Abstract
The single-stranded adeno-associated virus type 2 (AAV) genome is flanked by terminal repeats (TRs) that fold back on themselves to form hairpinned structures. During AAV DNA replication, the TRs are nicked by the virus-encoded Rep proteins at the terminal resolution site (trs). This origin function apparently requires three sequence elements, the Rep binding element (RBE), a small palindrome that comprises a single tip of an internal hairpin within the TR (RBE′), and the trs. Previously, we determined the sequences at the trs required for Rep-mediated cleavage and demonstrated that the trs endonuclease reaction occurs in two discrete steps. In the first step, the Rep DNA helicase activity unwinds the TR, thereby extruding a stem-loop structure at the trs. In the second step, Rep transesterification activity cleaves the trs. Here we investigate the contribution of the RBE and RBE′ during this process. Our data indicate that Rep is tethered to the RBE in a specific orientation during trs nicking. This orientation appears to align Rep on the AAV TR, allowing specific nucleotide contacts with the RBE′ and directing nicking to the trs. Accordingly, alterations in the polarity or position of the RBE relative to the trs greatly inhibit Rep nicking. Substitutions within the RBE′ also reduce Rep specific activity, but to a lesser extent. Interestingly, Rep interactions with the RBE and RBE′ during nicking seem to be functionally distinct. Rep contacts with the RBE appear necessary for both the DNA helicase and trs cleavage steps of the endonuclease reaction. On the other hand, RBE′ contacts seem to be required primarily for TR unwinding and formation of the trs stem-loop structure, not cleavage. Together, these results suggest a model of Rep interaction with the AAV TR during origin nicking through a tripartite cleavage signal comprised of the RBE, the RBE′, and the trs.
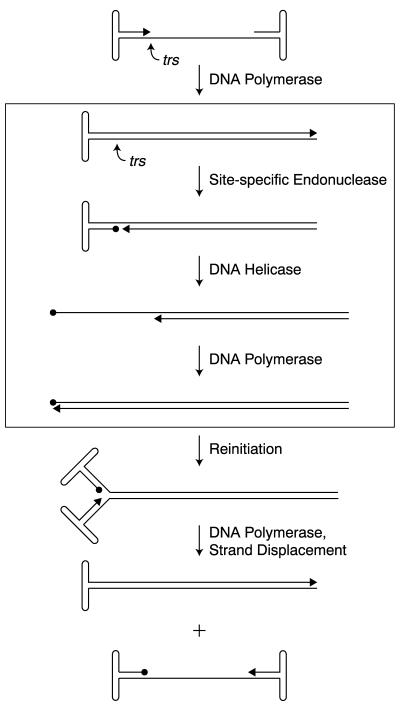
The single-stranded DNA, adeno-associated virus (AAV) genome is flanked by terminal repeats (TRs). Internal palindromes allow each TR to fold back on itself, forming terminal hairpinned structures that function as origins for AAV DNA replication, as well as integration and packaging signals (12, 21, 28). During infection, synthesis of the AAV genome is initiated by an unidentified host cell DNA polymerase using the 3′-hydroxyl primer of the hairpinned TR. This second-strand synthesis results in the replication of internal genes, allowing production of viral proteins (Fig. 1). Yet, continued AAV DNA synthesis requires the introduction of a site-specific, single-stranded nick into the TRs by the virus-encoded, nonstructural Rep proteins, Rep78 and Rep68 (14, 22, 33, 35). In our current model of AAV DNA replication, Rep origin nicking and subsequent Rep-mediated unwinding of the TR generate a 3′-hydroxyl primer for repair synthesis of the TR. During this process of terminal resolution, Rep induces a single-stranded nick at the terminal resolution site (trs), forming a 5′-phosphotyrosyl linkage between Rep and the nicking site (Fig. 1) (14, 31, 33). Current evidence suggests that this Rep origin nicking activity requires three functional elements within the AAV TR, the canonical Rep binding element (RBE), a portion of the small internal palindromes within the terminal hairpin (RBE′), and the trs (see Fig. 2A) (8, 19, 20, 27, 32, 36). The secondary structure of the internal palindromes may also play a role (17).
FIG. 1.
Model of AAV DNA replication. The boxed region illustrates the steps involved in the terminal resolution of AAV viral ends. In vitro, Rep68 is necessary and sufficient for both the site-specific endonuclease and helicase activities required for terminal resolution. The viral 3′ end is indicated with an arrow. Circles depict Rep covalently attached to the viral 5′ end at the terminal resolution site (trs).
FIG. 2.
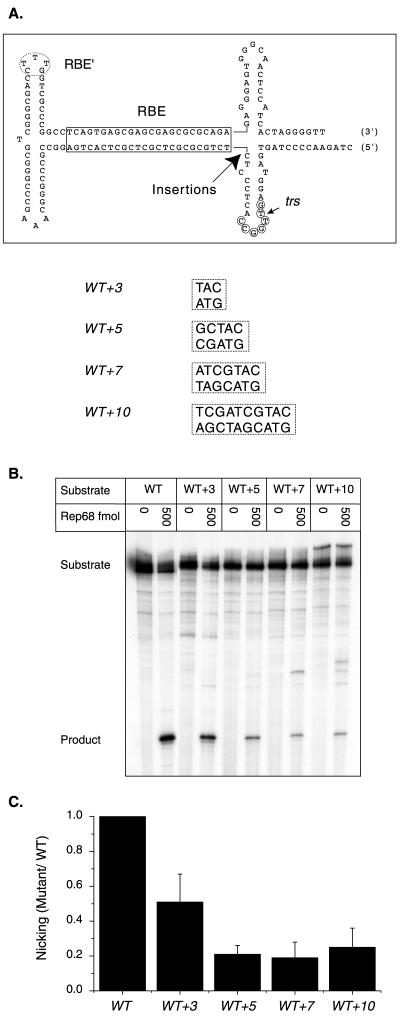
Rep nicking activity on RBE insertion mutants. (A) The wt AAV TR is depicted after extrusion of trs stem-loop structure. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. The position of insertions is indicated with a large arrow. The inserted sequences are given next to the mutant identifier. (B) Rep68 endonuclease reactions were performed on wt and insertion substrates in the presence of 0.5 mM ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. A representative gel is shown. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. The positions of substrates and products are indicated. (C) Nicking data were obtained from two independent trials. The relative Rep specific activity for each mutant was expressed as the fraction of mutant substrate nicked divided by the fraction of wt substrate nicked at the same Rep concentration. Ratios obtained at different Rep concentrations in the two trials were then averaged and graphed for each mutant (n = 4 for all substrates except WT+10, where n = 7). Bars indicate the standard deviations from means.
