Abstract
Introduction
Mounting evidence indicates that an individual's humoral adaptive immune response plays a critical role in the setting of SARS-CoV-2 infection, and that the efficiency of the response correlates with disease severity. The relationship between the adaptive immune dynamics in the lower airways with those in the systemic circulation, and how these relate to an individual's clinical response to SARS-CoV-2 infection, are less understood and are the focus of this study.
Material and methods
We investigated the adaptive immune response to SARS-CoV-2 in paired samples from the lower airways and blood from 27 critically ill patients during the first wave of the pandemic (median time from symptom onset to intubation 11 days). Measurements included clinical outcomes (mortality), bronchoalveolar lavage fluid (BALF) and blood specimen antibody levels, and BALF viral load.
Results
While there was heterogeneity in the levels of the SARS-CoV-2-specific antibodies, we unexpectedly found that some BALF specimens displayed higher levels than the paired concurrent plasma samples, despite the known dilutional effects common in BALF samples. We found that survivors had higher levels of anti-spike, anti-spike-N-terminal domain and anti-spike-receptor-binding domain IgG antibodies in their BALF (p<0.05), while there was no such association with antibody levels in the systemic circulation.
Discussion
Our data highlight the critical role of local adaptive immunity in the airways as a key defence mechanism against primary SARS-CoV-2 infection.
Shareable abstract
Among critically ill COVID-19 patients, low levels of anti-SARS-CoV-2 antibodies in the lower airways, more than those levels in blood, correlate with poor outcome. This suggests the local mucosal adaptive immune system is critical to SARS-CoV-2 response. https://bit.ly/49CNazp
Introduction
The COVID-19 pandemic has had unprecedented effects on human health [1]. Some individuals with SARS-CoV-2 infection remain asymptomatic; others develop severe illness with acute respiratory distress syndrome and multi-organ failure. While new COVID-19 infections remain common, over the past 3 years the proportion that becomes critically ill has decreased significantly, presumably owing to increasing population immunity to SARS-CoV-2 from prior infections and/or COVID-19 vaccination, or as a result of the improved availability of therapeutics such as antivirals [2]. However, among people with severe disease, the immunological determinants of fatal outcomes are not well known. To understand what contributes to disease severity, progression and mortality, there has been much focus on the roles of the innate and adaptive immune responses in determining clinical outcomes.
Several investigations have detailed circulating levels of IgM, IgA and IgG antibodies directed against different domains of the SARS-CoV-2 spike protein, and how these change over the course of the disease [1–5]. Other studies have aimed to correlate an individual's adaptive systemic immune response to SARS-CoV-2 infection with their clinical course and disease severity [3–8]. Generally, this has been done by comparing SARS-CoV-2-specific antibody levels in the blood of asymptomatic infected individuals to those with mild, moderate or severe COVID-19 disease. Overall, higher levels of IgG and IgA antibodies have been detected in those with severe disease while IgM levels are not significantly associated with severity of illness [3, 7, 9–11]. Furthermore, a failure to mount an early anti-SARS-CoV-2 immunoglobulin response in blood is associated with poor outcome [6, 8].
What remains unexplored is an evaluation of immunoglobulin levels in the blood of patients with similar disease severity (e.g. all intubated, or all critically ill) or the quantification of antibodies in the lower airways [11, 12]. In a prior investigation, we showed that in cross-sectional sampling of the lower airways of critically ill COVID-19 patients obtained ∼10 days after intubation, lower levels of anti-SARS-CoV-2 antibodies were associated with increased mortality [12]. In the current study, we evaluated in parallel the dynamics of local (lower airway) and systemic (blood) adaptive immune signatures and clinical outcomes among hospitalised, intubated and critically ill COVID-19 adult patients. We hypothesised that anti-SARS-CoV-2 antibody levels within the lower airways, rather than those in systemic circulation, would be associated with mortality among critically ill COVID-19 patients.
Material and methods
Study population
This observational longitudinal cohort study included patients 18 years or older admitted to the intensive care units at NYU Langone Health (New York, NY, USA) from 10 March to 10 May 2020 with a nasal swab-confirmed diagnosis of SARS-CoV-2 infection by reverse transcriptase PCR (RT-PCR) assay and respiratory failure requiring invasive mechanical ventilation. Clinical specimens were obtained during clinically indicated bronchoscopy performed for airway clearance or for percutaneous tracheostomy placement. Surviving subjects signed informed consent to participate in this study. Specimens and metadata from deceased patients were de-identified and included in this study. Comprehensive demographic and clinical data were collected from the electronic medical record. All protocols were carried out in accordance with the Declaration of Helsinki and the study protocol was approved by the Institutional Review Board of New York University (IRB#s16-00122).
