Abstract
Although prior studies have investigated cellular infection by dengue virus (DV), many have used highly passaged strains. We have reassessed cellular infection by DV type 2 (DV2) using prototype and low-passage isolates representing genotypes from different geographic areas. We observed marked variation in the susceptibility to infection among cell types by different DV2 strains. HepG2 hepatoma cells were susceptible to infection by all DV2 strains assayed. Although the prototype strain generated higher titers of secreted virus than the low-passage isolates, this difference did not correspond to positive- or negative-strand viral RNA levels and thus may reflect variation in efficiency among DV2 isolates to translate viral proteins or package and/or secrete virus. In contrast, human foreskin fibroblasts were susceptible to the prototype and low-passage Thai isolates but not to five Nicaraguan strains tested, as reflected by the absence of accumulation of negative-strand viral RNA, viral antigen, and infectious virus. A similar pattern was observed with the antibody-dependent pathway of infection. U937 and THP-1 myeloid cells and peripheral blood monocytes were infected in the presence of enhancing antibodies by the prototype strain but not by low-passage Nicaraguan isolates. Again, the barrier appeared to be prior to negative-strand accumulation. Thus, depending on the cell type and viral isolate, blocks that limit the production of infectious virus in vitro may occur at distinct steps in the pathway of cellular infection.
Dengue virus (DV) is a single-stranded positive-polarity enveloped RNA flavivirus that causes dengue fever (DF), the most prevalent arthropod-borne viral illness in humans. Four DV serotypes are transmitted by mosquitoes, and infection results in a clinical spectrum ranging from an acute, self-limited febrile illness (DF) to a life-threatening syndrome (dengue hemorrhagic fever/dengue shock syndrome [DHF/DSS]). Globally, DV causes an estimated 100 million new cases of DF and 250,000 cases of DHF/DSS per year, with 2.5 billion people at risk (40). Despite the worldwide morbidity associated with DV infection, neither the molecular virology nor the pathogenesis of DV is well characterized.
In primary DV infection, DV enters target cells after the envelope protein E attaches to an uncharacterized receptor that may display highly sulfated glycosaminoglycans (7). Secondary infection occurs after inoculation with a different DV serotype. In this case, the virus enters cells through a primary receptor but also may form immune complexes with preexisting nonneutralizing antibodies and interact with alternate receptors (9) such as Fcγ receptors I and II (32), resulting in antibody-dependent enhancement of infection (ADE) (14, 16). ADE is hypothesized to contribute to the pathogenesis of severe dengue illness (16, 23), as epidemiological studies have identified secondary infection as a risk factor for DHF and have shown that the presence of preexisting anti-DV antibodies correlates with DHF (6, 54). Nonetheless, despite the large number of secondary infections in endemic areas, only a small percentage progress to DHF. Environmental, host, and viral factors are hypothesized to contribute to the progression of DHF (16, 40). In support of this, distinct DV strains show disparate abilities to induce DHF (48, 49, 58). Particular structural differences in several viral proteins and the 5′ and 3′ untranslated regions between DV type 2 (DV2) genotypes have been found to correlate with disease severity (30, 35). How host and viral factors interplay to cause DHF remains uncertain, although T-lymphocyte activation and an exuberant production of inflammatory cytokines are hypothesized to play critical roles (52).
Studies of pathologic specimens from patients with DHF suggest that many tissues may be involved, as viral antigens are expressed in liver, lymph node, spleen, and bone marrow (8, 29, 51). Monocytes and macrophages are reported to display DV antigens in pathologic specimens from patients with DHF (14). Many cell types, including epithelial and endothelial cells and fibroblasts, have been shown to support viral replication in the absence of enhancing antibodies (1, 2, 4, 27, 28, 36, 39); however, many of these studies have used laboratory-adapted DV strains. Results obtained with high-passage DV strains may differ from those obtained with low-passage isolates, as dominant mutations that confer phenotypes that may not be physiologically relevant are acquired in vitro (25, 48).
In this paper, we reassess antibody-dependent and antibody-independent infection of cells of multiple lineages using a prototype DV2 strain and recent isolates. In a subset of cells, asymmetric competitive reverse transcriptase-PCR (RT-PCR), flow cytometry, and plaque assays were used to quantitate the steady-state levels of positive and negative viral RNA strands, the percentage of cells that express viral antigen, and the amount of secreted virus, respectively. Dose-response studies were conducted to assess the relative susceptibilities of particular cells to individual viral isolates. Overall, we find significant variation in the ability of DV2 isolates to productively infect different cells. Depending on the cell type and viral strain, productive infection may be limited by barriers to the accumulation of negative-strand viral RNA, the production of viral antigen, or, possibly, the packaging and secretion of infectious virus.
MATERIALS AND METHODS
Cell culture.
Human umbilical vein endothelial cells (HUVEC) were purchased commercially (Clonetics Corporation, San Diego, Calif.), maintained according to the manufacturer's instructions in endothelial cell culture media (EGM Bullet kit; Clonetics Corporation), and used from passages two to five. The following human cells were obtained from the American Type Culture Collection (Manassas, Va.), unless otherwise noted, and were cultured in RPMI medium (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS) (Sigma Chemical Co., St. Louis, Mo.), 100 U of penicillin G/ml, and 100 μg of streptomycin/ml at 37°C in 5% CO2: human foreskin fibroblasts (HFF; gift from M. Grigg and J. Boothroyd, Palo Alto, Calif.; used through passage 16), U937 myelomonocytes, Monomac-6 myelomonocytes (gift from J. Ernst, San Francisco, Calif.), HepG2 hepatoma cells, HeLa cervical epithelioid carcinoma cells (gift from L. Riley, Berkeley, Calif.), HL-60 promyelocytic leukemic cells, SW13 adrenal cortex adenocarcinoma cells (gift from R. Andino, San Francisco, Calif.), JY Epstein-Barr virus-transformed B cells (gift from L. Petruzzelli, Ann Arbor, Mich.), SKW3 T-lymphoma cells (gift from L. Petruzzelli), and K562 erythroleukemic cells (gift from L. Petruzzelli), COS-7 African green monkey transformed kidney fibroblasts (gift from L. Klickstein, Boston, Mass.), and L929 murine fibroblasts (gift from L. Riley) were also maintained in RPMI medium. THP-1 monocyte leukemic cells (gift from S. Goth and R. Stephens, Berkeley, Calif.) and 293 embryonal kidney fibroblasts were maintained in Dulbecco modified Eagle medium (DMEM) (Gibco BRL)–10% FBS–100 U of penicillin G and 100 μg of streptomycin/ml at 37°C in 5% CO2. BHK21-15 hamster kidney cells (gift from S. Kliks, San Francisco, Calif.) were maintained in α-modified Eagle medium (α-MEM) (Gibco BRL)–10% FBS–100 U of penicillin G and 100 μg of streptomycin/ml at 37°C in 5% CO2. C6/36, an Aedes albopictus cell line, was cultured in Leibovitz's L-15 medium (Gibco BRL)–10% FBS–100 U of penicillin G and 100 μg of streptomycin/ml at 28°C in the absence of CO2. Human peripheral blood monocytes and monocyte-derived macrophages were maintained in endotoxin-free RPMI medium–2.5% human AB serum (Sigma Chemical Co.) in the absence of antibiotics. U937 and HL-60 cells were differentiated toward the granulocyte or macrophage lineage by the addition of dimethyl sulfoxide (DMSO; 1.25% [vol/vol]) or phorbol-myristate acetate (PMA; 16 nM; Sigma Chemical Co.) for 72 h prior to infection.
Antibodies.
