ABSTRACT
While the Plasmodium falciparum malaria parasite continues to cause severe disease globally, Mozambique is disproportionally represented in malaria case totals. Acquisition of copy number variations (CNVs) in the parasite genome contributes to antimalarial drug resistance through overexpression of drug targets. Of interest, piperaquine resistance is associated with plasmepsin 2 and 3 CNVs (pfpmp2 and pfpmp3, respectively), while CNVs in the multidrug efflux pump, multidrug resistance-1 (pfmdr1), increase resistance to amodiaquine and lumefantrine. These antimalarials are partner drugs in artemisinin combination therapies (ACTs) and therefore, CNV detection with accurate and efficient tools is necessary to track ACT resistance risk. Here, we evaluated ~300 clinically derived samples collected from three sites in Mozambique for resistance-associated CNVs. We developed a novel, medium-throughput, quadruplex droplet digital PCR (ddPCR) assay to simultaneously quantify the copy number of pfpmp3, pfpmp2, and pfmdr1 loci in these clinical samples. By using DNA from laboratory parasite lines, we show that this nanodroplet-based method is capable of detecting picogram levels of parasite DNA, which facilitates its application for low yield and human host-contaminated clinical surveillance samples. Following ddPCR and the application of quality control standards, we detected CNVs in 13 of 229 high-quality samples (prevalence of 5.7%). Overall, our study revealed a low number of resistance CNVs present in the parasite population across all three collection sites, including various combinations of pfmdr1, pfpmp2, and pfpmp3 CNVs. The potential for future ACT resistance across Mozambique emphasizes the need for continued molecular surveillance across the region.
KEYWORDS: malaria, drug resistance, copy number variation, droplet digital PCR, plasmepsin, multidrug resistance
INTRODUCTION
Despite decades of research, the Plasmodium malaria parasite continues to cause severe disease in humans (1). The global burden of malaria cases reached 247 million and 619,000 deaths in 2021, and the African country of Mozambique accounted for 4.7% of these cases (2). P. falciparum is the world’s most deadly malarial parasite and causes infections across this region (3). Research on drug resistance is particularly important since there is no widely employed vaccine and antimalarial drugs are a major tool for control of malaria. Due to Mozambique’s sizable contribution to annual malaria cases, it is crucial to investigate the potential rise of antimalarial resistance in this area.
Artemisinin combination therapies (ACTs) are the most common treatment for P. falciparum malaria infections (4). Globally, ACTs are administered with several different partner drugs; in Mozambique, the first-line treatment for uncomplicated malaria is ACT with artemether–lumefantrine (AL), dihydroartemisinin–piperaquine (DHAP), or artesunate–amodiaquine (AS–AQ) (5). Due to reports of low ACT efficacy in Africa (3, 6–9), we aim to determine the risk of AL, AS-AQ, and DHAP resistance emergence in Mozambique.
Mutations in the pfkelch13 gene are the primary markers of artemisinin partial resistance and are actively being monitored globally. However, the high efficacy of ACTs is in large part due to the inclusion of partner drugs (10, 11). Unfortunately, resistance to the majority of ACT partner drugs has been reported in Africa and Asia (7, 11, 12). Parasite genotypes associated with this resistance predominantly involve copy number variations (CNVs), which increase the transcription of genes encoding for proteins that confer resistance. Key resistance-associated CNVs include pfmdr1 (for lumefantrine and amodiaquine) as well as pfpmp2 and pfpmp3 genes (for piperaquine) encoding a multidrug efflux pump and digestive proteases, respectively (12–17). It is important to actively monitor these CNVs, particularly in countries like Mozambique where resistance may only just be emerging (18).