Lower airway bronchoscopic sampling procedure
A total of 27 unvaccinated and mechanically ventilated COVID-19-infected adult patients with longitudinal collection of lower airway bronchoalveolar lavage fluid (BALF) and blood specimens were included. At least two BALF specimens were obtained per patient (median 2 specimens, interquartile range (IQR) 2–3 specimens). Paired BALF and blood specimens were collected at the same time points for 16 patients. There were 11 patients with BALF samples who did not have blood specimens collected. The median time from symptom onset to the first (baseline) BALF specimen collection was 17 days (IQR 14–20 days). Most baseline BALF specimens were obtained within the first week of intubation (median 5 days, IQR 3–7 days from intubation).
Both background and supraglottic (buccal) specimens were obtained prior to the procedure, as previously described [13]. The background specimens were obtained by passing sterile saline through the suctioning channel of the bronchoscope prior to the procedure. BALF specimens were obtained from one lung segment at the discretion of the treating physician as clinically indicated. Specimens were then transferred to a biosafety level 3 (BSL3) laboratory for processing. Once there, 2 mL of whole BALF was stored in a tube prefilled with 2 mL of Zymo Research's DNA/RNA Shield (R1100-250, Zymo Research Corp.) for RNA/DNA preservation and virus inactivation. In addition, background control specimens (saline passed through the bronchoscope prior to procedure) and supraglottic aspirates were stored in the same RNA/DNA shield. A subset of specimens underwent BALF cell separation by centrifugation and cells were cryopreserved in DMSO while acellular BALF was aliquoted for cytokine measurements. A paired blood specimen was also obtained in EDTA tubes (catalogue no. 355450; Becton Dickinson) and PAXgene Blood RNA tubes (catalogue no. 762165; PreAnalytiX).
SARS-CoV-2 antibody profiling in BALF and blood specimens
The New York University proprietary custom Multiepitope Bead-based Immunoassay was used to measure antibody responses to three SARS-CoV-2 recombinant proteins (SARS-CoV-2 (2019-nCoV) spike S2 ECD-His, catalogue no. 40590-V08B; SARS-CoV-2 (2019-nCoV) spike receptor binding domain (RBD)-His, catalogue no. 40592-V08B; and SARS-CoV-2 (2019-nCoV) spike S1 N-terminal domain (NTD)-His & AVI, catalogue no. 40591-V49H-B; all Sino Biological), using control analytes of human serum albumin, tetanus toxoid (TT), staphylococcal leucocidin (LukS) and anti-human IgG (Jackson Immunoresearch) coupled to commercial paramagnetic beads (MagPix, Luminex), as adapted from the manufacturer's instructions [11, 12]. Antibody reactivity was evaluated by comparing levels in plasma and BALF among SARS-CoV-2-infected patients and among smokers recruited for a pre-pandemic investigation in which research bronchoscopy was performed [14]. The immunoglobulin levels are qualitatively presented as the median fluorescence intensity (MFI) units. BALF dilution was 1:200 and plasma dilution was 1:1000.
Viral load detection targeting the N gene
Digital droplet PCR was performed using the Bio-Rad SARS-CoV-2 Droplet Digital PCR Kit following the manufacturer's instructions. The kit uses primers and probes for three targets from the Centers for Disease Control and Prevention 2019-nCoV assay. The system generates droplets for every sample tested and the PCR amplification occurs within each droplet for three targets: N1 and N2 target the N gene of the SARS-CoV-2 and RP targets the human RNAse P gene for sample integrity and as an internal control. Following PCR amplification, the positive and negative droplets are designated as a dot-plot on the QuantaSoft Analysis Pro software (BioRad). Once the data from the run is loaded onto the software, the triplex probe mix assay is selected for visualising the two-dimensional dot-plot graph. On the graph, the positive and negative dots appear as clusters. The different clusters were defined using the colour-coded guide in the software. These clusters can be defined as positive or negative reactions for the different targets (i.e. N1, N2 and RP, or a combination of any of these). The clusters were designated as positive for N1, N2 or RP or a combination (N1+N2, N1+RP etc.). After applying the cluster designation, each individual well was selected separately to manually inspect the cluster designations. Quantifications of the three targets were generated by the software as copies·μL−1 once the cluster designation was complete. The calculations were then performed to factor in the dilutions made for the PCR run to generate the RNA copies·μL−1. The absolute viral load was calculated using the average of the N1 and N2 genes.
Statistical analysis
To assess the significance of the observed differences in antibody levels between paired blood and BALF specimens, a Wilcoxon signed-rank test was used. A Mann–Whitney U test was employed to examine the differences between antibody levels in surviving versus deceased patients. These nonparametric tests were selected owing to the non-normal distribution of the immunoglobulin data (supplementary table S1), the relatively small sample size and the independent nature of the two groups. A Pearson correlation was used for the correlation analysis between viral load and BALF levels of SARS-CoV-2-specific immunoglobulins. Study sample sizes were not predetermined through formal power analysis and the investigation was conducted based on the opportunity to study such samples.
