Significance
Controlling the primary African malaria vector, Anopheles gambiae mosquitoes, is crucial for reducing malaria transmission. Conventional methods relying on insecticides are losing effectiveness. The sterile insect technique (SIT) has successfully eradicated pests, but implementing it for A. gambiae is hindered by technological gaps. Our precision-guided SIT (pgSIT) uses CRISPR to induce male sterilization and female elimination for use in SIT. Through engineered Cas9 and gRNA strains, we achieve over 99.5% male sterility and over 99.9% female lethality. Genetically sterilized males display longevity, induce population suppression, and, according to models, can eliminate wild A. gambiae populations. This finding enhances the malaria genetic biocontrol toolkit, allowing scalable, confined suppression in the species.
Keywords: pgSIT, malaria, suppression
Abstract
Controlling the principal African malaria vector, the mosquito Anopheles gambiae, is considered essential to curtail malaria transmission. However, existing vector control technologies rely on insecticides, which are becoming increasingly ineffective. Sterile insect technique (SIT) is a powerful suppression approach that has successfully eradicated a number of insect pests, yet the A. gambiae toolkit lacks the requisite technologies for its implementation. SIT relies on iterative mass releases of nonbiting, nondriving, sterile males which seek out and mate with monandrous wild females. Once mated, females are permanently sterilized due to mating-induced refractoriness, which results in population suppression of the subsequent generation. However, sterilization by traditional methods renders males unfit, making the creation of precise genetic sterilization methods imperative. Here, we introduce a vector control technology termed precision-guided sterile insect technique (pgSIT), in A. gambiae for inducible, programmed male sterilization and female elimination for wide-scale use in SIT campaigns. Using a binary CRISPR strategy, we cross separate engineered Cas9 and gRNA strains to disrupt male-fertility and female-essential genes, yielding >99.5% male sterility and >99.9% female lethality in hybrid progeny. We demonstrate that these genetically sterilized males have good longevity, are able to induce sustained population suppression in cage trials, and are predicted to eliminate wild A. gambiae populations using mathematical models, making them ideal candidates for release. This work provides a valuable addition to the malaria genetic biocontrol toolkit, enabling scalable SIT-like confinable, species-specific, and safe suppression in the species.
Malaria is a deadly parasitic disease that kills a child every minute (1). While widespread vaccine distribution recently began to avert the worst disease outcomes (2, 3), eradication remains elusive. Controlling the principal African malaria mosquito vector, Anopheles gambiae, promises to facilitate control and perhaps even elimination of disease transmission in the most highly infected areas. However, currently implemented control methods including insecticide-based technologies, and environmental controls, are increasingly less effective with the list of resistant populations growing yearly (4). Therefore, novel noninsecticidal control measures are needed to curb the spread of disease.
To fill this critical niche, genetic vector control technologies are being developed in Anopheles gambiae. In this species, the technology farthest down the developmental pipeline is gene drives—selfish genetic elements capable of unilaterally engineering entire wild populations (5–7). However, they have a propensity for breakage via generation of resistant alleles (8, 9), though this is not guaranteed (5). This, coupled with their self-autonomous spread (10), has unsurprisingly spurred scientific, social, ethical, economic, ecological, practical, and political concerns hindering their potential roll-out (11–17). To expedite approval and save lives and to provide more durable, and controllable, immediate options, it is imperative we develop alternative vector control measures that have safe track records. The sterile insect technique (SIT) is one such potential technology, as it has been used to eliminate the tsetse fly, screwworm, melon fly, medfly, and Aedes pest populations to great effect (18–23). Requiring male sterilization and benefitting from a female-removal component (24), SIT acts through the mass releases of infertile males which naturally locate, copulate with, and sterilize their monandrous female mates. Sterilization can be achieved either by transfer of radio-sterilized “defective” sperm (25), or following copulation with a spermless male in highly monandrous species, as is the case for A. gambiae (26). Because the control agent is an insect rather than a traditional pesticide, it has minimal if any off-target effects. Furthermore, SIT males can seek out and sterilize females in cryptic locations that insecticides may miss and are the sex that does not drink blood nor spread disease. As a result, SIT acts as a species-specific and chemical-free “organic” insecticide that has the potential to enable automated, safe, scalable, and controllable suppression when adapted to A. gambiae.
Building a scalable genetic SIT system in anophelines requires creating and combining precise male-sterilization and female-elimination systems, a process not yet successfully undertaken in the species. Sterilization by traditional chemo- or radio- sterilization methods unfortunately impairs male fitness (27–31). Oxidation of somatic DNA, lipids, and proteins (32) causes reduced emergence (18, 30), longevity (28, 33), sperm production (34), and ability to compete during copulation (28, 29, 35, 36)—a lekking-based process where competition is fierce (37). Only partially sterilizing radiation doses generate sufficiently competitive males in trials; however, they have compromised population suppression efficacy (38, 39). For these reasons, researchers have sought to develop male sterilization technologies using more precise genetic methods. For example, A. gambiae lines have been developed which shred the embryonic X-chromosome (40, 41), or express proapoptotic factors in the testis (42), resulting in sterility or offspring killing. However, these lines are difficult to rear in mass because they lack the ability to induce, or repress, the sterility phenotype. In a step toward a more scalable technology, a binary CRISPR system was recently demonstrated in A. gambiae which could generate spermless males in a more inducible manner (43). However, it caused incomplete genetic sterilization (95%) and lacked a sex-sorting component—a hindrance shared by all A. gambiae sterilization technologies to date.
Efforts to develop efficient genetic sexing systems (GSSs) in A. gambiae have been fruitful but limited. Historically, to achieve male-only lines, scientists employed a genetic sexing strain (44) reliant on Y chromosome–linked resistance to dieldrin. However, many of these lines are now extinct (45), and use of dieldrin is banned as it is highly neurotoxic (46, 47), impeding implementation. Therefore, safe genetic sex separation systems in A. gambiae have thus far been limited to optical sorting (48–50) or transgenic expression of sex-specific fluorophores (51–53) followed by fluorescent sorting of neonate larvae via COPAS (54). However, these fluorescence-based technologies require larval sorting of the released generation directly prior to release, making an egg-based distribution modality impossible, a desirable feature if scaling to cover vast distances. Systems which shred the X-chromosome have also been generated which yield highly male-biased populations. However, these lines unfortunately lack inducibility, or repressibility, making them exceedingly difficult to scale (55, 56). Fortunately, the genetic sexing system Ifegenia (Inherited Female Elimination by Genetically Encoded Nucleases to Interrupt Alleles) was recently developed, which permits egg distribution due to automatic genetic sexing. Ifegenia remarkably kills >99.9% females using a binary CRISPR system to target the female-essential gene femaleless (fle), producing a robust and inducible GSS through genetic crosses (57). Taken together, there remains high demand for a scalable SIT system that encompasses both highly penetrant male sterilization and female elimination.
One complete genetic SIT system which combines female elimination and male sterilization is termed the precision-guided sterile insect technique (pgSIT). It has been successfully developed in Aedes and Drosophila (58–61), but not yet in an anopheline species. PgSIT induces male sterilization and female elimination in the offspring of a cross between separate Cas9 and gRNA lines that target male-fertility and female-essential genes during development, resulting in an “inducible” system suited to large-scale releases. In this work, we develop a pgSIT system in Anopheles gambiae that is highly efficient at sterilizing males and eliminating females, providing a vector control option for eliminating the deadly African malaria mosquito. We develop a multi-gRNA line targeting the well-characterized female-essential locus, doublesexF (dsxF) (5), and male-fertility genes Zero population growth (zpg) (26) and β2-tubulin (β2) (53). We demonstrate that crossing this gRNA line to Cas9 yields female androgenization and robust male sterilization in the resulting hybrid F1 offspring. We then improve female elimination by introducing the GSS Ifegenia (57), which targets the female-essential gene, fle, enabling an egg-based distribution modality. In this more complete pgSIT system, we demonstrate complete female killing (>99.9%), near complete male sterility (>99.5%), efficient population suppression in cage trials, and model-predicted elimination of A. gambiae populations in the wild, demonstrating proof-of-principle that pgSIT is a candidate system for confinable vector control of the deadly A. gambiae malaria vector.
Results
Founding and Characterizing gZBD: Females Are Incompletely Androgenized; Males Are Highly Sterile.
To develop a pgSIT system in A. gambiae, we built a gRNA-expressing transgene, gZBD, that encodes an Actin5c-m2Turquoise marker and five gRNAs: one gRNA targeting the germline-essential gap-junction gene zpg (gRNAzpg.1)(43), two gRNAs targeting the sperm motility gene β2-tubulin (gRNAβ2.1,gRNAβ2.2), and two gRNAs targeting the female differentiation gene dsxF (gRNAdsxF.2 expressed twice) (Fig. 1A). We established two distinct gZBD families, gZBDA18 and gZBDD15, with different transgene insertion sites and expression profiles, and identified their chromosomal insertion positions by inverse PCR (62). For the Cas9 line, we utilized the Vasa2-Cas9 line (63), VZC, (henceforth referred to as Cas9) characterized by a 3xP3-DsRed selectable marker. It was selected due to its robust germline expression profile and ability to deposit Cas9 in the embryo, which promotes desired F1 mosaic mutagenesis during development resulting in a phenomenon we coined lethal mosaicism (58).
Fig. 1.
