Abstract
Mouse hepatitis virus (MHV) is a 31-kb positive-strand RNA virus that is replicated in the cytoplasm of infected cells by a viral RNA-dependent RNA polymerase, termed the replicase. The replicase is encoded in the 5′-most 22 kb of the genomic RNA, which is translated to produce a polyprotein of >800 kDa. The replicase polyprotein is extensively processed by viral and perhaps cellular proteinases to give rise to a functional replicase complex. To date, two of the MHV replicase-encoded proteinases, papain-like proteinase 1 (PLP1) and the poliovirus 3C-like proteinase (3CLpro), have been shown to process the replicase polyprotein. In this report, we describe the cloning, expression, and activity of the third MHV proteinase domain, PLP2. We show that PLP2 cleaves a substrate encoding the first predicted membrane-spanning domain (MP1) of the replicase polyprotein. Cleavage of MP1 and release of a 150-kDa intermediate, p150, are likely to be important for embedding the replicase complex in cellular membranes. Using an antiserum (anti-D11) directed against the C terminus of the MP1 domain, we verified that p150 encompasses the MP1 domain and identified a 44-kDa protein (p44) as a processed product of p150. Pulse-chase experiments showed that p150 is rapidly generated in MHV-infected cells and that p44 is processed from the p150 precursor. Protease inhibitor studies revealed that unlike 3CLpro activity, PLP2 activity is not sensitive to cysteine protease inhibitor E64d. Furthermore, coexpression studies using the PLP2 domain and a substrate encoding the MP1 cleavage site showed that PLP2 acts efficiently in trans. Site-directed mutagenesis studies confirmed the identification of cysteine 1715 as a catalytic residue of PLP2. This study is the first to report enzymatic activity of the PLP2 domain and to demonstrate that three distinct viral proteinase activities process the MHV replicase polyprotein.
The RNA-dependent RNA polymerase or replicase of most positive-strand RNA viruses is translated from the genomic RNA as a large precursor polyprotein. The precursor polyprotein is then processed by viral and/or cellular proteinases to generate functional replicase enzymes. The processing of the replicase is critical for the appropriate localization, assembly, and function of the replicase complex. Understanding replicase processing is a key element in elucidating the mechanisms that control RNA virus replication.
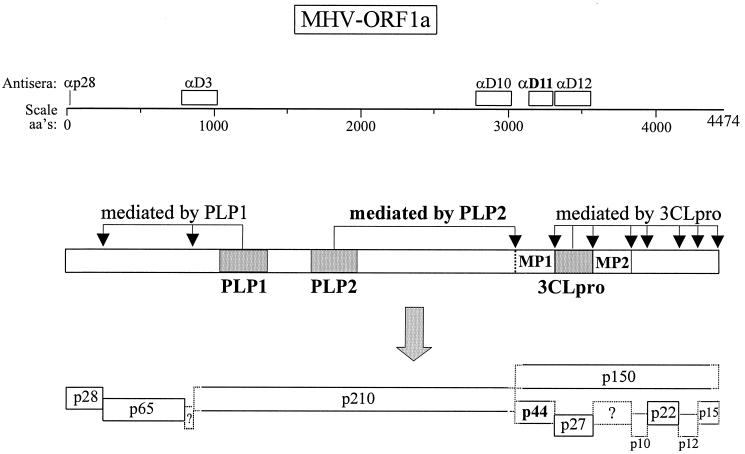
The replicase of the murine coronavirus mouse hepatitis virus (MHV) is the largest and most complex viral RNA replicase yet identified. The gene encoding the replicase, gene 1, encompasses 22 kb of the 31-kb positive-strand RNA genome (4, 27). Gene 1 contains two open reading frames (ORF1a and ORF1b) which overlap and have the capacity to generate a >800-kDa ORF1ab polyprotein (7, 27). Gene 1 is the 5′-most gene in the viral genome and is translated immediately upon entry of the virus into the cell (9, 10). Similar to other RNA viruses, MHV encodes proteinase domains in the replicase polyprotein. These proteinases mediate processing of the polyprotein into products. Sequence analysis of MHV genomic RNA led to the prediction of three proteinase domains: two papain-like cysteine proteinases, termed PLP1 and PLP2, and a domain similar to the poliovirus 3C proteinase and therefore designated 3C-like, 3CLpro (18, 19, 27). Both PLP1 and 3CLpro have been shown to function during viral replication and help drive the processing of the ORF1ab replicase polyprotein into at least 15 products (2, 3, 5, 6, 8, 11, 12, 16, 17, 31–34, 37, 38).
Processing of the MHV replicase polyprotein by viral proteinases is required for ongoing viral replication. This was shown in studies using protease inhibitor E64d, a cysteine protease inhibitor that blocks trans processing by PLP1 and 3CLpro (25, 31, 38). In the presence of E64d, specific steps in replicase processing are inhibited and viral RNA synthesis ceases (31, 38). These experiments clearly demonstrated the critical role of ongoing proteolytic processing during MHV viral RNA synthesis.
As a step toward understanding why proteolytic processing is critical for generating a functional MHV replicase complex, we are working to identify the replicase gene products and enzymes responsible for their release from the precursor polyprotein. Previously, we identified a 150-kDa ORF1a replicase product, p150, that is an intermediate between the nascent ORF1a replicase polyprotein and one ultimate product, 3CLpro (p27) (37). We detected p150 by using a polyclonal antiserum to 3CLpro (anti-D12) and demonstrated that p150 is a precursor to p27. We speculated that the p150 intermediate extended upstream of the 3CLpro domain and included the putative membrane-spanning domain, MP1. The MP1 domain is thought to be critical for embedding the replicase complex into cellular membranes. In the study presented here, we show that p150 does indeed encompass MP1. We generated a polyclonal antiserum (anti-D11) that detected p150 and the MP1-containing cleavage product, p44, from MHV-infected cells. To identify the proteinase responsible for generating p150, we generated expression constructs encoding one of the three possible MHV proteinase domains and the substrate cleavage site. We report that the MHV PLP2 domain efficiently processes the putative MP1 cleavage site and that PLP2 can act in trans. This is the first report of enzymatic activity for MHV PLP2.
MATERIALS AND METHODS
Virus and cells.
Plaque-cloned MHV strain JHM-X (35) was propagated on 17Cl-1 cells (43) maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS), 5% tryptone phosphate broth (TPB), 2% penicillin–streptomycin (P/S), and 2% 200 mM l-glutamine (l-glu). All tissue culture reagents were purchased from Gibco-BRL, Gaithersburg, Md. The infectivity of the virus stock was determined by plaque assay with murine delayed brain tumor (DBT) cells (23) as indicator cells. The DBT cells were grown in minimal essential medium supplemented with 5% FCS, 2% l-glu, 2% P/S, and 10% TPB. HeLa-MHVR cells (15), which are HeLa cells stably transfected with the MHV receptor, were kindly provided by T. Gallagher, Loyola University, Chicago, Ill. HeLa-MHVR cells were grown in DMEM supplemented with 10% FCS, 0.5% P/S, 0.5% l-glu, and 5 mM sodium HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.4).
Generation of anti-GST-D11 serum.