Results
Cohort/study design
Table 1 shows demographic and clinical characteristics of the included patients based on survival (21 survivors versus six deceased). The majority of patients were male (26 patients, 96%) and younger than 51 years (19 patients, 70%). A history of asthma, chronic kidney disease and organ transplant was significantly associated with mortality (chi-square p-value <0.001). Overall, 33% of the cohort required dialysis and 63% of the cohort were placed on veno-venous extracorporeal membrane oxygenation.
TABLE 1.
Baseline demographics and clinical data
| Totalbold> | Survived | Deceased | p-value | |
|---|---|---|---|---|
| Patients (n) | 27 | 21 (77.8) | 6 (22.2) | |
| Age (years) | 47 (39–54) | 46 (33–49) | 51 (49–56) | 0.08 |
| Age category (years) | 0.13 | |||
| <51 | 19 (70.4) | 16 (76.2) | 3 (50) | |
| 51–60 | 3 (11.1) | 1 (4.8) | 2 (33.3) | |
| 61–70 | 3 (11.1) | 3 (14.3) | 0 (0) | |
| >70 | 2 (7.4) | 1 (4.8) | 1 (16.7) | |
| Sex (male) | 26 (96.3) | 20 (95.2) | 6 (100) | 0.59 |
| Race | 0.46 | |||
| White | 10 (37) | 7 (33.3) | 3 (50) | |
| Other/unknown | 17 (63) | 14 (66.7) | 3 (50) | |
| Ethnicity | 0.48 | |||
| Hispanic or Latino | 9 (33.3) | 7 (33.3) | 2 (33.3) | |
| Not Hispanic or Latino | 14 (51.9) | 10 (47.6) | 4 (66.7) | |
| Unknown | 4 (14.8) | 4 (19) | 0 (0) | |
| BMI (kg·m−2) | 29 (24–33) | 27 (24–30) | 33 (29.75–35.5) | 0.16 |
| Charlson Comorbidity Index | 3 (2–4.5) | 3 (2.25–3.75) | 3 (1–5) | 0.18 |
| Comorbidities | ||||
| Asthma | 2 (7.4) | 0 (0) | 2 (33.3) | <0.001 |
| Chronic kidney disease | 2 (7.4) | 0 (0) | 2 (33.3) | <0.001 |
| Prior stroke | 4 (14.8) | 2 (9.5) | 2 (33.3) | 0.15 |
| Diabetes | 10 (37) | 9 (42.9) | 1 (16.7) | 0.24 |
| Hyperlipidaemia | 10 (37) | 7 (33.3) | 3 (50) | 0.46 |
| Hypothyroidism | 2 (7.4) | 1 (4.8) | 1 (16.7) | 0.33 |
| Hypertension | 13 (48.1) | 9 (42.9) | 4 (66.7) | 0.30 |
| Prior solid organ or stem cell transplant | 2 (7.4) | 0 (0) | 2 (33.3) | <0.001 |
| Smoking status | 0.87 | |||
| Current | 4 (14.8) | 3 (14.3) | 1 (16.7) | |
| Ex-smoker | 3 (11.1) | 2 (9.5) | 1 (16.7) | |
| Never-smoker | 20 (74.1) | 16 (76.2) | 4 (66.7) | |
| Early hospitalisation details | ||||
| Symptom onset to intubation | 11 (7.5–14.5) | 11 (7–14) | 11.5 (9.5–16.5) | 0.46 |
| Hospital admission to intubation | 3.5 (2–6.5) | 2 (1–4) | 7 (5–10) | 0.09 |
| APACHE score (day of intubation) | 19 (16–25) | 17 (15–23) | 26 (20.5–29.25) | 0.03 |
| Outcomes (days) | ||||
| Hospital length of stay | 60 (39–83.5) | 60 (41–83) | 57 (34.75–122.75) | 0.66 |
| ICU length of stay | 51 (34.5–65.5) | 51 (35–64) | 51 (35–76) | 0.84 |
| Ventilator# | 48 (24.5–62) | 48 (24–62) | 46.5 (28–76.25) | 0.56 |
Data are expressed as n (%) or median (interquartile range), unless otherwise indicated. Chi square and Mann–Whitney Wilcoxon tests were performed for categorical and continuous variables, respectively. Bold text indicated statistical significance. BMI: body mass index; APACHE: Acute Physiology and Chronic Health Evaluation; ICU: intensive care unit. # : total number of days on mechanical ventilation.
Most patients (25 out of 27, 93%) were hospitalised during March and April 2020, before the availability of COVID-19 vaccines and before clinical management was standardised. Nearly all patients received substantial doses of corticosteroids during their hospitalisation, defined as equivalent to daily prednisone dose ≥40 mg (22 patients, 82%). The median time spent on prednisone was 21 days (IQR 9–40 days), with median daily prednisone equivalent of 37 mg (IQR 12–75 mg). There was no difference in the median number of days on prednisone within the first 2 weeks of hospitalisation between those who survived (8 days, IQR 5–9 days) and those who died (8 days, IQR 5–10 days) (p=0.78). There was also no statistical difference in the average of each individual's average daily dose of steroids (prednisone equivalent 50 mg, IQR 41–74 mg versus 50 mg, IQR 35–104 mg; p=0.91). Only two patients (7.4%) received remdesivir treatment during their hospitalisation, and 17 patients (63%) received tocilizumab. No patients in this cohort received monoclonal antibodies.
