Abstract
Background
DNA repair plays a crucial role in the development and progression of different types of cancers. Nevertheless, little is known about the role of DNA repair-related genes (DRRGs) in esophageal cancer (EC). The present study aimed to identify a novel DRRGs prognostic signature in EC.
Methods
Gene set enrichment analysis (GSEA) was performed to screen 150 genes related to DNA repair, which is the most important enrichment gene set in EC. Univariate and multivariate Cox regression analyses were used to screen DRRGs closely associated with prognosis. The difference in the expression of hub DRRGs between tumor and normal tissues was analyzed. Combined with clinical indicators (including age, gender, and tumor stage), we evaluated whether the 4-DRRGs signature was an independent prognostic factor. In addition, we evaluated the prediction accuracy using a receiver operating characteristic (ROC) curve and visualized the model’s performance via a nomogram.
Results
Four-DRRGs (NT5C3A, TAF9, BCAP31, and NUDT21) were selected by Cox regression analysis to establish a prognostic signature to effectively classify patients into high- and low-risk groups. The area under the curve (AUC) of the time-dependent ROC of the prognostic signature for 1- and 3-year was 0.769 and 0.720, respectively. Compared with other clinical characteristics, the risk score showed a robust ability to predict the prognosis in EC, especially in the early stage of EC. Furthermore, we constructed a nomogram to interpret the clinical application of the 4-DRRGs signature.
Conclusions
In conclusion, we identified a prognostic signature based on the DRRGs for patients with EC, which can contribute independent value in identifying clinical outcomes that complement the TNM system in EC.
Keywords: DNA repair, prognostic model, esophageal cancer (EC), The Cancer Genome Atlas (TCGA)
Highlight box.
Key findings
• Our study identified a 4-gene DNA repair-related signature that stratified esophageal cancer patients into high and low-risk groups, demonstrating high prognostic accuracy and independence from traditional clinical indicators.
What is known and what is new?
• DNA repair mechanisms are critical for preventing carcinogenesis by managing genomic stability. Malfunctions in these pathways could drive cancer development and contribute to treatment responses in esophageal cancer.
• Our study revealed that certain DNA repair-related genes significantly influenced the prognosis of esophageal cancer and a 4-gene prognostic signature was identified to independently predict clinical outcomes in esophageal cancer patients.
What is the implication, and what should change now?
• Understanding and targeting DNA repair pathways could have significant impacts on the management of esophageal cancer. Personalized treatment strategies based on DNA repair-related genes might improve survival for patients with esophageal cancer.
Introduction
As one of the most invasive malignancies, esophageal cancer (EC) ranks seventh among the leading causes of cancer-related deaths worldwide (1). Most patients are diagnosed at an advanced stage, resulting in a poor 5-year overall survival (OS) rate of around 20% (1). Therefore, it is imperative to explore novel biomarkers and therapeutic targets for EC. EC is a complex and heterogeneous disease. It has been reported that DNA repair pathways play a role in regulating the response to chemotherapy and radiotherapy in EC (2-4).
DNA repair plays a vital role in maintaining cell and tissue homeostasis, as it responds to both endogenous and exogenous DNA insults. Defects in DNA repair can lead to an accumulation of genomic changes, increasing the risk of carcinogenesis and cancer development (5). Specific genes in the DNA repair pathways were found to be associated with the risk of esophageal squamous cell carcinoma (ESCC) and gastric cancer (6). Moreover, studies have shown that cancer cells exhibit mutations and aberrant expression in genes associated with DNA repair responses (7,8). Alterations in DNA repair pathways not only drive cancer but also contribute to its development. However, the understanding of the roles of DNA repair-related genes (DRRGs) in EC remains limited (9).
In this study, we analyzed the mRNA expression dataset from The Cancer Genome Atlas (TCGA) database, profiling hallmark gene sets in 158 cases of EC. Interestingly, we found that the DNA_REPAIR pathway ranked highest in terms of its function in EC. Subsequently, we conducted a comprehensive functional study of DRRGs to investigate their significance in EC. Through this analysis, we identified and validated an individualized prognostic model based on DRRGs for OS in EC patients. Our findings also revealed several EC-related RBPs, shedding light on the molecular mechanisms underlying EC progression. These DRRGs hold promise as potential biomarkers for diagnosis and prognosis in EC patients. We present this article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-262/rc).
