Abstract
A-MYB (MYBL1) is a transcription factor with a role in meiosis in spermatocytes. The related B-MYB protein is a key oncogene and a master regulator activating late cell cycle genes. To activate genes, B-MYB forms a complex with MuvB and is recruited indirectly to cell cycle genes homology region (CHR) promoter sites of target genes. Activation through the B-MYB-MuvB (MMB) complex is essential for successful mitosis. Here, we discover that A-MYB has a function in transcriptional regulation of the mitotic cell cycle and can substitute for B-MYB. Knockdown experiments in cells not related to spermatogenesis show that B-MYB loss alone merely delays cell cycle progression. Only dual knockdown of B-MYB and A-MYB causes G2/M cell cycle arrest, endoreduplication, and apoptosis. A-MYB can substitute for B-MYB in binding to MuvB. The resulting A-MYB-MuvB complex activates genes through CHR sites. We find that A-MYB activates the same target genes as B-MYB. Many of the corresponding proteins are central regulators of the cell division cycle. In summary, we demonstrate that A-MYB is an activator of the mitotic cell cycle by activating late cell cycle genes.
Graphical Abstract
Graphical Abstract.
Introduction
B-MYB (MYBL2) is an activating transcription factor and a prominent oncogene (1,2). It is ubiquitously expressed in essentially all cell types. The protein is often found overexpressed and its gene is frequently amplified in tumors (3). The central function of B-MYB as a transcriptional activator has been linked to stimulating the cell cycle by forming B-MYB-MuvB activator complexes (4).
Progression through the cell cycle is a tightly regulated process involving the sequential transcriptional activation of cell cycle genes during the appropriate phases. Cell cycle-dependent transcription largely depends on two regulatory systems, the RB-E2F and DREAM/MuvB protein complexes (5–12). RB-E2F protein complexes only regulate early cell cycle genes with maximal expression from late G1 to S phase through binding to E2F promoter sites. In contrast, the DREAM/MuvB protein complex controls transcription of early and late cell cycle genes through two distinct mechanisms employing E2F sites and cell cycle genes homology region (CHR) promoter sites (8,12,13). As DREAM also acts through E2F sites, it functionally overlaps with RB-E2F in repressing gene transcription. However, DREAM exerts additional functions separate from RB-E2F when binding CHR elements.
To form the DREAM repressor, the MuvB complex binds the RB-like proteins p107 or p130 as well as the E2F proteins E2F4 or E2F5 and DP (6–8). The MuvB core complex with its subunits LIN9, LIN37, LIN52, LIN54 and RBBP4 serves as a scaffold, and by alternating binding of different factors causes a switch from the repressing DREAM to the activating B-MYB/FOXM1-MuvB complexes (8,12,14). DREAM represses transcription of late cell cycle genes through CHR elements. To downregulate early cell cycle genes, DREAM binds E2F sites (7,8,12,15–17). Through hyperphosphorylation of p107 and p130 these RB-like proteins and the E2F components dissociate from the MuvB core complex causing the loss of the DREAM/MuvB repressor function. In order to activate late cell cycle genes beginning in late G1, MuvB binds the activating transcription factors B-MYB and later FOXM1. The resulting B-MYB-MuvB (MMB) and FOXM1-MuvB complexes activate a large number of cell cycle genes, thereby acting as strong oncogenes (4,14). A fundamental property of the MMB and FOXM1-MuvB complexes is that they are unable to bind E2F sites. They exclusively bind through CHR promoter elements to activate genes (8,10,12,14,18).
An important mechanism is the unusual mode by which these MuvB-derived complexes contact the DNA of their target genes. B-MYB and FOXM1 do not bind the DNA in target promoters directly. Rather, B-MYB and FOXM1 contact MuvB proteins, and MuvB then binds directly with its subunit LIN54 to CHR sites as specific transcription factor binding sites (6,8,10,14,17).
The role of B-MYB in proliferation is also highlighted by its critical function for the recruitment of FOXM1 and YAP1 to the MuvB complex, leading to increased activation of late cell cycle genes (4,14,18). The YAP-MMB, FOXM1-MuvB and MMB complexes have been shown to induce cell cycle progression and proliferation (4,14,18,19). Moreover, the p53–p21–DREAM axis regulates expression of MMB target genes in response to DNA damage, further emphasizing the importance of antagonizing B-MYB function in order to arrest the cell cycle (13,15,20–24).
In contrast to the general function of B-MYB in all proliferating cells, already early results from mice had indicated that A-MYB (MYBL1) has a specific function in male germ cells. A-Myb loss in mice causes deficits in cell proliferation and development of mammary glands as well as a block in spermatogenesis (25). Consistent with its function in testes, A-MYB is expressed in meiotic spermatocytes, is required for the progression of meiosis, and is considered a transcriptional master regulator of meiosis (26–31). However, in contrast to this specific function in spermatocytes, A-MYB is expressed also in proliferating cells and was found to be differentially phosphorylated throughout the cell cycle already in early studies (32). Additionally, several more recent studies suggested an oncogenic function for A-MYB (33–35).
Comparison of A-MYB and B-MYB binding to MuvB as well as their roles in transcription also reveals diverging properties of the two factors. Although both have been found to interact with the LIN9 and LIN52 subunits of the MuvB complex through their C-terminal regions, in vitro binding of B-MYB with truncated protein variants indicated a higher affinity of B-MYB compared to A-MYB (7,8,36). Moreover, only B-MYB together with other MuvB proteins was described to bind to a large number of genes in regions harboring conserved CHR sites (10). In addition, only B-MYB was shown to activate late cell cycle genes, including Cyclin B1 (CCNB1), Cyclin B2 (CCNB2), and Ki-67 (MKI67) as part of MMB (4,8,37). However, B-MYB knockdown did not lead to a full loss of activation through CHR sites in CCNB2 and MKI67 promoters (8,37). This yields the hypothesis that other factors than B-MYB—possibly A-MYB—can activate transcription through CHR sites.
Taken together, published results leave open the questions whether A-MYB functions in CHR-dependent transcriptional regulation and substantially affects control of the cell division cycle. Here, we report that A-MYB can substitute for B-MYB in binding to MuvB. The resulting A-MYB-MuvB complex activates expression of the same set of genes that are activated by the B-MYB-MuvB complex. Most importantly, also functionally A-MYB substitutes for B-MYB because only the combined loss of A-MYB and B-MYB causes a block of the cell cycle, leads to endoreduplication, and induces apoptosis.
Materials and methods
Cell culture and cell count analysis
Human fibroblast BJ cells (DSMZ, Braunschweig, Germany), human colorectal carcinoma HCT116 cells (provided by Bert Vogelstein), human embryonic kidney HEK293 cells (DSMZ), human papillomavirus-related cervical adenocarcinoma HeLa cells (DSMZ), human pediatric hepatocellular carcinoma Hep3B cells (DSMZ), human retinal pigment epithelial hTERT immortalized RPE-1 cells (DSMZ), human glioblastoma T98G cells (DSMZ), and human bone osteosarcoma U2OS cells (DSMZ) were cultivated in Dulbecco's modified Eagle's medium (DMEM; Lonza). For standard growth medium, DMEM was supplemented with 5% fetal calf serum (FBS Good; PAN Biotech), 5% Serum Substitute (Panexin NTA, PAN Biotech), and 1% penicillin/streptomycin (PAN Biotech). All cell lines were cultivated under standard growth conditions with 37°C and 10% CO2.
For density-arrest, cells were grown to 100% confluence and cultivated for three more days with a medium change every day. For a synchronized release from density-arrest, cells were washed with PBS, trypsinized, and seeded in a ratio 1 to 10 in a 20% FCS-containing growth medium.
For growth arrest by starvation, RPE-1 cells were seeded in 15 cm dishes with 1 · 106 cells per dish, and cultivated for 24 h under standard conditions. Then cells were washed twice with PBS and cultivated in FCS- and serum-substitute-free medium for three days to induce a G0 cell cycle arrest. To restimulate a synchronized cell cycle entry, cells were cultivated in a 20% FCS-containing growth medium for 20 h.
For cell count analysis, cells were trypsinized and collected in a defined volume of standard growth medium and counted twice with the Countstar BioTech module (Countstar). PCR-based tests for mycoplasma contamination were performed using the Mycoplasma PCR Detection Kit (Applied Biological Materials).
siRNA transfection and luciferase promoter reporter assays
siRNA transfection for RNA and protein analyses as well as cell count and flow cytometry was performed with HCT116 and U2OS cells by reverse transfection of 1 × 106 cells in a 5 cm dish and 5 ml growth medium with 20 nM total siRNA and 5 μl Lipofectamine™ RNAiMAX (Thermo Fisher). For persistent knockdowns over more than three days, cells were trypsinized and reverse transfected in 6-well plates (4 · 105 cells per well) with 20 nM total siRNA and 2 μl Lipofectamine™ RNAiMAX. HEK293, HeLa, RPE-1, BJ, and Hep3B cells were reverse-transfected in 6-well plates with 20 nM siRNA and 2 μl Lipofectamine™ RNAiMAX per well. For siRNA knockdowns prior to luciferase assays, HCT116 and U2OS cells were trypsinized and seeded in 24-well plates with 50 000 cells per well and reverse transfected with 20 nM siRNA and 0.5 μl Lipofectamine™ RNAiMAX. As double knockdowns were compared to single knockdowns, combinations of targeting and non-targeting control siRNAs with total constant siRNA concentrations were used (Supplementary Table S1). Luciferase reporter assays were performed with extracts of siRNA and plasmid-transfected cells. Twenty-four hours post siRNA transfection cells were transfected with 150 ng of a pGL4.10 promoter plasmid (Cyclin B2-wt or Cyclin B2-CHRmut) and 100 ng of a pGL4.70-Renilla control plasmid using 1.25 μg linear Polyethylenimine (764965, Sigma-Aldrich) per well (8). Cells were lyzed and analyzed 24 h after plasmid transfection.
For rescue luciferase assays, HCT116 and Hep3B cells were trypsinized and seeded in 24-well plates with 5 × 104 cells per well and reverse-transfected with 20 nM siRNA, 100 ng empty expression vector pEGFP (pEGFP-C1) or mouse A-Myb expression plasmid mAmyb-GFP (mAmyb-pEGFP-C1), 75 ng of a pGL4.10 promoter plasmid (Cyclin B2-wt or Cyclin B2-CHRmut), 75 ng of a pGL4.70-Renilla control plasmid, and 1 μl DharmaFECT Duo (Dharmacon/Horizon).
For overexpression luciferase assay, HCT116, Hep3B and HeLa cells were trypsinized and seeded in 24-well plates with 5 · 104 cells per well. RPE-1 cells were trypsinized and seeded in 24-well plates with 1 × 105 cells per well. All cells were reverse-transfected with 75 ng of a pGL4.10 promoter reporter constructs (Cyclin B2-wt or Cyclin B2-CHRmut), 50 ng of a pGL4.70 Renilla control plasmid, and 125 ng expression plasmid (pEGFP-C1 or mA-Myb-pEGFP-C1) using 1.25 μg linear polyethylenimine per well.
Live cell staining, DNA content staining, and flow cytometry
For combined analysis of cell division, cell death, and apoptosis via flow cytometry, cells were stained with CFSE stain CellTrace™ Violet (Thermo Fisher) 48 h prior to flow cytometry according to the manufacturer's protocol. For staining of apoptotic and dead cells, the supernatant of the cell culture was collected, cells were trypsinized, added to the supernatant, and centrifuged at 500g for 5 min. Cell pellets were resuspended in PBS containing 400 nM Apotracker™ Green (Biolegend), incubated for 15 min, and washed twice with PBS. Immediately before flow cytometric analysis, propidium iodide (Sigma-Aldrich) was added to a final concentration of 0.08 μg/ml. For DNA content analysis, cells were fixed in one volume PBS/1 mM EDTA and three volumes of absolute ethanol overnight at 4°C, centrifuged for 8 min at 500g, and resuspended in PBS/1 mM EDTA. DNA was stained with propidium iodide at a final concentration of 20 μg/ml. Combined live cell staining of CFSE, propidium iodide, and the apoptosis tracker as well as DNA content of propidium iodide-stained ethanol-fixed cells were analyzed by flow cytometry (LSR II, Becton Dickinson). The online tool floreada.io was used for data analysis.
Reverse-transcription semi-quantitative PCR and next generation RNA sequencing
Total RNA was isolated using Direct-zol RNA Miniprep (Zymo) according to the manufacturer's protocol. For RNA semi-quantification, reverse transcription and semi-quantitative PCR were combined in a one-step semi-quantitative PCR (RT-qPCR) reaction using the GoTaq® 1-Step RT-qPCR System (Promega). Samples were analyzed on an ABI 7300 Real-Time PCR System (Applied Biosystems). All primer sequences are listed in Supplementary Table S2.
