Abstract
Analyses of a collection of full-length TT virus genomes showed nearly half of them to be recombinant. The results were highly significant and revealed homologous recombination both within and among genotypes, often involving extremely divergent lineages. Recombination breakpoints were significantly more common in the noncoding region of the TT virus genome than in the coding region.
TT virus (TTV) is a newly discovered nonenveloped DNA virus that was initially considered to be a possible agent of viral hepatitis, since it was first recovered from a patient with posttransfusion hepatitis of unknown etiology (12, 15). Subsequent studies, however, have shown it to be very widespread and to occur at an extremely high prevalence even in healthy populations (1, 9, 18, 20), casting doubt on its causal role in human disease. Its circular, single-stranded, negative-sense DNA genome, approximately 3,850 nucleotides in length (10, 11), bears little or no identifiable similarity to other known viruses, and TTV appears to represent a new virus family, tentatively designated Circinoviridae (11).
For a DNA virus, TTV exhibits an astonishingly large amount of genetic diversity. To date, more than 16 genotypes separated by more than 30% divergence at the nucleotide level have been described (6, 7, 14, 16, 18, 22), incorporating three hypervariable regions (13). Understanding the origins of such diversity is a fundamental problem in virology. While the role played by mutation has long been considered, it is becoming increasingly apparent that recombination also plays a key role in the evolution of many virus groups (19, 24). The recent availability of several full-length TTV sequences (3, 6, 10, 11, 16), along with evidence for mixed infection by multiple genetic types (2, 4, 17, 21), prompted this investigation into whether recombination might also play a role in the evolution of TTV.
A total of 15 full-length or near-full-length TTV genomes with the following isolate names (accession numbers) were collected from GenBank: TA278 (AB017610) (16); GH1 (AF122913) (11); TUS01 (AB017613) (16); SANBAN (AB025946) (6); JA20 (AF122914), JA9 (AF122915), JA10 (AF122919), JA4 (AF122917), JA1 (AF122916), JA2B (AF122918), US32 (AF122921), and US35 (AF122920) (3); and BDH1 (AF116842), TTVCHN1 (AF079173), and TTVCHN2 (AF129887). The sequences were aligned using CLUSTAL W (23) and adjusted by hand. The resulting 3,853-nucleotide full-length TTV alignment is available from the author on request.
The aligned data set was analyzed by various methods to identify possible recombinant isolates, to characterize their putative recombination breakpoints, and to test results suggestive of recombination for statistical significance. First, an exploratory tree analysis (5) was performed by sliding a 400nucleotide window down the sequence alignment in 200-nucleotide increments, generating a series of trees for the different regions. All trees were reconstructed with the PAUP* program (version 4; Sinauer Associates, Sunderland, Mass.) using the neighbor-joining algorithm with distances estimated under the HKY85 model of DNA substitution. Seven isolates clearly changed topological position over different regions of their genomes, an indication of possible mosaic structure, and were earmarked as putative recombinants (data not shown).
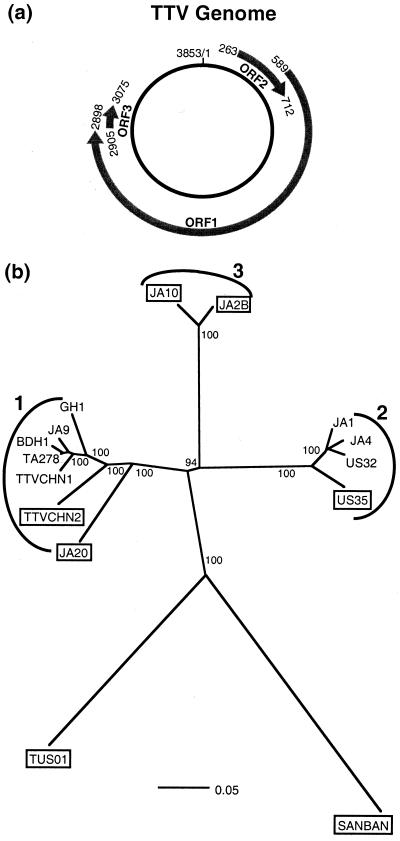
Each putative recombinant isolate was subsequently examined using sliding window diversity plots to determine which of the other sequences in the data set it most closely resembled over the conflicting regions of its genome (5, 25). A neighbor-joining tree of the full-length data set, with bootstrap percentages based on 1,000 replicates, was also reconstructed and is shown along with a diagram of the TTV genome in Fig. 1. Surprisingly, although almost half of the isolates appeared to contain sequence regions from more than one genotype, analysis of the full-length alignment gave no indication of this, with all putative recombinants supported by high bootstrap scores within distinct clades (Fig. 1b).