Methods
Data extraction from the TCGA database
The transcriptome data and clinical information of EC (including squamous cell carcinoma and adenocarcinoma) were retrieved from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Patients with incomplete follow-up information and survival time <30 days were excluded to reduce statistical bias in the following analysis. To identify differentially expressed genes between tumor samples and normal samples in EC patients, the “limma” package in R software was utilized. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene set enrichment analysis (GSEA)
The GSEA tool (http://www.broadinstitute.org/gsea/index.jsp) was implemented to assess the significance of the identified gene sets between adjacent and tumor tissues. A total of 168 mRNA expressions profiles, consisting of 10 mRNAs from adjacent tissues and 158 mRNAs from primary EC tissues, were extracted from the TCGA dataset and subjected to analysis. Further investigation of functions was based on the normalized P value (P<0.05) and the normalized enrichment score (NES).
Construction and validation of a DRRGs-related prognostic signature in predicting survival
We employed univariate Cox regression analysis to identify DRRGs that exhibited significant associations with the OS of EC patients. Subsequently, multivariate Cox regression analysis was performed to construct a risk score that could effectively evaluate patient prognosis. The risk score was determined using the following formula: risk scores = Vi × Ci, summed for each signature gene, where Vi represents the expression value of a gene and Ci represents the regression coefficient of the gene. Based on the median risk cutoff value, all patients were categorized into high- and low-risk groups. The prognostic model’s predictive performance was assessed using the area under the curve (AUC) value of the receiver operating characteristic (ROC) curve at 1 and 3 years, with evaluation facilitated by the “survivalROC” package in R. Additionally, we employed the log-rank method and Kaplan-Meier curves to further validate the predictive ability of the risk score.
Statistical analysis
The Cox regression analysis was performed using the “survival” package in R software (Version 4.3.1). For the normalization and differential expression analysis, the “limma” package was utilized. A significance threshold of P<0.05 was applied in all statistical analyses.
Results
Screen of the top-ranking function genes in EC
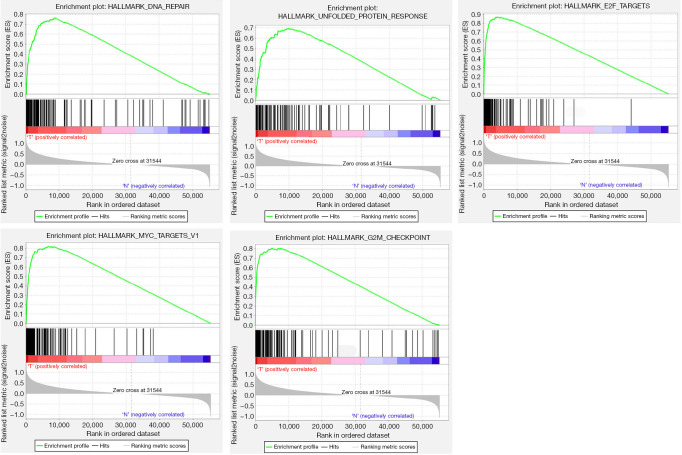
The flow chart of this study is shown as Figure 1. Fifty specific hallmark gene sets were derived by aggregating MSigDB version 7.2 gene sets to summarize and represent specific, well-defined biological states or processes. GSEA was further utilized to explore and identify significantly different gene sets between the EC tissues and adjacent normal tissues. The top five gene sets that exhibited significant enrichment were DNA_REPAIR, UNFOLDED_PROTEIN_RESPONSE, E2F_TARGETS, MYC_TARGETS_V1, and G2M_CHECKPOINT (Table 1, Figure 2). For further analysis, we selected the DNA_REPAIR gene set, consisting of 150 genes, which had the lowest P value (P<0.001) and highest NES-value (NES =2.2617).
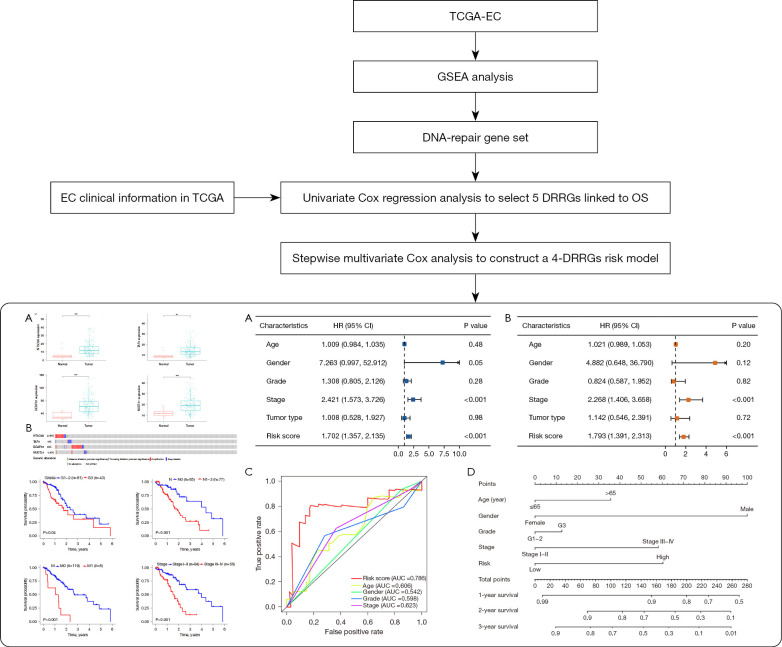
Figure 1.
