Abstract
The RNA genome of Borna disease virus (BDV) shows extraordinary stability in persistently infected cell cultures. We performed bottleneck experiments in which virus populations from single infected cells were allowed to spread through cultures of uninfected cells and in which RNase protection assays were used to identify virus variants with mutations in a 535-nucleotide fragment of the M-G open reading frames. In one of the cell cultures, the major virus species (designated 2/1) was a variant with two point mutations in the G open reading frame. When fresh cells were infected with a low dose of a virus stock prepared from 2/1-containing cells, only a minority of the resulting persistently infected cultures contained detectable levels of the variant, whereas the others all seemed to contain wild-type virus. The BDV variant 2/1 remained stable in the various persistently infected cell cultures, indicating that the cells were resistant to superinfection by wild-type virus. Indeed, cells persistently infected with prototype BDV He/80 were also found to resist superinfection with strain V and vice versa. Our screen for mutations in the viral M and G genes of different rat-derived BDV virus stocks revealed that only one of four stocks believed to contain He/80 harbored virus with the original sequence. Two stocks mainly contained a novel virus variant with about 3% sequence divergence, whereas the fourth stock contained a mixture of both viruses. When the mixture was inoculated into the brains of newborn mice, the novel variant was preferentially amplified. These results provide evidence that the BDV genome is mutating more frequently than estimated from its invariant appearance in persistently infected cell cultures and that resistance to superinfection might strongly select against novel variants.
Genome replication of RNA viruses is error prone, because RNA-dependent RNA polymerases lack proofreading activity (10, 18). Nevertheless, field isolates of certain RNA viruses exhibit a high degree of genetic stability over many decades (10). The selective forces which restrict virus variability are presumably complex and nonuniform. In most cases, the mechanisms of restriction are largely unknown.
Borna disease virus (BDV) is a newly classified nonsegmented negative-strand RNA virus that can persistently infect the central nervous systems of a broad range of warm-blooded animals and possibly humans without destruction of its host cells (11, 13, 21, 29). Natural and experimental infections with BDV usually result in immune-system-mediated neurological disease and behavioral abnormalities (5, 13, 14, 34). Sequence comparisons between old and recent BDV isolates of diseased animals from regions of endemicity in Central Europe revealed viral genome conservation of greater than 95%, in spite of the fact that some of these viruses were passaged many times in experimental animals or in cell culture (3, 28). Recent evidence indicates that BDV strains from outside the classical regions of endemicity (e.g., Japan, Sweden, and the United States) (1, 2, 7, 11, 19) are virtually identical (95 to 98% identity) to the Central European strains (3, 28). However, we recently characterized a novel field isolate from a diseased horse in eastern Austria whose genome was only about 85% identical to that of classical BDV strains, demonstrating that BDV can easily tolerate more nucleotide exchanges (22). Intriguingly, the nucleotide exchanges of the novel BDV isolate have little effect on the primary structures of most viral proteins (22).
To elucidate the molecular basis of the high stability of the BDV genome in persistently infected cells, we tested the hypothesis that virus mutants are constantly present but fail to emerge under standard conditions. We report the successful amplification of a novel virus mutant by creating bottleneck conditions under which virus populations of single infected cells were allowed to spread to uninfected cells that were provided in large excess. We propose that the observed resistance of BDV-infected cells to superinfection with BDV variants contributes to viral genome stability. Analysis of laboratory strains further showed that they can undergo dynamic changes during passage in the brains of animals and that new virus variants can eventually become dominant in the population.
MATERIALS AND METHODS
Infection of cell cultures.
Single rat astroglia cells (C6 cells) from a cell culture persistently infected with BDV strain He/80 (6) were seeded in 96-well dishes. After adherence, approximately 400 uninfected C6 cells were added. After the cells had grown to confluency, passages were done at a ratio of 1:5 for 3 to 4 weeks. The degree of virus infection of the cultures was determined by indirect immunofluorescence assay (IFA) as described previously (16) using a polyclonal serum directed against the viral phosphoprotein (31). Standard virus infections were carried out with 105 C6 cells and various amounts of virus stocks in the presence of culture medium containing 10% fetal calf serum (FCS). Cells infected with high doses (approximately 100, 500, 2,500, and 10,000 focus-forming units [FFU]) of BDV virus stock derived from 2/1-containing cells were passaged at a ratio of 1:5 for 3 to 4 weeks.
Preparation of total RNA.
Total RNA of either BDV-infected C6 cells or the brains of neonatally infected rats and mice harvested at 4 weeks postinfection were prepared using peqGOLD TriFast (Peqlab) according to the manufacturer's recommendations. The precipitated RNA samples were dissolved in water.
In vitro transcription and RPA.
