Significance
Despite their importance in neuronal and cardiac control, Hyperpolarization-activated Cyclic Nucleotide-gated (HCN) channels have only one approved drug: ivabradine, an open-pore blocker that lacks subtype specificity. Our cryogenic Electron Microscopy (cryo-EM) structure of the HCN4 channel in complex with ivabradine is of broad interest because we obtained the open-pore conformation of HCN channels in the absence of hyperpolarization, explained the mechanism of block in terms of electrostatic repulsion between the drug and the ions, and, most importantly, identified the unpredicted contribution to ivabradine binding of residues outside the pore, opening up the perspective of subtype-specific drug design.
Keywords: HCN, ivabradine, structure
Abstract
HCN1-4 channels are the molecular determinants of the If/Ih current that crucially regulates cardiac and neuronal cell excitability. HCN dysfunctions lead to sinoatrial block (HCN4), epilepsy (HCN1), and chronic pain (HCN2), widespread medical conditions awaiting subtype-specific treatments. Here, we address the problem by solving the cryo-EM structure of HCN4 in complex with ivabradine, to date the only HCN-specific drug on the market. Our data show ivabradine bound inside the open pore at 3 Å resolution. The structure unambiguously proves that Y507 and I511 on S6 are the molecular determinants of ivabradine binding to the inner cavity, while F510, pointing outside the pore, indirectly contributes to the block by controlling Y507. Cysteine 479, unique to the HCN selectivity filter (SF), accelerates the kinetics of block. Molecular dynamics simulations further reveal that ivabradine blocks the permeating ion inside the SF by electrostatic repulsion, a mechanism previously proposed for quaternary ammonium ions.
The era of cryo-EM has provided high-resolution information on the structure of many ion channels with physiological importance. This paved the way for understanding structure/function correlates of these proteins in physiological and pathophysiological conditions. A great expectation from cryo-EM-resolved structures is that they will also provide insight into the function of important channel-interacting small molecules, thus allowing the design of more efficient and specific drugs. Motivated by this goal, we have solved the structure of the open HCN4 channel with its prototypical blocking drug, ivabradine, in the pore. We cross-correlate this high-resolution structural information with a bulk of functional data on ivabradine action as an open-pore blocker to detail the mode of channel block at an atomistic level.
HCN4 is predominantly expressed in the human cardiac sinoatrial node (1) and is considered the molecular determinant of the If current (2, 3). We had previously set the experimental protocol to purify HCN4 channels in the closed and in the open-pore configuration and solved the structures by single-particle cryo-EM (4). Molecular dynamic (MD) simulations (4–6) confirm that the open pore is conductive and displays the functional features of HCN4 channels as originally described for the native f-channels, revealing molecular mechanisms for the low single-channel conductance (7), mixed Na+-K+ permeability (8), and block by cesium (9). The high resolution of cryo-EM-generated structures now provides the unique opportunity to further study the interaction between HCN4 open channels and ivabradine, the prototypical HCN blocker.
Ivabradine is the only HCN-specific drug approved for clinical use and the sole member of Class 0 antiarrhythmic agents (10). Commercialized as Procoralan or Corlentor (11), ivabradine reduces the heart rate and is approved for use in case of stable angina pectoris and symptomatic heart failure (12). In contrast to beta blockers and calcium antagonists, ivabradine has no inotropic action but is a purely bradycardic agent, acting on the pacemaker current If that drives the diastolic depolarization phase of the sinoatrial node action potential (13). From the chemical point of view, ivabradine is a phenylalkylamine and a derivative of zatebradine (Thomae ULFS49), a heart rate–reducing molecule developed earlier (14). The ivabradine molecule is composed of two hydrophobic moieties, benzazepinone and benzocyclobutane, connected by a linker region that contains a positively charged, tertiary nitrogen atom (Fig. 1A).
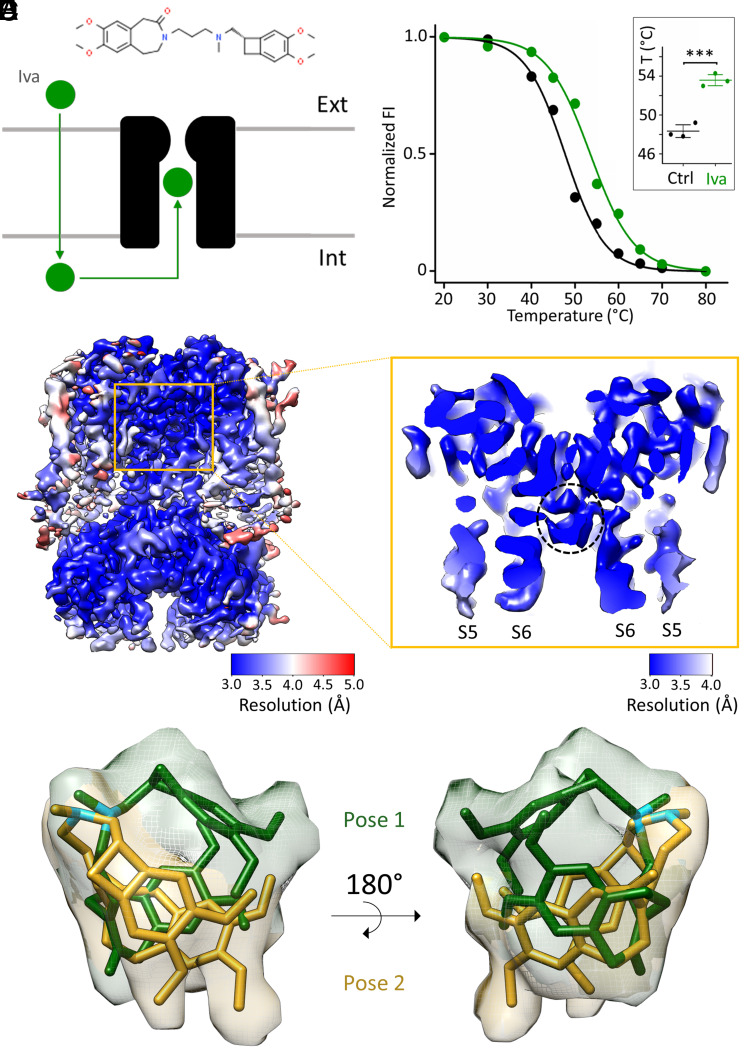
Fig. 1.