Flow diagram of this study. **, P<0.01, ***, P<0.001. TCGA, The Cancer Genome Atlas; EC, esophageal cancer; GSEA, Gene Set Enrichment Analysis; DRRGs, DNA repair-related genes; OS, overall survival; AUC, area under the curve; HR, hazard ratio; CI, confidence interval.
Table 1. Gene sets enriched in EC (158 mRNA samples).
| GS follow link to MSigDB | Size | NES | NOM, P value | FDR q-value |
|---|---|---|---|---|
| HALLMARK_DNA_REPAIR | 150 | 2.2617 | <0.001 | 0 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 110 | 2.2048 | <0.001 | 0 |
| HALLMARK_E2F_TARGETS | 198 | 2.1889 | <0.001 | 0 |
| HALLMARK_MYC_TARGETS_V1 | 196 | 2.1749 | <0.001 | 0 |
| HALLMARK_G2M_CHECKPOINT | 196 | 2.1644 | <0.001 | 0 |
EC, esophageal cancer; GS, gene set; NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate.
Figure 2.
GSEA results for the top five enrichment hallmark gene sets in EC. GSEA, Gene Set Enrichment Analysis; EC, esophageal cancer.
The DRRGs signature for OS prediction of EC patients
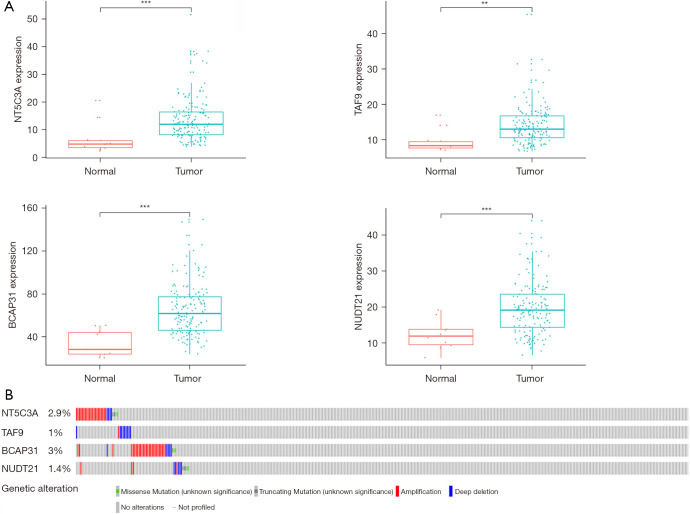
To explore the novel biomarkers that correlate with the outcome of patients with EC, we used the data from the TCGA EC dataset, which includes both RNA sequencing and adequate follow-up information. The general clinical features are shown in Table 2. After applying univariate Cox proportional hazard regression, 5 DRRGs that were found to be significantly associated with OS (P<0.05) were selected into a stepwise multivariate Cox regression analysis (Table S1). Finally, a total of four hub DRRGs were selected to establish the prognostic signature (NT5C3A, TAF9, BCAP31, NUDT21) (Table S2). The risk score of each patient was evaluated by applying the Vi × Ci formula (as reported in Methods), as follows: Risk score = (0.0346× ExpNT5C3A) + (0.0454× ExpTAF9) + (0.0192× ExpBCAP31) + (0.0486× ExpNUDT21). Additionally, we assessed the expression levels of these four hub genes in both EC tumor tissues and adjacent normal tissues. The results showed significant upregulation of all four hub genes in the EC tumor tissues (P<0.01, Figure 3A). Finally, an additional analysis of clinical EC samples from the cBioPortal database revealed that the 4 hub DRRGs were altered in 14 out of 158 sequenced cases, accounting for 9% of the samples (Figure 3B), highlighting the importance of these four DRRGs in esophageal carcinogenesis.