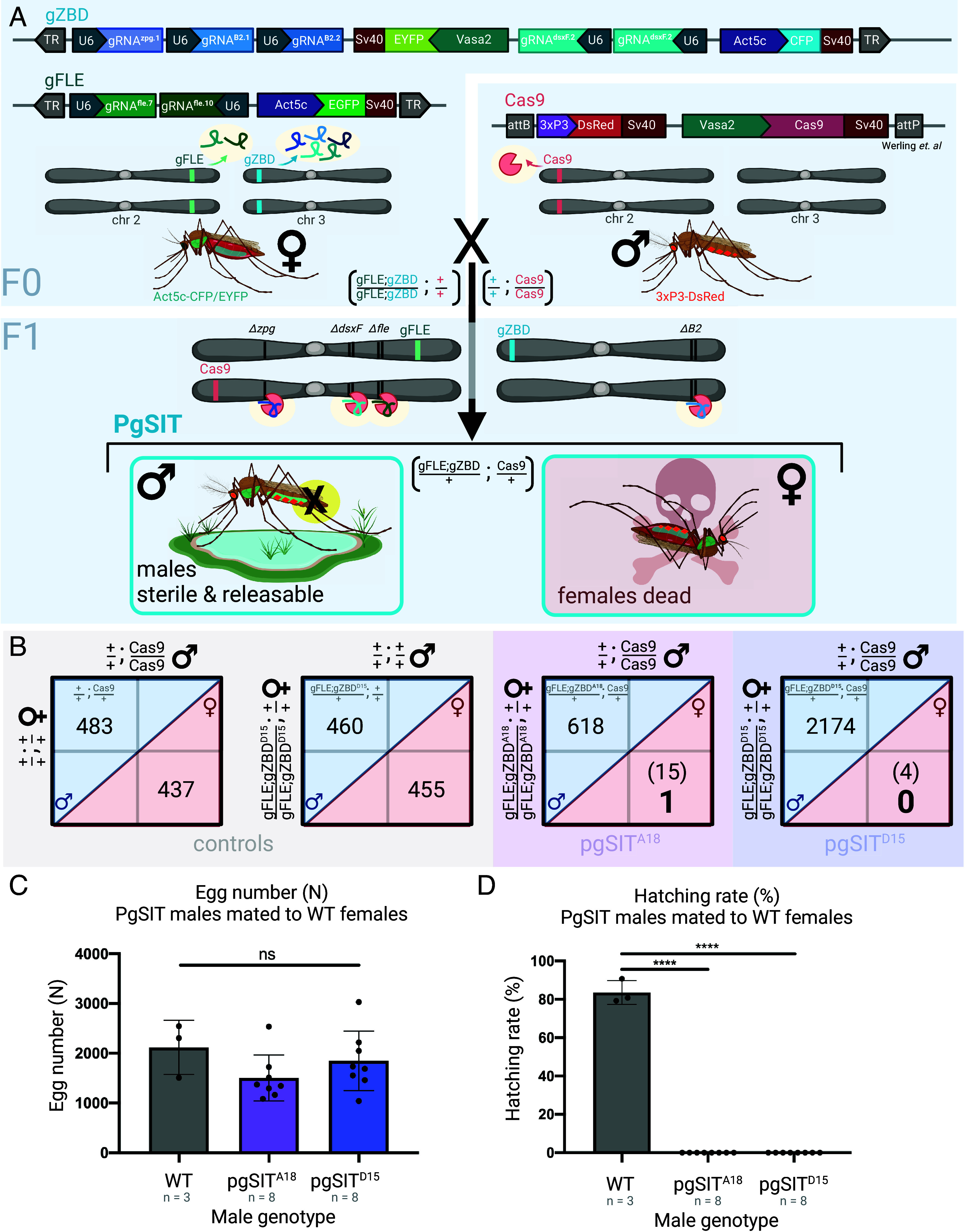
Homozygous pgSIT gRNA females crossed to Cas9 males produce nearly exclusively sterile male F1 offspring. (A) gZBD transgenics express one gRNA targeting zero population growth (zpg) (lavender), two gRNAs targeting β2-tubulin (periwinkle), and two gRNAs targeting the female-specific exon 5 of doublesex (dsxF) (teal) under the expression of individual PolIII U6 promoters, some carrying modified scaffolds (Methods). Also included is a whole-body fluorescent selectable marker, Actin5c-m2turquoise (denoted CFP for brevity), as well as a Vasa2-EYFP marker to aid in germline visualization, a marker which was not visible in these lines. gZBD transgenic lines were individually crossed to a second line, gFLE, to generate double homozygous transgenic lines termed (gFLE;gZBD). gFLE targets femaleless (fle) via two gRNAs also under the expression of the Pol III U6 promoter, and includes an Actin5c-EGFP cassette for selection by whole-body fluorescence. A third line, Cas9, expresses Cas9 in the germline under the Vasa2 promoter and includes a 3xP3-DsRed cassette for selection by central nervous system fluorescence. Crossing (gFLE;gZBD) females to Cas9 males yields transheterozygous pgSIT individuals who bear all three transgenes, resulting in active mosaic mutagenesis, and causing female killing and male sterilization. (B) Among control and pgSIT test crosses, the female-killing phenotype was quantified in the F1 generation, reported as male and female sibling pupa counts. Male and female counts are delineated within blue and red diagonal areas, respectively. Control crosses of Cas9 or (gFLE;gZBDD15) homozygotes to wild type result in approximately equal F1 male and female pupa counts. Crosses between (gFLE;gZBDA18) or (gFLE;gZBDD15) homozygous females and Cas9 homozygous males result in significantly reduced F1 female pupa numbers (in parentheses) (P < 0.0001 for both groups, Binomial test). The number of pupae which survived to adulthood to fly are denoted in large bold font. (C) Crossing 50 pgSIT males to 50 wild type females results in statistically the same number of eggs being laid. Three cage replicates and eight cage replicates shown for wild type control and pgSIT test genotypes respectively. Raw egg counts shown (ns, one-way ANOVA, Dunnett's multiple comparisons test). Mean and SD shown. (D) Crossing 50 pgSIT males to 50 wild type females results in complete sterilization of females when assayed by hatching rate (n% = n 1-d-old larvae/n eggs laid), with high significance compared to the wild type control group (P < 0.0001 for each group, one-way ANOVA, Dunnett's multiple comparisons test). Three cage replicates and eight cage replicates shown for wild type control and pgSIT test genotypes respectively. Created with Biorender.com.
We hypothesized that crossing the gZBD and Cas9 lines would yield (+/gZBD; +/Cas9) F1 hybrid offspring with the desired female androgenization and male sterilization mosaic knockout phenotypes. Among the hybrid F1 offspring, we identified some intersex (+/gZBD; +/Cas9) females with male claspers on female genitalia indicative of dsxF mutagenesis (SI Appendix, Fig. S1) (5). When assaying gZBDA18 and gZBDD15 individually, we observed only 77% and 68% intersex phenotype penetrance respectively among F1 hybrid offspring (Dataset S1), with some females able to initiate blood feeding (n = 3/26). While this assay involved examining genital claspers and not internal reproductive morphology, it still indicates incomplete androgenization making them generally unacceptable for vector control in this species.
To determine whether (+/gZBD; +/Cas9) males are sterile, we performed crosses of 50 wild type females to 50 F1 (+/gZBDA18; +/Cas9) or (+/gZBDD15; +/Cas9) males and assayed the hatching rates of their F2 offspring. We observed sterilization of all females mated to (+/gZBDA18; +/Cas9) males, with a hatch rate of 0%, and most females mated to (+/gZBDD15; +/Cas9) males with an F2 offspring hatch rate of 5.1%, compared to 82.3% hatching rate in wild type controls (SI Appendix, Fig. S2 and Dataset S2). Hatched F2 larvae were verified to express the transgenic fluorescence ratios indicative of (+/gZBD; +/Cas9) paternity, verifying the presence of an “escapee” fertile male. Sequencing these F2 larvae revealed no mutations at the target sites, suggesting incomplete germline mutagenesis in their (+/gZBD; +/Cas9) father. Cumulatively, our data show that the preliminary pgSIT design robustly sterilizes males with efficiency dependent upon genomic insertion site but fails to sufficiently incapacitate females.
Improving Female Killing by Combining with gFLE.
We hypothesized that we could improve the female elimination properties of our system by additionally targeting the recently discovered female-essential gene fle through the introgression of the Ifegenia GSS gRNA line (57). To do this, we separately crossed both gZBD lines into the previously published gFLEG transgenic line (hereon shortened to gFLE) to produce the doubly homozygous gRNA lines (gFLE;gZBDA18) and (gFLE;gZBDD15). The gFLE line expresses two gRNAs targeting fle and an Actin5c-EGFP selectable marker (57) (Fig. 1A), making it distinguishable from the Actin5c-m2Turquoise on gZBD. We previously showed that crossing gFLE males to Cas9 females results in complete female death in the F1 transheterozygous offspring before the pupal stage. Therefore, we hypothesized that (gFLE;gZBD) crossed to Cas9 would produce a robust pgSIT system with the male-sterilizing properties of gZBD and the female-killing properties of gFLE.
pgSIT (+/gFLE;gZBD; +/Cas9) Individuals Are Mosaic Mutants, but Some Crosses Are Lethal.
Prior pgSIT and Ifegenia systems generated F1 hybrids using F0 Cas9 females (as opposed to males) because they are capable of maternal deposition of Cas9 into F1 embryos, resulting in stronger mosaic mutagenesis and more penetrant phenotypes. For initial verification of mutagenesis, we crossed homozygous (gFLE;gZBD) males to homozygous Cas9 females and confirmed mutations in zpg, β2-tubulin, dsxF, and fle in F1 embryos (SI Appendix, Fig. S3). However, we observed these crosses resulted in severe F1 mortality at the early larval stage, even though separate F0 crosses between gFLE or gZBD males to Cas9 females were viable. Fortunately, the reciprocal F0 cross using Cas9 males generated viable F1 offspring and was used to generate the (+/gFLE;gZBD; +/Cas9) genotype used in all subsequent experiments. For simplicity, the hybrid F1 (+/gFLE;gZBD; +/Cas9) genotype is henceforth abbreviated to “pgSIT” genotype, with (+/gFLE;gZBDA18; +/Cas9) and (+/gFLE;gZBDD15; +/Cas9) shortened to pgSITA18 and pgSITD15 respectively.
The pgSIT System Induces Robust Female Elimination and Produces Fit Sterile Males.
We next determined whether this pgSIT system is capable of robustly eliminating females and sterilizing males. We observed almost exclusively male pupae among both pgSITA18 and pgSITD15 individuals, indicating robust female elimination (Fig. 1B). Specifically, for 618 pgSITA18 male pupae scored, 15 female sibling pupae were identified, out of which only one survived to fly; and for 2,174 pgSITD15 male pupae scored, four female pupae siblings were identified, out of which none survived to fly (Fig. 1B and Dataset S3). Consistent with prior work (57), both pgSITA18 and pgSITD15 lines exhibited robust female elimination, 99.8% and 100% respectively, sufficient to be candidates for field releases. To determine whether pgSIT males have high sterility, we crossed 50 pgSITA18 or pgSITD15 males to 50 wild type females and calculated percent fertility of the resulting broods. Out of 16 total cages assayed (800 males total, 400 for each family), zero larvae hatched, demonstrating 100% sterility of both-pgSITA18 and pgSITD15 males in these assays (Fig. 1 C and D and Dataset S4). A more accurate sterility measurement for the population as a whole is >99.5% for each line assuming half of the males were represented in the assay, well above the 98% considered by some to be adequate for SIT campaigns (64).
Moving forward, we selected pgSITD15 for further characterization, crosses, and analysis due to its strong female-killing and male sterility phenotypes, as well as husbandry considerations. To characterize pgSITD15, we performed Nanopore sequencing on pooled pgSITD15 males and confirmed the single insertion site of gZBDD15 to be within a noncoding region of chr. 3L (NT_078267.5:4828892-4828896). To verify the sterility phenotype, we performed dissections on male pgSITD15 lower reproductive tracts, which revealed the absence of normal testicular tissue (SI Appendix, Fig. S4A). As expected, we observed atrophied testes within (+/gZBD; +/Cas9) controls due to zpg and B2-tubulin targeting. However, we also observed atrophied and occasionally absent testes among (+/gFLE; +/Cas9) controls, a phenotype not noticed in prior work due to the fertility of the (+/gFLE; +/Cas9) male population as a whole (57). This suggests that targeting all of these genes together may have an additive or synergistic effect, causing the complete sterility observed in Fig. 1C, as opposed to the partial fertility observed in SI Appendix, Fig. S2C. Taken together, these results demonstrate that pgSITD15 males are sufficiently sterilized to be candidates for SIT by most measures.
For the most effective population suppression, males must be able to mate with and induce mating refractoriness in females, in addition to having high fitness. In A. gambiae, refractoriness is induced following the transfer of a gelatinous mating plug to the female during copulation, a structure originally produced by the male accessory glands (MAGs) (65, 66). Dissections revealed that despite lacking testes, pgSITD15 males still developed other important tissues for reproduction such as claspers, an aedegus, and MAGs (SI Appendix, Fig. S4A). In line with the development of MAGs, we confirmed that pgSITD15 males were able to successfully transfer a mating plug during copulation (SI Appendix, Fig. S4B), indicating females should be refractory to further mating (66, 67). To quantify general pgSIT male fitness, we determined their longevity through survival curve assays (SI Appendix, Fig. S4C and Dataset S5). These revealed that pgSITD15 males have approximately the same longevity as wild type males (p = ns Mantel–Cox test), living slightly but not significantly longer than controls. In summary, these results suggest that pgSITD15 males do not have significant fitness costs that could curtail their longevity and develop all structures critical for reproduction, suggesting they have high fitness overall.