Runoff transcriptions were performed with 0.2 μg of EcoRI-linearized plasmid pRPAwt (20) in a total volume of 20 μl of T7 transcription buffer (Roche Molecular Biochemicals) containing 28 U of RNasin (Amersham); 0.5 μM (each) ATP, GTP, and CTP; 0.05 μM UTP (Pharmacia); 3 U of T7 RNA polymerase (MBI Fermentas); and 10 μCi of 32P-labeled UTP (3,000 Ci/mmol; Amersham). After incubation at 37°C for 90 min, the total volume was raised to a final volume of 30 μl with transcription buffer including 4 U of RNase-free DNase (Ambion) and incubated for a further 30 min at 37°C to digest plasmid DNA. Purification of the 32P-labeled RNA and subsequent RNase protection assay (RPA) reactions were carried out as described previously (32) using approximately 1.4 × 105 cpm of 32P-labeled RPA probe and 10 μg of total RNA per sample. The digestion of unhybridized RNA involved 16 U of RNase T1 (Ambion)/sample and either 0.02 or 4 μg of RNase A (Roche Molecular Biochemicals).
Virus stocks.
Virus stocks from C6 cells persistently infected with BDV strain He/80 (6) were prepared as described previously (4) with slight modifications. Briefly, 25 confluent 90-mm-diameter plates were washed with 20 mM HEPES (pH 7.4) and incubated with 10 ml of 20 mM HEPES (pH 7.4) containing 250 mM MgCl2 and 1% FCS for 1.5 h at 37°C. Subsequently, the virus-containing supernatants were harvested and centrifuged twice at 2,500 × g for 5 min to remove cell debris. The virus particles were concentrated by ultracentrifugation for 1 h at 20°C and 80,000 × g onto a 20% sucrose cushion containing 10 mM HEPES (pH 7.4) and 0.5% FCS. The virus-containing pellets were finally resuspended in phosphate-buffered saline. Titration of BDV using Vero cells was performed as described previously (17).
The following virus stocks from He/80-infected rats were used in this study: no. 66, fifth passage in brains of newborn Lewis rats (a gift from L. Stitz, Tübingen, Germany); no. 62, fifth passage in brains of adult Lewis rats (a gift from L. Stitz); no. 61, fifth passage in brains of newborn Lewis rats (the fourth passage of this virus was a gift from L. Stitz, whereas the fifth passage was done locally as described below); and no. 102, third brain passage in newborn Lewis rats (a gift of K. Carbone, Rockville, Md.), passaged once in the brain of an adult Lewis rat. The last virus stock was only available as an archival RNA sample (27).
Animal infections.
Newborn β2m0/0 MRL mice were infected by the intracerebral route with 10 μl of rat brain-derived virus stock no. 61, corresponding to approximately 500 FFU, in the thalamic region of the left hemisphere using a Hamilton syringe as described previously (15). Four weeks postinfection, successful infections of the mice were confirmed by analyzing the total RNA of the brains by RPA. Virus stock no. 61 was prepared from the brains of 4-week-old Lewis rats that were intracerebrally infected neonatally with about 103 FFU of BDV.
Direct sequencing of RT-PCR products.
First-strand cDNA synthesis was carried out with 4 μg of total RNA and He/80-specific primer 1908F (5′-TCCTATGTGGAGCTCAAGGAC-3′) in a final volume of 20 μl using Superscript RT (Gibco Life Technologies) as recommended by the manufacturer. Subsequent nested PCR (two series of 30 cycles as follows: 95°C, 1 min; 55°C, 1 min; 72°C, 1 min) was carried out in a total volume of 100 μl containing 5 U of Taq polymerase (Roche Molecular Biochemicals) using primers 2550R (5′-CTTAACAGTACCAGTGTACCG-3′) and 1908F as the first primer set and 2530R (5′-GATTGATGATCGGTCAGCG-3′) and 1925F (5′-GACAAGGTAATCGTCCCTGG-3′) as the second primer set. The first PCR mixture contained 2 μl of the reverse transcription (RT) reaction mixture, whereas the second PCR mixture contained 5 μl of the first PCR mixture. Amplification products were gel purified and directly sequenced (Toplab).
Nucleotide sequence accession numbers.
Partial nucleotide sequences of variant 2/1 and virus stocks no. 102 and 62 were deposited in GenBank under accession no. AJ271120, AJ271118, and AJ250177, respectively.
RESULTS
Detection of sequence variations among different BDV strains by RPA.
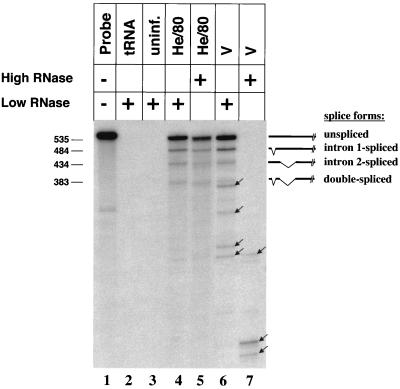
To ensure that RPA is a reliable method to identify small nucleotide differences between closely related BDV strains, total RNA of C6 cells infected with BDV strains He/80 and V were analyzed using an RNA probe complementary to the coding sequence of the M and G proteins (nucleotides [nt] 1975 to 2510) of He/80. Within this region, the two strains differ at 22 nucleotide positions. At low concentrations of RNase A (0.02 μg), the expected unspliced and three spliced viral transcripts of He/80 were detected (Fig. 1, lane 4). When RNA of cells infected with strain V was used, additional bands appeared (Fig. 1, lane 6). A 200-fold-increased RNase A concentration (4 μg) did not significantly change the RPA signal pattern observed with He/80 transcripts (Fig. 1, lane 5). However, pronounced degradation of the RPA probe was evident when strain V-derived RNA was analyzed (Fig. 1, lane 7), indicating that the mismatches of strain V transcripts are efficiently recognized. Thus, under stringent conditions, RPA can be used as a diagnostic tool to identify sequence variations among BDV strains. All further RPA experiments were therefore carried out with high concentrations of RNase A. It should be noted that due to the nucleotide specificities of the RNases used in the assay, unpaired A residues were not recognized.
