Abstract
All enveloped viruses must bud through a cellular membrane in order to acquire their lipid bilayer, but little is known about this important stage in virus biogenesis. We have developed a quantitative biochemical assay to monitor the budding of Semliki Forest virus (SFV), an enveloped alphavirus that buds from the plasma membrane in a reaction requiring both viral spike proteins and nucleocapsid. The assay was based on cell surface biotinylation of newly synthesized virus spike proteins and retrieval of biotinylated virions using streptavidin-conjugated magnetic particles. Budding of biotin-tagged SFV was continuous for at least 2 h, independent of microfilaments and microtubules, strongly temperature dependent, and relatively independent of continued exocytic transport. Studies of cell surface spike proteins at early times of infection showed that these spikes did not efficiently bud into virus particles and were rapidly degraded. In contrast, at later times of infection, spike protein degradation was markedly reduced and efficient budding was then observed. The previously described cholesterol requirement in SFV exit was shown to be due to a block in budding in the absence of cholesterol and correlated with the continued degradation of spike proteins at all times of virus infection in sterol-deficient cells.
Virus budding is a critical step in the life cycle of all enveloped viruses. Budding may be defined as the progressive envelopment of the virus core by a cellular membrane enriched with viral membrane proteins, culminating in a membrane fission reaction to release the completed virus particle. Different viruses use different host cell membranes as budding sites, including the plasma membrane and various membranes of the exocytic pathway. Viruses also differ in their requirements for virus proteins to drive the budding reaction (14). Viruses such as the alphaviruses and hepadnaviruses have a strict requirement for both nucleocapsids and spike proteins to permit virus budding (31, 57, 59). In contrast, the budding reactions of various other viruses can be driven by the capsid or core protein, by the matrix protein, or by the membrane proteins, without obligatory involvement of the other viral protein subunits (14). Virus budding reactions are an important area of research because of their key roles in virus replication, their potential as therapeutic targets, and their relevance to cellular membrane budding reactions. However, quantitative experimental methods to specifically assay budding have been limited. In the absence of such focused systems, broader studies of infectious particle production by necessity measure a wide range of reactions during the virus life cycle, including the biosynthesis of viral components and the traffic of viral membrane proteins through the exocytic pathway. The lack of more effective model systems has made it difficult to address fundamental questions about viral budding reactions, such as requirements for cellular components and energy sources.
Alphaviruses such as Semliki Forest virus (SFV) are simple, well-characterized enveloped animal viruses (see references 20 and 56 for a review). Each SFV particle contains 240 copies of four structural proteins: the capsid protein, which packages the single plus-stranded RNA genome into nucleocapsids, and three envelope proteins, the type I transmembrane polypeptides E1 and E2 (each about 50 kDa) and the peripheral E3 polypeptide (∼10 kDa). The envelope proteins assemble into 80 spikes, each consisting of a trimer, (E1/E2/E3)3. Both the spike protein layer and the viral nucleocapsid are arranged as T=4 icosohedral structures, which associate with each other via a one-to-one interaction of the E2 internal domain and the capsid protein (7, 13). The virus lipid bilayer is derived from the host cell plasma membrane during budding.
The life cycle of SFV and other alphaviruses has been studied in detail (20, 56). SFV enters host cells via receptor-mediated endocytosis, and virus membrane fusion is mediated by the spike protein and triggered by the low pH present in the endosome (15, 20). In addition to its low pH requirement, fusion of alphaviruses such as SFV is also strongly dependent on the presence of cholesterol and sphingolipid in the target membrane (21, 25, 37, 38, 65). Following fusion between the viral and endosome membranes, nucleocapsids are released into the cytoplasm and viral replication is initiated. Progeny RNA molecules associate with capsid protein in the cytoplasm to form new nucleocapsids. The spike protein E1 subunit and the E2 precursor, p62, are translated and translocated into the rough endoplasmic reticulum, where they are glycosylated and form a stable but noncovalently associated heterodimer. The dimer is transported through the secretory pathway and processing of p62 to mature E2 and E3 is carried out in the late secretory pathway by furin, a cellular protease. E1, E2, and E3 are then transported to the plasma membrane, where virus budding occurs.
Alphavirus budding is a clear example of a budding reaction that requires both nucleocapsids and spike proteins. This is due to a specific interaction between the capsid protein and a key tyrosine-containing motif in the cytoplasmic tail of E2 (56, 67). Structural studies indicate that this region of E2 binds to a hydrophobic pocket on the surface of the nucleocapsid (28, 52). Expression studies demonstrated that, although p62 could be transported to the cell surface in the absence of E1, stable association with nucleocapsids did not occur, suggesting that the dimeric interaction of E1 and E2 is important to maintain the correct conformation of the E2 tail for nucleocapsid binding (1). Further studies showed that lateral interactions between heterodimers are also required for efficient budding (12). In keeping with the known structure of the alphavirus particle (7, 13), it appears that the E1/E2 heterodimers assemble into trimers which associate via lateral interactions and that the resultant multivalency of E2-nucleocapsid binding promotes virus budding.
In addition to these requirements for viral proteins in budding, studies of alphavirus infection in cholesterol-depleted cells have revealed a role for cholesterol in efficient virus exit. Along with their well-characterized requirement for cholesterol in the membrane fusion reaction, both SFV and Sindbis virus (SIN) show strong requirements for cholesterol in exit (32, 34). In contrast, srf-3 (sterol requirement in function), an SFV mutant selected for more efficient growth in cholesterol-depleted cells, has markedly increased levels of both fusion and exit in the absence of cholesterol compared to those of wild-type (wt) virus (34, 62). The increased cholesterol independence of srf-3 fusion and exit were demonstrated to be caused by a single point mutation in the srf-3 E1 subunit, proline 226→serine (P226S) (62). Mutagenesis studies of SIN demonstrated that the E1 226 region is also involved in the cholesterol dependence of its fusion and exit (32). However, although these studies reveal a role for cholesterol in the alphavirus exit pathway, the stage of the viral exit pathway affected and the mechanism of the cholesterol effect remain unclear.
We have here established a quantitative biochemical assay for SFV budding that is generally applicable to viruses that bud from the plasma membrane. The assay was based on biotin derivatization of radiolabeled virus spike proteins at the cell surface and subsequent retrieval of biotin-tagged virions using magnetic streptavidan particles. We have used the assay to define several basic properties of SFV budding, including kinetics, temperature dependence, and the role of cytoskeletal elements and exocytic transport. Using wt virus and the srf-3 mutant, we demonstrated that the previously observed cholesterol requirement for production of wt SFV was due to a block in spike protein budding from cholesterol-depleted cells. The block in wt virus budding correlated with the rapid degradation of the wt spike proteins throughout the infectious cycle in sterol-deficient cells.
(This research was conducted by Y.E.L. in partial fulfillment of the requirements for a Ph.D. from the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells were cultured at 37°C in Dulbecco's modified Eagle's (DME) medium containing 100 U of penicillin/ml and 100 μg of streptomycin/ml (P/S), 5% fetal calf serum (FCS), and 10% tryptose phosphate broth (38). Control cholesterol-containing C6/36 mosquito cells were maintained at 28°C in DME medium with P/S and 10% heat-inactivated FCS (34). Cholesterol-depleted C6/36 cells were prepared by four passages at 28°C in DME medium containing P/S and 10% delipidated heat-inactivated FCS (32, 38) and used for experiments between passages 5 and 15 (32, 33). The levels of both free and esterified cholesterol in the cholesterol-depleted cells were less than 2% of those of control cells (38).
The wt SFV stock was a well-characterized plaque-purified SFV isolate (16), and was used to infect BHK cells for budding experiments. wt SFV stock from the pSP6-SFV-4 infectious clone (29, 62) was prepared as described below and used to infect cholesterol-containing or depleted C6/36 cells. DNA sequence analysis of both stocks at the structural protein region showed identical amino acid sequences (16). srf-3 was an SFV mutant selected for efficient growth in cholesterol-depleted C6/36 cells. The srf-3 phenotype is conferred by a single point mutation in the E1 subunit, which changes the proline at position 226 to serine (62). To prepare wt and srf-3 stocks from infectious clones, infectious RNAs were transcribed in vitro from the wt and mutant SFV infectious clones and introduced into BHK cells by electroporation, and the cells were cultured for 12 to 18 h before the collection of the virus stock, as previously described (62). [35S]methionine- and [35S]cysteine-labeled SFV or unlabeled, purified SFV was prepared by growth in BHK cells and purified by banding on a discontinuous sucrose gradient or tartrate gradient, respectively (26).
Virus infection, metabolic labeling, and cell surface budding assay.
BHK cells, control C6/36 cells, or cholesterol-depleted C6/36 cells were cultured on 35-mm plates for 1 to 3 days until just confluent and then infected by the following protocols. BHK cells (ca. 1 × 106 to 2 × 106 cells/plate) were infected with wt SFV at 10 PFU/cell in 0.6 ml of minimal essential medium (MEM) containing P/S, 10 mM HEPES (pH 7.4), and 0.2% bovine serum albumin (BSA) for 1 h at 37°C. Cells were then washed to remove the input virus, and the incubation was continued for another 4 to 5 h (a total of 5 to 6 h) in 1 ml of the same medium. Control C6/36 cells (ca. 2 × 106 to 6 × 106 cells/plate) were infected with wt or srf-3 at the same multiplicities of infection (either 10 or 100 PFU/cell) in 0.7 ml of Opti-MEM containing 0.2% BSA (O/B) for 2 h at 28°C, washed with O/B, and incubated in 1.5 ml of O/B for the indicated times (1.5 to 5 h). To overcome the inhibition of wt virus fusion in cholesterol-depleted C6/36 cells and permit viral protein expression, depleted cells (ca. 3 × 106 to 6 × 106 cells/plate) were infected with wt SFV at 1,000 PFU/cell or srf-3 at 100 PFU/cell in 0.7 ml of O/B for 2 h at 28°C. The cells were then washed twice with O/B and incubated in 1.5 ml of O/B with or without 20 mM NH4Cl for the indicated times.
