Abstract
The synaptonemal complex (SC) is a meiotic interface that assembles between parental chromosomes and is essential for the formation of gametes. While the dimensions and ultrastructure of the SC are conserved across eukaryotes, its protein components are highly divergent. Recently, an unexpected component of the SC has been described in the nematode C. elegans: the Skp1-related proteins SKR-1/2, which are components of the Skp1, Cullin, F-box (SCF) ubiquitin ligase. Here, we find that the role of SKR-1 in the SC is conserved in nematodes. The P. pacificus Skp1 ortholog, Ppa-SKR-1, colocalizes with other SC proteins throughout meiotic prophase, where it occupies the middle of the SC. Like in C. elegans, the dimerization interface of Ppa-SKR-1 is required for its SC function. A dimerization mutant, Ppa-skr-1F105E, fails to assemble SC and is almost completely sterile. Interestingly, the evolutionary trajectory of SKR-1 contrasts with other SC proteins. Unlike most SC proteins, SKR-1 is highly conserved in nematodes. Our results suggest that the structural role of SKR-1 in the SC has been conserved since the common ancestor of C. elegans and P. pacificus, and that rapidly evolving SC proteins have maintained the ability to interact with SKR-1 for at least 100 million years.
Keywords: meiosis, synaptonemal complex, SCF, evolution, nematodes, Skp1, P. pacificus
Summary
During sexual reproduction, the parental chromosomes align along their length and exchange genetic information. These processes depend on a chromosomal interface called the synaptonemal complex. The structure of the synaptonemal complex is conserved across eukaryotes, but, surprisingly, the components that make it up are dramatically different in different organisms. Here we find that a protein well known for its role in regulating protein degradation has been moonlighting as a structural component of the synaptonemal complex in the nematode Pristionchus pacificus, and that it has likely carried out both of these functions for more than 100 million years.
Introduction
The synaptonemal complex (SC) is a conserved interface that facilitates chromosome organization during meiosis. The SC aligns parental chromosomes end-to-end and regulates genetic exchanges between them, ultimately allowing for the proper segregation of chromosomes during the meiotic divisions. First identified by electron microscopy over 60 years ago, the SC is made up of two parallel axes (also called lateral or axial elements) separated by repeating striations that make up the central region of the SC (throughout, we refer to the central region of the SC simply as ‘the SC’ (Page and Hawley 2004; Zickler and Kleckner 2015)).
Despite its essential role in reproduction and its conserved ultrastructure across sexually reproducing organisms, SC components have diverged beyond recognition in multiple eukaryotic clades (Kursel, Cope, and Rog 2021; Hemmer and Blumenstiel 2016). Indeed, new SC components are still being identified, and we likely still lack the full complement of SC components in most model organisms. Further complicating molecular studies, SC components exhibit near-complete co-dependence for assembly onto chromosomes, in worms and in other organisms (Colaiácovo et al. 2003; MacQueen et al. 2002; Smolikov et al. 2007; Smolikov, Schild-Prüfert, and Colaiácovo 2009; Collins et al. 2014; Page et al. 2008; Schramm et al. 2011). Recently, co-expression of SC components allowed their purification from bacteria (Blundon et al. 2024). This suggests that SC subunits intimately associate with one another to form the repeating building blocks of an assembled SC. However, only a few intra-SC interaction interfaces have been defined (Dunce et al. 2018; Dunne and Davies 2019; Sánchez-Sáez et al. 2020; Dunce, Salmon, and Davies 2021; Kursel, Martinez, and Rog 2023), and, due to sequence divergence, it is unclear whether any of them constitute a conserved feature of the SC.
Recently, two unexpected SC proteins were identified in C. elegans: the Skp1-related proteins SKR-1 and SKR-2 (due to their functional redundancy we refer to them throughout as SKR-1/2; (Blundon et al. 2024)). SKR-1/2 are essential members of the Skp1, Cullin, F-box (SCF) ubiquitin ligase complex, which plays a part in virtually all eukaryotic cellular processes including germline designation (DeRenzo, Reese, and Seydoux 2003), sex determination (Clifford et al. 2000), transcriptional regulation (Ouni, Flick, and Kaiser 2010), circadian oscillation (Han et al. 2004) and hormone signaling in plants (Gray et al. 1999), to name a few (Willems, Schwab, and Tyers 2004). Within the SCF complex, Skp1 acts as an adapter by binding the N-terminus of Cul1 and the F-box motif in the F-box protein, linking the core scaffold to the substrate of the ubiquitin ligase machinery. SKR-1/2 co-purify with all other C. elegans SC proteins, localize to the SC, and are required for SC assembly in vivo. Notably, the SCF Cullin subunit CUL-1 does not localize to the SC and is not required for SC assembly. These data support the conclusion that SKR-1/2 are bona fide SC proteins in C. elegans (Blundon et al. 2024).
Here we address two outstanding questions regarding the role of SKR-1 in the SC. 1) Is the structural role of SKR-1 in the SC conserved in other nematodes? And 2) Does SKR-1 share a similar evolutionary signature to other SC proteins? We identify a single SKR-1 ortholog in the distantly related nematode Pristionchus pacificus, Ppa-SKR-1, and find that it localizes to the middle of the SC. Like in C. elegans, the predicted dimerization interface in Ppa-SKR-1 is necessary for SC assembly. Our results indicate that Ppa-SKR-1 is a structural component of the SC in P. pacificus, suggesting that its role in the SC originated at least 100 million years ago, in the common ancestor of Pristionchus and Caenorhabditis nematodes. Interestingly, we find that the primary sequence of SKR-1 is conserved, setting it apart from other SC proteins and shedding light on the evolutionary pressures that shape the SC.
Results
Identifying P. pacificus SKR-1
C. elegans and P. pacificus are a useful species pair for comparative studies. Like C. elegans, P. pacificus is a free-living, hermaphroditic nematode that has six pairs of chromosomes. Previous studies of meiosis in P. pacificus identified two SC proteins; Ppa-SYP-1 (Kursel, Cope, and Rog 2021) and Ppa-SYP-4 (Rillo-Bohn et al. 2021). Consistent with the rapid divergence of SC proteins, Ppa-SYP-4 and Ppa-SYP-1 exhibit little to no sequence homology, respectively, with their C. elegans counterparts. Given the recent identification of SKR-1/2 as a structural component of the SC in C. elegans (Blundon et al. 2024), we wondered whether SKR-1 plays a similar SC role in P. pacificus.
