by interacting with enzymes involved in RyR regulation, e.g., CaMKII via Calmodulin (25), PKA via S100A1 (25, 26), and SERCA2a via S100A1 (27). The effects of S100A1- or CaM-binding to RyR have been analyzed at the single-channel level using planar lipid bilayers or by measuring Ca2+ release from SR vesicles. These functional data indicate that Ca2+-free CaM (apo-CaM) activates RyR1 and inhibits RyR2, whereas Ca2+-bound CaM (Ca-CaM) stabilizes the closed state of RyR1 and RyR2 (28–30). Interestingly, a recent report revealed that CaM-binding to RyR1 abolishes a pathological conformation of the MH mutation R614C (30) suggesting that CaM can have alleviating effects on dysfunctional RyR channels under certain pathologic conditions. The effects of S100A1 on RyR channels resemble those of CaM to a certain extent. RyR1-activation by recombinant S100A1 has been reported for SR vesicles isolated from rat skeletal muscle (31) and for purified RyR1 in planar lipid bilayer experiments at submicromolar free [Ca2+], at which S100A1 is predicted to exist in the Ca2+-free apo-form (apo-S100A1) (24). Moreover, S100A1 decreased the activity of RyR2 channels in planar lipid bilayers at similar conditions (32). Hence, like apo-CaM, apo-S100A1 shows opposite effects on RyR1 and RyR2 activity. In line with apo-S100A1 stabilizing the closed state of RyR2, S100A1 added exogenously to permeabilized quiescent cardiomyocytes decreased Ca2+ spark frequency, amplitude, and duration (33). Under systolic conditions, S100A1 provoked increased fractional SR Ca2+ release in voltage-clamped rabbit cardiomyocytes, implying that Ca-S100A1, unlike Ca-CaM, can improve E-C coupling gain presumably by RyR2 activation (34). Similarly, it has been suggested that Ca-S100A1 activates RyR1 since SKO in mice impaired Ca2+ transients and the maximal isometric force during tetanic stimulation (11, 12, 23).
Previously reported single particle cryoelectron microscopy (cryo-EM) analyses of RyR1 at near atomic-resolution granted insights into its domain architecture and provided atomic models for nearly the complete protein. The homotetrameric Ca2+-channel exhibits an evolutionarily conserved mushroom-like shape with a fourfold symmetry along the central pore axis (35–39). The C-terminal portion of RyR forms the ion-conducting transmembrane domain (stalk), while the massive cytoplasmic shell (head) allosterically regulates the RyR channel function and presents a large scaffold for multiple protein–protein interactions (22). The cytosolic shell consists of extended α-solenoid scaffold regions interspersed by globular domains such as two β-trefoil domains at the N terminus (NTD-A and NTD-B), three SPRY (SPIa kinase and ryanodine receptor) domains, and two tandem repeat domains (RY1&2 and RY3&4). The flexible armadillo-like repeat network of the cytoplasmic shell comprises the N-terminal (NSol), junctional (JSol), and bridging (BSol) solenoids, and continues as core solenoid (CSol) in the central core region of RyR channels. The BSol domain has an exposed position at the periphery of the channel forming a large part of the surface and, hence, is thought to be important for protein–protein interactions in RyR macromolecular complexes. The BSol conformation allosterically affects pore gating of RyR channels by forming intra- and interprotomer contacts with neighboring domains including BSol with NSol, BSol with NTD-A (of adjacent protomer), and BSol with SPRY2 (of adjacent protomer) (40, 41). Stimulatory or inhibitory signals are transmitted from the cytosolic shell to the pore via CSol. The BSol and CSol domains are directly connected by the shell-core linker (SCL) bridging the cleft between the C-terminal BSol region and the JSol/CSol interface. The α-solenoid network in RyR channels may enable protein interaction partners of the cytosolic shell to allosterically modulate RyR activity (40, 41). Structural data of RyR-heterocomplexes are only available for the small proteins Calstabin and CaM. Similar to S100A1, CaM binds to the JSol/CSol interface, SCL, and N-terminal BSol region of RyR1. Thereby, apo-CaM and Ca-CaM stabilize the BSol domain in activating and inhibitory conformational states, respectively (30, 36, 42).
The S100A1 structure has been solved by NMR and X-ray crystallography showing antiparallel homodimers in apo or Ca2+-bound states (43–45). Each monomeric subunit comprises two Ca2+ binding helix–loop–helix motifs (EF-hands) which are connected by a flexible loop (hinge region), forming an X-type four-helix bundle (incl. α-helices H1-H4). The N-terminal moiety (H1-H2) is considered a “pseudo” EF-hand (pEF) because it coordinates Ca2+ binding mainly through backbone carbonyl groups, whereas the canonical C-terminal EF-hand (cEF; H3-H4) tightly binds Ca2+ through acidic side chains. This is reflected in the weak Ca2+ affinity of pEF exhibiting a dissociation constant in the high micromolar range (KD: > 250 μM) that is about an order of magnitude larger compared to cEF (KD: ~27 to 50 μM), implying that the pEF-hand would not significantly bind Ca2+ at physiological conditions (44, 46). However, previous studies reported that the Ca2+-affinity of S100A1 increases after oxidation of S100A1 at cysteine C86 by S-nitrosylation or S-glutathionylation (46). Interactions with target proteins may also alter the Ca2+ affinity of S100A1 by stabilizing Ca-S100A1 (23, 47). Ca2+-binding to both pEF- and cEF-hands induces a conformational change of S100A1 resulting in the exposure of a hydrophobic pocket between helices H3 and H4 that mediates interactions with several regulatory protein targets. Interestingly, previous NMR data showed that a RyR1-derived peptide named RyRP12 (a.a. 3616-3627 in human RyR1) can bind into the hydrophobic pocket of S100A1 (12, 23). The RyRP12 sequence belongs to the SCL domain that is known to interact with CaM. A direct competition between S100A1 and CaM for binding to RyR has been implicated (12, 23) suggesting that S100A1-binding may indirectly modify RyR activity by displacing CaM. Such a competition would depend on the relative binding affinities and local protein concentrations of free CaM versus free S100A1. However, FRET studies comparing S100A1- and CaM-binding to RyR implied that S100A1 does not directly compete with CaM under physiological-like conditions. Instead, it was suggested that S100A1 would bind to RyR on a separate binding site altering allosterically the structure of the RyR–CaM complex (48). But structural data for the S100A1 interaction with full-length RyR have remained elusive until now. Here, we show cryo-EM structures of S100A1 bound to RyR1 in presence and absence of Ca2+. Our structural models of RyR1-S100A1 provide insights for the regulation of RyR1 channels by S100A1.
Results
Ca2+ Dependence of the Interaction between S100A1 and RyR1.
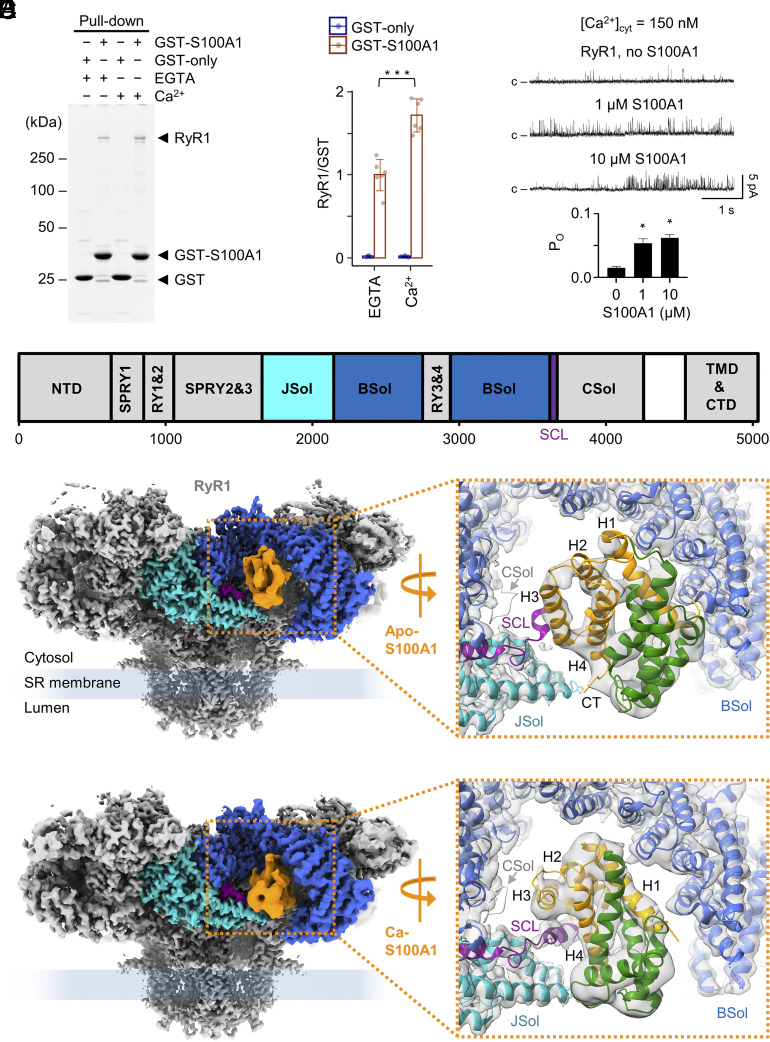
In order to confirm the interaction between S100A1 and RyR1, we performed Glutathione S-Transferase (GST) pull-down assays using recombinant GST-tagged S100A1 purified from Escherichia coli and endogenous RyR1 purified from mouse skeletal muscles. GST pull-downs were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining (Fig. 1A). Coomassie-stained protein bands specific for RyR1 were only detected after pull-down with GST-S100A1 but not with GST-only (negative control). Addition of 1 mM free Ca2+ to the GST pull-down assay resulted in a ~1.7-fold increase of the RyR1 signal compared to the EGTA-only condition (no free Ca2+) indicating that Ca2+ induces a stronger S100A1-binding mode (Fig. 1B).