FIG. 1.
RPA for the detection of sequence variations between different strains of BDV. Nucleotide differences between the BDV strains He/80 and V were visualized by RPA using a 535-nt RNA probe (lane 1) complementary to the M and G open reading frames of strain He/80. Analysis was carried out using 10 μg of tRNA (lane 2), RNA samples (10 μg) prepared from total cell lysates of uninfected (uninf.) C6 cells (lane 3), C6 cells infected with BDV strain He/80 (lanes 4 and 5), or C6 cells infected with BDV strain V (lanes 6 and 7). Low (0.02-μg) or high (4-μg) concentrations of RNase A were used (+) as indicated. The repertoire of RNA species and the expected gel positions of the corresponding RPA signals are indicated on the right. The arrows mark signals resulting from RNA species with slightly different sequences. The numbers to the left indicate the mobility of molecular size markers (in nucleotides).
Characterization of a new BDV variant from persistently infected cells.
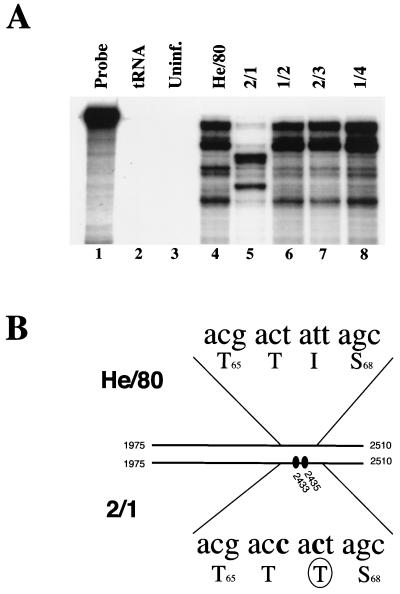
Based on the hypothesis that a certain percentage of cells within a population of BDV-infected cells may harbor virus variants in addition to wild-type virus, we speculated that cocultivation of uninfected cells with a single BDV-infected cell might allow the characterization of such variants. To test this hypothesis, we seeded uninfected C6 cells onto a single He/80-infected C6 cell to allow the spread of virus to neighboring cells. Monitoring of infection by indirect IFA indicated that after 3 to 4 weeks of passaging, 50 to 90% of the cells were infected with BDV (data not shown). Subsequent RPA screening using total RNA of 30 independent infected C6 cell cultures revealed that one culture contained a BDV variant (designated 2/1), as indicated by a dramatic change in the signal pattern (Fig. 2A, lane 5). To confirm this observation we determined the nucleotide sequence of the variant by sequencing nested RT-PCR amplification products corresponding to the M-G coding sequence (nt 1975 to 2510). Two nucleotide changes were identified in 2/1 at positions 2433 and 2435 compared to He/80 (Fig. 2B), one of which resulted in a nonconservative change of isoleucine to threonine at position 67 of the G protein.
FIG. 2.
Identification of a new variant of BDV He/80. (A) Samples of total RNA (10 μg) from a standard C6 cell culture persistently infected with BDV He/80 (lane 4) or from newly established persistently infected C6 cell cultures each initially infected with a single He/80-infected C6 cell (lanes 5 to 8) were analyzed by RPA as for Fig. 1, using high concentrations of RNase A. Note the novel signal pattern of the virus in culture 2/1. Undigested RPA probe (lane 1). No specific products were observed when the RPA was carried out with 10 μg of tRNA (lane 2) or 10 μg of total RNA from uninfected C6 cells (lane 3). (B) Fragments of the M-G open reading frames (nt 1975 to 2510) of standard virus (He/80) and variant 2/1 were amplified by RT-PCR, and their sequences were compared. The two T-to-C mutations found in 2/1 (shaded) and the resulting amino acid change in the G open reading frame (I67T [circled]) are indicated.