For RNA sequencing, total RNA quality was assessed using a 5200 Fragment Analyzer (Agilent). Only samples with an RQN ≥7.0 were used for further analysis. Library preparation and strand-specific RNA sequencing were performed by Genewiz/Azenta Life Sciences.
Analysis of RNA sequencing data
RNA-seq data of two biological replicates per condition and the two cell lines were deposited at GSE240799. Reads were trimmed using cutadapt v4.4 (38). Fastqc v0.12.0 available on https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ was used for quality control. Trimmed reads were mapped to the hg38 genome using Salmon v1.10.1 (39,40). Differential gene expression between samples was analyzed separately for HCT116 and U2OS cells with DEseq2 (41). Gene names were annotated using the Ensembl database v108 (42). Heatmaps of differentially expressed genes were generated with pheatmap v1.0.12 (43). Enrichment analyses based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed with clusterProfiler v4.0 (44–46). Genes were annotated as MMB target genes based on chromatin immunoprecipitation data of MMB components and cell cycle dependent expression behavior in the targetgenereg.org database v1.0 (47). Genes were considered upregulated upon p53 activation, if the p53 score was ≥30 on a scale from −50 to 50 in the targetgenereg.org database v2.0 (16). Association between gene regulation and enrichment of transcription factor binding sites as annotated by JASPAR 2022 in the corresponding promoter regions (−200 bp to +50 bp of the transcriptional start site) was analyzed by monaLisa v1.6.0 (48,49).
DNA affinity purification
DNA affinity purification assays were performed as described earlier (50). Briefly, biotinylated DNA probes were amplified via PCR from wild-type and mutant mCcnb2-210 bp promoter reporter pGL4.10 plasmids (8). Proteins binding to these DNA probes in vitro were purified from nuclear extracts of HCT116 or T98G cells. Purified proteins were analyzed by SDS-PAGE and Western blotting.
Co-immunoprecipitation
Native protein extracts of HCT116 or T98G cells were generated by scraping the adherent cells and collecting them in PBS, followed by centrifugation at 500g for 5 min and resuspension in native lysis buffer (50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.5% IGEPAL CA-630). The cells were fragmented by five strokes through a 27-gauge syringe. Cell fragments were separated from the protein extracts by centrifugation at 20 000g for 10 min. The supernatant was used as native protein extract. A volume containing 2 mg protein was then incubated with 2 μg antibody for 2 h at room temperature. BSA-blocked Protein G Dynabeads™ (Thermo Fisher) were added to the solution to bind antibody-bound protein complexes. Beads were washed seven times with 200 μl native lysis buffer. Purified proteins were eluted from beads by addition of 70 μl 1x Laemmli buffer and shaking for 5 min at 95°C. Antibodies used for immunoprecipitation: rabbit IgG isotype control (02-6102, Thermo Fisher), LIN9 (A300-BL2981, Bethyl Laboratories/Fortis), LIN37 (T1, custom-made at Pineda Antikörper-Service, Berlin, Germany), LIN54 (A303-799A, Bethyl Laboratories/Fortis), A-MYB (HPA008791, Sigma-Aldrich), and B-MYB (A 301–655A, Bethyl Laboratories/Fortis).
SDS-PAGE and western blotting
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described earlier (Krause et al., 2000). For protein detection, these antibodies were used: β-actin (A5441, Sigma-Aldrich), A-MYB (HPA008791, Sigma-Aldrich), B-MYB (A 301–655A, Bethyl Laboratories/Fortis), CDC25C (H-6, Santa Cruz Biotechnology), Cyclin B2 (A-2, Santa Cruz Biotechnology), E2F4 (E3G2G rabbit mAb, 40291, Cell Signaling Technology), histone 3 (D2B12 XP® rabbit mAb, 4620S, Cell Signaling Technology), LIN9 (A300-BL2981, Bethyl Laboratories/Fortis), LIN37 (T3, custom-made at Pineda Antikörper-Service, Berlin, Germany) (Müller et al., 2016), LIN54 (A303-799A, Bethyl Laboratories/Fortis), Survivin (71G4B7, Cell Signaling Technology). The monoclonal B-MYB LX015.1 antibody (hybridoma media 1:5) was a kind gift from Roger Watson. Western blot based protein quantification was performed with the LabImage 1D software (Kapelan Bio-Imaging).
Chromatin immunoprecipitations (ChIPs)
RPE-1 cells were cell cycle-synchronized by growth factor withdrawal for three days followed by restimulation with 20% FCS and 5% Panexin-NTA. The cells were subjected to cross-linking with 1% formaldehyde for 10 min at room temperature. ChIPs were performed as described earlier [8]. The following antibodies were used to precipitate DREAM, MuvB, and FOXM1-MMB complex components: E2F4 (E3G2G, Cell Signaling Technology), LIN9 (A300-BL2981, Bethyl Laboratories/Fortis), LIN37 (T3, custom-made at Pineda Antikörper-Service, Berlin, Germany), A-MYB (HPA008791, Sigma-Aldrich), B-MYB (A 301–655A, Bethyl Laboratories/Fortis), and FOXM1 (D3F2B, Cell Signaling Technology). A non-targeting rabbit antibody (IgG, Dako, Glostrup, Denmark) was used as a control for nonspecific signals. For all precipitations, 1–2 μg of antibody and 30 μl of Protein G Dynabead suspension (Invitrogen) were used. For FOXM1 precipitations, the FOXM1 antibody was used in a 1:50 dilution. Immunoprecipitated DNA was used as template for quantitative real-time PCR which was performed on an ABI 7300 qPCR System (Applied Biosystems, Forster City, CA) using the GoTaq® qPCR Kit (Promega). ChIP primer sequences are listed in Supplementary Table S2. All PCR results were normalized to input controls. For calculation of averages of biological replicates, all normalized values were normalized to the non-targeting IgG control after growth factor withdrawal.
Plasmids and DNA probes
The coding region of mouse A-Myb was amplified from genomic cDNA extracted from NIH3T3 cells by standard PCR and was cloned into pEGFP-C1 (Clontech). The 210 bp promoter region of the murine Cyclin B2 gene and its CHR mutant have been described before (8). DNA probes for affinity purification with the same sequence as the murine wt and CHR mutant Cyclin B2 gene were generated by PCR with a 3′-biotinylated primer (Invitrogen). Primer sequences are given in Supplementary Table S2.
Results
Only combined knockdown of A-MYB and B-MYB leads to a substantial decrease in expression of cell cycle genes.
In order to investigate the function of A-MYB and B-MYB in regulating the cell cycle, siRNA knockdowns of A-MYB (A-MYB KD), B-MYB (B-MYB KD) or both (A-MYB + B-MYB dKD) were conducted and compared to a control treatment (Ctrl KD). Knockdowns were performed in the human colon carcinoma HCT116 and the osteosarcoma U2OS cell lines to reduce the possibility of observing cell type-specific effects. Cyclin B2, CDC25C, and Survivin were chosen as model genes for late cell cycle genes, as their CHR promoter sites and regulation by the B-MYB-MuvB complex during the cell cycle are well characterized (20,23,51).
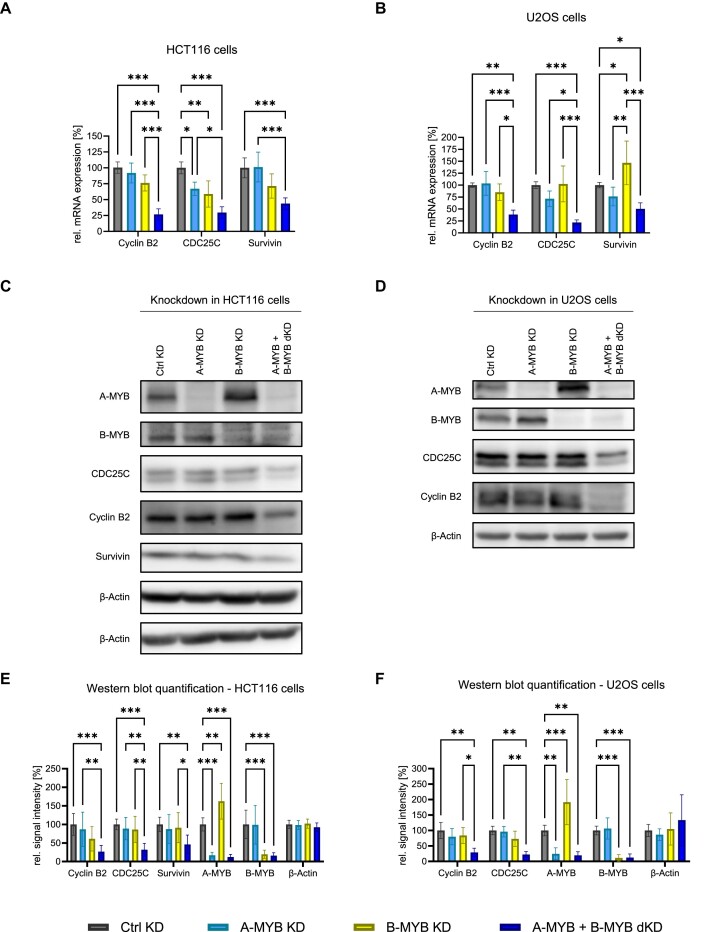
Unexpectedly, B-MYB KD did not lead to a consistently significant decrease of Cyclin B2, CDC25C and Survivin mRNA expression in both HCT116 and U2OS cells (Figure 1A, B). Furthermore, also A-MYB knockdown alone did not significantly influence Cyclin B2 and Survivin expression in both cell lines. CDC25C expression was only slightly decreased by A-MYB KD in HCT116 cells but not in U2OS cells. In contrast, an A-MYB + B-MYB dKD led to a substantial loss of Cyclin B2, CDC25C and Survivin mRNA expression in both cell lines (Figure 1A, B).
Figure 1.
Only combined knockdown of A-MYB and B-MYB leads to a substantial decrease of Cyclin B2, CDC25C, and SURVIVIN expression. Forty-eight hours after knockdown with targeting (KD, dKD) and non-targeting (neg. control, Ctrl KD) siRNAs, cells were harvested for protein and RNA analysis. Cyclin B2, CDC25C, and SURVIVIN mRNA expression after knockdown was determined by RT-qPCR in extracts from HCT116 (A) and U2OS (B) cells (n = 4). Following knockdown, HCT116 (C) and U2OS (D) protein extracts were analyzed by Western Blot. β-Actin was used as a loading control. The relative signal intensity of bands was quantified for HCT116 (E) (n = 6) and U2OS (F) cells (n = 5). Mean ± SD are given, and significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001).
Expression of Cyclin B2 and the other late cell cycle genes was downregulated in the same manner on the protein level (Figure 1C,D). While A-MYB or B-MYB knockdown alone had no significant effect on the expression of Cyclin B2, CDC25C, and Survivin proteins, A-MYB and B-MYB double knockdown significantly decreased expression of the three cell cycle regulators (Figure 1E, F).
Notably, A-MYB protein expression increased significantly upon B-MYB KD in both HCT116 and U2OS cells (Figure 1C-F). However, mRNA levels of A-MYB are not influenced by B-MYB KD, indicating that the A-MYB regulation by loss of B-MYB is controlled on the protein level (Supplementary Figure S1).