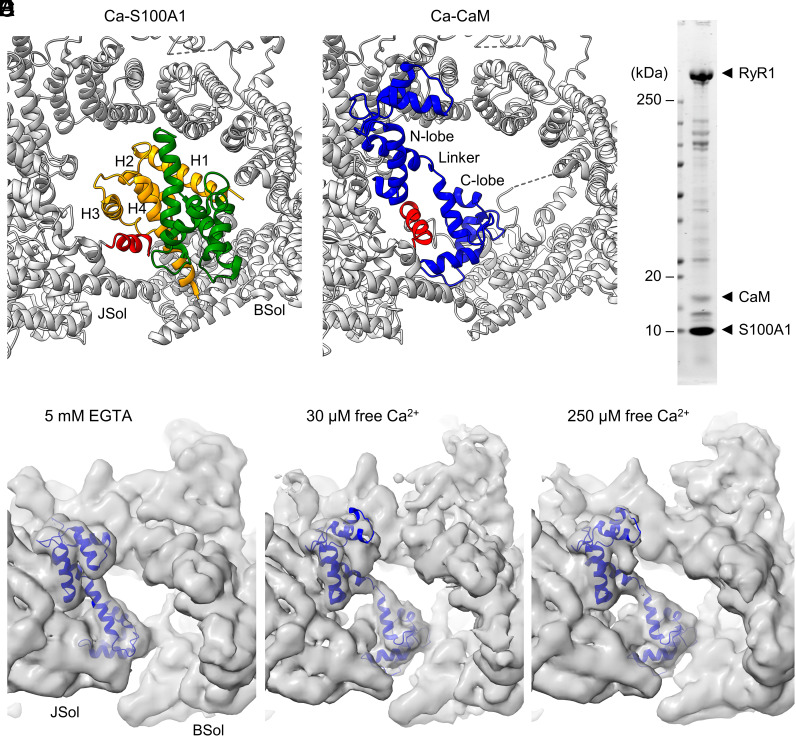
Fig. 1.
Interaction between RyR1 and S100A1 under EGTA and Ca2+-saturated conditions. (A) GST pull-down of GST-tagged S100A1 and RyR1. Eluted samples were subjected to SDS-PAGE followed by Coomassie staining. Co-pull-down of RyR1 with GST-S100A1 is observed under apo (1 mM EGTA) and Ca2+-saturated conditions (1 mM free Ca2+) confirming the interaction between both proteins. GST-only served as negative control. (B) Quantification of the Coomassie-stained protein bands. The ratios of the RyR1 signal versus GST-S100A1 or GST-only are plotted for the EGTA and Ca2+-saturated conditions. The quantification indicates that addition of Ca2+ results in a slightly stronger co-pull-down of RyR1 (by factor ~1.7). Two-tailed Student’s t test; ***P < 0.001. (C) Single channel recordings (Top) of purified RyR1 reconstituted in planar lipid bilayers at 150 nM free Ca2+ and quantification of the open probability (Bottom) of RyR1 with and without S100A1 (n = 5 per group). Channel openings in current traces are represented as upward deflections, while baseline currents correspond to the closed state (c) of RyR1. Upon addition of 1 µM or 10 µM S100A1, the open probability (PO) of RyR1 significantly increased from 0.01 to 0.05 or 0.07, respectively. Two-tailed Student’s t test; *P < 0.05. (D) Schematic view of the domain architecture of mouse RyR1 comprising the N-terminal domain (NTD), the junctional (JSol), bridging (BSol), and core (CSol) solenoids, three SPRY domains, two tandem repeat domains (RY1&2 and RY3&4), the SCL, as well as the transmembrane and C-terminal domains (TMD & CTD). (E and F) Side view on cryo-EM composite maps of RyR1 with apo-S100A1 at 5 mM EGTA only (E) and with Ca-S100A1 at 250 µM free Ca2+/caffeine/ATP (F). S100A1 is colored in orange and RyR1 according to the domain architecture scheme given above (D). In the magnifications, the conformational states of apo- and Ca-S100A1 are shown (PDBs: 8VK3 and 8VK4, respectively). The monomeric subunits 1 (orange) and 2 (green) of homodimeric S100A1 and the surrounding binding site of RyR1 are depicted as ribbon models. Both, apo-S100A1 and Ca-S100A1 bind into the cleft between the BSol (blue) and JSol (cyan) domains but exhibit different binding poses. As a result of Ca2+-induced conformational change, Ca-S100A1 exposes a hydrophobic pocket between helices H3-H4 and the hinge region that interacts with the central helix of SCL (magenta). Ca-S100A1 moves deeper beneath the BSol domain compared to apo-S100A1.
The interaction between purified RyR1 and S100A1 was assessed by cryo-EM single particle analysis (SI Appendix, Figs. S1 and S2). Following cryo-EM data collection and particle selection of RyR1–S100A1 complexes, the S100A1 signal was distinctive and differences in the S100A1 binding poses on RyR1 with and without Ca2+ were revealed as described in detail below (Fig. 1 D–F). Hence, our GST pull-down and cryo-EM data confirmed RyR1-S100A1 interactions and implied that S100A1 can bind to RyR1 not only in a Ca2+-bound mode (Ca-S100A1) (Fig. 1F) but also in a weaker Ca2+-free mode (apo-S100A1) (Fig. 1E).
To analyze the functional effect of S100A1 on RyR1 activity, we reconstituted purified RyR1 channels in planar lipid bilayers. Single-channel currents were recorded at 150 nM free Ca2+ (Fig. 1C and SI Appendix, Fig. S3A). The open probability (PO) of RyR1 determined from the single-channel recordings significantly increased upon adding 1 µM S100A1 (from PO = 0.01 to PO = 0.05, n = 5, P < 0.05) and continued to increase as S100A1 was added up to 10 µM (PO = 0.07, n = 5, P < 0.05). Hence, our single-channel recordings confirm that S100A1 can activate RyR1 at submicromolar Ca2+-concentrations as previously reported (24). At 150 nM free Ca2+, S100A1 presumably exists in the apo-form since the two EF hand motifs of S100A1 bind Ca2+ with low affinity in vitro (cEF, KD: ~27 to 50 μM; pEF, KD: > 250 μM) (44, 46), implying that the apo-form of S100A1 is mediating the activation of RyR1. To form Ca-S100A1, we increased the free Ca2+ concentration to 250 µM. Under the high Ca2+ condition, we did not detect a significant change in single-channel currents of RyR1 after adding 1 µM or 10 µM S100A1 (PO = ~0.2) (SI Appendix, Fig. S3B).
Structure of RyR1 in Complex with apo-S100A1.
To solve the protein complex structure of S100A1 bound to RyR1, we chromatographically purified endogenous RyR1 from mouse skeletal muscle and recombinant mouse S100A1 overexpressed in E. coli. Samples of RyR1–S100A1 complexes were prepared by adding a 10-fold molar excess of S100A1 homodimers (~150 µM final) to RyR1 protomers (~15 µM final) in presence of 5 mM EGTA to chelate Ca2+. After vitrification, the final samples of RyR1–S100A1 complexes were assessed by cryo-EM single particle analysis (SI Appendix, Fig. S1). Initially, homogeneous refinements (using CryoSPARC) of all RyR1 particles (defined as total number of high-quality particles identified as RyR1 in the cryo-EM dataset; N = 336,237) indicated a density for S100A1 located in the cleft between the JSol and BSol domains of RyR1. To enrich the S100A1 density, particles of RyR1 bound to S100A1 were selected by 3D classification followed by local refinements (using CryoSPARC). As a result, the S100A1 density signal took on the shape of homodimeric S100A1 and the secondary structural elements of S100A1 became clear. Only one of the four RyR1 protomers was occupied by S100A1 in a well-defined binding pose after 3D classification.
The structure of the mouse homolog of RyR1 was solved at an overall resolution of ~2.5 Å, reaching local resolutions of ~2.3 Å in the final composite map of all particles (N = 1,344,948 after C4 symmetry expansion). Importantly, depletion of the apo-S100A1 density by 3D variability did not alter the structural conformation of RyR1 compared to all particles (SI Appendix, Fig. S4 A and B). Moreover, the occupancy of the S100A1 binding site in all particles was negligible compared to the selected RyR1 particles with S100A1 (SI Appendix, Fig. S4B). Hence, we used the structure of all particles to derive the atomic model of mouse RyR1 (PDB: 8VJJ). The structure contained the endogenously bound small protein Calstabin but not Calmodulin as expected from previous rabbit RyR1 purifications using similar protocols (35, 49). The mouse and rabbit RyR1 homologs share a high degree of conservation in their sequences (95.9% identity, 97.4% similarity). In line with this, the mouse homolog exhibits the conserved RyR1 architecture known from the rabbit RyR1 structures (35, 37–39).
For the selected RyR1-S100A1 particles of the C4 symmetry expanded dataset (N = 36,507; ~2.7%), the apo-S100A1-binding site of RyR1 was determined at a local resolution of ~3.5 Å (SI Appendix, Fig. S5A), including apo-S100A1 solved at ~5 to 7 Å resolution (SI Appendix, Fig. S5B). A final composite map for RyR1-S100A1 was built from local refinements of the expanded particles at a global resolution of ~3.2 Å (Fig. 1 E, Left). The S100A1 density was not sufficient to model side chains but revealed the backbone architecture of the S100A1-dimer. This allowed us to unambiguously dock the atomic-resolution NMR structure of apo-S100A1 (PDB: 2L0P) into the cryo-EM density map by rigid-body fitting followed by flexible fitting and to derive an atomic model for the RyR1–S100A1 complex in absence of Ca2+ (PDB: 8VK3) (Fig. 1 E, Right).
In the RyR1–S100A1 complex, one monomeric subunit (H1-H4) of the S100A1-dimer is bound beneath the BSol domain while the second antiparallelly oriented subunit (H1′-H4′) protrudes laterally into the cytosolic periphery. Overall, apo-S100A1 adopts a compact conformation in which the regions around the N and C terminus of the first subunit participate in interactions with the BSol and JSol domains of RyR1, respectively (SI Appendix, Fig. S6 A–C). Next to JSol, the SCL of RyR1 forms contacts with the H3 helix of S100A1-subunit 1 (SI Appendix, Fig. S6B). At these local contact sites, several hydrogen bonds and salt bridges are predicted by the model to stabilize apo-S100A1 binding (SI Appendix, Fig. S6 B–D) but, given limitations in resolution and potential dynamic changes of S100A1–RyR1 interactions, these side chain interactions remain putative.
Compared to RyR1 particles depleted of S100A1, we did not detect any significant conformational change in the RyR1 structure upon binding of apo-S100A1 in cryo-EM maps (SI Appendix, Fig. S4B). Similarly, comparison between the atomic models of all RyR1 particles (PDB: 8VJJ) and selected RyR1 particles with apo-S100A1 (PDB: 8VK3) revealed no structural changes in RyR1 (SI Appendix, Fig. S4C) that would explain RyR1 activation by S100A1 at submicromolar Ca2+-concentrations as observed in planar lipid bilayers (Fig. 1C). In particular, the pore remained in a closed conformation. Hence, apo-S100A1 did not overcome the stabilization of the closed RyR1 state by Calstabin as expected for Ca2+-free RyR1 without additional activating ligands (such as ATP and caffeine) (35, 38–40).