The D11 region was generated by reverse transcription-PCR (RT-PCR) from RNA isolated from MHV-JHM-infected cells with primers FP30 and FP31 (Table 1) by using RNAzol B according to the manufacturer's instructions and as previously described (37). The D11 region was cloned in frame with gluthathione S-transferase (GST) in the pGEX-KG vector, and fusion protein was induced by addition of 150 μM IPTG (isopropyl β-d-thiogalactopyranoside) for 2 h, isolated and purified as previously described (37). Rabbits were injected intramuscularly and subcutaneously with 1 mg of GST-D11 fusion protein emulsified in 1 ml of adjuvant (Freund's complete adjuvant for the first immunization and incomplete adjuvant for two subsequent immunizations). Serum was isolated from blood collected 10 to 14 days after each injection. The titer of sera taken after the second and third immunizations to GST-D11 fusion protein was >250,000.
TABLE 1.
Primers used for amplification or mutagenesis of MHV-JHM sequences
| Primer | Oligonucleotide sequence (5′ to 3′)a | Nucleotide no.b | Polarity | Domain generated |
|---|---|---|---|---|
| FP30 | TATCTAGATGTTCGTAGTGATGGC | 9940–9955 | Forward | GST-D11 insert |
| FP31 | TGAAGCTTAGGCAGTTGGAGG | 10187–10199 | Reverse | |
| B165 | CCTCTAGAATAATGGGCAAGAAAGTCGTG | 2710–2725 | Forward | |
| B190 | TAACGCGTTCAGAGATCTGCCGCAGACAGACCACTATACTG | 5198–5227 | Reverse | PLP1/MPI insert |
| B195 | TTAGATCTTGTATTGTGGGCCCTTATG | 8790–8809 | Forward | |
| B194 | AACTTAAGTCATAAAAATGATGTAGTAACAGA | 10199–10219 | Reverse | |
| B162 | TATCTAGAATAATGGTGTGCTTCTACCCACTGTTTGTCCTTGTTGG | 7448–7479 | Forward | Cen-3CLpro insert |
| B170 | GAAGATCTTCACTCTTGTGCGCTTTGACTGTAGC | 11125–11148 | Reverse | |
| B204 | GGGGATCCATAATGGAAGCGCTGCGACATGAT | 4790–4807 | Forward | PLP2-Cen insert |
| B205 | CCCTCGAGCCAGGCTTACTACATCCATA | 7652–7671 | Reverse | |
| B206 | GGGGATCCAACATGGTGACAGATTTAGGCTATA | 7249–7268 | Forward | Cen-MP1 insert |
| B207 | TAATCTAGAAATGATGTAGTAACAGAGGCA | 10195–10216 | Reverse | |
| B212 | GCAGTCAAATAATAATGGCTATATAAATGTGGC | 5341–5373 | Forward | PLP2-C1715G |
| B213 | GCCACATTTATATAGCCATTATTATTTGACTGC | 5341–5373 | Reverse | |
| B215 | GCTATATAAATGTGGCAGGTTTAATGCTGCAACAC | 5358–5392 | Forward | PLP2-C1721G |
| B216 | GTGTTGCAGCATTAAACCTGCCACATTTATATAGC | 5358–5392 | Reverse |
Preparation of radiolabeled whole-cell lysates and immunoprecipitation.
MHV-JHM infection, radiolabeling of proteins, preparation of whole-cell lysates, and immunoprecipitation were performed by our laboratory as previously described (37), with two exceptions. First, HeLa-MHVR cells were used for all labeling experiments. Second, proteins were metabolically radiolabeled with Tran35S-label for 2 h, and infected cells were harvested at 7 h postinfection (hpi). For experiments involving proteinase inhibitor E64d (Matreya, Inc., Pleasant, Pa.), the drug was dissolved in dimethyl sulfoxide to generate a 100-μg/μl stock solution. E64d was added to the medium to a final concentration of 400 μg/ml for 1 h prior to labeling and was also added to the methionine-free media during the starvation and radiolabeling of the cells. The antibodies used in this study included rabbit anti-p28 (2), anti-D3, anti-D10, and anti-D12 (37) polyclonal antisera and mouse anti-V5 monoclonal antibody (Invitrogen).
Generating expression constructs for MHV-JHM ORF1a.
Constructs expressing specific regions of ORF1a were generated by RT-PCR amplification from RNA isolated from MHV-JHM-infected cells by using RNAzol B as previously described (37). Following amplification with specific primers listed in Table 1, products were purified, digested with the appropriate restriction enzymes, and then ligated into the polylinker region of specific vectors as follows: Cen-3CLpro was ligated into the XbaI-BglII site of pET-11d (Novagen), PLP2-Cen was ligated into the BamHI-XhoI site into pcDNA3.1/V5-His (Invitrogen), and Cen-MP1 was ligated into the BamHI-XbaI site into pcDNA3.1/V5-His. pPLP2-MP1 was generated by a two-step PCR procedure. PLP2-Cen and Cen-MP1 PCR products were digested with EcoRI, isolated, gel purified, and ligated together overnight in the presence of T4 ligase at 16°C. The ligated product was then as used a template for second-round PCR amplification with outside primers B204 and B207. The resulting product of 5,446 bp was digested with BamHI and XbaI, gel purified, and ligated into the BamHI-XbaI site of pcDNA3.1/V5-His. pPLP1/MP1 was also generated by a two-step PCR procedure. The PLP1 domain was amplified from pT7-NBgl DNA (2) by using primers B165 and B190. The MP1 domain was amplified from pCen-3CLpro DNA by using primers B194 and B195. The two PCR products were digested with BglII, gel purified, and allowed to ligate overnight in the presence of T4 ligase at 16°C. The ligated product was used as a template and amplified by using outside primers B165 and B194. The resulting product of 3,937 bp was gel purified and ligated directly into the pCR-XL-TOPO vector (Invitrogen). Three plasmids, pPLP2-Cen, pCen-MP1, and pPLP2-MP1, required special growth conditions. After ligation, these plasmids were transformed into Escherichia coli DH5α cells by heat shock at 42°C for 90 s. The transformed bacteria were maintained in S.O.C. broth (2% [wt/vol] Bacto Tryptone, 0.5% [wt/vol] yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) at room temperature (20 to 25°C) for 1.5 h and then plated on Luria-Bertani plates containing 50 μg of ampicillin per ml and incubated at room temperature for 4 to 5 days. Under these conditions, a majority of bacteria retained the full-length insert, but the transformed colonies grew slowly and were smaller than typical colonies. All subsequent incubation steps using these transformed bacteria (such as for minipreps and large-scale plasmid preparations) were also performed at room temperature.
Plasmid DNA purification.