In this study, we assessed the presence of resistance-associated CNVs in clinical samples from Mozambique using a novel “quadruplex” droplet digital PCR (ddPCR) assay. This nanodroplet-based method involves end-point PCR and fluorescent probes to amplify and quantify gene copies of interest (19). DdPCR presents an attractive tool for the rapid detection of resistance genotypes in malaria-endemic areas for several reasons. When compared with Sanger sequencing, ddPCR is able to detect minor variants with high accuracy, particularly at lower DNA concentrations (20, 21). When compared to standard quantitative PCR, ddPCR can reduce average costs per sample by 72% by requiring fewer runs to evaluate more loci (22). Furthermore, ddPCR requires less hands-on time per sample due to a lower requirement for replicates and serial dilutions, thus decreasing the chance of technical errors (23). Finally, ddPCR requires sub-nanogram levels of DNA that are easily attainable from minimally invasive collection methods like dried blood spots. While instrument access may limit use in some areas, droplet-based PCR assays are becoming more popular for molecular surveillance (22, 24).
To further extend the benefits of ddPCR, we developed a quadruplex assay, where the three CNV loci (pfmdr1, pfpmp2, and pfpmp3) and a control gene (pfhsp70) are evaluated simultaneously. This approach improves assay efficiency and technical consistency over duplex assays (21). Following optimization, the ddPCR assay proved accurate and sensitive and identified a low but appreciable level of resistance-conferring CNVs across three Mozambique provinces. This study supports the continued need for molecular surveillance across this region to improve the detection of ACT resistance before it becomes more widespread.
RESULTS
The optimized quadruplex ddPCR assay is accurate, specific, and sensitive
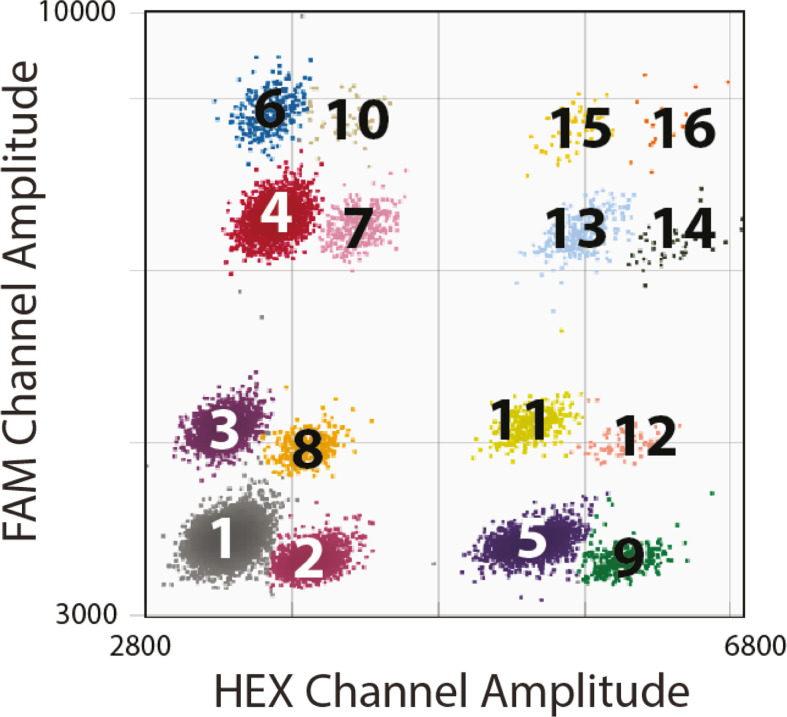
The development of a two-color “quadruplex” assay requires optimization of primer annealing temperature, dilutions, and cycling number (see Materials and Methods and Fig. S1 to S3). Copy number determination is based on comparing positive-droplet populations (Fig. 1), which represent the relative proportions of DNA targets in the sample. In our studies, P. falciparum hsp70 is used as a single copy “reference gene” to determine the copy number of the other “target genes” (i.e., target-CN/reference-CN ≠1 indicates a change in copy number status). With appropriate parasite DNA input (>0.05 ng), the resulting quadruplex assay has 16 distinguishable orthogonal droplet clusters (Fig. 1).
Fig 1.