Antibody profile in the lower airways and systemic circulation
To investigate antibody responses to COVID-19 and other antigens, we used a previously validated multiplex assay with >20-fold greater sensitivity than an index clinical anti-spike assay [11, 12]. We first compared the levels of several antibodies to SARS-CoV-2-specific (spike (total), spike-NTD, spike-RBD, nucleocapsid) and non-SARS-CoV-2-specific epitopes, with a control antimicrobial response to TT reflecting prior vaccination and to LukS reflecting past natural staphylococcal infection, in plasma and lower airway specimens from patients with COVID-19 and SARS-CoV-2-negative controls (supplementary figure S1). As anticipated at these post-infectious time points, SARS-CoV-2-specific antibody levels were significantly higher in COVID-19 patients compared to controls (p<0.001), both in the lower airways and in the systemic circulation (supplementary figure S1A). In contrast, circulating antibodies against non-SARS-CoV-2 antigens (TT and LukS) were similar in specimens from COVID-19 patients and controls. However, when the levels of these same antibodies against non-SARS-CoV-2 antigens (TT and LukS) were evaluated in the lower airways, we observed significantly higher levels among COVID-19 patients (p<0.001, supplementary figure S1B).
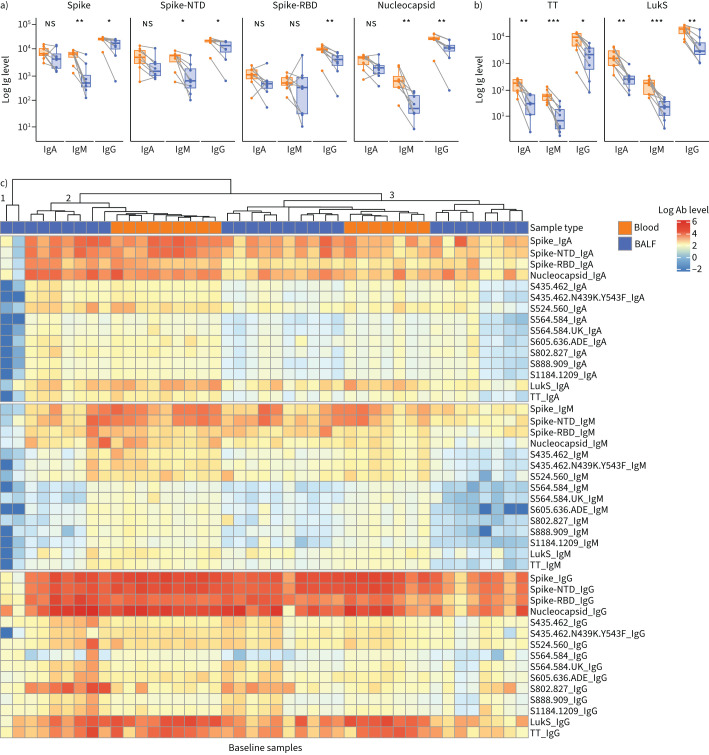
We then evaluated the levels of IgA, IgG and IgM antibodies in the BALF and paired blood specimens at baseline. As expected for a BALF specimen, the concentrations of antibodies in the lower airway specimens were overall lower than those in the systemic circulation (figure 1a, b). Notably, while this trend was consistent among the two non-SARS-CoV-2-related antibody responses (TT and LukS), for some patients certain SARS-CoV-2-related antibody types were higher in BALF specimens compared to blood.
FIGURE 1.
Baseline antibody levels in paired lower airway and plasma specimens. a, b) Paired blood and bronchoalveolar lavage fluid (BALF) specimens at baseline, with lines connecting specimens from the same subject (n=8), showing levels of antibodies to SARS-CoV-2-specific epitopes (a) and non-SARS-CoV-2-specific epitopes (b). c) Heatmap of levels of antibodies to both SARS-CoV-2-specific and non-SARS-CoV-2-specific epitopes in baseline blood and BALF specimens (blood, n=16 specimens; BALF, n=27). Statistical significance based on Wilcoxon signed rank test. Ig: immunoglobulin; NTD: N-terminal domain; RBD: receptor-binding domain; ns: nonsignificant; TT: tetanus toxoid; LukS: staphylococcal leucocidin; Ab: antibody. *: p≤0.05; **: p≤0.01; ***: p≤0.001.
Clustering analyses of baseline lower airway and blood specimen levels revealed three clusters (figure 1c). Cluster one included two lower airway specimens with low levels of all immunoglobulin isotypes. Clusters two and three contained both BALF and plasma specimens, distinguished by cluster two having higher levels of both IgA and IgM spike, spike-NTD and spike-RBD antibodies.