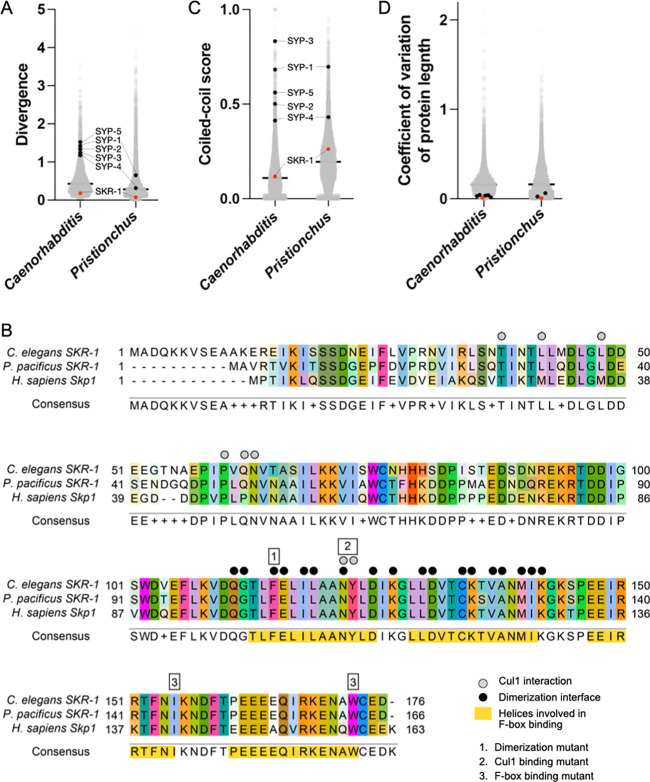
We used C. elegans SKR-1 as a BLASTp query against P. pacificus El Paco V3 predicted proteins. We identified a single strong hit which we refer to as Ppa-SKR-1. Ppa-SKR-1 clusters with C. elegans SKR-1/2 on a strongly supported branch to the exclusion of all other Skp1-related proteins in P. pacificus (Figure S1). While the C. elegans genome contains a recent duplication of SKR-1 called SKR-2 (Blundon et al. 2024), our phylogenetic analysis reveals that P. pacificus contains only one copy of SKR-1. We similarly queried seven additional Pristionchus proteomes and found that most species have a single SKR-1 ortholog (Figure S2)). We note that P. pacificus, like C. elegans, encodes many predicted Skp1-related proteins: 32 in P. pacificus and 21 in C. elegans (Figure S1; (Nayak et al. 2002)). While the expansion of the Skp1 family in nematodes complicates comprehensive tracing of their evolutionary history, SKR-1 orthologs appear to be the most conserved among Skp1-related proteins, and cluster together in a well-supported clade (Figure S2).
Ppa-SKR-1 localizes to the center of the SC
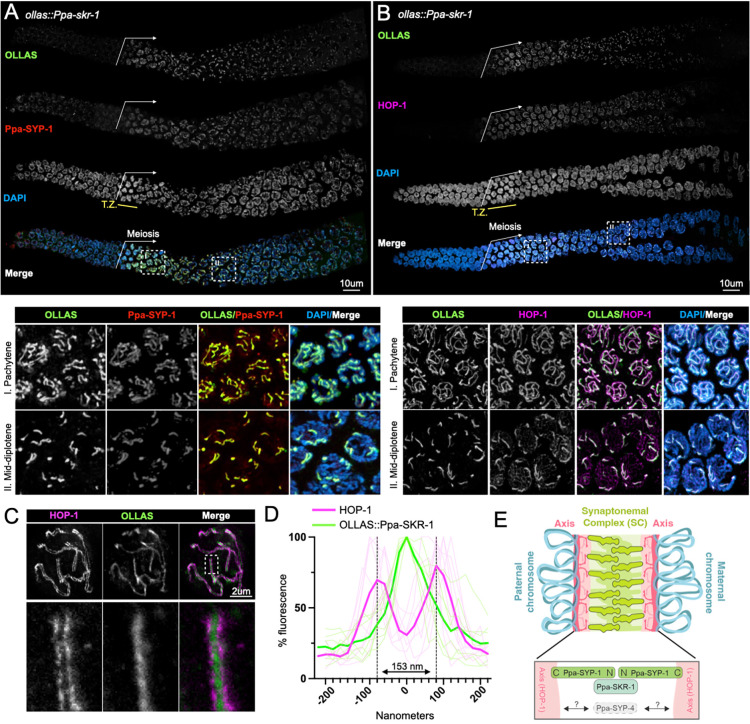
We used CRISPR/Cas9 to insert an OLLAS tag on the N-terminus of Ppa-SKR-1 and examined its localization during meiosis (Figure 1). OLLAS::Ppa-SKR-1 appears as threads on meiotic chromosomes from the time of SC assembly at meiotic entry, throughout pachytene (the stage when the SC is completely assembled on all chromosomes), and to diplotene (the extended stage of SC disassembly; Figure 1A). This pattern matches that of other SC proteins (Rillo-Bohn et al. 2021; Kursel, Cope, and Rog 2021). The axis component HOP-1 (Rillo-Bohn et al. 2021) localizes to meiotic chromosomes slightly before OLLAS::Ppa-SKR-1 as faint lines indicative of unpaired chromosomes (Figure 1B). As OLLAS::Ppa-SKR-1 signal begins to overlap with HOP-1, the lines of HOP-1 are brighter, reflecting paired, synapsed chromosomes. During diplotene, OLLAS::Ppa-SKR-1 remains on the bright-staining regions of HOP-1 until the SC fully disassembles.
Figure 1: Ppa-SKR-1 localizes to the middle of the SC.
(A) Top panel, confocal image of whole gonads from ollas::Ppa-skr-1 stained with anti-OLLAS, anti-SYP-1 and DAPI. Bottom panel, zoom in on pachytene (I) or mid-diplotene (II) nuclei. (B) Confocal image as in (A) except with HOP-1 staining. In (A) and (B), the beginning of the meiotic gonad is indicated with a white arrow and the transition zone is labeled below the DAPI channel in yellow (T.Z.). (C) Super-resolution STED image of a single pachytene nucleus from ollas::Ppa-skr-1 worms stained with anti-OLLAS and anti-HOP-1. Zoom-in panels show OLLAS::Ppa-SKR-1 between parallel HOP-1 tracks. (D) Plot of line scans of pixel intensity for anti-HOP-1 and anti-OLLAS across parallel axes in ollas::Ppa-skr-1 worms. The average distance between parallel axes is 153nm. (E) Cartoon of the P. pacificus synaptonemal complex with the orientation and position of Ppa-SYP-1 and Ppa-SKR-1 relative to HOP-1 indicated in the bottom panel. The relative position of Ppa-SYP-4 is not known (grey arrows and question marks). Adapted from (Kursel, Aguayo Martinez, and Rog 2023).
SC proteins occupy stereotypical positions in the ~150nm space separating the two parental chromosomes. Ppa-SYP-1, like its C. elegans counterpart, spans the 100nm width of the SC in a head-to-head manner (N-terminus in, C-terminus out) such that staining with a C-terminal epitope produces two parallel lines and N-terminal staining produces a single thread in the middle of the SC (Köhler et al. 2020; Kursel, Cope, and Rog 2021; Schild-Prüfert et al. 2011). Using STED super-resolution microscopy, we found that the axis protein HOP-1 formed parallel tracks that are 153nm wide on average (Figure 1C, D) and that Ppa-SKR-1 localized to the central region of the SC, midway between the parallel HOP-1 tracks. These cytological data indicate that, like in C. elegans, Ppa-SKR-1 occupies the middle of the SC ladder, where the N-terminus of SYP-1 is located (Figure 1E, (Blundon et al. 2024)).
The Ppa-SKR-1 dimerization interface is required for SC assembly
The essential functions of Skp1 make it challenging to study its role in the SC. C. elegans worms lacking both SKR-1 and −2 fail to hatch, reflecting the essential roles of SCF in embryogenesis and cell proliferation (Nayak et al. 2002; Blundon et al. 2024). Given that P. pacificus harbors a single Skp1 ortholog, we predicted that gene deletion would result in embryonic lethality. We therefore wished to generate a separation-of-function allele of Ppa-skr-1.
Previous studies found that Skp1 dimerizes via a conserved hydrophobic interface that is not essential for F-box binding (Kim et al. 2020; Henzl, Thalmann, and Thalmann 1998). In C. elegans, mutations that disrupt SKR-1/2’s ability to dimerize (skr-1F115E) cause a complete failure of SC assembly and prevent SKR-1/2 localization to an already formed SC. Importantly, these mutations do not abolish SCF activity, suggesting that SKR-1/2 dimerization is necessary specifically for SC function (Blundon et al. 2024).