Structure of RyR1 in Complex with Ca-S100A1.
To analyze whether Ca-S100A1 exhibits a different binding mode than the apo-form, we prepared cryo-EM samples of RyR1–S100A1 complexes in the same way as described for the Ca2+-free structure but added 0.25 mM free Ca2+, 5 mM caffeine, and 10 mM ATP instead of only EGTA. Following the same processing as before, the RyR1 structure of all particles (N = 292,757) was determined with an overall resolution of ~2.9 Å and local resolutions reaching ~2.7 Å (PDB: 8VJK) (SI Appendix, Fig. S2). For the selected RyR1-S100A1 expanded particles (N = 31,572; ~2.7%), we obtained a structure at ~3.6 Å global resolution (Fig. 1 F, Left). The S100A1 binding site of RyR1 was solved at ~3.8 Å (SI Appendix, Fig. S7A) and bound S100A1 at ~5 to 7 Å local resolution (SI Appendix, Fig. S7B) allowing unambiguously fitting of the atomic model of S100A1 in the Ca2+-bound conformation (PDB: 2K2F) previously obtained by NMR at atomic resolution (23). From the final composite map of local refinements, we were able to derive a structural model for the RyR1–S100A1 complex in presence of high [Ca2+] (PDB: 8VK4) (Fig. 1 F, Right).
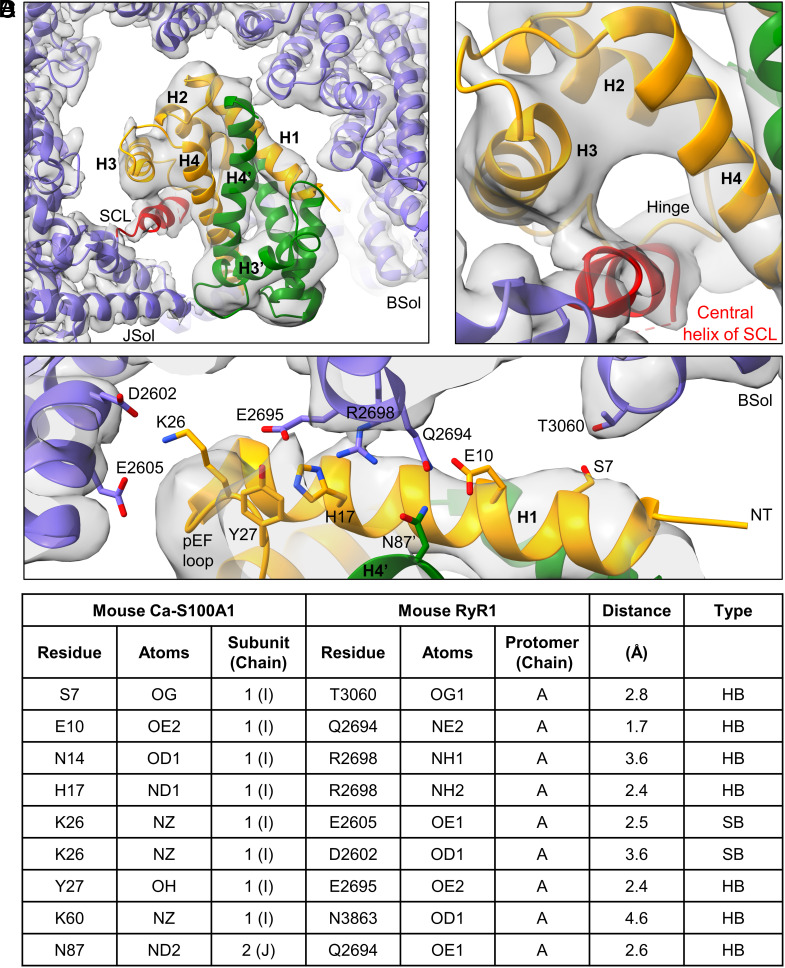
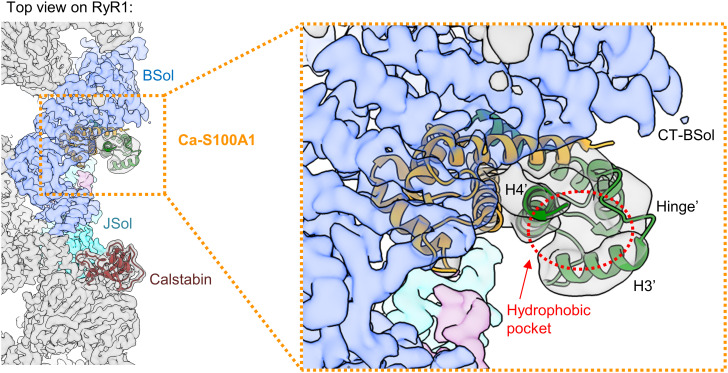
The structural model reveals that Ca-S100A1 remains in a similar binding orientation as apo-S100A1 but intrudes deeper into the RyR1 structure beneath the BSol domain (SI Appendix, Fig. S8 A and B). Subunit 1 of Ca-S100A1 mediates nearly all the interactions with RyR1 (Fig. 2 A–D). In particular, helix H1 of Ca-S100A1 closely interacts with the C-terminal BSol region forming putative hydrogen bonds and salt bridges (Fig. 2 C and D). Helix H3 of Ca-S100A1 may protrude to the flexible loop of the CSol domain at a.a. ~3860-3868, but the resolution of this loop is insufficient to detect contacts (Fig. 1 F, Right). Importantly, Ca-S100A1 exposes a hydrophobic pocket between helices H3 and H4 that serves as a major interaction site (23, 47). The hydrophobic pocket of subunit 1 is occupied by the central region of the SCL (a.a. ~3625-3639 in mouse RyR1) mediating tight interactions with helices H3, H4, and the hinge region of S100A1-subunit 1 (Fig. 2B). These Ca2+-dependent interactions with the SCL may explain why the affinity between S100A1 and RyR1 increases upon Ca2+ binding as shown by the GST pull-down assays (Fig. 1 A and B). However, compared to apo-S100A1, the more engaged binding mode of Ca-S100A1 did not lead to a substantially higher occupancy of the S100A1 binding site in our structural analyses (SI Appendix, Figs. S1 and S2). This may reflect dynamic changes in S100A1 binding potentially due to flexible BSol motions in single particles not stabilized by RyR1 clustering interactions as would be the case on the SR membrane in vivo where RyR1 is tightly packed in side-by-side arrays organized in checkerboard-like arrangements (50). Interestingly, the hydrophobic pocket of subunit 2 is exposed at the hydrophilic surface next to the BSol domain where it would be prone to interactions with the cytosolic environment (Fig. 3).
Fig. 2.
Local interaction hot spots between RyR1 and Ca-S100A1. (A) Overview of the Ca-S100A1 binding site of RyR1 (cryo-EM map in gray). The monomeric subunits 1 (orange) and 2 (green) of homodimeric S100A1 and the surrounding binding site of RyR1 (violet) are depicted as ribbon models (PDB: 8VK4). (B) Interaction site between the SCL of RyR1 and subunit 1 of Ca-S100A1. The central helix (red) of the SCL binds into the hydrophobic pocket formed by helices H3-H4 and the hinge region of subunit 1. (C) Interaction site between the BSol domain of RyR1 and subunit 1 of Ca-S100A1. Helix H1 of subunit 1 forms close contacts with the BSol domain. (D) Table of S100A1-RyR1 side chain interactions predicted by the atomic model under Ca2+-saturated condition. The listed H-bonds (HB) and salt bridges (SB) of side chains (depicted in C) are only putative, given limitations in resolution and potential dynamic changes of S100A1 binding.
Fig. 3.
Hydrophobic pocket of the second monomeric subunit of S100A1 exposed at the cytosolic surface of RyR1. Top view on the cryo-EM composite map of RyR1 (gray) with Ca-S100A1 (subunit 1 in orange, subunit 2 in green; PDB: 8VK4) and Calstabin (brown). The S100A1 binding site of RyR1 is formed by JSol (cyan), BSol (blue), and SCL (magenta). In the magnification, the hydrophobic pocket of subunit 2 (hollow red circle), formed by the hinge region and helices H3′-H4′, is shown to be largely exposed at the surface next to the C-terminal BSol region (CT-BSol) while subunit 1 is bound to RyR1 beneath the BSol domain.
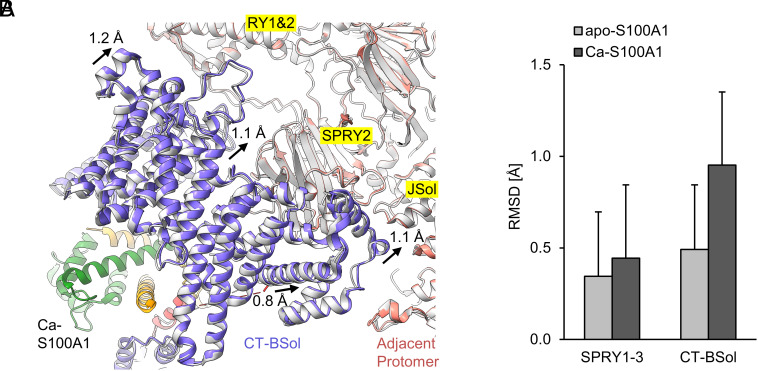
In comparison with all RyR1 particles, the selected RyR1-S100A1 particles underwent a conformational change. Ca-S100A1 binding induced a counterclockwise sideways motion of the C-terminal BSol region (CT-BSol) by ~1 Å, getting closer to the adjacent RyR1 protomer at the RY1&2, SPRY2, and JSol domains (Fig. 4A and SI Appendix, Table S1). Local RMSD values calculated for the superimposed structures of RyR1 before and after Ca-S100A1 binding confirmed that the CT-BSol motion (~1 Å) is substantially larger than the background motion at the SPRY1-3 region (~0.4 Å). In contrast, apo-S100A1 binding to RyR1 did not substantially increase the RMSD value for the CT-BSol region (Fig. 4B). The S100A1-induced CT-BSol motion may lead to tighter interactions between the adjacent protomers in the RyR1 structure, which in return could stabilize specific conformational states of RyR1 and, hence, represent the structural basis for the modulation of RyR1 activity by S100A1.
Fig. 4.
