Abstract
We have cloned the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Here, we have subjected the HCMV BAC to random transposon (Tn) mutagenesis using a Tn1721-derived insertion sequence and have provided the conditions for excision of the BAC cassette. We report on a fast and efficient screening procedure for a Tn insertion library. Bacterial clones containing randomly mutated full-length HCMV genomes were transferred into 96-well microtiter plates. A PCR screening method based on two Tn primers and one primer specific for the desired genomic position of the Tn insertion was established. Within three consecutive rounds of PCR a Tn insertion of interest can be assigned to a specific bacterial clone. We applied this method to retrieve mutants of HCMV envelope glycoprotein genes. To determine the infectivities of the mutant HCMV genomes, the DNA of the identified BACs was transfected into permissive fibroblasts. In contrast to BACs with mutations in the genes coding for gB, gH, gL, and gM, which did not yield infectious virus, BACs with disruptions of open reading frame UL4 (gp48) or UL74 (gO) were viable, although gO-deficient viruses showed a severe growth deficit. Thus, gO (UL74), a component of the glycoprotein complex III, is dispensable for viral growth. We conclude that our approach of PCR screening for Tn insertions will greatly facilitate the functional analysis of herpesvirus genomes.
Human cytomegalovirus (HCMV) infection is widespread and usually without symptoms in healthy adults, but it can cause severe disease in the immunologically immature or immunodeficient host and is a leading cause of birth abnormalities in industrialized countries (7). Its 230-kbp linear double-stranded DNA genome is among the largest in the herpesvirus family, encoding well over 150 proteins. The nucleic acid sequence predicts about 55 open reading frames (ORFs) coding for transmembrane glycoproteins (10). Some of these glycoproteins modulate the communication of the infected cell with the host's immune system (20, 53). Only a limited number of glycoproteins represent virion components (8, 23). The envelope of HCMV consists of at least three distinct types of covalently linked glycoprotein complexes (18). The homology to herpes simplex virus type 1 (HSV-1) virion glycoproteins indicates a certain degree of conservation among herpesvirus glycoproteins (8). While the 12 HSV-1 glycoprotein ORFs have already been subjected to extensive mutational analysis (43), their HCMV counterparts so far have escaped such studies in the context of viral infection (8).
F-factor-based bacterial artificial chromosomes (BACs) can efficiently be used for the propagation of the genomes of large recombinant DNA viruses in Escherichia coli (36). Cloning a herpesvirus genome, exemplified by the murine CMV (MCMV) genome, as an infectious BAC made herpesviruses generally accessible to the methods of bacterial genetics (38). By now, several other herpesvirus genomes have been cloned as BACs, representing members of alpha-, beta-, and gammaherpesviruses (1, 6, 14, 21, 44, 47, 51; G. Hahn, M. Mach, M. Messerle, and U. H. Koszinowski, Abstr. 24th Int. Herpesvirus Workshop, abstr. 13.013, 1999). Transposons (Tn) are well established tools for random insertion mutagenesis of bacterial genomes (4). Using the MCMV BAC as an example, we have introduced this method for the random mutagenesis of cloned herpesvirus genomes (9). More recently, the general feasibility of this approach has been independently confirmed by others with a different Tn system (47).
Here, we report on the application of this technique for the mutational analysis of the full-length infectious genome of HCMV. We have developed a fast screening procedure for a Tn insertion library of HCMV genomes. For retrieval of Tn insertions in a gene of interest, only three consecutive rounds of PCR analysis on hierarchically pooled DNA samples are required. To demonstrate the efficacy of the method, we retrieved and analyzed mutants of known HCMV envelope glycoprotein genes. This method should significantly speed up the access to genomes with mutations within specific ORFs and thus facilitate the assignment of specific functions to individual herpesvirus genes.
MATERIALS AND METHODS
Recombinant viruses and cells.
Virus propagation and viral DNA extraction were essentially performed as previously described (6). MRC-5 cells (human fetal lung fibroblasts; BioWhittaker, Verviers, Belgium) were used for transfection and propagation of reconstituted viruses. The original HCMV BAC, referred to as pHB-5, represents an infectious derivative of AD169 (American Type Culture Collection) lacking the genes US2 to US6 (nucleotides [nt] 193360 to 196045) (6). Nucleotide numbering and delineation of ORFs are given according to the sequence published by Chee et al. (10) (GenBank accession no. X17403), irrespective of the additional 929 bp contained in the EcoRI c′ fragment of the cloned virus (13, 39).
Plasmid construction.
To reinsert the deleted US2 to US6 genes and to make the BAC cassette, a fusion construct of pMBO131 (40) with a gpt selection marker (17) excisable from the genome, the following plasmids were used for homologous recombination in bacteria. For construction of pUH-16, one loxP site, excised as a PstI/SacI fragment (5′-ctc gag ctc cac cgc ggt ggc ggc cac gga tgc atc cgt ggc cgc GCA TAA CTT CGT ATA GCA TAC ATT ATA CGA AGT TAT cta gca gat ctg cag-3′) from pllNsi (M. Messerle, unpublished), a derivative of pP4 (12), was cloned into pSL301 (Invitrogen, San Diego, Calif.). The loxP site was flanked with homologous sequences from HCMV (a BssHII/NheI fragment spanning nt 191395 to 193360 of AD169) and the BAC cassette (an XbaI/SpeI fragment excised from pEB1097 [6]) (see Fig. 1C). From this construct BamHI and MscI fragments were excised and placed between the BamHI and SmaI sites of shuttle plasmid pST76K-SR (kindly supplied by G. Pósfai, Szeged, Hungary). pST76K-SR carries the recA and sacB genes and allows mutagenesis of BAC plasmids in the recombination-deficient E. coli strain DH10B to be performed essentially as described in reference 55. For construction of pUH-15, one loxP site was excised from pllNsi by digestion with SacI and EcoRI and inserted into an oligonucleotide linker (5′-cat gGA TCC GCG GCC GCT TTC TCG AGC TCA TGC ATT TTG AAT TCG GCG CGC CTT TTC TAG AGG ATC CAa gct-3′), replacing the multiple cloning site (NcoI-HindIII) of pSL301. Next, a PstI fragment excised from pMin-1 (26), which codes for the RP4 origin of transfer, was inserted into a unique PstI site imported with the loxP fragment. Subsequently, US2 to US6 (nt 193003 to 198197) were added as a XhoI/EagI fragment of pCM1052 (16). After addition of the BAC homology domain (AscI to XbaI of pEB1097) the whole insert was excised with BamHI and transferred into shuttle vector pST76K-SR. Allelic exchange (see below) of pHB-5 with pUH-16 and pUH-15 yielded HB-5loxP and AD169-BAC, respectively (Fig. 1C).