Previously, we identified the core trs sequence necessary for efficient Rep-catalyzed nicking, 3′-CCGGT/TG-5′ (6). This core sequence is strand specific in that it is required only on the nicked strand. Interestingly, the sequences flanking the trs contain an inverted repeat conserved among various AAV serotypes (6). Similar to rolling circle origins of DNA replication (23, 24), these inverted repeats appear to form a nicking site stem-loop structure. In the case of AAV, extrusion of this structure requires ATP-dependent, Rep helicase activity, but once this structure is formed, the actual endonuclease reaction is not dependent on ATP (6). Since the nicking site is within the single-stranded region of this origin stem-loop, it appears that the nicking intermediate is a single-stranded trs (6).
In addition to a specific sequence and structure at the trs, efficient Rep nicking requires two additional sequence recognition elements within the AAV TR, the RBE and RBE′. Mutational analysis has identified a core 22-bp sequence required for stable Rep binding to linear TR substrates (27). This RBE includes the tetrameric GAGC repeat identified by several groups as necessary for stable Rep binding to both linear and hairpinned TR substrates (4, 8, 19, 20, 32, 36). Moreover, chemical interference assays indicate that all major Rep contacts within the linear portion of the hairpinned TR fall within the RBE (1, 25, 27). Thus, the 22-bp RBE appears to be the primary sequence element promoting Rep binding to the AAV TR. Homologues of this RBE are present at the AAV p5 promoter, the preferential proviral integration site on human chromosome 19, and within several viral and cellular promoters (3, 13, 15, 16, 18, 26, 36–38).
Although the contribution of the RBE to Rep-catalyzed trs nicking has not been determined, mutant AAV genomes containing multiple transversions in the RBE replicate at much lower levels than do wild-type (wt) genomes (4). This observation has led to the conclusion that stable Rep binding to the AAV TR is necessary for efficient origin nicking and subsequent viral replication. There is one report of a Rep mutant that fails to bind the AAV TR and yet nicks TR substrates in vitro, albeit at lower levels than those of the wt. However, this Rep mutant does not cleave these substrates at the trs but 11 or 12 nucleotides downstream of the correct nicking site (2). Thus, our current model predicts that the RBE establishes the polarity of Rep interaction with the AAV TR, correctly aligning Rep over the trs, culminating in nicking of the correct strand at the correct site. This alignment over the trs is thought to be quite precise since the correct strand of the trs is nicked even when both strands contain the same sequence (32).
Rep interaction with the AAV TR is enhanced by sequences within the internal palindromes of the terminal hairpin. Rep binds the complete TR with 125- to 170-fold-greater affinity than linear TR substrates lacking the internal palindromes (20, 27). Moreover, Rep trs nicking on similar linear TR substrates is reduced 4- to 50-fold compared to hairpinned TR substrates (7, 19, 34, 39). Previously, it was thought that Rep made no specific contacts with the terminal hairpin, but recently, Ryan et al. (27) identified Rep sequence contacts with the CTTTG motif at one tip of the secondary structure element. Curiously, this short sequence, referred to here as the RBE′, has a constant position with respect to the trs regardless of the orientation of the TR (flip or flop). Deletion of the RBE′ and adjacent sequences reduces both Rep nicking in vitro and viral DNA replication in vivo (5, 32, 39).
Though many functions have been attributed to the interaction of Rep with the AAV TRs, the mechanics and architecture of this interaction remain undetermined. Here we investigate the functional roles of the RBE and RBE′ in an attempt to better characterize the mechanism of Rep-catalyzed trs nicking in vitro. We determine the contribution of the RBE by altering the polarity and distance of the RBE relative to the trs within mutant TR substrates. Increased spacing between the RBE and trs or a change in the polarity of the RBE dramatically reduces Rep specific activity, and only the wt orientation of the RBE is able to support efficient Rep nicking. These data indicate that association with the RBE is critical to the correct alignment of the Rep active site over the trs for efficient cleavage. Additionally, we characterize the contribution of the RBE′ to Rep-mediated trs nicking using a panel of substitution mutants. The mutants indicate that specific nucleotides within the RBE′ are required for efficient, Rep-mediated cleavage. These RBE′ contacts apparently contribute to Rep-mediated unwinding of sequences near the trs and formation of the correct nicking intermediate. Together, these results suggest a model for Rep interaction with the AAV TR during trs nicking.
MATERIALS AND METHODS
Purification of baculovirus-expressed Rep68.
Rep68 was purified to homogeneity from baculovirus-infected Sf9 cells as previously described (40). Rep68 was purified by sequential chromatography on phenyl-Sepharose, single-stranded DNA-cellulose, and DEAE-cellulose. Preparations were more than 99% pure as judged by sodium dodecyl sulfate-acrylamide gel electrophoresis followed by silver staining (40).