FIG. 1.
TTV circular genome and phylogenetic tree of the full-length alignment. (a) Schematic diagram of the TTV circular genome (isolate TA278) with open reading frames 2, 1, and 3 indicated. The remaining 1,048 nucleotides of the noncoding region account for approximately 27% of the TTV genome. (b) Unrooted neighbor-joining tree with associated bootstrap percentages from analysis of the full-length alignment. Branch lengths are drawn to scale, and genotypes 1 to 3 are indicated. The seven putative recombinants identified by the initial exploratory analyses are boxed.
The putative recombinants were next subjected to a maximum likelihood (ML) method for estimating recombination breakpoints and testing their significance (8). Briefly, this approach finds breakpoints by dividing an alignment of a putative recombinant and its two “parents” (those sequences it most resembles in different genomic regions) into two regions (using either one or two breakpoints) and finding a separate ML tree for each. All possible partitions (breakpoints) are tried, and the scores for the two trees in each case are combined to give a “recombination model” likelihood for that particular partitioning of the alignment. If recombination has occurred, the highest combined score is expected when the alignment is broken at the actual recombination breakpoint(s), since the two trees reconstructed in this case best reflect the true phylogenetic history of the separate recombinant regions (8, 25).
Statistical significance for the inferred breakpoints is determined by comparing the recombination model likelihood to the likelihood obtained from the unbroken alignment (i.e., the “no recombination model”) by using a likelihood ratio test and a Monte Carlo approach based on sequences simulated without recombination (8, 25). Significance is established if the observed improvement in likelihood under the recombination model (i.e., when more than one tree can fit the data) is greater than that expected by chance, as assessed by comparison with the null distribution generated from simulated data. These analyses were performed on four overlapping, 1,200-nucleotide regions of the full-length alignment, and all significance tests were based on 200 simulated data sets for each set of breakpoints evaluated.
Finally, confirmation that significant results reflected mosaic genomes containing regions with different evolutionary histories was provided by constructing bootstrap phylogenetic trees (1,000 replicates) for the different recombinant regions.
Table 1 shows the results of the breakpoint analysis of putative recombinants and their “parents,” those isolates most similar to them in different genomic regions. The breakpoints identified within every putative recombinant genome closely matched those expected from the exploratory tree and diversity analyses and proved to be highly statistically significant, with likelihood ratios in each case much higher than any generated by 200 simulated data sets (P < 0.005). The table also lists the percent similarity between putative recombinants and their parents, both over the full-length alignment and in specific recombinant fragments, showing that otherwise divergent isolates shared marked similarity in some genomic regions.
TABLE 1.
