Abstract
Although the isolation and counting of small extracellular vesicles (sEVs) are essential steps in sEV research, an integrated method with scalability and efficiency has not been developed. Here, we present a scalable and ready‐to‐use extracellular vesicle (EV) isolation and counting system (EVics) that simultaneously allows isolation and counting in one system. This novel system consists of (i) EVi, a simultaneous tandem tangential flow filtration (TFF)‐based EV isolation component by applying two different pore‐size TFF filters, and (ii) EVc, an EV counting component using light scattering that captures a large field‐of‐view (FOV). EVi efficiently isolated 50–200 nm‐size sEVs from 15 µL to 2 L samples, outperforming the current state‐of‐the‐art devices in purity and speed. EVc with a large FOV efficiently counted isolated sEVs. EVics enabled early observations of sEV secretion in various cell lines and reduced the cost of evaluating the inhibitory effect of sEV inhibitors by 20‐fold. Using EVics, sEVs concentrations and sEV PD‐L1 were monitored in a 23‐day cancer mouse model, and 160 clinical samples were prepared and successfully applied to diagnosis. These results demonstrate that EVics could become an innovative system for novel findings in basic and applied studies in sEV research.
Keywords: counting, isolation, light scattering, small extracellular vesicle, tangential flow filtration
1. INTRODUCTION
Small extracellular vesicles (sEVs) are lipid‐bilayered vesicles with diameters of approximately 50−200 nm that are secreted by most human cells and circulate in the body (Colombo et al., 2014; Kalluri & LeBleu, 2020; Théry et al., 2002). Numerous studies have focused on sEV biogenesis (Costa‐Silva et al., 2015; Greening et al., 2015; Kaiser, 2016), diagnosis (Melo et al., 2015; Moon et al., 2016; Thakur et al., 2014) and therapeutic applications (Chen et al., 2018; Im et al., 2019; Lee et al., 2022; Okoye et al., 2014). Efficient sEV isolation and counting are critical prerequisites for these studies and have a significant impact on molecular analyses of downstream targets (Théry et al., 2018; Witwer et al., 2013).
However, currently available isolation and counting methods have limitations that restrict their practical uses. Ultracentrifugation (UC) has scalability and pelleting issues, despite its wide usage (Gardiner et al., 2016; Royo et al., 2020). Especially, pelleting of sEVs can lead to misinterpretation of their physical/biological characteristics (Kalluri & LeBleu, 2020; Linares et al., 2015; Witwer et al., 2013). Recently, to preserve the characteristics of sEV, gentle isolation techniques using size‐exclusion chromatography (SEC) have been employed, enabling a more accurate and efficient sEV isolation (Gardiner et al., 2016; Royo et al., 2020; Tóth et al., 2021). However, SEC also has a limited capacity, which impedes high‐throughput or large‐scale sEV isolation (Baranyai et al., 2015; Burgess, 2018; Lobb et al., 2015). In addition, existing techniques, including precipitation, immunoaffinity, and microfluidics‐based methods, exhibit limited scalability (Chen et al., 2021; Gurunathan et al., 2019; Li et al., 2017; Liangsupree et al., 2021; Théry et al., 2006). However, membrane‐based filtration enables scalable sEV isolation through its flexible surface area manipulation (Konoshenko et al., 2018). Notably, tangential flow filtration (TFF) provides continuous filtration, clog prevention, and improved scalability. TFF is primarily used for medium‐ and large‐scale sEV isolation, especially in the mass production of industry‐scale and clinical‐grade sEVs in good manufacturing practice (GMP) facilities, but rarely used in a laboratory‐scale (Ahn et al., 2022; Heinemann et al., 2014; Heinemann et al., 2014; Watson et al., 2018). Thus, leveraging and optimizing the scalability of TFF has the potential to develop a new system applicable across all scales.
In sEV counting, nanoparticle tracking analysis (NTA) is a popular method of choice (Gardiner et al., 2016; Royo et al., 2020). NTA analyzes light scattered particles in a sample using laser illumination and camera capture to accurately determine particle size and concentration (Gardiner et al., 2013; Koritzinsky et al., 2017). The selection of the field‐of‐view (FOV) setting in NTA determines the particle size detection area, which affects analysis accuracy and reliability. A larger FOV detects more EV particles by capturing a wider area, whereas a smaller FOV provides better resolution for accurate size measurement (Kashkanova et al., 2022). Current NTA instruments are designed to provide acceptable measurements of both particle size and concentration. As a result, the FOV is a little bit compromised, resulting in inefficient counting. Consequently, a relatively high concentration of samples is required to measure the size and concentration, resulting in the consumption of a significant number of EVs (Bell et al., 2012; El Baradie et al., 2020; Filipe et al., 2010; Tian et al., 2016). Therefore, using a large FOV setting for sEV counting would reduce sample consumption, despite less accurate size measurement.
We hypothesized that coupling isolation and counting into one system could improve workflow efficiency, experiment repeatability, and research and application productivity with the conservation of valuable samples. Therefore, based on these assumptions, in the present study, we aimed to couple sEV isolation and counting using the TFF principle and FOV adjustments in the light scattering technique.
Here we present an EV isolation and counting system (EVics) that isolates sEVs within a specific size range using two pore‐size TFF filters and counts them using light scattering with a large FOV (Figure 1). The isolation component (EVi) enabled the preparation of highly purified sEVs from small to large volumes. The counting component (EVc) allowed particle counting using a small amount of sample. Moreover, we extensively tested EVics in practical applications in various systems of cells (investigation of the secretion dynamics of sEVs at early time points and screening for sEV secretion inhibitors), animal models (monitoring sEV concentration), and clinical samples (sEV PD‐L1 expression levels in individual subjects, n = 160). In summary, we present the utility of EVics as a scalable and ready‐to‐use sEV preparation system for basic research, therapeutic development, and diagnostic applications.
FIGURE 1.
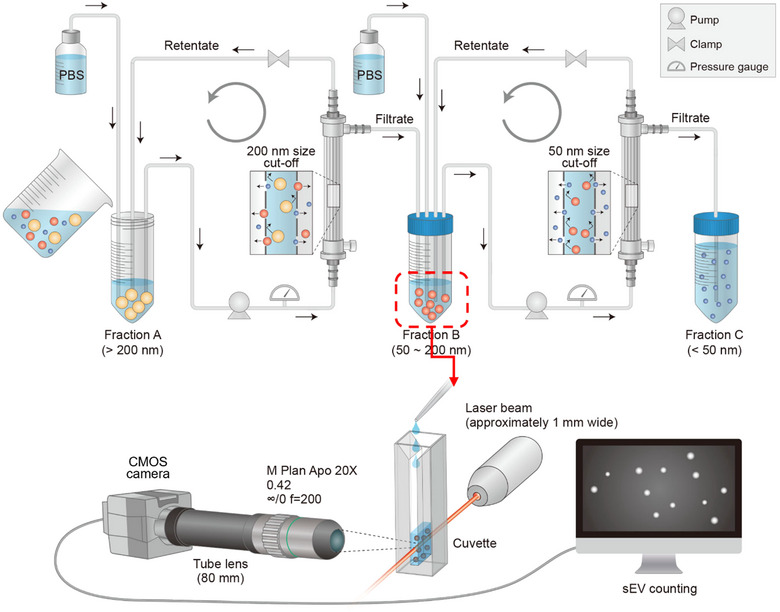
Proposed sEV preparation system: EVics. Schematic design of the EVics. Particles of 50–200 nm in size isolated from simultaneous tandem TFF (EVi) are immediately transferred to EVc for counting. Ready‐to‐use sEV samples with known concentrations can be used for subsequent experiments.
2. MATERIALS AND METHODS
2.1. Cell lines
All human cancer cells (American Type Culture Collection) were grown at 37°C under a humidified atmosphere of 5% CO2 and 95% air. MDA‐MB231, MCF7, and A375 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 1% antibiotic–antimycotic solution. MCF10A cells were grown in mammary epithelial cell growth medium (MEGM Lonza, Basel, Switzerland) with 5% FBS, 52 µg mL−1 bovine pituitary extract, 0.5 µg mL−1 hydrocortisone, 10 ng mL−1 EGF, and 5 µg mL−1 insulin. SK‐MEL‐28 cells were cultured in Minimum Essential Medium with Earle's Balanced Salts (MEM/EBSS; HyClone, Logan, UT, USA). A549 cells were cultured in Roswell Park Memorial Institute (RPMI) with 10% FBS and 1% antibiotic–antimycotic solution. All cell lines were tested for mycoplasma contamination using polymerase chain reaction (PCR).
2.2. Isolation of sEVs from conditioned media
Before the isolation of sEVs, clarified conditioned media (CCM) were first generated via serial centrifugation at 300×g at 4°C for 5 min and 2500×g at 4°C for 20 min to remove cells and debris.
