Abstract
Post-translational modifications (PTMs) endow proteins with new properties to respond to environmental changes or growth needs. With the development of advanced proteomics techniques, hundreds of distinct types of PTMs have been observed in a wide range of proteins from bacteria, archaea, and eukarya. To identify the roles of these PTMs, scientists have applied various approaches. However, high dynamics, low stoichiometry, and crosstalk between PTMs make it almost impossible to obtain homogeneously modified proteins for characterization of the site-specific effect of individual PTM on target proteins. To solve this problem, the genetic code expansion (GCE) strategy has been introduced into the field of PTM studies. Instead of modifying proteins after translation, GCE incorporates modified amino acids into proteins during translation, thus generating site-specifically modified proteins at target positions. In this review, we summarize the development of GCE systems for orthogonal translation for site-specific installation of PTMs.
Graphical Abstract

1. INTRODUCTION
In the first class of biochemistry, it is taught that proteins are composed of a set of 20 amino acids. Two additional amino acids, selenocysteine and pyrrolysine, have also been found as components of a limited number of proteins in specific organisms.1–3 At the very beginning, they were thought as modifications of cysteine and lysine residues respectively, but later scientists found that both amino acids are linked to their cognate tRNAs and then delivered to ribosomes for protein synthesis, sharing a similar process of translation with the set of 20 canonical amino acids. In a more accurate way, these 22 amino acids together are named as proteinogenic amino acids. On the other hand, protein side chains usually undergo modifications during different growth stages or after stimuli in living cells. Since these modifications occur after translation, they are named as post-translational modifications (PTMs).4,5
The first protein PTM was discovered 90 years ago. Fritz Lipmann and Phoebus Levene detected phosphoserine in casein in 1933.6 Later, lysine acetylation in histones was discovered by Vincent Allfrey in 1964.7 During the past decade, PTM studies have encountered a new era with the development of advanced proteomics techniques that have identified more than 600 distinct types of PTMs in thousands of proteins covering almost all the biological processes in bacteria, archaea, and eukarya.8–10 PTMs can change charges of protein side chains such as phosphorylation and acetylation to affect substrate binding or interactions with other biomolecules.11–14 PTMs can also add specific tags to proteins for triggering corresponding biological processes such as ubiquitination and glycosylation.15–18 More importantly, PTMs have been found to be closely associated with a variety of human diseases including cancers,19 cardiovascular diseases,20 diabetes,21 neurodegenerative diseases,22 and even coronavirus diseases in the recent COVID-19 pandemic.23 Thus, protein PTMs have been attracting attentions of scientists from different research fields for decades.
There are several major challenges in PTM studies. First, the same type of PTM can occur at multiple sites in a single protein. Second, different types of PTMs can occur at the same residue in proteins. Third, PTMs at different sites have crosstalk between each other, no matter if they are the same or distinct types of PTMs. Fourth, PTMs are high dynamic processes between modified and unmodified forms. Thus, it is necessary to generate a protein with a homogeneous modification at the PTM site to demonstrate its site-specific effect on the target protein, and this is essential for downstream studies such as functional determination and drug development. To achieve this goal, classic approaches usually use PTM mimicking amino acids to replace PTMs at the target site by site-directed mutagenesis. For example, glutamate is commonly used as the substitution for a phosphorylated serine and threonine or even tyrosine residue,24 while glutamine is usually used as the mimic for an acetylated lysine residue.25 The advantages of this strategy include the following: First, it is easy to implement with commercially available kits; Second, these PTM mimicking amino acids are stable in cells, which is especially important for in vivo studies. However, the differences in sizes and charges of these PTM mimicking amino acids from those of native PTMs sometimes make them not sufficient substitutions.26,27 Thus, the most rigorous way is to produce proteins with native PTMs at target sites.
Due to high dynamics, low stoichiometry, and existence of multiple PTMs, it is almost impossible to isolate homogeneously modified proteins from living cells. Scientists have applied various biological and chemical approaches to overcome those challenges.28,29 First, specific modifying enzymes such as kinases and acetyltransferases can be used to modify proteins at target sites.30,31 However, this approach only has confined applications due to limited knowledge in substrate specificities of modifying enzymes. Second, solid-phase peptide synthesis can be implemented to generate site-specifically modified peptides,32,33 but this method only applies for proteins or peptides with short lengths. To overcome the length limit of solid-phase peptide synthesis, the native chemical ligation reaction has been established to link PTM-containing peptide fragments to form a full-length protein with PTMs at target sites.34,35 However, such semisynthetic approach sometimes needs refolding of proteins after purification. Furthermore, all these above-mentioned methods only apply to in vitro studies, leaving gaps in installing desired PTMs into target proteins in living cells.
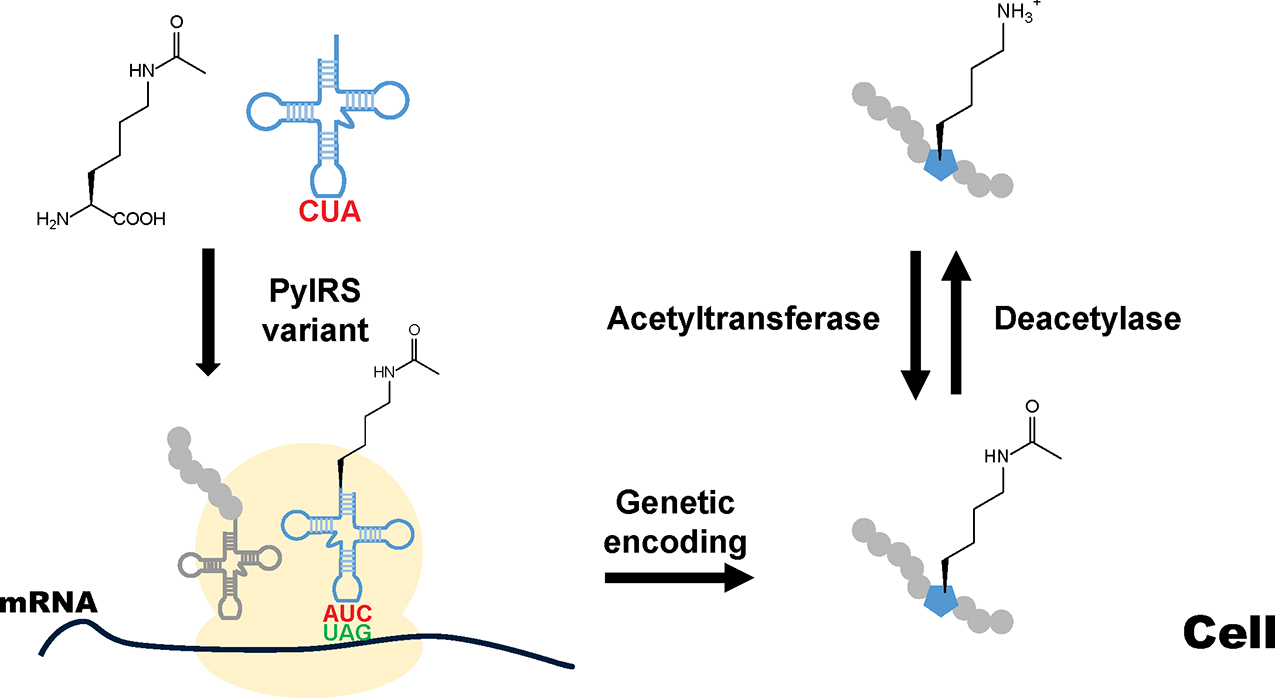
To fill in this gap, the genetic code expansion (GCE) strategy has been applied for PTM studies.36–40 It introduces an orthogonal translation system (OTS) into living cells to genetically encode a noncanonical amino acid (ncAA) at a chosen site of target proteins.41–43 The OTS is mainly composed of an engineered aminoacyl-tRNA synthetase (AARS), which recognizes the ncAA, and an engineered tRNA, which bears a specific anticodon to read an assigned codon in mRNA (Figure 1).44–46 The orthogonality of an OTS includes the following: First, the target ncAA cannot be recognized by host AARSs; Second, the engineered AARS cannot recognize canonical amino acids and cannot aminoacylate host tRNAs; Third, the engineered tRNA cannot be aminoacylated by host AARSs. To date, majority of established OTSs are derived from the pair of tyrosyl-tRNA synthetase (TyrRS) and tRNATyr from Methanococcus jannaschii or Escherichia coli and the pair of pyrrolysyl-tRNA synthetase (PylRS) and tRNAPyl from Methanosarcina species.47,48 There are also a series of “not-so-popular” orthogonal pairs, expanding the diversity of ncAAs that can be genetically encoded in living cells.49 It should be noted that there are several potential problems when applying the GCE method. First, since the introduced OTS can decode the assigned codon (commonly a UAG stop codon) as an ncAA not only in the target mRNA but also in mRNAs of native genes in host cells, the GCE method could lead to off-target ncAA incorporation at more sites than the desired target in living cells. Moreover, due to the competition from release factors or near cognate tRNAs for the assigned stop codon, the GCE approach could generate truncated or mistranslated target proteins, correspondingly. To overcome these issues, several strategies have been applied such as optimizing OTS efficiencies, eliminating release factors, and engineering elongation factor-Tu (EF-Tu) or ribosomes.50
Figure 1.

The scheme for the GCE strategy. The noncanonical amino acid (ncAA) is specifically recognized by an engineered aminoacyl-tRNA synthetase (AARS*) and attached to a cognate tRNA*. The pair of introduced AARS* and tRNA* is orthogonal in host cells and does not cross react with native pairs of AARS and tRNA in hosts. Then ncAA-charged tRNA* is delivered to the ribosome and reads an assigned codon in mRNA, allowing genetic encoding of an ncAA into the target protein at a controlled site.
Instead of modifying proteins post-translationally, the GCE method can install PTMs into proteins cotranslationally, producing proteins with homogeneous PTMs at target sites for further studies, especially for in vivo studies.51–53 In later sections of this review, we will describe common PTMs of individual amino acid in nature and summarize methods for site-specific installation of PTMs into proteins, focusing on the development of OTSs for genetically encoding various ncAAs into proteins as native PTMs, PTM analogs, or PTM precursors either in vitro or in vivo. We also discuss limitations of certain methods and propose future directions of OTS development for PTM studies.
2. ARGININE MODIFICATIONS
Arginine is a basic amino acid with a guanidinium group that has a pKa value of 13.8,54 making arginine residues positively charged under physiological conditions. The majority of arginine PTMs have been found in histones, including methylation and citrullination (deimination) of the guanidinium group.55,56 There are also some other arginine modifications such as phosphorylation and ADP-ribosylation.57,58 In this section, we summarize the development of OTSs for generating proteins with arginine methylation and citrullination.
2.1. Arginine Methylation
Arginine methylation has been found to regulate a wide range of biological processes, including DNA damage repair, transcription, splicing, cell signaling, and cell cycles.59–61 They are involved in many human diseases such as cancers and inflammatory-related diseases.62–64 Arginine methylation is catalyzed by protein arginine methyltransferases (PRMTs), which transfer the methyl group from S-adenosyl-l-methionine (SAM) to the guanidinium group. To date, humans are known to have nine PRMTs, which are further categorized into three types according to their methylation products (Figure 2). Type I PRMTs generate monomethylarginine and can add another methyl group to the same nitrogen atom to form asymmetric N,N′-dimethyl-arginine. Type II PRMTs also generate monomethylarginine but can add a methyl group to another nitrogen atom in the guanidinium group to form symmetric N,N-dimethylarginine. Type III PRMTs only generate monomethylarginine.65,66 For the reverse reaction, it is still controversial whether specific protein arginine demethylases exist to remove methyl groups from methylated arginine residues or not.67
Figure 2.

Three types of PRMTs. All three types of PRMTs can generate monomethylarginine. Type I PRMTs can further form asymmetric N,N′-dimethyl-arginine, while Type II PRMTs produce symmetric N,N-dimethyl-arginine. Type III PRMTs only generate monomethylarginine. SAM: S-Adenosyl-l-methionine; SAH: S-adenosyl-l-homocysteine.
To study site-specific effects of arginine methylation on protein function and structure, two chemical approaches have been developed to install methylated arginine or analogs into recombinant proteins in vitro. Both methods need to first apply site-directed mutagenesis to replace the target arginine residue with cysteine. The first reaction is based on cysteine alkylation with α,β-unsaturated amidine scaffold harboring different degrees of methylation (Figure 3A).68 Such methylarginine analogs have two variations from methylarginine: γ-methylene is replaced with a sulfur atom, and the secondary amine is substituted by ε-methylene. To install native methylarginine, C(sp3)–C(sp3) bond formation through carbon free radicals has been adopted (Figure 3B).69 The mutated cysteine residue fist undergoes elimination to form dehydroalanine (Dha) and then reacts with methylarginine-containing free radicals generated from alkyl halides via homolytic bond division in the presence of NaBH4.
Figure 3.

Chemical approaches to installing arginine methylation or analogs into proteins in vitro. A) The mutated cysteine residue is alkylated with α,β-unsaturated amidine scaffold harboring different degrees of methylation. Monomethylarginine analog: R1 = H, R2 = CH3, R3 = H; Asymmetric N,N′-dimethyl-arginine analog: R1 = H, R2 = CH3, R3 = CH3; Symmetric N,N-dimethyl-arginine analog: R1 = CH3, R2 = CH3, R3 = H. B) The mutated cysteine residue undergoes elimination to form dehydroalanine and then reacts with methylarginine-containing free radicals generated from alkyl halides in NaBH4. Monomethylarginine: R1 = CH3 R2 = H; Asymmetric N,N′-dimethyl-arginine: R1 = CH3, R2 = CH3.
To date, no OTS has been developed to incorporate methylarginine site-specifically into proteins in vivo. However, one study used the GCE strategy with cell-free translation systems to incorporate methylarginine into proteins, providing bases for further developing OTSs for genetic encoding methylarginine.70 In this work, monomethylarginine was first linked to a mutant yeast tRNAArg harboring a four-base anticodon CCCG by yeast arginyl-tRNA synthetase (ArgRS) through in vitro aminoacylation. Then methylarginine-charged tRNAArgCCCG was isolated and added into the cell-free translation system to decode the mutated CGGG codon at the target position of mRNA as methylarginine (Figure 4). This approach was only applicable for monomethylarginine but not for dimethyl-arginine, as it was shown that ArgRS cannot aminoacylate tRNAArg with neither asymmetric N,N′-dimethylarginine nor symmetric N,N-dimethyl-arginine.70 Another potential problem with this approach is that the cell-free translation system also produces arginine-charged tRNAArgCCG, which can compete with methylarginine-charged tRNAArgCCCG for the first three-base codon (CGG) in the CGGG four-base codon, thus forming a distinct read frame after the four-base codon. Indeed, truncated protein fragments were detected in this work due to a downstream stop codon if the CGGG codon is decoded as CGG.70 This problem could be solved by removing tRNAArgCCG in the cell-free translation system by biotin-tagged antisense DNA oligos and replacing all CGG codons in the mRNA with other Arg codons. Such a strategy has been used for other ncAA incorporation by cell-free translation systems.71
Figure 4.

The cell-free translation approach to incorporating monomethylarginine site-specifically into proteins. Yeast arginyl-tRNA synthetase (ArgRS) aminoacylates a mutant yeast tRNAArg bearing a four-base anticodon CCCG with monomethylarginine by in vitro aminoacylation. Then methylarginine-charged tRNAArgCCCG is added into the cell-free translation system to read an assigned CGGG codon in the mRNA as monomethylarginine to install arginine methylation at the target site.
2.2. Citrullination
Citrullination, or peptidyl arginine deimination, is a PTM of converting an arginine residue to a citrulline residue in proteins (Figure 5A). As the guanidinium group is hydrolyzed to become a ureido group, the arginine side chain losses its positive charge. The guanidinium group also losses two hydrogen-bond donors but obtains two hydrogen-bond acceptor sites.72 Protein citrullination regulates a variety of cellular processes, including gene expression, cell signaling, metabolism, and apoptosis.73,74 It is associated with a variety of human diseases, including cancers,75 inflammatory and autoimmune disorders,76 cardiovascular diseases,77 and neurodegeneration diseases.78 Citrullination is catalyzed by peptidyl arginine deiminases (PADs).79 There are 5 annotated PADs in the human genome (PAD1–4 and PAD6).80,81 So far, citrullination is still considered to be an irreversible PTM without known citrulline erasers, despite the existence of argininosuccinate synthetase and argininosuccinate lyase, which work together to convert free citrulline to arginine in the urea cycle.82–84
Figure 5.

Protein citrullination. A) The guanidinium group of the arginine residue is hydrolyzed by peptidyl arginine deiminases (PADs) to form a citrulline residue. B) The structures of citrulline and its mimicking amino acid, glutamine.
To study protein citrullination, glutamine has been used as a citrulline-mimicking amino acid.85 Glutamine has a carboxamide moiety that has similar numbers of hydrogen bond donors and acceptors to those in the ureido group, so it is considered as a feasible mimic of citrulline. However, the side chain of citrulline is ~2.7 Å longer than that of glutamine, making glutamine not an accurate substitution of citrulline (Figure 5B). To install native citrullination, the similar Dha-mediated approach to installing arginine methylation has been applied in vitro (Figure 3B).69 Instead, a citrulline moiety-containing free radical is generated from a corresponding alkyl halide in the presence of NaBH4 by homolytic bond fission and then reacts with Dha to form a C–C bond, thus changing the mutated cysteine residue to a citrulline residue.
Similar to installing arginine methylation (Figure 4), the cell-free translation system was also used to incorporate citrulline at the original arginine site by the GCE strategy.70 Yeast ArgRS charges tRNAArgCCCG with citrulline to form citullinyl-tRNAArgCCCG by in vitro aminoacylation that is then isolated and added into the cell-free translation system to decode the four-base codon CGGG in mRNA as citrulline. The fact that wild-type ArgRS can also recognize citrulline indicates the difficulty in engineering ArgRS to recognize citrulline but not arginine, making direct incorporation of citrulline challenging in vivo. Recently, an alternative strategy has been developed to overcome this challenge.86 Instead of citrulline, photocaged citrulline is first genetically encoded into proteins in mammalian cells. To develop an OTS that can incorporate o-nitrobenzyl photocaged citrulline, both the archaeal PylRS/tRNAPyl pair and the E. coli leucyl-tRNA synthetase (EcLeuRS)/tRNALeuCUA pair were selected for further engineering, because both pairs are known to be orthogonal in mammalian cells and can recognize ncAAs with long and bulky side chains.48,87 A selection based on GFP fluorescence was implemented and obtained an EcLeuRS variant (T252A, M40I, Y499I, Y527A, and H529G) that can incorporate photocaged citrulline site-specifically into proteins. After genetic encoding of photocaged citrulline into target proteins in living cells, the caging group can be removed by UV irradiation (365 nm), leaving native citrulline at the target position for downstream studies (Figure 6). By using this approach, it was shown that autocitrullination of R372 and R374 in protein arginine deiminase 4 deceases the enzyme activity by 181- and 9-fold, respectively.
Figure 6.

Site-specific incorporation of citrulline into proteins in mammalian cells. o-Nitrobenzyl photocaged citrulline is first genetically encoded in the target protein by an orthogonal pair of E. coli LeuRS (EcLeuRS) variant and tRNALeuCUA at an assigned position with a UAG stop codon. Then the caging group is removed by UV 365 nm exposure, and citrulline is formed at the desired site in the target protein.
3. ASPARAGINE MODIFICATIONS
Asparagine is a polar amino acid with a carboxamide group that has two hydrogen bond donors and two hydrogen bond acceptor sites. Asparagine residues have several types of PTMs such as hydroxylation to regulate gene transcription and signaling networks,88,89 spontaneous deamidation as a molecular marker for protein degradation,90,91 and methylation to mitigate protein damages from deamidation.92 A unique PTM for asparagine is N-glycosylation, though recent studies have found that arginine residues of few proteins in hosts are targets of N-glycosylation catalyzed by bacterial effector proteins for host–pathogen interactions.93 In this section, we focus on the development of OTSs for generating proteins with N-glycosylation analogs.
3.1. N-Glycosylation
N-Glycosylation can be found in about half of all eukaryotic proteins94 and affects protein structure, stability, solubility, folding, and quality control.95 It plays important roles in protein trafficking and cell signaling.96 N-Glycosylation is catalyzed by oligosaccharyl transferase that transfers oligosaccharide from a lipid-linked donor to the asparagine residue in a Asn-Xaa-Ser/Thr consensus sequence.97,98
Most challenges in glycosylation studies result from its high heterogeneity with varied numbers of saccharides, types of saccharides, and linkage between saccharides. To overcome these challenges, scientists have made remarkable progresses to generate glycoproteins with homogeneous saccharides through native chemical ligation, chemo-selective ligation, and enzymatic catalysis and remodeling.99–101 Take a chemical approach based on S-alkylation as an example: the target asparagine residue is first mutated to cysteine, which reacts with glycosyl-β-N-iodoacetamide to form a sulfur-containing analog of N-glycosylation in vitro (Figure 7).102
Figure 7.

A chem-selective ligation approach to generating N-glycosylation analogs. The target asparagine residue is first mutated to cysteine, which is then reacted with glycosyl-β-N-iodoacetamide by S-alkylation to form a sulfur-containing analog of N-glycosylation in vitro.
The direct incorporation of glycosylated asparagine into proteins by GCE has been challenging for decades due to the difficulty in engineering AARS binding pockets to accommodate large-sized glycans. Alternatively, several semisynthetic approaches combining GCE and chemical ligation have been developed. In an earlier study, an aspartate derived thioester was first genetically encoded into proteins by an Methanosarcina barkeri PylRS (MbPylRS) variant (A267Y, Y271A, N311T, C313G, and Y349F) in E. coli cells. After aspartate thioester-containing recombinant proteins were purified, the thioester moiety was linked to a cysteine-glycan conjugate by transthioesterification, followed by an S- to N-acyl transfer to form a cysteine-bridged N-glycosylation analog in vitro (Figure 8A).103 The advantage of this approach is that the free thiol group in the cysteine bridge can be further used as a handle to add tags to the target protein for purification, detection, and functional characterization. In a later study, aspartate β-benzyl ester was first genetically encoded into target proteins by another MbPylRS variant (N311C, C313S, and Y349F) in E. coli cells. After the target proteins were purified, the ester moiety was hydrazinolyzed to form alanine-β-hydrazide, followed by ligation with reducing carbohydrate in vitro (Figure 8B).104 By this approach, a variety of carbohydrates can be linked to target proteins such as N-acetylated monosaccharides, disaccharides, trisaccharide, and tetrasaccharide analogs. More importantly, the product is a close mimic of native N-glycosylation, facilitating further generation of complex glycans through enzymatic catalysis and remodeling.
Figure 8.