Two-dimensional plot of the quadruplex ddPCR assay. Sixteen clusters of droplets (maximum possible) represent (1): droplets containing no target DNA (negative population), (2): droplets containing at least one copy of pfhsp70, (3): droplets containing at least one copy of pfmdr1, (4): droplets containing at least one copy of pfpmp3, (5): droplets containing at least one copy of pfpmp2, (6): droplets with both pfpmp3 and pfmdr1, (7): droplets with both pfpmp3 and pfhsp70, (8): droplets with both pfmdr1 and pfhsp70, (9): droplets with both pfhsp70 and pfpmp2, (10): droplets with pfmdr1, pfhsp70, and pfpmp3, (11): droplets with pfpmp2 and pfmdr1, (12): droplets with pfhsp70, pfmdr1, and pfpmp2, (13): droplets with pfpmp2 and pfpmp3, (14): droplets with pfhsp70, pfpmp2, and pfpmp3, (15): droplets with pfmdr1, pfpmp2, and pfpmp3, (16): and droplets with pfmdr1, pfpmp2, pfpmp3, and pfhsp70.
When comparing duplex versus quadruplex assays, we observed no significant difference in copy numbers of pfmdr1, pfpmp2, and pfpmp3 using a laboratory parasite line with a known gene copy number (Fig. S4). Additionally, we confirmed that the assays are P. falciparum-specific (Fig. S5) and lack interference by human DNA (Fig. S6), which represents the majority of DNA in clinical surveillance samples.
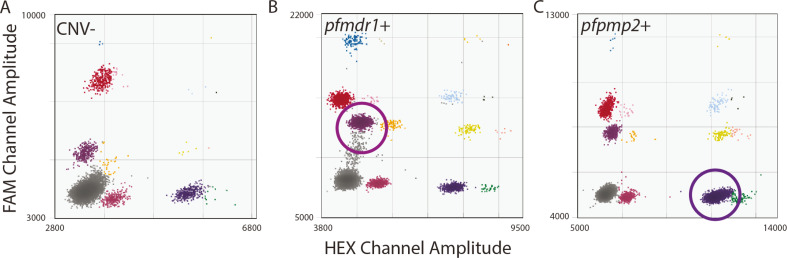
Assessments that used DNA from laboratory parasite lines also displayed the accuracy and sensitivity of the novel quadruplex assays. As expected, we observed single copies for all ddPCR gene targets for the NF54 parasite line (Fig. 2A), a three-copy pfmdr1 CNV for the Dd2 line (25, 26) (Fig. 2B), and a four-copy pfpmp2 CNV for the PM2GT clone F4 due to episomal expression (27) (Fig. 2C). When we evaluated the sensitivity of the quadruplex assay across a 5-log range (Fig. S7 and S8A, R2 of 0.9766), we detected positive droplets down to 0.08 pg of total parasite DNA (0.00008 ng, Fig. S7A).
Fig 2.
Parasite lines with known CNVs show the accuracy of the quadruplex ddPCR assay. All samples were run at 60 cycles. Clusters of droplets are colored as in Fig. 1. (A) Sample: laboratory-cultured P. falciparum NF54 DNA with no known CNVs in loci of interest (0.08 ng DNA, 17,739 total droplets, CN: 0.94/1.01/1.09; pfpmp3, pfpmp2, and mdr1, respectively). Note: this sample is also part of the dilution series represented in Fig S7. (B) Sample: laboratory-cultured P. falciparum Dd2 DNA with three copies of pfmdr1 (0.02 ng DNA, 18,400 total droplets, CN: 2.94; mdr1). (C) Sample: laboratory-cultured P. falciparum PM2GT clone F4 DNA with four copies of pfpmp2 (0.02 ng DNA, 9,690 total droplets, CN: 4.43; pfpmp2).
To explore the impact of genotype mixtures on CNV detection using the novel quadruplex assay, we evaluated mixtures of P. falciparum laboratory parasite lines with different genotypes. We were able to detect copy number changes in assays where the CNV+ sample is diluted at various proportions with a CNV- sample (Fig. S9). Based on these data, we can reliably detect CNVs when they are the major genotype (>75%). We also determined that a copy number of >1.5 represents a duplication event in at least 75% of the parasite genomes within the sample. Based on confidence intervals (CIs) from individual experiments, we may be able to detect CNVs at lower levels (i.e., ~50% of the population, Fig. S9A). Therefore, in high-quality samples (low 95% CI), copy numbers between 1.2 and 1.5 were noted as potentially CNV+; this threshold is in line with previous studies that used 1.2 as a cutoff for CNV detection (22).