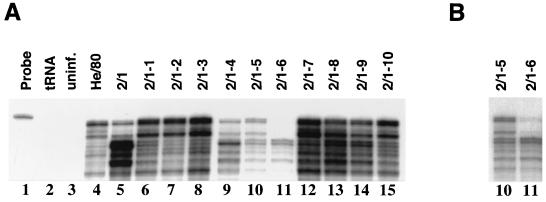
Closer inspection of the RPA signal pattern of 2/1-infected cells (Fig. 2A, lane 5) showed a weak pattern of protected bands similar to that of wild-type He/80, in addition to 2/1-specific signals. To determine whether a He/80-like virus was still present in the cell culture, 10 C6 cell cultures were independently infected with less than 10 FFU of a virus stock prepared from 2/1-infected cells as described in Materials and Methods. Screening for BDV by RPA 3 weeks postinfection revealed that 7 out of 10 independent cultures contained predominantly the wild-type-like virus (Fig. 3A, lanes 6 to 8 and 12 to 15). Two cultures were persistently infected with a mixture of the He/80-like strain and 2/1, as judged by the intensities of the various signals (Fig. 3A, lanes 9 and 10), whereas one culture seemed to be infected with the 2/1 variant only (Fig. 3A, lane 11). However, longer exposure of the film showed that the He/80-like strain was still present in this culture (Fig. 3B, lane 11). The signal intensities observed by RPA differed strongly between 2/1- (Fig. 3A, lanes 9 to 11) and He/80-infected cells (Fig. 3A, lanes 6 to 8 and 12 to 15), although equal amounts of total RNA were used for analysis, indicating that the absolute amounts of viral transcripts were lower in cell cultures infected with 2/1. Consistent with these findings, the percentages of infected cells at 3 weeks postinfection, as determined by IFA, were higher than 50% in cell cultures predominantly infected with the He/80-like virus and less than 20% in cell cultures predominantly infected with 2/1 (data not shown), indicating that the efficiencies of virus spread in these cultures differed.
FIG. 3.
Nonuniform nature of BDV variant 2/1. (A) Samples of total RNA (10 μg) from a standard C6 cell culture persistently infected with BDV He/80 (lane 4), cell culture 2/1 (lane 5), and newly established persistently infected C6 cell cultures infected with a low dose (<10 FFU) of a BDV stock prepared from cell culture 2/1 (lanes 6 to 15) were analyzed by RPA as for Fig. 1. Note that the signal patterns of most cultures differed from that of cell culture 2/1, indicating that several virus variants were present. Undigested RPA probe (lane 1). No specific products were observed when the RPA was carried out with 10 μg of tRNA (lane 2) or 10 μg of total RNA from uninfected C6 cells (lane 3). (B) Longer exposure of lanes 10 and 11 shown in panel A.
Cells persistently infected with BDV are resistant to superinfection with other BDV strains.
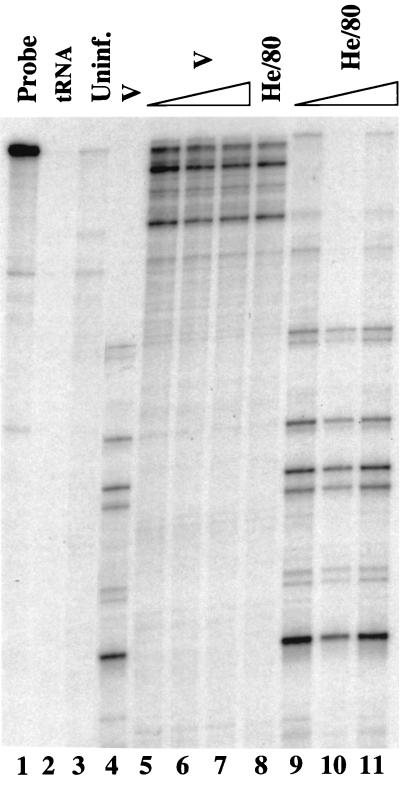
The virus populations in cell cultures derived from the bottleneck experiment remained stable after persistent infection was established, as judged from the unchanged RPA signal patterns (data not shown). Resistance of BDV-infected cells to superinfection by other BDV variants could explain this behavior. To test this hypothesis, C6 cells persistently infected with the BDV strain He/80 were incubated with increasing doses of strain V virus stocks. In parallel, uninfected C6 cells were infected with the lowest dose of strain V virus used for the superinfection experiment. Three weeks postinfection, almost all cells of the entire control culture contained viral antigen, as determined by indirect IFA (data not shown). RPA confirmed the infection of these cells by BDV strain V (Fig. 4, lane 4). Since strain V-specific RPA signals were absent from the other cultures, we concluded that cells persistently infected with He/80 could not be infected with strain V under these conditions, independent of the initial dose of virus used (Fig. 4, lanes 5 to 7). A similar resistance to superinfection with BDV strain He/80 was observed in C6 cells persistently infected with strain V (Fig. 4, lanes 9 to 11) or 2/1 (data not shown).
FIG. 4.
BDV-infected cells are resistent to superinfection. C6 cell lines persistently infected with either strain He/80 (lanes 5 to 7) or strain V (lanes 9 to 11) were incubated with various doses (100, 500, and 2,500 FFU) of partially purified BDV strain V (lanes 5 to 7) or with various doses (40, 200, and 4,000 FFU) of partially purified BDV strain He/80 (lanes 9 to 11). To control for the quality of the virus stocks, uninfected C6 cells were infected with 100 FFU of strain V (lane 4) or with 40 FFU of strain He/80 (lane 8). Three weeks postinfection, when almost all cells in the control cultures were expressing viral antigen, samples of total RNA (10 μg) from the various cultures were analyzed by RPA as for Fig. 1, using high concentrations of RNase A. Undigested RPA probe (lane 1). No specific products were observed when the RPA was carried out with 10 μg of tRNA (lane 2) or 10 μg of total RNA from uninfected C6 cells (lane 3).