Following completion of the above virus infection protocols, the cells were metabolically labeled to monitor the newly synthesized spike proteins. Cells were starved in methionine- and cysteine-free DME medium for 15 min and labeled for 5 or 15 min in 0.6 ml of this medium containing 40 to 200 μCi of [35S] methionine-cysteine/ml (Pro-Mix Cell Labeling Mix; Amersham Pharmacia Biotech, Arlington Heights, Ill.). Cells were chased for various times in 1 ml of medium containing a 10-fold excess of methionine and cysteine, either MEM containing 10 mM HEPES (pH 8.0) plus 0.2% BSA (for BHK cells) or O/B medium (for C6/36 cells). The cells were then placed on ice, washed twice with ice-cold phosphate-buffered saline (PBS; pH 8.0) containing 1 mM glucose, and derivatized by incubating for 15 min with 0.6 ml of fresh biotin solution on ice on a reciprocal shaker platform. Biotin solution was freshly prepared before each use and consisted of 0.5 mg of EZ-Link Sulfo-NHS-LC-Biotin/ml (Pierce Chemical Co., Rockford, Ill.) in PBS (pH 8.0) plus 1 mM glucose. A single derivatization was used, since one or two additional treatments did not increase the cell surface signal and led to a slight decrease in subsequent virus budding (data not shown). Free biotin was then quenched by washing the cells four times with ice-cold MEM containing 10 mM HEPES (pH 8.0), 0.2% BSA, and 10 mM glycine, followed by further analysis as described below.
To allow budding and release of biotin-derivatized viruses, derivatized cells were shifted from ice to a water bath at either 37°C (for BHK cells) or 28°C (for C6/36 cells) and incubated in 1 ml of post-biotin incubation medium (MEM containing P/S and 10 mM HEPES [pH 8.0] without bicarbonate but supplemented with the equivalent concentration of NaCl). The post-biotin incubation medium was collected after the indicated time, and cell debris was removed by centrifugation in a microfuge for 10 min at 12,000 rpm. Protease inhibitors were added to final concentrations of 1 mM PMSF (phenylmethylsulfonyl fluoride), 1 μg of pepstatin per ml, and 1 μg of leupeptin per ml. Samples were then aliquoted and stored on ice for analysis the following day. Alternatively, samples were diluted with an equal volume of 50% (wt/wt) sucrose (in buffer containing 50 mM Tris [pH 7.4] and 100 mM NaCl) and frozen at −80°C. Either storage condition preserved intact, protease-impermeable virus membranes (data not shown). To quantitate the cell-associated biotinylated spike proteins at various incubation times, cells were lysed in 0.5 ml of lysis buffer containing protease inhibitors and 1% Triton X-100 (23), the nuclei were removed by centrifugation, and the biotinylated spike proteins were quantitatively retrieved as described below.
Quantitative retrieval of biotinylated virus particles and spike proteins.
One-quarter of the post-biotin incubation medium samples and one-quarter of the cell lysate samples were used for analysis. The retrieval method and quantitation of budding efficiency were essentially the same for BHK cells, control C6/36 cells, or cholesterol-depleted C6/36 cells. Medium samples were incubated with 25 μl of BioMag Streptavidin Ultra-Load Particles (mag-SA; PerSeptive Biosystems, Framingham, Mass.) for 30 min at 4°C on a nutator. The mag-SA particles were collected using a magnetic tube holder (Magnetic Particle Concentrator MPC-E; Dynal, Oslo, Norway), and washed three times with 0.5 ml of cold PBS containing 0.2% BSA and 1 mM PMSF and one time with cold PBS containing 1 mM PMSF. Retrieval of biotinylated spike proteins from the cell lysate samples was similar except that 30 μl of mag-SA was used and wash solutions contained 1% Triton X-100. Specific retrieval of biotinylated spike proteins from the medium was performed by adding 1% Triton X-100 to medium samples, followed by retrieval with 25 μl of mag-SA and washing as for cell lysate samples. mag-SA-bound material was then released by heating the samples to 95°C in 1× sodium dodecyl sulfate (SDS) sample buffer for 5 min. Samples were analyzed by electrophoresis on 10% acrylamide gels followed by fluorography (23). In order to accurately recover radiolabeled capsid protein from budded virus for SDS-polyacrylamide gel electrophoresis (PAGE) analysis, 3 μg of purified nonradiolabeled SFV per sample was included as carrier in the 1× SDS sample buffer. Gels were quantitated by phosphorimaging (ImageQuant, v. 1.2; Molecular Dynamics, Inc., Sunnyvale, Calif.).
Preliminary experiments showed that different lots of mag-SA had variable retrieval efficiencies. To overcome this batch-to-batch variation, 200 μl of mag-SA was incubated with 0.5 ml of PBS containing 5% dialyzed FCS (biotin-free) at 4°C for half an hour, washed three times with 1 ml of PBS containing 0.1% BSA, and then resuspended in this buffer to the original volume. This prewash of mag-SA significantly improved the specific binding of suboptimal batches (maximally ∼6-fold) and resulted in similar binding capacities for different batches of mag-SA (data not shown). Both capsid protection assays and electron microscopy (as described below) demonstrated that the increased binding was not due to virus aggregation. Therefore, mag-SA batches of low binding capacities were treated using the procedure described above immediately before use.
Capsid protease protection assay.
To assay virus particle and virus membrane integrity, biotinylated virus was retrieved from the post-biotin incubation medium using mag-SA as described above. After the final PBS wash, the mag-SA was resuspended in 25 μl of 10 mM Tris (pH 7.4)–50 mM NaCl. The samples were digested with 50 μg of trypsin XIII (Sigma) per ml for 10 min on ice in the presence or absence of 1% Triton X-100. Trypsin was then inhibited by the addition of soybean trypsin inhibitor and PMSF to final concentrations of 150 μg/ml. Then, 3 μg of cold purified SFV was added as carrier to the samples before boiling them in SDS gel buffer, followed by SDS-PAGE analysis. As a control, parallel trypsin digestion and SDS-PAGE analysis were performed using gradient-purified radiolabeled SFV.
Electron microscopy.
Electron microscopic analysis was used to evaluate the morphology of biotinylated viruses retrieved by mag-SA. Two 35-mm plates of BHK cells were infected as described above, mock pulse-labeled, chased, and either derivatized with biotin or mock treated. Both samples were then postincubated for 1 h at 37°C, and one-half of the medium was retrieved by using 50 μl of mag-SA. After the final PBS wash, the mag-SA pellets were fixed in 1 ml of 2.5% glutaraldehyde in 0.1 M cacodylate for 30 min, dehydrated, embedded, processed, and evaluated by transmission electron microscopy, all as previously described for cell pellets (34). Parallel samples from radiolabeled cells were evaluated by retrieval and SDS-PAGE and showed efficient budding and specific mag-SA binding.
Electron microscopy was also used to localize biotin at the cell surface. Cells were infected and biotin-conjugated using our standard procedure and fixed before or after a post-biotin incubation step of 45 min at 37°C. Biotin was detected by incubating cells with streptavidin conjugated to horseradish peroxidase, followed by gold-conjugated antibody to horseradish peroxidase (Carolyn Machamer, personal communication). Cells were then fixed and processed as described above.
Assay of E1/E2 dimer association in C6/36 cells.
Control or cholesterol-depleted C6/36 cells were infected with either wt SFV or the srf-3 mutant as described above. The cells were pulse-labeled, chased for 30 min, and lysed in 0.5 ml of lysis buffer containing 1% NP-40, 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.9 mM CaCl2, 0.5 mM MgCl2, 1 μg of leupeptin/ml, 1% aprotinin, 1 mM PMSF, and 1 mg of BSA/ml (19, 63). Monoclonal antibodies (MAb) that recognize either the E1 subunit (E1-1) or the E2 subunit (E2-3) (23) were used to coprecipitate the spike dimer, essentially as previously described (19, 63).
Analysis of spike protein transport to the cell surface of cholesterol-depleted cells.
Due to the fusion block in cholesterol-depleted cells, only about 10 to 30% of the cells could be infected even at high multiplicities, and mosquito cells do not efficiently shut off host protein synthesis. Thus, to accurately measure the rate of spike protein transport to the cell surface, the spike proteins expressed in the cells were first quantitatively immunoprecipitated with an anti-spike protein antibody (23). Half of the immunoprecipitate was directly analyzed by SDS-PAGE to determine the amount of total radiolabeled spike protein in the cells. To quantitate the biotinylated spike proteins derivatized at the cell surface, the other half of the immunoprecipitate was boiled in SDS-containing buffer to release the spike proteins from the zysorbin (reference 10 and as described below). The released spike proteins were then retrieved by mag-SA binding and analyzed by SDS-PAGE.
Quantitation of virus budding and biotinylated spike protein degradation.