Murine hybridomas against DV envelope (E protein; 3H5-1, anti-DV2; 5D4-11, anti-DV3) or membrane (prM protein; 2H2-9, anti-DV) proteins or flavivirus antigens (4G2) were obtained (American Type Culture Collection) and grown in DMEM (Gibco BRL) supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, and 100 U of penicillin G and 100 μg of streptomycin/ml at 37°C in 5% CO2. Supernatants were collected from cell cultures that had reached greater than 50% cell death, centrifuged, filtered, and stored at −20 or 4°C. For some investigations, monoclonal antibodies (MAbs) were purified after 45% NH4SO4 precipitation and protein A affinity chromatography and directly conjugated to fluorescein isothiocyanate (FITC; Molecular Probes, Eugene, Oreg.) (19). Unless otherwise noted, in functional assays and immunostaining, purified immunoglobulin G was used at 10 to 20 μg/ml and tissue culture supernatants were used at a 1/4 final dilution. FITC-labeled goat anti-mouse antibodies (Sigma Chemical Co.) were used at a 1/250 dilution after centrifugation (14,000 × g for 5 min) to remove insoluble debris.
Virus stocks.
DV2 strains used in this study include a prototype DHF strain from Thailand (16681; kindly provided by the Centers for Disease Control and Prevention, Fort Collins, Colo.) (53), two recent DHF isolates from Thailand (C0477 and K0049; a gift from R. Rico-Hesse, San Antonio, Tex.) (48), and recent DF isolates from Nicaragua (N9622, N1042, N1043, N1047, and N1064; a gift from A. Balmaseda, Managua, Nicaragua) (3). All experiments used viral stocks from tissue culture passage two except those with strain 16681. Viral stocks were obtained by inoculating monolayers of C6/36 cells in 75-cm2 tissue culture flasks with virus diluted 1:5 to 1:10 in 1 ml of L15 containing 2% FBS. After 1 h, 14 ml of L15 supplemented with 10% FBS was added, and the cells were cultured for 7 days. Cells and supernatant were then harvested by gentle pipetting. Cell debris was removed by centrifugation (2,000 × g for 5 min), and the viral supernatant was adjusted to 20% FBS, aliquoted, and stored at −80°C.
Virus titration by plaque assay.
Virus production was titered by plaque assays using BHK21-15 cells. BHK21 cells were seeded in 6-well (6 × 105 cells/well) or 12-well (3 × 105 cells/well) plates in α-MEM with 10% FBS for 3 h at 37°C. Medium was then removed, serial dilutions of virus supernatants in α-MEM with 2% FBS were added (0.30 ml/well for 6-well plates and 0.15 ml/well for 12-well plates), and the cells were incubated for 2 h at 37°C. Subsequently, α-MEM containing 5% FBS and 1% low-melting-point agarose (SeaPlaque; FMC Bioproducts, Rockland, Maine; 3 ml/well for 6-well plates and 1.5 ml/well for 12-well plates) was added, and the plates were incubated at 37°C for 5 days. The plaques were visualized after 10% formaldehyde fixation (>1 h at room temperature) and removal of the agarose plug by staining briefly (15 to 30 s) with a 1% crystal violet solution in 20% ethanol. Virus concentrations were determined as PFU per milliliter and used to calculate the multiplicity of infection (MOI) in infection experiments.
Monocytes and monocyte-derived macrophages.
Monocytes were isolated from the whole blood of healthy volunteers by Ficoll-Hypaque gradient centrifugation (12). In brief, peripheral blood mononuclear cells were isolated from the interface of a Ficoll gradient, washed four times in endotoxin-free phosphate-buffered saline (PBS) at 4°C, quantitated, and resuspended in endotoxin-free RPMI medium supplemented with 1% human AB serum (Sigma Chemical Co.). Monocytes were selected by adherence after adding cells (2.5 × 106 cells/well in 1 ml) to individual wells of a tissue culture-treated 12-well plate and incubating for 1 h at 37°C. Unattached cells (e.g., platelets and lymphocytes) were removed by eight washes with RPMI medium. The monocyte phenotype was confirmed by immunostaining with phycoerythrin-conjugated anti-CD14 (Becton Dickinson, Franklin Lakes, N.J.). The percentage of monocytes after adherence ranged from 88 to 95%. To generate monocyte-derived macrophages, monocytes were cultured in RPMI medium supplemented with 2.5% human AB serum for 6 days at 37°C.
Cell infection (antibody-independent).
Cells were infected after adherence to tissue culture plastic (293, SW13, HUVEC, HFF, HepG2, COS-7, L929, U937 plus PMA, monocytes, monocyte-derived macrophages) or in suspension (U937, HL-60, Monomac-6, THP-1, JY, SKW3, K562). Experiments with SW13 cells demonstrated that the state of the cells (adherent or suspended) had little effect on the efficiency of infection (data not shown). Adherent cells (105 to 2 × 105 cells/well) were seeded in 12- or 24-well plates. At the time of infection, medium was removed and the virus was diluted in α-MEM with 2% FBS, added to monolayers or suspensions of cells at a given MOI, and incubated at 37°C for 2 h. The viral supernatants were removed, and the cells were washed six times to remove residual virus and then incubated at 37°C for 72 h prior to harvest. For cells infected in suspension, cells were washed in α-MEM with 2% FBS, exposed to viral supernatants (total volume, 200 μl), and incubated at 37°C for 2 h with agitation every 20 min to prevent cell sedimentation. Cells were then washed six times by centrifugation (900 × g for 3 min) and reseeded in 6- or 12-well plates for 72 h at 37°C. Supernatants were collected for plaque assay, and cells were harvested for flow cytometry and RNA determination.
Cell infection (antibody-dependent).
For antibody-dependent enhancement of DV infection studies, cells bearing Fcγ receptors (U937, THP-1, Monomac-6, monocytes, monocyte-derived macrophages) were subjected to infection in the presence of subneutralizing concentrations (less than 1 μg/ml) of antibody using a modification of a previously published protocol (5). Cells (2.5 × 105) were resuspended in 100 μl of α-MEM with 2% FBS. MAb (200 ng of antiflavivirus antibody 4G2 or anti-DV2 antibody 3H5-1 in 50 μl) was mixed with 50 μl of diluted DV2 virus and added to cells, and the resulting mixture was incubated for 2 h at 37°C. Cells were then extensively washed (eight times) to remove residual free virus, growth medium was replaced, and antibody (200 ng of 4G2 or 3H5) was added back as previously described (5). Cells were incubated for 96 h at 37°C prior to supernatant and cell harvest for plaque, flow cytometry, and RNA determination assays.
Flow-cytometric analysis.
For antibody-independent or antibody-dependent infection, DV2-infected and control cells were harvested at 72 or 96 h after infection, respectively. An aliquot (125 μl) of supernatant was removed for storage at −80°C for plaque assay, and the cells were divided into two pools, one for flow-cytometric analysis and one for RNA quantitation. In addition, an aliquot of cells was removed for quantitation of the total number of cells with a hemocytometer. For flow-cytometric determination of DV infection, harvested cells were transferred into individual wells of a 96-well U-bottom non-tissue culture plate. Cells were washed three times in PBS by centrifugation, fixed in PBS with 4% paraformaldehyde for 10 min at room temperature, washed twice in PBS, and permeabilized in Hank's balanced salt solution (Sigma Chemical Co.) containing 10 mM HEPES (pH 7.3), 0.1% saponin (Aldrich Chemical, St. Louis, Mo.), and 0.02% NaN3 (HHSN). For indirect immunofluorescence experiments, cells were resuspended in 100 μl of HHSN and 25 μl of MAb (anti-DV2 for detection of positive samples and anti-DV3 as a negative control), incubated for 1 to 2 h on ice, washed three times in HHSN (4°C), resuspended in a 1/250 dilution of FITC-labeled goat anti-mouse immunoglobulin G (50 μl), and incubated for 1 h on ice in the dark. Cells were subsequently washed three times in HHSN (4°C), fixed in 0.5% paraformaldehyde, and stored in the dark prior to flow cytometry. To avoid background staining associated with the presence of enhancing antibodies, immunofluorescence detection was performed with directly conjugated MAbs for the antibody-dependent DV2 infection studies. For these experiments, permeabilized cells were resuspended in a mixture of HHSN (100 μl) and antibody (a 20-μg/ml concentration of FITC-labeled anti-DV for detection of positive samples or FITC-labeled anti-DV3 as a negative control) supplemented with 5% human serum and incubated for 1 to 2 h on ice in the dark. Subsequently, cells were washed and fixed as described above for indirect immunofluorescence. Additional controls demonstrated that the background binding of the anti-DV2 or anti-DV MAb to uninfected cells was equivalent to the background binding of the anti-DV3 MAb used as a negative control (data not shown). Samples were analyzed on a FACSCAN flow cytometer using Cellquest software (Becton Dickinson).