Sideways motion of the C-terminal BSol region upon binding of Ca-S100A1. (A) View from below the S100A1 binding site of RyR1. The atomic models of RyR1-S100A1 (colored; PDB: 8VK4) and all RyR1 particles (gray; PDB: 8VJK) are superimposed under Ca2+-saturated conditions (250 µM free Ca2+/caffeine/ATP). Upon Ca-S100A1 binding (subunit 1 in orange, subunit 2 in green), the CT-BSol region (violet) moved sideways by ~1 Å toward the RY1&2, SPRY2, and JSol domains of the adjacent protomer (salmon). (B) Local C-alpha RMSD were calculated for the superimposed structures at the SPRY1-3 region (a.a. 629-1657 in mouse RyR1) and CT-BSol region (a.a. 2950-3613). Compared to SPRY1-3, CT-BSol showed a substantially increased RMSD value upon binding of Ca-S100A1 but not for apo-S100A1.
Compared to the Ca2+-free RyR1 structure under EGTA condition, the addition of the small activating ligands caffeine, ATP, and Ca2+ to RyR1 induced a downward motion of CT-BSol that differs from the sideways motion of CT-BSol induced by Ca-S100A1 (SI Appendix, Fig. S9). However, the RyR1 pore adopted only closed conformational states due to the high [Ca2+] inactivating RyR1 as reported previously (51).
Steric Overlap between the CaM and S100A1 Binding Sites on RyR1 in Presence and Absence of Ca2+.
Previous reports suggested direct competition between S100A1 and CaM for the same binding site on RyR1 (12, 23). To show putatively clashing regions of sterically overlapping S100A1 and CaM binding sites on RyR1, we compared the RyR1-S100A1 structures with known RyR1-CaM structures (30, 49). Under EGTA conditions, the flexible central linker of apo-CaM (PDB: 6X32) and the helix H3 of apo-S100A1 show similar positions on RyR1 (SI Appendix, Fig. S10 A and B) and would sterically overlap to a large extent (SI Appendix, Fig. S10 E, Left). Next to helix H3, a steric overlap would occur between the cEF-loop of apo-S100A1 and the N-lobe of apo-CaM as well as between the hinge region of apo-S100A1 and the C-lobe of apo-CaM (SI Appendix, Fig. S10 E, Left). Under Ca2+-saturated conditions, the C-lobe of Ca-CaM (PDB: 7TZC) overlaps with helices H2–H4 of Ca-S100A1 (SI Appendix, Fig. S10 C–E). Additionally, both Ca-CaM and Ca-S100A1 show tight interactions with the SCL region of RyR1 (Fig. 5 A and B). Compared to RyR1-CaM structures, the S100A1 binding induces a conformational change of the SCL. The structurally resolved central part of the core-shell linker changes its orientation and rotates by around 90° toward the CT-BSol region upon binding of S100A1 (Fig. 5A). Moreover, apo-S100A1 and Ca-CaM may coexist under physiological conditions, since the pEF-hand of S100A1 has a low Ca2+-affinity (KD > 250 μM) and the Ca2+-binding to the pEF-hand is a prerequisite for the conformational change to Ca-S100A1 (44, 46). Interestingly, the C-lobe of Ca-CaM would only slightly intersect with the flexible hinge region of apo-S100A1, while the N-lobe would slightly collide with the helix H3 and cEF-loop (SI Appendix, Fig. S10 E, Middle). Thus, the binding sites of apo-S100A1 and Ca-CaM show less overlap compared to the other states. Taken together, the CaM and S100A1 binding events onto RyR1 are predicted to sterically clash in their Ca2+-free and Ca2+-bound states.
Fig. 5.
Steric displacement of S100A1 by CaM from RyR1 under Ca2+-free and Ca2+-bound conditions. (A and B) Atomic model of Ca-S100A1 (subunit 1 in orange, subunit 2 in green; PDB: 8VK4) (A) and Ca-CaM (blue; PDB: 7TZC) (B) bound to RyR1 (gray). The helices H1-H4 of S100A1-subunit 1 (A) and the N- and C-lobe of CaM (B) are indicated. The central helix (red) of the SCL of RyR1 changes its orientation if bound to Ca-S100A1 (A) compared to Ca-CaM (B). (C) Coomassie-stained SDS-gel of the protein sample prepared for cryo-EM consisting of RyR1 (~1.6 µM protomer), CaM (~7.8 µM), S100A1 (~40.6 µM dimer). (D–F) Cryo-EM screenings of the RyR1-S100A1-CaM sample at 5 mM EGTA (D), 30 µM free Ca2+ (E), or 250 µM free Ca2+ (F). Under these experimental conditions, the cryo-EM maps showed densities for CaM (blue ribbon model) but not for S100A1, implying that apo-CaM displaces apo-S100A1 from RyR1 (under EGTA condition) while Ca-CaM displaces apo- and Ca-S100A1 (at 30 µM and 250 µM free Ca2+, respectively).
To investigate whether CaM and S100A1 compete for the overlapping binding sites on RyR1 or if an induced fit mechanism allows both small proteins to bind simultaneously, we prepared cryo-EM samples consisting of purified RyR1 (~1.6 µM protomer), CaM (~7.8 µM), and S100A1 (~40.6 µM dimer) under apo- and Ca2+-conditions (Fig. 5C). Thus, a 5X molar excess of S100A1-dimer versus CaM was concurrently added to RyR1. Cryo-EM screenings of samples at 5 mM EGTA, 30 µM free Ca2+, or 250 µM free Ca2+ showed distinct density signals for CaM but not for S100A1 implying that apo-CaM displaces apo-S100A1 (EGTA condition) (Fig. 5D) while Ca-CaM displaces apo- and Ca-S100A1 (at 30 µM and 250 µM free Ca2+, respectively) (Fig. 5 E and F). The CaM densities under Ca2+-free and Ca2+-bound conditions exhibited binding poses on mouse RyR1 similar to previously published atomic models for apo-CaM bound to porcine RyR1 (PDB: 6X32) (30) and Ca-CaM bound to rabbit RyR1 (PDB: 7TZC) (49), respectively (Fig. 5 D–F). This suggests evolutionary conservation of the RyR1-CaM interaction across mammalian species.
Hence at least under experimental in vitro conditions, our cryo-EM data reveal steric clashes between S100A1 and CaM binding sites and exclude simultaneous binding of CaM and S100A1 to RyR1 protomers.
Interaction between Endogenous S100A1 and RyR in Skeletal Muscle, Heart, and Cerebellum.
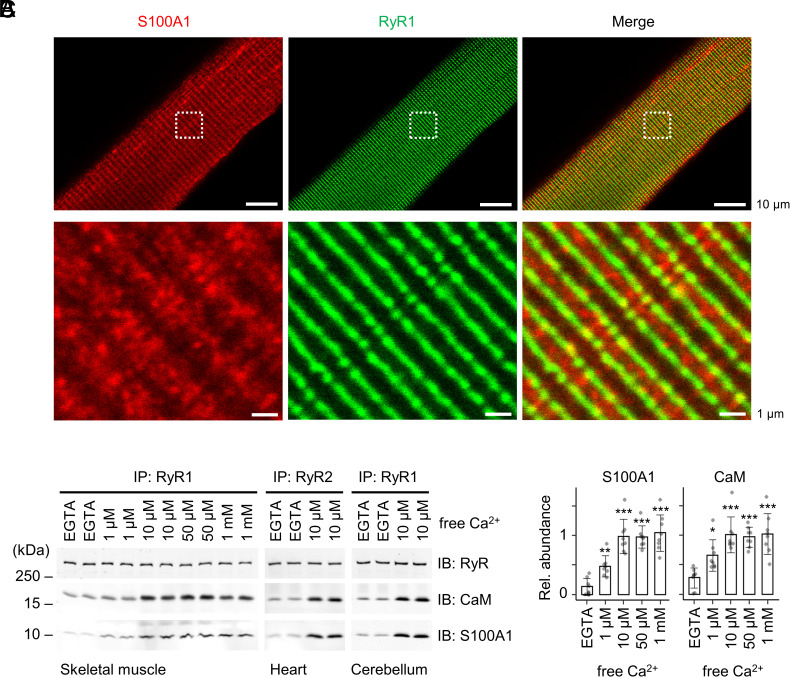
To investigate whether intracellular RyR clusters are associated with endogenous S100A1, we isolated ventricular myocytes (VMs) and immunostained them with S100A1 and RyR2 antibodies followed by stimulated emission depletion (STED) superresolution microscopy (SI Appendix, Fig. S12 A and B). Compared to skeletal myocytes, VMs exhibited higher expression levels of S100A1 (3–5) while the RyR1 and RyR2 isoforms are evolutionarily conserved sharing similar structural architectures and protein–protein interactions (22, 36, 37) including S100A1 (SI Appendix, Fig. S11). The S100A1-antibody specificity was verified by a negative control consisting of VMs isolated from S100A1-knockout (SKO) mice (10). As expected, the double-stained SKO-VMs showed only immunosignals for RyR2 but not for S100A1 (SI Appendix, Fig. S12 A, Right). Similar to WT-VMs (SI Appendix, Fig. S12 A, Left), RyR2 in SKO-VMs was arranged in a distribution pattern typical for terminal cisternae of the SR network (SI Appendix, Fig. S12 A, Right). Hence, the clustering and nano-domain organization of RyR2 was apparently unaffected by SKO. In WT-VMs, the immunosignal for the soluble protein S100A1 was enriched in a cross-striated pattern typical for SR membrane proteins (SI Appendix, Fig. S12 A, Left). Based on a nominal ~50 nm lateral STED resolution and established image analysis workflows (52–54), the immunosignals of S100A1 and RyR2 partially overlapped in WT-VMs indicating colocalization (SI Appendix, Fig. S12 B–D). Similarly, S100A1 and RyR1 analyzed by confocal imaging in skeletal myofibers isolated from mouse extensor digitorum longus (EDL) muscles showed partially overlapping immunosignals (Fig. 6A). The partial signal associations of S100A1 with the SR striation pattern of junctional RyR1 clusters in EDL myofibers resemble those of S100A1 with RyR2 in VMs. Taken together, our coimmunofluorescence imaging results imply partial colocalization between S100A1 and RyR channels that may relate to dynamic localization patterns of the soluble protein S100A1 known to interact with several other proteins besides RyR including SERCA situated on the longitudinal SR (1, 2, 7).
Fig. 6.