DNA substrates.
The TR substrates used in this study were constructed from gel-purified, synthetic oligonucleotides (Genosys) as previously described (6). However, construction methods were scaled up to increase yields. Accordingly, 200 pmol of two annealed oligonucleotides containing the RBE and the trs sequences were ligated to 1,000 pmol of a third oligonucleotide containing the terminal hairpin. Oligonucleotides were ligated together at 32°C for 2 h in a 100-μl reaction volume containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg of bovine serum albumin per ml, and 1,600 U of T4 DNA ligase (New England Biolabs). Complete 163-nucleotide TR constructs were purified from ethidium bromide-stained, 10% denaturing polyacrylamide gels containing 50% urea. DNA concentrations of purified substrates were determined using the Pico Green fluorometric reagent (Molecular Probes). Each panel of mutant and wt constructs was assayed together to ensure accurate relative DNA concentrations. TR substrates were 5′ end labeled at 37°C in a 10-μl reaction mixture containing 200 fmol of substrate, 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol, 20 μCi of [γ-32P]ATP, and 20 U of T4 polynucleotide kinase (New England Biolabs). Final concentrations of labeled substrates were determined on the basis of specific activity and confirmed with the Pico Green fluorometric reagent (Molecular Probes).
trs endonuclease assay.
The trs endonuclease reactions were performed as described previously (6, 14). The 20-μl reaction mixtures contained 25 mM HEPES-KOH (pH 7.5), 20 mM NaCl, 5.5 mM MgCl2, 10 ng of bovine serum albumin per ml, 0.25 pM 5′-end-labeled TR substrate (104 cpm/fmol), and 0.5 mM ATP, unless otherwise indicated. The reaction mixtures were incubated at 37°C for 1 h. Proteinase K-digested reaction products were phenol-chloroform extracted, ethanol precipitated, washed in 70% ethanol, and fractionated on 10% denaturing polyacrylamide gels containing 50% urea. The amount of product formed was determined with a phosphorimager (Fuji). To confirm that we were in the linear range of the phosphorimager, we experimentally compared radioactive standards by phosphorimager and scintillation counting.
Unless otherwise indicated, each mutant was assayed at three or four Rep concentrations, giving several data points for each substrate. Since the kinetics of Rep nicking are sigmoidal with respect to enzyme concentration (40), this approach provided the opportunity to measure Rep activity within a linear range for nicking and to repeat the nicking assay multiple times for each substrate. Only the correct-sized product resulting from Rep cleavage at the trs was counted for analysis. Minor cuts at less favored sites or nicks present in the starting substrate were not included in phosphorimager analysis.
RESULTS
Spacing between the RBE and trs is critical during Rep nicking.
Previously, we determined that Rep makes sequence-specific contacts at the trs and that sequences flanking the trs include a conserved inverted repeat which apparently forms a nicking site stem-loop structure (6). Substrates in which this stem-loop was extruded (NOSTEM) abolished the ATP requirement for Rep68-mediated trs nicking in vitro, indicating that the actual Rep endonuclease reaction did not require ATP. Moreover, Rep nicked the NOSTEM substrate with a two- to threefold-greater specific activity than that for wt. Since the specific activity of Rep nicking on this substrate was essentially the same in the presence and in the absence of ATP, we concluded that the DNA helicase activity of Rep was not required once the trs stem-loop was formed (6). Although these observations helped to clarify the nature of Rep interaction with the trs, the functional contribution of Rep interaction with the RBE during trs nicking remained unclear.
To assess the importance of spacing between the RBE and the trs during Rep nicking, a panel of insertion mutants was constructed. These mutant TRs contained 3, 5, 7, or 10 bp of heterologous sequence inserted directly between the RBE and the trs stem-loop structure (Fig. 2A) in a region that does not overlap with either element. TRs were constructed from three synthetic oligonucleotides that were ligated together as described in Materials and Methods. Complete TR constructs were then purified from ethidium bromide-stained, 10% denaturing polyacrylamide gels and 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Rep trs cleavage was assayed on mutant and wt substrates in vitro using homogeneously pure Rep68 as described in Materials and Methods.
Rep68 nicking was reduced on all of the insertion mutants compared to wt and decreased as the spacing between the RBE and the trs increased (Fig. 2B and C). Furthermore, no significant improvement was seen in the WT+10 mutant, in which the trs site was expected to be on approximately the same side of the DNA helix as the wt substrate. This trend suggests that the actual spacing between the RBE and trs is important during Rep nicking and not just the relative position of the trs on the surface of the DNA helix. Although most of the Rep68-mediated cleavage on the mutant substrates occurred at the trs, some non-trs Rep68 nicking was observed on the 7- and 10-bp insertion mutants. This secondary site nicking occurred on the correct strand but internally with respect to the trs, suggesting that the increased spacing between the RBE and trs was changing the specificity of Rep68 nicking. Together, these observations indicate that the spacing between the RBE and the nicking site is critical for both accurate and efficient Rep68 trs cleavage.
Rep remains bound to the RBE during trs nicking.
Although the spacing between the RBE and trs appeared important, the reason for the spacing requirement was unclear. One possibility was that the increased distance of our spacer mutants prevented formation of the nicking site stem-loop structure by endogenous Rep DNA helicase activity. To investigate this possibility, we constructed a 10-bp insertion mutant that included a preferentially extruded trs stem-loop structure (Fig. 3A, NOSTEM+10 substrate). Rep nicking on this substrate and its wt counterpart, NOSTEM, should no longer require ATP or DNA helicase activity (6). Thus, we reasoned that if increased spacing inhibited trs stem-loop formation, then preferentially extruding this structure should result in efficient Rep68 trs nicking of the 10-bp insertion mutant in the absence of ATP.