Table 2. The summary information of patients with EC.
| Characteristic | Patients, n (%) |
|---|---|
| Age (years) | |
| ≤65 | 98 (62.03) |
| >65 | 60 (37.97) |
| Gender | |
| Male | 135 (85.44) |
| Female | 23 (14.56) |
| Grade | |
| G1 | 16 (10.13) |
| G2 | 65 (41.14) |
| G3 | 43 (27.22) |
| Unknown | 34 (21.51) |
| T | |
| T1 | 27 (17.09) |
| T2 | 37 (23.42) |
| T3 | 75 (47.47) |
| T4 | 4 (2.53) |
| Unknown | 15 (9.49) |
| N | |
| N0 | 65 (41.14) |
| N1 | 62 (39.24) |
| N2 | 9 (5.70) |
| N3 | 6 (3.80) |
| Unknown | 16 (10.12) |
| M | |
| M0 | 119 (75.32) |
| M1 | 8 (5.06) |
| Unknown | 31 (19.62) |
| Stage | |
| I | 16 (10.13) |
| II | 68 (43.04) |
| III | 48 (30.38) |
| IV | 8 (5.06) |
| Unknown | 18 (11.39) |
EC, esophageal cancer.
Figure 3.
Identification of mRNAs associated with patient survival. (A) Different mRNA expression of four genes between the tumor and non-tumor tissues based on The Cancer Genome Atlas; (B) genetic alterations for the four selected genes of esophageal cancer in the cBioPortal database. **, P<0.01, ***, P<0.001.
Prognostic analysis of survival signature based on DRRGs
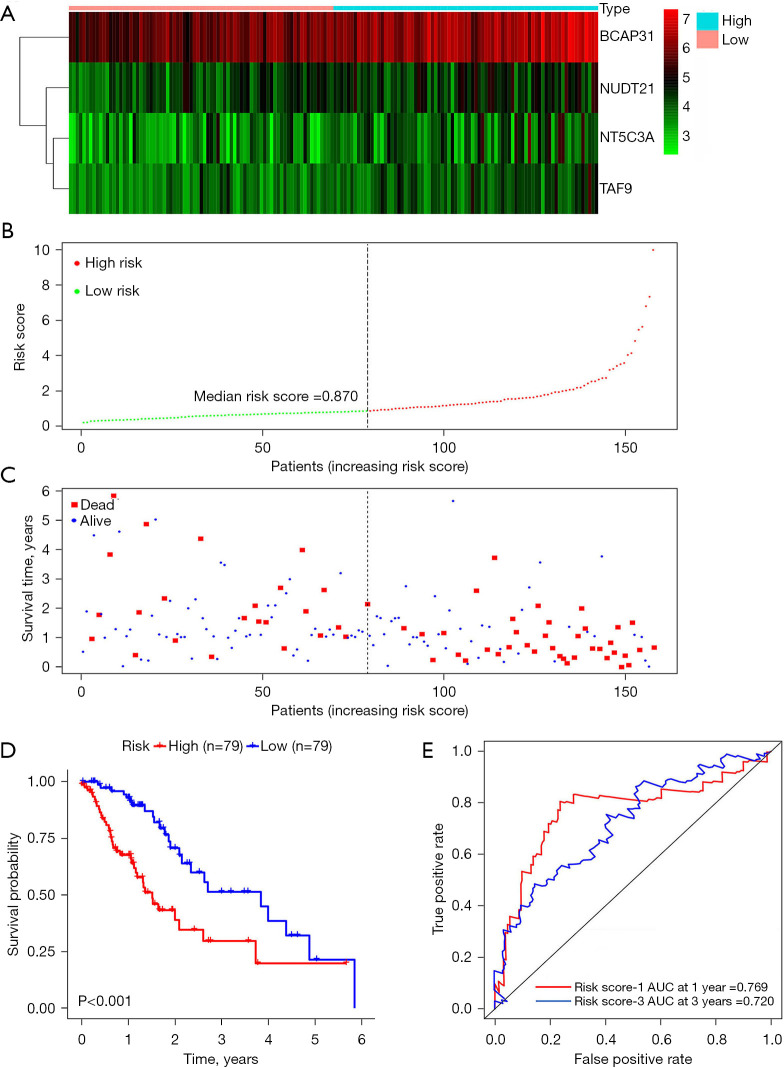
To evaluate the reliability of the risk score, we then dichotomized the EC patients into high- and low-risk groups according to the median value of risk score. The expression heat map, the distribution of patients’ risk score, and survival status are presented in Figure 4A-4C. Kaplan-Meier curves were further employed to clarify the relationship between the risk score and OS. The results indicated that the high-risk group had a worse OS compared to the low-risk group (P<0.001, Figure 4D). To estimate the prognostic ability of the 4-DRRGs biomarker signature, a time-dependent ROC analysis was performed. The ROC curves of the DDRGs signature model are shown in Figure 4E, with an AUC of 0.769 and 0.720 for 1- and 3-year survival respectively, which indicated that it has a moderate diagnostic performance.