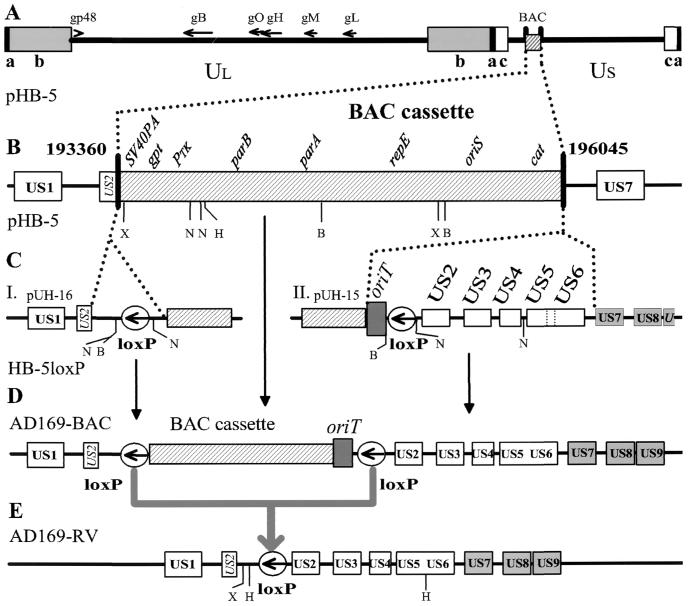
FIG. 1.
Completion of the full-length HCMV BAC and excision of the BAC cassette. (A) In HCMV BAC plasmid pHB-5, the BAC cassette replaces genes US2 through US6 (nt 193360 to 196045). (B) Details of the region of BAC insertion. X, XbaI; N, NsiI; H, HpaI; B, BglII. (C) For construction of HB-5loxP, a loxP site flanked with 2.0-kbp fragments homologous to HCMV and BAC sequences was cloned into pST76K-SR. Site-directed introduction into the HCMV genome by shuttle mutagenesis yielded an additional BglII site and two additional NsiI sites (I). Subsequently, the genes US2 to US6 were introduced by shuttle mutagenesis together with a loxP site and an RP4 origin of transfer (oriT) (II). (D) Cotransfection of AD169-BAC with a plasmid expressing recombinase Cre into permissive cells leads to the removal of the BAC cassette from the HCMV genome. (E) The virus progeny AD169-RV is distinguishable from AD169-wt by XbaI and HpaI restriction sites located next to the loxP site.
Tn mutagenesis.
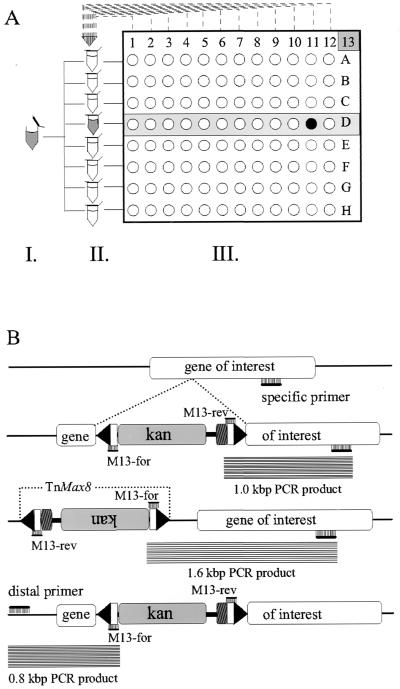
Insertion mutagenesis was performed as previously described (9). Briefly, the temperature-sensitive Tn donor plasmid pTsTM8 was electroporated into E. coli strain DH10B harboring AD169-BAC and plated at 30°C on Luria-Bertani (LB) agar plates containing chloramphenicol (13.6 μg/ml) and ampicillin (100 μg/ml). Bacterial clones containing both the HCMV BAC plasmid and the Tn donor plasmid were grown as liquid cultures at 30°C in the presence of both antibiotics. Small aliquots (approximately 2 μl per plate) were spread on LB agar plates at 43°C and selected with chloramphenicol and kanamycin (50 μg/ml) for transposition events. Bacterial colonies (about 200 per plate) were replated for another round of purification at 43°C in the presence of chloramphenicol and kanamycin and were then grown as liquid cultures in 96-well microtiter plates. Aliquots of the liquid culture of individual bacterial clones were pooled by rows, and DNA was prepared by following the alkaline lysis protocol (45) to generate DNA pools. DNA pools of the microtiter plate were generated by combining samples of the eight DNA row pools (see Fig. 3A).
FIG. 3.
Schematic outline for the PCR-based screening procedure for Tn insertions. (A) Three rounds of PCR were performed on the hierarchically pooled aliquots of the Tn library to locate a candidate mutant to a specific well in one of the 96-well plates. (B) A primer binding specifically to a genomic position of interest and the Tn-specific primers M13-for and M13-rev were included in the reaction. If an insertion event occurred close to the position of the specific primer binding site, a PCR product was generated irrespective of the orientation of the Tn. kan, ORF encoding the kanamycin resistance marker; upside down lettering indicates orientation opposite to that of the two other ORFs shown. The size of the PCR amplificate indicates whether the insertion lies within or outside the area of interest. In a confirmatory PCR with a distal primer of opposite orientation, mutants were checked for the absence of deletions at the Tn insertion site.
PCR screening for Tn insertions within specific genes.
For the detection of Tn insertions at positions of interest, three rounds of PCR were performed (PCR conditions: 7 min at 94°C, followed by 40 cycles of 16 at 95°C, 11 s at 55°C, and 1 to 2 min at 72°C, using AmpliTaq Gold and the GeneAmp 9700 PCR cycler [Perkin-Elmer, Darmstadt, Germany]). The Tn can insert in both orientations into the BAC (see Fig. 3A). Therefore, two Tn-specific primers (M13-for and M13-rev) were included in the reaction. The position-specific search primers used in this study are listed in Table 1. In the first round of PCR, the DNA pools, each representing all mutants contained in one microtiter plate, were examined. In the second round, the DNA pools representing the mutants stored in specific rows of those individual plates that tested positive in the first round were screened. From the individual wells of the rows identified as positive, crude DNA extracts were made by boiling aliquots of the frozen bacteria briefly in 0.1% Triton X-100 (45). These DNA samples were subjected to a third round of PCR screening in order to determine the exact location of the desired mutant within the Tn library. With a second distal primer oriented in the opposite direction (listed in Table 1) and located about 2 kbp from the search primer, a confirmatory PCR was performed to exclude illegitimate deletion events at the Tn integration site. The exact location of the Tn insertion was determined by restriction enzyme analysis and direct sequencing of BAC DNA with primers M13-for (5′-GCC GCT GTA AAA CGA CGG CCA GT-3′) and M13-rev (5′-GGC CGC AGG AAA CAG CTA TGA CC-3′) as described previously (9).
TABLE 1.
