Abstract
Recent drug regimens have had much success in the treatment of human immunodeficiency virus (HIV)-infected individuals; however, the incidence of resistance to such drugs has become a problem that is likely to increase in importance with long-term therapy of this chronic illness. An analysis and understanding of the molecular interactions between the drug(s) and the mutated viral target(s) is crucial for further progress in the field of AIDS therapy. The protease inhibitor amprenavir (APV) generates a signature set of HIV type 1 (HIV-1) protease mutations associated with in vitro resistance (M46I/L, I47V, and I50V [triple mutant]). Passage of the triple-mutant APV-resistant HIV-1 strain in MT4 cells, in the presence of increasing concentrations of saquinavir (SQV), gave rise to a new variant containing M46I, G48V, I50V, and I84L mutations in the protease and a resulting phenotype that was resistant to SQV and, unexpectedly, resensitized to APV. This phenotype was consistent with a subsequent kinetic analysis of the mutant protease, together with X-ray crystallographic analysis and computational modeling which elucidated the structural basis of these observations. The switch in protease inhibitor sensitivities resulted from (i) the I50V mutation, which reduced the area of contact with APV and SQV; (ii) the compensating I84L mutation, which improved hydrophobic packing with APV; and (iii) the G-to-V mutation at residue 48, which introduced steric repulsion with the P3 group of SQV. This analysis establishes the fine detail necessary for understanding the loss of protease binding for SQV in the quadruple mutant and gain in binding for APV, demonstrating the powerful combination of virology, molecular biology, enzymology, and protein structural and modeling studies in the elucidation and understanding of viral drug resistance.
The aim of this work was to establish an understanding, at the atomic and molecular level, of the shifting human immunodeficiency virus (HIV) protease resistance phenotype established when two protease inhibitors (amprenavir [APV] and saquinavir [SQV]) were sequentially added in an in vitro viral resistance experiment. APV is a novel arylsulfononyl diaminopropane peptidomimetic of the HIV protease inhibitor (PI) class. This compound is a potent, low-molecular-weight, orally bioavailable drug with excellent pharmacokinetic characteristics and a distinct in vitro resistance pattern (8, 14). The field of work relating to HIV replication and PI-associated resistance is large and ever increasing (for reviews, see references 1, 2, and 13 and references therein). The signature set of HIV type 1 (HIV-1) protease mutations associated with in vitro resistance to APV are M46I/L, I47V, and I50V, the last of which distinguishes APV from other PIs in terms of its resistance profile. In general, in vitro experiments involving pairs of PIs have demonstrated increases in phenotypic resistance to both agents associated with alterations in the protease sequence consistent with those associated with each individual PI (data not shown). In contrast, when a clonal APV-resistant HIV-1 strain (containing mutations at positions 46, 47 and 50 [14]) was passaged in the presence of increasing concentrations of SQV, the triple-mutant virus (M46I, I47V, I50V) further mutated. The resulting predominant HIV-1 variant encoded a protease with the mutations M46I, G48V, I50V, and I84L (Table 1). Phenotypic analysis of a clonally pure SQV-selected variant demonstrated an expected increase in the 50% inhibitory concentration (IC50) of SQV from 14 to 1,655 nM but an unexpected 40-fold resensitization to APV (with the IC50 decreasing from 2,135 to 55 nM), resulting in a return to wild-type virus sensitivity to APV.
TABLE 1.
In vitro passaging of APV-resistant HIV-1 in the presence of SQVa
| Passage no.b | IC50 (nM)c of:
|
Mutations in protease | |
|---|---|---|---|
| APV | SQV | ||
| 0 | 2,135 | 14 | 46I,47V,50V |
| 6 | 55 | 1655 | 46I,48V,50V,84L |
The method is detailed in reference 14. MT4 cells were infected with HIVHXB2 containing the APV resistance-conferring triple mutation in the protease gene (46I, 47V, and 50V) at a multiplicity of infection of <0.01 50% tissue culture infective dose per cell, and virus was passaged at increasing concentrations of SQV (40 nM for three passages, 80 nM for one passage, and 160 nM for two passages). Cultures were examined daily for viral cytopathic effect, with fresh compound and medium being added every 7 days. Samples were collected for viral titer and IC50 determination and viral DNA sequencing.
The SQV passaging experiment took 87 days.