Control or cholesterol-depleted C6/36 cells were infected with wt or srf-3 and tested at various times of infection using the budding assay. Due to the lower expression level of viral spike proteins early in the infection cycle and the lack of host protein shutoff, the analysis of both medium and cell lysate samples was modified by direct mag-SA retrieval of samples, followed by immunoprecipitation with an anti-spike protein antibody to remove non-spike protein background (10). The biotinylated proteins from one-quarter of the cell lysate or medium were first retrieved using 30 μl of mag-SA in the presence of detergent and then released from mag-SA by heating them twice at 95°C for 5 min in 50 μl of buffer containing 3% SDS, 50 mM NaCl, and 10 mM Tris (pH 7.4). The mag-SA was removed, the supernatant was diluted with 450 μl of buffer (50 mM NaCl; 10 mM Tris, pH 7.4; 1% Triton X-100) and immunoprecipitated with anti-spike protein antibody, and the samples were analyzed by SDS-PAGE. The biotinylated viral proteins released in the medium and the biotinylated spike proteins remaining in the cells were quantitated and used to determine the efficiency of budding and the spike protein degradation rate.
RESULTS
Development of an SFV budding assay.
In order to measure SFV budding, a method to specifically monitor the incorporation of plasma membrane spike proteins into virus particles was required. During the initial phases of this work, we explored a variety of possible assays for SFV budding. We tested the use of temperature-sensitive SFV or SIN mutants (43, 53) or ionic conditions (64) to reversibly accumulate budding-competent virus spike proteins at the plasma membrane. None of these conditions gave a clearcut exit block that could be synchronously released to monitor budding (data not shown). We therefore developed a direct biochemical method to specifically tag SFV spike proteins at the plasma membrane and monitor their incorporation into virus particles.
In this assay, BHK cells were infected with SFV at a multiplicity of 10 PFU/cell for 5 to 6 h, pulsed-labeled with [35S] methionine-cysteine, and chased for 45 min to permit delivery of the newly labeled spike proteins to the plasma membrane. The cells were then placed on ice, and the cell surface proteins were covalently derivatized with biotin. The cells were incubated at 37°C to permit budding of radioactive virus containing biotinylated spike proteins, which was then retrieved using streptavidan coupled to magnetic particles, and analyzed by SDS-PAGE (see Materials and Methods for details). A number of preliminary experiments were performed to determine optimal assay conditions (data not shown). Growth curve studies demonstrated that the assay interval occurred during a period of efficient production of infectious progeny virus, i.e., approximately 103 PFU/cell/h, and prior to significant cytopathic effects. The chase time was selected to permit maximal accumulation of radiolabeled spike proteins at the plasma membrane in the absence of significant release of radiolabeled virus. Control experiments demonstrated that the biotinylation conditions were specific for proteins at the cell surface and gave negligible labeling of internal proteins (see also reference 10). Both E1 and E2 were efficiently biotin derivatized under these conditions (cf. Fig. 6).
FIG. 6.
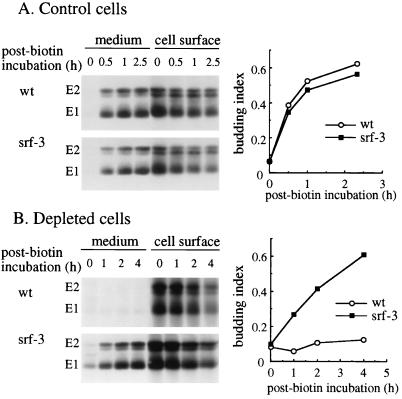
Effect of cholesterol depletion on SFV budding. (A) Cholesterol-containing C6/36 mosquito cells were infected with wt SFV or srf-3 at 100 PFU/cell for 6 h, pulse-labeled for 15 min, chased for 30 min, and derivatized with biotin. The cells were then incubated at 28°C for the indicated times. The 0-h sample was incubated on ice for the duration of the experiment. Biotinylated virus particles in the medium and biotinylated spike proteins present in the cells were collected by mag-SA retrieval at each time point. Samples were analyzed by SDS-PAGE and phosphorimaging, and the data are expressed as a budding index, defined as the spike protein radioactivity retrieved from the medium divided by the spike protein radioactivity retrieved from both the medium and the cell lysate. (B) Cholesterol-depleted C6/36 cells were infected with wt SFV at 1,000 PFU/cell or with srf-3 at 100 PFU/cell for 24 h, pulse-labeled for 15 min, chased for 90 min, derivatized with biotin, and incubated at 28°C for the indicated times. Mag-SA retrieval and quantitation were performed as in panel A. Panels A and B show representative examples of three experiments in each cell type.
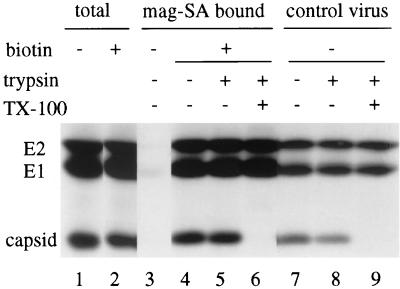
Results of a typical budding assay are shown in Fig. 1. Infected, radiolabeled BHK cells were either biotin derivatized (lanes 2, 4, 5, and 6) or mock derivatized (lanes 1 and 3) and then incubated at 37°C for 60 min to permit virus budding, a step that is referred to here as “post-biotin incubation.” The total radiolabeled spike proteins released in the post-biotin incubation media were recovered by acid precipitation of the media (lanes 1 and 2). Similar amounts of radiolabeled spike proteins were released from the biotinylated (lane 2) and mock-treated (lane 1) samples, indicating that biotin derivatization did not significantly affect virus budding and release. The post-biotin incubation media were then treated with mag-SA in the absence of detergent, conditions that should preserve intact virions. Although abundant radiolabeled viral proteins were present in both biotin and mock-treated samples (lanes 1 and 2), mag-SA retrieval was specific for biotin-derivatized virus (lanes 4 to 6 versus lane 3). Control experiments established that retrieval of biotin-containing virus was quantitative under our standard experimental conditions (see Materials and Methods; also data not shown). The presence of capsid protein in the retrieved sample (lane 4) and in a gradient-purified radiolabeled control virus sample (lane 7) suggested mag-SA retrieval of complete viral particles. In order to address the integrity of the retrieved virus, we took advantage of capsid protein's sensitivity to trypsin digestion and its protection from proteolysis when enveloped by the virus lipid bilayer (24). Digestion by exogenous trypsin demonstrated that the capsid protein present in the retrieved samples was protected from proteolysis (lane 5), similar to capsid protein in gradient-purified radiolabeled SFV (lane 8). Protection was lost when the virus membrane in either sample was disrupted by the addition of Triton X-100 (lanes 6 and 9). Under these digestion conditions the spike protein E1 and E2 subunits are resistant to trypsin (lanes 5, 6, 8, and 9), in keeping with previous results indicating the relative trypsin resistance of the native E1/E2 heterodimer (15a, 22).
FIG. 1.
Characterization of retrieval and integrity of biotinylated virus particles. BHK cells were infected with SFV at 10 PFU/cell for 5 h, labeled with [35S] methionine-cysteine at 100 μCi/ml for 15 min, and chased for 45 min. The cells were next either derivatized with biotin or mock derivatized and then incubated for 60 min at 37°C; the media were then collected for analysis. The total viral spike proteins in an aliquot of the media were determined by trichloroacetic acid precipitation (lanes 1 and 2). Equivalent aliquots of the media were retrieved with mag-SA in the absence of detergent and either directly analyzed by SDS-PAGE (lanes 3 and 4) or digested with exogenous trypsin before SDS-PAGE analysis (lanes 5 and 6) in the presence or absence of 1% Triton X-100 as indicated. Capsid protein is trypsin-sensitive and fully digested when the virus membrane is disrupted by detergent. [35S]methionine-[35S]cysteine-labeled gradient purified SFV was similarly processed in parallel as a control (lanes 7 to 9). A representative example of three experiments is shown, with all lanes exposed for the same length of time.
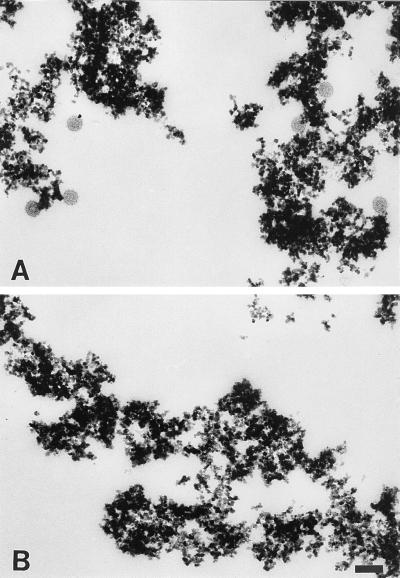
Electron microscopy was used to observe the morphology of the biotinylated virus bound to mag-SA. Samples were prepared from virus-infected cells following either standard biotin conjugation or mock biotin treatment and then retrieved as described above using mag-SA. The mag-SA appeared as electron-dense particles (Fig. 2). The biotin-derivatized sample contained a number of individual virions bound to the mag-SA (Fig. 2A), while no virus particles were observed in the mock-treated sample (Fig. 2B), a finding in agreement with the biochemical results presented above. The retrieved virus appeared to be morphologically normal and contained spike proteins, an intact-appearing lipid bilayer, and an electron-dense nucleocapsid core (Fig. 2A), in agreement with the biochemical analysis of the retrieved radiolabeled virus samples presented in Fig. 1.
FIG. 2.
Electron microscopy of retrieved virus particles. Similar to Fig. 1, BHK cells were infected with SFV for 6 h, biotinylated (A) or mock treated (B), and further incubated at 37°C for 60 min. Virus particles in the medium were retrieved by mag-SA and processed for electron microscopy. Note that virus particles are only observed in the biotinylated sample. Bar, 0.1 μm.
