Abstract
The Microphthalmia-associated Transcription Factor (MITF) governs numerous cellular and developmental processes. In mice, it promotes specification and differentiation of the retinal pigmented epithelium (RPE), and in humans, some mutations in MITF induce congenital eye malformations. Herein, we explore the function and regulation of Mitf in Drosophila eye development and uncover two roles. We find that knockdown of Mitf results in retinal displacement (RDis), a phenotype associated with abnormal eye formation. Mitf functions in the peripodial epithelium (PE), a retinal support tissue akin to the RPE, to suppress RDis, via the Hippo pathway effector Yorkie (Yki). Yki physically interacts with Mitf and can modify its transcriptional activity in vitro. Severe loss of Mitf, instead, results in the de-repression of retinogenesis in the PE, precluding its development. This activity of Mitf requires the protein phosphatase 2A holoenzyme STRIPAK-PP2A, but not Yki; Mitf transcriptional activity is potentiated by STRIPAK-PP2A in vitro and in vivo. Knockdown of STRIPAK-PP2A results in cytoplasmic retention of Mitf in vivo and in its decreased stability in vitro, highlighting two potential mechanisms for the control of Mitf function by STRIPAK-PP2A. Thus, Mitf functions in a context-dependent manner as a key determinant of form and fate in the Drosophila eye progenitor epithelium.
Keywords: Striatin, YAP, PP2A, Retina, fate, epithelial morphology
Graphical Abstract
Summary Statement
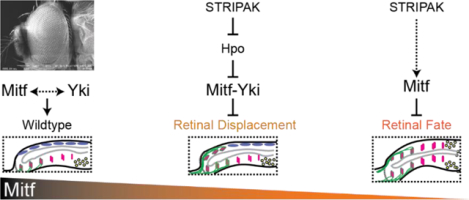
Mitf is a key determinant of peripodial fate and of retinal morphology in the developing fly eye. STRIPAK-PP2A, but not Yki, is required for Mitf to suppress retinal fate, whereas Mitf acts with Yki to prevent retinal displacement.
Introduction
Mitf is the sole Drosophila representative of the MITF/TFE basic helix-loop-helix transcription factor family, which includes vertebrate MITF, TFEB, TFEC, and TFE3 (Hallsson et al., 2007; Hallsson et al., 2004; La Spina et al., 2020). Mitf is a master regulator of autophagy where it controls the expression of numerous genes essential for lysosomal function (Bouche et al., 2016; Tognon et al., 2016; Zhang et al., 2015). This conserved role is orchestrated by TFEB, TFE3 and MITF in vertebrates (Martina et al., 2014; Palmieri et al., 2011; Ploper et al., 2015; Sardiello et al., 2009; Zhang et al., 2015). MITF is also a key factor in both fate specification and differentiation of the melanocytic cell lineage and of the retinal pigmented epithelium (RPE) of the vertebrate eye – a non-neuronal tissue that supports development of the retina at the embryonic stage and photoreceptor function in the adult (reviewed in (Goding and Arnheiter, 2019)). Dysfunction of MITF in humans is associated with various syndromic diseases (see OMIM 156845), including COMMAD (coloboma, osteopetrosis, microphthalmia, macrocephaly, albinism and deafness), resulting in congenital malformations of the eye (George et al., 2016). In the fruit fly, we previously reported that Mitf is expressed in the eye progenitor epithelium and that overexpression of exogenous Mitf can disrupt eye development (Hallsson et al., 2004). However, no loss-of-function (LOF) data were reported as the gene lies on the intractable chromosome IV of Drosophila. Given the association between MITF and multiple ocular phenotypes in vertebrates, and armed with new reagents for fly Mitf and chromosome IV, we sought to establish whether Mitf is required for development of the compound eye.
The Drosophila compound eye develops during the larval and pupal stages (reviewed in (Chen and Desplan, 2020)). Its primordial tissue, the eye imaginal disc, undergoes eye fate specification during larval stages 1 and 2 (L1 and L2). By early L2, the eye disc is comprised of two epithelial layers that are discernable by their physical properties and molecular markers – the retinal (neural) and the peripodial (PE; non-neural) epithelia (Fig. 1A–A’). The PE plays a support role in the proper development of the retina but does not give rise to any part of the compound eye (Cho et al., 2000; Gibson and Schubiger, 2000, 2001; Weasner et al., 2020). Retinogenesis begins early in L3 at the posterior of the retinal epithelium, and progresses anteriorly, as rows of progenitor cell clusters acquire neuronal fate and then differentiate into photoreceptors (Fig. 1B–C). By the end of L3, about 2/3 of the retinal array is formed and can be visualized as a crystalline lattice of differentiating photoreceptor cell clusters surrounded by accessory cells. Perturbations of these stages of eye development often result in aberrant compound eyes incompatible with normal vision.
Yorkie (Yki; vertebrate family members YAP1 and WWTR1/TAZ) is known to regulate numerous cellular and developmental processes as the principal effector of the Hippo pathway. Its activity is negatively regulated when Hippo kinase (Hpo; vertebrate STK3/MST2 and STK4/MST1) phosphorylates Warts kinase (Wts; vertebrate LATS1/2) which, in turn, phosphorylates Yki causing its cytoplasmic retention (reviewed in (Misra and Irvine, 2018)). Our recent work has identified recurrent roles for the Hippo-Yki pathway during Drosophila eye development. Disruption of Yki function up to mid-L2, in the presumptive PE, results in a PE-to-Retina fate change and yields eye discs with double retinae and no PE (Fig. 1D) (Neal et al., 2022). Disruption of Yki activity in the PE in late-L2 through early-L3 leads instead to a striking morphological defect named Retinal Displacement (RDis). RDis is categorized as either mild (mRDis) or severe (sRDis), based on its expressivity (see Methods for definition), and is so-called because the developmentally older posterior portion of the retinal epithelium shifts onto the PE plane of the disc (Fig. 1E) (DeSantis et al., 2023; Neal et al., 2022). While Yki itself functions in both RDis and fate, we have also identified adherens junctions (AJs) and two Protein Phosphatase 2A holoenzymes (the STRiatin Interacting Phosphatase And Kinase – STRIPAK – and PP2A(B’) complexes) as general or context-specific regulators of Yki (DeSantis et al., 2023; Neal et al., 2020, 2022). These factors function through the Hpo-Yki axis at multiple stages of development to promote normal compound eye formation.
In this work, we investigated the potential role of Mitf in Drosophila eye development. We find that Mitf is required to preserve eye disc morphology by preventing RDis, as well as to promote PE fate and prevent double-retina formation. Mitf preserves disc morphology through a Yki-dependent process. We show that Mitf can physically interact with Yki – as we demonstrate also for the human MITF and YAP1 proteins – and that Yki can modify Mitf-driven transcription in vitro. In contrast, we find that Mitf activity in PE fate is dependent on STRIPAK-PP2A, but not Yki. The heterotrimeric PP2A holoenzyme comprises constitutive A (scaffold) and C (catalytic) subunits, and a variable B-type subunit that confers substrate-specificity; a total of 5 genes encode the four classes of B subunits (B, B’, B”, B”’) in the fly. Depleting cells of the sole STRIPAK-specific B”’ subunit, Connector of Kinase to AP-1 (Cka), attenuates Mitf transcriptional activity on directly regulated targets, both in vivo and in vitro. In addition, Cka knock-down (KD) can reduce Mitf stability in vitro and diminish the nuclear/cytoplasmic distribution ratio of an inactivated form of Mitf in vivo. Thus, we have uncovered multiple roles for Mitf in Drosophila eye development – controlling disc morphology and PE fate – and have identified Yki and STRIPAK-PP2A as context-specific factors that interact with Mitf and contribute to its activity during these two distinct phases of eye development, respectively.
Materials and Methods
Drosophila genetics and fly lines
See Table S1 for a list of stocks used and their sources, and Table S2 for precise genotypes and experimental conditions for all reported crosses. Constitutive transgene expression in the eye imaginal disc was driven by the “flip-out” method, using ey-FLP with Act5Cp>CD2>GAL4 (ActeyFLP>GAL4; (Pignoni and Zipursky, 1997)). Briefly, flippase (FLP) is expressed throughout the developing eye disc via the ey enhancer. This catalyzes the excision of the CD2 interruption cassette from the Act5Cp>CD2>GAL4 transgene and results in constitutive GAL4 expression under the control of the Act5C promoter. A UAS-Dcr-2 transgene was used, as indicated. Dcr-2 promotes processing of long-hairpin RNAi reagents and was used to enhance the penetrance of MitfRNAi-induced phenotypes. Experimental larvae were dissected at the late L3 stage, unless otherwise indicated.
The Vienna Drosophila Resource Center (VDRC) UAS-MitfRNAi stock (v108519) used in this study (Dietzl et al., 2007) contains insertions at two landing sites (30B3 and 40D3), a possibility reported by Green et al. (2014), and validated by the VDRC and ourselves. Chromosome II (ChrII) recombinants with UAS-MitfRNAi were molecularly confirmed to maintain both insertions. Our reference to “homozygous” UAS-MitfRNAi (or 4XMitfRNAi) refers to experimental animals with two insertions on each of the maternal and paternal chromosomes. Spurious genetic interactions with the Hippo signaling cascade have been observed with dual-insert RNAi reagents from the VDRC, due to transgenesis into the tio locus at 40D3 (Green et al., 2014; Vissers et al., 2016). To preclude this possibility, we combined one copy of the MitfRNAi chromosome (2XMitfRNAi) with the “KK-TK” empty vector insertion line (VDRC_ID 60101). We did not observe increased RDis penetrance over that induced by the 2XMitfRNAi alone (Fig. S1A). These data suggest that upregulation of tio does not contribute to the results described herein.
Mosaic analysis with a repressible cell marker (MARCM) experiments were carried out essentially as reported by Lee et al. (Lee and Luo, 1999). FLP was expressed under the control of the ey enhancer (ey-FLP) and catalyzed the excision of a y+ interruption cassette from (Act5Cp>y+>GAL4) (Ito et al., 1997), to yield constitutive GAL4 expression. GAL80, expressed from a tub-GAL80 transgene, represses GAL4 activity in FLP-FRT mediated mitotic clones homozygous for this allele and in heterozygous tissue, while GAL4 activity was present to drive Mitf (as indicated) and GFP expression (UAS-GFP) in positively marked clones lacking GAL80.
Herein, we employed multiple wt UAS-Mitf transgenes, including those derived from P-element insertions (MitfCDS3 and MitfCDS6) and those targeted to specific genomic sites (Mitf96E). As shown in Figure 4, differential expressivity of the transgenes was leveraged to obtain phenotypes of different severities. Alternatively, different transgene insertions were used to facilitate the generation of specific experimental genotypes; for instance, the Chr II insertion UAS-MitfCDS6 was used with the dpp-GAL4 reagent (inserted on Chr III). To the extent tested, all wt UAS-Mitf transgenes elicit similar phenotypes.
MitfEA, also known as dominant-negative Mitf or MitfDN, has been previously reported (Hallsson et al., 2004). The derivative, MitfEAΔR, has a single arginine deleted from the 4R repeat in the basic region of the protein (residues 14–17 of bHLHzip_MITF_like consensus domain; NCBI accession cd11397); it is dimerization competent, but is defective in DNA binding and transcriptionally inactive.
Retinal displacement (RDis) and PE-to-Retina fate change were scored as per our prior work (DeSantis et al., 2023; Neal et al., 2020, 2022). Mild (m) RDis refers to eye discs where posterior, developmentally older, neuronal clusters are displaced into the disc margin and those where up to one row of displaced neuronal clusters are observed in the PE plane. Severe (s) RDis refers to discs where multiple rows of displaced neuronal clusters are observed in the PE plane. Briefly, differentiating photoreceptor clusters (positive for the pan-neural marker Elav and photoreceptor marker chaoptin (Chp, or 24B10 antigen) are normally restricted to the posterior retinal epithelium, but come to lie within the PE plane of the disc in both RDis and PE-to-Retina fate change (Fig. 1D–E). In PE-to-Retina fate change, these differentiating neuronal clusters are flanked anteriorly, in both cell layers, by developmentally younger clusters (Elav-positive and 24B10-negative) and a stripe of retinal progenitor cells (positive for the retinal determination factor Dac but negative for Elav and 24B10) (compare Fig. 1D to wt in Fig. 1C). This is not the case for RDIs, where the displaced Elav+24B10-positive clusters on the PE side abut PE cells, and Elav-only and Dac-positive cells are seen only in the retinal plane (compare Fig. 1E to wt in Fig. 1C).
Generation of the Mitf129 allele
CRISPR gRNA targets 5′- TAAGCGGGTACAGACGTTAC-3′ (exon 1b) and 5′- GGTTCTTGCATCTGACACAT-3′ (exon 9) were selected using the CRISPR Optimal Target Finder website (http://targetfinder.flycrispr.neuro.brown.edu/). Both gRNA sequences were cloned into plasmid pCFD4 based on the standard protocol (http://www.crisprflydesign.org/wp-content/uploads/2014/06/Cloning-with-pCFD4.pdf) and transgenesis of the attP2 locus was carried out (BestGene Inc.). The double-gRNA stock was crossed with the germline-expressing vas-Cas9 stock from the Bloomington Drosophila Stock Center (BDSC#51323) and F1 progeny were screened to identify the Mitf129 allele. This allele was induced on a 2FRT-RFP102D chromosome to facilitate screening, and was balanced over P{Tb1-RFP}IV to facilitate experimental analysis (see (Pina and Pignoni, 2012)). P{Tb1-RFP}IV dominantly confers a strong larval Tubby (Tb) phenotype (and weak red cuticle fluorescence), thus permitting easy identification of larvae that carry at least one copy of it. Furthermore, chromosome IV does not undergo meiotic recombination in Drosophila (Goldsmith et al., 2022; Hartmann and Sekelsky, 2017), making P{Tb1-RFP}IV effectively function as a balancer chromosome, although organisms carrying two copies of this chromosome are viable and fertile.