Assays of virus infectivity and susceptibility were performed with MT4 cells using the MTT cell viability reagent. For each assay, 10 concentrations of each compound were used over a twofold dilution range, in quadruplicate. The IC50 is the concentration of compound that protected 50% of the cells from virus killing.
Mutant protease generation, crystallization, and enzymatic analysis.
A synthetic HIV-1 protease gene encoding the 46I, 48V, 50V, 84L mutations was generated using standard methods of PCR mutagenesis (4). Essentially, internal mutagenic primers 46/48/50 top (5′ CCGAAGATCATTGTTGGCGTTGGTGGT 3′) and 46/48/50 bot (5′ ACCACCAACGCCAACAATGATCTTCGG 3′) or 84 top 5′ (CCTGTAAACCTGATTGGTCGTAAC 3′) and 84 bot 5′ (GTTACGACCAATCAGGTTTACAGG 3′) were mixed with appropriate external primers, the HIV-1 protease gene, nucleotides, and Taq polymerase. PCRs were undertaken to generate two mutant protease genes containing the 46I, 48V, and 50V mutations together and the 84L single mutation separately, which were then recombined via an intervening unique restriction enzyme site (KpnI). Complete sequencing of the fully mutated gene demonstrated that only the desired mutations were present. An NdeI-EcoRI fragment containing the open reading frame for the mutated protease was recloned into the expression vector pSPC27 (10). The quadruple-mutant protease was expressed in Escherichia coli BL21DE3 in a 10-liter fermentor and purified from inclusion bodies by the modified method of Hui et al. (5).
The quadruple-mutant and wild-type protease values for Km (275 and 115 μM, respectively) and kcat (5.6 and 12.2 s−1, respectively) are similar, giving catalytic efficiencies of 20 and 106 mM−1s−1, respectively (Table 2) (see references 8 to 10 for methodological details). Although the catalytic efficiency for the mutant is approximately fivefold poorer overall, the mutant product is still a relatively efficient protease. A comparison of the inhibitory constants for each of the marketed HIV-1 protease inhibitors (APV, SQV, indinavir, ritonavir, and nelfinavir) demonstrated that the quadruple-mutant protease was effectively inhibited by all of the PIs except SQV (Table 3). The Ki value for SQV was 300-fold higher in the quadruple-mutant protease than in the wild-type protease and is consistent with the viral phenotype seen in tissue culture assays. Table 4 shows the additive effects of the individual mutations that make up the quadruple mutant. A comparison of the Ki mutant/wild-type ratio for each of the individual mutant proteases with that of the combined quadruple-mutant protease for APV or SQV demonstrates approximate additivity compared to the final kinetic character of the quadruple-mutant product. These data also indicate that the I50V mutation negatively affects the inhibitory affinity of APV or SQV for mutant protease while the M46I and I84L mutations are likely to compensate for this mutation (especially for the APV-protease interaction). It can also be seen that the G48V mutation has the most significant effect on the Ki ratio for SQV.
TABLE 2.
Kinetic analysis of the HIV-1 protease mutant (M46I, G48V, I50V, I84L)a
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Relative efficiency |
|---|---|---|---|---|
| Wild type | 115 | 12.2 | 106 | 5.3 |
| Mutant | 275 | 5.6 | 20 | 1.0 |
For details, see reference 10. The substrate His-Lys-Ala-Arg-Val-Leu/p(NO2) Phe-Glu-Ala-Nle-Ser-NH2 is referred to as substrate 1 in reference 10. The assay conditions are 1 mM dithiothreitol–1 mM EDTA in 50 mM citrate (pH 4.5). Vials containing this buffer and various amounts of the substrate were preincubated at 37°C for 10 min before initiating reactions by the addition of wild-type or mutant HIV-1 protease. After sufficient time to allow about 15% hydrolysis of the substrate (15 to 30 min), trifluoroacetic acid was added to a final concentration of 5%. Products were analyzed by high-pressure liquid chromatography, and the data were fitted to the Michaelis-Menten equation to evaluate kcat and Km.
TABLE 3.