2.3. Ultracentrifugation
CCM were centrifuged at 10,000×g at 4°C for 30 min to remove large vesicles and apoptotic bodies. Next, the supernatants were passed through a 0.2 µm membrane syringe filter (#S6534‐FMOSK; Sartorius, Göttingen, Germany) or 0.2 µm bottle top vacuum filter (#43118, Corning Inc., Corning, NY, USA). Then, the filtrate was centrifuged at 120,000×g at 4°C for 90 min (SW32‐Ti rotor, Beckman Optima XE‐100 Ultracentrifuge; Beckman Coulter, Brea, CA, USA) to pellet sEVs. The sEV pellets were rinsed with phosphate‐buffered saline (PBS) and centrifuged again at 120,000×g at 4°C for 90 min. Purified sEV pellets were resuspended in PBS for further analysis.
2.4. EVi
EVi was used to isolate sEVs from CCM. To purify sEVs from small volumes of CCM (2−100 mL), small scale TFF filters (200 nm [surface area (SA) of 20 cm2, C02‐P20U‐05‐N] and 50 nm [SA 28 cm2, C02‐S05U‐05‐N]; Spectrum Labs, San Fransico, CA, USA) were used. For large volumes (100 mL−2 L), large scale TFF filters (200 nm [SA 140 cm2, D02‐P20U‐05‐N] and 50 nm [SA 190 cm2, D02‐S05U‐05‐N]; Spectrum Labs) were used. The two different pore size TFF modules were equipped with two independent peristaltic pumps and tubing circuits. The entire system was equilibrated with PBS, and the CCM were loaded into a sterile feed reservoir. The CCM were continuously circulated through the TFF filter module and processed at 15−35 mL min−1 for optimization. Throughout the process, the pressure in the TFF device was maintained below 20 psi. For large volumes (>100 mL), large filters were operated at a flow rate corresponding to the optimized shear rate (the 200 nm filter at 3500 s−1 (116 mL min−1) and the 50 nm filter at 3800 s−1 (170 mL min−1). We minimized the final volume by adjusting the filter position and tube length for maximum concentration. The final volume of the product concentrated was approximately 700 µL for the small scale TFF filters and approximately 9 mL for the large scale TFF filters.
2.5. Conventional dead‐end filtration (DEF)/TFF process
CCM were passed through a 0.2 µm membrane syringe filter (#S6534‐FMOSK, Sartorius) or 0.22 µm bottle top vacuum filter (#43118, Corning) according to the sample volume. Then, the filtrate was circulated through the 50 nm pore size TFF module. The samples were concentrated to approximately 1 mL and diafiltrated 10 times with PBS. Finally, the sample was concentrated to a volume of ∼700 µL and analyzed.
2.6. SEC: qEV
A qEV2/35 nm SEC column (Izon) was prepared by washing with 10 mL of PBS. CCM (30 mL) were preconcentrated to a volume of 2 mL using an Amicon Ultra 15 centrifugal filter. The preconcentrated CCM were loaded onto the qEV column and collected every 2 mL via gravitational flow. Fractions 1 to 7, corresponding to the void volume (approximately 14 mL), were discarded, and the sEVs fractions (8 to 11) were pooled and analyzed.
2.7. Exoquick precipitation (EQ)
ExoQuick precipitation (System Biosciences, Palo Alto, CA, USA) was performed according to the manufacturer's instructions. Briefly, 30 mL of CCM were mixed with the ExoQuick‐TC solution (7.5 mL) by inverting the tube several times. The sample was incubated overnight at 4°C and then centrifuged at 1500×g for 30 min to remove the supernatant. The supernatant was discarded, and the pellets were resuspended in 1 mL of PBS.
2.8. Isolation of sEVs from plasma
Plasma samples from human or mouse were centrifuged at 2500×g for 15 min to remove debris and again at 10,000×g for 30 min to pellet and remove large particles. Then, the plasma was diluted to 2 mL, and the sEVs were isolated as described in the previous EVi section.
2.9. Transmission electron microscope (TEM) analysis
TEM analysis was performed to detect the sEVs and nanobeads (NIST Traceable, Thermo Fisher Scientific). Samples were fixed with 2% paraformaldehyde (PFA) for 5 min and deposited on a pure carbon‐coated grid for 2 min. After staining with 2% uranyl acetate solution for 1 min, the grid was washed and dried at room temperature. The grid was imaged at a range of magnification from ×2k to ×20k depending on the sample, using a transmission electron microscope (HT7700, Hitachi) operated at 120 kV.
2.10. NTA
The size distribution and concentration of sEVs were measured using a violet laser (405 nm; Nanosight LM10; Malvern Panalytical, Malvern, UK). Each sample was either undiluted or diluted (1:1000) in PBS to the measurement range (∼200 particles per frame) depending on the initial sample concentration. The software (Nanosight version 3.2) settings for the analysis were as follows: camera level, 9−10; detection threshold, 2−3; and measurement time, 30 s (30 frames s−1 [fps]).
2.11. Determination of sEV protein concentration
The concentration of sEV proteins was determined using a Bicinchoninic acid (BCA) (#23225; Thermo Fisher Scientific, Waltham, MA, USA) or microBCA assay (#23235; Thermo Fisher Scientific, Waltham, MA, USA). To measure protein concentrations, sEVs were extracted using 1× RIPA buffer (#50−188; Merck Millipore, Burlington, MA, USA). BCA assays or microBCA assays were performed according to the manufacturer's instructions.
2.12. Western blot analysis
Equal amounts (or equal volumes of samples) of cellular or sEV proteins were separated using 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (GE Healthcare, Chicago, IL, USA). The membranes were blocked for 2 h at room temperature with 5% non‐fat dry milk in Tris‐Buffered Saline with 0.1 % Tween 20 (TBS‐T). and then the membrane was probed with primary antibodies overnight at 4°C. The following antibodies were used for western blot analysis: anti‐CD63 (ab68418; Abcam, Cambridge, UK), anti‐TSG101 (ab125011; Abcam), anti‐Flotillin‐1 (3253; Cell Signaling Technology, Danvers, NA, USA) and anti‐ALIX (2171S; Cell Signaling Technology). All primary antibodies were used at a 1:1000 dilution. The membranes were washed three times with TBS‐T for 15 min and incubated for 2 h at room temperature with a horseradish peroxidase‐linked secondary antibody (7076S or 7074S; Cell Signaling Technology). Images were visualized using a FUSION Solo S imaging system (Vilber) with enhanced chemiluminescence detection reagents (#34580; Thermo Fisher Scientific).
2.13. Cellular assay
Transwell migration assays were performed using a Costar Transwell system (CLS3364; Corning). Briefly, MCF7 cells (5 × 104 cells) were suspended in 200 µL serum‐free medium and seeded in the upper insert chamber, and 600 µL 1% FBS‐containing medium was added to the lower chamber. Four hours after the MCF7 cells were seeded, the media in both the upper and lower chambers were removed. Cells that had migrated into the lower chamber through the 8 µm pore membrane were stained with crystal violet solution and then visualized using a microscope (×4 magnification). Transwell invasion assays were also performed using the Costar Transwell system. Briefly, MDA‐MB231 cells (2 × 103 cells) were suspended in 200 µL serum‐free medium and seeded in the upper insert chamber, and 500 µL medium was added to the lower chamber. Four hours after the MDA‐MB231 cells were seeded, the media in both the upper and lower chambers were removed. Cells that had invaded the lower chamber through the 8 µm pore membrane were stained with crystal violet solution and then visualized using a microscope (×4 magnification).
2.14. Measurement setup of EVc
A low‐coherence light source with a wavelength of 661 nm (COHERENT) was used, and the power of the laser was 30 mW. Laser passes through the cuvette and scatter the particles. The scattered light was imaged through a complementary metal‐oxide‐semiconductor (CMOS) camera (Allied Vision, Alvium 1800U ‐ 319 m) positioned at a 90 degree angle to the laser in the lowest‐noise mode (12‐bit) with 2.1 e‐ temporal dark noise. The infinity corrected 20× lens with 0.42 numerical aperture and ±1.6 depth of focus (Mitutoyo) was mounted on a camera equipped with extension tube. FOV was by changing the length of extension tube. We typically use a FOV of 1544 pixels × 2064 pixels, equivalent to 670 × 900 µm2. Images were taken in the absence of significant particle movement (fluid flow, not a Brownian motion) after the sample was loaded into the cuvette because if the particle movement is large, traces of the particle movement appear in the image following the exposure time. Images were captured following optimized conditions: 50 ms exposure time, 1.5 gamma, 15 gain. By taking six images with an interval of 5 s, we extracted particles per frame using ImageJ software (https://imagej.net/) and calculated the concentration of the sample.