Semisynthetic approaches to installing N-glycosylation analogs into proteins. A) An aspartate derived thioester is first incorporated into proteins by GCE and then linked to a cysteine-glycan conjugate by transthioesterification in vitro, followed by an S- to N-acyl transfer to form a cysteine-bridged N-glycosylation analog. B) Aspartate β-benzyl ester is first encoded into proteins by GCE and then hydrazinolyzed to form alanine-β-hydrazide in vitro, followed by ligation with reducing carbohydrate.
4. CYSTEINE MODIFICATIONS
Cysteine is a polar amino acid bearing a thiol group with a pKa value of 8.3, making cysteine residues usually involved in enzymatic reactions as nucleophiles. Disulfide bonds between cysteine residues are essential for protein folding and stability.105,106 Common PTMs of cysteine residues include oxidation for redox signaling pathways,107 methylation for membrane protein functions,108 S-nitrosylation for protein localization,109 and lipidation for cell signaling.110 In this section, we focus on the development of OTSs for generating proteins with analogs of S-palmitoylation, which is one type of protein lipidation unique to cysteine residues.
4.1. S-Palmitoylation
Protein lipidation is to link long-chain fatty acids to proteins, regulating membrane protein localization, trafficking, and interactions with other biomolecules.111 Abnormalities of protein lipidation have been shown to be associated with human diseases such as metabolic disorders, neurological diseases, and cancers.112–114 Common types of protein lipidation include N-myristoylation, S-prenylation, and S-palmitoylation, while C-phosphatidylethanolaminylation and N-palmitoylation are rare (Figure 9).115 Among them, S-palmitoylation is the only reversible modification, while others are irreversible and commonly linked to termini of proteins. S-Palmitoylation adds palmitate to cysteine residues by protein S-acyltransferases that contain a cysteine-rich domain with a consensus Asp-His-His-Cys (DHHC) motif.116
Figure 9.
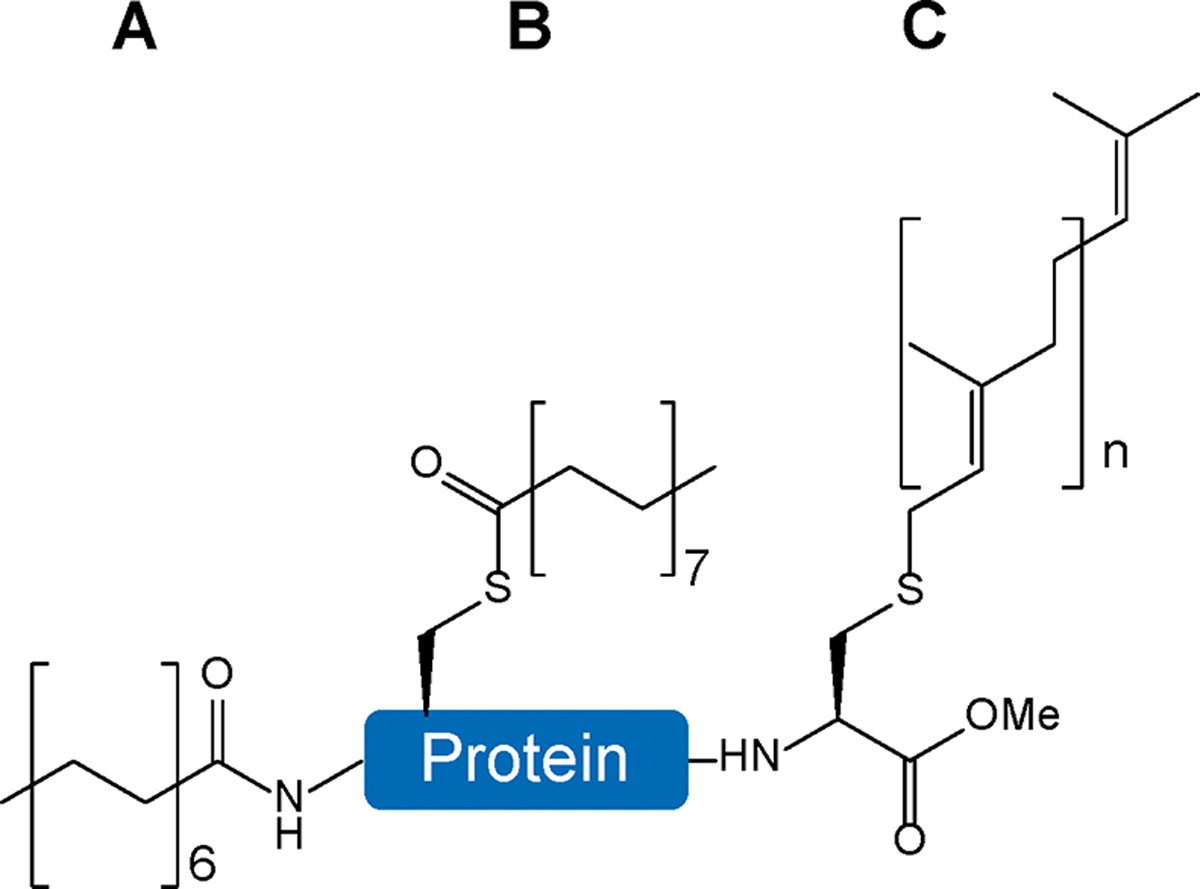
Common types of protein lipidation. A) N-Myristoylation is an irreversible modification linked to the N-termini of proteins. B) S-Palmitoylation is a reversible modification attached to internal cysteine residues. C) S-Prenylation is an irreversible modification linked to cysteine residues at C-termini of proteins (n = 2: farnesylation; n = 3: geranylgeranylation).
To study protein lipidation, classic chemical approaches such as solid-phase peptide synthesis and native chemical ligation have been used to generate proteins with lipid moieties.113 Semisynthetic methods combining GCE and chemical ligation have also been developed. In an earlier study, p-ethynylphenylalanine, an alkyne group-containing phenylalanine analog, was first genetically encoded into proteins by a yeast phenylalanyl-tRNA synthetase (ScPheRS) variant (T415A) in E. coli cells. After protein purification, it was linked to palmitic acid-azide by copper-catalyzed azide–alkyne cycloaddition to form an S-palmitoylation mimic in vitro (Figure 10A).117 In a later study, p-azido-phenylalanine (pAzF), an azide-containing phenylalanine analog, was first site-specifically incorporated into proteins by an engineered M. jannaschii TyrRS (MjTyrRS) variant (Y34T, E107N, D158P, I159L, and L162Q) in E. coli cells. Then the pAzF-containing protein was purified and reacted with dibenzocyclooctyne group-containing palmitic acid by strain-promoted azide–alkyne cycloaddition in vitro (Figure 10B).118
Figure 10.

Semisynthetic approaches by GCE and chemical ligation to installing S-palmitoylation mimics into proteins. A) p-Ethynylphenylalanine is first site-specifically incorporated into proteins and then linked to 15-azido-pentadecanoic acid by copper-catalyzed azide–alkyne cycloaddition in vitro. B) p-Azido-phenylalanine is first genetically encoded into proteins and then reacts with dibenzocyclooctyne-palmitic acid conjugate by strain-promoted azide–alkyne cycloaddition in vitro.
Both above-mentioned ncAA-mediated semisynthetic methods only apply to in vitro studies. The long length of fatty acids makes it impossible to engineer AARSs for direct incorporation of S-palmitoylation in living cells. Recently, an alternative approach based on GCE has been developed to generate proteins with an S-palmitoylation mimic in mammalian cells.119 In this work, a carbon–carbon triple bond containing compound, bicyclo[6.1.0]nonyne-lysine, was first genetically encoded into proteins by an Methanosarcina mazei PylRS (MmPylRS) variant (Y306A and Y384F) in mammalian cells and reacted with tetrazine-containing palmitic acid derivatives by the biocompatible inverse-electron-demand Diels–Alder reaction to form S-palmitoylation mimics in living cells (Figure 11). This work also demonstrated that such S-palmitoylation mimics can activate downstream signaling transduction, demonstrating promising applications in protein lipidation studies in vivo.120
Figure 11.

Site-specific incorporation of S-palmitoylation mimics into proteins in mammalian cells. Bicyclo[6.1.0]nonyne-lysine is first genetically encoded into proteins by an orthogonal pair of engineered PylRS/tRNAPyl at an assigned position with a UAG stop codon and then reacts with a tetrazine-containing palmitic acid analog by the inverse-electron-demand Diels–Alder reaction to form an S-palmitoylation mimic in living cells.
5. GLUTAMINE MODIFICATIONS
Glutamine is another carboxamide-containing amino acid. Glutamine residues also undergo spontaneous deamidation, which triggers protein aggregation,121,122 and methylation, which regulates transcription,123 translation,124 and methanogenesis.125 In this section, we focus on the development of OTSs for generating proteins with glutamine methylation.
5.1. Glutamine Methylation
Glutamine methylation is relatively rare compared to lysine methylation and arginine methylation. To date, only a few proteins have been identified as glutamine methylation targets, including release factors,124,126 ribosomal proteins,127 histones,123 and methanogenic enzymes.125 Glutamine methylation is catalyzed by glutamine methyltransferases, which are also SAM-dependent enzymes like arginine methyltransferase.125 Interestingly, the enzyme HEMK2-TRMT112 catalyzes methylation of both lysine residues in histones and glutamine residues in release factors.128 Recently, an ncAA-mediated approach has been developed to generate proteins with glutamine methylation.129 First, L-glutamic acid γ-benzyl ester is site-specifically incorporated into proteins by an MbPylRS variant (N311S, C313A, and Y349F) in E. coli cells. After protein purification, it is converted into acyl hydrazide, followed by a reaction with acetyl acetone to form a Knorr pyrazole at pH 3. Finally, methylamine is added into the mixture with adjusted pH 9 to generate proteins with native glutamine methylation at the target position in vitro (Figure 12). To date, there is no established OTS to site-specifically incorporate methylated glutamine into proteins in living cells, which will be the next milestone in the field of glutamine methylation.
Figure 12.

An ncAA-mediated approach to installing glutamine methylation into target proteins. L-Glutamic acid γ-benzyl ester is first site-specifically incorporated into proteins by GCE, then it is converted to glutamyl hydrazide in vitro, followed by a reaction with acetyl acetone to form a Knorr pyrazole intermediate. In the last step, methylamine is added to generate proteins with glutamine methylation at the target position.
6. HISTIDINE MODIFICATIONS
Histidine is a basic amino acid with an imidazole group. The pKa value of imidazole in water is about 6, but protein local environments can change the pKa value of imidazole, making histidine residues wobble between positive and neutral under physiology conditions. Histidine residues are involved in catalytic mechanisms of many enzymes such as proteases. PTMs of histidine residues include phosphorylation involved in cellular signal transduction,130,131 hydroxylation found in ribosomal proteins,132,133 oxidation associated with redox sensing,134 and methylation related to regulation of cytoskeletal dynamics and protein translation.135,136 In this section, we focus on the development of OTSs for generating proteins with histidine methylation.
6.1. Histidine Methylation
Histidine methylation is an emerging field, even though it was first discovered in actin in 1967.137,138 A recent study has identified 299 histidine methylation sites in 254 human proteins, which is less abundant compared to 788 lysine methylation sites in 584 human proteins and 1154 arginine methylation sites in 764 human proteins.139 Histidine methylation is catalyzed by protein histidine methyltransferases (PHMTs), which are also SAM-dependent enzymes (Figure 13).136 Only three proteins have been assigned as PHMTs in humans to date. SETD3 and METTL18 can generate 3-methyl-histidine,140,141 while METTL9 produces 1-methyl-histidine.142
Figure 13.

Histidine methylation by protein histidine methyltransferases (PHMTs). There are two kinds of PHMTs depending on the methylation site in the imidazole group. In humans, SETD3 and METTL18 generate 3-methyl-histidine, while METTL9 produces 1-methyl-histidine. SAM: S-adenosyl-l-methionine. SAH: S-adenosyl-l-homocysteine.
Before human PHMTs were identified, an earlier study has already applied GCE to genetically encode 3-methyl-histidine and other histidine analogs in both E. coli and mammalian cells.143 PylRS was engineered to recognize histidine analogs because of its high plasticity for amino acid recognition.144 An MbPylRS variant (L270I, Y271F, L274G, C313F, and Y349F) was obtained after selection for 3-methyl-histidine specifically. In this work, 3-methyl-histidine was site-specifically incorporated into a fluorescent protein and changed its spectral properties. Now, with the knowledge in PHMTs, it is promising to generate proteins with naturally existing histidine methylation to study their biological functions.
7. LYSINE MODIFICATIONS
Lysine is another basic amino acid with an amine group at the ε-end. The pKa value of the ε-amine group is 10.8, making lysine residues bring positive charges under most physiological conditions. Lysine is one of the most frequent PTM-targeting amino acids. About 30 distinct types of lysine PTMs have been identified in eukaryotic and bacterial cells.145 The most abundant lysine PTMs include ubiquitination, acetylation, SUMOylation, succinylation, crotonylation, methylation, malonylation, and 2-hydroxy-isobutyrylation. In this section, we summarize the development of OTSs to generate proteins with lysine PTMs and their analogs.
7.1. Lysine Acetylation
Lysine acetylation was first discovered in histones in 1964.7 During many years after its discovery, studies on lysine acetylation have focused on its regulatory roles in transcription.146,147 With the development of advanced proteomics techniques, now it is known that lysine acetylation is not only existing in histones but also in nonhistone proteins.148,149 Moreover, lysine acetylation is not unique for eukaryotic cells but also occurs in bacterial cells.150–152 The biological processes in which lysine acetylation plays roles range from gene expression to cell singling and metabolism.13,153 Lysine acetylation is involved in various human diseases including cancers,154 neurodegenerative diseases,155 kidney diseases,156 and bacterial infections.157 Lysine acetylation is catalyzed by protein acetyltransferases, which have 5 major families: the Gcn5-related N-acetyltransferase family, the CBP/p300 coactivators, the TAFII group of transcription factors, the MYST family, and the SRC family of coactivators.158 Lysine acetylation is a reversible PTM, and the removal of Nε-acetyl amide moieties is catalyzed by lysine deacetylases, which can be divided into two main families: hydrolytic deacetylases and NAD+-dependent deacetylases (sirtuins).159
To study the functions of lysine acetylation, OTSs for direct incorporation of acetyllysine into target proteins in living cells have been developed (Figure 14). The first OTS for acetyllysine incorporation was established in E. coli cells and was derived from the pair of MbPylRS and tRNAPyl.160 The residues (Leu266, Leu270, Tyr271, Leu274, Cys313, and Trp383) around the binding pocket of pyrrolysine were randomized for selections, and two variants recognizing acetyllysine were obtained, MbAcKRS-1 (D76G, L266 V, L270I, Y271F, L274A, and C313F) and MbAcKRS-2 (L270I, Y271L, L274A, and C313F). By further engineering MbAcKRS-1, a better MbAcKRS-3 (D76G, L266M, L270I, Y271F, L274A, and C313F) was shown to improve acetyllysine incorporation.161 Later, another OTS for acetyllysine was developed based on MmPylRS.162 Two MmPylRS variants, MmAcKRS-1 (L301M, Y306L, L309A, and C348F) and MmAcKRS-2 (L301M, Y306L, and C348S), were demonstrated to have higher acetyllysine incorporation efficiency. However, even with those optimized AcK-OTSs, the suppression of stop codons by acetyllysine-charged tRNAPyl is still relatively low compared to OTSs for other ncAAs. To further increase acetyllysine incorporation efficiency, several approaches to engineering components in AcK-OTSs have been implemented. First, because tRNAPyl is originally from methanogenic archaea, its binding to host EF-Tu is not optimal. Thus, the interacting nucleotides of tRNAPyl with EF-Tu, C6-G67:C7-G66, were mutated to C6-G67:G7-C66 for higher binding affinity.163 Second, due to the size limit of mutant libraries, earlier studies only randomized residues around the amino acid binding pocket, while residues in other parts of the enzyme could also benefit the incorporation. To overcome this challenge, phage-assisted continuous evolution (PACE) was performed for the full-length chimeric PylRS (chPylRS, the chimera of residues 1–149 of MbPylRS and residues 185–454 of MmPylRS).164 Indeed, several mutations at other parts of chPylRS (V31I, T56P, H62Y, and A100E) further enhanced acetyllysine incorporation. Third, the stop codon is commonly used for ncAA incorporation, while release factors can compete with ncAA-charged tRNAs for those assigned stop codons, thus decreasing ncAA incorporation efficiencies. To solve this problem, the C-terminal domain of ribosomal protein L11 was overexpressed to decrease release factor-mediated termination, showing the ability to incorporate acetyllysine at three distinct sites in a single protein.165 Furthermore, because of the orthogonality of PylRS-derived OTSs in both bacteria and eukaryotes, genetic encoding of acetyllysine into proteins has been established in different organisms, including E. coli,160,162 Salmonella,166 yeast,167,168 mammalian cells,169,170 stem cells,171 and animals.172 The applications of these AcK-OTSs in determining the roles of lysine acetylation in regulating specific proteins, from histones to nonhistone proteins, have been nicely reviewed recently.36,38,40,173
Figure 14.
Direct installation of lysine acetylation into proteins in living cells by GCE. Nε-Acetyllysine is genetically encoded into the target protein by an engineered PylRS variant and tRNAPyl at a designed position with a UAG stop codon in living cells. The acetylation of lysine residues is a reversible process, regulated by acetyltransferases and deacetylases.
There is one potential problem when using AcK-OTSs for in vivo studies. Due to existence of deacetylases, site-specifically incorporated acetyllysine residues by GCE undergo deacetylation, especially in yeast or mammalian cells, and the stoichiometry of acetylation at target sites varies from batch to batch to make inconsistence among replicates. Thus, it is necessary to use nondeacetylatable analogs of acetyllysine, especially when long-time observation is needed for in vivo studies. Several lysine derivatives can be used for this purpose (Figure 15). First, 2-amino-8-oxononanoicacid (KetoK) has the same length of side chains as acetyllysine but replaces the nitrogen atom with carbon, so it cannot be hydrolyzed. The site-specific incorporation of KetoK into proteins has been established using MbAcKRS-1 in E. coli cells.174 Second, thioacetyllysine (TAcK) replaces the oxygen atom with sulfur, and TAcK-containing small peptides have been shown to inhibit sirtuin deacetylases.175 The OTS for TAcK has been developed by further engineering MbAcKRS-1 with two additional mutations (F271L and F313C).176 In this work, TAcK-containing proteins were shown to be recognized by acetyllysine-antibodies and had similar enzyme activities to acetyllysine-containing proteins. Moreover, TAcK-containing proteins were shown to resist CobB, the sirtuin deacetylase in E. coli. All these facts indicate TAcK is an ideal non-deacetylatable mimic of acetyllysine for in vivo studies. Later, trifluoro-acetyllysine (TFAcK) was genetically encoded into proteins by an MbPylRS variant (L266M, L270I, L274A, and C313F) as a probe to detect protein conformational changes by 19F NMR.177 In this study, TFAcK-containing proteins were also demonstrated to be resistant to sirtuin deacetylases. Recently, another stable mimic of acetyllysine, 1,2,4-triazole amino acid (ApmTri), has been site-specifically incorporated into proteins by WT MmPylRS in mammalian cells.178 The ApmTri-containing proteins were shown to have similar binding affinity with other proteins to that of acetyllysine-containing proteins. All these above-mentioned acetyllysine mimics offer alternative approaches when acetyllysine residues undergo rapid deacetylation in living cells.
Figure 15.

Acetyllysine and its nondeacetylatable analogs. AcK: acetyllysine; KetoK: 2-amino-8-oxononanoicacid; TAcK: thio-acetyllysine; TFAcK: trifluoro-acetyllysine; ApmTri: 1,2,4-triazole amino acid.
7.2. Lysine Methylation
Lysine methylation was first discovered in a bacterial flagellar protein in 1959, earlier than its discovery in histones.179 Besides its key role in regulating gene expression, lysine methylation of nonhistone proteins is also involved in a wide range of biological processes such as DNA damage repair, cellular signaling, metabolism, cell cycle, and cell differentiation.180–182 Abnormalities of lysine methylation has been found in patients of cancers,183,184 nervous system diseases,185,186 diabetes,187 cardiovascular diseases,188 and developmental disorders.189 Methylation of lysine residues is catalyzed by protein lysine methyltransferases (PKMTs). Humans have more than 60 PKMTs, which can be mainly classified as SET-domain-containing PKMTs and non-SET domain PKMTs.190,191 The reverse process, demethylation, is catalyzed by protein lysine demethylases (KDMs) through two distinct mechanisms: aminooxidation or hydroxylation.192 Different from methylation of arginine, glutamine, and histidine residues, lysine methylation has three degrees (mono-, di-, and trimethylation), and its regulatory role depends on methylation degrees.182,193 Another important feature of lysine methylation is that it keeps positive charges of lysine residues under physiological conditions, which is distinct from other lysine PTMs.
To generate proteins with degree- and site-specific lysine methylation, several semisynthetic approaches have been developed (Figure 16). Sulfur-containing lysine methylation mimics can be generated by alkylation chemistry of cysteine residues in vitro.194 Sulfur-containing analogs of lysine methylation can also be obtained by Michael addition reactions of Dha tags with thiol-containing compounds.195,196 It should be noted that those thioether bonds are sensitive to oxidation. To generate proteins with native lysine methylation, C–C bond formation reactions by free radical chemistry69 or single-electron transfer197 have been adopted. However, proteins generated by these approaches lose native stereo-chemistry at target lysine residues.
Figure 16.

Semisynthetic approaches to generating proteins with lysine methylation with different degrees. The target lysine residue is first mutated to cysteine. A) Alkylation of the cysteine residue forms sulfur-containing lysine methylation mimics. Then cysteine can be eliminated to form dehydroalanine (Dha). B) Michael addition of Dha produces sulfur-containing lysine methylation mimics. C) The Free radical reaction or D) single-electron transfer by Bpin (alkylboronic acid pinacol ester) with Dha forms C–C bond to generate proteins with native lysine methylation. Monomethylation: R1 = CH3, R2 = H, R3 = H; dimethylation: R1 = CH3, R2 = CH3, R3 = H; trimethylation: R1 = CH3, R2 = CH3, R3 = CH3.
Direct installation of lysine methylation into proteins in vivo has not been established by GCE yet, largely due to the high structural similarity between methylated lysine and unmodified lysine. However, several approaches based on ncAA-mediated chemical or physical reactions have been developed to install lysine methylation with different degrees. Monomethylation can be achieved by acid treatment of tert-butyloxycarbonyl-methyllysine incorporated by OTSs with WT MbPylRS or an MmPylRS variant (Y384F),198,199 Ru-mediated elimination of allylcarbamoyl-methyllysine encoded by the OTS with an MbPylRS variant (Y349F),200 reductive decaging of p-nitrobenzyloxycarbonyl-methyllysine installed by the OTS with an MbPylRS variant (Y271G and C313 V),201 and UV decaging of o-nitrobenzylcarbamoyl-methyllysine encoded by OTSs with an MbPylRS variant (Y271I, L274M, and C313A) or an MmPylRS variant (Y306M, L309A, and C348T) (Figure 17).202,203 More importantly, the approach by UV decaging was demonstrated to generate proteins with monomethyllysine in mammalian cells, providing a powerful tool for in vivo studies on lysine methylation.202
Figure 17.

The generation of monomethyllysine-containing proteins by ncAA-mediated approaches. A) 2% trifluoroacetic acid (TFA) treatment of tert-butyloxycarbonyl-methyllysine. B) Chloro-pentamethylcyclopentadienyl-cyclooctadiene-ruthenium(II) ([Cp*Ru(cod)Cl])-catalyzed cleavage of allylcarbamoyl-methyllysine. C) Reductive decaging of p-nitrobenzyloxycarbonyl-methyllysine. D) UV decaging of o-nitrobenzylcarbamoyl-methyllysine in living cells.
For dimethylation, two ncAA-mediated approaches have been established in vitro (Figure 18). In the earlier work, tert-butyloxycarbonyl (Boc)-lysine was first site-specifically incorporated into proteins by WT MbPylRS in E. coli cells. After purification, other free lysine residues in the protein were protected by the benzyloxycarbonyl (Cbz) group. Next, the Boc group was removed by trifluoroacetic acid, followed by the installation of dimethylation by reductive alkylation with formaldehyde and a dimethylamine borane complex. Finally, Cbz-protected lysine residues were deprotected by trifluoromethanesulfonic acid, trifluoroacetic acid, and dimethylsulfide mixtures.204 Later, an easier method was developed.205 Nε-(4-azidobenzoxycarbonyl)-δ,ε-dehydrolysine was genetically encoded into proteins by an MmPylRS variant (L309T, C348G, and Y384F) in E. coli cells. Then proteins were purified, and the precursor residue was reduced to allysine by Staudinger reduction with tris(2-carboxyethyl)phosphine, followed by conversion to dimethyllysine by reductive amination with dimethylamine in the presence of NaCNBH3.
Figure 18.

The generation of dimethyllysine-containing proteins by ncAA-mediated approaches. A) tert-Butyloxycarbonyl (Boc)-lysine is first genetically encoded into proteins at the target site 1, and all free lysine residues at other positions 2 are protected with benzyloxycarbonyl (Cbz) group by benzyloxycarbonyloxy-succinimide (Cbz-OSu). Then the Boc-protected lysine residue is deprotected by trifluoroacetic acid (TFA) and methylated by reductive alkylation using formaldehyde and a dimethylamine borane complex. Finally, Cbz groups at other lysine resides are removed by a mixture of trifluoromethanesulfonic acid (TFMSA)/trifluoroacetic acid (TFA)/dimethylsulfide (DMS). B) Nε-(4-azidobenzoxycarbonyl)-δ,ε-dehydrolysine is first site-specifically incorporated into proteins by GCE, followed by Staudinger reduction with tris(2-carboxyethyl)phosphine (TCEP) to form allysine and reductive amination with dimethylamine in the presence of NaCNBH3.
For trimethylation, instead of being derived from cysteine residues, two ncAAs were used to generate Dha tags, followed by Michael addition or metal-catalyzed C–C bond formation (Figure 19).206,207 Moreover, both methods can generate target proteins with lysine methylation at all three degrees. In the earlier study, phenylselenocysteine (PhSeCys) was first genetically encoded into proteins by the OTS with an MjTyrRS variant (Y32W, L65H, H70G, F108N, Q109S, D158S, and L162E). Then PhSeCys was converted to Dha with H2O2, followed by Michael addition with corresponding thiols to form sulfur-containing methyllysine mimics.206 In the later work, phosphoserine was first site-specifically incorporated into proteins by the OTS for phosphoserine (discussed in section 9.1) and converted to Dha under mild alkali conditions, followed by metal-mediated C–C bond formation with corresponding iodides to form native methyllysine modifications.207 Both methods can only be implemented in vitro, so the next breakthrough in this field is to develop methods to generate di- and trimethyllysine-containing proteins by cell-friendly and bio-orthogonal reactions.
Figure 19.