Inhibition coefficients of HIV-1 protease mutant (M46I, G48V, I50V, I84L)a
| Protease inhibitorb |
ki (nM) for:
|
Mutant/ wild-type ratio | |
|---|---|---|---|
| Wild type | Mutant | ||
| APV | 0.6 | 1.3 | 2 |
| SQV | 0.8 | 240 | 300 |
| IDV | 1 | 4 | 4 |
| RTV | 0.3 | 0.5 | 2 |
| NFV | 1.1 | 1.4 | 1 |
For details, see reference 10. The substrate His-Lys-Ala-Arg-Val-Leu/p(NO2) Phe-Glu-Ala-Nle-Ser-NH2 is referred to as substrate 1 in reference 10. Assay conditions were as in the experiment in Table 2, except that the various PIs were allowed to preincubate with the wild-type or mutant protease and the reactions were initiated by addition of the substrate. Ki values were determined as described in the footnote to Table 2.
IDV, indinavir; RTV, ritonavir; NFV, nelfinavir.
TABLE 4.
Kinetic analysis of HIV-1 protease with individual mutationsa
X-ray diffraction studies and molecular modeling of the mutant protease-PI complexes.
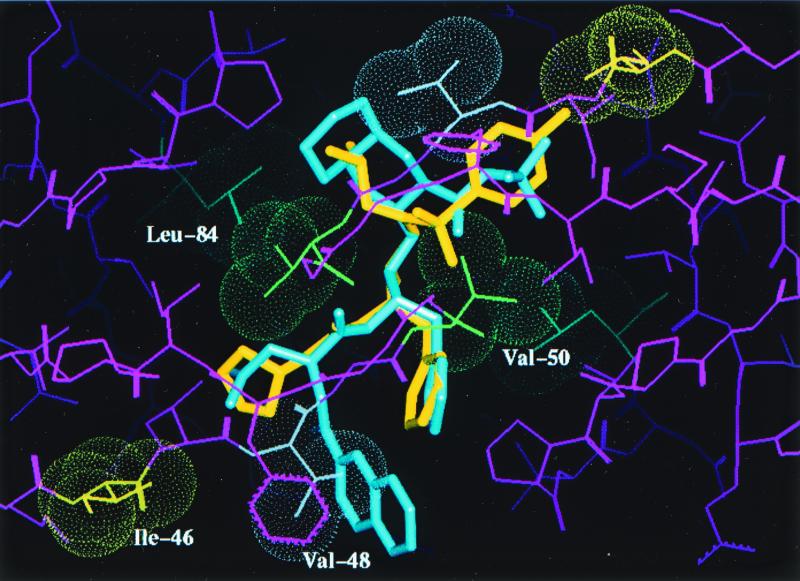
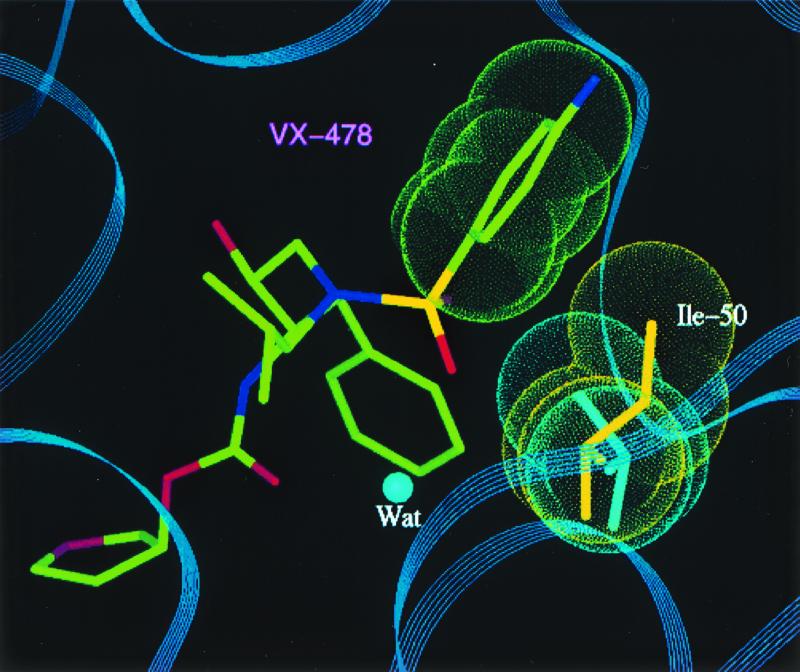
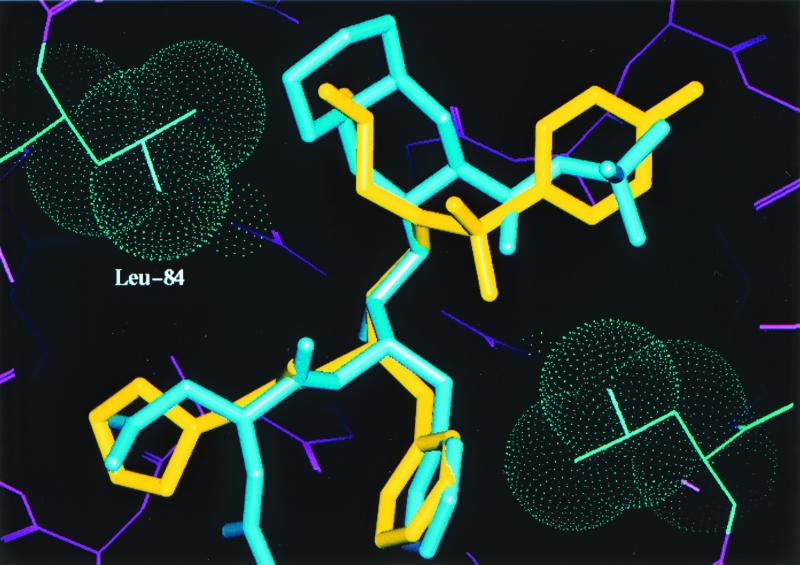
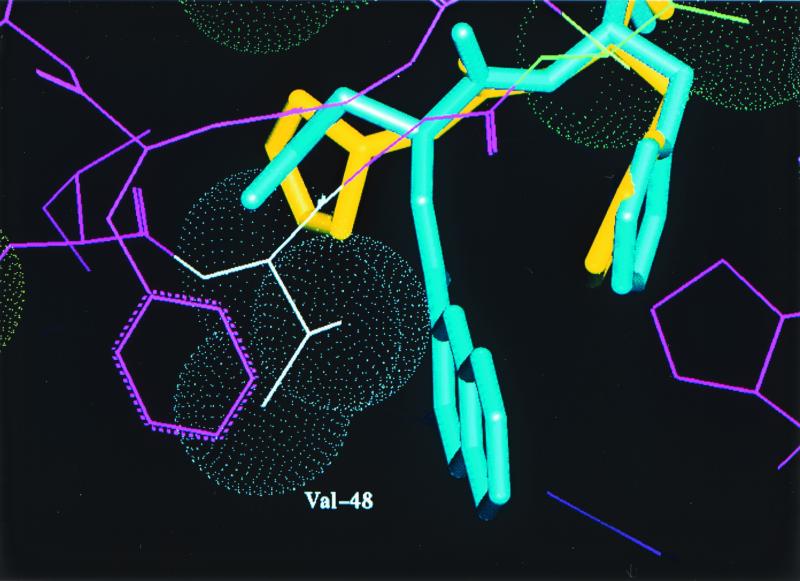
Crystals of the mutant protease-PI complexes were generated by refolding the protease, using the rapid dilution method, in the presence of APV by using the hanging-drop method (detailed in reference 6). Data collection, refinement, and model building were performed as detailed in reference 6. SQV was modeled into the active site of the mutant protease by superposing the backbone coordinates of the mutant and the HIV-1 protease-SQV complex structure (modeling details described previously). A model based on the X-ray diffraction data collected (data not shown) with the quadruple-mutant HIV-1 