Search primers used for the identification of Tn insertions into envelope glycoprotein genes
| Primer | Sequence |
|---|---|
| UL55-for | 5′-AAA ACA TAG CGG ACC GTG AG-3′ |
| UL55-afo | 5′-GCA AGG CAT CAA GCA AAA ATC-3′ |
| UL55-are | 5′-CAC CGA TTC CAT GCT GGA C-3′ |
| UL55-rev | 5′-AGT GGC GAC GTG CCA ACA GC-3′ |
| UL75-for | 5′-AGA CCC ATA ACA GTA CCT CG-3′ |
| UL75-rev | 5′-ATC TCC GCA GAG CGT TCC CC-3′ |
| UL100-are | 5′-TTT GTG TGT GTT TGC GCC-3′ |
| UL100-rev | 5′-CCT CTA GAA GGC CGT ACC AG-3′ |
| UL115-are | 5′-GGA CAC AGA TAG CTC CAG-3′ |
| UL115-rev | 5′-AGG ACG ACG ACG AGT ACG AC-3′ |
| UL4-for | 5′-GCG ATG AGT CCA TAA AGC ACC-3′ |
| UL4-rev | 5′-CCA ACC ACA CAA AAG ACA ACA-3′ |
| UL74-for | 5′-CTA CGA CAT TGC TGC TTC-3′ |
| UL74-rev | 5′-TCG TTG TAA TAA AGT ACA CGC C-3′ |
Allelic exchange.
For the generation of revertant BACs, 4 to 5 kbp of viral DNA was excised from BAC pHB-5 using appropriate restriction enzymes and cloned into plasmid pST76A-SR, a derivative of pST76A (41) carrying the recA and sacB genes from pST76K-SR (M. Wagner and C. Ménard, unpublished data). Plasmids pUH-25 to pUH-34, which were used for the construction of revertants, are listed in Table 2. The principles of recA-mediated allelic replacement in E. coli have been described elsewhere (38, 40, 55). Resolution of cointegrates was significantly improved by counterselecting against sacB (6, 22). Briefly, temperature-sensitive shuttle plasmids (pUH-15, -16, and -25 to -34) were cotransformed with the HCMV BAC plasmid into electrocompetent E. coli DH10B, and transformants were selected at 30°C on agar plates containing chloramphenicol, ampicillin, and/or kanamycin. Cultivation at nonpermissive 43°C in the presence of chloramphenicol and ampicillin or kanamycin led to the selection of clones containing cointegrates. By recA-mediated recombination, cointegrates are resolved with an ∼50% chance to either the initial BAC or the BAC variant carrying the desired mutation (38). Resolved cointegrates were selected for in the presence of 5% sucrose at 30°C by making use of the sacB gene carried on the shuttle plasmid (5, 6). To confirm allelic replacement and loss of the Tn insertion, potentially positive colonies were replica plated on chloramphenicol and kanamycin plates and screened for kanamycin-sensitive clones.
TABLE 2.
Plasmids used for the generation of revertant genomes
| Plasmid | Enzyme 1a | Nucleotide position | Enzyme 2a | Nucleotide position |
|---|---|---|---|---|
| pUH-25 | EcoRV (SmaI) | 105127 | PstI (NsiI) | 110694 |
| pUH-26 | SpeI (XbaI) | 80099 | XhoI (XhoI) | 84790 |
| pUH-27 | SphI (SphI) | 159669 | NheI (NheI) | 168330 |
| pUH-28 | SphI (SphI) | 107919 | SalI (SalI) | 112129 |
| pUH-29 | KpnI (KpnI) | 144435 | KpnI (KpnI) | 147618 |
| pUH-30 | XhoI (XhoI) | 79366 | SstI (SstI) | 83906 |
| pUH-31 | NsiI (NsiI) | 81000 | Eco47III (SmaI) | 85583 |
| pUH-32 | SnaBI (HpaI) | 11427 | SstI (SstI) | 15544 |
| pUH-33 | SnaBI (HpaI) | 105042 | SnaBI (HpaI) | 109189 |
| pUH-34 | XbaI (XbaI) | 106258 | PmlI (HpaI) | 110389 |
Enzymes used for cleaving shuttle vector pST76A-SR are in parentheses.
Reconstitution of BAC cassette-free mutant viruses.
One microgram of BAC DNA purified on Nucleobond columns (Macherey Nagel, Düren, Germany) was cotransfected with 0.3 μg of pcDNApp71tag, a plasmid expressing pp71, for enhancement of infectivity (3, 6, 35) (kindly provided by B. Plachter, Mainz, Germany) and 0.5 μg of plasmid pBRep-Cre using 12 μl of Superfect according to the manufacturer's instructions (Qiagen, Hilden, Germany). pBRep-Cre contains an XhoI fragment of pMC-Cre (19) fused to pBRep (W. Brune, unpublished) expressing recombinase Cre for excision of the BAC cassette. In parallel, transfections were performed with complementing plasmids carrying the respective HCMV DNA fragment that spans the Tn insertion site (pUH-25 to pUH-34; listed above). Five days posttransfection cells were split 1:3. A marked cytopathic effect usually became apparent 10 to 12 days posttransfection. When no plaques became visible, cells were split 1:3 at day 15 after transfection. Cells were monitored for 1 month after transfection. When after repeated transfection experiments no plaques could be obtained unless the complementing plasmid was included, the continuity of the sequence interrupted by Tn insertion was classified as essential for virus growth.
RESULTS
Completion of the HCMV BAC.
We wanted to construct a Tn insertion library with a full-length HCMV AD169 genome. Published infectious BAC clone pHB-5 was constructed by deleting genes US2 to US6 to accommodate the BAC cassette and to restrict the overlength of the pHB-5 BAC genome to about 5 kbp (6). Tn insertion adds an additional 2 kbp of overlength, and the reintroduction of the missing US2-to-US6 sequence would result in a propensity for random deletions imposed by the packaging restraints of the virion during virus reconstitution (49) (data not shown). Therefore, in order to minimize genome instability, the BAC cassette was made excisable by attached loxP sites, similar to the completion of the MCMV BAC and the pseudorabies virus (PrV) BAC (48, 54).
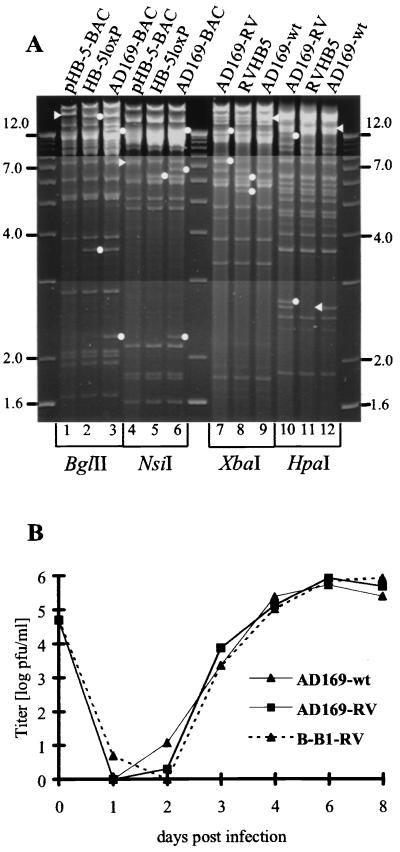
Homologous recombination of pHB-5 with pUH-16 and pUH-15 in E. coli yielded HB-5loxP and AD169-BAC, respectively (Fig. 1). Recombinase Cre-mediated excision of the BAC cassette yielded AD169-RV, a viral genome reduced to only a small amount of overlength with respect to the wild-type (wt) genome (Fig. 1E). To avoid the disruption of upstream regulatory sequences of US1 by the retained loxP site, a small duplication surrounding the loxP site was introduced. The intermediate construct HB-5loxP (Fig. 1C), containing a single loxP site, and AD169-BAC were digested with BglII and NsiI and compared with plasmid pHB-5 (Fig. 2A, lanes 1 to 6). Upon introduction of a loxP site into pHB-5, a 25.5-kbp BglII fragment (Fig. 2A, lane 1) was cleaved into two subfragments of 3.6 and 22.1 kbp (Fig. 2A, lane 2). Subsequent introduction of genes US2 to US6 resulted in additional bands at 13.7 and 2.2 kbp (Fig. 2A, lane 3). The constructions outlined in Fig. 1 converted a 7.1-kbp NsiI fragment of pHB-5 (Fig. 2A, lane 4) into two new fragments of 6.2 and 1.1 kbp (Fig. 2A, lane 5). Further manipulation of HB-5loxP as depicted in Fig. 1C yielded additional NsiI fragments of 2.2, 6.7, and 13.0 kbp (Fig. 2A, lane 6).