Immunohistochemistry
Standard protocols were used to dissect and immunostain eye and wing discs and salivary glands. Primary antibodies were guinea pig α-Mitf (1:500, (Zhang et al., 2015)), rabbit α-LacZ (1:1000, Cappel), rabbit α-Ey (1:2000, (Halder et al., 1998)), Invitrogen rabbit α-GFP (1:104; catalog # A-6455) and from the Developmental Studies Hybridoma Bank (DSHB) - 1:200 rat α-Elav (clone 7E8A10, deposited by G.M. Rubin), 1:500 mouse α-Dac (clone mAbdac2-3, deposited by G.M. Rubin), 1:50 rat α-DE-Cad (clone DCAD2, deposited by T. Uemura), 1:200 mouse α-Eya (clone eya10H6, deposited by S. Benzer and N.M. Bonini), 1:200 mouse α-Chp (clone 24B10, deposited by S. Benzer and N. Colley), 1:200 mouse α-Arm (clone N2 7A1, deposited by E. Wieschaus), 1:200 mouse α-LacZ (clone 40–1a, deposited by J.R. Sanes), 1:100 mouse α-Ey (deposited by P. Callaerts). Cy2-, Cy3-, and Cy5-conjugated goat α-mouse, rat, guinea pig or rabbit secondary antibodies were used at 1:400 (Jackson ImmunoResearch). Phalloidin Alexa-546 conjugate (1:50; Molecular Probes) and/or Hoechst 33342 (1:105; Invitrogen) was included in either the secondary antibody solution or in the mounting medium as counterstain, where indicated. Discs were mounted in medium consisting of 65% glycerol and 2.5% n-propyl gallate (Sigma) in 1xPBS, or Vectashield (Vector Labs).
Image acquisition and processing
Confocal stacks (minimum 3 per genotype) were recorded in Leica LASX software using a Leica DM5500Q microscope with SPEII confocal head. Color micrographs of wing imaginal discs were acquired using a Leica DFC300FX color camera. Color images of adult heads and wings were acquired on a Nikon E600 microscope using an Accu-scope Excelis camera. Images were processed in LASX and/or in Adobe Photoshop; only global manipulations were applied to images. Tissue was imaged under identical and non-saturating conditions for quantification of fluorescence signals; the subcellular distribution of Mitf was determined by calculating the ratio of area-corrected fluorescence intensity of the nuclear (defined by DAPI) and cytoplasmic (defined by DE-Cad cell boundary, minus nucleus) compartments of a cell. Quantification was made from a single central plane in each cell (30 cells/gland). For representative images, fluorescence intensities were adjusted both pre- and post-imaging for best presentation and no quantitative inferences should be made from these data. Colorimetric images were acquired with fixed exposures such that maximal signals were not saturated. LacZ activity was quantified by comparing log-transformed background-subtracted signals from a hand-drawn region of interest, using FIJI (NIH) and Microsoft Excel. Figures were assembled in Adobe Illustrator.
Preparation of dsRNA
dsRNA was prepared using the MEGAscript T7 kit (Ambion). Briefly, plasmids containing the verified full-length cDNAs of target genes were ordered from the Drosophila Genomics Resource Center (DGRC). dsRNA target regions were identified from the Drosophila RNAi Screening Center (DRSC) database (Mitf: DRSC21056; yki: DRSC28304; RagA/B: DRSC14450; RagC/D: DRSC35563; Cka: DRSC34541). Target regions were amplified by PCR using corresponding T7-tagged primers and 1 μg of PCR template was used for cDNA synthesis. Negative control dsRNA targeting GFP (cDNA nucleotides 1–435) was as described previously (Neal et al., 2019). RNA was pelleted by centrifugation at >18,000 × g at 4°C for 15 min and resuspended in 100–200 μL of nuclease free water (Invitrogen).
Cell culture
Drosophila S2-DRSC (S2) cells were purchased from the DGRC (RRID:CVCL_Z992). Cells were grown at 25°C in M3 Medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (SH30070.02, HyClone), 50 units/mL penicillin G + 50 μg/mL streptomycin sulfate (Gibco). All experiments were performed between cell passages 3–30. S2 cells were plated at a density of 1–3 × 106 cells/mL in 6- or 24-well plates (1 mL or 0.5 mL/well, respectively) and allowed to adhere to the bottom of the well. For dsRNA treatments in 24-well plates, growth medium was removed after 24 hrs and serum-free M3 medium containing dsRNA (2.5 μg) was exchanged daily, for 3 days. Cells were transfected using TransIT-Insect (MIR 6100, Mirus) according to the manufacturer’s instructions. Plasmids used were pΔTub-hRluc, pGL3–8Mbox-hsp70, pAcpA-Mitf-myc, pAc-Yki-FLAG (modified from pAc-HA-Yki; gift from D. Pan), and typically 50–200 ng of each plasmid was used. The pΔTub promoter was subcloned from pΔTubHA4C, a gift from D. Rincon-Limas (Zhang et al., 2013), into pGL4.70 (Promega). pGL3–8Mbox-hsp70 was constructed in our lab, by concatenating 2 copies of the 4Mbox sequence reported in (Zhang et al., 2015), where Mbox = “tcatgtg” (Supplemental File 1 – Complete and annotated sequence of pGL3–8Mbox-hsp70).
HeLa cells (gift of the Viapiano Lab, Upstate Medical University) were grown at 37°C and 5% CO2 in 1X Modified IMEM (A10489–01, Gibco) supplemented with 10% heat-inactivated fetal bovine serum, 50 units/mL penicillin G + 50 μg/mL streptomycin sulfate. This cell line was authenticated through a commercial service (https://azgc.arizona.edu/services/complete-solutions/cell-line-authentication). Experiments were performed between cell passage 2–20. Plasmids were obtained from AddGene – pFLAG-YAP1 (66853; (Bao et al., 2011)), pEGFP-N1-MITF-A (38132; (Roczniak-Ferguson et al., 2012)); empty pEGFP-N1 was obtained from the Viapiano Lab (Upstate Medical University).
Validation of gene expression and KD in S2 cells
Using standard protocols, total RNA was isolated from S2 cells treated with GFP, Mitf, RagA/B+RagC/D, yki, or Cka dsRNA, as above, using RNAzol (Molecular Research Center, Inc.). 1 μg of total RNA was reverse-transcribed using oligo(dT)20 and SuperScript III (Invitrogen). Quantitative PCR was performed on a Bio-Rad CFX 384 Real Time PCR System, using LightCycler 480 SYBR Green I Master Mix (Roche) and 10 ng of cDNA template, using the following DRSC primer pairs (5’ –> 3’): Mitf_F – AGTATCGGAGTAGATGTGCCAC + Mitf_R – CGCTGAGATATTGCCTCACTTG; yki_F – ATCAGCCCCATTCAGTTGAAC + yki_R – CCTCCCACTGCGTAGATTTTGTA; Cka_F – AATACCGGGCATCTTGCACTT + Cka_R – CAAGCAGCATAGCAATGCGAG; RagA/B_F – ACTATATCGCCCGTGATACGA + RagA/B_R – CCCAGAGATTAAGCACCAGGT; RagC/D_F – CATCCAAAAAGTGGTCTTCCACA + RagC/D_R – CGTCCTTCACGATCTTACTGGTC and published control primers Act5C_F – GGCGCAGAGCAAGCGTGGTA + Act5C_R – GGGTGCCACACGCAGCTCAT (Nevil et al., 2017). Gene expression was normalized to Act5c as a control, and differential gene expression under KD conditions was calculated by the 2−ΔΔCq method. Expression of all genes discussed was readily detected in S2 cells and is reported, relative to Act5C expression (Fig. S2A–A”’). Data were analyzed in Microsoft Excel.
Luciferase assays
RNAi-treated S2 cells were transfected 24 hrs after the third RNAi treatment and were lysed 72 hours later at room temperature in passive lysis buffer (Promega). Firefly and Renilla luciferase activities were immediately assessed using the Dual Luciferase Reporter Assay System (Promega) using a Synergy 2 plate reader (BioTek). Raw luminescence data were exported to Microsoft Excel for analysis.
Western blots and immunoprecipitation
S2 and HeLa cells were lysed 72 hrs post-transfection by rotation for 1 hr at 4°C in buffer containing 150 mM NaCl, 50 mM Tris, 1% NP-40, 1X cOmplete Mini EDTA-free Protease Inhibitor (04693159001, Roche), 1X Pierce Phosphatase Inhibitor (A32957, Thermo), Phosphatase Inhibitor Cocktail I (10 μL/mL, P2850, Sigma), 5 mM NaF, 1 mM Na3VaO3, 1 mM β-glycerophosphate, and 1 mM phenylmethylsulfonyl fluoride. Total protein was quantified using the Pierce BCA Protein Assay Kit (Thermo). 500 μg total protein was used for each overnight immunoprecipitation (at 4°C, with rotation), with either Pierce α-C-myc (88842, Thermo) or Pierce α-DYKDDDDK-FLAG (A36797, Thermo) antibody-conjugated magnetic beads. Sample processing continued using standard protocols.
Fixed volumes of IP (10 μL) and luciferase (20 μL) lysates, or 15–20 μg of input protein were electrophoresed on acrylamide gels (4% stacking, 8% separating) in a Bio-Rad Mini-PROTEAN tank at 100V and 120–150V, respectively. Protein was transferred to nitrocellulose membrane (1620112, Bio-Rad) using a Bio-Rad semi-dry transfer system for 30–35 min at 15V. Standard western blotting methods were used. Primary antibodies used were 1:4000 rabbit α-Myc at (71D10, Cell Signaling) and 1:500 mouse α- DYKDDDDK (1:500) (DSHB clone 12C6c, deposited by D. Alfandari). Total protein was detected on the membrane using Revert 700 (926–11021, LiCor) and specific immune complexes were detected with near-IR secondary antibodies (1:10,000 goat-α-mouse or goat α-rabbit IRDye 680RD or IRDye 800 (LiCor), 1:10,000 IRDye 800 goat-α-Mouse (Rockland), or 1:10,000 AlexaFluor680 goat-α-mouse Light Chain (Thermo)). Imaging was done using the LiCor Odyssey Imaging System (CLX-0642) on auto exposure for both 700 and 800 channels. Images were taken at 169 μm resolution and medium quality. Quantification of signal intensity was performed in LiCor ImageStudio (v5.2).
Statistical analyses
For genetic interaction analyses, Fisher’s Exact test of 2×3 or 2×4 contingency tables were used for pairwise comparisons (PA is reported). An online implementation of this tool was used (http://vassarstats.net/fisher2x3.html; http://vassarstats.net/fisher2x4.html; accessed throughout September and October, 2023). LacZ, luciferase, western blot and nuclear localization data were analyzed using one-way ANOVA with Dunnett’s or Šidák’s test for multiple comparisons (adjusted P values reported) or using Student’s t-test for pairwise analyses (precise P values reported), using GraphPad Prism (v10.0.3).
Reproducibility
Representative images and blots are shown in all Figures. All experiments were repeated at least once. For categorical observations, precise numbers of discs observed are given in the Figures. Multiple investigators independently observed the key phenotypes. Representative full blots are shown in Supplemental Figures.
Results
Mitf functions to preserve eye disc morphology.
As a first step to investigate a possible role for Mitf in Drosophila eye development, we induced constitutive KD of Mitf expression throughout the larval eye disc by RNA-interference (RNAi), using the compound driver ey-FLP, Act5Cp>IC>GAL4 (Actey-FLP>GAL4; see Methods). Either one or two copies of a chromosome carrying two UAS-MitfRNAi transgenes (2XMitfRNAi chromosome, see Methods) were combined with this driver and with a UAS-Dcr-2 transgene. Thus, we generated Dicer-enhanced 2XMitfRNAi and 4XMitfRNAi KD conditions.
In discs with 2XMitfRNAi, we observed a partially penetrant RDis phenotype (34% RDis). The phenotype presented as mild RDis (10/29 mRDis), with one row of clusters displaced into the margin or onto PE plane (see also Methods). Severe RDis (sRDis), with multiple rows of developing photoreceptor clusters displaced onto the PE plane, was not observed in 2XMitfRNAi discs (0/29) (Fig. 1F–J; Fig. S1A). Notably, the severity and penetrance of RDis was enhanced by using this chromosome in the homozygous state (4XMitfRNAi; 78% RDis: 27/36 sRDis, 1/36 mRDis) (Fig. 1G–J). Control L3 eye discs lacking the 2XMitfRNAi chromosome showed the normal pattern of neurogenesis; Elav-positive cells (neurons) were present only in the retinal epithelium, and a single band of Dac-positive cells (Dac-positive retinal progenitors) was located anterior to the emerging neuronal array (Fig. 1F–F’). In 4XMitfRNAi discs, several rows of neuronal clusters (Elav-positive) were mislocalized to the PE plane (Fig. 1G–G’). However, as is characteristic for the RDis phenotype (DeSantis et al., 2023; Neal et al., 2022), no Dac-positive cells were observed in the PE (Fig. 1G’) and all neurons in the PE were developmentally older as shown by co-expression of the photoreceptor-specific antigen 24B10 (Chp) along with Elav (Fig. 1I–I’, compare to wt in H–H’). Importantly, an RNAi-resistant Mitf rescue construct (Zhang et al., 2015) significantly reduced the severity and penetrance of the observed phenotype (Fig. 1K; 20% RDis: 1/25 sRDis, 4/25 mRDis). Altogether, these data confirm that the observed phenotype is RDis and that it results from Mitf KD in the eye disc.
Figure 1. Mitf is required in the PE to suppress RDis, and functions via Yki.