protease-APV complex is shown in Fig. 1. This figure shows the active site of the protease (a homodimer), with each pair of mutated residues (M46I, G48V, I50V, and I84L) indicated by van der Waals surfaces and the drug interactant from each pair highlighted. APV is shown in yellow, and SQV is shown in blue. Figure 2 is a close-up of the hydrophobic contact between the side chain of I50 (gold) and the P2′ phenyl ring (green) of the benzenesulfonamide moiety of APV in the S2′ pocket. A detailed description of this interaction and the effect of the valine mutation has been given elsewhere (10); essentially, there is a reduction in the interaction between APV and the mutant protease. A similar reduction in the interaction occurs between the mutant protease and the iso-butyl group of SQV. The I84L mutation within the protease compensates for this loss of hydrophobic packing, at least for APV, by reintroducing a comparable area of hydrophobic contact (Fig. 3). Modeling suggests that this would also be the case for SQV if it could line up in the pocket as normal, but the G48V mutation precludes this. The major change leading to SQV resistance is the G48V mutation, which introduces a bulky side chain into the S3 pocket. This leads to steric hindrance of the P3 quinoline group of SQV (bicyclic blue ring in Fig. 4) but not of the P2 furanyl group of APV (yellow five-membered ring in Fig. 4). Residue 46 (containing the M-to-I mutation) was demonstrated to be exposed to solvent and not directly involved in interactions with either inhibitor; instead, it is likely to be compensatory for the mutations within the protease-active site for reasons such as domain flexibility (3, 12).