Assessment of clinical samples reveals a low level of CNVs across the Mozambique parasite population
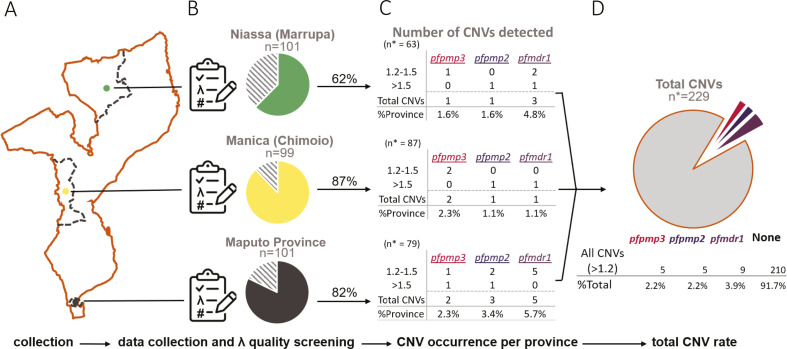
Of the 297 clinical P. falciparum samples collected from Mozambique (Fig. 3A), we were able to assess 296 samples (99%, Table S1). The majority of samples (229/279, 77%) passed quality screening (λ = 0.005–1.1) due to low 95% CI (Fig. 3B; Table S2; Fig. S8B). When separated by province, we considered 87% of Manica samples, 82% of Maputo samples, and 62% of Niassa samples as high-quality samples (Fig. 3B). Clinical samples with measurable DNA levels fell within the ddPCR positive droplet range of our standards (Fig. S8A), confirming the presence of P. falciparum DNA in these samples. During these studies, we detected high-quality clinical samples with as few as 80 positive droplets per target (with a mean of ~16,000 total droplets), which equates to ~0.02 ng of parasite DNA input based on our standard curve (Table S2; Fig. S8A).
Fig 3.
Summary of sample processing and results by province. (A) Depiction of Mozambique sites where samples were collected. Provinces are depicted in dashed lines. The city of collection within the province is depicted with a colored dot. (B) Sample quality control preformed from ddPCR data. Names of the province and city of collection (in parentheses) above pie charts representing the pass rate of quality control using λ (see Materials and Methods); the dashed portion represents failed quality control. n = total number of samples collected. (C) Results of ddPCR CNV analysis per-target in each province. n* = total number of samples passing quality screening in each province; 1.2–1.5 represents “potential CNVs”; >1.5 represents “called CNVs”; total CNVs are the sum of the CNVs > 1.2; %Province = Total CNVs/n*. (D) The cumulative total of all CNVs (>1.2) observed from all provinces. n* = combined total samples that passed quality screening.
Together, we measured a mean copy number for high-quality samples across all loci of 0.95 copies (mean of 0.92, 0.95, and 0.99 for pfpmp3, pfpmp2, and pfmdr1, respectively; Fig. S10A; Table S2), indicating that the assay did not exhibit locus-specific detection bias. We identified four different high-quality clinical samples that harbored definitive CNVs (>1.5 copies) with six total CNVs (Fig. 3C; Fig. S10A); the samples were spread across the three provinces (one from Maputo, two from Niassa, and one from Manica). Three of the CNVs included the pfpmp2 gene, two included pfmdr1, and one included pfpmp3. When we manually inspected ddPCR plots, all of these samples showed high-quality droplet formation and locus detection (Fig. S10B through E). By including samples with copy numbers between 1.2 and 1.5, we identified nine additional high-quality clinical samples across all three provinces that were potentially positive for CNVs in at least one of the three loci (nine for pfmdr1, five for pfpmp3, and five for pfpmp2). Some samples were positive for two CNVs; one sample showed both pfmdr1 and pfpmp2 CNVs (Fig. S10C) and four samples with both pfpmp2 and pfpmp3 CNVs (Fig. S10D, Table S2).