In separate experiments, we also examined the production of infectious virus particles under the experimental conditions used for the budding assay. Similar titers of virus were produced per hour at 37°C from cells that were continuously maintained under 37°C incubation conditions and from cells that were first preincubated for ∼15 min on ice and then returned to 37°C incubation conditions (data not shown). Thus, exposure to low temperature did not significantly affect subsequent virus production. Infected cells were also biotin derivatized using the standard budding protocol, and the release of infectious virus during the post-biotin incubation was evaluated by plaque assay. Virus titers were within the range observed for nonbiotinylated cells (data not shown), suggesting that biotinylation of the cells did not significantly affect infectious virus production.
Taken together, these results demonstrate that the budding assay was specific for virions containing cell surface-labeled spike proteins. The biotinylation procedure did not appreciably affect virus budding as assayed either biochemically or by infectivity. The retrieved samples were sealed, intact virions with the morphology typical of alphavirus particles.
Specificity of the assay for SFV budding from the plasma membrane.
A key issue in the validation of this assay was its ability to specifically monitor the budding of newly formed progeny virus. Since SFV is known to bind to cell surface receptors (56), it was important to determine if the dissociation of newly budded virus particles from plasma membrane receptors played an important role in the kinetics of exit. In addition, it was also important to prove that the retrieved virus resulted from de novo budding of cell surface spike proteins during the post-biotin incubation rather than from the release of receptor-bound virus particles formed prior to biotinylation. We addressed these points both morphologically and biochemically. Electron microscopy was performed using streptavidin and immunogold to follow cell surface biotin (see Materials and Methods). Immediately after biotin derivatization, infected BHK cells showed abundant gold labeling at the plasma membrane (data not shown). Label was associated with numerous linear regions of the membrane and also with some regions overlying nucleocapsids or containing forming virus buds. Negligible gold labeling was observed in the absence of biotinylation. After post-biotin incubation of the biotinylated cells at 37°C for 45 min, a markedly increased number of gold-labeled virus particles and forming virus buds were observed. These morphological results suggested that the assay was detecting newly formed virus particles.
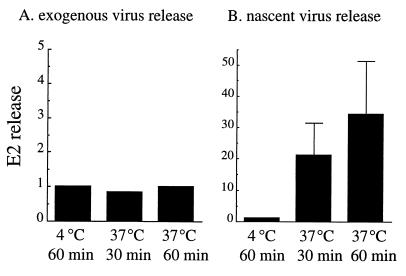
In order to directly test the contribution of preformed virus particles to the assay, we determined the properties of virus receptor release under the conditions used for the standard budding assay. Gradient-purified radiolabeled SFV was allowed to bind to virus-infected, unlabeled cells on ice. Unbound virus was washed away, and the cells with bound virus were biotin conjugated using our standard assay conditions. The samples were then incubated either on ice or at 37°C, and the released biotin-derivatized virus was retrieved by mag-SA. Samples were analyzed by SDS-PAGE, and the release of exogenous virus was compared to that of parallel cultures in which a standard budding assay was performed. Samples were quantitated and compared using phosphorimaging of the retrieved virus E2 protein since, unlike E2, the E1 protein can be released from BHK cells as a soluble, non-virus-associated protein under some experimental conditions (10, 66) and since infected cells contain a larger pool of capsid protein than spike protein, resulting in a lower specific activity of capsid after pulse-labeling (47), (Y. E. Lu and M. Kielian, unpublished data).
The release of exogenous bound virus was temperature independent, with comparable amounts of release occurring at either 4 or 37°C and after 30 or 60 min (Fig. 3A). In contrast, very little newly synthesized virus was retrieved when the cells were incubated at 4°C (Fig. 3B). The amount of nascent virus retrieved increased dramatically when the cells were incubated for increasing times at 37°C. Thus, budding was strongly time and temperature dependent and was the principal step in virus exit measured by our assay. The assay primarily detected virus particles formed de novo during the post-biotin incubation period. Prior experiments had demonstrated that virus receptor binding is substantially decreased in infected cells (51), a result in agreement with our results demonstrating the lack of a significant role of receptor dissociation in the kinetics of virus exit.
FIG. 3.
Comparison of the properties of release of prebound virus and nascent virus. (A) Release of prebound SFV. [35S]methionine-[35S]cysteine-labeled gradient purified SFV (∼3 × 106 cpm/plate) was bound to unlabeled SFV-infected BHK cells on ice for 1 h. Using the standard protocol, unbound virus was removed by washing, and the cells with bound exogenous virus were biotin derivatized and quenched. (B) Release of nascent virus. BHK cells were infected with SFV for 5 h, radiolabeled, and biotin derivatized as in Fig. 1. Both the panel A and panel B samples were then incubated in post-biotin incubation medium (pH 8.0) at 4 or 37°C for 30 or 60 min to permit budding or release of radiolabeled virus. The media were then collected, and the biotinylated virus was retrieved using mag-SA. The samples were analyzed by SDS-PAGE and quantitation of the viral E2 protein. The amount of E2 release at 4°C was set to 1 for each panel. Note the different scales on the y axis of the A and B panels. Panel A is a representative example of two experiments; panel B shows averaged data and standard deviations from four separate experiments.
Kinetics and temperature dependence of SFV budding.
We next used the assay to characterize basic features of virus budding in BHK cells. Retrieval was performed in the absence of detergent in order to recover virus particles, as described above. Such biotin-tagged virus samples would contain both biotinylated and nonbiotinylated radiolabeled spike proteins. Alternatively, retrieval could also be performed in the presence of detergent, and under these conditions the assay could specifically characterize the efficiency of cell surface spike protein budding, in the absence of retrieval of nonbiotinylated spike proteins.
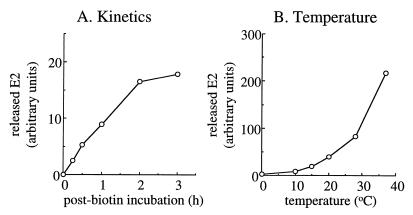
The kinetics of SFV budding were measured by incubation of BHK cells in post-biotin incubation medium for various times, followed by retrieval in the absence of detergent and quantitation of the radiolabeled E2 released in biotin-tagged virus (Fig. 4A). Biotinylated virus budding was continuous for at least 2 h at 37°C, indicating that a pool of biotin-tagged spike proteins remained at the cell surface during this time and continued to be incorporated into virus particles. In separate experiments, we also characterized the efficiency of budding of biotin-tagged spike proteins by specifically quantitating the total biotinylated spike proteins present in the cell lysate at the start of the incubation and the biotinylated spike proteins retrieved from the medium in the presence of detergent. These analyses demonstrated that ca. 13 to 36% of the biotin-tagged spike proteins were released within a 2-h incubation period at 37°C.
FIG. 4.
Properties of SFV budding. (A) Kinetics. BHK cells were infected with SFV for 5 h, radiolabeled, and biotin derivatized as in Fig. 1. Post-biotin incubation was carried out at 37°C for the indicated times, and biotinylated virus particles in the medium were retrieved by mag-SA and analyzed by SDS-PAGE and phosphorimaging to quantitate the E2 subunit, graphed in arbitrary units. A representative example of five experiments is shown. (B) Temperature dependence. BHK cells were prepared as in panel A, the post-biotin incubation was carried out at the indicated temperature for 2 h, and budding was quantitated as in panel A. A representative example of four experiments is shown.
The temperature dependence of SFV budding was characterized by incubating biotin-tagged BHK cells at different temperatures for 2 h and then quantitating biotinylated virus particles released into the medium (Fig. 4B). SFV budding was most efficient at 37°C, but measurable budding occurred even at temperatures as low as 10°C. Thus, although budding was strongly temperature dependent, it did not appear to be as extensively blocked at reduced temperatures as some cellular membrane trafficking events, such as endocytic traffic to late endosomes or protein transport from the trans-Golgi network (TGN) to the plasma membrane, both of which are inhibited at temperatures below 19°C (36, 42).
The role of membrane transport and cytoskeletal elements in SFV budding.
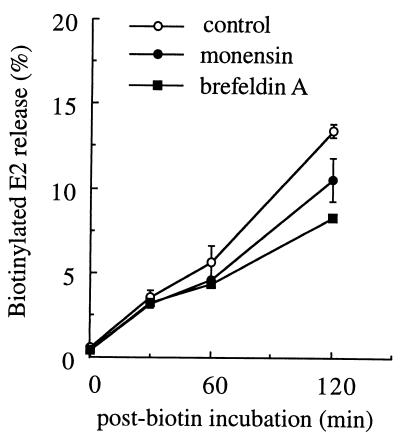
The experiments described above indicated that a population of biotin-tagged spike proteins at the cell surface continued to be incorporated into virus particles for at least 2 h at 37°C. During this time, virus budding would cause the net loss of spike proteins, nucleocapsids, and membrane lipids from the cellular pool. We used the membrane transport inhibitors monensin and brefeldin A to test the importance of replenishment of plasma membrane components to continued budding (Fig. 5). Monensin inhibits exocytic transport from the medial Golgi to the plasma membrane (18), while brefeldin A inhibits transport from the endoplasmic reticulum to the Golgi and from the TGN to the plasma membrane (27, 44). The transport effects of both inhibitors are known to occur rapidly after their addition to cells, and control experiments demonstrated that the addition of monensin or brefeldin A to the chase medium blocked 90 to 95% of the transport of newly synthesized spike proteins to the cell surface during the 45-min chase (data not shown). The kinetics of inhibition thus made it possible to add the inhibitors at the start of the post-biotin incubation and observe their effects on subsequent virus budding. Budding of biotin-tagged spike proteins was specifically monitored in order to observe the effects of the transport inhibitors on virus budding, rather than on the delivery of additional radiolabeled spike proteins to the plasma membrane. SFV budding was unaffected by monensin or brefeldin A during the first 30 min of incubation, during which time about 3.5% of the total biotin-labeled E2 protein was released in virus particles (Fig. 5). A slight decrease was observed for both of the drugs after a 60-min incubation, from ∼5.6% budding efficiency in untreated cells to ∼4.5% in treated cells. Even after 2 h of incubation of cells in the presence of monensin or brefeldin A, SFV budding occurred at reasonable efficiency, although it was reduced (ca. 20 to 40%) compared to that in untreated cells. Thus, budding of spike proteins from the cell surface was relatively independent of continued exocytic transport to the plasma membrane. This result emphasizes that the assay focuses on the late steps of budding from the plasma membrane rather than on the general exocytic pathway involved in spike protein processing and transport.
