Abstract
In an attempt to generate broadly cross-reactive, neutralizing monoclonal antibodies (MAbs) to simian immunodeficiency virus (SIV), we compared two immunization protocols using different preparations of oligomeric SIV envelope (Env) glycoproteins. In the first protocol, mice were immunized with soluble gp140 (sgp140) from CP-MAC, a laboratory-adapted variant of SIVmacBK28. Hybridomas were screened by enzyme-linked immunosorbent assay, and a panel of 65 MAbs that recognized epitopes throughout the Env protein was generated. In general, these MAbs detected Env by Western blotting, were at least weakly positive in fluorescence-activated cell sorting (FACS) analysis of Env-expressing cells, and preferentially recognized monomeric Env protein. A subset of these antibodies directed toward the V1/V2 loop, the V3 loop, or nonlinear epitopes were capable of neutralizing CP-MAC, a closely related isolate (SIVmac1A11), and/or two more divergent strains (SIVsmΔB670 CL3 and SIVsm543-3E). In the second protocol, mice were immunized with unfixed CP-MAC-infected cells and MAbs were screened for the ability to inhibit cell-cell fusion. In contrast to MAbs generated against sgp140, the seven MAbs produced using this protocol did not react with Env by Western blotting and were strongly positive by FACS analysis, and several reacted preferentially with oligomeric Env. All seven MAbs potently neutralized SIVmac1A11, and several neutralized SIVsmΔB670 CL3 and/or SIVsm543-3E. MAbs that inhibited gp120 binding to CD4, CCR5, or both were identified in both groups. MAbs to the V3 loop and one MAb reactive with the V1/V2 loop interfered with CCR5 binding, indicating that these regions of Env play similar roles for SIV and human immunodeficiency virus. Remarkably, several of the MAbs generated against infected cells blocked CCR5 binding in a V3-independent manner, suggesting that they may recognize a region analogous to the conserved coreceptor binding site in gp120. Finally, all neutralizing MAbs blocked infection through the alternate coreceptor STRL33 much more efficiently than infection through CCR5, a finding that has important implications for SIV neutralization assays using CCR5-negative human T-cell lines.
Human and simian immunodeficiency viruses (HIV and SIV) are closely related retroviruses that produce AIDS in humans and related immunodeficiency syndromes in some species of macaques, respectively. SIV infection of rhesus macaques has become an important animal model for HIV infection and AIDS in humans and for the development of an effective HIV vaccine (20). Several reports have shown that the humoral immune response can, under some circumstances, protect nonhuman primates from infection by HIV, SIV, or SHIVs (SIVs that are engineered to contain an HIV type 1 [HIV-1] Env protein) (28, 41, 57, 72, 79). In addition, infections by SIVs with partially deglycosylated Envs have generated neutralizing antibodies that can efficiently neutralize wild-type virus in vitro (73), while immunization of mice with cells expressing fusion-competent HIV-1 Env elicited humoral responses that could neutralize numerous primary virus isolates in vitro (52). Finally, recent findings have shown that the passive administration of neutralizing monoclonal antibodies (MAbs) could prevent mucosal and in utero transmission of pathogenic SHIVs (3, 58). Collectively, these findings raise hope that an appropriately designed Env-based immunogen will generate a protective humoral response to HIV.
A key feature of any effective vaccine against HIV will be the ability to protect against infection with multiple, divergent isolates. Unfortunately, the humoral response elicited by monomeric gp120 is not broadly cross-neutralizing, making it unlikely that vaccination with this form of Env will prevent infection by the heterogeneous viruses circulating in the general population (10, 12). HIV and SIV Env glycoprotein is expressed on the surface of the virus as a noncovalently linked oligomer, and immunization with oligomeric Env preparations has been shown to generate antibodies that preferentially recognize oligomeric Env (8, 24). A correlation between antibody reactivity with oligomeric Env and neutralization ability has been noted in several reports (30, 64, 69, 76). With these studies in mind, we immunized mice with cell-associated or soluble forms of oligomeric SIV Env in an attempt to elicit broadly cross-reactive, neutralizing antibodies. A secondary goal was to create a large panel of well-characterized MAbs directed toward diverse epitopes throughout SIV Env; while many antibodies to HIV have been described and their binding sites have been determined, much less is known about the antigenic structure of SIV Env. As will be described, a number of MAbs reactive with the V3 or V1/V2 loops or less well-defined conformational determinants on gp120 derived from both protocols were capable of neutralizing related and more divergent isolates. Several of these MAbs have been shown to interfere with Env binding to CD4 and/or the CCR5 coreceptor. A large number of nonneutralizing MAbs with epitopes distributed throughout the Env protein have also been generated. This panel of well-characterized MAbs should be highly useful for future structure-function and immunologic studies of SIV Env.
MATERIALS AND METHODS
Preparation of purified SIV Env.
The CP-MAC env gene (49) was cloned into pSC65, and a premature stop codon was introduced just N terminal to the membrane-spanning (transmembrane [TM]) domain using PCR-based mutagenesis. A recombinant vaccinia virus (vAE1) was made using this plasmid and the Western Reserve vaccinia virus strain, using standard techniques (23). To generate recombinant protein, BHK cells were infected with vAE1 at a multiplicity of infection of 10. Four hours post infection, cells were washed twice with phosphate-buffered saline (PBS), and serum-free Dulbecco modified Eagle medium (DMEM; Gibco-BRL) was added. Cell supernatant was harvested 24 h postinfection, cleared by centrifugation, and filtered through a 0.45-μm-pore-size filter. Triton X-100 was added to a final concentration of 0.05% to inactivate vaccinia virus and sodium azide was added to 0.05% to prevent microbial growth. Env protein was purified using wheat germ agglutinin coupled to agarose beads (Vector Laboratories) for lectin affinity chromatography. Env was eluted in 1 M N-acetyl-d-glucosamine (Sigma) in morpholineethanesulfonic acid (MES) buffer (20 mM MES [pH 7.0], 0.13 M NaCl, 10 mM CaCl2), the sugar was removed by buffer exchange with an Amicon stir cell fitted with a YM30 membrane (Amicon), and the remaining volume was concentrated to produce a purified protein stock of 0.5 to 1 mg of soluble gp40 (sgp140) per ml. SIVmacCP-MAC gp120 was produced by infecting 293T cells with the recombinant vaccinia virus vTF1.1 (which produces T7 polymerase [1]) at a multiplicity of infection of 10 for 1 h at 37°C and then transiently transfecting with a plasmid encoding CP-MAC gp120 under the control of the T7 promoter (pCR3.1; Invitrogen). This gp120 construct was generated using a Quickchange mutagenesis kit (Stratagene) to introduce a premature stop codon just N terminal to the gp120/gp41 cleavage site. The SIVmac17E-Fr gp120 plasmid has been described elsewhere (25).
MAb production.
For immunization protocols using soluble oligomeric Env, BALB/c mice were immunized intradermally with 50 μg of CP-MAC sgp140 three times at 4-week intervals, with a final boost 4 days before hybridoma fusion protocols were performed. The first immunization was done in Freund's complete adjuvant; incomplete Freund's adjuvant was used for subsequent immunizations. Splenocytes from a one animal were harvested and cells were fused with SP2 myeloma cells as previously described (91). Of 200 uncloned hybridomas that were initially reactive with sgp140 by enzyme-linked immunosorbent assay (ELISA), 65 were derived from a single cell based on cloning efficiency and were characterized further. For immunization protocols that utilized cell-associated Env, mice received four intraperitoneal inoculations at 1-week intervals of 106 living SupT1 cells (in PBS) chronically infected with CP-MAC. Four days following the final inoculation, fusions were performed. Hybridomas were screened in 96-well plates for the ability to inhibit syncytium formation of a 1:10 mix of CP-MAC-infected to uninfected SupT1 cells at 24 h. Eight stable hybridoma cell lines were obtained from two fusions. Of these, one, termed 12G5, was shown to react with CXCR4 and has been described elsewhere (29); the other seven were specific for SIV Env.
Velocity gradient centrifugation.
Supernatants containing sgp140 or gp120 were concentrated 10-fold in a Centricon spin column (MWCO 50) and loaded onto a 5 to 20% (wt/vol) continuous sucrose gradient. Samples were centrifuged in an SW40 rotor at 40,000 rpm for 20 h at 4°C. Fractions were collected from the bottom, and aliquots were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with the anti-SIV gp120 antibody DA6. This MAb was generated in our lab to monomeric gp120 from HIV-2/ST (provided by Ray Sweet, Smith Kline Beecham, King of Prussia, Pa.) and has been shown to cross-react with many heterologous HIV-2 and SIV gp120s. The DA6 epitope has been mapped to amino acids 76 to 99 of the SIVmac251 gp120 (J. A. Hoxie, unpublished data).
Immunoblotting and immunoprecipitation.
The ability of each MAb to detect denatured, unpurified CP-MAC sgp140 was evaluated in Western blots of gels after SDS-PAGE run under reducing conditions. Blots were blocked in BLOTTO (PBS with 50 g of nonfat dry milk and 0.1% Tween 20), cut into strips, and incubated with MAbs diluted in BLOTTO to 1 μg/ml. Bound antibody was detected with goat anti-mouse conjugated to horseradish peroxidase (GAM-HRP; Promega) and enhanced chemiluminescence (Amersham). Immunoprecipitations were performed using 50 μl of supernatant from cells infected with vAE1 (harvested and used within 24 h), 50 μl of hybridoma supernatant, 25 μl of protein A-agarose beads (diluted to a binding capacity of 4 mg/ml; Gibco-BRL), and 350 μl of PBS. After 1 h of binding at room temperature (RT), the beads were washed twice with cold PBS and 50 μl of 1× sample buffer with β-mercaptoethanol was added. Samples were analyzed by SDS-PAGE and Western blotted with DA6.
Flow cytometry.