Biochemical and structural characterization of ivabradine binding to HCN4. (A) Cartoon representation of ivabradine (green sphere) crossing the plasma membrane and entering the intracellular pore vestibule of HCN4 (black). The chemical structure of ivabradine is shown on Top. (B) Representative denaturing curves of purified GFP-tagged HCN4 protein without (black) and with 0.5 mM ivabradine (green). Normalized fluorescence intensity (FI) is plotted against preconditioning temperature (°C). Solid lines show a sigmoidal dose–response equation fitting to the data (SI Appendix). Inset: Mean Tm values for control (48.3 ± 0.4 °C, black) and + ivabradine (53.6 ± 0.4 °C, green) (n = 3 experiments ± SEM). Statistical analysis was performed with one-way ANOVA, followed by Fisher's test (***P < 0.001). (C) Density map and (D) enlargement of the pore region (S5 and S6 helices are marked) of HCN4 with ivabradine. (D) The pore is open, and the intracellular pore vestibule is occupied by a nonprotein density circled with a dotted black line. Maps are color-coded by local resolution (Å), as indicated by the two bars. (E and F) Ivabradine molecule modeled within the nonprotein density in two different poses: Pose 1 (yellow) and pose 2 (green) (PDB: 8OFI) displaying similar position of the tertiary amine of the aliphatic linker (N atoms colored in light blue) and different orientation of the hydrophobic benzazepinone and benzocyclobutane moieties. Density is zoned around poses 1 and 2 and is colored accordingly.
The blocking action of ivabradine has been extensively described as being use and current dependent in native f-channels as well as in HCN channels (15–17). As an overall lipophilic molecule, ivabradine crosses the lipid membrane and enters the HCN pore cavity from the cytosolic side (Fig. 1A). For accessing the binding region in the pore cavity, it needs the channel to be opened by hyperpolarization before it is driven by depolarization into its blocking site. A special feature of ivabradine block is its current dependence, resulting from the interaction of block with the flow of permeating ions through the pore (15, 18). The block is also use dependent, as it requires several cycles of hyperpolarization/depolarization to reach full block. The blocking action of ivabradine is thus incremental and accrues gradually in small additions at each cycle of hyperpolarization, during which the pore opens, and depolarization, during which the molecule is driven inside the pore. This mode of action favors the clinical aspect in that ivabradine blocks more efficiently at faster (tachycardic) heart rates (19). Indeed, while indicated for coronary heart disease and heart failure, off-label experiments have shown that ivabradine is also efficient for the treatment of inappropriate sinus tachycardia (20).
Although ivabradine is highly selective for blocking the If current (17, 21), it does not show marked specificity among the HCN subtypes (18). Such subtype-specific blockers, however, are urgently needed to treat pathologies in which one of the subtypes prevails. While, for example, HCN4 is more prevalent in cardiac disorders, other subtypes play more important roles in neurological diseases. Intractable forms of HCN1-related early infantile epileptic encephalopathies (EIEE) (22, 23) or HCN2-driven neuropathic pain (24) are just two examples.
Since advancements in the field of drug development are favored by high-resolution structural information, we solved the structure of the HCN4 protein in complex with ivabradine. This study confirms unambiguously that the drug is an open-channel blocker and identifies the residues that interact with the molecule inside the cavity.
Validation by mutational analysis and MD simulations further provides clear indications on the modality of block that is based on the electrostatic interaction between the blocker and the cation in the selectivity filter (SF). Our study sheds light on the inhibitory mechanism of ammonium ions on HCN in particular, and on Kv channels in general (25), and further explains the HCN preference for ivabradine.
Results
Prior to grid preparation for cryo-EM analysis, we tested whether ivabradine binds to the purified HCN4 protein. This was done with a thermostability assay based on fluorescence-detection size-exclusion chromatography (26). By comparing the melting curve of Green Fluorescence Protein (GFP)-tagged HCN4 without (black) and with 0.5 mM ivabradine (green), we found that the drug stabilizes the protein, increasing the melting temperature (Tm) by about 5 °C (Fig. 1B). The Inset in panel B shows the mean values (±SEM) of Tm in the absence (48.3 ± 0.4 °C) and presence of ivabradine (53.6 ± 0.4 °C) from three independent experiments.
We interpreted stabilization as proof of ivabradine binding to the open pore and further subjected the protein sample to single-particle cryo-EM studies. A total of 31,670 micrograph movies were collected, and data were processed without imposing symmetry. We reconstructed a density map from 107,523 particles of the tetrameric HCN4 protein with an overall resolution of 3.6 Å (Fig. 1C and SI Appendix, Fig. S1 A–E) and a local resolution of the pore of 3.2 Å (Fig. 1D). The pore is open as indicated by the position of the S6 helices at the cytosolic side [Fig. 1D, (4)] and contains a nonprotein density (circled with a black dotted line in Fig. 1D) in the intracellular vestibule. In this density, larger than expected for a single molecule, we could model ivabradine in two partially overlapping and mutually exclusive poses, identified as pose 1 (green) and pose 2 (yellow) in Fig. 1 E and F. Interesting to note, the position of the tertiary amine (N atoms in light blue) is the same in both poses, while the orientation of the bulky hydrophobic groups benzazepinone and benzocyclobutane varies between the two poses. Important to mention here is that ivabradine is in both cases coordinated by the hydrophobic residues Y507 and I511 from the S6 helix (see below, s). As discussed later, this interaction of the drug with the protein is consistent with a previous hypothesis based on functional data and in silico docking experiments (16, 27). It is not possible to exactly calculate the percentage of occupancy of the two poses. An estimation was obtained by performing B-factor refinements of two atomic models of HCN4, each containing one of the two ivabradine poses. We obtained B-factors of 20.27 Å2 and 23.15 Å2 for poses 1 and 2, respectively. Since the B-factors are similar, we argue that ivabradine can equally populate both poses.
