Abstract
We have characterized a temperate phage (MM1) from a clinical isolate of the multiply antibiotic-resistant Spanish/American 23F Streptococcus pneumoniae clone (Spain23F-1 strain). The 40-kb double-stranded genome of MM1 has been isolated as a DNA-protein complex. The use of MM1 DNA as a probe revealed that the phage genome is integrated in the host chromosome. The host and phage attachment sites, attB and attP, respectively, have been determined. Nucleotide sequencing of the attachment sites identified a 15-bp core site (5′-TTATAATTCATCCGC-3′) that has not been found in any bacterial genome described so far. Sequence information revealed the presence of an integrase gene (int), which represents the first identification of an integrase in the pneumococcal system. A 1.5-kb DNA fragment embracing attP and the int gene contained all of the genetic information needed for stable integration of a nonreplicative plasmid into the attB site of a pneumococcal strain. This vector will facilitate the introduction of foreign genes into the pneumococcal chromosome. Interestingly, DNAs highly similar to that of MM1 have been detected in several clinical pneumococcal isolates of different capsular types, suggesting a widespread distribution of these phages in relevant pathogenic strains.
Streptococcus pneumoniae is an important human pathogen and is presently the leading cause of pneumonia, meningitis, and bloodstream infections in the elderly and one of the main causes of middle ear infections in children. In addition, pneumococcal resistance to β-lactam antibiotics has been as high as 33.5% in the United States (54). Most of these resistances have been achieved as the result of interspecific gene transfers of DNA fragments between pneumococci and phylogenetically close species that colonize the same ecological niche (i.e., the nasopharynx), leading to acquisition of low-affinity penicillin-binding proteins (26). It has been reported that among the mechanisms of DNA transfer, lysogenic conversion by bacteriophages appears to be advantageous in several bacterial systems (36). The role of phages in the evolution and transfer of bacterial virulence determinants is a topic of increasing research (10, 58), and the potential use of bacteriophages for therapy and prophylaxis for antibiotic-resistant bacteria has been suggested (3, 37).
Pneumococcal phages have been a subject of continuous interest in our laboratory since the isolation of these phages was first reported (34, 55). The biological properties of several lytic and temperate phages infecting S. pneumoniae have been recently reviewed (24). The presence of temperate phages in fresh clinical isolates of S. pneumoniae was reported many years ago (4, 5). The outstanding similarity between the lytA gene, coding for the major lytic enzyme of pneumococcus, and the corresponding lytic genes coding for several pneumococcal phage amidases (19, 46) has led to the preparation of a probe, pCE3, based in the use of the 5′-end moiety of the lytA gene (16). This probe has been used to detect lysogenic strains of pneumococcus (A. Fenoll, personal communication). A recent survey carried out on clinical isolates of pneumococci by using the whole lytA as a probe confirmed and extended previous observations on the high incidence (about 75%) of prophage carriage among natural isolates (44). However, these two procedures have severe limitations, since strains containing remnants of the lytA gene in the genome might provide erroneous data on the real presence of pneumococcal phages in clinical samples (45). Nevertheless, the lysis of fresh isolates after treatment with mitomycin C and the observation of phage-like particles in the crude supernatants of these lysates also suggested the presence of a high proportion of temperate phages in clinical strains of pneumococcus. Due to the well-documented difficulties in isolation and purification of pneumococcal phages (24), none of these interesting observations provides an easy way to carry out detailed molecular characterization of some of the temperate phages in order to develop reliable studies that might document the real value of these phages as vehicles of virulence genes. Furthermore, the abundant presence of temperate phages in pneumococcus might influence genetic variation in natural populations of S. pneumoniae. That is, the bacterium-phage coevolution might result in several attributes in pathogenic microorganisms. For example, it has been suggested that phage infection may be a requirement in the pathogenesis of Shiga-like toxin-producing Escherichia coli-associated diseases (58). Currently, microbial pathogens such as S. pneumoniae are developing a great variety of strategies to guarantee their own survival and expansion. As already documented for many other bacteria, phages might be important vehicles to introduce new factors that microbes can eventually use to cause infection and disease (38).
The multiresistant 23F Spanish clone (Spain23F-1) is the best example to illustrate the rapid spread of drug resistance, in this case originally detected in Spain and then rapidly disseminated to other parts of the world (40). A recent study conducted in 38 states of the United States revealed that of 328 isolates highly resistant to penicillin (≥2.0 μg/ml), about 40% belonged to the Spain23F-1 clone (35). In this study, we have purified and characterized a temperate phage, named MM1, isolated from the 23F strain 949. Moreover, we have also determined the attP and attB attachment sites, as well as the phage integrase gene required for site-specific recombination. A nonreplicative vector based on phage integration elements has been constructed and shown to be able to integrate in a specific attB site in the S. pneumoniae chromosome. To our knowledge, a detailed analysis of the phage integration system in pneumococcus had not been previously documented.
MATERIALS AND METHODS
Bacteria, bacteriophages, plasmids, and growth conditions.
The bacterial strains, bacteriophages, and plasmids used in this study are listed in Table 1. S. pneumoniae was grown in C medium (29) supplemented with yeast extract (0.8 mg/ml) (Difco Laboratories) at 37°C without shaking and the growth was monitored with a Hach 2100N nephelometer. E. coli was grown in Luria-Bertani medium at 37°C with shaking. Phage MM1 was induced from the lysogenic strain 949. At a cell concentration of 1.2 × 108 CFU/ml, mitomycin C was added to a final concentration of 75 ng/ml, and the culture was incubated in the dark at 37°C until lysis occurred. The phages were precipitated with NaCl (0.5 M) and polyethylene glycol 6000 (10%) and purified in a two-step CsCl gradient procedure as previously described (21).
TABLE 1.
Bacterial strains, plasmids, phages, and primers
| Material | Description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| S. pneumoniae | ||
| 949 | Lysogenic for phage MM1; serotype 23F | 40 |
| 496 | Serotype 23F | 40 |
| 499 | Serotype 23F | 40 |
| 622 | Serotype 23F | 40 |
| 8249 | Serotype 19A | 32 |
| CSUB 3409 | Serotype 9V | J. Liñares |
| CSUB 4086 | Serotype 14 | J. Liñares |
| SSISP33C/1 | Serotype 33C | Statens Seruminstitut |
| SSISP33F/1 | Serotype 33F | Statens Seruminstitut |
| 746 | Lysogenic for phage HB-746; serotype 8 | 47 |
| 708 | Hex− | 53 |
| M24 | S3−lytA Hex− | 20 |
| M222 | Hex− | 17 |
| PM11 | 708 pIAPU1 integrant strain | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 50 |
| Plasmids | ||
| pUCE191 | pUC derivative; Lnr; 4.1 kb | 1 |
| pCE3 | pBR325 derivative; Cmr; harbors the 5′ moiety of lytA | 16 |
| pIAPU1 | pUCE191::EcoRI-PstI MM1 int-attP; 5.6 kb | This study |
| Phages | ||
| MM1 | Temperate phage from strain 949 | This study |
| HB-746 | Temperate phage from strain 746 | 47 |
| Dp-1 | Virulent phage | 34 |
| Cp-1 | Virulent phage | 48 |
| EJ-1 | Temperate phage from strain 101/87 | 13 |
| ω2 | Virulent phage | 55 |
| Primers | ||
| EGP2 | 5′-GCAATTATATTCATTTTCTCTCC-3′ | This study |
| EGP4 | 5′-GAAGATAGGAGGATAAACTGG-3′ | This study |
| EGP8 | 5′-GGAATTCCCCACACTCAAATTTTGGC-3′ | This study |
| EGP9 | 5′-AACTGCAGAAATTGTTCTTTCACCGCAGG-3′ | This study |
| EGP14 | 5′-CCATCAAGACACCATTCGCC-3′ | This study |
| EGP15 | 5′-CATATTGTAGACCATCGAGGC-3′ | This study |
SDS-PAGE.