293T cells were transiently transfected with plasmids encoding full-length CP-MAC env and the cDNA version of rev (kindly supplied by Mike Malim, University of Pennsylvania) by calcium phosphate precipitation; medium containing 7.5 mM sodium butyrate was substituted 6 to 8 h posttransfection. The next day, the cells were lifted with PBS, transferred to V-bottom 96-well plates, and washed once with FACS (fluorescence-activated cell sorting) staining buffer (PBS plus 2% fetal calf serum and 0.05% sodium azide). Cells were incubated with 100 μl of hybridoma supernatant for 30 min at 4°C. Cells were then washed twice with FACS staining buffer and incubated with horse anti-mouse phycoerythrin-conjugated secondary (Vector Laboratories) at 1 μg/ml for 30 min at 4°C in the dark. Cells were washed twice with FACS staining buffer, transferred to FACS tubes, and analyzed with a Becton Dickinson FACScanner immediately following staining. Purified anti-CP-MAC MAb 17A11 and the anti-HIV-1 IIIB MAb D47 were used as positive and negative controls, respectively, in each experiment, and mean channel fluorescence (MCF) values were normalized to that of 17A11 for comparisons between experiments.
ELISAs.
SIV Env ELISAs were performed in 96-well plates to which concanavalin A (ConA; Vector Laboratories) had been adsorbed in capture buffer (20 mM Tris, 100 mM NaCl, 0.05% NaN3 [pH 8.5]) by incubation for 1 h at RT. Plates were washed with PBS containing 0.05% Tween 20 and incubated with sgp140- or gp120-containing supernatant diluted in PBS for 2 h at RT or overnight at 4°C. Plates were washed and blocked with BLOTTO for 1 to 2 h at RT. Primary antibodies were added as indicated for each experiment, and plates were incubated for 1.5 to 2 h at RT. Plates were washed, and GAM-HRP secondary antibody (Promega) was added at a dilution of 1:2,500 in BLOTTO without azide. Following a final wash, 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS; Pierce) dissolved in substrate buffer (0.1 M sodium acetate, 0.1% Tween 20 [pH 4.2]) was added, and the optical density at 405 nm (OD405) was measured. Competition ELISAs were carried out in the above manner on plates with ConA-bound CP-MAC gp120 except that following incubation with competing antibody (100 μl of hybridoma supernatant), the indicated biotinylated antibody was added without washing and the plate was incubated for an additional 1.5 h at RT. In competition ELISAs, streptavidin conjugated to HRP (Sigma) was used for detection of the biotinylated antibody; binding of the competing antibodies to gp120 was verified on a separate plate via detection with GAM-HRP.
Antipeptide ELISAs were performed by adsorbing the indicated peptides (obtained from the EVA Project or generously provided by Hermann Staats at Duke Medical Center) diluted to 1 μg/ml in capture buffer for 1 h at RT or overnight at 4°C. Plates were then blocked with BLOTTO for 1 to 2 h at RT and incubated with hybridoma supernatants diluted in BLOTTO to approximately 10 μg/ml for 1.5 h at RT. Plates were washed, and bound MAb detected with GAM-HRP as described above.
ELISAs to detect antibodies that blocked soluble CD4 (sCD4) binding were done using CP-MAC gp120 captured to 96-well plates coated with the 31C7 antibody recognizing the N terminus of SIV gp120. Plates were next blocked with BLOTTO and incubated with MAb at 10 μg/ml for 1.5 h; then sCD4 (100 μl of supernatant from cells infected with vCB5; kindly provided by Chris Broder) was added without washing, and incubation continued for another 1.5 h. Bound sCD4 was detected using rabbit serum R1168 raised against sCD4 at a 1:500 dilution for 30 min followed by a goat anti-rabbit detection antibody (Boehringer Mannheim) at 1:500 for 15 min. ABTS substrate was added, and OD405 values were recorded.
ELISAs for the quantification of mouse immunoglobulin G (IgG) were performed by adsorbing anti-mouse Ig antibody (Boehringer Mannheim) in capture buffer (1.2 μg/ml) to 96-well plates for 1 h at RT or overnight at 4°C. Plates were blocked with BLOTTO for at least 15 min, and serial dilutions of a mouse IgG (Sigma) standard curve and unknowns were incubated for 1.5 h at RT. Bound IgG was detected with GAM-HRP, and the linear portion of the standard curve was used to calculate the mouse IgG concentration in the samples.
Reporter virus neutralization assays.
Luciferase reporter viruses were produced by cotransfecting 293T cells with plasmids encoding SIV Env proteins under the control of the cytomegalovirus promoter and with the NL-Luc-R−-E− backbone plasmid (18, 19); fresh medium containing 7.5 mM sodium butyrate was added 4 to 6 h posttransfection. Supernatant containing pseudotyped virus was harvested 2 days posttransfection, preincubated for 1 h at 37°C with anti-Env MAbs diluted in DMEM-10 (DMEM with 10% fetal calf serum [HyClone] and 1% penicillin-streptomycin [Gibco-BRL]), and used to infect 96-well plates of GHOST target cells stably transduced with CD4 only (GHOST parent), CD4 and human CCR5, or CD4 and human STRL33 (generously provided by Dan Littman, Skirball Institute; low-passage GHOST cells were used for these experiments). Infections were performed in DMEM-10 without supplements. Cells were lysed in 0.5% Triton X-100 in PBS 3 to 4 days postinfection, and luciferase in the cell lysate was quantified in a luminometer using luciferase reagent (Promega). Infections were done in duplicate; the duplicates were averaged for each antibody dilution, and these averages were used to generate neutralization curves from which 50 and 90% inhibitory concentrations (IC50 and IC90) were determined.
Direct Env binding assays.
A 500-μl aliquot of unpurified SIVmac17E-Fr gp120 (0.5 to 1 μg) was preincubated with 10 μg of MAb for 30 min at RT and then incubated with 293T cells stably expressing high levels of rhesus CCR5 or with parental 293T cells for 1 h at 37°C. The cells were washed once with cold, serum-free DMEM and lysed in 0.5% Triton X-100 in PBS with protease inhibitors (Complete; Boehringer Mannheim). Lysates were evaluated for bound Env by SDS-PAGE and Western blotting with DA6.
RESULTS
Production of MAbs to soluble or cell-associated Env oligomers.
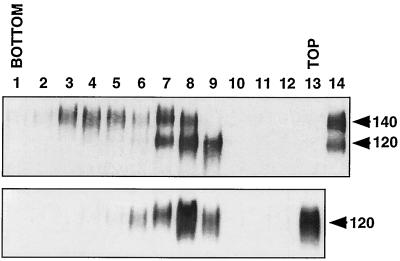
In general, antibody binding to oligomeric HIV-1 Env correlates with virus neutralization better than binding to monomeric gp120 (30, 64, 69, 76), and immunization with oligomeric rather than monomeric Env results in more efficient generation of antibodies to discontinuous epitopes (8, 24). With these results in mind, we used two different forms of oligomeric SIV Env protein as immunogens with the hope of generating broadly cross-neutralizing MAbs. The Env protein of CP-MAC, a lab-adapted variant of SIVmacBK28, was chosen both because of its highly stable gp120/gp41 association and because high levels of Env protein are expressed on the surface of infected cells (49, 50). In the first protocol, mice were immunized with purified, soluble CP-MAC gp140 generated by introducing a premature stop codon prior to the TM domain of gp41. The cleavage site between gp120 and gp41 was left intact with the expectation that the strong SU-TM association between CP-MAC gp120 and gp41 would stabilize Env oligomers (49). Purified sgp140 was subjected to sucrose velocity gradient centrifugation to confirm the oligomeric status of this construct. A separate gradient that contained only monomeric gp120 was run in parallel. As shown in Fig. 1, the sgp140 preparation included cleaved, monomeric gp120 as well as uncleaved gp140, which could be separated into both oligomeric and monomeric fractions. The predominance of uncleaved Env in this preparation likely resulted from saturation of the cellular protease responsible for Env cleavage due to the high levels of protein expression obtained in this vaccinia virus-driven system. Stable oligomers containing cleaved gp120 and gp41 either were not present in these preparations or were not stable to ultracentrifugation. Hybridomas produced from immunizations with this sgp140 preparation were screened by ELISA for reactivity with the immunogen, and a total of 65 stable, clonal hybridoma cell lines were produced and evaluated.
FIG. 1.
Oligomeric nature of CP-MAC sgp140. Unpurified CP-MAC sgp140 (top) and gp120 (bottom) were concentrated 10-fold by low-speed centrifugation, loaded onto a 5 to 20% continuous sucrose gradient, and centrifuged at 40,000 rpm in an SW40 rotor at 4°C for 20 h. The gradient was separated into approximately 1-ml fractions, and an aliquot of each was subjected to SDS-PAGE under reducing conditions and Western blotting with an anti-gp120 MAb (DA6). Samples of the input sgp140 and gp120 proteins were run in the far right lanes as controls.
Given that high levels of uncleaved SIV Env oligomers were present in the sgp140 immunogen and because Env cleavage is important for coreceptor binding (25), we also immunized mice with unfixed SupT1 cells that were chronically infected with CP-MAC. As previously shown, these cells express high levels of Env protein on the cell surface due to a Tyr-to-Cys mutation in the gp41 cytoplasmic domain that eliminates an Env endocytosis signal (50, 77). We screened these hybridomas for the ability to inhibit CP-MAC-induced syncytium formation of SupT1 cells and obtained a total of 7 hybridomas that potently inhibited syncytium formation. All seven MAbs reacted strongly with CP-MAC-infected, but not uninfected, SupT1 cells by FACS analysis (data not shown).
Identification of MAbs recognizing linear epitopes.