The generation of trimethyllysine-containing proteins by ncAA-mediated approaches. A) The dehydroalanine (Dha) tag is first formed by oxidation of genetically encoded phenylselenocysteine and then reacted with trimethylamine-containing thoil to form a mimic of trimethyllysine. B) The incorporated phosphoserine residue is converted to Dha by mild alkali treatment, followed by a metal-mediated coupling reaction with trimethylamine-containing iodide.
7.3. Lysine Acylation
Besides acetylation, lysine residues also undergo various types of acylation that have been recently identified by a series of proteomics studies.208 Among them, succinylation, crotonylation, malonylation, and 2-hydroxy-isobutyrylation have higher abundance than other types of acylation such as formylation and propionylation.145 Although the function, mechanism, and regulation of lysine acylation have not been as well studied as lysine acetylation, it is known that they are distributed as widely as lysine acetylation, from histones to nonhistone proteins and from eukaryotes to prokayotes.209 They are playing important roles in gene expression, cell signaling, and metabolism, and are involved in many human diseases.210 Here, we summarize established OTSs for genetically encoding lysine acylation or analogs in vivo (Figure 20).
Figure 20.

Genetically encoded acylated lysine or analogs. A) Nonpolar groups; B) polar groups; C) charged groups; D) other acylation; E) acylation analogs.
7.3.1. Nonpolar Acyl Groups.
Lysine propionylation has been found in histones and a number of bacterial proteins.211–213 The direct incorporation of propionyl-lysine into recombinant histones has been established in E. coli cells by WT MmPylRS214 or an MbPylRS variant (Y271F and C313T).215
Lysine butyrylation exists in histones and is involved in the onset and progression of cancer, diabetes, and cardiovascular diseases as well as growth, development, and metabolism of rice.216,217 OTSs for genetic encoding butyryl-lysine into proteins have been established in E. coli cells with WT MmPylRS214 or MbPylRS.215
Lysine crotonylation can be found in both histones and nonhistone proteins.218,219 Its unique C=C double bond forms a rigid planar conformation, indicating distinct regulatory mechanisms from other lysine acylation.220 OTSs based on WT MmPylRS214 or an MbPylRS variant (C313 V and M315Y)215 succeeded in site-specifically incorporating crotonyl-lysine into recombinant histones in E. coli cells. The protein yield was several milligrams per liter of growth media. Another OTS with an MbPylRS variant (L274A, C313F, and Y349F) was shown to genetically encode crotonyl-lysine into histones in human HEK293T cells, showing a promising application for in vivo studies.221
Lysine formylation occurs in histones and other nuclear proteins for regulating chromatin conformation and gene activity and functions as a secondary modification responding to oxidative DNA damage.222 MbPylRS was engineered to charge tRNAPyl with formyl-lysine, and a variant with five mutations (L266M, L270I, Y271F, L274A, and C313F) was able to generate homogeneously formylated proteins in both E. coli and mammalian cells.223 This work also demonstrated that the antibody for acetyllysine cannot recognize incorporated formyl-lysine residues, implying distinct regulatory functions between formylation and acetyllysine.
Lysine benzoylation has only been found in histones so far.208 It regulates gene expression of a series of proteins in metabolism and cell signaling.224,225 Several OTSs for benzoyllysine incorporation have been established. An MbPylRS variant (Y349W),226 an MmPylRS variant (Y384F),227 and an Methanomethylophilus alvus PylRS (MaPylRS) variant (Y206W)228 were shown to site-specifically incorporate benzoyl-lysine into target proteins in both E. coli and mammalian cells.
Due to the structural similarity among members of the nonpolar acylation group, those evolved PylRS variants have cross activities for different acylated lysine. For example, the OTS for propionyl-lysine can also encode crotonyl-lysine, while the OTS for crotonyl-lysine can incorporate both propionyl-lysine and butyryl-lysine.215 This phenomenon raises a question of how cells distinguish these highly similar modifications to implement different regulatory mechanisms.
7.3.2. Polar Acyl Groups.
Lysine 2-hydroxyisobutyrylation is widely distributed in both bacteria and eukaryotes, playing roles in protein synthesis, protein degradation, and cellular metabolism.229,230 The direct incorporation of 2-hydroxyisobutyryl-lysine into target protein has been successful in both E. coli and mammalian cells by an MbPylRS variant (C313T and Y349F).231
Lysine β-hydroxybutyrylation is a newly identified PTM in histones and nonhistone proteins in eukaryotes,232 which links cellular metabolism to gene expression.233 Emerging evidence shows its association with metabolic diseases, cardiovascular diseases, diabetes, neuropsychiatric disorders, and cancers.234,235 A recent study showed that lysine β-hydroxybutyrylation could improve stability of COVID-19 antibodies in cells.236 The OTS for genetic encoding of lysine β-hydroxybutyrylation into proteins has been established in both E. coli and mammalian cells with an MbPylRS variant (C313 V and Y349F).237
Lysine lactylation is also a newly discovered PTM and can be found in both bacteria and eukaryotes.238,239 The same as β-hydroxybutyrate, lactate is an important metabolite in cells, so lysine lactylation also play roles in cellular metabolism and is associated with a series of diseases related to metabolism such as cancers, diabetes, and metabolic disorders.240 An MbPylRS variant (Y271 M and C313T) was evolved to site-specifically incorporate lactyl-lysine into proteins in both E. coli and mammalian cells.237
7.3.3. Charged Acyl Groups.
Lysine succinylation, malonylation, and glutarylation are three PTMs found in both histones and nonhistone proteins from eukaryotic and bacterial cells.241–243 They are mostly involved in cellular metabolism and stress responses.244 A special feature of these three lysine PTMs is that they change lysine residues from positively charged to negatively charged, making them distinct from other lysine acylation. This feature also challenges the development of OTSs for direct incorporation them into proteins by GCE due to their bulky sizes and negative charges. Alternatively, an indirect method was developed to generate proteins with site-specific lysine succinylation, malonylation, and glutarylation.245 The precursor azidonorleucine was first site-specifically incorporated into proteins by an MmPylRS variant (Y306L, C348I, and Y384F), followed by traceless Staudinger ligation with phosphinothioester with corresponding acyl moieties. Theoretically, this approach can be used to generate proteins with all kinds of lysine acylation.
7.3.4. Other Acylation.
Lysine lipoylation has been known to exist in a limited number of protein complexes such as pyruvate dehydrogenase and α-ketoglutarate dehydrogenase as a handle to transfer reaction intermediates between subunits.246 A recent proteomic study identified a series of new protein targets for lysine lipoylation, indicating its possible roles in other biological processes.247 The OTS for installing lysine lipoylation has been developed in both E. coli and mammalian cells with an MbPylRS variant (Y271A, L274M, and C313V).237
Lysine fatty acylation with long chain fatty acyl groups such as myristoylation (C14) and palmitoylation (C16) has been identified in nonhistone proteins and is mostly involved in changing membrane protein affinity and localization for cell signaling.248 The long chain of fatty acids makes the direct incorporation of lysine myristoylation or palmitoylation into proteins almost impossible by GCE. A recent study developed an OTS for site-specifically installing lysine heptanoylation with a seven-carbon fatty acyl group into proteins with an MmPylRS variant (L301M, Y306A, L309A, and C348F) in E. coli cells.249 Although lysine heptanoylation has not been found in nature yet, the OTS could still provide a tool for studying native lysine fatty acylation.
Lysine aminoacylation is a newly identified PTM catalyzed by corresponding aminoacyl-tRNA synthetases to sense and transmit intracellular amino acid signals.250 As one type of lysine aminoacylation, lysine threonylation has been found in more than 100 proteins. The OTS for genetic encoding threonyl-lysine into proteins has been recently established in both E. coli and mammalian cells with an MmPylRS variant (Y384F).251 Later, another lysine aminoacylation, methionyllysine, was site-specifically incorporated into proteins by an MbPylRS variant (L274A, C313S, and Y349F) in both E. coli and mammalian cells. Using such a protein as a probe, a new PTM aminoacylated lysine ubiquitination was identified in human cell lines.252
7.3.5. Acylation Analogs.
Besides the above-mentioned lysine acylation, which exists in nature, a series of lysine acylation analogs have also been genetically encoded into proteins: A) Stable mimics for in vivo studies. Pent-4-ynyloxycarbonyl-lysine was used as a stable mimic of lysine butyrylation to study Salmonella infection, which can be genetically encoded into proteins by the OTS with an MbPylRS variant (L274A, C313S, and Y349F);253 B) Probes for biophysical studies. 2-Fluorobenzoyl-lysine and 2,5-difluorobenzoyl-lysine were site-specifically incorporated into proteins by an MmPylRS variant (Y384F) as probes for 19F NMR to monitor protein structure and interaction;227 C) Chemical handles for detectable tags. 7-Octenoyl-lysine and 7-azidoheptanoyl-lysine were site-specifically incorporated into proteins by an MmPylRS variant (L309A and C348S) first and then reacted with a fluorogenic tetrazine dye through the alkene-tetrazine cycloaddition reaction or a dibenzocyclooctyne (DBCO) dye through click chemistry to detect deacetylation by sirtuins;254,255 D) Photo-cross-linkers for mapping interaction networks. Diazirine-containing photoaffinity crotonyl-lysine was first genetically encoded into proteins by an MmPylRS variant (L309A, C348F, Y384W). After exposure with UV 365 nm, biomolecules that interact with target proteins were photo-cross-linked and identified by mass spectrometry.256 Recently, this strategy was extended to other lysine acylation such as propionylation, butyrylation, 2-hydroxyisobutyrylation,and β-hydroxybutyrylation.257
7.4. Lysine Ubiquitination/SUMOylation
Lysine ubiquitination is the most abundant lysine PTM in nature and can be found in proteins involved in a wide range of biological processes, not only confined to its well-known role in directing protein degradation.258,259 Ubiquitin (Ub) is a small protein with 76 amino acid. The C-terminal glycine residue of Ub is covalently linked to specific lysine residues in substrate proteins through an isopeptide bond by a successive process catalyzed by three enzymes: E1 Ub-activating enzyme, E2 Ub-conjugating enzyme, and E3 Ub-ligase.260 Beyond monoubiquitination, Ub can also attach to one of seven lysine residues or the N-terminal methionine residue in other Ub units to form polyubiquitination.15 Besides Ub, there are also some Ub-like proteins such as SUMO and NEDD8 that have similar properties to Ub, making the profile of ubiquitination in cells more complicated.261 Thus, it is necessary to generate proteins with homogeneous ubiquitination or SUMOylation to determine its site-specific roles. Although direct incorporation of ubiquitinated/SUMOylated lysine into protein is impossible for the GCE strategy due to their large sizes, numerous efforts have been made to generate homogeneously ubiquitinated/SUMOylated proteins by ncAA-mediated approaches.
In an earlier study, a thiol-containing lysine, D-Cys-ε-lysine, was first genetically encoded into proteins by WT PylRS, then reacted with a Ub analog with a C-terminal thioester by thiol exchange, followed by intramolecular S- to N-acyl transfer to form a ubiquitinated protein in vitro (Figure 21A).262 However, this method cannot generate the native isopeptide bond between Ub and lysine residues. To achieve this goal, a method called GOPAL (genetically encoded orthogonal protection and activated ligation) was developed with a similar procedure of generating lysine methylation shown in Figure 18A. Instead, it used a Cbz-protected Ub with a C-terminal thioester as the Ub donor.263 Although this approach generates ubiquitinated proteins with native isopeptide linkage, it is a relatively laborious process. To address this concern, a more facile method was established later.264 A thiol-lysine precursor, p-nitrocarbobenzyloxy-thiol-lysine, was first site-specifically incorporated into proteins by an OTS with an MbPylRS variant (Y271M, L274G, C313A, and Y349W) and converted to thiol-lysine by reductive decaging, followed by native chemical ligation with a Ub thioester. The thiol group was then removed by desulfurization reagent to generate ubiquitinated proteins in vitro (Figure 21B).
Figure 21.

The generation of lysine ubiquitination in proteins by ncAA-mediated approaches. The ubiquitin moiety is linked to lysine residues by A) native chemical ligation with D-Cys-ε-lysine; B) native chemical ligation with thiol-lysine; C) click chemistry with 2-propynyloxycarbonyl-lysine; D) oxime ligation with aminooxy-lysine; E) sortase-catalyzed transpeptidation with glycyl-glycyl-lysine.
Lysine ubiquitination is a reversible PTM, and the removal of ubiquitin moieties is catalyzed by deubiquitinases.265 To generate stable ubiquitinated proteins, nonhydrolyzable ubiquitination analogs have been incorporated into proteins by ncAA-mediated approaches. By using click chemistry, an alkynyl-containing lysine derivative, 2-propynyloxycarbonyllysine, was first genetically encoded into protein by WT MbPylRS and then reacted with azide-containing Ub (Figure 21C).266 In another work, the biorthogonal oxime ligation reaction was used to link aldehyde-containing Ub and aminooxy-lysine-containing proteins (Figure 21D). The aminooxy-lysine was first protected by a Boc group, then site-specifically incorporated into protein by an MbPylRS variant (Y349W), and finally deprotected by TFA treatment.267
All these methods mentioned above only apply to in vitro studies with purified proteins. To overcome this limitation, an ncAA-mediated approach combines biocompatible chemical and enzymatic reactions to generate proteins with site-specific lysine ubiquitination mimics in vivo (Figure 21E).268 Instead of linking Ub moiety to lysine residues by chemical reactions in above-mentioned approaches, an enzyme called sortase A was used for this step. Sortase A is an enzyme from Staphylococcus aureus and covalently links proteins to cell wall.269 It recognizes a specific motif LPXTG, cuts between T and G, and attaches T to peptides containing N-terminal glycine residues. To link Ub to a lysine residue, the C-terminal sequence of Ub was mutated from LRLRGG to LPLTGG. Then the remaining GG moiety need to attach to a lysine residue in the target protein. To achieve the goal, the GCE strategy was applied to site-specifically incorporate the GG moiety containing-lysine precursor, 2-azidoacetyl-glycyl-lysine, by an MbPylRS variant (L274A, N311Q and C313S), which was then converted to glycyl–glycyl-lysine by Staudinger reduction with the cell-permeable phosphine 2-(diphenylphosphino)benzoic acid in living cells. The final protein product has the native isopeptide linkage between Ub and lysine but also bears two mutations at the C-terminal of Ub (R72P and R74T), making the ubiquitinated protein resistant to deubiquitinases. This work also used the same strategy to install lysine SUMOylation into proteins in living mammalian cells. Later, a series of orthogonal sortases and asparaginyl endopeptidases were engineered to provide more flexible motif selections.270,271
8. SERINE/THREONINE MODIFICATIONS
Serine and threonine are two polar amino acids with hydroxyl groups, making them favorable targets of PTMs. Besides well-known phosphorylation and O-glycosylation, both of them also undergo several relatively rare PTMs such as acetylation, which can compete with other PTMs to have antagonized effects,272 sulfonation, which is involved in protein assembly and signal transduction,273 and palmitoylation, which regulates membrane protein functions.274 In this section, we focus on the development of OTSs for generating proteins with homogeneous phosphorylation or O-glycosylation on serine or threonine residues.
8.1. Serine Phosphorylation
Serine phosphorylation is the most abundant PTM in nature. The addition of the phosphate group brings two negative charges to the serine residue, affecting protein structure, function, and interactions.275 Due to the transferable property of the phosphate group, phosphorylation plays essential roles in cell signaling pathways, regulating a variety of processes including cellular metabolism, cell cycle, cell differentiation, and stress responses276,277 Because of its tight association with human diseases, phosphorylation is a common therapeutical target.278,279 Protein phosphorylation is regulated by kinases and phosphatases. In humans, there are more than 500 kinases and approximately 200 phosphatases,280,281 making phosphorylation involved in a huge cell signaling network. To illustrate the effects of phosphorylation on target proteins, enzymatic, chemical, and biological approaches have been developed to generate phosphorylated proteins.12,282 Among these methods, GCE is able to generate site-specifically phosphorylated proteins in living cells, providing a powerful tool to study protein phosphorylation in vivo. The applications of GCE in characterizing specific phosphorylation events have been nicely reviewed recently.51,283
Phosphoserine (Sep) is not a synthetic amino acid, but a native amino acid in cells, which is an important intermediate in serine biosynthesis.284 In methanogenic archaea, phosphoseryl-tRNA synthetase (SepRS) charges tRNACys with Sep for cysteine biosynthesis.285 Inspired by this fact, the pair of Methanococcus maripaludis SepRS (MmSepRS) and a modified MjtRNACys with CUA as the anticodon (tRNASep) was used to generate Sep-tRNASep, which is later delivered to ribosomes by EF-Tu to decode the UAG codon in mRNA as Sep. However, ET-Tu has a relatively weaker binding with negatively charged amino acids due to two acidic amino acid residues in the amino acid binding pocket.286 Thus, E. coli EF-Tu was engineered to accommodate Sep-charged tRNASep (EF-Sep) with mutations of H67R, D216G, E217N, F219Y, T229S, and N274W. MmSepRS, tRNASep, and EF-Sep forms the first OTS for Sep incorporation in E. coli cells (Figure 22).287 To date, Sep-OTSs have been introduced into Samonella,166 mammalian cells,288 and cell-free translation systems.71,289
Figure 22.

Genetic encoding phosphoserine into proteins in living cells and further optimization. Phosphoserine (Sep) is linked to tRNACysCUA by phosphoseryl-tRNA synthetase (SepRS) to generate Sep-tRNA, which is then delivered to the ribosome by an EF-Tu variant, EF-Sep, to decode UAG codon in mRNA as Sep, thus generating the target protein with site-specific serine phosphorylation. To improve Sep-incorporation, several strategies have been adopted, including inactivating serine phosphatase to increase intracellular Sep concentration, inactivating release factors to eliminate early termination, using nonhydrolyzable Sep mimics to prevent hydrolysis by protein phosphatases, and further engineering SepRS, tRNACysCUA, and EF-Sep.
The first Sep-OTS could only produce phosphorylated proteins at the microgram level. To improve Sep incorporation efficiencies, components in the whole process of Sep-encoding have been optimized (Figure 22). A) Serine phosphatase inactivation. Inactivation of serine phosphatase (SerB in E. coli or PSPH in mammalian cells) can increase the intracellular Sep concentration to provide sufficient substrate for generating Sep-charged tRNA.288,290–292 B) Removal of release factor-1 (RF1). One major factor to affect UAG codon-mediated ncAA incorporation is the competition from RF1 in E. coli cells. To eliminate such competition, RF1 has been inactivated in E. coli strains. However, one problem with RF1-deficient strains is that there are hundreds of native genes in E. coli using TAG as the termination signal. Inactivation of RF1 will add unwanted C-terminal extension to those proteins, and some of them are essential for E. coli cells. Thus, partial or complete replacement of those TAG codons with TAA codons was implemented to recover cell growth and to increase Sep-incorporation in E. coli cells, including the EcAR7.ΔA strain in which stop codons of 7 essential genes in E. coli were modified from TAG to TAA,290,293 the C321.ΔA strain in which all 321 TAG codons in E. coli MG1655 cells were changed to TAA codons,289,294,295 and the B-95.ΔA stain in which 95 TAG codons in E. coli BL21(DE3) cells were mutated to TAA.296 Another strategy to overcome the drawback of RF-1 deficient strains is to remove near cognate tRNAs for UAG codons to prevent the decoding of UAG codons as canonical amino acids such as tyrosine and glutamine. This strategy has been used in cell-free translation systems for phosphoprotein synthesis.71 C) Further engineering SepRS, tRNASep, and EF-Sep. Further engineering each component in the Sep-OTS has been implemented by generating larger mutation libraries, optimizing selection strategies, or hybridizing components from different species, which includes the SepRS9-EFSep21 system with an optimized MmSepRS variant (K347E, N352D, E412S, E414I, P495R, I496R and L512I) and an optimized E. coli EF-Tu variant (H67R, D216 V, E217N, F219Y, T229S, and N274W),297 the SepRSv1.0/tRNAv1.0CUA pair with an improved MmSepRS variant (E412P, E414F, K417 K, P495M, I496W, and F529) and an enhanced MjtRNASep mutant (A29G, U31A, A39U, U40C, and U41C) without the need of EF-Tu variants,292 as well as an OTS that contains SepRSv1.0/tRNAv1.0CUA, an EF-1α variant (EF-1α-Sep, L77R, Q251N, D252G, V264S, andN307W), and eRF1(E55D) for Sep-incorporation in mammalian cells.288 D) Nonhydrolyzable Sep analogs. Protein phosphatases in cells can dephosphorylate phosphoproteins generated by GCE. Thus, a nonhydrolyzable Sep analog, phosphonomethylene alanine (Pma), was genetically encoded into proteins in both E. coli and mammalian cells.288,292 Due to the high similarity between Pma and Sep, Pma can be recognized by SepRS. However, endogenous Sep in cells can also compete with Pma for the Sep-OTS. To increase Pma-incorporation, endogenous Sep concentration was decreased by overexpressing serine phosphatase (SerB in E. coli or PSPH in mammalian cells) to hydrolyze free Sep and inactivating phosphoserine aminotransferase (SerC in E. coli or PSAT in mammalian cells) to block the production of Sep.288,292
8.2. Threonine Phosphorylation
Threonine is different from serine only by one methyl group, but that methyl group matters. That is why nature keeps both serine and threonine as basic components for protein synthesis after billions of years of evolution. The abundance of serine phosphorylation in nature is about 4.5-fold that of threonine phosphorylation.298 The structure and function of serine phosphorylation and threonine phosphorylation also differ from each other.299
Because of the similarity between Sep and phosphothreonine (pThr), the established Sep-OTS is a good start for pThr-OTS development. To achieve this goal, there are two concerns to be addressed first. pThr does not exist as a free amino acid in most species such as E. coli and mammalian cells, which are the most common hosts for GCE, and negatively charged compounds commonly have low cellular uptake. Thus, the first problem is how to increase intracellular pThr concentration for pThr-tRNA generation. PduX, a threonine kinase in Salmonella, generates pThr as an intermediate for coenzyme B12 biosynthesis.300,301 Overexpressing PduX can increase pThr concentration in E. coli cells.302 The second problem to be solved is how to engineer SepRS to recognize pThr but not Sep. Combining parallel selection and deep sequencing, an MmSepRS variant (G320A and L321Y) was shown to distinguish Sep and pThr, and site-specifically incorporated pThr into target proteins in E. coli cells.302 A recent study further optimized the pThr-OTS with several modifications such as inactivating phosphoserine aminotransferase SerC to decrease Sep concentration and utilizing RF-1 deficient strains to generate proteins with site-specific threonine phosphorylation.303 Next steps for pThr-OTS development will be introducing it into mammalian cells and genetic encoding nonhydrolyzable pThr analogs.
8.3. O-Glycosylation
Besides N-glycosylation discussed in section 4.1, O-glycosylation is another type of protein glycosylation with O-linkage between glycans and hydroxyl group-containing amino acid residues, including serine, threonine, hydroxyproline, hydroxylysine, and rarely tyrosine.304 In addition to the roles in protein localization and cell signaling, emerging evidence has demonstrated the involvement of O-glycosylation in protein aggregation and phase separation.305 O-glycosylation has been found to be associated with genetic disorders, infectious diseases, immune-related diseases, neurodegenerative diseases, and cancers.306,307
Chemical and enzymatic approaches have been implemented to generate proteins with O-glycosylation mimetics. The site-directed mutated cysteine residue is transformed into Dha by alkylating reagents and reacted with active glycosylation derivatives.308,309 Furthermore, thioglycoligase engineered from Streptomyces plicatus hexosaminidase is able to transfer N-acetylglucosamine (GlcNAc) to a site-directed mutated cysteine residue to form a protein with S-GlcNAc, which is a physiological mimetic of O-glycosylation.310 However, both methods only apply to in vitro studies with purified proteins.
Although direct incorporation of O-glycosylated serine/threonine into proteins in living cells has not been established yet, the applications of GCE in cell-free translation systems to produce glycosylated proteins have been reported earlier.311,312 Instead of charging tRNA with glycosylated amino acids by engineered AARSs, chemical or T4 ligase-catalyzed aminoacylation was used to aminoacylate tRNA, which later decodes the assigned codon as glycosylated amino acids. For cell-based approaches, several OTSs have been utilized to genetically encode ncAAs bearing functional groups, which can be reacted with glycan moiety-containing compounds to form glycosylated proteins. For example, a similar approach shown in Figure 11 was applied. First, bicyclo[6.1.0]nonyne-lysine was site-specifically incorporated into proteins by an MmPylRS variant (Y306A and Y384F) in E. coli cells. Then purified proteins were reacted with a tetrazine-containing glucosamine analog by the inverse-electron-demand Diels–Alder reaction to form glycosylation mimic-containing proteins in vitro.313 Since the inverse-electron-demand Diels–Alder reaction can be applied in living cells, this approach has the potential for in vivo studies. In another work, like the method demonstrated in Figure 21C, click chemistry was utilized to link alkyne-containing residues with azido-containing glycans to produce target proteins with glycosylation mimics in vitro.314
9. TRYPTOPHAN MODIFICATIONS
Tryptophan is an aromatic amino acid with an indole ring as the side chain and is commonly found inside of globular proteins. Its intrinsic fluorescence makes itself a native probe for detecting protein conformational changes.315 Tryptophan residues in proteins mainly undergo oxidation and nitration, which is involved in stress responses to reactive oxygen species (ROS) and reactive nitrogen species (RNS) (Figure 23).316,317 In this section, we focus on the development of OTSs for genetically encoding tryptophan hydroxylation, one type of tryptophan oxidation.
Figure 23.