Purified phage virions were boiled for 10 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and loaded in gels containing SDS and 12.5% (wt/vol) polyacrylamide as described previously (50). The gels were stained using Coomassie blue.
Recombinant DNA techniques.
The preparation of pneumococcal DNA has been described elsewhere (56). Protein-free phage DNAs were obtained by treatment of purified phage preparations with SDS and proteinase K as described previously (47). DNA-protein complexes were isolated as previously described (21). Plasmid DNA was extracted from E. coli by the rapid alkaline method (6). DNA restriction fragments or amplified fragments for cloning, probe preparation, and sequencing were isolated from 0.7% (wt/vol) agarose gels with a GeneClean II kit (Bio101, La Jolla, Calif.). Restriction endonucleases (New England Biolabs, Beverly, Mass.) and T4 DNA ligase and Klenow DNA polymerase (Amersham Pharmacia Biotech., Uppsala, Sweden) were used as recommended by the suppliers. Transformation of E. coli DH5α was carried out by the RbCl method (50). Transformants were selected on Luria-Bertani plates with ampicillin (100 μg/ml). The transformation procedure for S. pneumoniae has been described elsewhere (56). S. pneumoniae clones obtained upon transformation with the integrative vector were scored on blood agar plates containing lincomycin (0.6 μg/ml).
Southern hybridization.
Restricted DNA fragments were separated on a 0.7% (wt/vol) agarose gel and transferred to Hybond N+ membranes (Amersham Pharmacia Biotech) by vacuum blotting with blotter model 785 (Bio-Rad Laboratories) as described by the supplier. For determination of the attB chromosomal location, DNA from the S. pneumoniae strain M24 was restricted with ApaI, SacII, or SmaI enzyme and run in a 1% agarose gel by the pulsed-field gel electrophoresis (PFGE) technique as previously described (2), using a contour-clamped homogeneous electric field DRII apparatus (Bio-Rad). DNA fragments were then blotted as described by Southern (52). DNA probes were digoxigenin labeled with a DNA labeling and detection kit (Roche, Mannheim, Germany). Hybridizations were carried out at 65°C, and detections were performed as recommended by the supplier.
Electron microscopy.
Phage particles purified as described above were dialyzed against 0.1 M ammonium acetate (pH 7.0) and negatively stained with 1% sodium phosphotungstate. Samples were examined at 80 kV in a Philips EM 300 electron microscope.
Preparation of MM1 antiserum.
Purified MM1 phage particles were mixed with an equal volume of Freund's complete adjuvant. The preparation was injected subcutaneously, and the rabbit was reinoculated four more times, at 15-day intervals, with Freund's incomplete adjuvant. Each inoculation was done using 15 μg of phage proteins. The serum was collected 15 days after the last inoculation.
DNA sequencing.
DNA sequencing was carried out by using an ABI Prism 377 DNA sequencer (Applied Biosystems, Inc.). DNA and protein sequences were analyzed with the PC/GENE software package version 6.85 (Intelligenetics, Mountain View, Calif.) or using the programs present in the Deambulum (http: //www.infobiogen.fr) and National Center for Biotechnology Information (http: //www.ncbi.nlm.nih.gov) sites. Sequence similarity searches were performed using the EMBL/GenBank, SWISS-PROT, and PIR databases.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the attP-, attR-, attB-, and attL-containing fragments have been deposited in GenBank under accession numbers AJ400629, AJ400630, AJ400631, and AJ400632, respectively.
RESULTS
Isolation and characterization of phage MM1.
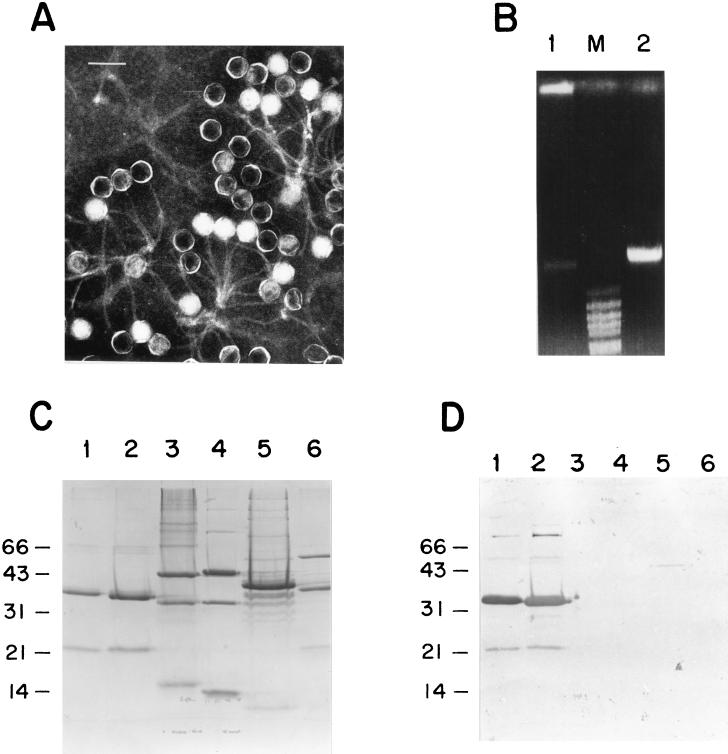
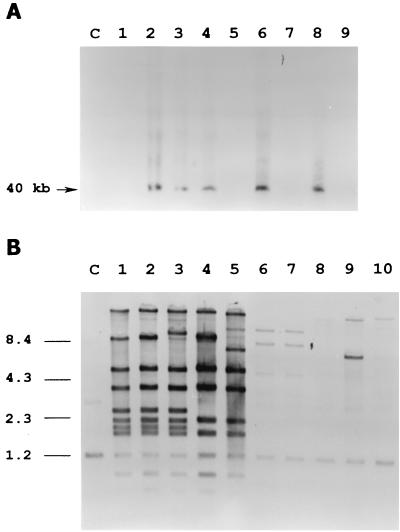
In a survey to look for the presence of temperate phages in freshly clinical isolates of pneumococcus, using plasmid pCE3 as a probe, we paid special attention to the pneumococcal strain 949, which belongs to the Spain23F-1 clone. The presence of two hybridization bands was the first hint suggesting that this strain contained a temperate phage. The lysis of the pneumococcal strain 949 when the culture was treated with mitomycin C gave additional support to the hypothesis of the presence of a temperate phage. Moreover, purification of the lysed culture revealed a bluish band after two CsCl gradients. Electron microscopy of the purified particles showed that this phage, named MM1, belongs to the Siphoviridae family, with an icosahedral head (60 nm in diameter), and a long tail (160 nm in length) (Fig. 1A). SDS-PAGE of MM1 and five different pneumococcal phages showed that MM1 virions contained two main bands of 36 and 22 kDa (Fig. 1C). A Western blot analysis using a polyclonal antiserum raised against the MM1 virion revealed common bands between phages MM1 and HB-746, and a faint band with a protein of phage Cp-1, whereas no signal was detected with any other phages in our collection (Fig. 1D). It should be mentioned that HB-746 is a phage that was originally isolated from a type 8 strain (5). From this preliminary characterization, it was concluded that phage MM1 might share several traits with HB-746, a temperate phage that has the peculiarity of having a protein covalently bound to the 5′ ends of its DNA (47). To test whether phage MM1 also has this characteristic, we prepared DNA from purified virions of MM1 that had been treated or not with proteinase K before phenol extraction. As clearly illustrated in Fig. 1B, the DNA remained at the top of the gel in the sample that was not treated with proteinase K, a peculiarity attributed to the presence of a DNA-protein complex (47), whereas the proteinase K-treated DNA migrated normally into the gel. The stability of the DNA complex was also tested by treatment with different chaotropic agents and conditions that affect ionic and hydrophobic associations (21); e.g., MM1 DNA treated with 2% SDS and 2% mercaptoethanol at 65°C for 10 min or with 6 M urea at 37°C for 30 min did not penetrate the agarose gel (data not shown). In spite of these common traits between the MM1 and HB-746 phages, restriction enzyme digestions with HindIII, PstI, and PvuII revealed that they are different phages. A molecular size of about 40 kb for MM1 DNA was determined from the sum of the sizes of DNA fragments obtained with several restriction enzymes. Furthermore, PFGE analysis of the entire DNA also indicated that the molecular size was ca. 40 kb (see Fig. 8A).