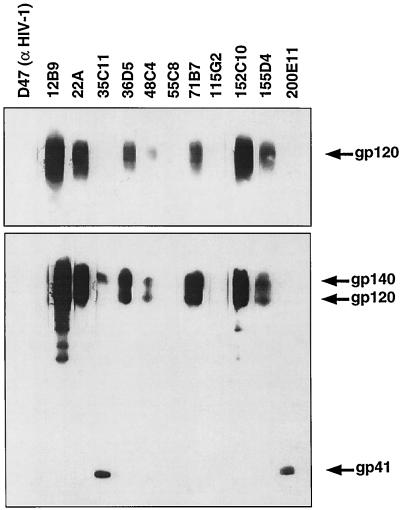
To determine subunit reactivity and to discriminate between antibodies recognizing linear and discontinuous epitopes, all MAbs were screened for the ability to detect CP-MAC gp120 and sgp140 Env protein by Western blotting. Most of the MAbs raised against sgp140 were at least weakly reactive with Env by Western blotting. A subset of these MAbs are shown in Fig. 2. The majority recognized epitopes in gp120, but 12 of 65 detected the truncated gp41 subunit. Interestingly, anti-gp41 MAbs displayed two distinct phenotypes. While 35C11 detected both sgp140 and gp41, 200E11 recognized only the gp41 subunit by Western blotting (Fig. 2). MAb 200E11, but not 35C11, also detected gp41 homodimers under nonreducing conditions, indicating that its epitope was exposed on oligomeric gp41 (data not shown). A small number of MAbs raised against sgp140, including 55C8 and 115G2, were unable to recognize CP-MAC gp140 or gp120 by Western blotting (Fig. 2 and Table 1). As both of these MAbs reacted strongly with CP-MAC sgp140 by ELISA, this finding suggests that their epitopes are lost when Env is subjected to SDS-PAGE and Western blotting. Interestingly, the seven MAbs derived from mice immunized with infected cells and selected for the ability to inhibit syncytium formation did not detect Env by Western blot under reducing conditions. However, under nonreducing conditions, several reacted weakly with gp140 and gp120 (data not shown) and all immunoprecipitated gp120 at least weakly in the absence of gp41, indicating that their epitopes lie in the gp120 subunit. Thus, as a group, these MAbs were strongly dependent on Env conformation.
FIG. 2.
Ability of the MAbs raised against sgp140 to detect Env by Western blotting. Supernatant from vAE1-infected cells (bottom) or from cells expressing only monomeric gp120 (top) was subjected to SDS-PAGE under reducing conditions, blotted onto polyvinylidene difluoride membranes, blocked in BLOTTO, and cut into strips which were incubated with the indicated MAbs at 1 μg/ml in BLOTTO. Bound antibodies were detected with GAM-HRP and enhanced chemiluminescence. An exposure is presented that allows Env detection with the less effective antibodies. The anti-HIV-1 V3 loop antibody D47 is included as a negative control.
TABLE 1.
Summary of MAb propertiesa
| Antibody | Epitope | No. of clones | Reactivity
|
||
|---|---|---|---|---|---|
| Western | IP | FACS | |||
| Soluble gp140 | |||||
| 47G6 | 21-40 | 3 | ++ | + | + |
| 136B10 | 31-50 | 1 | ++ | + | ++ |
| 179F10 | 41-50 | 2 | ++ | + | + |
| 115G2 | 41-60 | 3 | − | + | + |
| 12B9 | 76-99 | 6 | ++ | + | + |
| 152C10 | 81-99 | 3 | ++ | +/− | − |
| 163B9 | 81-100 | 1 | ++ | − | − |
| 22A | 111-130 (V1) | 3 | ++ | + | ++ |
| 55C8 | 161-180 (V1/V2) | 4 | − | +/− | + |
| 171C2 | 171-190 (V1/V2) | 2 | ++ | + | ++ |
| 71B7 | 271-290 | 1 | + | + | − |
| 48C4 | 281-290 | 2 | + | + | + |
| 36D5 | 321-340 (V3) | 2 | ++ | + | +++ |
| 28E6 | 361-380 | 1 | + | + | + |
| 31C7 | 501-510 | 3 | ++ | + | + |
| 35C11 | 601-620 (TM) | 11 | + | + | ++/+ |
| 200E11 | 611-630 (TM) | 1 | + | + | − |
| 6B8 | Group 1 (N terminal) | 6 | + | − | +/− |
| 8H1 | Group 2 (V1/V2) | 1 | ++ | + | ++ |
| 189D5 | Group 3 (N terminal | 1 | ++ | + | + |
| 77D6 | Group 4 (76-99) | 6 | ++ | + | + |
| 123A2 | Group 5 | 2 | + | + | − |
| Infected cells | |||||
| 3E9 | Conformational | NA | − | ++ | ++ |
| 4B11 | CD4/CCR5 BS | NA | − | ++ | ++ |
| 5B11 | CD4 BS | NA | − | ++ | ++ |
| 7D3 | CCR5 BS | NA | − | ++ | ++ |
| 8C7 | CD4/CCR5 BS | NA | − | ++ | ++ |
| 11F2 | CCR5 BS | NA | − | ++ | ++ |
| 17A11 | CD4/CCR5 BS | NA | − | ++ | ++ |
One antibody from each epitope group is listed. The number of clones mapping to the same epitope (clones) and the reactivities of these clones in Western blots, immunoprecipitations (IP), and FACS analysis are shown. Western and IP rankings are subjective relative to reference antibodies (DA6 and 8C7, respectively), FACS rankings are relative to the MCF obtained with the 17A11 antibody (10 to 25% [+], 26 to 50% [++], or 50 to 100% [+++] of the MCF obtained with 17A11 on that day, minimum of two assays per antibody). Data for neutralizing MAbs are in boldface. NA, not applicable; BS, binding site.
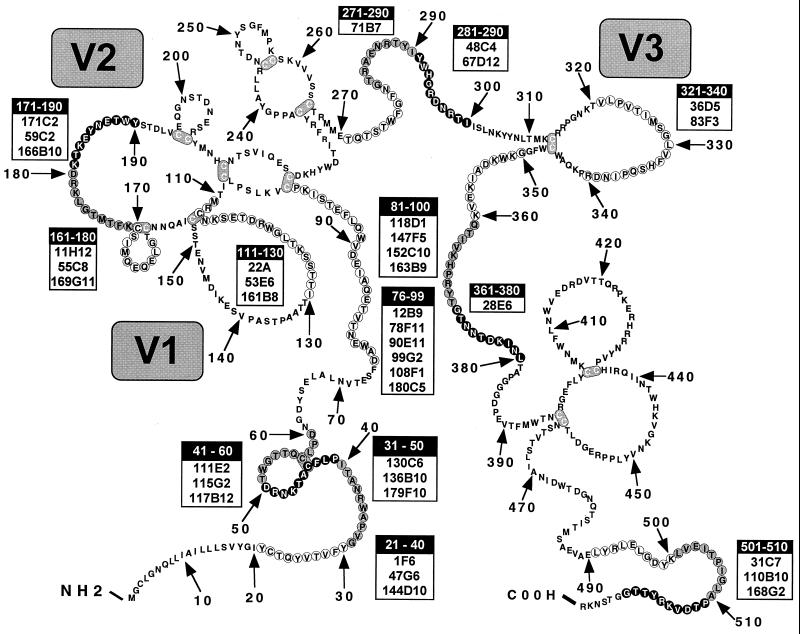
To further define the epitopes recognized by the panel of MAbs, each was tested by ELISA against a series of overlapping, 20-mer peptides covering the entire ectodomain of SIVmac32H, as well as several peptides based on SIVmac251 sequences. As CP-MAC was derived from a molecular clone of SIVmac251 (BK28) (49) and the 32H isolate is very closely related to SIVmac251 (75), we expected that antibodies to linear determinants in CP-MAC would recognize these peptides. As shown in Fig. 3, three immunodominant regions in gp120 were identified: (i) the N terminus (residues 21 to 70), (ii) residues 81 to 100, and (iii) the V1/V2 region (residues 111 to 190). Antibodies that recognized epitopes in four other regions were also identified (Fig. 3). Surprisingly, two MAbs that did not recognize Env in Western blots reacted with peptides: 55C8 reacted with a V1/V2 peptide (residues 161 to 180), and 115G2 recognized a peptide from the gp120 N terminus (residues 41 to 60). MAbs reactive with the gp41 ectodomain by Western blotting were screened against overlapping peptides from this subunit. Eleven of 12 anti-gp41 MAbs recognized the previously identified immunodominant region (residues 601 to 620) containing the highly conserved disulfide bond in gp41 (11, 33, 42, 45) (data not shown). The epitope of the one anti-gp41 MAb not reactive with this peptide was shifted slightly toward the C terminus (residues 611 to 630). 200E11 had a phenotype distinct from those of the other anti-gp41 MAbs in it that failed to react with Env-expressing cells by FACS (Table 1) and that it detected gp41 but not gp140 by Western blotting (see above).
FIG. 3.
Epitope mapping of MAbs using overlapping peptides in gp120. Antibodies produced from immunization with sgp140 were screened by ELISA for reactivity with overlapping 20-mers representing almost the complete gp120 ectodomain. Antibodies that reached a signal-to-noise ratio of 3 or greater on a peptide were scored as positive; most positive results represent signal/noise ratios of 10 or greater. Grey scale represents areas of overlap between adjacent peptides. The amino acid sequence of BK28, not of the peptides, is shown. Mutations between BK28 and CP-MAC are E84K, R112K, R120K, M327R, and T475I. Disulfide bonds (loops) are hypothetical and are based on the HIV-1 disulfide bonding pattern (39, 54).
Determination of surface exposure of antibody epitopes.
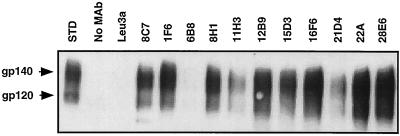
To determine which MAbs recognized nondenatured Env protein, immunoprecipitations were performed with fresh preparations of sgp140 (containing a mixture of gp140, gp120, and gp41), and Env was detected by Western blotting with the anti-gp120 MAb DA6. A representative blot showing several of these MAbs is presented in Fig. 4, and the results for all antibodies are summarized in Table 1. As expected, the majority of the antibodies immunoprecipitated CP-MAC Env. However, several antibodies immunoprecipitated freshly prepared sgp140 only weakly or not at all, suggesting that their epitopes are not exposed on the folded Env protein (Table 1). MAb 8C7 (Fig. 4) and the other six MAbs generated against chronically infected cells (not shown) immunoprecipitated CP-MAC gp140 and gp120.
FIG. 4.
Immunoprecipitation of sgp140. Supernatant collected from vAE1-infected cells was incubated with the indicated MAbs and protein A-beads, and the immunoprecipitates were analyzed by Western blotting. Input supernatant is shown as a standard (STD); negative controls are precipitations to which no antibody (No MAb) or the irrelevant anti-CD4 MAb (Leu3A) was added. An exposure is shown which allows the detection of Env precipitated by less efficient antibodies.