We used structural homology to predict the dimerization interface in Ppa-SKR-1 (Figure 2A). We found that a residue critical for dimerization in Dictyostelium Skp1, F97 (Kim et al. 2020), aligns closely with F105 in Ppa-SKR-1 (Figure 2A). To test the function of the putative dimerization interface, we used CRISPR/Cas9 to make ollas::Ppa-skr-1F105E. Gratifyingly, we easily obtained ollas::Ppa-skr-1F105E homozygous animals. Out of 46 F2s singled from heterozygous ollas::Ppa-skr-1F105E F1 parents, 12 were homozygous wildtype, 22 were heterozygous and 12 were homozygous for ollas::Ppa-skr-1F105E, matching expected Mendelian ratios. This suggests that the F105E mutation does not disrupt SCF functions during development.
Figure 2: Conserved dimerization interface in SKR-1 is required for P. pacificus meiosis.
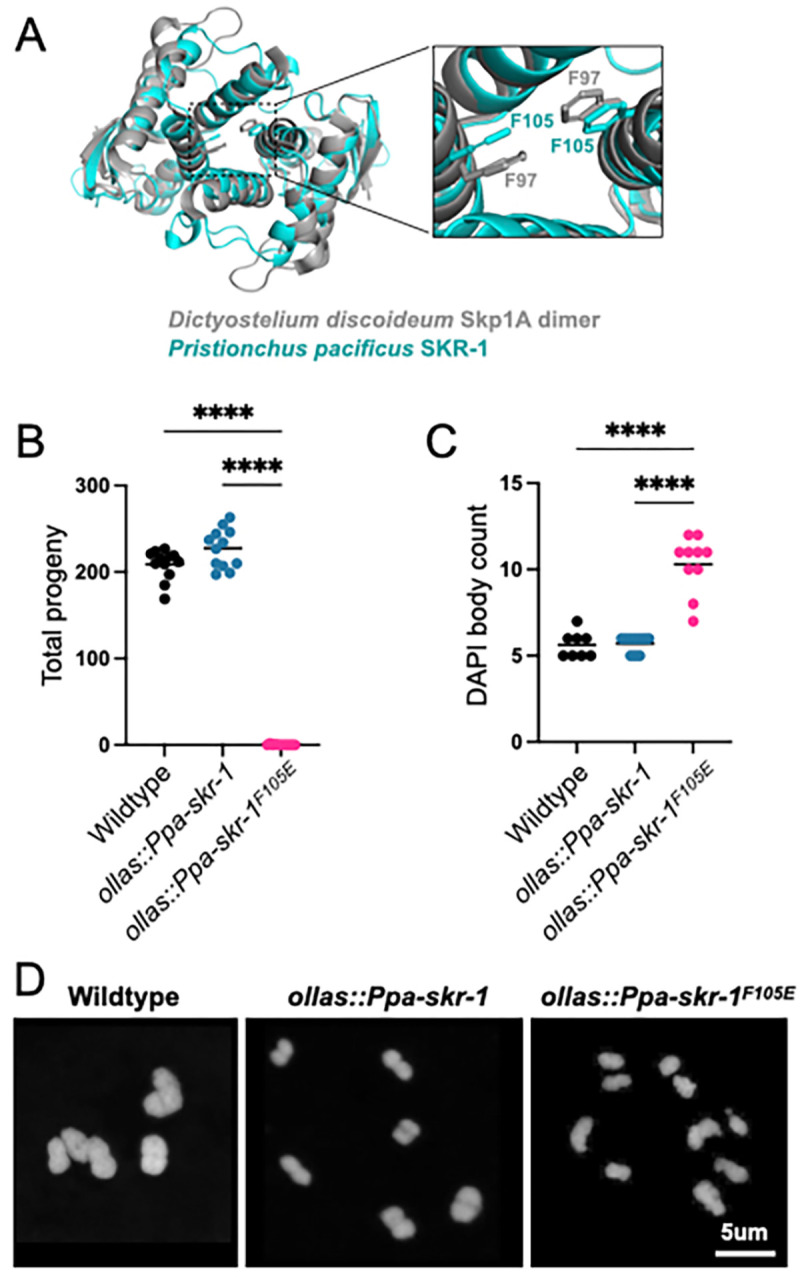
(A) Alignment of P. pacificus SKR-1 AlphaFold model (cyan) to Dictyostelium Skp1A dimer NMR structure (PDB structure 6V88, gray). Conserved phenylalanines required for dimerization are labeled in zoom. Dot plot depicting total progeny (B) and DAPI body count (C) for wild-type P. pacificus, ollas::Ppa-skr-1 and ollas::Ppa-skr-1F105E. Asterisks reflect P-values from Tukey’s multiple comparison test where **** indicates P < 0.0001. (D) Representative images of DAPI-stained Meiosis I bivalents (DAPI bodies) from the indicated genotypes.
To evaluate successful completion of meiosis, we counted total progeny in wild-type, ollas::Ppa-skr-1 and ollas::Ppa-skr-1F105E worms. Total progeny produced by ollas::Ppa-skr-1 worms were comparable to that of the wild-type P. pacificus, indicating that the OLLAS insertion did not interfere with meiosis. In contrast, ollas::Ppa-skr-1F105E worms were almost sterile, mimicking other SC null mutants (Figure 2B). Notably, several homozygous hermaphrodites produced one to two progeny, further indicating that OLLAS::Ppa-SKR-1F105E can carry out the non-meiotic functions of Skp1 proteins. Together, this analysis indicated that Ppa-SKR-1 dimerization is necessary for reproduction.
To examine meiotic dysfunction in more detail, we monitored successful formation of crossovers in meiotic prophase. Chromosomes that form a crossover are joined at metaphase of Meiosis I, forming so-called “bivalents” that can be visualized by staining DNA with DAPI. Since P. pacificus has six chromosome pairs, successful generation of a crossover on each pair yields six DAPI-staining bodies. We found no significant difference in DAPI body counts between wild-type and ollas::Ppa-skr-1 worms. They averaged 5.6 and 5.7 DAPI bodies, respectively (Figure 2C). However, ollas::Ppa-skr-1F105E worms had a significantly elevated DAPI body count (mean = 10.5) suggesting that failure of chromosome pairing or crossover formation underlies the reduced progeny count in ollas::Ppa-skr-1F105E worms (Figure 2D).