FIG. 3.
Rep nicking activity on RBE insertion mutants in the NOSTEM background. (A) The two TR substrates containing preferentially extruded trs stem-loop structure are illustrated. NOSTEM and NOSTEM+10 TRs are depicted after formation of trs stem-loop structure. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. The position and sequence of the NOSTEM+10 insertion are indicated with boldface italics. (B) Rep68 endonuclease reactions were performed on wt and the WT+10 insertion substrates in the presence of 0.5 mM ATP as described in Materials and Methods. Rep68 endonuclease reactions were performed on NOSTEM and NOSTEM+10 insertion substrates in the absence of ATP. Products were resolved on a 10% denaturing polyacrylamide gel. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. (C) The gel from panel B was phosphorimaged, and the amounts of substrate and product were determined. The fraction of nicked substrate for wt and mutant TRs was then calculated at each Rep concentration and plotted. Closed squares, wt; closed triangles, NOSTEM; open squares, WT+10; open triangles, NOSTEM+10.
In fact, preferential extrusion of the nicking site stem-loop structure in the 10-bp insertion mutant did not result in efficient trs nicking. Rep68 nicking on this substrate was barely detectable (Fig. 3B and C, NOSTEM+10 substrate). In contrast, preferential extrusion of the trs stem-loop structure in the wt background increased Rep68 specific activity as previously reported (Fig. 3, NOSTEM substrate) (6). This result indicated that Rep68 is unable to make functional contacts with the trs, in the absence of ATP, when the spacing between the RBE and trs has been increased. Since our previous study indicated that Rep helicase activity is not required for cleavage once the trs stem-loop is formed, these data strongly suggested that Rep is unable to recognize and nick the trs efficiently unless Rep is also physically interacting with the RBE.
The RBE aligns Rep over the trs.
If Rep must maintain contact with the RBE during nicking, then the polarity of the RBE within the TR should have a strong effect on nicking efficiency. To test this prediction, we constructed three additional mutants in which the 22-bp RBE was replaced with its complement, inverse, or inverse complement (Fig. 4A). If any of these polarity changes still supported Rep trs nicking, then this would suggest a possible model of Rep interaction with the AAV TR.
FIG. 4.
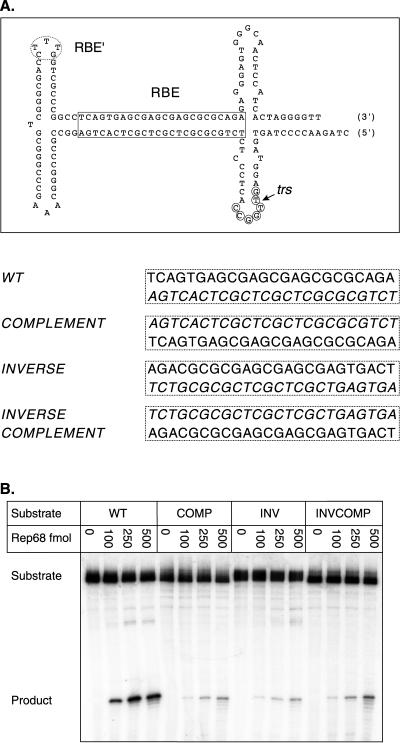
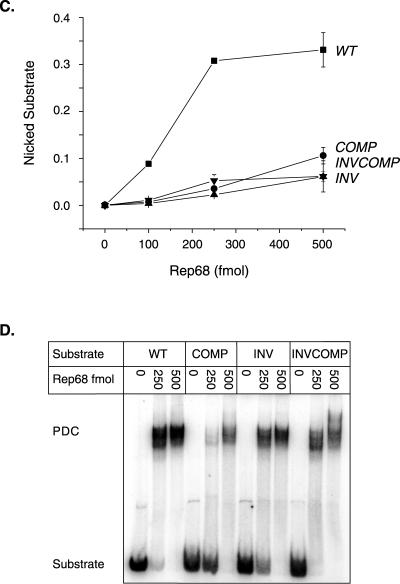
Rep endonuclease activity on RBE polarity mutants. (A) The wt AAV TR is depicted after extrusion of the trs stem-loop structure. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. The wt RBE was replaced with alternative orientations of this sequence. The sequences of the various RBE orientations are given next to the mutant identifier. Note that the integrity of the RBE base composition is maintained. Only the polarity of the nucleic acid sequence has been altered. (B) Rep68 endonuclease reactions were performed on wt and insertion substrates in the presence of 0.5 mM ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. A representative gel is shown. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. The positions of substrates and products are indicated. (C) The gels from two independent trials were phosphorimaged, and the amounts of substrate and product were determined. The fractions of nicked substrate for wt and mutant TRs were calculated at each Rep concentration, averaged between trials, and plotted. Closed squares, wt; closed circles, complement; closed triangles, inverse; closed inverted triangles, inverse complement (n = 2 for all data points). Bars indicate the range at each data point in the two independent trials. (D) Rep binding to wt and mutant TRs was assayed under endonuclease conditions in the absence of ATP (see Materials and Methods). Reactions were resolved on a 4% native polyacrylamide gel to separate substrate from protein-bound DNA complexes (PDCs). The positions of substrate and protein-bound DNA complexes are indicated.