PgSIT Can Induce Population Suppression.
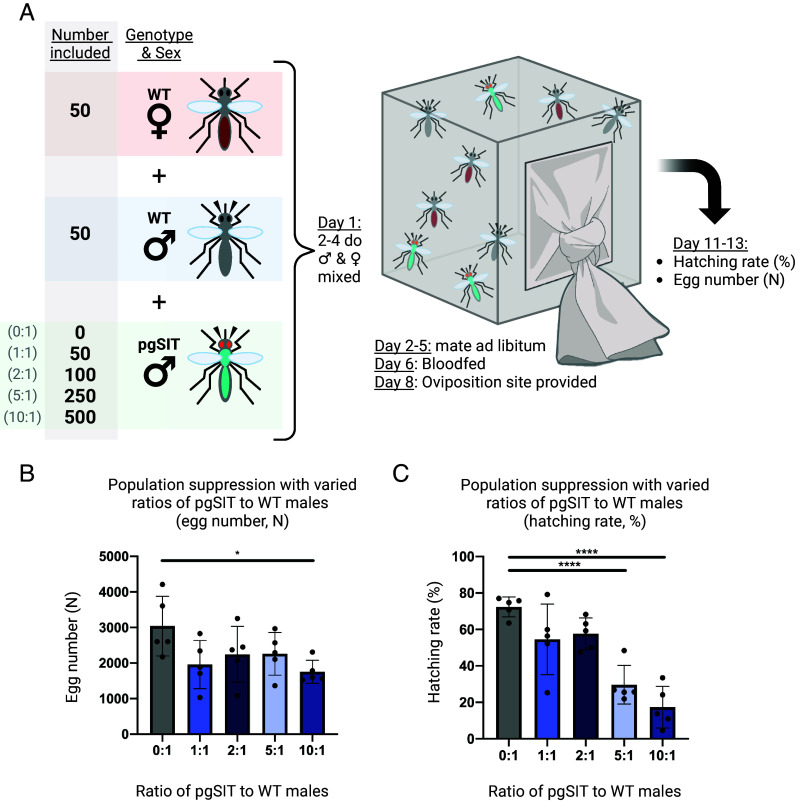
We next set out to determine whether pgSITD15 males could cause population suppression in cage trials. For this, we established competition cages of pgSITD15 males against 50 wild type males at 0:1, 1:1, 2:1, 5:1, or 10:1 ratios and added 50 wild type females as potential mates. The resulting broods were assayed for percent fertility (Fig. 2A). Consistent with release ratios required to suppress Aedes populations (61), we observed significant population suppression from the 10:1 and 5:1 release ratios (17.4% and 29.7% mean hatching rate, both P < 0.0001), and nonsignificant, less pronounced, suppression from the 2:1 and 1:1 releases (57.6% and 54.6% mean hatching rate respectively, nonsignificant), compared to the 0:1 control (72.4% mean hatching rate) (Fig. 2 B and C). In line with other GM vector control systems, pgSIT males are unsurprisingly less fit than wild type by nature of their high transgene load, a parameter taken into account when determining high ratios for release. Specifically transgenic males can be calculated to be 49.2% as fit as wild type males as the difference between the observed and theoretical suppression percentages of the 1:1 versus 2:1 release groups assayed in Fig. 2B. Hence, release ratios of 10:1 or higher are common (68) for other sterile transgenic mosquito control campaigns, demonstrating that A. gambiae pgSIT males achieve sufficient suppression to be considered candidates for SIT releases, but larger trials are needed.
Fig. 2.
Population suppression following release of pgSIT males at different ratios to wild type. (A) Test suppression cages were established with 50 wild type males, 50 wild type virgin females, and either 0, 50,100, 250, or 500 pgSITD15 males (for the 0:1, 1:1, 2:1, 5:1, and 10:1 pgSIT:wild type male ratios respectively). After mating and blood feeding, the hatching rate was calculated for each cage. (B) The egg counts from population suppression assay cages. Groups 0:1 and 10:1 are significantly different (P < 0.05, Dunnett’s multiple comparisons). Mean and SD shown. (C) Population suppression as measured by the hatching rate (%) from cages suppressed by different ratios of pgSIT males to wild type males. Hatching rate is reported as the percent of eggs which hatched (n% = n 1-d-old larvae/n eggs laid). The 0:1 control group differs significantly with both the 5:1 (P < 0.001) and 10:1 (P < 0.001) groups (one-way ANOVA, Dunnett’s multiple comparisons test). Mean and SD shown. Created with Biorender.com.
The broods from population suppression assays were also monitored for the presence of fluorescent transgenic F2 offspring which would indicate a fertile pgSITD15 father. Among the 20 cages tested containing pgSIT males (Fig. 2 B and C and Dataset S6), only a single brood yielded transgenic larvae (n = 43 transgenic larvae total, from the 2:1 suppression group), suggesting the presence of a single fertile male which escaped the sterilization phenotype, providing evidence of the only fertile pgSIT male observed throughout the course of this work.
Modeling pgSIT As a Suppression Technology.
We next modeled hypothetical releases of pgSIT A. gambiae eggs to explore their potential to eliminate a local A. gambiae population resembling that of the Upper River region of The Gambia using the MGDrivE 3 framework (69) with parameters listed in Dataset S7. The modeling framework was calibrated to malaria prevalence data from a randomized controlled trial conducted in the Upper River region (70), and informed by local entomological data (71) and rainfall data sourced from Climate Hazards Group InfraRed Precipitation with Station data (CHIRPS, https://www.chc.ucsb.edu/data/chirps). Parameters describing the pgSIT system were based on results from this paper suggesting the pgSIT system in A. gambiae induces near complete male sterility (>99.5%) and female inviability (>99.9%), with offspring of sterile males being unviable at the egg stage. Semi-field data for anophelines suggest that mating competitiveness of sterile males from SIT programs is either the same as or 20 to 55% that of wild males (Bouyer and Vreysen, 2020). To be conservative, we chose 37.5% as our estimate for this parameter, as it falls in the middle of the lower range. We assumed a 25% reduction in pgSIT male lifespan compared to wild-type males, despite no reductions in lifespan being observed in this work, as fitness costs sometimes emerge in the field (68).
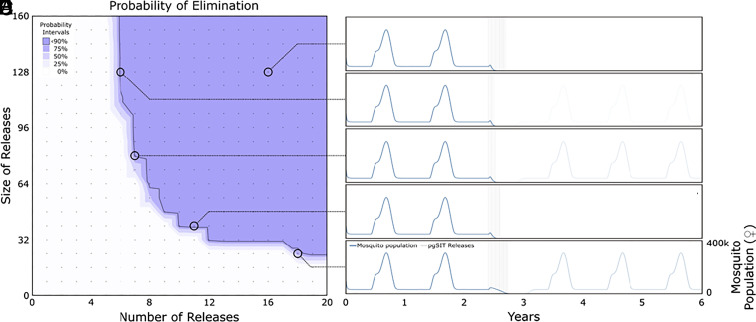
With the parameterized modeling framework in place, we simulated 0 to 20 consecutive weekly releases of pgSIT eggs at a ratio of 0 to 160 pgSIT eggs (female and male) per wild A. gambiae adult (female and male) (Fig. 3). Previous pgSIT modeling studies (58, 61) suggested that Aedes aegypti populations could potentially be eliminated by 10 to 24 consecutive weekly releases of 40 to 400 pgSIT eggs per wild adult; however, we focused on release schemes having smaller weekly release sizes as a more cost-effective option. The mosquito population in the Upper River region is highly seasonal, as reflected in the first 2 y of the time-series dynamics (prerelease), so we simulated pgSIT eggs released from the beginning of the rainy season (June 1st), just as the A. gambiae population begins to grow—a timing determined optimal for several genetic control systems (68, 72). Simulation output predicts local A. gambiae elimination for achievable release schemes —≥10 weekly releases of 40 pgSIT eggs per adult mosquito, ≥7 weekly releases of 80 pgSIT eggs per adult mosquito, and ≥6 weekly releases of 128 pgSIT eggs per adult mosquito. In absolute numbers, this represents ≥10 weekly releases of ~75 million pgSIT eggs throughout inhabited areas of the Upper River region (an area of 2,070 km2), ≥7 weekly releases of ~150 million pgSIT eggs, or ≥6 weekly releases of ~240 million pgSIT eggs. In many cases where elimination is not achieved, significant population suppression still occurs and is maintained for >6 mo, which would be expected to have a significant epidemiological impact.
Fig. 3.
Modeling population suppression and elimination by release of pgSIT A. gambiae. Weekly egg releases were simulated in a randomly mixing A. gambiae population resembling the Upper River region of The Gambia using the MGDrivE 3 simulation framework (69) with parameters described in Dataset S7. Probability of A. gambiae population elimination is depicted for a range of release schemes described by the number of consecutive weekly releases and number of pgSIT eggs released per wild-type adult. The contour plot (Left) depicts regions of parameter space for which the local mosquito population is eliminated with probabilities ≥0, 25, 50, 75, or 90% (as measured by the proportion of 100 simulations that lead to elimination). 90% elimination probability is depicted by a solid line. Time-series mosquito population dynamics (Right) are depicted for a selection of scenarios from the contour plot. Releases of pgSIT eggs (female and male) are modeled beginning June 1st (beginning of the Upper River rainy season) in the third year of the simulation. Scenario (A) depicts a large release very likely to achieve elimination (16 weekly releases at a 128:1 ratio of pgSIT eggs to wild adults); scenario (B) depicts a release scheme that achieves transient suppression but not elimination (5 weekly releases at a 128:1 ratio); scenarios (C and D) depict release schemes ~90% likely to achieve elimination (7 weekly releases at an 80:1 ratio, and 10 weekly releases at a 40:1 ratio, respectively); and scenario (E) represents a release scheme with a ~75% elimination probability (18 weekly releases at a 24:1 ratio), with the population rebounding in ~25% of simulations (faint lines in years 4 to 6).
Discussion
In this work, we develop the genetic SIT technology, pgSIT, in the malaria vector A. gambiae, meeting the demand for a confinable and proven mosquito suppression technology in this species. Overall, we demonstrate that our pgSIT system exhibits remarkable male sterilization (100% in assays, >99.5% for the population as a whole), and female elimination (100% in assays, >99.9% for the population as a whole), and strong population suppression effects in cage trials, yielding a system amenable to high-throughput safe SIT releases of presterilized and pre-sex-sorted eggs. In detail, we generated a gRNA-expressing transgene, gZBD, targeting zpg, dsxF, and β2-tubulin for CRISPR cleavage. When crossed to a Cas9 transgenic line, we observed in the hybrid progeny significant but incomplete female androgenization due to dsxF targeting. We also observed complete or nearly complete sterility of-(+/gZBDA18; +/Cas9) or (+/gZBDD15; +/Cas9) respectively, due to zpg and β2-tubulin targeting. To improve female-elimination, we crossed the gZBD line to the Ifegenia Genetic Sexing System line, gFLE, and established double homozygous gRNA-expressing lines (gFLE;gZBDA18) and (gFLE;gZBDD15). When crossed to Cas9 we confirmed the presence of mutations within each gene in the hybrid progeny, however, crosses generated with the Cas9 transgene derived maternally were lethal. While further elucidation of this phenotype was beyond the scope of this work, we postulate this is due to an overabundance of on- and off-target cleavage because maternal Cas9 is expected to yield stronger embryonic mutagenesis due to maternal deposition by the Vasa promoter (73).