To determine reactivity with native Env oligomers, MAbs were evaluated by FACS analysis of 293T cells transiently expressing CP-MAC Env. The majority of the cell-associated Env in this case should be cleaved and oligomeric, although some gp120 shedding likely occurs despite the tight gp120-gp41 association in CP-MAC, and some uncleaved Env may be present. Most MAbs were at least weakly reactive with CP-MAC Env by FACS compared to a pcDNA3-transfected control (Table 1). Of note, several epitopes were highly exposed on the native Env protein, including residues 31 to 50, the V1/V2 loop, and particularly the V3 loop. These findings are consistent with those previously reported for HIV-1 (64, 76, 80, 88). With the exception of 200E11, MAbs to gp41 were also reactive by FACS, which may reflect binding to gp41 subunits from which gp120 has been shed. All seven of the MAbs raised to CP-MAC-infected cells were strongly positive by FACS, giving MCF values equivalent to or greater than those obtained with the V3 loop MAbs (data not shown).
Epitope mapping of MAbs directed against discontinuous epitopes.
A subset of the MAbs that did not react with SIV Env peptides (8H1, 51G3, 77D6, 143C11, 155B4, and 189D5) were biotinylated and used in competition ELISAs. Five competition groups were identified (Table 2). Group 1 included MAbs that competed with each other and 136B10, which binds to residues 31 to 50. Interestingly, N-terminal antibodies other than 136B10 did not compete with group 1 antibodies for Env binding (136B10 is the only MAb that recognized both the 21-40 and 31-50 peptides). Competition group 2 included only 8H1, which was blocked by MAbs reactive with residues in the V1/V2 region. Groups 3 and 4 defined additional competition groups within the gp120 N terminus. The binding of group 4 MAbs and those directed to residues 76 to 99 enhanced the binding of the group 3 MAb 189D5 (approximately 120 and 150% of control, respectively), while antibodies to the N terminus (residues 21 to 40) competed with MAb 189D5. MAbs from an overlapping region in the N terminus (residues 21 to 70) enhanced binding of group 4 MAbs. Two antibodies that did not fit into any of these groups were designated group 5. The seven MAbs raised against infected cells competed for Env binding with 189D5 (group 3, 40 to 60% of control) and 8H1 (group 2, 65 to 80% of control) but not 51G3 or 155B4 (group 1) or the anti-V3 loop MAbs 36D5 and 83F3.
TABLE 2.
Competition grouping of MAbs raised against sgp140 that do not map to peptidesa
| Group | MAb | Competing MAb | Enhancing MAb |
|---|---|---|---|
| 1 | 6B8 | ||
| 51G3 | |||
| 86F4 | 136B10 only (31-50) | NA | |
| 139A | |||
| 155B4 | |||
| 2 | 8H1 | 161-180 (V1/V2) | NA |
| 3 | 189D5 | 21-40 | Group 4, 76-99 |
| 4 | 77D6 | ||
| 107E5 | |||
| 113A | 76-99 | 21-70 | |
| 143C11 | |||
| 174D4 | |||
| 5 | 123A2 | ND | ND |
| 181H8 |
Supernatants from vAE1-infected cells were captured to 96-well plates with ConA; after blocking with BLOTTO, competing MAbs were added. Following incubation for 1.5 h, biotinylated antibody (which did not recognize any peptides) was added without washing. Bound biotinylated antibodies were detected with streptavidin conjugated to HRP. Antibodies were classified as competing (reduced biotinylated antibody binding by at least 50%) or enhancing (increased biotinylated antibody binding to 150% or greater) as indicated. NA, not applicable; ND, no data.
Oligomer sensitivity of MAbs.
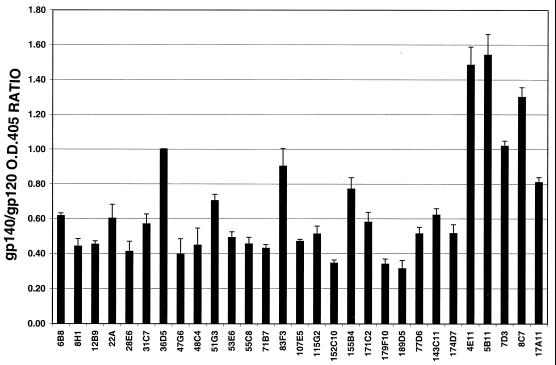
To assess whether any of the MAbs preferentially recognized oligomeric Env, ELISAs were performed in parallel 96-well plates coated with ConA-captured CP-MAC gp120 or sgp140 (which, as shown in Fig. 1, contains oligomeric, uncleaved gp140 and possibly gp120-gp41 oligomers prior to ultracentrifugation). The ratio of antibody reactivity to plates coated with gp120 and with the sgp140-containing preparation was calculated (Fig. 5). As the V3 loop is predicted to be a highly exposed epitope and thus less likely to be affected by oligomerization status, we set the average OD for V3 loop MAbs 36D5 and 83F3 against gp120 and sgp140 at a value of 1 and calculated the reactivity of the other MAbs relative to this value. In this manner, differences in the amount of gp120 or sgp140 captured to the plates were accounted for. All of the antibodies generated against sgp140 reacted more strongly with gp120 than with the oligomer-containing preparation, suggesting that these MAbs preferentially recognize monomeric Env. Strikingly, of the five MAbs tested that were raised to chronically infected cells, three (4E11, 5B11, and 8C7) were more reactive with the oligomer-containing Env than with the monomeric gp120 preparation. Two MAbs from this group, 3E9 and 11F2, did not react with gp120 by ELISA and could not be analyzed in this manner. However, the finding that these antibodies reacted with sgp140 by ELISA (not shown) indicates that they also reacted preferentially with oligomeric Env.
FIG. 5.
MAb oligomer sensitivity. ELISAs using the indicated antibodies (at least one antibody from each category shown in Table 1) were conducted side-by-side on plates coated with ConA-captured gp120 and oligomer-containing sgp140 (vAE1 supernatant). Background was subtracted, and the ratio of the OD405s on each plate was calculated; values are standardized to the signal with the V3 loop MAb 36D5. Error bars represent the standard error of the mean. Two MAbs, 3E9 and 11F2, were not tested in this assay due to their weak binding to monomeric gp120.
Neutralization profiles of the MAbs on target cells expressing CCR5 or alternate coreceptors.
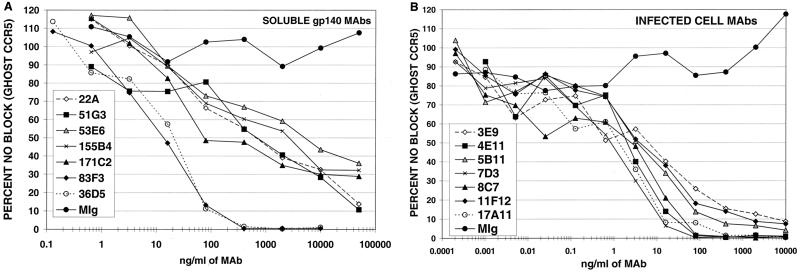
At least one MAb from each epitope group listed in Table 1 was tested for the ability to neutralize luciferase reporter viruses pseudotyped with CP-MAC Env used to infect GHOST target cells expressing human CD4 and CCR5 (Table 3 and Fig. 6). Among MAbs generated against sgp140, the most potent neutralizing MAbs recognized the V3 loop (36D5 and 83F3), with IC90s of 71 and 84 ng/ml, respectively (Fig. 6A and Table 3). In addition, many of the MAbs that mapped to the V1/V2 loops neutralized homologous virus under these conditions, although only one (22A) reached an IC90 of <50 μg/ml. In general, the IC50s of anti-V1/V2 antibodies were 2 to 3 logs greater than those of the anti-V3 loop MAbs. Weak neutralization activity was also seen with group 1 MAbs 6B8, 51G3, and 155B4 (Table 3 and Fig. 6A). As a group, these MAbs reacted weakly with Env by Western blotting, failed to or inefficiently immunoprecipitated gp120 and gp140, and were poorly reactive with Env-expressing cells in FACS assays. Although group 1 antibodies competed with a single N-terminal antibody, 136B10, this N-terminal antibody did not have an IC50 under 50 μg/ml. Taken together, these results suggest that group 1 antibodies recognize a conformational epitope that is not well exposed on the native Env oligomer. No neutralizing activity was observed with MAbs generated against sgp140 that reacted with other epitopes (Table 3).
TABLE 3.
Neutralization profiles of MAbs raised against sgp140a
| MAb | Epitope | CCR5 (ng/ml)
|
STRL33 (ng/ml)
|
||
|---|---|---|---|---|---|
| IC50 | IC90 | IC50 | IC90 | ||
| 47G6 | 21-40 | >50,000 | >50,000 | >50,000 | >50,000 |
| 179F10 | 41-50 | >50,000 | >50,000 | >50,000 | >50,000 |
| 115G2 | 41-60 | >50,000 | >50,000 | >50,000 | >50,000 |
| 12B9 | 76-99 | >50,000 | >50,000 | >50,000 | >50,000 |
| 152C10 | 81-100 | >50,000 | >50,000 | >50,000 | >50,000 |
| 22A | 111-130(V1) | 1,143 | 41,333 | 21 | 1,600 |
| 53E6 | 111-130(V1) | 12,600 | >50,000 | 125 | 24,000 |
| 55C8 | 161-180(V1/V2) | >50,000 | >50,000 | >50,000 | >50,000 |
| 171C2 | 171-190(V1/V2) | 467 | >50,000 | 32 | 4,000 |
| 71B7 | 271-290 | >50,000 | >50,000 | >50,000 | >50,000 |
| 48C4 | 281-290 | >50,000 | >50,000 | >50,000 | >50,000 |
| 36D5 | 321-340(V3) | 16 | 71 | 3 | 22 |
| 83F3 | 321-340(V3) | 16 | 84 | 4 | 25 |
| 28E6 | 361-380 | >50,000 | >50,000 | >50,000 | >50,000 |
| 31C7 | 501-510 | >50,000 | >50,000 | >50,000 | >50,000 |
| 35C11 | 601-620 (TM) | >50,000 | >50,000 | >50,000 | >50,000 |
| 200E11 | 611-630 (TM) | >50,000 | >50,000 | >50,000 | >50,000 |
| 6B8 | Group 1 | 7,000 | >50,000 | 220 | 3,000 |
| 51G3 | Group 1 | 850 | 50,000 | 95 | 3,375 |
| 155B4 | Group 1 | 822 | >50,000 | 95 | 13,000 |
| 8H1 | Group 2 | 50,000 | >50,000 | 3,667 | >50,000 |
| 189D5 | Group 3 | >50,000 | >50,000 | >50,000 | >50,000 |
| 77D6 | Group 4 | >50,000 | >50,000 | >50,000 | >50,000 |
| 107E5 | Group 4 | >50,000 | >50,000 | >50,000 | >50,000 |
| 143C11 | Group 4 | >50,000 | >50,000 | >50,000 | >50,000 |
At least one antibody from each epitope group was tested for the ability to neutralize luciferase reporter viruses pseudotyped with the CP-MAC Env protein when used to infect GHOST-CCR5 or GHOST-STRL33 target cells. Each antibody was evaluated in a minimum of three or as many as seven experiments; IC50s and IC90s were calculated for each experiment, and the average values are presented. Data for antibodies that were found to neutralize are in boldface.