FIG. 5.
Role of membrane transport in SFV budding. BHK cells were infected and biotinylated as in Fig. 4 and then incubated on ice for 10 min in post-biotin incubation medium containing 10 μM monensin or 5 μg of brefeldin A per ml. The cells were then shifted to 37°C in the same medium, and the incubation was continued until the indicated times. The medium was then treated with mag-SA in the presence of detergent to specifically retrieve biotinylated spike proteins. Samples were analyzed by SDS-PAGE and phosphorimaging to quantitate the E2 subunit. The biotinylated E2 released in the medium at each time point was expressed as the percentage of the total biotinylated E2 present on the cell surface at time zero. The points are the average of duplicate samples, and the bars show the range. A representative example of three experiments is shown.
In an experiment similar to that described in Fig. 5, SFV budding from the cell surface was measured in the presence of cytochalasin D or nocodazole, drugs that specifically disrupt actin microfilaments or microtubules, respectively. SFV budding was unaffected by the inclusion of 10 μg of cytochalasin D/ml in the post-biotin incubation medium (data not shown). Inclusion of nocodazole at concentrations of 10 μM yielded a slight inhibition of SFV budding (data not shown), which appeared to be primarily due to the inhibition of protein transport by the acute addition of nocodazole (3). This effect on Golgi transport is known to dissipate if the cells are cultured in the continued presence of nocodazole, while microtubule function remains blocked. In a separate experiment, we therefore measured the production of infectious progeny SFV by infecting BHK cells for 1 h at 37°C and then incubating the cells in the presence of 0 to 10 μg of cytochalasin D/ml or 0 to 100 μM nocodazole for 11 h. Titers of 1010 PFU/ml resulted by this time point, and no effect of either cytochalasin D or nocodazole on the yield of infectious virus was observed (data not shown), further indicating that SFV budding was independent of microfilaments and microtubules.
Localization of the cholesterol requirement in SFV exit.
Having established and characterized the SFV budding assay, we wished to use it to analyze the previously described block in the exit pathway of wt SFV from cholesterol-depleted cells (34). As a control, we used the SFV mutant srf-3 which, although not completely sterol independent, is more efficient at both fusion with and exit from cholesterol-depleted cells (6, 62). We optimized the infection and incubation conditions for the budding assay in control or sterol-depleted C6/36 mosquito cells and used these conditions to test wt and srf-3 budding. Spike proteins in both wt- and srf-3-infected control cells were efficiently biotin labeled at the cell surface, and biotin-tagged viruses budded from control cells infected with either virus (Fig. 6A). Quantitation showed comparable levels of budding for both viruses, with a budding index of about 0.6 after 2.5 h (graph in Fig. 6A). When the same assay was performed in cholesterol-depleted cells, efficient biotin labeling of the E1 and E2 spike protein subunits was observed for either wt or mutant-infected cells (Fig. 6B). However, the efficiency of the budding of wt virus was reduced to background levels, while abundant budding of srf-3 was observed, with a budding index of ∼0.6 after 4 h of post-biotin incubation. These results thus suggested that the cholesterol requirement for the exit of wt SFV resided in a late stage of virus assembly, budding from the plasma membrane, and demonstrated that the budding of the srf-3 mutant was significantly less cholesterol dependent than that of wt virus.
Characterization of spike protein dimerization and transport in sterol-depleted cells.
Although the results presented above demonstrated that both the wt and the srf-3 spike proteins were accessible to biotin labeling at the plasma membrane of depleted cells, it was possible that in the absence of cholesterol the conformation or quantity of the wt spike protein was inadequate to support efficient budding. A decrease in wt spike protein transport might alter the plasma membrane spike protein pool and thus affect virus budding. Alternatively, the wt E1/E2 dimer interaction might be altered, similar to an SFV spike protein mutant that has a striking reduction in the stability of the E1/E2 dimer and greatly reduced budding but unimpaired transport (10). Thus, it was important to compare wt and srf-3 spike protein properties in cholesterol-depleted cells and to determine if the wt spike protein was altered in either dimer stability or delivery rate to the plasma membrane.
We first tested the dimer stability of wt and srf-3 spike proteins in control versus cholesterol-depleted cells. Cells were infected with either virus, pulse-labeled, and chased, and the presence of the E1/E2 dimer was assayed by coimmunoprecipitation with MAb against the E1 or E2 subunits (Table 1). The efficiency of coimmunoprecipitation, as previously reported (63), was dependent on the MAb, with the anti-E2 MAb showing greater efficiency than the anti-E1 MAb. However, the ability of each MAb to precipitate the other subunit in the dimer was not affected by the presence or absence of cellular cholesterol and did not differ between the wt virus and the srf-3 mutant. The dimer interaction of E1 and E2 is stabilized by basic pH and dissociates upon acid pH treatment (63). We found that pretreatment of the cell lysates at pH values between 6.5 and 8.0 altered the coprecipitation efficiencies as predicted, but no differences were observed between wt and mutant or control versus sterol-depleted samples (data not shown). Similar results were obtained when the cell surface spike proteins were biotin derivatized and analyzed for dimer stability by coimmunoprecipitation (data not shown).
TABLE 1.
Association of the E1/E2 dimer in control and sterol-depleted cellsa
| Virus type | E2/E1 or E1/E2 ratio ± SDb for:
|
|||
|---|---|---|---|---|
| Anti-E1 MAb in:
|
Anti-E2 MAb in:
|
|||
| Control cells | Depleted cells | Control cells | Depleted cells | |
| wt | 0.28 ± 0.07 | 0.24 ± 0.11 | 1.02 ± 0.13 | 0.84 ± 0.15 |
| srf-3 | 0.24 ± 0.02 | 0.26 ± 0.10 | 0.96 ± 0.10 | 1.00 ± 0.09 |
Cholesterol-containing or depleted C6/36 cells were infected with wt and srf-3 as in Fig. 6, pulse-labeled for 15 min, chased for 30 min (control cells) or 60 min (depleted cells), and lysed in NP-40-containing buffer at pH 7.4. Lysates were immunoprecipitated with subunit-specific MAb and analyzed by SDS-PAGE and phosphorimaging.
Ratio of E2/E1 (for anti-E1 MAb) or E1/E2 (for anti-E2 MAb). An average of three or four independent experiments is given.
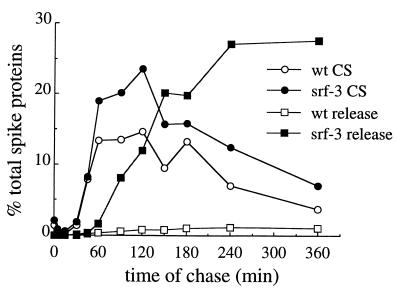
We then assayed the kinetics of wt and srf-3 spike protein transport to the surface of cholesterol-depleted cells. Depleted cells were infected with wt or mutant virus, pulse-labeled for 5 min, and chased for the indicated time, and the arrival of radiolabeled proteins at the cell surface was detected by their accessibility to biotin labeling (Fig. 7, CS samples). In parallel, the release of radioactive spike proteins in virus particles in the medium was quantitated (Fig. 7, release samples). The initial rates of wt and srf-3 spike protein transport to the plasma membrane of depleted cells were equivalent, with ∼8% of the total radiolabeled spike proteins detected at the cell surface after a 45-min chase. The proportion of radiolabeled spike proteins at the plasma membrane reached a plateau level between 60 and 120 min, and at this time the cell surface distribution of wt spike proteins (13 to 15% of the total) was consistently lower than that of srf-3 (19 to 24% of the total). As expected, the release of radioactive wt virus from the cholesterol-depleted cells was dramatically reduced, ∼1% of the total radiolabeled spike proteins after a 6-h chase compared to ∼28% for srf-3. A lag of 30 to 45 min was observed between the initial appearance of radiolabeled srf-3 spike proteins on the cell surface and the release of radiolabeled virus into the medium. This suggests that, following its arrival at the cell surface, the newly synthesized spike protein undergoes further interactions or rearrangements before its final incorporation into a completed virus particle.
FIG. 7.
Transport of wt and mutant spike proteins to the plasma membrane in cholesterol-depleted cells. Cholesterol-depleted C6/36 cells were infected with wt SFV (1,000 PFU/cell) or srf-3 (100 PFU/cell) for 24 h. Transport of newly synthesized spike proteins to the plasma membrane was then measured by pulse-labeling for 5 min and chasing for the indicated times at 28°C. At each time point, the medium was collected, the cell surface spike proteins were derivatized with biotin, and the cells were harvested by detergent lysis. The radiolabeled spike proteins present on the cell surface at each time point were assessed by quantitative immunoprecipitation of an aliquot of the cell lysate using a polyclonal antibody to the SFV spike protein, followed by mag-SA retrieval. The amount of spike proteins derivatized by cell surface biotinylation, referred to as cell surface (CS) samples, was expressed as a percentage of the total radiolabeled spike proteins immunoprecipitated at the 5-min chase time. For comparison, at each time point the amount of radiolabeled spike proteins released into the medium due to virus budding was also determined by quantitative immunoprecipitation using the same antibody and is expressed as a percentage of the total spike proteins at the 5-min chase time (release samples). A representative example of two experiments is shown.