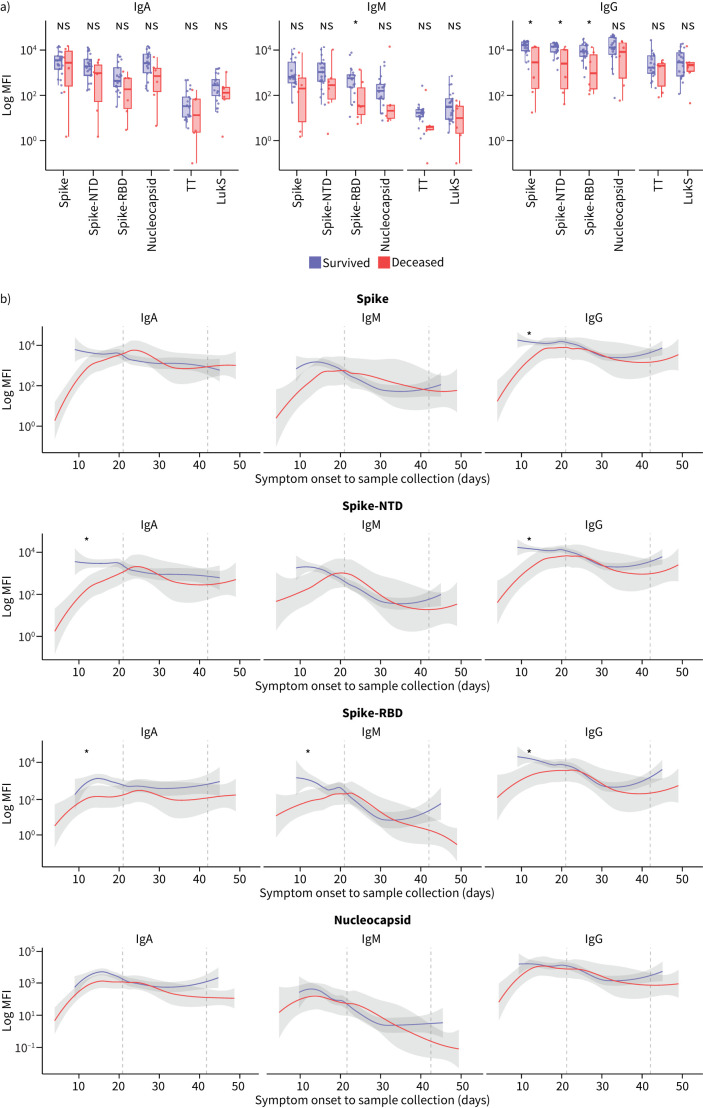
Impaired adaptive lower airway immune responses to SARS-CoV-2 among deceased patients
We examined the association of baseline levels of antibodies and clinical prognosis. In blood, neither levels of SARS-CoV-2-specific antibodies nor levels of non-SARS-CoV-2-specific antibodies were significantly different between the deceased and survivor subgroups (supplementary figure S2A, p>0.05). In contrast, baseline levels of many anti-SARS-CoV-2 antibodies in BALF specimens, especially for the IgG isotype, were lower among the deceased patients compared to the survivors (figure 2a). We modelled the dynamic change in the levels of SARS-CoV-2-related antibodies over time. For the time points available, the anti-SARS-CoV-2-specific immunoglobulin levels in blood did not follow a stable pattern and no clear differences were found when comparing the deceased and survivor groups (supplementary figure S2B, p>0.05). In contrast, in the deceased group, levels of many anti-SARS-CoV-2-specific antibodies in the lower airway showed a blunted response within the first 21 days from symptom onset compared to the survivor group (figure 2b). More specifically, this trend was notable among anti-spike-RBD IgM antibody levels, anti-spike-NTD and anti-spike-RBD IgA antibody levels, and anti-spike, anti-spike-NTD and anti-spike-RBD IgG antibody levels, but not among anti-nucleocapsid antibodies (figure 2b, p≤0.05 for those deemed significant). This was not the case after the first 21 days from symptom onset, where we did not find a significant difference in antibody levels in the lower airway in deceased patients compared to survivors (p>0.05).
FIGURE 2.
Lower airway levels of SARS-CoV-2-specific and non-SARS-CoV-2-specific antibodies by clinical outcome. a) Baseline levels of antibodies to SARS-CoV-2-specific epitopes (spike, spike-N-terminal domain (NTD), spike-receptor-binding domain (RBD), nucleocapsid) and non-SARS-CoV-2-specific epitopes (tetanus toxoid (TT), staphylococcal leucocidin (LukS)) by isotope in bronchoalveolar lavage fluid (BALF) specimens in the survivor versus deceased groups. b) Longitudinal levels of antibodies to SARS-CoV-2-specific epitopes in BALF over time by clinical outcome from day of symptom onset to sample collection. Statistical significance tested within the first 3 weeks, weeks 4–6 and >6 weeks by Mann–Whitney Wilcoxon test. Ig: immunoglobulin; MFI: median fluorescence intensity; ns: nonsignificant. *: p≤0.05.