Tryptophan oxidation and nitration modifications found in proteins.
9.1. Tryptophan Hydroxylation
Tryptophan hydroxylation has been found in a series of proteins such as α-crystallin and apolipoprotein. α-crystallin is a major lens protein, and oxidation of tryptophan residues in crystallin leads to darkening of lens and a potential link to cataractogenesis.318 Oxidation of tryptophan residues in apolipoprotein can decrease cholesterol efflux capacity.319 It is commonly thought that protein oxidation is harmful. However, oxidation of tryptophan residues does not always have negative effects and sometimes is essential for protein functions.316 For example, copper-binding protein MopE in Methylococcus capsulatus needs oxidation of tryptophan residues to facilitate the binding of copper.320 Thus, it is necessary to characterize site-specifically hydroxylated tryptophan residues.
5-Hydroxy-tryptophan (5HTP), one type of tryptophan hydroxylation, has been genetically encoded into protein in both E. coli and mammalian cells. The OTSs for 5HTP incorporation utilize orthogonal pairs of tryptophanyl-tRNA synthetase (TrpRS) and tRNATrp instead of PylRS and TyrRS systems, which are commonly used for ncAA incorporation. The 5HTP-OTS for using in E. coli cells was derived from Saccharomyces cerevisiae TrpRS/tRNATrp with an ScTrpRS variant (T107C, P254T, and C255A), which was obtained by directed evolution with compartmentalized partnered replication.321 The suppressor SctRNATrpCUA could be mischarged by E. coli LysRS, so an SctRNATrpCUA mutant (U16G, G43U, and U58G) was obtained by engineering to eliminate misacylation by EcLysRS.321 Becasue AARS/tRNA pairs are usually not orthogonal in organisms from the same domain, the ScTrpRS/tRNATrp pair cannot be used in mammalian cells. To achieve the goal of developing a 5HTP-OTS for mammalian cells, the pair of TrpRS/tRNATrp from E. coli was used as the scaffold for further engineering. To take the advantage of the facile E. coli-based AARS evolution system, the genes of EcTrpRS and EctRNATrp were removed from the E. coli genome, and the pair of ScTrpRS/tRNATrp was introduced into E. coli to decode native tryptophan codons. Then the pair of EcTrpRS/tRNATrp was evolved in this altered translational machinery E. coli strain. The final 5HTP-OTS for using in mammalian cells contains an EcTrpRS variant (V144G and V146C) and an amber suppressor EctRNATrpCUA. This 5HTP-OTS was also able to site-specifically incorporate a series of tryptophan analogs such as 5-methoxy-tryptophan, 5-bromo-tryptophan, and 5-azidotryptophan, which could be useful for studies on other tryptophan PTMs.322
10. TYROSINE MODIFICATIONS
Tyrosine is an aromatic amino acid with a hydroxyl group, making itself a special amino acid containing both nonpolar and polar moieties. As an aromatic amino acid, tyrosine residues also undergo oxidation such as hydroxylation and nitration like tryptophan. On the other end, the hydroxyl group makes tyrosine a favorable target of phosphorylation. Besides these modifications, tyrosine sulfation and halogenation also occur naturally. In this section, we summarize the development of OTSs for tyrosine PTMs or analogs (Figure 24).
Figure 24.