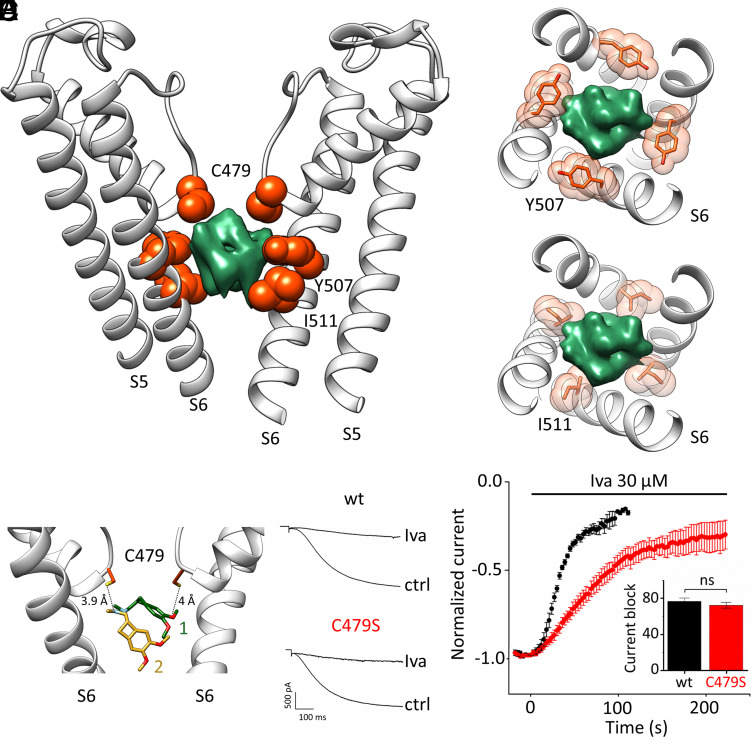
Fig. 3.
Molecular determinants of ivabradine binding to HCN4. (A) Cross-membrane view of the HCN4 pore (S5–S6) bound to ivabradine (PDB: 8OFI, SI Appendix, Table S1). For clarity, only two opposite subunits of the tetramer are shown. Residues contacting ivabradine (green density) are shown as orange spheres and labeled: C479 from the SF; Y507, and I511 from S6 (RbHCN4 numbering). Spheres represent the van der Waals radii of the atoms. (B and C) Transverse views of the intracellular pore cavity (S6) showing the four Y507 (B) and I511 (C) residues (orange sticks and spheres) that surround ivabradine (green density). (D) Distances between two opposite C479 residues (orange sticks, S atoms in yellow) and ivabradine pose 1 (green sticks) and pose 2 (yellow sticks). N atoms of the tertiary amine and methoxyl groups are colored in light blue and red, respectively. (E) Representative current traces of HCN4 wt (Upper) and C479S mutant (Lower), in control (ctrl) and after superfusion with 30 µM ivabradine (Iva) at steady-state block. (F) Mean time courses of current amplitudes of HCN4 wt (black) and C479S mutant (red), recorded at −140 mV, before and after perfusion at time = 0 with 30 µM ivabradine. (Inset) mean maximal block by 30 µM ivabradine of HCN4 wt and C479S mutant channel: 76.8 ± 3.7% (wt, black); 72.3 ± 3.2% (C479S, red). There is no statistical difference between the two values (t test). Values are mean of n = 3 experiments ± SEM.
We eventually generated the final atomic model of the protein bound to the drug, hereafter named HCN4–IVA (SI Appendix, Fig. S2 and Table S1). Fig. 2A shows the HCN4 protein as a dimer, after removal of two monomers from the tetramer for clarity. The S4–S5 linker and the pore domain, consisting of the pore helix (PH), SF, and the transmembrane helices S5 and S6, are shown in light gray and tagged. Ivabradine is shown in the two poses (green and yellow) in the nonprotein density inside the pore.
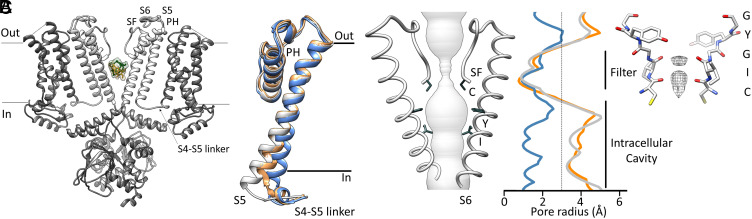
Fig. 2.
Structure of the open pore of HCN4 in complex with ivabradine. (A) HCN4 bound to ivabradine (green/yellow sticks and densities), in a cross-membrane view. Only two opposite subunits are shown with S5, S6, PH, SF, and the linker between the S4 and S5 (S4–S5 linker) in light gray and labeled in one subunit. (B) Superposition, in a cross-membrane view, of the S5 helix in the closed HCN4 structure [blue, PDB: 7NP4, (4), open orange, PDB: 7NP3, (4), and open with ivabradine (gray, PDB: 8OFI, SI Appendix, Table S1)]. The S5 movement is most pronounced in the presence of ivabradine (gray). The helical turn in the S4–S5 linker is unfolded in the HCN4–IVA protein (gray). (C, Left) HCN4–IVA pore diameter shown as a gray volume. Helix S6 and SF lining the pore are labeled. Residues involved in ivabradine binding are shown as sticks and labeled. (Middle) Plot of the pore radii of HCN4 closed (blue, PDB: 7NP4), open (orange, PDB: 7NP3), and open in the presence of ivabradine (gray, PDB: 8OFI, SI Appendix, Table S1). The dotted line marks the radius of hydrated K+. Corresponding portions of the SF and intracellular cavity are indicated by black vertical bars. (Right) SF of HCN4 in the presence of ivabradine. The sequence 479CIGYG481 (RbHCN4 numbering) is marked. Densities within the filter are represented as meshes.
The Open Pore in Complex with Ivabradine.
The open-pore structure of HCN4-IVA differs from the HCN4 open-pore structure previously solved in the absence of ivabradine (4), PDB 7NP3), hereafter mentioned as open HCN4. Superposition of the S4–S5 linker and S5 helix of HCN4-IVA (gray) with those of open HCN4 (orange) and, for reference, closed HCN4 pore [blue, (4), PDB 7NP4] shows that the presence of ivabradine inside the pore markedly accentuates structural features previously associated with pore opening in HCN channels: S4–S5 linker unfolding and uplifting of the cytosolic end of the S5 helix (4), (28). Indeed, in HCN4-IVA (gray), the unfolding of the S4–S5 linker is completed, leading to a larger displacement of S5, if compared with open HCN4 (orange). Note that these features have been already seen in the HCN4 open-pore structure solved in the presence of the membrane mimetic amphipols, which have chemically stabilized S5 splayed out [(4), PDB 7NMN].
Fig. 2C shows as a gray volume the ion permeation pathway of the HCN4-IVA pore, lined by the S6 helix and SF. The side chains of residues C479, Y507, and I511 forming the ivabradine binding site (see below) are shown and labeled. The corresponding pore radius is plotted on the right (gray), together with those of the previously published open HCN4 (orange) and closed HCN4 pore (blue) (4). The comparison with the radius of open HCN4 highlights that ivabradine has no impact on the widening of the S6 helix, while it stabilizes other features of the open pore such as the unfolding of S4–S5 and the full movement of S5 (Fig. 2B). Fig. 2C also shows that the SF is in the conductive conformation (4). Nonprotein densities, which are interpreted as ions (shown in gray), are visible at the carbonyl plane of cysteines and isoleucines, residues forming the ion-binding sites of HCN channels (4–6).