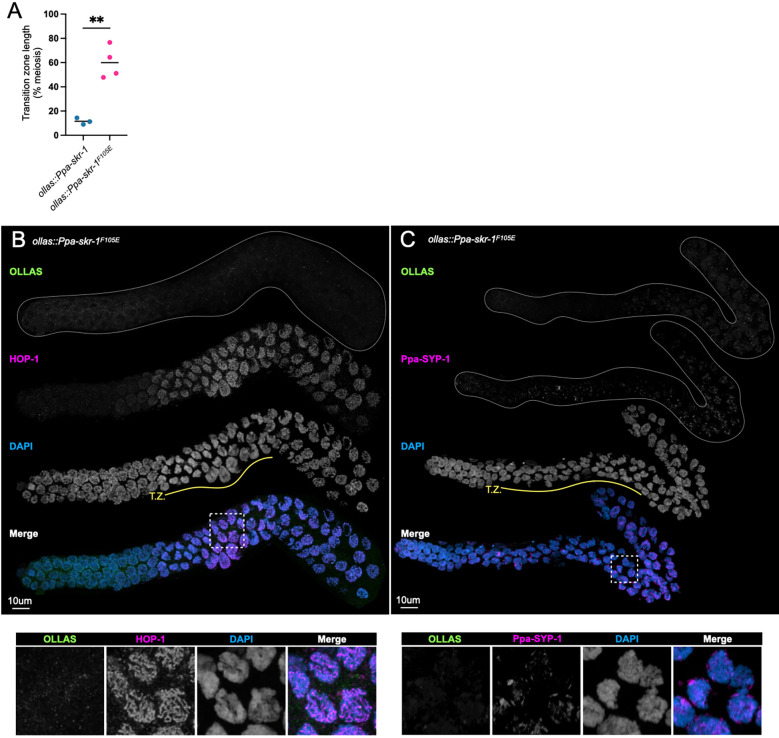
Cytological examination established that Ppa-skr-1F105E worms lack an SC. Meiotic nuclei in the mutant spent an extended duration in the transition zone - the region of the gonad where the SC assembles, marked by crescent-shaped nuclei (Figure 3, compare to Figure 1A, B). An increase in transition zone length is seen in other SC mutants (MacQueen et al. 2002; Colaiácovo et al. 2003; Smolikov et al. 2007; Smolikov, Schild-Prüfert, and Colaiácovo 2009) and is thought to reflect a synapsis checkpoint (Harper et al. 2011). HOP-1 appeared as thin tracks throughout the gonad in Ppa-skr-1F105E worms, indicative of chromosomes that were unable to assemble an SC (Figure 3B). Furthermore, Ppa-SYP-1 staining revealed complete lack of SC assembly (Figure 3C). In C. elegans and other species, SC components seem to be required for each other’s stability (Colaiácovo et al. 2003; Hurlock et al. 2020; Smolikov et al. 2007; Smolikov, Schild-Prüfert, and Colaiácovo 2009; Blundon et al. 2024; Z. Zhang et al. 2020). Indeed, Ppa-SYP-1 staining was almost completely absent in ollas::Ppa-skr-1F105E worms. Moreover, when SC components are present but cannot load onto chromosomes, SC material forms large aggregates called polycomplexes (Page and Hawley 2004). Notably, polycomplexes are absent in ollas::Ppa-skr-1F105E worms (Figure 3) and in C. elegans skr-1F115E worms (Blundon et al. 2024), suggesting other SC component are not able to assemble in the dimerization mutant. These data indicate that, like in C. elegans, SC formation in P. pacificus depends on Ppa-SKR-1 dimerization. Taken together with Ppa-SKR-1 localization (Figure 1), our data indicate that Ppa-SKR-1 is a structural component of the P. pacificus SC.
Figure 3: Ppa-SKR-1F105E fails to assemble the SC.
(A) Dot plot showing transition zone length as percent of meiosis. Asterisks reflect the P-value from an unpaired T-test where ** indicates P < 0.01. (B) and (C), Confocal images of whole gonads from P. pacificus ollas::skr-1F105E stained with anti-OLLAS, anti-HOP-1 (B) or anti-SYP-1 (C), and DAPI. Lower panels in (B) and (C) show zoom-in on regions indicated by white, dashed boxes and the transition zone is labeled below the DAPI channel in yellow (T.Z.).
Unlike other SC components, SKR-1 sequence is conserved in nematodes
We previously found that SC proteins in nematodes, Drosophila and mammals have a unique evolutionary signature; diverged protein sequence but conserved length and position of coiled-coil domains and conserved overall protein length (Kursel, Cope, and Rog 2021). We hypothesized that this evolutionary signature could be explained by the SC mode of assembly, which likely relies on weak multi-valent interactions mediated by coiled-coil domains. Since the sequence requirements for coiled-coil domains are flexible (typically defined as a heptad repeat where the first and fourth residues are hydrophobic and the fifth and seventh are charged or polar), selection to maintain coiled-coil domains could allow for significant sequence divergence. At the time of our analysis, SKR-1 had not been identified as an SC protein. Therefore, we wished to compare the evolutionary signature of SKR-1 to the other SC proteins.
Unlike the other SC proteins in Caenorhabditis and Pristionchus, the sequence of SKR-1 is conserved in both clades, ranking in the bottom one percentile for amino acid substitutions per site (Figure 4A). Unsurprisingly, residues involved in CUL-1 binding, F-box protein binding, and the dimerization interface are highly conserved, even between C. elegans, P. pacificus and H. sapiens (Figure 4B). We also found that SKR-1 does not contain conserved coiled-coil domains (Figure 4C, S3A). Pristionchus SKR-1 does have a low-scoring predicted coiled-coil domain from amino acids 20 – 47 (Figure S3A). However, AlphaFold does not predict a coiled-coil formed by Ppa-SKR-1 and this coiled-coil signature is not conserved in Caenorhabditis (Figure S3A) or in Dictyostelium, where the corresponding residues are mostly disordered in the NMR structure (Kim et al. 2020). Together, this argues against the functional importance of coiled-coil domains in SKR-1 (Figure S3B). Lastly, the length of SKR-1 is conserved, like other SC proteins (Figure 4D). Taken together, our analysis indicates that the evolutionary trajectory of SKR-1 is distinct from other SC proteins in Caenorhabditis and Pristionchus and suggests that its interaction with other SC proteins is mediated by domains other than coiled-coils.
Figure 4: SKR-1 has an evolutionary signature distinct from other SC proteins.
(A) Dot plot showing protein divergence for the Caenorhabditis and Pristionchus proteomes. SYP proteins and SKR-1 are indicated (black and pink, respectively). (B) Alignment of Skp1 orthologs from C. elegans and P. pacificus, and H. sapiens with Cul1 interaction, dimerization and F-box binding sites labeled (Zheng et al. 2002; Kim et al. 2020). Additionally, three mutants generated by Blundon and Caesar et al. are indicated by numbered boxes (Blundon et al. 2024). (C) and (D), dot plots showing coiled-coil conservaBon and coefficient of variaBon of protein length for the Caenorhabdi+s and Pris+onchus proteomes. SYP proteins and SKR-1 are indicated as in (A).
Discussion
We found that SKR-1 is a structural member of the SC in P. pacificus. Ppa-SKR-1 dynamically localizes to meiotic chromosomes in a manner that is indistinguishable from that of other SC proteins. Like other SC proteins, Ppa-SKR-1 exhibits stereotypic localization relative to the axes: it localizes to the middle of the SC, placing it near the N-terminus of Ppa-SYP-1 (Figure 1E). Finally, like in C. elegans, the dimerization interface of Ppa-SKR-1 is necessary for SC assembly but not for other essential functions. Taken together, our cytological, functional and phylogenetic data suggest that the function of SKR-1 as a structural component of the SC has been conserved since the common ancestor of C. elegans and P. pacificus, at least 100 million years ago.
Our work on the conservation of an SC role for SKR-1 in nematodes raises the possibility that it extends to Skp1 proteins in other clades. Unsurprisingly, proteasome-mediated degradation regulates multiple key steps in meiosis (Ahuja et al. 2017; Rao et al. 2017; Guan et al. 2022) and the proteasome itself localizes to the SC in C. elegans and mice (Rao et al. 2017; Ahuja et al. 2017). Skp1 also localizes to the SC in male and female mice (Guan et al. 2020), and in Arabidopsis plants where it is called ASK1 (Wang et al. 2004). In both cases, its disruption leads to meiotic defects (Yang et al. 2006). However, the essential functions of the proteosome and Skp1, and the consequent far-ranging effects of their disruption, has made it difficult to parse their role in the protein degradation from any potential structural role in the SC.
C. elegans has proved to be an especially valuable system for studying the role of Skp1 in the SC because it contains two partially redundant paralogs, SKR-1 and SKR-2. Having two SKR-1 paralogs allowed Blundon and Cesar et al. to identify the separation-of-function dimerization mutant. We similarly found that a mutation predicted to disrupt Ppa-SKR-1 dimerization results in separation of function; worms are viable and have no obvious growth defects indicating SCF functions are intact, but they are sterile due to failure of SC assembly. It will be interesting to explore whether the corresponding Skp1 dimerization interface - which is conserved at the protein sequence level in mammals and plants - would help to generate separation-of-function alleles in other model organisms.