Tyrosine modifications and analogs. 1: phosphotyrosine; 2: photocage phosphotyrosine; 3: bis(dimethylamino)phosphoryloxy-phenylalanine; 4: 4-phosphomethyl-phenylalanine; 5: p-carboxymethyl-phenylalanine; 6: p-(phosphonoamino)-phenylalanine; 7: sulfotryosine; 8: 3,4-dihydroxy-phenylalanine (DOPA); 9: o-nitrobenzyl DOPA; 10: 3-nitro-tyrosne; 11: 3-chloro-tyrosine; 12: 3,5-dichloro-tyrosine; 13: 3-bromo-tyrosine; 14: 3,5-dibromo-tyrosine; 15: 3-iodo-tyrosine; 16: 3,5-diiodo-tyrosine.
10.1. Tyrosine Phosphorylation
Although tyrosine phosphorylation is not as abundant as serine/threonine phosphorylation in nature, it plays important roles in cell cycle, cell differentiation, cell shape and movement, synaptic transmission, and bacterial virulence.323,324 In humans, there are at least 90 protein tyrosine kinases and 107 protein tyrosine phosphatases that function coordinately to control tyrosine phosphorylation levels in different proteins.325,326 Thus, abnormalities of those enzymes lead to a series of human diseases such as cancers, Alzheimer’s disease, diabetes, skeletal diseases, and cardiovascular diseases.327
To generate proteins with site-specific phosphorylation in living cells, a big challenge is to provide sufficient phosphotyrosine (pTyr) (1 of Figure 24) for tRNA aminoacylation. This is difficult because there are no known biosynthesis pathways for pTyr, and pTyr has low cell permeability due to its negative charges. Thus, production of proteins with homogeneous tyrosine phosphorylation was first established by applying GCE in cell-free translation systems in which pTyr was linked to tRNA by chemical aminoacylation.328 Later, this method was optimized by adopting a photocaged pTyr analog (2 of Figure 24) to overcome the weak recognition of pTyr by EF-Tu329 or with modified ribosomes to improve pTyr incorporation.330 Another approach to bypassing the low concentration of pTyr in cells is to use a neutral charged pTyr analog, bis(dimethylamino)phosphoryloxy-phenylalanine (3 of Figure 24), as a precursor that was genetically encoded into proteins by an MmPylRS variant (A302S, L309M, I322L, N346A, W348G, and W417T) in E. coli cells. Then purified proteins were treated by acid to remove the protecting group to form native tyrosine phosphorylation in vitro.331
To develop pTyr-OTSs in living cells, the first question is how to increase intracellular pTyr concentration. Two studies utilized different methods to solve this problem. In the earlier work, five phosphatases were removed from E. coli cells, including AphA (nonspecific acidic phosphatase), PgpA (phosphatidyl-glycerophosphatase in membrane), PhoA (nonspecific alkaline phosphatase), PphB (phosphatase involved in the misfolded protein stress), and SerB (phosphoserine phosphatase). These five phosphatases were selected by screening all possible pTyr phosphatases in the E. coli genome. Removal of phosphatases has also been shown to increase cellular uptake of phosphate-containing compounds to respond to phosphate-limiting conditions.291 Combining with an MjTyrRS variant (Y32L, L65R, D158G, I159C, L162 K, D286R) to charge MjtRNATyrCUA with pTyr and an EF-Tu variant (E216 V, D217G, F219G) to facilitate the binding of negatively charged pTyr, the first pTyr-OTS was established in E. coli cells, through the incorporation efficiency is relatively low.332 In a later study, synthesized dipeptide (Lys-pTyr) was transported into E. coli cells by the dipeptide transporter DppA and then hydrolyzed by nonspecific peptidases in cells to provide sufficient pTyr for aminoacylation. An MjTyrRS variant (Y32S, L65A, F108 K, Q109H, D158G and L162 K) was used to charge MjtRNATyrCUA with pTyr and site-specifically incorporate pTyr into proteins in E. coli cells. However, dephosphorylated protein products were observed by this method resulting from phosphatases in cells. Thus, a nonhydrolyzable pTyr analog, 4-phosphomethyl-phenylalanine (4 of Figure 24), was genetically encoded into proteins using the same dipeptide transport strategy and pTyr-OTS in E. coli cells.333
Several OTSs for genetic encoding of stable pTyr analogs have also been developed, including p-carboxymethyl-phenylalanine (5 of Figure 24), which can be site-specifically incorporated into proteins by the OTS with an MjTyrRS variant (Y32S, L65A, F108 K, Q109H, D158G, and L162 K) for E. coli cells334 or the OTS with an EcTyrRS variant (Y37H, L71 V, and D182G) or another EcTyrRS variant (Y37H, L71 V, D182G, L186M, and D265R) for mammalian cells,335,336 p-(phosphonoamino)phenylalanine (6 of Figure 24), which is generated by the Staudinger-phosphite reaction from a genetically encoded p-azidophenyalanine residue by an MjTyrRS variant (Y32T, E107N, D158P, I159L, and L162Q),337 and sulfotyrosine (sTyr, 7 of Figure 24), which is also a tyrosine PTM in nature. The next breakthrough of pTyr-OTS development will be establishing pTyr-OTSs in mammalian cells.
10.2. Tyrosine Sulfation
Tyrosine sulfation was first discovered in fibrinogen 70 years ago.338 However, research on its functions and significance in human health has emerged recently. Tyrosine sulfation has been identified mostly in secretory and membrane-bound proteins of higher eukaryotes so far.339 It mainly affects protein–protein interactions and is involved in hemostasis, receptor–ligand recognition, visual functions, and viral infections.340 Tyrosine sulfation is catalyzed by tyrosylprotein sulfotransferase, which transfers an activated sulfate from 3′-phosphoadenosine-5′-phosphosulfate to tyrosine residues.341 Different from most PTMs, tyrosine sulfation is thought to be an irreversible process, and this is also the reason why it can be used as a stable analog of tyrosine phosphorylation.
The OTSs for genetically encoding sTyr into proteins have been established in both E. coli and mammalian cells. The first sTyr-OTS for E. coli cells contains an MjTyrRS variant (Y32L, L65P, D158G, I159C, and L162 K).342 Later, this sTyr-OTS was optimized by increasing copy numbers of the MjTyrRS variant and MjtRNATyrCUA and using an improved MjTyrRS variant (Y32L, L65P, D158G, I159T, and L162 K)343 or adopting the RF-1 deficient E. coli strain (C321.ΔA).344 Because the pair of MjTyrRS and MjtRNATyr is not orthogonal in mammalian cells, the pair of EcTyrRS and EctRNATyr is commonly used for incorporation of tyrosine analogs in mammalian cells. However, the facile E. coli-based selection system cannot be used to engineer E. coli AARSs. To solve this problem, one study utilized a yeast selection system to engineer EcTyrRS and obtained an EcTyrRS variant (L71 V, W129F, and D182G) that can recognize sTyr for site-specific incorporation of sTyr into proteins in mammalian cells.345 To take the advantage of E. coli selection systems, another study developed an altered translational machinery E. coli strain in which the native EcTyrRS/tRNA pair is replaced by MjTyrRS/tRNA pair to decode tyrosine codons, and then EcTyrRS was evolved in this engineered E. coli strain. Finally, an EcTryRS variant (L71 V, D182G, and L196M) was obtained to site-specifically incorporate sTyr into proteins in mammalian cells.346 Recently, a biosynthetic reaction for sTyr by tyrosine sulfotransferase was introduced into both E. coli and mammalian cells. Combined with established sTyr-OTSs, this approach generated proteins with higher sTyr-incorporation efficiency.347
10.3. Tyrosine Hydroxylation
Hydroxylation of free tyrosine by tyrosine hydroxylase forms 3,4-dihydroxy-phenylalanine (DOPA, 8 of Figure 24), which is the precursor of neurotransmitters dopamine, norepinephrine, and epinephrine.348 Hydroxylation of tyrosine residues in proteins has been found in mussel proteins to increase adhesivity349 and identified as a new histone maker associated with transcription.350
Site-specific incorporation of DOPA into proteins has been established in both E. coli and mammalian cells. In E. coli cells, an OTS with an MjTyrRS variant (Y32L, A67S, H70N, and A167Q) was developed to genetically encoding DOPA into proteins.351 However, due to the structural similarity between DOPA and tyrosine, this MjTyrRS variant could also recognize tyrosine and bring it to the target site. To circumvent this issue, a photocaged DOPA, o-nitrobenzyl DOPA (9 of Figure 24), was first site-specifically incorporated into proteins by an MjTyrRS variant (Y32A, L65I, H70A, Q109A, D158A, I159A, L162M, A167N, and A180Q) in E. coli cells, and then decaged by UV irradiation in vitro.352 Later, a biosynthetic reaction for DOPA by tyrosine-phenol lyase was introduced into E. coli cells. Instead of feeding cells with DOPA, this method could provide sufficient DOPA for protein synthesis and was shown to increase DOPA-incorporation efficiency.353 To select MjTyrRS variants that can discriminate DOPA and tyrosine, a recent study combined classic selection methods and a selection approach based on the biochemical properties of DOPA to provide additional stringency for selection. MjtRNATyrCUA in the OTS has a mutated anticodon CUA from GUA, and MjTyrRS contains an anticodon binding domain for recognizing the GUA anticodon in the original MjtRNATyr, so the anticodon binding region of MjTyrRS (F261, H283, P284, M285, and R286) was engineered for better CUA anticodon recognition. Together with an evolved DOPA binding pocket, the final MjTyrRS variant (L65E, H70A, Y119F, D158A, I159A, F261S, H283P, P284L, M285R, and R286A) was able to eliminate tyrosine incorporation at target positions.354 To develop an DOPA-OTS for mammalian cells, a chimeric AARS that contains a human mitochondrial PheRS variant (Q356N, T467S, and A507S) and the tRNA binding domain of MbPylRS was created. Such chimeric design of PylRS and other AARSs was also able to genetically encode various aromatic amino acid derivatives into proteins in both E. coli and mammalian cells.355
10.4. Tyrosine Nitration
Distinct from most PTMs mentioned above, which are catalyzed by enzymes, nitration of tyrosine residues in proteins is triggered by various RNS such as peroxynitrite (ONOO−) and nitrogen dioxide radicals (·NO2).356 Tyrosine nitration is involved in several RNS-related biological processes such as oxidative stress responses, cell signaling, and cell death.357 It is known to be associated with aging, cancers, atherosclerosis, and neurodegenerative diseases.358–360
As the major product of tyrosine nitration, 3-nitro-tyrosne (3NY, 10 of Figure 24) has been genetically encoded into proteins in both E. coli and mammalian cells. The first 3NY-OTS for E. coli cells was established with an engineered MjTyrRS variant (Y32R, H70L, Q155M, D158G, I159L, and L162H).361 This 3NY-OTS was optimized in several later studies. First, an altered selection scheme was implemented to modify the screening stringency and obtained an improved MjTyrRS variant (Y32H, H70C, D158S, I159A, and L162R) that can increase 3NY incorporation by 7-fold.362 Later, MjtRNATyr in the 3NY-OTS was engineered with two mutations in the anticodon loop (G34C/G37A), which further improved 3NY incorporation in E. coli cells.363 Recently, the RF-1 deficient stain B-95.ΔA was used for genetic encoding of 3-NY with an enhanced MjTyrRS variant (Y32H, H70T, D158H, I159A, and L162R) by computational screening.364 To develop 3NY-OTSs for mammalian cells, MbPylRS was engineered to recognize 3NY but not tyrosine, and an MbPylRS variant (N311S and C313S) showed the highest incorporation efficiency.
10.5. Tyrosine Halogenation
Tyrosine halogenation is another type of nonenzymatic PTM and commonly occurs under oxidative stress conditions. Halogenation of proteins by reactive halogen species (RHS) mostly impairs protein structure and function, thus leading to aging, cancers, and other long-term diseases.365 Varied with types and numbers of halogen atoms, there are several halogenated tyrosine moieties identified in cells and tissues, including 3-chloro-tyrosine (ClY), 3,5-dichloro-tyrosine (Cl2Y), 3-bromo-tyrosine (BrY), 3,5-dibromo-tyrosine (Br2Y), 3-iodo-tyrosine (IY), and 3,5-diiodo-tyrosine (I2Y) (11 to 16 of Figure 24).366 Although tyrosine halogenation can be formed by treating proteins with RHS in vitro, it usually generates halogenated proteins with multiple halogenated residues and various types of halogenation.367
To demonstrate site-specific effects of tyrosine halogenation on protein functions, a recent study developed OTSs to genetically encode halogenated tyrosine into proteins in E. coli cells.368 Due to the structural similarity among those halogenated tyrosine moieties, one MjTyrRS variant (Y32G, L65E, H70G, F108Q, Q109C, D158A, L162N) could recognize ClY, Cl2Y, BrY, Br2Y, and IY, while another MjTyrRS variant (Y32V, L65E, H70G, F108N, D158A, L162C) was selected for I2Y. This study showed that halogenation of tyrosine residues changes the phenolic pKa of tyrosine residues significantly (Y: 9.90, ClY: 8.31, Cl2Y: 6.52, BrY: 8.29, Br2Y: 6.42, IY: 8.30, I2Y: 6.45) and affects protein assembly and dynamics. For example, halogenation of Tyr222 in E. coli filamentous cell-division protein FtsZ decreases its GTPase activity and polymerization rates, thus affecting ring formation in living cells. As tyrosine halogenation is a typical format of protein oxidation, the information on site-specific effects of halogenation on protein structures and functions will contribute to our understanding of oxidative damage-related human diseases such as cancers and aging.
11. MODIFICATIONS OF OTHER AMINO ACIDS
Besides the above-mentioned amino acids, other amino acids also undergo various PTMs.10 Alanine residues can be modified by methylation and acetylation; aspartate and glutamate residues undergo acetylation, phosphorylation, and hydroxylation; phenylalanine, leucine, and isoleucine residues can be modified by methylation and hydroxylation; glycine residues undergo myristoylation and palmitoylation; methionine and valine residues can be modified by acetylation and oxidation; proline residues undergo acetylation, hydroxylation, and methylation. The OTSs for these PTMs, analogs, or precursors have not been established yet.
12. SUMMARY AND PERSPECTIVES
In this review, we summarize currently available OTSs for genetically encoding various ncAAs into proteins as native PTMs, PTM analogs, or PTM precursors either in living cells or in vitro (Table 1). Although plenty of OTSs have been developed for PTM studies, there are still many gaps to fill in, such as developing OTSs for mammalian cells or model animals, replacing in vitro harsh chemical reactions with in vivo biocompatible reactions to generate proteins with native PTMs or PTM analogs from precursors, designing nonhydrolyzable but functional PTM mimics for in vivo studies, and installing multiple PTMs into a single protein simultaneously to study crosstalk between PTMs.
Table 1.
Availability of OTSs for Generating Proteins with PTMs*
| Acetylation | Citrullination | Glycosylation | Hydroxylation | Lipidation | |
|---|---|---|---|---|---|
| Arg | xa | ++b | x | x | |
| Asn | +c | x | |||
| Cys | x | ++ | |||
| Gin | |||||
| His | x | x | x | ||
| Lys | +++d | x | x | ++ | |
| Ser | x | + | x | ||
| Thr | x | + | x | ||
| Trp | +++ | ||||
| Tyr | x | x | +++ | ||
| Methylation | Nitration | Phosphorylation | Sulfation | Ubiquitination | |
| Arg | + | x | |||
| Asn | x | ||||
| Cys | x | x | x | ||
| Gin | + | ||||
| His | +++ | x | |||
| Lys | ++ | x | ++ | ||
| Ser | x | +++ | X | ||
| Thr | x | +++ | x | ||
| Trp | x | x | x | ||
| Tyr | +++ | +++ | +++ |
Shading: The PTM has not been identified in nature to date.
x indicates no OTS has been established for the native PTM, PTM analogs, or PTM precursors yet.
++ indicates OTSs have been established for genetic encoding of precursors that can be converted to PTMs or analogs in living cells.
+ indicates OTSs have been established for genetic encoding of precursors that can be converted to PTMs or analogs in vitro.
+++ indicates OTSs have been established for genetic encoding of native PTMs in living cells.
Protein methylation occurs mostly on arginine and lysine residues, less commonly on histidine residues. Unfortunately, no OTS has been established to genetically encode methylarginine into proteins in living cells. The previous study showed that WT yeast ArgRS can charge tRNAArg with monomethylarginine,70 so it should be difficult to engineer ArgRS to discriminate monomethylarginine and arginine. However, the same study also showed that WT yeast ArgRS cannot charge tRNAArg with dimethyl-arginine, indicating that the amino acid binding pocket of ArgRS is sufficient to distinguish the difference between arginine and dimethyl-arginine. Thus, it is possible to further engineer ArgRS to recognize dimethyl-arginine but not arginine and establish ArgRS-based OTSs for dimethyl-arginine incorporation in living cells. As for another common protein methylation, there are also no established OTSs for installing lysine methylation directly in living cells. Monomethyllysine residues could be formed by photo decaging of a methyllysine precursor in living cells, while di- and trimethyllysine residues could only be generated by in vitro chemical reactions from precursors (Figures 17–19). To date, classic methods of engineering AARSs for ncAA incorporation commonly randomize residues around the amino acid binding pockets. Due to the limited size of mutant libraries (<1010), the number of randomized residues cannot be more than 7 to make good coverage of all possible mutations. This number could be good enough for most ncAAs but may not be sufficient to distinguish small structural differences such as lysine and methyllysine. Thus, approaches to engineering AARSs on the full-length scale such as phage-assisted continuous evolution164 or predicting AARS variants for specific ncAAs more accurately by computational screening364 could be adopted to establish OTSs for methyllysine in living cells by further engineering existing OTSs based on PylRS or LeuRS, which have flexible amino acid binding pockets.48,87
Protein phosphorylation mostly occurs on serine, threonine, and tyrosine residues and less frequently on histidine, aspartate, and arginine residues. OTSs for Sep, pThr, and pTyr have been established in E. coli cells, but only the Sep-OTS has been introduced into mammalian cells and animals. Thus, developing OTSs for pThr and pTyr in mammalian cells is the next milestone in this field. The pThr-OTS for E. coli cells was derived from the Sep-OTS, which is also orthogonal in mammalian cells, so the major challenge is how to generate pThr in mammalian cells. Threonine kinase PduX from Salmonella produces pThr in E. coli cells302 but may not function in mammalian cells, which is the first thing to determine before developing pThr-OTS for mammalian cells. The pTyr-OTS for E. coli cells was derived from the pair of MjTyrRS/tRNATyr, which is not orthogonal in mammalian cells. Instead, the pair of EcTyrRS/tRNATyr has been used to genetically encode several tyrosine analogs in mammalian cells,336,345 so it is feasible to evolve EcTyrRS for site-specific incorporation of pTyr. However, the same as pThr-OTS development, the first problem to solve is how to transport or generate pTyr in mammalian cells. The dipeptide transporter was used for pTyr generation in E. coli cells.333 This approach has also been used to transport ncAAs into Caenorhabditis elegans.369 A promising fact is that humans also have dipeptide transporters,370 but it needs to first test whether pTyr-containing dipeptide can be transported and then hydrolyzed to release free pTyr inside mammalian cells. For other less common phosphorylations such as arginine and histidine phosphorylation, a big challenge is that they are unstable. Thus, nonhydrolyzable phosphorylation mimics should be ideal alternatives for them. To engineer AARSs for phosphorylated arginine or histidine, PylRS could be a feasible start due to its permissiveness for substrates. Moreover, the PylRS-based OTS has been used to genetically encode methyl-histidine in living cells.143
Protein oxidation is another field to which GCE can make great contributions. Sulfur-containing amino acids (cysteine and methionine) and aromatic amino acids (tyrosine, tryptophan, and histidine) undergo oxidation by ROS, nitration or nitrosylation by RNS, or halogenation by RHS in cells.371 Most of these modifications are not catalyzed by enzymes and nonspecific. Thus, the heterogeneity is a major challenge to determine which modification sites are essential. The GCE approach can generate site-specifically modified proteins to solve this problem. The OTS development for these oxidative modifications on aromatic residues is feasible and promising because products of these modifications are mostly stable and there are well-developed OTSs for tyrosine, tryptophan and histidine analogs.372 Some OTSs such as hydroxylation of tyrosine and tryptophan have already been established (sections 10.1 and 10.3–10.5). On the other end, although cysteine and methionine oxidation has been extensively studied, the input from GCE is limited. One issue is the stability of sulfur oxidation products. For example, cysteine sulfenic acid (Cys-SOH) commonly undergoes further oxidation to cysteine sulfinic acid (Cys-SOOH) in cells.373 Thus, nonoxidizable mimics or caged precursors are options for OTS development. Another issue is there are no well-established OTSs for cysteine or methionine derivatives. To solve this problem, the orthogonal pair of MmSerRS/tRNASer for E. coli cells could be a candidate for Cysox-OTS development,374 while LeuRS and PylRS could be further engineered for Metox-OTS.48,87
PTMs do not work alone but function coordinately to regulate protein structure and function.375 To install multiple PTMs into a single protein simultaneously, several GCE-based methods have been developed. Combining the Sep-OTS and site-specific phosphorylation by a threonine kinase, one study generated a protein with both serine phosphorylation and threonine phosphorylation.376
In another work, two mutually orthogonal OTSs, MbPylRS-based AcK-OTS and MmSepRS-based Sep-OTS, were introduced into E. coli cells simultaneously to site-specifically incorporated acetyllysine and phosphoserine at two sites in malate dehydrogenase to study crosstalk between acetylation and phosphorylation.377 In the same work, another frequently used orthogonal pair, the MjTyrRS/tRNATyr pair, was shown to be orthogonal to both pairs of MbPylRS/tRNAPyl and MmSepRS/tRNASep, indicating the possibility of installing three distinct PTMs into a single protein. Using a similar strategy, acetyllysine and selenocysteine have been genetically encoded into proteins simultaneously.378,379 Recently, several approaches to site-specifically incorporating three distinct ncAAs into a single protein have been reported. For example, OTSs based on EcTyrRS, EcTrpRS, and MbPylRS are mutually orthogonal to each other and are able to decode UAG, UGA, and UAA codons as three distinct ncAAs in mammalian cells respectively,380 while OTSs derived from EcLeuRS, MmPylRS and EcTyrRS are also mutually orthogonal to each other and can read UAG, UGA, and UAA codons as three different ncAAs in mammalian cells separately.381 However, suppressing all three stop codons could lead to issues of cell growth, so sense codons or quadruplet codons have been selected for multiple ncAA encoding. One study generated 12 triply orthogonal pairs of PylRS/tRNAPyl by screening PylRS sequences from different species. As PylRS is relatively flexible for anticodon, such triply orthogonal pairs successfully incorporated three distinct ncAAs at UAG, AGGA or AGUA codons, individually.382 In another work, a programmed initiator was used to genetically encode an ncAA at the N-terminus of proteins toward a UAU codon. Combining with two mutually orthogonal PylRS/tRNAPyl pairs, three ncAAs were site-specifically incorporated into a single protein at the N-terminal and two internal sites toward UAU, UAG, and UAA codons, respectively.383
The GCE approach has a variety of applications in PTM studies. With site-specifically installed PTMs, the effects of PTMs on the functions and structures of target proteins can be characterized site-by-stie in vitro. With incorporated nonhydrolyzable PTM analogs, the impacts of PTMs on biological processes can be determined in living cells without the concern of endogenous demodifying enzymes. Besides studying proteins harboring PTMs, GCE methods have also been used to study modifying and demodifying enzymes. For example, GCE has been applied in studying protein kinases, including spatiotemporal control of kinase activity, regulation of protein kinase phosphorylation states, and bioorthogonal manipulation of kinase isoforms.283 On the other hand, using site-specifically modified proteins as substrates, the specificity of demodifying enzymes such as phosphatases and deacetylases can be precisely determined. Besides PTMs discussed in this review, some other PTMs such as ADP-ribosylation,384 neddylation (NEDD8, a Ub-like protein),385 and S-nitrosylation386 have also been found in nature. Further developing OTSs for them or their analogs will enlarge the existing library of PTM-OTSs, providing sufficient tools for studies on emerging PTMs.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R15GM140433 and P20GM139768) and Arkansas Biosciences Institute (the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000).
ABBREVIATIONS
- 3NY
3-nitro-tyrosne
- 5HTP
5-Hydroxy-tryptophan
- AARS
aminoacyl-tRNA synthetase
- ApmTri
1,2,4-triazole amino acid
- ArgRS
arginyl-tRNA synthetase
- Boc
tert-butyloxycarbonyl
- Cbz
benzyloxycarbonyl
- DBCO
dibenzocyclooctyne
- Dha
dehydroalanine
- DMS
dimethylsulfide
- DOPA
3,4-dihydroxy-phenylalanine
- EF-Tu
elongation factor-Tu
- GCE
genetic code expansion
- GlcNAc
N-Acetylglucosamine
- KDM
protein lysine demethylase
- KetoK
2-amino-8-oxononanoicacid
- LeuRS
leucyl-tRNA synthetase
- ncAA
noncanonical amino acid
- OTS
orthogonal translation system
- PAD
peptidyl arginine deiminase
- PheRS
phenylalanyl-tRNA synthetase
- PHMT
protein histidine methyltransferase
- PKMT
protein lysine methyltransferase
- Pma
phosphonomethylene alanine
- PRMT
protein arginine methyltransferase
- pThr
phosphothreonine
- PTM
Posttranslational modification
- pTyr
phosphotyrosine
- PylRS
pyrrolysyl-tRNA synthetase
- RHS
reactive halogen species
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- SAH
S-adenosyl-l-homocysteine
- SAM
S-adenosyl-l-methionine
- Sep
Phosphoserine
- SepRS
phosphoseryl-tRNA synthetase
- sTyr
sulfotyrosine
- TAcK
thio-acetyllysine
- TCEP
tris(2-carboxyethyl)phosphine
- TFA
trifluoroacetic acid
- TFAcK
trifluoro-acetyllysine
- TFMSA
trifluoromethanesulfonic acid
- TrpRS
tryptophanyl-tRNA synthetase
- TyrRS
tyrosyl-tRNA synthetase
- Ub
Ubiquitin
Biographies
Qinglei Gan received her B.S. degree at Nanjing University and M.S. degree at Iowa State University under the mentorship of Professor Alan Myers. Then she worked in the Plant Transformation Facility at Iowa State University. She joined Dr. Chenguang Fan’s laboratory in 2016 as a research associate. Her research mainly focuses on developing and applying the genetic code expansion strategy for post-translational modification studies.