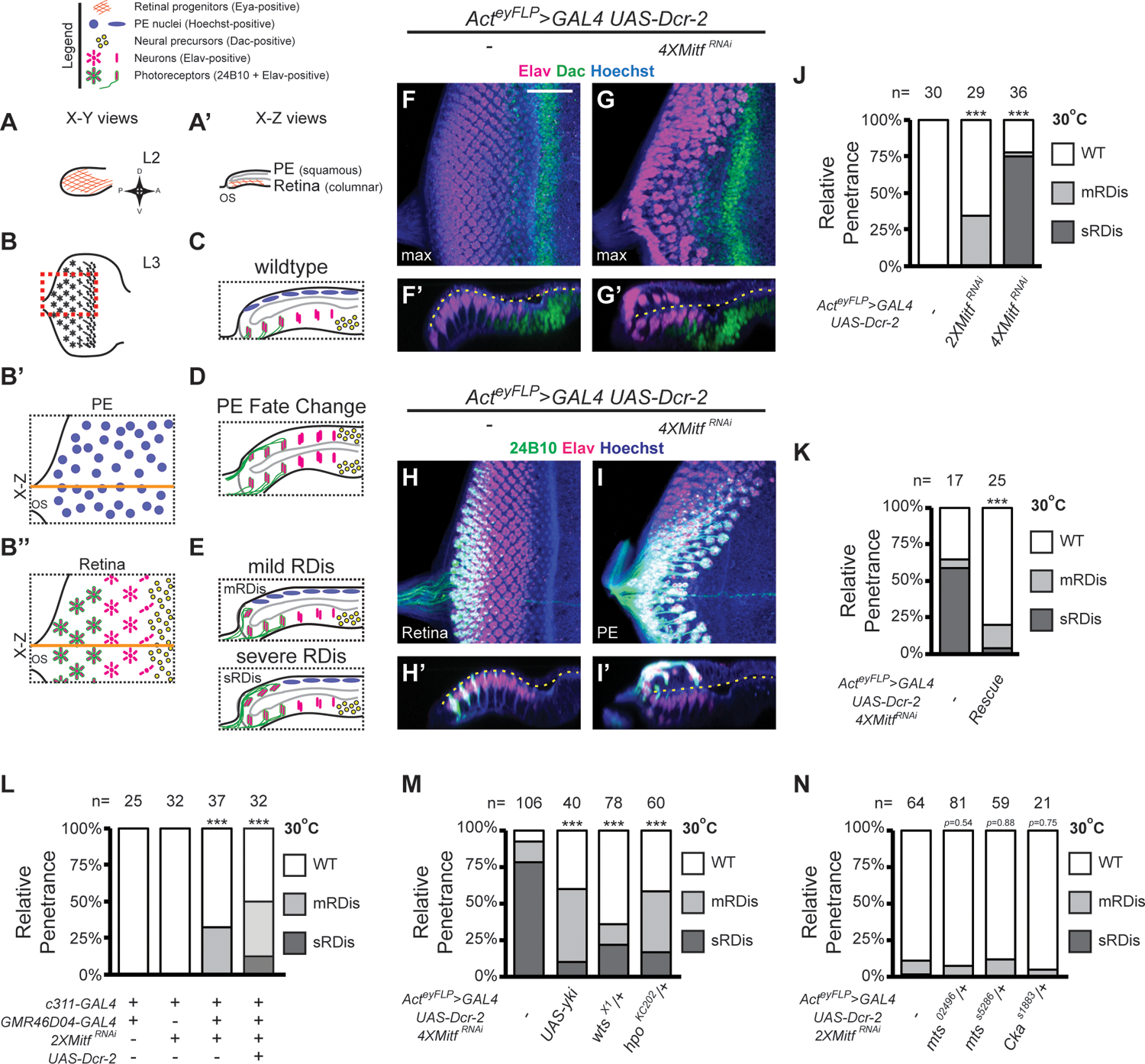
(A-E) Diagrams depicting normal and aberrant development of eye imaginal discs. Orthogonal views of Eya expression in wt L2 eye discs (A-A’). L3 eye disc schematic with region of interest outlined (red dashed box) (B). PE (B’) and retinal (B”) planes of a late L3 wt eye disc. Orange line: plane of optical X-Z section, shown in (C), where grey outline defines disc lumen. X-Z optical sections illustrating PE-to-retinal fate change (D) and mild or severe retinal displacement (mRDis or sRDis, respectively; E). (F-I’) L3 eye discs stained for Dac (green) and Elav (magenta) (F-G’), or Elav (magenta) and 24B10 (green) (H-I’), shown as X-Y (F, G, H, I) and X-Z (F’, G’, H’, I’) limited projections. Hoechst 33342 (DNA; blue); disc lumen (dashed yellow line). (J-K) Quantification of RDis phenotype (J: 2XMitfRNAi p=3.19 × 10−4; 4XMitfRNAi p=1.40 × 10−11; 2XMitfRNAi vs 4XMitfRNAi p=5.16 × 10−11); (K: “Rescue” = UAS-MitfRes; p=1.90 × 10−4). (L) Quantification of RDis phenotype in dual PE-driver experiments (relative to control: 2XMitfRNAi p=2.36 × 10−4; 2XMitfRNAi+Dcr-2 p=2.46 × 10−6; relative to 2XMitfRNAi: 2XMitfRNAi+Dcr-2 p=0.0605). (M-N) Quantification of RDis (M: UAS-yki, p=6.34 × 10−15; wtsX1/+, p=6.31 × 10−6; hpoKC202/+, p=5.92 × 10−10); (N: mts02496/+, p=0.54; mtsS5286/+, p=0.88; CkaS1883/+, p=0.75. Scale Bar (in F) = 25 μm; *** p<0.001
To assess the function of PE-expressed Mitf, we used the PE-restricted GAL4 drivers c311-GAL4 (expressed from L1 to L3) and hth46D04-GAL4 (broadly expressed from late L2; Neal et al., 2022). Expression of 2XMitfRNAi using c311-GAL4 did not perturb eye development (Fig. 1L; 32/32 wt discs). However, in eye discs where 2XMitfRNAi expression is driven by both c311-GAL4 and hth46D04-GAL4, we observed the mRDis phenotype (Fig. 1L; 32% RDis: 0/37 sRDis, 12/37 mRDis). Including UAS-Dcr-2 further increased the penetrance and expressivity of RDis in dual-driver 2XMitfRNAi discs (Fig. 1L; 50% RDis: 12/32 sRDis, 4/32 mRDis). The penetrance and expressivity of RDis in these discs exceeded that observed in the Actey-FLP>GAL4 UAS-Dcr-2 2XMitfRNAi genotype (34% mRDis, 0% sRDis; Fig. 1J). Notably, the temporal overlap of the two PE GAL4s drives broad expression in the PE starting in late L2, extending into L3 (Neal et al., 2022). This time frame corresponds well with the late-L2 to early-L3 activity period identified for previously studied RDis-associated factors, such as Yki, Cka and Armadillo (Arm, β-Catenin) (genetic pathways summarized in Fig. S1B) (DeSantis et al., 2023; Neal et al., 2022).
Given the genetic evidence of Mitf activity in the PE, we next sought to confirm that it is indeed expressed in this tissue and, particularly, at the late-L2 stage. Using in situ hybridization, we were able to detect Mitf transcripts in the PE at the late L2 stage (Fig. S1C–C’). In addition, using a synthetic reporter that combines the proximal 2.2 kb of the Mitf promoter with the coding sequence of enhanced green fluorescent protein (eGFP) (Zhang et al., 2015), we observed GFP-positive cells throughout the eye disc at the L2 stage, in both the PE and the retinal cell layers (Fig. S1D–D’). Broad expression in the PE domain persisted into the L3 stage (Fig. S1E–E’), but may in part reflect GFP persistence. Nonetheless, both lines of evidence suggest that Mitf is expressed in the PE of the eye disc throughout the developmental window previously associated with RDis (late L2) and with the broad overlap of c311-GAL4 and hth46D04-GAL4 expression.
Altogether, these data suggested that Mitf activity is required to preserve disc morphology and prevent RDis, through its action in the PE. This result also raised the question of Mitf’s relationship with Yki, given the similarity of RDis induced by Mitf KD and Yki KD.
MitfRNAi-induced RDis is Yki dependent.
To gain mechanistic insight into the roles of Mitf and Yki in RDis suppression, we relied on genetic interaction experiments. In particular, we considered that overexpression (OE) of Yki can rescue RDis induced by disruption of AJs or PP2A(B’) (DeSantis et al., 2023; Neal et al., 2022). Hence, we asked whether RDis induced by 4XMitfRNAi could be rescued by overexpression of wt Yki, using a UAS-Yki transgene that does not perturb eye development on its own. We observed a robust decrease in the penetrance and expressivity of RDis in such Yki OE + Mitf KD discs (Fig. 1M; control – 92% RDis: 83/106 sRDis, 15/106 mRDis; UAS-Yki – 60% RDis: 4/40 sRDis, 20/40 mRDis).
We next considered a more stringent assay. Specifically, we tested whether increasing the activity of endogenous Yki, through heterozygosity for its negative regulators Hpo or Wts (hpo/+ or wts/+), was sufficient to rescue 4XMitfRNAi-induced RDis. Indeed, we observed significantly reduced penetrance and expressivity of MitfRNAi-induced RDis in such larvae (Fig. 1M; wtsX1/+ – 36% RDis: 17/78 sRDis, 11/78 mRDis; hpoKC202/+ – 58% RDis: 10/60 sRDis, 25/60 mRDis); control larvae, hemizygous for wtsX1 or hpo KC202, are phenotypically normal (not shown). These findings suggest that discs with reduced Mitf are highly sensitized to changes in Yki activity, and altogether, support the interpretation that reduced Yki activity is a significant cause of RDis in Mitf KD discs.
Since Mitf might exercise its effect on Yki by acting upstream of the core Hippo kinase cascade, we next focused on STRIPAK-PP2A, which negatively regulates Hippo signaling by directly modifying Hpo kinase activity (Ribeiro et al., 2010). We thus tested for phenotypic enhancement of 2XMitfRNAi RDis (Fig. 1N; 11% RDis: 1/64 sRDis, 6/64 mRDis) by decreased STRIPAK-PP2A activity, using reagents previously shown to work in other RDis paradigms (Neal et al., 2022). Specifically, we used LOF alleles of the PP2A catalytic subunit Mts, encoded by microtubule star (mts), and of the STRIPAK-specific PP2A B”’ subunit Cka. Relative to Mitf KD controls, we did not observe enhanced expressivity or increased penetrance of the RDis phenotype in 2XMitfRNAi mts/+ discs (Fig. 1N; mts02496/+, 7% RDis; mtsS5286/+, 12% RDis), nor in 2XMitfRNAi Cka/+ discs (Fig. 1N; CkaS1883/+, 5% RDis); animal hemizygous for mts or Cka are phenotypically normal (not shown). Hence, we concluded that aberrant Yki activity in Mitf KD discs is unlikely to result from perturbation of the STRIPAK-Hpo regulatory node.
AJs are also known to function as a regulatory hub for Yki activity (Fig. S1B) (Rauskolb et al., 2014) and we previously established that RDis derived via loss of Arm/β-catenin in the PE (armRNAi-induced RDis) could be rescued by exogenous Yki expression (DeSantis et al., 2023). To assess whether AJs were at play in the MitfRNAi RDis paradigm, we observed AJs in 4xMitfRNAi discs via immunohistochemical detection of Arm and DE-Cadherin. We found the AJs to be indistinguishable from those in wt discs (Fig. S1F–I’). Hence, we concluded that decreased Yki activity in Mitf KD discs is unlikely to be due to AJ-dependent mechanisms.
In summary, we did not observe genetic interactions between Mitf and STRIPAK-PP2A, or AJs. Thus, while Yki-loss appears to be an important factor in MitfRNAi-induced RDis, we found no evidence to suggest that Mitf acts congruently with more distant regulators of Yki, with respect to this phenotype. Nonetheless, the strong evidence of a genetic interaction between Mitf and Yki prompted us to consider whether these factors might interact more directly to suppress RDis.
Yki binds Mitf and potentiates Mitf transcriptional activity in vitro
As a transcription co-factor, Yki has the potential to modulate the activity of DNA-binding transcription regulators, such as Mitf. Thus, we sought to interrogate a potential functional interaction between Yki and Mitf by perturbing their activity in Drosophila S2 cells. We began by using 8MBox-lucifearse (8MBox-luc), a reporter regulated by a synthetic array of 8 Mitf binding sites (Mbox, see Methods). Baseline reporter expression was modest under GFP dsRNA (control) conditions, and was significantly reduced upon treatment of cells with Mitf dsRNA (Fig. 2A); the specificity and degree of RNA KD by dsRNA treatment was assessed by qPCR (Fig. S2A–A”’). To increase the activity of endogenous Mitf protein, we used RNAi-mediated KD of Rag GTPases, an approach previously reported to work in mammalian cells (Ikeda et al., 2021). Rag GTPases, encoded by the RagA/B and RagC/D loci in flies, are required for mTORC1 activation on the surface of the lysosome (Kim et al., 2008; Sancak et al., 2008), and thus for MITF/TFE factor inactivation. As expected, Rag KD resulted in an increase in 8MBox-Luc reporter activity in S2 cells; specifically, activity increased by 35-fold (Fig. 2A’; normalized to 1). Under this condition of Rags KD-induced 8MBox-luc expression, treatment of cells with Mitf dsRNA returned activity to baseline levels (Fig. 2A’), confirming that Rags KD induces Mitf activation in S2 cells. Using the Rag depletion paradigm, we then co-depleted cells of yki using dsRNA. Strikingly, this resulted in a 46% reduction in reporter activation, down from 22-fold to 12-fold above baseline (Fig. 2A”). These results strongly suggest that Yki is required to potentiate the transcriptional activity of Mitf in S2 cells.
Figure 2. Yki physically interacts with Mitf and potentiates its transcriptional output.