Interactions between endogenous S100A1 and RyR channels. (A) Confocal coimmunofluorescence images of S100A1 (red) and RyR2 (green) in skeletal myofibers (green) isolated from mouse EDL muscles. RyR1 exhibited a cross-striated pattern typical for the junctional sarcoplasmic reticulum and partially overlapping with S100A1. Dashed boxes indicate magnification shown below. Scale bar, 10 µm (Top) and 1 µm (Bottom). (B) Ca2+ dependency of coimmunoprecipitations (IP) between endogenous RyR channels and S100A1 versus CaM. RyR complexes were immunoprecipitated from skeletal muscle, heart muscle, and cerebellar lysates at indicated free [Ca2+] followed by immunoblotting against RyR, S100A1, and CaM. (C) Quantification of bound S100A1 and CaM in the IPs of RyR1 from the skeletal muscle. The relative abundance of S100A1/RyR1 (Left) and CaM/RyR1 (Right) is plotted against free [Ca2+] (mean ± SD; n = 8 per group) showing a tendentiously stronger Ca2+ dependence of the endogenous S100A1–RyR1 interaction compared to CaM. Two-tailed Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate interactions between endogenous S100A1 and RyR channels, we immunoprecipitated RyR complexes from skeletal muscle, cardiac muscle, and cerebellar tissue lysates under EGTA conditions or with 10 µM free Ca2+ (Fig. 6B). In the RyR immunoprecipitations (IPs) from the different tissues, addition of 10 µM free Ca2+ increased the amount of RyR-bound S100A1 compared to EGTA conditions. Similar Ca2+-dependent interactions between RyR1 and S100A1 were observed in skeletal muscle and cerebellum, as well as between RyR2 and S100A1 in cardiac muscle tissues (Fig. 6B).
To analyze the Ca2+ dependence of endogenous RyR1–S100A1 complexes in more detail, IPs of RyR1 complexes from skeletal muscle lysates were exposed to Ca2+ at the following concentrations: 1 µM, 10 µM, 50 µM, 1,000 µM free Ca2+ and EGTA-only as negative control. Compared to the EGTA condition, the S100A1 associated with RyR1 increased in presence of 1 µM free Ca2+ and reached a plateau at ~10 µM free Ca2+ (Fig. 6 B and C).
Importantly, these results revealed interactions between S100A1 and RyR1 in the cellular lysates at low micromolar [Ca2+] under physiologic-like conditions. Hence in endogenous RyR1-heterocomplexes, Ca-S100A1 is likely stabilized by interactions of its hydrophobic pocket with the SCL at lower [Ca2+] than expected based on the weak Ca2+-affinity reported for recombinantly purified S100A1 (pEF-hand: KD > 250 μM) (44, 46).
Similar to S100A1, CaM in the IPs of RyR complexes significantly increased in presence of low micromolar Ca2+ reaching a plateau at ~10 µM free Ca2+ (Fig. 6 B and C). Comparison of EGTA conditions versus ~10 µM free Ca2+ showed similar Ca2+-dependent increases of RyR-bound CaM in the skeletal muscle, heart muscle, and cerebellar tissue samples but not as strong as for S100A1 (Fig. 6 B and C). Hence, at least under EGTA conditions, the RyR complexes in the co-IPs were not saturated with CaM and S100A1 while the RyR-binding affinity of both CaM and S100A1 increased at low micromolar Ca2+. These results suggest a dynamic interplay between CaM and S100A1 binding to RyR channels presumably regulated by local [Ca2+], other CaM/S100A1 interaction partners, and posttranslational modifications.
Discussion
We report here the structures of S100A1 bound to RyR1 under Ca2+-free and Ca2+-saturated conditions solved by cryo-EM single particle analysis. Until now, structural data of RyR1 heterocomplexes were only available for Calstabin and CaM (30, 37, 49) albeit several other interaction partners of RyR1 (including S100A1) have been reported (22). Previous approaches to identify and characterize the S100A1 binding site of RyR channels mainly relied on methods involving RyR-truncation and -peptide binding (12, 24). However, limitations of truncation and peptide approaches can arise from nonspecific hydrophobic interactions of artificially exposed surfaces that would be otherwise buried in the RyR1 structure or SR membrane resulting in misleading binding site predictions. Wright et al. reported the solution NMR structure of Ca-S100A1 bound to the 12-a.a. long RyR1-derived peptide named RyRP12 (human a.a. 3616-3627) (23). Importantly, the NMR structure revealed how the helical peptide RyRP12 interacts with the hydrophobic pocket of Ca-S100A1 via favorable hydrophobic and electrostatic interactions stabilizing the Ca2+-bound conformational state of S100A1. The RyRP12 sequence belongs to the SCL domain that is also known to interact with Ca-CaM (12, 36, 49). Considering RyRP12 as pars pro toto for full-length RyR1, it was suggested that Ca-CaM and Ca-S100A1 compete for overlapping binding sites on RyR1 (12, 23). Interestingly, tight interactions between Ca-S100A1 and the SCL of RyR1 are consistent with our cryo-EM data. However, in the full-length RyR1–S100A1 complex, the hydrophobic pocket of Ca-S100A1 is involved in interactions with the central helix of the SCL (Fig. 2B) but not with the N-terminal part including the RyRP12 sequence. Moreover, the central helix of the SCL and peptide RyRP12 occupy the hydrophobic pocket in antiparallel orientations. The central helix shares a similar binding pose with peptide TRTK12, derived from the actin capping protein CapZ, in the TRTK12-S100A1 NMR structure (47). Similarly, the cryo-EM structure of Ca-CaM bound to RyR1 (PDB: 7TZC) indicates that the middle region of the SCL forms the central α-helix implicated in interactions with the N-lobe, flexible linker, and C-lobe of Ca-CaM, while the N-terminal region of the SCL wraps around the C-lobe (Fig. 5B).
Ca-S100A1 binding to RyR1 induces a conformational change in the SCL. Due to the conformational change, the central α-helix of the SCL changes its orientation and rotates toward the CT-BSol region (red helix in Fig. 5 A and B). Moreover, the CT-BSol region moves sideways (by ~1 Å) in a counterclockwise direction relevant to the adjacent RyR1 protomer near the RY1&2, SPRY2, and JSol domains, resulting in slightly tighter interprotomer contacts (Fig. 4 A and B). Hence, S100A1 may regulate RyR1 activity by altering the conformation of CT-BSol. However, we did not observe any conformational change in the pore domain. Interestingly, a similar sideways motion of CT-BSol has been reported for the CPVT-mutant RyR2-R2474S upon binding of the Rycal drug ARM210 that stabilizes the closed conformation of the leaky Ca2+-channel (36). Alleviating effects on dysfunctional RyR1 channels carrying the MH mutation R614C have also been associated with CaM binding since it abolishes a pathological conformation of the MH mutant (30). Hence, S100A1 interactions stabilizing CT-BSol could have similar beneficial effects on leaky RyR channels. This may underpin how restoring S100A1 protein levels by cardiac viral gene transfer in animal heart failure models can reduce pathological SR Ca2+ leak (55).
Since RyR1 activity is directly regulated by CaM, the ability of S100A1 to compete with CaM-binding to RyR1 may contribute to the modulation of RyR1 activity. However, as shown by cryo-EM particle screenings, CaM completely displaced S100A1 from the RyR1-particles in presence and absence of Ca2+, even when a fivefold molar excess of homodimeric S100A1 over CaM was added (Fig. 5). Hence, at least in vitro, the RyR1 affinity for CaM is substantially stronger than for S100A1 under both Ca2+-free and Ca2+-bound conditions.
Similarly, using in vitro FRET-based assays, Rebbeck et al. reported a lack of steric competition between S100A1 and CaM for RyR binding since S100A1 did not efficiently interfere with the FRET sensor pair consisting of donor-labeled Calstabin and acceptor-labeled CaM bound to RyR1 or RyR2 channels of SR vesicles (48). In time-resolved FRET measurements, RyR-bound CaM exhibited two detectable states that were perturbed in their distribution after adding high micromolar [S100A1]. The authors suggested that S100A1 would bind to RyR at a separate binding site allosterically altering the structure of the RyR–CaM complex (48). However, in our cryo-EM structures, we could not detect a second S100A1 binding site on RyR1. Interestingly, time-resolved FRET data at 30 µM free Ca2+ indicated that S100A1 only altered the conformation of the N- but not the C-lobe of CaM bound to RyR1, implying that S100A1 may interfere with the CaM-RyR1 interaction at the N-lobe while residual C-lobe interactions stabilize CaM binding to RyR1 (48).
At 30 µM free Ca2+, Ca-CaM and apo-S100A1 may coexist since the conformational change to Ca-S100A1 requires higher micromolar [Ca2+] in vitro (pEF-hand: KD > 250 μM) (44, 46). The structural comparison of the S100A1 versus CaM binding sites of RyR1 indicated only a small steric overlap between apo-S100A1 and Ca-CaM (SI Appendix, Fig. S10E). Thus, simultaneous binding of both small proteins by an induced fit mechanism seems plausible. However, using cryo-EM, we did not observe any indication of simultaneous CaM and S100A1 binding events on RyR1 protomers at 5 mM EGTA only, 30 µM free Ca2+, or 250 µM free Ca2+ (Fig. 5 D–F).
Intracellularly, the local availability of free CaM is regulated by CaM sequestering proteins (56) that may indirectly facilitate S100A1 binding to RyR1. It is well documented that proteins of the calpacitin family (e.g., neurogranin/RC3 or neuromodulin/GAP43) can bind to CaM with high affinity and mask available CaM, i.e., sequestering CaM away from its regulatory downstream targets such as RyR1 (56). Similar to CaM, S100A1 is highly abundant in cardiomyocytes and slow-twitch skeletal myocytes (3–5, 57) and can interact with numerous proteins (1, 2). The total concentration in adult VMs has been estimated to be ~10 μM for S100A1 (10) and ~5 µM for CaM (57) but the free concentrations available for RyR interactions are unknown. At the subcellular level, S100A1 is locally enriched at the SR where S100A1 has been reported to interact with RyR and SERCA2a–phospholamban complexes regulating SR Ca2+ release and uptake, respectively (7, 9, 27). The intracellular S100A1 distribution overlaps with RyR clusters as shown by coimmunofluorescence imaging of EDL myofibers (Fig. 6A) and VMs (SI Appendix, Fig. S12). This suggests that S100A1 exhibits high local concentrations at the SR near RyR clusters facilitating S100A1–RyR interactions. Besides local availability and steric accessibility, the RyR–S100A1 binding affinity could be directly affected by posttranslational modifications, lipid–protein interactions, or protein–protein interactions in vivo.