Chenguang Fan is an associate professor in the Department of Chemistry and Biochemistry at University of Arkansas. In 2009, he received his Ph.D. degree at Iowa State University under the mentorship of Professor Thomas Bobik. He stayed in the same laboratory as a postdoc to continue his Ph.D. studies on coenzyme B12 metabolism. In 2012, he pursued his second postdoctoral studies with Professor Dieter Söll at Yale University, working on development and applications of the genetic code expansion technique. He joined University of Arkansas as an assistant professor in 2016. His research is mainly at the interface of chemistry and biology, focusing on post-translational modifications and protein engineering. He was a recipient of the Ralph E. Powe Junior Faculty Enhancement Award, Robert C. and Sandra Connor Endowed Faculty Fellowship, and Arkansas Biosciences Institute New Investigator of the Year 2018.
Footnotes
Author Contributions
CRediT: Qinglei Gan visualization, writing-original draft, writing-review & editing; Chenguang Fan conceptualization, funding acquisition, writing-original draft, writing-review & editing.
The authors declare no competing financial interest.
Contributor Information
Qinglei Gan, Department of Chemistry and Biochemistry, University of Arkansas, Fayetteville, Arkansas 72701, United States.
Chenguang Fan, Department of Chemistry and Biochemistry and Cell and Molecular Biology Program, University of Arkansas, Fayetteville, Arkansas 72701, United States.
REFERENCES
- (1).Cone JE; Del Rio RM; Davis JN; Stadtman TC Chemical Characterization of the Selenoprotein Component of Clostridial Glycine Reductase: Identification of Selenocysteine as the Organoselenium Moiety. Proc. Natl. Acad. Sci. U.S.A. 1976, 73, 2659–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kryukov GV; Castellano S; Novoselov SV; Lobanov AV; Zehtab O; Guigo R; Gladyshev VN Characterization of Mammalian Selenoproteomes. Science 2003, 300, 1439–1443. [DOI] [PubMed] [Google Scholar]
- (3).Srinivasan G; James CM; Krzycki JA Pyrrolysine Encoded by Uag in Archaea: Charging of a Uag-Decoding Specialized Trna. Science 2002, 296, 1459–1462. [DOI] [PubMed] [Google Scholar]
- (4).Dreyfus JC; Kahn A; Schapira F Posttranslational Modifications of Enzymes. Curr. Top. Cell Regul. 1978, 14, 243–297. [DOI] [PubMed] [Google Scholar]
- (5).Harding JJ Nonenzymatic Covalent Posttranslational Modification of Proteins in Vivo. Adv. Protein Chem. 1985, 37, 247–334. [DOI] [PubMed] [Google Scholar]
- (6).Lipmann FA; Levene PA Serinephosphoric Acid Obtained on Hydrolysis of Vitellinic Acid. J. Biol. Chem. 1932, 98, 109–114. [Google Scholar]
- (7).Allfrey VG; Faulkner R; Mirsky AE Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. U.S.A. 1964, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Aslebagh R; Wormwood KL; Channaveerappa D; Wetie AGN; Woods AG; Darie CC Identification of Posttranslational Modifications (Ptms) of Proteins by Mass Spectrometry. Adv. Exp. Med. Biol. 2019, 1140, 199–224. [DOI] [PubMed] [Google Scholar]
- (9).Doll S; Burlingame AL Mass Spectrometry-Based Detection and Assignment of Protein Posttranslational Modifications. ACS Chem. Biol. 2015, 10, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ramazi S; Zahiri J Posttranslational Modifications in Proteins: Resources, Tools and Prediction Methods. Database (Oxford) 2021, 2021, baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rubin CS; Rosen OM Protein Phosphorylation. Annu. Rev. Biochem. 1975, 44, 831–887. [DOI] [PubMed] [Google Scholar]
- (12).Bilbrough T; Piemontese E; Seitz O Dissecting the Role of Protein Phosphorylation: A Chemical Biology Toolbox. Chem. Soc. Rev. 2022, 51, 5691–5730. [DOI] [PubMed] [Google Scholar]
- (13).Verdin E; Ott M 50 Years of Protein Acetylation: From Gene Regulation to Epigenetics, Metabolism and Beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [DOI] [PubMed] [Google Scholar]
- (14).Drazic A; Myklebust LM; Ree R; Arnesen T The World of Protein Acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [DOI] [PubMed] [Google Scholar]
- (15).Pickart CM Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [DOI] [PubMed] [Google Scholar]
- (16).Squair DR; Virdee S A New Dawn Beyond Lysine Ubiquitination. Nat. Chem. Biol. 2022, 18, 802–811. [DOI] [PubMed] [Google Scholar]
- (17).Illiano A; Pinto G; Melchiorre C; Carpentieri A; Faraco V; Amoresano A Protein Glycosylation Investigated by Mass Spectrometry: An Overview. Cells 2020, 9, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).West CM Current Ideas on the Significance of Protein Glycosylation. Mol. Cell. Biochem. 1986, 72, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang H; Yang L; Liu M; Luo J Protein Post-Translational Modifications in the Regulation of Cancer Hallmarks. Cancer Gene Ther. 2023, 30, 529–547. [DOI] [PubMed] [Google Scholar]
- (20).Liu YP; Zhang TN; Wen R; Liu CF; Yang N Role of Posttranslational Modifications of Proteins in Cardiovascular Disease. Oxid. Med. Cell Longev. 2022, 2022, 3137329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hu A; Zou H; Chen B; Zhong J Posttranslational Modifications in Diabetes: Mechanisms and Functions. Rev. Endocr. Metab. Disord. 2022, 23, 1011–1033. [DOI] [PubMed] [Google Scholar]
- (22).Baskakov IV From Posttranslational Modifications to Disease Phenotype: A Substrate Selection Hypothesis in Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Mishra S; Bassi G; Nyomba BG Inter-Proteomic Posttranslational Modifications of the Sars-Cov-2 and the Host Proteins – a New Frontier. Exp. Biol. Med. (Maywood) 2021, 246, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Otto NM; McDowell WG; Dickey DM; Potter LR A Glutamate-Substituted Mutant Mimics the Phosphorylated and Active Form of Guanylyl Cyclase-A. Mol. Pharmacol. 2017, 92, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gorsky MK; Burnouf S; Dols J; Mandelkow E; Partridge L Acetylation Mimic of Lysine 280 Exacerbates Human Tau Neurotoxicity in Vivo. Sci. Rep. 2016, 6, 22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kozelekova A; Naplavova A; Brom T; Gasparik N; Simek J; Houser J; Hritz J Phosphorylated and Phosphomimicking Variants May Differ-a Case Study of 14–3–3 Protein. Front. Chem. 2022, 10, 835733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Venkat S; Chen H; Stahman A; Hudson D; McGuire P; Gan Q; Fan C Characterizing Lysine Acetylation of Isocitrate Dehydrogenase in Escherichia Coli. J. Mol. Biol. 2018, 430, 1901–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yang A; Cho K; Park HS Chemical Biology Approaches for Studying Posttranslational Modifications. RNA Biol. 2018, 15, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chuh KN; Batt AR; Pratt MR Chemical Methods for Encoding and Decoding of Posttranslational Modifications. Cell Chem. Biol. 2016, 23, 86–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang Y; Liu Z; Cheng H; Gao T; Pan Z; Yang Q; Guo A; Xue Y Ekpd: A Hierarchical Database of Eukaryotic Protein Kinases and Protein Phosphatases. Nucleic Acids Res. 2014, 42, D496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Xu Y; Zhang S; Lin S; Guo Y; Deng W; Zhang Y; Xue Y Weram: A Database of Writers, Erasers and Readers of Histone Acetylation and Methylation in Eukaryotes. Nucleic Acids Res. 2017, 45, D264–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wade JD; Tregear GW Solid Phase Peptide Synthesis: Recent Advances and Applications. Australas. Biotechnol. 1993, 3, 332–336. [PubMed] [Google Scholar]
- (33).Merrifield RB Solid-Phase Peptide Synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1969, 32, 221–296. [DOI] [PubMed] [Google Scholar]
- (34).Dawson PE; Muir TW; Clark-Lewis I; Kent SB Synthesis of Proteins by Native Chemical Ligation. Science 1994, 266, 776–779. [DOI] [PubMed] [Google Scholar]
- (35).Muir TW Semisynthesis of Proteins by Expressed Protein Ligation. Annu. Rev. Biochem. 2003, 72, 249–289. [DOI] [PubMed] [Google Scholar]
- (36).Peng T; Das T; Ding K; Hang HC Functional Analysis of Protein Post-Translational Modifications Using Genetic Codon Expansion. Protein Sci. 2023, 32, e4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Liu WR; Wang YS; Wan W Synthesis of Proteins with Defined Posttranslational Modifications Using the Genetic Non-canonical Amino Acid Incorporation Approach. Mol. Biosyst. 2011, 7, 38–47. [DOI] [PubMed] [Google Scholar]
- (38).Niu W; Guo J Co-Translational Installation of Posttranslational Modifications by Non-Canonical Amino Acid Mutagenesis. Chembiochem 2023, 24, e202300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wu D; Liu T Studying Reversible Protein Post-Translational Modification through Co-Translational Modification. Chembiochem 2023, 24, e202200716. [DOI] [PubMed] [Google Scholar]
- (40).Chen H; Venkat S; McGuire P; Gan Q; Fan C Recent Development of Genetic Code Expansion for Posttranslational Modification Studies. Molecules 2018, 23, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).de la Torre D; Chin JW Reprogramming the Genetic Code. Nat. Rev. Genet. 2021, 22, 169–184. [DOI] [PubMed] [Google Scholar]
- (42).Mukai T; Lajoie MJ; Englert M; Soll D Rewriting the Genetic Code. Annu. Rev. Microbiol. 2017, 71, 557–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Liu CC; Schultz PG Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [DOI] [PubMed] [Google Scholar]
- (44).Kim S; Yi H; Kim YT; Lee HS Engineering Translation Components for Genetic Code Expansion. J. Mol. Biol. 2022, 434, 167302. [DOI] [PubMed] [Google Scholar]
- (45).Vargas-Rodriguez O; Sevostyanova A; Soll D; Crnkovic A Upgrading Aminoacyl-Trna Synthetases for Genetic Code Expansion. Curr. Opin. Chem. Biol. 2018, 46, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Reynolds NM; Vargas-Rodriguez O; Soll D; Crnkovic A The Central Role of Trna in Genetic Code Expansion. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang L; Brock A; Herberich B; Schultz PG Expanding the Genetic Code of Escherichia Coli. Science 2001, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- (48).Wan W; Tharp JM; Liu WR Pyrrolysyl-Trna Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta. Proteins Proteomics 2014, 1844, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Andrews J; Gan Q; Fan C ″Not-So-Popular″ Orthogonal Pairs in Genetic Code Expansion. Protein Sci. 2023, 32, e4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Fu X; Huang Y; Shen Y Improving the Efficiency and Orthogonality of Genetic Code Expansion. Biodes. Res. 2022, 2022, 9896125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen J; Tsai YH Applications of Genetic Code Expansion in Studying Protein Post-Translational Modification. J. Mol. Biol. 2022, 434, 167424. [DOI] [PubMed] [Google Scholar]
- (52).Ding W; Zhao H; Chen Y; Lin S New Strategies for Probing the Biological Functions of Protein Post-Translational Modifications in Mammalian Cells with Genetic Code Expansion. Acc. Chem. Res. 2023, 56, 2827–2837. [DOI] [PubMed] [Google Scholar]
- (53).Zhou W; Deiters A Chemogenetic and Optogenetic Control of Post-Translational Modifications through Genetic Code Expansion. Curr. Opin. Chem. Biol. 2021, 63, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Fitch CA; Platzer G; Okon M; Garcia-Moreno BE; McIntosh LP Arginine: Its Pka Value Revisited. Protein Sci. 2015, 24, 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Fuhrmann J; Clancy KW; Thompson PR Chemical Biology of Protein Arginine Modifications in Epigenetic Regulation. Chem. Rev. 2015, 115, 5413–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Smith BC; Denu JM Chemical Mechanisms of Histone Lysine and Arginine Modifications. Biochim. Biophys. Acta 2009, 1789, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Fuhrmann J; Schmidt A; Spiess S; Lehner A; Turgay K; Mechtler K; Charpentier E; Clausen T Mcsb Is a Protein Arginine Kinase That Phosphorylates and Inhibits the Heat-Shock Regulator Ctsr. Science 2009, 324, 1323–1327. [DOI] [PubMed] [Google Scholar]
- (58).Hawse WF; Wolberger C Structure-Based Mechanism of Adp-Ribosylation by Sirtuins. J. Biol. Chem. 2009, 284, 33654–33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Lorton BM; Shechter D Cellular Consequences of Arginine Methylation. Cell. Mol. Life Sci. 2019, 76, 2933–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Raposo AE; Piller SC Protein Arginine Methylation: An Emerging Regulator of the Cell Cycle. Cell Div. 2018, 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wu Q; Schapira M; Arrowsmith CH; Barsyte-Lovejoy D Protein Arginine Methylation: From Enigmatic Functions to Therapeutic Targeting. Nat. Rev. Drug Discovery 2021, 20, 509–530. [DOI] [PubMed] [Google Scholar]
- (62).Xu J; Richard S Cellular Pathways Influenced by Protein Arginine Methylation: Implications for Cancer. Mol. Cell 2021, 81, 4357–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Srour N; Khan S; Richard S The Influence of Arginine Methylation in Immunity and Inflammation. J. Inflamm. Res. 2022, 15, 2939–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Chang K; Gao D; Yan J; Lin L; Cui T; Lu S Critical Roles of Protein Arginine Methylation in the Central Nervous System. Mol. Neurobiol. 2023, 60, 6060–6091. [DOI] [PubMed] [Google Scholar]
- (65).Zhang J; Jing L; Li M; He L; Guo Z Regulation of Histone Arginine Methylation/Demethylation by Methylase and Demethylase (Review). Mol. Med. Rep. 2019, 19, 3963–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wolf SS The Protein Arginine Methyltransferase Family: An Update About Function, New Perspectives and the Physiological Role in Humans. Cell. Mol. Life Sci. 2009, 66, 2109–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Wesche J; Kuhn S; Kessler BM; Salton M; Wolf A Protein Arginine Methylation: A Prominent Modification and Its Demethylation. Cell. Mol. Life Sci. 2017, 74, 3305–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Le DD; Cortesi AT; Myers SA; Burlingame AL; Fujimori DG Site-Specific and Regiospecific Installation of Methylarginine Analogues into Recombinant Histones and Insights into Effector Protein Binding. J. Am. Chem. Soc. 2013, 135, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wright TH; Bower BJ; Chalker JM; Bernardes GJ; Wiewiora R; Ng WL; Raj R; Faulkner S; Vallee MR; Phanumartwiwath A; et al. Posttranslational Mutagenesis: A Chemical Strategy for Exploring Protein Side-Chain Diversity. Science 2016, 354, aag1465. [DOI] [PubMed] [Google Scholar]
- (70).Akahoshi A; Suzue Y; Kitamatsu M; Sisido M; Ohtsuki T Site-Specific Incorporation of Arginine Analogs into Proteins Using Arginyl-Trna Synthetase. Biochem. Biophys. Res. Commun. 2011, 414, 625–630. [DOI] [PubMed] [Google Scholar]
- (71).Gan Q; Fan C Increasing the Fidelity of Noncanonical Amino Acid Incorporation in Cell-Free Protein Synthesis. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Christophorou MA The Virtues and Vices of Protein Citrullination. R. Soc. Open Sci. 2022, 9, 220125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Fuhrmann J; Thompson PR Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem. Biol. 2016, 11, 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Sarnik J; Makowska JS Highlighting the Versatility of the Citrullination Process. Immunobiology 2022, 227, 152233. [DOI] [PubMed] [Google Scholar]
- (75).Harada K; Carr SM; Shrestha A; La Thangue NB Citrullination and the Protein Code: Crosstalk between Post-Translational Modifications in Cancer. Philos. Trans. R Soc. London B Biol. Sci. 2023, 378, 20220243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ciesielski O; Biesiekierska M; Panthu B; Soszynski M; Pirola L; Balcerczyk A Citrullination in the Pathology of Inflammatory and Autoimmune Disorders: Recent Advances and Future Perspectives. Cell. Mol. Life Sci. 2022, 79, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Fert-Bober J; Sokolove J Proteomics of Citrullination in Cardiovascular Disease. Proteomics Clin. Appl. 2014, 8, 522–533. [DOI] [PubMed] [Google Scholar]
- (78).Jang B; Ishigami A; Maruyama N; Carp RI; Kim YS; Choi EK Peptidylarginine Deiminase and Protein Citrullination in Prion Diseases: Strong Evidence of Neurodegeneration. Prion 2013, 7, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Das K; Butler GH; Kwiatkowski V; Clark AD Jr.; Yadav P; Arnold E Crystal Structures of Arginine Deiminase with Covalent Reaction Intermediates; Implications for Catalytic Mechanism. Structure 2004, 12, 657–667. [DOI] [PubMed] [Google Scholar]
- (80).Mondal S; Thompson PR Chemical Biology of Protein Citrullination by the Protein a Arginine Deiminases. Curr. Opin. Chem. Biol. 2021, 63, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Mondal S; Thompson PR Protein Arginine Deiminases (Pads): Biochemistry and Chemical Biology of Protein Citrullination. Acc. Chem. Res. 2019, 52, 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Morris SM Jr. Regulation of Enzymes of the Urea Cycle and Arginine Metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [DOI] [PubMed] [Google Scholar]
- (83).Yu B; Howell PL Intragenic Complementation and the Structure and Function of Argininosuccinate Lyase. Cell. Mol. Life Sci. 2000, 57, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Husson A; Brasse-Lagnel C; Fairand A; Renouf S; Lavoinne A Argininosuccinate Synthetase from the Urea Cycle to the Citrulline-No Cycle. Eur. J. Biochem. 2003, 270, 1887–1899. [DOI] [PubMed] [Google Scholar]
- (85).Slack JL; Jones LE Jr.; Bhatia MM; Thompson PR Autodeimination of Protein Arginine Deiminase 4 Alters Protein–Protein Interactions but Not Activity. Biochemistry 2011, 50, 3997–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Mondal S; Wang S; Zheng Y; Sen S; Chatterjee A; Thompson PR Site-Specific Incorporation of Citrulline into Proteins in Mammalian Cells. Nat. Commun. 2021, 12, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wu N; Deiters A; Cropp TA; King D; Schultz PG A Genetically Encoded Photocaged Amino Acid. J. Am. Chem. Soc. 2004, 126, 14306–14307. [DOI] [PubMed] [Google Scholar]
- (88).Greve JM; Pinkham AM; Cowan JA Human Aspartyl (Asparaginyl) Hydroxylase. A Multifaceted Enzyme with Broad Intra- and Extra-Cellular Activity. Metallomics 2021, 13, mfab044. [DOI] [PubMed] [Google Scholar]
- (89).Lando D; Peet DJ; Gorman JJ; Whelan DA; Whitelaw ML; Bruick RK Fih-1 Is an Asparaginyl Hydroxylase Enzyme That Regulates the Transcriptional Activity of Hypoxia-Inducible Factor. Genes Dev. 2002, 16, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Geiger T; Clarke S Deamidation, Isomerization, and Racemization at Asparaginyl and Aspartyl Residues in Peptides. Succinimide-Linked Reactions That Contribute to Protein Degradation. J. Biol. Chem. 1987, 262, 785–794. [PubMed] [Google Scholar]
- (91).Lindner H; Helliger W Age-Dependent Deamidation of Asparagine Residues in Proteins. Exp. Gerontol. 2001, 36, 1551–1563. [DOI] [PubMed] [Google Scholar]
- (92).Lowenson J; Clarke S Does the Chemical Instability of Aspartyl and Asparaginyl Residues in Proteins Contribute to Erythrocyte Aging? The Role of Protein Carboxyl Methylation Reactions. Blood Cells 1988, 14, 103–118. [PubMed] [Google Scholar]
- (93).Pan X; Luo J; Li S Bacteria-Catalyzed Arginine Glycosylation in Pathogens and Host. Front. Cell Infect. Microbiol. 2020, 10, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Apweiler R; Hermjakob H; Sharon N On the Frequency of Protein Glycosylation, as Deduced from Analysis of the Swiss-Prot Database. Biochim. Biophys. Acta 1999, 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- (95).Roth J; Zuber C; Park S; Jang I; Lee Y; Kysela KG; Le Fourn V; Santimaria R; Guhl B; Cho JW Protein N-Glycosylation, Protein Folding, and Protein Quality Control. Mol. Cells 2010, 30, 497–506. [DOI] [PubMed] [Google Scholar]
- (96).Varki A Biological Roles of Glycans. Glycobiology 2017, 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Schwarz F; Aebi M Mechanisms and Principles of N-Linked Protein Glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [DOI] [PubMed] [Google Scholar]
- (98).Shrimal S; Cherepanova NA; Gilmore R Cotranslational and Posttranslocational N-Glycosylation of Proteins in the Endoplasmic Reticulum. Semin. Cell Dev. Biol. 2015, 41, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Li C; Wang LX Chemoenzymatic Methods for the Synthesis of Glycoproteins. Chem. Rev. 2018, 118, 8359–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Nomura K; Liu Y; Kajihara Y Synthesis of Homogeneous Glycoproteins with Diverse N-Glycans. Adv. Carbohydr. Chem. Biochem. 2022, 81, 57–93. [DOI] [PubMed] [Google Scholar]
- (101).Machida K; Imataka H In Glycoscience Protocols (Glycopodv2); Nishihara S, Angata K, Aoki-Kinoshita KF, Hirabayashi J, Eds.; Saitama (JP), 2021. [PubMed] [Google Scholar]
- (102).Macmillan D; Bill RM; Sage KA; Fern D; Flitsch SL Selective in Vitro Glycosylation of Recombinant Proteins: Semi-Synthesis of Novel Homogeneous Glycoforms of Human Erythropoietin. Chem. Biol. 2001, 8, 133–145. [DOI] [PubMed] [Google Scholar]
- (103).Holloran N; Collins D; Rathnayake U; Zhang B; Koh M; Kang C; Garner P Site-Specific Synthesis of Cysteine-Bridged Glycoproteins Via Expressed Protein Glycoligation. Bioconjugate Chem. 2020, 31, 2362–2366. [DOI] [PubMed] [Google Scholar]
- (104).Wu Q; Dong W; Miao H; Wang Q; Dong S; Xuan W Site-Specific Protein Modification with Reducing Carbohydrates. Angew. Chem., Int. Ed. 2022, 61, e202116545. [DOI] [PubMed] [Google Scholar]
- (105).Wedemeyer WJ; Welker E; Narayan M; Scheraga HA Disulfide Bonds and Protein Folding. Biochemistry 2000, 39, 4207–4216. [DOI] [PubMed] [Google Scholar]
- (106).Darby N; Creighton TE Disulfide Bonds in Protein Folding and Stability. Methods Mol. Biol. 1995, 40, 219–252. [DOI] [PubMed] [Google Scholar]
- (107).Garrido Ruiz D; Sandoval-Perez A; Rangarajan AV; Gunderson EL; Jacobson MP Cysteine Oxidation in Proteins: Structure, Biophysics, and Simulation. Biochemistry 2022, 61, 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Cansado J To Finish Things Well: Cysteine Methylation Ensures Selective Gtpase Membrane Localization and Signalling. Curr. Genet. 2018, 64, 341–344. [DOI] [PubMed] [Google Scholar]
- (109).Gould N; Doulias PT; Tenopoulou M; Raju K; Ischiropoulos H Regulation of Protein Function and Signaling by Reversible Cysteine S-Nitrosylation. J. Biol. Chem. 2013, 288, 26473–26479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Casey PJ Protein Lipidation in Cell Signaling. Science 1995, 268, 221–225. [DOI] [PubMed] [Google Scholar]
- (111).Hang HC; Linder ME Exploring Protein Lipidation with Chemical Biology. Chem. Rev. 2011, 111, 6341–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Fhu CW; Ali A Protein Lipidation by Palmitoylation and Myristoylation in Cancer. Front. Cell Dev. Biol. 2021, 9, 673647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Chen B; Sun Y; Niu J; Jarugumilli GK; Wu X Protein Lipidation in Cell Signaling and Diseases: Function, Regulation, and Therapeutic Opportunities. Cell Chem. Biol. 2018, 25, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Resh MD Targeting Protein Lipidation in Disease. Trends Mol. Med. 2012, 18, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Wang R; Chen YQ Protein Lipidation Types: Current Strategies for Enrichment and Characterization. Int. J. Mol. Sci. 2022, 23, 2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Mitchell DA; Vasudevan A; Linder ME; Deschenes RJ Protein Palmitoylation by a Family of Dhhc Protein S-Acyltransferases. J. Lipid Res. 2006, 47, 1118–1127. [DOI] [PubMed] [Google Scholar]
- (117).Lim SI; Mizuta Y; Takasu A; Hahn YS; Kim YH; Kwon I Site-Specific Fatty Acid-Conjugation to Prolong Protein Half-Life in Vivo. J. Controlled Release 2013, 170, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Cho J; Lim SI; Yang BS; Hahn YS; Kwon I Generation of Therapeutic Protein Variants with the Human Serum Albumin Binding Capacity Via Site-Specific Fatty Acid Conjugation. Sci. Rep. 2017, 7, 18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Li Y; Wang S; Chen Y; Li M; Dong X; Hang HC; Peng T Site-Specific Chemical Fatty-Acylation for Gain-of-Function Analysis of Protein S-Palmitoylation in Live Cells. Chem. Commun. (Camb) 2020, 56, 13880–13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Garst EH; Lee H; Das T; Bhattacharya S; Percher A; Wiewiora R; Witte IP; Li Y; Peng T; Im W; et al. Site-Specific Lipidation Enhances Ifitm3Membrane Interactions and Antiviral Activity. ACS Chem. Biol. 2021, 16, 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Lampi KJ; Wilmarth PA; Murray MR; David LL Lens Beta-Crystallins: The Role of Deamidation and Related Modifications in Aging and Cataract. Prog. Biophys. Mol. Biol. 2014, 115, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Robinson NE Protein Deamidation. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Tessarz P; Santos-Rosa H; Robson SC; Sylvestersen KB; Nelson CJ; Nielsen ML; Kouzarides T Glutamine Methylation in Histone H2a Is an Rna-Polymerase-I-Dedicated Modification. Nature 2014, 505, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Heurgue-Hamard V; Champ S; Mora L; Merkoulova-Rainon T; Kisselev LL; Buckingham RH The Glutamine Residue of the Conserved Ggq Motif in Saccharomyces Cerevisiae Release Factor Erf1 Is Methylated by the Product of the Ydr140w Gene. J. Biol. Chem. 2005, 280, 2439–2445. [DOI] [PubMed] [Google Scholar]
- (125).Gagsteiger J; Jahn S; Heidinger L; Gericke L; Andexer JN; Friedrich T; Loenarz C; Layer G A Cobalamin-Dependent Radical Sam Enzyme Catalyzes the Unique C(Alpha) -Methylation of Glutamine in Methyl-Coenzyme M Reductase. Angew. Chem., Int. Ed. 2022, 61, e202204198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Nakahigashi K; Kubo N; Narita S; Shimaoka T; Goto S; Oshima T; Mori H; Maeda M; Wada C; Inokuchi H Hemk, a Class of Protein Methyl Transferase with Similarity to DNA Methyl Transferases, Methylates Polypeptide Chain Release Factors, and Hemk Knockout Induces Defects in Translational Termination. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Lhoest J; Colson C Genetics of Ribosomal Protein Methylation in Escherichia Coli. Ii. A Mutant Lacking a New Type of Methylated Amino Acid, N5-Methylglutamine, in Protein L3. Mol. Gen. Genet. 1977, 154, 175–180. [DOI] [PubMed] [Google Scholar]
- (128).Gao J; Wang B; Yu H; Wu G; Wan C; Liu W; Liao S; Cheng L; Zhu Z Structural Insight into Hemk2-Trmt112-Mediated Glutamine Methylation. Biochem. J. 2020, 477, 3833–3838. [DOI] [PubMed] [Google Scholar]
- (129).Yang X; Miao H; Xiao R; Wang L; Zhao Y; Wu Q; Ji Y; Du J; Qin H; Xuan W Diverse Protein Manipulations with Genetically Encoded Glutamic Acid Benzyl Ester. Chem. Sci. 2021, 12, 9778–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Kowluru A Emerging Roles for Protein Histidine Phosphorylation in Cellular Signal Transduction: Lessons from the Islet Beta-Cell. J. Cell Mol. Med. 2008, 12, 1885–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Saito H Histidine Phosphorylation and Two-Component Signaling in Eukaryotic Cells. Chem. Rev. 2001, 101, 2497–2509. [DOI] [PubMed] [Google Scholar]