Small-scale purification of plasmid DNAs was performed with the Wizard purification system according to the manufacturer's instructions (Promega). Large-scale purification of plasmid DNAs was prepared by using an ammonium acetate precipitation method (36) with some modifications. Briefly, transformed bacteria from 250 ml of liquid culture (optical density at 600 nm of 0.6 to 1.0) were resuspended in 10 ml of cell resuspension solution (50 mM Tris-HCl, [pH 7.5], 10 mM EDTA, 100 μg of RNase A per ml). The cells were lysed by adding 10 ml of cell lysis solution (0.2 M NaOH, 1% sodium dodecyl sulfate [SDS]) and neutralized with 10 ml of neutralization solution (1.32 M potassium acetate [pH 4.8]). The mixture was centrifuged at 12,000 × g for 10 min at 4°C. Supernatant was transferred to a new tube, and 0.6 volume of isopropanol was added. Samples were mixed by inversion and incubated at room temperature for 10 min prior to centrifugation at 12,000 × g for 10 min at 4°C. After centrifugation, the pellet containing crude plasmid DNA was resuspended in 4 ml of 2 M ammonium acetate (pH 7.4) by gentle rocking and incubated on ice for 10 min. After centrifugation (10 min at 12,000 × g), the supernatant was transferred to a fresh tube, 4 ml of isopropanol was added, and the combination was mixed by inversion and incubated at room temperature for 10 min. Centrifugation was performed at 12,000 × g for 10 min at 4°C. The final DNA pellet was resuspended in 2 ml of sterile, deionized water, and 5 μl of 10-mg/ml RNAase was added for 20 min at 37°C. Following RNase treatment, 1 ml of ice-cold 7.5 M ammonium acetate (pH 7.6) was added, and the sample was mixed by inversion. The sample was then incubated at room temperature for 5 min followed by centrifugation at 12,000 × g for 10 min at room temperature. The supernatant was transferred to a new tube, and 3 ml of isopropanol was added, mixed by inversion, and incubated at room temperature for 10 min. Centrifugation was performed at 12,000 × g for 10 min at room temperature. The resulting pellet was washed with 70% ethanol and resuspended in sterile, deionized water for future use.
Gel purification of digested plasmid DNAs.
All plasmid DNAs digested with the appropriate restriction enzymes were gel purified by the crystal violet (CV) method according to the manufacturer's instructions (Invitrogen) with some modifications. Briefly, 40 μl of digested DNA (0.2 to 2 μg) was mixed with 8 μl of 6× CV loading buffer (100 μg of CV per ml, 20 mM EDTA, 30% glycerol) and loaded onto a 0.8% agarose gel containing CV (20 to 40 μl of 2-mg/ml CV stock solution in 50 ml of 0.8% agarose gel in 1× TAE buffer [0.04 M Tris-acetate, 0.001 MEDTA]). The DNA was subjected to electrophoresis at 80 V in 1× TAE buffer for 30 min (or until the DNA band of interest was approximately halfway down the gel). The DNA was visible as a blue band, excised from the gel, and transferred to a new tube containing 2.5 volumes of sodium iodide solution (6.6 M sodium iodide, 16 mM sodium sulfite). The mixture was incubated at 50°C for 2 min or until the gel had completely melted. The tube containing the mixture was placed at room temperature, 1 volume of Wizard miniprep binding resin (Promega) was added, and the combination was mixed by vortexing. The DNA-resin mixture was poured into a Promega Wizard Plus SV miniprep spin column, fitted into a 2-ml microcentrifuge tube, and subjected to centrifugation at 3,000 × g for 30 s. The DNA-resin complex was trapped on the filter and washed twice with 400 μl of washing solution (1 mM NaCl, 75% ethanol). After the washing solution was discarded, the column was spun at maximum speed to dry the DNA-resin for 3 min. The DNA was eluted from the resin with 40 μl of sterile, deionized water. The DNA concentration was estimated by running 10 μl of the eluted DNA on a 1% agarose gel containing 0.5 μg of ethidium bromide per ml.
Transfection and vaccinia virus T7 expression of MHV-ORF1a products.
Plasmid DNAs encoding specific regions of MHV-JHM ORF1a were transfected into HeLa-MHVR cells infected with vaccinia virus expressing T7 polymerase (vTF7.3; kindly provided by B. Moss, National Institutes of Health, Bethesda, Md.) (14). Briefly, monolayer cultures of 50 to 80% HeLa-MHVR cells in 60-mm-diameter petri dishes were infected with vTF7.3 at a multiplicity of infection of 10 for 1 h. The cells were then rinsed with phosphate-buffered saline (PBS) and transfected with 1 μg of plasmid DNA in 20 μl of 2-mg/ml Lipofectamine (Gibco-BRL, Bethesda, Md.) for 3 h, following the manufacturer's instructions. At 4 hpi, the media were replaced with serum-containing DMEM to allow the cells to recover from transfection. At 5 hpi, cells were washed with PBS and the media were replaced with methionine-free medium. At 5.5 hpi, Tran35S-label (50 μCi/ml) was added to the medium to radiolabel newly synthesized proteins. Whole-cell lysates were prepared at 10.5 hpi and subjected to immunoprecipitation as described above.
Mutagenesis of pPLP2-Cen.
Plasmid DNA pPLP2-Cen was subjected to site-directed mutagenesis by using synthetic oligonucleotides with single-nucleotide mismatches and amplification with high-fidelity Pfu DNA polymerase according to the manufacturer's instructions (QuickChange Site-Directed Mutagenesis; Stratagene). Briefly, polyacrylamide gel electrophoresis (PAGE)-purified oligonucleotides B212 and B213 (Table 1) were annealed and extended on the pPLP2-Cen template DNA by using Pfu DNA polymerase. The template DNA was subjected to 12 cycles of amplification with denaturation at 95°C for 30 s, annealing of oligonucleotides at 55°C for 1 min, and DNA extension at 68°C for 17 min. The template DNA was digested with DpnI, and the amplified DNA was transformed into Epicurian coli XL1-Blue supercompetent cells (Stratagene). Plasmid DNA was isolated from the bacteria, and the PLP2-Cen coding region was sequenced to confirm the point mutation and to ensure that no other mutations were introduced during the mutagenesis procedure. The resulting plasmid was designated pPLP2-C1715G. By the same procedure, PAGE-purified oligonucleotides B215 and B216 (Table 1) were used to change the PLP2 nucleotide sequence such that cysteine 1721 was converted to a glycine, and the resulting plasmid was designated pPLP2-C1721G.
RESULTS
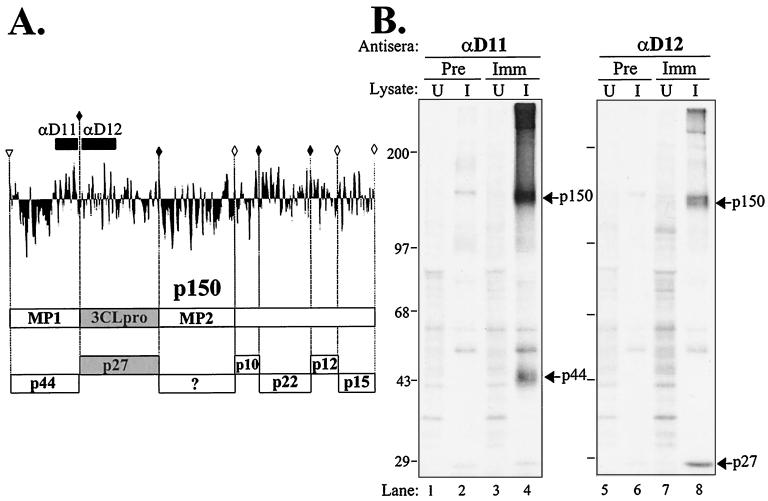
Previously, we identified a 150-kDa replicase intermediate from MHV-infected cells by using antibodies to the 3CLpro domain (37). We proposed that p150 extended from a putative PLP cleavage site upstream of the MP1 domain to the end of ORF1a. To determine if p150 did extend into the MP1 domain and to identify putative p150 cleavage products, we generated an antiserum to an 86-amino-acid region of the MP1 region that encoded a small, predicted hydrophilic domain. This domain was designated D11 (Fig. 1A). As described in Materials and Methods, this D11 region was expressed as a GST fusion protein in bacteria. The fusion protein was purified and used to immunize rabbits to generate a polyclonal antiserum, anti-D11. Upon testing the specificity of this antiserum with radiolabeled cell lysates from uninfected and MHV-infected cells, the p150 protein and a newly identified MHV product, p44, were detected (Fig. 1B, lane 4). The p150 replicase product migrates slightly faster on the gel than a nonspecific product, which may represent small amounts of uncleaved spike glycoprotein that bind to the protein A-Sepharose beads, which is detected with all tested preimmune and immune antisera (Fig. 1B, lanes 2, 4, 6, and 8). The p150 replicase product precipitated with anti-D11 appears to be identical to that detected by immunoprecipitation with anti-D12 (Fig. 1B, compare lanes 4 and 8). These results indicate that p150 contains both the MP1 domain and the 3CLpro region.