The Ivabradine Contact Sites in the Pore Cavity.
Three residues facing the intracellular pore cavity are in proximity with ivabradine: C479 from the SF and Y507 and I511 from the S6 helix (Fig. 3A). In the tetrameric arrangement of the pore, Y507 and I511 form two hydrophobic rings spaced by a helix turn, surrounding the likewise bulky hydrophobic groups of the drug (Fig. 3 B and C). This large hydrophobic contact surface between the molecule and the protein provides a structural explanation for the finding that substitution of Y and I with alanine residues abolishes the blocking effect of the drug by reducing the affinity by 100-fold (16, 27).
As for C479, this residue forms the innermost ion-binding site of the SF. This is a peculiarity of HCN channels since all other K+ channels, including CNG and NaK channels, have a highly conserved Ser/Thr in this position (29). Two of the four cysteines are 3.9 and 4 Å apart from the protonated tertiary amine and a methoxy group of the drug, respectively (Fig. 3 D, Left), distances compatible with weak hydrogen bonds (HBs). Given the debated and controversial ability of cysteine sulfur to form HBs (30–35), we decided to test the hydrophilic nature of the interaction with ivabradine by substituting C479 with a serine, whose side chain is of similar length but with a clear hydrophilic nature.
We measured the ivabradine block of wt and C479S HCN4 currents by patch-clamp electrophysiology in HEK293T cells. Ivabradine was perfused extracellularly at 30 µM. Recordings were performed in whole-cell configuration by means of a standard voltage clamp protocol that repetitively imposes voltage jumps from −140 mV to +5 mV (see Methods for details). Mutation C479S did not alter the properties of HCN4 (Fig. 3E), as previously reported for HCN2 (36). The degree of block by ivabradine at steady state was the same (Fig. 3 E and F, histogram): 76.8 ± 3.7% for the wt and 72.3 ± 3.2% for the C479S mutant. However, the mutation almost doubled the half time of block onset (Fig. 3F): The half time (T1/2) was 30.5 ± 1 s (n = 5) for the wt and 54 ± 2 s (n = 6) for the C479S mutant. We conclude that the cysteine side chains contribute to the dynamics of ivabradine block via apolar interactions.
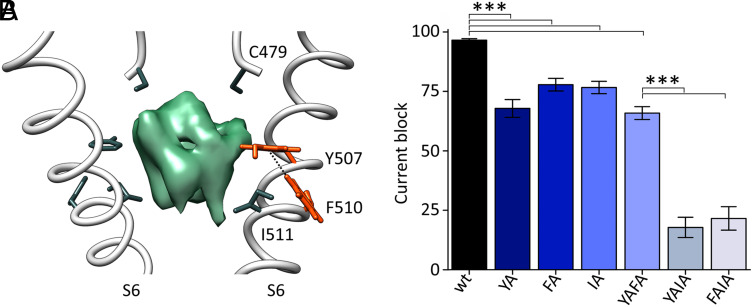
It was previously shown that mutation of F509 in human HCN4 reduces ivabradine binding. These data were interpreted as evidence of the direct interaction of F509 with the drug inside the pore (27). At first glance, we were surprised to find that in the RbHCN4 structure, the corresponding phenylalanine (F510) does not point inside the pore (Fig. 4A); hence, it exhibits no direct contact with the blocker (green density, Fig. 4A). A closer scrutiny reveals, however, that F510 is involved in an edge-to-face π-interaction with Y507 (Fig. 4A and SI Appendix, Fig. S3), suggesting that F510 indirectly affects ivabradine binding by controlling the orientation of the Y507 side chain. We tested this hypothesis experimentally by introducing single and double mutations (Fig. 4B and SI Appendix, Fig. S4). Indeed, in agreement with what was previously reported in human HCN4 (27), Y507A and I511A single mutations cause a mild reduction of ivabradine block (70 to 80% of wt, Fig. 4B), while their combination abolishes, almost completely, the blocking effect of the drug (18% of wt, Fig. 4B). This indicates that Y507 and I511 equally contribute to the ivabradine binding site inside the cavity (Fig. 3 A–C). The effect of F510A is similar to the single Y507A and I511A mutations (80% of wt, Fig. 4B), while in the context of double mutants, it depends on whether F510A is associated with Y507A or I511A. If coupled with I511A, it reduces the block almost completely (20% of wt, Fig. 4B), while in combination with Y507A, it has no effects (70% of wt, Fig. 4B). These findings confirm the hypothesis that F510 is chemically linked to Y507 (π-interaction, Fig. 4A) and indirectly affects IVA block via this residue.
Fig. 4.
Role of F510 in ivabradine block. (A) Cross-membrane view of the HCN4 pore (S6) bound to ivabradine (PDB: 8OFI, SI Appendix, Table S1). For clarity, only two opposite subunits of the tetramer are shown. Residues directly or indirectly involved in ivabradine (green density) binding are shown as sticks and labeled: C479 from the SF and Y507 and I511 from S6 (RbHCN4 numbering). F510 develops a π-interaction (edge-to-face, 5.8 Å) with Y507. Side chains of both residues are shown as orange sticks, and their hydrogen atoms are shown. (B) Mean maximal block by 300 µM ivabradine of HCN4 wt and Y507A (YA), I511A (IA), Y507A-I511A (YAIA), Y507A-F510A (YAFA), and F510A-I511A (FAIA) mutant channels: 97.8 ± 0.6% (wt); 68.7 ± 3.8% (YA); 77.6 ± 2.6% (IA); 18.0 ± 4.3% (YAIA); 67.7 ± 2.2% (YAFA); 21.8 ± 5.0% (FAIA). Values are mean of n ≥ 3 experiments ± SEM. Statistical analysis was performed with one-way ANOVA, followed by Fisher’s test (***P < 0.001). The percentage of current block of YA, IA, and YAFA mutants is not statistically different, as well as the one of YAIA and FAIA.
Modality of Block by Ivabradine.