Remarkably, pgSIT individuals of both lines exhibit strong female elimination, 99.8% and 100% respectively for pgSITA18 and pgSITD15, in addition to high levels of sterility, >99.5% each. From these two lines, we selected pgSITD15 for more in-depth characterization. In line with the sterility phenotype, pgSITD15 males lacked testes but maintained otherwise normal lower reproductive tracts (SI Appendix, Fig. S4A). Interestingly, (+/gFLE; +/Cas9) controls also displayed aberrant and occasionally absent testes, a phenotype not noticed in prior work due to population fertility as a whole (57). Though further elucidation is beyond the scope of this work, we postulate that it is mimicking the function of fle’s closest well-characterized homolog, Transformer2 (tra2), whose misregulation during fly spermatogenesis causes infertility and defective sperm (74). This finding potentially explains why the pgSITD15 individuals in Fig. 1D had higher rates of sterility than the (+/gZBDD15; +/Cas9) males in SI Appendix, Fig. S2C, if this phenotype has an additional sterilization effect. In line with having an otherwise normal reproductive tract, we confirmed that pgSIT males were able to transfer a mating plug, a key requirement for induction of refractoriness in females (SI Appendix, Fig. S4B) (66, 67).
We further confirmed that pgSITD15 males are long-lived by the survival curve assay and validated their genomic integration locus by Nanopore. In competition cage trial assays, we demonstrated that pgSITD15 males are capable of causing significant population suppression when competing against wild type males at 10:1 and 5:1 ratios (both P < 0.0001, Dunnett’s multiple comparison) yielding a 4.2× and 2.4× fold reduction in average hatching rate respectively, a strong suppression phenotype similar to those observed in pgSIT in other organisms (58, 61). Finally, modeling suggests this system is capable of eliminating local A. gambiae populations, and hence interrupting malaria transmission, for achievable release schemes of ≥10 releases of 40 pgSIT eggs per adult mosquito or ≥7 weekly releases of 80 pgSIT eggs per adult mosquito. In total, this work demonstrates that this pgSIT system exhibits all of the necessary properties for consideration as a line for SIT-like vector control of A. gambiae.
The pgSIT system outlined here may also enable suppression of the adjacent species within the A. gambiae complex: A. arabiensis, A. quadriannulatus, A. melas, and A. merus. Not only are the target sites for transgenic gRNAs, gRNAzpg.1, gRNAB2.2, gRNAdsxF.2, gRNAfle.7, and gRNAfle.10 conserved making introgression into these species possible (75, 76), but an overabundance of released A. gambiae pgSIT males may breach natural mating barriers to directly suppress these species as well (77–80). With gene drives being proposed to spread beyond target species assuming the drive target site is conserved and not mutated (81), the possibility of this occurring with other vector control strategies such as pgSIT should be explored as well.
pgSIT could be most directly compared to the nondriving suppression technologies fsRIDL, X-shredders, and Ifegenia. fsRIDL is a GM technology wherein a flightless female transgene is chemically repressed in the laboratory setting by addition of tetracycline or an analog (68). In the absence of tetracycline, flight muscle toxicity is caused by uncontrolled protein overproduction expressed by the female flight muscle-specific Actin-4 promoter (Labbé et al. 2012 (82); Marinotti et al. 2013 (83)). When males with this transgene are released, they mate with wild females, and their offspring have the transgene derepressed, resulting in transgenic daughters incapable of flight. These daughters are nonbiting as they persist on the ground, failing to ever fly beyond the initial oviposition site. Releases of fsRIDL males can cause population suppression and have been field-proven following trials in the Cayman Islands, Mexico, Brazil, and Florida (68, 84). Such a technology would show great potential in A. gambiae, and has already been developed in the neighboring species A. stephensi (83). However, no female-specific flight muscle promoters have yet been identified and published in A. gambiae, and the closest putative Actin-4 homologs do not exhibit female-specific RNA expression (AGAP011515, AGAP001676, AGAP011516, AGAP005095, AGAP000651, AGAP011514) (85), making development of this technology in this species potentially problematic. Indeed, a successful fsRIDL system in A. gambiae has not yet been published. Once published, fsRIDL could present a more scalable alternative to pgSIT as the phenotype is chemically repressible, making unnecessary any sorting steps to induce the phenotype. If circumventing sorting steps is desired, temperature-inducible pgSIT (TIpgSIT) could be developed in which Cas9 is temperature inducible, making pgSIT inducible sans crossing more akin to fsRIDL (86). However, with future iterations of pgSIT having the potential to produce 2 million males per COPAS sort-hour (see below, SI Appendix, Text S1) it is unclear how significant this difference in scalability will be. A further important distinction between fsRIDL and pgSIT is that pgSIT is designed not to produce significant GM offspring which persist in the environment (though GM offspring from a single fertile male was observed in this study). Though no technology is perfect, this is a property where fsRIDL is lacking, potentially making pgSIT more alluring to locales where anti-GM sentiments persist. In all, until fsRIDL is developed in A. gambiae, we propose pgSIT to be a viable alternative nondriving vector control technology in this space, one which may be superior in some respects.
On the other end of the technological spectrum are X-shredders which have been developed in the species for over a decade (55, 56). These technologies are endonuclease-expressing transgenes which target the male-derived X chromosome, resulting in male sterility or male bias depending on whether they target the X-chromosome in the offspring or in the testis respectively. Sterilizing X-shredders developed to date lack an inducible sex separation component and are dominantly sterile. Therefore, cumbersome addition of nontransgenic individuals each generation is a necessity to maintain the line, and makes manual sorting prior to releases a requirement (87). While male-bias X-shredders are capable of over 95% male bias, they do not achieve the >99.9% levels achieved by pgSIT and are similarly noninducible, making scale-up problematic. Though mathematically fewer X-shredder males compared SIT need to be released to achieve suppression (88), their lack of phenotype inducibility makes the currently published lines difficult to scale. While systems to induce the X-shredder phenotype could theoretically be developed, they have yet to be published, again making pgSIT a theoretically better alternative.
Ifegenia is a sister technology to pgSIT, relying on the same type of crossing mechanism for phenotype induction. Ifegenia relies on the crossing together of Cas9 and female-killing gRNA lines to develop male-only populations for release. These males however are not sterile as in pgSIT, but instead transmit the female-killing CRISPR transgenes and female-killing mutant femaleless alleles on to the next generation, causing population suppression over time. Though somewhat similar to fsRIDL, Ifegenia has been modeled to be less effective at suppressing wild populations than pgSIT (57). This, coupled with the release of GM offspring designed to persist in the environment, makes Ifegenia less alluring than pgSIT in multiple respects. In all, among the nondriving technologies for targeting A. gambiae, they are either not yet developed (fsRIDL), not scalable (X-shredders), or less effective (Ifegenia) than pgSIT, making pgSIT a leading candidate technology for confinable suppression of this deadly pest.
In contrast to these technologies are gene drives, nonconfinable, self-spreading selfish genetic elements capable of unilaterally engineering entire populations. These self-propagating genetic strains are the most advanced and scalable technology already developed in the species (5, 89), but their uncontrollability has raised political, ethical, ecological, and socioeconomic concerns, hindering their release (13). The long-term durability of gene drives is also unclear as many generate their own resistance mutations often due to the constitutive coexpression of CRISPR components, ultimately hindering their spread. Following release of population suppression drives, selection pressures for resistance mutations which evade extinction will be very high making the system prone to breakage, as they rely on the integrity of a single gRNA target site. That said, pgSIT is comparably more robust as the two CRISPR strains are maintained separately, preventing resistance allele generation, and only producing the released “predominantly dead end” males following a controlled cross in the production facility. During facility rearing, lines could be regularly validated to verify the absence of surprise resistance polymorphisms, preventing the emergence of a resistance allele during the final cross. Even if a resistance allele were to arise in pgSIT, for example in a female, it could be eliminated by mating with a sterile male, preventing transmission of the allele to further offspring, overall making this technology resistant to resistance. In essence, pgSIT acts to inhibit the process of reproduction whereas gene drives act on the underlying genetics, so they are not subject to the same resistance mechanisms. Therefore in this vein, allele pumps are of little concern for pgSIT, as carrying a resistance allele does not make a female less likely to be sterilized by a pgSIT male (90). Among those published in A. gambiae, pgSIT fills a unique niche between these technologies; it is more high-throughput than X-shredders, more confineable and controllable than gene drives, and because it is almost exclusively a “genetic dead end” (releasing orders of magnitude fewer transgenes into the population than technologies like Ifegenia) it promises to be a powerful tool in the A. gambiae vector control toolkit.
Compared to some other vector control methods, pgSIT is more scalable and can be released during all life stages, notably eggs. For GM vector control campaigns except fsRIDL and sex-biasing gene drives (5, 91), the rate-limiting step for releases is sorting males from the undesirable, disease-transmitting, females in the released generation. If not performed manually, which is limited to 500 pupae per hour per operator (92), this is achieved by optical sorting or by fluorescence-based sex sorters (48–51, 53). The former utilizes an AI-trained camera to distinguish between sexes as adults (49, 50), while the latter relies on transgenic sex-specific fluorescence to enable sorting of nascent larvae by a COPAS fluorescence sorter machine (54). For some other vector control measures, sorting occurs on the individuals directly to be released, yielding a fairly low 2:1 sort:release ratio (two mosquitoes sorted per one released, SI Appendix, Text S1) (41, 42, 55). In pgSIT, however, sex sorting occurs the generation prior to release (F0 generation), and the released generation is automatically genetically sex-sorted (F1 generation), giving pgSIT, and the sister technology Ifegenia a sort:release ratio closer to 1:50 due to the high reproductive output of a single sorted F0 female (SI Appendix, Text S1). These features not only make pgSIT higher-throughput by orders of magnitude, but also enable delivery of eggs via drone (93, 94). It is important to note however that systems such as fsRIDL, which rely on chemical suppression/induction require no sorting step, and are therefore maximally scalable having no sort:release ratio. While manual F0 sorting was performed in this study, a technique effective for small-scale field trials (87), F0 sorting by optical sorters could be adapted to the current iteration of pgSIT to immediately enable larger-scale trials and possibly even release campaigns. Future iterations of pgSIT, termed pgSIT2.0, could consolidate the two gRNA transgenes and introduce sex-specific fluorescent reporters to enable higher-throughput F0 sorting by COPAS (54, 95–97). With a COPAS sorting capacity at conservatively 40,000 larvae per hour (54), F0 sorting a pgSIT2.0 system could yield 2 million F1 sterilized males in the next generation with one machine (SI Appendix, Text S1), facilitating production required for releases on a continental scale.
While pgSIT does not aim to release transgenes into the population, our observation of rare fertile escapee males indicates that release of some CRISPR transgenes into the population will likely occur. It has been shown that population eradication by pgSIT does not require complete (100%) sterility penetrance, as appreciable levels of suppression can be achieved by incompletely penetrant systems (60). The released transgenes would still separately express Cas9 and gRNAs, and they are incapable of gene drive given their dislinkage and genomic position. Such transgenes would be expected to be lost from the population given their inherent fitness defects. Importantly, these alleles may include rare resistance alleles (defined as functional mutant alleles at the gRNA target site), however because fresh pgSIT individuals would be released iteratively, wild females carrying these alleles would be sterilized and prevented from transmitting it further to their offspring. Therefore while both fsRIDL and pgSIT do technically release GM carrying offspring into the environment, the latter does so at an order of magnitude lower rate, potentially making it more acceptable for locales with strong anti-GM sentiment.