FIG. 6.
Neutralization of SIVmacCP-MAC. Luciferase reporter viruses pseudotyped with the CP-MAC Env protein were preincubated for 1 h at 37°C with serial dilutions of the indicated MAbs and used to infect GHOST-CCR5 cells. Cells were lysed 3 days postinfection, and luciferase activity in the cell lysate was quantified. Mouse Ig (MIg) is included as a negative control. One representative experiment is shown for neutralizing antibodies raised against sgp140 (A) and antibodies raised against chronically infected cells (B). Each data point represents averaged duplicates.
MAbs produced by immunization with chronically infected cells were originally screened for the ability to block syncytium formation and were thus predicted to possess neutralizing activity against cell-free virus. As shown in Fig. 6B, all seven of these MAbs potently neutralized reporter viruses pseudotyped with the CP-MAC Env. The most potent neutralizing antibody was 7D3, with an IC90 of 6 ng/ml (Table 4). 4E11 and 8C7 were next in potency, displaying IC90s of 31 and 20 ng/ml, respectively.
TABLE 4.
Neutralization profiles of MAbs raised against chronically infected cellsa
| MAb | CCR5 (ng/ml)
|
STRL33 (ng/ml)
|
||
|---|---|---|---|---|
| IC50 | IC90 | IC50 | IC90 | |
| 3E9 | 12 | 600 | 2 | 27 |
| 4E11 | 2 | 31 | 6 | 60 |
| 5B11 | 3 | 130 | 2 | 23 |
| 7D3 | 1 | 6 | 0.3 | 3 |
| 8C7 | 2 | 20 | 1 | 7 |
| 11F2 | 3 | 500 | 2 | 28 |
| 17A11 | 4 | 50 | 1 | 8 |
The seven antibodies that inhibited syncytium formation were tested for neutralization as described in the footnote to Table 3, and IC50s and IC90s were calculated in the same manner.
Both sets of MAbs were also screened for the ability to inhibit infection of target cells expressing CD4 and STRL33 (Tables 3 and 4). As we have previously reported (26), although the most efficient coreceptor for CP-MAC is CCR5, STRL33 also supports infection by CP-MAC-pseudotyped reporter viruses. No additional neutralizing antibodies were identified using the STRL33 target cells. However, eight of nine neutralizing MAbs generated against sgp140 reached an IC90 below 50 μg/ml on GHOST STRL33 cells, while only three of these MAbs reached an IC90 below this level on GHOST CCR5 target cells. Furthermore, all neutralizing MAbs had lower IC50s when tested on STRL33-expressing cells; in some cases, IC50s were 1 to 2 logs lower. This increase in neutralization potency on STRL33-expressing target cells was less apparent with MAbs that potently neutralized infection through CCR5. The rank orders of neutralization ability were similar on both CCR5- and STRL33-expressing target cells, although some variation was observed among antibodies generated against chronically infected cells (Table 4). These results suggest that infection through alternate coreceptors, such as STRL33, is more readily neutralized than infection using the principal SIV coreceptor, CCR5 (21).
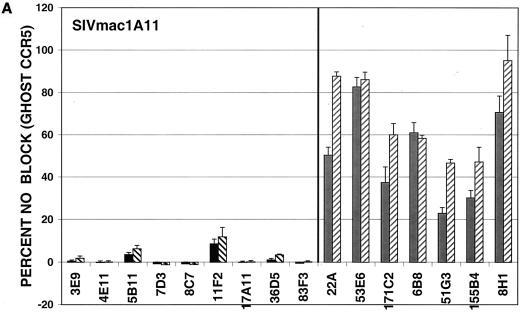
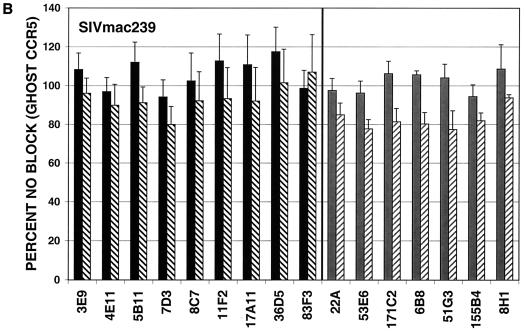
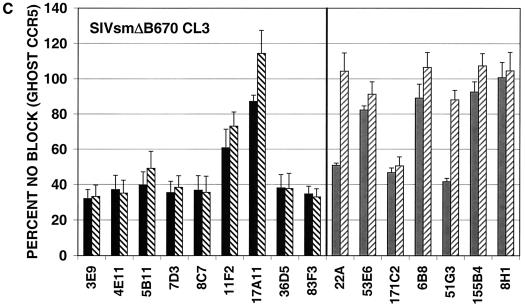
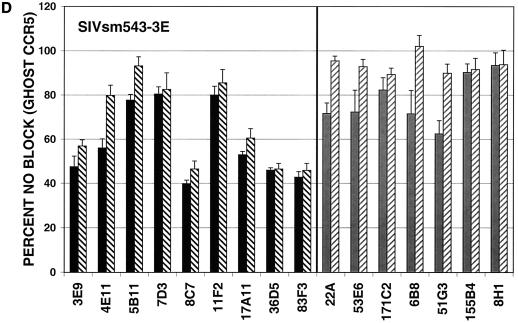
MAbs capable of neutralizing the CP-MAC reporter virus were next assayed for the ability to neutralize reporter viruses pseudotyped with more divergent Envs. GHOST cells expressing human CD4 and CCR5 were used for these experiments, and antibodies were tested at two concentrations, 50 or 10 μg/ml and 5 or 1 μg/ml, depending on their potency against viruses pseudotyped with the homologous CP-MAC Env. In general, MAbs raised against infected cells more efficiently neutralized divergent isolates than those raised against purified sgp140, with the exception of the two anti-V3 MAbs (36D5 and 83F3), which were similar to the infected cell MAbs in neutralizing potency (Fig. 7). The typically neutralization-sensitive isolate SIVmac1A11 (55), which is relatively closely related to CP-MAC (93.1% similarity to CP-MAC in the Env protein), was most sensitive to neutralization by these MAbs. SIVmac239, a related (94.2% similarity in Env) but classically neutralization-resistant isolate, was resistant to all MAbs tested (Fig. 7B). Infection by more divergent Envs, SIVsmΔB670 CL3 (2) (83.8% similarity in Env) and SIVsm543-3E (37) (81.5% similarity in Env), was inhibited by subsets of these MAbs (Fig. 7C and D). None of the MAbs reached an IC90 against SIVsmΔB670 CL3 or SIVsm543-3E at concentrations tested. In general, the relative potency of the MAbs against CP-MAC reporter viruses correlated with their ability to neutralize more divergent isolates.
FIG. 7.
Neutralization profiles of antibodies using heterologous SIV isolates. Neutralization assays were conducted as described for SIVmacCP-MAC but using viruses pseudotyped with SIV Env clones SIVmac1A11 (A), SIVmac239 (B), SIVsmΔB670 CL3 (C), and SIVsm543-3E (D). Antibodies were tested at two concentrations depending on their potency: black bars, 10 μg/ml; black hatched bars, 1 μg/ml; grey bars, 50 μg/ml; grey hatched bars, 5 μg/ml. Values represent means of averaged duplicates from three independent experiments; error bars represent standard error of the mean.
Effect of neutralizing MAbs on Env binding to CD4 and CCR5.
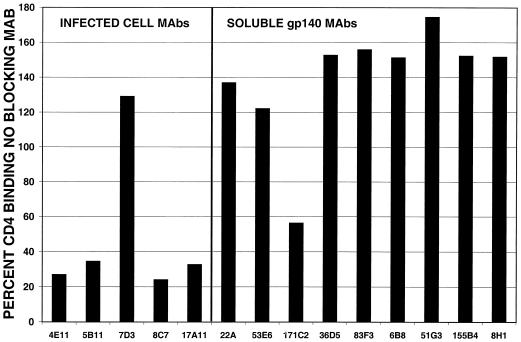
The ability of an antibody to prevent viral attachment to target cells is a major determinant of its neutralization potency. For HIV-1, most neutralizing antibodies either bind to the V3 loop and thereby disrupt coreceptor binding or interfere with CD4 binding (9, 17, 62, 63, 82–84, 89). Both of these neutralization mechanisms were evaluated for the subset of anti-SIV Env MAbs shown to neutralize homologous virus (Table 3). The ability of MAbs to inhibit gp120 binding to sCD4 was evaluated by ELISA using unpurified, monomeric CP-MAC gp120 (the sgp140 preparation was not used because it contained significant amounts of uncleaved protein which binds CD4 less efficiently [25]). Two neutralizing MAbs, 3E9 and 11F2, were not tested in this assay, as they did not bind CP-MAC gp120 in this format. A representative experiment is shown in Fig. 8. Several of the neutralizing MAbs blocked binding of sCD4 to gp120, including four of the MAbs generated against chronically infected cells. Interestingly, the most potent neutralizing antibody, 7D3, did not affect sCD4 binding. Only one of the MAbs produced by immunization with sgp140, 171C2, interfered with CD4 binding. This MAb maps to the V1/V2 loop and may be similar to a MAb to the SIVcpz V2 loop that has also been shown to block sCD4 binding (88).