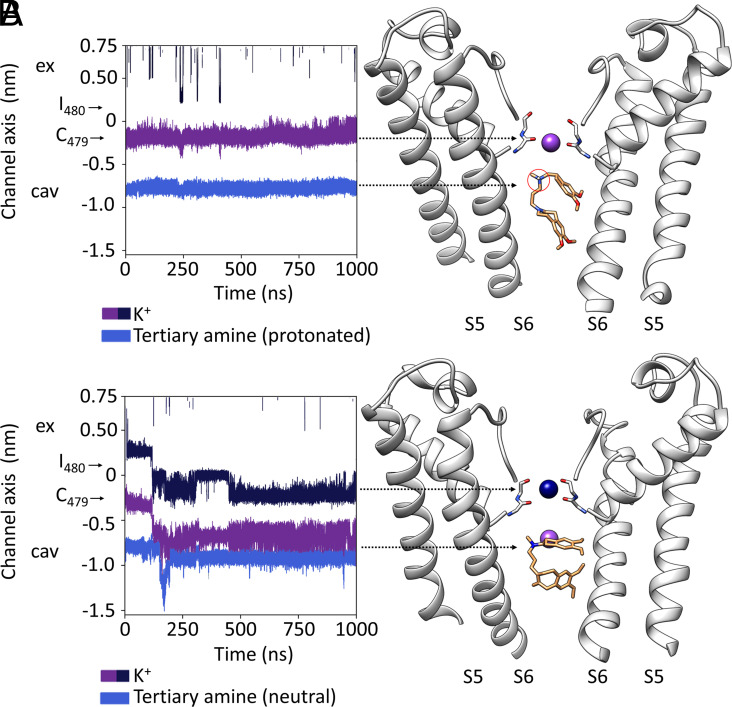
At first glance, it seems possible that ivabradine blocks ion permeation by steric occlusion of the cavity. Nonetheless, it should be considered that ivabradine contains in the aliphatic linker a weak basic tertiary amine (pKa = 8.6) (Fig. 1A) that is largely protonated (about 95%) at the physiological pH 7.4 (18). Therefore, it is possible that the positively charged N atom interacts with the permeating cation, as previously postulated to explain the “current dependency” of ivabradine block (15, 18). In both poses (1 and 2), the protonated amine is right below the SF (Fig. 3D) and can exert an electrostatic repulsion on the cation in the binding site. To test this hypothesis, we performed 17 MD simulations for a total length of 13 µs with ivabradine in a protonated or neutral form and in both poses (Fig. 1 E and F and SI Appendix, Table S2). For completeness, we have included also a third pose of ivabradine —“docked IVA”—previously identified by molecular docking as top ranking for energetic and scoring function (4). Given the extremely low (about 1 pS) single-channel conductance of HCN channels (7), we applied extreme conditions, high voltage (−500 to −700 mV) and high ion concentrations (1 M KCl) to increase the rate of ion permeation (4–6). Fig. 5 shows sample simulations performed with pose 1, but similar results were obtained with pose 2 and docked-IVA. The blocker, both in the protonated (Fig. 5A) and neutral state (Fig. 5B), stably occupies the cavity with the tertiary amine pointing toward the SF. The two hydrophobic moieties are more dynamic but constantly develop extensive hydrophobic interactions with Y507 and I511 in the cavity. Hence, the simulations further support both structural (Figs. 1 and 3) and functional results (Fig. 4). The positively charged amine (Fig. 5A) occupies the position normally occupied by the permeating ion when it leaves the filter and enters the vestibule. In this position, the protonated amine exerts a constant repulsive force that blocks the K+ inside the filter with the effect that no ion passages through SF were detected (two to four independent simulations per pose, with a total simulation time of 5.5 µs, SI Appendix, Table S2).
Fig. 5.
Mechanism of ivabradine block: (A and B) For simplicity, the present figure shows MD simulations of ivabradine pose 1, protonated (A) and neutral (B). MDs of pose 2 are, indeed, equivalent. (A, Left) Trajectory for a K+ ion (purple) in the SF and for the protonated tertiary amine of ivabradine (blue) in the pore cavity in a MD simulation performed with HCN4 pore only, in KCl solution. Positions of the carbonyl plane of C479 and I480 are indicated by black arrows. Ex (extracellular side of the pore); cav (intracellular pore cavity). (Right) Representative snapshot of the MD simulation shown in the Left panel, cross-referenced with the latter by arrowheads. For clarity, only two opposite subunits of the pore are shown. Over the entire simulation, the ion (purple) is stably located in the SF. Ivabradine (orange sticks) stably occupies the intracellular cavity. This localizes the protonated tertiary amine (circled in red) just below the SF for the entire simulation. (B, Left) Trajectories as in A, for K+ ions (purple and blue) in the SF and for the neutral tertiary amine of ivabradine (light blue). Positions of the carbonyl plane of C479 and I480 are indicated by black arrows. Ex (extracellular side of the pore); cav (intracellular pore cavity). (Right) Representative snapshot of the MD simulation shown in the left panel, cross-referenced with the latter by arrows. Entrance of a K+ ion (blue) from the extracellular side of the SF moves the second ion (purple) from the FS to the intracellular pore cavity. Neutralized ivabradine (orange sticks) stably occupies the intracellular cavity even when a K+ (purple) permeates the pore.
To further validate the “electrostatic repulsion” as the mode of block, we repeated simulations with neutral ivabradine (two independent simulations per pose, with a total simulation time of 7.5 µs, SI Appendix, Table S2). In five out of six simulations, we observed the transition of ions within the filter and in the vestibule, despite the presence of the drug. Therefore, distances between the N atom of the tertiary amine and K+ have a lower median and a wider distribution in MDs with the neutral ivabradine compared to the protonated one (SI Appendix, Fig. S5). Fig. 5B shows the simulation that started with ivabradine in pose 1. The arrival of the new ion (dark blue) inside the SF displaces the second ion (purple) into the wide vestibule, where it occupies a position approximately at the same height along the main channel axis as the noncharged amine.
By increasing the driving force for K+ (higher voltage), we observed a full transition of the ion through the pore, from the vestibule to the extracellular solution (SI Appendix, Fig. S6 and Table S2).
The results of MD simulations, together with the data obtained with the mutational analysis, underpin that 1) the presence of ivabradine per se does not prevent, in a steric manner, the passage of ions in the cavity; 2) the protonated amine group is necessary and sufficient to block K+ ions in the filter; and 3) the hydrophobic contacts with Y507 and I511 stabilize ivabradine with the tertiary amine group facing upside, toward the SF.
Discussion
Here, we present the structure of rabbit HCN4 in complex with its blocker ivabradine at 3 Å resolution. The molecule binds inside the open pore of HCN channels. This finding validates, on an atomistic level, a large set of functional data that described ivabradine block as use and state dependent (15, 18). Not least, the good agreement between structural and functional data once again confirms that the open-pore structure of HCN4 obtained by our experimental conditions—in the absence of an imposed voltage—is a reliable and powerful model to understand permeation and block of these channels.