Unexpected diversity of He/80 virus stocks with different passage histories.
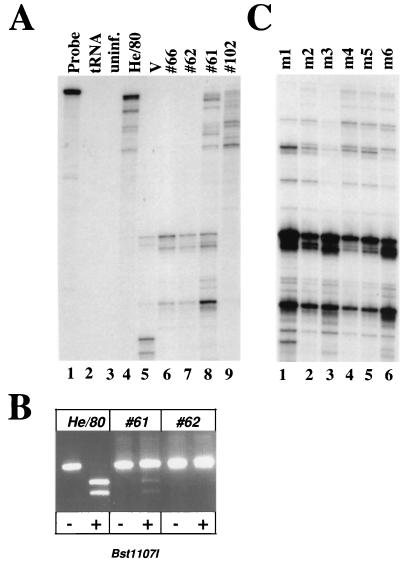
To evaluate the genome stability of BDV in vivo, we used RPA to screen stocks of He/80 with different passage histories in rat brains for mutations in the viral M and G genes. RNA of C6 cells infected with BDV stock no. 66 or no. 62 (Fig. 5A, lanes 6 and 7) yielded RPA signal patterns that were clearly distinct from those of the prototype strains He/80 and V (Fig. 5A, lanes 4 and 5). Intriguingly, analysis of stock no. 61 revealed two overlapping RPA signal patterns (Fig. 5A, lane 8), one similar to that of prototype He/80 and one similar to those of no. 62 and no. 66, suggesting coinfection with at least two viruses. Analysis of virus stock no. 102 (Fig. 5A, lane 9) revealed an RPA signal pattern similar to that of prototype He/80 (Fig. 5A, lane 4) but with distinct signal intensities, suggesting high sequence similarity between the two strains. Direct sequencing of corresponding RT-PCR amplification product indeed confirmed the complete identity of the two viruses in the region of interest. The differences observed in the RPA thus probably resulted from partial degradation of the archival RNA from virus stock no. 102. The sequence of the M-G gene region in BDV stock no. 62 revealed 17 nucleotide changes compared to that of the prototype He/80 sequence. The deduced partial amino acid sequence of the M protein in virus no. 62 was identical to that in He/80, whereas the G protein of no. 62 harbored two amino acid substitutions (I143L and I144L).
FIG. 5.
RPA analysis of a collection of standard stocks of BDV He/80. (A) Samples of total RNA (10 μg) were prepared from rat brains or C6 cell cultures infected with the following BDV He/80 stocks: no. 66, fifth passage in brains of newborn Lewis rats (lane 6); no. 62, fifth passage in brains of adult Lewis rats (lane 7); no. 61, fifth passage in brains of newborn Lewis rats (lane 8); and no. 102, three passages in brains of newborn Lewis rats and one subsequent passage in the brain of an adult Lewis rat (lane 9). RNAs were analyzed by RPA using high concentrations of RNase A, as described in the legend to Fig. 1. Undigested RPA probe (lane 1). No specific products were observed when the RPA was carried out with 10 μg of tRNA (lane 2) or 10 μg of total RNA from uninfected C6 cells (lane 3). RNA from C6 cells infected with BDV strains He/80 (lane 4) and V (lane 5) yielded the expected characteristic signal patterns. (B) Corresponding RT-PCR products of standard BDV He/80, virus stock no. 61, and virus stock no. 62 were incubated with (+) or without (−) restriction enzyme Bst11071, and products were visualized by agarose gel electrophoresis and ethidium bromide staining. (C) Analysis of stock no. 61 after one passage in the brains of six individual newborn β2m0/0 MRL mice (m1 to m6).
Despite the complex signal pattern observed with RNA of stock no. 61-infected rat brains, direct sequencing of corresponding RT-PCR amplification products from the M-G gene region revealed complete identity to stock no. 62 (data not shown), indicating that viruses with the latter sequence were predominant. To demonstrate the existence of a mixed virus population in stock no. 61, we digested RT-PCR amplification products of the M-G gene regions of prototype He/80, stock no. 62, and stock no. 61 with the restriction enzyme Bst1107I, which cuts prototype He/80 but not virus no. 62 (Fig. 5B). The RT-PCR product derived from stock no. 61 was partially cleaved by Bst1107I (Fig. 5B), supporting our previous RPA results, which had suggested the presence of small amounts of He/80-like virus and large amounts of no. 62-like virus.
To investigate whether one of the two virus variants might eventually overgrow the other, we infected newborn mice with BDV stock no. 61 by the intracerebral route. Since stock no. 61 had been generated by five sequential passages in brains of newborn rats, we assumed that this deliberate change in animal hosts might create a bottleneck situation, which may favor replication of the fitter virus variant. In fact, RPA analysis of RNA from the brains of the infected mice sacrificed at 4 weeks postinfection revealed major changes in the signal patterns: the no. 62-specific RPA signals were now clearly dominant in all six mice, whereas the He/80-like signals showed markedly reduced intensities (Fig. 5C).