Breakpoint analysis results for the seven putative TT virus recombinants
| Isolate | Region | Length (nucleotides) | Parent | Genotypea | % Similarity: genome/fragmentd | P value |
|---|---|---|---|---|---|---|
| TTVCHN2b | 490–867 | 378 | JA2B | 3 | 76.2/85.1 | <0.005 |
| 868–2004 | 1137 | TTVCHN1 | 1 | 90.0/98.3 | <0.005 | |
| 2005–2730 | 726 | JA20 | 1 | 83.7/98.8 | <0.005 | |
| 2731–2900 | 170 | TTVCHN1 | 1 | 90.0/90.0 | <0.005 | |
| 2901–3288 | 388 | JA2B | 3 | 76.2/90.4 | <0.005 | |
| 3289–3739; | 451 | TTVCHN1 | 1 | 90.0/89.3 | ||
| 1–489c | 489 | TTVCHN1 | 1 | 90.0/90.0 | <0.005 | |
| TUS01 | 3083–3334 | 252 | US32 | 2 | 63.2/93.6 | <0.005 |
| 3335–3658 | 324 | GH1 | 1 | 65.0/97.2 | <0.005 | |
| 3659–3082 | 3,277 | SANBAN | 61.8/59.4 | <0.005 | ||
| SANBAN | 3175–3282 | 108 | TA278 | 1 | 61.6/100.0 | <0.005 |
| 3283–3174 | 3,745 | TUS01 | 61.8/61.3 | <0.005 | ||
| JA20 | 3426–3641 | 216 | JA2B | 3 | 74.7/87.0 | <0.005 |
| 3642–3425 | 3,637 | TA278 | 1 | 85.0/85.8 | <0.005 | |
| JA10b | 94–259 | 166 | JA20 | 1 | 73.2/98.8 | <0.005 |
| 260–3489 | 3,230 | JA2B | 3 | 94.8/94.5 | <0.005 | |
| 3490–3617 | 128 | SANBAN | 59.4/67.4 | <0.005 | ||
| JA2Bb | 94–259 | 166 | JA20 | 1 | 74.7/98.2 | <0.005 |
| 260–3641 | 3,382 | JA10 | 3 | 94.8/94.5 | <0.005 | |
| US35 | 2329–3713 | 1,385 | US32 | 2 | 92.6/97.6 | <0.005 |
| 3714–2328 | 2,468 | JA1 | 2 | 91.8/89.6 | <0.005 |
Only genotypes 1 to 3 are listed.
Near-full-length genome. For TTVCHN2, nucleotides 3740 to 3853 in the aligned data set were not available, and for JA10 and JA2B, nucleotides 3642 to 93 were not available.
3289 to 3739 and 1 to 489 were treated as a single fragment for this analysis since the intervening region was not available.
Comparison is between each putative recombinant isolate and its parent.
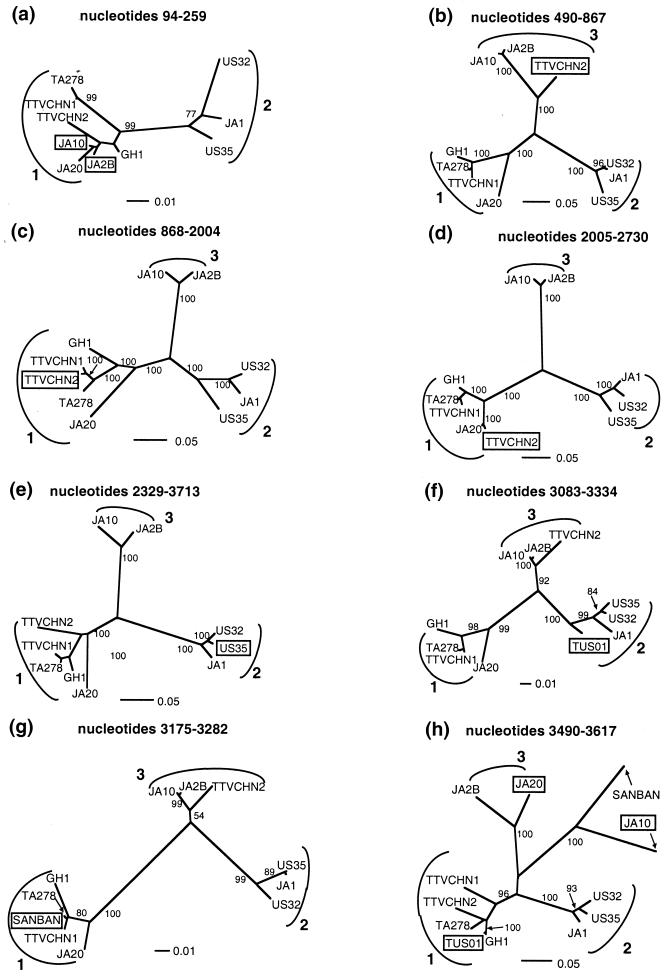
Strongly supported bootstrap trees (Fig. 2) confirmed the presence within single genomes of sequence from different genotypes or lineages: JA10 and JA2B (Fig. 2a) contained nearly identical stretches of genotype 1 sequence, most similar to that of JA20, and appeared to be descendants of a single, recombinant common ancestor. TTVCHN2 (Fig. 2b to d) grouped alternately with genotypes 1 and 3 in various parts of its genome. US35 (Fig. 2e) moved from a position basal to US32 and JA1 to group with US32. TUS01 (Fig. 2f), otherwise extremely divergent (see Fig. 1b), contained some genotype 2 sequence. Likewise, SANBAN (Fig. 2g) contained some genotype 1 sequence. Finally, over a similar region (Fig. 2h; see Table 1 for the precise breakpoints), JA20 exhibited genotype 3 sequence, TUS01 contained genotype 1 sequence, and JA10 grouped with the extremely divergent isolate SANBAN. For clarity, three of the isolates shown in Fig. 1b (BDH1, JA9, and JA4) were not included here because they were very similar to other isolates across the full-length alignment. Also for clarity, the two very divergent isolates TUS01 and SANBAN were only included for regions where they were recombinants or parents of other recombinants. Neither the alignment nor the subsequent analysis was significantly affected by the presence or absence of these two isolates, and bootstrap trees produced using all 15 isolates produced virtually identical findings in all cases. These results indicate that TTV undergoes relatively frequent recombination.