Taken together, these data suggested that decreased wt virus budding from sterol-depleted cells was not due to an alteration in E1/E2 dimer interaction or a decreased rate of spike protein transport to the cell surface. However, the steady-state distribution of wt spike proteins on the plasma membrane was lower than that of srf-3, even though their initial transport rates to the cell surface were similar. The decrease in wt spike proteins at the plasma membrane suggested a possible increase in the degradation of wt spike proteins in cholesterol-depleted cells. Such increased degradation might be a consequence of reduced budding or might itself lead to reduced budding from sterol-depleted cells. In order to test for alterations in spike protein turnover and to assess their importance in budding, we compared the rate of spike protein degradation at various points in the infectious cycle in control and depleted cells.
Comparison of spike protein budding and degradation.
Little was known about the turnover of the cell surface spike protein in either control or sterol-depleted cells. Previous studies in BHK cells demonstrated that under conditions in which budding is blocked, such as the expression of spike proteins in the absence of capsid protein, spike proteins are rapidly degraded (10, 66). We examined the question of cell surface spike protein turnover first in control C6/36 cells, using biotin derivatization to mark spike proteins that had reached the plasma membrane and comparing cells at various times postinfection.
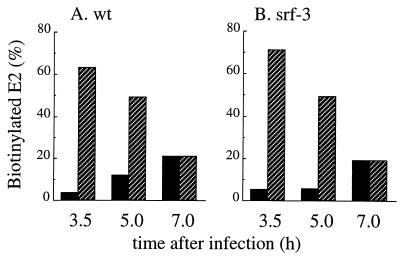
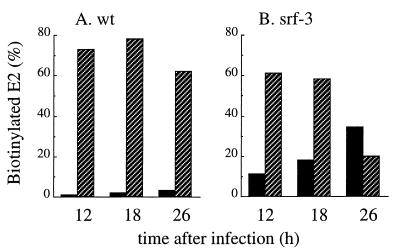
Control C6/36 cells were infected with wt and srf-3 for 3.5, 5, or 7 h, time points chosen to be prior to or during the logarithmic period of infectious progeny virus production (data not shown; see also reference 62). The cells were then pulse-labeled, chased, and derivatized with biotin using our standard procedure. The cells were next incubated for 2 h, and the degradation of the biotinylated spike proteins was quantitated by comparing the amounts of biotin-labeled spike proteins present at the start and at the end of the 2-h incubation period (Fig. 8, hatched bars). In parallel, the release of biotinylated spike proteins into the medium in virus particles was measured by mag-SA retrieval (Fig. 8, solid bars). The results showed a striking difference in the fate of the cell surface spike proteins at different times of infection. Early in the infection cycle, almost none of the biotinylated spike proteins were released as budded virus particles. Instead, the cell surface spike proteins of both wt and srf-3 were rapidly turned over (60 to 70% degradation within the 2-h incubation period). In contrast, later in the infection cycle, efficient budding of both wt and srf-3 spike proteins was observed (∼20% budding of the biotinylated spike proteins at the 7-h infection time), in keeping with our previous results (Fig. 6A). The degradation of biotinylated cell spike proteins was dramatically reduced, to ∼20% within the 2-h incubation period. The results with wt and srf-3 were similar at all times of infection in cholesterol-containing cells. Thus, during the infection cycle increased incorporation of cell surface spike proteins into virus particles was observed at infection times when the degradation of cell surface spike proteins was suppressed. These results suggested that efficient budding correlated with the accumulation of a critical concentration of spike proteins at the plasma membrane, which was achieved at later times of infection.
FIG. 8.
Degradation and budding of wt and srf-3 spike proteins in control cells. Cholesterol-containing C6/36 cells were infected with wt SFV (A) or srf-3 (B) at 10 PFU/cell for the indicated times. At each time of infection, the cells were pulse-labeled for 15 min with [35S]methionine-cysteine, using concentrations of 200 μCi/ml for the 3.5-h time point and 80 μCi/ml for the remaining time points. The cells were then chased for 30 min, derivatized with biotin (time zero), and incubated at 28°C for 2 h. The biotinylated spike proteins present in cell lysates and media were quantitated using mag-SA retrieval and immunoprecipitation as described in Materials and Methods. The amount of budding after 2 h (solid bars) was determined by comparing the biotinylated E2 subunit in the medium to the biotinylated E2 present in the cell lysate at time zero. The degradation of cell surface spike proteins after 2 h (hatched bars) was determined by subtracting the biotinylated E2 recovered in the cell lysate and medium from the biotinylated E2 in the cell lysate at time zero and is expressed as the percentage of the biotinylated E2 in the cell lysate at time zero. A representative example of two experiments is shown.
We next investigated if such a shift in the lifetime of the spike protein occurred in cholesterol-depleted C6/36 cells (Fig. 9). Again, time points of infection were chosen to be prior to or during the logarithmic period of infectious progeny virus production (12 to 26 h for srf-3 [62]). Since srf-3 can produce progeny virus and reinfect cells in the absence of cholesterol, infection would result in mixed cell populations at various stages of the infection cycle. To synchronize the infection cycle and prevent secondary infection, both wt- and srf-3-infected cells were incubated in the presence of 20 mM NH4Cl from 2 h postinfection until 30 min prior to pulse-labeling. These conditions did not affect the efficient release of srf-3 from depleted cells (Fig. 9B and data not shown). The infected cells were then pulse-labeled, chased, biotin derivatized, and incubated for 5 h to permit virus budding. Biotin-derivatized spike protein turnover and release in virus particles were analyzed as in Fig. 8.
FIG. 9.
Degradation and budding of wt and srf-3 spike proteins in cholesterol-depleted cells. Cholesterol-depleted C6/36 cells were infected with wt SFV at 1,000 PFU/cell or with srf-3 at 100 PFU/cell for 2 h and then incubated in the presence of 20 mM NH4Cl to inhibit secondary infection. Thirty minutes before the completion of the indicated infection time, the cells were washed and the incubation was completed in the absence of NH4Cl. The cells were then pulse-labeled for 15 min with 35S-labeled methionine-cysteine using concentrations of 200 μCi/ml for wt infected cells and 40 μCi/ml for srf-3-infected cells. The cells were then chased for 90 min, derivatized with biotin, incubated at 28°C for 5 h, and analyzed for spike protein degradation (hatched bars) and budding (solid bars) as in Fig. 8. A representative example of two experiments is shown.
Early in the infection cycle in depleted cells (12 h), although wt and srf-3 spike proteins reached the plasma membrane, the biotinylated spike proteins were efficiently degraded during the subsequent incubation (∼60 to 70% degradation during the 5-h incubation period [hatched bars]). Very little biotin-tagged spike protein was released by budding of either virus at this time of infection (Fig. 9, solid bars). Similar high levels of spike protein degradation were observed after 6 or 8 h of infection by either virus (data not shown). As infection progressed (26 h), efficient virus release was observed for srf-3, with 35% of total biotinylated spike proteins released in virus particles within a 5-h incubation period (Fig. 9B, solid bars). Degradation of biotinylated srf-3 spike proteins was markedly decreased by this point in infection. In contrast, wt biotin-tagged spike proteins were efficiently degraded throughout the infection cycle (60 to 80%), and little spike protein budding was observed (<4%) (Fig. 9A). Further incubation of wt infected cells for 48 h gave similar budding and degradation results, indicating that a longer infection time did not permit rescue of the wt spike protein (data not shown). Differential spike protein degradation thus appears to be responsible for the decrease we observed between the plasma membrane distribution of srf-3 and wt spike proteins in depleted cells (Fig. 7).
These results suggested that during virus infection the cell surface spike protein population converts from rapid degradation and inefficient budding early in the infection cycle to efficient virus release and reduced spike protein degradation later in infection. The srf-3 mutant spike protein showed this phenotype in either control or cholesterol-depleted cells, even under conditions in which secondary infection was blocked. However, the wt cell surface spike protein, although efficiently budded from control cells, was rapidly degraded at all points of the infection cycle in sterol-depleted cells and did not bud efficiently in the absence of cholesterol.
DISCUSSION
We have developed a reliable biochemical method to measure the incorporation of cell surface viral spike proteins into budded virions. This assay quantitatively retrieved intact, morphologically normal virus particles. Our studies showed that the assay was specific for budding, defined as the final steps in the assembly and pinching off of a completed virus particle from the plasma membrane. In the case of SFV, such a budding reaction presumably would include the lateral association of the spike polypeptides into trimers and higher-order complexes, the formation of circumferential spike protein-nucleocapsid interactions, and the fission of the virus membrane to produce the enveloped virion. Biotin tagging of plasma membrane spike proteins permitted the collective evaluation of these steps in the budding reaction separate from the protein biosynthetic and secretory pathways.
Our studies characterized several aspects of virus budding, including kinetics, temperature dependence, and the role of membrane transport and cytoskeletal elements. The biotin-tagged spike protein pool was continuously incorporated into virus particles for at least 2 h at 37°C. Budding, although strongly temperature dependent, still occurred at temperatures such as 20°C which inhibit some membrane trafficking events. Our results with monensin and brefeldin A demonstrated that once the nascent spike protein was transported to the plasma membrane pool, budding during a 2-h incubation period at 37°C was largely independent of further exocytic delivery to the cell surface. Although we assume that at some point treatment with membrane transport inhibitors would limit budding, our data indicate that the amount of virus budding during a 2-h incubation time (estimated from growth curves as ∼4 × 103 PFU/cell) did not sufficiently deplete the cells of membrane components to produce a block in budding. This result also provides further evidence that the budding assay monitors incorporation of cell surface spike proteins into virus particles.