A-MYB regulates Cyclin B2 by indirectly binding to its CHR promoter element through MuvB
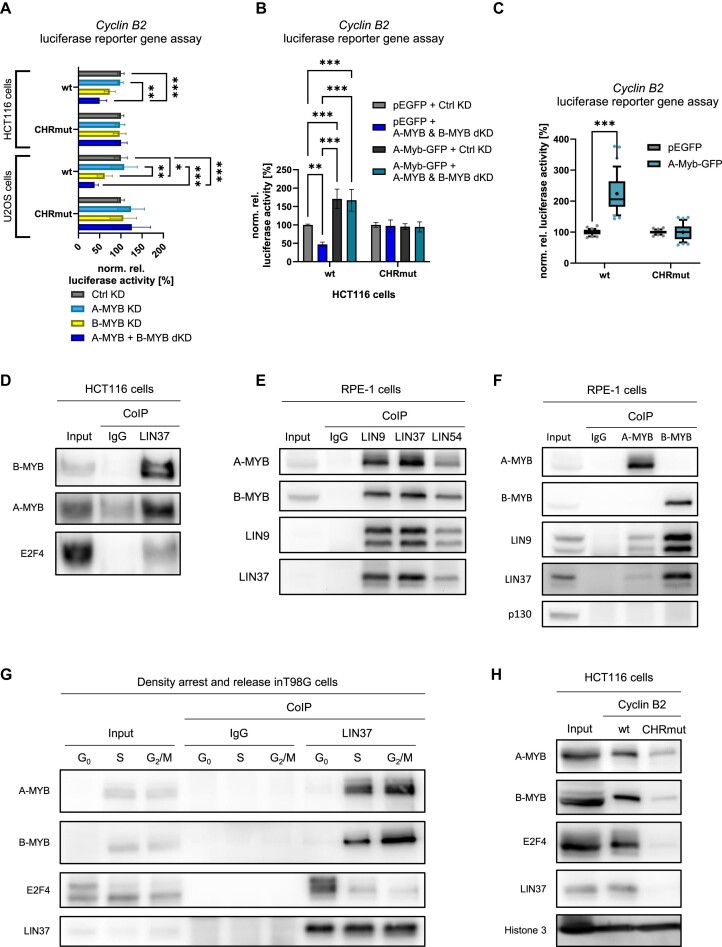
Next, we examined whether A-MYB exerts its function through CHR-dependent transcriptional regulation. As an example for regulation by the CHR, we employed reporter assays with Cyclin B2 wt and CHR-mutated (CHRmut) promoter constructs (Figure 2A). The Cyclin B2 wt promoter was downregulated in both HCT116 and U2OS cells after A-MYB + B-MYB dKD. After B-MYB knockdown alone, a significant Cyclin B2 reporter downregulation was observed in U2OS but not in HCT116. Importantly, downregulation was completely lost when the CHR promoter site was mutated when using the Cyclin B2 CHRmut promoter reporter. The effect of the A-MYB + B-MYB dKD was rescued by mouse A-Myb overexpression in HCT116 cells (Figure 2B). In Hep3B cells, which show very low A-MYB expression, B-MYB KD alone already leads to a Cyclin B2 reporter downregulation in a CHR-dependent manner (Supplementary Figure S2). However, recombinant expression of mouse A-Myb reverses the effect of B-MYB KD in Hep3B cells. In line with these results, mouse A-Myb overexpression also induces Cyclin B2 reporter expression dependent on the CHR element (Figure 2C). These data suggest that A-MYB as well as B-MYB exert their functions through CHR sites in their target promoters.
Figure 2.
A-MYB regulates Cyclin B2 by indirectly binding to its CHR promoter element through MuvB. (A) siRNA knockdown was performed in HCT116 (n = 5) and U2OS (n = 4) cells. Twenty-four hours after knockdown, cells were transfected with luciferase reporter constructs carrying the wild-type (wt) or CHR-mutated (CHRmut) Cyclin B2 promoter. Forty-eight hours after knockdown, luciferase activity was quantified from cell lysates. Differences between B-MYB KD and A-MYB + B-MYB dKD are statistically not significant. (B) To rescue HCT116 cells from siRNA knockdown of A-MYB and B-MYB, cells were transfected with the mouse A-Myb-overexpressing plasmid mA-Myb-GFP or the empty vector pEGFP as well as luciferase reporter constructs carrying the wild-type (wt) or CHR-mutated (CHRmut) Cyclin B2 promoter in parallel to the siRNA transfections. Forty-eight hours after knockdown, luciferase activity was quantified from cell lysates (n = 3). (C) For luciferase assays after mouse A-Myb overexpression, HCT116, RPE-1, Hep3B or HeLa cells were transfected with the mAmyb-GFP overexpression vector as well as luciferase reporter constructs carrying the wild-type (wt) or CHR-mutated (CHRmut) Cyclin B2 promoter. Forty-eight hours after knockdown, luciferase activity was quantified from cell lysates (n = 12). (D) Binding of A-MYB to LIN37 was tested in CoIP assays with HCT116 native protein extracts. As controls, E2F4 representing the DREAM complex and B-MYB as MMB component were precipitated (representative experiment of n = 4). (E) Binding of A-MYB to the MuvB core components LIN9, LIN37 and LIN54 was tested in CoIP assays with RPE-1 native protein extracts. As controls, LIN9 and LIN37 representing the MuvB core complex and B-MYB as an MMB component were precipitated (representative experiment of n = 3). (F) Binding of the MuvB core to A-MYB was tested in CoIP assays with RPE-1 native protein extracts. As negative control, p130 representing the DREAM complex was precipitated (representative experiment of n = 3). (G) T98G cells were synchronized in the cell cycle by density arrest and released for 15 h or 24 h into the cell cycle to obtain cell populations enriched in G0, S and G2/M phase, respectively. From each time point, native protein extracts were isolated and subjected to a LIN37 CoIP. Binding of DREAM and MMB components B-MYB, E2F4, and LIN37 as well as A-MYB was analyzed by Western blot (representative from n = 3). (H) Binding of A-MYB to wild-type (wt) and CHR mutant (CHRmut) Cyclin B2 promoter probes in DNA-affinity purification was analyzed by Western blot (representative of n = 3). Mean ± SD are given, and significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001).
These results imply that A-MYB binds the MuvB complex, which attaches through its LIN54 component to CHR promoter sites. Therefore, we examined this complex formation in co-immunoprecipitation (CoIP) experiments (Figure 2D–F). CoIPs of the MuvB core component LIN37 resulted in enrichment of A-MYB in HCT116 cells (Figure 2D). The established LIN37 interactors B-MYB, as part of the B-MYB-MuvB complex, and E2F4, a DREAM complex component, served as positive controls (Figure 2D). The same result could be observed in CoIPs of the MuvB core components LIN9, LIN37 and LIN54 in RPE-1 cells, with even stronger A-MYB binding due to the fact that RPE-1 cells express A-MYB at increased levels (Figure 2E, Supplementary Figure S7A, B). Additionally, LIN9 and LIN37 were bound in A-MYB CoIPs in RPE-1 cells, though with a lower yield than in B-MYB CoIPs (Figure 2F). In contrast, the DREAM component p130 was not bound by A-MYB or B-MYB. These results are consistent with data from large-scale database entries also detecting binding of A-MYB to MuvB complex components (52–54).
Another important aspect of cell cycle-dependent transcription through MuvB is the switch from repressive DREAM to activating B-MYB-MuvB complex binding at CHR sites during cell cycle progression from G0 to mitosis. In order to investigate A-MYB binding to MuvB in different cell cycle phases, we performed CoIPs of LIN37 with E2F4, A-MYB and B-MYB from extracts of synchronized T98G cells (Figure 2G). Cell cycle phase synchronization was validated by flow cytometry (Supplementary Figure S3A). We observe that LIN37 was expressed and precipitated throughout the cell cycle (Figure 2G). E2F4 binding decreased from G0 to S phase and even further in G2/M. In a reciprocal trend, B-MYB bound LIN37 only from S and G2/M-enriched extracts. Importantly, A-MYB binding to LIN37/MuvB was found reciprocal to that of E2F4 and in parallel to that of B-MYB, suggesting that A-MYB is a factor participating in the switch from DREAM to A/B-MYB-MuvB (Figure 2G). Consequently, these results also suggest that an A-MYB-MuvB complex can be formed in parallel to the established B-MYB-MuvB complex, with A-MYB likely competing successfully with B-MYB for binding to MuvB.
As it is well established that the MuvB complex binds through CHR sites, we examined whether A-MYB, with its association to MuvB that we have shown above, binds differentially to CHR sites. We performed DNA affinity purification assays from HCT116 nuclear extracts with a wild-type Cyclin B2 promoter probe containing its conserved CHR site (wt) and a Cyclin B2 promoter variant harboring a mutated CHR (CHRmut), resembling the same promoter sequences as employed earlier in the reporter promoter assays. The results show that A-MYB protein binding to the Cyclin B2 promoter was dependent on the CHR site. Binding of the MuvB core component LIN37, the B-MYB-MuvB component B-MYB, and the DREAM component E2F4 served as positive controls (Figure 2H). Histone bound to the DNA probe independent of the CHR site and therefore served as a negative control for differential binding.
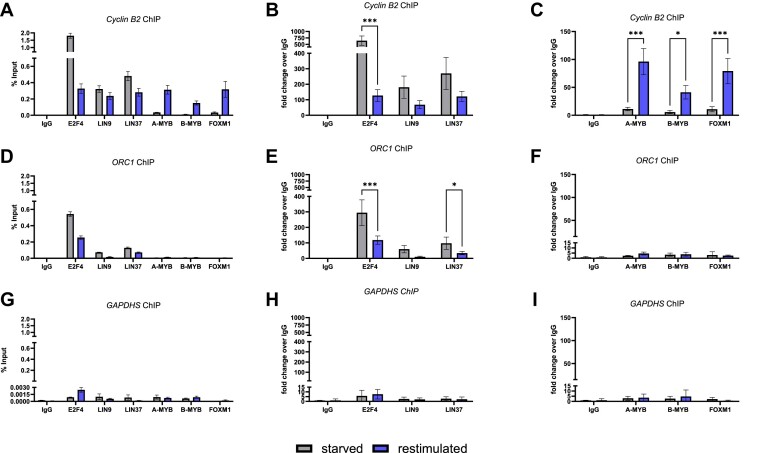
As A-MYB binds in vitro to the MuvB core only in S, G2, and M phases, we investigated the cell cycle-dependent binding of A-MYB in living cells by ChIP of cell cycle-synchronized RPE-1 cells (Figure 3; Supplementary Figure S3B). We observed that A-MYB binds to the Cyclin B2 promoter in late cell cycle phases but not in resting cells. A-MYB binding displayed the same pattern as the binding of B-MYB and FOXM1 (Figure 3A, C). No significant binding of A-MYB was found to the control promoters of ORC1 and GAPDHS, which served as negative controls for B-MYB binding (Figure 3). The unrelated GAPDHS promoter was not bound by any of the DREAM and MuvB components. The ORC1 promoter carries an E2F site and can bind DREAM. However, because ORC1 does not have a CHR site, it is incapable of binding the MMB complex (12). In contrast to A-MYB, the DREAM component E2F4 bound to the Cyclin B2 promoter predominantly in growth factor-starved cells, while the MuvB components LIN9 and LIN37 bound throughout the cell cycle (Figure 3B). Thus, A-MYB can bind promoters in the same manner as B-MYB in living cells.
Figure 3.
A-MYB binds the Cyclin B2 promotor dependent on the cell cycle. RPE-1 cells were cultivated under growth factor withdrawal (starved) und restimulated by growth factor addition to distinguish between a G0- and an S/G2/M-enriched condition. Promoter binding of DREAM and MMB components was analyzed by ChIP. The relative enrichment of the Cyclin B2 promoter (A), the ORC1 promoter (early cell cycle gene) (D), and the negative control GAPDHS promoter (G) is visualized relative to the input DNA (representative experiment of n = 3). Binding of DREAM components to the Cyclin B2 promoter (B), the ORC1 promoter (E), and the GAPDHS promoter (H) was analyzed relative to the IgG negative control of the starved condition (n = 3). Binding of the activating MMB components B-MYB and FOXM1 as well as A-MYB to the Cyclin B2 promoter (C), the ORC1 promoter (F), and the GAPDHS promoter (I) was analyzed relative to the IgG negative control of the starved condition (n = 3).). Mean ± SD are given, and significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001).
Taken together, these results are consistent with the model that A-MYB binds to CHR sites indirectly through the MuvB complex even in presence of B-MYB. Additionally, the data suggest that either A-MYB or B-MYB are required to transcriptionally activate cell cycle genes through their promoter CHR sites in late cell cycle phases.
Only A-MYB and B-MYB double knockdown substantially influences mRNA expression. Affected target genes are associated with mitosis
After establishing a model of A-MYB- and B-MYB-dependent gene regulation by indirect recruitment to CHR sites through MuvB, next-generation RNA sequencing (RNA-seq) of RNA from the HCT116 and U2OS cells after knockdown was performed to broaden the perspective from the model gene Cyclin B2 to a transcriptome-wide level.