FIG. 2.
Neighbor-joining trees reconstructed from the recombinant regions revealed by the breakpoint analysis. Branch lengths are drawn to scale, associated bootstrap values are shown, and genotypes 1 to 3 are indicated. The region of the full-length alignment used to reconstruct each tree is included above it, and the recombinant isolate(s) for that region are boxed. Other trees not shown produced similarly high bootstrap support for phylogenetically conflicting regions. These results illustrate that many of the individual full-length sequences in the TTV data set contain regions with conflicting evolutionary histories.
Of the seven recombinants, five (TUS01, SANBAN, JA20, JA10, and JA2B) were identified as intergenotype homologous recombinants; one (US35) was an intragenotype homologous recombinant; and one (TTVCHN2) showed evidence of both inter- and intragenotype homologous recombination. Three isolates (TTVCHN2, TUS01, and JA10) contained gene sequences from at least three different sources, indicating a history of multiple recombination events. This observation was not surprising, given the extremely high percentage of recombinants among these natural TTV isolates (>46%). One of these, TTVCHN2, contained two distinct regions of genotype 3 sequence (Fig. 2c and d).
A comparison of all the recombinant regions identified in this study (Table 1) with the structure of the TTV genome (Fig. 1a) revealed a surprising preponderance of breakpoints in the relatively short noncoding region. After correcting for nonindependent comparisons, 13 out of 19 breakpoints fell within the contiguous 1,048-nucleotide noncoding region, while only 6 out of the 19 were found within the 2,805 nucleotides spanning open reading frames 2, 1, and 3. The difference in the numbers of breakpoints falling in noncoding versus coding sequence was highly significant based on a chi-square test (χ2 = 16.3; df = 1; P < 0.0001). It is not clear whether this reflects a higher rate of recombination events in the noncoding region or the enhanced fitness of noncoding compared to coding region recombinants. While neither the coding nor the noncoding region breakpoints appeared to map to especially similar sequence regions, such as direct or inverted repeats, they did not generally correspond to insertions or deletions either. The evident pattern of homologous crossing over may thus reflect a copy choice mechanism for recombination in these novel DNA viruses. Because information about different levels of recombination in different regions of viral genomes is extremely rare, these results are particularly interesting.