FIG. 1.
Characteristics of phage MM1. (A) Electron micrograph of a negatively stained preparation of purified MM1 virions. Bar, 100 nm. (B) Agarose gel electrophoresis of MM1 DNA. Lane 1, untreated DNA-protein complex; lane 2, DNA-protein complex digested with proteinase K (50 μg/ml, final concentration) at 37°C for 30 min; lane M, molecular size markers from BstEII-digested λ DNA. (C) Structural polypeptides of pneumococcal phages analyzed by SDS–12.5% PAGE. Lane 1, MM1; lane 2, HB-746, lane 3, Dp-1; lane 4, ω2; lane 5, Cp-1; lane 6, EJ-1. (D) Western blot analysis of the gel shown in panel C. The blotted gel was tested with a polyclonal antiserum raised against phage MM1, used at a dilution of 1/1,000. Molecular size markers (in kilodaltons) are indicated on the left.
FIG. 8.
Comparative analysis of phage DNAs from several lysogenic S. pneumoniae clinical isolates. (A) Southern blot of total DNA extracted from mitomycin C-induced cultures, separated by PFGE and hybridized with MM1 DNA as a probe. The arrow indicates the extrachromosomal band. Lane C, uninduced strain 949; lanes 1 to 9, respectively, induced cultures of strains 496, 622, 499, 949, CSUB 4086, 8249, CSUB 3409, SSISP33C/1, and SSIS33F/1. (B) Southern blot of chromosomal DNAs of lysogenic strains digested with HindIII, run on an agarose gel, and hybridized with MM1 DNA as a probe. Lane C, nonlysogenic strain M222; lanes 1 to 10, respectively, strains 949, 622, 499, 746, 8249, CSUB 3409, CSUB 4086, 496, SSISP33C/1, and SSISP33F/1. The positions of molecular size standards are indicated on the left in kilobases.
Identification of the attachment sites.
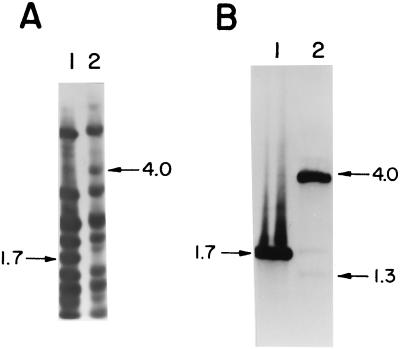
To locate the phage attachment site of MM1, attP, DNAs from the lysogenic strain 949 and the MM1 phage were digested with different restriction enzymes, the resulting fragments were separated by electrophoresis and blotted, and the membrane was hybridized with MM1 DNA as a probe. When we compared the DraI digestions, all of the hybridization bands of MM1 were present in the lysogenic strain DNA, except for a 1.7-kb fragment and a new 4-kb band found in the prophage pattern (Fig. 2A). This finding was consistent with a recombination event occurring between the attP site, located in the 1.7-kb fragment, and the bacterial attachment site, attB, resulting in the splitting of attP and the formation of a new 4-kb junction fragment. The second junction fragment is probably overlapped by one of the other restriction fragments. The phage 1.7-kb DraI fragment was isolated, cloned into SmaI-digested pUC19, and used as a probe in the same membrane. Following this procedure, a second junction fragment (1.3-kb band) was detected (Fig. 2B). The faintness of the hybridization signal was due to the small portion of phage DNA present in this junction site (see below).
FIG. 2.
Identification of the attP, attL, and attR sites. Southern blots of phage MM1 (lanes 1) and strain 949 (lanes 2) DNAs digested with DraI and hybridized with MM1 DNA (A) or with the 1.7-kb DraI MM1 DNA fragment containing the attP site (B) are shown. Arrows indicate the sizes of relevant bands, in kilobases.
We used an inverse PCR strategy to clone the junction fragments. attR was amplified as follows. Fifteen micrograms of DraI-digested S. pneumoniae 949 DNA was self-ligated in a 400-μl reaction volume, precipitated, and used for PCR amplification with oligonucleotides EGP2 and EGP4. The resulting 2.5-kb fragment was purified from an agarose gel and sequenced. Despite several attempts, attL could not be amplified by this method. We then took advantage of the partial S. pneumoniae genome sequence (http://www.tigr.org) and our sequence of attP and attR to look for a locus of identity, probably located in the attB zone. This region was found in the contig sp_100, and an oligonucleotide, EGP14, deduced from the bacterial sequence was designed. The attL sequence was amplified, using oligonucleotides EGP9 and EGP14, as a 0.6-kb fragment that was purified and sequenced. Finally, taking into account that most lysogenic bacteria suffer spontaneous phage excision from the bacterial chromosome resulting in attB site restoration, we could amplify attB with oligonucleotides EGP14 and EGP15, deduced from the bacterial parts of attL and attR, respectively, using DNA from strain 949 as the template. The resulting 1,852-bp amplicon was sequenced.