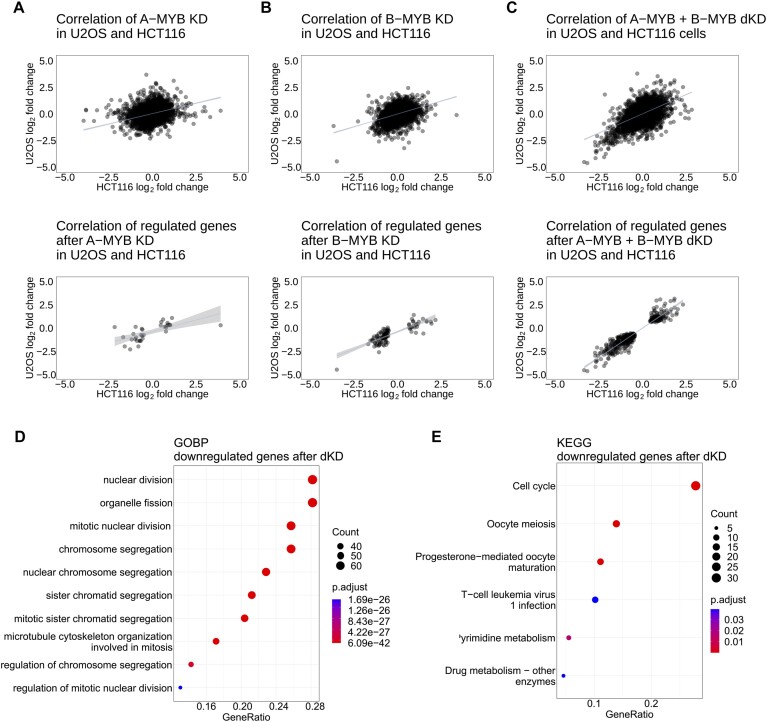
Figure 4 summarizes the impact of A-MYB KD, B-MYB KD and A-MYB + B-MYB dKD compared to the Ctrl KD in both cell lines. After A-MYB KD, only very few mRNAs were expressed differentially (Figure 4A). In addition, the gene set downregulated after B-MYB KD showed only a minor increase (Figure 4B). In contrast, A-MYB + B-MYB dKD yielded a large number of downregulated mRNAs (Figure 4C). Furthermore, a considerable number of genes was upregulated by A-MYB + B-MYB dKD. Again, these data further strengthen the hypothesis that A-MYB substitutes for major transcriptional functions of B-MYB. The dominant effects of A-MYB + B-MYB dKD can also be seen in heatmap analyses for both HCT116 and U2OS cells (Suppl. Data 1 and 2).
Figure 4.
A-MYB and B-MYB double knockdown but not single knockdowns influence RNA expression. Affected target genes are associated with mitosis. The correlation of expression of all RNA-seq hits (upper diagrams) and differentially expressed genes (lower diagrams) in HCT116 (log2 fold-change on the x-axis) and U2OS cells (log2 fold-change on the y-axis) was generated for (A) A-MYB KD, (B) B-MYB KD and (C) A-MYB + B-MYB dKD relative to the control (Ctrl KD). Genes were considered differentially expressed, if the log2 fold-change adjusted P-value was <0.01. For each scatter plot a linear regression with a 95% confidence interval of all data points was calculated. For A-MYB + B-MYB dKD a GOBP (D) and KEGG (E) analysis of genes significantly downregulated in HCT116 and U2OS cells were performed. For linear regression values see Supplementary Table S3.
In fact, the sets of genes up- and downregulated after A-MYB KD and upregulated after B-MYB KD were too small to find enriched pathways by Gene Ontology Biological Pathways (GOBP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Only after knockdown of both, A-MYB and B-MYB, the set of regulated genes was large enough to achieve reliable GOBP and KEGG pathway enrichment analyses of downregulated genes (Figure 4D and E). Genes downregulated after A-MYB and B-MYB double knockdown were associated in GOBP and KEGG analyses with the cell cycle, in the GOBP analysis more specifically with mitosis and mitotic processes. Previously, A-MYB had only been associated with meiosis. Thus, it is of great interest which gene groups are enriched in the knockdown experiments beside the group of cell cycle genes. At first glance, the KEGG analysis of the double knockdown identifies an enrichment of oocyte meiosis. However, when analyzing the specific genes included in oocyte meiosis, all enriched genes, e.g. CDC25C, PLK1, CCNB1/2 or AURKA, are actually mitotic genes as well. A GOBP analysis of genes downregulated after B-MYB KD also led to an enrichment of mitotic pathways, however, with much lower enrichment score and a smaller number of genes found when compared to the GOBP analysis of genes downregulated after A-MYB + B-MYB dKD (Supplementary Figure S4B). This suggests that the additional loss of A-MYB increases the impact on the expression of genes associated with mitosis.
As the set of genes upregulated after A-MYB and B-MYB double knockdown was substantially smaller than the downregulated gene set (Figure 4C, E), GOBP analysis yielded no significant enrichment (Supplementary Figure S4A). However, the p53 signaling pathway was enriched in the KEGG analysis.
A-MYB and B-MYB double knockdown affects B-MYB-MuvB targets and leads to p53 target induction without affecting RB targets
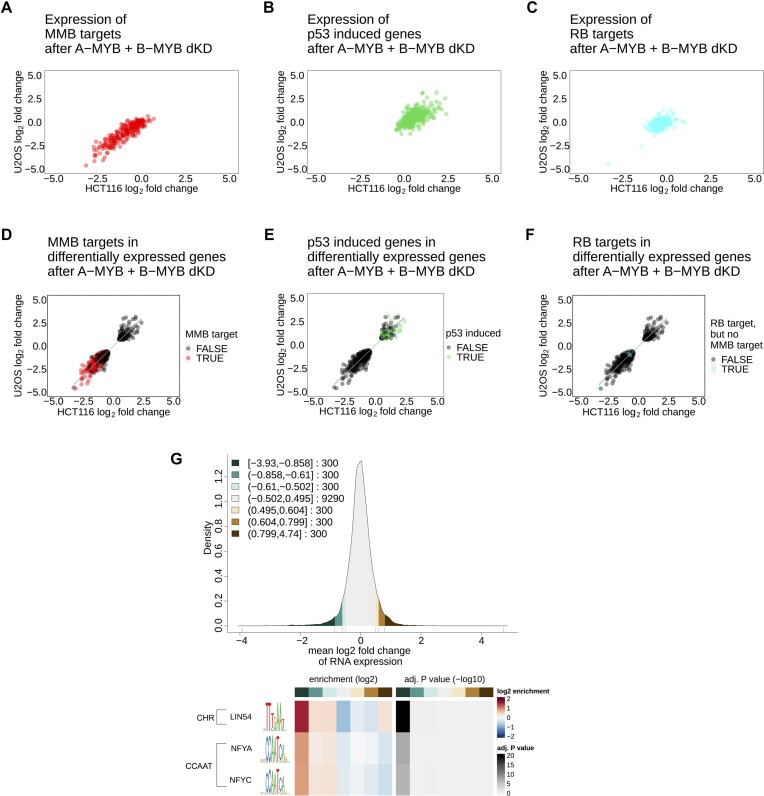
To investigate whether B-MYB-MuvB targets are also consistently changed in their expression by the A-MYB and B-MYB double knockdown, the targetgenereg.org database was employed (16,47). In this database, B-MYB-MuvB targets are defined by their cell cycle-dependent expression and the binding of B-MYB, FOXM1 and MuvB components to their promoters in ChIP experiments. We used our RNA-seq data and compared it to ChIP and transcription factor binding site data deposited in the database to identify groups of target genes. After A-MYB KD or B-MYB KD alone, the RNA expression of B-MYB-MuvB targets listed in targetgenereg.org remained unaltered (Supplementary Figure S5A-F). Consistent with the GOBP enrichment analysis, their expression was downregulated upon A-MYB + B-MYB dKD (Figure 5A). Importantly, B-MYB-MuvB targets represent the vast majority of downregulated genes after A-MYB + B-MYB dKD (Figure 5D).
Figure 5.
A-MYB + B-MYB double knockdown affects B-MYB-MuvB targets and leads to p53 target induction without affecting RB targets. The correlation of RNA-seq hits after A-MYB and B-MYB double knockdown in HCT116 (x-axis) and U2OS cells (y-axis) was filtered for (A) MYB-MuvB targets from targetgenereg.org in red, (B) p53 upregulated genes with a score ≥30 from targetgenereg.org in green, or (C) RB targets but not MYB-MuvB targets in targetgenereg.org in blue. To show the portion of each gene set within all regulated genes, the correlation of genes differentially expressed in HCT116 (x-axis) and U2OS cells (y-axis) was highlighted with (D) MYB-MuvB targets from targetgenereg.org in red, (E) p53 upregulated genes with a score ≥30 from targetgenereg.org in green, or (F) RB targets but not MYB-MuvB targets in targetgenereg.org in blue. For scatter plots (D, E and F) a linear regression with 95% confidence interval of all data points was calculated. To analyze the enrichment of motifs in the promoter regions of regulated genes after A-MYB + B-MYB dKD, the gene regulation in HCT116 and U2OS cells was averaged to a mean log2 fold-change and down- and upregulated genes were clustered in sets of 300 genes. The distribution of gene regulation after A-MYB + B-MYB dKD is shown as a density plot (G) with green scales for downregulated genes and brown scales for upregulated genes. The resulting seven clusters (3 downregulated gene clusters, 1 unregulated gene cluster, 3 upregulated gene clusters) were analyzed for motif enrichment from the JASPAR database in their core promoter region (−200 bp to +50 bp from the transcriptional start site). The enrichment and the significance of the enrichment are depicted as a heatmap (H) for each motif that was significantly (P < 0.05) enriched with a false discovery rate <10−3 in the up- or downregulated clusters. For linear regression values see Supplementary Table S3.
In contrast, genes induced by p53 according to the targetgenereg.org database were upregulated in cells with the double knockdown, but to a lesser extent and with a higher proportion of genes not regulated at all (Figure 5B). Even though only a relatively small subset of p53 targets is consistently upregulated, these genes still represent a considerable proportion of all upregulated genes (Figure 5E).
The fact that p53 target genes were upregulated and B-MYB-MuvB target genes were downregulated raised the question whether MMB targets were possibly only downregulated as a secondary effect after induction of p53 through the p53-p21-DREAM pathway (24). Therefore, a set of genes regulated by RB-E2F complexes but not by the B-MYB-MuvB complex was analyzed for regulation upon A-MYB + B-MYB dKD (Figure 5C). This gene set resembles early cell cycle genes, regulated by E2F motifs in their promoters. These genes are repressed through the p53-p21-RB/DREAM pathway in a similar way as are B-MYB-MuvB target genes upon strong p53 induction (13,24,55). If p53 induction was actually indirectly causing B-MYB-MuvB target gene downregulation, RB targets would be downregulated as well. However, RB target genes were not regulated after A-MYB + B-MYB dKD (Figure 5F). Therefore, B-MYB-MuvB target regulation must be caused directly by A-MYB and B-MYB loss and not indirectly via a p53 response.
Consistent with the MMB target regulation, promoter motif analysis categorized by mean gene regulation after A-MYB + B-MYB dKD in HCT116 and U2OS cells (Figure 5G) yielded enrichment of the LIN54 motif, equal to the CHR, and a correlation between enrichment of the motif and stronger downregulation of the corresponding RNA expression after A-MYB + B-MYB dKD (Figure 5H). Also, CCAAT-boxes, the NF-Y binding motifs, were enriched in sets of downregulated genes after A-MYB + B-MYB dKD. In contrast, A-MYB KD data sets showed no enrichment of CHR or CCAAT-box motifs (Supplementary Figure S5G) and genes downregulated after single B-MYB KD were substantially enriched only for the CHR (LIN54 binding motif), but still with a much lower enrichment factor than after A-MYB + B-MYB dKD (Supplementary Figure S5H).
DepMap data reveal an association between increased A-MYB expression and low impact of B-MYB loss on proliferation and survival
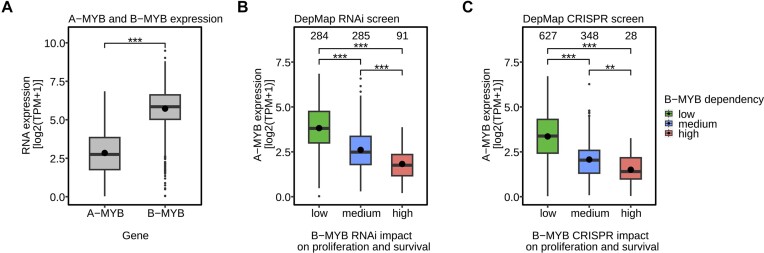
The finding that A-MYB can substitute for B-MYB in cell cycle gene regulation on a transcriptome-wide level prompted us to investigate whether this regulation also affects proliferation and cell survival. To obtain an initial overview, the DepMap database was employed (56,57). Analyzing the dependency of cancer cells on A-MYB or B-MYB (Supplementary Figure S6A), B-MYB loss has a higher negative impact on cell survival and proliferation on average. This is consistent with the gene regulation analysis. However, the effect of B-MYB loss comprises a wide spectrum, including even no impact by B-MYB loss in many cell lines (Supplementary Figure S6C, D).