2.15. Animal study
In the syngeneic tumor models, EMT6 breast tumor cells (2 × 105 cells in 100 µL of PBS with 50% Matrigel) were orthotopically injected into the left fat pad of female BALB/c (BALB/cAnNCrl) mice aged 6−7 weeks, purchased from Orient Bio (Seongnam, Korea). The mice were bred and maintained in a specific pathogen‐free barrier facility. Macitentan (MAC, an sEV inhibitor, purchased from MedChemExpress, Monmouth Junction, NJ, USA) was then orally administered at 50 mg kg−1 day −1 for 5−23 days. Tumor volumes were recorded until they reached the maximum value defined by the Institutional Animal Care and Use Committee (IACUC) guidelines. When the tumors reached an average size of 50−100 mm3, the mice were randomized into groups with a comparable distribution of starting tumor volumes. The mice were euthanized when their tumor volume reached 1500 mm3. Every 3−4 days, tumor growth was measured using calipers, and blood (∼15 µL) was collected using a capillary tube via retro‐orbital blood collection. Tumor size was estimated using the following equation: volume (cm3) = width2 × length × 0.5. Plasma was prepared from the blood samples for sEV isolation, then sEVs were purified and counted using EVics, and plasma sEV PD‐L1 (anti‐PD‐L1, 14‐5983‐82; eBioscience, San Diego, CA USA) levels were measured using Enzyme‐linked immunosorbent assay (ELISA). All animal experiments were performed according to the protocol approved by the Kyungpook National University IACUC (approval number: 2020‐0016).
2.16. Clinical samples
Plasma was obtained from freshly drawn blood by centrifugation at 2500 × g for 15 min using a separate plasma tube. Then, sEVs were isolated and counted using EVics to measure plasma sEV concentrations. This study was approved by the Institutional Review Board (IRB, approval number: 2018‐01‐033) of Kyungpook National University Medical Center, where all clinical samples were collected. Informed consent was obtained from all the participants.
2.17. ELISA
For quantification of proteins on sEVs, 96‐well plates were coated with capture antibodies against various markers at 100 µL well−1 in 0.2 mol L−1 sodium phosphate buffer (pH 6.5) and incubated overnight at 4°C. The following capture antibodies were used: anti‐EGFR (1:500, MA5‐13269; Invitrogen, Carlsbad, CA, USA), anti‐EpCAM (1:500; 14‐9326‐82, eBioscience), anti‐PD‐L1 (1:500, 14‐5983‐82; eBioscience) and anti‐CD63 (1:500; NBP2‐42225; Novus Biologicals, Littleton, CO, USA). The plates were blocked for 1 h at 37°C with 200 mL of PBS containing 2% bovine serum albumin (BSA) and washed three times with PBS containing 0.05% Tween 20 (PBS‐T). The sEV samples (100 µL) were added to the plates and incubated for 2 h at room temperature. Following three washes with PBS‐T, a biotinylated detection antibody cocktail was added to each well (100 µL well−1) and incubated for 1 h at room temperature. The following antibodies for the detection antibody cocktail were used: anti‐CD63 (ab8219; Abcam), anti‐CD9 (14‐0098‐82; eBioscience), and anti‐CD81 (ab7955; Abcam). After three washes with PBS‐T, streptavidin‐horseradish peroxidase was added to each well, and the plate was incubated for 1 h. The reagent mixture (3,30,5,50‐tetramethylbenzidine‐containing hydrogen peroxide) was added to each well, the reaction was stopped with 1 mol L−1 phosphoric acid, and optical density was measured at 450 nm using an automated plate reader.
2.18. Statistical analysis
Statistical analyses were performed using GraphPad Prism version 9.5.1 (GraphPad Software). Unpaired two‐tailed Student's t‐tests were used to compare two sets of data. Error bars in graphs represent means ± standard deviations (SDs). Tumor volume and ELISA data are presented as means ± standard errors of mean (SEMs). All in vitro experiments were performed in triplicate unless otherwise stated. For all statistical tests, p‐values less than 0.05 were considered statistical significance; *, **, ***, and **** represent p values less than 0.05, 0.01, 0.001, and 0.0001, respectively.
3. RESULTS
3.1. System design and optimization of EVi
Our study aimed to develop a scalable, ready‐to‐use sEV preparation and counting system with low consumption of valuable EV samples. To achieve scalable and effective sEV isolation, we chose TFF membrane filtration, which allows EV particle isolation based on their sizes. The conventional TFF process usually involves a preliminary step of DEF to remove large particles (Comşa et al., 2015; Heinemann et al., 2014; Lee et al., 2020; Lee et al., 2020; Watson et al., 2018; Woo et al., 2020). However, DEF has been reported to have some limitations, including membrane clogging and the risk of external contamination. To overcome these critical limitations, we designed an improved system by replacing the DEF with TFF. We configured a simultaneous tandem TFF system (EVi) by running two different pore‐size (50 and 200 nm) TFF filters to simultaneously remove large particles and soluble proteins, thereby obtaining sEVs within the 50−200 nm size range (Figure S1).
To optimize the process variables, small scale TFF filters were used to establish the flow rate (shear rate) and diafiltration volume (DV). The optimal flow rate is determined based on the membrane diameter and the number of fibers in the filter. Thus, in this study, we referred to the flow rate (L min−1)‐shear (s−1) information provided by the filter manufacturer and experimentally identified the optimal conditions. We used BSA and 100 and 300 nm standard nanobeads. In this pilot experiment, the optimal conditions were selected by considering a balance between the yield of 100 nm beads, the exclusion of 300 nm beads, and the efficiency of BSA removal. Before optimizing the variables for EVi, we optimized each filter individually. First, to optimize the 200 nm pore size TFF filter, we filtered 100 and 300 nm beads suspended in 20 mL of DMEM at three different flow rates (15‐, 25‐, and 35‐mL min−1) and three different DVs (10, 20, and 30 DVs). The filtrate (fraction B in Figure 1) was analyzed using NTA (Figure 2a). Subsequently, for the 200 nm pore size TFF filter, operating at 25 mL min−1 (equivalent to a shear rate of 3500 s−1) with a 10 or 20 DV setting resulted in the optimal condition for efficient separation. Second, to optimize the 50 nm pore size TFF filter, we filtered 100 nm beads and BSA in 20 mL of DMEM at three different flow rates (25‐, 30‐, and 35‐mL min−1) and the same three DVs as those mentioned previously (Figure 2b). The retentate (fraction B in Figure 1) was analyzed using NTA and BCA protein assays. Although the performance at 35 mL min−1 appears superior in terms of BSA removal, there is a concurrent decrease in the yield of 100 nm beads. In addition, the reduction in 100 nm bead yield suggests membrane clogging, which could impact the filter's lifespan. Consequently, we deemed the 35 mL min−1 condition unsuitable and excluded it. The optimal conditions for the 50 nm pore size TFF filter were 25 mL min−1 (equivalent to a shear rate of 3800 s−1) and 30 mL min−1 (4400 s−1).
FIGURE 2.
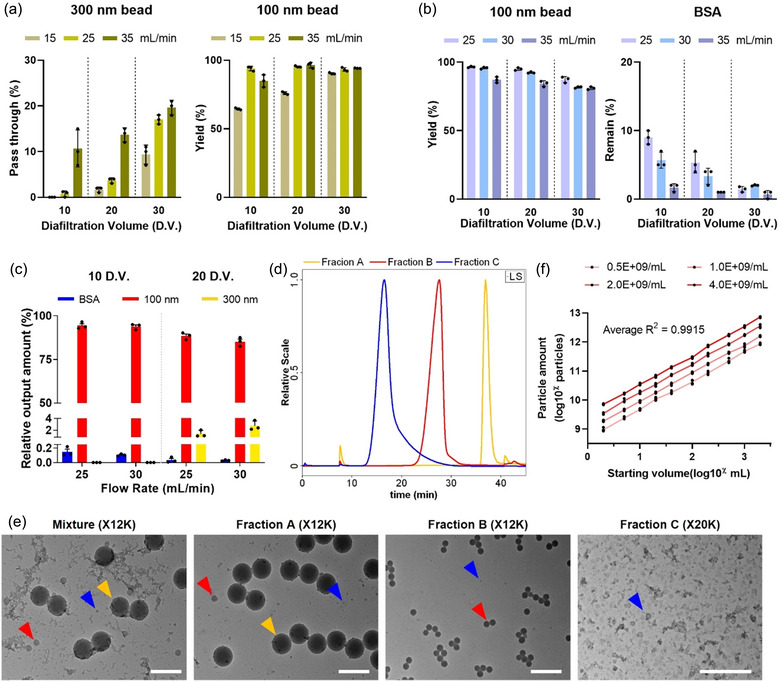
Optimization of EVi. (a) Optimization of 200 nm pore size filter process variables. Fraction B of EVi was measured using NTA after operation (n = 3, independent experiments using 100 and 300 nm beads spiked into DMEM). (b) Optimization of 50 nm pore size filter process variables. Fraction B of EVi was measured using NTA and BCA assay after operation (n = 3, independent experiments using 100 nm beads and BSA spiked into DMEM). (c) Optimization of EVi process variables (n = 3, independent experiments using 100 and 300 nm beads and BSA spiked into DMEM). (d) Elution profiles of Fraction A‐C on HF5‐MALS‐DLS‐UV system. Fractions A (yellow line), B (red line), and C (blue line) were individually injected after EVi processing. Bead mixture (30 nm, 100 nm, and 300 nm) in DMEM was processed using EVi. (e) TEM image of starting bead mixture and each fraction using EVi. Scale bar, 500 nm. (f) Characterization of the particle amount as a function of the sample processing volume at different bead concentrations (n = 4, independent experiments using 100 nm beads spiked into DMEM). The data are presented as means ± SD.