RNA extraction and competitive RT-PCR.
Total RNA was extracted from infected cells using the RNEasy minikit (Qiagen, Valencia, Calif.) according to the manufacturers' instructions and was eluted in 35 to 100 μl of RNase-free double-distilled water. Positive- and negative-strand DV RNA was quantitated using a recently developed competitive RT-PCR assay (10). In brief, competitors for both the positive and negative strand were designed by fusing sequences from the nonstructural 3 gene of DV2 to sequences of the green fluorescent protein gene. The positive-strand (324 nucleotides) and negative-strand (332 nucleotides) competitors were synthesized by T7 RNA polymerase, purified after DNase treatment by passage over an RNEasy spin column, and quantitated by spectrophotometry. For quantitative, asymmetric, competitive RT-PCR, serial dilutions of the competitor RNA were mixed with a fixed amount of DV RNA harvested from infected cells and subjected to RT-PCR. cDNA was synthesized with the RT Rav-2 (Amersham Pharmacia, Piscataway, N.J.) and either the antisense (to measure the positive strand) or sense (to measure the negative strand) primer and amplified with Taq polymerase (Perkin-Elmer, Foster City, Calif.) using a previously published protocol (20). PCR products were separated by 1.5% agarose gel electrophoresis. The amount of viral RNA was determined from the competitor concentration that produced competitor and DV bands of equal intensities. RNA per cell was calculated as follows: RNA per cell = [(competitor concentration in copies per microliter) × (total volume of RNA/volume of RNA in RT-PCR)]/(total number of cells).
RESULTS
Infection of cell lines of different lineages with DV2.
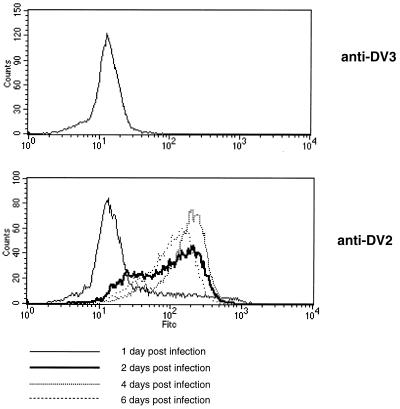
Experiments were undertaken initially to determine which cells were infected by DV2 in vitro in the absence of enhancing antibodies using a flow-cytometric assay that permitted quantitative, objective, and reproducible analysis. Because most prior studies have been performed by immunofluorescence microscopy with manual quantitation, few dose-response studies were available to show the relative susceptibilities to infection among cell types. In our experiments, flow-cytometric analysis was used to measure the intracellular accumulation of viral proteins as a marker of cell infection after labeling with anti-E or anti-prM MAbs. Permeabilized cells were used because the levels of E and prM proteins on the surfaces of intact infected cells were variable and did not consistently correspond to the levels observed within cells. We analyzed DV2-positive cells relative to two negative controls: uninfected cells (data not shown) and infected cells stained with a MAb to the E protein of DV3. A representative example is shown (Fig. 1); the SW13 human carcinoma cell line expressed significant amounts of E protein intracellularly over time (17% of cells at 1 day postinfection, 68% at 2 days postinfection, 90% at 4 days postinfection, and 79% at 6 days postinfection) after infection with the prototype 16681 DV2 strain.
FIG. 1.
Time course of infection of SW13 cells by DV2. SW13 cells were exposed to DV2 (strain 16681; MOI of 1) and incubated for 1, 2, 4, and 6 days. Cells (2 × 105) were processed for flow cytometry after fixation and detergent permeabilization and incubated with a MAb that recognizes the E protein of DV3 (top) or DV2 (bottom). The flow-cytometric data are expressed as the log of fluorescence intensity. One representative experiment of two is shown.
Subsequent studies were undertaken with 15 cell types of multiple lineages, including tumor cell lines, hematopoietic cells (monocytes, monocyte-derived macrophages), and primary untransformed cells (HFF and HUVEC). The cells assessed were of human origin with the exception of BHK21 (hamster), COS-7 (monkey), and L929 (murine) cells. Several of the cell lines were exposed to the 16681 prototype virus over a range of input concentrations (MOI, 0.01 to 10) to obtain a dose-response curve so that the relative susceptibilities to infection could be determined. Other cells (HL-60, U937) were exposed to pharmacological agents to assess how the state of differentiation affected infection. Overall, in the absence of antibody, adherent cells of epithelial (SW13, HepG2, BHK21) and mesenchymal (HFF, 293, L929, COS-7) lineage showed the greatest susceptibility to and highest percentage of cell infection (Table 1). Most cell lines of hematopoietic origin, with the exception of the erythroleukemic cell line K562, were relatively resistant to infection with the prototype strain of DV2 in the absence of enhancing antibodies, as judged by flow cytometry. Peripheral blood monocytes and monocyte-derived macrophages, which have been suggested to play a role in infection in vivo in DHF, showed no significant infection by flow cytometry. A subset of HUVEC was susceptible to DV2 infection but only at high MOI.
TABLE 1.
Antibody-independent infection of cells with DV2
| Cell line | Cell type | % Positive cellsa | MOI50b |
|---|---|---|---|
| Epithelial lineage | |||
| BHK21-15 | Hamster kidney | 72.6 | |
| SW13 | Adrenal adenocarcinoma | 73.6 | |
| HepG2 | Hepatoma | 78.0 | 0.05 |
| 293 | Embryonal kidney | 90.5 | 0.05 |
| HeLa | Epithelioid carcinoma | 62.1 | 1.0 |
| Myeloid lineage | |||
| U937 | Histiocytic lymphoma | <0.2 | |
| U937 + DMSO (1.25%) | 3.7 | ||
| U937 + PMA (16 nM) | 2.1 | ||
| HL-60 | Promyelocytic leukemia | 0.3 | |
| HL-60 + DMSO (1.25%) | 2.4 | ||
| HL-60 + PMA (16 nM) | 1.0 | ||
| Monomac-6 | Myelomonocytic leukemia | <0.2 | |
| Monocyte-derived macrophages | <0.2 | ||
| Hematopoietic lineage | |||
| JY | EBVc B-cell lymphoma | 1.3 | |
| SKW3 | T-cell leukemia | <0.2 | |
| K562 | Erythroleukemia | 45.0 | 0.05 |
| Fibroblast lineage | |||
| HFF | HFF | 65.0 | 0.7 |
| L929 | Murine fibroblast | 78.0 | 0.5 |
| COS-7 | Monkey kidney fibroblast | 81.0 | 0.3 |
| Endothelial cell lineage | |||
| HUVEC | Human umbilical vein endothelial cell | 12.0 | 5.0 |
Human cells (unless specified) were exposed to DV2 (strain 16681), cultured for 72 h, harvested, and subjected to flow cytometry with MAbs against the E protein. The percentage of positive cells reflects expression of DV2 antigens after infection at the maximal input virus concentration (MOI, 10). Data are expressed as the average percentages of cells infected and are from at least two experiments per cell type. The detection of 0.2% positive cells approaches the sensitivity of this assay.