A portion of samples were rerun to assess the reproducibility of copy number estimates using the quadruplex assay (17 CNV- samples and 13 CNV+ samples). All CNV- samples yielded reproducible calls (Table S1). Despite an additional 18 months of DNA storage, the majority of high-quality samples with potential CNVs (>1.2 copies) also yielded reproducible calls (83% for pfmdr1, 67% of pfpmp3, and 83% for pfpmp2, Table S1).
DISCUSSION
We identified optimal ddPCR conditions through extensive experimentation including evaluations of cycle number, dilution range, quality control metrics, and potential effects from contaminating DNA from the human host or non-falciparum species (Fig. S1 to S7). We established that our novel assay is accurate (Fig. 2; Fig. S4 and S9), specific (Fig. S5 and S6), and sensitive (Fig. S7). We employed metrics to determine sample quality and identified a copy number range for CNV detection in clinical samples. Using this refined approach, we identified a low level of CNVs in clinical parasite populations in Mozambique.
The clinical samples evaluated in our study were prepared from dried blood spots collected after screening for P. falciparum. Despite the high sensitivity and accuracy of the ddPCR assay, some samples proved unquantifiable due to low DNA content or quality (Fig. 3B; Table S1). These assay failures were likely due to the age of the purified DNA; in the current study, there was a minimum of 8 months between DNA extraction and ddPCR analysis (maximum of 28 months), which in addition to degradation from freeze–thaw and shipping temperatures (see Materials and Methods) could contribute to declining sample quality. Indeed, when samples were repeated ~20 months following the initial runs, we had a failure rate of >50% (Table S1); most of these samples failed due to low positive droplet numbers indicating DNA degradation as the cause.
The P. falciparum CNV frequency that we observed in this study was in the range found in previous studies from Mozambique (7, 18). Although we only detected potential CNVs in a total of 13 samples (Table S2), this is appreciative in a small sampling of clinical infections (13/229, 5.7% total; pfpmp3: 2.2%; pfpmp2: 2.2%; and pfmdr1: 3.9%, Fig. 3). For comparison, one study estimated 1.1% (4/350) and 1.4% (5/350) pfpmp2 and pfmdr1 CNV prevalence, respectively (18). Another Mozambican study identified multiple copies of pfpmp2 and pfmdr1 in 11.3% (8/71) and 16.7% (2/12) of samples, respectively (7).
The incidence of pfmdr1 CNVs (copy number of >1.2) in our study was approximately twice as common as the pfpmp2 or pfpmp3 loci (Fig. 3; Fig. S10A). Additionally, of the nine pfmdr1 CNV+ samples, three samples also harbored pfpmp2 or pfpmp3 CNVs (Table S2). Concomitant CNVs may reflect an increase in parasite adaptation to DHAP, which has been directly linked with all three CNVs in genome-wide association studies (8, 16). Interestingly, previous studies from Mozambique did not detect pfpmp3 CNVs in clinical samples. Although at a very low rate, we detected samples with CNVs (copy number of >1.2) that encompassed both pfpmp2 and pfpmp3 genes (3/229, Fig. S10D; Table S2). The report of pfpmp2/3 co-amplification is consistent with studies from Cambodia, where the CNV region spanned the two neighboring genes (28).
Our data are consistent with the observation that the piperaquine partner drug has not yet been widely used to treat uncomplicated malaria in Mozambique. However, its use during complicated malaria and MDA may select resistant parasite populations. As far as we are aware, this drug has been used for MDA in three Mozambique provinces; DHAP MDA was applied between 2015 and 2017 in the Magude district (29, 30), which is approximately 100 km from the current study health facility (Hospital provincial da Matola) in the Maputo province, and in 2021 and 2023 in Cabo Delgado, which is outside of the region of testing in this study. Although a recent study that used the same clinical samples as our study did not detect validated mutations in pfkelch13 (31), the use of MDA in the region and our identification of a low rate of pfmdr1 and pfpmp2/3 CNVs necessitate continued molecular surveillance to better understand genotype dynamics across this region. Fortunately, the prevalence of resistance CNVs in Mozambique remains far below that reported in other countries such as Cambodia (23%) (17), Vietnam (54.3%) (32), and other African countries (average of 21.1% for pfmdr1 and >30% for pfpmp2) (7).