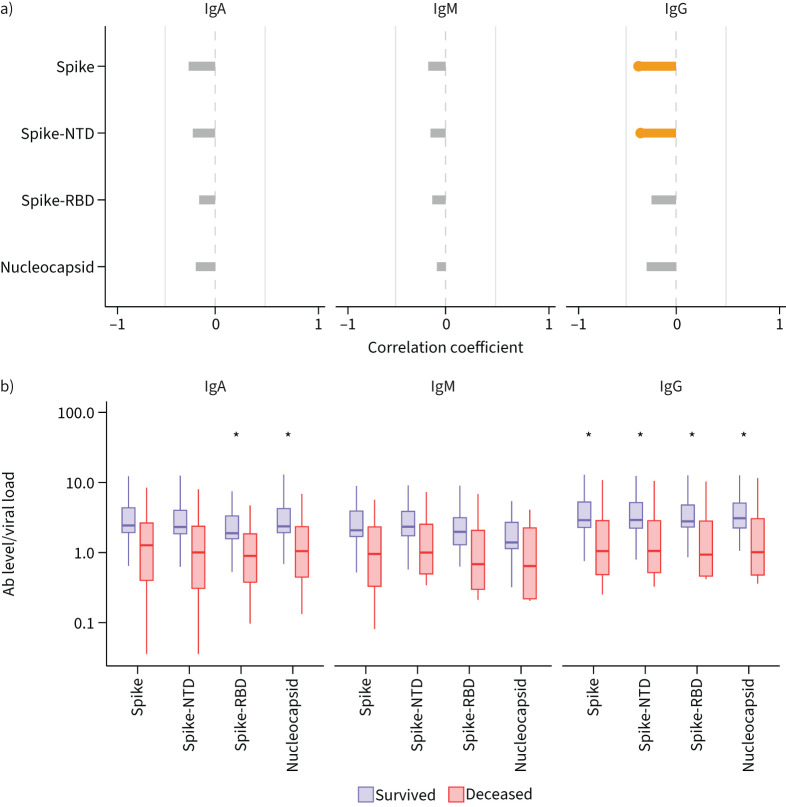
We then investigated whether anti-SARS-CoV-2 antibody responses in the lung and systemic circulation were associated with viral control. We assessed the relationship of SARS-CoV-2 levels (as measured by quantitative RT-PCR testing of BALF specimens) in the lower airways with levels of anti-SARS-CoV-2 antibodies in both the blood and lower airways. In blood, the levels of anti-spike IgM, anti-spike-RBD IgA and anti-nucleocapsid IgA were positively correlated with lower airway SARS-CoV-2 viral load (supplementary figure S3A, Pearson p<0.05). In contrast, BALF levels of anti-spike IgG and anti-spike-NTD IgG were negatively correlated with lower airway SARS-CoV-2 viral load levels (figure 3a, Pearson p<0.05). In addition, we calculated the ratio of anti-SARS-CoV-2 antibodies to the absolute SARS-CoV-2 levels in the BALF at baseline (figure 3b, p≤0.05). The ratio was significantly lower in the deceased group compared to those that survived for all anti-SARS-CoV-2 IgG antibodies as well as for anti-spike-RBD IgA and anti-nucleocapsid IgA (figure 3b, p≤0.05). In comparison, we did not find statistically significant differences in the ratio of blood anti-SARS-CoV-2 antibodies to absolute SARS-CoV-2 levels between the deceased and survivor groups (supplementary figure 3B). These data further support the hypothesis that an inability to mount an early local pulmonary anti-SARS-CoV-2-specific antibody response is a key step in viral replication and COVID-19 pneumonitis.
FIGURE 3.
Correlation between viral load and SARS-CoV-2-specific immunoglobulin (Ig) levels in the lower airways. a) Correlation between bronchoalveolar lavage fluid (BALF) viral load and BALF levels of SARS-CoV-2-specific immunoglobulins. Orange colour indicates statistically significant correlation (Pearson r<0.05). b) Ratio of the log10 anti-SARS-CoV-2 antibodies (Ab) to the log10 viral load in the BALF at baseline. Statistical significance based on Mann–Whitney Wilcoxon test. NTD: N-terminal domain; RBD: receptor-binding domain. *: p≤0.05.
Discussion
The systemic host adaptive immune system plays a critical role in the defence against and control of SARS-CoV-2 infection, but the immune response in the mucosae of the lower airways remains poorly understood. Our cohort of 27 mechanically ventilated COVID-19 patients demonstrated heterogeneity in the levels of anti-SARS-CoV-2 antibodies. A proportion of the paired samples showed similar antibody levels in the BALF and plasma specimens despite the known dilution expected from BALF sampling. Strikingly, reduced levels of anti-spike, anti-spike-NTD and anti-spike-RDB IgG antibodies in lower airways, but not in systemic circulation, correlated with fatal outcome. Furthermore, this decreased immunological response in the lower airway was inversely correlated with SARS-CoV-2 levels.