For cells with appreciable levels of infection, a dose-response curve was conducted over a range of virus concentrations (MOI, 0.1 to 10.0). MOI50, relative virus concentration that caused infection at 50% of the maximum rate.
EBV, Epstein-Barr virus.
Infection of HepG2 and HFF with distinct DV2 strains.
Although the flow cytometry data provided information as to the relative susceptibilities to infection of several cell types, we wanted to extend the analysis to low-passage clinical isolates. The majority of prior infection studies with DV2 have been performed with two prototype strains (New Guinea C [5, 18, 27, 28, 32–34, 39] and 16681 [2, 15, 17, 18, 56]). Therefore, two cell types, one tumor cell line (HepG2) and one untransformed primary cell line (HFF), were tested for infection with three recent DV2 isolates with documented passage histories. Cells of the hepatocyte lineage are implicated in the pathogenesis of DV infection in vivo since patients with DF and DHF show evidence of liver injury. HFF cells were chosen as an example of dermal fibroblasts that may play a role in primary DV infection as the initial site of viral replication after mosquito inoculation (27). C0477 and K0049 are strains from Thailand (southeast Asian genotype) (48), and N9622 is a strain from Nicaragua (3) (Jamaican genotype). Experiments were conducted from viral stocks generated from the second passage in mosquito cells. In addition to the assessment of infection by flow cytometry, parallel plaque assay experiments were performed.
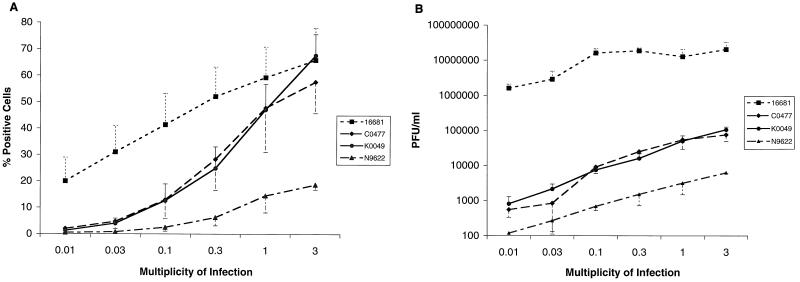
HepG2 cells were exposed to the four DV2 strains over a range of viral concentrations (MOI, 0.01 to 3). Flow cytometric analysis revealed that, at an MOI of 3, HepG2 cells were infected on average (n = 3) 66% ± 12% by 16681, 58% ± 12% by C0477, 68% ± 8% by K0049, and 18% ± 2% by N9622. Dose-response curves demonstrated that the prototype 16681 strain had the lowest threshold of infection and required the least input virus to reach maximal infection (Fig. 2A). Furthermore, the Nicaraguan strain required the most input virus and plateaued at the lowest percentage of infection. Plaque assay studies showed that, at the highest MOI used, infectious virus was produced in the supernatant at a concentration of 2.0 × 107 ± 1.2 × 107 PFU/ml by 16681, 7.6 × 104 ± 2.6 × 104 PFU/ml by C0477, 1.1 × 105 ± 2.1 × 104 PFU/ml by K0049, and 6.3 × 103 ± 2.4 × 102 PFU/ml by N9622. The dose-response curves confirm the disparity between the prototype strain and the recent DV isolates (Fig. 2B). Even at low levels of input virus (MOI, 0.01 to 0.03), cells infected with 16681 produced abundant infectious virus at levels 1,000- to 10,000-fold greater than those produced by the other strains.
FIG. 2.
Infection of HepG2 cells by different strains of DV2. HepG2 cells were exposed to the prototype 16681 strain or recent Thai (K0049 and C0477) and Nicaraguan (N9622) isolates over a range of MOIs and incubated for 72 h. Cells (106) were processed for flow cytometry (A), and supernatants were harvested for plaque assays (B). The flow-cytometric data are expressed as the percentages of positive cells. The plaque assay data are expressed as the numbers of PFU per milliliter based on cytopathic effect in BHK21 cells. The data are the averages of three experiments, and the error bars represent the standard errors of the means.
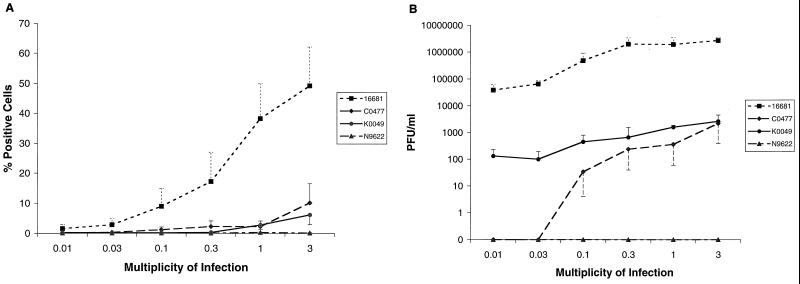
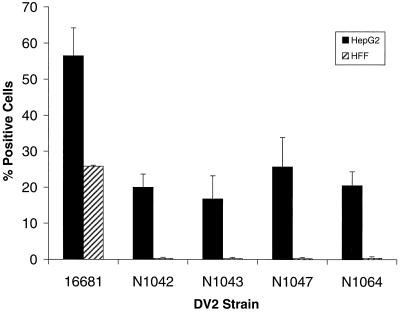
Similar dose-response studies were performed with HFF cells. Analysis of intracellular viral antigen by flow cytometry suggested that HFF cells were less susceptible than HepG2 cells to DV infection (Fig. 3A). At low MOI, infection of HFF cells with the prototype 16681 strain was notably lower than that of HepG2 cells. For example, an MOI of 0.03, which infected 31% of HepG2 cells, infected only 3% of HFF. Furthermore, C0047 and K0049 infected HFF cells significantly (6 to 10%) only at the highest MOI assayed. We were unable to show intracellular antigen accumulation after infection with Nicaraguan strain N9622, even at comparable MOIs. This pattern was confirmed in plaque assays (Fig. 3B), which demonstrated no infectious virus in the supernatants of HFF cells exposed to N9622. Because strain N9622 did not infect HFF cells productively despite accumulating antigen and generating infectious virus in HepG2 cells, four additional Nicaraguan strains were tested. The DV2 strains N1042, N1043, N1047, and N1064 were isolated from DF patients from Nicaragua in 1999, have the same restriction site-specific PCR subtype as N9622 (“Jamaica;” data not shown), and were passaged only twice. Although all four Nicaraguan strains infected HepG2 cells (17 to 26% positive cells, 3 × 103 to 5 × 103 PFU/ml), they did not accumulate significant amounts of antigen or infectious virus in HFF cells at the highest MOI tested (Fig. 4 and data not shown).
FIG. 3.
Infection of HFF cells by different strains of DV2. HFF cells were exposed to the prototype 16681 strain or recent Thai (K0049 and C0477) and Nicaraguan (N9622) isolates over a range of MOIs and incubated for 72 h. Cells (2 × 105) were processed for flow cytometry (A), and supernatants were harvested for plaque assays (B). The data are expressed as described for Fig. 2.
FIG. 4.
Infection of HepG2 and HFF cells by Nicaraguan DV2 isolates. HFF cells and HepG2 cells were exposed to the 16681 prototype Thai strain or recent Nicaraguan isolates (N1042, N1043, N1047, N1064) at an MOI of 3 and incubated for 72 h. Cells (HepG2, 6 × 105; HFF, 2 × 105) were processed for flow cytometry. The data are expressed as described for Fig. 2.
Antibody-dependent enhancement of infection: myeloid cells.
Flow cytometry and plaque assays were performed on cells of myeloid lineage in the presence and absence of enhancing antibodies using prototype and low-passage DV2 isolates. Prior studies quantitated levels of secreted virus in supernatants (5, 17, 24, 28, 45) but did not directly measure antibody-dependent and antibody-independent infection rates of myeloid cells with recent DV isolates.