DISCUSSION
In this study, we demonstrate that although BDV exhibits extraordinary genetic stability under standard cell culture conditions, it is possible to isolate stable genetic virus variants from these cultures. Simple bottleneck situations under which the virus content of single persistently infected cells was allowed to spread in uninfected cell cultures yielded 1 culture out of 30 in which a virus variant (designated 2/1) with two nucleotide exchanges in the G gene was stably propagated. Since screening for new BDV variants was performed using a diagnostic RPA that covered only about 1/18 of the viral genome, it is reasonable to assume that additional cultures containing mutant viruses with alterations outside the examined genome region were present. This view is supported by the observation that virus spread was slow not only in culture 2/1 but also in several other cultures of the bottleneck experiment that propagated viruses with wild-type He/80 sequence in the examined M-G gene region. Because mutations in various locations of the viral genome can presumably contribute to such phenotypic changes, it remains unclear whether the mutations found in the G gene of BDV variant 2/1, which result in a single-amino-acid change, are indeed responsible for the attenuated phenotype.
More than one virus was present in the original cell culture containing BDV variant 2/1. As a consequence, most persistently infected cell cultures resulting from infection with cell-free virus from 2/1 cells did not propagate this mutant virus but rather viruses with the wild-type genotype in the M-G gene region. A possible explanation for the failure of virus variant 2/1 to propagate efficiently under these experimental conditions is that its replication might be helper virus dependent. Indeed, all efforts to obtain cell cultures propagating exclusively virus variant 2/1 were unsuccessful. Since infection experiments with high doses of cell-free virus from 2/1 cells did not result in a preferential replication of the mutant virus, it seems not to have properties of defective interfering particles.
We find it surprising that replication of virus variant 2/1 in the original cell culture was apparently not affected by the wild-type-like virus, which was simultaneously present. Since our experiments with cell-free virus stocks clearly showed that the fitness of variant 2/1 is reduced, it remains to be explained why this handicap was not disadvantageous under the conditions used in the bottleneck experiment. It is possible that the cell-to-cell spread of BDV is mechanistically distinct from the entry of cell-free virus into host cells and that the defect of virus mutant 2/1 might selectively affect the latter pathway. Supernatants of BDV-infected C6 cells contain only minute amounts of cell-free infectious virus (23) (data not shown), suggesting that direct cell-to-cell spread is the preferred mode of virus propagation in this cell line.
An important result of this study was the finding that persistently infected C6 cells are highly resistant to superinfection by other strains of BDV. We believe that this biological property of BDV contributes significantly to its genome stability, in addition to the counterselection of newly generated variants with lower fitness within the cell. Because BDV is noncytolytic and because it has no growth inhibitory effect on persistently infected cells, resistance to superinfection generates an ideal ecological niche for resident viruses to produce progeny without competition by genetically distinct viruses entering the cell from outside. The drawback of this situation is that due to the lack of susceptible cells, novel BDV variants with increased fitness cannot outgrow the resident virus population unless plenty of uninfected cells are available. Such conditions can be generated experimentally by enforcing bottleneck situations. In vivo, optimal conditions for novel virus variants to outgrow the parental viruses might exist during the first few weeks after infection of the central nervous system of a new host.
Superinfection interference was originally described for avian retroviruses (33, 35) and was later also found to affect most noncytolytic mammalian retroviruses (36). Resistance to superinfection by retroviruses results from interaction of the env gene product of the endogenous virus with cellular components that function as virus receptors, resulting in reduced availability of virus entry mediators at the cell surface (9). Superinfection interference has also been described for other viruses, including noncytolytic variants of foot-and-mouth disease virus (8) and measles virus (12). The mechanisms of these restriction phenomena have not yet been elucidated. Similarly, the mechanism of the BDV interference phenomenon that we describe here is presently unknown. Preliminary studies showed that it is not mediated by interferons or other soluble factors (M. Schwemmle and P. Staeheli, unpublished observation). When we used the RPA to determine whether stocks of BDV He/80 with different passage histories in brains of adult or newborn rats might contain viruses with mutations in the M-G gene region, we found that only one of four stocks yielded the expected RPA signal pattern. Two stocks contained a BDV variant that was about 2 to 3% different from prototype He/80. Interestingly, the fourth virus stock contained a mixture of prototype He/80 and the variant. Since all four virus stocks were generated from the same starting material, it seems likely that the variant was already present in that material and that it was selected from the virus mixture with variable efficacy. In support of this view, we found that one additional passage of the mixed virus stock in mouse brains resulted in a preferential loss of the prototype He/80 strain. It is possible that the new variant is a direct descendent of He/80 that arose by mutation. Alternatively, the diseased horse from which He/80 was originally isolated might have been infected with two strains of BDV. Because the sequence of the new virus variant closely resembles that of prototype He/80 in some parts of the genome and that of prototype strain V in others, it seems more likely that it represents a wild-type virus that evolved from a common ancestor of the Central European strains of BDV. In fact, Binz and coworkers (3) presented evidence that simultaneous infection of horses with two different genotypes of BDV is possible. It is tempting to speculate that the various rat brain-derived He/80 virus stocks that contain different concentrations of “mouse-pathogenic” variant viruses may have caused the reported variability of neurological symptoms in experimentally infected mice (15, 26). This could explain, at least in part, the observation of Rubin et al. (26) that efficient replication of BDV in adult mice was only achieved after multiple passages of the initial rat-derived virus in mouse brains.