Figure 4.
DRRGs prognostic signature of EC patients based on TCGA. (A) Expression heat map; (B) risk score distribution; (C) survival status; (D) Kaplan-Meier curves showed that patients in the high-risk group had significantly poorer OS than those in the low-risk group; (E) ROC curve of OS-related prognostic signature. AUC, area under the curve; DRRGs, DNA repair-related genes; EC, esophageal cancer; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic; OS, overall survival.
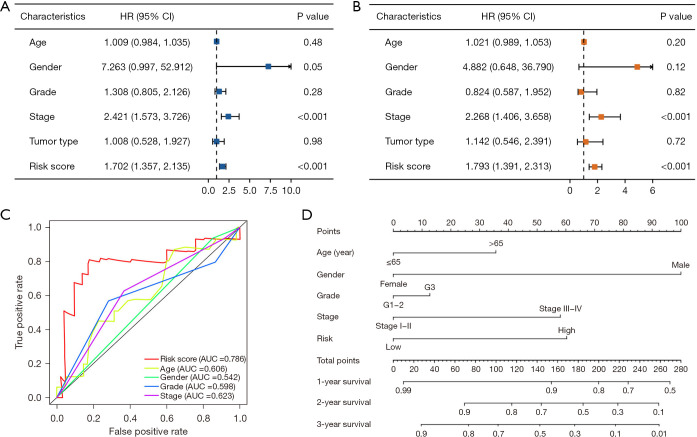
Evaluation of independent prognostic factors in EC
Univariate and multivariate Cox hazard analyses were conducted to assess the risk score obtained from the prognostic signature, along with conventional clinical parameters such as age, gender, tumor grade, and tumor stage. The results demonstrated significant associations between the risk score and tumor stage with OS in both univariate [hazard ratio (HR): 1.702; 95% confidence interval (CI): 1.357−2.135; P<0.001; and HR: 2.421; 95% CI: 1.573−3.726; P<0.001; respectively] (Figure 5A) and multivariate (HR: 1.793; 95% CI: 1.391−2.313; P<0.001; and HR: 2.268; 95% CI: 1.406−3.658; P<0.001; respectively) (Figure 5B) analysis. The risk score was an independent predictor of survival and exhibited the highest value of AUC compared to other clinical factors (Figure 5C). To facilitate its practical application in clinical settings, a nomogram was developed to predict the survival probability of patients (Figure 5D). Every value of each indicator corresponds to a distinct point. For instance, a male patient under 65 years old diagnosed with EC, with G1–2 tumor grade, I–II stage, and classified as low risk based on the calculated risk score, would be assigned a total score of 100 points. Consequently, the corresponding 1-year survival rate ranges between 90% and 99%, the 2-year survival rate is approximately 82%, and the 3-year survival rate is estimated to be around 70%.
Figure 5.
Evaluation of independent prognostic factors in EC. (A) Univariate Cox regression analysis results; (B) multivariate Cox regression analysis results; (C) ROC curve of risk score and clinical features in predicting the accuracy of patients’ survival prognosis; (D) nomogram plotted by the independent factors of patients’ overall survival. HR, hazard ratio; CI, confidence interval; AUC, area under the curve; EC, esophageal cancer; ROC, receiver operating characteristic.