The molecular details of SKR-1 interaction with other SC components remain unknown in both C. elegans and P. pacificus. SKR-1 proteins are not merely recruited to the SC like other so-called ‘client’ proteins, including the crossover regulator family ZHP-1/2/3/4 (Jantsch et al. 2004) and the polo-like kinase PLK-2 (L. Zhang et al. 2018; Harper et al. 2011; Labella et al. 2011). For example, the localization pattern of ZHP-1/2/3/4 is distinct from SC proteins and the SC can still assemble in the absence of the ZHPs. In contrast, SKR-1 is essential for SC assembly in both C. elegans and P. pacificus, and it contributes to the stability of SC components in vivo and in vitro. Such intimate co-dependence suggests the existence of underlying protein-protein interactions that provide specificity and stability.
The protein surfaces that mediate interactions between SC proteins must co-evolve to maintain compatibility. In this light, the high conservation of SKR-1 versus the high divergence of other SC components might seem surprising since proteins in complex often have homogenous evolutionary rates (Wong et al. 2008) and genes whose evolutionary rates covary tend to be functionally related (Clark, Alani, and Aquadro 2012). However, a more recent study reported that direct physical interaction is only a weak driver of evolutionary rate covariation (Little, Chikina, and Clark 2024). Moreover, moonlighting proteins that function in multiple complexes can confound such analyses. Taking these factors into account, SKR-1’s role in the highly conserved SCF complex might overwhelm any signal of shared evolutionary rates with other SC proteins. In addition, we note that the SC is a condensate (Rog, Köhler, and Dernburg 2017), and that many condensates rely on weak, multivalent interactions to recruit and exclude member and non-member components, respectively (Shin and Brangwynne 2017) . SC proteins might have multiple, redundant interaction interfaces with SKR-1, each too weak to pose a strong constraint on the primary sequence.
The recent duplication of SKR-1 in the lineage leading to C. elegans (Blundon et al. 2024) could suggest that gene duplication has allowed Skp1 proteins to adopt a novel function - a structural component of the SC. However, our findings suggest that the role of SKR-1 in the SC is more ancient and that a single SKR-1 protein has likely performed both functions in the common ancestor of C. elegans and P. pacificus. An ancestral dual-function protein suggests that SKR-1 has been subjected to evolutionary pressures to maintain both functions for at least 100 million years. Interestingly, SKR-1’s dual roles in SCF and the SC entail that mutations in skr-1 might have pleiotropic effects in development (SCF) versus reproduction (SC). If so, C. elegans may represent a lineage where such intralocus conflict is resolving by gene duplication and specialization (Castellanos, Wickramasinghe, and Betrán 2024). In this scenario, the different structural and functional requirements of the SC versus the SCF complex could be divided between SKR-1 and SKR-2, allowing them to eventually differentiate into an SC-dedicated protein and an SCF-dedicated one. Such specialization has likely taken place throughout the broader Skp1-related gene family, which has massively expanded in nematodes (Nayak et al. 2002). Intralocus conflict and related processes provide a leading framework in the evolution of aging (Adler and Bonduriansky 2014), suggesting that the evolutionary trajectory of SKR-1 in nematodes could shed light on the evolution of aging more broadly.
Materials and Methods
Worm strains and maintenance
We used Pristionchus pacificus strain PS312 for the wildtype control and for injections to make ollas::Ppa-skr-1. To make ollas::Ppa-skr-1F105E, we injected into ollas::Ppa-skr-1. All strains were grown at 20°C on NGM agar with OP50 bacteria. We maintained PS312 and ollas::Ppa-skr-1 in a homozygous state but since ollas::Ppa-skr-1F105E was sterile, we maintained it as a heterozygous line by singling animals and genotyping each generation. We consistently observed severe SC defects in one-quarter of the progeny from a heterozygous parent and never observed severe defects in progeny from ollas::Ppa-skr-1 or PS312 parents. For DAPI body counts, we identified gonads with SC defects in progeny of ollas::Ppa-skr-1F105E heterozygous animals, and considered those gonads with severe SC defects to be homozygous. To perform progeny counts of ollas::Ppa-skr-1F105E, we singled from a heterozygous parent, counted progeny and genotyped by PCR (see below) after the complete brood was laid.
Identification of P. pacificus SKR-1
We used C. elegans SKR-1 as a query in a BLASTp search, implemented on pristionchus.org, of the P. pacificus El Paco V3 genome (Dieterich et al. 2007). The top hit was ppa_stranded_DN29817_c0_g1_i2, a 166 amino acid protein. We also performed a tBLASTn search using C. elegans SKR-1 as a query against the El Paco V4 genome (GCA_000180635.4) implemented on ncbi.nlm.nih.gov. This identified the coding sequence KAF8362560.1, which encodes a 166 amino acid protein identical to ppa_stranded_DN29817_c0_g1_i2. When we used the 166 amino acid protein as a query in a BLASTp search of the C. elegans proteome, the top his was C. elegans SKR-1 (F46A9.5).
We note that performing the same BLASTp search against the P. pacificus genome on wormbase.org (Sternberg et al. 2024) produces a top hit to PPA23980, a protein with 1443 amino acids that contains a predicted ABC transporter transmembrane domain in its N-terminus and homology to SKR-1 in its C-terminus. We suspect that this is due to an annotation error that merges two genes since wormbase.org also hosts the El Paco V4 genome assembly and the start codon of the 166 amino acid version of SKR-1 is preserved in PPA23980.
To confirm that ppa_stranded_DN29817_c0_g1_i2 is indeed the SKR-1 ortholog in P. pacificus, we generated a neighbor-joining phylogenetic tree with all hits that resulted from BLASTp search of P. pacificus with C. elegans SKR-1 (File S1, S2, S3). Since P. pacificus ppa_stranded_DN29817_c0_g1_i2 groups closest with C. elegans SKR-1/2 (Figure S1, File S3), it is most likely to be the SKR-1 ortholog. Thus, we refer to ppa_stranded_DN29817_c0_g1_i2 as Ppa-SKR-1.
Sequence collection, alignment and phylogenetic analysis
We identified Caenorhabditis SKR-1 orthologs using the EnsEMBL Compara pipeline implemented on wormbase.org (Harris et al. 2010). We only kept sequences from species with a single predicted ortholog, with the exception of C. elegans, which has an SKR-1 paralog, SKR-2, leaving 16 SKR-1 sequences for analysis. We identified Pristionchus SKR-1 orthologs by performing BLASTp with C. elegans SKR-1 against the eight Pristionchus genomes available on Pristionchus.org (Dieterich et al. 2007). We saved the top hit from each search. We used Clustal Omega for all protein alignments and Geneious Tree Builder (neighbor-joining method, Geneious Prime version 2023.2.1) with 100x bootstrap resampling to generate the phylogenies in supplementary Figures 1 and 2. All protein sequences, alignments and trees are available as supplemental data (File S4 – S9).
CRISPR genome editing
We aimed to insert an OLLAS tag in the N-terminus of Ppa-SKR-1, immediately following the start methionine. We made an injection mix containing 1ul Cas9 (IDR, Alt-R S.p. Cas9 Nuclease V3, 10ug/ul), 3.5ul repair template (200uM), 3.5ul annealed tracr/crRNA mix and 0.5ul duplex buffer (IDT). We injected the gonads of wildtype (PS312) young adult hermaphrodite P. pacificus and singled each injected worm to its own plate. We extracted DNA from ~16 combined F1 worms from each plate and genotyped with primers that span the OLLAS insertion site (LEK1094 GTTTCACAACAACGGCCCTC and LEK1095 CTTGATGACGTCACGGGGAA) to identify “jackpot plates” (i.e., plates with high rates of OLLAS insertion). We singled as many F1s as possible from the jackpot plates and genotyped again to identify individual insertion events.