Importantly, the monomeric subunit 1 of homodimeric S100A1 mediates most of the interactions with RyR1 (Fig. 1 E and F), while subunit 2 of S100A1 is largely exposed to the surface (Fig. 3), suggesting that subunit 2 could be substituted by another S100-isoform without affecting the binding to RyR1. In the Ca2+-saturated state, the second hydrophobic pocket of S100A1 faces outward to the aqueous milieu and would be prone to interactions with the cytoplasmic environment (Fig. 3). The importance of the hydrophobic pocket as a major interaction site of S100A1 is pointed out by the fact that co-IPs of endogenous S100A1 and RyR1 in skeletal lysates require micromolar [Ca2+] (Fig. 6C). Hence, Ca-S100A1 could be involved in formation of larger RyR-heterocomplexes that, in turn, could stabilize interactions between RyR1 and Ca-S100A1 leading to a higher Ca2+ affinity of S100A1 in vivo.
Taken together, the structural data reveal how S100A1 interacts with RyR1 in presence and absence of Ca2+. Future studies are warranted to elucidate in detail the structural and functional role of homo- and, potentially, heterodimeric S100A1 in endogenous RyR-heterocomplexes under physiological and pathological conditions. Given the previously implicated regulatory role of S100A1 in Ca2+ release by RyR1 (11, 12) and RyR2 channels (32–34, 55), the S100A1 binding site conserved between the RyR isoforms (SI Appendix, Fig. S11) may provide the structural basis for the development of therapeutic strategies regarding treatments of RyR-related disorders.
Methods
Antibodies.
Primary antibodies: anti-S100A1 (rabbit polyclonal; Abcam, ab11428), anti-CaM (recombinant monoclonal; Abcam, ab45689), anti-RyR [mouse monoclonal (C3-33); Thermo Fisher Scientific, MA3-916], anti-RyR [mouse monoclonal (34C); Thermo Fisher Scientific, MA3-925], anti-RyR (rabbit polyclonal; homemade antibody #5029). Secondary antibodies used for immunoblotting: anti-mouse IRDye 680RD (donkey polyclonal; LI-COR, #926-68072), anti-rabbit IRDye 800CW (donkey polyclonal; LI-COR, #926-32213). Secondary antibodies used for immunofluorescence: anti-mouse-STAR635P (goat polyclonal; Abberior, 200020075), anti-rabbit-STAR580 (goat polyclonal; Abberior, 200120058).
Purification of Recombinant S100A1.
Mouse S100A1 cDNA (NP_035439.1) was amplified by PCR and cloned into the pGEX-6P-1 vector (GE Healthcare). The inserted S100A1 cDNA sequence was confirmed by DNA sequencing. Recombinant S100A1 was expressed as N-terminally GST-tagged fusion protein in E. coli BL21 (DE3) induced by addition of 1 mM IPTG at OD 0.6 followed by incubation at 30 °C for 4 h. After centrifugation for 10 min at 6,500 g, the cell pellets were resuspended in buffer A [phosphate-buffered saline (PBS) pH 7.4, 0.5 mM AEBSF, 1 mM 1,4-dithiothreitol (DTT)] and lysed using an emulsiflex (Avestin EmulsiFlex-C3). To remove cell debris, the lysate was centrifugated for 10 min at 100,000 g. The GST-tagged S100A1 protein in the supernatant was affinity purified on Glutathione Sepharose 4B prepacked columns (GE Healthcare). After washing with 5 column volumes (CV) of buffer A, the GST affinity tag was removed using on-column cleavage with PreScission Protease (GE Healthcare) resulting in the elution of untagged S100A1. S100A1 fractions were pooled, concentrated, and dialyzed overnight at 4 °C into storage buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT).
Purification of Recombinant CaM and Tobacco Etch Virus (TEV) Protease.
Recombinant CaM and TEV protease were purified by affinity chromatography as previously described (36, 49). In brief, human CaM was recombinantly expressed in E. coli BL21 (DE3) with an N-terminal 6-histidine affinity tag and a TEV protease cleavage site. Protein expression was induced by addition of 0.2 mM IPTG at OD 0.6 followed by overnight incubation at room temperature. After centrifugation (10 min 6,500 g), the cell pellets were resuspended in buffer A (PBS pH 7.4, 20 mM imidazole, 0.5 mM AEBSF, 5 mM 2-mercaptoethanol) and lysed using an emulsiflex (Avestin EmulsiFlex-C3). To remove cell debris, the lysate was centrifugated for 10 min at 100,000 g. The His-tagged CaM protein in the supernatant was purified via immobilized metal affinity chromatography (IMAC) using HisTrap columns (GE Healthcare). After washing with 5 column volumes of buffer A, the protein was eluted using a linear gradient from buffer A to buffer B (buffer A containing 500 mM imidazole). CaM fractions were pooled. To cleave off the 6-histidine affinity tag, TEV protease (~1 mg) was added to CaM and the mixture was dialyzed overnight at 4 °C into storage buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT). In a second IMAC after TEV proteolysis, the dialysate was loaded onto a HisTrap column. The flow-through fractions containing CaM were collected and concentrated using 10,000 kDa cutoff centrifugation filters (MilliporeSigma).
TEV protease was purified using IMAC in the same manner as CaM except for an uncleavable his-tag. After a one-step IMAC purification, elution fractions containing the TEV protease were pooled, concentrated, and dialyzed overnight at 4 °C into storage buffer including 10% glycerol.
Purification of Endogenous RyR1 from Mouse Skeletal Muscle.
RyR1 was purified from mouse skeletal muscle using a modified protocol previously reported for RyR1 purification from rabbit skeletal muscle (35, 49). All purification steps were performed in ice-cold buffers unless otherwise stated. Back and thigh muscle tissue was dissected from C57BL6 mice (8 to 20 wk old). ~30 g of skeletal muscle tissue was resuspended in ~200 mL buffer A (10 mM tris maleate pH 6.8, 1 mM EGTA, 1 mM benzamidine hydrochloride, 0.5 mM AEBSF) and lysed in a Waring blender. The suspension was centrifugated for 10 min at 11,000 g and the supernatant was filtered through cheesecloth to remove debris. After centrifugation for 30 min at 36,000 g, the pelleted membrane fraction was solubilized in buffer B [10 mM HEPES pH 7.4, 0.8 M NaCl, 1% CHAPS, 0.1% phosphatidylcholine, 1 mM EGTA, 2 mM DTT, 0.5 mM AEBSF, 1 mM benzamidine hydrochloride, one protease inhibitor tablet (Pierce)] followed by homogenization using a glass tissue grinder (Kontes). The solute was diluted with buffer C (buffer B without NaCl) at a 1:1 ratio and the homogenization was repeated. After centrifugation for 30 min at 100,000 g to remove CHAPS-insoluble material, the supernatant was filtered and then loaded onto a HiTrap Q HP column (5 mL, GE Healthcare) previously equilibrated with buffer D [10 mM HEPES pH 7.4, 400 mM NaCl, 0.25% CHAPS, 1 mM EGTA, 0.5 mM TCEP, 0.01% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti)]. After washing with six column volumes of buffer D, the immobilized protein on the HiTrap Q HP column was eluted using a linear gradient from 480 to 550 mM NaCl with buffers D and E (buffer D with 600 mM NaCl). RyR1-containing fractions were pooled and concentrated to 0.4 mL using 100,000 kDa cutoff centrifugation filters (MilliporeSigma) followed by size-exclusion chromatography using a TSKgel G4SWXL column (TOSOH Biosciences) with buffer F (buffer D with 150 mM NaCl). RyR1 fractions were pooled and concentrated to >10 g/L using 100,000 kDa cutoff centrifugation filters (MilliporeSigma).
Cryo-EM samples were prepared by adding a 10-fold molar excess of homodimeric S100A1 (final: ~150 µM S100A1-dimer) per RyR1-protomer (final: ~15 µM RyR1-protomer) in presence of either 5 mM EGTA or 10 mM NaATP, 5 mM caffeine, and 30 μM free Ca2+. Free Ca2+ concentrations were calculated using MaxChelator (https://somapp.ucdmc.ucdavis.edu/pharmacology/bers/maxchelator/webmaxc/webmaxcE.htm).
Cryo-EM Sample Preparation and Data Collection.
Cryo-EM grids were prepared using a Vitrobot Mark IV (Thermo Fisher Scientific) as described previously (36, 37). In brief, 3 µL samples of purified proteins were transferred onto UltrAuFoil holey gold grids (Quantifoil R 0.6/1.0, Au 300) cleaned with EasiGlow (PELCO). Grids were blotted for 8 s at force 10 with ashless filter paper (Whatman) at 4 °C with 100% relative humidity and vitrified by plunge-freezing into liquid ethane chilled by liquid nitrogen.
Cryo-EM grids were screened in-house on a Glacios Cryo-TEM (Thermo Fisher Scientific) microscope with a 200-kV x-FEG source and a Falcon 3EC direct electron detector (Thermo Fisher Scientific) using EPU software (Thermo Fisher Scientific) to operate the microscope and collect data. High-resolution cryo-EM data were obtained at Columbia University on a Titan Krios 300-kV (Thermo Fisher Scientific) microscope equipped with an energy filter (slit width 20 eV) and a K3 direct electron detector (Gatan). Cryo-EM data were collected using Leginon software for automated data acquisition (58). The nominal magnification was 105,000× in electron counting mode, corresponding to a pixel size of 0.83 Å. The electron dose rate was set to 16 e−/pixel/sec with 2.5 s exposures for a total dose of 50 to 60 e/Å2.
Cryo-EM data were processed in CryoSPARC (59). Image stacks were aligned using Patch motion and defocus values estimated by Patch CTF estimation. RyR1 particles were automatically picked using the template picker with preexisting templates. Following 2D classification with 150 classes, RyR1 particles from the highest-resolution classes were pooled. An initial molecular volume of the RyR1 particles was generated by ab initio 3D reconstruction and subjected to homogeneous refinement with C4 symmetry imposed followed by heterogeneous refinement with two classes to further select the best particles. The particle stack was quadrupled by C4 symmetry expansion. Masks generated for subsequent local refinements were composed as follows: The first mask comprised the N-terminal domain, the three SPRY domains, the tandem repeat domain RY1&2, and Calstabin; the second mask the CSol and JSol domains with S100A1; the third mask the BSol domain with S100A1; the fourth mask the pore region including the thumb-and-forefinger (TaF), transmembrane (TM), and C-terminal (CTD) domains of RyR1. C4 symmetry was only imposed on the mask of the pore region. Smaller masks focused on the more variable density signal of only S100A1 or the tandem repeat domains RY1&2-only and RY3&4-only. To enrich the S100A1 density, particles with S100A1 were selected by three rounds of 3D classification with the small mask focused on S100A1. After completing homogeneous and local refinements, composite maps were independently generated for the selected RyR1-S100A1 particles or all RyR1 particles under EGTA-only condition or with Ca2+, caffeine, and ATP (SI Appendix, Figs. S1 and S2). To assemble composite maps, the focused maps from local refinements were aligned to the global consensus map from the corresponding homogeneous refinement and combined in UCSF ChimeraX (60). The pixel size was calibrated using correlation coefficients with a map generated from the crystal structure of the N-terminal domain (61). Atomic models were manually built in Coot (62) starting with a cryo-EM structural model of RyR1 (PDB: 7TZC), while models of S100A1 in the apo-conformation and Ca2+-bound conformation were obtained from NMR structures (PDB: 2L0P and PDB: 2K2F, respectively). Atomic models were refined using real-space refinement in Phenix (63). Figures of RyR1-S100A1 structures were prepared with UCSF ChimeraX (60).