FIG. 2.
Recombinase Cre-mediated excision of the BAC vector sequences results in a nearly wt AD169-RV. (A) Lanes 1 to 6, restriction enzyme digests of HCMV BAC plasmids pHB-5, HB-5loxP, and AD169-BAC isolated from bacteria; lanes 7 to 12, digestion of viral DNA extracted from cells infected with viruses reconstituted from BAC plasmids AD169-BAC (AD169-RV) and pHB-5 (RVHB5), or the parental strain AD169-wt. Relevant restriction sites are depicted in Fig. 1. Arrowheads, restriction endonuclease fragments; dots, additional fragments resulting from manipulations described in the legend for Fig. 1. (B) Single-step growth curve of recombinant viruses. AD169-RV is compared to parental HCMV strain AD169-wt and B-B1-RV, an AD169-BAC-based mutant virus with the Tn sequences stably integrated at the 3′-terminal end of UL78, nt 114124. MRC-5 cells were infected at an MOI of 0.1. Virus titers from cells and supernatant were determined in duplicate by a standard plaque assay at the indicated time points.
Transfection of the completed AD169-BAC into human embryonic fibroblast cells followed by recombinase Cre-mediated excision of the BAC cassette should yield recombinant virus AD169-RV (Fig. 1E). Comparison of the DNAs of the recombinant viruses AD169-RV and RVHB5 (6) with that of the parental strain, AD169 (Fig. 2A, lanes 7 to 12) displayed the alterations of the restriction patterns as predicted in Fig. 1C. A 18.9-kbp XbaI fragment present in prototype AD169 (Fig. 2A, lane 9) was cleaved into two subfragments of 7.5 and 11.8 kbp (Fig. 2A, lane 7). In RVHB5 these fragments were absent, and 6.3- and 5.7-kbp fragments of the BAC cassette were retained in the viral DNA (Fig. 2A, lane 8). The single 13.3-kbp HpaI fragment of wt AD169 (AD 169-wt) (Fig. 2A, lane 12) yielded, due to the insertion of an HpaI site, the 2.7- and 11.1-kbp HpaI fragments of AD169-RV (Fig. 2A, lane 10), whereas the 2.6-kbp HpaI fragment carrying US6 was absent from recombinant virus RVHB5 (lane 11). Recombinant virus AD169-RV showed growth properties indistinguishable from those of AD169-wt (Fig. 2B). Therefore, the completed AD169-BAC provided a useful substrate for Tn mutagenesis. Single Tn insertions into BAC vector-deficient AD169-RV resulted in an overlength of the genome well below that of the 235 kbp of RVHB5 or conventional lacZ insertion mutants, a genome size that is still packaged and that yields intact viral progeny (6, 29). As shown exemplarily with mutant B-B1 containing a sequence disruption at the extreme end of ORF UL78 (nt 114124), insertion mutagenesis with a Tn-derived mobile DNA sequence has no apparent effect on viral growth (Fig. 2B).
Generation and screening of a library of genomic HCMV Tn mutants.
The Tn previously described (9, 26) has a high preference for insertion into plasmids or BACs. In more than 90% of the transposition events the BAC plasmid represented the target, while insertions into the bacterial genome occurred with a frequency of less than 10%. Therefore, a positive selection of Tn-inserted BACs via bacterial conjugation was not required. Bacterial clones with Tn insertions were generated and stored as glycerol stocks in 96-well microtiter plates. These, together with a series of DNA pools of the mutant genomes, constitute a library of Tn mutants (Fig. 3A). Assuming a completely random distribution of Tn insertions over the entire genomic BAC, such a library would have to comprise 5,294 individual clones to have a 99% chance of finding an insertion every 200 nt. About 1,050 mutants would suffice for a Tn insertion somewhere within every 1.0-kb fragment. In a first approach, our library included 2,000 random mutant bacterial clones.
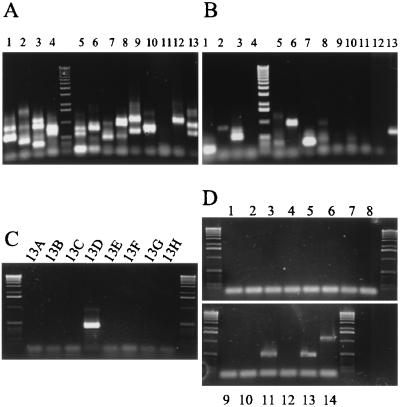
To test the hypothesis of random insertions, half of this library of genomic HCMV mutants was screened for insertion events located in the ORFs coding for known virion glycoproteins. DNA pools of each plate were tested by a PCR using three oligonucleotide primers as outlined in Fig. 3B. Since the AD169 genome has been completely sequenced (10), search primers can be designed to bind selectively to any position of interest, whereas the Tn-specific primers M13-for and M13-rev hybridize to sites near the inverted repeat structures of the transposed element (26). A PCR product should be generated wherever a Tn has inserted near the search primer position, irrespective of the Tn orientation (Fig. 3B). The observed fragment sizes of the PCR products obtained from the first round of search should permit a choice among suitable candidates. Figure 4A and B show two representative experiments for a first round of PCR search scanning DNA plate pools. In Fig. 4A 13 plates were tested with a primer specific for gB (UL55-rev; listed in Table 1). The detection of numerous PCR products indicated Tn insertion events at various positions close to the search primer position, suggesting that the library comprises a large number of insertions into gene UL55 (gB) (Fig. 4A). However, when this library of about 1,000 mutants was probed with a gM-specific primer (UL100-for), candidate mutants were detected much less frequently (Fig. 4B). This difference suggested that insertions are not equally distributed over the entire viral genome.
FIG. 4.