To make ollas::Ppa-skr-1F105E we followed a similar strategy as above except we injected into ollas::Ppa-skr-1. We screened the pooled F1s by doing PCR with primers LEK1111 (GAGAAGGGAACAACGTGGGT) and LEK1112 (CGCGCGTCTCATTCAACAAA) and digesting with MboI. The predicted Cas9 cut site is near an MboI site in ollas::skr-1, so CRISPR repair events could destroy the MboI site. In this scenario, wildtype plates will have bands that are 259, 241 and 92 base pairs in length after MboI digest but plates that contain CRISPR mutants will also have a 351 base pair band. We singled F1s from plates with the 351 base pair band and did a second round of genotyping with LEK1111 and LEK1112, this time followed by digest with SalI. Animals that contain CRISPR repair events from the injected homology template will gain an SalI site. PCR from wildtype animals will remain undigested (592 base pairs) whereas PCR from a mutant animal will get cut (336 and 256 base pair bands). See Table S1 for a list of primers, crRNAs and repair templates used for CRISPR.
Immunofluorescence and confocal microscopy
We prepared gonads for immunofluorescence and confocal microscopy as we have done previously (Kursel, Cope, and Rog 2021; Phillips, McDonald, and Dernburg 2009). Briefly, we dissected age-matched adult worms in egg buffer with 0.01% Tween-20 and fixed in a final concentration of 1% formaldehyde. We transferred samples to a HistoBond microscope slide, froze for 1 minute on dry ice and quickly immersed the slide in −20°C methanol for one minute. Slides were washed in PBST and blocked in Roche Block (Cat # 11096176001) for 30 minutes at room temperature. We incubated the slides in 80 μl of primary antibody overnight at 4°C. Primary antibody concentrations were as follows: Rabbit anti-PPA-SYP-1 1:500 (Kursel, Cope, and Rog 2021), Rat anti-OLLAS 1:200 (Invitrogen Catalog # MA5–16125), Rabbit anti-PPA-HOP-1 1:300 (Rillo-Bohn et al. 2021). The following day, slides were washed for three rounds of 10 minutes in PBST, then incubated in secondary antibody. Secondary antibody concentrations were as follows: Donkey anti-rabbit Cy3 1:500 and Donkey anti-rat Alexa488 1:500 (Jackson ImmunoResearch). Finally, we washed slides in PBST and DAPI and mounted them in NPG-Glycerol. Slides were imaged on a Zeiss LSM880 confocal microscope with Airyscan and a 63 × 1.4 NA oil objective. Confocal images presented in this manuscript are maximum intensity projections.
STED super-resolution microscopy
Gonads for STED microscopy were prepared as for confocal microscopy with the following changes: 1) we omitted DAPI staining, 2) we used Goat anti-Rabbit STAR RED 1:200 (Abberior # STRED-1002–500UG) and Goat anti-Rat Alexa 594 1:200 (Jackson ImmunoResearch) as secondaries, and 3) we mounted the samples in Abberior Mount Liquid Antifade (Abberior # MM-2009–2X15ML). Samples were imaged on Aberrior STEDYCON mounted on a Nikon Eclipse Ti microscope with a 100 × 1.45 NA oil objective. Line scans were performed in FIJI (Schindelin et al. 2012).
Structural modeling and alignment
We used AlphaFold (Jumper et al. 2021), implemented in ColabFold (Mirdita et al. 2022), to model the structure of full-length Ppa-SKR-1. We used Pymol ((Schrodinger 2015), version 2.5.7) to visualize Ppa-SKR-1 and to align it to the Dictyostelium Skp1A dimer NMR structure ((Kim et al. 2020), PDB structure 6V88).
Progeny counts
We singled twelve L4s from each genotype and grew them at 20°C. We moved the parents to a fresh plate every day for four days and counted the progeny after allowing them to mature for up to five days. For the ollas::Ppa-skr-1F105E genotype, we singled 50 F1s from a heterozygous animal. We moved the F1s to fresh plates daily as described. At the end of the fourth day of egg laying, we identified the homozygous animals among the F1s by genotyping the parent with LEK1111/LEK1112 PCR primers followed by SalI digest as described above. We counted progeny from those animals confirmed to be homozygous mutants.
Calculating divergence, coiled-coil conservation and length conservation
The Caenorhabditis and Pristionchus proteome values (Figure 4A, 4C and 4D) were published previously (Kursel, Cope, and Rog 2021). We calculated divergence values, coiled-coil conservation scores and coefficient of variation of protein length for SKR-1 as we have done previously for SC proteins (Kursel, Cope, and Rog 2021) using SKR-1 orthologs from Caenorhabditis or Pristionchus collected as described above.
Statistical analysis
We used an ordinary one-way ANOVA with Tukey’s multiple comparisons test to test for differences in total progeny and DAPI body counts between genotypes (Figure 2B and 2C). In Figure 3A, we used an unpaired t test to test for differences in transition zone length.
Supplementary Material
Acknowledgements
We would like to thank the Rog Lab for discussions, Abby Dernburg for antibodies and Yumi Kim for discussions and for sharing data prior to publication. Some strains used in this work were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We acknowledge the HSC Cell Imaging Core at the University of Utah for use of the STED microscope and The University of Utah Center for High Performance Computing for computational resources. KG was supported by the Undergraduate Research Opportunity Program at the University of Utah and by the Biology Research Scholar Award from the School of Biological Sciences. This work was supported by grants R35GM128804 from NIGMS and 2219605 from NSF.
Data availability
Worm strains generated in this study are available by request. All sequence alignments and phylogenies are included as supplementary data files. Proteome-wide analysis of divergence, coiled-coil scores and protein length variation in Caenorhabditis and Pristionchus was published previously (Kursel, Cope, and Rog 2021).