Initially, we asked if the RBE polarity mutations prevented Rep binding to the AAV TR. Although the inverse complement RBE was expected to bind with approximately the same efficiency as that of wt, it was not clear whether the more severe alterations in strand polarity in the inverse and complement mutants would affect binding. To assess Rep binding to these mutants, steady-state binding assays were done under the same reaction conditions as for the nicking assays. However, to prevent nicking and the subsequent accumulation of covalent complexes between Rep and the TR substrates, ATP was omitted from the binding reactions. These reaction mixtures were then resolved on a native polyacrylamide gel to separate the Rep-bound TR complexes from the starting substrates. As shown in Fig. 4D, all of the RBE polarity mutants were bound by Rep at the enzyme concentrations used in the nicking assays (Fig. 4B and C). This result indicates that the strand polarity of the RBE sequence does not significantly affect Rep binding.
In contrast, the results from Rep68 nicking assays on the three polarity mutants indicated that only the wt RBE was capable of directing efficient Rep trs nicking (Fig. 4B and C). Thus, changes in the polarity of the RBE dramatically inhibited the association of the Rep endonuclease active site with the trs. This is consistent with the notion that the RBE aligns Rep or a Rep complex in an orientation that is favorable for subsequent trs cleavage.
Curiously, none of our polarity mutations completely prevented Rep68 trs nicking. The small amount of cleavage observed on these mutants may arise from at least two possibilities. First, Rep may be capable of recognizing and cleaving the trs in the presence of ATP, outside the context of other elements. It should be noted, however, that the amount of Rep trs cleavage observed on these mutant substrates is quite small (5- to 10-fold less than that wt) and similar to levels observed on nicking site mutants that we previously reported (6). Second, the low level of cleavage with the polarity mutants may indicate that other sequence elements, like the RBE′, are also contributing to the alignment of Rep along the AAV TR and directing nicking to the trs (see below). In either case, the RBE appears to align Rep along the TR and direct nicking to the trs.
The RBE′ is required for efficient Rep-mediated nicking.
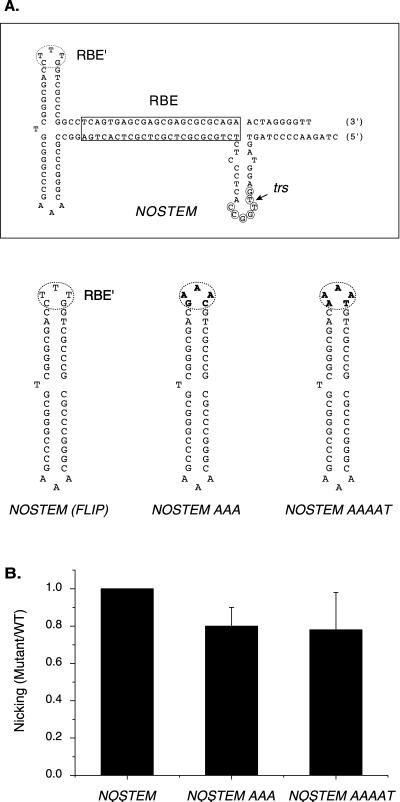
Despite the importance of the RBE in Rep-catalyzed nicking, previous binding assays have detected Rep contacts with a single tip of the internal palindromes of the AAV TR (27). This sequence, referred to as the RBE′, has a constant position with respect to the trs regardless of the orientation of the TR (flip or flop) (see Fig. 6). The functional importance of RBE′ sequences during Rep trs nicking was recently confirmed by Wu et al. (39). This group observed a three- to eightfold reduction in Rep nicking activity on TR substrates in which the RBE′ had been deleted.
FIG. 6.
Rep endonuclease activity on RBE′ substitution mutants in the NOSTEM background. (A) The NOSTEM substrate is depicted. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. The terminal hairpins of the RBE′ substitution mutants are also depicted. Mutated sequences are indicated in boldface. (B) Rep68 endonuclease reactions were performed on NOSTEM and NOSTEM RBE′ substitution substrates in the absence of ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. The gel was phosphorimaged, and the amounts of substrate and product were determined. The relative Rep specific activity for each mutant was expressed as the fraction of mutant substrate nicked divided by the fraction of wt substrate nicked at the same Rep concentration. Ratios obtained at different Rep concentrations were then averaged and graphed for each mutant (n = 4 for all mutants). Bars indicate standard deviations from the means.
The data from Rep68 nicking assays conducted on these RBE′ substitution substrates were consistent with those from previous binding and nicking assays (27, 39). Rep68 cleavage was reduced on all substrates containing substitutions in the CTTTG motif by two- to threefold [Fig. 5B and C, compare AAA, AAAAT, or SWITCH with WT (FLIP)]. Furthermore, substitutions in the complementary sequence that comprises the other tip of the internal palindromes had no effect on nicking [Fig. 5B and C; compare TTT with WT (FLIP) or AAA with SWITCH]. To determine whether other sequences within the internal palindromes affected nicking, we made a mutant in which all of the internal palindromic sequence was deleted in the context of a covalently closed end (Fig. 5A and C, LINEAR). Rep specific activity on the LINEAR substrate was only moderately reduced compared to the specific activity observed on the RBE′ substitution mutants (Fig. 5, compare AAA, SWITCH, and AAAAT to LINEAR). Together, these nicking results supported the hypothesis that Rep makes specific nucleotide contacts with the RBE′ during trs nicking and the most important contacts within the internal palindromes are within RBE′. If internal palindrome sequences outside the RBE′ contributed significantly to Rep nicking, then we would have expected a greater reduction in Rep68 nicking on the LINEAR substrate.
FIG. 5.