Strikingly, the effect of B-MYB loss on cell proliferation and survival can however best be predicted by A-MYB expression, accounting for about 30% of all model predictors (Supplementary Figure S6B), even though A-MYB mRNA expression is considerably lower than B-MYB mRNA expression (Figure 6A). Increased A-MYB mRNA expression correlates very well with a decreased effect of B-MYB loss either by knockout (Figure 6B) or knockdown (Figure 6C). These observations support the notion of a cell type-independent function of A-MYB as a substitute for B-MYB.
Figure 6.
DepMap data reveal an association between increased A-MYB expression and low impact of B-MYB loss on proliferation and survival. (A) A cell line screen from the DepMap database compares the mRNA expression level of A-MYB and B-MYB in 2900 cell lines in a boxplot. Two additional boxplots based on the DepMap database correlate the mRNA expression of A-MYB (y-axis) with the impact of (B) B-MYB loss by siRNA knockdown (B-MYB RNAi gene effect on the x-axis) in 660 cell lines and (C) B-MYB knockout (B-MYB CRISPR gene effect on the x-axis) in 1003 cell lines on cell proliferation and survival. The impact of B-MYB loss was categorized into low (gene effect > −0.5), medium (gene effect ≤ −0.5 and > −1), or high (gene effect ≤ −1) negative effect on proliferation and survival. For these analyses the effects are considered relative to a set of genes, which cells are fully dependent on. The effects of those genes are set to −1. Significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001).
With these strong correlation data, it became imperative to assess whether A-MYB functionally compensates for B-MYB in regulating proliferation, as also suggested by our results on A-MYB- and B-MYB-dependent cell cycle gene regulation.
A-MYB compensates for B-MYB loss in proliferation and causes a lower rate of G2/M-arrested cells as well as a lower rate of DNA aberrations
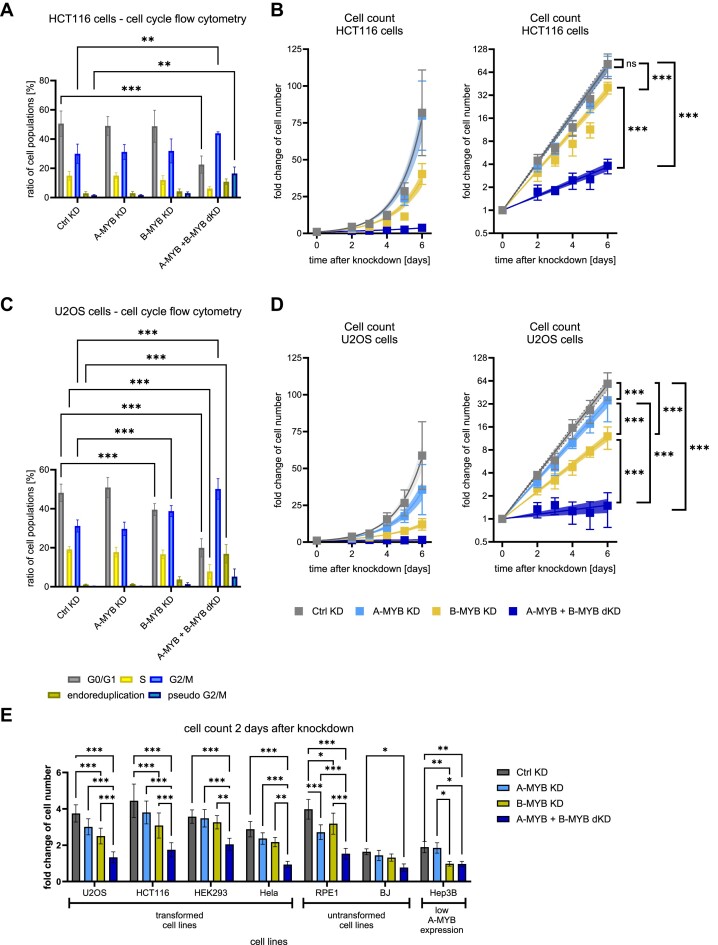
In order to investigate the effect of A-MYB and B-MYB on cell proliferation, siRNA knockdowns were performed and cell cycle distribution two days after knockdown as well as cell count at time points from two to six days after knockdown were analyzed in HCT116 and U2OS cells (Figure 7).
Figure 7.
A-MYB compensates for B-MYB loss in proliferation and causes a lower rate of G2/M-arrested cells as well as a lower rate of DNA aberrations. siRNA knockdown was performed on HCT116 (A, B) and U2OS cells (C, D). For flow cytometric analysis of DNA content to evaluate cell cycle distribution, (A) HCT116 (n = 3) and (C) U2OS (n = 5) cells were harvested 48 h after knockdown. Significance tests were performed for the change of the cell population ratio between Ctrl KD and the other conditions. To evaluate the change in cell numbers after knockdown, (B) HCT116 (n = 3) and (D) U2OS cells (n = 3) were counted every 24 h beginning from the second day after siRNA transfection. To assure a stable knockdown throughout the experiment, cells were re-transfected with siRNA after 72 h. Graphs on the left show exponential regression on a linear scale and graphs on the right display the same regression on a logarithmic scale. (E) siRNA knockdown was performed on the transformed cell lines U2OS, HCT116, HEK293 and HeLa, the untransformed cell lines RPE-1, and BJ, as well as the transformed cell line Hep3B with reportedly low A-MYB expression. Cells were counted two days after knockdown. Mean ± SD are given, and significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001). Exponential regression curves are displayed with 95% confidence intervals. Significances between exponential regression curves were calculated by Extra sum-of-squares F test (ns, not significant; ***P < 0.001).
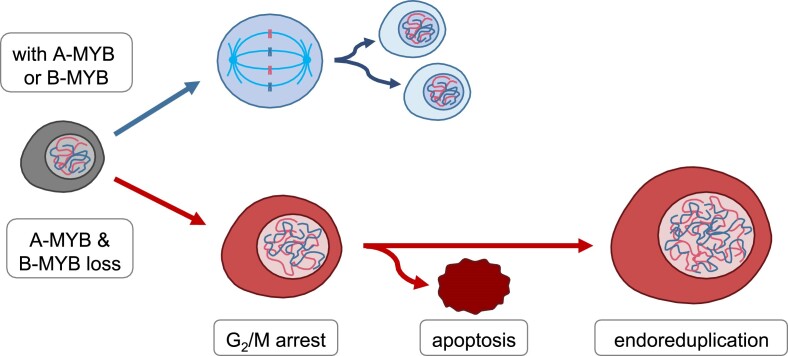
A-MYB knockdown alone did not influence cell cycle distribution in HCT116 (Figure 7A) or U2OS cells (Figure 7C). In addition, knockdown of B-MYB alone did not change cell cycle distribution in HCT116 cells and caused only minor changes in U2OS cells. In contrast, A-MYB + B-MYB dKD, however, led to a considerable decrease of cell populations in G0/G1 as well as in S phase and to an increase in G2/M populations in both HCT116 and U2OS cells. This indicates that combined loss of A-MYB and B-MYB causes cell cycle arrest in G2/M.
Of note, following A-MYB + B-MYB dKD, two cell populations drastically increased that were very low in control knockdown experiments (Figure 7A, C). These cells contained a DNA content higher than 2-fold from the content before treatment. These observations indicate that HCT116 as well as U2OS cells after G2/M arrest re-entered the cell cycle before completion of cell division, leading to endoreduplication followed by pseudo G2/M phases. Therefore, we further distinguished between cells containing approximately a 4-fold DNA content, corresponding to pseudo G2/M phases, and cells with a DNA content between 2-fold and 4-fold, resembling cells in the process of endoreduplication. Interestingly, in HCT116 and U2OS cells, populations displaying a DNA content of larger than 2-fold increased significantly upon double knockdown, reflecting a more stable G2/M arrest and additional DNA aberrations through endoreduplication (Figure 7A, C).
Importantly, we also counted the cells over a period of six days following knockdown in these experiments. A-MYB KD had no major impact on cell proliferation in any of the two cell lines and B-MYB KD displayed diminished proliferation rates (Figure 7B,D). Remarkably, A-MYB and B-MYB double knockdown resulted in the loss of nearly all proliferative activity. Thus, the response pattern in these analyses is consistent with our earlier results.
In order to rule out effects that are restricted to specific cell types, a panel of four transformed cell lines (HCT116, U2OS, HEK293, and HeLa cells), two cell lines from untransformed origin (RPE-1 and BJ cells) as well as a transformed cell line with low A-MYB expression based on DepMap RNA expression data sets (Hep3B) was analyzed. Decreased A-MYB and B-MYB expression upon siRNA knockdown was validated on the mRNA level (Supplementary Figure S7). Importantly, A-MYB + B-MYB dKD led to the strongest growth limitation in all cell types (Figure 7E). Interestingly, in all cell lines except in Hep3B cells, a further drop from B-MYB single knockdown to A-MYB + B-MYB dKD was observed. These observations imply that A-MYB has a general function in controlling proliferation. As Hep3B cells were chosen for their very low A-MYB expression, the lack of a further drop is explained by the low capacity to substitute for B-MYB before A-MYB is knocked down. This also suggests that this siRNA does not exert nonspecific stress on the cells when added to the MYBL2 knockdown. On the other end of the A-MYB expression spectrum, single knockdown of A-MYB in RPE-1 cells, which display a high A-MYB protein expression, caused already a substantial reduction of proliferation compared to the lower effect following single knockdown of B-MYB (Figure 7E; Supplementary Figure S7). Taken together, these data suggest that A-MYB has a function in controlling the cell cycle and can substitute for B-MYB in driving cell proliferation.
Combined loss of A-MYB and B-MYB causes a decrease in cell division and an increase in apoptosis rates
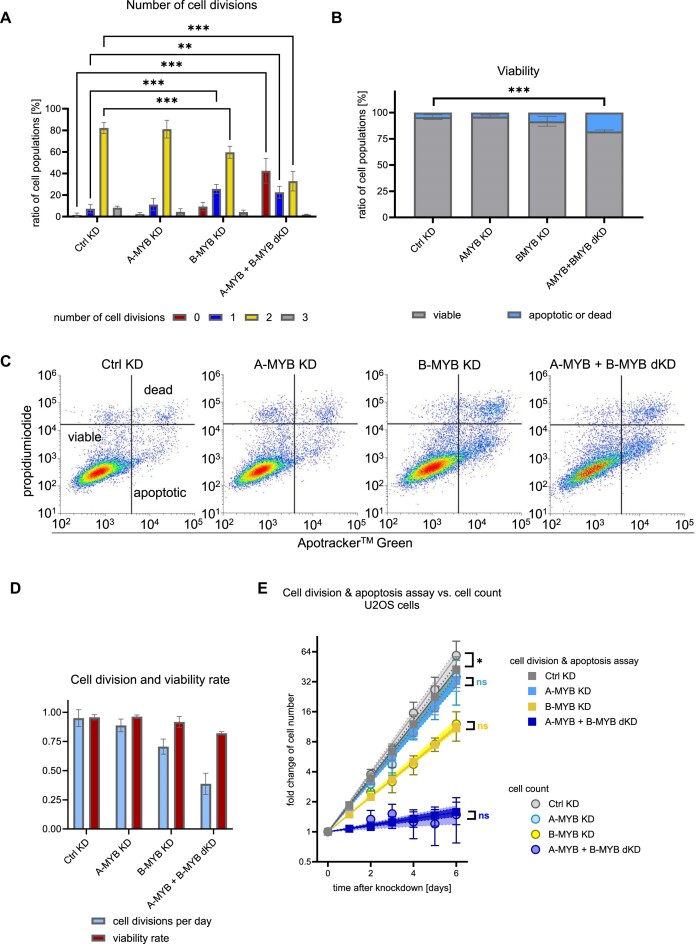
Cell proliferation is the product of two opposing processes, cell division and cell death. Particularly as the RNA-seq data indicated that p53 target genes were induced as a secondary effect of A-MYB and B-MYB double knockdown, we quantified cell division rates and cell viability in a single assay. To this end, a flow cytometry-based apoptosis assay was combined with a CFSE cell division assay on U2OS cells after knockdown (Figure 8).
Figure 8.