In the absence of enhancing antibodies, the flow-cytometric assay revealed little evidence of DV2 (strain 16681) antigen expression in HL-60 promyelocytes, U937 myelomonocytes, THP-1 monocyte leukemic cells, peripheral blood monocytes, and monocyte-derived macrophages even at the highest MOI tested (Tables 1 and 2). However, infectious virus was detected at low levels in supernatants from U937 cells and some monocyte donors that were infected with the prototype DV2 strain at high MOIs (MOI of 10; Table 2 and data not shown). The disparity may reflect the lower sensitivity of flow cytometry for detecting infrequent (i.e., <1 in 100) positive cells or may be due to the presence of a significant number of cells infected at levels below immunofluorescence detection. When enhancing MAbs were added, reproducible increases in the percentage of U937 cells expressing viral antigen when infected with the Thai strains were observed (16681, from 0.1% ± 0.09% to 23.0% ± 8%; K0049, from 0.09% ± 0.01% to 2.6% ± 0.7%) but not with a Nicaraguan strain (N9622, from 0.07% ± 0.07% to 0.08% ± 0.07%). Antibody addition also augmented the percentage of THP-1 cells that expressed antigen after infection with 16681 (from 0.06% ± 0.05% to 1.4% ± 0.3%) but not with other DV2 strains. Moreover, even at the highest MOI tested, DV2 in the presence of enhancing MAbs did not infect enough peripheral blood monocytes or monocyte-derived macrophages to be detected by flow cytometry (data not shown). In comparison, enhancing MAbs significantly increased infectious virus production of the 16681 prototype strain. Increases of several log units in viral titer were observed in supernatants from THP-1 and U937 cells infected with 16681 in the presence of enhancing MAbs; however, the effect in monocytes was variable among donors and between experiments (Table 2 and data not shown). When recent DV isolates were used (K0049 and N9622), only U937 cells showed an increase in virus production; no appreciable titers were detected in THP-1 cells, monocytes, or macrophages despite the presence of antibody, the use of 106 cells per assay, and the addition of virus at the highest possible MOI.
TABLE 2.
Antibody-dependent DV2 infection of myeloid cells
| Cells | DV2 strain | % Positive cellsa ± SEM with:
|
PFU/mlb ± SEM with:
|
||
|---|---|---|---|---|---|
| No MAb | 4G2 MAb | No MAb | 4G2 MAb | ||
| U937 | 16681 | <0.2 | 23 ± 8 | 4.4 × 103 ± 1.6 × 103 | 3.6 × 106 ± 1.7 × 106 |
| C0477 | <0.2 | 0.8 ± 0.3 | 0 | 1.8 × 102 ± 1.2 × 102 | |
| K0049 | <0.2 | 2.6 ± 0.7 | 0 | 8.9 × 103 ± 2.2 × 103 | |
| N9622 | <0.2 | <0.2 | 0 | 0 | |
| THP-1 | 16681 | <0.2 | 1.4 ± 0.3 | 0 | 8.1 × 103 ± 1.3 × 103 |
| C0477 | <0.2 | 0.2 ± 0.1 | 0 | 0 | |
| K0049 | <0.2 | <0.2 | 0 | 3.3 × 101 ± 2.6 × 101 | |
| N9622 | <0.2 | <0.2 | 0 | 0 | |
U937 or THP-1 cells were exposed to DV2 strains (16681, MOI of 10; K0049, MOI of 4.5; N9622, MOI of 3) in the absence (no MAb) or presence (4G2) of an enhancing MAb. After incubation for 96 h, cells were processed for flow cytometry. The data are the averages of three experiments. The detection of 0.2% positive cells approaches the sensitivity of this assay.
After exposure of U937 or THP-1 cells to DV2 strains, supernatants were harvested for plaque assays. The data are the averages of three experiments. A value of zero indicates no plaques were detected in any of the experiments.
Competitive RT-PCR: quantitation of RNA levels.
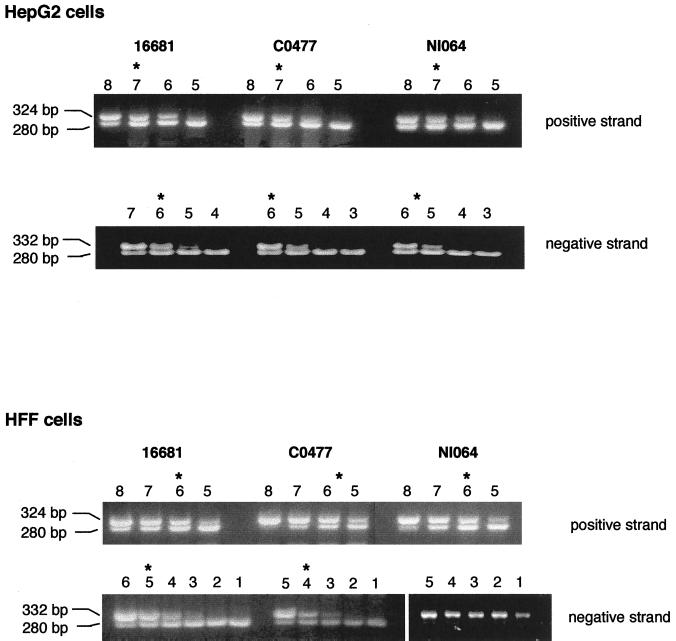
The flow-cytometric and plaque assays demonstrated that, among recent isolates, the Thai strains consistently showed greater levels of infection than the Nicaraguan strains. Because our previous work suggested that the accumulation of negative-strand viral RNA was critical for productive cell infection by DV (10), we hypothesized that the disparity in levels of infection among isolates might be explained at a viral RNA level. To address this, we utilized an asymmetric, competitive RT-PCR to measure positive and negative viral RNA strands. HepG2 and HFF cells were infected with the prototype and recently isolated DV2 strains and analyzed 72 h after infection for steady-state levels of viral RNA (Fig. 5 and Table 3). In HepG2 cells, there was little significant difference in levels of positive and negative viral RNA strands among DV2 strains. Although additional studies that assess the kinetics of viral RNA production are required, the disparity in virus production among the recent isolates in HepG2 cells did not correlate with differences in the levels of viral RNA. In HFF cells, although positive-strand levels were lower than those in HepG2 cells, again there was little difference in the amounts of positive-strand RNA among viral isolates. In contrast, we observed significant differences in negative-strand viral RNA levels in HFF cells infected with the different viral isolates. At 72 h after exposure to virus, there was a 100-fold reduction between the prototype strain and a recent Thai isolate and exceedingly low levels of negative-strand viral RNA in HFF cells that were exposed to the Nicaraguan strains. In HFF cells, these differences in negative-strand RNA correlated with the differences in antigen accumulation and virus production.
FIG. 5.
Asymmetric, competitive RT-PCR quantification of positive- and negative-strand viral RNA. HepG2 and HFF cells were exposed to Thai (16681, C0477) or Nicaraguan (N1064) strains and incubated for 72 h. Cells (HepG2, 6 × 105; HFF, 6 × 105) were harvested, total RNA was isolated, and quantitative asymmetric RT-PCR was performed with fixed amounts of cellular RNA in the presence of 10-fold-decreasing concentrations of positive- or negative-strand competitor. The RT-PCR product was subjected to agarose gel electrophoresis. The number above each lane represents the log of the number of copies of competitor used. The amount of viral RNA was determined from the competitor concentration that produces competitor and DV bands of equal intensities (asterisk). For the positive strand, the equivalence points and numbers of RNA copies per cell were as follows. HepG2 cells: 16681, 107, 1,666 RNA copies/cell; C0477, 107, 1,666 RNA copies/cell; N1064, 107, 1,666 RNA copies/cell; HFF cells: 16681, 106, 167 RNA copies/cell; C0477, 5 × 105, 83 RNA copies/cell; N1064, 106, 167 RNA copies/cell. For the negative strand, the equivalence points and numbers of RNA copies per cell were as follows. HepG2 cells: 16681, 106, 83 RNA copies/cell; C0477, 106, 83 RNA copies/cell; N1064, 5 × 105, 42 RNA copies/cell; HFF cells: 16681, 105, 16 RNA copies/cell; C0477, 104, 0.8 RNA copies/cell; N1064, <101, <0.01 RNA copies/cell. The limit of sensitivity of the assay is 0.01 under these conditions.