Rep endonuclease activity on RBE′ substitution mutants. (A) The wt AAV TR is depicted after extrusion of trs stem-loop structure. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. Additionally, the position of the SmaI endonuclease site is indicated with a line. The terminal hairpins of flop and RBE′ substitution mutants are also depicted. Mutated sequences are indicated in boldface. (B) Rep68 endonuclease reactions were performed on wt and substitution substrates in the presence of 0.5 mM ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. The positions of substrates and products are indicated. (C) The gel from panel B and a second gel containing reaction products from wt and LINEAR substrates were phosphorimaged, and the amounts of substrate and product were determined. The relative Rep specific activity for each mutant was expressed as the fraction of mutant substrate nicked divided by the fraction of wt substrate nicked at the same Rep concentration. Ratios obtained at different Rep concentrations were then averaged and graphed for each mutant (n = 2 for all mutants except for LINEAR, where n = 4). Bars indicate ranges between the different Rep concentrations.
To confirm the importance of the RBE′ to nicking, we also tested Rep68 nicking on the wt flop substrate. We expected this substrate to nick at approximately wt levels because all three components of the TR (trs, RBE, and RBE′) had the correct sequence and orientation. However, Rep68 nicked the flop substrate with about half the efficiency for the flip substrate, suggesting that other factors influenced Rep trs nicking activity. Since both the flip and flop orientations of the AAV TR maintain the RBE and the RBE′ in the same position relative to the trs, the difference in Rep nicking activity must be due to the dissimilar sequences flanking the RBE′ in these two substrates [Fig. 5A; compare WT (FLIP) with WT (FLOP)]. Indeed, our previous analysis indicated that Rep made additional base contacts within the terminal hairpin sequences flanking the RBE′ when bound to the flop substrates compared to when it was bound to flip substrates (27).
RBE′ facilitates DNA helicase activity and trs cruciform extrusion.
Although it was clear that Rep contacts with RBE′ were important for efficient nicking, it was not clear whether they affected the DNA helicase or endonuclease activity of Rep. We reasoned that, if contacts with RBE′ were important for trs transesterification activity, then RBE′ mutations should inhibit Rep nicking, even after extrusion of the trs stem-loop structure. To clarify this issue, a panel of RBE′ substitution mutants was constructed in the NOSTEM background (Fig. 6A). As discussed earlier, the NOSTEM substrate contains a preformed trs stem-loop structure and nicking of this substrate does not require ATP-dependent Rep helicase activity, allowing nicking assays to be done in the absence of ATP. Interestingly, both of our NOSTEM RBE′ mutants were nicked at nearly wt levels in the absence of ATP (Fig. 6B). Moreover, Rep nicked these NOSTEM RBE′ substitutions about twofold more efficiently than the same RBE′ mutations in the wt TR background (compare Fig. 5C, AAA and AAAAT, with Fig. 6B, NOSTEM AAA and NOSTEM AAAAT). This result indicates that Rep interaction with RBE′ is not necessary for the Rep transesterification reaction. Rather, it appears that RBE′ is required primarily for efficient, Rep-mediated unwinding of the AAV TR and formation of the nicking intermediate.
DISCUSSION
Previous binding and chemical interference studies in the absence of ATP have determined that Rep makes contact with two distinct elements within the AAV TR, the linear RBE and the CTTTG motif at one tip of one of the internal palindromes, the RBE′ (8, 19, 20, 27). In the presence of ATP, Rep makes additional contacts with the trs that lead to transesterification (6, 14, 32, 33). Regardless of the orientation of the internal palindromes, flip or flop, these three elements are maintained in a constant position relative to the trs during viral DNA replication. During the course of this study, we have analyzed the contribution of Rep binding contacts along the AAV TR to Rep-mediated trs nicking. Using synthetic AAV TR substrates, we have altered the position and polarity of the RBE relative to the trs and mutated the primary sequence of the RBE′. In vitro Rep trs nicking assays on these mutant substrates indicate that both the RBE and RBE′ are required for efficient Rep-catalyzed trs nicking.
The RBE is required both for origin unwinding and for trs nicking.
Rep nicking activity decreased dramatically when the spacing between the RBE and the trs was altered, indicating that the position of the RBE relative to the trs is critical for efficient cleavage. This observation is consistent with previous in vivo studies of AAV DNA replication, in which AAV genomes harboring mutant RBEs replicated at lower levels than those of wt genomes (4), and in vitro studies, which indicated that the RBE was necessary for Rep binding (8, 19, 20, 27). Recently, we showed that the trs endonuclease reaction occurs in two steps, an initial unwinding of the TR by the Rep-associated DNA helicase that leads to the extrusion of the trs stem-loop structure and the subsequent transesterification reaction that leads to cleavage of the trs (6). We also demonstrated that Rep has a site-specific DNA helicase activity that unwinds DNA containing an RBE (40). Our data from the RBE polarity and spacing mutants in this report indicate that Rep must maintain contact with the RBE during both the TR unwinding and the trs cleavage steps of the endonuclease reaction.
At least two models could describe Rep interaction with the TR during trs transesterification. For example, it is possible that Rep initially binds the RBE and then translocates along the nicked strand to the downstream nicking site, in a manner similar to the restriction endonuclease EcoKI (9–11). Once at the trs, Rep would recognize the nicking site and initiate the transesterification reaction. In this model, the trs stem-loop structure may function as a helicase pause site, allowing prolonged contact between Rep and the nicking site. Alternatively, Rep may be tethered to the RBE during nicking. Endogenous Rep helicase activity would allow downstream contact with the trs, melting of the duplex nicking site, and formation of the nicking site stem-loop structure. In this second model (illustrated in Fig. 7), the nicking site stem-loop structure would effectively reposition the trs closer to the RBE-bound Rep, allowing efficient cleavage.
FIG. 7.