Sequence comparisons of the M-G gene region with database entries showed that the new virus variant was almost identical to BDV isolate RW98, which is believed to have originated from the blood of a psychiatric patient (24, 25). This view was confirmed when other genome regions of the variant were sequenced and compared to RW98 (30). This surprising finding, together with the fact that the various BDV stocks analyzed in this study originated from the laboratory in which RW98 was isolated, raises serious questions about a human origin of this virus.
The results of this study are important for the correct interpretation of epidemiological data. Because the genome stability of BDV in persistently infected cell cultures is extraordinarily high, minor sequence variations in virus isolates from human tissues were previously regarded as solid proof that contamination with laboratory strains had not occurred. Since accidental contamination presumably occurs only at extremely low virus doses, it represents a situation which strongly resembles the bottleneck condition applied in this study. We have shown here that such conditions favor the outgrowth of variant viruses.
ACKNOWLEDGMENTS
We thank Georg Kochs, Jürgen Hausmann, Juan Carlos de la Torre, and Otto Haller for helpful comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Zentrum für Klinische Forschung I of the Universitätsklinikum Freiburg. C.S. is a fellow of the German Stipendienprogramm Infektionsforschung, DKFZ, Heidelberg.
REFERENCES
- 1.Berg A L, Berg M. A variant form of feline Borna disease. J Comp Pathol. 1998;119:323–331. doi: 10.1016/s0021-9975(98)80054-6. [DOI] [PubMed] [Google Scholar]
- 2.Berg A L, Dorries R, Berg M. Borna disease virus infection in racing horses with behavioral and movement disorders. Arch Virol. 1999;144:547–559. doi: 10.1007/s007050050524. [DOI] [PubMed] [Google Scholar]
- 3.Binz T, Lebelt J, Niemann H, Hagenau K. Sequence analyses of the p24 gene of Borna disease virus in naturally infected horse, donkey and sheep. Virus Res. 1994;34:281–289. doi: 10.1016/0168-1702(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, de la Torre J C, Lewis A, Ludwig H, Lipkin W I. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci USA. 1992;89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briese T, Hornig M, Lipkin W I. Bornavirus immunopathogenesis in rodents: models for human neurological diseases. J Neurovirol. 1999;5:604–612. doi: 10.3109/13550289909021289. [DOI] [PubMed] [Google Scholar]
- 6.Cubitt B, Oldstone C, de la Torre J C. Sequence and genome organization of Borna disease virus. J Virol. 1994;68:1382–1396. doi: 10.1128/jvi.68.3.1382-1396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czygan M, Hallensleben W, Hofer M, Pollak S, Sauder C, Bilzer T, Blumcke I, Riederer P, Bogerts B, Falkai P, Schwarz M J, Masliah E, Staeheli P, Hufert F T, Lieb K. Borna disease virus in human brains with a rare form of hippocampal degeneration but not in brains of patients with common neuropsychiatric disorders. J Infect Dis. 1999;180:1695–1699. doi: 10.1086/315068. [DOI] [PubMed] [Google Scholar]
- 8.de la Torre J C, Davila M, Sobrino F, Ortin J, Domingo E. Establishment of cell lines persistently infected with foot-and-mouth disease virus. Virology. 1985;145:24–35. doi: 10.1016/0042-6822(85)90198-9. [DOI] [PubMed] [Google Scholar]
- 9.Delwart E L, Panganiban A T. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J Virol. 1989;63:273–280. doi: 10.1128/jvi.63.1.273-280.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 11.Dürrwald R, Ludwig H. Borna disease virus (BDV), a (zoonotic?) worldwide pathogen. A review of the history of the disease and the virus infection with comprehensive bibliography. Zentbl Veterinarmed B. 1997;44:147–184. doi: 10.1111/j.1439-0450.1997.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Munoz R, Celma M L. Measles virus from a long-term persistently infected human T lymphoblastoid cell line, in contrast to the cytocidal parental virus, establishes an immediate persistence in the original cell line. J Gen Virol. 1992;73:2195–2202. doi: 10.1099/0022-1317-73-9-2195. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Dunia D, Sauder C, de la Torre J C. Borna disease virus and the brain. Brain Res Bull. 1997;44:647–664. doi: 10.1016/S0361-9230(97)00276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosztonyi G, Ludwig H. Borna disease-neuropathology and pathogenesis. Curr Top Microbiol Immunol. 1995;190:39–73. [PubMed] [Google Scholar]