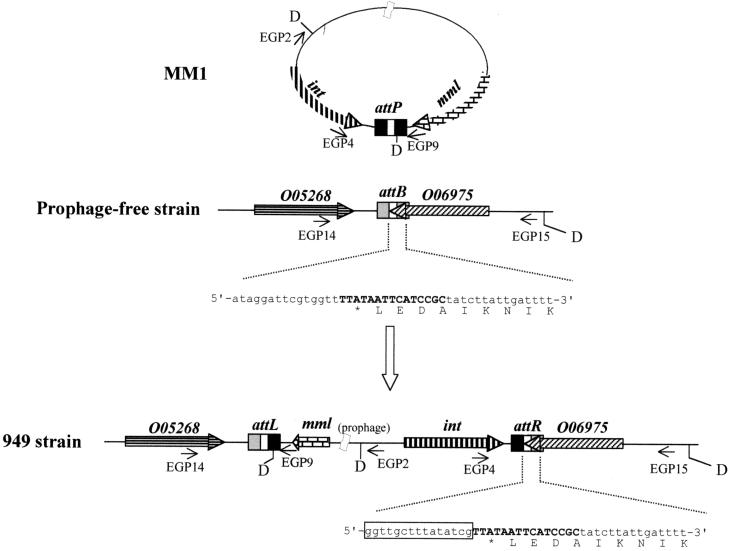
Alignment of the sequence obtained from the att sites revealed a 15-bp core site (5′-TTATAATTCATCCGC-3′) where the site-specific recombination process presumably takes place (Fig. 3 and 4). Searches in the databases revealed a single site in the S. pneumoniae genome but not in the other bacterial genomes already sequenced. Furthermore, comparison of the bacterial sequences from the attB, attL, and attR sites showed that strain 949 displays 99.6% identity at the nucleotide level with the corresponding region of the S. pneumoniae type 4 genome. The observed point mutations do not affect the genomic arrangement of the attB-containing region.
FIG. 3.
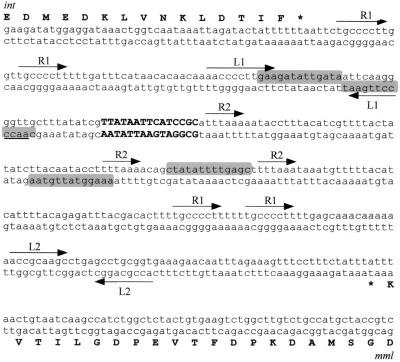
Schematic representation of the site-specific integration of phage MM1 DNA into S. pneumoniae chromosome. The central region of the att sites represent the core and is shown as boldface capital letters. Oligonucleotides (EGP series) used for the att site amplification are represented by thin arrows. The nucleotide sequence and the deduced C-terminal amino acid sequence of O06975 are indicated. The phage sequence in the attR site is boxed. D, DraI sites mentioned in the text; int, integrase gene; mml, phage lytic gene.
FIG. 4.
Nucleotide sequence of the attP site. The core is indicated in boldface capital letters. Direct repeats are marked R1 and R2. Facing arrows L1 and L2 represent putative transcription terminators. Putative integration host factor binding sites are shaded. int and mml, integrase and MM1 lytic enzyme genes, respectively.
Analysis of the 1.7-kb DraI fragment, which contains the attP site, showed that the core was present in this fragment but that the entire attP region probably was not, since a DraI site lies 4 bp downstream of the 3′ end of the core. This observation explains the low intensity of the hybridization signal obtained with the 1.3-kb junction site (Fig. 2B), as this fragment overlaps with the probe only in the core region. The complete sequence of the attP region was obtained from a cloned 3.2-kb HaeIII fragment that contains the lytic enzyme gene of phage MM1, mml, and overlaps with the 1.7-kb DraI fragment (unpublished results). Sequence analysis of the attP-containing region showed the presence of several direct repeats, including the 8-bp-long sequence 5′-TGCCCCTT-3′, which is repeated four times in the core surrounding region. Furthermore, four putative integration host factor binding sites, very similar to the consensus sequence deduced from attP sites of lambdoid phages [5′-(C/T)AANNNNTTGAT(A/T)-3′] (30), were also found. Two hairpin structures (L1 and L2) with free energies of −19.4 and −7.8 kcal/mol, respectively, are present in this region (Fig. 4). They could behave as rho-independent terminators for the two open reading frames (ORFs) that flank the core region.
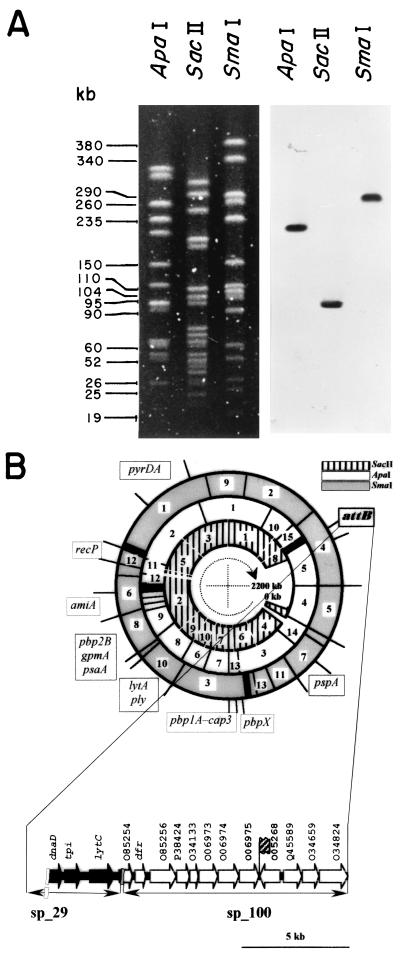
Analysis of the attB region showed that the core overlaps the 3′ end of an ORF (O06975) coding for a 303-amino-acid-long protein which has 42% identity and 78% similarity with a protein of unknown function of Bacillus subtilis (accession no. O06975). Remarkably, integration of the phage DNA in the bacterial chromosome does not disrupt the sequence of this ORF, as its stop codon is present in the core. Sequence analysis of this region also showed the presence of the 3′ end of an ORF (O05268) lying 34 bp to the left of the attB core (Fig. 3) and oriented in the opposite direction compared to O06975. The amino acid sequence of O05268 was deduced from the contig sp_100 and showed that O05268 could code for a 321-amino-acid-long protein displaying 54% identity and 80% similarity with the thioredoxine reductase from B. subtilis (accession no. O05268). To locate the attB site on the pneumococcal chromosomal map (25), we used an attB-specific probe amplified with oligonucleotides EGP14 and EGP15, deduced from the bacterial regions of attL and attR, respectively. Restriction fragments from the pneumococcal DNA isolated from strain M24 that were Southern blotted, subjected to PFGE, and hybridized with this probe revealed a single band in each case, corresponding to ApaI fragment 5, SacII fragment 8, and SmaI number 4 (Fig. 5A). We also show in Fig. 5B a fragment of the contig sp_29. We have observed that contigs sp_100 and sp_29 contained partial regions of O85254, and the gene lytC, recently demonstrated to code for the first identified pneumococcal lysozyme (23), is located in the 3′ end of contig sp_29.
FIG. 5.