Model of Rep interaction with the AAV TR during trs nicking. The various steps involved in Rep-catalyzed trs nicking are illustrated. The gray ovals each depict separate Rep molecules. The black circles represent the Rep catalytic site. The trs is indicated with an arrow. See the text for details.
Rep nicking data from our insertion mutants are not consistent with a translocation model of Rep-mediated trs nicking. Rep is a fairly strong helicase capable of unwinding 345 bp per min (40). If Rep was initially binding the AAV TR through the RBE and then actively being translocated downstream analogous to EcoKI, then we would not expect small increases in spacing between the RBE and the trs to affect the specific activity of Rep nicking. Yet, Rep had a lowered specific activity on all spacer mutants compared to that of wt (Fig. 2). Moreover, the specific activity of Rep nicking decreased rapidly as spacing between the RBE and the trs increased. Since it is unlikely that 5, 7, or 10 bp of intervening sequence would prevent translocation of Rep from the RBE toward the trs, it appears that the mechanism of Rep-mediated trs cleavage does not include helicase-stimulated translocation. Furthermore, artificially fixing the trs stem-loop structure in the extruded configuration should remove the need for Rep DNA helicase activity and thus contact with the RBE. However, Rep nicking on our 10-bp insertion mutant was barely detectable even after extrusion of the trs stem-loop structure (Fig. 3). Thus, it appears that Rep maintains contact with the RBE during both DNA helicase and trs cleavage activities.
We note that none of our RBE spacer or polarity mutations completely prevented Rep-mediated trs nicking. Apparently, Rep is able to recognize and nick the trs regardless of RBE position, albeit at much decreased levels. Thus, other elements within the AAV TR such as the RBE′ and trs must also contribute to Rep nicking. In the case of the trs, this is not surprising, because our previous study indicated that Rep makes sequence specific contacts at the trs during nicking. Mutation of sequences within the 7-base core trs sequence site reduced Rep cleavage 6- to 10-fold compared to wt substrates, suggesting that Rep specificity for the trs is quite stringent (6). Indeed, Smith and Kotin (29) recently showed directly that Rep can cleave a single-stranded, trs-containing oligonucleotide in the absence of RBE or RBE′ sequences.
Contribution of the RBE′ to Rep trs nicking.
The importance of the RBE′ to Rep-mediated nicking was anticipated by viral DNA replication assays as well as Rep binding and nicking assays (5, 8, 17, 19, 27, 39). Previous AAV DNA replication assays demonstrated that the internal palindromes of the TR are necessary for efficient viral DNA replication. AAV plasmid constructs with deletions of the RBE′ and adjacent sequences replicated at lower levels than did wt AAV constructs (5, 17). Furthermore, in vitro studies indicated that Rep requires the internal palindromes for efficient TR binding and nicking (8, 19, 27, 32). During TR binding, Rep appears to make limited sequence contacts with the internal palindromes, and the most prominent of these contacts occur within RBE′ (27).
Our data confirmed that Rep is making sequence-specific contacts with the CTTTG motif of the RBE′. Substitutions within this sequence significantly reduced Rep nicking activity (Fig. 5). However, when we examined these same RBE′ mutations in the context of the NOSTEM background, the reduction in Rep nicking activity was very small (Fig. 6). This suggested that Rep no longer requires contact with the RBE′ once the stem-loop has been formed and implied that interaction with the RBE′ was important primarily for Rep TR unwinding activity, rather than the transesterification reaction.
The RBE′ sequences appear to be the most significant Rep contacts with the internal palindromes during trs nicking. Rep nicking activity on our LINEAR substrate was only slightly reduced compared to our RBE′ substitution mutants, supporting this conclusion. However, the slight reduction in Rep specific activity observed on our LINEAR substrate does imply that either sequences flanking the CTTTG motif or the internal palindrome structure itself contributes to efficient Rep cleavage. Additionally, Rep nicking activity was reduced on our flop substrate compared to that on the flip substrate. Although this alternative orientation of the AAV TR maintains the CTTTG motif in the same position relative to the trs, the sequences flanking this motif are different from the flip orientation. Indeed, Ryan et al. (27) observed differences in Rep binding contacts between the flip and flop orientations within these flanking internal palindrome sequences. Perhaps this indicates that Rep association with the flop orientation is fundamentally different from that with the flip orientation. This concept is supported by chemical interference assays that reveal differences between Rep contacts within the RBEs of the two substrates. Although Rep makes many discrete contacts within the RBEs of both flip and flop, the strength of individual base contacts is different in the two TR orientations (27). Finally, the reduction in nicking activity seen with our RBE′ substitution mutants and the LINEAR mutant (about threefold) was less than we and others had previously seen on substrates that were missing portions of the internal palindromes (5- to 100-fold) (8, 19, 32, 39). This was most likely due to the fact that the substrates used in this study were covalently closed at one end, whereas previous studies had used SmaI-cut or linear oligonucleotide substrates. Thus, previously used substrates would likely be unwound by the Rep helicase activity to generate single-stranded DNA molecules. In contrast, the substrates used in this study would rapidly reanneal to duplex molecules.
The RBE appears to align Rep asymmetrically on the TR.
When we examined all three possible polarity changes of the RBE sequence (Fig. 4, INVERSE, COMPLEMENT, and INVERSE COMPLEMENT), only the wt polarity retained significant nicking activity. Yet, all of these polarity mutants bound Rep with affinities that were comparable to the wt substrate. This suggested that RBE binding is not particularly sensitive to strand polarity. It also suggested that Rep interaction with the RBE during nicking is inherently asymmetric and serves to align the Rep nicking complex in the appropriate orientation on the TR for the subsequent helicase and transesterification reactions.