It is important to note that detailed information on the strategy of sequencing was obtained for all seven recombinants (3, 6, 11, 16; C.-H. Huang, personal communication) and provided convincing evidence that the mosaic genomes identified here are natural recombinants and not laboratory artifacts. First, the successful amplification of long, overlapping sequence regions depended upon the presence of intact, circular TTV genomes, not fragmented ones. Crucially, for each recombinant, a comparison of the endpoints of the amplified PCR products with the inferred breakpoints showed that they did not correspond: the breakpoints listed in Table 1 could not be explained by the regions amplified and sequenced for each isolate. Furthermore, laboratory artifacts cannot readily explain the common recombinant region shared by JA10 and JA2B (Fig. 2a): it seems much more plausible that they diverged from a recombinant ancestor than that they arose by independent but identical errors (8, 25).
It is worth considering in this context that Taq polymerase has been shown to produce recombinant molecules reminiscent of the products of rolling circle DNA synthesis (26). However, since the recombination events described in that study were nonhomologous, it is not clear that such a process could underlie the homologous recombination detected here. Significantly, the majority of the genomes in the present data set were isolated from carefully collected and stored patient serum, most of which were not observed to be multiply infected, were the result of extremely clean, single, intense PCR products of the expected size, and were confirmed by evaluation of multiple PCR products and found to represent single sequences (J. C. Erker, personal communication). Taken together, these observations strongly suggest that these chimeric genomes were generated by natural recombination, not laboratory error.
The full-length tree with its associated bootstrap values (Fig. 1b) is quite instructive in light of the detailed evidence for recombination among the isolates depicted. For instance, not only does each recombinant fall within a distinct clade of this tree, but every one is supported by a deceptive 100% bootstrap score even though it contains extensive regions of alternative ancestry. At the same time, the highly structured tree clearly indicates that a good deal of phylogenetic signal remains among the isolates analyzed, despite the effects of recombination. Except, for instance, in the case of JA10 and JA2B (Fig. 2a), much of the evidence for recombination described here could reflect fairly recent events between different TTV lineages. The often high degree of similarity between recombinants and their parents (Table 1) suggests that in some cases little independent evolution has occurred since recombination.
In conclusion, the findings reported here imply that short sequence regions, particularly from the noncoding regions of TTV genomes, may be inadequate markers for identifying and typing isolates and for reconstructing the evolutionary history of this group. Furthermore, although long sequence regions or full-length genomes will be preferable for studies of TTV, even these must be scrutinized with care to reveal the fingerprint of recombination. The detection of recombination in TTV underscores the notion that for any group of viruses the assumption of clonality should be validated by explicit tests whenever phylogenies are used for virological inference. The evidence for recombination revealed by these analyses of full-length TTV genomes is remarkable because it shows for the first time that recombination not only occurs but is widespread in this newly discovered group and so is probably an important force in its evolution. Perhaps most intriguingly, the results demonstrate that extremely divergent variants of this novel DNA virus are linked, by recombination, into a single gene pool.
Acknowledgments
This work was supported by The Rhodes Trust and by the Natural Sciences and Engineering Research Council of Canada.
I thank James Erker and Cheng-hui Huang for helpful correspondence and Eddie Holmes and two anonymous reviewers for comments on the manuscript.