In all pgSIT presents an advancement for vector control of A. gambiae. It exhibits most of the criteria required for use in SIT releases; it is capable of producing highly sterile males and in mass, it causes robust population suppression, it displays advantageous fitness parameters, and it is more confineable and more scalable than alternative GM technologies developed to date in this species. In all, pgSIT presents a powerful tool in the toolkit for control of this deadly malaria vector, potentially enabling control of this deadly pest.
Methods
Mosquito Rearing and Maintenance.
A. gambiae used in this work was derived from the stock G3 strain. Mosquitoes were reared in an ACL-2 insectary under 12 h light/ dark cycles at 27 °C with a water source provided for drinking and ambient humidity. Adult mosquitoes were placed in Bugdorm, 17.5 × 17.5 × 17.5 cm cages. Adults were fed 0.3 M aqueous sucrose ad libitum. Males and females were allowed to mate for 4 to 7 d prior to being provided with a blood meal on anesthetized mice for about 15 min. Egg dishes, composed of urinalysis cups filled with water and lined with a filter paper cone, were provided to cages 48 h after a blood meal. Eggs were allowed to melanize and hatch unperturbed for 3 d in the egg dish before being floated into trays filled with DI water. Larvae were reared and fed, and pupae were screened and sexed, in accordance with established protocols (98).
Cloning and Plasmids.
Cloning and molecular biology work was undertaken using established cloning protocols. Plasmid 1114H (gZBD) is available at Addgene (200640) (99). All other transgenes used in this work were previously published (57, 63)
gRNA Design.
The target gene reference sequences for zpg (AGAP006241), B2-tubulin (AGAP008622), dsxF (AGAP004050) were retrieved from VectorBase (100), and sequences were confirmed by PCR. gRNA’s were designed using software available at https://crispor.tefor.net. One gRNA targeting dsxF was expressed twice designed to target the extreme 3’ end of the dsxF exon 5 coding sequence, termed gRNAdsxF.2, (5′ TTATCATCCACTCTGACGGG 3′). It was designed to carry a standard S. pyogenes RNA scaffold sequence containing a SNP of (5′ GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACTGAGTCGGTGCTTTTTT 3′). Two gRNAs targeting the first exon of B2-tubulin were designed, gRNAB2.1, (5′ GCTCGATATCGTGCGCAAGG 3′), and gRNAB2.2, (5′ CCAAATAGGCGCTAAGTTCT 3′). To minimize repetitiveness in the plasmid, both gRNAs were designed to carry a variant gRNA scaffold sequence (courtesy of the George Church lab) of (5′ TTCCAGAGCTATGGAAACATAGCAAGTTGGAATAAGGCTTAGTCCGTACTCAACTTGAAAAAGTGGCACCGAGTCGGTGCATTTTTT 3′). A single gRNA targeting the first exon of zpg previously shown to cause robust germline mutagenesis (43, 63, 101) was used, gRNAzpg.1 (5′ GATCCGATCACGCAGTCGAT 3′). This gRNA harbored a standard S. pyogenes gRNA scaffold (5′ GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT 3′). The gRNAs gRNAfle.7 (5′ CGACGGCTCGTTCATCGCTG 3′) and gRNAfle.10 (5′ ATCGAGCGCGTCGCCTGGTA 3′) targeting fle were previously described (57, 102).
Embryonic Microinjections.
Injections were carried out as described previously (103–105). In brief, the gZBD plasmid injection mix was prepared by maxiprep and was diluted to 350 ng/µL solution in diH2O. 45 m to 2.5 h old embryos were harvested from a stock cage of the G3 line and aligned on a glass slide, posterior end up, along the edge of a dampened Millipore mixed cellulose esters membrane (CAT No. HAWP04700F1) covered with a cut-to-size Whatman filter paper (CAT No. 1001-150), as diagrammed (103). The posterior end of embryos was injected with a quartz needle filled with injection mix and controlled by an Eppendorf FemtoJet4x injection system (CAT No. 5253000025). Injected embryos remained on the slide, and the slides were placed in a water dish with the end of the Whatman filter paper submerged to permit capillary action to prevent the eggs from drying. Neonate F0 larvae were removed beginning 48 h post injection and were reared separately.
Fluorescent Sorting, Sexing, and Imaging.
All imaging of A. gambiae was carried out under a Leica M165FC fluorescent stereomicroscope outfitted with a Leica DMC2900 camera. Fluorescence was visualized using the CFP/YFP/mCherry triple filter, and pupal sex was determined by examining the pupal genital terminalia (106).
gZBD Family Establishment.
The nomenclature for the F0, F1, and F2 generation demarcations within this section of the methods follows the more traditional use of these generational markers within the field. It differs from the use of F0 and F1 in the main text which is in reference to stock parental (F0) and hybrid (F1) generations for study of pgSIT. They are in reference to different experiments and genotypes, and are not to be confused.
To establish gZBD transgenics, embryonic microinjection of the gZBD transgene was carried out into F0 individuals essentially as described above. F0’s were reared to adulthood, outcrossed to wild type G3 stock line of the opposite sex, and blood fed. The resulting F1 offspring yielded multiple F1 “founder” transgenic larvae which were identified by fluorescence. Female F1 individuals were isolated individually and used to found the gZBDA and gZBDD families. The gZBDA and gZBDD families both exhibited fluorescence patterns indicative of multiple insertion sites. Therefore, to generate subfamilies with single insertion sites, gZBDA and gZBDD female transgenics were outcrossed to G3 wild type males in bulk (over 100 individuals of each sex) for five generations, selecting for female transgenics each generation. Single females from the “diluted” gZBDA and gZBDD lines were isolated and allowed to lay separately, and a single brood that had uniform fluorescent patterns suggesting a single insertion site from each family was used to found the gZBDA18 and gZBDD15 subfamilies.
Identifying and Validating Genomic Insertion Site of gZBD Transgenes.
To identify the genomic insertion site of gZBD, genomic samples were taken from crushed gZBDA18 and gZBDD15 adults, and inverse PCR was performed (107). In short, 1 to 3 μg of genomic DNA was treated with TaqI restriction enzyme for 4 h, then circularized with ligase in a dilute 100 μL reaction. The sample was reconcentrated by Sodium Acetate precipitation followed by resuspension in 10 μL water, of which 1 μL was used to template the inverse PCR. PCR was carried out with the primers 1114H.S3 and 1114H.S4, (5′ CTGTGCATTTAGGACATCTCAGTC 3′) and (5′ GACGGATTCGCGCTATTTAGAAAG 3′) respectively, the latter of which amplifies outward beyond the piggyBac terminal repeat of the gZBD plasmid and into adjacent genomic sequences. PCR amplicons were gel extracted, cloned into pJET (Thermo Scientific, Cat. No./ID: K1231), and colonies were individually sequenced. Reads were aligned against the piggyBac terminal repeat of the gZBD transgene with all sequencing beyond the repeat terminus corresponding to the locus of integration. Through this method, gZBDA18 and gZBDD15 were found to be integrated in the (chr3L:34188038) and (chr3L:828896) loci respectively. Primers for standard PCR were designed to confirm the genomic integration, and used to homozygous the transgenic lines used throughout this work. To identify the transgenic gZBDA18 allele, the primers 1114H.S4 and 1114H.ipS1; (5′ GACGGATTCGCGCTATTTAGAAAG 3′) and (5′ CATTGAACGGTCTATGCTGTCATGTAC 3′) respectively, were used for PCR amplification. To identify the presence of the wild type (unintegrated) gZBDA18 allele, the primers 1114H.ipS1 and 1114H.S31, (5′ CATTGAACGGTCTATGCTGTCATGTAC 3′) and (5′ CGTTCTTGCGAAAAGGTGAAAAGTG 3′) respectively, were used. To identify the transgenic gZBDD15 allele, primers 1114H.S17 and 1114H.S29, (5′ GACTGAGATGTCCTAAATGCAC 3′) and (5′ CTCGTGACCCTCGTTATAG 3′) respectively, were used, while the primers 1114H.S30 and 1114H.S29, (5′ CATGTTGTTCTTTTGGAAAGC 3′) and (5′ CTCGTGACCCTCGTTATAG 3′) respectively, were used to identify the presence of a wild type (unintegrated) gZBDD15 allele.
Δdsx Knockout Phenotype Characterization of gZBD Families.
Following embryonic microinjections of gZBD into F0 embryos, the F1 generation yielded transgenic “founder” larvae. While female F1 founders were used to establish clonal isofemale lines for study, the male F1 founders—with mixed uncharacterized and unknown insertion sites—were crossed to Cas9 females in bulk. The resulting F2 transheterozygous hybrids (+/gZBD; +/Cas9) were imaged for genital androgenization (SI Appendix, Fig. S1).
Male Sterility Characterization of +/gZBD and +/gFLE;gZBD Families.
For crosses assaying male sterility, we established cages of 50 transgenic hybrid sterile males —(+/gZBD; +/Cas9) or pgSIT (+/gFLE;gZBD; +/Cas9)—to 50 virgin wild type females on day 1, and allowed them to mate ad libitum. On day 6 females were fed a mouse blood meal, and an oviposition site (egg dish) was provided on day 8. Larvae were counted on days 11, 12, and 13 and checked for the presence of fluorescence. If F2 larvae were fluorescent at transgene ratios expected of progeny from a hybrid transgenic father, they were counted and presumed to belong to an escapee fertile male. These F2 offspring for (+/gZBD; +/Cas9) sterility experiments were collected and sequenced for mutations at the gRNA target sites following sequencing protocols listed above. If F2 larvae were completely nonfluorescent, then a contamination was presumed to have occurred via inclusion of a nontransgenic male, and the replicate discarded (one replicate). Eggs and eggshells were counted on days 13 and 14. Hatching rate was calculated as the number of larvae over the number of eggs (Fig. 1C and SI Appendix, Fig. S2B); the number of eggs is also reported (Fig. 1D and SI Appendix, Fig. S2C).
Establishing Homozygous pgSITA18 and pgSITD15 Lines.
To establish the doubly homozygous gFLE;gZBDA18 and gFLE;gZBDD15 lines, we began by crossing gZBDA18 and gZBDD15 separately to gFLE. For five generations brightly fluorescent individuals with an ‘aqua’ fluorescence color, indicative of dual EGFP and m2Turquoise fluorescence, were sorted for as pupae and allowed to mate ad libitum. Then, after a number of generations, individuals were fluorescently sorted and allowed to mate ad libitum as described above, but following blood feeding females were isolated into single oviposition cups to lay egg clutches in isolation. From each resulting brood, a small pool of individuals were taken as L1 larvae to check for gFLE and gZBD homozygosity via PCR. Primers 1154A.S32 and 1154A.S3, (5′ CTTTCTAACGGTACGCAGCAG 3′) and (5′ AACAGCCACAACGTCTATATCATG 3′) respectively, were used to identify the presence of the transgene in the gFLE transgenic locus, while the primers 1154A.S32 and 1154A.S34, (5′ CTTTCTAACGGTACGCAGCAG 3′) and (5′ GCTCCAGTTCATGTCGATAGAC 3′) respectively, were used to identify the presence of a wild type gFLE locus. Primers for analysis of gZBDA18 and gZBDD15 loci are listed above (see Identifying and Validating Genomic Insertion Site of gZBD Transgenes in Methods).