Localization of candidate mutants to individual wells of the Tn library. (A) First-round PCR search for mutants within the gene coding for gB using the UL55-rev primer. Each lane on the gel shows PCR products generated on secondary DNA pools representing all Tn mutants in one plate. A size marker (1-kbp ladder) is shown between lanes 4 and 5. (B) The PCR search was performed on the same DNA samples as in panel A but a primer specific for the gM gene (UL100-for) was used. (C) Pooled DNA samples representing the mutant clones stored in lanes A to H of plate 13 were subjected to PCR analysis with primers UL100-for, M13-for, and M13-rev. (D) All 12 individual wells (lanes 1 to 12) of row D identified in panel C were subjected to a third round of PCR screening. The desired mutant viral genome was detected in position D-11 on plate 13 of the Tn library. Pooled DNA from row D was included as a positive control (lane 13). Lane 14, PCR performed with the same DNA sample as in lane 13 using the distal primer UL100-rev with M13-for and M13-rev. The detection of the expected 1.2-kbp band was indicative of the absence of irregular deletion events.
When the plate pool tested positive for the gene of interest, two more rounds of PCR analysis were necessary to identify the bacterial clone carrying the respective BAC viral mutant (Fig. 3A). In plate 13 (Fig. 4B, lane 13) the size of the amplified PCR product predicted a Tn insertion in the gene encoding the C-terminal part of gM. In a second round of PCR screening, the mutant was traced by examining the DNA pools representing all mutants stored in rows A to H of plate 13 (Fig. 4C). After the mutant was localized to row D, individual wells of that row were subjected to a third round of PCR analysis in order to identify the well containing the clone of interest (Fig. 4D, lanes 1 to 12). According to this protocol only the mutant clones that carried an insertion close to the position of interest were analyzed, while all other clones were left uncharacterized. This method of mutant isolation is fast and involves only three steps of PCR screening of hierarchically pooled samples of DNA and bacterial clones.
Characterization of the Tn mutants.
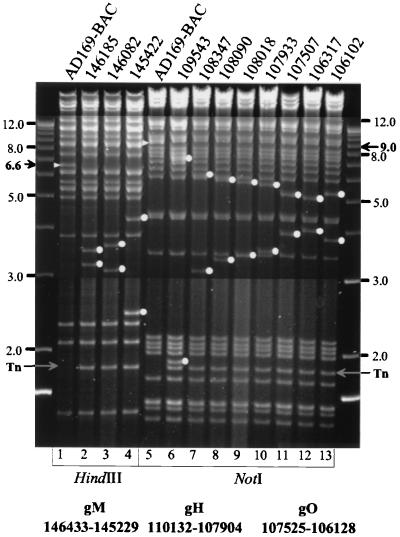
A first estimate of the distance between the Tn insertion site and the binding site of the search primer can be achieved by determining the size of the observed PCR product. In rare instances, Tn mutagenesis can result in adventitious deletions at the Tn insertion site (9), which probably occur via an illegitimate resolution of nearby double Tn insertions or by intramolecular transposition (4). To exclude such deletion mutants, a confirmatory PCR with an oligonucleotide primer (e.g., UL100-rev) located about 2 kbp downstream from the search primer and distal from the Tn integration site was performed (Fig. 4D, lane 14). When the resulting PCR fragment matched the expected size, the mutant genome was further analyzed by restriction enzyme digestions with several enzymes. Figure 5 shows examples of how Tn insertions result in new restriction enzyme cleavage products. The gene encoding the integral membrane protein gM is located in the 6.6-kbp HindIII R fragment of HCMV (32) (Fig. 5, lane 1). In the Tn mutants the 6.6-kbp HindIII R fragment is cleaved into two subfragments of corresponding sizes (Fig. 5, lanes 2 to 4) and an additional Tn-specific 1.8-kbp fragment appears due to a HindIII site located within each of the inverted repeats of the Tn (26). In lanes 6 to 13 Tn insertions into gene UL75 (gH) and the immediately adjacent gene UL74 (gO) are shown. These genes are located in a 9.0-kbp NotI fragment of AD169-BAC (Fig. 5, lane 5). This fragment is cleaved into two subfragments of corresponding sizes (Fig. 5, lanes 6 to 13). A Tn-specific 1.8-kbp fragment results from two NotI sites located near the Tn ends and is present in all mutated HCMV BACs but not in the parental clone, AD169-BAC (Fig. 5, lane 5). Finally, the exact nucleotide position of the Tn insertion was determined by direct sequencing of the BAC using the M13 primers.
FIG. 5.
Tn insertion mutants show random insertions within chosen genes. Lane 1, HindIII digest of the AD169-BAC DNA isolated from bacteria; UL100 (gM) is located on the 6.6-kbp HindIII R fragment (black arrow, white arrowhead); lanes 2 to 4, BAC mutants with insertions into UL100 digested with HindIII; the exact nucleotide positions of the Tn insertion are indicated above the lanes; lane 5, NotI digest of parental AD169-BAC; genes UL74 (gO) and UL75 (gH) are located on a 9.0-kbp NotI fragment (black arrow, white arrowhead); lanes 6 to 13, NotI digest of AD169-BAC-derived genomic HCMV mutants carrying Tn insertions in the genes coding for gH and gO. DNA fragments were separated on a 0.6% agarose gel, and molecular size markers are given in kilobase pairs on either side. Grey arrows, Tn-specific 1.8-kbp fragments; dots, subfragments resulting from cleavage of restriction endonuclease fragments.
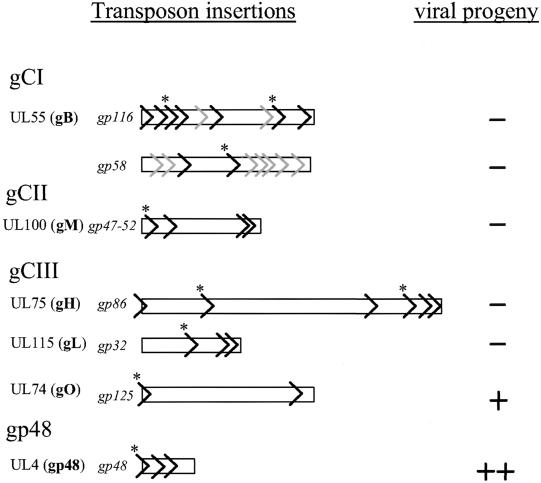
Our library was large enough to isolate at least two mutants of each glycoprotein gene we were interested in. The individual BAC clones characterized are summarized in Fig. 6. For the gB gene the insertions were located at nt 83389, 83171, 83108*, 83012, 82743, 82305*, 82166, and 81329*; disruptions of the gH gene were found at nt 110107, 109543*, 108347, 108090*, 108018, and 107933; gL gene insertions were found at nt 164159*, 163840, and 163712; insertions into the gM gene were detected at nt 146185*, 146082, 145422, and 145420; gp48 gene insertions were found at nt 13470*, 13614, and 13798; those disrupting the gO gene mapped to nt 107507* and 106317. For mutants corresponding to nucleotides denoted with an asterisk, a revertant genome was also created by allelic replacement in E. coli.
FIG. 6.