Actin microfilaments appear to be involved in the maturation and assembly of several enveloped viruses, such as measles virus (55, 61) and human immunodeficiency virus (30, 45), but not in the production of vesicular stomatitis virus and influenza virus (17). Our results from budding assays and growth curve analyses suggested that actin filaments and microtubules were not required for either SFV budding or for the overall production of infectious progeny virus, including the transport of spike proteins and nucleocapsid to the site of budding.
Our previous work has described a role for cellular cholesterol in the efficient production of progeny SFV and SIN, demonstrable either by infection at high multiplicity or by RNA transfection (32–34, 62). Cholesterol-depleted infected cells express the virus genome, translate the capsid and spike proteins, and process the p62 subunit but show inefficient virus exit. Virus mutants such as srf-3 that are less cholesterol dependent for fusion and entry are also less sterol dependent for virus exit. Using the budding assay developed in these studies, we were here able to demonstrate that the block in wt virus exit from cholesterol-depleted cells was due to an inhibition of virus budding from the plasma membrane. These studies thus localized the site of the cholesterol-dependent block to a late stage in virus exit at the cell surface and showed that budding from sterol-depleted cells was increased in the srf-3 mutant. Pulse-chase analysis of depleted infected cells demonstrated that the wt and mutant SFV spike proteins were equivalently dimerized and transported through the exocytic pathway to the plasma membrane. However, the wt spike protein was rapidly degraded (t1/2 of <2 h) and showed a lower steady-state distribution on the plasma membrane compared to the more efficiently budding srf-3 mutant. Rapid turnover of cell surface spike proteins was not simply due to the absence of cholesterol, since at early times of infection both wt and srf-3 spike proteins were rapidly degraded in either control or depleted cells. As infection progressed, the spike protein degradation rate markedly decreased for wt virus in control cells and for srf-3 in either control or depleted cells, and abundant virus budding was observed. Thus, the behavior of the wt spike protein in depleted cells was unusual in that rapid spike protein degradation was maintained for the duration of infection. These results demonstrated a strong inverse correlation between the spike protein degradation rate and virus budding. Although the relationship between these two events was striking, it is not yet clear whether rapid degradation of cell surface spike proteins is a cause or an effect of decreased virus budding.
Rapid turnover of SFV spike proteins has been previously described under conditions in which budding is impaired, such as expression of the spike in the absence of capsid protein (10, 66). Although at least a proportion of the degradation occurs after the spike protein is delivered to the plasma membrane (10, 66), efficient endocytic uptake of the cell surface E2 subunit was not detected (66). Thus, the site of spike protein degradation in these experiments is not clear. In contrast, the gp160 envelope proteins of human and simian immunodeficiency viruses are efficiently endocytosed from the plasma membrane pool via interaction of a highly conserved tyrosine in the cytoplasmic tail with adapter complexes and clathrin-coated pits (2). The internalization of gp160 decreases the spike protein concentration at the plasma membrane and in the budded virus particle, reduces syncytia formation (46), and may be important in decreasing the susceptibility of virus-infected cells to the humoral immune response (46). Interestingly, at least one report suggests that gp160 internalization is blocked by association with the Gag precursor protein (11), raising the possibility that Gag interaction may suppress endocytosis and redirect gp160 into budding virions. In contrast, the wt form of the influenza virus hemagglutinin (HA) is normally endocytosed at a very low rate and has a half-life of ca. 8 to 10 h (40, 68), similar to that of typical plasma membrane proteins. Insertion of a tyrosine-based internalization motif into the HA cytoplasmic tail causes the protein to be very rapidly endocytosed, and further signals mediate the rapid intracellular degradation of HA in late endosomes and/or lysosomes (t1/2 of ∼2 to 3 h) (68). Thus, it is clear that viral spike proteins can go through several different pathways at the cell surface, including rapid degradation through endocytic clearance, and long-lived protein maintenance in the absence of endocytosis.
Importantly, our data demonstrated that even when the entire SFV wt virus genome was expressed, wt cell surface spike proteins were rapidly degraded at early times of infection or in sterol-depleted cells. This result was unexpected and differs from previous experiments in which SFV spikes were degraded when expressed in the absence of nucleocapsid (66) or in which mutant virus spike proteins were degraded (10). To address the relationship between the degradation of cell surface SFV spike proteins and virus budding, we attempted to block turnover of the wt spike protein in sterol-depleted cells. Cells were treated with the weak bases NH4Cl or chloroquine or a protease inhibitor cocktail containing leupeptin, pepstatin, and aprotinin. No significant increase in the half-life of the spike protein or in virus budding was observed (Lu and Kielian, unpublished data). This experiment was complicated by the fact that, even if SFV spike proteins were being cleared from the cell surface by endocytosis, blocking degradation alone might not return the spike protein to the plasma membrane in quantities sufficient to promote budding. Thus, it is not yet clear what role endocytic uptake of the SFV spike protein may play in its degradation or whether degradation controls the budding-competent concentration of the spike protein at the plasma membrane.
How might cholesterol influence the budding of the SFV spike protein? One possibility is that the spike protein requires cholesterol-enriched regions of the membrane for budding and that the srf-3 mutation makes the spike less dependent on such regions. Within cellular membranes, cholesterol plays an important role in the formation of cholesterol and sphingolipid-enriched lipid domains termed “rafts,” operationally assayed by their resistance to solubilization with the detergent Triton X-100 at 4°C (4, 5, 50). A variety of experiments have demonstrated that the influenza virus HA associates preferentially with rafts via the HA transmembrane domain (49) and that influenza virus buds from and is enriched in raft domains (48). In contrast, although SFV is dependent on cellular cholesterol for efficient budding, the SFV spike protein is not associated with detergent-resistant rafts and these domains are not enriched in the SFV particle (48; Lu and Kielian, unpublished data). Thus, the reason for the alphavirus cholesterol requirement in budding does not appear to be due to a requirement for raft domains, and a suggestion for the role of cholesterol may come from studies of the alphavirus srf mutants.
To date, all of the SFV and SIN mutants that display increased fusion and infection of cholesterol-depleted cells also show increased exit from depleted cells (32, 34, 62; P. K. Chatterjee and M. Kielian, unpublished data). These data suggest a connection between the sterol dependence of membrane fusion and that of virus exit. The SFV E1 protein undergoes several distinct conformational changes during the low pH-triggered fusion reaction, and the kinetics and efficiency of these changes are increased by the presence of cholesterol in the target membrane (6, 8). Studies of srf-3 have shown that the cholesterol independence of its membrane fusion reaction is controlled by the increased cholesterol independence of these E1 conformational changes (6). Thus, the presence of cholesterol in the target membrane during fusion functionally affects the conformation of the virus E1 subunit. It is possible that during exit cholesterol promotes the formation of a budding-competent conformation of the SFV spike protein and that the srf-3 mutant is less cholesterol dependent for production of such active spike proteins. This budding-competent, cholesterol-enhanced conformation of the spike protein could represent the spike protein trimer or higher-order, laterally interacting spike protein oligomers. The formation of such lateral interactions would be expected to be critical for the productive association of the spike with the nucleocapsid during budding. The rapid degradation of the wt and srf-3 spike proteins at early times of infection, or of wt spike protein in cholesterol-depleted cells, could be a secondary effect produced by the lack of such lateral interactions. In this scenario, decreased lateral interactions would be caused by either the low level of spike protein expression at the plasma membrane early in infection or by the absence of cholesterol in the case of the wt spike protein. Alternatively, it is also possible that the formation of lateral interactions is directly controlled by the degradation rate of the spike protein via its regulation of the amount of spikes at the plasma membrane.
Our studies on virus budding also offer insights into cellular membrane budding processes, including the role of lipids in the budding reaction. Vesicular trafficking mediates the transport and sorting of cargo and membrane proteins during endocytosis and exocytosis in eukaryotic cells. A key step in this traffic is the formation of vesicles through the process of vesicle budding (see reference 54 for review). Although our understanding of vesicle budding is incomplete, formation of exocytic transport vesicles has been shown to involve the formation of a complex containing coat components, priming membrane proteins, and small GTPases. Polymerization of the vesicle coat appears to lead to membrane deformation and release of a budded vesicle. Studies to date also suggest that specific lipids carry out important functions during the formation of membrane vesicles (9, 35). For example, the formation of clathrin-coated endocytic vesicles has been shown to be inhibited by depletion of cholesterol from the plasma membrane, the site of this budding reaction (39, 58). A recent study reported that the membrane protein synaptophysin specifically interacts with cholesterol and suggested that the generation of neurosecretory vesicles may be driven by lateral associations of cholesterol and synaptophysin in the forming membrane bud (60). Thus, a precedent for a role of cholesterol-protein interactions in the formation of highly curved membrane structures is found in the budding reactions of several types of cellular vesicles. As our mechanistic knowledge of these processes evolves, it will be interesting to follow the similarities and differences between the functions of cholesterol in alphavirus budding and in cellular membrane budding reactions.