Localization of the attB site on the physical and genetic maps of the S. pneumoniae M24 DNA. (A) PFGE of the DNA obtained from strain M24 digested with ApaI, SacII, or SmaI was performed, and the fragments were blotted and hybridized with a DNA probe containing the attB site (see text). (B) The localizations of most restriction fragments and the genetic markers are taken from reference (25), and the attB site is shown in boldface. At the bottom, the location of attB is denoted by a hatched flag in contig sp_100 as deduced from the preliminary sequence of the genome of S. pneumoniae already released. A fragment of contig sp_29 located upstream of sp_100 is also shown. ORFs that have been described in previous publications are designated by their gene names, and the rest of the genes are identified by the designation of their most similar homologues.
Identification of the integrase.
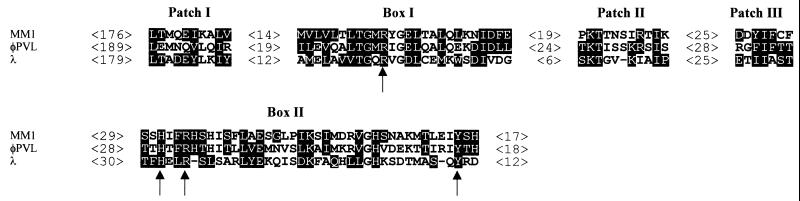
The complete sequencing of the 1.7-kb DraI fragment reported above allowed the identification of a first ORF coding for a 116-amino-acid protein and a second ORF, named int, which coded for a 375-amino-acid protein. It is preceded by a Shine-Dalgarno sequence (5′-GAGGT-3′) located 8 bp upstream of the start codon. The stop codon of the int gene is located 87 bp upstream of the attP core. BLAST searches performed with the Int sequence identified similar site-specific recombinases belonging to the λ integrase family. The best score, 33% identity and 70% similarity, was obtained with the Staphylococcus aureus phage φPVL integrase. An alignment performed with the MM1, φPVL, and λ phage integrases is shown in Fig. 6, where only the regions corresponding to the conserved boxes and patches defined by Esposito and Scocca (15) and Nunes-Düby et al. (41) are shown. Box I contains the conserved Arg residue, and the triad His-Arg-Tyr is present in box II. These four amino acids, which are involved in the recombinase activity and considered a hallmark of the λ Int family recombinases, are present at the expected positions in the MM1 Int. In addition, the three patches containing charged amino acids and highly conserved, precisely spaced, hydrophobic residues could be found in the MM1 Int sequence. The presence of the previously identified boxes and patches, the global sequence similarity with other λ integrases, and the location of int close to the attP site strongly suggested that this ORF encodes the MM1 integrase. A schematic representation of the site-specific integration of MM1 DNA into the pneumococcal chromosome is depicted in Fig. 3.
FIG. 6.
Sequence alignment of integrases of S. pneumoniae phage MM1, S. aureus phage φPVL, and coliphage λ. Amino acids matching the consensus sequence deduced from alignment of Int family integrases (41) are boxed. The arrows indicate the four invariant amino acids that are key for the recombinase activity. The number of amino acids between each motif is indicated.
Construction of an integrative vector.
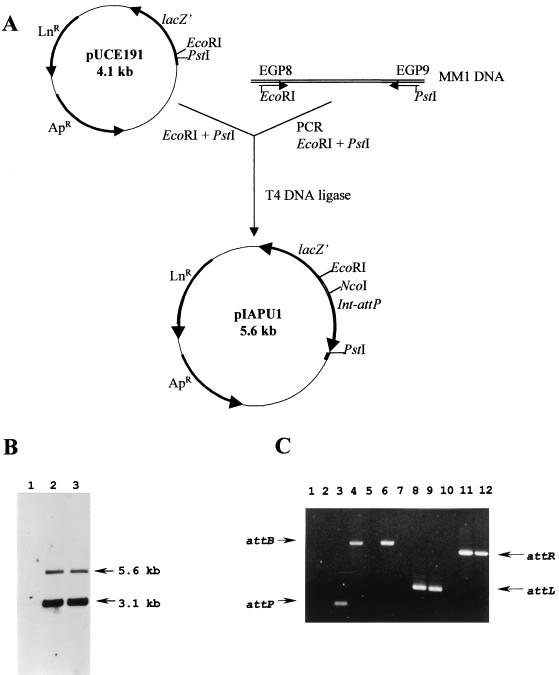
To demonstrate that attP and the int gene were actually sufficient to mediate site-specific integration into the pneumococcal chromosome, a 1,527-bp DNA fragment, embracing the int-attP cassette, was PCR amplified using oligonucleotides EGP8 and EGP9. These oligonucleotides are located 112 bp upstream from the int start codon and 186 bp downstream from the 3′ end of the attP core, respectively. This fragment contains the putative promoter, the structural gene of the integrase, and 186 bp downstream of the core of the attP site. The 1,527-bp amplified product was restricted with the enzymes EcoRI and PstI and ligated into the EcoRI- and PstI-digested pUCE191 plasmid (Fig. 7A). The 5,599-bp recombinant plasmid (pIAPU1) was first introduced into E. coli DH5α and then transferred into S. pneumoniae 708 by transformation and selected for lincomycin resistance. Since pIAPU1 is a nonreplicative plasmid in S. pneumoniae, lincomycin resistance is expressed upon chromosomal integration. Two clones, named PM11 and PM12, were chosen, although the amplification analysis revealed the same pattern for both transformants. Site-specific integration of a single copy of pIAPU1 in the attB site should lead to the detection of two fragments of 5,600 and 3,150 bp when chromosomal DNA is cut with NcoI. In fact, this was the case when the DNA prepared from PM11 was restricted with this enzyme, electrophoresed, and blotted and the membrane was probed with the 1,527-bp int-attP-containing fragment (Fig. 7B).
FIG. 7.
Construction of an integrative vector for S. pneumoniae. (A) Schematic representation of the pIAPU1 integrative vector. Ap, ampicillin; Ln, lincomycin. (B) Site-specific integration of a single copy of pIAPU1 into the S. pneumoniae chromosome. Strain 708 was transformed with pIAPU1. DNAs extracted from the parental strain 708 (lane 1) and from two transformants, PM11 and PM12 (lanes 2 and 3), were cut with NcoI, run on an agarose gel, Southern blotted, and hybridized with the int-attP cassette as a probe. The sizes of hybridizing fragments are indicated. (C) Attachment site detection using PCR. The positions of the different attachment sites are indicated. The DNAs used as templates were from strain 708 (lanes 1, 4, 7, and 10), from transformant PM11 (lanes 2, 5, 8, and 11), and from lysogenic strain 949 (lanes 3, 6, 9, and 12). The oligonucleotides used were EGP4 and EGP9 for attP, EGP14 and EGP15 for attB, EGP9 and EGP14 for attL, and EGP4 and EGP15 for attR.