FIG. 8.
Ability of neutralizing MAbs to interfere with sCD4 binding to gp120. CP-MAC gp120 was attached to 96-well plates using an antibody directed against the Env C terminus, and the plates were blocked by incubation in BLOTTO. The indicated antibodies were added followed 1.5 h later by sCD4 added without washing. sCD4 binding was detected using a rabbit serum generated against sCD4 and goat anti-rabbit-HRP conjugate. Results are expressed relative to OD values obtained when no anti-Env antibody was added (100%). A representative experiment is shown; identical results were obtained in a second assay.
To determine whether any of the neutralizing MAbs interfered with coreceptor binding, their ability to block CD4-independent gp120 binding to target cells expressing rhesus CCR5 was evaluated. SIVmac17E-Fr gp120 was used in this assay, as this Env binds rhesus CCR5 very efficiently in the absence of CD4 (25), thus allowing CCR5 binding to be measured without the confounding effects of Env-CD4 binding. All MAbs used in this assay recognized SIVmac17E-Fr gp120 by ELISA or immunoprecipitation, although as with the CP-MAC gp120, 3E9 and 11F2 were minimally reactive with this monomeric protein (data not shown).
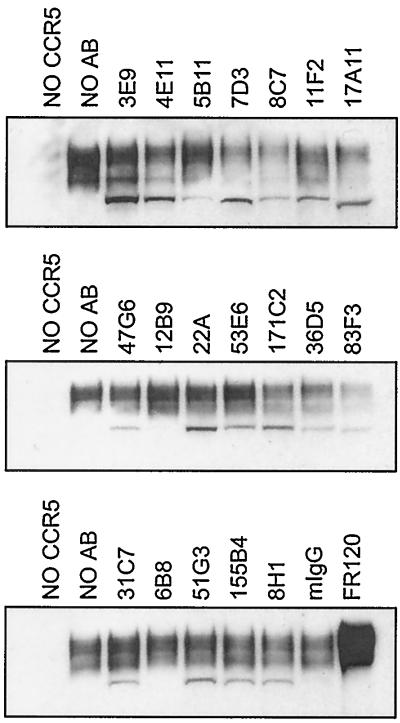
Remarkably, most of the neutralizing antibodies generated against chronically infected cells efficiently blocked Env-CCR5 binding (Fig. 9); 8C7, 7D3, and 17A11 were the most potent MAbs in this assay. In addition, the V3 loop antibodies 36D5 and 83F3, which did not affect Env-CD4 binding, also inhibited gp120 binding to CCR5. 171C2, which recognizes the V1/V2 loop and partially inhibited CD4 binding, also decreased Env binding to CCR5 in the absence of CD4, suggesting that the V1/V2 loops contribute to CCR5 binding by SIV Env consistent with their described role in HIV-1 tropism (7, 15, 34, 48). Although 11F2 bound SIVmac17E-Fr gp120 poorly, this MAb clearly interfered with Env-CCR5 binding. It is important to note that none of the antibodies generated against chronically infected cells competed with the anti-V3 loop MAbs 36D5 and 83F3 for sgp140 binding by ELISA (data not shown). This result suggests that these MAbs do not block CCR5 binding simply by masking a conformation-dependent epitope in the V3 loop, but rather that they may interact with a conserved chemokine receptor binding site similarly to the anti-HIV-1 MAb 17b (74). The coreceptor binding domain is likely to be more exposed in CD4-independent isolates (38), and this may have allowed for more efficient generation of antibodies against this conserved domain, as CP-MAC is a CD4-independent virus (25). Further studies will be required to test this hypothesis.
FIG. 9.
Ability of neutralizing MAbs to block Env-CCR5 binding. Unpurified SIV-17E-Fr gp120 was preincubated with no antibody (NO AB) or the indicated MAbs for 30 min at RT, added to 293T cells stably expressing rhesus CCR5 or parental 293T cells, and incubated for 1 h at 37°C. Cells were washed once and lysed, and bound Env was detected by Western blotting with DA6. mIgG, mouse IgG.
DISCUSSION
Several studies have shown that the humoral immune response can, under some circumstances, protect nonhuman primates from HIV or SIV infection (3, 28, 41, 57, 58, 72, 79). In addition, recent immunization studies with partially deglycosylated forms of SIV Env (73) and cell-associated fusion-competent HIV-1 Env (52) have produced antibodies which are effective against neutralization-resistant isolates. Collectively, these findings suggest that an appropriately designed subunit or noninfectious vaccine immunogen could produce a protective humoral immune response and encourage further studies towards this goal (13).
Recent studies have helped identify features in Env that should probably be recapitulated in Env-based immunogens in order to engender an effective immune response. The HIV and SIV Env protein, like those of most other enveloped viruses, is expressed as a noncovalently associated oligomer. The binding of antibodies to oligomeric HIV Env protein has been shown to correlate with neutralization ability (30, 64, 69, 76), and immunizations of mice or rabbits with various oligomeric forms of HIV Env have indicated that, relative to immunizations with monomeric gp120, this approach results in an antibody response which more efficiently recognizes oligomeric Env (8, 24). Several cross-reactive, neutralizing anti-HIV MAbs have been obtained that react with functionally important domains in the context of oligomeric Env, including IgG1b12, which has a rare CD4 binding site specificity (14), 2G12, which binds to conserved carbohydrate moieties on gp120 (87), and 2F5, which binds to a determinant in the ectodomain of gp41 (11, 66, 71). Recent insights into the structure of HIV Env have helped to identify highly conserved domains, including binding sites for CD4 and chemokine receptors, that are likely to be important targets for neutralizing antibodies (74, 90). It will be important to delineate the structure and ultimately the immunogenicity of these critical functional domains in the context of the native oligomeric Env protein.
While many neutralizing antibodies to HIV-1 have been reported and their mechanisms of action have been determined (see reference 70 for a review), less is known about the antigenic structure of SIV Env proteins and the mechanisms of antibody-mediated neutralization (43). Neutralizing murine and simian MAbs that react with conformational epitopes within the SIV V1/V2, V3, and V4 variable loops (4–6, 32, 44, 45, 59) and that characteristically inhibit homologous but not heterologous strains (32) have been described. However, while one MAb that may interfere with CD4 binding has been reported (5), antibodies that inhibit SIV-coreceptor interactions have not been described, and the role of the V3 loop as a target for neutralizing antibodies to SIV remains unclear (32, 40, 43, 45, 92). In addition, although humoral immune responses have been characterized in infected macaques and in macaques immunized with SIV gp120 and sgp140 (81), little is known about how the oligomeric structure of the SIV Env impacts the humoral immune response. As with HIV-1, it is unclear what types of immunogens will be required to generate broadly neutralizing antibody responses. To begin to address these questions, we produced and characterized a panel of murine MAbs to two different forms of oligomeric SIV Env from CP-MAC, a laboratory-passaged variant of SIVmacBK28 remarkable for its stable gp120-gp41 interaction and high surface Env expression (49). A panel of MAbs was produced which recognize diverse epitopes throughout the SIV Env that should be useful for further studies of SIV antigenic structure. Several neutralization-competent MAbs were also identified.
Immunization with sgp140 yielded antibodies to SIV Env that recognized 14 distinct peptide epitopes, 12 in gp120 and 2 in gp41, as well as several conformation-dependent MAbs that were mapped into five competition groups. Most of the MAbs generated from sgp140 immunization reacted at least weakly with denatured SIV Env by Western blotting, and only a few of these MAbs had potent neutralizing activity. Two MAbs directed against the V3 loop (36D5 and 83F3) potently neutralized viruses pseudotypes with Env derived from CP-MAC or the related isolate, SIVmac1A11. Several neutralizing MAbs with epitopes in the V1/V2 loops and less well-defined conformationally sensitive binding sites were also identified. The low proportion of the MAbs generated against sgp140 that neutralized virus infection and recognized strictly conformation-dependent epitopes could be due to several factors. Although the CP-MAC molecular clone exhibits a tight gp120-gp41 association (49), only uncleaved Env sedimented in the oligomeric fractions during velocity gradient sedimentation (Fig. 1). Since gp120-gp41 cleavage appears to be a prerequisite for efficient SIV Env CD4 and coreceptor binding (25), the failure to generate stable, cleaved SIV Env oligomers could have accounted, at least in part for, the low frequency of neutralizing antibodies generated using this immunogen. Furthermore, a significant fraction of the sgp140 preparation may be monomeric (Fig. 1). Finally, secreted gp140 could differ from native oligomeric Env in less obvious ways, as only half of the 72 MAbs produced against sgp140 recognized cell surface Env by FACS analysis although all MAbs were reactive with sgp140 by ELISA (Table 1 and data not shown). In summary, the high fraction of uncleaved sgp140, the presence of monomeric protein, and/or conformational differences between sgp140 and cell-associated oligomeric Env could have contributed to the failure of this immunogen to elicit broadly cross-neutralizing antibodies.