Literature cited
- Adler Margo I., and Bonduriansky Russell. 2014. “Sexual Conflict, Life Span, and Aging.” Cold Spring Harbor Perspectives in Biology 6 (8). 10.1101/cshperspect.a017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja Jasvinder S., Sandhu Rima, Mainpal Rana, Lawson Crystal, Henley Hanna, Hunt Patricia A., Yanowitz Judith L., and Valentin Börner G.. 2017. “Control of Meiotic Pairing and Recombination by Chromosomally Tethered 26S Proteasome.” Science 355 (6323): 408–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundon Joshua M., Cesar Brenda I., Bae Jung Woo, Čavka Ivana, Haversat Jocelyn, Ries Jonas, Köhler Simone, and Kim Yumi. 2024. “Skp1 Proteins Are Structural Components of the Synaptonemal Complex in C. Elegans.” Science Advances 10 (7): eadl4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos María Del Pilar, Wickramasinghe Chathuri Devmika, and Betrán Esther. 2024. “The Roles of Gene Duplications in the Dynamics of Evolutionary Conflicts.” Proceedings. Biological Sciences / The Royal Society 291 (2024): 20240555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark Nathan L., Alani Eric, and Aquadro Charles F.. 2012. “Evolutionary Rate Covariation Reveals Shared Functionality and Coexpression of Genes.” Genome Research 22 (4): 714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R., Lee M. H., Nayak S., Ohmachi M., Giorgini F., and Schedl T.. 2000. “FOG-2, a Novel F-Box Containing Protein, Associates with the GLD-1 RNA Binding Protein and Directs Male Sex Determination in the C. Elegans Hermaphrodite Germline.” Development 127 (24): 5265–76. [DOI] [PubMed] [Google Scholar]
- Colaiácovo Mónica P., MacQueen Amy J., Martinez-Perez Enrique, McDonald Kent, Adamo Adele, Volpe Adriana La, and Villeneuve Anne M.. 2003. “Synaptonemal Complex Assembly in C. Elegans Is Dispensable for Loading Strand-Exchange Proteins but Critical for Proper Completion of Recombination.” Developmental Cell 5 (3): 463–74. [DOI] [PubMed] [Google Scholar]
- Collins Kimberly A., Unruh Jay R., Slaughter Brian D., Yu Zulin, Lake Cathleen M., Nielsen Rachel J., Box Kimberly S., et al. 2014. “Corolla Is a Novel Protein That Contributes to the Architecture of the Synaptonemal Complex of Drosophila.” Genetics 198 (1): 219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo Cynthia, Reese Kimberly J., and Seydoux Geraldine. 2003. “Exclusion of Germ Plasm Proteins from Somatic Lineages by Cullin-Dependent Degradation.” Nature 424 (6949): 685–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich Christoph, Roeseler Waltraud, Sobetzko Patrick, and Sommer Ralf J.. 2007. “Pristionchus.org: A Genome-Centric Database of the Nematode Satellite Species Pristionchus Pacificus.” Nucleic Acids Research 35 (Database issue): D498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunce James M., Dunne Orla M., Ratcliff Matthew, Millán Claudia, Madgwick Suzanne, Usón Isabel, and Davies Owen R.. 2018. “Structural Basis of Meiotic Chromosome Synapsis through SYCP1 Self-Assembly.” Nature Structural & Molecular Biology 25 (7): 557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunce James M., Salmon Lucy J., and Davies Owen R.. 2021. “Structural Basis of Meiotic Chromosome Synaptic Elongation through Hierarchical Fibrous Assembly of SYCE2-TEX12.” Nature Structural & Molecular Biology 28 (8): 681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne Orla M., and Davies Owen R.. 2019. “Molecular Structure of Human Synaptonemal Complex Protein SYCE1.” Chromosoma 128 (3): 223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray William M., Carlos del Pozo J., Walker Loni, Ho bbie Lawrence, Risseeuw Eddy, Banks Travis, Crosby William L., Yang Ming, Ma Hong, and Estelle Mark. 1999. “Identification of an SCF Ubiquitin–Ligase Complex Required for Auxin Response in Arabidopsis Thaliana.” Genes & Development 13 (13): 1678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Yongjuan, Adrian Leu N., Ma Jun, Chmátal Lukáš, Ruthel Gordon, Bloom Jordana C., Lampson Michael A., Schimenti John C., Luo Mengcheng, and Jeremy Wang P.. 2020. “SKP1 Drives the Prophase I to Metaphase I Transition during Male Meiosis.” Science Advances 6 (13): eaaz2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Yongjuan, Lin Huijuan, Adrian Leu N., Ruthel Gordon, Fuchs Serge Y., Busino Luca, Luo Mengcheng, and Jeremy Wang P.. 2022. “SCF Ubiquitin E3 Ligase Regulates DNA Double-Strand Breaks in Early Meiotic Recombination.” Nucleic Acids Research 50 (9): 5129–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Linqu, Mason Mary, Risseeuw Eddy P., Crosby William L., and Somers David E.. 2004. “Formation of an SCF(ZTL) Complex Is Required for Proper Regulation of Circadian Timing.” The Plant Journal: For Cell and Molecular Biology 40 (2): 291–301. [DOI] [PubMed] [Google Scholar]
- Harper Nicola C., Rillo Regina, Jover-Gil Sara, Assaf Zoe June, Bhalla Needhi, and Dernburg Abby F.. 2011. “Pairing Centers Recruit a Polo-like Kinase to Orchestrate Meiotic Chromosome Dynamics in C. Elegans.” Developmental Cell 21 (5): 934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris Todd W., Antoshechkin Igor, Bieri Tamberlyn, Blasiar Darin, Chan Juancarlos, Chen Wen J., Cruz Norie De La, et al. 2010. “WormBase: A Comprehensive Resource for Nematode Research.” Nucleic Acids Research 38 (Database issue): D463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer Lucas W., and Blumenstiel Justin P.. 2016. “Holding It Together: Rapid Evolution and Positive Selection in the Synaptonemal Complex of Drosophila.” BMC Evolutionary Biology 16 (May): 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzl M. T., Thalmann I., and Thalmann R.. 1998. “OCP2 Exists as a Dimer in the Organ of Corti.” Hearing Research 126 (1–2): 37–46. [DOI] [PubMed] [Google Scholar]
- Hurlock Matthew E., Čavka Ivana, Kursel Lisa E., Haversat Jocelyn, Wooten Matthew, Nizami Zehra, Turniansky Rashi, et al. 2020. “Identification of Novel Synaptonemal Complex Components in C. Elegans.” The Journal of Cell Biology 219 (5). 10.1083/jcb.201910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch Verena, Pasierbek Pawel, Mueller Michael M., Schweizer Dieter, Jantsch Michael, and Loidl Josef. 2004. “Targeted Gene Knockout Reveals a Role in Meiotic Recombination for ZHP-3, a Zip3-Related Protein in Caenorhabditis Elegans.” Molecular and Cellular Biology 24 (18): 7998–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper John, Evans Richard, Pritzel Alexander, Green Tim, Figurnov Michael, Ronneberger Olaf, Tunyasuvunakool Kathryn, et al. 2021. “Highly Accurate Protein Structure Prediction with AlphaFold.” Nature 596 (7873): 583–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Hyun W., Eletsky Alexander, Gonzalez Karen J., Wel Hanke van der, Strauch Eva-Maria, Prestegard James H., and West Christopher M.. 2020. “Skp1 Dimerization Conceals Its F-Box Protein Binding Site.” Biochemistry 59 (15): 1527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler Simone, Wojcik Michal, Xu Ke, and Dernburg Abby F.. 2020. “Dynamic Molecular Architecture of the Synaptonemal Complex.” BioRxiv. bioRxiv. 10.1101/2020.02.16.947804. [DOI] [Google Scholar]
- Kursel Lisa E., Aguayo Martinez Jesus E., and Rog Ofer E.. 2023. “A Suppressor Screen in C. Elegans Identifies a Multi-Protein Interaction Interface That Stabilizes the Synaptonemal Complex.” BioRxiv. 10.1101/2023.08.21.554166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel Lisa E., Cope Henry D., and Rog Ofer. 2021. “Unconventional Conservation Reveals Structure-Function Relationships in the Synaptonemal Complex.” ELife 10 (November). 10.7554/eLife.72061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursel Lisa E., Aguayo Martinez Jesus E., and Rog Ofer. 2023. “A Suppressor Screen in C. Elegans Identifies a Multiprotein Interaction That Stabilizes the Synaptonemal Complex.” Proceedings of the National Academy of Sciences of the United States of America 120 (50): e2314335120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella Sara, Woglar Alexander, Jantsch Verena, and Zetka Monique. 2011. “Polo Kinases Establish Links between Meiotic Chromosomes and Cytoskeletal Forces Essential for Homolog Pairing.” Developmental Cell 21 (5): 948–58. [DOI] [PubMed] [Google Scholar]