- (132).Yanshina DD; Bulygin KN; Malygin AA; Karpova GG Hydroxylated Histidine of Human Ribosomal Protein Ul2 Is Involved in Maintaining the Local Structure of 28s Rrna in the Ribosomal Peptidyl Transferase Center. FEBS J. 2015, 282, 1554–1566. [DOI] [PubMed] [Google Scholar]
- (133).Bundred JR; Hendrix E; Coleman ML The Emerging Roles of Ribosomal Histidyl Hydroxylases in Cell Biology, Physiology and Disease. Cell. Mol. Life Sci. 2018, 75, 4093–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Moye-Rowley WS Redox Sensing and Histidine Oxidation: No Longer Perr-Fect Strangers. Nat. Chem. Biol. 2006, 2, 234–235. [DOI] [PubMed] [Google Scholar]
- (135).Kwiatkowski S; Drozak J Protein Histidine Methylation. Curr. Protein Pept. Sci. 2020, 21, 675–689. [DOI] [PubMed] [Google Scholar]
- (136).Jakobsson ME Enzymology and Significance of Protein Histidine Methylation. J. Biol. Chem. 2021, 297, 101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Asatoor AM; Armstrong MD 3-Methylhistidine, a Component of Actin. Biochem. Biophys. Res. Commun. 1967, 26, 168–174. [DOI] [PubMed] [Google Scholar]
- (138).Johnson P; Harris CI; Perry SV 3-Methylhistidine in Actin and Other Muscle Proteins. Biochem. J. 1967, 105, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Kapell S; Jakobsson ME Large-Scale Identification of Protein Histidine Methylation in Human Cells. NAR Genom. Bioinform. 2021, 3, lqab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Wilkinson AW; Diep J; Dai S; Liu S; Ooi YS; Song D; Li TM; Horton JR; Zhang X; Liu C; et al. Setd3 Is an Actin Histidine Methyltransferase That Prevents Primary Dystocia. Nature 2019, 565, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Malecki JM; Odonohue MF; Kim Y; Jakobsson ME; Gessa L; Pinto R; Wu J; Davydova E; Moen A; Olsen JV; et al. Human Mettl18 Is a Histidine-Specific Methyltransferase That Targets Rpl3 and Affects Ribosome Biogenesis and Function. Nucleic Acids Res. 2021, 49, 3185–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Davydova E; Shimazu T; Schuhmacher MK; Jakobsson ME; Willemen H; Liu T; Moen A; Ho AYY; Malecki J; Schroer L; et al. The Methyltransferase Mettl9Mediates Pervasive 1-Methylhistidine Modification in Mammalian Proteomes. Nat. Commun. 2021, 12, 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Xiao H; Peters FB; Yang PY; Reed S; Chittuluru JR; Schultz PG Genetic Incorporation of Histidine Derivatives Using an Engineered Pyrrolysyl-Trna Synthetase. ACS Chem. Biol. 2014, 9, 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Crnković A; Suzuki T; Söll D; Reynolds NM Pyrrolysyl-Trna Synthetase, an Aminoacyl-Trna Synthetase for Genetic Code Expansion. Croat. Chem. Acta. 2016, 89, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Zhang W; Tan X; Lin S; Gou Y; Han C; Zhang C; Ning W; Wang C; Xue Y Cplm 4.0: An Updated Database with Rich Annotations for Protein Lysine Modifications. Nucleic Acids Res. 2022, 50, D451–D459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Grunstein M Histone Acetylation in Chromatin Structure and Transcription. Nature 1997, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- (147).Sterner DE; Berger SL Acetylation of Histones and Transcription-Related Factors. Microbiology and molecular biology reviews: MMBR 2000, 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Narita T; Weinert BT; Choudhary C Functions and Mechanisms of Non-Histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [DOI] [PubMed] [Google Scholar]
- (149).Shvedunova M; Akhtar A Modulation of Cellular Processes by Histone and Non-Histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349. [DOI] [PubMed] [Google Scholar]
- (150).Ouidir T; Kentache T; Hardouin J Protein Lysine Acetylation in Bacteria: Current State of the Art. Proteomics 2016, 16, 301–309. [DOI] [PubMed] [Google Scholar]
- (151).Christensen DG; Baumgartner JT; Xie X; Jew KM; Basisty N; Schilling B; Kuhn ML; Wolfe AJ Mechanisms, Detection, and Relevance of Protein Acetylation in Prokaryotes. mBio 2019, 10, e02708–02718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).VanDrisse CM; Escalante-Semerena JC Protein Acetylation in Bacteria. Annu. Rev. Microbiol. 2019, 73, 111–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Menzies KJ; Zhang HB; Katsyuba E; Auwerx J Protein Acetylation in Metabolism - Metabolites and Cofactors. Nat. Rev. Endocrinol. 2016, 12, 43–60. [DOI] [PubMed] [Google Scholar]
- (154).Yang J; Song C; Zhan X The Role of Protein Acetylation in Carcinogenesis and Targeted Drug Discovery. Front. Endocrinol. (Lausanne) 2022, 13, 972312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Kabir F; Atkinson R; Cook AL; Phipps AJ; King AE The Role of Altered Protein Acetylation in Neurodegenerative Disease. Front. Aging Neurosci. 2023, 14, 1025473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Li XY; Yu JT; Dong YH; Shen XY; Hou R; Xie MM; Wei J; Hu XW; Dong ZH; Shan RR; et al. Protein Acetylation and Related Potential Therapeutic Strategies in Kidney Disease. Pharmacol. Res. 2023, 197, 106950. [DOI] [PubMed] [Google Scholar]
- (157).Ren J; Sang Y; Lu J; Yao YF Protein Acetylation and Its Role in Bacterial Virulence. Trends Microbiol. 2017, 25, 768–779. [DOI] [PubMed] [Google Scholar]
- (158).Allis CD; Berger SL; Cote J; Dent S; Jenuwien T; Kouzarides T; Pillus L; Reinberg D; Shi Y; Shiekhattar R; et al. New Nomenclature for Chromatin-Modifying Enzymes. Cell 2007, 131, 633–636. [DOI] [PubMed] [Google Scholar]
- (159).Gregoretti IV; Lee YM; Goodson HV Molecular Evolution of the Histone Deacetylase Family: Functional Implications of Phylogenetic Analysis. J. Mol. Biol. 2004, 338, 17–31. [DOI] [PubMed] [Google Scholar]
- (160).Neumann H; Peak-Chew SY; Chin JW Genetically Encoding Nε-Acetyllysine in Recombinant Proteins. Nat. Chem. Biol. 2008, 4, 232–234. [DOI] [PubMed] [Google Scholar]
- (161).Neumann H; Hancock SM; Buning R; Routh A; Chapman L; Somers J; Owen-Hughes T; van Noort J; Rhodes D; Chin JW A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histones Defines the Effects of H3 K56 Acetylation. Mol. Cell 2009, 36, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Umehara T; Kim J; Lee S; Guo LT; Soll D; Park HS N-Acetyl Lysyl-Trna Synthetases Evolved by a Ccdb-Based Selection Possess N-Acetyl Lysine Specificity in Vitro and in Vivo. FEBS Lett. 2012, 586, 729–733. [DOI] [PubMed] [Google Scholar]
- (163).Fan C; Xiong H; Reynolds NM; Soll D Rationally Evolving Trnapyl for Efficient Incorporation of Noncanonical Amino Acids. Nucleic Acids Res. 2015, 43, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Bryson DI; Fan C; Guo LT; Miller C; Soll D; Liu DR Continuous Directed Evolution of Aminoacyl-Trna Synthetases. Nat. Chem. Biol. 2017, 13, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Huang Y; Russell WK; Wan W; Pai P-J; Russell DH; Liu W A Convenient Method for Genetic Incorporation of Multiple Noncanonical Amino Acids into One Protein in Escherichia Coli. Mol. Biosyst. 2010, 6, 683–686. [DOI] [PubMed] [Google Scholar]
- (166).Gan Q; Lehman BP; Bobik TA; Fan C Expanding the Genetic Code of Salmonella with Non-Canonical Amino Acids. Sci. Rep. 2016, 6, 39920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Kim SW; Lee KJ; Kim S; Kim J; Cho K; Ro HS; Park HS Genetic Incorporation of N(Epsilon)-Acetyllysine Reveals a Novel Acetylation-Sumoylation Switch in Yeast. Biochim. Biophys. Acta 2017, 1861, 3030–3037. [DOI] [PubMed] [Google Scholar]
- (168).Hancock SM; Uprety R; Deiters A; Chin JW Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids Via a Pyrrolysyl-Trna Synthetase/Trna Pair. J. Am. Chem. Soc. 2010, 132, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Mukai T; Kobayashi T; Hino N; Yanagisawa T; Sakamoto K; Yokoyama S Adding L-Lysine Derivatives to the Genetic Code of Mammalian Cells with Engineered Pyrrolysyl-Trna Synthetases. Biochem. Biophys. Res. Commun. 2008, 371, 818–822. [DOI] [PubMed] [Google Scholar]
- (170).Cohen S; Arbely E Single-Plasmid-Based System for Efficient Noncanonical Amino Acid Mutagenesis in Cultured Mammalian Cells. Chembiochem 2016, 17, 1008–1011. [DOI] [PubMed] [Google Scholar]
- (171).Elsasser SJ; Ernst RJ; Walker OS; Chin JW Genetic Code Expansion in Stable Cell Lines Enables Encoded Chromatin Modification. Nat. Methods 2016, 13, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Han S; Yang A; Lee S; Lee H-W; Park CB; Park H-S Expanding the Genetic Code of Mus Musculus. Nat. Commun. 2017, 8, 14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Neumann-Staubitz P; Lammers M; Neumann H Genetic Code Expansion Tools to Study Lysine Acylation. Adv. Biol. (Weinh) 2021, 5, e2100926. [DOI] [PubMed] [Google Scholar]
- (174).Huang Y; Wan W; Russell WK; Pai PJ; Wang Z; Russell DH; Liu W Genetic Incorporation of an Aliphatic Keto-Containing Amino Acid into Proteins for Their Site-Specific Modifications. Bioorg. Med. Chem. Lett. 2010, 20, 878–880. [DOI] [PubMed] [Google Scholar]
- (175).Smith BC; Denu JM Mechanism-Based Inhibition of Sir2 Deacetylases by Thioacetyl-Lysine Peptide. Biochemistry 2007, 46, 14478–14486. [DOI] [PubMed] [Google Scholar]
- (176).Venkat S; Nannapaneni DT; Gregory C; Gan Q; McIntosh M; Fan C Genetically Encoding Thioacetyl-Lysine as a Non-Deacetylatable Analog of Lysine Acetylation in Escherichia Coli. FEBS Open Bio. 2017, 7, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Zhang F; Zhou Q; Yang G; An L; Li F; Wang J A Genetically Encoded (19)F Nmr Probe for Lysine Acetylation. Chem. Commun. (Camb) 2018, 54, 3879–3882. [DOI] [PubMed] [Google Scholar]
- (178).Kirchgassner S; Braun MB; Bartlick N; Koc C; Reinkemeier CD; Lemke EA; Stehle T; Schwarzer D Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the Bet Family. Angew. Chem., Int. Ed. 2023, 62, e202215460. [DOI] [PubMed] [Google Scholar]
- (179).Ambler RP; Rees MW Epsilon-N-Methyl-Lysine in Bacterial Flagellar Protein. Nature 1959, 184, 56–57. [DOI] [PubMed] [Google Scholar]
- (180).Wu Z; Connolly J; Biggar KK Beyond Histones - the Expanding Roles of Protein Lysine Methylation. FEBS J. 2017, 284, 2732–2744. [DOI] [PubMed] [Google Scholar]
- (181).Zhang X; Huang Y; Shi X Emerging Roles of Lysine Methylation on Non-Histone Proteins. Cell. Mol. Life Sci. 2015, 72, 4257–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Lanouette S; Mongeon V; Figeys D; Couture JF The Functional Diversity of Protein Lysine Methylation. Mol. Syst. Biol. 2014, 10, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Han D; Huang M; Wang T; Li Z; Chen Y; Liu C; Lei Z; Chu X Lysine Methylation of Transcription Factors in Cancer. Cell Death Dis. 2019, 10, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Stark GR; Wang Y; Lu T Lysine Methylation of Promoter-Bound Transcription Factors and Relevance to Cancer. Cell Res. 2011, 21, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Wang Z; Liu H Lysine Methylation Regulates Nervous System Diseases. Neuropeptides 2019, 76, 101929. [DOI] [PubMed] [Google Scholar]
- (186).Rowe EM; Xing V; Biggar KK Lysine Methylation: Implications in Neurodegenerative Disease. Brain Res. 2019, 1707, 164–171. [DOI] [PubMed] [Google Scholar]
- (187).Sun GD; Cui WP; Guo QY; Miao LN Histone Lysine Methylation in Diabetic Nephropathy. J. Diabetes Res. 2014, 2014, 654148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Yi X; Jiang X; Li X; Jiang DS Histone Lysine Methylation and Congenital Heart Disease: From Bench to Bedside (Review). Int. J. Mol. Med. 2017, 40, 953–964. [DOI] [PubMed] [Google Scholar]
- (189).Bhat KP; Umit Kaniskan H; Jin J; Gozani O Epigenetics and Beyond: Targeting Writers of Protein Lysine Methylation to Treat Disease. Nat. Rev. Drug Discovery 2021, 20, 265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Luo M Chemical and Biochemical Perspectives of Protein Lysine Methylation. Chem. Rev. 2018, 118, 6656–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Black JC; Van Rechem C; Whetstine JR Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Kaniskan HU; Martini ML; Jin J Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2018, 118, 989–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Martin C; Zhang Y The Diverse Functions of Histone Lysine Methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [DOI] [PubMed] [Google Scholar]
- (194).Simon MD; Chu F; Racki LR; de la Cruz CC; Burlingame AL; Panning B; Narlikar GJ; Shokat KM The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell 2007, 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Chalker JM; Lercher L; Rose NR; Schofield CJ; Davis BG Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications. Angew. Chem., Int. Ed. Engl. 2012, 51, 1835–1839. [DOI] [PubMed] [Google Scholar]
- (196).Bernardes GJ; Chalker JM; Errey JC; Davis BG Facile Conversion of Cysteine and Alkyl Cysteines to Dehydroalanine on Protein Surfaces: Versatile and Switchable Access to Functionalized Proteins. J. Am. Chem. Soc. 2008, 130, 5052–5053. [DOI] [PubMed] [Google Scholar]
- (197).Josephson B; Fehl C; Isenegger PG; Nadal S; Wright TH; Poh AWJ; Bower BJ; Giltrap AM; Chen L; Batchelor-McAuley C; et al. Light-Driven Post-Translational Installation of Reactive Protein Side Chains. Nature 2020, 585, 530–537. [DOI] [PubMed] [Google Scholar]
- (198).Nguyen DP; Garcia Alai MM; Kapadnis PB; Neumann H; Chin JW Genetically Encoding N(Epsilon)-Methyl-L-Lysine in Recombinant Histones. J. Am. Chem. Soc. 2009, 131, 14194–14195. [DOI] [PubMed] [Google Scholar]
- (199).Yanagisawa T; Takahashi M; Mukai T; Sato S; Wakamori M; Shirouzu M; Sakamoto K; Umehara T; Yokoyama S Multiple Site-Specific Installations of Nepsilon-Monomethyl-L-Lysine into Histone Proteins by Cell-Based and Cell-Free Protein Synthesis. Chembiochem 2014, 15, 1830–1838. [DOI] [PubMed] [Google Scholar]
- (200).Ai HW; Lee JW; Schultz PG A Method to Site-Specifically Introduce Methyllysine into Proteins in E. Coli. Chem. Commun. (Camb) 2010, 46, 5506–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Rehn A; Lawatscheck J; Jokisch ML; Mader SL; Luo Q; Tippel F; Blank B; Richter K; Lang K; Kaila VRI; et al. A Methylated Lysine Is a Switch Point for Conformational Communication in the Chaperone Hsp90. Nat. Commun. 2020, 11, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Groff D; Chen PR; Peters FB; Schultz PG A Genetically Encoded Ε-N-Methyl Lysine in Mammalian Cells. ChemBioChem. 2010, 11, 1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Wang Y-S; Wu B; Wang Z; Huang Y; Wan W; Russell WK; Pai P-J; Moe YN; Russell DH; Liu WR A Genetically Encoded Photocaged Nε-Methyl-L-Lysine. Mol. Biosyst. 2010, 6, 1557–1560. [DOI] [PubMed] [Google Scholar]
- (204).Nguyen DP; Alai MMG; Virdee S; Chin JW Genetically Directing ε-N,N-Dimethyl-L-Lysine in Recombinant Histones. Chem. Biol. 2010, 17, 1072–1076. [DOI] [PubMed] [Google Scholar]
- (205).Wang ZA; Zeng Y; Kurra Y; Wang X; Tharp JM; Vatansever EC; Hsu WW; Dai S; Fang X; Liu WR A Genetically Encoded Allysine for the Synthesis of Proteins with Site-Specific Lysine Dimethylation. Angew. Chem., Int. Ed. Engl. 2017, 56, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Guo J; Wang J; Lee JS; Schultz PG Site-Specific Incorporation of Methyl- and Acetyl-Lysine Analogues into Recombinant Proteins. Angew. Chem., Int. Ed. Engl. 2008, 47, 6399–6401. [DOI] [PubMed] [Google Scholar]
- (207).Yang A; Ha S; Ahn J; Kim R; Kim S; Lee Y; Kim J; Söll D; Lee H-Y; Park H-S A Chemical Biology Route to Site-Specific Authentic Protein Modifications. Science 2016, 354, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Xu Y; Shi Z; Bao L An Expanding Repertoire of Protein Acylations. Mol. Cell Proteomics 2022, 21, 100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Shang S; Liu J; Hua F Protein Acylation: Mechanisms, Biological Functions and Therapeutic Targets. Signal Transduct. Target Ther. 2022, 7, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Blasl AT; Schulze S; Qin C; Graf LG; Vogt R; Lammers M Post-Translational Lysine Ac(Et)Ylation in Health, Ageing and Disease. Biol. Chem. 2022, 403, 151–194. [DOI] [PubMed] [Google Scholar]
- (211).Chen Y; Sprung R; Tang Y; Ball H; Sangras B; Kim SC; Falck JR; Peng J; Gu W; Zhao Y Lysine Propionylation and Butyrylation Are Novel Post-Translational Modifications in Histones. Mol. Cell Proteomics 2007, 6, 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Miao Y; Wang Y; Huang D; Lin X; Lin Z; Lin X Profile of Protein Lysine Propionylation in Aeromonas Hydrophila and Its Role in Enzymatic Regulation. Biochem. Biophys. Res. Commun. 2021, 562, 1–8. [DOI] [PubMed] [Google Scholar]
- (213).Sun M; Xu J; Wu Z; Zhai L; Liu C; Cheng Z; Xu G; Tao S; Ye BC; Zhao Y; et al. Characterization of Protein Lysine Propionylation in Escherichia Coli: Global Profiling, Dynamic Change, and Enzymatic Regulation. J. Proteome Res. 2016, 15, 4696–4708. [DOI] [PubMed] [Google Scholar]
- (214).Gattner MJ; Vrabel M; Carell T Synthesis of Ε-N-Propionyl-, Ε-N-Butyryl-, and Ε-N-Crotonyl-Lysine Containing Histone H3 Using the Pyrrolysine System. Chem. Commun. (Camb) 2013, 49, 379–381. [DOI] [PubMed] [Google Scholar]
- (215).Wilkins BJ; Hahn LE; Heitmüller S; Frauendorf H; Valerius O; Braus GH; Neumann H Genetically Encoding Lysine Modifications on Histone H4. ACS Chem. Biol. 2015, 10, 939–944. [DOI] [PubMed] [Google Scholar]
- (216).Xue Q; Yang Y; Li H; Li X; Zou L; Li T; Ma H; Qi H; Wang J; Yu T Functions and Mechanisms of Protein Lysine Butyrylation (Kbu): Therapeutic Implications in Human Diseases. Genes Dis. 2023, 10, 2479–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Liu S; Liu G; Cheng P; Xue C; Zhou Y; Chen X; Ye L; Qiao Z; Zhang T; Gong Z Genome-Wide Profiling of Histone Lysine Butyrylation Reveals Its Role in the Positive Regulation of Gene Transcription in Rice. Rice (N Y) 2019, 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Wu Q; Li W; Wang C; Fan P; Cao L; Wu Z; Wang F Ultradeep Lysine Crotonylome Reveals the Crotonylation Enhancement on Both Histones and Nonhistone Proteins by Saha Treatment. J. Proteome Res. 2017, 16, 3664–3671. [DOI] [PubMed] [Google Scholar]
- (219).Wan J; Liu H; Chu J; Zhang H Functions and Mechanisms of Lysine Crotonylation. J. Cell Mol. Med. 2019, 23, 7163–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Jiang G; Li C; Lu M; Lu K; Li H Protein Lysine Crotonylation: Past, Present, Perspective. Cell Death Dis. 2021, 12, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Kim CH; Kang M; Kim HJ; Chatterjee A; Schultz PG Site-Specific Incorporation of Ε-N-Crotonyllysine into Histones. Angew. Chem., Int. Ed. Engl. 2012, 51, 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Wisniewski JR; Zougman A; Mann M Nepsilon-Formylation of Lysine Is a Widespread Post-Translational Modification of Nuclear Proteins Occurring at Residues Involved in Regulation of Chromatin Function. Nucleic Acids Res. 2008, 36, 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Wang T; Zhou Q; Li F; Yu Y; Yin X; Wang J Genetic Incorporation of N(Epsilon)-Formyllysine, a New Histone Post-Translational Modification. Chembiochem 2015, 16, 1440–1442. [DOI] [PubMed] [Google Scholar]
- (224).Huang H; Zhang D; Wang Y; Perez-Neut M; Han Z; Zheng YG; Hao Q; Zhao Y Lysine Benzoylation Is a Histone Mark Regulated by Sirt2. Nat. Commun. 2018, 9, 3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Tan D; Wei W; Han Z; Ren X; Yan C; Qi S; Song X; Zheng YG; Wong J; Huang H Hbo1 Catalyzes Lysine Benzoylation in Mammalian Cells. iScience 2022, 25, 105443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Ji Y; Ren C; Miao H; Pang Z; Xiao R; Yang X; Xuan W Genetically Encoding Epsilon-N-Benzoyllysine in Proteins. Chem. Commun. (Camb) 2021, 57, 1798–1801. [DOI] [PubMed] [Google Scholar]
- (227).Tian H; Yang J; Guo AD; Ran Y; Yang YZ; Yang B; Huang R; Liu H; Chen XH Genetically Encoded Benzoyllysines Serve as Versatile Probes for Interrogating Histone Benzoylation and Interactions in Living Cells. ACS Chem. Biol. 2021, 16, 2560–2569. [DOI] [PubMed] [Google Scholar]
- (228).Cao L; Liu J; Ghelichkhani F; Rozovsky S; Wang L Genetic Incorporation of ϵ-N-Benzoyllysine by Engineering Methanomethylophilus Alvus Pyrrolysyl-Trna Synthetase. Chembiochem 2021, 22, 2530–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Huang S; Tang D; Dai Y Metabolic Functions of Lysine 2-Hydroxyisobutyrylation. Cureus 2020, 12, e9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Zhang N; Wang S; Tian H; Li S; Liu L; Li J; Chen D; Zhao S; Yan X; Niaz M; et al. Functions of Lysine 2-Hydroxyisobutyrylation and Future Perspectives on Plants. Proteomics 2023, 23, e2300045. [DOI] [PubMed] [Google Scholar]
- (231).Xiao H; Xuan W; Shao S; Liu T; Schultz PG Genetic Incorporation of Ε-N-2-Hydroxyisobutyryl-Lysine into Recombinant Histones. ACS Chem. Biol. 2015, 10, 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Hou W; Liu G; Ren X; Liu X; He L; Huang H Quantitative Proteomics Analysis Expands the Roles of Lysine Beta-Hydroxybutyrylation Pathway in Response to Environmental Beta-Hydroxybutyrate. Oxid. Med. Cell Longev. 2022, 2022, 4592170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Xie Z; Zhang D; Chung D; Tang Z; Huang H; Dai L; Qi S; Li J; Colak G; Chen Y; et al. Metabolic Regulation of Gene Expression by Histone Lysine Beta-Hydroxybutyrylation. Mol. Cell 2016, 62, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Zhou T; Cheng X; He Y; Xie Y; Xu F; Xu Y; Huang W Function and Mechanism of Histone Beta-Hydroxybutyrylation in Health and Disease. Front. Immunol. 2022, 13, 981285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Xu H; Wu M; Ma X; Huang W; Xu Y Function and Mechanism of Novel Histone Posttranslational Modifications in Health and Disease. Biomed. Res. Int. 2021, 2021, 6635225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Li Z; Zhang Y; Han M; Deng H; Wu F; Liu G; Chen GQ Lysine Beta-Hydroxybutyrylation Improves Stability of Covid-19 Antibody. Biomacromolecules 2022, 23, 454–463. [DOI] [PubMed] [Google Scholar]
- (237).Ren C; Wu Q; Xiao R; Ji Y; Yang X; Zhang Z; Qin H; Ma JA; Xuan W Expanding the Scope of Genetically Encoded Lysine Post-Translational Modifications with Lactylation, Beta-Hydroxybutyrylation and Lipoylation. Chembiochem 2022, 23, e202200302. [DOI] [PubMed] [Google Scholar]
- (238).Li Z; Gong T; Wu Q; Zhang Y; Zheng X; Li Y; Ren B; Peng X; Zhou X Lysine Lactylation Regulates Metabolic Pathways and Biofilm Formation in Streptococcus Mutans. Sci. Signal. 2023, 16, eadg1849. [DOI] [PubMed] [Google Scholar]
- (239).Chen AN; Luo Y; Yang YH; Fu JT; Geng XM; Shi JP; Yang J Lactylation, a Novel Metabolic Reprogramming Code: Current Status and Prospects. Front. Immunol. 2021, 12, 688910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Xie Y; Hu H; Liu M; Zhou T; Cheng X; Huang W; Cao L The Role and Mechanism of Histone Lactylation in Health and Diseases. Front. Genet. 2022, 13, 949252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Zhao G; Zhen J; Liu X; Guo J; Li D; Xie J; Xie L Protein Post-Translational Modification by Lysine Succinylation: Biochemistry, Biological Implications, and Therapeutic Opportunities. Genes Dis. 2023, 10, 1242–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (242).Zou L; Yang Y; Wang Z; Fu X; He X; Song J; Li T; Ma H; Yu T Lysine Malonylation and Its Links to Metabolism and Diseases. Aging Dis. 2023, 14, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Xie L; Xiao Y; Meng F; Li Y; Shi Z; Qian K Functions and Mechanisms of Lysine Glutarylation in Eukaryotes. Front. Cell Dev. Biol. 2021, 9, 667684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (244).Hirschey MD; Zhao Y Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol. Cell Proteomics 2015, 14, 2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Wang ZA; Kurra Y; Wang X; Zeng Y; Lee YJ; Sharma V; Lin H; Dai SY; Liu WR A Versatile Approach for Site-Specific Lysine Acylation in Proteins. Angew. Chem., Int. Ed. Engl. 2017, 56, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Rowland EA; Snowden CK; Cristea IM Protein Lipoylation: An Evolutionarily Conserved Metabolic Regulator of Health and Disease. Curr. Opin. Chem. Biol. 2018, 42, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).Tang Q; Guo Y; Meng L; Chen X Chemical Tagging of Protein Lipoylation. Angew. Chem., Int. Ed. Engl. 2021, 60, 4028–4033. [DOI] [PubMed] [Google Scholar]
- (248).Komaniecki G; Lin H Lysine Fatty Acylation: Regulatory Enzymes, Research Tools, and Biological Function. Front. Cell Dev. Biol. 2021, 9, 717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Fu C; Chen Q; Zheng F; Yang L; Li H; Zhao Q; Wang X; Wang L; Wang Q Genetically Encoding a Lipidated Amino Acid for Extension of Protein Half-Life in Vivo. Angew. Chem., Int. Ed. Engl. 2019, 58, 1392–1396. [DOI] [PubMed] [Google Scholar]
- (250).He XD; Gong W; Zhang JN; Nie J; Yao CF; Guo FS; Lin Y; Wu XH; Li F; Li J; et al. Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations. Cell Metab. 2018, 27, 151–166. [DOI] [PubMed] [Google Scholar]
- (251).Zang J; Chen Y; Liu C; Lin S Probing the Role of Aurora Kinase a Threonylation with Site-Specific Lysine Threonylation. ACS Chem. Biol. 2023, 18, 674–678. [DOI] [PubMed] [Google Scholar]
- (252).Zang J; Chen Y; Liu C; Hu L; Zhao H; Ding W; Yuan Y; Lin S Genetic Code Expansion Reveals Aminoacylated Lysine Ubiquitination Mediated by Ube2w. Nat. Struct. Mol. Biol. 2023, 30, 62–71. [DOI] [PubMed] [Google Scholar]
- (253).Zhang ZJ; Pedicord VA; Peng T; Hang HC Site-Specific Acylation of a Bacterial Virulence Regulator Attenuates Infection. Nat. Chem. Biol. 2020, 16, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Wang WW; Zeng Y; Wu B; Deiters A; Liu WR A Chemical Biology Approach to Reveal Sirt6-Targeted Histone H3 Sites in Nucleosomes. ACS Chem. Biol. 2016, 11, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Wang WW; Angulo-Ibanez M; Lyu J; Kurra Y; Tong Z; Wu B; Zhang L; Sharma V; Zhou J; Lin H; et al. A Click Chemistry Approach Reveals the Chromatin-Dependent Histone H3k36 Deacylase Nature of Sirt7. J. Am. Chem. Soc. 2019, 141, 2462–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (256).Xie X; Li XM; Qin F; Lin J; Zhang G; Zhao J; Bao X; Zhu R; Song H; Li XD; et al. Genetically Encoded Photoaffinity Histone Marks. J. Am. Chem. Soc. 2017, 139, 6522–6525. [DOI] [PubMed] [Google Scholar]
- (257).Qin F; Li B; Wang H; Ma S; Li J; Liu S; Kong L; Zheng H; Zhu R; Han Y; et al. Linking Chromatin Acylation Mark-Defined Proteome and Genome in Living Cells. Cell 2023, 186, 1066–1085. [DOI] [PubMed] [Google Scholar]
- (258).Callis J The Ubiquitination Machinery of the Ubiquitin System. Arabidopsis Book 2014, 12, No. e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (259).Swatek KN; Komander D Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Hershko A; Ciechanover A The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [DOI] [PubMed] [Google Scholar]
- (261).van der Veen AG; Ploegh HL Ubiquitin-Like Proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [DOI] [PubMed] [Google Scholar]
- (262).Li X; Fekner T; Ottesen JJ; Chan MK A Pyrrolysine Analogue for Site-Specific Protein Ubiquitination. Angew. Chem., Int. Ed. Engl. 2009, 48, 9184–9187. [DOI] [PubMed] [Google Scholar]
- (263).Virdee S; Ye Y; Nguyen DP; Komander D; Chin JW Engineered Diubiquitin Synthesis Reveals Lys29-Isopeptide Specificity of an Otu Deubiquitinase. Nat. Chem. Biol. 2010, 6, 750–757. [DOI] [PubMed] [Google Scholar]