Schematic representation of Tn insertions into the HCMV virion glycoprotein genes. Mutants were characterized by appropriate restriction enzyme digests and direct sequencing on BACs prior to transfection. >, insertion of a Tn sequence resulting in the disruption of that gene. For gB many more candidates are present in the library, but these have not been confirmed by sequencing (grey symbols). Transfection results for insertion mutants are summarized at the right. When upon repeated transfection no viral progeny could be obtained (−), plaque formation could reproducibly be rescued by cotransfection of subgenomic fragments of HCMV DNA (4 to 5 kbp) spanning the insertion site. ∗, mutant with revertant genome constructed by allelic exchange in bacteria and found to be infectious.
Identification of essential glycoprotein genes by reconstitution of mutant viruses.
We tested whether insertions in genes coding for envelope glycoproteins still allow virus replication in cultured fibroblasts. For this purpose, each of the mutant genomes depicted in Fig. 6 was transfected into MRC-5 cells. Mutant genomes that did not give rise to plaques were complemented by cotransfection with plasmids carrying 4 to 5 kb of viral DNA overlapping the Tn insertion site (pUH-25 to pUH-34). In all cases, complementation in vitro reproducibly rescued plaque formation, proving that the Tn insertion was the reason for the failure to replicate. In addition, a number of revertant genomes were constructed by allelic exchange in E. coli (13470-Rev, 81329-Rev, 82305-Rev, 83108-Rev, 107507-Rev, 108090-Rev, 109543-Rev, 146185-Rev, and 164159-Rev; numbers indicate the nucleotide position of the Tn insertion). Both types of revertants (obtained either by recombination in eukaryotic cells or by allelic exchange in bacteria) showed wt growth properties along with wt restriction patterns (data not shown). This suggested that mutant genomes with Tn insertions within the genes encoding gB, gH, gM, and gL are not viable. As reported previously (42, 52) the UL4 gene product, gp48, is a nonessential component of the viral envelope. Our data confirm these findings. Mutated BACs with a Tn integrated into gene UL4 gave rise to a mutant viral progeny without significantly impaired replication kinetics, irrespective of the multiplicity of infection (MOI) applied (data not shown). The results of the transfection experiments are summarized in Fig. 6.
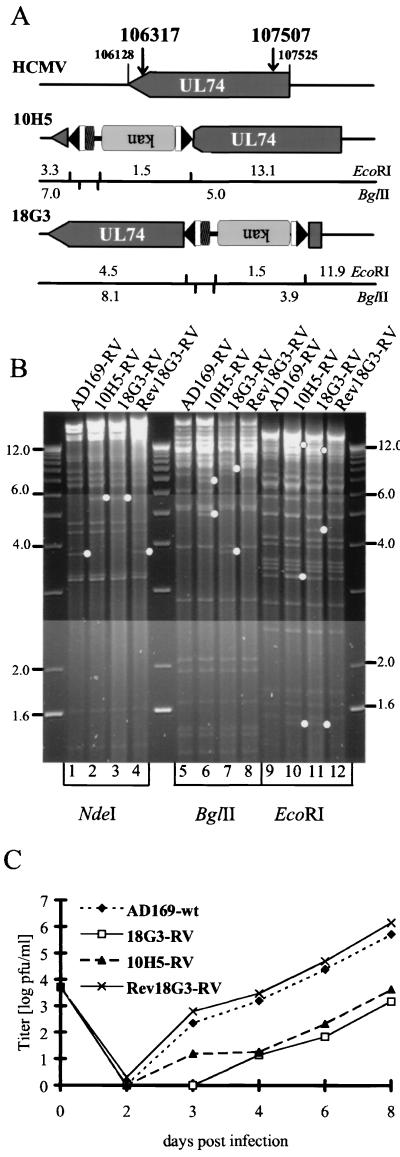
Remarkably, the mutant BACs with disruptions of gene UL74 (gO) also yielded viable virus, suggesting that the HCMV gO is not essential for the infectious cycle of HCMV. Still, all genomes with an insertion into UL74 led to mutant viruses with an attenuated growth phenotype in cell culture (Fig. 7C), which remained conserved over several rounds of replication. The gO-defective viruses exhibited a severely reduced plaque size, which was observed irrespective of whether the Tn insertion was at the beginning (18G3: insertion at nt 107507), corresponding to an alteration in the coding sequence at amino acid position 8 leading to a premature stop 11 amino acids later, or at the end (10H5: insertion at nt 106317), corresponding to a truncation of the protein at amino acid 404, of the UL74 gene (nt 107525 to 106128) (Fig. 7A). These data do not yet allow the conclusion that a mutated protein with a stop at this position is associated with a loss of gO function. An inserted Tn sequence can also destabilize the mRNA transcript and thus result in a functional gene knockout (56). If a truncated gene product should be expressed, a set of insertion mutants would help to identify functionally important domains. Figure 7B shows the DNA extracted from the mutant viruses with a Tn stably integrated into the gene encoding gO compared to that from a virus reconstituted from the parental AD169-BAC, which is indistinguishable from revertant virus Rev18G3-RV, obtained by allelic replacement. Repair of the mutated UL74 gene with an intact sequence restored wt growth properties (Fig. 7C).
FIG. 7.
Mutant viruses defective in gene UL74 (gO) exhibit a reduction in growth kinetics. (A) Schematic representation of the Tn insertions. kan, ORF encoding the kanamycin resistance marker; upside down lettering indicates opposite orientation. Boldface numbers, exact nucleotide positions of the interruption of the ORF; lightface numbers, expected fragment sizes in kilobase pairs. (B) Viral DNA isolated from cells infected with AD169-RV (lanes 1, 5, and 9) compared to that from the UL74-deficient mutant viruses reconstituted from clones 10H5 (lanes 2, 6, and 10) and 18G3 (lanes 3, 7, and 11), together with that from a revertant genome generated by allelic exchange in bacteria and reconstituted to yield Rev18G3-RV (lanes 4, 8, and 12). Viral DNA was digested with NdeI (lanes 1 to 4). The UL74 gene is located on a 3.8-kbp NdeI fragment (lane 1), which, due to the Tn insertion in variants 10H5 and 18G3, shifts to a 5.6-kbp fragment (lanes 2 and 3, dots). In lanes 5 to 8 (BglII digests) and lanes 9 to 12 (EcoRI digests) the 18G3 revertant has a pattern indistinguishable from that of AD169-RV, whereas both gO-deficient viruses show the expected altered fragment sizes depicted in panel A (dots). (C) Single-step growth curve of gO mutants compared to revertant virus Rev18G3-RV. Confluent monolayers of MRC-5 cells were infected at an MOI of 0.01. Virus was harvested from both media and infected cells at the indicated time points and titered by plaque assay. Values shown are the results of duplicate assays from duplicate infections.