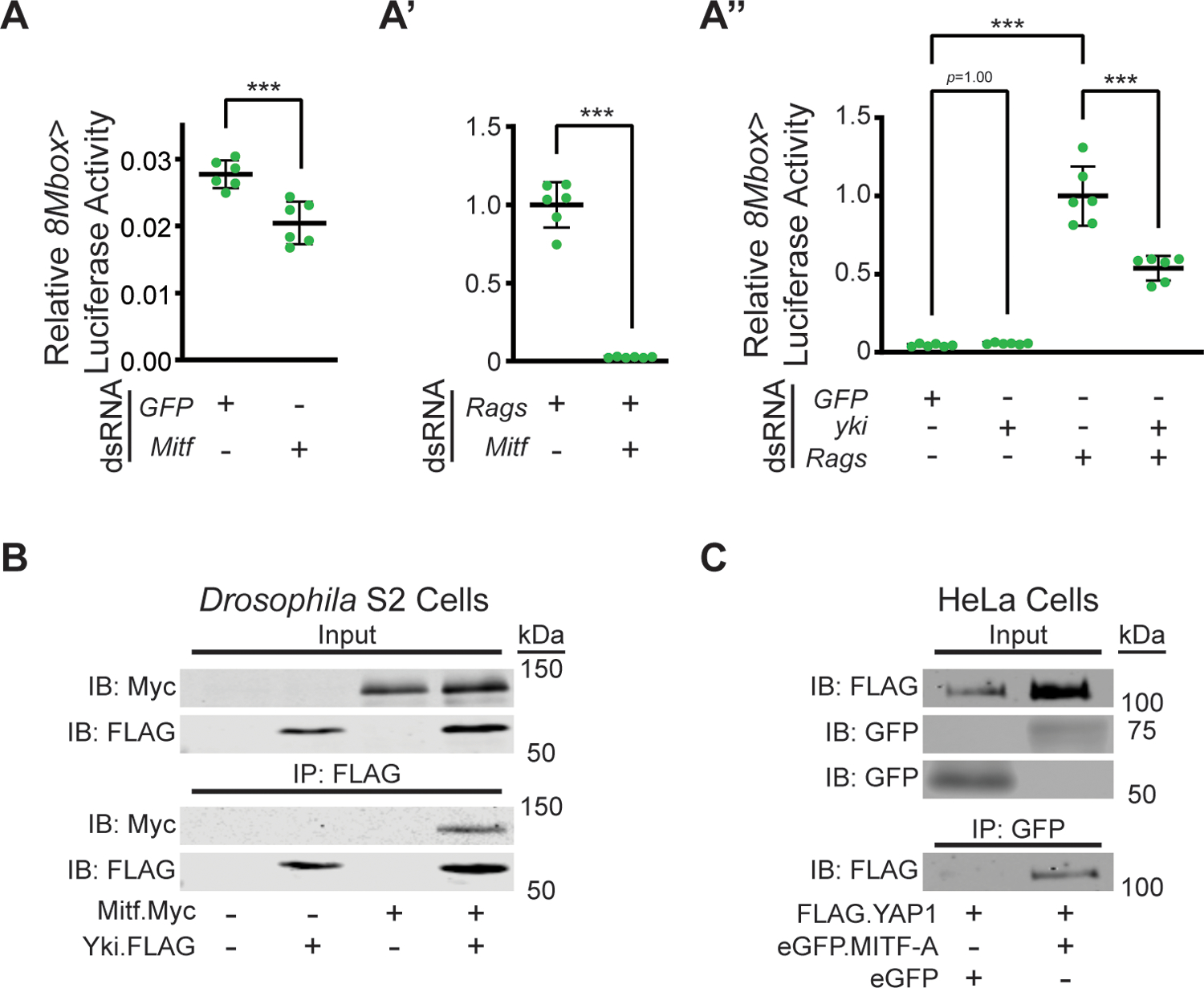
(A-A”) 8Mbox>Luciferase activity in S2 cells. Mitf KD decreases basal 8Mbox>Luciferase activity (A, p=0.0009). Rags KD increases basal 8Mbox>Luciferase activity (A’, note scale relative to panel A), in an Mitf-dependent manner (A’, p<0.0001). Rags KD-induced 8Mbox>Luciferase activity (p<0.0001, relative to basal activity), but not basal 8Mbox>Luciferase activity, is attenuated by yki dsRNA (p<0.0001 and p=1.00, respectively). *** p<0.001 (B) Co-IP of Mitf.Myc with Yki.FLAG from Drosophila S2 DRSC cell lysates. (C) Co-IP of FLAG.YAP1 with eGFP.MITF-A, but not eGFP alone, from HeLa cell lysates.
Next, we hypothesized that if the observed potentiation occurred via formation of a Yki-Mitf transcription complex, we should be able to detect a physical interaction between the two proteins. To test for binding, we relied on co-immunoprecipitation (co-IP) of tagged versions of the two proteins. Thus, we transiently transfected S2 cells with Mitf.Myc and Yki.FLAG expression vectors and used α-FLAG magnetic beads for co-IP (Fig. 2B; Fig. S2B). The Myc epitope (Mitf) was detected only in precipitates from cells expressing both Mitf and Yki, and not in those from cells expressing either factor alone (Fig. 2B), thus providing evidence of such an interaction. To investigate whether this interaction was conserved among the orthologous human proteins, MITF and YAP1, we performed co-IP assays from transiently-transfected HeLa cells expressing FLAG.YAP1 and either eGFP or eGFP.MITF-A. We then used α-GFP-conjugated magnetic beads for IP. Consistent with our findings in Drosophila cells, the FLAG epitope (YAP1) was detected only in precipitates from eGFP.MITF-A-expressing cells, and not from those expressing eGFP alone (Fig. 2C; Fig. S2C), providing strong evidence of a physical interaction between mammalian YAP1 and MITF.
In summary, our analyses demonstrate that Drosophila Yki and Mitf interact functionally to affect Mitf transcriptional activity, and that this may reflect a physical interaction between the two Drosophila proteins. Moreover, this interaction appears to be evolutionarily conserved, as human YAP1 and MITF can also bind. Whether this is the mechanisms at work in RDis will require further testing, but the high sensitivity of MitfRNAi-induced RDis to varying levels of Yki, Wts and Hpo (Fig. 1M) is consistent with the activity of a Yki-Mitf complex.
Mitf is required for PE fate.
In addition to its role in preserving eye disc morphology, Yki also acts prior to mid-L2 to control PE fate specification. Yki KD prior and up to mid-L2 results in the transformation of the PE into a second mirror image retina (Fig. 1D) (Neal et al., 2022). Hence, our interest was raised as to whether Mitf also played a role in PE fate specification, given the evidence of its functional interaction in vitro and genetic interaction in the RDis paradigm, with Yki. Despite using a constitutive and early driver, Actey-FLP>GAL4, that is sufficient to induce PE-to-Retina fate change with UAS-ykiRNAi or UAS-CkaRNAi (Neal et al., 2020; Zhang et al., 2011), we saw no evidence of a perturbation of PE fate in Actey-FLP>GAL4 UAS-Dcr-2 MitfRNAi discs, even under the 4XMitfRNAi condition (see Fig. 1G–G’ and Fig. 1I–I’).
The fact that we observed very low penetrance RDis in 2XMitfRNAi discs, and incompletely penetrant RDis in 4XMitfRNAi discs, led us to consider that Mitf KD might be incomplete. Hence, we pursued alternate experimental strategies to generate more severe Mitf LOF conditions in which to investigate a possible role for Mitf in PE fate specification.
We first generated a null allele of Mitf using CRISPR-CAS9 genome editing (Fig. S3A). This allele, Mitf129, is characterized by the deletion of most of the coding sequences of Mitf and is predicted to result in very early termination of all isoforms (Fig. S3B). Working with this allele proved challenging, because of its recessive lethality and the paucity of genetic tools for Drosophila chromosome IV. To facilitate our studies, we developed a chromosome IV that is marked with the Tubby-inducing Tb1 dominant allele, encoded via a Tb1-RFP fusion transgene, P{Tb1-RFP}IV. Using this tool, we ascertained that the majority of Mitf129/Mitf129 homozygous (non-Tb) larvae died at or before the L2 stage. Some “escapers” survived into the L3 stage (Fig. S3C), allowing us to observe their eye discs, which appeared to be developmentally arrested and similar in size to early L1 discs (Fig. 3A–A’). Although neurons were not observed (no Elav-positive cells were detected; not shown), cells throughout these discs were immunoreactive for the retinal fate determinant Eyes absent (Eya) (Fig. 3A–A’). Eya expression initiates in the emerging retinal epithelium of the wt disc very early in L2 (Fig. 1A–A’); it is, therefore, the earliest marker of retinal primordium formation (Kenyon et al., 2003). Striking to us was that there appeared to be Eya positive cells throughout the small Mitf129/Mitf129 L3 eye discs, suggesting a failure of PE specification. Whereas this phenotype was suggestive of a possible PE-to-Retinal fate change, it was not possible to distinguish two juxtaposed epithelia within these small discs.
Figure 3. Mitf is required for PE fate specification.
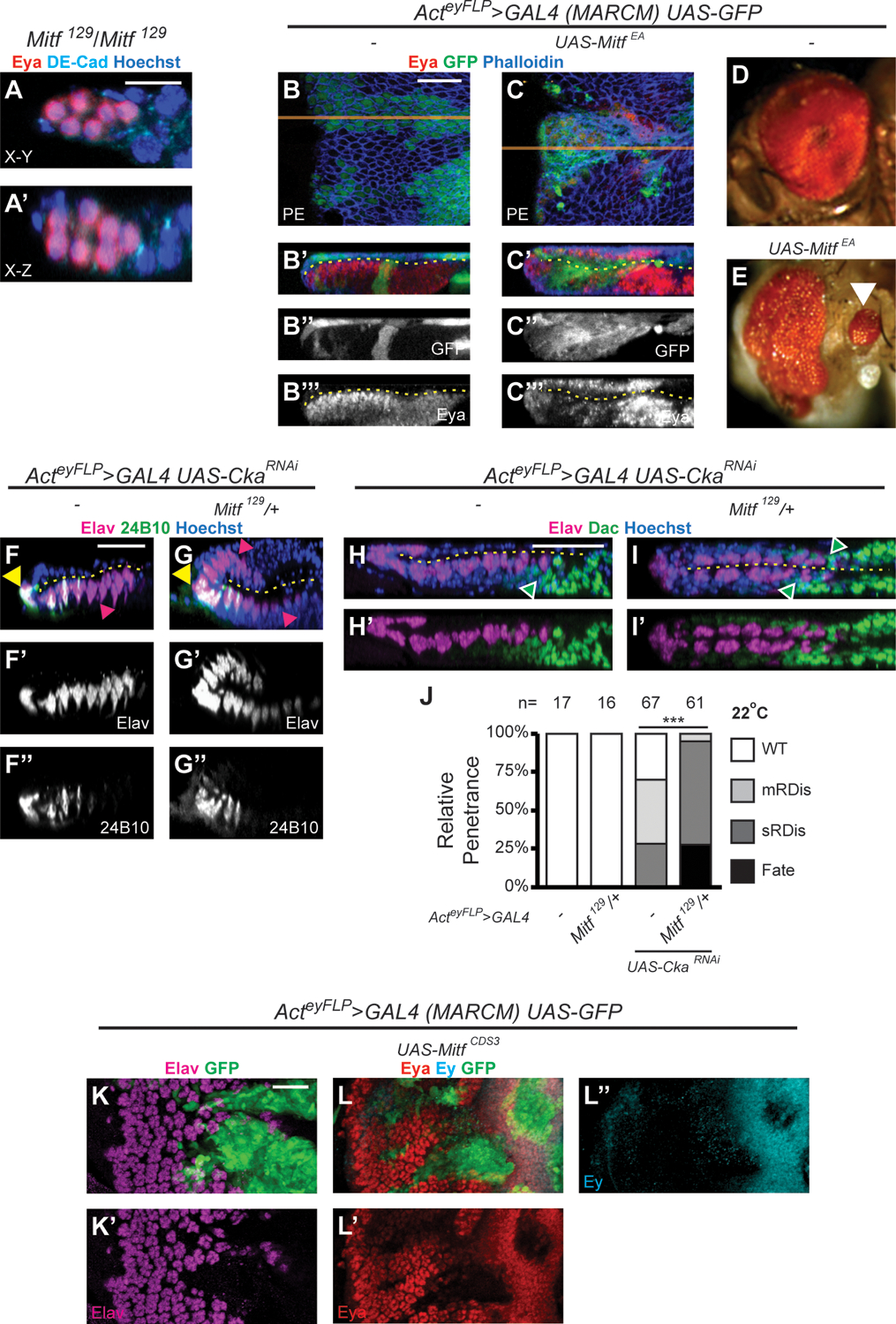
(A-A’) X-Y (A) and X-Z (A’) projections of Mitf129/Mitf129 eye disc (n=12). Discs stained for Eya (red), DE-Cad (cyan) and Hoechst 33342 (DNA, blue); Scale Bar in A = 10 μm. (B-C”’) MARCM clones overexpressing GFP alone (green, UAS-GFP, B-B”’) or in combination with MitfEA (UAS-MitfEA; C-C”’); X-Y projections of PE clones (B, C; orange line = plane of X-Z projection); X-Z projections (B’-B”’, C’-C”’). Discs stained for Eya (red) and F-actin (Phalloidin, blue); disc lumen (dashed yellow line); Scale Bar in B = 25 μm. (D-E) Adults developing from MARCM UAS-GFP larvae have wt eyes (D), whereas rare adults from MARCM UAS-MitfEA larvae exhibited malformed compound eyes with ectopic secondary compound eyes (white arrowhead) (E). (F-J) Genetic interaction assay between Cka and Mitf. At 22°C, CkaRNAi discs exhibit RDis (F-F” and H-H’) and CkaRNAi discs heterozygous for Mitf (Mitf129/+) exhibit PE-to-retina fate change (G-G” and I-I’). Discs are stained for Elav (magenta) and 24B10 (green) (F-G”) or Elav (magenta) and Dac (green) (H-I’), with Hoechst 33342 (DNA, blue); disc lumen (dashed yellow line); Scale Bars in F, H = 25 μm. Magenta arrowheads (F, G) indicate cells expressing Elav only localized anterior to cells expressing Elav + 24B10, in the retina (F) or in both cell layers (G). Green arrowheads (H, I) indicate Dac-positive cells anterior to Elav-positive cells in the retina (H) or in both cell layers (I). Quantification of phenotypes (J). Heterozygosity for Mitf129 increases phenotypic penetrance in CkaRNAi discs (p=1.01 × 10−14). (I-J”) MARCM clones overexpressing GFP (UAS-GFP, green) in combination with wt Mitf (UAS-MitfCDS3); X-Y projections of retinal clones are shown; discs stained for Elav (magenta, I-I’) or Eya (red, J-J’) and Ey (cyan, J, J”); Scale Bar in K = 25 μm.
As an alternative approach to generating LOF conditions, we relied on a dominant negative form of Mitf (UAS-MitfEA, also known as “MitfDN”) (Hallsson et al., 2004). In cell culture, MitfEA fails to induce expression of a synthetic reporter of Mitf transcriptional activity and strongly reduces reporter expression when co-transfected with wt Mitf (Hallsson et al., 2004). Using Mosaic Analysis with a Repressible Cell Marker (MARCM), we induced UAS-MitfEA-expressing clones in the eye disc. Relative to wt controls expressing GFP alone (Fig. 3B–B”’), few MitfEA-expressing larvae reached the L3 stage. In those that did, we detected multiple MitfEA-expressing clones in the eye-disc, including in the PE (GFP-marked). Interestingly, some of the PE clones showed co-expression of the retinal marker Eya (Fig. 3C–C”’). Moreover, in very rare eclosed adults, we observed ectopic small eyes in the post-gena, a cuticular region derived from the PE (Haynie and Bryant, 1986), along with dysmorphic compound eyes (Fig. 3D–E; white arrowhead in E).