TABLE 3.
Summary of competitive RT-PCR data
| DV2 straina | No. of copies of RNA/cellb
|
|
|---|---|---|
| Positive strand | Negative strand | |
| HepG2 cells | ||
| 16681 | 657 ± 303 | 31 ± 17 |
| C0477 | 875 ± 175 | 25 ± 11 |
| K0049 | 735 ± 385 | 9 ± 5 |
| N9622 | 875 ± 525 | 9 ± 6 |
| N1042 | 1,666 ± 125 | 12 ± 4 |
| N1043 | 1,500 (ND) | 16 (ND) |
| N1047 | 1,500 (ND) | 16 (ND) |
| N1064 | 1,583 ± 117 | 26 ± 18 |
| HFF cells | ||
| 16681 | 81 ± 37 | 16 ± 11 |
| C0477 | 78 ± 55 | 0.5 ± 0.4 |
| K0049 | 110 ± 90 | 0.2 ± 0.5 |
| N9622 | 166 (ND) | 0.02 |
| N1042 | 44 ± 10 | ND |
| N1043 | 23 (ND) | <0.01c |
| N1047 | 131 ± 91 | 0.01 ± 0.02 |
| N1064 | 103 ± 63 | 0.01 ± 0.01 |
Cells (4 × 105) were exposed to Thai strains (16681, C0477, and K0049) or Nicaraguan strains (N9622, N1042, N1043, N1047, and N1064), incubated for 72 h, and harvested.
Total RNA was isolated and subjected to asymmetric, competitive RT-PCR using an antisense primer for reverse transcription to measure the positive strand and a sense primer to measure the negative strand. The data are averages ± standard errors of the means. ND, not determined.
The limit of sensitivity of the assay is 0.01 under these conditions.
Parallel studies were performed with myeloid cells (U937, THP-1, and Monomac-6) to determine the levels of positive and negative viral RNA strands after antibody-dependent infection with the prototype (16681), and low-passage (C0477 and N9622) strains. Overall, among all cell types and viral strains, at 72 h after infection there was a 10- to 100-fold-lower level of positive-strand RNA present relative to the levels observed in HFF and HepG2 cells. The average amounts of positive strand viral RNA per cell for different DV2 strains ranged from 0.4 to 3.6 for U937 cells, 0.6 to 5.6 for THP-1 cells, and 0.5 to 4.5 for Monomac-6 cells. This variation did not correlate with productive infection, as no significant difference in positive-strand RNA levels between the prototype strain and the recent isolates was observed in any of the cells tested. In contrast, significant differences in the levels of negative-strand RNA, which corresponded to the degree of productive infection, were observed. In each of the myeloid cells, the Nicaraguan isolate demonstrated a 100-fold reduction and the Thai isolate showed a 10- to 50-fold reduction in the level of negative-strand viral RNA relative to the quantity of negative-strand RNA generated by the prototype strain (0.03 negative-strand RNA copies per cell).
DISCUSSION
In this paper, we have reassessed antibody-dependent and antibody-independent DV2 infection using both prototype and low-passage viral isolates. Even though equivalent MOIs were used, a wide variation in the abilities of different DV2 isolates to infect cells productively was observed, as judged by the accumulation of viral RNA and antigen and the generation of infectious virus in cell supernatants. In certain cell types (e.g., HFF and myeloid cells), the disparities in productive infection correlated with the accumulation of negative-strand viral RNA. In other cell types (e.g., HepG2), productive infection occurred with all strains, but with marked differences in the amounts of viral antigen and the numbers of infectious virions detected. With each DV2 isolate, bound virus penetrated all cell types tested efficiently, as the level of positive-strand viral RNA changed little after exposure to proteinase K or alkaline high-ionic-strength solutions, treatments that detach surface-bound virus (M. Diamond and E. Harris, unpublished results). We surmise that factors subsequent to viral entry play a role in regulating the cellular tropism of productive DV infection. This could explain why the broad expression of heparan sulfate, a molecule proposed as a receptor for DV (7), does not correlate with the restriction of infection observed in vivo. Depending on the viral isolate and target cell, an interplay between virus- and cell-specific determinants may govern the level of initial translation of input virus, negative-strand viral RNA synthesis, viral protein production, and virion secretion.
The spectrum of DV infection in humans includes subclinical infection, mild or severe febrile illnesses, and the life-threatening DHF/DSS. Epidemiological and clinical studies have suggested that both viral and host factors influence the severity of disease (16, 40, 57). However, the lack of an adequate animal model and in vitro correlates has hindered the characterization of viral determinants of virulence. Molecular epidemiological studies have supported the importance of viral factors, as genetic analyses correlate specific DV2 genotypes with more severe disease (47, 50). The DV2 genotype native to Central and South America associates clinically with only DF despite widespread secondary infection (58), and the African DV2 genotype has a sylvatic cycle but does not cause appreciable disease in humans (47), whereas DV2 genotypes of southeast Asian origin are often associated with DHF/DSS (48, 49). Full-length sequencing studies of viral isolates with different genotypes and distinct clinical phenotypes correlate disease severity with particular sequence differences in several DV genes and in the 5′ and 3′ untranslated regions (30, 35). Individual DV2 genotypes may contain or lack particular sequences that influence disease transmission and virulence.
Prior functional studies in vitro have suggested that different viral genotypes exhibit different cellular infection characteristics. For example, DV2 strains isolated from patients with various disease severities produced different levels of infectious virus in LLC-MK2 cells (35), peripheral blood leukocytes, and C6/36 cells (41). In this paper, we demonstrate that, despite the use of similar MOI, DV2 isolates varied in their capacities to infect the same cell type in vitro depending on the virus genotype and passage history. We found that in HFF and myeloid cells, the degree of productive infection correlated with the accumulation of negative-strand viral RNA; strains with high levels of negative-strand viral RNA produced greater levels of viral antigen and secreted more infectious virus. In contrast, even 48 h after infection, HFF cells that were infected with different DV2 isolates and that generated various amounts of progeny virus contained comparable levels of positive-strand viral RNA. At present, it is unclear why negative-strand viral RNA levels should correlate with viral antigen production since significant amounts of positive-strand RNA are present within the cell. Nonetheless, this observation agrees with our prior studies with interferon-treated HepG2 cells, where antigen accumulation and virus production corresponded to the quantity of negative-strand RNA present (10). It is also consistent with reports of tight coupling between translation and replication (43) and between replication and viral assembly (44) in poliovirus, another positive strand RNA virus. One explanation that we are currently investigating is that the majority of input positive-strand RNA cannot be translated either because it has been modified by host antiviral effector molecules or targeted away from the translation machinery. Particular genetic determinants among DV2 strains may influence the rate of input virus translation or negative-strand viral RNA production and/or stability.
Viral determinants may also influence steps in cellular infection that are downstream of the synthesis and accumulation of negative-strand DV RNA. Our data showed a wide variation in the ability of HepG2 cells to produce infectious virus after infection with eight DV2 strains despite the presence of equivalent amounts of negative- and positive-strand viral RNA. For example, infection of HepG2 cells with the recent Thai isolates (C0477 and K0049) and the prototype DV2 strain (16681) resulted in a 3-log-unit difference in viral titer despite similar levels of RNA and similar percentages of cells expressing viral antigen. Presumably, sequence-specific determinants can modulate the rate of antigen accumulation or viral packaging or secretion. Analysis of the DV genome suggests that alterations in the downstream loop of the 5′ untranslated region may affect translation efficiency (30). Mutations in this region modify RNA-ribosome interactions and attenuate poliovirus (21) and Venezuelan equine encephalitis virus (22). Alternatively, viral proteins may be involved, as mutations in the NS1 protein of DV and the related Kunjin flavivirus affect dimerization and reduce the ability to produce infectious virus by more than 100-fold (13, 46). Future studies are required to identify the rate-limiting steps of virus production with DV strains that have well-defined clinical histories and known sequences.