Combined loss of A-MYB and B-MYB causes a decrease in cell division and an increase in apoptosis rates. A combined analysis of apoptosis and cell division by flow cytometric analysis of stained live cells was performed on U2OS cells 72 h after knockdown. (A) For cell division analysis, U2OS cells were stained with CFSE from 24 h until 72 h after knockdown (n = 3). (B) Cell viability of U2OS cells after knockdown (n = 3) was based on ApoTracker and (C) propidium iodide staining (representative experiment from n = 3). In significance tests in (A) and (B) cell population ratios were compared between Ctrl KD and the other conditions. (D) Based on cell division and viability data, cell division and viability rates can be calculated as parameters to create an exponential regression function of cell growth. (E) A prognosis of cell growth based on flow cytometric analysis after 72 h was compared to the time course analysis of U2OS cell growth based on cell count. Mean ± SD are given, and significances were calculated by two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001). Exponential regression curves are displayed with 95% confidence intervals. Significances between exponential regression curves of cell division and apoptosis assay compared to cell count were calculated by extra sum-of-squares F test (ns, not significant; *P < 0.05).
The CFSE staining from the start until three days after knockdown demonstrated that the vast majority of cells underwent two cell divisions under the control knockdown (Ctrl KD) as well as under the A-MYB KD conditions, as expected for unaffected cell proliferation (Figure 8A). Following B-MYB KD, the population of cells that had divided twice significantly decreased and the fraction that divided once significantly increased. Strikingly, the most drastic effects were observed when both A-MYB and B-MYB were knocked down. The largest cell population did not divide at all (Figure 8A).
In the same set of experiments, the viability measured in apoptosis assays decreased only significantly after A-MYB + B-MYB dKD (Figure 8B), which is based on the apoptosis and cell death staining in flow cytometry (Figure 8C). Taken together, strong effects on cell division and viability are observed only with a combined knockdown of A-MYB and B-MYB. Of note, the effects on cell division appear stronger than the effects on cell viability.
As cell viability and cell division should be the only factors that determine proliferation of a cell culture, we developed two equations to calculate the proliferation rate of a cell population from the results of the combined apoptosis and cell division assay (Suppl. Data 3). For this, CFSE data were simplified into a single cell division rate, reflecting the average number of cell divisions per day, whereas the relative population of viable cells in the apoptosis assay indicated the viability rate (Figure 8D). Based on these two parameters, a cell growth prognosis can be calculated (Supplementary Data 3). To evaluate the quality of this indirect measurement of cell growth based on the combined apoptosis and cell division assay, these results were compared with the actual cell count of U2OS cells during time progression following knockdown already introduced in Figure 7D.
For each knockdown the cell division and apoptosis assay correlated very well with the cell count data (Figure 8E), proving that the loss of growth potential upon A-MYB + B-MYB dKD is caused by both a substantial decrease in cell division as well as a gain in apoptosis. Similar results were obtained with HCT116 cells (Supplementary Figure S8).
In summary, the results provide evidence that A-MYB can functionally substitute for B-MYB in various cell types and that it contributes to drive the cell cycle and to inhibit apoptosis.
Discussion
B-MYB is a key cell cycle regulator and is well known for its oncogenic function (2,19). Its proliferation-supporting role and elevated expression associated with adverse prognosis for cancer patients has been well established in many tumor types, for example in breast cancer (58), renal cell carcinoma (59), and colorectal cancer (60).
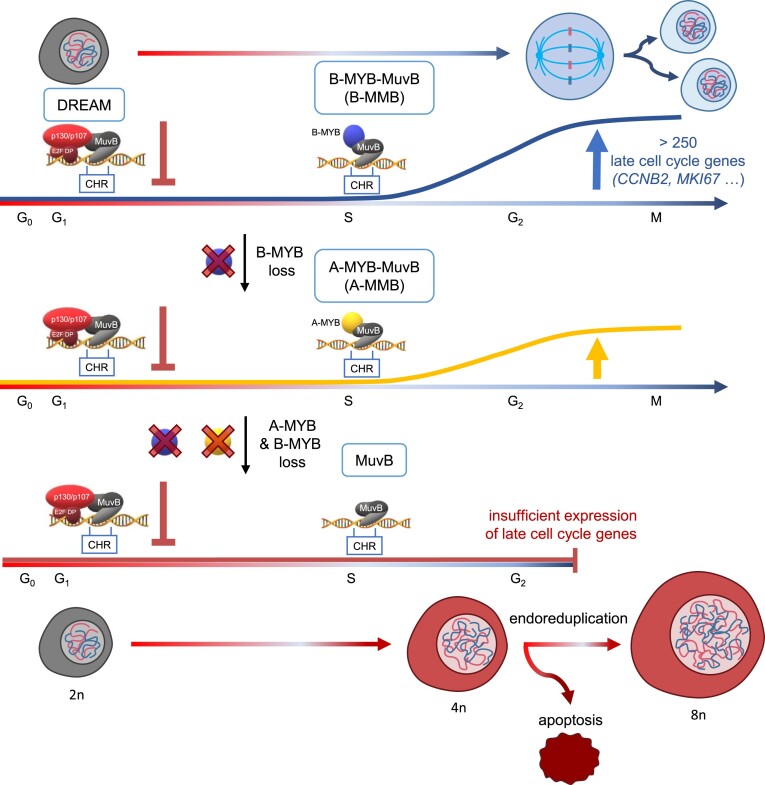
B-MYB exerts this function by binding the MuvB complex to activate genes that support progression through the cell cycle (7,61). Interestingly, B-MYB employs a noncanonical mode of DNA binding (Figure 9). It attaches not directly to the DNA but binds to the MuvB complex, which contacts the DNA directly through its LIN54 subunit at CHR promoter sites of target genes (8,24,62). The resulting MMB complex binds and activates the same genes that are downregulated by DREAM repressor complex. A switch in complex composition from the repressing DREAM to the activating MMB complex can be reversed by p53 activation via the p53–p21 axis. This switch causes cell cycle arrest (15,24). Since MMB target genes largely stimulate cell cycle progression, B-MYB activity causes an increase in proliferation (Figure 9).
Figure 9.
A-MYB substitutes for B-MYB in cell cycle gene regulation and cell proliferation. Loss of A-MYB and B-MYB leads to a G2/M arrest and subsequent endoreduplication as well as apoptosis. Ploidy is specified for resting diploid cells as examples.
Although A-MYB was observed to also bind MuvB, albeit with a lower affinity than B-MYB, its pro-proliferative role by activation of late cell cycle genes has not been thoroughly investigated (7,52–54). Here, we show that A-MYB can substitute for B-MYB as a transcription factor, cell cycle regulator, and protein counteracting induction of apoptosis.
Two central questions arise from this functional substitution. How relevant is the regulation by A-MYB in non-testicular cell types and does A-MYB—similar to B-MYB - display oncogenic properties?
Up to now, A-MYB has mostly been associated with spermatogenesis with high expression levels in testis. When examining tissue-wide expression data, it becomes evident that A-MYB is expressed also in several other tissues, including proliferative tissues such as lymphoid tissues (63,64). These results indicate a more general function for A-MYB, which is supported by results from various cell types presented here. In one central experiment, we find that proliferation can be further reduced by A-MYB knockdown even when cells had been deprived of B-MYB. This effect is also observed in cells that had been shown to depend on B-MYB expression, thus serving as model systems for B-MYB function, e.g. HeLa and HEK293 cells (4,65). Such experiments suggest that A-MYB is not only expressed but also possess important functions in most tissues.
Importantly, MYBL1 has also been implicated in the development of tumors. Overexpression and mutation of MYBL1 has been reported for several tumor types, implying that MYBL1 has properties of a proto-oncogene and an oncogene (2). For example, in a large fraction of pediatric low-grade gliomas, rearrangements including duplications and truncated mutants of the MYBL1 gene were observed. The A-MYB mutants in these astrocytomas had lost their negative regulatory domain at the C-terminus, which caused anchorage-independent growth of 3T3 cells and tumor formation in nude mice (33,66). Analyses in adenoid cystic carcinomas (ACC) led to the conclusion that MYBL1, similar to the related MYB gene, acts as an oncogenic driver through fusion translocations with NFIB and RAD51B (67). In related studies also in ACC, translocations to fusion genes of MYBL1 with NFIB and their expression correlated with poor prognosis for patients (68,69). In another example from hepatocellular carcinoma (HCC), it was described that MYBL1 is often overexpressed in HCC and high MYBL1 expression correlated with advanced disease and poor patient prognosis. Notably, A-MYB-overexpressing cells showed pro-metastatic properties in vitro, and ectopic expression of A-MYB stimulated cell proliferation in cell culture as well as in xenograft tumor models (34). Furthermore, MYBL1 was found overexpressed in subclasses of cells from triple-negative breast cancer (TNBC), implicating MYBL1 as a potential prognostic marker (70).
Generally, the pro-proliferative properties suggested by these reports are in accordance with results from mice lacking A-Myb. Mybl1-deficient animals do not develop increased numbers of tumors but are overall smaller in size compared to wild-type mice, with prominent hypoplasia of B cells and some tissues such as breast alveolar structures, seminiferous tubules in testes, and spleen (25,71). Conversely, Mybl1 transgene overexpression causes hyperproliferation of B lymphocytes as well as hyperplasia of spleen and lymph nodes (72). These results are in accordance with high MYBL1 expression levels found in Burkitt lymphomas (73). Importantly, all these reports analyzing MYBL1 expression, mutations, and functions in cells from various tissues as well as studies describing A-Myb function in mouse models are consistent with results described here. We show functional results for A-MYB in a spectrum of non-testicular cell types originating from cancer tissues of colon, bone, brain, liver, kidney and cervix as well as untransformed epithelial cells and fibroblasts. Thus, in regard to pro-proliferative and anti-apoptotic properties of A-MYB and its function in various cell types, results are consistent with observations from the earlier reports. Of note, A-MYB functions in the experiments described here were analyzed with endogenous expression levels in these cell lines.
Our analyses show that A-MYB binds MuvB in a cell cycle-dependent manner. The resulting A-MYB-MuvB complex is recruited to CHR promoter sites in a cell cycle-dependent manner analogous to B-MYB-MuvB (Figure 9). CHR promoter sites are indicative for late cell cycle genes (10,12). Thus, even though A-MYB expression is lower than B-MYB expression, A-MYB at endogenous levels in non-testicular cells can bind indirectly to late cell cycle gene promoters as part of an A-MYB-MuvB complex. When trying to estimate relative affinities, we find that A-MYB and B-MYB are bound from the input of nuclear extract with similar efficiency to the CHR promoter sites. This observation is in contrast to published affinity estimates (36). The discrepancy can be explained by the use of truncated versions of overexpressed proteins in the previous report compared to utilization of full-length proteins at endogenous levels in experiments presented here.
The binding of A-MYB to the CHR site led us to investigate the functional relevance of A-MYB association with MuvB proteins and CHR sites. The central question was how this binding affects the functional overlap of A-MYB with B-MYB in regulating cell division genes and controlling the cell cycle in general. The analysis of model genes as well as transcriptome-wide RNA-seq analysis demonstrated that A-MYB has major impact on MuvB- and CHR-dependent gene regulation as a substitute for B-MYB. Only a combined loss of A-MYB and B-MYB causes a substantial loss of gene activation. The genes downregulated after A-MYB and B-MYB loss are associated with mitosis and a majority represents late cell cycle genes reported as targets of B-MYB through the MMB complex (16). Only in a subset of cell lines expressing A-MYB at exceptionally low levels, represented by Hep3B cells in our study, the A-MYB-MuvB complex cannot substitute for B-MYB-MuvB in regulating the Cyclin B2 gene as an example. However, recombinant expression of mouse A-Myb can control a Cyclin B2 gene reporter dependent on the CHR site also in Hep3B cells. In addition, when performing a motif enrichment analysis of the core promoter regions of the downregulated genes after A-MYB + B-MYB dKD, the CHR site as the LIN54/MuvB binding site was highly enriched. This shows that A-MYB and B-MYB can substitute for each other in CHR-dependent transcriptional activation of late cell cycle genes through the MuvB complex by forming either an A-MYB-MuvB or B-MYB-MuvB complex. We therefore recommend redefining the commonly used term MYB-MuvB (MMB) to represent both alternative complexes.
These results also explain our earlier observations on B-MYB-dependent transcription. When we knocked down expression of B-MYB, expression of B-MYB targets CCNB2 (Cyclin B2) and MKI67 (Ki-67) did not drop to the expected low levels (8,37). In this setting, the unaffected A-MYB protein levels could replace the lost B-MYB transcriptional cofactor. Furthermore, the integration of A-MYB into the MYB-MuvB complex can also explain the increased A-MYB protein levels upon B-MYB knockdown, as A-MYB proteins are expected to be protected from degradation when integrated into complexes.