Crosses to Generate F1 gRNA/Cas9 Hybrids.
For all crosses, pupae were fluorescently sorted and sexed, and allowed to emerge as adults in separate cages to ensure female virginity before crossing. Unless otherwise indicated, crosses of 50 males × 50 females were set up on Day 1 with 2- to 4-d-old adults, allowed to mate ad libitum, then blood fed on day 6. The crosses to generate the (+/gZBD; +/Cas9) genotype in SI Appendix, Fig. S1 were generated with maternal Cas9 paternal gRNA F0 directionality, while the crosses to generate the same genotype of males in SI Appendix, Fig. S2 used the reciprocal cross. The crosses to generate the F1 pgSIT (+/gFLE;gZBD; +/Cas9) genotype were performed with F0 Cas9 females and gRNA males for mutation analysis in SI Appendix, Fig. S3, but following identification of the lethal phenotype of this cross directionality, all subsequent crosses to generate this genotype used the Cas9 male and gRNA female directionality (Figs. 1 C and D and 2 B and C).
Quantifying Female Elimination of F1 (+/gFLE;gZBD; +/Cas9).
F1 (+/gRNA; +/Cas9) hybrids generated with maternal gRNA and paternal Cas9 were sex-sorted daily as pupae. Counts of males and females were recorded starting the first day pupation is observed until the day all larvae had become pupae (Fig. 1B and Dataset S4), typically a 4 to 6 day timespan. Male and female pupae were placed in separate cages to emerge as adults, and the survival of female pupae was closely monitored. Females who emerged as adults and were able to fly were crossed to 50 wild type adult males and allowed to mate ad libitum, they were observed during blood feeding for their ability to take blood meal, and given an egg dish 48 h post blood feed.
Testes Dissections.
Four-day-old adult virgin males were immobilized on ice for <1 h, and the lower reproductive tract was dissected into PBS by pulling slowly from the claspers. Images were taken with a Leica M165FC fluorescent stereomicroscope outfitted with a Leica DMC2900 camera under 6.5× magnification. Lighting orientation, brightness, exposure time, and white balance were not controlled for, so no conclusions about tissue color, brightness, or tone should be made from these images (SI Appendix, Fig. S4A).
Mating Plug Transfer Assay.
To determine whether or not pgSIT males could transfer a mating plug, we crossed 100 pgSITD15 or wild type 5 to 7 d-old virgin males to 100 wild type virgin 5- to 7-d-old females at dusk when males were swarming. We allowed them to mate ad libitum for 45 min, during which we verified the presence of copulating pairs at the bottom of the cage. After 45 min, many females were removed onto ice. The terminal abdominal segments of females were imaged, ventral side up, with a Leica M165FC fluorescent stereomicroscope and a CFP/YFP/mCherry triple filter (SI Appendix, Fig. S4B). The presence of the mating plug could be seen through the female cuticle by autofluorescence within the female atrium—a previously established assay for verifying mating plug transfer (66, 108). Lighting orientation, brightness, exposure time, and white balance were not controlled for, so conclusions about plug brightness should not be made from these images.
Male Adult Survival Assay.
Seventeen male pupae were put into each small Bugdorm cage on day 0. This number of males was selected to minimize crowding and competition between males. On day 1, the number of dead pupae or drowned adults was counted and removed, but were not included in survival curve counts. From day 2 onward the number of adult dead males were counted, removed, and recorded each day. All cages within a replicate were summed for the final survival curve analysis, yielding a total of 110 wild type males and 81 pgSITD15 males analyzed. At the end of the assay, for cages that had no more living mosquitoes but had individuals that were unaccounted for (4 cages out of 12 cages total), the unaccounted individuals were censored on the final day of the survival curve analysis and marked as censorship notches in SI Appendix, Fig. S4C. Raw survival counts broken down by cage can be found in Dataset S6.
Insertion Site Mapping by Nanopore.
Insertion sites for gFLE and Cas9 transgenes were previously determined to be located at 2R(NT_078266.2): 23,279,556-23,279,559 and 2L(NT_078265.2):10,326,500-10,326,503, respectively (57). To determine the insertion site for the gZBD transgene, we performed Oxford Nanopore sequencing of genomic DNA from adult transheterozygous pgSITD15 males harboring the +/gZBD transgene in addition to the +/Cas9 and +/gFLE transgenes. DNA was extracted in pools of 6 to 8 adult mosquitoes using the Blood & Cell Culture DNA Midi Kit (Qiagen, Cat# 13343) following the manufacturer’s protocol. The sequencing library was prepared using the Oxford Nanopore SQK-LSK110 genomic library kit and sequenced on a single MinION flowcell (R9.4.1) for 72 h. Basecalling was performed with ONT Guppy basecalling software version 6.4.6 using dna_r9.4.1_450bps_sup model generating 3.92 million reads above the quality threshold of Q≧10 with N50 of 6,608 bp and total yield of 14.43 Gb.
To identify transgene insertion sites, nanopore reads were aligned to the gZBD plasmid sequence (Plasmid #1114H, Addgene #200640 (99)) using minimap2 (109). Reads mapped to the plasmids were extracted and mapped to the A. gambiae genome (GCF_000005575.2_AgamP3). Exact insertion sites were determined by examining read alignments in Interactive Genomics Viewer (IGV). The gZBDD15 transgene is integrated between positions 4,828,892 and 4,828,896 on chromosome 3L (NT_078267.5). The site is located in the intergenic region between AGAP010485 and AGAP010486. The previously determined integration sites for gFLE and Cas9 transgenes were confirmed with the nanopore data. The nanopore sequencing data have been deposited to the NCBI sequence read archive (PRJNA978105).
Sequencing of gRNA Expression Cassettes.
gDNA from gZBDD15 and pgSITD15 were extracted (Qiagen, DNeasy Blood & Tissue Kits, Cat. No./ID: 69504) from pools of three adults, PCR amplified (Q5 HotStart DNA polymerase (NEB, Cat. No./ID: M0493L)), and Sanger sequenced for the 7 gRNA expression cassettes. The gRNAdsxF.2 cassette was amplified and sequenced with the 1114H.S7 (5′ CGGTTTTGTTTGCAGCGAGTTGTG 3′) and aa151 (5′ GGTAATCGATTTTTTCAGTGCAG 3′) primers. The gRNAzpg.1, gRNAB2.1, gRNAB2.2, gRNAfle.7, and gRNAfle.10 expression cassettes were amplified and sequenced all together with the 1114H.S1 (5′ CTCAAAATTTCTTCTATAAAGTAACAAAAC 3′) and 1114G.C2 (5′ CGAGGTTCTCCTTATGCTCTGTG 3′) primers. PCR amplicons were run on 1% agarose gel at 120 V for 20 min, and then gel was extracted with the Zymoclean Gel DNA Recovery Kit (Zymo Research, Cat. No./ID: D4007).
Target Site Mutation Analysis.
Mutations under the gRNA target sites were identified in F1 (+/gFLE;gZBDD15; +/Cas9) hybrid offspring resulting from a cross between 50 (gFLE;gZBDD15) males and 50 Cas9 females. The male–female directionality of this F0 cross was chosen because Cas9 females provide maternal deposits of Cas9 protein into the embryo, producing F1 hybrid offspring with a high mosaic mutation load and allowing for sequencing of many mutant alleles. F1 hybrid offspring were collected in bulk as late-stage embryos and were DNA extracted (Qiagen, DNeasy Blood & Tissue Kits, Cat. No./ID: 69504) and PCR amplified [Q5 HotStart DNA polymerase (NEB, Cat. No./ID: M0493L)]. The zpg locus was amplified with the 114H.S34 and 1114H.S37; 1114H.S34 (5′ GTAGAAAGAGCAAGGAAAGAAACG 3′) and 1114H.S37 (5′ GTTCCGAATTTCCAAGTGCTTC 3′) primers respectively. The β2-tubulin locus was amplified with the 1114H.S38 (5′ GCTAAATATCAGACGGCTTTC 3′) and 1114H.S39 (5′ GCGAATTTTCGAAATCAGCAG 3′) primers. The dsxF locus was amplified with the 1114E.S33 (5′ CTTGCCATCCTATGGAACTGC 3′) and 1114E.S32 (5′ GGTGAAAATATTGTTGATGCGC 3′) primers. The fle locus was amplified with the aa174 (5′ CGACTCACTATAGGGAGAGCGGC 3′) and aa175 (5′ AAGAACATCGATTTTCCATGGCAG 3′) primers (57). PCR amplicons were run on 1% agarose gel at 120 V for 20 min, and then gel was extracted with the Zymoclean Gel DNA Recovery Kit (Zymo Research, Cat. No./ID: D4007). Purified amplicons were then cloned into the pJET vector (Thermo Scientific, Cat. No. / ID: K1231), transformed into chemically competent Escherichia coli (Promega, JM109), and plated on LB-Ampicillin plates. Plates were sent for Sanger Colony sequencing with universal primers PJET1-2F (5′ CGACTCACTATAGGGAGAGCGGC 3′) and/or PJET 1-2R (5′ AAGAACATCGATTTTCCATGGCAG 3′), with each colony representing a single PCR amplicon from an individual mutant allele.
Because their mutation frequency was qualitatively weaker, to enrich for mutant alleles under gRNAB2.1, gRNAB2.2, and gRNAfle.7, their genomic target sites were PCR-amplified, and these PCR amplicons were digested with a restriction enzyme whose recognition site overlaps the expected gRNA cut sites, such that an undigestible PCR product indicates a likely CRISPR mutation (SI Appendix, Fig. S3) (110). The β2-tubulin locus was amplified with the 1114A.S43 (5′ GAGAGCAACACTCGTGCG 3′) and 1114A.S44 (5′CAGGGTGGCATTGTACG 3′) primers and the amplicon was digested with FspI (NEB cat#R0135S) or DdeI (NEB cat#R0175S) to identify mutations by gRNAB2.1 and gRNAB2.2 respectively. To identify mutations by gRNAfle.7, the fle locus was amplified with the 1154A.S23 (5′ CTCAGCAAGCAGTATGCCAAC 3′) and 1154A.S8 (5′ GTTGAACGCTTCGTCGTACG 3′) primers, and the amplicon was digested with BseYI (NEB cat# R0635S). All PCRs were performed using Q5 HotStart DNA polymerase (NEB, Cat. No./ID: M0493L). Digestions were performed at 37 °C for 1 h, then run on 1% agarose gel at 120 V for 25 min. Undigested bands corresponding to mutant PCR products were gel extracted with the Zymoclean Gel DNA Recovery Kit (Zymo Research, Cat. No./ID: D4007), then cloned into pJET vectors (Thermo Scientific, Cat. No./ID: K1231), transformed into chemically competent E. coli (Promega, JM109), and plated on LB-Ampicillin plates. Plates were sent for Sanger Colony sequencing with universal primers PJET1-2F (5′ CGACTCACTATAGGGAGAGCGGC 3′) and/or PJET 1-2R (5′ AAGAACATCGATTTTCCATGGCAG 3′), with each colony representing a single PCR amplicon from an individual mutant allele. Sequences were compared to the reference genome sequences of AGAP006241, AGAP008622, AGAP004050, AGAP013051 for zpg, β2-tubulin, dsxF, and fle respectively (SI Appendix, Fig. S3).
Population Suppression Assays.