To establish the process for variables of EVi, we adjusted EVi based on the experimental results shown in Figure 2(a, b) using a mixture of BSA and 100 and 300 nm beads in 20 mL of DMEM. Separation was performed at 25 mL min−1 for the 200 nm pore size filter and at 25 and 30 mL min−1 for the 50 nm pore size filter, with 10 and 20 DVs for fractions A and B, respectively. After processing, fraction B was analyzed (Figure 2c). We found that optimal separation conditions could be established for the 200 nm filter at 25 mL min−1 (3500 s−1) and the 50 nm filter at 30 mL min−1 (3800 s−1) that produced yields of 94.38% for 100 nm beads, 0% for 300 nm beads, and 99.996% for BSA removal. To compare the performance of EVi with that of conventional TFF processing (DEF/TFF; DEF followed by TFF), the particle recovery of the two methods was evaluated for input sample volume. At low sample volumes, there was no significant difference between DEF/TFF and EVi, but the recovery yield and reproducibility of DEF/TFF decreased with increased sample volume. In contrast, EVi showed a relatively linear increase in recovery with greater sample volumes, and their results were superior to those of DEF/TFF (Figure S2).
We next conducted particle separation using an EVi system with a nanobead mixture (30, 100, and 300 nm) spiked into 20 mL of DMEM and analyzed each fraction (fraction A, B, and C). Each fraction was transferred to the HF5‐MALS‐DLS‐UV system, and the hydrodynamic radius was analyzed (Figures 2d and S3). We confirmed that the elution profile of fraction A overlapped with a 300 nm peak, fraction B coincided with a 100 nm peak, and fraction C overlapped with a 30 nm peak. The distinct elution profiles of each fraction indicated that EVi effectively separated the different‐sized particles based on the pore size of the filter. In addition, TEM and nanoflow cytometry analysis of each separated fraction confirmed that EVi effectively separated the different‐size particles (Figures 2e and S4).
To evaluate the scalability and reliability of the EVi system, we prepared four different concentrations of 100 nm bead samples in DMEM and performed EVi of serially diluted concentrations in incremental volumes ranging from 2 to 2000 mL (Figure 2f). The results showed linearity with the input concentration and volume, indicating that EVi can successfully isolate particles over a wide dynamic range. Thus, the various experimental validations show that EVi is a reliable system capable of effectively separating particles of different sizes and volumes based on filter pore size.
3.2. Characterization of sEVs obtained by EVi
We investigated whether EVi could separate 50−200 nm‐sized sEVs from cell culture media. The MDA‐MB‐231 cell culture media were processed following the EVi protocol and the sEV fraction was analyzed using TEM and NTA (Figure 3a). Lipid bilayered vesicles ranging from 50 to 200 nm sizes were visualized via TEM images. We also isolated human and mouse plasma using EVi, and NTA results exhibited particles ranging from 50 to 200 nm in size (Figure S5), indicating that clinical samples or animal plasma samples could be applicable to EVi. Western blot analysis showed the presence of the well‐established sEV markers CD63, Alix, Flotillin‐1, and TSG101 enriched in the EVi extracts compared with those in cell lysates while the marker for the endoplasmic reticulum (calnexin) was undetectable (Figure 3b). To confirm the scalability and reliability, we isolated sEVs from MDA‐MB‐231 cell culture media across a range of volumes (from 2 mL to 1 L) using EVi (Figure 3c). As expected, our results demonstrate that the particle yield increased linearly in proportion to the input volume, further confirming EVi scalability and reliability.
FIGURE 3.
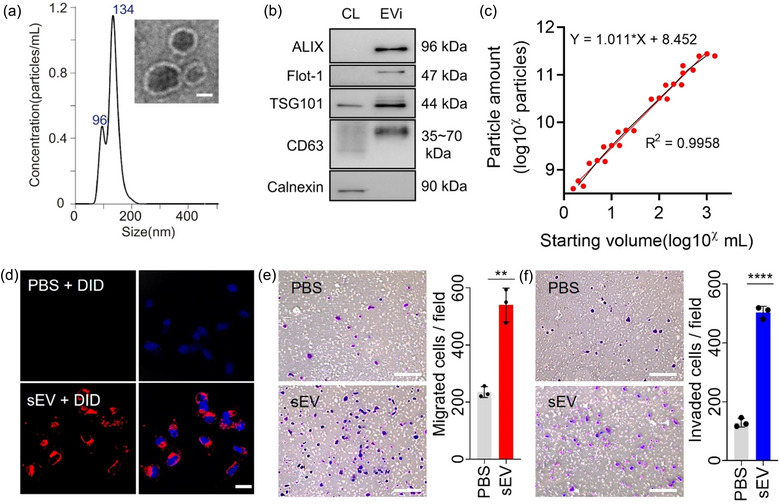
Characterization of EVi. (a) Size distribution (using NTA) and TEM image (inset) of MDA‐MB231 cell‐derived sEVs prepared using EVi system. Scale bar, 50 nm. (b) Immunoblot of various proteins in sEVs and whole‐cell lysates (CL) from MDA‐MB231 cells. One µg of protein was loaded in each lane. (c) Characterization of the particle yield as a function of the sample processing volume (n = 3). (d) Uptake of DID‐labeled sEVs isolated using EVi was detected using a confocal microscope. Scale bar, 20 µm. (e, f) Effect of MDA‐MB‐231 cell‐derived sEVs on MCF7 cells. Cell migration (e) and invasion assays (f) were performed (n = 3). Scale bar, 200 µm. Data are presented as means ± SD. *P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001, respectively; ns, not significant; unpaired two‐sided t‐test.
To investigate whether sEVs obtained through EVi are taken up by other cells, we performed an sEV uptake assay. MDA‐MB‐231 cell‐derived sEVs isolated using EVi were fluorescence‐labeled and then treated to MCF7 cells, and the images were analyzed using confocal microscopy (Figure 3d). We confirmed that sEVs from MDA‐MB‐231 cells were taken up by MCF7 cells, indicating that sEVs obtained through EVi retained their property to be taken up by other cells. To evaluate the maintenance of functionality in the sEVs obtained by EVi, migration and invasion assays were performed in Transwell chambers. Less‐malignant MCF7 cells were treated with malignant MDA‐MB‐231 cell‐derived sEVs and we found that MDA‐MB‐231 cell‐derived sEVs increased the mobility and invasiveness of MCF cells (Figure 3e, f). Furthermore, a wound‐healing assay also demonstrated an enhanced wound closure effect of MCF7 cells by MDA‐MB‐231 cell‐derived sEVs (Figure S6), as previously reported (Amaro et al., 2016; Amaro et al., 2016; Comşa et al., 2015). These results indicate that EVi allows successful isolation of sEVs from a wide range of cell culture medium volumes (2 mL−1 L) and that the isolated sEVs maintain the property to be taken up by other cells and retain their functionality.
3.3. EVi outperforms other isolation methods
First, we compared the EVi with the conventional TFF process, DEF/TFF. Equal volume of MDA‐MB‐231 cell culture media (200 mL) were processed following each isolation methods and final volume was adjusted equally (700 µL). Immunoblot of equal‐sample‐volume or equal‐protein‐amount analysis showed higher yield and purity of EVi compared to DEF/TFF (Figure S7a, Sb). While conventional TFF process DEF does not involve diafiltration, EVi incorporates a diafiltration step using a 200 nm pore size TFF filter. This step enables EVi to obtain more particles and remove impurities, resulting in higher purity. We also compared total particle yields, total protein amount and purity using NTA and BCA assay (Figure S7c‐e). As a result, it was confirmed that EVi is superior to DEF/TFF in terms of yield and purity. Total processing time was approximately 30 min for EVi, whereas approximately 35 min for conventional TFF process (data not shown).