Our data show that knockdown of B-MYB alone already has a somewhat stronger effect on target gene expression than knockdown of A-MYB. Related results were obtained in knockout mouse models. Mybl1 deletion led to a general growth retardation in mice, indicating a reduced proliferative capacity of A-Myb-negative mouse embryos (25). The phenotype in Mybl2-negative mouse embryos is more severe as they display reduced proliferation of the blastocyst inner cell mass and die around E4.5-E6.5, likely caused by deficits from perturbed S phase regulation in the mice lacking B-Myb (74–77). The results from the two knockout models likely can be explained by the relative expression of A-Myb and B-Myb proteins with the resulting ability to substitute for each other as A-MYB is not detected in embryonic stem cells (74).
However, also in case of similar relative expression levels, particularly in adult tissues, several observations may explain the dominant role in transactivation of B-MYB over A-MYB. First, both A-MYB and B-MYB can also be bound and activated for transactivation by CBP/p300 binding to specific domains (78,79). CCAAT-boxes, the DNA motifs that can recruit CBP/p300 indirectly, are enriched in late cell cycle genes as well as in the genes downregulated by a combined loss of A-MYB and B-MYB (80–84). This highlights a potential role of CBP/p300 for A-MYB- and B-MYB-MuvB-dependent transactivation. Whether transactivation mechanisms by CBP/p300 differ for A-MYB and B-MYB is currently unclear. Second, also direct DNA binding of A-MYB and B-MYB, e.g. in enhancer regions, may affect their activity. Along these lines, it was shown for B-MYB that direct DNA binding employing its DNA binding domain is affected also by cell cycle-dependent phosphorylation (85). Third, the regulation of the phosphorylation status of A-MYB and B-MYB may vary. For example, it has been shown that Cyclin F binds B-MYB upon induction of DNA damage, thereby suppressing Cyclin A-dependent B-MYB phosphorylation, which is required for B-MYB transcriptional activity (86). Whether such mechanisms also apply to A-MYB and are responsible for differing modulation of transcription by the two factors has not been investigated. Fourth, the in vivo affinity of A-MYB versus B-MYB to MuvB may differ. The affinity of the MuvB-interacting C-terminal region of A-MYB towards its binding pocket in the MuvB core complex was found to be lower than the affinity of B-MYB to MuvB (36). The smaller affinity could yield a lower transactivation capacity for the A-MYB-MuvB complex. In our experiments, a comparison of A-MYB and B-MYB CoIPs implies a smaller capacity of A-MYB to coprecipitate MuvB core complex components. Inversely, CoIPs of MuvB core components, however, showed very clear A-MYB binding, similar to B-MYB. This proves that the potentially decreased affinity of A-MYB towards the MuvB core does not prevent formation of functional A-MYB-MuvB complexes. Finally, as mentioned before, A-MYB expression levels in most cell types appear lower than B-MYB expression levels, which may limit the formation of A-MYB-MuvB complexes. For example, the RPE-1 cell line, which displays a high A-MYB expression level, was the only among the tested cell lines in which A-MYB KD alone had a highly significant impact on Cyclin B2 expression, highlighting the importance of elevated A-MYB expression levels for its function as a B-MYB substitute.
When evaluating the functional overlap of A-MYB with B-MYB, the cellular processes such as cell cycle progression and apoptosis induction are of particular interest. We observe a G2/M arrest after combined knockdown of A-MYB and B-MYB. This block of the cell cycle results from the loss of expression of many genes that are required for the transition from G2 phase to mitosis (87). As seen from our experiments, a crucial set of these genes depends on transcriptional activation by A-MYB or B-MYB. G2/M arrest can be abrogated by single expression of either A-MYB or B-MYB, further supporting the notion that A-MYB and B-MYB functionally substitute for each other. When analyzing proliferation, no apparent difference was found between A-MYB function in transformed and untransformed cell lines. However, in one transformed cell line, namely Hep3B, A-MYB expression was too low to substitute for B-MYB in late cell cycle gene regulation and proliferation. This result is in line with the findings in the DepMap database that indicated a subset of cells displaying a low A-MYB transcript expression and thus being sensitive to B-MYB loss. Hep3B cells are part of this subset. Based on our exemplary analyses, this subset of transformed cells could potentially be addressed selectively by targeting only B-MYB but not A-MYB in a therapeutic setting, as A-MYB might be able to substitute for B-MYB in the untransformed cells. However, further tests on untransformed cells and animal models are required to assess this hypothesis. Generally, these results imply that the prognostic value of B-MYB expression depends also on A-MYB expression levels, and therefore expression of both factors must be considered in clinical prognosis and therapeutical approaches.
G2/M arrest in the context of A-MYB/B-MYB loss led to endoreduplication and thereby to chromosomal aberrations, as reported for other conditions that led to G2/M arrest (88–90). Of note, the increase in DNA content is independent of the ploidy in the starting cell population. As an example, we examined U2OS cells, which are near-triploid (91). Upon A-MYB and B-MYB double knockdown, U2OS cells also largely went through pseudo G2/M phase resulting in near-hexaploid cells. In addition, our findings are consistent with another study in Drosophila that found a signaling pathway involving Cyclin A, MMB, and Aurora B to play a crucial role regulating the preference between a mitotic cell cycle and endoreduplication, leading to polyploidy (92). Of note, myb, the only homologue of the MYB family in Drosophila, was found to suppress endoreduplication as well (93). An explanation for the rapid re-entry into DNA replication may be the constant expression of early cell cycle genes regulated by RB-E2F complexes even after cells arrest at G2/M because of A-MYB and B-MYB loss. Early cell cycle genes are required for DNA replication in S phase, but may also lead to a fast entry into endoreduplication.
Loss of A-MYB and B-MYB function also results in apoptosis. Cell cycle arrest upon A-MYB/B-MYB loss leads to cellular stress, as evidenced by the p53-like pattern of upregulated genes in RNA-seq analyses. This cellular stress ultimately causes apoptosis. The role of endoreduplication in this context remains elusive. Whether the induction of endoreduplication increases cellular stress and thereby apoptosis or whether it counteracts apoptotic signaling by diminishing cell cycle arrest, is currently unknown. However, evidence from treatment attempts targeting the mitotic machinery suggests that endoreduplication may rather be a mechanism to escape from cell death and acquire resistance against mitotic arrest (90,94). Elucidation of mechanistic details on induction of endoreduplication and apoptosis by A-MYB/B-MYB loss will require further studies on the signaling factors affected downstream of the MYB-MuvB/CHR regulatory system.
In summary, we show that A-MYB binds the MuvB complex and transcriptionally activates a plethora of cell cycle genes driving cell cycle progression. A-MYB substitutes for the B-MYB oncogene as a subunit of the MYB-MuvB complex and compensates for B-MYB loss in preventing G2/M arrest, endoreduplication, polyploidy, and cell death. These results provide evidence that A-MYB has a function beyond spermatogenesis and displays pro-proliferative properties of a proto-oncogene and oncogene. It needs to be elucidated how the interplay of A-MYB with B-MYB and particularly their relative abundance in cells influences their activity in supporting proliferation and preventing apoptosis.
Supplementary Material
Acknowledgements
The authors thank Kathrin Jäger at the core unit for performing flow cytometry, Bert Vogelstein for the kind gift of HCT116 cells, Roger Watson for generously providing the anti-B-MYB monoclonal antibody, and Christine Engeland for critically reading the manuscript.
Author contributions: R.K. and K.E. developed the study. R.K. performed the experiments. R.K. and K.E. wrote the manuscript.
Contributor Information
Robin Kohler, Molecular Oncology, Medical School, University of Leipzig, Semmelweisstr. 14, 04103 Leipzig, Germany.
Kurt Engeland, Molecular Oncology, Medical School, University of Leipzig, Semmelweisstr. 14, 04103 Leipzig, Germany.
Data availability
RNA-seq data underlying this article are available in the Gene Expression Omnibus and can be accessed under accession code GSE240799.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [424870812 to K.E.]; Roland-Ernst-Stiftung [04/22 to K.E.]. Funding for open access charge: Deutsche Forschungsgemeinschaft and University of Leipzig.
Conflicts of interest statement
None declared.
References
- 1. Musa J., Aynaud M.-M., Mirabeau O., Delattre O., Grünewald T.G.. MYBL2 (B-Myb). A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death. Dis. 2017; 8:e2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cicirò Y., Sala A.. MYB oncoproteins. Emerging players and potential therapeutic targets in human cancer. Oncogenesis. 2021; 10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen X., Lu Y., Yu H., Du K., Zhang Y., Nan Y., Huang Q.. Pan-cancer analysis indicates that MYBL2 is associated with the prognosis and immunotherapy of multiple cancers as an oncogene. Cell Cycle. 2021; 20:2291–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sadasivam S., Duan S., DeCaprio J.A.. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012; 26:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinberg R.A. The retinoblastoma protein and cell cycle control. Cell. 1995; 81:323–330. [DOI] [PubMed] [Google Scholar]
- 6. Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., Eyss B.v., Gagrica S., Hänel F., Brehm A., Gaubatz S.. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007; 6:1903–1913. [DOI] [PubMed] [Google Scholar]
- 7. Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A.. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007; 26:539–551. [DOI] [PubMed] [Google Scholar]
- 8. Müller G.A., Quaas M., Schümann M., Krause E., Padi M., Fischer M., Litovchick L., DeCaprio J.A., Engeland K.. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012; 40:1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadasivam S., DeCaprio J.A.. The DREAM complex. Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer. 2013; 13:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller G.A., Wintsche A., Stangner K., Prohaska S.J., Stadler P.F., Engeland K.. The CHR site. Definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014; 42:10331–10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dyson N.J. RB1. A prototype tumor suppressor and an enigma. Genes Dev. 2016; 30:1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Müller G.A., Stangner K., Schmitt T., Wintsche A., Engeland K.. Timing of transcription during the cell cycle. Protein complexes binding to E2F, E2F/CLE, CDE/CHR, or CHR promoter elements define early and late cell cycle gene expression. Oncotarget. 2017; 8:97736–97748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uxa S., Bernhart S.H., Mages C.F.S., Fischer M., Kohler R., Hoffmann S., Stadler P.F., Engeland K., Müller G.A.. DREAM and RB cooperate to induce gene repression and cell-cycle arrest in response to p53 activation. Nucleic Acids Res. 2019; 47:9087–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X., Müller G.A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A.D., Engeland K.. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell Biol. 2013; 33:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer M., Quaas M., Steiner L., Engeland K.. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016; 44:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischer M., Schwarz R., Riege K., DeCaprio J.A., Hoffmann S.. TargetGeneReg 2.0. A comprehensive web-atlas for p53, p63, and cell cycle-dependent gene regulation. NAR Cancer. 2022; 4:zcac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer M., Schade A.E., Branigan T.B., Müller G.A., DeCaprio J.A.. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 2022; 47:1009–1022. [DOI] [PubMed] [Google Scholar]
- 18. Pattschull G., Walz S., Gründl M., Schwab M., Rühl E., Baluapuri A., Cindric-Vranesic A., Kneitz S., Wolf E., Ade C.P.et al.. The Myb-MuvB complex is required for YAP-dependent transcription of mitotic genes. Cell Rep. 2019; 27:3533–3546. [DOI] [PubMed] [Google Scholar]
- 19. Iness A.N., Felthousen J., Ananthapadmanabhan V., Sesay F., Saini S., Guiley K.Z., Rubin S.M., Dozmorov M., Litovchick L.. The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B-Myb. Oncogene. 2019; 38:1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quaas M., Müller G.A., Engeland K.. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012; 11:4661–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fischer M., Grundke I., Sohr S., Quaas M., Hoffmann S., Knörck A., Gumhold C., Rother K.. p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes. PLoS One. 2013; 8:e63187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fischer M., Steiner L., Engeland K.. The transcription factor p53. Not a repressor, solely an activator. Cell Cycle. 2014; 13:3037–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fischer M., Quaas M., Nickel A., Engeland K.. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget. 2015; 6:41402–41417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53. I have a DREAM. Cell Death Differ. 2018; 25:114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toscani A., Mettus R.V., Coupland R., Simpkins H., Litvin J., Orth J., Hatton K.S., Reddy E.P.. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997; 386:713–717. [DOI] [PubMed] [Google Scholar]