Single-particle cryo-EM analysis pinpoints in this study two similar poses, equally probable, of ivabradine inside the pore that are very stable also in MD simulations. The finding of two poses is not surprising, given the results from molecular docking performed on the open pore of HCN4 (4) or on a homology model of the HCN1 open pore derived from the closely related CNG channel TAX-4 (16).
In both poses, the bulky hydrophobic groups of the drug display extensive hydrophobic interactions with residues Y507 and I511 in the pore vestibule and with C479 in the SF. The hydrophobic nature of the interaction with C479 is further confirmed by mutation C479S that slows down the block. A fourth residue, F510, controls ivabradine block indirectly by π-stack with Y507. The fact that the side chain of F510 points toward the S6–S5 interface that contains a lipid moiety in the closed HCN4 pore structure (4) raises the question whether the presence of the lipid can—in some indirect way—impact ivabradine binding.
The above-described features provide a structural framework for an atomistic understanding of the ivabradine block of HCN channels. Ivabradine is not the only known blocker of these channels but the most important one. It has a much higher selectivity for HCN channels compared to other less specific If channel blockers such as, for instance, ZD7288. The latter blocker also acts in the intracellular pore cavity, but the sites of interaction are different from the ones used by ivabradine (37, 38). Consequently, also the inhibitory action of the two drugs and, most important, their selectivity for HCN channels differ markedly (15, 39, 40).
Our results also provide a coherent explanation of the modality by which ivabradine blocks ion conductivity in HCN channels. Two possible modalities of block were considered, pore hindrance and electrostatic interaction, given that the charged tertiary amine of ivabradine is, in both poses, stably found under the SF. MD simulations provided the unique opportunity to discriminate between these hypotheses confirming that the block is electrostatic and depends on the mutual repulsion between the amine nitrogen of ivabradine and the cation in the SF. These data are in perfect agreement with, and provide an elegant molecular interpretation of, the “current-dependent” HCN4 block model proposed previously (18, 27). In studies of native If and HCN4 (15, 18), voltage dependence of ivabradine block was shown to derive from the direction and amplitude of current flow, rather than from voltage per se, according to a current-dependent block mechanism. This was supported by evidence that i) block is strengthened by outward and relieved by inward current; ii) its voltage dependence shifts in accordance with changes of If reversal potential; and iii) block removal is lost when external Cs+ stops inward current flow. These and other observations led to the proposal that ivabradine block is exerted at the intracellular side and involves competition of the tertiary ammonium ion with permeating ions for a binding site along the permeation pathway. Our combinatory structural and in silico analysis emphasizes that ivabradine differs markedly from that of other HCN channel blockers such as ULFS49 (zatebradine) and ZD7288. For example, block of pacemaker channels by these latter drugs is not current dependent (15).
The mechanism of ivabradine block based on electrostatic repulsion can be easily extended to the block of K+ channels by quaternary ammonium ions, that, similarly to ivabradine, bind to the intracellular pore cavity, just below the SF. X-ray crystallography and simulation studies (25) have shown that, in KcsA, the position of the Quaternary Ammonium (QA) blocker in the vestibule depends on that of K+ in the filter, giving rise to the hypothesis that their interaction is electrostatic, model recently supported by a functional study (41). Comparison of the open-pore structure with and without ivabradine underscores that the drug exerts a stabilizing effect beyond the pore vestibule on the S5 and the S4–S5 linker. Two movements associated with pore opening in HCN4 (4) and in HCN1 (28) appear very pronounced in the structure with ivabradine: i) the up-movement of the cytosolic end of S5 and ii) the unfolding of the S4–S5 linker. The structure of the open pore of HCN4 bound to ivabradine further confirms that these two movements are mandatory steps in the channel opening pathway.
Subtype specificity of HCN blockers is a much-desired property as these channels are involved in diverse human pathologies. HCN4 dysfunctions, for example, lead to cardiac arrhythmias (12), HCN2 leads to chronic neuropathic pain (24), and HCN1 to severe EIEE (22, 23). However, HCN1 and HCN2-based diseases cannot be treated with ivabradine, as its bradycardic impact on the heart, due to HCN4, is unavoidable.
The crucial ivabradine-interacting residues, C479, Y507, and I511, identified here, are identical in all HCN subtypes, apart from a conservative substitution in HCN1 (Iso to Val), which has no impact on IVA block (16). This has two important implications: First, it predicts that the basic mode of ivabradine block should be the same in all HCN subtypes and, therefore, also in the physiologically relevant HCN heteromers (3); second, the sequence identity of the pore cavity among all HCN subtypes predicts that subtype-specific drugs will unlikely emerge from chemical modifications of ivabradine interaction inside the pore.
Materials and Methods
Protein Expression, Purification, and fSEC-TS Assay.
HCN4 protein with an N-terminal His6x-eGFP tag was expressed in HEK293F cells as detailed in SI Appendix. Membranes were isolated and HCN4 was extracted and purified in buffer containing lauryl maltose neopentyl glycol (LMNG), and cholesteryl hemisuccinate (CHS) by a Nickel resin chromatography and then by size-exclusion chromatography, according to the protocol detailed in SI Appendix. Fluorescence Size Exclusion Chromatography-based Thermostability (fSEC-TS) assay was performed using a Superose 6 increase 10/300 GL SEC column (GE Healthcare Life Sciences) connected to a Prominence UFLC system (Shimadzu) possessing a RF-10AXL fluorescence detector (Shimadzu) to monitor the emission signal (509 nm) of the excited eGFP fused at the N terminus of HCN4. A detailed description of sample preparation and data analysis is provided in SI Appendix.
CryoEM Sample Preparation and Data Collection.
Purified HCN4 in presence of ivabradine was applied to glow-discharged UltrAu R0.6/1.0 300-mesh gold holey grids (Quantifoil). Vitrification was performed by using a Vitrobot Mk IV (Thermo Fischer Scientific), and data collection was performed on a Titan Krios electron microscope (FEI) operating at 300kV acceleration voltage. A detailed description of sample preparation and data collection is provided in SI Appendix.
CryoEM Data Processing, Model Building, and Refinement.
Initial steps of data processing were performed using RELION-3.1, as detailed in SI Appendix. A total of 31670 micrograph movies were selected for data processing. As described in SI Appendix, particle picking was performed using both RELION-3.1 and WARP, while 2D/3D classification and 3D refinement were carried out by using RELION-3.1. A final dataset of 444873 particles were selected, extracted with recentered coordinates and an unbinned box size of 400 pixels and moved into CryoSPARCv3.3.1 for a further 3D classification (heterogeneous refinement with four classes) without imposing symmetry. 107523 particles from to the best class were subjected to a non-uniform refinement, again without imposing symmetry, to obtain the final map. Details are reported in SI Appendix. HCN4 – IVA model was rigid body fitted with UCSF CHIMERA and real-space refinement was performed with PHENIX using the previous HCN4 (PDB: 7NMN) coordinates as template.