Next, we compared the efficiency of EVi with the current main isolation approach, UC and SEC. We isolated sEVs from MDA‐MB‐231 cell culture medium using each method and analyzed the presence of sEV proteins using immunoblot analysis (Figure 4a). EVi produced much higher intensities for sEV proteins than UC and SEC, indicating an increased yield of sEV proteins via the EVi method. To compare the purity of sEVs isolated using EVi and other isolation methods (UC, SEC, and EQ), we also isolated sEVs from culture media from three different cancer cells and calculated their purities (Figures 4b, c, and S8). We found that EVi‐isolated sEVs from all three cell lines showed the highest purity, followed by sEVs isolated by SEC, UC, and EQ. Comparison of the intensity of the CD63 protein of MDA‐MB‐231‐derived sEVs isolated by the four methods showed that the signal intensity tendency matched the purity analysis results (Figure S9). In addition, we compared the processing time required for each isolation method and found that EVi required less processing time for conditioned media or plasma samples than any of the other isolation methods (Figure 4d).
FIGURE 4.
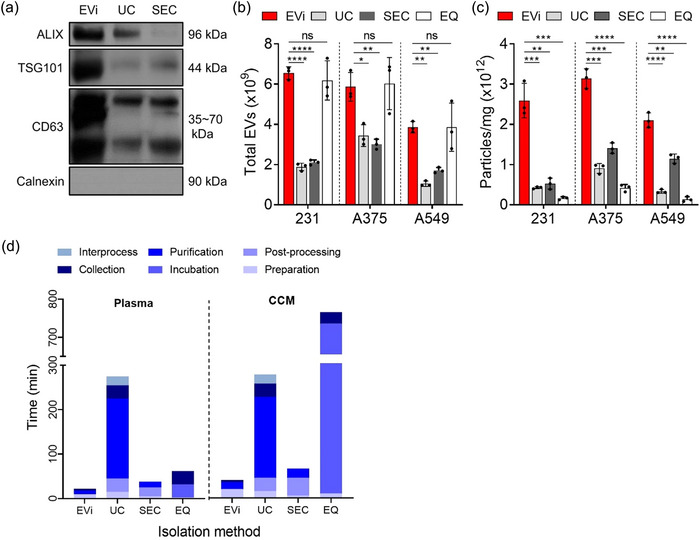
Comparison of EVi with other isolation methods. (a) Quantitative immunoblot of sEV markers for MDA‐MB231 cell‐derived sEVs obtained using EVi, UC and SEC. 4 µg of proteins were loaded in each lane. (b, c) Total yields (b) and purities (c) were characterized using NTA analysis and BCA assay for each isolation method in three different cancer cell lines (n = 3), as indicated. 231, MDA‐MB‐231 breast cancer cell line; A375, A375 melanoma cell line; and A549, A549 lung cancer cell line. An equal volume of CCM (30 mL) was used to isolate sEVs according to each isolation method. (d) Comparison of the sample processing time by EVi and other isolation methods. Processed volumes were 500 µL for plasma and 200 mL for CCM, respectively. CCM, clarified conditioned media. Data are presented as means ± SD. *P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001, respectively; ns, not significant; unpaired two‐sided t‐test.
3.4. System design and optimization of EVc
To efficiently count sEV, we developed an EV counting system by utilizing the light scattering‐based measurement principle and adjusting the FOV to count EVs in the size range of 50−200 nm obtained by EVi.
First, we arranged the components of the counting system to facilitate easy loading and measurement of sEV samples in small quantities. We positioned the laser module and camera perpendicularly around the cuvette (Figures 5a and S10). The particles suspended in the sample cuvette scatter the incident laser light, which is then visualized using a CMOS camera equipped with a 20× objective with a long working distance. Next, to minimize the sample amount used for sEV counting, the sensitivity to concentration should be increased by capturing a wider area of view. Therefore, we optimized the tube lens length to configure a large FOV (Figure S11). This resulted in optimized parameters of a tube lens length of 80 mm, a FOV of ∼670 × 900 × 3.2 µm, and a scattering volume of 1.93 nL, which is 24.1 times larger than that of a conventional NTA instrument (0.08 nL).
FIGURE 5.
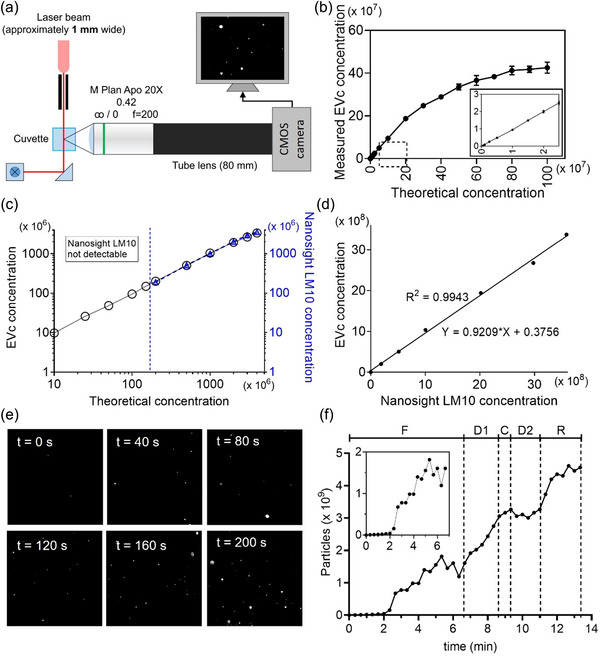
Characterization of EVc. (a) Schematic diagram of EVc system. (b) Calibration curve of EVc system (n = 3). The inset shows the magnified plots for the dashed box area. (c, d) Comparison of measured concentrations by EVc and Nanosight LM10 instrument obtained from the same particle concentration (n = 3). Linear regression analysis from c was presented in (d). (e) Representative time‐lapse images of every 40 s during particle purification using EVi. (f) Changes in particles extracted from the data (e). F, filtration; D1, first diafiltration; C, concentration; D2, second diafiltration; and R, recovery. Inset plot represents the magnified plots for the F step. Data are presented as means ± SD.
Finally, for accurate and reproducible measurement of sEV concentration, we optimized the image acquisition parameter using 100 nm standard nanobead (Figure S12). The optimal image acquisition parameters were found to be 1.5 gamma, 50 ms exposure, and 15 gain. We also observed a change in the measured concentration of particles over time to identify the appropriate measurement time (Figure S13). For the tracking time shorter than 20 min, no significant difference in the number of particles was observed. To verify the performance of our counting system, a calibration curve was plotted by comparing the actual concentration with the measured concentration using 100 nm standard beads (Figure 5b). Furthermore, to investigate the practical application of our system, MDA‐MB‐231 cell derived sEVs were counted using EVc and compared to those of NTA (Figure 5c, d). The results demonstrated that EVc was capable of measuring particles at approximately 1/100th of the concentration required by NTA instruments (Figure 5c), and the calculated concentrations from EVc and Nanosight LM10 exhibited good concordance, demonstrating the reliability of EVc (Figure 5d).
To investigate the compatibility of EVi and EVc, we used standard nanoparticles to test whether EVc could detect the changes in the concentration of nanoparticles during the EVi purification process over time. Nanoparticles were processed using EVi, and the concentration of purified nanoparticles in fraction B of EVi was measured by EVc every 20 s; representative time‐lapse images are displayed (Figure 5e). The results suggest that EVi and EVc could be coupled effectively. In addition, detailed information on how EVi operates over time could be obtained and the acquired information can then be used to operate EVi efficiently (Figure 5f). As expected, the concentration of the particles gradually increased during the filtration step (F), and the increased concentration during the first diafiltration step (D1) was a direct result of diafiltration. No particle loss occurred in the second diafiltration (D2), indicating that the buffer exchange performed well. Circulation in the recovery step (R) had a significant effect on particle yield, suggesting that the recovery step must be performed during the TFF process.
3.5. Utilization of various in vitro research using EVics
We tested the applicability of EVics in various types of studies (in vitro, in vivo, and clinical settings) conducted in the EV field. As EVics performed effective purification and sensitive counting, we were able to accurately determine the number of secreted sEVs at early time points (Figure 6a). Following sEV purification from various cell‐derived cultures grown in 100 mm culture dishes using EVi at 6‐h intervals for up to 24 h, we measured the total number of sEVs using EVc and calculated the secreted sEVs per cell (Figures 6b and S14). The results showed that sEV secretion differed depending on the types of cells and that all cell lines secreted more than 50% of the total amount of sEVs within 0–6 h, after which the number of secreted sEVs per cell increased slowly until 24 h. In addition, among the breast cancer cells, malignant cells (MDA‐MB‐231) secreted more sEVs than less malignant cells (MCF7 > MCF10A). Furthermore, we evaluated several drugs that inhibit sEV secretion (Im et al., 2019; Trajkovic et al., 2008) using EVics and examined material consumption (Figure 6c). The sEV inhibitor exhibited a statistically significant effect on sEV secretion from cells grown in both 150 and 35 mm (6‐well) culture dishes. Using EVics, the amount of material used was reduced by 20 times, and the time required for purification and analysis was decreased by approximately 4.7 times (∼180 to 40 min).
FIGURE 6.