Prior investigations have primarily focused on the systemic adaptive immune response following acute infection with SARS-CoV-2, with little emphasis on mucosal immunity. These studies found that blood levels of virus-specific antibodies inversely correlate with disease severity, with delayed systemic antibody production associated with poor outcome [4, 7]. Given that the site of immunological exposure to viral antigens can significantly influence the immune response and protect against subsequent infections, understanding the dynamic antibody response in key mucosal sites is essential. While we are not the first to explore mucosal immunity, most other studies have focused on quantifying SARS-CoV-2-specific antibodies in the saliva and nares [6, 15–18]. Levels of viral-specific antibodies in the upper airways significantly correlate with levels in the blood, suggesting that antibody levels in the mucosa may be derived from transudation from blood rather than local production [15, 18]. Our study seeks to explore the interplay between lower airway and systemic immunity.
We rationalised that higher levels of non-SARS-CoV-2-related antibodies in the lower airways of mechanically ventilated COVID-19 patients, compared to controls, are likely due to higher protein concentrations in the alveolar space of individuals with acute respiratory distress syndrome as a consequence of capillary leak and/or high local antibody production by mucosal lymphoid tissue associated with the lower airway [19]. Here, we have shown that, while blood levels of non-SARS-CoV-2-specific antibodies are similar in COVID-19 patients and controls, antibody levels in the lower airways are significantly higher in COVID-19 patients. When evaluating paired BALF and blood specimens among our COVID-19 patients, we found that for some subjects SARS-CoV-2-related antibodies were higher in BALF specimens than in blood. This finding was unexpected because the technique for BALF sample collection dilutes epithelial lining fluid ∼50–100 times using sterile saline. Hence, higher BALF antibody levels in these individuals likely reflects high local anti-COVID-19 pulmonary production. This strongly supports a role for active pulmonary mucosal antibody production during SARS-CoV-2 infection.
With only 27 patients included in this analysis, this investigation is limited by its small sample size, with the majority of subjects being male. The study was focused on patients who were mechanically ventilated, with an emphasis on collecting specimens longitudinally and whenever a clinically indicated bronchoscopy was performed. For this reason, it was not always possible to collect specimens at precise intervals and not every BALF and blood specimen was collected on the same day. This limited the number of specimens we could include in our paired analysis. Because we used the first clinically indicated bronchoscopy to collect our baseline sample, we do not have specimens collected from these patients prior to the onset of their severe illness; consequently, we were unable to evaluate immunoglobulin responses at earlier time points. However, by including patients with similar degrees of disease severity, we established an association with poor clinical outcome that may aid future risk-stratification of critically ill patients. Finally, these data are limited to unvaccinated subjects during the first wave of the COVID-19 pandemic. Because COVID-19 vaccination has reduced the risk of severe COVID-19, further investigations are needed to understand the lack of sufficient immune response among patients experiencing breakthrough SARS-CoV-2 infections.
Future investigations should include larger cohorts of patients with a wider range of disease severity, with an emphasis on prospective serial collection of paired lower airway and blood specimens before and after progression to severe disease. Further work is also needed to assess other factors that may influence the adaptive immune responses, including gene expression profiling and in-depth assessments of the regulation of T-cell and B-cell functions.
In summary, these data support not only the importance of local adaptive immune responses in the lower airways of mechanically ventilated COVID-19 adults, but also the lower airway's contribution to disease outcomes. An improved understanding of the immune response in the respiratory tract might lead to identification of prognostic biomarkers and development of new approaches to treat viral pathogens such as SARS-CoV-2. This study serves to highlight and add to evidence supporting the key role of local antibody production in the mucosal tissues as a mechanism of defence against poor outcomes of SARS-CoV-2 infection.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00789-2023.SUPPLEMENT2 (73.8KB, pdf)
Supplementary figures 00789-2023.SUPPLEMENT (393KB, pdf)
Acknowledgements
We thank Pravesh Regmi and Jung-Ho Youn from the National Institutes of Health's Department of Laboratory Medicine, Clinical Center, for help with viral load detection. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Provenance: Submitted article, peer reviewed.
Ethics statement: Informed consent was obtained from all subjects and the study was approved by our Institutional Review Board.
Code used for the analyses presented in the current manuscript are available at https://github.com/segalmicrobiomelab/SARS-CoV-2.airway.adaptive.immune.response.
Conflict of interest: Other than the funding disclosed in the Support Statement, there are no further conflicts of interest to disclose.