On day 1 of experimentation, cages were seeded with 0, 50, 100, 250, or 500 virgin 2- to 4-d-old pgSIT males (for release ratios of 0:1, 1:1, 2:1, 5:1, and 10:1, respectively) intermixed with 50 virgin 2- to 4-d-old wild type males. Then, 50 2- to 4-d-old virgin wild type females were aspirated into the cage. Adults were allowed to mate ad libitum, then blood-fed on a mouse on day 6. A wet filter paper (oviposition site) was provided on day 8, and eggs were allowed to develop and hatch undisturbed. Hatched larvae were counted on days 11, 12, and 13 and screened for fluorescence, which would indicate a fertile pgSIT father. Egg shells were counted on days 13 and 14. Only replicates which yielded >1,000 eggs were included to guarantee ample representation of male contribution. Each data point represents the counts from a single distinct cross cage; individual cages were not scored multiple times.
Mathematical Modeling.
We used the MGDrivE 3 framework (111) to simulate releases of A. gambiae pgSIT eggs to suppress mosquitoes in the Upper River region of The Gambia. MGDrivE 3 is a modular framework for simulating releases of genetic control systems in spatially structured mosquito populations which includes modules for inheritance, life history, and epidemiology. The inheritance pattern of the pgSIT system was modeled within the inheritance module of MGDrivE (112). Based on laboratory data, we assumed the pgSIT system in A. gambiae would induce complete male sterility and female inviability, with inviability being manifest at hatching. We assumed that pgSIT eggs would be introduced into the environment in cups with sufficient water volume and larval resources such that larval mortality would be density-independent. Survival of eggs released in cups was determined by expected juvenile life stage durations and daily mortality rates (Dataset S7) leading to a viable emergence rate of 26% for male eggs. Offspring of pgSIT sterile males are unviable at the egg stage. Based on semi-field data for SIT anophelines, we assumed a 62.5% reduction in pgSIT male mating competitiveness (113). To be conservative, we also assumed a 25% reduction in pgSIT male lifespan compared to wild-type males, despite no reductions in lifespan being observed in this work (58, 61).
The MGDrivE 3 framework (111) models the development of mosquitoes from egg to larvae to pupae to adult with overlapping generations, larval mortality increasing with larval density (114), and a mating structure in which females retain the genetic material of the adult male with whom they mate for the duration of their adult lifespan. Life history of A. gambiae was modeled using standard bionomic parameters (Dataset S7) and seasonality in larval carrying capacity driven by rainfall data from the Upper River region of The Gambia (https://www.chc.ucsb.edu/data/chirps). To smooth the seasonal profile of the raw rainfall data, we leveraged a Fourier analysis-based approach that involves fitting a mixture of sinusoids to the raw data (https://github.com/mrc-ide/umbrella). Entomological data from the Upper River region (71) suggested vector breeding sites in this region are substantially more abundant in the rainy season than in the dry season, suggesting larval carrying capacity in the dry season was ~10% that of the peak rainy season. We calibrated the model to malaria prevalence data from a randomized-controlled trial of mass drug intervention in the Upper River region (70) by linking MGDrivE 3 (111) to the Imperial College London (ICL) malaria model (115, 116) by allowing forces of infection (i.e., the probability of infection from mosquito-to-human and human-to-mosquito per individual per unit time) to be exchanged between the two models. Weekly releases of pgSIT A. gambiae eggs were simulated from the beginning of the rainy season (June 1st), for a variable number of weeks and release sizes.
Ethical Conduct of Research.
All animals were handled in accordance with the Guide for the Care and Use of Laboratory Animals as recommended by the NIH and approved by the UCSD Institutional Animal Care and Use Committee (IACUC, Animal Use Protocol #S17187) and UCSD Biological Use Authorization (BUA #R2401).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Acknowledgments
We thank Judy Ishikawa for helping with mosquito husbandry and Akshay Bharadwaj for laboratory assistance. This work was supported in part by funding from an NIH award (R01AI151004), EPA (Award #RD84020401), and an Open Philanthropy award (309937-0001) awarded to O.S.A.; and by funds from the Bill & Melinda Gates Foundation (INV-017683) awarded to J.M.M. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the U.S. government. Figures were made with https://Biorender.com.
Author contributions
J.M.M. and O.S.A. designed research; and R.A.A., A.L.S., J.J.P., M.L.C., S.C., A.M., H.M.S.C., I.A., and J.M.M. performed research.
Competing interests
O.S.A. is a founder of Agragene, Inc. and Synvect, Inc. with equity interest. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. All other authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission. M.J.S. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Complete sequence maps and plasmids are deposited at https://Addgene.org (200640) (99). All Nanopore sequencing data have been deposited to the NCBI sequence read archive (PRJNA978105) (117). All data used to generate figures are provided in supporting information. A. gambiae transgenic lines are available upon request to O.S.A.
Supporting Information
References
- 1.World Health Organization, “World Malaria Report 2021” (WHO, 2021).
- 2.Willyard C., The slow roll-out of the world’s first malaria vaccine. Nature 612, S48–S49 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Egbewande O. M., The RTS, S malaria vaccine: Journey from conception to recommendation. Public Health Pract. 4, 100283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranson H., Lissenden N., Insecticide resistance in African anopheles mosquitoes: A worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 32, 187–196 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kyrou K., et al. , A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champer J., Buchman A., Akbari O. S., Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Emerging technology: concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs S., et al. , Resistance to a CRISPR-based gene drive at an evolutionarily conserved site is revealed by mimicking genotype fixation. PLoS Genet. 17, e1009740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond A. M., et al. , The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13, e1007039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noble C., Adlam B., Church G. M., Esvelt K. M., Nowak M. A., Current CRISPR gene drive systems are likely to be highly invasive in wild populations. Elife 7, e33423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James S., et al. , Pathway to Deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of malaria in Sub-Saharan Africa: Recommendations of a scientific working group. Am. J. Trop. Med. Hyg. 98, 1–49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kofler N., et al. , Editing nature: Local roots of global governance. Science 362, 527–529 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Long K. C., et al. , Core commitments for field trials of gene drive organisms. Science 370, 1417–1419 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Oye K. A., et al. , Biotechnology. Regulating gene drives. Science 345, 626–628 (2014). [DOI] [PubMed] [Google Scholar]
- 15.National Academies of Sciences, Engineering, and Medicine, Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values (The National Academies Press, Washington, DC, 2016). [PubMed] [Google Scholar]
- 16.Taitingfong R. I., et al. , Exploring the value of a global gene drive project registry. Nat. Biotechnol. 41, 9–13 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Raban R., Marshall J. M., Hay B. A., Akbari O. S., Manipulating the destiny of wild populations using CRISPR. Annu. Rev. Genet. 57, 361–390 (2023), 10.1146/annurev-genet-031623-105059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyck V. A., Hendrichs J., Robinson A. S., Sterile Insect Technique: Principles and Practice in Area-wide Integrated Pest Management (Taylor & Francis, 2021). [Google Scholar]
- 19.Krafsur E. S., Whitten C. J., Novy J. E., Screwworm eradication in North and Central America. Parasitol. Today 3, 131–137 (1987). [DOI] [PubMed] [Google Scholar]
- 20.Vreysen M. J., et al. , Glossina austeni (Diptera: Glossinidae) eradicated on the island of Unguja, Zanzibar, using the sterile insect technique. J. Econ. Entomol. 93, 123–135 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Koyama J., Kakinohana H., Miyatake T., Eradication of the melon fly, Bactrocera cucurbitae, in Japan: Importance of behavior, ecology, genetics, and evolution. Annu. Rev. Entomol. 49, 331–349 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Castanera P., Control integrado de la mosca mediterránea de la fruta, Ceratitis capitata (Wiedemann) (Diptera: Tephritidae) en cítricos. Phytoma España 153, 131–133 (2003). [Google Scholar]
- 23.Zheng X., et al. , Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572, 56–61 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Rendón P., McInnis D., Lance D., Stewart J., Medfly (Diptera: Tephritidae) genetic sexing: Large-scale field comparison of males-only and bisexual sterile fly releases in Guatemala. J. Econ. Entomol. 97, 1547–1553 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Robinson A. S., Mutations and their use in insect control. Mutat. Res. 511, 113–132 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Thailayil J., Magnusson K., Godfray H. C. J., Crisanti A., Catteruccia F., Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 108, 13677–13681 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdel-Malek A. A., Tantawy A. O., Wakid A. M., Studies on the eradication of Anopheles pharoensis by the sterile-male technique using cobalt-60. 3. Determination of the sterile dose and its biological effects on different characters related to “fitness” components. J. Econ. Entomol. 60, 20–23 (1967). [DOI] [PubMed] [Google Scholar]
- 28.Sharma V. P., Razdan R. K., Ansari M. A., Anopheles stephensi: Effect of gamma-radiation and chemosterilants on the fertility and fitness of males for sterile male releases. J. Econ. Entomol. 71, 449–450 (1978). [DOI] [PubMed] [Google Scholar]
- 29.Ageep T. B., et al. , Participation of irradiated Anopheles arabiensis males in swarms following field release in Sudan. Malar. J. 13, 484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreasen M. H., Curtis C. F., Optimal life stage for radiation sterilization of Anopheles males and their fitness for release. Med. Vet. Entomol. 19, 238–244 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Maïga H., et al. , Mating competitiveness of sterile male Anopheles coluzzii in large cages. Malar. J. 13, 460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monaghan P., Metcalfe N. B., Torres R., Oxidative stress as a mediator of life history trade-offs: Mechanisms, measurements and interpretation. Ecol. Lett. 12, 75–92 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Proverbs M. D., Induced sterilization and control of insects. Annu. Rev. Entomol. 14, 81–102 (1969). [DOI] [PubMed] [Google Scholar]
- 34.Damiens D., Vreysen M. J. B., Gilles J. R. L., Anopheles arabiensis sperm production after genetic manipulation, dieldrin treatment, and irradiation. J. Med. Entomol. 50, 314–316 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Abdeu-malek A. A., Tantawy A. O., Wakid A. M., Others, Studies on the eradication of Anopheles pharoensis by the sterile-male technique using cobalt-60. III. Determination of the sterile dose and its biological effects on different characters related to “fitness” components. J. Econ. Entomol. 60, 20–23 (1967). [DOI] [PubMed] [Google Scholar]
- 36.Helinski M. E. H., Parker A. G., Knols B. G. J., Radiation biology of mosquitoes. Malar. J. 8, S6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manoukis N. C., et al. , Structure and dynamics of male swarms of Anopheles gambiae. J. Med. Entomol. 46, 227–235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helinski M. E. H., Knols B. G. J., Mating competitiveness of male Anopheles arabiensis mosquitoes irradiated with a partially or fully sterilizing dose in small and large laboratory cages. J. Med. Entomol. 45, 698–705 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Munhenga G., et al. , Mating competitiveness of sterile genetic sexing strain males (GAMA) under laboratory and semi-field conditions: Steps towards the use of the Sterile Insect Technique to control the major malaria vector Anopheles arabiensis in South Africa. Parasit. Vectors 9, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein T. A., Windbichler N., Deredec A., Burt A., Benedict M. Q., Infertility resulting from transgenic I-PpoI male Anopheles gambiae in large cage trials. Pathog. Glob. Health 106, 20–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Windbichler N., Papathanos P. A., Crisanti A., Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 4, e1000291 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto D. S., et al. , A synthetic male-specific sterilization system using the mammalian pro-apoptotic factor in a malaria vector mosquito. Sci. Rep. 9, 8160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smidler A. L., et al. , CRISPR-mediated germline mutagenesis for genetic sterilization of Anopheles gambiae males. Sci. Rep. 14, 4057 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dandalo L. C., Munhenga G., Kaiser M. L., Koekemoer L. L., Development of a genetic sexing strain of Anopheles arabiensis for KwaZulu-Natal, South Africa. Med. Vet. Entomol. 32, 61–69 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Yamada H., et al. , The Anopheles arabiensis genetic sexing strain ANO IPCL1 and its application potential for the sterile insect technique in integrated vector management programmes. Acta Trop. 142, 138–144 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Baselt R., Encyclopedia of toxicology. J. Anal. Toxicol. 38, 464–464 (2014). [Google Scholar]
- 47.Curtis C. F., Genetic sex separation in Anopheles arabiensis and the production of sterile hybrids. Bull. World Health Organ. 56, 453–454 (1978). [PMC free article] [PubMed] [Google Scholar]
- 48.Crawford J. E., et al. , Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 38, 482–492 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Zha T., et al. , “Predictive models for visually classifying insects.” US Patent 17863802.9 (2021).