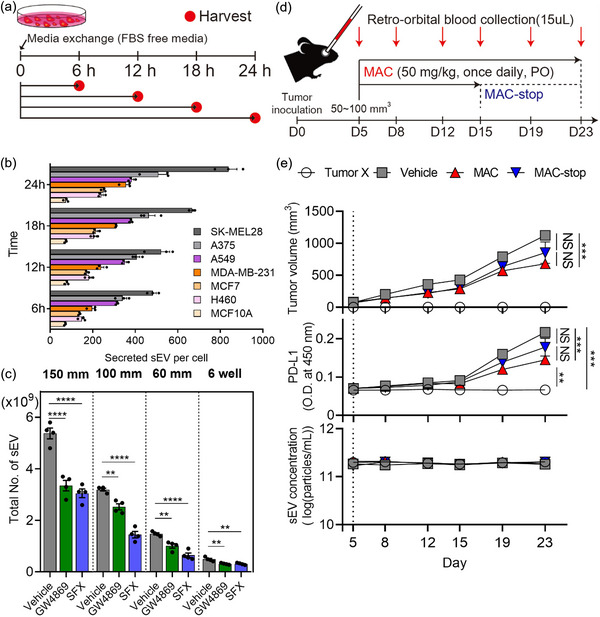
Application of EVics in vitro and in vivo studies. (a) Experimental design of the cellular sEV secretion investigation. (b) Investigation of sEV secretion behavior from various cell lines over time (n = 3). (c) Inhibitory effect of sEV inhibitors (GW4869, SFX) on MDA‐MB‐231 breast cancer cells (n = 4). SFX, sulfisoxazole. (d) Experimental design of syngeneic cancer models. (e) Growth curves of EMT6 (top), levels of plasma sEV PD‐L1 (middle), and plasma sEV concentrations (bottom) of immunocompetent mice subjected to the indicated treatments (n = 5−6). Vertical dotted lines represent the beginning of macitentan (MAC) administration. Data are presented as mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001, respectively; NS, not significant; unpaired two‐sided t‐test.
3.6. EVics allows monitoring the effects of an sEV inhibitor in mice
Next, we devised an experiment to test the applicability of EVics in vivo. In general, it is difficult to monitor plasma sEV concentrations and the amounts of sEV proteins in individual mice, mainly owing to the considerably limited amount of blood collected from each mouse. Therefore, there have been limited reports on the pharmacokinetics related to sEVs, and only differences at the endpoint of experiments were reported. Therefore, we designed experiments to investigate the feasibility of monitoring sEVs in animal models by using the advantages of EVics (Figure 6d). In the syngeneic tumor models, EMT6 breast tumor cells were orthotopically injected into the left fat pad of the female BALB/c mice and macitentan (MAC, a sEV inhibitor (Lee et al., 2022)) was orally administered. Approximately 15 µL of blood was obtained by orbital blood collection once every 3 or 4 days and plasma was prepared for sEV isolation. Then sEVs were purified and counted using EVics, and the plasma sEV PD‐L1 levels were determined by using ELISA (Figure 6e). The level of plasma sEV PD‐L1 increased with the growth of the tumor in syngeneic mice with EMT6 breast tumor cells. In contrast, the tumor grew more slowly and the level of plasma sEV PD‐L1 was lower in MAC‐treated mice than in those of the vehicle group. When MAC treatment was stopped (MAC‐stop) from day 15, tumor volume and the level of plasma sEV PD‐L1 tended to increase again, although statistical significance was not observed. However, sEV concentrations in the blood were maintained similarly over time regardless of the presence of tumors or treatment with MAC as a sEV inhibitor.
3.7. Clinical application of EVics
To determine the practical clinical applicability of EVics, 100 µL of plasma samples were collected from patients of breast cancer (n = 40), lung cancer (n = 40), and colorectal cancer (n = 40) and healthy individuals (n = 40). Plasma sEVs from these individuals were isolated and counted using EVics to measure sEV concentrations (Figure 7a). The plasma sEV concentrations were increased in all cancer types compared with those in healthy controls, despite no significant differences between cancer types. Furthermore, to investigate the efficiency of EVi in removing plasma‐derived lipoproteins, the levels of LDL/VLDL and HDL were measured before and after sEV isolation (Figure S15). The results indicated that our system removed 91.53% of LDL/VLDL and 95.96% of HDL, indicating effective removal of lipoproteins. Although lipoproteins were not completely eliminated, the substantial enrichment of sEVs in the 50−200 nm size range suggests that our system is applicable to plasma samples. Next, we randomly selected six samples from each group to compare the yields of sEVs isolated using EVi and UC (Figure 7b). Overall, EVi isolated significantly more sEVs from the plasma in all samples than did UC, indicating the superior ability of EVi for efficiently handling the small volumes of clinical samples.
FIGURE 7.
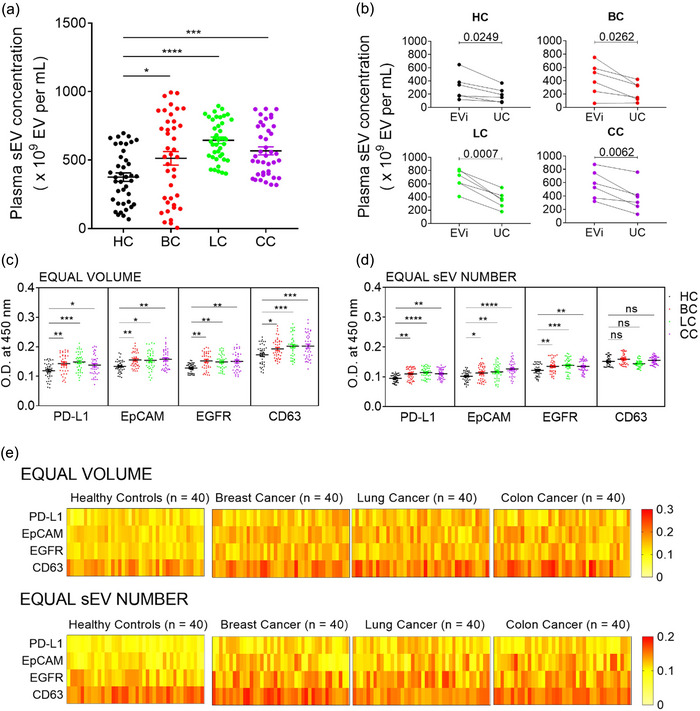
Diagnostic application of EVics. (a) Investigation of human plasma sEV concentrations in various cancer types. We used 100 µL of plasma to isolate sEVs. (b) Comparison of sEV concentrations measured using EVc and different isolation methods. sEVs of six randomly selected plasma samples from each group were isolated by different methods and counted using EVc. (c, d) ELISA for cancer diagnosis. Plasma sEV samples from healthy controls (n = 40) and patients with cancer (breast cancer, lung cancer, or colon cancer, n = 40 for each group) were analyzed. The relative expressions of three cancer‐related markers (PD‐L1, EpCAM, and EGFR) and CD63 were measured using ELISA. Equal volumes (c) of sEVs (equivalent to 25 µL of plasma sEVs) or equal numbers (d) of sEVs (1 × 10 (Moon et al., 2016) sEVs) were loaded into a 96‐well plate and analyzed. (e) Heatmap of analyzed data from (c) and (d). Data are presented as mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001, respectively; ns, not significant; Dunnet's multiple comparisons test.
To evaluate whether sEVs prepared using EVics could be used for diagnostic applications, we performed ELISA to determine the expression of sEV markers and cancer‐related markers (PD‐L1, EpCAM, and EGFR) in equal volumes Figure 7c) and equal numbers of sEVs (Figure 7d). In the equal‐volume analysis, the level of each marker increased differently compared to that in healthy controls, depending on the cancer type. The level of sEV PD‐L1 was the highest in lung cancer, followed by colon and breast cancers. Moreover, sEV EpCAM levels were the highest in colon cancer, followed by breast and lung cancers. Lastly, sEV EGFR levels were the highest in breast cancer, followed by colon and lung cancers. In addition, the tendency in sEV CD63 levels was consistent with that of the plasma sEV concentration shown in Figure 7(a), suggesting that sEV CD63 levels may reflect the sEV concentrations. In the analysis with equal‐number sEVs, the statistical significance of each marker changed when the number of sEVs was adjusted based on EVc counts. Unlike the equal volume analysis, differences in sEV CD63 levels between each group disappeared in the equal sEV number analysis, indicating that the EVics‐based quantity correction worked effectively. Figure 7(e) shows the expression of the four markers in the overall cohort.
4. DISCUSSION
Numerous methods for EV isolation and counting have been developed in recent years, highlighting their importance. However, currently reported problems in isolation and counting methods have limited their practical usage in sEV research, suggesting an urgent need for improvements. Thus, we have developed a novel system by coupling isolation and counting technologies into one system to produce ready‐to‐use sEV by improving TFF for isolation and light scattering technology for counting. Our results demonstrated that EVics is a system useful for scalable sEV isolation and efficient sEV counting within a single workflow system. EVi is a system for isolating sEVs through simultaneously operating two TFF filters with 50 and 200 nm pore sizes. EVc is a light scattering‐based counting system with a large FOV to count the sEVs isolated using EVi.