Support statement: This study was supported by grants R37 CA244775 (to L.N. Segal; from the National Cancer Institute/National Institutes of Health (NIH)) and R21 GM147800 (to L.N. Segal; from the National Institute of General Medical Sciences/NIH); COVID-19 and Respiratory Virus Research Award number COVID-917496 (to L.N. Segal; from the American Lung Association); a Colton Pilot Project Grant (to L.N. Segal, L. Angel and K. Krolikowski); grant UWSC1085.1 (to L.N. Segal, R. Postelnicu and L. Evans; from the Centers for Disease Prevention and Control Foundation); a PACT grant (to L.N. Segal; from the Foundation for the NIH); grants R01 AI143861 (to K. Khanna; from the National Institute of Allergy and Infectious Disease (NIAID)/NIH) and R01 AI143861-02S1 (to K. Khanna; from NIAID/NIH); and a Stony Wold-Herbert Fund Grant-in-Aid/Fellowship (to I. Sulaiman and C.R. Barnett). This work was supported in part by the Division of Intramural Research of the NIAID/NIH (to E. Ghedin and M. Chung) and a Chest Research Grant in COVID-19 (to C.R. Barnett). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Msemburi W, Karlinsky A, Knutson V, et al. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2023; 613: 130–137. doi: 10.1038/s41586-022-05522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narayanan SA, Jamison DA, Jr, Guarnieri JW, et al. A comprehensive SARS-CoV-2 and COVID-19 review, part 2: host extracellular to systemic effects of SARS-CoV-2 infection. Eur J Hum Genet 2024; 32: 10–20. doi: 10.1038/s41431-023-01462-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carsetti R, Zaffina S, Piano Mortari E, et al. Different innate and adaptive immune responses to SARS-CoV-2 infection of asymptomatic, mild, and severe cases. Front Immunol 2020; 11: 610300. DOI: 10.3389/fimmu.2020.610300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng X, Yin J, Zhang J, et al. Longitudinal profiling of antibody response in patients with COVID-19 in a tertiary care hospital in Beijing, China. Front Immunol 2021; 12: 614436. DOI: 10.3389/fimmu.2021.614436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li K, Huang B, Wu M, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun 2020; 11: 6044. DOI: 10.1038/s41467-020-19943-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas C, Klein J, Sundaram ME, et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat Med 2021; 27: 1178–1186. DOI: 10.1038/s41591-021-01355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravichandran S, Lee Y, Grubbs G, et al. Longitudinal antibody repertoire in “mild” versus “severe” COVID-19 patients reveals immune markers associated with disease severity and resolution. Sci Adv 2021; 7: eabf2467. DOI: 10.1126/sciadv.abf2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozonoff A, Schaenman J, Jayavelu ND, et al. Phenotypes of disease severity in a cohort of hospitalized COVID-19 patients: results from the IMPACC study. EBioMedicine 2022; 83: 104208. DOI: 10.1016/j.ebiom.2022.104208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis 2020; 71: 2027–2034. DOI: 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang NY, Pang AS, Chow VT, et al. Understanding neutralising antibodies against SARS-CoV-2 and their implications in clinical practice. Mil Med Res 2021; 8: 47. DOI: 10.1186/s40779-021-00342-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cagigi A, Yu M, Osterberg B, et al. Airway antibodies emerge according to COVID-19 severity and wane rapidly but reappear after SARS-CoV-2 vaccination. JCI Insight 2021; 6: e151463. DOI: 10.1172/jci.insight.151463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulaiman I, Chung M, Angel L, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol 2021; 6: 1245–1258. DOI: 10.1038/s41564-021-00961-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsay JJ, Wu BG, Badri MH, et al. Airway microbiota is associated with upregulation of the PI3 K pathway in lung cancer. Am J Respir Crit Care Med 2018; 198: 1188–1198. DOI: 10.1164/rccm.201710-2118OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulaiman I, Wu BG, Li Y, et al. Functional lower airways genomic profiling of the microbiome to capture active microbial metabolism. Eur Respir J 2021; 58: 2003434. DOI: 10.1183/13993003.03434-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen JI, Dropulic L, Wang K, et al. Comparison of levels of nasal, salivary, and plasma antibody to severe acute respiratory syndrome coronavirus 2 during natural infection and after vaccination. Clin Infect Dis 2023; 76: 1391–1399. doi: 10.1093/cid/ciac934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Chua BY, Selva KJ, et al. SARS-CoV-2 infection results in immune responses in the respiratory tract and peripheral blood that suggest mechanisms of disease severity. Nat Commun 2022; 13: 2774. DOI: 10.1038/s41467-022-30088-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervia C, Nilsson J, Zurbuchen Y, et al. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J Allergy Clin Immunol 2021; 147: 545–557. doi: 10.1016/j.jaci.2020.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 2020; 5: eabe5511. DOI: 10.1126/sciimmunol.abe5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fein A, Grossman RF, Jones JG, et al. The value of edema fluid protein measurement in patients with pulmonary edema. Am J Med 1979; 67: 32–38. DOI: 10.1016/0002-9343(79)90066-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00789-2023.SUPPLEMENT2 (73.8KB, pdf)
Supplementary figures 00789-2023.SUPPLEMENT (393KB, pdf)