MD simulations and trajectory analysis.
As detailed in SI Appendix, simulations were conducted using GROMACS with the Amber99sb*-ILDN force field for the protein together with the GAFF2 parameters for IVA+/neutral IVA. All trajectories were analysed using Biotite versions 0.35-0.37 and GROMACS.
Electrophysiology.
Whole-cell recordings were performed using patch-clamp on HEK293T cells or HEK293F cells cultured and transfected as detailed in SI Appendix. HCN4 currents were recorded at room temperature using an ePatch (Elements srl) or a Dagan 3900A amplifier (Dagan Corporation). Signals acquired with the Dagan 3900A were digitized using a Digidata 1550B (Moelcular Devices). A detailed description of current recordings and analysis is provided in SI Appendix. Data were analyzed with Clampfit (Molecular devices) and Origin (OriginLab) softwares.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of beam time on CM01, and we would like to thank Dr. Gregory Effantin for assistance. We also gratefully acknowledge the computing time provided to them on the high-performance computer Lichtenberg at the NHR Centers NHR4CES at TU Darmstadt. This is funded by the Federal Ministry of Education and Research and the state governments participating on the basis of the resolutions of the GWK for national high-performance computing at universities. The work was partially funded by Telethon grant no. GGP20021, Prin 2022 grant no. 2022EMA8FA, and Fondation Leducq Research Grant no. 19CVD03 to D.D. and A.M., by Deutsche Forschungsgemeinschaft under grant no. HA5261/6-1 to J.H.K. and K.H., by Deutsche Forschungsgemeinschaft under grant no. TH558/34-1 to G.T., grant NIH 5 R01 NS 109366 04 for partial support of O.B.C. and by Cariplo Young Investigator grant no. 2018–0231 to A.S. We are grateful to the University of Milano NOLIMITS Center and to Fondazione Romeo and Enrica Invernizzi, Pediatric Research Center, for continuous support.
Author contributions
A.S., A.P., D.D., O.B.C., G.T., and A.M. designed research; A.S., J.H.K., A.C.-S., A.S.S., A.P., R.C., and K.H. performed research; A.S., J.H.K., A.C.-S., A.S.S., A.P., R.C., K.H., M.B., O.B.C., G.T., and A.M. analyzed data; and A.S., M.B., D.D., G.T., and A.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Cryo-EM map was deposited in the Electron Microscopy Data Bank under accession number EMD-16860 (42). Atomic coordinates were deposited into the PDB under accession number 8OFI (43). Required input files to run MD simulations described in this work are available at Zenodo (44). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Chandler N. J., et al. , Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation 119, 1562–1575 (2009), 10.1161/CIRCULATIONAHA.108.804369. [DOI] [PubMed] [Google Scholar]
- 2.Shi W., et al. , Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circulat. Res. 85, e1–e6 (1999), 10.1161/01.res.85.1.e1. [DOI] [PubMed] [Google Scholar]
- 3.Altomare C., et al. , Heteromeric HCN1-HCN4 channels: A comparison with native pacemaker channels from the rabbit sinoatrial node. J. Physiol. 549, 347–359 (2003), 10.1113/jphysiol.2002.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saponaro A., et al. , Gating movements and ion permeation in HCN4 pacemaker channels. Mol. Cell 81, 2929–2943.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer D., Wissmann J., Moroni A., Thiel G., Hamacher K., Weak cation selectivity in HCN channels results from K(+)-Mediated release of Na(+) from selectivity filter binding sites. Function (Oxford England) 3, zqac019 (2022), 10.1093/function/zqac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krumbach J. H., et al. , Alkali metal cations modulate the geometry of different binding sites in HCN4 selectivity filter for permeation or block. J. General Physiol. 155, e202313364 (2023), 10.1085/jgp.202313364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiFrancesco D., Characterization of single pacemaker channels in cardiac sino-atrial node cells. Nature 324, 470–473 (1986), 10.1038/324470a0. [DOI] [PubMed] [Google Scholar]
- 8.DiFrancesco D., A study of the ionic nature of the pace-maker current in calf Purkinje fibres. J. Physiol. 314, 377–393 (1981), 10.1113/jphysiol.1981.sp013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiFrancesco D., Block and activation of the pace-maker channel in calf Purkinje fibres: Effects of potassium, caesium and rubidium. J. Physiol. 329, 485–507 (1982), 10.1113/jphysiol.1982.sp014315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei M., Wu L., Terrar D. A., Huang C.L.-H., Modernized classification of cardiac antiarrhythmic drugs. Circulation 138, 1879–1896 (2018), 10.1161/CIRCULATIONAHA.118.035455. [DOI] [PubMed] [Google Scholar]
- 11.Diaz A., Tardif J.-C., Heart rate slowing versus other pharmacological antianginal strategies. Adv. Cardiol. 43, 65–78 (2006), 10.1159/000095429. [DOI] [PubMed] [Google Scholar]
- 12.DiFrancesco D., Camm J. A., Heart rate lowering by specific and selective I(f) current inhibition with ivabradine: A new therapeutic perspective in cardiovascular disease. Drugs 64, 1757–1765 (2004), 10.2165/00003495-200464160-00003. [DOI] [PubMed] [Google Scholar]
- 13.DiFrancesco D., The role of the funny current in pacemaker activity. Circulat. Res. 106, 434–446 (2010), 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- 14.DiFrancesco D., Some properties of the UL-FS 49 block of the hyperpolarization-activated current (i(f)) in sino-atrial node myocytes. Pflugers Archiv. Eur. J. Physiol. 427, 64–70 (1994), 10.1007/BF00585943. [DOI] [PubMed] [Google Scholar]
- 15.Bucchi A., Baruscotti M., DiFrancesco D., Current-dependent block of rabbit sino-atrial node I(f) channels by ivabradine. J. General Physiol. 120, 1–13 (2002), 10.1085/jgp.20028593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanguay J., Callahan K. M., Avanzo D’N., Characterization of drug binding within the HCN1 channel pore. Sci. Rep. 9, 465 (2019), 10.1038/s41598-018-37116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thollon C., et al. , Electrophysiological effects of S 16257, a novel sino-atrial node modulator, on rabbit and guinea-pig cardiac preparations: Comparison with UL-FS 49. Br. J. Pharmacol. 112, 37–42 (1994), 10.1111/j.1476-5381.1994.tb13025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucchi A., Tognati A., Milanesi R., Baruscotti M., DiFrancesco D., Properties of ivabradine-induced block of HCN1 and HCN4 pacemaker channels. J. Physiol. 572, 335–346 (2006), 10.1113/jphysiol.2005.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiFrancesco D., Borer J. S., The funny current: Cellular basis for the control of heart rate. Drugs 67, 15–24 (2007), 10.2165/00003495-200767002-00003. [DOI] [PubMed] [Google Scholar]