Systematic monitoring of resistance markers from malaria-infected patients using sensitive and accurate assays can assist in the prevention of treatment failure due to resistance (4, 33–35). Our novel quadruplex ddPCR assay is specific for P. falciparum loci (Fig. S5), and not suitable for molecular surveillance of P. ovale infections, which cause malaria infections in this region of Africa (36, 37). The assay is capable of detecting P. falciparum CNVs that are not yet prominent in a parasite population but may be emerging over time (potential CNVs in high-quality samples, >1.2 copies). Therefore this tool, and others capable of detecting minor parasite genotypes, are an important component of malaria resistance surveillance programs; malaria-endemic regions with emerging resistance CNVs can increase malaria prevention efforts to protect from future widespread antimalarial resistance.
MATERIALS AND METHODS
Laboratory & control parasites, culturing, and genomic DNA extraction
Laboratory parasite DNA from P. falciparum NF54 and Dd2 (MRA1000 and 156, respectively) were used to design and validate assay efficacy before clinical sample use. Parasite culture was carried out as previously (38). Briefly, P. falciparum parasites were grown in vitro at 37°C at 3% hematocrit (serotype A-positive human erythrocytes) in RPMI 1640 medium (Invitrogen, Waltham, MA, USA) containing 28 mM NaHCO3 and 25 mM HEPES and supplemented with AlbuMAX (TM) II Lipid-Rich BSA (Invitrogen, 11021029) in sterile, sealed flasks, flushed with 5% O2, 5% CO2, and 90% N2 (39, 40). Cultures were maintained with media changes three times each week and sub-cultured as necessary to keep parasitemia below 3%.
DNA from laboratory parasite lines was isolated as previously (41). Parasites were lysed with 0.15% saponin (Sigma Life Science) and washed with 1 x PBS to remove RBC membranes. Then, cells were treated with 200 ug/mL proteinase K (Thermo Fisher Scientific) and 0.1% L-loril sarkosil (Teknova Inc, Hollister, CA, USA) at 37° overnight. DNA was extracted using phenol/chloroform/isoamyl alcohol (25:24:1), pH 7.8–8.1 (Invitrogen), and MaXtract high-density gel tubes (Qiagen) twice, and twice more with only chloroform (Sigma-Aldrich) using standard methods. A final precipitation step with 100% ethanol removed the remaining salts and phenol. The resulting DNA was quantified using the Qubit double-stranded DNA High-Sensitivity kit as per the manufacturer’s recommendations (Invitrogen).
DNA from a laboratory parasite line harboring ~4 copies of pfpmp2 maintained on the chromosome and episomes was provided by the Michael Klemba (PM2GT clone F4) (27). Clinical samples obtained for this study came from the University of Virginia Medical Center from patients with clinical malaria, as determined by rapid diagnostic tests and peripheral blood smears (see ethical approval above). Blood was obtained from these patients within 24 hours of phlebotomy and used to extract DNA from malaria parasites using the method described above.
Area of study, clinical isolates, and genomic DNA preparation
Mozambique clinical samples used in this study were obtained from patients admitted in health facilities from three settings, namely, Northern (Hospital distrital de Marrupa in Niassa), Central (Centro de Saúde Eduardo Mondlhane and Centro de Saúde 7 de abril in Manica), and Southern (Hospital provincial da Matola in Maputo) areas of Mozambique, between April and August of 2021, as previously described (31). The choice of Niassa, Manica, and Maputo provinces not only aligns with our scientific goals but also with the objectives of Instituto Nacional de Saúde and National Malaria Control Programme Mozambique, contributing to the systematic mapping of malaria cases at the national level.
A total of 450 participants of all ages with malaria-positive rapid diagnostic test (RDT) were recruited and provided 100 µL of blood samples on filter paper (Whatman FTA cards), after written informed consent. All dried blood spot samples were then stored under −20°C until they were used for genotyping.