DISCUSSION
We report on the generation of a library of Tn insertion mutants of the reconstituted full-length AD169 genome cloned as an infectious BAC in E. coli. Pooled DNA samples of this library were used to screen for single Tn insertions in various genes of interest. As several individual candidates can be handled in parallel, a whole set of mutants can be identified for any position of interest in less than a week's time. For the six genes encoding glycoproteins reported here, a library of approximately 2,000 clones proved large enough to contain at least two independent Tn insertions per gene studied. Transfection of mutant genomes gave rise to infectious viral progeny only for insertions into UL4 and UL74. Insertions within the genes coding for gB, gH, gL, and, interestingly, gM were not compatible with virus replication in fibroblasts.
Our studies show that any genomic region can be rapidly screened for insertion events by a sequential three-step PCR analysis of the Tn insertion library. BACs usually are present only as a single copy per bacterial cell (46). Exposure of the BAC-containing bacteria to the Tn donor plasmid for a restricted period of time for mutagenic conditions led to rare transposition events resulting in mainly single Tn insertions (9). The known preference of this minimal Tn1721-derived Tn for supercoiled plasmids versus bacterial genomes (4) was also valid for BACs. We found a Tn insertion into the MCMV (9) and HCMV BAC plasmids in about 90% of the clones analyzed. Thus, for the generation of a library with single Tn insertions, the bacteria with insertions into the bacterial chromosome did not require practical consideration. However, during the setup of the experimental system the choice of the antibiotic resistance gene was not without impact on Tn mutagenesis. A single copy of a tetracycline resistance gene apparently confers only a weak resistance against the antibiotic. Bacteria with two or more Tn insertions in AD169-BAC were predominantly selected when TnMax13, encoding a tetracycline resistance gene, was used together with a tetracycline concentration of 10 μg/ml (data not shown). A switch to the aphA3 gene conferring resistance to kanamycin reproducibly resulted in single Tn insertions. However, by reducing the tetracycline concentration single Tn insertions should also be obtained.
Single insertions of the Tn selectively into the viral BAC are important for establishing useful libraries for fast screening. Some Tn, such as the Tn5 or Tn10 derivatives, have a considerable propensity to integrate into the bacterial genome thus requiring discrimination between insertion events into the viral BAC and into the E. coli genome. Such a separation has been achieved either by DNA extraction and retransformation of BACs (47) or, alternatively, via bacterial conjugation using an oriT element inserted into the BAC cassette. However, for efficient transfer into E. coli the library had to be amplified, which increases the risk of a repeated selection of identical clones that probably did not arise from independent insertion events (47).
The Tn mutagenesis which we have carried out previously for the MCMV BAC and here for AD169-BAC is rapid and direct, but is it also random? Whereas for Tn7 a strict sequence specificity has been reported (4), Tn1721, a member of the Tn3 family, and Tn5 display an almost random distribution of integration sites (4). Tn insertions were observed at random locations throughout the AD169-BAC genome. With about 250 Tn mutants of MCMV and HCMV analyzed so far, we never found a repetition of an identical Tn insertion. Instead, we have detected independent clonal insertion events 20 nt or less from each other within the same gene. Thus, random insertion mutagenesis could perhaps be used to generate a series of gene truncation mutants to map functional domains of a target protein. However, for unknown reasons the insertions are not entirely equally distributed over the genome, as indicated by an apparent accumulation of insertions in the UL55 region. Nevertheless, our library of about 2,000 individual mutant clones has so far proved large enough to identify at least two genomic HCMV mutants per gene studied.
A targeted deletion of a complete ORF in the context of the viral genome is considered a potent means to functionally analyze an HCMV gene. This approach, however, may also affect regulatory sequences controlling the function of other genes, with the consequence that the phenotype of a mutant cannot be attributed with certainty to the deleted ORF. This problem is only partially addressed by the construction of a revertant and is not excluded by studying the directly neighboring genes. In principle, each individual Tn insertion mutant, as the product of a random sequence disruption, suffers from the same ambiguity. Therefore, a consistent phenotype of several insertion mutants from different positions within one ORF probably defines the property of the targeted gene with a higher degree of confidence. A variability of phenotypes would either indicate effects on other genes or suggest the presence of functionally independent domains in the targeted gene. By studying the properties of several independent Tn mutants with insertions dispersed all over the area of interest, a rapid and reliable assignment of functions may be achieved even for overlapping ORFs and spliced genes. Therefore, we conclude that, collectively, each set of gene mutants, although not necessarily an individual mutant, describes the properties of the targeted gene.
Products of six ORFs have been described as constituents of the HCMV virion envelope, most of them being organized in three distinct glycoprotein complexes numbered I to III (8, 18, 23). UL33 codes for a G protein-coupled receptor homolog that has also been detected in the envelopes of purified virus particles (37) but that is much less abundant than the glycoproteins mentioned above and thus may result from random incorporation into budding particles. It had to be presumed that, by analogy to HSV-1 (43) and all other herpesviruses studied, deletions within UL55 (gB) and UL75 (gH) would not be viable (8). For HCMV, the essential viral functions of attachment and fusion have been assigned to gB and gH, respectively (11). Proof for this assumption can only be derived from defined genetic constructs and is now provided by this study. This type of construction was not possible through virus mutant construction by recombination in eukaryotic cells due to the lack of complementing helper cells. Work on essential virus genes requires two components, the defined viral gene mutants and cell lines expressing the gene under study for complementation. Our defined sets of genomic mutants will be a useful tool to evaluate transfected cell lines with regard to their helper function. This should provide a basis for the subsequent functional analysis of gene mutants.
The results obtained in this study provide at least two new pieces of information. First, HCMV gM (UL100) was shown to be essential for growth in fibroblasts. The function of the integral membrane protein gM is largely unknown. This extremely hydrophobic protein is associated with at least one antigenically distinct component in a multimeric complex (28). Apart from a diffuse heparin binding potential (27) no molecular functions have been proposed for glycoprotein complex II (gCII). The requirement for HCMV gM contrasts with findings for the alphaherpesviruses, where, despite its high conservation, the gM gene homolog UL10 in HSV-1 has been deleted without severe consequences (2). Deletion of the gM gene homolog UL10 in PrV also resulted in attenuated but viable viral progeny (15). Data from HSV-1 and PrV suggest an involvement in penetration of their respective gM homologs (15).
Second, the HCMV gO function is not required for propagation of the virus in fibroblasts. Along with two other protein components, gH and gL, gO forms a trimeric, covalently linked complex called gCIII (24, 33). Alphaherpesviruses do not contain a gene homologous to the gO gene. Moreover, in Epstein-Barr virus, which so far contains the only other example of a heterotrimeric gH-related complex, the BZLF2 gene product, a gO-related protein, can be omitted for infection in a cell-type-specific manner (34). Likewise, for HCMV, which in vivo infects multiple cell types, gO might act as a coreceptor binding partner cooperating with the fusion-competent gH. It will be of interest to see, whether gO is essential for the infection of some target tissues by HCMV. The other two components of the gCIII complex, gH and gL, are necessary for the formation of infectious viruses. This is corroborated by similar findings throughout the herpesvirus family (31, 43). HCMV gH (UL75) has been proposed to be involved in attachment to a ubiquitous cell surface receptor (8), whereas gL is essential for the transfer of the essential component gH from the endoplasmic reticulum to the cell surface (25, 30, 50). Careful analysis of the molar ratios of the envelope components of HCMV suggested that, in addition to a heterotrimeric disulfide-linked form of gH (gCIII), additional noncomplexed gH molecules are present on the surface of the virion (33).