- 26. Bolcun-Filas E., Bannister L.A., Barash A., Schimenti K.J., Hartford S.A., Eppig J.J., Handel M.A., Shen L., Schimenti J.C.. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development. 2011; 138:3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li X.Z., Roy C.K., Dong X., Bolcun-Filas E., Wang J., Han B.W., Xu J., Moore M.J., Schimenti J.C., Weng Z.et al.. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell. 2013; 50:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maezawa S., Sakashita A., Yukawa M., Chen X., Takahashi K., Alavattam K.G., Nakata I., Weirauch M.T., Barski A., Namekawa S.H.. Super-enhancer switching drives a burst in gene expression at the mitosis-to-meiosis transition. Nat. Struct. Mol. Biol. 2020; 27:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu T., Biasini A., Cecchini K., Saflund M., Mou H., Arif A., Eghbali A., Rooij D.d., Weng Z., Zamore P.D.et al.. A-MYB/TCFL5 regulatory architecture ensures the production of pachytene piRNAs in placental mammals. RNA. 2022; 29:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander A.K., Rice E.J., Lujic J., Simon L.E., Tanis S., Barshad G., Zhu L., Lama J., Cohen P.E., Danko C.G.. A-MYB and BRDT-dependent RNA Polymerase II pause release orchestrates transcriptional regulation in mammalian meiosis. Nat. Commun. 2023; 14:1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cecchini K., Biasini A., Yu T., Säflund M., Mou H., Arif A., Eghbali A., Colpan C., Gainetdinov I., Rooij D.G.d.et al.. The transcription factor TCFL5 responds to A-MYB to elaborate the male meiotic program in mice. Reproduction. 2023; 165:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ziebold U., Klempnauer K.H.. Linking Myb to the cell cycle. Cyclin-dependent phosphorylation and regulation of A-Myb activity. Oncogene. 1997; 15:1011–1019. [DOI] [PubMed] [Google Scholar]
- 33. Ramkissoon L.A., Horowitz P.M., Craig J.M., Ramkissoon S.H., Rich B.E., Schumacher S.E., McKenna A., Lawrence M.S., Bergthold G., Brastianos P.K.et al.. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8188–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie B., Liu Y., Zhao Z., Liu Q., Wang X., Xie Y., Liu Y., Liu Y., Yang Y., Long J.et al.. MYB proto-oncogene-like 1-TWIST1 axis promotes growth and metastasis of hepatocellular carcinoma cells. Mol. Ther. Oncolytics. 2020; 18:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Zhu J., Wu Y., Yu Y., Li Y., Shen J., Zhang R.. MYBL1 induces transcriptional activation of ANGPT2 to promote tumor angiogenesis and confer sorafenib resistance in human hepatocellular carcinoma. Cell Death. Dis. 2022; 13:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guiley K.Z., Iness A.N., Saini S., Tripathi S., Lipsick J.S., Litovchick L., Rubin S.M.. Structural mechanism of Myb-MuvB assembly. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:10016–10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uxa S., Castillo-Binder P., Kohler R., Stangner K., Müller G.A., Engeland K.. Ki-67 gene expression. Cell Death Differ. 2021; 28:3357–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:10. [Google Scholar]
- 39. Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.-C., Kitts P.A., Murphy T.D., Pruitt K.D., Thibaud-Nissen F., Albracht D.et al.. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017; 27:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R.et al.. Ensembl 2022. Nucleic Acids Res. 2022; 50:D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kolde R. Pheatmap. Pretty heatmaps. R Package Version. 2012; 1:726. [Google Scholar]
- 44. Kanehisa M., Goto S.. KEGG. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.et al.. clusterProfiler 4.0. A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L., Hill D.P.et al.. The Gene Ontology knowledgebase in 2023. Genetics. 2023; 224:iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fischer M., Grossmann P., Padi M., DeCaprio J.A.. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016; 44:6070–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Machlab D., Burger L., Soneson C., Rijli F.M., Schübeler D., Stadler M.B.. monaLisa. An R/Bioconductor package for identifying regulatory motifs. Bioinformatics. 2022; 38:2624–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Lemma R.B., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Pérez N.et al.. JASPAR 2022. The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022; 50:D165–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Müller G.A., Engeland K.. DNA Affinity Purification. A Pulldown Assay for Identifying and Analyzing Proteins Binding to Nucleic Acids. Methods Mol. Biol. 2021; 2267:81–90. [DOI] [PubMed] [Google Scholar]
- 51. Lange-zu Dohna C., Brandeis M., Berr F., Mössner J., Engeland K.. A CDE/CHR tandem element regulates cell cycle-dependent repression of cyclin B2 transcription. FEBS Lett. 2000; 484:77–81. [DOI] [PubMed] [Google Scholar]
- 52. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N.et al.. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014; 42:D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P.et al.. STRING v11. Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oughtred R., Rust J., Chang C., Breitkreutz B.-J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F.et al.. The BioGRID database. A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021; 30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Engeland K. Cell cycle regulation. P53-p21-RB signaling. Cell Death Differ. 2022; 29:946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M.et al.. Defining a cancer dependency map. Cell. 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dempster J.M., Boyle I., Vazquez F., Root D.E., Boehm J.S., Hahn W.C., Tsherniak A., McFarland J.M.. Chronos. A cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021; 22:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bayley R., Ward C., Garcia P.. MYBL2 amplification in breast cancer. Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta Rev. Cancer. 2020; 1874:188407. [DOI] [PubMed] [Google Scholar]
- 59. Sun S.-S., Fu Y., Lin J.-Y.. Upregulation of MYBL2 independently predicts a poorer prognosis in patients with clear cell renal cell carcinoma. Oncol. Lett. 2020; 19:2765–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ren F., Wang L., Shen X., Xiao X., Liu Z., Wei P., Wang Y., Qi P., Shen C., Sheng W.et al.. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am. J. Cancer Res. 2015; 5:1542–1552. [PMC free article] [PubMed] [Google Scholar]
- 61. Osterloh L., Eyss B.v., Schmit F., Rein L., Hübner D., Samans B., Hauser S., Gaubatz S.. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007; 26:144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmit F., Cremer S., Gaubatz S.. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009; 276:5703–5716. [DOI] [PubMed] [Google Scholar]
- 63. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A.et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 64. Karlsson M., Zhang C., Méar L., Zhong W., Digre A., Katona B., Sjöstedt E., Butler L., Odeberg J., Dusart P.et al.. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021; 7:eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Werwein E., Cibis H., Hess D., Klempnauer K.-H.. Activation of the oncogenic transcription factor B-Myb via multisite phosphorylation and prolyl cis/trans isomerization. Nucleic Acids Res. 2019; 47:103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang J., Wu G., Miller C.P., Tatevossian R.G., Dalton J.D., Tang B., Orisme W., Punchihewa C., Parker M., Qaddoumi I.et al.. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013; 45:602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brayer K.J., Frerich C.A., Kang H., Ness S.A.. Recurrent fusions in MYB and MYBL1 define a common, transcription factor-driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 2016; 6:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitani Y., Liu B., Rao P.H., Borra V.J., Zafereo M., Weber R.S., Kies M., Lozano G., Futreal P.A., Caulin C.et al.. Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin. Cancer Res. 2016; 22:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Togashi Y., Dobashi A., Sakata S., Sato Y., Baba S., Seto A., Mitani H., Kawabata K., Takeuchi K.. MYB and MYBL1 in adenoid cystic carcinoma. Diversity in the mode of genomic rearrangement and transcripts. Mod. Pathol. 2018; 31:934–946. [DOI] [PubMed] [Google Scholar]
- 70. Player A., Abraham N., Burrell K., Bengone I.O., Harris A., Nunez L., Willaims T., Kwende S., Walls W.. Identification of candidate genes associated with triple negative breast cancer. Genes Cancer. 2017; 8:659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vora K.A., Lentz V.M., Monsell W., Rao S.P., Mettus R., Toscani A., Reddy E.P., Manser T.. The T cell-dependent B cell immune response and germinal center reaction are intact in A-myb-deficient mice. J. Immunol. 2001; 166:3226–3230. [DOI] [PubMed] [Google Scholar]
- 72. DeRocco S.E., Iozzo R., Ma X.P., Schwarting R., Peterson D., Calabretta B.. Ectopic expression of A-myb in transgenic mice causes follicular hyperplasia and enhanced B lymphocyte proliferation. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Golay J., Erba E., Bernasconi S., Peri G., Introna M.. The A-myb gene is preferentially expressed in tonsillar CD38+, CD39-, and sIgM- B lymphocytes and in Burkitt's lymphoma cell lines. J. Immunol. 1994; 153:543–553. [PubMed] [Google Scholar]
- 74. Tanaka Y., Patestos N.P., Maekawa T., Ishii S.. B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem. 1999; 274:28067–28070. [DOI] [PubMed] [Google Scholar]
- 75. Lorvellec M., Dumon S., Maya-Mendoza A., Jackson D., Frampton J., García P.. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem. Cells. 2010; 28:1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. García P., Berlanga O., Watson R., Frampton J.. Generation of a conditional allele of the B- myb gene. Genesis. 2005; 43:189–195. [DOI] [PubMed] [Google Scholar]
- 77. García P., Frampton J.. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J. Cell Sci. 2006; 119:1483–1493. [DOI] [PubMed] [Google Scholar]
- 78. Facchinetti V., Loffarelli L., Schreek S., Oelgeschläger M., Lüscher B., Introna M., Golay J.. Regulatory domains of the A-Myb transcription factor and its interaction with the CBP/p300 adaptor molecules. Biochem. J. 1997; 324:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schubert S., Horstmann S., Bartusel T., Klempnauer K.-H.. The cooperation of B-Myb with the coactivator p300 is orchestrated by cyclins A and D1. Oncogene. 2004; 23:1392–1404. [DOI] [PubMed] [Google Scholar]
- 80. Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999; 239:15–27. [DOI] [PubMed] [Google Scholar]
- 81. Chan H.M., La Thangue N.B.. p300/CBP proteins. HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001; 114:2363–2373. [DOI] [PubMed] [Google Scholar]
- 82. Wasner M., Tschöp K., Spiesbach K., Haugwitz U., Johne C., Mössner J., Mantovani R., Engeland K.. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003; 536:66–70. [DOI] [PubMed] [Google Scholar]
- 83. Müller G.A., Engeland K.. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010; 277:877–893. [DOI] [PubMed] [Google Scholar]
- 84. Fischer M., Quaas M., Wintsche A., Müller G.A., Engeland K.. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res. 2014; 42:163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wijeratne T.U., Guiley K.Z., Lee H.-W., Müller G.A., Rubin S.M.. Cyclin-dependent kinase-mediated phosphorylation and the negative regulatory domain of transcription factor B-Myb modulate its DNA binding. J. Biol. Chem. 2022; 298:102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Klein D.K., Hoffmann S., Ahlskog J.K., O’Hanlon K., Quaas M., Larsen B.D., Rolland B., Rösner H.I., Walter D., Kousholt A.N.et al.. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat. Commun. 2015; 6:5800. [DOI] [PubMed] [Google Scholar]
- 87. Viner-Breuer R., Yilmaz A., Benvenisty N., Goldberg M.. The essentiality landscape of cell cycle related genes in human pluripotent and cancer cells. Cell Div. 2019; 14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Moon D.O., Kim M.O., Kang C.H., Lee J.D., Choi Y.H., Kim G.Y.. JNK inhibitor SP600125 promotes the formation of polymerized tubulin, leading to G2/M phase arrest, endoreduplication, and delayed apoptosis. Exp. Mol. Med. 2009; 41:665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mili D., Abid K., Rjiba I., Kenani A.. Effect of SP600125 on the mitotic spindle in HeLa Cells, leading to mitotic arrest, endoreduplication and apoptosis. Mol. Cytogenet. 2016; 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sinha D., Duijf P.H.G., Khanna K.K.. Mitotic slippage. An old tale with a new twist. Cell Cycle. 2019; 18:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ben-Shoshan S.O., Simon A.J., Jacob-Hirsch J., Shaklai S., Paz-Yaacov N., Amariglio N., Rechavi G., Trakhtenbrot L.. Induction of polyploidy by nuclear fusion mechanism upon decreased expression of the nuclear envelope protein LAP2β in the human osteosarcoma cell line U2OS. Mol. Cytogenet. 2014; 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rotelli M.D., Policastro R.A., Bolling A.M., Killion A.W., Weinberg A.J., Dixon M.J., Zentner G.E., Walczak C.E., Lilly M.A., Calvi B.R.. A Cyclin A-Myb-MuvB-Aurora B network regulates the choice between mitotic cycles and polyploid endoreplication cycles. PLoS Genet. 2019; 15:e1008253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fitzpatrick C.A., Sharkov N.V., Ramsay G., Katzen A.L.. Drosophila myb exerts opposing effects on S phase, promoting proliferation and suppressing endoreduplication. Development. 2002; 129:4497–4507. [DOI] [PubMed] [Google Scholar]
- 94. Cheng B., Crasta K.. Consequences of mitotic slippage for antimicrotubule drug therapy. Endocr. Relat. Cancer. 2017; 24:T97–T106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data underlying this article are available in the Gene Expression Omnibus and can be accessed under accession code GSE240799.