Various reports have documented the application of TFF for isolating EVs. However, a notable issue in this technique, which we aim to overcome, lies not within TFF itself but rather in the preceding DEF step used for large particle removal. During DEF, significant particles accumulate and form a cake layer on the membrane surface, leading to a substantial reduction in processing speed and yield. The present study demonstrated significantly improved results in terms of yield, purity, and process time compared to the conventional TFF process commonly used for sEV isolation. Additionally, the single‐step isolation process is expected to alleviate concerns about external contamination and result in cost savings owing to the semi‐permanent TFF filter.
Notably, EVi successfully isolated sEVs in the 50−200 nm range with proven scalability and reliability. Unexpectedly, simultaneous filtration using EVi improved BSA removal compared to sequential filtration. This could be attributed to fraction B of EVi becoming less diluted during the process, causing BSA molecules present in the sample to pass through the filters more frequently. In addition, EVi can produce biologically active sEVs and outperform the other methods in terms of purity, sample volume, and isolation time. In a recent study, a combination of multiple steps was employed to obtain a high‐purity sEV pool (Comşa et al., 2015). In contrast, we were able to obtain sEVs of similar purity using only EVi, suggesting that EVi is a convenient system for high‐purity sEV isolation in a single step. Therefore, EVi is expected to offer biologically active sEV not only for laboratory‐scale research but also for large‐scale production of clinical‐grade sEVs in industries requiring GMP facilities.
We achieved a reduction in sEV consumption by approximately 100 times less than that of Nanosight LM10 by optimizing the FOV and simplifying its use by employing a cuvette. This suggests that EVc provides a convenient EV counting system with low sEV consumption.
By utilizing EVics, we investigated, for the first time, sEV secretion behavior at early time points in various cell lines. Furthermore, we were able to study the inhibitory effects of sEV secretion inhibitors using only 2 mL (6‐well plates) of culture supernatant, thereby saving the cost and time. These results indicate that EVics can be utilized in various studies on sEV secretion in vitro. Moreover, by using EVics, we were able to monitor changes in blood sEV concentrations and sEV PD‐L1 levels during tumor progression in individual mice. Remarkably, our sEV concentration monitoring results provided the first direct evidence that sEV concentration in the blood might be maintained via cellular homeostasis. This finding highlights how EVics can contribute to understanding the kinetics of sEV concentrations or sEV derived surface proteins in the blood in response to drug treatment, thus indicating the applicability of EVics for in vivo studies.
In addition, to demonstrate the clinical utility of EVics, we examined 160 clinical samples. The blood sEV concentrations were significantly higher in patients with various types of cancer than in healthy controls. Moreover, our comparison of UC and EVi in Figure 7(b) demonstrates the superior advantages of the EVics system for sEV‐based research using clinical samples. Furthermore, the statistically significant differences in biomarkers before and after adjusting for sEV number at diagnosis highlight the importance of considering sEV numbers in sEV‐based diagnostic research. Although sEVs were successfully prepared from clinical samples using EVics in this study, there is a limitation in its clinical application because EVi can process only one sample at a time. Thus, the development of a system that can process multiple samples and features a filter with a scaled‐down size for small volumes would be more useful for application to clinical samples.
In this study, we proposed a ready‐to‐use sEV preparation system that also provides particle concentration for the first time. By coupling isolation and counting into one preparation process, the amount of sample required for counting could be significantly reduced. This ability is important when working with limited amounts of clinical specimens. EVics provides purified sEV containing particle concentration information, and various approaches conducted in this study revealed that the particle concentration itself could also be used as information. Although isolation and counting parts were not integrated into one device in this study, further optimizations are needed to achieve this integration.
In conclusion, we have expanded the conceptual scope of sEV research preparation by combining sEV isolation and counting, thereby proposing a scalable and ready‐to‐use sEV preparation system, which has versatile applications in the sEV research field.
AUTHOR CONTRIBUTIONS
J.H.B. and M.C.B. conceived idea and designed the study. J.H.B. performed experiments and analyses. C.H.L. conducted mouse experiments and collected clinical samples. J.H.B., C.H.L., D.J., and M.C.B. wrote the manuscript. M.C.B. supervised the project. B.J.S., H.L., K.Y., and M.C.B. discussed the results and edited the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea, funded by the Ministry of Science & ICT (2021R1A5A2021614, 2023R1A2C3005553), DGIST Program of the Ministry of Science and ICT (21‐DGRIP‐01).
Bae, J.‐H. , Lee, C.‐H. , Jung, D. , Yea, K. , Song, B.‐J. , Lee, H. , & Baek, M.‐C. (2024). Extracellular vesicle isolation and counting system (EVics) based on simultaneous tandem tangential flow filtration and large field‐of‐view light scattering. Journal of Extracellular Vesicles, 13, e12479. 10.1002/jev2.12479
REFERENCES
- Ahn, S. H. , Ryu, S. W. , Choi, H. , You, S. , Park, J. , & Choi, C. (2022). Manufacturing therapeutic exosomes: From bench to industry. Molecules and Cells, 45(5), 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro, A. , Angelini, G. , Mirisola, V. , Esposito, A. I. , Reverberi, D. , Matis, S. , Maffei, M. , Giaretti, W. , Viale, M. , Gangemi, R. , Emionite, L. , Astigiano, S. , Cilli, M. , Bachmeier, B. E. , Killian, P. H. , Albini, A. , & Pfeffer, U. (2016). A highly invasive subpopulation of MDA‐MB‐231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo. Oncotarget, 7(42), 68803–68820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, W. , Kozak, D. , Coleman, V. A. , Jämting, Å. K. , & Trau, M. (2013). A comparative study of submicron particle sizing platforms: accuracy, precision and resolution analysis of polydisperse particle size distributions. Journal of Colloid and Interface Science, 405, 322–330. [DOI] [PubMed] [Google Scholar]
- Baranyai, T. , Herczeg, K. , Onódi, Z. , Voszka, I. , Módos, K. , Marton, N. , Nagy, G. , Mäger, I. , Wood, M. J. , El Andaloussi, S. , Pálinkás, Z. , Kumar, V. , Nagy, P. , Kittel, Á. , Buzás, E. I. , Ferdinandy, P. , & Giricz, Z. (2015). Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE, 10(12), e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, N. C. , Minelli, C. , Tompkins, J. , Stevens, M. M. , & Shard, A. G. (2012). Emerging techniques for submicrometer particle sizing applied to Stöber silica. Langmuir: The ACS Journal of Surfaces and Colloids, 28(29), 10860–10872. [DOI] [PubMed] [Google Scholar]
- Burgess, R. R. (2018). A brief practical review of size exclusion chromatography: Rules of thumb, limitations, and troubleshooting. Protein Expression and Purification, 150, 81–85. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Huang, A. C. , Zhang, W. , Zhang, G. , Wu, M. , Xu, W. , Yu, Z. , Yang, J. , Wang, B. , Sun, H. , Xia, H. , Man, Q. , Zhong, W. , Antelo, L. F. , Wu, B. , Xiong, X. , Liu, X. , Guan, L. , Li, T. , … Guo, W. (2018). Exosomal PD‐L1 contributes to immunosuppression and is associated with anti‐PD‐1 response. Nature, 560(7718), 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Zhu, Q. , Cheng, L. , Wang, Y. , Li, M. , Yang, Q. , Hu, L. , Lou, D. , Li, J. , Dong, X. , Lee, L. P. , & Liu, F. (2021). Exosome detection via the ultrafast‐isolation system: EXODUS. Nature Methods, 18(2), 212–218. [DOI] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Comşa, Ş. , Cîmpean, A. M. , & Raica, M. (2015). The story of MCF‐7 breast cancer cell line: 40 years of experience in research. Anticancer Research, 35(6), 3147–3154. [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T. M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T. L. , … Lyden, D. (2015). Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology, 17(6), 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baradie, K. B. Y. , Nouh, M. , O'Brien Iii, F. , Liu, Y. , Fulzele, S. , Eroglu, A. , & Hamrick, M. W. (2020). Freeze‐dried extracellular vesicles from adipose‐derived stem cells prevent hypoxia‐induced muscle cell injury. Frontiers in Cell and Developmental Biology, 8, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe, V. , Hawe, A. , & Jiskoot, W. (2010). Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharmaceutical Research, 27(5), 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, C. , Di Vizio, D. , Sahoo, S. , Théry, C. , Witwer, K. W. , Wauben, M. , & Hill, A. F. (2016). Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. Journal of Extracellular Vesicles, 5, 32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, C. , Ferreira, Y. J. , Dragovic, R. A. , Redman, C. W. , & Sargent, I. L. (2013). Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. Journal of Extracellular Vesicles, 2, 19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest, C. , Joimel, U. , Huang, L. , Pritchard, L. L. , Petit, A. , Dulong, C. , Buquet, C. , Hu, C. Q. , Mirshahi, P. , Laurent, M. , Fauvel‐Lafève, F. , Cazin, L. , Vannier, J. P. , Lu, H. , Soria, J. , Li, H. , Varin, R. , & Soria, C. (2013). Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: differences in MDA‐MB‐231 and MCF‐7 breast cancer cell lines. BMC Cancer, 13, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening, D. W. , Gopal, S. K. , Xu, R. , Simpson, R. J. , & Chen, W. (2015). Exosomes and their roles in immune regulation and cancer. Seminars in Cell & Developmental Biology, 40, 72–81. [DOI] [PubMed] [Google Scholar]
- Gurunathan, S. , Kang, M. H. , Jeyaraj, M. , Qasim, M. , & Kim, J. H. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells, 8(4), 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, M. L. , Ilmer, M. , Silva, L. P. , Hawke, D. H. , Recio, A. , Vorontsova, M. A. , Alt, E. , & Vykoukal, J. (2014). Benchtop isolation and characterization of functional exosomes by sequential filtration. Journal of Chromatography. A, 1371, 125–135. [DOI] [PubMed] [Google Scholar]
- Im, E. J. , Lee, C. H. , Moon, P. G. , Rangaswamy, G. G. , Lee, B. , Lee, J. M. , Lee, J. C. , Jee, J. G. , Bae, J. S. , Kwon, T. K. , Kang, K. W. , Jeong, M. S. , Lee, J. E. , Jung, H. S. , Ro, H. J. , Jun, S. , Kang, W. , Seo, S. Y. , Cho, Y. E. , … Baek, M. C. (2019). Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nature Communications, 10(1), 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, J. (2016). Malignant messengers. Science (New York, N.Y.), 352(6282), 164–166. [DOI] [PubMed] [Google Scholar]
- Kalluri, R. , & LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science (New York, N.Y.), 367(6478), eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkanova, A. D. , Blessing, M. , Gemeinhardt, A. , Soulat, D. , & Sandoghdar, V. (2022). Precision size and refractive index analysis of weakly scattering nanoparticles in polydispersions. Nature Methods, 19(5), 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko, M. Y. , Lekchnov, E. A. , Vlassov, A. V. , & Laktionov, P. P. (2018). Isolation of extracellular vesicles: General methodologies and latest trends. BioMed Research International, 2018, 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky, E. H. , Street, J. M. , Star, R. A. , & Yuen, P. S. (2017). Quantification of exosomes. Journal of Cellular Physiology, 232(7), 1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. H. , Bae, J. H. , Choe, E. J. , Park, J. M. , Park, S. S. , Cho, H. J. , Song, B. J. , & Baek, M. C. (2022). Macitentan improves antitumor immune responses by inhibiting the secretion of tumor‐derived extracellular vesicle PD‐L1. Theranostics, 12(5), 1971–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. H. , Ha, D. H. , Go, H. K. , Youn, J. , Kim, H. K. , Jin, R. C. , Miller, R. B. , Kim, D. H. , Cho, B. S. , & Yi, Y. W. (2020). Reproducible large‐scale isolation of exosomes from adipose tissue‐derived mesenchymal stem/stromal cells and their application in acute kidney injury. International Journal of Molecular Sciences, 21(13), 4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Kaslan, M. , Lee, S. H. , Yao, J. , & Gao, Z. (2017). Progress in exosome isolation techniques. Theranostics, 7(3), 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liangsupree, T. , Multia, E. , & Riekkola, M. L. (2021). Modern isolation and separation techniques for extracellular vesicles. Journal of Chromatography. A, 1636, 461773. [DOI] [PubMed] [Google Scholar]
- Linares, R. , Tan, S. , Gounou, C. , Arraud, N. , & Brisson, A. R. (2015). High‐speed centrifugation induces aggregation of extracellular vesicles. Journal of Extracellular Vesicles, 4, 29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb, R. J. , Becker, M. , Wen, S. W. , Wong, C. S. , Wiegmans, A. P. , Leimgruber, A. , & Möller, A. (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of Extracellular Vesicles, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, R. P. , Caro‐Vegas, C. P. , Costantini, L. M. , Landis, J. T. , Griffith, J. D. , Damania, B. A. , & Dittmer, D. P. (2018). Large‐scale, cross‐flow based isolation of highly pure and endocytosis‐competent extracellular vesicles. Journal of Extracellular Vesicles, 7(1), 1541396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, S. A. , Luecke, L. B. , Kahlert, C. , Fernandez, A. F. , Gammon, S. T. , Kaye, J. , LeBleu, V. S. , Mittendorf, E. A. , Weitz, J. , Rahbari, N. , Reissfelder, C. , Pilarsky, C. , Fraga, M. F. , Piwnica‐Worms, D. , & Kalluri, R. (2015). Glypican‐1 identifies cancer exosomes and detects early pancreatic cancer. Nature, 523(7559), 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, P. G. , Lee, J. E. , Cho, Y. E. , Lee, S. J. , Jung, J. H. , Chae, Y. S. , Bae, H. I. , Kim, Y. B. , Kim, I. S. , Park, H. Y. , & Baek, M. C. (2016). Identification of developmental endothelial locus‐1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 22(7), 1757–1766. [DOI] [PubMed] [Google Scholar]
- Okoye, I. S. , Coomes, S. M. , Pelly, V. S. , Czieso, S. , Papayannopoulos, V. , Tolmachova, T. , Seabra, M. C. , & Wilson, M. S. (2014). MicroRNA‐containing T‐regulatory‐cell‐derived exosomes suppress pathogenic T helper 1 cells. Immunity, 41(1), 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo, F. , Théry, C. , Falcón‐Pérez, J. M. , Nieuwland, R. , & Witwer, K. W. (2020). Methods for separation and characterization of extracellular vesicles: Results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells, 9(9), 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, B. K. , Zhang, H. , Becker, A. , Matei, I. , Huang, Y. , Costa‐Silva, B. , Zheng, Y. , Hoshino, A. , Brazier, H. , Xiang, J. , Williams, C. , Rodriguez‐Barrueco, R. , Silva, J. M. , Zhang, W. , Hearn, S. , Elemento, O. , Paknejad, N. , Manova‐Todorova, K. , Welte, K. , … Lyden, D. (2014). Double‐stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Research, 24(6), 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, 30, 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Zitvogel, L. , & Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nature Reviews. Immunology, 2(8), 569–579. [DOI] [PubMed] [Google Scholar]
- Tian, X. , Nejadnik, M. R. , Baunsgaard, D. , Henriksen, A. , Rischel, C. , & Jiskoot, W. (2016). A comprehensive evaluation of nanoparticle tracking analysis (NanoSight) for characterization of proteinaceous submicron particles. Journal of Pharmaceutical Sciences, 105(11), 3366–3375. [DOI] [PubMed] [Google Scholar]
- Tóth, E. Á. , Turiák, L. , Visnovitz, T. , Cserép, C. , Mázló, A. , Sódar, B. W. , Försönits, A. I. , Petővári, G. , Sebestyén, A. , Komlósi, Z. , Drahos, L. , Kittel, Á. , Nagy, G. , Bácsi, A. , Dénes, Á. , Gho, Y. S. , Szabó‐Taylor, K. É. , & Buzás, E. I. (2021). Formation of a protein corona on the surface of extracellular vesicles in blood plasma. Journal of Extracellular Vesicles, 10(11), e12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic, K. , Hsu, C. , Chiantia, S. , Rajendran, L. , Wenzel, D. , Wieland, F. , Schwille, P. , Brügger, B. , & Simons, M. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York, N.Y.), 319(5867), 1244–1247. [DOI] [PubMed] [Google Scholar]
- Watson, D. C. , Yung, B. C. , Bergamaschi, C. , Chowdhury, B. , Bear, J. , Stellas, D. , Morales‐Kastresana, A. , Jones, J. C. , Felber, B. K. , Chen, X. , & Pavlakis, G. N. (2018). Scalable, cGMP‐compatible purification of extracellular vesicles carrying bioactive human heterodimeric IL‐15/lactadherin complexes. Journal of Extracellular Vesicles, 7(1), 1442088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐’t Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2, 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, C. H. , Kim, H. K. , Jung, G. Y. , Jung, Y. J. , Lee, K. S. , Yun, Y. E. , Han, J. , Lee, J. , Kim, W. S. , Choi, J. S. , Yang, S. , Park, J. H. , Jo, D. G. , & Cho, Y. W. (2020). Small extracellular vesicles from human adipose‐derived stem cells attenuate cartilage degeneration. Journal of Extracellular Vesicles, 9(1), 1735249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