To confirm the site specificity and to evaluate the stability of the integration event of PM11, we also performed PCR analysis. attL and attR sites could be amplified using genomic DNAs from the lysogenic parental strain 949 and from the transformant PM11 strain as templates (Fig. 7C, lanes 8, 9, 11, and 12), demonstrating the specificity of the integration of the plasmid in the bacterial chromosome. attB-containing amplicons could be detected using DNAs from 949 and 708 (Fig. 7C, lanes 4, and 6), and attP was amplified when using DNA from 949 (Fig. 7C, lane 3). The fact that attB and attP were amplified when using DNA from 949 as a template clearly reflects a spontaneous induction event leading to the release of phage progeny in the lysogenic culture and then restoring intact attB and attP sites. These two sites could not be amplified when using DNA from PM11, showing the stability of the integrated copy of the integrative plasmid pIAPU1 in the bacterial chromosome.
Lysogeny in different pneumococcal strains.
To test the incidence of this particular type of phage among different clinical isolates, we analyzed three other strains belonging to the Spain23F-1 clone, as well as other isolates from serotypes 19A (strain 8249, an important multiresistant strain, originally isolated in South Africa), 14 (CSUB 3409), 9V (CSUB 4086), 33C (SSISP33C/1), and 33F (SSISP33F/1). A first experimental approach was based on the comparison of the growth curves of several strains treated with mitomycin C or not. Later, total DNAs prepared from these pneumococcal cultures were subjected to PFGE and Southern blotting using MM1 DNA as a probe. As shown in Fig. 8A, three isolates belonging to the 23F serotype, as well as strains 8249 and SSISP33C/1, showed an extra chromosomal hybridization band of about 40 kb, whereas in the case of the 496, CSUB 3409, CSUB 4086, and SSISP33F/1 strains there were no visible bands. These results might suggest that some of the phages released after induction had a close relationship. Nevertheless, since we had reported the high similarity between the lytic genes from pneumococcal phages and their host (19), the common hybridization bands could be simply attributed to the presence of lytA-like genes in phages that otherwise could be very different when more accurate analysis are performed. Hence, we digested the bacterial DNAs of the strains mentioned above, along with 746 and M222 as controls, with HindIII, and the fragments generated by these digestions were probed with MM1 DNA. The result showed three different patterns (Fig. 8B): (i) strains 949, 499, 622, 746, and 8249 had very similar, but not identical, hybridization profiles (lanes 1 to 5); (ii) strain SSISP33C/1 showed one strong and several weak hybridization bands (lane 9); (iii) strains CSUB 3409 and CSUB 4086 only had two weak bands at different positions from the other ones (lanes 6 and 7). Strains 496 and SSISP33F/1 gave no bands, except the common 1.2-kb fragment containing the host lytic gene, lytA (22). The simplest explanation for these findings could be that, in the first case, the temperate phages are very similar, despite the distinct strains from which they were isolated and different geographic origins of the corresponding strains. Besides, the phage of strain SSISP33C/1 is more distant from these phages in terms of genome similarity, and strains CSUB 3409 and CSUB 4086 harbor the most different phages of the strains tested here, which most likely are defective phages because there were no bands visible in the PFGE-Southern blot analysis (Fig. 8A). Another possibility was that strains CSUB 3409 and CSUB 4086 contained some phage remnants in their chromosomes, as has already demonstrated for other organisms (33), including pneumococcus (45).
DISCUSSION
Double-stranded-DNA-containing bacteriophages infect a large diversity of bacterial hosts and probably are the most abundant group of similar organisms in the biosphere (27). In S. pneumoniae only a few examples of well-characterized phages have been described, but they have revealed a striking morphological and physiological variety (24). In two recent reports, it has been claimed that 76% of the pneumococcal clinical isolates carried a prophage in the chromosome (44, 51). This was based on the presence of two or more chromosomal SmaI fragments that hybridized with a lytA probe in lysogenic strains, with one of these bands corresponding to the gene coding for the major host autolysin and the other corresponding to the gene present in the temperate phage coding for the phage lysin (16). Nevertheless, in these reports there was no indication of phage purification, which is a limiting factor to establish any relationship among these temperate phages. To investigate a precise biological role that provides clues about the ubiquitous presence of phages in clinical isolates of virulent pneumococci, we decided to investigate several clones of the multiresistant 23F serotype and tried to purify one phage (MM1) from the Spain23F-1 clone. This clone was selected because strains resistant to penicillin, tetracycline, and chloramphenicol and variably resistant to erythromycin have been spread worldwide (35). The genome of phage MM1 appears to be quite similar, but not identical, to that of phage HB-746 isolated from strain 746, a type 8 pneumococcus. In fact, from preliminary sequence data of the HB-746 genome obtained from PhageTech, Inc. (Montreal, Canada), we know that the DNA fragment encompassing the integrase gene of MM1 does not display significant similarity to the HB-746 genome, although at the protein level, the corresponding ORF of HB-746 has 41% identity with the integrase of MM1. These two phages also share the peculiar characteristic of having DNA-protein complexes that are capable of becoming integrated into the host chromosome. The determination of the precise biological role of the proteins covalently linked to the DNAs of these pneumococcal temperate phages still remains a mystery. We have previously suggested a protective role for incoming DNA after the phage has injected the DNA into the host bacteria or a function during integration, assuming that the bound protein has retained an enzymatic activity (47). Similar mechanisms have been postulated to explain the integrative process of the T-DNA molecules generated in the Agrobacterium tumefaciens system for transferring the DNA into the genome of the host plant (28). In the case of these pneumococcal phages there must be an additional mechanism allowing the phage genome to regain the terminal protein when these peculiar temperate phages enter the lytic cycle. Furthermore, the study of the genome of MM1 might provide a reasonable way to investigate genes involved in the mechanism leading to the programmed loss and recovery of the terminal protein when shifting from the lytic to the lysogenic cycle and vice versa.
In work on the molecular characterization of MM1, we have analyzed the genetic determinants required for phage DNA integration. Temperate bacteriophages integrate their DNAs into the host chromosome by a site-specific recombination process following the Campbell model (9). Two specific attachment sites, one on the bacterial chromosome (attB) and the other on the phage genome (attP), are recombined by the activity of a phage-encoded integrase. There are well-characterized examples of site-specific recombination in gram-negative bacteria, especially that of bacteriophage λ (30). Although the integration system of phages of gram-positive bacteria is less well documented, data are available for several phages of S. aureus (11, 31, 59), for bacteriophage T12 of Streptococcus pyogenes (36), for the actinophage RP3 (18), and for several lactic acid bacterial phages (8, 14, 42, 57). We have now identified the attP-containing phage DNA, the bacterial attachment site attB, and the host-phage junctions attL and attR of the MM1 prophage. These sequences share a 15-bp identity region, a typical size for the cores of other phages that integrate through a site-specific recombination mechanism. The attP region, 313 bp long, is located between two ORFs, coding for the integrase and the lytic enzyme, that are convergently transcribed. The attP site has several traits in common with other attP sites, such as a high percent A+T (67%) (although not significantly higher than that of the host DNA) (49) and a complex array of direct and inverted repeats. These repeat sequences have been postulated to be the binding or recognition sites for phage-encoded proteins such as integrase or excisionase or for host factors analogous to the E. coli integration host factor (12), although precise assignment of these sites in phage MM1 must await the purification of these proteins. From the nucleotide sequence of the MM1 integration region we deduced an ORF encoding a polypeptide of 375 amino acids located adjacent to attP and transcribed towards it. Evidence suggesting that this ORF encoded the MM1 integrase came from the location, size, and similarity to site-specific recombinases of the λ integrase family. This was confirmed by the construction of a functional vector promoting site-specific integration. This report represents the first demonstration of such a mechanism carried out for a pneumococcal phage, since with the other two temperate phages previously studied, we did not succeed in accurately sequencing the attachment sites (13, 45).