To better understand this phenotype, we quantified the proportion and position of Eya-expressing clones in the PE (Fig. S3D). Only 3 of 35 GFP+MitfEA-expressing clones displayed Eya induction, and all 3 extended to the posterior margin of the disc (3/15 posterior clones; 0/20 internal clones). Importantly, BMP signaling (specifically, the ligand Decapentaplegic or Dpp) is required along the disc posterior to promote eye field formation in the wt disc (Chen et al., 1999; Kenyon et al., 2003). We thus hypothesized that enhancing Dpp signaling in MitfEA-expressing PE clones would increase the penetrance of retinal induction in these clones. To test this hypothesis, we co-expressed an activated form of the type I BMP receptor Thickveins (TkvQD; UAS-tkvQD) along with MitfEA and once again scored GFP-marked MitfEA+TkvQD-expressing clones based on their location within the PE. Under these conditions, we observed Eya expression in internal clones, far from the disc margin (27%; 3/11 clones), and of 10 PE clones extending to the disc posterior margin, 50% expressed Eya (versus 20% in clones without TkvQD). Importantly, Eya expression was never detected in PE clones expressing GFP alone (n>20), nor in those expressing TkvQD alone (0/36). Altogether, these findings strongly suggest that Mitf plays a role in suppressing retinal fate within the PE.
We next considered a genetic-interaction paradigm with Cka to gain further evidence of Mitf’s role in PE fate. We previously established that KD of Cka at 22°C using Actey-FLP>GAL4 generates a sensitized genetic background in which PE-to-retina fate change is not observed but can be readily induced by partially compromising the activity of other fate-associated factors, such as the PP2A core subunits Mts and Pp2A-29B (Neal et al., 2020). We therefore reasoned that if Mitf truly contributed to PE fate and the suppression of retina fate, a partial loss of Mitf should result in PE-to-Retina fate change in CkaRNAi discs, under these assay conditions. As expected, CkaRNAi-only control-discs from larvae reared at 22°C did not show fate transformation (Fig. 3F–F”). This was in stark contrast to when we introduced one copy of the null Mitf129 allele into this genetic background (Mitf129/+), where we observed Elav-positive cells anterior to Elav+24B10 positive cells in both layers of the eye disc (Fig. 3G–G”; magenta arrowheads). Dac-positive retinal progenitor cells were only observed in the retinal layer of CkaRNAi discs (Fig. 3H–H’; green arrowhead), but were observed in both cell layers in discs from CkaRNAi Mitf129/+ larvae (Fig. 3I–I’; green arrowheads). The penetrance of RDis versus fate phenotypes in larvae of these genotypes was quantified (Fig. 3J). In CkaRNAi-only controls, 28% of discs (19/67) exhibited sRDis, 42% (28/67) mRDis and remaining discs were wt, whereas in CkaRNAi Mitf129/+ larvae, 28% of the eye discs (17/61) showed PE-to-retinal fate change and the remainder exhibited RDis (Fig. 3J; 3/61 mRDis, 41/61 sRDis). Control discs from wt and Mitf129/+ larvae were phenotypically normal (Fig. 3J; n=17 and n=16, respectively). Hence, depleting Mitf within an eye disc with reduced Cka compromised PE-fate specification and resulted in retinogenesis in the presumptive PE, thus providing additional evidence for the contribution of Mitf to the establishment of PE fate.
As a final inquiry into the role of Mitf in PE fate, we implemented a gain-of-function paradigm. Selector genes couple their promotion of a specific fate with their ability to suppress an alternate fate. For instance, yki is required not only to promote PE fate and suppress retina fate within the presumptive PE, but can also suppress retina identity when overexpressed throughout the eye disc (Zhang et al., 2011). To investigate whether Mitf shared the ability to suppress eye identity, we induced MARCM clones overexpressing wt Mitf (UAS-MitfCDS3; see Methods) within the eye disc. Compared with control clones expressing GFP only (Fig. S3E–F’), Mitf-OE clones in the developing retinal epithelium showed loss of neurons, as detected by loss of Elav immunoreactivity (Fig. 3K–K’). Importantly, Mitf-OE clones also failed to express the eye specification proteins eyeless (Ey; vertebrate PAX6) and Eya (Fig. 3L–L”), two factors of the retinal determination cascade that begin expressing early and persist in eye progenitors (Ey) or in progenitors and developing retinal cells (Eya) throughout the larval stages of eye development. Restoring Ey or Eya expression in such Mitf-OE clones, through exogenous transgenes, did not result in the restoration of retinal development (Fig. S3G–H”). These results show that Mitf can antagonize eye identity in the retinal epithelium and that it most likely does so at multiple levels of the retinal determination cascade.
In summary, four lines of evidence, including analysis of homozygous mutant Mitf129 discs, dominant-negative MitfEA expression, Mitf129-CkaRNAi genetic interactions, and Mitf OE, support the conclusion that Mitf is a determinant of PE fate. That more severe LOF conditions were required to uncover Mitf’s role in fate, relative to RDis, suggests that there may be different thresholds of Mitf activity required in these different developmental contexts.
Retinal Suppression by Mitf requires STRIPAK, but not Yki.
Having established that Yki is a mediator of Mitf’s role in RDis, we were curious as to whether it also mediated Mitf’s role in PE fate. We were further motivated by the fact that both factors individually promote PE fate and can suppress retina identity. Therefore, we performed additional experiments to investigate this potential mechanism. To facilitate these studies, we adopted a “global” overexpression approach, whereby a wt Mitf transgene (UAS-Mitf96E or UAS-MitfCDS3) was constitutively overexpressed throughout the eye disc using Actey-FLP>GAL4. This resulted in a robust but variable suppression of retina phenotype that we scored as Category 1, 2, or 3, with Category 3 being the most severe (Fig. 4A–A”; see caption for descriptions). Data are reported as % Category 1; % Category 2; % Category 3. For discs expressing Mitf alone, we observed 21%; 19%; 60% penetrance with UAS-Mitf96E (n=47; Fig. 4B) and 59%; 33%; 8% penetrance with UAS-MitfCSD3 (n=39; Fig. 4B’, Fig. S4A–A”); no wt discs were observed in either experiment. Using these two different transgenic insertions presented us with more severe (UAS-Mitf96E) and less severe (UAS-MitfCDS3) genetic backgrounds in which to assay genetic interactions.
Figure 4. Retinal suppression by Mitf requires STRIPAK-PP2A, but not Yki.
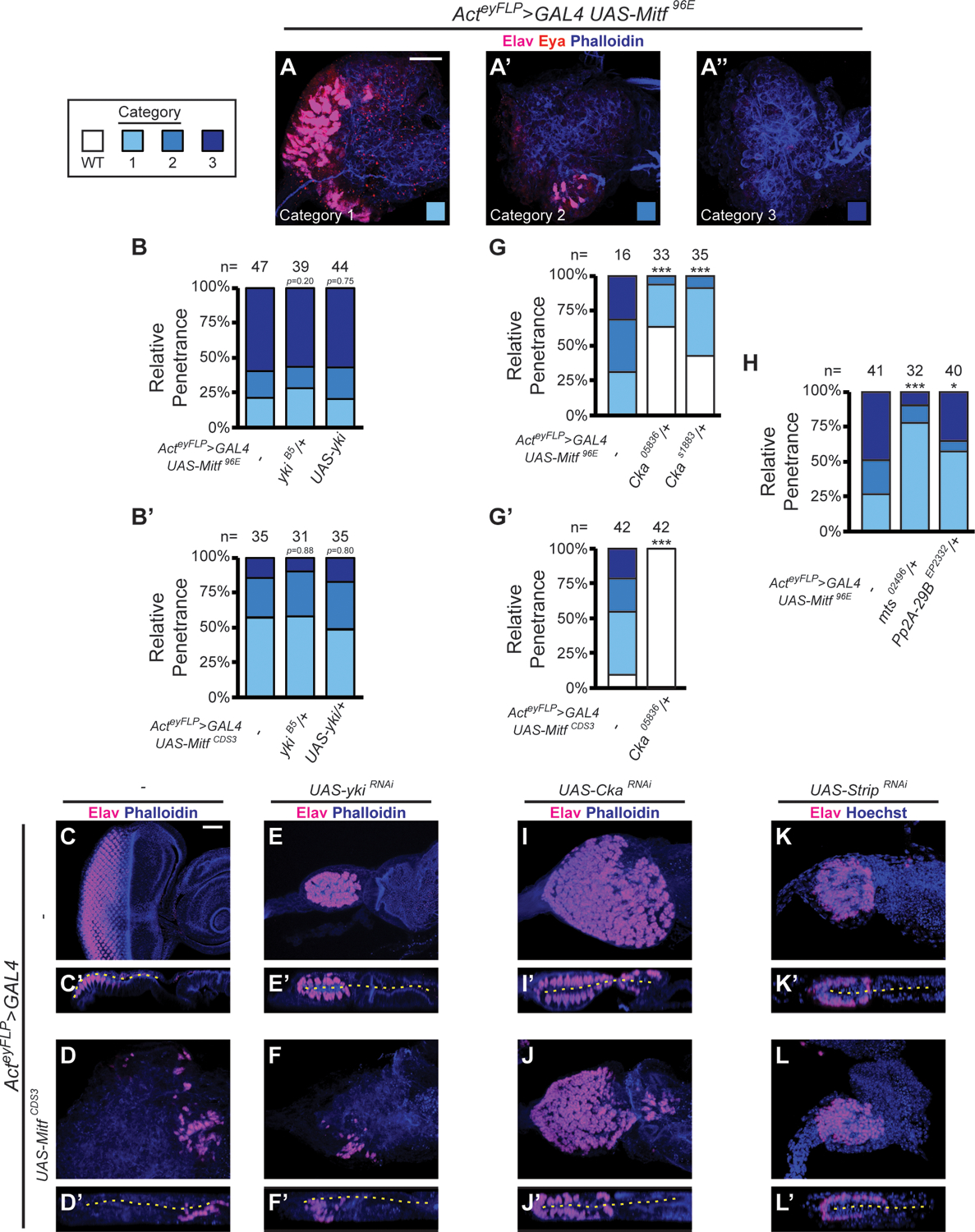
(A-A”) Example images of categorization scheme for retinal suppression phenotype, upon disc-wide expression of wt Mitf (UAS-Mitf96E); discs stained for Elav (magenta), Eya (red), and F-actin (Phalloidin, blue); Scale Bar in A = 25 μm. Category 1 – many neurons in disorganized pattern (A), Category 2 – few neurons (A’) and Category 3 – no neurons (A”). (B-B’) Quantification of retinal suppression phenotype in discs expressing wt Mitf96E (B: ykiB5/+, p=0.20; UAS-yki; p=0.75) or MitfCDS3 (B’: ykiB5/+, p=0.88; UAS-yki; p=0.80). (C-F) Genetic epistasis experiments between Mitf and yki. Discs stained for Elav (magenta) and F-actin (Phalloidin, blue); disc lumen (dashed yellow line); Scale Bar in C = 25 μm. (G-H) Quantification of retinal suppression phenotype in discs expressing wt Mitf96E (G: Cka05836/+, p=2.95 × 10−7; CkaS1883/+, p=7.36 × 10−6; H: mts02496/+: p=3.18 × 10−5; Pp2A-29BEP2332/+: p=0.0111), or MitfCDS3 (G’: Cka05836/+, p=1.94 × 10−19). (I-L’) Genetic epistasis experiments among Mitf and Cka, or Strip. Discs stained for Elav (magenta) and F-actin (Phalloidin, blue), or Hoechst 33342 (DNA, blue), as indicated; disc lumen (dashed yellow line); scaled as in C-F’. * p< 0.05, *** p<0.001
We used this paradigm to investigate the effects of directly altering Yki levels on Mitf OE-induced retinal suppression. For these experiments, we used the null ykiB5 allele and the same UAS-yki transgene that rescues MitfRNAi-induced RDis but does not lead to dominant retinal suppression on its own. We predicted that, if Yki plays a role in Mitf-induced retinal suppression, we would observe a decrease in retinal suppression in the presence of ykiB5/+ and an increase with UAS-yki. To our surprise, in the more severe UAS-Mitf96E context, neither heterozygosity for yki (ykiB5/+; 28%; 15%; 56%; n=39), nor OE of wt Yki (UAS-yki; 20%; 23%; 57%; n=44), significantly altered phenotypic penetrance (Fig. 4B); ykiB5/+ larvae are phenotypically normal (not shown). We thus repeated the experiment in the less severe UAS-MitfCDS3 background, one perhaps more sensitive to phenotypic modification. Yet again, neither heterozygosity for the null allele ykiB5 (ykiB5/+; 49%; 34%; 17%; n=35), nor OE of wt Yki (UAS-yki; 58%; 32%; 10%; n=31), significantly altered phenotypic penetrance (Fig. 4B’). Thus, Yki does not appear to be required for Mitf to suppress retinal fate.
To conclusively establish the independence of Mitf-induced retinal suppression from Yki, we considered an alternate paradigm (Fig. 4C–F’). For this line of inquiry, we used a strong Yki KD reagent, UAS-ykiRNAi, under conditions that result in fully penetrant PE-to-retina fate change (compare ykiRNAi disc in Fig. 4E–E’ with wt control in Fig. 4C–C’). Importantly, this is an opposing phenotype to that of MitfCDS3-induced retinal suppression (Fig. 4D–D’). Hence, we reasoned that, if retina suppression by Mitf is independent of Yki (Fig. 4B–B’), we would not see retinal duplications in MitfCDS3 OE + ykiRNAi discs. The experimental results emerged as predicted (Fig. 4F–F’). In contrast to the double-retina phenotype of discs expressing ykiRNAi alone (Fig. 4E–E’), discs co-expressing MitfCDS3 + ykiRNAi showed retinal suppression similar to that of MitfCDS3-only discs (compare Fig. 4F–F’ with Fig. 4D–D’). Neurons, when present, were restricted to the retinal plane of these discs (Figs. 4D’, 4F’). Hence, even a drastic reduction in Yki in the weaker of the two Mitf OE models, via ykiRNAi expression, was unable to prevent retinal suppression by Mitf. Altogether, these data support our assessment that Mitf suppresses retina independently of Yki.
Beyond Yki, PP2A complexes are also prominent actors in the PE fate and RDis paradigms, with STRIPAK-PP2A (containing the fly PP2A B”’ subunit Cka) playing essential roles in both processes (Neal et al., 2020, 2022). Although Mitf’s role in PE fate appeared to be independent of Yki, we hypothesized that it may yet depend on STRIPAK. In particular, the strong genetic interaction between Mitf129 and CkaRNAi described above – Mitf129/+ induced PE-to-retinal fate transformation in CkaRNAi larvae reared at 22°C (Fig. 3F–J) – hinted at an interdependence of Mitf and STRIPAK-PP2A activities in suppressing retinal identity.