In contrast to MAbs induced by immunization with sgp140, MAbs induced by live, infected cells were notable for their conformational dependence, high reactivity with cell surface Env, and potency in neutralization assays, including the ability to neutralize at least some heterologous isolates. Differences in the screening protocols clearly account for some of these findings. While hybridomas derived using sgp140 were screened by ELISA, hybridomas raised against infected cells were screened for the ability to inhibit syncytium formation. However, all seven infected cell MAbs reacted with CP-MAC sgp140 by ELISA, indicating that had antibodies with these properties been generated by immunization with sgp140, they could have been identified by ELISA screening. It is possible that although epitopes recognized by MAbs to cell-associated Env were retained in CP-MAC sgp140, they were not immunogenic in the context of a soluble protein. In addition, Env expressed on the cell surface is likely to be fully cleaved, and oligomers may be more stable in this context. Finally, although the CP-MAC-infected SupT1 cells used in this protocol have undetectable cell surface CD4 by FACS analysis (data not shown), it is possible that some Env-CD4 complexes are formed in the endoplasmic reticulum or on the cell surface at very low levels. Since CD4 binding triggers conformational changes in SIV Env that enable it to interact with CCR5 more efficiently (25, 27, 36, 56), such complexes may expose a conserved CCR5 binding domain analogous to that shown for the HIV-1 gp120 (74); immunization with fusion-competent Env has been previously shown to induce potent, cross-reactive neutralizing antibodies to HIV-1 (52). In summary, cell-associated Env induced efficient, neutralizing MAbs at higher frequency than sgp140, and these MAbs recognized distinct, conformational epitopes important for Env-CD4 and/or Env-CCR5 binding.
At least five classes of neutralizing MAbs were identified following immunization with two different forms of oligomeric SIV Env: (i) MAbs to the V3 loop (36D5 and 83F3), (ii) MAbs recognizing the V1/V2 loop (8H1, 22A, 53E6, and 171C2), (iii) group 1 conformational antibodies which overlap the N terminus (6B8, 51G3, and 155B4), (iv) MAbs which interfere with CD4 binding site (171C2, 4E11, 5B11, 8C7, and 17A11), and (v) MAbs that disrupt CCR5 binding (36D5, 83F3, 171C2, 4E11, 7D3, 8C7, 11F12, and 17A11). These findings are in agreement with previously reported results for SIV and HIV-1 with the exception of the MAbs reacting with the V3 loop. For HIV-1, V3 loop antibodies are among the most potent neutralizing antibodies (10, 76) and have been shown to inhibit HIV-1 gp120-chemokine receptor binding (35, 61, 85, 89). In contrast, previous studies have reported that SIV MAbs that recognize the V3 loop are nonneutralizing, suggesting that the function of the V3 loop may be different for SIV and HIV-1 (32, 40, 45, 68), although there is evidence that the SIV V3 loop is involved in cell tropism (46). In our study, however, anti-V3 loop antibodies potently neutralized homologous and some heterologous viruses and blocked gp120 binding to CCR5, indicating that the V3 loop plays identical roles in chemokine receptor binding for SIV and HIV-1. The potency of V3 loop MAbs against some heterologous SIV isolates may be explained in part by the lesser variability in the SIV V3 loop than in the V3 loop of HIV-1 (67).
Antibodies directed toward the V1/V2 loops have been identified in HIV-1-infected patients and in animals immunized with a variety of HIV-1 Env preparations. In many but not all cases these antibodies neutralized homologous isolates, although as a group they were less potent than antibodies directed to the V3 loop (31, 60, 65, 80, 88). Similarly, anti-V1/V2 antibodies have been previously reported to neutralize SIV (6, 22, 45, 47, 59), consistent with the results reported here for MAbs 22A, 171C2, and 8H1. In general, antibodies to the N and C termini of HIV-1 are not neutralizing, and our finding that group 1 antibodies neutralize homologous virus (albeit weakly) was somewhat surprising. It is possible that these MAbs inhibit a conformational change in Env that is important for fusion. With the exception of a single MAb described by Babas et al. (5), few SIV antibodies directed against the CD4 binding site have been reported to date. Our MAbs that inhibit CD4-binding (4E11, 5B11, 8C7, and 17A11, and 171C2) efficiently neutralized CP-MAC reporter virus infection of CD4-positive cells, as would be predicted from studies using HIV-1 (17, 63, 82, 84). Finally, several MAbs blocked Env-CCR5 binding; interestingly, these were the most potent neutralizing antibodies against homologous virus and some heterologous strains. Many of the MAbs that blocked CCR5 binding did so independently of the V3 loop. This group may represent a novel subset of neutralizing SIV MAbs that block CCR5 binding through interactions with the conserved coreceptor binding site and should prove useful for a variety of future studies of SIV Env structure.
Finally, an important conclusion from this work is that the target cells used in neutralization assays can have a significant impact on the observed potency of a neutralizing antibody. Studies with HIV-1 have shown that chemokine receptor use does not significantly affect neutralization sensitivity (16, 51, 86). However, these studies evaluated only infection of cells expressing the major HIV coreceptors, CXCR4 and CCR5. While CCR5 is the most efficient coreceptor for the majority of SIV isolates, many strains are able to use alternative coreceptors such as STRL33 and GPR15 which are, in general, poorly utilized by HIV-1 strains (21). When identical preparations of CP-MAC-pseudotyped reporter viruses were used to infect target GHOST cells expressing CD4 and either CCR5 or STRL33, infection through STRL33 was blocked much more efficiently than infection through CCR5. It is important to consider that our neutralization studies were performed with cells expressing high levels of coreceptor, and it is unclear how well this in vitro neutralization protocol reflects conditions in vivo. However, it is not surprising that virus entry through STRL33 would be more easily blocked than infection through CCR5 given that the SIV Envs that we have examined clearly have lower affinities for STRL33 than CCR5 (25) and that higher concentrations of STRL33 than of CCR5 are needed to support virus infection (78). Thus, SIV neutralization protocols using human T-cell lines that do not express CCR5 may be far less stringent assays than protocols that employ target cells expressing CCR5. This conclusion is supported by earlier findings that several macaque seras capable of low-level neutralization of SIVmac251 on CEMx174 target cells (a CCR5-negative cell line) were unable to prevent the infection of human or rhesus peripheral blood mononuclear cells (which express CCR5) even at low dilutions (53).
In summary, this panel of well-characterized anti-SIV Env MAbs should be useful for future structural and immunologic studies of SIV Env proteins. In addition, results using the two immunization protocols suggest that Env-based immunogens that maintain the native structure of the oligomeric Env protein can elicit MAbs that are broadly neutralizing. Future immunization protocols with alternate preparations of Env that maintain this structure will be of considerable interest.
ACKNOWLEDGMENTS
We thank the members of the Doms laboratory for helpful discussions and advice.
A.L.E. was supported by NIH Medical Scientist Training Program grant 2T32GM07170. R.W.D. and J.A.H. were supported by NIH grants R01 AI45378, AI40880, and AI38225, and R.W.D. was supported by NIH grant A1-35383 and by an Elizabeth Glaser Scientist award from the Pediatric AIDS Foundation.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amedee A M, Lacour N, Gierman J L, Martin L N, Clements J E, Bohm R, Harrison R M, Murphey-Corb M. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J Virol. 1995;69:7982–7990. doi: 10.1128/jvi.69.12.7982-7990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba T W, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini L A, Posner M R, Katinger H, Stiegler G, Bernacky B J, Rizvi T A, Schmidt R, Hill L R, Keeling M E, Lu Y, Wright J E, Chou T C, Ruprecht R M. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 4.Babas T, Belhadj-Jrad B, Le Grand R, Dormont D, Montagnier L, Bahraoui E. Specificity and neutralizing capacity of three monoclonal antibodies produced against the envelope glycoprotein of simian immunodeficiency virus isolate 251. Virology. 1995;211:339–344. doi: 10.1006/viro.1995.1414. [DOI] [PubMed] [Google Scholar]
- 5.Babas T, Le Grand R, Dormont D, Bahraoui E. Production and characterization of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS Res Hum Retroviruses. 1997;13:1109–1119. doi: 10.1089/aid.1997.13.1109. [DOI] [PubMed] [Google Scholar]
- 6.Benichou S, Legrand R, Nakagawa N, Faure T, Traincard F, Vogt G, Dormont D, Tiollais P, Kieny M P, Madaule P. Identification of a neutralizing domain in the external envelope glycoprotein of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1992;8:1165–1170. doi: 10.1089/aid.1992.8.1165. [DOI] [PubMed] [Google Scholar]
- 7.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broder C C, Earl P L, Long D, Abedon S T, Moss B, Doms R W. Antigenic implications of human immunodeficiency virus type-1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc Natl Acad Sci USA. 1994;91:11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broliden P A, Makitalo B, Akerblom L, Rosen J, Broliden K, Utter G, Jondal M, Norrby E, Wahren B. Identification of amino acids in the V3 region of gp120 critical for virus neutralization by human HIV-1-specific antibodies. Immunology. 1991;73:371–376. [PMC free article] [PubMed] [Google Scholar]
- 10.Burton D R. A vaccine for HIV type 1: the antibody perspective. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11:S87–S98. [PubMed] [Google Scholar]
- 12.Burton D R, Moore J P. Why do we not have an HIV vaccine and how can we make one? Nat Med. 1998;4:495–498. doi: 10.1038/nm0598supp-495. [DOI] [PubMed] [Google Scholar]
- 13.Burton D R, Parren P W. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat Med. 2000;6:123–125. doi: 10.1038/72200. [DOI] [PubMed] [Google Scholar]
- 14.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W H I, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cecilia D, KewalRamani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamat S, Nara P, Berquist L, Whalley A, Morrow W J W, Köhler H, Kang C-Y. Two major groups of neutralizing anti-gp120 antibodies exist in HIV-infected individuals. J Immunol. 1992;149:649–654. [PubMed] [Google Scholar]
- 18.Chen B K, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 20.Desrosiers R C. The simian immunodeficiency viruses. Annu Rev Immunol. 1990;8:557–578. doi: 10.1146/annurev.iy.08.040190.003013. [DOI] [PubMed] [Google Scholar]
- 21.Doms R W, Edinger A L, Moore J P. Coreceptor use by primate lentiviruses. Los Alamos National HIV Sequence Database, part 3. Los Alamos, N.Mex: Los Alamos National Laboratory; 1999. pp. 1–12. [Google Scholar]