FIG. 1.
Structure of the active site of the HIV-1 protease dimer shown complexed with APV (yellow) and SQV (blue). Van der Waals surfaces are indicated for each pair of amino acids in the quadruple mutant, with the drug-interacting residue of each pair highlighted.
FIG. 2.
Close-up view of the interaction between I50 (side chain in gold) and the phenyl ring (green) of APV (also called VX-478) in the S2′ pocket of the enzyme. The valine mutant side chain is colored turquoise. The flap water molecule is also indicated.
FIG. 3.
Close-up view of the interaction between mutant residue L84 and both APV and SQV. This interaction partially compensates for the loss of hydrophobic interaction between 50V and either APV or SQV.
FIG. 4.
Close-up view of the steric repulsion of the P3 group of SQV induced by the introduction of the V48 side chain into the S3 pocket.
One assumes that the smallest number of mutations that enabled the efficient survival of the APV-resistant mutated virus in the presence of increasing amounts of SQV was the conversion to the observed quadruple-mutant SQV-resistant virus. Questions that arise from such an observation include the following. Given the association between SQV resistance and the pair of mutations G48V and L90M, why did the latter mutation not appear? What does this say about the role of residue 47 in APV resistance? And why should APV sensitivity reappear when the selection was for SQV resistance? There are likely to be multiple and potentially complicated pathways toward the generation of PI resistance in HIV, even in vitro. This will depend on the starting sequence of the virus, the particular PI used, and the intensity of the selection pressure applied to the replicating virus. Speculation regarding the shift from the 46I, 47V, and 50V mutations to the 46I, 48V, 50V, and 84L mutations during the SQV selection is as follows. The starting point in the studies described here was the APV-resistant viral strain (with mutations at residues 46, 47, and 50), which had remained sensitive to SQV. M46I and I47V as single mutations have no effect on inhibitor binding for APV or SQV (9) but appear to be compensatory for the I50V mutation in the triple mutant, since the catalytic efficiency of the triple-mutant protease is greater than that of the I50V protease alone. This, together with the marked increase in Ki (270-fold) for APV, allows for an increased vitality or fitness of such a triply mutated HIV and hence survival in the presence of APV. The M46I mutation may result in the locking of the flap portion of the protease in the closed position (3, 12), while residue 47, also in the flap, is part of the S2/S2′ binding pocket but does not interact directly with either APV or SQV. Viral survival of the APV-resistant triple mutant in the presence of SQV necessitated further mutation. The principal mutations associated with SQV resistance have been reported to be G48V and L90M (7, 11), although in these and our studies (data not shown) the G48V mutation appears to be the dominant change in the enzymological mechanism of resistance. The L90M mutation indirectly affects the interaction between the catalytic D25 side chains and the central OH group of all the PIs, including APV, while the G48V mutation has a direct steric effect on SQV, as described above. In fact, the presence of L90M together with I50V in HIV protease leads to a marked increase in Ki for SQV (data not shown), while addition of L90M to I47V plus I50V in the protease causes a reversal in this trend (data not shown). A comparison of the Ki mutant to Ki wild type ratio for the various mutant proteases with an added L90M mutation demonstrated the following ranking with respect to SQV: (L90M alone) < (I47V + L90M) < (M46I + I47V + I50V) < (M46I + I47V + I50V + L90M) < (M46I + M48V + I50V + I84L) < (G48V + L90M) (data not shown). From the above, it appears reasonable that the G48V mutation would be favored when converting an APV-resistant strain to one resistant to SQV. However, a protease with consecutive mutations at residues 46, 47, and 48 would probably be affected negatively, requiring residue 46 or 47 to change, probably a reversion. We suggest that residue 47 is most likely to revert, since the 46I/L mutation is a relatively common change associated with PI resistance and is often linked with other protease mutations, probably playing a compensatory role in proteases resistant to other PIs as well as APV. This does not necessarily require a reassessment of residue 47 in APV resistance, since in the context of the triple mutation it appears to be compensatory for the I50V mutation in terms of the catalytic efficiency of the mutated protease. We assume that the I84L change recompensates, possibly in terms of substrate affinity, or that M46I provides sufficient compensation within the context of the quadruple mutation. Residues 50 and 84 are at the center of the active site, with their side chains at the interface of subsites 1 and 2 and capable of interacting with residues P2 through P2′ (10). 84L as a single mutation has a minor effect on the interaction of the protease with SQV (fourfold increase in Ki). Why, then, does the 84L mutation appear? In the absence of data for the G48V-plus-I84L combination, we can only speculate that this combination, either within the quadruple background or as a pair, has an effect on the affinity of the protease for SQV or the catalytic efficiency of the protease containing the quadruple mutation. The fact that this combination of mutations elicited a resensitization to APV, while understandable mechanistically as described here, is obviously coincidental, since the selection pressure of the experiment was for SQV resistance alone. Such resensitization was not observed with any other PI combinations tested at the same time (data not shown), hence the unexpected phenotype described here.