- 15.Hallensleben W, Schwemmle M, Hausmann J, Stitz L, Volk B, Pagenstecher A, Staeheli P. Borna disease virus-induced neurological disorder in mice: infection of neonates results in immunopathology. J Virol. 1998;72:4379–4386. doi: 10.1128/jvi.72.5.4379-4386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallensleben W, Staeheli P. Inhibition of Borna disease virus multiplication by interferon: cell line differences in susceptibility. Arch Virol. 1999;144:1209–1216. doi: 10.1007/s007050050580. [DOI] [PubMed] [Google Scholar]
- 17.Herzog S, Rott R. Replication of Borna disease virus in cell cultures. Med Microbiol Immunol. 1980;168:153–158. doi: 10.1007/BF02122849. [DOI] [PubMed] [Google Scholar]
- 18.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 19.Iwata Y, Takahashi K, Peng X, Fukuda K, Ohno K, Ogawa T, Gonda K, Mori N, Niwa S, Shigeta S. Detection and sequence analysis of borna disease virus p24 RNA from peripheral blood mononuclear cells of patients with mood disorders or schizophrenia and of blood donors. J Virol. 1998;72:10044–10049. doi: 10.1128/jvi.72.12.10044-10049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jehle C, Lipkin W I, Staeheli P, Marion R M, Schwemmle M. Authentic Borna disease virus transcripts are spliced less efficiently than cDNA-derived viral RNAs. J Gen Virol. 2000;81:1947–1954. doi: 10.1099/0022-1317-81-8-1947. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig H, Bode L, Gosztonyi G. Borna disease: a persistent virus infection of the central nervous system. Prog Med Virol. 1988;35:107–151. [PubMed] [Google Scholar]
- 22.Nowonty N, Kolodziejek J, Jehle C, Suchy A, Staeheli P, Schwemmle M. Isolation of a new subtype of Borna disease virus. J Virol. 2000;74:5655–5658. doi: 10.1128/jvi.74.12.5655-5658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pauli G, Ludwig H. Increase of virus yields and releases of Borna disease virus from persistently infected cells. Virus Res. 1985;2:29–33. doi: 10.1016/0168-1702(85)90057-7. [DOI] [PubMed] [Google Scholar]
- 24.Planz O, Rentzsch C, Batra A, Rziha H J, Stitz L. Persistence of Borna disease virus-specific nucleic acid in blood of psychiatric patient. Lancet. 1998;352:623. doi: 10.1016/S0140-6736(05)79577-5. [DOI] [PubMed] [Google Scholar]
- 25.Planz O, Rentzsch C, Batra A, Winkler T, Buttner M, Rziha H J, Stitz L. Pathogenesis of borna disease virus: granulocyte fractions of psychiatric patients harbor infectious virus in the absence of antiviral antibodies. J Virol. 1999;73:6251–6256. doi: 10.1128/jvi.73.8.6251-6256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin S A, Waltrip R W D, Bautista J R, Carbone K M. Borna disease virus in mice: host-specific differences in disease expression. J Virol. 1993;67:548–552. doi: 10.1128/jvi.67.1.548-552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauder C, de la Torre J C. Cytokine expression in the rat central nervous system following perinatal Borna disease virus infection. J Neuroimmunol. 1999;96:29–45. doi: 10.1016/s0165-5728(98)00272-0. [DOI] [PubMed] [Google Scholar]
- 28.Schneider P A, Briese T, Zimmermann W, Ludwig H, Lipkin W I. Sequence conservation in field and experimental isolates of Borna disease virus. J Virol. 1994;68:63–68. doi: 10.1128/jvi.68.1.63-68.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwemmle M, Hatalski C G, Lewis A J, Lipkin W I. Borna Virus. In: Ahmed R, Chen I, editors. Persistent disease virus infection of the nervous system. New York, N.Y: John Wiley & Sons; 1999. pp. 559–573. [Google Scholar]
- 30.Schwemmle M, Jehle C, Formella S, Staeheli P. Sequence similarities between human bornavirus isolates and laboratory strains question human origin. Lancet. 1999;354:1973–1974. doi: 10.1016/S0140-6736(99)04703-0. [DOI] [PubMed] [Google Scholar]
- 31.Schwemmle M, Salvatore M, Shi L, Richt J, Lee C, Lipkin W. Interactions of the Borna disease virus P, N, and X proteins and their functional implications. J Biol Chem. 1998;273:9007–9012. doi: 10.1074/jbc.273.15.9007. [DOI] [PubMed] [Google Scholar]
- 32.Stalder A, Pagenstecher A, Kincaid C, Campbell I. Analysis of gene expression by multiprobe RNase protection assay. Methods Mol Med. 1998;22:53–66. doi: 10.1385/0-89603-612-X:53. [DOI] [PubMed] [Google Scholar]
- 33.Steck F T, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966;29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- 34.Stitz L, Dietzschold B, Carbone K M. Immunopathogenesis of Borna disease. Curr Top Microbiol Immunol. 1995;190:75–92. doi: 10.1007/978-3-642-78618-1_5. [DOI] [PubMed] [Google Scholar]
- 35.Temin H M, Kassner V K. Replication of reticuloendotheliosis viruses in cell culture: chronic infection. J Gen Virol. 1975;27:267–274. doi: 10.1099/0022-1317-27-3-267. [DOI] [PubMed] [Google Scholar]
- 36.Weiss R A, Clapham P, Nagy K, Hoshino H. Envelope properties of human T-cell leukemia viruses. Curr Top Microbiol Immunol. 1985;115:235–346. doi: 10.1007/978-3-642-70113-9_15. [DOI] [PubMed] [Google Scholar]