- 22.D'Souza M P, Kent K A, Thiriart C, Collignon C, Milman G. International collaboration comparing neutralization and binding assays for monoclonal antibodies to simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:415–422. doi: 10.1089/aid.1993.9.415. [DOI] [PubMed] [Google Scholar]
- 23.Earl P, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 16.17.1–16.17.16. [Google Scholar]
- 24.Earl P L, Broder C C, Long D, Lee S, Peterson J, Chakrabarti S, Doms R W, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edinger A L, Hoffman T L, Sharron M, Lee B, O'Dowd B, Doms R W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 27.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emini E A, Schleif W A, Nunberg J H, Conley A J, Eda Y, Tokiyoshi S, Putney S D, Matsushita S, Cobb K E, Jett C M, et al. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 29.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 30.Fouts T R, Binley J R, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung M S C, Sun C R Y, Gordon W L, Liou R-S, Chang T W, Sun W N C, Daar E S, Ho D D. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J Virol. 1992;66:848–856. doi: 10.1128/jvi.66.2.848-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glamann J, Burton D R, Parren P W, Ditzel H J, Kent K A, Arnold C, Montefiori D, Hirsch V M. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J Virol. 1998;72:585–592. doi: 10.1128/jvi.72.1.585-592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnann J W J, Nelson J A, Oldstone M B A. Fine mapping of an immunodominant domain in the transmembrane glycoprotein of human immunodeficiency virus. J Virol. 1987;61:2639–2641. doi: 10.1128/jvi.61.8.2639-2641.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van't Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 35.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill C M, Kwon D, Jones M, Davis C B, Marmon S, Daugherty B L, DeMartino J A, Springer M S, Unutmaz D, Littman D R. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. J Virol. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 37.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins W R, Monterfiori D C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoxie J A. Hypothetical assignment of intrachain disulfide bonds for HIV-2 and SIV envelope glycoproteins. AIDS Res Hum Retroviruses. 1991;7:495–499. doi: 10.1089/aid.1991.7.495. [DOI] [PubMed] [Google Scholar]
- 40.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P M, Meloen R H, Desrosiers R C, Burns D P W, Bolognesi D P, LaRosa G J, Putney S D. The principle neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson P R, Lifson J D, Czajak S C, Cole K S, Manson K H, Glickman R, Yang J, Montefiori D C, Montelaro R, Wyand M S, Desrosiers R C. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang C-Y, Nara P, Chamat S, Caralli V, Ryskamp T, Haigwood N, Newman R, Köhler H. Evidence for non-V3-specific neutralizing antibodies that interfere with gp120/CD4 binding in human immunodeficiency virus 1-infected humans. Proc Natl Acad Sci USA. 1991;88:6171–6175. doi: 10.1073/pnas.88.14.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent K A. Neutralising epitopes of simian immunodeficiency virus envelope glycoprotein. J Med Primatol. 1995;24:145–149. doi: 10.1111/j.1600-0684.1995.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 44.Kent K A, Gritz L, Stallard G, Cranage M P, Collignon C, Thiriart C, Corcoran T, Silvera P, Stott E J. Production and characterization of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS. 1991;5:829–836. doi: 10.1097/00002030-199107000-00006. [DOI] [PubMed] [Google Scholar]
- 45.Kent K A, Rud E, Corcoran T, Powell C, Thiriart C, Collignon C, Stott E J. Identification of two neutralizing and 8 non-neutralizing epitopes on simian immunodeficiency virus envelope using monoclonal antibodies. AIDS Res Hum Retroviruses. 1992;8:1147–1151. doi: 10.1089/aid.1992.8.1147. [DOI] [PubMed] [Google Scholar]
- 46.Kirchhoff F, Mori K, Desrosiers R C. The “V3” domain is a determinant of simian immunodeficiency virus cell tropism. J Virol. 1994;68:3682–3692. doi: 10.1128/jvi.68.6.3682-3692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kodama T, Burns D P W, Silva D P, Veronese F D, Derosiers R C. Strain-specific neutralizing determinant in the transmembrane protein of simian immunodeficiency virus. J Virol. 1991;65:2010–2018. doi: 10.1128/jvi.65.4.2010-2018.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Hoxie J A. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J Virol. 1994;68:5509–5522. doi: 10.1128/jvi.68.9.5509-5522.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of SIVmac transmembrane molecule increases envelope glycoprotein expression on cells and virions. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LaCasse R A, Follis K E, Moudgil T, Trahey M, Binley J M, Planelles V, Zolla-Pazner S, Nunberg J H. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J Virol. 1998;72:2491–2495. doi: 10.1128/jvi.72.3.2491-2495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 53.Langlois A J, Desrosiers R C, Lewis M G, KewalRamani V N, Littman D R, Zhou J Y, Manson K, Wyand M S, Bolognesi D P, Montefiori D C. Neutralizing antibodies in sera from macaques immunized with attenuated simian immunodeficiency virus. J Virol. 1998;72:6950–6955. doi: 10.1128/jvi.72.8.6950-6955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leonard C K, Spellman M W, Riddle L, Harris R J, Thomas J N, Gregory T J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 55.Luciw P A, Shaw K E, Unger R E, Planelles V, Stout M W, Lackner J E, Pratt-Lowe E, Leung N J, Banapour B, Marthas M L. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res Hum Retroviruses. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- 56.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 57.Mascola J R, Lewis M G, Stiegler G, Harris D, VanCott T C, Hayes D, Louder M, Brown C R, Sapan C V, Frankel S S, Lu Y, Robb M L, Katinger H, Birx D L. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4090–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mascola J R, Stiegler G, VanCott T C, Katinger H, Carpenter C B, Hanson C E, Beary H, Hayes D, Frankel S S, Birx D L, Lewis M G. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 59.Matsumi S, Matsushita S, Yoshimura K, Javaherian K, Takatsuki K. Neutralizing monoclonal antibody against an external envelope glycoprotein (gp110) of SIVmac251. AIDS Res Hum Retroviruses. 1995;11:501–508. doi: 10.1089/aid.1995.11.501. [DOI] [PubMed] [Google Scholar]
- 60.McKeating J A, Shotton C, Cordell J, Graham S, Balfe P, Sullivan N, Charles M, Page M, Bolmstedt A, Olofsson S, et al. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:4932–4944. doi: 10.1128/jvi.67.8.4932-4944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mondor I, Moulard M, Ugolini S, Klasse P J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 62.Moore J P, Burton D R. HIV-1 neutralizing antibodies: how full is the bottle? Nat Med. 1999;5:142–144. doi: 10.1038/5502. [DOI] [PubMed] [Google Scholar]
- 63.Moore J P, Ho D D. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore J P, Sattentau Q J, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon S, Fung M S, Traincard F, Pinkus M, Robey G, Robinson J E, Ho D D, Sodroski J. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J Virol. 1993;67:6136–6151. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Overbaugh J, Rudensey L M, Papenhausen M D, Beneviste R E, Morton W R. Variation in simian immunodeficiency virus Env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palker T J, Muir A J, Spragion D E, Staats H F, Langlois A, Montefiori D C. The V3 domain of SIVmac251 gp120 contains a linear neutralizing epitope. Virology. 1996;224:415–426. doi: 10.1006/viro.1996.0548. [DOI] [PubMed] [Google Scholar]
- 69.Parren P W, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parren P W H I, Moore J P, Burton D R, Sattentau Q J. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13(Suppl. A):S137–S162. [PubMed] [Google Scholar]
- 71.Poignard P, Klasse P J, Sattentau Q J. Antibody neutralization of HIV-1. Immunol Today. 1996;17:239–246. doi: 10.1016/0167-5699(96)10007-4. [DOI] [PubMed] [Google Scholar]
- 72.Putkonen P, Thorstensson R, Ghavamzadeh L, Albert J, Hild K, Biberfeld G, Norrby E. Prevention of HIV-2 and SIVsm infection by passive immunization in cynomlgus monkeys. Nature. 1991;352:436–438. doi: 10.1038/352436a0. [DOI] [PubMed] [Google Scholar]
- 73.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 74.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 75.Rud E W, Cranage M, Yon J, Quirk J, Ogilvie L, Cook N, Webster S, Dennis M, Clarke B E. Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J Gen Virol. 1994;75:529–543. doi: 10.1099/0022-1317-75-3-529. [DOI] [PubMed] [Google Scholar]
- 76.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M F, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharron M, Pohlmann S, Price K, Lolis E, Tsang M, Kirchhoff F, Doms R W, Lee B. Expression and coreceptor activity of STRL33 on primary peripheral blood lymphocytes. Blood. 2000;96:41–49. [PubMed] [Google Scholar]
- 79.Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho M W, Martin M A. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 80.Shotton C, Arnold C, Sattentau Q, Sodroski J, McKeating J A. Identification and characterization of monoclonal antibodies specific for polymorphic antigenic determinants within the V2 region of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1995;69:222–230. doi: 10.1128/jvi.69.1.222-230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silvera P, Flanagan B, Kent K, Rud E, Powell C, Corcoran T, Bruck C, Thiriart C, Haigwood N L, Stott E J. Fine analysis of humoral antibody response to envelope glycoprotein of SIV in infected and vaccinated macaques. AIDS Res Hum Retroviruses. 1994;10:1295–1304. doi: 10.1089/aid.1994.10.1295. [DOI] [PubMed] [Google Scholar]
- 82.Thali M, Furman C, Ho D D, Robinson J, Tilley S, Pinter A, Sodroski J. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 1992;66:5635–5641. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thali M, Olshevsky U, Furman C, Gabuzda D, Posner M, Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly neutralizing human monoclonal antibody. J Virol. 1991;65:6188–6193. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 86.Trkola A, Ketas T, KewalRamani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warrier S V, Pinter A, Honnen W J, Girard M, Muchmore E, Tilley S A. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J Virol. 1994;68:4636–4642. doi: 10.1128/jvi.68.7.4636-4642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 90.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 91.Yokoyama W. Production of monoclonal antibodies. In: Coligan J, Kruisbeek A, Marguiles D, Shevach E M, Strober W, editors. Current protocols in immunology. Vol. 1. New York, N.Y: John Wiley & Sons; 1994. pp. 2.5.2–2.5.17. [Google Scholar]
- 92.Zhuge W, Gao Q, Liu Z Q, Narayan O, Stephens E B. Neutralization of SIVmac239/17E in lymphocyte cultures involves virus strain-specific linear and conformational epitopes encoded by different regions of the env gene including the “V3” domain. Virology. 1996;222:184–192. doi: 10.1006/viro.1996.0409. [DOI] [PubMed] [Google Scholar]