VM Isolation from Adult Mouse Hearts.
VMs were isolated from 8- to 20-wk-old C57BL6/N mouse hearts as previously described (52). Mouse hearts were excised and cannulated at the aorta. The aortic cannula was mounted on a modified Langendorff apparatus to retrogradely perfuse the heart with a modified Ca2+-depleted Tyrode buffer (10 mM HEPES, 0.6 mM Na2HPO4, 0.6 mM KH2PO4, 4.6 mM NaHCO3, 120.4 mM NaCl, 14.7 mM KCl, 1.2 mM MgSO4, 30 mM Taurin, 10 mM 2,3-butanedione monoxime, 5.5 mM Glucose, pH 7.4) at 37 °C. 2 mg/mL collagenase type II (Sigma) and 40 µM CaCl2 were added to the Tyrode perfusion buffer to induce enzymatic digestion. After 9 min at 37 °C, the ventricles were dissected and the isolated VMs were transferred into Tyrode buffer containing 10% FBS (Sigma) to terminate the enzymatic digestion. Isolated VMs were fixed, permeabilized, and stained with RyR1 and S100A1 antibodies followed by immunofluorescence confocal microscopy and STED nanoscopy.
Myofiber Isolation from Adult Mouse EDL Muscles.
Myofibers were isolated from EDL muscles of 8- to 12-wk-old C57BL6/J mice as previously described (64). In brief, EDL muscles were dissected from mouse hindlimbs followed by digestion with 2 mg/mL collagenase type I (Sigma) in DMEM-F12 (Thermo Fisher Scientific) at 37 °C for ~1 h. Single myofibers were liberated by gentle trituration in DMEM with 2% horse serum (Sigma) to terminate the enzymatic digestion. Isolated EDL myofibers were fixed, permeabilized, stained with RyR1 and S100A1 antibodies, and analyzed by immunofluorescence confocal microscopy.
Immunofluorescence Confocal Microscopy and Superresolution STED Nanoscopy.
For immunofluorescence imaging, EDL myofibers or VMs were plated on glass coverslips covered with Matrigel (BD Biosciences) or laminin (BD Biosciences), respectively. EDL myofibers or VMs were fixed with 4% PFA (Merck Millipore) in PBS at room temperature for 10 min, permeabilized, and blocked with 0.2% Triton-X100 (Carl Roth) and 10% FBS (Merck Millipore) in PBS (Gibco) at 4 °C overnight. Primary antibodies diluted in blocking solution (0.2% Triton-X100 and 10% FBS in PBS) were added to EDL myofibers or VMs followed by incubation overnight at 4 °C. EDL myofibers or VMs were washed thrice with blocking solution and then incubated with secondary antibodies anti-mouse-STAR635P (Abberior Instruments) or anti-rabbit-STAR580 (Abberior Instruments) diluted 1:200 in blocking solution at room temperature for 2 h. After washing, the fluorescently labeled coverslips were mounted onto microscope slides using Prolong Gold Antifade Mountant (Thermo Fisher Scientific), dried in air protected from light, and sealed with nail polish. Images were collected by confocal microscopy (for EDL) using a Leica STELLARIS 5 microscope system or confocal microscopy/STED nanoscopy (for VMs) using a Leica TCS SP8 system microscope system equipped with a HC PL APO C2S 100x/1.40 oil objective. The STED workflow was optimized for the STAR635P and STAR580 fluorophores using the following parameters: pixel size 16.23 x 16.23 nm, pixel dwell time 400 ns, scanning speed 600 Hz, 32x line averaging, excitation by a white light laser at 635 and 580 nm, STED at 775 nm, and fluorescence detection between 650 to 700 nm and 600 to 630 nm, respectively. The 775 synchronized depletion laser was aligned at the start of each imaging session. In STED mode, a time gating of 0.5 to 6 μs was used. The pinhole size was set to one airy unit in all recordings. Adjustment of the STED laser power to maximize resolution and raw image processing in Fiji (https://imagej.net/Fiji) were carried out according to previously established workflows (52–54).
Single Channel Data Acquisition and Analysis.
Single channel recordings of purified RyR1 reconstituted in planar lipid bilayers were conducted as described previously (36, 49, 65). In brief, a 3:1 mixture of phosphatidylethanolamine and phosphatidylcholine (Avanti Polar Lipids) was dissolved in decane (30 mg/mL). The lipid mixture was applied across a 200-µm aperture in a polysulfonate cup (Warner Instruments). The planar lipid bilayer formed in the aperture partitioned two 1 mL chambers referred to as cis and trans representing cytoplasmic and SR/ER luminal compartments, respectively. The cis chamber was held at virtual ground, while the trans chamber was connected to the headstage input of a bilayer voltage clamp amplifier (BC-525D, Warner Instruments). The buffers in the two chambers were composed as follows: 250/125 mM HEPES/Tris, 50 mM KCl, 1 mM EGTA, 0.64 mM CaCl2, pH 7.35 on cis and 250 mM HEPES, 50 mM KCL, 53 mM Ca(OH)2, pH 7.35 on trans. Free Ca2+ concentrations in the cis chamber were calculated using MaxChelator. Purified RyR1 channels were added to the cis side of the planar lipid bilayer. To facilitate channel incorporation into the bilayer lipid membrane, the cis side was made hyperosmotic with KCl added up to 400 to 500 mM. After channel incorporation, the cis chamber was perfused with cis buffer. Single channel currents of RyR1 were recorded at room temperature before and after adding recombinant S100A1 to the cis chamber. The voltage across the planar lipid bilayer was set to 0 mV by using a Bilayer Clamp BC-525D amplifier (Warner Instruments). Single channel currents were filtered at 1 kHz and digitized at 4 kHz. Software used for data acquisition: Digidata 1440A and Axoscope 10.2. Software used for data analysis: Clampfit 10.2 (Molecular Devices), GraphPad Prism, and Microsoft Excel 2010.
IP of Endogenous RyR Heterocomplexes.
To immunoprecipitate endogenous RyR heterocomplexes from different tissue types, lysates of skeletal muscle, cardiac muscle, or cerebellar tissues (100 µg) were incubated with a RyR-specific antibody (4 μg of Ab #5029) in a modified RIPA buffer (0.5 mL; 50 mM Tris-HCl pH 7.4, 0.9% NaCl, 5.0 mM NaF, 1 mM DTT, 1.0 mM Na3VO4, 0.5% Triton X-100, and protease inhibitors) for 1 h at 4 °C. To assess the Ca2+ dependence of RyR heterocomplexes, the free Ca2+ concentration in the RIPA buffer was adjusted to 1 µM, 10 µM, 50 µM, or 1,000 µM in presence of 1 mM EGTA, while EGTA-only served as Ca2+-depleted control condition. Free Ca2+ concentrations were calculated using MaxChelator. The immune complexes were captured on protein A sepharose beads (Sigma) at 4 °C for 1 h followed by washing three times with buffer. Proteins were eluted with 2x Laemmli SDS-sample buffer and subjected to SDS-PAGE followed by immunoblotting.
Immunoblot Analysis.
Protein samples were denatured in 1x Laemmli SDS-sample buffer at 70 °C for 5 min. Equal amounts of protein were loaded and separated on 4 to 20% gradient SDS-PAGE gels (Thermo Fisher Scientific) by electrophoresis. Proteins were transferred onto nitrocellulose membranes (0.45 µm, Protran Premium NC, Cytivia). Membranes were immunoblotted with primary antibodies overnight at 4 °C using S100A1, CaM, or RyR antibodies diluted 1:1,000. Primary antibodies were probed with secondary antibodies overnight at 4 °C using anti-mouse IRDye 680RD (donkey polyclonal; LI-COR, #926-68072) or anti-rabbit IRDye 800CW (donkey polyclonal; LI-COR, #926-32213) diluted 1:10,000. Membranes were scanned by an Odyssey CLx infrared imaging station (LI-COR). Protein bands of interest were quantified using Image Studio software (LI-COR).
Statistical Data Analysis.
Statistical significance was determined by the unpaired two-tailed t test or one-way-ANOVA using mathematical analysis software GraphPad Prism 8.0, CRAN R version 4.0.2, and Microsoft Excel 2010. A P-value < 0.05 was considered statistically significant.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Cryo-EM data were collected at the Columbia University Cryo-Electron Microscopy Center. We thank R. Grassucci and Z. Zhang from the cryo-EM facility for their essential services. We thank P. Most and J. Ritterhoff for providing S100A1-related materials. We would also like to acknowledge the molecular graphics and analyses performed using UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the NIH R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. These studies were supported by NIH grants R01HL145473, R01DK118240, R01HL142903, R01HL140934, R01NS114570, R01AR070194, R25HL156002R25, R25NS076445, P01 HL164319, and T32 HL120826 (to A.R.M.). Deutsche Forschungsgemeinschaft supported these studies under Germany’s Excellence Strategy (EXC2067/1-390729940) (to S.E.L.).
Author contributions
G.W. and A.R.M. designed research; G.W., M.C.M., C.T., H.D., S.B., G.C.R., Q.Y., and Y.L. performed research; G.W., M.C.M., S.R., S.B., Q.Y., A.C., A.W., S.E.L., and A.R.M. analyzed data; G.W., M.C.M., S.B., S.E.L., and A.R.M. edited and revised manuscript; and G.W. wrote the paper.
Competing interests
Columbia University and A.R.M. own stock inARMGO, a company developing compounds targeting RyR and have patents on Rycals.The remaining authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission. S.R. is a guest editor invited by the Editorial Board.
Contributor Information
Gunnar Weninger, Email: gw2424@cumc.columbia.edu.
Andrew R. Marks, Email: arm42@cumc.columbia.edu.