- Little Jordan, Chikina Maria, and Clark Nathan L.. 2024. “Evolutionary Rate Covariation Is a Reliable Predictor of Co-Functional Interactions but Not Necessarily Physical Interactions.” ELife 12 (February). 10.7554/eLife.93333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen Amy J., Colaiácovo Mónica P., McDonald Kent, and Villeneuve Anne M.. 2002. “Synapsis-Dependent and -Independent Mechanisms Stabilize Homolog Pairing during Meiotic Prophase in C. Elegans.” Genes & Development 16 (18): 2428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita Milot, Schütze Konstantin, Moriwaki Yoshitaka, Heo Lim, Ovchinnikov Sergey, and Steinegger Martin. 2022. “ColabFold: Making Protein Folding Accessible to All.” Nature Methods 19 (6): 679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak Sudhir, Santiago Fernando E., Jin Hui, Lin Debbie, Schedl Tim, and Kipreos Edward T.. 2002. “The Caenorhabditis Elegans Skp1-Related Gene Family: Diverse Functions in Cell Proliferation, Morphogenesis, and Meiosis.” Current Biology: CB 12 (4): 277–87. [DOI] [PubMed] [Google Scholar]
- Ouni Ikram, Flick Karin, and Kaiser Peter. 2010. “A Transcriptional Activator Is Part of an SCF Ubiquitin Ligase to Control Degradation of Its Cofactors.” Molecular Cell 40 (6): 954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page Scott L., and Scott Hawley R.. 2004. “The Genetics and Molecular Biology of the Synaptonemal Complex.” Annual Review of Cell and Developmental Biology 20: 525–58. [DOI] [PubMed] [Google Scholar]
- Page Scott L., Khetani Radhika S., Lake Cathleen M., Nielsen Rachel J., Jeffress Jennifer K., Warren William D., Bickel Sharon E., and Scott Hawley R.. 2008. “Corona Is Required for Higher-Order Assembly of Transverse Filaments into Full-Length Synaptonemal Complex in Drosophila Oocytes.” PLoS Genetics 4 (9): e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips Carolyn M., McDonald Kent L., and Dernburg Abby F.. 2009. “Cytological Analysis of Meiosis in Caenorhabditis Elegans.” Methods in Molecular Biology 558: 171–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H. B. D. Prasada, Qiao Huanyu, Bhatt Shubhang K., Bailey Logan R. J., Tran Hung D., Bourne Sarah L., Qiu Wendy, et al. 2017. “A SUMO-Ubiquitin Relay Recruits Proteasomes to Chromosome Axes to Regulate Meiotic Recombination.” Science 355 (6323): 403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillo-Bohn Regina, Adilardi Renzo, Mitros Therese, Avşaroğlu Barış, Stevens Lewis, Köhler Simone, Bayes Joshua, et al. 2021. “Analysis of Meiosis in Pristionchus Pacificus Reveals Plasticity in Homolog Pairing and Synapsis in the Nematode Lineage.” ELife 10 (August): e70990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog Ofer, Köhler Simone, and Dernburg Abby F.. 2017. “The Synaptonemal Complex Has Liquid Crystalline Properties and Spatially Regulates Meiotic Recombination Factors.” ELife 6 (January). 10.7554/eLife.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Sáez Fernando, Laura Gómez-H Orla M. Dunne, Cristina Gallego-Páramo Natalia Felipe-Medina, Manuel Sánchez-Martín Elena Llano, Pendas Alberto M., and Davies Owen R.. 2020. “Meiotic Chromosome Synapsis Depends on Multivalent SYCE1-SIX6OS1 Interactions That Are Disrupted in Cases of Human Infertility.” Science Advances 6 (36). 10.1126/sciadv.abb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert Kristina, Saito Takamune T., Smolikov Sarit, Gu Yanjie, Hincapie Marina, Hill David E., Vidal Marc, McDonald Kent, and Colaiácovo Monica P.. 2011. “Organization of the Synaptonemal Complex during Meiosis in Caenorhabditis Elegans.” Genetics 189 (2): 411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin Johannes, Arganda-Carreras Ignacio, Frise Erwin, Kaynig Verena, Longair Mark, Pietzsch Tobias, Preibisch Stephan, et al. 2012. “Fiji: An Open-Source Platform for Biological-Image Analysis.” Nature Methods 9 (7): 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm Sabine, Fraune Johanna, Naumann Ronald, Hernandez-Hernandez Abrahan, Höög Christer, Cooke Howard J., Alsheimer Manfred, and Benavente Ricardo. 2011. “A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility.” PLoS Genetics 7 (5): e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. L. C. 2015. “The PyMOL Molecular Graphics System.” Version 1: 8. [Google Scholar]
- Shin Yongdae, and Brangwynne Clifford P.. 2017. “Liquid Phase Condensation in Cell Physiology and Disease.” Science 357 (6357). 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Smolikov Sarit, Eizinger Andreas, Schild-Prufert Kristina, Hurlburt Allison, McDonald Kent, Engebrecht Joanne, Villeneuve Anne M., and Colaiácovo Mónica P.. 2007. “SYP-3 Restricts Synaptonemal Complex Assembly to Bridge Paired Chromosome Axes during Meiosis in Caenorhabditis Elegans.” Genetics 176 (4): 2015–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov Sarit, Schild-Prüfert Kristina, and Colaiácovo Mónica P.. 2009. “A Yeast Two-Hybrid Screen for SYP-3 Interactors Identifies SYP-4, a Component Required for Synaptonemal Complex Assembly and Chiasma Formation in Caenorhabditis Elegans Meiosis.” PLoS Genetics 5 (10): e1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg Paul W., Auken Kimberly Van, Wang Qinghua, Wright Adam, Yook Karen, Zarowiecki Magdalena, Arnaboldi Valerio, et al. 2024. “WormBase 2024: Status and Transitioning to Alliance Infrastructure.” Genetics 227 (1). 10.1093/genetics/iyae050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Yixing, Wu Hong, Liang Genqing, and Yang Ming. 2004. “Defects in Nucleolar Migration and Synapsis in Male Prophase I in the Ask1–1 Mutant of Arabidopsis.” Sexual Plant Reproduction 16 (6): 273–82. [Google Scholar]
- Willems Andrew R., Schwab Michael, and Tyers Mike. 2004. “A Hitchhiker’s Guide to the Cullin Ubiquitin Ligases: SCF and Its Kin.” Biochimica et Biophysica Acta 1695 (1–3): 133–70. [DOI] [PubMed] [Google Scholar]
- Wong Philip, Althammer Sonja, Hildebrand Andrea, Kirschner Andreas, Pagel Philipp, Geissler Bernd, Smialowski Pawel, et al. 2008. “An Evolutionary and Structural Characterization of Mammalian Protein Complex Organization.” BMC Genomics 9 (December): 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Xiaohui, Timofejeva Ljudmilla, Ma Hong, and Makaroff Christopher A.. 2006. “The Arabidopsis SKP1 Homolog ASK1 Controls Meiotic Chromosome Remodeling and Release of Chromatin from the Nuclear Membrane and Nucleolus.” Journal of Cell Science 119 (Pt 18): 3754–63. [DOI] [PubMed] [Google Scholar]
- Zhang Liangyu, Köhler Simone, Rillo-Bohn Regina, and Dernburg Abby F.. 2018. “A Compartmentalized Signaling Network Mediates Crossover Control in Meiosis.” ELife 7 (March). 10.7554/eLife.30789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Zhenguo, Xie Songbo, Wang Ruoxi, Guo Shuqun, Zhao Qiuchen, Nie Hui, Liu Yuanyuan, et al. 2020. “Multivalent Weak Interactions between Assembly Units Drive Synaptonemal Complex Formation.” The Journal of Cell Biology 219 (5). 10.1083/jcb.201910086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Ning, Schulman Brenda A., Song Langzhou, Miller Julie J., Jeffrey Philip D., Wang Ping, Chu Claire, et al. 2002. “Structure of the Cul1–Rbx1–Skp1–F BoxSkp2 SCF Ubiquitin Ligase Complex.” Nature 416 (6882): 703–9. [DOI] [PubMed] [Google Scholar]
- Zickler Denise, and Kleckner Nancy. 2015. “Recombination, Pairing, and Synapsis of Homologs during Meiosis.” Cold Spring Harbor Perspectives in Biology 7 (6). 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Worm strains generated in this study are available by request. All sequence alignments and phylogenies are included as supplementary data files. Proteome-wide analysis of divergence, coiled-coil scores and protein length variation in Caenorhabditis and Pristionchus was published previously (Kursel, Cope, and Rog 2021).