- (264).Virdee S; Kapadnis PB; Elliott T; Lang K; Madrzak J; Nguyen DP; Riechmann L; Chin JW Traceless and Site-Specific Ubiquitination of Recombinant Proteins. J. Am. Chem. Soc. 2011, 133, 10708–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (265).Komander D; Clague MJ; Urbe S Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [DOI] [PubMed] [Google Scholar]
- (266).Eger S; Scheffner M; Marx A; Rubini M Synthesis of Defined Ubiquitin Dimers. J. Am. Chem. Soc. 2010, 132, 16337–16339. [DOI] [PubMed] [Google Scholar]
- (267).Stanley M; Virdee S Genetically Directed Production of Recombinant, Isosteric and Nonhydrolysable Ubiquitin Conjugates. Chembiochem 2016, 17, 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Fottner M; Brunner AD; Bittl V; Horn-Ghetko D; Jussupow A; Kaila VRI; Bremm A; Lang K Site-Specific Ubiquitylation and Sumoylation Using Genetic-Code Expansion and Sortase. Nat. Chem. Biol. 2019, 15, 276–284. [DOI] [PubMed] [Google Scholar]
- (269).Mazmanian SK; Liu G; Ton-That H; Schneewind O Staphylococcus Aureus Sortase, an Enzyme That Anchors Surface Proteins to the Cell Wall. Science 1999, 285, 760–763. [DOI] [PubMed] [Google Scholar]
- (270).Fottner M; Weyh M; Gaussmann S; Schwarz D; Sattler M; Lang K A Modular Toolbox to Generate Complex Polymeric Ubiquitin Architectures Using Orthogonal Sortase Enzymes. Nat. Commun. 2021, 12, 6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (271).Fottner M; Heimgartner J; Gantz M; Muhlhofer R; Nast-Kolb T; Lang K Site-Specific Protein Labeling and Generation of Defined Ubiquitin-Protein Conjugates Using an Asparaginyl Endopeptidase. J. Am. Chem. Soc. 2022, 144, 13118–13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Mukherjee S; Hao YH; Orth K A Newly Discovered Post-Translational Modification–the Acetylation of Serine and Threonine Residues. Trends Biochem. Sci. 2007, 32, 210–216. [DOI] [PubMed] [Google Scholar]
- (273).Medzihradszky KF; Darula Z; Perlson E; Fainzilber M; Chalkley RJ; Ball H; Greenbaum D; Bogyo M; Tyson DR; Bradshaw RA; et al. O-Sulfonation of Serine and Threonine: Mass Spectrometric Detection and Characterization of a New Posttranslational Modification in Diverse Proteins Throughout the Eukaryotes. Mol. Cell Proteomics 2004, 3, 429–440. [DOI] [PubMed] [Google Scholar]
- (274).Guan X; Fierke CA Understanding Protein Palmitoylation: Biological Significance and Enzymology. Sci. China Chem. 2011, 54, 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Tarrant MK; Cole PA The Chemical Biology of Protein Phosphorylation. Annu. Rev. Biochem. 2009, 78, 797–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Pawson T; Scott JD Protein Phosphorylation in Signaling–50 Years and Counting. Trends Biochem. Sci. 2005, 30, 286–290. [DOI] [PubMed] [Google Scholar]
- (277).Day EK; Sosale NG; Lazzara MJ Cell Signaling Regulation by Protein Phosphorylation: A Multivariate, Heterogeneous, and Context-Dependent Process. Curr. Opin. Biotechnol. 2016, 40, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (278).Ardito F; Giuliani M; Perrone D; Troiano G; Lo Muzio L The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Pang K; Wang W; Qin JX; Shi ZD; Hao L; Ma YY; Xu H; Wu ZX; Pan D; Chen ZS; et al. Role of Protein Phosphorylation in Cell Signaling, Disease, and the Intervention Therapy. MedComm (2020) 2022, 3, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (280).Zhang H; Cao X; Tang M; Zhong G; Si Y; Li H; Zhu F; Liao Q; Li L; Zhao J; et al. A Subcellular Map of the Human Kinome. Elife 2021, 10, e64943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Sacco F; Perfetto L; Castagnoli L; Cesareni G The Human Phosphatase Interactome: An Intricate Family Portrait. FEBS Lett. 2012, 586, 2732–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Duan HZ; Nie ZK; Li Y; Chen YX Unremitting Progresses for Phosphoprotein Synthesis. Curr. Opin. Chem. Biol. 2020, 58, 96–111. [DOI] [PubMed] [Google Scholar]
- (283).Qin X; Liu T Recent Advances in Genetic Code Expansion Techniques for Protein Phosphorylation Studies. J. Mol. Biol. 2022, 434, 167406. [DOI] [PubMed] [Google Scholar]
- (284).Ros R; Cascales-Minana B; Segura J; Anoman AD; Toujani W; Flores-Tornero M; Rosa-Tellez S; Munoz-Bertomeu J Serine Biosynthesis by Photorespiratory and Non-Photorespiratory Pathways: An Interesting Interplay with Unknown Regulatory Networks. Plant Biol. (Stuttg) 2013, 15, 707–712. [DOI] [PubMed] [Google Scholar]
- (285).Sauerwald A; Zhu W; Major TA; Roy H; Palioura S; Jahn D; Whitman WB; Yates JR; Ibba M; Söll D Rna-Dependent Cysteine Biosynthesis in Archaea. Science 2005, 307, 1969–1972. [DOI] [PubMed] [Google Scholar]
- (286).Dale T; Sanderson LE; Uhlenbeck OC The Affinity of Elongation Factor Tu for an Aminoacyl-Trna Is Modulated by the Esterified Amino Acid. Biochemistry 2004, 43, 6159–6166. [DOI] [PubMed] [Google Scholar]
- (287).Park H-S; Hohn MJ; Umehara T; Guo L-T; Osborne EM; Benner J; Noren CJ; Rinehart J; Söll D Expanding the Genetic Code of Escherichia Coli with Phosphoserine. Science 2011, 333, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Beranek V; Reinkemeier CD; Zhang MS; Liang AD; Kym G; Chin JW Genetically Encoded Protein Phosphorylation in Mammalian Cells. Cell Chem. Biol. 2018, 25, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Oza JP; Aerni HR; Pirman NL; Barber KW; ter Haar CM; Rogulina S; Amrofell MB; Isaacs FJ; Rinehart J; Jewett MC Robust Production of Recombinant Phosphoproteins Using Cell-Free Protein Synthesis. Nat. Commun. 2015, 6, 8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (290).Heinemann IU; Rovner AJ; Aerni HR; Rogulina S; Cheng L; Olds W; Fischer JT; Söll D; Isaacs FJ; Rinehart J Enhanced Phosphoserine Insertion During Escherichia Coli Protein Synthesis Via Partial Uag Codon Reassignment and Release Factor 1 Deletion. FEBS Lett. 2012, 586, 3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (291).Steinfeld JB; Aerni HR; Rogulina S; Liu Y; Rinehart J Expanded Cellular Amino Acid Pools Containing Phosphoserine, Phosphothreonine, and Phosphotyrosine. ACS Chem. Biol. 2014, 9, 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (292).Rogerson DT; Sachdeva A; Wang K; Haq T; Kazlauskaite A; Hancock SM; Huguenin-Dezot N; Muqit MM; Fry AM; Bayliss R; et al. Efficient Genetic Encoding of Phosphoserine and Its Nonhydrolyzable Analog. Nat. Chem. Biol. 2015, 11, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (293).Aerni HR; Shifman MA; Rogulina S; O’Donoghue P; Rinehart J Revealing the Amino Acid Composition of Proteins within an Expanded Genetic Code. Nucleic Acids Res. 2015, 43, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (294).Pirman NL; Barber KW; Aerni HR; Ma NJ; Haimovich AD; Rogulina S; Isaacs FJ; Rinehart J A Flexible Codon in Genomically Recoded Escherichia Coli Permits Programmable Protein Phosphorylation. Nat. Commun. 2015, 6, 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (295).George S; Aguirre JD; Spratt DE; Bi Y; Jeffery M; Shaw GS; O’Donoghue P Generation of Phospho-Ubiquitin Variants by Orthogonal Translation Reveals Codon Skipping. FEBS Lett. 2016, 590, 1530–1542. [DOI] [PubMed] [Google Scholar]
- (296).Zhu P; Gafken PR; Mehl RA; Cooley RB A Highly Versatile Expression System for the Production of Multiply Phosphorylated Proteins. ACS Chem. Biol. 2019, 14, 1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (297).Lee S; Oh S; Yang A; Kim J; Söll D; Lee D; Park HS A Facile Strategy for Selective Incorporation of Phosphoserine into Histones. Angew. Chem., Int. Ed. Engl. 2013, 52, 5771–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (298).Khoury GA; Baliban RC; Floudas CA Proteome-Wide Post-Translational Modification Statistics: Frequency Analysis and Curation of the Swiss-Prot Database. Sci. Rep. 2011, 1, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (299).Barchi JJ Jr.; Strain CN The Effect of a Methyl Group on Structure and Function: Serine Vs. Threonine Glycosylation and Phosphorylation. Front. Mol. Biosci. 2023, 10, 1117850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (300).Fan C; Bobik TA The Pdux Enzyme of Salmonella Enterica Is an L-Threonine Kinase Used for Coenzyme B12 Synthesis. J. Biol. Chem. 2008, 283, 11322–11329. [DOI] [PubMed] [Google Scholar]
- (301).Fan C; Fromm HJ; Bobik TA Kinetic and Functional Analysis of L-Threonine Kinase, the Pdux Enzyme of Salmonella Enterica. J. Biol. Chem. 2009, 284, 20240–20248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (302).Zhang MS; Brunner SF; Huguenin-Dezot N; Liang AD; Schmied WH; Rogerson DT; Chin JW Biosynthesis and Genetic Encoding of Phosphothreonine through Parallel Selection and Deep Sequencing. Nat. Methods 2017, 14, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (303).Moen JM; Mohler K; Rogulina S; Shi X; Shen H; Rinehart J Enhanced Access to the Human Phosphoproteome with Genetically Encoded Phosphothreonine. Nat. Commun. 2022, 13, 7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (304).Zarschler K; Janesch B; Pabst M; Altmann F; Messner P; Schaffer C Protein Tyrosine O-Glycosylation–a Rather Unexplored Prokaryotic Glycosylation System. Glycobiology 2010, 20, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (305).Li X; Pinou L; Du Y; Chen X; Liu C Emerging Roles of O-Glycosylation in Regulating Protein Aggregation, Phase Separation, and Functions. Curr. Opin. Chem. Biol. 2023, 75, 102314. [DOI] [PubMed] [Google Scholar]
- (306).Magalhaes A; Duarte HO; Reis CA The Role of O-Glycosylation in Human Disease. Mol. Aspects Med. 2021, 79, 100964. [DOI] [PubMed] [Google Scholar]
- (307).Li J; Guo B; Zhang W; Yue S; Huang S; Gao S; Ma J; Cipollo JF; Yang S Recent Advances in Demystifying O-Glycosylation in Health and Disease. Proteomics 2022, 22, e2200156. [DOI] [PubMed] [Google Scholar]
- (308).Gorelik A; van Aalten DMF Tools for Functional Dissection of Site-Specific O-Glcnacylation. RSC Chem. Biol. 2020, 1, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Harel O; Jbara M Posttranslational Chemical Mutagenesis Methods to Insert Posttranslational Modifications into Recombinant Proteins. Molecules 2022, 27, 4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (310).Tegl G; Hanson J; Chen HM; Kwan DH; Santana AG; Withers SG Facile Formation of Beta-Thioglcnac Linkages to Thiol-Containing Sugars, Peptides, and Proteins Using a Mutant Gh20 Hexosaminidase. Angew. Chem., Int. Ed. Engl. 2019, 58, 1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (311).Fahmi NE; Dedkova L; Wang B; Golovine S; Hecht SM Site-Specific Incorporation of Glycosylated Serine and Tyrosine Derivatives into Proteins. J. Am. Chem. Soc. 2007, 129, 3586–3597. [DOI] [PubMed] [Google Scholar]
- (312).Matsubara T; Iijima K; Watanabe T; Hohsaka T; Sato T Incorporation of Glycosylated Amino Acid into Protein by an in Vitro Translation System. Bioorg. Med. Chem. Lett. 2013, 23, 5634–5636. [DOI] [PubMed] [Google Scholar]
- (313).Machida T; Lang K; Xue L; Chin JW; Winssinger N Site-Specific Glycoconjugation of Protein Via Bioorthogonal Tetrazine Cycloaddition with a Genetically Encoded Trans-Cyclooctene or Bicyclononyne. Bioconjugate Chem. 2015, 26, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (314).Hyun JY; Kim S; Lee CH; Lee HS; Shin I Efficient Preparation and Bioactivity Evaluation of Glycan-Defined Glycoproteins. ACS Chem. Biol. 2021, 16, 1930–1940. [DOI] [PubMed] [Google Scholar]
- (315).Ghisaidoobe AB; Chung SJ Intrinsic Tryptophan Fluorescence in the Detection and Analysis of Proteins: A Focus on Forster Resonance Energy Transfer Techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (316).Ehrenshaft M; Deterding LJ; Mason RP Tripping up Trp: Modification of Protein Tryptophan Residues by Reactive Oxygen Species, Modes of Detection, and Biological Consequences. Free Radic. Biol. Med. 2015, 89, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Nuriel T; Hansler A; Gross SS Protein Nitrotryptophan: Formation, Significance and Identification. J. Proteome Res. 2011, 74, 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (318).Hains PG; Truscott RJ Post-Translational Modifications in the Nuclear Region of Young, Aged, and Cataract Human Lenses. J. Proteome Res. 2007, 6, 3935–3943. [DOI] [PubMed] [Google Scholar]
- (319).Hadfield KA; Pattison DI; Brown BE; Hou L; Rye KA; Davies MJ; Hawkins CL Myeloperoxidase-Derived Oxidants Modify Apolipoprotein a-I and Generate Dysfunctional High-Density Lipoproteins: Comparison of Hypothiocyanous Acid (Hoscn) with Hypochlorous Acid (Hocl). Biochem. J. 2013, 449, 531–542. [DOI] [PubMed] [Google Scholar]
- (320).Helland R; Fjellbirkeland A; Karlsen OA; Ve T; Lillehaug JR; Jensen HB An Oxidized Tryptophan Facilitates Copper Binding in Methylococcus Capsulatus-Secreted Protein Mope. J. Biol. Chem. 2008, 283, 13897–13904. [DOI] [PubMed] [Google Scholar]
- (321).Ellefson JW; Meyer AJ; Hughes RA; Cannon JR; Brodbelt JS; Ellington AD Directed Evolution of Genetic Parts and Circuits by Compartmentalized Partnered Replication. Nat. Biotechnol. 2014, 32, 97–101. [DOI] [PubMed] [Google Scholar]
- (322).Italia JS; Addy PS; Wrobel CJ; Crawford LA; Lajoie MJ; Zheng Y; Chatterjee A An Orthogonalized Platform for Genetic Code Expansion in Both Bacteria and Eukaryotes. Nat. Chem. Biol. 2017, 13, 446–450. [DOI] [PubMed] [Google Scholar]
- (323).Whitmore SE; Lamont RJ Tyrosine Phosphorylation and Bacterial Virulence. Int. J. Oral Sci. 2012, 4, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (324).Hunter T Tyrosine Phosphorylation: Thirty Years and Counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (325).Alonso A; Sasin J; Bottini N; Friedberg I; Friedberg I; Osterman A; Godzik A; Hunter T; Dixon J; Mustelin T Protein Tyrosine Phosphatases in the Human Genome. Cell 2004, 117, 699–711. [DOI] [PubMed] [Google Scholar]
- (326).Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- (327).Hunter T The Croonian Lecture 1997. The Phosphorylation of Proteins on Tyrosine: Its Role in Cell Growth and Disease. Philos. Trans. R Soc. London B Biol. Sci. 1998, 353, 583–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (328).Arslan T; Mamaev SV; Mamaeva NV; Hecht SM Structurally Modified Firefly Luciferase. Effects of Amino Acid Substitution at Position 286. J. Am. Chem. Soc. 1997, 119, 10877–10887. [Google Scholar]
- (329).Rothman DM; Petersson EJ; Vázquez ME; Brandt GS; Dougherty DA; Imperiali B Caged Phosphoproteins. J. Am. Chem. Soc. 2005, 127, 846–847. [DOI] [PubMed] [Google Scholar]
- (330).Chen S; Maini R; Bai X; Nangreave RC; Dedkova LM; Hecht SM Incorporation of Phosphorylated Tyrosine into Proteins: In Vitro Translation and Study of Phosphorylated Ikappab-Alpha and Its Interaction with Nf-Kappab. J. Am. Chem. Soc. 2017, 139, 14098–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (331).Hoppmann C; Wong A; Yang B; Li S; Hunter T; Shokat KM; Wang L Site-Specific Incorporation of Phosphotyrosine Using an Expanded Genetic Code. Nat. Chem. Biol. 2017, 13, 842–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (332).Fan C; Ip K; Söll D Expanding the Genetic Code of Escherichia Coli with Phosphotyrosine. FEBS Lett. 2016, 590, 3040–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (333).Luo X; Fu G; Wang RE; Zhu X; Zambaldo C; Liu R; Liu T; Lyu X; Du J; Xuan W; et al. Genetically Encoding Phosphotyrosine and Its Nonhydrolyzable Analog in Bacteria. Nat. Chem. Biol. 2017, 13, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (334).Xie J; Supekova L; Schultz PG A Genetically Encoded Metabolically Stable Analogue of Phosphotyrosine in Escherichia Coli. ACS Chem. Biol. 2007, 2, 474–478. [DOI] [PubMed] [Google Scholar]
- (335).He X; Ma B; Chen Y; Guo J; Niu W Genetic Encoding of a Nonhydrolyzable Phosphotyrosine Analog in Mammalian Cells. Chem. Commun. (Camb) 2022, 58, 5897–5900. [DOI] [PubMed] [Google Scholar]
- (336).Grasso KT; Singha Roy SJ; Osgood AO; Yeo MJR; Soni C; Hillenbrand CM; Ficaretta ED; Chatterjee A A Facile Platform to Engineer Escherichia Coli Tyrosyl-Trna Synthetase Adds New Chemistries to the Eukaryotic Genetic Code, Including a Phosphotyrosine Mimic. ACS Cent. Sci. 2022, 8, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (337).Serwa R; Wilkening I; Del Signore G; Muhlberg M; Claussnitzer I; Weise C; Gerrits M; Hackenberger CP Chemoselective Staudinger-Phosphite Reaction of Azides for the Phosphorylation of Proteins. Angew. Chem., Int. Ed. Engl. 2009, 48, 8234–8239. [DOI] [PubMed] [Google Scholar]
- (338).Bettelheim FR Tyrosine-O-Sulfate in a Peptide from Fibrinogen. J. Am. Chem. Soc. 1954, 76, 2838–2839. [Google Scholar]
- (339).Stone MJ; Chuang S; Hou X; Shoham M; Zhu JZ Tyrosine Sulfation: An Increasingly Recognised Post-Translational Modification of Secreted Proteins. New Biotechnol. 2009, 25, 299–317. [DOI] [PubMed] [Google Scholar]
- (340).Yang YS; Wang CC; Chen BH; Hou YH; Hung KS; Mao YC Tyrosine Sulfation as a Protein Post-Translational Modification. Molecules 2015, 20, 2138–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (341).Huttner WB Protein Tyrosine Sulfation. Trends Biochem. Sci. 1987, 12, 361–363. [Google Scholar]
- (342).Liu CC; Schultz PG Recombinant Expression of Selectively Sulfated Proteins in Escherichia Coli. Nat. Biotechnol. 2006, 24, 1436–1440. [DOI] [PubMed] [Google Scholar]
- (343).Liu CC; Cellitti SE; Geierstanger BH; Schultz PG Efficient Expression of Tyrosine-Sulfated Proteins in E. Coli Using an Expanded Genetic Code. Nat. Protoc. 2009, 4, 1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (344).Schwessinger B; Li X; Ellinghaus TL; Chan LJ; Wei T; Joe A; Thomas N; Pruitt R; Adams PD; Chern MS; et al. A Second-Generation Expression System for Tyrosine-Sulfated Proteins and Its Application in Crop Protection. Integr. Biol. (Camb) 2016, 8, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (345).He X; Chen Y; Beltran DG; Kelly M; Ma B; Lawrie J; Wang F; Dodds E; Zhang L; Guo J; et al. Functional Genetic Encoding of Sulfotyrosine in Mammalian Cells. Nat. Commun. 2020, 11, 4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (346).Italia JS; Peeler JC; Hillenbrand CM; Latour C; Weerapana E; Chatterjee A Genetically Encoded Protein Sulfation in Mammalian Cells. Nat. Chem. Biol. 2020, 16, 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (347).Chen Y; Jin S; Zhang M; Hu Y; Wu KL; Chung A; Wang S; Tian Z; Wang Y; Wolynes PG; et al. Unleashing the Potential of Noncanonical Amino Acid Biosynthesis to Create Cells with Precision Tyrosine Sulfation. Nat. Commun. 2022, 13, 5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (348).Fitzpatrick PF The Aromatic Amino Acid Hydroxylases: Structures, Catalysis, and Regulation of Phenylalanine Hydroxylase, Tyrosine Hydroxylase, and Tryptophan Hydroxylase. Arch. Biochem. Biophys. 2023, 735, 109518. [DOI] [PubMed] [Google Scholar]
- (349).Waite JH; Tanzer ML Polyphenolic Substance of Mytilus Edulis: Novel Adhesive Containing L-Dopa and Hydroxyproline. Science 1981, 212, 1038–1040. [DOI] [PubMed] [Google Scholar]
- (350).Tan M; Luo H; Lee S; Jin F; Yang JS; Montellier E; Buchou T; Cheng Z; Rousseaux S; Rajagopal N; et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 2011, 146, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (351).Alfonta L; Zhang Z; Uryu S; Loo JA; Schultz PG Site-Specific Incorporation of a Redox-Active Amino Acid into Proteins. J. Am. Chem. Soc. 2003, 125, 14662–14663. [DOI] [PubMed] [Google Scholar]
- (352).Hauf M; Richter F; Schneider T; Faidt T; Martins BM; Baumann T; Durkin P; Dobbek H; Jacobs K; Moglich A; et al. Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code. Chembiochem 2017, 18, 1819–1823. [DOI] [PubMed] [Google Scholar]
- (353).Kim S; Sung BH; Kim SC; Lee HS Genetic Incorporation of L-Dihydroxyphenylalanine (Dopa) Biosynthesized by a Tyrosine Phenol-Lyase. Chem. Commun. (Camb) 2018, 54, 3002–3005. [DOI] [PubMed] [Google Scholar]
- (354).Thyer R; d’Oelsnitz S; Blevins MS; Klein DR; Brodbelt JS; Ellington AD Directed Evolution of an Improved Aminoacyl-Trna Synthetase for Incorporation of L-3,4-Dihydroxyphenylalanine (L-Dopa). Angew. Chem., Int. Ed. Engl. 2021, 60, 14811–14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (355).Ding W; Zhao H; Chen Y; Zhang B; Yang Y; Zang J; Wu J; Lin S Chimeric Design of Pyrrolysyl-Trna Synthetase/Trna Pairs and Canonical Synthetase/Trna Pairs for Genetic Code Expansion. Nat. Commun. 2020, 11, 3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (356).Radi R Nitric Oxide, Oxidants, and Protein Tyrosine Nitration. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 4003–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (357).Sabadashka M; Nagalievska M; Sybirna N Tyrosine Nitration as a Key Event of Signal Transduction That Regulates Functional State of the Cell. Cell Biol. Int. 2021, 45, 481–497. [DOI] [PubMed] [Google Scholar]
- (358).Chakravarti B; Chakravarti DN Protein Tyrosine Nitration: Role in Aging. Curr. Aging Sci. 2017, 10, 246–262. [DOI] [PubMed] [Google Scholar]
- (359).Griswold-Prenner I; Kashyap AK; Mazhar S; Hall ZW; Fazelinia H; Ischiropoulos H Unveiling the Human Nitroproteome: Protein Tyrosine Nitration in Cell Signaling and Cancer. J. Biol. Chem. 2023, 299, 105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (360).Lee JR; Kim JK; Lee SJ; Kim KP Role of Protein Tyrosine Nitration in Neurodegenerative Diseases and Atherosclerosis. Arch. Pharm. Res. 2009, 32, 1109–1118. [DOI] [PubMed] [Google Scholar]
- (361).Neumann H; Hazen JL; Weinstein J; Mehl RA; Chin JW Genetically Encoding Protein Oxidative Damage. J. Am. Chem. Soc. 2008, 130, 4028–4033. [DOI] [PubMed] [Google Scholar]
- (362).Cooley RB; Feldman JL; Driggers CM; Bundy TA; Stokes AL; Karplus PA; Mehl RA Structural Basis of Improved Second-Generation 3-Nitro-Tyrosine Trna Synthetases. Biochemistry 2014, 53, 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (363).Rauch BJ; Porter JJ; Mehl RA; Perona JJ Improved Incorporation of Noncanonical Amino Acids by an Engineered Trnatyr Suppressor. Biochemistry 2016, 55, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (364).Beyer JN; Hosseinzadeh P; Gottfried-Lee I; Van Fossen EM; Zhu P; Bednar RM; Karplus PA; Mehl RA; Cooley RB Overcoming near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine Trna Synthetase. J. Mol. Biol. 2020, 432, 4690–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (365).Feeney MB; Schoneich C Tyrosine Modifications in Aging. Antioxid. Redox Signal. 2012, 17, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (366).Desiderio DM; Nibbering NM Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases; John Wiley & Sons, 2006. [Google Scholar]
- (367).Santos AL; Lindner AB Protein Posttranslational Modifications: Roles in Aging and Age-Related Disease. Oxid. Med. Cell Longev. 2017, 2017, 5716409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (368).Sun H; Jia H; Kendall O; Dragelj J; Kubyshkin V; Baumann T; Mroginski MA; Schwille P; Budisa N Halogenation of Tyrosine Perturbs Large-Scale Protein Self-Organization. Nat. Commun. 2022, 13, 4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (369).Parrish AR; She X; Xiang Z; Coin I; Shen Z; Briggs SP; Dillin A; Wang L Expanding the Genetic Code of Caenorhabditis Elegans Using Bacterial Aminoacyl-Trna Synthetase/Trna Pairs. ACS Chem. Biol. 2012, 7, 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (370).Anderle P; Nielsen CU; Pinsonneault J; Krog PL; Brodin B; Sadee W Genetic Variants of the Human Dipeptide Transporter Pept1. J. Pharmacol. Exp. Ther. 2006, 316, 636–646. [DOI] [PubMed] [Google Scholar]
- (371).Spickett CM; Pitt AR Protein Oxidation: Role in Signalling and Detection by Mass Spectrometry. Amino Acids 2012, 42, 5–21. [DOI] [PubMed] [Google Scholar]
- (372).Porter JJ; Mehl RA Genetic Code Expansion: A Powerful Tool for Understanding the Physiological Consequences of Oxidative Stress Protein Modifications. Oxid. Med. Cell Longev. 2018, 2018, 7607463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (373).Hamann M; Zhang T; Hendrich S; Thomas JA Quantitation of Protein Sulfinic and Sulfonic Acid, Irreversibly Oxidized Protein Cysteine Sites in Cellular Proteins. Methods Enzymol. 2002, 348, 146–156. [DOI] [PubMed] [Google Scholar]
- (374).Zambaldo C; Koh M; Nasertorabi F; Han GW; Chatterjee A; Stevens RC; Schultz PG An Orthogonal Seryl-Trna Synthetase/Trna Pair for Noncanonical Amino Acid Mutagenesis in Escherichia Coli. Bioorg. Med. Chem. 2020, 28, 115662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (375).Beltrao P; Bork P; Krogan NJ; van Noort V Evolution and Functional Cross-Talk of Protein Post-Translational Modifications. Mol. Syst. Biol. 2013, 9, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (376).Balasuriya N; Kunkel MT; Liu X; Biggar KK; Li SSC; Newton AC; O’Donoghue P Genetic Code Expansion and Live Cell Imaging Reveal That Thr308 Phosphorylation Is Irreplaceable and Sufficient for Akt1 Activity. J. Biol. Chem. 2018, 293, 10744–10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (377).Venkat S; Sturges J; Stahman A; Gregory C; Gan Q; Fan C Genetically Incorporating Two Distinct Post-Translational Modifications into One Protein Simultaneously. ACS Synth. Biol. 2018, 7, 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (378).Wright DE; Altaany Z; Bi Y; Alperstein Z; O’Donoghue P Acetylation Regulates Thioredoxin Reductase Oligomerization and Activity. Antioxid. Redox Signal. 2018, 29, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (379).Morosky P; Comyns C; Nunes LGA; Chung CZ; Hoffmann PR; Soll D; Vargas-Rodriguez O; Krahn N Dual Incorporation of Non-Canonical Amino Acids Enables Production of Post-Translationally Modified Selenoproteins. Front. Mol. Biosci. 2023, 10, 1096261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (380).Osgood AO; Zheng Y; Roy SJS; Biris N; Hussain M; Loynd C; Jewel D; Italia JS; Chatterjee A An Efficient Opal-Suppressor Tryptophanyl Pair Creates New Routes for Simultaneously Incorporating up to Three Distinct Noncanonical Amino Acids into Proteins in Mammalian Cells. Angew. Chem., Int. Ed. 2023, 62, e202219269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (381).Shi N; Tong L; Lin H; Zheng Z; Zhang H; Dong L; Yang Y; Shen Y; Xia Q Optimizing Erf1 to Enable the Genetic Encoding of Three Distinct Noncanonical Amino Acids in Mammalian Cells. Adv. Biol. (Weinh) 2022, 6, e2200092. [DOI] [PubMed] [Google Scholar]
- (382).Dunkelmann DL; Willis JCW; Beattie AT; Chin JW Engineered Triply Orthogonal Pyrrolysyl-Trna Synthetase/Trna Pairs Enable the Genetic Encoding of Three Distinct Non-Canonical Amino Acids. Nat. Chem. 2020, 12, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (383).Tharp JM; Vargas-Rodriguez O; Schepartz A; Soll D Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons. ACS Chem. Biol. 2021, 16, 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (384).Suskiewicz MJ; Prokhorova E; Rack JGM; Ahel I Adp-Ribosylation from Molecular Mechanisms to Therapeutic Implications. Cell 2023, 186, 4475–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (385).Enchev RI; Schulman BA; Peter M Protein Neddylation: Beyond Cullin-Ring Ligases. Nat. Rev. Mol. Cell Biol. 2015, 16, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (386).Fernando V; Zheng X; Walia Y; Sharma V; Letson J; Furuta S S-Nitrosylation: An Emerging Paradigm of Redox Signaling. Antioxidants (Basel) 2019, 8, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]