The elucidation of the determinants required for the integration of MM1 has allowed the construction of a site-specific integration vector for S. pneumoniae. Although this gram-positive bacterium possesses a remarkable and well-characterized mechanism for incorporating foreign DNA, because of its natural competence, a site-specific vector like pIAPU1 described here presents some new advantages over the alternative methods developed to take up genetic material. As an example, this vector will help in the specific insertion of any heterologous gene into the attB site in a single copy, which could eventually be useful for gene expression studies with this important human pathogen.
We do not know whether orf116, which precedes the integrase gene, could function as an excisionase, since the typical traits of this kind of protein have not been identified in the orf116 product. On the other hand, 208 bp downstream of the attP core region (Fig. 4) we could locate the 3′ end of the lytic gene, which, together with the holin gene, forms part of the lytic cassette of this phage (unpublished observations). Concerning attB, it is interesting that MM1 integrates into a gene of unknown function with the peculiarity that the stop codon of this gene is included in the core region of attP, which implies that lysogenization of the host does not inactivate this gene.
The unexpected similarities found between phage MM1 and the HB family of phages prompted us to undertake a broader examination of the presence of this phage among different multiresistant isolates belonging to other capsular serotypes. From the examples presented here we can conclude that an MM1-like phage appears to be spread among several of the most abundant pneumococcal strains studied. Moreover, the presence in strains SSISP33C/1, CSUB 3409, and CSUB 4086 of another phage(s) very different from MM1 can be expected (Fig. 8). On the other hand, we do not favor the idea, suggested by Bernheimer (4, 5), that lysogeny is associated with only certain pneumococcal capsular types, since in a more general context, we have found that the presence of fully functional defective or remnant prophages in the chromosome is indeed a general trait among pneumococcal isolates, including some atypical pneumococci that are deoxycholate insensitive using a classical taxonomic test (13). Botstein has proposed that evolution of lambdoid phages happens by exchange of genes organized in functional modules (7). The biological and functional similarities between phage-encoded enzymes and the host pneumococcal amidase have been well documented (46). One of the highest levels of identity between bacterial and phage genes (87.1%) was observed when comparing the host lytA and hbl3 from the HB-3 temperate phage (46). This nucleotide sequence similarity allows recombination between both genomes that permits restructuring and evolutionary adaptation in both organisms. In addition, it has been established that lactococcal phages are able to acquire pieces of the host chromosome (39).
The high incidence of lysogeny among clinical strains has raised the possibility that part of the exchange of genetic information found in pneumococcus in vivo is carried out through transduction or is facilitated by phage functions (44). Actually, it is well documented that some temperate phages bear virulence-related genes in many bacterial systems (reference 27 and references therein), and a process similar to transduction but requiring competence development was described previously for pneumococci (43). The interchange of capsular polysaccharides has been revealed to be a quite common process in nature as a mechanism to improve serotype replacement, which can provide to pathogenic species like pneumococcus an excellent and profitable way to escape from a vaccine prepared against a limited number of capsular types, like the newly developed heptavalent vaccine. The possibility of in vivo DNA exchange by a transfection-like mechanism is attractive, since the higher efficiency of pseudotransduction of large fragments of DNA compared to transformation could give this mechanism an advantage over transformation for the observed in vitro capsular switch events between the cassette-like organization of the genes coding for capsules (35). Phages capable of lysing unencapsulated (nonlysogenic) indicator strains have been readily isolated from carriers or patients (34, 48, 55), and the high incidence of temperate phage carriage in S. pneumoniae could strongly influence the structure of natural populations of pneumococci in their ecological niche. Incidentally, it has been suggested that defense against phage infection may be another selective pressure that facilitates the structure and maintenance of capsular polysaccharide in this species (44). In addition to all of these relevant biological roles of pneumococcal phages, the observation that a high proportion of isolates with clinical relevance carry phages (e.g., Spain23F-1) also invites speculation that these phages might contribute to transfer of antibiotic resistance markers between strains through a generalized transduction-like mechanism. We now have available the possibility of sequencing the complete genomes of temperate phages isolated and purified from very relevant pneumococcal strains. This approach together with the development of an integrative phage vector will be an important tool to facilitate the study of potential virulence genes.
ACKNOWLEDGMENTS
We acknowledge E. García for encouragement and many fruitful discussions and J. L. García for critical reading of the manuscript. We thank M. Carrasco and E. Cano for technical assistance. V. Muñoz, M. Fontenla, and A. Hurtado are also acknowledged for the art work.
This work was supported by grants from Dirección General de Investigación Científica y Técnica (PB96-0809) and the Comunidad Autónoma de Madrid (08.2/0014.2/98). E.G. was the recipient of a Marie Curie Research Training Grant.
REFERENCES
- 1.Arrecubieta C, García E, López R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 2.Arrecubieta C, López R, García E. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol. 1994;176:6375–6383. doi: 10.1128/jb.176.20.6375-6383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrow P A, Soothill J S. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol. 1997;5:268–271. doi: 10.1016/S0966-842X(97)01054-8. [DOI] [PubMed] [Google Scholar]
- 4.Bernheimer H P. Lysogeny in pneumococci freshly isolated from man. Science. 1977;195:66–68. doi: 10.1126/science.12565. [DOI] [PubMed] [Google Scholar]
- 5.Bernheimer H P. Lysogenic pneumococci and their bacteriophages. J Bacteriol. 1979;138:618–624. doi: 10.1128/jb.138.2.618-624.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botstein D. A theory of modular evolution for bacteriophages. Ann NY Acad Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 8.Bruttin A, Foley S, Brüssow H. The site-specific integration system of the temperate Streptococcus thermophilus bacteriophage φSfi21. Virology. 1997;237:148–158. doi: 10.1006/viro.1997.8769. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 11.Coleman C, Knights J, Russel R, Shanley D, Birkbeck T H, Dougan G, Charles I. Insertional inactivation of the Staphylococcus aureus β-toxin by bacteriophage phi-13 occurs by site- and orientation-specific integration of the phi-13 genome. Mol Microbiol. 1991;5:933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 12.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 13.Díaz E, López R, García J L. EJ-1, a temperate bacteriophage of Streptococcus pneumoniae with a Myoviridae morphotype. J Bacteriol. 1992;174:5516–5525. doi: 10.1128/jb.174.17.5516-5525.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont L, Boizet-Bonhoure B, Coddeville M, Auvray F, Rizenthaler P. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J Bacteriol. 1995;174:5516–5525. doi: 10.1128/jb.177.3.586-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esposito D, Scocca J J. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenoll A, Martínez-Suárez J V, Muñoz R, Casal J, García J L. Identification of atypical strains of Streptococcus pneumoniae by a specific DNA probe. Eur J Clin Microbiol Infect Dis. 1990;9:396–401. doi: 10.1007/BF01979468. [DOI] [PubMed] [Google Scholar]