Host factors also account for some of the variation in the clinical manifestations of DV infection. However, few cellular factors that modulate DV infection have been characterized. In this study, we show that, although eight different DV2 strains infect HepG2 cells productively, only three of them (the Thai strains) infect HFF cells. Although genetic differences among DV2 strains may account for the disparity in infection within a given cell type, they do not explain why infections of distinct cell types by the same virus are different. One hypothesis that could explain this is the existence of qualitative or quantitative differences in the viral receptors on HepG2 and HFF cells. Although the hypothesis is plausible, some experimental evidence argues against it. First, adherent cells in vitro (both HepG2 and HFF cells) express high levels of heparan sulfate (11, 55), a putative receptor for DV attachment (7). Second, although only a subset of DV2 strains generate negative-strand viral RNA and infectious virions in HFF cells, comparable levels of positive-strand viral RNA are detected several days after exposure to all DV2 strains. This suggests that entry alone does not determine cellular tropism, although the route of entry may influence the degree of productive infection achieved. We suggest that the presence or absence of as yet uncharacterized cellular factors modulates the permissiveness of a particular cell type to DV infection.
Although infection of cells of myeloid lineage by DV in the absence or presence of enhancing antibodies has been extensively documented (5, 15, 17, 18, 24, 26, 28, 32, 33, 45), few studies have used low-passage isolates (31, 37, 38, 42). Our data with the prototype, highly passaged 16681 strain are consistent with the results of prior studies. Infection in the absence of enhancing antibodies resulted in low-level virus production that was detectable only by plaque assay from supernatants of U937 cells and of monocytes from a subset of donors. The addition of enhancing antibodies augmented 16681 infection in U937 and THP-1 cells such that a significant percentage of infected cells were detected by flow cytometry. In contrast, when low-passage isolates were used, productive infection was detected only in U937 cells. Even at the highest MOI possible and in the presence of enhancing antibodies, we and others (31) were unable to detect infection consistently by either flow-cytometric or viral plaque assays of THP-1 cells, peripheral blood monocytes, or monocyte-derived macrophages. Some of this disparity was reflected at the negative-strand viral RNA level, as very small amounts were detected in myeloid cells infected with recent isolates.
By using multiple cell types and DV2 strains, we have shown that the degree of viral infection in vitro reflects an interplay between viral and cell-specific factors. Moreover, the results from our in vitro DV infection assays correlated with the DV2 genotype, as the three Thai (southeast Asian genotype) strains tested productively infected HFF cells whereas the five Nicaraguan (Jamaican genotype) isolates showed no measurable evidence of productive infection. In addition, there was a preliminary correlation between the in vitro infection pattern and in vivo phenotype. Although more viruses need to be analyzed to sustain this correlation, it is likely that viral determinants influence disease expression in the context of host and environmental factors. Future studies with an extended panel of Asian, African, and American DV2 isolates, as well as other serotypes, are under way to assess whether an in vitro phenotype correlates with DV genotype. If this observation is borne out, genetic analyses will be incorporated to identify the viral determinants that confer virulence in DV infection.
ACKNOWLEDGMENTS
We thank R. Rico-Hesse for providing recent DV isolates, L. Petruzzelli, L. Klickstein, S. Goth, R. Stephens, L. Riley, M. Grigg, J. Boothroyd, J. Ernst, and S. Kliks for providing cell lines and vectors, and P. Dazin for assistance with flow cytometry. We are grateful to J. Ernst and R. Beatty for constructive editorial comments and to S. Halstead and S. Kliks for helpful discussions and advice.
The work was supported by a National Institutes of Health grant to E. Harris (AI-42052) and by fellowships from the Giannini Foundation of the Bank of America and the Infectious Diseases Society of America to M. S. Diamond.
REFERENCES
- 1.Anderson R, King A D, Innis B L. Correlation of E protein binding with cell susceptibility to dengue 4 virus infection. J Gen Virol. 1992;73:2155–2159. doi: 10.1099/0022-1317-73-8-2155. [DOI] [PubMed] [Google Scholar]
- 2.Avirutnan P, Malasit P, Seliger B, Bhakdi S, Husmann M. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J Immunol. 1998;161:6338–6346. [PubMed] [Google Scholar]
- 3.Balmaseda A, Sandoval E, Perez L, Gutierrez C M, Harris E. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg. 1999;61:893–897. doi: 10.4269/ajtmh.1999.61.893. [DOI] [PubMed] [Google Scholar]
- 4.Bonner S M, O'Sullivan M A. Endothelial cell monolayers as a model system to investigate dengue shock syndrome. J Virol Methods. 1998;71:159–167. doi: 10.1016/s0166-0934(97)00211-5. [DOI] [PubMed] [Google Scholar]
- 5.Brandt W E, McCown J M, Gentry M K, Russell P K. Infection enhancement of dengue type 2 virus in the U-937 human monocyte cell line by antibodies to flavivirus cross-reactive determinants. Infect Immun. 1982;36:1036–1041. doi: 10.1128/iai.36.3.1036-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke D S, Nisalak A, Johnson D E, Scott R M. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 8.Couvelard A, Marianneau P, Bedel C, Drouet M T, Vachon F, Henin D, Deubel V. Report of a fatal case of dengue infection with hepatitis: demonstration of dengue antigens in hepatocytes and liver apoptosis. Hum Pathol. 1999;30:1106–1110. doi: 10.1016/s0046-8177(99)90230-7. [DOI] [PubMed] [Google Scholar]
- 9.Daughaday C C, Brandt W E, McCown J M, Russell P K. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptors and trypsin-resistant immune complex receptors. Infect Immun. 1981;32:469–473. doi: 10.1128/iai.32.2.469-473.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond M, Roberts T G, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich C P, Montes de Oca H. Surface sulfated mucopolysaccharides of primary and permanent mammalian cell lines. Biochem Biophys Res Commun. 1978;80:805–812. doi: 10.1016/0006-291x(78)91316-5. [DOI] [PubMed] [Google Scholar]
- 12.English D, Anderson B R. Single-step separation of red blood cells. granulocytes, and mononuclear cells on discontinuous density gradients of ficoll-hypaque. J Immunol Methods. 1974;5:249–253. doi: 10.1016/0022-1759(74)90109-4. [DOI] [PubMed] [Google Scholar]
- 13.Hall R A, Khromykh A A, Mackenzie J M, Scherret J H, Khromykh T I, Mackenzie J S. Loss of dimerisation of the nonstructural protein NS1 of Kunjin virus delays viral replication and reduces virulence in mice, but still allows secretion of NS1. Virology. 1999;264:66–75. doi: 10.1006/viro.1999.9956. [DOI] [PubMed] [Google Scholar]
- 14.Halstead S B. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis. 1989;11(Suppl. 4):S830–S839. doi: 10.1093/clinids/11.supplement_4.s830. [DOI] [PubMed] [Google Scholar]