In conclusion, an interesting drug-resistant phenotype was observed during the in vitro passage of APV-resistant HIV-1 in the presence of SQV, resulting in SQV resistance and APV resensitization. A detailed dissection of the protease from this virus at the enzymological and structural levels demonstrated facets of the PI-protease interaction that were perfectly understandable as a gain or loss of individual interatomic interactions within the context of the combined quadruple-mutant active site. We are optimistic that the establishment of such a detailed understanding of the HIV-1 protease-PI interaction will lead to the design of more efficacious drugs with less resistance potential.
Acknowledgments
We thank Jim Griffith, John Fulghum, and Sam Pazhanisamy for work related to this project and Ann Kwong and Vicki Sato for careful reading of the manuscript and suggested improvements that were gratefully received.
REFERENCES
- 1.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condra J H. Virological and clinical implications of resistance to HIV-1 protease inhibitors. Drug Resistance Updates. 1998;1:292–299. doi: 10.1016/s1368-7646(98)80045-9. [DOI] [PubMed] [Google Scholar]
- 3.Erickson J W, Burt S K. Structural mechanisms of HIV drug resistance. Annu Rev Pharmacol Toxicol. 1996;36:545–571. doi: 10.1146/annurev.pa.36.040196.002553. [DOI] [PubMed] [Google Scholar]
- 4.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis. A practical approach. Oxford, United Kingdom: IRL Press and Oxford University Press; 1991. pp. 217–247. [Google Scholar]
- 5.Hui J O, Tomasselli A G, Reardon I M, Lull J M, Brunner D P, Tomich C S, Heinrikson R L. Large scale purification and refolding of HIV-1 protease from Escherichia coli inclusion bodies. J Protein Chem. 1993;12:323–327. doi: 10.1007/BF01028194. [DOI] [PubMed] [Google Scholar]
- 6.Kim E E, Baker C T, Dwyer M D, Murcko M A, Tung R D, Navia M A. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J Am Chem Soc. 1995;117:1181–1182. [Google Scholar]
- 7.Maschera B, Darby G, Palu G, Wright L L, Tisdale M, Myers R, Blair E D, Furfine E S. Human immunodeficiency virus: mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996;271:33231–33235. doi: 10.1074/jbc.271.52.33231. [DOI] [PubMed] [Google Scholar]
- 8.Partaledis J A, Yamaguchi K, Tisdale M, Blair E E, Falcione C, Maschera B, Myers R E, Pazhanisamy S, Futer O, Cullinan A B, Stuver C M, Byrn R A, Livingston D J. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pazhanisamy S, Partaledis J A, Rao B G, Livingston D J. In vitro selection and characterization of VX-478 resistant HIV-1 variants. Adv Exp Med Biol. 1998;436:75–83. doi: 10.1007/978-1-4615-5373-1_10. [DOI] [PubMed] [Google Scholar]
- 10.Pazhanisamy S, Stuver C M, Cullinan A B, Margolin N, Rao B G, Livingston D J. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 11.Roberts N A. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS. 1995;9:527–532. [PubMed] [Google Scholar]
- 12.Rose R B, Craik C S, Stroud R M. Domain flexibility in retroviral proteases: structural implications for drug resistant mutations. Biochemistry. 1998;37:2607–2621. doi: 10.1021/bi9716074. [DOI] [PubMed] [Google Scholar]
- 13.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antiviral Newsl. 1997;5:129–142. [Google Scholar]
- 14.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]