Once again, we reasoned that if Mitf works through/with STRIPAK-PP2A in PE fate, then partial loss of Cka should alleviate Mitf-induced retina suppression. Indeed, the severe Mitf96E OE phenotype was significantly suppressed by either of two hypomorphic alleles of Cka (Fig. 4G). As compared to the UAS-Mitf96E control with 31%; 38%; 31% penetrance (n=16), the two LOF alleles of Cka showed a strong reduction of the more severe phenotypic categories. For Cka05836/+, penetrance decreased to 30%; 6%; 0% (n=33), and for CkaS1883/+, to 49%; 9%; 0% (n=35) (Fig. 4G). Furthermore, heterozygosity for Cka05836 completely suppressed the phenotype in the less severe UAS-MitfCDS3 background, such that all discs were wt (N=42; Fig. 4G’). These data are consistent with the observed PE-to-retina fate change in CkaRNAi Mitf129/+ discs, and argue that Mitf requires Cka activity to suppress retina. Furthermore, it suggests that this might reflect an involvement of the STRIPAK-PP2A complex.
To substantiate the connection to PP2A, we also tested for genetic interactions with the PP2A catalytic component gene mts and the core structural component gene Pp2A-29B, again looking for suppression of the severe UAS-Mitf96E OE phenotype (27%; 24%; 49%; n=41) (Fig. 4H). Significant suppression of the phenotype was observed with a reduction in either factor, mts (mts02496/+: 78%; 13%; 9%; n=32) or Pp2A-29B (Pp2A-29BEP2332/+: 58%; 8%; 35%; n=40) (Fig. 4H; animals hemizygous for either mts or Pp2A-29B are phenotypically normal (not shown). These data confirm that Mitf is dependent on STRIPAK-PP2A activity to antagonize retinal fate.
As a final test of this conclusion, we used the strong KD reagent CkaRNAi under conditions that result in fully penetrant PE-to-retina fate change (Fig. 4I–I’), the opposing phenotype to that of MitfCDS3-induced retinal suppression (Fig. 4D–D’). Based on the evidence that Mitf-induced retina suppression requires Cka (Fig. 4G–G’), we predicted that MitfCDS3 OE + CkaRNAi discs would show the CkaRNAi phenotype. This was indeed what we observed in this experiment. Combining MitfCDS3 OE with CkaRNAi yielded discs with neurons in both layers (Fig. 4J–J’), similar to those expressing CkaRNAi alone (Fig. 4I–I’). To substantiate this finding with respect to STRIPAK, we additionally examined Strip. Strip encodes a core STRIPAK component, a scaffold protein, and StripRNAi also induces retinal duplication (Neal et al., 2020) (Fig. 4K–K’). Combining MitfCDS3 OE with StripRNAi yielded discs with neurons in both layers (Fig. 4L–L’), like those expressing StripRNAi alone (Fig. 4K–K’). Thus, StripRNAi recapitulated what we had observed with CkaRNAi, identifying STRIPAK as a factor indispensable for Mitf function in PE fate.
From this series of experiments, despite the convergent phenotype induced by both factors, we conclude that Mitf antagonizes retinogenesis independently of Yki. Furthermore, our data regarding mts, Pp2A-29B, Cka and Strip provide strong evidence for a context-dependent regulation of Mitf, in PE-versus-Retinal fate specification, by STRIPAK-PP2A. It remains unresolved how Yki and Mitf might independently regulate the PE-Retina fate choice.
Regulation of Mitf protein activity by STRIPAK-PP2A.
To further explore the dependence of Mitf function on STRIPAK-PP2A, we leveraged the Drosophila wing disc, a tissue that, unlike the head, is dispensable not only for larval development but also for fly eclosion from the pupal case. For instance, using dpp-GAL4 to overexpress wt Mitf (UAS-MitfCDS6) resulted in flies with a very robust dominant phenotype, severe malformation of the wings (Fig. 5A). This provided us with an alternate paradigm in which to test for genetic interactions.
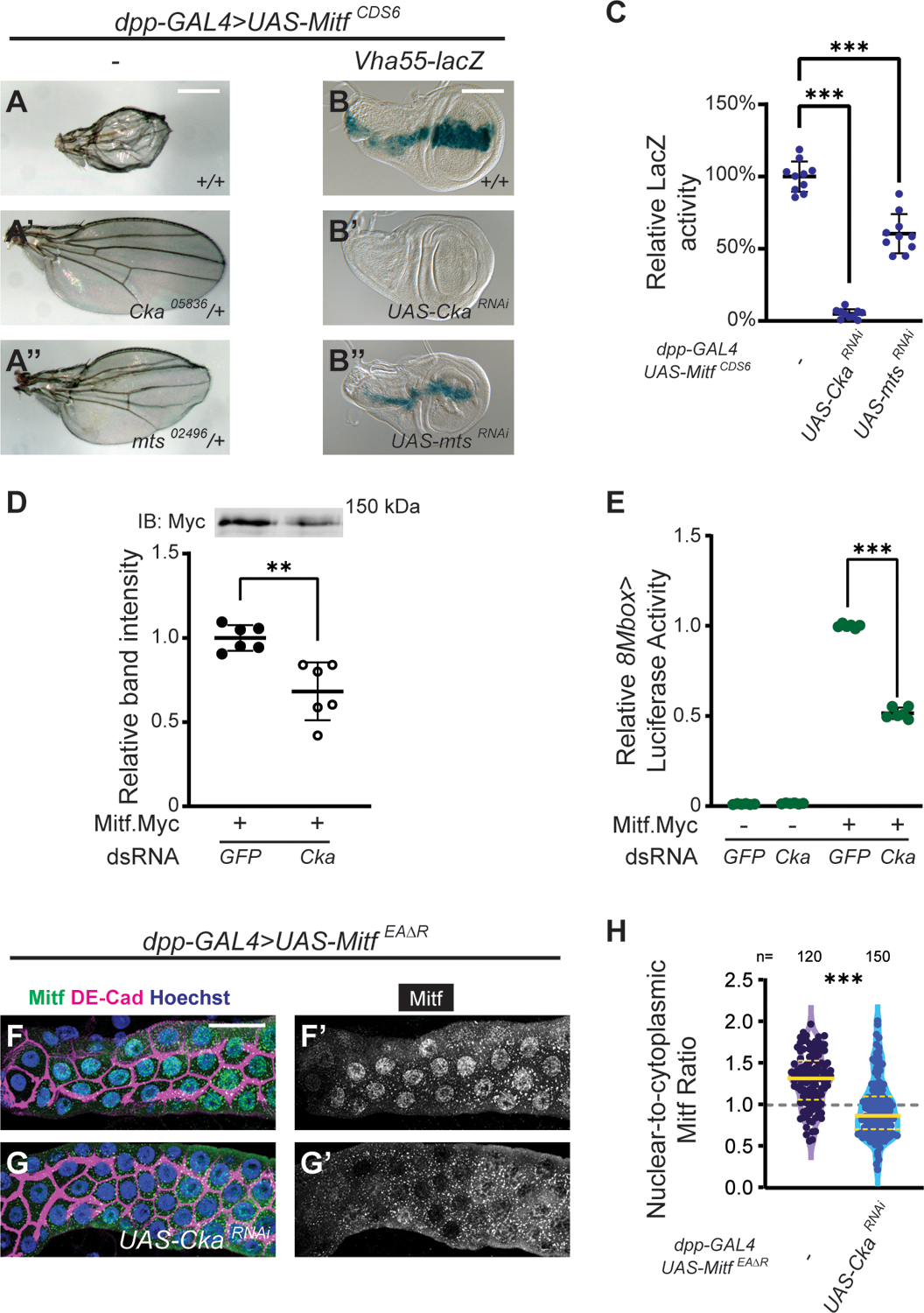
Figure 5. Loss of Cka attenuates Mitf transcriptional activity and suppresses Mitf nuclear accumulation.
(A-A”) Adult wing phenotypes; genotypes as indicated; Scale Bar in A = 250 μm. (B-B”) Vha55-LacZ expression, visualized by β-galactosidase activity (blue); Scale Bar in B = 100 μm. (C) Quantification of β-galactosidase activity across 10 discs per genotype (CkaRNAi, p<0.0001; mtsRNAi, p<0.0001). (D) Representative western blot and quantification of the relative abundance of Mitf.Myc in S2 cell luciferase lysates (3 experiments, each with 2 technical replicates). Mitf.Myc abundance was reduced by treatment with Cka dsRNA (p=0.002). (E) 8Mbox>Luciferase activity in Drosophila S2 cells was strongly induced in the presence of Mitf (approximately 100-fold baseline activity, normalized to 1) and was attenuated by Cka dsRNA (p<0.0001); control GFP dsRNA data are repeated from Fig. 2A. (F-G’) Expression of DNA-binding-defective MitfEAΔR in salivary glands (dpp>UAS-MitfEAΔR, green); glands were co-stained for DE-Cad (magenta) and Hoechst 33342 (DNA, blue); Scale Bar in F = 50 μm. (H) Quantification of nuclear-to-cytoplasmic Mitf signal ratio (see Methods; control – 4 glands, 120 cells; CkaRNAi – 5 glands, 150 cells; p<0.0001). ** p<0.01, *** p<0.001
Based on our results in the eye, we hypothesized that reduction of STRIPAK-PP2A activity would reduce or abrogate the ability of Mitf to compromise wing development. We therefore introduced mutant alleles of STRIPAK-associated genes, Cka and mts, into this paradigm to assess their effects. Similar to what we observed with retinal suppression, heterozygosity for Cka (CkaS1883/+) completely suppressed the Mitf OE wing phenotype (Fig. 5A’). Moreover, we also observed partial restoration of wing morphology when Mitf OE was combined with heterozygosity for the PP2A catalytic subunit gene mts (mts02496/+) (Fig. 5A”). These results identify STRIPAK-PP2A as a regulator of Mitf activity in a second developmental paradigm.
To gain mechanistic insight into this genetic interaction, we assessed the transcriptional activity of Mitf in the wing via an enhancer trap reporter of the Mitf target gene vha55, vha55-lacZ (Zhang et al., 2015). In the larval wing disc, vha55-LacZ activity is strongly induced by ectopic expression of Mitf (dpp>MitfCDS6) (expression normalized to 100%; Fig. 5B–C). Given our above results (Fig. 5A’–A”), we hypothesized that reduced STRIPAK-PP2A function would decrease or abrogate Mitf target gene expression. Indeed, CkaRNAi essentially eliminated, and mtsRNAi decreased, Mitf-induced vha55-LacZ expression (Fig. 5B’–C; 4.9% and 60% of control, respectively). This effect was also confirmed using alleles of Cka (CkaS1883/+) and mts (mts02496/+), in place of RNAi KD (Fig. S5A–B; 51% and 58% of control, respectively). These results mirrored, at the transcriptional level, the genetic interactions observed in the adult wing assay.
Next, we investigated two potential mechanisms by which STRIPAK-PP2A might attenuate Mitf transcriptional activity: a) decrease in Mitf protein or b) cytoplasmic sequestration of Mitf. Both are well-known mechanisms for the regulation of MITF transcriptional activity (Goding and Arnheiter, 2019). To investigate the first mechanism (protein stability), we tested the effect of Cka KD in a Mitf.Myc OE paradigm in S2 cells, thus permitting us to assess protein levels via the Myc tag (Fig. 5D). Relative to cells treated with GFP dsRNA, the abundance of Mitf.Myc was reduced to 68% of control in cells treated with Cka dsRNA (Fig. 5D and Fig. S5C; Cka KD to 14.5% of control). To explore the functional consequence of this reduction in Mitf, we used luciferase assays. Whereas treating S2 cells with Cka dsRNA in the absence of Mitf.Myc had a negligible effect on basal 8Mbox-Luc activity, it reduced Mitf.Myc-induced 8Mbox-Luc activity from 100% to 52% (Fig. 5E). These in vitro data support the hypothesis that a STRIPAK-PP2A-mediated decrease in Mitf abundance contributes to the decrease in Mitf transcriptional activity that we observed both in vivo (Fig. 5B–B”) and here, in vitro.
Notwithstanding the above results, we continued to investigate the second mechanism (nuclear/cytoplasmic localization), by adapting an in vivo paradigm previously used to study the subcellular localization of Mitf (Zhang et al., 2015). This assay takes advantage of the giant cells of the Drosophila salivary glands, and makes use of a non-functional Mitf protein in order to bypass Mitf’s promotion of its own sequestration, through a negative feedback loop (Zhang et al., 2015). Thus, we examined the subcellular distribution of overexpressed MitfEAΔR, a variant that is unable to bind DNA, with and without Cka KD. In control glands, MitfEAΔR is both nuclear and cytoplasmic (Fig. 5F–F’; 1.31 mean nuclear:cytoplasmic ratio), yet upon KD of Cka (CkaRNAi) the nuclear enrichment of Mitf is strongly reduced (Fig. 5G–H; 0.94 mean nuclear:cytoplasmic ratio). This result offers an alternate explanation for the reduced transcriptional activity of Mitf observed upon depletion of Cka. Specifically, this finding points to the cytoplasmic sequestration of Mitf, resulting in its physical segregation from its transcriptional targets.
The above data demonstrate that reducing the function of STRIPAK-PP2A negatively affects Mitf target gene expression. Since vha55-lacZ and 8Mbox-luc are both direct transcriptional targets of Mitf, these results imply that STRIPAK-PP2A is directly or indirectly regulating the activity of Mitf itself. Furthermore, our quantification of Mitf abundance in vitro and of its localization in vivo suggest two possible and non-mutually-exclusive mechanisms for the effect of STRIPAK-PP2A on Mitf – changes in stability and subcellular distribution.
Discussion
Eye disc morphology is regulated by the concerted actions of Mitf and Yki.
We recently described the RDis phenotype, in which the interface between the retinal epithelium and PE becomes displaced from the posterior margin of the L3 eye disc into the PE plane, resulting in dysmorphism of the compound eye (DeSantis et al., 2023; Neal et al., 2022). In these prior studies, we identified Yki as the key regulator of this aspect of disc morphology, and demonstrated its dual regulation via a PP2A(B’)/PP2A(B”’)-Hpo-Wts signaling axis and an AJ-Wts signaling axis (DeSantis et al., 2023; Neal et al., 2022). Herein, we have shown that Mitf also functions to suppress RDis (Fig. 1F–K), and Mitf similarly exerts this function from within the PE (Fig. 1L). While we found that Mitf’s suppression of RDis appears to be mediated by Yki (Fig. 1M), we did not observe robust genetic interactions between Mitf and either the PP2A-Hpo or the AJ-Wts regulatory axes (Fig. 1N; Fig. S1F–I’). The latter observation suggested that a closer, possibly direct, interaction occurred between Mitf and Yki. Indeed, we found that Yki can modify transcriptional output from an Mitf reporter in vitro (Fig. 2A”), and that Yki and Mitf can physically interact (Fig. 2B). This interaction appears to be evolutionarily conserved, as it also occurs between the human proteins YAP1 and MITF (Fig. 2C). Thus, Yki and Mitf may function jointly in the preservation of disc morphology.