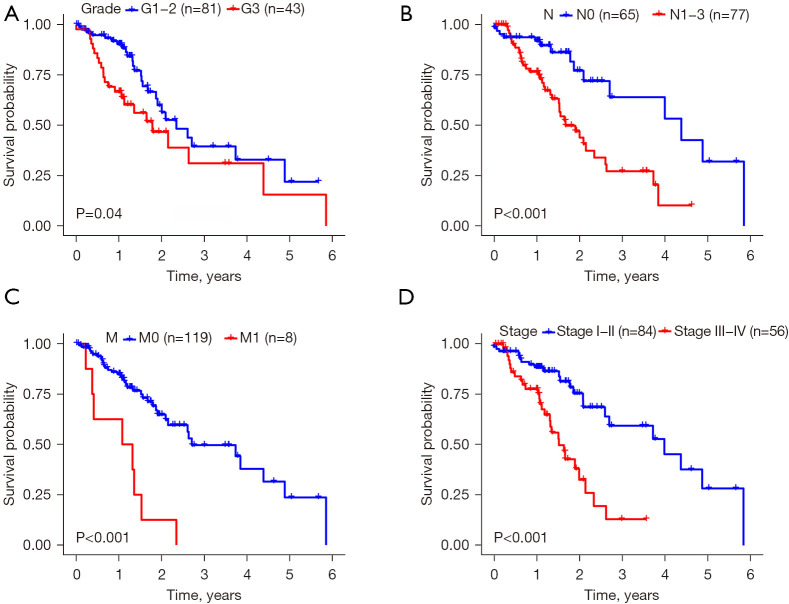
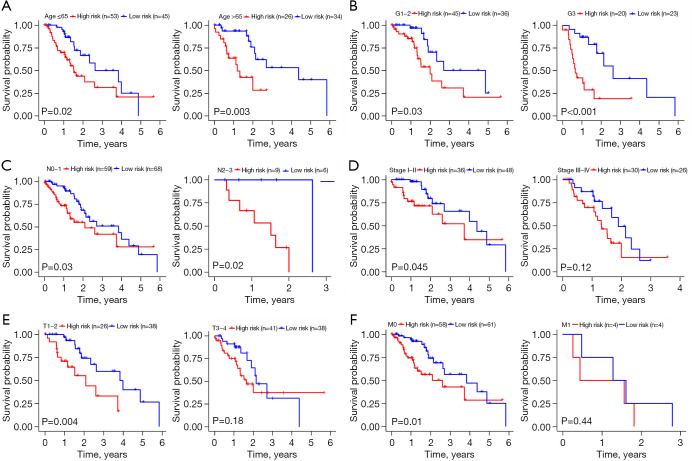
Validation of the 4-DRRGs signature in predicting survival by Kaplan-Meier curves
We first validated the impact of different clinical characteristics on survival and then conducted comprehensive subgroup analysis to determine the applicability of the established 4-DRRGs signature in EC patients of different genders, grades, and stages. The Kaplan-Meier curves demonstrated that certain clinical characteristics, including tumor grade (G3) (P=0.04), N stage (N1−3) (P<0.001), M stage (M1) (P<0.001), and tumor stage (III−IV) (P<0.001), were significantly associated with poorer OS (Figure 6). Furthermore, our results indicated that the risk score served as a reliable prognostic predictor for EC patients when stratified by age (≤65 or >65 years), tumor grade (G1−2 or G3), and N stage (N0−1 or N2−3) (Figure 7A-7C). However, the prognostic value of the risk score varied when patients were stratified by tumor stage, T stage, and M stage. Specifically, within the stage I−II subgroups, patients in the low-risk group exhibited significantly better OS than those in the high-risk group (P=0.045), whereas no significant difference was observed within the stage III−IV subgroups (P=0.12). Notably, the prognostic power of the risk score was more pronounced in individuals with T1–2 stage disease (P=0.004) compared to those with T3−4 stage disease (P=0.18). Similar trends were observed in relation to the M stage (M0, P=0.01; M1, P=0.44) (Figure 7D-7F). These findings suggest that the risk score derived from the DRRGs signature may serve as a more effective prognostic predictor for early and intermediate stages of EC.
Figure 6.
Different clinical features that can predict patient survival via Kaplan-Meier curves. (A) Tumor grade; (B) N stage; (C) M stage; (D) tumor stage.
Figure 7.
Stratified analysis for associations between the risk score and survival of EC. Kaplan-Meier curve for (A) age ≤65 years (n=98) and age >65 years (n=60) patients group; (B) tumor grade for G1−2 (n=81) and G3 (n=43) patients group; (C) N stage for N0−1 (n=127) and N2−3 (n=15) patients group; (D) tumor stage for stage I−II and stage III−IV patients group; (E) T stage for T1−2 and T3−4 patients group; (F) M stage for M0 and M1 patient group. EC, esophageal cancer.