FIG. 1.
Detection of p150 and p44 from MHV-infected cells by using anti-D11 serum. (A) Hydrophobicity plot of the predicted p150 region and schematic diagram of reported and predicted products (17, 37). The D11 and D12 domains, to which antisera were generated, are depicted. Solid diamonds indicate confirmed MHV 3CLpro cleavage sites, and open diamonds indicate predicted 3CLpro sites. The putative p150 cleavage site is indicated by the open arrowhead. MP1 and MP2, the predicted membrane-spanning domains (27), are indicated. (B) Immunoprecipitation of replicase products from MHV-infected cells. Radiolabeled lysates were prepared from uninfected (U) or MHV-infected (I) HeLa-MHVR cells and subjected to immunoprecipitation with preimmune serum (Pre) or serum from rabbits immunized with GST-D11 or GST-D12 protein (Imm). Products were analyzed by electrophoresis on SDS–5 to 10% polyacrylamide gels. The gels were processed and subjected to autoradiography. MHV-specific proteins p150, p44, and p27 are indicated.
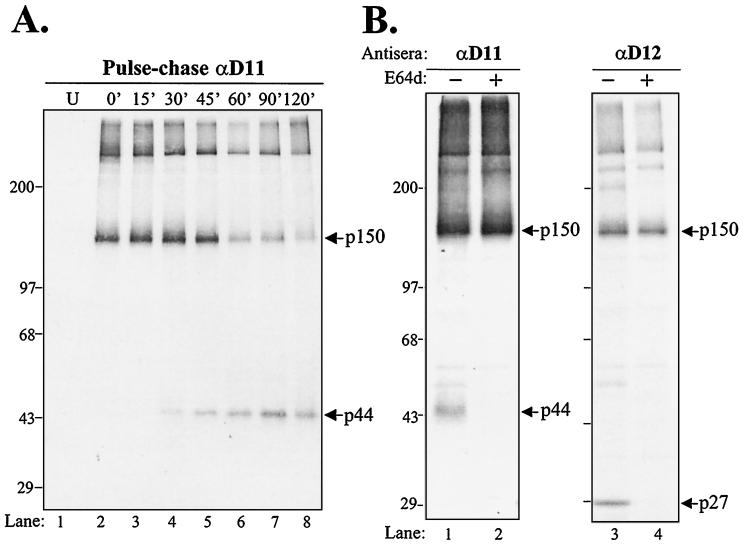
To determine if p150 and p44 exhibited a precursor-product relationship, we performed pulse-chase analysis. MHV-infected cells were radiolabeled for 30 min, and cells were washed with PBS and incubated in medium containing excess cold methionine. Cell lysates were prepared at the times indicated and subjected to immunoprecipitation with anti-D11 serum. At early times (0, 15, and 30 min of chase), immunoprecipitation with anti-D11 revealed the p150 product (Fig. 2A). This suggests that processing of p150 is very rapid and likely occurs cotranslationally. With increasing periods of chase, the amount of p150 diminished with a concomitant increase in p44. The kinetics of the processing of p150 into products was the same as we previously reported for processing of p150 to generate p27 (37).
FIG. 2.
Analysis of precursors and products detected in MHV-infected cells by using replicase-specific antisera anti-D11 and anti-D12. (A) Pulse-chase analysis. Lysates prepared from MHV-infected cells were pulse-labeled with Tran35S-label from 4.5 to 5.0 hpi and chased with medium containing excess methionine for the times indicated. Lysates were prepared and subjected to immunoprecipitation with anti-D11 serum and analyzed as described in the legend to Fig. 1. The p150 and p44 products are indicated by arrows. (B) Inhibition of 3CLpro-mediated proteolytic processing by E64d. MHV-infected HeLa-MHVR cells were labeled with Tran35S-label from 5 to 7 hpi in either the absence (−) or presence (+) of 400 μg of protease inhibitor E64d per ml and subjected to immunoprecipitation with anti-D11 and anti-D12 sera.
It has previously been shown that MHV proteinases are sensitive to the cysteine protease inhibitor E64d. Lu and coworkers demonstrated that E64d inhibits the autoproteolytic processing of 3CLpro (p27) (31). In addition, E64d has been shown to inhibit PLP1 processing of p65, but not the rapid cis cleavage by PLP1 to generate p28 (25). To determine if processing of p150 was sensitive to E64d, we radiolabeled MHV proteins in the presence and absence of E64d and subjected the lysates to immunoprecipitation with specific antisera. We found that processing of p44 and p27 was inhibited by E64d (Fig. 2B, lanes 2 and 4). However, the processing event required to generate p150 was not inhibited by E64d. As shown in Fig. 1A, the cleavage site between p44 and p27 is a conserved LQ/S site recognized by the 3CLpro (33). When 3CLpro activity is inhibited by E64d, only the precursor p150 is detected. These results indicate that a proteinase other than 3CLpro likely mediates the processing of p150.