Taken together, our studies have established an assay for the budding of SFV spike proteins from the cell surface into virus particles. We have also demonstrated that this assay can be adapted to other alphaviruses, such as SIN, as well as to other viruses, such as the rhabdovirus vesicular stomatitis virus (Lu and Kielian, unpublished data). Our results localized the inhibition of SFV exit in cholesterol-depleted cells to the virus budding step and demonstrated that a relatively cholesterol-independent SFV mutant is more permissive for budding from sterol-depleted cells. A striking correlation of efficient budding with decreased spike protein turnover was observed. Future experiments will address spike protein endocytic traffic and the mechanism of rapid spike protein turnover and their importance in the regulation of virus budding. In addition, the sustained budding of biotin-tagged spike proteins from SFV-infected cells and its relative insensitivity to continued cellular secretion suggest the possibility of adapting this assay for use in a semipermeabilized cell system. In vitro systems based on semipermeabilized cells have been used to reconstitute and characterize many cellular reactions involving the budding and fusion of vesicular carriers (reviewed in reference 41). Such an assay would be invaluable for the experimental manipulation and study of virus budding and has not yet been reported for any enveloped animal virus.
ACKNOWLEDGMENTS
We thank Anna Ahn and Christina Eng for technical assistance and Frank Macaluso and the members of the Analytical Imaging Facility of The Albert Einstein College of Medicine for assistance with electron microscopy. We thank Marianne Marquardt for her contributions to exploring the role of spike protein transport and dimer formation in SFV budding. We also thank the members of our lab for helpful discussions and suggestions and Duncan Wilson and the members of our lab for critical reading of the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM57454), by the Jack K. and Helen B. Lazar fellowship in Cell Biology, and by Cancer Center Core Support Grant NIH/NCI P30-CA13330.
REFERENCES
- 1.Barth B U, Garoff H. The nucleocapsid-binding spike subunit E2 of Semliki Forest virus requires complex formation with the E1 subunit for activity. J Virol. 1997;71:7857–7865. doi: 10.1128/jvi.71.10.7857-7865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom G S, Goldstein L S B. Cruising along microtubule highways: how membranes move through the secretory pathway. J Cell Biol. 1998;140:1277–1280. doi: 10.1083/jcb.140.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D A, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 5.Brown D A, Rose J K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee P K, Vashishtha M, Kielian M. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J Virol. 2000;74:1623–1631. doi: 10.1128/jvi.74.4.1623-1631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng R H, Kuhn R J, Olson N H, Rossman M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corver J. Membrane fusion activity of Semliki Forest virus. Ph.D. thesis. Groningen, The Netherlands: Groningen University; 1998. [Google Scholar]
- 9.De Camilli P, Emr S D, McPherson P S, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 10.Duffus W A, Levy-Mintz P, Klimjack M R, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekstrom M, Liljeström P, Garoff H. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 1994;13:1058–1064. doi: 10.1002/j.1460-2075.1994.tb06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 14.Garoff H, Hewson R, Opstelten D-J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garoff H, Wilschut J, Liljeström P, Wahlberg J M, Bron R, Suomalainen M, Smyth J, Salminen A, Barth B U, Zhao H. Assembly and entry mechanisms of Semliki Forest virus. Arch Virol. 1994;9:329–338. doi: 10.1007/978-3-7091-9326-6_33. [DOI] [PubMed] [Google Scholar]
- 15a.Gibbons D L, Ahn A, Chatterjee P K, Kielian M. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J Virol. 2000;74:7772–7780. doi: 10.1128/jvi.74.17.7772-7780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glomb-Reinmund S, Kielian M. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J Virol. 1998;72:4281–4287. doi: 10.1128/jvi.72.5.4281-4287.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin J A, Compans R W. Effect of cytochalasin B on the maturation of enveloped viruses. J Exp Med. 1979;150:379–391. doi: 10.1084/jem.150.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths G, Quinn P, Warren G. Dissection of the Golgi complex. I. Monensin inhibits the transport of viral membrane proteins from medial to trans Golgi cisternae in baby hamster kidney cells infected with Semliki forest virus. J Cell Biol. 1983;96:835–850. doi: 10.1083/jcb.96.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Justman J, Klimjack M R, Kielian M. Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol. 1993;67:7597–7607. doi: 10.1128/jvi.67.12.7597-7607.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 21.Kielian M, Chatterjee P K, Gibbons D L, Lu Y E. Specific roles for lipids in virus fusion and exit: Examples from the alphaviruses. In: Hilderson H, Fuller S, editors. Subcellular biochemistry. 34. Fusion of biological membranes and related problems. New York, N.Y: Plenum Publishers; 2000. pp. 409–455. [DOI] [PubMed] [Google Scholar]
- 22.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian M, Jungerwirth S, Sayad K U, DeCandido S. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian M C, Helenius A. The role of cholesterol in the fusion of Semliki Forest virus with membranes. J Virol. 1984;52:281–283. doi: 10.1128/jvi.52.1.281-283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielian M C, Keränen S, Kääriäinen L, Helenius A. Membrane fusion mutants of Semliki Forest virus. J Cell Biol. 1984;98:139–145. doi: 10.1083/jcb.98.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klausner R D, Donaldson J G, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Owen K E, Choi H-K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 29.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Dai R, Tian C J, Dawson L, Gorelick R, Yu X F. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez S, Yao J-S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y E, Cassese T, Kielian M. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J Virol. 1999;73:4272–4278. doi: 10.1128/jvi.73.5.4272-4278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquardt M T, Kielian M. Cholesterol-depleted cells that are relatively permissive for Semliki Forest virus infection. Virology. 1996;224:198–205. doi: 10.1006/viro.1996.0521. [DOI] [PubMed] [Google Scholar]
- 34.Marquardt M T, Phalen T, Kielian M. Cholesterol is required in the exit pathway of Semliki Forest virus. J Cell Biol. 1993;123:57–65. doi: 10.1083/jcb.123.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin T F J. Greasing the Golgi budding machine. Nature. 1997;387:21–22. doi: 10.1038/387021a0. [DOI] [PubMed] [Google Scholar]
- 36.Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 37.Nieva J L, Bron R, Corver J, Wilschut J. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 1994;13:2797–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodal S D, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated vesicles. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth M G, Henis Y I, Brewer C B, Ktistakis N T, Shia S-P, Lazarovits J, Fire E, Thomas D, Zwart D E. Sorting of membrane proteins in the endocytic and exocytic pathways. In: Rupp R, Oka M, editors. Cell biology and biotechnology. New York, N.Y: Springer-Verlag; 1993. pp. 137–156. [Google Scholar]
- 41.Rothman J E, editor. Reconstitution of intracellular transport. Methods Enzymol. 1992;219:1–438. [Google Scholar]
- 42.Saraste J, Kuismanen E. Pre-Golgi and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- 43.Saraste J, von Bonsdorff C-H, Hashimoto K, Kääriäinen L, Keränen S. Semliki Forest virus mutants with temperature-sensitive defect of envelope proteins. Virology. 1980;100:229–245. doi: 10.1016/0042-6822(80)90516-4. [DOI] [PubMed] [Google Scholar]
- 44.Sariola M, Saraste J, Kuismanen E. Communication of post-Golgi elements with early endocytic pathway: regulation of endoproteolytic cleavage of Semliki Forest virus p62 precursor. J Cell Sci. 1995;108:2465–2475. doi: 10.1242/jcs.108.6.2465. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Biol Chem. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheele C M, Pfefferkorn E R. Kinetics of incorporation of structural proteins into Sindbis virions. J Virol. 1969;3:369–375. doi: 10.1128/jvi.3.4.369-375.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 49.Scheiffele P, Roth M G, Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 51.Singh I R, Suomalainen M, Varadarajan S, Garoff H, Helenius A. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology. 1997;231:59–71. doi: 10.1006/viro.1997.8492. [DOI] [PubMed] [Google Scholar]
- 52.Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 53.Smith J F, Brown D T. Envelopment of Sindbis virus: synthesis and organization of proteins in cells infected with wild-type and maturation-defective mutants. J Virol. 1977;22:662–678. doi: 10.1128/jvi.22.3.662-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Springer S, Spang A, Schekman R. A primer on vesicle budding. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 55.Stallcup K C, Raine C S, Fields B N. Cytochalasin B inhibits the maturation of measles virus. Virology. 1983;124:59–74. doi: 10.1016/0042-6822(83)90290-8. [DOI] [PubMed] [Google Scholar]
- 56.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strauss J H, Strauss E G, Kuhn R J. Budding of alphaviruses. Trends Microbiol. 1995;3:346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 58.Subtil A, Gaidarov I, Kobylarz K, Lampson M A, Keen J H, McGraw T E. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci USA. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiele C, Hannah M J, Fahrenholz F, Huttner W B. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- 61.Tyrrell D L, Ehrnst A. Transmembrane communication in cells chronically infected with measles virus. J Cell Biol. 1979;81:396–402. doi: 10.1083/jcb.81.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vashishtha M, Phalen T, Marquardt M T, Ryu J S, Ng A C, Kielian M. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J Cell Biol. 1998;140:91–99. doi: 10.1083/jcb.140.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wahlberg J M, Boere W A M, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waite N R F, Brown D T, Pfefferkorn E R. Inhibition of Sindbis virus release by media of low ionic strength: intracellular events and requirements for reversal. J Virol. 1972;10:537–544. doi: 10.1128/jvi.10.3.537-544.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White J, Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci USA. 1980;77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H, Garoff H. Role of cell surface spikes in alphavirus budding. J Virol. 1992;66:7089–7095. doi: 10.1128/jvi.66.12.7089-7095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C-H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwart D E, Brewer C B, Lazarovits J, Henis Y I, Roth M G. Degradation of mutant influenza virus hemagglutinins is influenced by cytoplasmic sequences independent of internalization signals. J Biol Chem. 1996;271:907–917. doi: 10.1074/jbc.271.2.907. [DOI] [PubMed] [Google Scholar]