Discussion
Accumulating evidence supports the involvement of DNA repair deficiencies in various cancers, including EC (10,11). DRRGs exhibit distinct expression patterns between adjacent and tumor tissues, which have been shown to be closely associated with patient prognosis (12-14). Genomic instability is considered a fundamental characteristic of cancer, and defective DNA repair mechanisms can contribute to oncogenic genomic instability. Recent research has demonstrated that altered DNA damage response pathways are closely linked to an increased vulnerability to cancer and significantly impact the effectiveness of cancer treatments, including therapy response and resistance (15). Consequently, targeting DRRGs has emerged as a promising approach in anticancer therapies (16,17). In this study, we utilized the hallmark gene sets, which encompass 50 well-defined biological processes relevant to a wide range of potential cellular responses (18). Our findings revealed that genes related to DNA repair pathways ranked as the top-enriched gene set in EC, suggesting that the abundance of DRRGs may serve as a potential indicator of malignant transformation in the development of EC.
The heterogeneity of EC determines that the prognostic value of a single gene is limited. Therefore, establishing a multi-gene prognostic signature instead of a single gene biomarker potentially provides more optimal feasibility in predicting prognosis for patients with EC. To date, no research has been reported on the transcriptional patterns of DRRGs in EC and its prospective prognostic value. Our study developed a prognostic signature based on 4-DRRGs which could predict the survival outcomes of patients with EC. Among the 4-DRRGs biomarkers detected in the present study, BCAP31 is reported to regulate proliferation, migration, and invasion and promote cancer progression (19). NUDT21, a newly identified post-transcriptional regulator, controls cell fate by connecting alternative polyadenylation to chromatin signaling (20,21). NUDT21 may also serve as an oncogene and promote cell growth and proliferation while inhibiting apoptosis through EIF2 signaling in pancreatic cancer (22). It is reported that p53 sequesters TAF9 from GLI1, which modulates the GLI1 oncogene activity (23). These DRRGs are potential prognostic markers and might be new therapeutic targets for EC in the future. However, the molecular mechanism of how these DRRGs contribute to EC progression requires further exploration.
To date, several studies have investigated the relationship between DNA repair and the initiation, progression, and invasion of cancer (14,24,25). However, only a limited number of prognostic signatures based on DRRGs have been developed, primarily in hepatocellular carcinoma (26), colon cancer and glioblastoma (27,28). In our present study, we identified a prognostic signature based on DRRGs in EC, to investigate the role of this signature in this particular cancer type. Our findings demonstrated that the risk score derived from this signature outperformed other clinical factors, functioning as both an independent risk predictor and a significant prognostic indicator. These results support the robustness of our risk score in predicting the prognosis of EC patients, especially for early and intermediate stages. Additionally, we developed a nomogram that can potentially aid clinicians in making informed treatment decisions for patients with EC. The present study has a certain limitation. EC is generally classified into squamous cell carcinoma and adenocarcinoma, which encompass distinct pathological subtypes with different etiological mechanisms. Nonetheless, the restricted number of EC patients within the TCGA dataset precluded the construction of individual models for each subtype. Consequently, this study conducted analysis on EC as a unified entity. It is anticipated that future research, with a more extensive sample size, will facilitate specific analyses of diverse EC patient cohorts.
Conclusions
In summary, our study successfully developed a novel prognostic signature consisting of four DRRGs for patients with EC. This signature holds promise as an independent prognostic predictor that can provide valuable insights into clinical outcomes, potentially complementing the traditional TNM system in EC.
Supplementary
The article’s supplementary files as
Acknowledgments
The results of this study are based on the data from TCGA (https://www.cancer.gov/tcga). We thank all contributors who provided the data for this study.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-262/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-262/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-262/coif). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med 2015;66:129-43. 10.1146/annurev-med-081313-121208 [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Hua Y, Jiang Z, et al. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res 2019;25:1989-2000. 10.1158/1078-0432.CCR-18-0773 [DOI] [PubMed] [Google Scholar]
- 4.Wang G, Guo S, Zhang W, et al. A Comprehensive Analysis of Alterations in DNA Damage Repair Pathways Reveals a Potential Way to Enhance the Radio-Sensitivity of Esophageal Squamous Cell Cancer. Front Oncol 2020;10:575711. 10.3389/fonc.2020.575711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 2016;16:35-42. 10.1038/nrc.2015.4 [DOI] [PubMed] [Google Scholar]