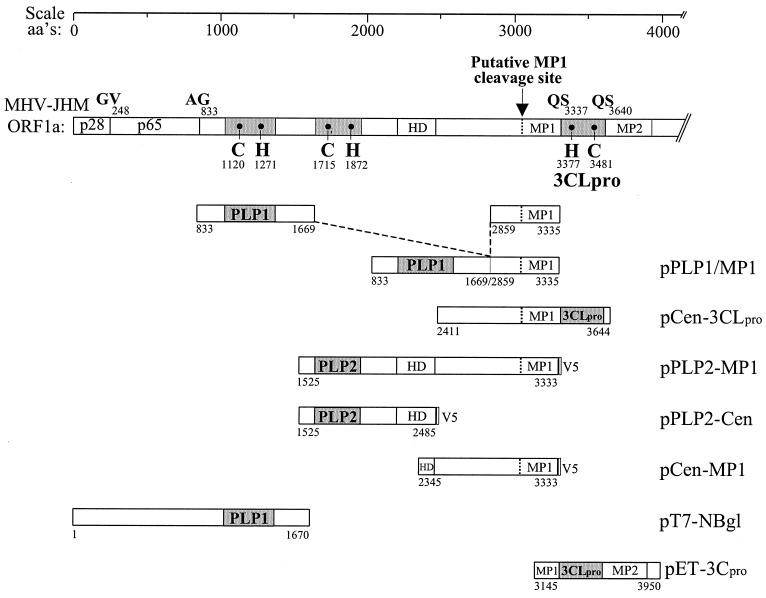
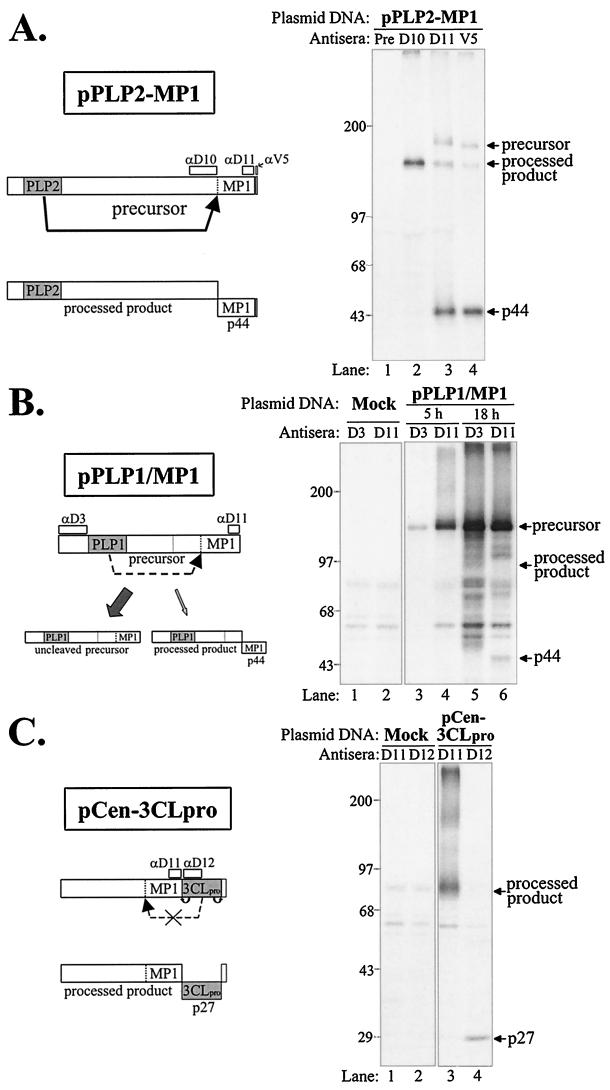
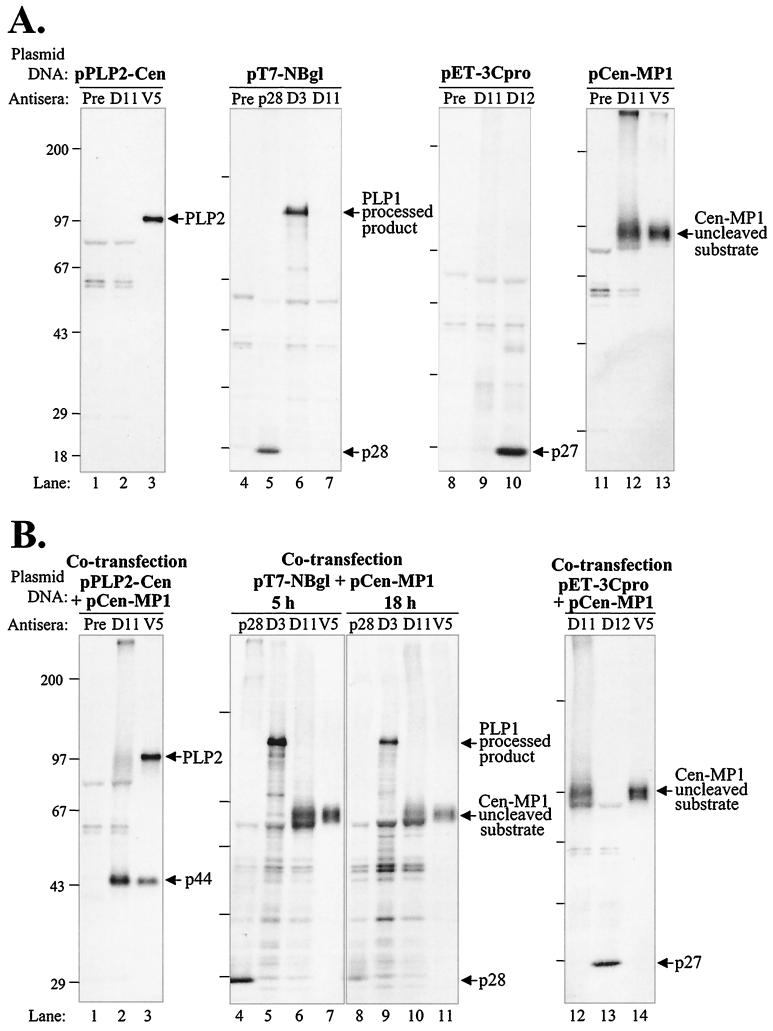
To identify the proteinase responsible for generating p150, we designed and generated expression constructs encoding the region predicted to contain the cleavage site with a region encoding one of the three possible viral proteinase domains (Fig. 3). The MHV replicase ORFs depicted were generated by RT-PCR and cloned into a plasmid vector downstream of a T7 promoter as described in Materials and Methods. Plasmid DNA was transfected into vTF7.3-infected cells, newly synthesized proteins were labeled with Tran35S-label, and products were detected by immunoprecipitation with specific antibodies. We found that the construct encoding the PLP2 domain expressed a polyprotein of 199 kDa that was efficiently processed to generate p44 (Fig. 4A). The processed product p44 was detected by the anti-D11 antiserum and with a monoclonal antibody (anti-V5) directed against the epitope tag expressed as part of this construct. This is the first evidence of proteinase activity of the MHV PLP2 domain.
FIG. 3.
Schematic diagram of constructs generated to assess MHV proteinase activity. The ORF of MHV-JHM ORF1a is shown. The numbering of the amino acids (aa's) is according to Lee et al. (27) as corrected by Bonilla et al. (4). Catalytic cysteine (C) and histidine (H) residues of PLP1 (3) and 3CLpro (34) are indicated. The predicted catalytic residues of PLP2 are C-1715 and H-1872 (27). The putative p150 site is located just upstream of the MP1 domain. Constructs that incorporate the V5 epitope tag are indicated (V5). See Materials and Methods for a description of the generation of each expression construct.
FIG. 4.
Analysis of proteolytic activity of PLP1, PLP2, and 3CLpro-containing polyproteins for the ability to cleave the predicted p150 cleavage site and release p44. HeLa-MHVR cells were infected with vaccinia virus vTF7.3 encoding the T7 polymerase and transfected with the indicated plasmid DNA. Proteins were labeled by the addition of Tran35S-label to the medium from 5.5 to 10.5 or 5.5 to 23.5 hpi, as indicated in the diagram. At the end of the labeling, cell lysates were prepared and subjected to immunoprecipitation with either MHV-specific polyclonal antisera (anti-D3, -D10, -D11, and -D12) or a mouse monoclonal antibody to an epitope tag included in the construct (anti-V5). Immunoprecipitated products were analyzed by electrophoresis on SDS–5 to 10% polyacrylamide gels, processed, and subjected to autoradiography. A schematic diagram of the expected products is on the left, and arrows on the right indicate the plasmid DNA-expressed products detected. (A to C) Expression products generated from pPLP2-MP1 DNA (A), pPLP1/MP1 DNA (B), and pCen-3CLpro DNA (C).