- 20.Cappato R., et al. , Clinical efficacy of ivabradine in patients with inappropriate sinus tachycardia: A prospective, randomized, placebo-controlled, double-blind, crossover evaluation. J. Am. College Cardiol. 60, 1323–1329 (2012), 10.1016/j.jacc.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 21.Bois P., Bescond J., Renaudon B., Lenfant J., Mode of action of bradycardic agent, S 16257, on ionic currents of rabbit sinoatrial node cells. Br. J. Pharmacol. 118, 1051–1057 (1996), 10.1111/j.1476-5381.1996.tb15505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marini C., et al. , HCN1 mutation spectrum: From neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain A J. Neurol. 141, 3160–3178 (2018), 10.1093/brain/awy263. [DOI] [PubMed] [Google Scholar]
- 23.Nava C., et al. , De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 46, 640–645 (2014), 10.1038/ng.2952. [DOI] [PubMed] [Google Scholar]
- 24.Lee M. C., et al. , A randomised, double-blind, placebo-controlled crossover trial of the influence of the HCN channel blocker ivabradine in a healthy volunteer pain model: An enriched population trial. Pain 160, 2554–2565 (2019), 10.1097/j.pain.0000000000001638. [DOI] [PubMed] [Google Scholar]
- 25.Faraldo-Gómez J. D., et al. , Mechanism of intracellular block of the KcsA K+ channel by tetrabutylammonium: Insights from X-ray crystallography, electrophysiology and replica-exchange molecular dynamics simulations. J. Mol. Biol. 365, 649–662 (2007), 10.1016/j.jmb.2006.09.069. [DOI] [PubMed] [Google Scholar]
- 26.Saponaro A., Sharifzadeh A. S., Moroni A., Detection of ligand binding to purified HCN channels using fluorescence-based size exclusion chromatography. Methods Enzymol. 652, 105–123 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Bucchi A., et al. , Identification of the molecular site of ivabradine binding to HCN4 channels. PLoS One 8, e53132 (2013), 10.1371/journal.pone.0053132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burtscher V., et al. , Structural basis for hyperpolarization-dependent opening of the human HCN1 Channel. bioRxiv [Preprint] (2023), 10.1101/2023.08.17.553623 (Accessed 17 August 2023). [DOI] [PMC free article] [PubMed]
- 29.Liu S., Lockless S. W., Equilibrium selectivity alone does not create K+-selective ion conduction in K+ channels Nat. Commun. 4, 2746 (2013), 10.1038/ncomms3746. [DOI] [PubMed] [Google Scholar]
- 30.Iyer B. R., Mahalakshmi R., Hydrophobic characteristic is energetically preferred for cysteine in a model membrane protein. Biophys. J. 117, 25–35 (2019), 10.1016/j.bpj.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyte J., Doolittle R. F., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982), 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 32.Marino S. M., Gladyshev V. N., Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 404, 902–916 (2010), 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazmanian K., Sargsyan K., Grauffel C., Dudev T., Lim C., Preferred hydrogen-bonding partners of cysteine: Implications for regulating cys functions. J. Phys. Chem. B 120, 10288–10296 (2016), 10.1021/acs.jpcb.6b08109. [DOI] [PubMed] [Google Scholar]
- 34.Nagano N., Ota M., Nishikawa K., Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett. 458, 69–71 (1999), 10.1016/s0014-5793(99)01122-9. [DOI] [PubMed] [Google Scholar]
- 35.Wolfenden R., Andersson L., Cullis P. M., Southgate C. C., Affinities of amino acid side chains for solvent water. Biochemistry 20, 849–855 (1981), 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 36.Macri V., Angoli D., Accili E. A., Architecture of the HCN selectivity filter and control of cation permeation. Sci. Rep. 2, 894 (2012), 10.1038/srep00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng L., Kinard K., Rajamani R., Sanguinetti M. C., Molecular mapping of the binding site for a blocker of hyperpolarization-activated, cyclic nucleotide-modulated pacemaker channels. J. Pharmacol. Exp. Therapeutics 322, 931–939 (2007), 10.1124/jpet.107.121467. [DOI] [PubMed] [Google Scholar]
- 38.Shin K. S., Rothberg B. S., Yellen G., Blocker state dependence and trapping in hyperpolarization-activated cation channels: Evidence for an intracellular activation gate. J. General Physiol. 117, 91–101 (2001), 10.1085/jgp.117.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.BoSmith R. E., Briggs I., Sturgess N. C., Inhibitory actions of ZENECA ZD7288 on whole-cell hyperpolarization activated inward current (If) in guinea-pig dissociated sinoatrial node cells. Br. J. Pharmacol. 110, 343–349 (1993), 10.1111/j.1476-5381.1993.tb13815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall P. W., et al. , ICI D7288, a novel sinoatrial node modulator. J. Cardiovasc. Pharmacol. 21, 902–906 (1993), 10.1097/00005344-199306000-00008. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel T. S., et al. , Asymmetric interplay between K(+) and blocker and atomistic parameters from physiological experiments quantify K(+) channel blocker release. Front. Physiol. 12, 737834 (2021), 10.3389/fphys.2021.737834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saponaro A., et al. , Ivabradine bound to HCN4 channel. EMDB. https://www.ebi.ac.uk/emdb/EMD-16860. Deposited 14 February 2023.
- 43.Saponaro A., et al. , Ivabradine bound to HCN4 channel. PDB. https://www.rcsb.org/structure/8OFI. Deposited 14 February 2023.
- 44.Saponaro A., et al. , Structural determinants of ivabradine block of the open pore of HCN4. Zenodo. 10.5281/zenodo.10889976. Deposited 10 April 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Cryo-EM map was deposited in the Electron Microscopy Data Bank under accession number EMD-16860 (42). Atomic coordinates were deposited into the PDB under accession number 8OFI (43). Required input files to run MD simulations described in this work are available at Zenodo (44). All other data are included in the manuscript and/or SI Appendix.