- 6.Li WQ, Hu N, Hyland PL, et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis 2013;34:1536-42. 10.1093/carcin/bgt094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouw KW, Goldberg MS, Konstantinopoulos PA, et al. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov 2017;7:675-93. 10.1158/2159-8290.CD-17-0226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qing T, Jun T, Lindblad KE, et al. Diverse immune response of DNA damage repair-deficient tumors. Cell Rep Med 2021;2:100276. 10.1016/j.xcrm.2021.100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Li X, Zhao L, et al. Identification of DNA-Repair-Related Five-Gene Signature to Predict Prognosis in Patients with Esophageal Cancer. Pathol Oncol Res 2021;27:596899. 10.3389/pore.2021.596899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørensen SG, Shrikhande A, Poulsgaard GA, et al. Pan-cancer association of DNA repair deficiencies with whole-genome mutational patterns. Elife 2023;12:e81224. 10.7554/eLife.81224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo IY, Huang YL, Lin CY, et al. SOX17 overexpression sensitizes chemoradiation response in esophageal cancer by transcriptional down-regulation of DNA repair and damage response genes. J Biomed Sci 2019;26:20. 10.1186/s12929-019-0510-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groelly FJ, Fawkes M, Dagg RA, et al. Targeting DNA damage response pathways in cancer. Nat Rev Cancer 2023;23:78-94. 10.1038/s41568-022-00535-5 [DOI] [PubMed] [Google Scholar]
- 13.Jinjia C, Xiaoyu W, Hui S, et al. The use of DNA repair genes as prognostic indicators of gastric cancer. J Cancer 2019;10:4866-75. 10.7150/jca.31062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateo J, Boysen G, Barbieri CE, et al. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol 2017;71:417-25. 10.1016/j.eururo.2016.08.037 [DOI] [PubMed] [Google Scholar]
- 15.Kheyrandish MR, Mir SM, Sheikh Arabi M. DNA repair pathways as a novel therapeutic strategy in esophageal cancer: A review study. Cancer Rep (Hoboken) 2022;5:e1716. 10.1002/cnr2.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awwad SW, Serrano-Benitez A, Thomas JC, et al. Revolutionizing DNA repair research and cancer therapy with CRISPR-Cas screens. Nat Rev Mol Cell Biol 2023;24:477-94. 10.1038/s41580-022-00571-x [DOI] [PubMed] [Google Scholar]
- 17.Pilié PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 2019;16:81-104. 10.1038/s41571-018-0114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yothers G, Song N, George TJ, Jr. Cancer Hallmark-Based Gene Sets and Personalized Medicine for Patients With Stage II Colon Cancer. JAMA Oncol 2016;2:23-4. 10.1001/jamaoncol.2015.3614 [DOI] [PubMed] [Google Scholar]
- 19.Dang E, Yang S, Song C, et al. BAP31, a newly defined cancer/testis antigen, regulates proliferation, migration, and invasion to promote cervical cancer progression. Cell Death Dis 2018;9:791. 10.1038/s41419-018-0824-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Y, Zheng T, Su Z, et al. Alternative polyadenylation reprogramming of MORC2 induced by NUDT21 loss promotes KIRC carcinogenesis. JCI Insight 2023;8:e162893. 10.1172/jci.insight.162893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brumbaugh J, Di Stefano B, Wang X, et al. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell 2018;172:106-120.e21. 10.1016/j.cell.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng YS, Chen ML, Lei WD, et al. NUDT21 knockdown inhibits proliferation and promotes apoptosis of pancreatic ductal adenocarcinoma through EIF2 signaling. Exp Cell Res 2020;395:112182. 10.1016/j.yexcr.2020.112182 [DOI] [PubMed] [Google Scholar]
- 23.Yoon JW, Lamm M, Iannaccone S, et al. p53 modulates the activity of the GLI1 oncogene through interactions with the shared coactivator TAF9. DNA Repair (Amst) 2015;34:9-17. 10.1016/j.dnarep.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majidinia M, Yousefi B. DNA repair and damage pathways in breast cancer development and therapy. DNA Repair (Amst) 2017;54:22-9. 10.1016/j.dnarep.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 25.Pardini B, Corrado A, Paolicchi E, et al. DNA repair and cancer in colon and rectum: Novel players in genetic susceptibility. Int J Cancer 2020;146:363-72. 10.1002/ijc.32516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Zhao L, Guo C, et al. Identification of a novel DNA repair-related prognostic signature predicting survival of patients with hepatocellular carcinoma. Cancer Manag Res 2019;11:7473-84. 10.2147/CMAR.S204864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Tan C, Ye M, et al. Development and validation of a DNA repair gene signature for prognosis prediction in Colon Cancer. J Cancer 2020;11:5918-28. 10.7150/jca.46328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S, Qian Z, Liang T, et al. Identification of a DNA Repair-Related Multigene Signature as a Novel Prognostic Predictor of Glioblastoma. World Neurosurg 2018;117:e34-41. 10.1016/j.wneu.2018.05.122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as