Interestingly, a chimeric construct encoding the MHV PLP1 domain fused with the MP1 domain exhibited a low level of processing (Fig. 4B, lane 6). A small amount of processed product of approximately 44 kDa was detected after overnight labeling, suggesting that PLP1 may recognize the MP1 cleavage site very inefficiently. Alternatively, there may be conformational requirements for PLP1 recognition of the MP1 cleavage site.
We also tested 3CLpro for its ability to cleave the MP1 cleavage site, since it had been previously suggested that 3CLpro may recognize an LK/R cleavage site and process the MP1 domain (see Table 1 in reference 27). In Fig. 4C, lane 4, we show that the 3CLpro domain is active in this expression construct and efficiently cleaves itself to release p27. However, the upstream cleavage site for the release of p44 is not recognized and processed. The upstream product has a predicted mass of 104 kDa, but the protein migrates to approximately 80 kDa under the SDS-PAGE conditions, likely due to the MP1 hydrophobic domain.
The analysis of the first three constructs, pPLP2-MP1, pPLP1/MP1, and pCen-3CLpro, demonstrated that a polyprotein containing the PLP2 domain generated a proteinase activity that was capable of cleaving the MP1 cleavage site and generating p44. To determine if the PLP2 domain itself could act in trans to mediate the cleavage of p44, we generated a construct expressing the substrate cleavage site (pCen-MP1) and cotransfected it with plasmids encoding individual proteinase domains (see Fig. 3 for diagrams of each construct). As a first step, we tested each construct for the expression of the appropriate product. Cells infected with vaccinia virus encoding the T7 polymerase were transfected with plasmid DNA encoding the domain of interest under the control of the T7 promoter as described above. Newly synthesized proteins were labeled with Tran35S-label and detected by immunoprecipitation with specific antibodies. We found that each plasmid encoded the expected products (Fig. 5A). pPLP2-Cen encoded a protein predicted to be 107 kDa (Fig. 5A, lane 3). The plasmid DNAs encoding the PLP1 domain (pT7-NBgl [2]) and 3CLpro domain (pET-3Cpro [37]) were shown to encode active proteinases, as demonstrated by their ability to process their respective polyproteins and produce p28 and p27 (Fig. 5A, lanes 5 and 10). The substrate encoding the putative MP1 cleavage site was also expressed and detected with either the anti-D11 antibody or the anti-V5 antibody that was directed to the epitope tag (Fig. 5A, lanes 12 and 13).
FIG. 5.
Analysis of trans-acting proteinase activity of PLP1, PLP2, and 3CLpro. HeLa-MHVR cells were infected and transfected with the indicated plasmid DNA and subjected to labeling and immunoprecipitation as described in Materials and Methods. (A) Protein products detected after transfection with plasmid DNA encoding one of the MHV proteinase domains or the putative p150 cleavage site, as indicated above each gel. (B) Protein products detected after cotransfection of pCen-MP1 DNA encoding the MHV p150 cleavage site substrate and one of the MHV proteinase domains, as indicated above the gel.
Next, plasmid DNA encoding the substrate (pCen-MP1) was cotransfected with plasmid DNA encoding a proteinase domain into vTF7.3-infected cells, and proteolytic processing was monitored by immunoprecipitation of radiolabeled proteins as described above. We found that the protein encoded by the PLP2 domain mediated efficient processing of the substrate to release p44 (Fig. 5B, lane 2). Using the anti-V5 antibody, we found that both the PLP2 protein and the processed product p44 were detected (Fig. 5B, lane 3). This result shows that the PLP2 proteinase can act in trans to generate p44. In contrast, neither PLP1 nor 3CLpro was able to act in trans to release p44 (Fig. 5B, lanes 6, 7, 10 to 12, and 14). We verified the activity of PLP1 and 3CLpro by detecting their autoproteolysis products p28 and p27, respectively (Fig. 5B, lanes 4, 8, and 13). We noted that the anti-D11 serum did not detect either the precursor or a small amino-terminal product of the pET-3Cpro translation product (Fig. 5, lane 12) (data not shown). The absence of the precursor reflects the rapid autoproteolytic processing by 3CLpro to generate p27. The amino-terminal product of pET-3Cpro is likely rapidly degraded or is folded in such a way that the anti-D11 serum cannot precipitate the truncated product. Overall, we concluded that enzymatically active forms of PLP1 and 3CLpro were unable to recognize and process the Cen-MP1 substrate in trans. Only protein products containing the PLP2 domain efficiently recognized the MP1 cleavage site to generate p44.
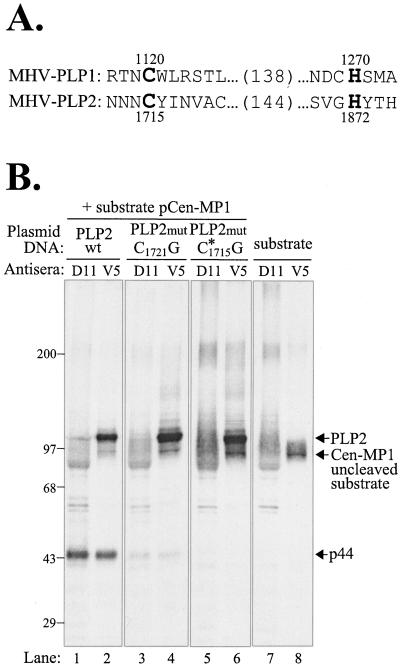
To determine if the predicted catalytic cysteine residue of PLP2 is required for proteinase activity, we performed site-directed mutagenesis on the pPLP2-Cen plasmid DNA and tested PLP2 mutant products for proteinase activity. The predicted catalytic cysteine and histidine residues, cysteine 1715 and histidine 1872, are shown aligned with the essential catalytic residues of MHV PLP1 (Fig. 6A). (Note that the amino acid numbering has been modified from the original published sequence of Lee et al. [27] by using the data of Bonilla et al. [4].) The predicted catalytic cysteine residue 1715 and downstream cysteine residue 1721 were mutated to glycine by oligonucleotide-mediated site-directed mutagenesis as described in Materials and Methods. Plasmid DNAs encoding the wild-type or mutant PLP2 domain were cotransfected with the pCen-MP1 DNA into HeLa-MHVR cells and analyzed for trans-cleavage activity as described above. The PLP2 protein with a cysteine-to-glycine substitution at position 1721 was able to process the pCen-MP1-encoded protein and generate p44 (Fig. 6, lanes 3 and 4). The amount of p44 is reduced compared to that generated by the wild-type PLP2 product (lanes 1 and 2), perhaps due to inefficient folding or altered conformation of the proteinase, but enzymatic activity is detected. In contrast, PLP2 containing a glycine substitution of the predicted catalytic cysteine 1715 residue does not cleave the Cen-MP1 substrate, and no p44 is detected (lanes 5 and 6). This result provides experimental evidence for the predicted activity and essential catalytic cysteine residue of the second papain-like cysteine proteinase domain of MHV (27).
FIG. 6.