- 15.Halstead S B. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis. 1979;140:527–533. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- 16.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 17.Halstead S B, O'Rourke E J. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265:739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- 18.Halstead S B, Venkateshan C N, Gentry M K, Larsen L K. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J Immunol. 1984;132:1529–1532. [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 20.Harris E, Roberts T G, Smith L, Selle J, Kramer L D, Valle S, Sandoval E, Balmaseda A. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36:2634–2639. doi: 10.1128/jcm.36.9.2634-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iizuka N, Kohara M, Hagino-Yamagishi K, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. Construction of less neurovirulent polioviruses by introducing deletions into the 5′ noncoding sequence of the genome. J Virol. 1989;63:5354–5363. doi: 10.1128/jvi.63.12.5354-5363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinney R M, Chang G J, Tsuchiya K R, Sneider J M, Roehrig J T, Woodward T M, Trent D W. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol. 1993;67:1269–1277. doi: 10.1128/jvi.67.3.1269-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliks S. Antibody-enhanced infection of monocytes as the pathogenetic mechanism for severe dengue illness. AIDS Res Hum Retroviruses. 1990;6:993–998. doi: 10.1089/aid.1990.6.993. [DOI] [PubMed] [Google Scholar]
- 24.Kliks S C, Nisalak A, Brandt W E, Wahl L, Burke D S. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 25.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontny U, Kurane I, Ennis F A. Gamma interferon augments Fc gamma receptor-mediated dengue virus infection of human monocytic cells. J Virol. 1988;62:3928–3933. doi: 10.1128/jvi.62.11.3928-3933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurane I, Janus J, Ennis F A. Dengue virus infection of human skin fibroblasts in vitro production of IFN-beta, IL-6 and GM-CSF. Arch Virol. 1992;124:21–30. doi: 10.1007/BF01314622. [DOI] [PubMed] [Google Scholar]
- 28.Kurane I, Kontny U, Janus J, Ennis F A. Dengue-2 virus infection of human mononuclear cell lines and establishment of persistent infections. Arch Virol. 1990;110:91–101. doi: 10.1007/BF01310705. [DOI] [PubMed] [Google Scholar]
- 29.Kurane I, Rothman A L, Livingston P G, Green S, Gagnon S J, Janus J, Innis B L, Nimmannitya S, Nisalak A, Ennis F A. Immunopathologic mechanisms of dengue hemorrhagic fever and dengue shock syndrome. Arch Virol Suppl. 1994;9:59–64. doi: 10.1007/978-3-7091-9326-6_7. [DOI] [PubMed] [Google Scholar]
- 30.Leitmeyer K C, Vaughn D W, Watts D M, Salas R, Villalobos de Chacon I, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y L, Liao C L, Chen L K, Yeh C T, Liu C I, Ma S H, Huang Y Y, Huang Y L, Kao C L, King C C. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Littaua R, Kurane I, Ennis F A. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990;144:3183–3186. [PubMed] [Google Scholar]
- 33.Mady B J, Erbe D V, Kurane I, Fanger M W, Ennis F A. Antibody-dependent enhancement of dengue virus infection mediated by bispecific antibodies against cell surface molecules other than Fc gamma receptors. J Immunol. 1991;147:3139–3144. [PubMed] [Google Scholar]
- 34.Mady B J, Kurane I, Erbe D V, Fanger M W, Ennis F A. Neuraminidase augments Fc gamma receptor II-mediated antibody-dependent enhancement of dengue virus infection. J Gen Virol. 1993;74:839–844. doi: 10.1099/0022-1317-74-5-839. [DOI] [PubMed] [Google Scholar]
- 35.Mangada M N, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244:458–466. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- 36.Marianneau P, Megret F, Olivier R, Morens D M, Deubel V. Dengue 1 virus binding to human hepatoma HepG2 and simian Vero cell surfaces differs. J Gen Virol. 1996;77:2547–2554. doi: 10.1099/0022-1317-77-10-2547. [DOI] [PubMed] [Google Scholar]
- 37.Marianneau P, Steffan A M, Royer C, Drouet M T, Jaeck D, Kirn A, Deubel V. Infection of primary cultures of human Kupffer cells by dengue virus: no viral progeny synthesis, but cytokine production is evident. J Virol. 1999;73:5201–5206. doi: 10.1128/jvi.73.6.5201-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marianneau P, Steffan A M, Royer C, Drouet M T, Kirn A, Deubel V. Differing infection patterns of dengue and yellow fever viruses in a human hepatoma cell line. J Infect Dis. 1998;178:1270–1278. doi: 10.1086/314466. [DOI] [PubMed] [Google Scholar]
- 39.Mentor N A, Kurane I. Dengue virus infection of human T lymphocytes. Acta Virol. 1997;41:175–176. [PubMed] [Google Scholar]
- 40.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morens D M, Marchette N J, Chu M C, Halstead S B. Growth of dengue type 2 virus isolates in human peripheral blood leukocytes correlates with severe and mild dengue disease. Am J Trop Med Hyg. 1991;45:644–651. doi: 10.4269/ajtmh.1991.45.644. [DOI] [PubMed] [Google Scholar]
- 42.Murgue B, Cassar O, Guigon M, Chungue E. Dengue virus inhibits human hematopoietic progenitor growth in vitro. J Infect Dis. 1997;175:1497–1501. doi: 10.1086/516486. [DOI] [PubMed] [Google Scholar]
- 43.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 44.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Sullivan M A, Killen H M. The differentiation state of monocytic cells affects their susceptibility to infection and the effects of infection by dengue virus. J Gen Virol. 1994;75:2387–2392. doi: 10.1099/0022-1317-75-9-2387. [DOI] [PubMed] [Google Scholar]
- 46.Pryor M J, Gualano R C, Lin B, Davidson A D, Wright P J. Growth restriction of dengue virus type 2 by site-specific mutagenesis of virus-encoded glycoproteins. J Gen Virol. 1998;79:2631–2639. doi: 10.1099/0022-1317-79-11-2631. [DOI] [PubMed] [Google Scholar]
- 47.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- 48.Rico-Hesse R, Harrison L M, Nisalak A, Vaughn D W, Kalayanarooj S, Green S, Rothman A L, Ennis F A. Molecular evolution of dengue type 2 virus in Thailand. Am J Trop Med Hyg. 1998;58:96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- 49.Rico-Hesse R, Harrison L M, Salas R A, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa M T, Nogueira R M, da Rosa A T. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 50.Rosen L. The Emperor's New Clothes revisited, or reflections on the pathogenesis of dengue hemorrhagic fever. Am J Trop Med Hyg. 1977;26:337–343. doi: 10.4269/ajtmh.1977.26.337. [DOI] [PubMed] [Google Scholar]
- 51.Rosen L, Drouet M T, Deubel V. Detection of dengue virus RNA by reverse transcription-polymerase chain reaction in the liver and lymphoid organs but not in the brain in fatal human infection. Am J Trop Med Hyg. 1999;61:720–724. doi: 10.4269/ajtmh.1999.61.720. [DOI] [PubMed] [Google Scholar]
- 52.Rothman A L, Ennis F A. Immunopathogenesis of dengue hemorrhagic fever. Virology. 1999;257:1–6. doi: 10.1006/viro.1999.9656. [DOI] [PubMed] [Google Scholar]
- 53.Russell P K, Udomsakdi S, Halstead S B. Antibody response in dengue and dengue hemorrhagic fever. Jpn J Med Sci Biol. 1967;20:103–108. [PubMed] [Google Scholar]
- 54.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead S B. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 55.Satoh C, Duff R, Rapp F, Davidson E A. Production of mucopolysaccharides by normal and transformed cells. Proc Natl Acad Sci USA. 1973;70:54–56. doi: 10.1073/pnas.70.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sittisombut N, Maneekarn N, Kanjanahaluethai A, Kasinrerk W, Viputtikul K, Supawadee J. Lack of augmenting effect of interferon-gamma on dengue virus multiplication in human peripheral blood monocytes. J Med Virol. 1995;45:43–49. doi: 10.1002/jmv.1890450109. [DOI] [PubMed] [Google Scholar]
- 57.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Endy T P, Raengsakulrach B, Rothman A L, Ennis F A, Nisalak A. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 58.Watts D M, Porter K R, Putvatana P, Vazquez B, Calampa C, Hayes C G, Halstead S B. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354:1431–1434. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]