These results are particularly intriguing because the Mitf and Yki pathways have been associated with alternative biological contexts – MITF and TFEB promote autophagy and cell survival under nutrient-restricted growth conditions (Di Malta et al., 2019; Goding and Arnheiter, 2019; Theodosakis et al., 2021), while Yki/YAP1 signaling is typically associated with proliferation under nutrient replete conditions (Misra and Irvine, 2018). The mechanisms underlying these alternate roles remains to be characterized, but it likely involves recently discovered crosstalk between these conserved pathways (Honda et al., 2023). YAP1 is known to act upstream of mTORC1 to promote its activity (Liu et al., 2019; Tumaneng et al., 2012), which consequently negatively regulates MITF/TFE factors. Conversely, YAP1 is itself regulated by autophagy, and is stabilized when MITF/TFE factors are inhibited by active mTORC1 (Liang et al., 2014). Thus, the emerging picture is that of two largely antagonistic pathways.
In contrast, our data point to a collaborative function of Yki and Mitf such that their combined action may lead to an “anti-RDis” transcriptional output that is distinct from that driven separately by either factor. Thus far, the only other instance of cooperative action between MITF/TFE and YAP1/TAZ factors has been reported in a RagA/B conditional KO mouse model of cardiac hypertrophy. In this model, a critical accretion of YAP1 and an activation of TFEB simultaneously occur such that cardiomyopathy results from an excessive accumulation of autophagosomes (Ikeda et al., 2021). This study provides compelling evidence that this phenotype comes about through a closer interaction of the nuclear co-factor YAP1 with the transcription factor TFEB, resulting in altered transcriptional activity as compared to TFEB alone (Ikeda et al., 2021). Notably, while this example is associated with a pathological condition, our RDis paradigm suggests that joint action of Mitf and Yki can also occur under normal conditions.
Additional studies will undoubtedly reveal other contexts in which MITF/TFE factors and YAP1/WWTR1 factors act in concert, rather than antagonistically, and further research is required to understand what underlies this mechanism and the nature of the modified transcriptomes that drive each biological process.
Multiple lines of evidence support the role of Mitf in suppressing retinal fate.
In the early stages of eye disc development, a critical binary fate decision determines the neural retina and the non-neural PE domains within the eye disc epithelium. We have previously shown that the transcription cofactor Yki is a key player in this process (Neal et al., 2020). Loss of Yki results in transformation of the PE into a mirror image retina and OE of Yki in the retinal epithelium leads to loss of retinal identity factors and thus failure of eye development (Zhang et al., 2011). Using four independent lines of evidence, we show here that Mitf activity also promotes PE fate and suppresses retina fate in the PE of the wt eye disc (Fig. 3A–J). Moreover, Mitf can also suppress retina identity when over-expressed within the retinal epithelium (Fig. 3K–L”).
In these roles, Mitf functions in a strikingly similar manner to Yki. Hence, we were very surprised to find that the retina suppression activity of Mitf OE is independent of Yki, as neither partial/severe loss of Yki nor Yki OE affected the penetrance or expressivity of this activity (Fig. 4B–B’). Yet, we observed significant genetic interactions between Mitf and the STRIPAK-PP2A components Cka, mts, and Pp2A-29B (Fig. 4G–H). Furthermore, strong KD of the STRIPAK components Cka and Strip, but not Yki, was epistatic to retinal suppression by Mitf and resulted in retinal duplication (Fig. 4C–F’, Fig. 4I–L’). Thus, our data are consistent with a model in which STRIPAK-PP2A is necessary to potentiate Mitf activity. In agreement with this model, in wing discs and in S2 cells, we found evidence that the transcriptional activity of Mitf was reduced when Cka was depleted (Fig. 5B–C; Fig. 5E). Further analyses identified two potential mechanisms for this regulation. In cell culture, we observed a decrease in the relative abundance of Mitf protein in Cka-depleted lysates (Fig. 5D), and in salivary glands, we found that nuclear enrichment of a non-functional form of Mitf was dependent on Cka (Fig. 5F–H). Hence, STRIPAK-PP2A might exert its effects on Mitf by promoting its nuclear entry and/or its protein stability. Indeed, these potential mechanisms are not mutually exclusive and are deserving of further analysis.
Phosphorylation of MITF/TFE factors has long been implicated in the control of both their stability and nuclear localization (Goding and Arnheiter, 2019; Nardone et al., 2023; Puertollano et al., 2018). As a phosphatase, it is plausible that STRIPAK-PP2A might regulate MITF/TFE factors. In fact, STRIPAK has been shown to regulate the activity of other transcription factors, such as CLOCK/CYCLE (circadian rhythms), NF-κB (immune response, cell growth and survival), and the glucocorticoid receptor (development, metabolism, and immune response) (Andreazza et al., 2015; Petta et al., 2017; Tsuchiya et al., 2017). However, Calcineurin, rather than STRIPAK or other PP2A, is thought to be the principal phosphatase that regulates MITF/TFE nuclear entry (Fock et al., 2019; Medina et al., 2015). Only in two specific contexts has PP2A-mediated dephosphorylation been shown to promote nuclear translocation of TFEB or TFE3 – specifically upon induction of acute oxidative stress by sodium arsenite (Martina and Puertollano, 2018), or upon inhibition of the phosphoinositide kinase PIKfyve (Hasegawa et al., 2022), with PP2A(B) identified as the holoenzyme active in the former (Martina and Puertollano, 2018).
To our knowledge, this work is the first to report a potential role for STRIPAK-PP2A in the post-translational regulation of an MITF/TFE factor. Though our data does not identify the specific molecular mechanism of this functional interaction, the retinal suppression paradigm and biochemical approaches could be used to do so in future studies.
Mitf and fate in visual system development.
The compound eye of Drosophila and the mammalian camera eye did not evolve from a common ancestral visual organ, yet the study of eye development has uncovered considerable conservation in the genetic networks that drive the development and function of the neural retina of distant species. This evidence led to much debate and ultimately the proposal of a shared evolutionary origin of the fly and mammalian eyes from simple photosensitive organs of a common bilaterian ancestor (Arendt, 2003; Fernald, 2004; Gehring, 2004). Since then, evidence has also slowly emerged about a striking conservation of function, and underlying molecular players, in the accessory cells/tissues of diverse fully formed eyes – such as the Semper cells and the pigment cells of the Drosophila compound eye, and the Müller glia and RPE of vertebrate eyes, respectively (Charlton-Perkins et al., 2021; Rathore et al., 2023). Here, we propose for this parallel to be extended to the PE and RPE as accessory tissues that support retinal development in both fly and vertebrates.
During vertebrate eye development, the optic vesicle invaginates to form an optic cup with juxtaposed cell layers that develop one into retina and the other into RPE. This topology is similar to that of the eye imaginal disc, where the flattened disc vesicle develops into retina on one side and PE on the other. Strikingly, in Drosophila and vertebrates (zebrafish, mouse), Yki/YAP1/TAZ have been shown to play a role in the fate decision that drives this partitioning into retina versus PE/RPE. Moreover, these factors promote the establishment and maintenance of the PE/RPE, at least in part, by suppressing an underlying program for retinogenesis (Kim et al., 2016; Miesfeld et al., 2015; Neal et al., 2020; Zhang et al., 2011). MITF has been implicated in this same process in the mouse eye (Bharti et al., 2012; Nguyen and Arnheiter, 2000), and with this paper, we offer evidence that Drosophila Mitf is critical for PE formation in the eye imaginal disc.
Thus, Mitf/MITF and Yki/YAP1 appear to play analogous roles in the establishment of the PE/RPE during development of fly and mouse eyes (Neal et al., 2020; Zhang et al., 2011); this paper). Whether this conservation extends to STRIPAK-PP2A remains to be seen, as the function(s) of STRIPAK in vertebrate eye development remains largely unexplored.
Supplementary Material
Figure S1. Data pertaining to the role of Mitf in RDis. (A) The presence of the empty KK-TK transgene in the tio locus did not increase the penetrance of RDis in 2XMitfRNAi discs (p=0.15). (B) Schematic summarizing the genetic pathways known to affect Yki function with respect to RDis (DeSantis et al., 2023; Neal et al., 2022). (C-C’) Differential interference contrast micrographs of colorimetric in situ hybridization showing Mitf expression in the L2 eye disc. Mitf is detected in the PE plane (en face in A and in cross-section in A’). (D-E’) Mitf2.2p::GFP expression (green) in L2 (D-D’) and L3 (E-E’) eye discs; PE (D, E) and retinal (D’, E’) planes are shown. (F-I’) Detection of adherens junction (AJ) components (Arm, F-G’; DE-Cad, H-I’) in control (F-F’ and H-H’) and 4XMitfRNAi-expressing (G-G’ and I-I’) discs; Scale Bar in F = 10 μm. In control discs, AJ markers were detected at cell-cell boundaries in the PE (F, H) and at the vertices of developing photoreceptor clusters in the retina (F’, H’; yellow arrows). Despite disc-wide expression of UAS-MitfRNAi and a highly penetrant sRDis phenotype, neither pattern of AJ marker expression was perturbed in these eye discs (G-G’, I-I’).
Figure S2. Validation of dsRNA KD and representative full western blots related to Figure 2. (A-A”’) Expression level of indicated transcripts, relative to Act5C in S2 cells treated with dsRNA, as indicated. *** p<0.001 (B) Full images of western blot shown in Fig. 2B. (C) Full images of western blot shown in Fig. 2C. *NS* - non-specific band detected using 12C6c antibody in input but not immunoprecipitated samples. *HC* Residual mouse IgG heavy chain detected in 12C6c immunoprecipitates.
Figure S3. Supporting data related to the Mitf129 loss-of-function allele. (A) The Mitf129 allele was generated using CRISPR technology (see Methods); sequencing of a genomic DNA amplicon of the Mitf locus revealed a large deletion, encompassing most of the coding exons (indicated by red box on a schematic exported from UCSC Genome Browser). Predicted transcripts (RA-RD) are labeled. (B) Primary amino acid sequence translations (PA-PD) from predicted transcripts affected by the Mitf129 deletion. While the N-terminal 53 amino acids are preserved in all cases, transcripts RA, RB, and RD are expected to encode a frameshift resulting in 6 novel amino acids (red) before the termination codon in the respective translations (PA, PB and PD). The reading frame of the RC transcript is preserved in Mitf129 but has a predicted deletion of 781 amino acids (indicated in blue) before the 4 native residues preceding the termination codon in PC. (C) Micrograph of larvae of the indicated genotypes, including rare homozygous Mitf129/ Mitf129 larvae that develop to at most early L3 size, while retaining L2-like mouth hooks. (D) Quantification of Eya-expressing MARCM clones in the PE, relative to the posterior disc margin. (E-F’) Eye discs with MARCM clones overexpressing GFP (UAS-GFP, green); discs stained for Elav (magenta, A-A’) or Eya (red, B-B’); Scale Bar in E = 25 μm. (G-H”) Eye discs with MARCM clones overexpressing GFP (UAS-GFP, green), Mitf (UAS-MitfCDS3), and either Ey (UAS-ey, G-G”) or Eya (UAS-eya, H-H”); XY projections of retinal clones are shown. Cells in posterior clones expressing Mitf and Ey show upregulation of Ey (cyan, G’) but not restoration of Eya (red, G”). Cells in posterior clones expressing Mitf and Eya show upregulation of Eya (red, H’) but not restoration of Elav (magenta, H”).
Figure S4. Representative Images of MitfCDS3 retina suppression phenotypes. (A-A”) Example images of categorization scheme for retinal suppression phenotype, upon disc-wide expression of wt Mitf (UAS-MitfCDS3); discs stained for Elav (magenta) and F-actin (Phalloidin, blue); Scale Bar in A = 25 μm. Category 1 – many neurons in disorganized pattern (A), Category 2 – few neurons (A’) and Category 3 – no neurons (A”).
Figure S5. Data supporting the effects of Cka and Mts reduction on Mitf transcriptional activity. (A-B) Mitf expression in the larval wing disc (dpp>UAS-MitfCDS6) resulted in upregulation of a reporter gene (Vha55-lacZ), as visualized by β-galactosidase activity (blue) (A’, compare with negative control in A); Scale Bar in A = 100 μm. Reporter expression was reduced in discs heterozygous for a hypomorphic Cka allele (A”; Ckas1883/+) and in discs heterozygous for a null allele of mts (A”’; mts02496/+). Quantification of β-galactosidase activity across 10 discs per genotype (CkaS1883/+, p=1.85 × 10−9; mts02496/+, p=4.60 × 10−8) (B). *** p<0.001 (C) Full images of western blot shown in Fig. 5D.
Highlights.
Mitf functions to preserve eye disc epithelium morphology during eye morphogenesis
Mitf works with Yki to maintain eye disc morphology
Mitf is a selector factor that promotes peripodial (PE) fate over retinal fate
Mitf works with STRIPAK-PP2A, not Yki, in PE development and retina suppression
Acknowledgements
Stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila Resource Center (VDRC, www.vdrc.at), or kindly provided by K. Irvine (Rutgers U.). We are grateful to many labs, cited in Methods, for their gifts of other reagents. We thank Dana DeSantis and Ryan Palumbo for critical comments on this manuscript.
Funding
This work was supported by a grant from the NIH to FP (NEI 5R01EY028221).
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
All data necessary to support the conclusions of this study are included. The authors will make all unique reagents available upon reasonable requests to the corresponding authors.