- 50.Lepek H., et al. , “Method for sex sorting of mosquitoes and apparatus therefor.” World Patent WO2018134829 (2018).
- 51.Magnusson K., et al. , Transcription regulation of sex-biased genes during ontogeny in the malaria vector Anopheles gambiae. PLoS One 6, e21572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernardini F., et al. , Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc. Natl. Acad. Sci. U.S.A. 111, 7600–7605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catteruccia F., Benton J. P., Crisanti A., An Anopheles transgenic sexing strain for vector control. Nat. Biotechnol. 23, 1414–1417 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Marois E., et al. , High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malaria J. 11, 302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galizi R., et al. , A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci. Rep. 6, 31139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galizi R., et al. , A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5, 3977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smidler A. L., et al. , A confinable female-lethal population suppression system in the malaria vector. Sci. Adv. 9, eade8903 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kandul N. P., et al. , Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun. 10, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kandul N. P., et al. , Precision guided sterile males suppress populations of an invasive crop pest. GEN Biotechnol. 1, 372–385 (2022). [Google Scholar]
- 60.Li M., et al. , Targeting sex determination to suppress mosquito populations. bioRxiv [Preprint] (2023). 10.1101/2023.04.18.537404 (Accessed 30 May 2023). [DOI] [PMC free article] [PubMed]
- 61.Li M., et al. , Suppressing mosquito populations with precision guided sterile males. Nat. Commun. 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lloyd V. K., Dyment D., Sinclair D. A. R., Grigliatti T. A., Different patterns of gene silencing in position-effect variegation. Genome 46, 1104–1117 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Werling K., et al. , Steroid hormone function controls non-competitive plasmodium development in Anopheles. Cell 177, 315–325.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alphey L., et al. , Sterile-insect methods for control of mosquito-borne diseases: An analysis. Vector Borne Zoonotic Dis. 10, 295–311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers D. W., et al. , Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLoS Biol. 7, e1000272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gabrieli P., et al. , Sexual transfer of the steroid hormone 20E induces the postmating switch in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 111, 16353–16358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shutt B., Stables L., Aboagye-Antwi F., Moran J., Tripet F., Male accessory gland proteins induce female monogamy in anopheline mosquitoes. Med. Vet. Entomol. 24, 91–94 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Carvalho D. O., et al. , Suppression of a field population of aedes aegypti in brazil by sustained release of transgenic male mosquitoes. PLoS Negl. Trop. Dis. 9, e0003864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mondal A., Sánchez C. H. M., Marshall J. M., MGDrivE 3: A decoupled vector-human framework for epidemiological simulation of mosquito genetic control tools and their surveillance. bioRxiv [Preprint] (2023). 10.1101/2023.09.09.556958 (Accessed 30 May 2023). [DOI] [PMC free article] [PubMed]
- 70.Dabira E. D., et al. , Mass drug administration of ivermectin and dihydroartemisinin-piperaquine against malaria in settings with high coverage of standard control interventions: a cluster-randomised controlled trial in The Gambia. Lancet Infect. Dis. 22, 519–528 (2022). [DOI] [PubMed] [Google Scholar]
- 71.Soumare H. M., et al. , Entomological impact of mass administration of ivermectin and dihydroartemisinin-piperaquine in The Gambia: A cluster-randomized controlled trial. Parasit. Vectors 15, 435 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eckhoff P. A., Wenger E. A., Godfray H. C. J., Burt A., Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl. Acad. Sci. U.S.A. 114, E255–E264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kandul N. P., et al. , Assessment of a split homing based gene drive for efficient knockout of multiple genes. G3 10, 827–837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belote J. M., Baker B. S., The dual functions of a sex determination gene in Drosophila melanogaster. Dev. Biol. 95, 512–517 (1983). [DOI] [PubMed] [Google Scholar]
- 75.Neafsey D. E., et al. , Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernardini F., Kriezis A., Galizi R., Nolan T., Crisanti A., Introgression of a synthetic sex ratio distortion system from Anopheles gambiae into Anopheles arabiensis. Sci. Rep. 9, 5158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Besansky N. J., et al. , Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. U.S.A. 100, 10818–10823 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weill M., et al. , The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol. Biol. 9, 451–455 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Caputo B., et al. , The “far-west” of Anopheles gambiae molecular forms. PLoS One 6, e16415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mancini E., et al. , Adaptive potential of hybridization among malaria vectors: Introgression at the immune locus TEP1 between Anopheles coluzzii and A. gambiae in “far-west” Africa. PLoS One 10, e0127804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Courtier-Orgogozo V., Danchin A., Gouyon P.-H., Boëte C., Evaluating the probability of CRISPR-based gene drive contaminating another species. Evol. Appl. 13, 1888–1905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Labbé G. M. C., Scaife S., Morgan S. A., Curtis Z. H., Alphey L., Female-specific flightless (fsRIDL) phenotype for control of Aedes albopictus. PLoS Negl. Trop. Dis. 6, e1724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marinotti O., et al. , Development of a population suppression strain of the human malaria vector mosquito, Anopheles stephensi. Malar. J. 12, 142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Halliday S., Oxitec Successfully Completes First Field Deployment of 2nd Generation FriendlyTM Aedes aegypti Technology. https://www.oxitec.com/en/news/oxitec-successfully-completes-first-field-deployment-of-2nd-generation-friendly-aedes-aegypti-technology. Accessed 30 January 2023.
- 85.Giraldo-Calderón G. I., et al. , VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kandul N. P., Liu J., Akbari O. S., Temperature-inducible precision-guided sterile insect technique. CRISPR J. 4, 822–835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yao F. A., et al. , Mark-release-recapture experiment in Burkina Faso demonstrates reduced fitness and dispersal of genetically-modified sterile malaria mosquitoes. Nat. Commun. 13, 796 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schliekelman P., Ellner S., Gould F., Pest control by genetic manipulation of sex ratio. J. Econ. Entomol. 98, 18–34 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Hammond A., et al. , A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gantz V. M., Bier E., The dawn of active genetics. Bioessays 38, 50–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas D. D., Donnelly C. A., Wood R. J., Alphey L. S., Insect population control using a dominant, repressible, lethal genetic system. Science 287, 2474–2476 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Lutrat C., et al. , Sex sorting for pest control: It’s raining men!. Trends Parasitol. 35, 649–662 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Marina C. F., et al. , Comparison of ground release and drone-mediated aerial release of Aedes aegypti sterile males in southern Mexico: Efficacy and challenges. Insects 13, 347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Virginio J. F., Gómez M., Almeida F., Pedrosa M. C., Drone-based aerial release mechanism for mosquitoes. WeRobotics: Geneva.
- 95.Weng S.-C., et al. , Efficient sex separation by exploiting differential alternative splicing of a dominant marker in Aedes aegypti. PLoS genetics 19, e1011065 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davydova S., et al. , Next-generation genetic sexing strain establishment in the agricultural pest Ceratitis capitata. Sci. Rep. 13, 19866 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J., Rayes D., Akbari O. S., A fluorescent sex-sorting technique for insects with the demonstration in Drosophila melanogaster. GEN Biotechnol. 3, 35–44 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benedict M. Q., The MR4 methods in Anopheles research laboratory manual. (Atlanta, CDC, 2018).
- 99.Smidler A., Li T., Plasmid 1114H. AddGene. https://www.addgene.org/200640/. Deposited 28 August 2023.
- 100.Ho N., Gesing S., Consortium, VectorBase: an updated bioinformatics rerefrefsource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Mol. Biol. 43, D707–D713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smidler A., “CRISPR-based innovative genetic tools for control of Anopheles gambiae mosquitoes,” Thesis, Harvard University; (2019) (July 25, 2022). [Google Scholar]
- 102.Pai J., Development of PgSIT and Gynecider, confinable population suppression systems in Anopheles Gambiae (2022).
- 103.Volohonsky G., et al. , Tools for Anopheles gambiae transgenesis. G3 5, 1151–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fuchs S., Nolan T., Crisanti A., Mosquito transgenic technologies to reduce Plasmodium transmission. Methods Mol. Biol. 923, 601–622 (2013). [DOI] [PubMed] [Google Scholar]
- 105.Pondeville E., et al. , Efficient ΦC31 integrase–mediated site-specific germline transformation of Anopheles gambiae. Nat. Protoc. 9, 1698–1712 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Staff M. R., “Anopheles laboratory biology and culture” in Methods in Anopheles Research, ed. 1. Atlanta. [Google Scholar]
- 107.Handler A. M., McCombs S. D., Fraser M. J., Saul S. H., The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc. Natl. Acad. Sci. U.S.A. 95, 7520–7525 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smidler A. L., Scott S. N., Mameli E., Shaw W. R., Catteruccia F., A transgenic tool to assess Anopheles mating competitiveness in the field. Parasit. Vectors 11, 651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smidler A. L., Terenzi O., Soichot J., Levashina E. A., Marois E., Targeted mutagenesis in the malaria mosquito using TALE nucleases. PLoS One 8, e74511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu S. L., et al. , MGDrivE 2: A simulation framework for gene drive systems incorporating seasonality and epidemiological dynamics. PLoS Comput. Biol. 17, e1009030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sánchez C H. M., Wu S. L., Bennett J. B., Marshall J. M., MGDrivE: A modular simulation framework for the spread of gene drives through spatially explicit mosquito populations. Methods Ecol. Evol. 11, 229–239 (2020). [Google Scholar]
- 113.Bouyer J., Vreysen M. J. B., Yes, irradiated sterile male mosquitoes can be sexually competitive! Trends Parasitol. 36, 877–880 (2020). [DOI] [PubMed] [Google Scholar]
- 114.White M. T., et al. , Modelling the impact of vector control interventions on Anopheles gambiae population dynamics. Parasit. Vectors 4, 153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Griffin J. T., et al. , Reducing Plasmodium falciparum malaria transmission in Africa: A model-based evaluation of intervention strategies. PLoS Med. 7, e1000324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Griffin J. T., et al. , Potential for reduction of burden and local elimination of malaria by reducing Plasmodium falciparum malaria transmission: A mathematical modelling study. Lancet Infect. Dis. 16, 465–472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Apte R., et al. , gZBD insertion in A. gambiae. NCBI. https://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA978105. Deposited 31 May 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Data Availability Statement
Complete sequence maps and plasmids are deposited at https://Addgene.org (200640) (99). All Nanopore sequencing data have been deposited to the NCBI sequence read archive (PRJNA978105) (117). All data used to generate figures are provided in supporting information. A. gambiae transgenic lines are available upon request to O.S.A.