Data, Materials, and Software Availability
Atomic coordinates have been deposited in the Protein Data Bank (PDB) and Cryo-EM density maps in the Electron Microscopy Data Bank (EMDB). The PDB accession codes are 8VK3 (66), 8VK4 (67), 8VJJ (68), and 8VJK (69). The corresponding EMDB accession codes are EMD-43299 (66), EMD-43304 (67), EMD-43283 (68), and EMD-43284 (69).
Supporting Information
References
- 1.Schafer B. W., Heizmann C. W., The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem. Sci. 21, 134–140 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez L. L., Garrie K., Turner M. D., Role of S100 proteins in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118677 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Engelkamp D., Schafer B. W., Erne P., Heizmann C. W., S100 alpha, CAPL, and CACY: Molecular cloning and expression analysis of three calcium-binding proteins from human heart. Biochemistry 31, 10258–10264 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Sleczka B. G., Levesque P. C., Adam L. P., Olah T. V., Shipkova P., LC/MS/MS-based quantitation of pig and human S100A1 protein in cardiac tissues: Application to gene therapy. Anal. Biochem. 602, 113766 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Kato K., Kimura S., S100ao (alpha alpha) protein is mainly located in the heart and striated muscles. Biochim. Biophys. Acta 842, 146–150 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Maco B., et al. , Ultrastructural distribution of the S100A1 Ca2+-binding protein in the human heart. Physiol. Res. 50, 567–574 (2001). [PubMed] [Google Scholar]
- 7.Volkers M., Rohde D., Goodman C., Most P., S100A1: A regulator of striated muscle sarcoplasmic reticulum Ca2+ handling, sarcomeric, and mitochondrial function. J. Biomed. Biotechnol. 2010, 178614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Most P., et al. , Transgenic overexpression of the Ca2+-binding protein S100A1 in the heart leads to increased in vivo myocardial contractile performance. J. Biol. Chem. 278, 33809–33817 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Remppis A., et al. , The small EF-hand Ca2+ binding protein S100A1 increases contractility and Ca2+ cycling in rat cardiac myocytes. Basic Res. Cardiol. 97 (suppl. 1), I56–I62 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Du X. J., et al. , Impaired cardiac contractility response to hemodynamic stress in S100A1-deficient mice. Mol. Cell Biol. 22, 2821–2829 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prosser B. L., et al. , S100A1 promotes action potential-initiated calcium release flux and force production in skeletal muscle. Am. J. Physiol. Cell Physiol. 299, C891–C902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prosser B. L., et al. , S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. J. Biol. Chem. 283, 5046–5057 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remppis A., et al. , Altered expression of the Ca(2+)-binding protein S100A1 in human cardiomyopathy. Biochim. Biophys. Acta 1313, 253–257 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Bennett M. K., et al. , S100A1 in human heart failure: Lack of recovery following left ventricular assist device support. Circ. Heart Fail 7, 612–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleger S. T., et al. , Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci. Transl. Med. 3, 92ra64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohde D., et al. , S100A1 gene therapy for heart failure: A novel strategy on the verge of clinical trials. J. Mol. Cell Cardiol. 50, 777–784 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Wehrens X. H., Lehnart S. E., Marks A. R., Intracellular calcium release and cardiac disease. Annu. Rev. Physiol. 67, 69–98 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kushnir A., Wajsberg B., Marks A. R., Ryanodine receptor dysfunction in human disorders. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1687–1697 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Betzenhauser M. J., Marks A. R., Ryanodine receptor channelopathies. Pflugers Arch. 460, 467–480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks A. R., Targeting ryanodine receptors to treat human diseases. J. Clin. Invest. 133, e162891 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bousova K., et al. , Shared CaM- and S100A1-binding epitopes in the distal TRPM4 N terminus. FEBS J. 285, 599–613 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Zalk R., Lehnart S. E., Marks A. R., Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 76, 367–385 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Wright N. T., et al. , S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 283, 26676–26683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treves S., et al. , Interaction of S100A1 with the Ca2+ release channel (ryanodine receptor) of skeletal muscle. Biochemistry 36, 11496–11503 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Marx S. O., Marks A. R., Dysfunctional ryanodine receptors in the heart: New insights into complex cardiovascular diseases. J. Mol. Cell Cardiol. 58, 225–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melville Z., et al. , The activation of protein kinase A by the calcium-binding protein S100A1 is independent of cyclic AMP. Biochemistry 56, 2328–2337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiewitz R., et al. , Ca2+ -dependent interaction of S100A1 with the sarcoplasmic reticulum Ca2+ -ATPase2a and phospholamban in the human heart. Biochem. Biophys. Res. Commun. 306, 550–557 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Balshaw D. M., Xu L., Yamaguchi N., Pasek D. A., Meissner G., Calmodulin binding and inhibition of cardiac muscle calcium release channel (ryanodine receptor). J. Biol. Chem. 276, 20144–20153 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Tripathy A., Xu L., Mann G., Meissner G., Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69, 106–119 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woll K. A., Haji-Ghassemi O., Van Petegem F., Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM. Nat. Commun. 12, 807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fano G., et al. , S-100a0 protein stimulates Ca2+-induced Ca2+ release from isolated sarcoplasmic reticulum vesicles. FEBS Lett. 255, 381–384 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi N., et al. , Cardiac hypertrophy associated with impaired regulation of cardiac ryanodine receptor by calmodulin and S100A1. Am. J. Physiol. Heart Circ. Physiol. 305, H86–H94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volkers M., et al. , S100A1 decreases calcium spark frequency and alters their spatial characteristics in permeabilized adult ventricular cardiomyocytes. Cell Calcium 41, 135–143 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Kettlewell S., Most P., Currie S., Koch W. J., Smith G. L., S100A1 increases the gain of excitation-contraction coupling in isolated rabbit ventricular cardiomyocytes. J. Mol. Cell Cardiol. 39, 900–910 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Zalk R., et al. , Structure of a mammalian ryanodine receptor. Nature 517, 44–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miotto M. C., et al. , Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment. Sci. Adv. 8, eabo1272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melville Z., Kim K., Clarke O. B., Marks A. R., High-resolution structure of the membrane-embedded skeletal muscle ryanodine receptor. Structure 30, 172–180.e3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Efremov R. G., Leitner A., Aebersold R., Raunser S., Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39–43 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Yan Z., et al. , Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Georges A., et al. , Structural basis for gating and activation of RyR1. Cell 167, 145–157.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santulli G., Lewis D., de Georges A., Marks A. R., Frank J., Ryanodine receptor structure and function in health and disease. Subcell Biochem. 87, 329–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi X., et al. , Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators. Proc. Natl. Acad. Sci. U.S.A. 116, 25575–25582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rustandi R. R., et al. , Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR. Biochemistry 41, 788–796 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Wright N. T., et al. , The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J. Mol. Biol. 353, 410–426 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Melville Z., et al. , X-ray crystal structure of human calcium-bound S100A1. Acta Crystallogr. F Struct. Biol. Commun. 73, 215–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goch G., Vdovenko S., Kozlowska H., Bierzynski A., Affinity of S100A1 protein for calcium increases dramatically upon glutathionylation. FEBS J. 272, 2557–2565 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Wright N. T., et al. , Solution structure of S100A1 bound to the CapZ peptide (TRTK12). J. Mol. Biol. 386, 1265–1277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebbeck R. T., et al. , S100A1 protein does not compete with calmodulin for ryanodine receptor binding but structurally alters the ryanodine receptor. Calmodulin Complex. J. Biol. Chem. 291, 15896–15907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melville Z., et al. , A drug and ATP binding site in type 1 ryanodine receptor. Structure 30, 1025–1034.e24 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Ferguson D. G., Schwartz H. W., Franzini-Armstrong C., Subunit structure of junctional feet in triads of skeletal muscle: A freeze-drying, rotary-shadowing study. J. Cell Biol. 99, 1735–1742 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak A. R., Samso M., Ca(2+) inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM. Elife 11, e75568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weninger G., et al. , Calpain cleavage of Junctophilin-2 generates a spectrum of calcium-dependent cleavage products and DNA-rich NT(1)-fragment domains in cardiomyocytes. Sci. Rep. 12, 10387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hebisch E., Wagner E., Westphal V., Sieber J. J., Lehnart S. E., A protocol for registration and correction of multicolour STED superresolution images. J. Microsc. 267, 160–175 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Brandenburg S., et al. , Direct proteomic and high-resolution microscopy biopsy analysis identifies distinct ventricular fates in severe aortic stenosis. J. Mol. Cell Cardiol. 173, 1–15 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Most P., et al. , Cardiac adenoviral S100A1 gene delivery rescues failing myocardium. J. Clin. Invest. 114, 1550–1563 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moradi F., et al. , Calmodulin-binding proteins in muscle: A minireview on nuclear receptor interacting protein, neurogranin, and growth-associated protein 43. Int. J. Mol. Sci. 21, 1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maier L. S., et al. , Dynamic changes in free Ca-calmodulin levels in adult cardiac myocytes. J. Mol. Cell Cardiol. 41, 451–458 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Suloway C., et al. , Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tung C. C., Lobo P. A., Kimlicka L., Van Petegem F., The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature 468, 585–588 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersson D. C., et al. , Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 14, 196–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dridi H., et al. , Heart failure-induced cognitive dysfunction is mediated by intracellular Ca(2+) leak through ryanodine receptor type 2. Nat. Neurosci. 26, 1365–1378 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weninger G., Marks A. R., Structure of mouse RyR1 in complex with S100A1 (EGTA-only dataset). Protein Data Bank (PDB). https://www.rcsb.org/structure/8VK3. Deposited 8 January 2024.
- 67.Weninger G., Marks A. R., Structure of mouse RyR1 in complex with S100A1 (high-Ca2+/CFF/ATP dataset). Protein Data Bank (PDB). https://www.rcsb.org/structure/8VK4. Deposited 8 January 2024.
- 68.Weninger G., Marks A. R., Structure of mouse RyR1 (EGTA-only dataset). Protein Data Bank (PDB). https://www.rcsb.org/structure/8VJJ. Deposited 7 January 2024.
- 69.Weninger G., Marks A. R., Structure of mouse RyR1 (high-Ca2+/CFF/ATP dataset). Protein Data Bank (PDB). https://www.rcsb.org/structure/8VJK. Deposited 7 January 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Atomic coordinates have been deposited in the Protein Data Bank (PDB) and Cryo-EM density maps in the Electron Microscopy Data Bank (EMDB). The PDB accession codes are 8VK3 (66), 8VK4 (67), 8VJJ (68), and 8VJK (69). The corresponding EMDB accession codes are EMD-43299 (66), EMD-43304 (67), EMD-43283 (68), and EMD-43284 (69).