Analysis of trans-acting proteinase activity of wild-type and mutant forms of PLP2. (A) Alignment of the MHV PLP1 and PLP2 domain amino acid sequences. PLP1 catalytic cysteine and histidine residues and PLP2 predicted catalytic cysteine and histidine residues are shown in boldface. (B) Protein products detected after cotransfection of DNAs encoding the MHV p150 cleavage site substrate and either the wild-type (wt) or a mutant (mut) form of the PLP2 domain, as indicated above the gel.
DISCUSSION
In this report, we show that the predicted second papain-like proteinase domain of the MHV replicase polyprotein encodes an active enzyme. PLP2 cleaves a site located just upstream of the first predicted membrane-spanning domain in the replicase polyprotein. Processing the replicase polyprotein at this site generates the p150 replicase intermediate that is likely critical for embedding the replicase complex into cellular membranes. p150 itself is extensively processed by MHV 3CLpro (37). In addition, the region upstream of p150 is processed by the PLP1 domain (2, 3, 5, 6, 8, 16). Therefore, at least three distinct proteinase activities, PLP1, PLP2, and 3CLpro, are required for the maturation of the MHV replicase complex.
Previously, we identified the p150 intermediate as a precursor to the MHV 3C-like proteinase, p27 (37). We proposed that the p150 intermediate extended upstream of p27 (3CLpro) and encompassed MP1. In this study, we generated an antiserum to a region of MP1 and used it to immunoprecipitate MHV replicase products from virus-infected cells. The new antiserum, anti-D11, detected p150 and a product of 44 kDa, p44 (Fig. 1B). Using pulse-chase analysis, we showed that there is a precursor-product relationship between p150 and p44 (Fig. 2A). Furthermore, p150 is rapidly generated in MHV-infected cells. The p150 intermediate is processed by the action of 3CLpro to generate p44, p27, and additional products. These 3CLpro-mediated cleavages can be inhibited by proteinase inhibitor E64d, but the cleavage of p150 is not inhibited (Fig. 2B). These results suggested that another proteinase, not 3CLpro, is responsible for processing the replicase polyprotein upstream of the MP1 domain and generating p150.
By assessing the ability of each of the three MHV proteinase domains to cleave a polyprotein substrate representing the region upstream of the MP1 domain (the putative MP1 cleavage site), we determined that only PLP2 efficiently cleaves the substrate (Fig. 4A). Furthermore, the PLP2 proteinase can process the MP1 cleavage site in trans (Fig. 5B). In contrast, catalytically active PLP1 and 3CLpro enzymes are unable to efficiently process the MP1 cleavage site. Finally, mutagenesis of the predicted catalytic residues of PLP2 knocked out the proteinase activity (Fig. 6), indicating that PLP2 does indeed encode the enzymatic activity.
PLP2 activity has been postulated for 9 years (27), but has eluded analysis for at least two reasons. First, we found that plasmid DNAs encoding the PLP2-MP1 region are unstable and may undergo deletion if the transformed bacteria are maintained at 37°C. We encountered this instability regardless of the vector or bacterial strain that was used to propagate the plasmid. However, if PLP2-MP1-encoding plasmids are maintained in transformed bacteria grown at room temperature, the plasmid DNA was retained and amplified intact. Once we established conditions to maintain and propagate plasmids encoding the PLP2-MP1 domain, we could isolate sufficient quantities of plasmid DNA for transfection experiments and monitor the enzymatic activity of the expressed protein products (see Materials and Methods). The second reason that PLP2 activity has been difficult to assess is because the putative substrate cleavage site was only recently identified (37). In previous studies using expression constructs or a recombinant fusion protein of maltose binding protein (MBP) and PLP2, the investigators were unable to demonstrate PLP2 enzymatic activity (5, 44). However, it should be noted that the PLP2 activity was tested on PLP1 cleavage sites. Therefore, it will be interesting to reexamine the activities of these PLP2 expression constructs and the MBP-PLP2 fusion protein when they are presented with the appropriate substrate.
One of the most important results from our studies is the demonstration that PLP1 and PLP2 proteinases recognize and process cleavage sites with different levels of efficiency. Expression studies using constructs encoding the PLP1 proteinase domain demonstrated that PLP1 efficiently processes the amino-terminal product of the replicase polyprotein to release p28 (Fig. 5A) (2, 3). In addition, PLP1 has also been shown to act in trans to process the p65 cleavage site (6). Site-directed mutagenesis studies of the determinants of the p28 cleavage sites revealed that the P5 (arginine or lysine), P2 (arginine), and P1 (glycine or alanine) positions are critical for recognition and processing of p28 by PLP1 (13, 24). The p65 cleavage site showed similar dependence on glycine or alanine in the P1 position and arginine or lysine in the P5 position. Furthermore, the p28 and p65 cleave sites could be interchanged and processed by PLP1 (6). In contrast, PLP1 recognizes the MP1 cleavage site very inefficiently in cis (Fig. 4B, lane 6) and does not process this site in trans (Fig. 5B, lanes 6, 7, 10, and 11). Do these differences in processing efficiency reflect distinct substrate specificity (i.e., that PLP2 will recognize different cleavage site determinants from those of PLP1) or show that the overall conformation of the polyprotein and the presentation of the cleavage site to the proteinase are the most critical factor for cleavage site recognition? Our future studies will be directed at identifying the critical determinants of the PLP2 enzymatic activity (such as the catalytic histidine residue and putative zinc-binding domain) and the cleavage site recognized by this proteinase. Comparison of the catalytic region and the determinants of the cleavage sites should help us understand the factors that control the specificity of the PLP1 and PLP2 enzymes.
Recent advances in the analysis of the viral cysteine proteinases will help direct these studies. A defined crystal structure has been solved for the leader cysteine proteinase of foot and mouth disease virus (FMDV Lpro) (20). This study revealed that FMDV Lpro adopts a compact version of the characteristic papain fold, with catalytic cysteine and histidine residues opposite each other in the groove where the polypeptide resides. Furthermore, a computer-generated structural analysis of the PLP1 domain human coronavirus 229E predicts similar spacing of catalytic cysteine and histidine residues, as seen in the crystal structure of papain and FMDV (22). This study goes on to propose and provide some experimental evidence that the coronavirus cysteine proteinases (PLP1 and PLP2) depend upon a zinc-binding finger to connect and stabilize the two domains that make up the papain-like fold of the enzyme. Such zinc-binding domains have been shown to be essential for the viral cysteine proteinases of human rhinovirus (40, 41) and hepatitis C virus (30, 42). It will be interesting to determine if MHV PLP1 and PLP2 bind zinc and if the predicted zinc-binding domain is required for enzymatic function.
A working model of how the MHV replicase polyprotein is processed is presented in Fig. 7. The model extends and modifies previous models for processing MHV ORF1a (32, 37). As shown in this study, the anti-D11 serum detects the p150 intermediate and the p44 processed product of MHV ORF1a. PLP2 likely acts on the nascent polyprotein and rapidly generates p150. This working model shows that three distinct proteinases are required for processing the MHV replicase. Processing of the replicase of another member of the order Nidovirales, Equine arteritis virus (EAV), also requires three distinct proteinase activities. The EAV replicase polyprotein 1ab is processed into 12 products, and the ORF1a products are involved in the membrane association of the complex (45). Indeed, the second cysteine proteinase of EAV is responsible for generating the hydrophobic nsp3 replicase product, which is analogous to the MHV p44 product. EAV nsp3 is thought to be an important scaffolding component of the replicase complex (45). The avian coronavirus infectious bronchitis virus (IBV) also releases a hydrophobic ORF1a replicase product, termed p41 (29). Interestingly, IBV encodes only one PLP activity that is responsible for processing glycine/glycine cleavage sites both upstream and downstream of the proteinase domain (29). Other coronaviruses, such as human coronavirus 229E, have been shown to encode PLP1 and PLP2 domains, but to date, only PLP1 has been shown to encode enzymatic activity (21).