REFERENCES
- 1.Abe K, Inami T, Asano K, Miyoshi C, Masaki N, Hayashi S, Ishikawa K I, Takebe Y, Win K M, El-Zayadi A R, Han K H, Zhang D Y. TT virus infection is widespread in the general populations from different geographic regions. J Clin Microbiol. 1999;37:2703–2705. doi: 10.1128/jcm.37.8.2703-2705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J K, Curran R, Berridge S, Grabowska A M, Jameson C L, Thomson B J, Irving W L, Sharp P M. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J Gen Virol. 1999;80:1759–1768. doi: 10.1099/0022-1317-80-7-1759. [DOI] [PubMed] [Google Scholar]
- 3.Erker J C, Leary T P, Desai S M, Chalmers M L, Mushahwar I K. Analyses of TT virus full-length genomic sequences. J Gen Virol. 1999;80:1743–1750. doi: 10.1099/0022-1317-80-7-1743. [DOI] [PubMed] [Google Scholar]
- 4.Forns X, Hegerich P, Darnell A, Emerson S U, Purcell R H, Bukh J. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J Med Virol. 1999;59:313–317. [PubMed] [Google Scholar]
- 5.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B X, Chen Y L, Barré-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hijikata M, Takahashi K, Mishiro S. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORF2 sequences implicating a great degree of diversity beyond genotypes. Virology. 1999;260:17–22. doi: 10.1006/viro.1999.9797. [DOI] [PubMed] [Google Scholar]
- 7.Hohne M, Berg T, Muller A R, Schreier E. Detection of sequences of TT virus, a novel DNA virus, in German patients. J Gen Virol. 1998;79:2761–2764. doi: 10.1099/0022-1317-79-11-2761. [DOI] [PubMed] [Google Scholar]
- 8.Holmes E C, Worobey M, Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol. 1999;16:405–409. doi: 10.1093/oxfordjournals.molbev.a026121. [DOI] [PubMed] [Google Scholar]
- 9.Leary T P, Erker J C, Chalmers M L, Desai S M, Mushahwar I K. Improved detection systems for TT virus reveal high prevalence in humans, nonhuman primates and farm animals. J Gen Virol. 1999;80:2115–2120. doi: 10.1099/0022-1317-80-8-2115. [DOI] [PubMed] [Google Scholar]
- 10.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Dexai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 13.Nishizawa T, Okamoto H, Tsuda F, Aikawa T, Sugai Y, Konishi K, Akahane Y, Ukita M, Tanaka T, Miyakawa Y, Mayumi M. Quasispecies of TT virus (TTV) with sequence divergence in hypervariable regions of the capsid protein in chronic TTV infection. J Virol. 1999;73:9604–9608. doi: 10.1128/jvi.73.11.9604-9608.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto H, Kato N, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. Distinct genotypes of a nonenveloped DNA virus associated with posttransfusion non-A to G hepatitis (TT virus) in plasma and peripheral blood mononuclear cells. J Med Virol. 1999;57:252–258. doi: 10.1002/(sici)1096-9071(199903)57:3<252::aid-jmv7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 16.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUSO1): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 18.Prescott L E, MacDonald D M, Davidson F, Mokili J, Pritchard D I, Arnot D E, Riley E M, Greenwood B M, Hamid S, Saeed A A, McClure M O, Smith D B, Simmonds P. Sequence diversity of TT virus in geographically dispersed human populations. J Gen Virol. 1999;80:1751–1758. doi: 10.1099/0022-1317-80-7-1751. [DOI] [PubMed] [Google Scholar]
- 19.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of “AIDS” viruses. Philos Trans R Soc Lond Ser B Biol Sci. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Hoshino H, Ohta Y, Yoshida N, Mishiro S. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol Res. 1998;12:233–239. [Google Scholar]
- 21.Takayama S, Yamazaki S, Matsuo S, Sugii S. Multiple infection of TT virus (TTV) with different genotypes in Japanese hemophiliacs. Biochem Biophys Res Commun. 1999;256:208–211. doi: 10.1006/bbrc.1999.0270. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka Y, Mizokami M, Orito E, Nakano T, Kato T, Ding X, Ohno T, Ueda R, Sonoda S, Tajima K, Miura T, Hayami M. A new genotype of TT virus (TTV) infection among Colombian native Indians. J Med Virol. 1999;57:264–268. doi: 10.1002/(sici)1096-9071(199903)57:3<264::aid-jmv9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worobey M, Holmes E C. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 25.Worobey M, Rambaut A, Holmes E C. Widespread intra-serotype recombination in natural populations of dengue virus. Proc Natl Acad Sci USA. 1999;96:7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaphiropoulos P G. Non-homologous recombination mediated by Thermus aquaticus DNA polymerase I. Evidence supporting a copy choice mechanism. Nucleic Acids Res. 1998;26:2843–2848. doi: 10.1093/nar/26.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]