- 17.Fenoll A, Muñoz R, García E, de la Campa A G. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the F0 complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol Microbiol. 1994;12:587–598. doi: 10.1111/j.1365-2958.1994.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel K, Schmid H, Schmidt U, Rausch H. The actinophage RP3 DNA integrates site-specifically into the putative tRNAArg (AGG) gene of Streptomyces rimosus. Nucleic Acids Res. 1995;23:58–63. doi: 10.1093/nar/23.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García E, García J L, García P, Arrarás A, Sánchez-Puelles J M, López R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci USA. 1988;85:914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García E, García P, López R. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniae type 3. Mol Gen Genet. 1993;239:188–195. doi: 10.1007/BF00281617. [DOI] [PubMed] [Google Scholar]
- 21.García E, Gómez A, Ronda C, Escarmís C, López R. Pneumococcal bacteriophage Cp-1 contains a protein bound to the 5′ termini of its DNA. Virology. 1983;128:92–104. doi: 10.1016/0042-6822(83)90321-5. [DOI] [PubMed] [Google Scholar]
- 22.García P, García J L, García E, López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 23.García P, González M P, García E, García J L, López R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol Microbiol. 1999;33:128–138. doi: 10.1046/j.1365-2958.1999.01455.x. [DOI] [PubMed] [Google Scholar]
- 24.García P, Martín A C, López R. Bacteriophages of Streptococcus pneumoniae: a molecular approach. Microb Drug Resist. 1997;3:165–176. doi: 10.1089/mdr.1997.3.165. [DOI] [PubMed] [Google Scholar]
- 25.Gasc A M, Kauc L, Barraille P, Sicard M, Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991;173:7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakenbeck R, Grebe T, Zănher D, Stock J B. β-Lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol Microbiol. 1999;33:673–678. doi: 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 27.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrera-Estrella A, Chen Z M, Van Montagu M, Wang K. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 1988;7:4055–4062. doi: 10.1002/j.1460-2075.1988.tb03299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in Pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 30.Landy A. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 31.Lee C Y, Iandolo J J. Structural analysis of staphylococcal bacteriophage φ11 attachment sites. J Bacteriol. 1988;170:2409–2411. doi: 10.1128/jb.170.5.2409-2411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H H, Tomasz A. Penicillin tolerance in multiply drug-resistant natural isolates of Streptococcus pneumoniae. J Infect Dis. 1985;152:365–372. doi: 10.1093/infdis/152.2.365. [DOI] [PubMed] [Google Scholar]
- 33.Longchamp P F, Mauël C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 34.McDonnell M, Ronda-Laín C, Tomasz A. “Diplophage”: a bacteriophage of Diplococcus pneumoniae. Virology. 1975;63:577–582. doi: 10.1016/0042-6822(75)90329-3. [DOI] [PubMed] [Google Scholar]
- 35.McGee L K, Klugman K P, Tomasz A. Serotypes and clones of antibiotic-resistant pneumococci. In: Tomasz A, editor. Streptococcus pneumoniae. Molecular biology and mechanism of disease. Larchmont, N.Y: Mary Ann Liebert, Inc. Pub.; 2000. pp. 375–379. [Google Scholar]
- 36.McShan W M, Tang Y F, Ferretti J J. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol Microbiol. 1997;23:719–728. doi: 10.1046/j.1365-2958.1997.2591616.x. [DOI] [PubMed] [Google Scholar]
- 37.Merril C R, Biswas B, Carlton R, Jensen N C, Creed G J, Zullo S, Adhya S. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA. 1996;93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao E A, Miller S M. Bacteriophages in the evolution of pathogen-host interaction. Proc Natl Acad Sci USA. 1999;96:9452–9454. doi: 10.1073/pnas.96.17.9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moineau S, Pandian S, Klaenhammer T R. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol. 1994;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 41.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen A, Josephsen J, Johnsen M G. TPW22, a lactococcal temperate phage with a site-specific integrase closely related to Streptococcus thermophilus phage integrases. J Bacteriol. 1999;181:7034–7042. doi: 10.1128/jb.181.22.7034-7042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter R D, Shoemaker N B, Ramper G, Guild W R. Bacteriophage-associated gene transfer in pneumococcus: transduction or pseudotransduction? J Bacteriol. 1979;137:556–567. doi: 10.1128/jb.137.1.556-567.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez M, Severina E, Tomasz A. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol. 1999;181:3618–3625. doi: 10.1128/jb.181.12.3618-3625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero A, López R, García P. The insertion site of the temperate phage HB-746 is located near the phage remnant in the pneumococcal host chromosome. J Virol. 1992;66:2860–2864. doi: 10.1128/jvi.66.5.2860-2864.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero A, López R, García P. Sequence of the Streptococcus pneumoniae bacteriophage HB-3 amidase reveals high homology with the major host autolysin. J Bacteriol. 1990;172:5064–5070. doi: 10.1128/jb.172.9.5064-5070.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero A, López R, Lurz R, García P. Temperate bacteriophages of Streptococcus pneumoniae that contain protein covalently linked to the 5′ ends of their DNA. J Virol. 1990;64:5149–5155. doi: 10.1128/jvi.64.10.5149-5155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronda C, López R, García E. Isolation and characterization of a new bacteriophage, Cp-1, infecting Streptococcus pneumoniae. J Virol. 1981;40:551–559. doi: 10.1128/jvi.40.2.551-559.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotta J. Pyogenic hemolytic streptococci. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams and Wilkins Co.; 1986. pp. 1047–1053. [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 51.Severina E, Ramirez M, Tomasz A. Prophage carriage as a molecular epidemiological marker in Streptococcus pneumoniae. J Clin Microbiol. 1999;37:3308–3315. doi: 10.1128/jcm.37.10.3308-3315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 53.Stassi D L, Lopez P, Espinosa M, Lacks S A. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1981;78:7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thornsberry C, Ogilvie P, Kahn J, Mauriz Y the Laboratory Investigator Group. Surveillance of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States in 1996–1997 respiratory season. Diagn Microbiol Infect Dis. 1997;29:249–257. doi: 10.1016/s0732-8893(97)00195-8. [DOI] [PubMed] [Google Scholar]
- 55.Tiraby J G, Tiraby E, Fox M S. Pneumococcal bacteriophages. Virology. 1975;68:566–569. doi: 10.1016/0042-6822(75)90300-1. [DOI] [PubMed] [Google Scholar]
- 56.Tomasz A. Requirement for protein synthesis during induction of competence. J Bacteriol. 1970;101:860–871. doi: 10.1128/jb.101.3.860-871.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Guchte M, Daly C, Fitzgerald G F, Arendt E K. Identification of int and attP on the genome of lactococcal bacteriophage Tuc2009 and their use for site-specific plasmid integration in the chromosome of Tuc2009-resistant Lactococcus lactis MG1363. Appl Environ Microbiol. 1994;60:2324–2329. doi: 10.1128/aem.60.7.2324-2329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 59.Ye Z H, Lee C Y. Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J Bacteriol. 1989;171:4146–4153. doi: 10.1128/jb.171.8.4146-4153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]