FIG. 7.
Schematic diagram of the proposed proteolytic processing scheme for MHV ORF1a. Domains of the MHV PLP1 and PLP2 and the 3CLpro and their respective cleavage sites are indicated. Synthetic peptide (p28) and fusion protein domains used to generate antisera are depicted above the amino acid (aa's) scale.
The processing of the MHV ORF1a polyprotein is complex and generates at least 12 distinct products. Why is the MHV replicase polyprotein (and indeed all nidovirus replicase polyproteins) so large and processed into so many distinct intermediates and products? Why are three distinct proteinases required for processing the MHV and EAV replicase polyproteins? The answers to these questions are currently unclear, but likely revolve around the requirements for accurate replication of a large (12 to 31 kb) genomic RNA, for mediating discontinuous transcription of a nested set of mRNAs, and for high-frequency RNA recombination, all features of nidovirus RNA synthesis (26). There may in fact be distinct replicase conformations or complexes that mediate specific activities, such as positive- and negative-strand RNA synthesis.
It has been elegantly demonstrated that the replicase of Sindbis virus (SV) requires only a single proteinase, nsp2, to sequentially process the SV replicase polyprotein into intermediates that function in negative-strand RNA synthesis. nsp2 then acts in trans to process the intermediates into four products that function in positive-strand RNA synthesis (28, 39). We hypothesize that the MHV replicase requires three distinct proteinase activities to generate the intermediates and products that function in the different stages of MHV RNA synthesis. Replicase intermediates such as p150, which contains two predicted membrane-spanning domains, may be critical for localizing the replicase complex to membranous sites in the cell. The size and complexity of the replicase may also be necessary to mediate functions that are uniquely required for the discontinuous transcription of mRNAs. In addition, some of the replicase products may function as a putative mismatch repair system that may be responsible for maintaining the genetic integrity of the 31-kb RNA genome (1). A more detailed understanding of the replicase intermediates and products is important so that individual replicase products (and uncleaved intermediates) can be expressed and assessed for their ability to localize within the cell, interact with host cell proteins, and complement RNA temperature-sensitive mutants. Such studies will help elucidate the mechanisms of MHV transcription and replication.
ACKNOWLEDGMENTS
We thank David Axtell for assistance with the production of GST fusion protein and John Zaryczny for assistance with rabbit injection and serum collection. We thank Nopporn Sittisombut, Chiang Mai University, Chiang Mai, Thailand, for helpful advice on cloning techniques and Anne Rowley, Northwestern University, Chicago, Ill., for helpful advice and comments on the manuscript.
This work was supported by Public Health Service research grant AI 32065 to S.C.B.
REFERENCES
- 1.Baker S C, Lai M M C. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990;9:4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S C, Shieh C-K, Soe L H, Chang M-F, Vannier D M, Lai M M C. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S C, Yokomori K, Dong S, Carlisle R, Gorbalenya A E, Koonin E V, Lai M M C. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonilla P J, Hughes S A, Pinon J D, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley I, Boursnell M E G, Binns M M, Bilimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denison M R, Hughes S A, Weiss S R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207:316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison M R, Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison M R, Perlman S. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157:565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison M R, Spaan W J M, van der Meer Y, Gibson C A, Sims A C, Prentice E, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Baker S C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher T M. Murine coronavirus membrane fusion is blocked by modification of thiols buried within the spike protein. J Virol. 1996;70:4683–4690. doi: 10.1128/jvi.70.7.4683-4690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H Q, Schiller J J, Baker S C. Identification of the polymerase polyprotein products p72 and p65 of the murine coronavirus MHV-JHM. Virus Res. 1996;145:101–109. doi: 10.1016/S0168-1702(96)01368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson Bost A, Carnahan R H, Lu X T, Denison M R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J Virol. 2000;74:3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya A E, Koonin E V, Lai M M C. Putative papain-related thiol proteases of positive-strand RNA viruses: identification of rubi- and aphthovirus proteinases and delineation of a novel conserved domain associated with proteinases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya A E, Snijder E J. Viral cysteine proteinases. Perspect Drug Discov Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarne A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herold J, Gorbalenya A E, Theil V, Schelle B, Siddell S G. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold J, Siddell S G, Gorbalenya A E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J Biol Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano N, Murakami T, Fujiwara K, Matsumoto M. Utility of mouse cell line DBT for propagation and assay of mouse hepatitis virus. Jpn J Exp Med. 1978;48:71–75. [PubMed] [Google Scholar]
- 24.Hughes S A, Bonilla P J, Weiss S R. Identification of the murine coronavirus p28 cleavage site. J Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J C, Spence R A, Currier P F, Lu X, Denison M R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, Monica N L, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemm J A, Rumenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim K P, Ng L F P, Liu D X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J Virol. 2000;74:1674–1685. doi: 10.1128/jvi.74.4.1674-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 31.Lu X, Lu Y, Denison M R. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology. 1996;222:375–382. doi: 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X T, Sims A C, Denison M R. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J Virol. 1998;72:2265–2271. doi: 10.1128/jvi.72.3.2265-2271.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Denison M R. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makino S, Taguchi F, Hirano N, Fujiwara K. Analysis of genomic and intracellular viral RNA and small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984;139:9–17. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saporito-Irwin S M, Todd Geist R, Gutmann D H. Ammonium acetate protocol for the preparation of plasmid DNA suitable for mammalian cell transfections. BioTechniques. 1997;23:424–427. doi: 10.2144/97233bm16. [DOI] [PubMed] [Google Scholar]
- 37.Schiller J J, Kanjanabaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi S T, Schiller J J, Kanjanahaluethai A, Baker S C, Oh J-W, Lai M M C. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommergruber W, Casari G, Fessl F, Seipelt J, Skern T. The 2A proteinase of human rhinovirus is a zinc containing enzyme. Virology. 1994;204:815–818. doi: 10.1006/viro.1994.1599. [DOI] [PubMed] [Google Scholar]
- 41.Sommergruber W, Seipelt J, Fessl F, Skern T, Liebig H-D, Casari G. Mutational analyses support a model for the HRV2 2A proteinase. Virology. 1997;234:203–214. doi: 10.1006/viro.1997.8595. [DOI] [PubMed] [Google Scholar]
- 42.Stempniak M, Hostomska Z, Nodes B R, Hostomsky Z. The NS3 proteinase domain of hepatitis C virus is a zinc-containing enzyme. J Virol. 1997;71:2881–2886. doi: 10.1128/jvi.71.4.2881-2886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturman L S, Takemoto K K. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect Immun. 1972;6:501–507. doi: 10.1128/iai.6.4.501-507.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng H, Piñón J D, Weiss S R. Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J Virol. 1999;73:2658–2666. doi: 10.1128/jvi.73.4.2658-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Meer Y, van Tol H, Locker J K, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]