References
- Andreazza S, Bouleau S, Martin B, Lamouroux A, Ponien P, Papin C, Chelot E, Jacquet E, Rouyer F, 2015. Daytime CLOCK Dephosphorylation Is Controlled by STRIPAK Complexes in Drosophila. Cell Rep 11, 1266–1279. [DOI] [PubMed] [Google Scholar]
- Arendt D, 2003. Evolution of eyes and photoreceptor cell types. Int J Dev Biol 47, 563–571. [PubMed] [Google Scholar]
- Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M, Okuno Y, Hata S, Nishina H, Hata Y, 2011. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochem 150, 199–208. [DOI] [PubMed] [Google Scholar]
- Bharti K, Gasper M, Ou J, Brucato M, Clore-Gronenborn K, Pickel J, Arnheiter H, 2012. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet 8, e1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche V, Espinosa AP, Leone L, Sardiello M, Ballabio A, Botas J, 2016. Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 12, 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins MA, Friedrich M, Cook TA, 2021. Semper’s cells in the insect compound eye: Insights into ocular form and function. Dev Biol 479, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Halder G, Zhang Z, Mardon G, 1999. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development 126, 935–943. [DOI] [PubMed] [Google Scholar]
- Chen YC, Desplan C, 2020. Gene regulatory networks during the development of the Drosophila visual system. Curr Top Dev Biol 139, 89–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KO, Chern J, Izaddoost S, Choi KW, 2000. Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell 103, 331–342. [DOI] [PubMed] [Google Scholar]
- DeSantis DF, Neal SJ, Zhou Q, Pignoni F, 2023. Peripodial adherens junctions regulate Ajuba-Yorkie signaling to preserve fly eye morphology. Biol Open 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Malta C, Cinque L, Settembre C, 2019. Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front Cell Dev Biol 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ, 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- Fernald RD, 2004. Eyes: variety, development and evolution. Brain Behav Evol 64, 141–147. [DOI] [PubMed] [Google Scholar]
- Fock V, Gudmundsson SR, Gunnlaugsson HO, Stefansson JA, Ionasz V, Schepsky A, Viarigi J, Reynisson IE, Pogenberg V, Wilmanns M, Ogmundsdottir MH, Steingrimsson E, 2019. Subcellular localization and stability of MITF are modulated by the bHLH-Zip domain. Pigment Cell Melanoma Res 32, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, 2004. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol 48, 707–717. [DOI] [PubMed] [Google Scholar]
- George A, Zand DJ, Hufnagel RB, Sharma R, Sergeev YV, Legare JM, Rice GM, Scott Schwoerer JA, Rius M, Tetri L, Gamm DM, Bharti K, Brooks BP, 2016. Biallelic Mutations in MITF Cause Coloboma, Osteopetrosis, Microphthalmia, Macrocephaly, Albinism, and Deafness. Am J Hum Genet 99, 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G, 2000. Peripodial cells regulate proliferation and patterning of Drosophila imaginal discs. Cell 103, 343–350. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Schubiger G, 2001. Drosophila peripodial cells, more than meets the eye? Bioessays 23, 691–697. [DOI] [PubMed] [Google Scholar]
- Goding CR, Arnheiter H, 2019. MITF-the first 25 years. Genes Dev 33, 983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith SL, Shimell M, Tauscher P, Daly SM, Shimmi O, O’Connor MB, Newfeld SJ, 2022. New resources for the Drosophila 4th chromosome: FRT101F enabled mitotic clones and Bloom syndrome helicase enabled meiotic recombination. G3 (Bethesda) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP, 2014. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods 11, 222–223. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ, 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125, 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Schepsky A, Arnheiter H, Steingrimsson E, 2007. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res 20, 185–200. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrimsson E, 2004. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA, Sekelsky J, 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: Imperfection or interesting exception? Fly (Austin) 11, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Tokuda E, Yao Y, Sasaki T, Inoki K, Weisman LS, 2022. PP2A-dependent TFEB activation is blocked by PIKfyve-induced mTORC1 activity. Mol Biol Cell 33, ar26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie JL, Bryant PJ, 1986. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J Exp Zool 237, 293–308. [DOI] [PubMed] [Google Scholar]
- Honda D, Okumura M, Chihara T, 2023. Crosstalk between the mTOR and Hippo pathways. Dev Growth Differ 65, 337–347. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Nah J, Shirakabe A, Zhai P, Oka SI, Sciarretta S, Guan KL, Shimokawa H, Sadoshima J, 2021. YAP plays a crucial role in the development of cardiomyopathy in lysosomal storage diseases. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D, 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F, 2003. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell 5, 403–414. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL, 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Park R, Lee JH, Shin J, Nickas J, Kim S, Cho SH, 2016. Yap is essential for retinal progenitor cell cycle progression and RPE cell fate acquisition in the developing mouse eye. Dev Biol 419, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spina M, Contreras PS, Rissone A, Meena NK, Jeong E, Martina JA, 2020. MiT/TFE Family of Transcription Factors: An Evolutionary Perspective. Front Cell Dev Biol 8, 609683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- Liang N, Zhang C, Dill P, Panasyuk G, Pion D, Koka V, Gallazzini M, Olson EN, Lam H, Henske EP, Dong Z, Apte U, Pallet N, Johnson RL, Terzi F, Kwiatkowski DJ, Scoazec JY, Martignoni G, Pende M, 2014. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J Exp Med 211, 2249–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ren H, Zhou Y, Shang L, Zhang Y, Yang F, Shi X, 2019. The hypoxia conditioned mesenchymal stem cells promote hepatocellular carcinoma progression through YAP mediated lipogenesis reprogramming. J Exp Clin Cancer Res 38, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R, 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Puertollano R, 2018. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J Biol Chem 293, 12525–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A, 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld JB, Gestri G, Clark BS, Flinn MA, Poole RJ, Bader JR, Besharse JC, Wilson SW, Link BA, 2015. Yap and Taz regulate retinal pigment epithelial cell fate. Development 142, 3021–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, Irvine KD, 2018. The Hippo Signaling Network and Its Biological Functions. Annu Rev Genet 52, 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardone C, Palanski BA, Scott DC, Timms RT, Barber KW, Gu X, Mao A, Leng Y, Watson EV, Schulman BA, Cole PA, Elledge SJ, 2023. A central role for regulated protein stability in the control of TFE3 and MITF by nutrients. Mol Cell 83, 57–73 e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal SJ, Dolezal D, Jusic N, Pignoni F, 2019. Drosophila ML-DmD17-c3 cells respond robustly to Dpp and exhibit complex transcriptional feedback on BMP signaling components. BMC Dev Biol 19, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal SJ, Zhou Q, Pignoni F, 2020. STRIPAK-PP2A regulates Hippo-Yorkie signaling to suppress retinal fate in the Drosophila eye disc peripodial epithelium. J Cell Sci 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal SJ, Zhou Q, Pignoni F, 2022. Protein Phosphatase 2A with B’ specificity subunits regulates the Hippo-Yorkie signaling axis in the Drosophila eye disc. J Cell Sci 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevil M, Bondra ER, Schulz KN, Kaplan T, Harrison MM, 2017. Stable Binding of the Conserved Transcription Factor Grainy Head to its Target Genes Throughout Drosophila melanogaster Development. Genetics 205, 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Arnheiter H, 2000. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development 127, 3581–3591. [DOI] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A, 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- Petta I, Bougarne N, Vandewalle J, Dejager L, Vandevyver S, Ballegeer M, Desmet S, Thommis J, De Cauwer L, Lievens S, Libert C, Tavernier J, De Bosscher K, 2017. Glucocorticoid Receptor-mediated transactivation is hampered by Striatin-3, a novel interaction partner of the receptor. Sci Rep 7, 8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL, 1997. Induction of Drosophila eye development by decapentaplegic. Development 124, 271–278. [DOI] [PubMed] [Google Scholar]
- Pina C, Pignoni F, 2012. Tubby-RFP balancers for developmental analysis: FM7c 2xTb-RFP, CyO 2xTb-RFP, and TM3 2xTb-RFP. Genesis 50, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM, 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A 112, E420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R, Ferguson SM, Brugarolas J, Ballabio A, 2018. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S, Stahl A, Benoit JB, Buschbeck EK, 2023. Exploring the molecular makeup of support cells in insect camera eyes. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD, 2014. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M, 2010. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell 39, 521–534. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM, 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM, 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A, 2009. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Theodosakis N, Pagan AD, Fisher DE, 2021. The role of MiT/TFE family members in autophagy regulation. Curr Top Biochem Res 22, 151–159. [PMC free article] [PubMed] [Google Scholar]
- Tognon E, Kobia F, Busi I, Fumagalli A, De Masi F, Vaccari T, 2016. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy 12, 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Osaki K, Kanamoto M, Nakao Y, Takahashi E, Higuchi T, Kamata H, 2017. Distinct B subunits of PP2A regulate the NF-kappaB signalling pathway through dephosphorylation of IKKbeta, IkappaBalpha and RelA. FEBS Lett 591, 4083–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL, 2012. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 14, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JH, Manning SA, Kulkarni A, Harvey KF, 2016. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat Commun 7, 10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner BP, Weasner BM, Kumar JP, 2020. Ghost in the machine: The peripodial epithelium, Molecular Genetics of Axial Patterning, Growth and Disease in Drosophila Eye. Springer, pp. 121–141. [Google Scholar]
- Zhang T, Zhou Q, Ogmundsdottir MH, Moller K, Siddaway R, Larue L, Hsing M, Kong SW, Goding CR, Palsson A, Steingrimsson E, Pignoni F, 2015. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J Cell Sci 128, 2938–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhou Q, Pignoni F, 2011. Yki/YAP, Sd/TEAD and Hth/MEIS control tissue specification in the Drosophila eye disc epithelium. PLoS One 6, e22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Arcia S, Perez B, Fernandez-Funez P, Rincon-Limas DE, 2013. pΔTubHA4C, a new versatile vector for constitutive expression in Drosophila. Mol Biol Rep 40, 5407–5415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Data pertaining to the role of Mitf in RDis. (A) The presence of the empty KK-TK transgene in the tio locus did not increase the penetrance of RDis in 2XMitfRNAi discs (p=0.15). (B) Schematic summarizing the genetic pathways known to affect Yki function with respect to RDis (DeSantis et al., 2023; Neal et al., 2022). (C-C’) Differential interference contrast micrographs of colorimetric in situ hybridization showing Mitf expression in the L2 eye disc. Mitf is detected in the PE plane (en face in A and in cross-section in A’). (D-E’) Mitf2.2p::GFP expression (green) in L2 (D-D’) and L3 (E-E’) eye discs; PE (D, E) and retinal (D’, E’) planes are shown. (F-I’) Detection of adherens junction (AJ) components (Arm, F-G’; DE-Cad, H-I’) in control (F-F’ and H-H’) and 4XMitfRNAi-expressing (G-G’ and I-I’) discs; Scale Bar in F = 10 μm. In control discs, AJ markers were detected at cell-cell boundaries in the PE (F, H) and at the vertices of developing photoreceptor clusters in the retina (F’, H’; yellow arrows). Despite disc-wide expression of UAS-MitfRNAi and a highly penetrant sRDis phenotype, neither pattern of AJ marker expression was perturbed in these eye discs (G-G’, I-I’).
Figure S2. Validation of dsRNA KD and representative full western blots related to Figure 2. (A-A”’) Expression level of indicated transcripts, relative to Act5C in S2 cells treated with dsRNA, as indicated. *** p<0.001 (B) Full images of western blot shown in Fig. 2B. (C) Full images of western blot shown in Fig. 2C. *NS* - non-specific band detected using 12C6c antibody in input but not immunoprecipitated samples. *HC* Residual mouse IgG heavy chain detected in 12C6c immunoprecipitates.
Figure S3. Supporting data related to the Mitf129 loss-of-function allele. (A) The Mitf129 allele was generated using CRISPR technology (see Methods); sequencing of a genomic DNA amplicon of the Mitf locus revealed a large deletion, encompassing most of the coding exons (indicated by red box on a schematic exported from UCSC Genome Browser). Predicted transcripts (RA-RD) are labeled. (B) Primary amino acid sequence translations (PA-PD) from predicted transcripts affected by the Mitf129 deletion. While the N-terminal 53 amino acids are preserved in all cases, transcripts RA, RB, and RD are expected to encode a frameshift resulting in 6 novel amino acids (red) before the termination codon in the respective translations (PA, PB and PD). The reading frame of the RC transcript is preserved in Mitf129 but has a predicted deletion of 781 amino acids (indicated in blue) before the 4 native residues preceding the termination codon in PC. (C) Micrograph of larvae of the indicated genotypes, including rare homozygous Mitf129/ Mitf129 larvae that develop to at most early L3 size, while retaining L2-like mouth hooks. (D) Quantification of Eya-expressing MARCM clones in the PE, relative to the posterior disc margin. (E-F’) Eye discs with MARCM clones overexpressing GFP (UAS-GFP, green); discs stained for Elav (magenta, A-A’) or Eya (red, B-B’); Scale Bar in E = 25 μm. (G-H”) Eye discs with MARCM clones overexpressing GFP (UAS-GFP, green), Mitf (UAS-MitfCDS3), and either Ey (UAS-ey, G-G”) or Eya (UAS-eya, H-H”); XY projections of retinal clones are shown. Cells in posterior clones expressing Mitf and Ey show upregulation of Ey (cyan, G’) but not restoration of Eya (red, G”). Cells in posterior clones expressing Mitf and Eya show upregulation of Eya (red, H’) but not restoration of Elav (magenta, H”).
Figure S4. Representative Images of MitfCDS3 retina suppression phenotypes. (A-A”) Example images of categorization scheme for retinal suppression phenotype, upon disc-wide expression of wt Mitf (UAS-MitfCDS3); discs stained for Elav (magenta) and F-actin (Phalloidin, blue); Scale Bar in A = 25 μm. Category 1 – many neurons in disorganized pattern (A), Category 2 – few neurons (A’) and Category 3 – no neurons (A”).
Figure S5. Data supporting the effects of Cka and Mts reduction on Mitf transcriptional activity. (A-B) Mitf expression in the larval wing disc (dpp>UAS-MitfCDS6) resulted in upregulation of a reporter gene (Vha55-lacZ), as visualized by β-galactosidase activity (blue) (A’, compare with negative control in A); Scale Bar in A = 100 μm. Reporter expression was reduced in discs heterozygous for a hypomorphic Cka allele (A”; Ckas1883/+) and in discs heterozygous for a null allele of mts (A”’; mts02496/+). Quantification of β-galactosidase activity across 10 discs per genotype (CkaS1883/+, p=1.85 × 10−9; mts02496/+, p=4.60 × 10−8) (B). *** p<0.001 (C) Full images of western blot shown in Fig. 5D.
Data Availability Statement
All data necessary to support the conclusions of this study are included. The authors will make all unique reagents available upon reasonable requests to the corresponding authors.