Although the RBE appears to play a central role in orienting Rep along the AAV TR during nicking, the architecture of this interaction is undefined. It is not yet clear what an active Rep complex looks like when it is bound to the TR. The kinetics of trs nicking are second order with respect to Rep and ATP concentration, suggesting that a dimer of Rep is sufficient for nicking activity (40). In contrast, Rep DNA helicase activity appears to be first order with respect to enzyme concentration. Furthermore, binding studies detect at least six different bound species, suggesting that Rep complexes can contain as many as six Rep molecules (20, 30). If a Rep dimer is the active nicking complex as implied by the kinetic data, then our data suggest that individual Rep monomers do not associate along a twofold axis of symmetry similar to the type II restriction endonucleases. Presumably such an arrangement of Rep molecules would be active on both our wt and inverse complement substrates. Indeed, our data imply that the Rep nicking complex is arranged asymmetrically along the RBE. This asymmetry may arise from the arrangement of Rep monomers within the homodimer or may reflect the involvement of higher-order complexes in the nicking reaction.
In conclusion, it appears that at least three discrete steps are involved in Rep-mediated AAV origin nicking (Fig. 7). First, Rep binds the TR through the RBE. The RBE aligns the Rep complex along the TR, allowing specific contacts with RBE′. These RBE′ contacts appear to stabilize the Rep complex and facilitate Rep-mediated DNA helicase activity. It is unclear if Rep maintains its original contacts and pulls unwound downstream sequences toward the RBE, allowing them to self-anneal into the trs stem-loop, or if stem-loop formation is more passive in nature. In either case, the net result of Rep helicase activity is the formation of the trs stem-loop. Once formed, this structure presents the single-stranded trs to the Rep transesterification active site in the proper position for nicking.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health: RO1 GM35723, P01 HL51811, and P01 NS 36302.
REFERENCES
- 1.Ashktorab H, Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989;63:3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batchu R B, Hermonat P L. Dissociation of conventional DNA binding and endonuclease activities by an adeno-associated virus Rep78 mutant. Biochem Biophys Res Commun. 1995;210:717–725. doi: 10.1006/bbrc.1995.1718. [DOI] [PubMed] [Google Scholar]
- 3.Batchu R B, Kotin R M, Hermonat P L. The regulatory rep protein of adeno-associated virus binds to sequences within the c-H-ras promoter. Cancer Lett. 1994;86:23–31. doi: 10.1016/0304-3835(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 4.Bishop B M, Santin A D, Quirk J G, Hermonat P L. Role of the terminal repeat GAGC trimer, the major Rep78 binding site, in adeno-associated virus DNA replication. FEBS Lett. 1996;397:97–100. doi: 10.1016/s0014-5793(96)01149-0. [DOI] [PubMed] [Google Scholar]
- 5.Bohenzky R A, LeFebvre R B, Berns K I. Sequence and symmetry requirements within the internal palindromic sequences of the adeno-associated virus terminal repeat. Virology. 1988;166:316–327. doi: 10.1016/0042-6822(88)90502-8. [DOI] [PubMed] [Google Scholar]
- 6.Brister J R, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini J A, Wiener S M, Owens R A, Kyostio S R, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies G P, Kemp P, Molineux I J, Murray N E. The DNA translocation and ATPase activities of restriction-deficient mutants of Eco KI. J Mol Biol. 1999;292:787–796. doi: 10.1006/jmbi.1999.3081. [DOI] [PubMed] [Google Scholar]
- 10.Ellis D J, Dryden D T, Berge T, Edwardson J M, Henderson R M. Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Nat Struct Biol. 1999;6:15–17. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]
- 11.Garcia L R, Molineux I J. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc Natl Acad Sci USA. 1999;96:12430–12435. doi: 10.1073/pnas.96.22.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauswirth W W, Berns K I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 13.Horer M, Weger S, Butz K, Hoppe Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 15.Kokorina N A, Santin A D, Li C, Hermonat P L. Involvement of protein-DNA interaction in adeno-associated virus Rep78-mediated inhibition of HIV-1. J Hum Virol. 1998;1:441–450. [PubMed] [Google Scholar]
- 16.Kyostio S R, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeFebvre R B, Riva S, Berns K I. Conformation takes precedence over sequence in adeno-associated virus DNA replication. Mol Cell Biol. 1984;4:1416–1419. doi: 10.1128/mcb.4.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noirot P, Bargonetti J, Novick R P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci USA. 1990;87:8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orozco B M, Hanley-Bowdoin L. A DNA structure is required for geminivirus replication origin function. J Virol. 1996;70:148–158. doi: 10.1128/jvi.70.1.148-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens R A, Weitzman M D, Kyostio S R, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 29.Smith R H, Kotin R M. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J Virol. 2000;74:3122–3129. doi: 10.1128/jvi.74.7.3122-3129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith R H, Spano A J, Kotin R M. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder R O, Im D S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder R O, Im D S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 34.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wonderling R S, Owens R A. Binding sites for adeno-associated virus Rep proteins within the human genome. J Virol. 1997;71:2528–2534. doi: 10.1128/jvi.71.3.2528-2534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wonderling R S, Owens R A. The Rep68 protein of adeno-associated virus type 2 stimulates expression of the platelet-derived growth factor B c-sis proto-oncogene. J Virol. 1996;70:4783–4786. doi: 10.1128/jvi.70.7.4783-4786.1996. . (Erratum, 70:9084.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, Davis M D, Owens R A. Factors affecting the terminal resolution site endonuclease, helicase, and ATPase activities of adeno-associated virus type 2 Rep proteins. J Virol. 1999;73:8235–8244. doi: 10.1128/jvi.73.10.8235-8244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Zolotukhin I, Im D S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]