The system described in this paper permits the rapid and targeted identification of entire sets of insertion mutants in any HCMV gene of interest. The number of mutants per gene depends on the size of the library and can be adjusted to suit one's need. Assessment of the functional importance of virion glycoproteins is just one application to demonstrate the potential use of a library of random Tn insertion mutants for a detailed genetic analysis of HCMV or any other BAC-cloned virus genome. By three consecutive steps of PCR screening on hierarchically pooled samples of the random Tn library, several independent mutant clones can be identified in less than a week and insertion mutants can be characterized prior to transfection. Sets of mutants with insertions at different locations in the gene help to faithfully attribute functional phenotypes to a gene product. This method might be particularly attractive for the analysis of viral genes that give rise to splice variants and that are composed of functionally distinguishable subdomains.
ACKNOWLEDGMENTS
We thank G. Pósfai, M. Wagner, and C. Ménard for providing plasmids. We are indebted to A. Hegele and A. Colomar for expert technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 455 and BR 1730/1-1) and from the Bundesministerium für Bildung und Forschung.
REFERENCES
- 1.Adler H, Messerle M, Wagner M, Koszinowski U H. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg C M, Berg D E, Groisman E A. Transposable elements and the genetic engineering of bacteria. In: Berg D E, Howe M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 879–925. [Google Scholar]
- 5.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 6.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 8.Britt W J, Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. doi: 10.1159/000150510. [DOI] [PubMed] [Google Scholar]
- 9.Brune W, Ménard C, Hobom U, Odenbreit S, Messerle M, Koszinowski U H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchinson III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Compton T, Nepomuceno R R, Nowlin D M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 12.Crnković-Mertens I, Messerle M, Milotić I, Szepan U, Kučić N, Krmpotić A, Jonjić S, Koszinowski U H. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol. 1998;72:1377–1382. doi: 10.1128/jvi.72.2.1377-1382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dargan D J, Jamieson F E, Maclean J, Dolan A, Addison C, McGeoch D J. The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J Virol. 1997;71:9833–9836. doi: 10.1128/jvi.71.12.9833-9836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delecluse H-J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleckenstein B, Müller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 17.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosin phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 18.Gretch D R, Kari B, Rasmussen L, Gehrz R C, Stinski M F. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J Virol. 1988;62:875–881. doi: 10.1128/jvi.62.3.875-881.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu H, Zou Y-R, Rajewski K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 20.Hengel H, Brune W, Koszinowski U H. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 1998;6:190–197. doi: 10.1016/s0966-842x(98)01255-4. [DOI] [PubMed] [Google Scholar]
- 21.Horsburgh B C, Hubinette M M, Qiang D, MacDonald M L, Tufaro F. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 1999;6:922–930. doi: 10.1038/sj.gt.3300887. [DOI] [PubMed] [Google Scholar]
- 22.Horsburgh B C, Hubinette M M, Tufaro F. Genetic manipulation of herpes simplex virus using bacterial artificial chromosomes. Methods Enzymol. 1999;306:337–352. doi: 10.1016/s0076-6879(99)06022-x. [DOI] [PubMed] [Google Scholar]
- 23.Huber M T, Compton T. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol. 1998;72:8191–8197. doi: 10.1128/jvi.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber M T, Compton T. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J Virol. 1999;73:3886–3892. doi: 10.1128/jvi.73.5.3886-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson A C, Johnson D C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and effects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahrs A F, Odenbreit S, Schmitt W, Heuermann D, Meyer T F, Haas R. An improved TnMax mini-transposon system suitable for sequencing, shuttle mutagenesis and gene fusions. Gene. 1995;167:53–57. doi: 10.1016/0378-1119(95)00671-0. [DOI] [PubMed] [Google Scholar]
- 27.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kari B, Li W, Cooper J, Goertz R, Radeke B. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J Gen Virol. 1994;75:3081–3086. doi: 10.1099/0022-1317-75-11-3081. [DOI] [PubMed] [Google Scholar]
- 29.Kaye J, Browne H, Stoffel M, Minson T. The UL16 gene of human cytomegalovirus encodes a glycoprotein that is dispensable for growth in vitro. J Virol. 1992;66:6609–6615. doi: 10.1128/jvi.66.11.6609-6615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaye J F, Gompels U A, Minson A C. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J Gen Virol. 1992;73:2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 31.Klupp B, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Nelson J A, Britt W J. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–3097. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luckow V A, Lee S C, Barry G F, Olins P O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margulies B J, Browne H, Gibson W. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology. 1996;225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messerle M, Crnković I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mocarski E S, Prichard M N, Tan C S, Brown J M. Reassessing the organization of the UL42-UL43 region of the human cytomegalovirus strain AD169 genome. Virology. 1997;239:169–175. doi: 10.1006/viro.1997.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 41.Pósfai G, Koob M D, Kirkpatrick H A, Blattner F R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripalti A, Mocarski E S. The products of human cytomegalovirus genes UL1-UL7, including gp48, are dispensable for growth in cell culture. In: Landini M P, editor. Progress in cytomegalovirus research: proceedings of the Third International Cytomegalovirus Workshop. Amsterdam, The Netherlands: Elsevier Publishers; 1991. pp. 57–62. [Google Scholar]
- 43.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 44.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith G A, Enquist L W. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith G A, Enquist L W. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97:4873–4878. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spaete R R, Perot K, Scott P I, Nelson J A, Stinski M F, Pachl C. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in transport of gH to the cell surface. Virology. 1993;193:853–861. doi: 10.1006/viro.1993.1194. [DOI] [PubMed] [Google Scholar]
- 51.Stavropoulos T A, Strathdee C A. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takekoshi M, Maeda-Takekoshi F, Ihara S, Sakuma S, Watanabe Y. Site-specific stable insertion into the human cytomegalovirus genome of a foreign gene under control of the SV40 promoter. Gene. 1991;101:209–213. doi: 10.1016/0378-1119(91)90413-6. [DOI] [PubMed] [Google Scholar]
- 53.Tomazin R, Boname J, Hegde N R, Lewinsohn D M, Altschuler Y, Jones T R, Cresswell P, Nelson J A, Riddell S R, Johnson D C. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat Med. 1999;5:1039–1043. doi: 10.1038/12478. [DOI] [PubMed] [Google Scholar]
- 54.Wagner M, Jonjić S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X W, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 56.Zhan X, Lee M, Abenes G, Reis I V, Kittinunvorakoon C, Ross-Macdonald P, Snyder M, Liu F. Mutagenesis of murine cytomegalovirus using a Tn3-based transposon. Virology. 2000;266:264–274. doi: 10.1006/viro.1999.0089. [DOI] [PubMed] [Google Scholar]