Parasite genomic DNA from dried blood spots was extracted using the Chelex method (42), and DNA was stored at −20°C. Real-time PCR for P. falciparum confirmation, targeting the 18S rRNA gene, was conducted as described in da Silva et al. (31). P. falciparum positive DNA samples (n = 297) from three provinces (Maputo n = 97, Niassa n = 100, and Manica n = 100) were assessed at the University of Virginia (shipped at room temperature, stored at −20°C). Clinical isolate DNA concentration ranged from 0.1 to 2.0ng/μL with an average concentration of ~0.4 ng/µL. Samples were coded at collection, and location details were blinded during experimentation.
Droplet digital PCR quadruplex assays and CNV analysis
To achieve optimal primer/probe concentrations, the two-color quadruplex assay was developed as two individual duplex assays on either fluorescence channel and then merged into a single quadruplex assay with small adjustments (Table 1: primer and probe concentrations). Because the ddPCR droplet-analyzer used in this study possesses two fluorescence channels (FAM and HEX), quadruplexing requires an intentional decrease in the fluorescence amplitude of one of the assays on each channel to distinguish the populations. The amplitude of these droplet clusters can be affected by several factors including the age of the primers and probes, purity of the input DNA, and interexperimental variables like UV exposure and droplet generation variance (43). Generally, when shifts in amplitude occur from interexperimental factors, they affect all clusters equally, allowing for accurate assignment of droplet clusters.
TABLE 1.
Optimized quadruplex assay reaction primer and probe concentrationsa
| Target | Probe concentration (nm) | Fluorophore | Primer concentration (nm) |
|---|---|---|---|
| pfpmp2a | 125 | HEX | 2,500 |
| pfhsp70b | 125 | HEX | 800 |
| pfpmp3a | 50 | FAM | 400 |
| pfmdr1 | 125 | FAM | 600 |
Bold targets indicate assays with higher fluorescence amplitudes on ddPCR plots.
Used as a single-copy reference gene.
Primer and probe sequences (Table 2) were BLASTed against Pf3D7 and human genomes to confirm the absence of potential secondary targets and assessed for potential dimer formation using the IDT OligoAnalyzer tool. The specificity for Plasmodium genes was also confirmed by the lack of amplification of human genomic DNA (Fig. S6). Optimal assay temperature was assessed by performing assays in duplex over a thermal gradient of annealing/extension temperatures and determining the condition with the highest average amplitude Fig. S1).
TABLE 2.
Primer and probe sequences for the quadruplex ddPCR reaction
| Target | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| pfpmp2 a | 5′-ATGGTGATGCAGAAGTTGGA-3′ | 5′-AACATCCTGCAGTTGTACATTTAAC-3′ | 5'-/5HEX/-CAGGATCTGCTAATTTATGGGTCCCA/3IABkFQ/-3' |
| pfhsp70 b | 5′-TGCTGTCATTACCGTTCCAG-3′ | 5′-AGCAGCTGCAGTAGGTTCATT-3′ | 5'-/5HEX/AGATGCTGGTACAATTGCAGGA/IABkFQ/-3' |
| pfpmp3 a | 5′-CCACTTGTGGTAACACGAAATTA-3′ | 5′-TGGTTCAAGGTATTGTTTAGGTTC-3′ | 5'-/56FAM/CCAACACTCGAATATCGTTCACCAA/IABkFQ/-3' |
| pfmdr1 b | 5′-TGCCCACAGAATTGCATCTA-3′ | 5′-TCGTGTGTTCCATGTGACTG-3′ | 5'-/56FAM/ACCCTGATCGAAATGGAACCT/IABkFQ/-3' |
Purified bulk DNA (~10 ng/µL) from laboratory parasite lines was digested with BamHI (New England BioLabs) for 12 hours at 37°C and then diluted to 0.004 ng/µL for input into ddPCR assays (total of 0.02 ng). Extracted DNA (5 µL) from clinical samples was directly digested with BamHI and diluted at a ratio of 1:2-1:10 into the ddPCR assay (Table S1; Fig. S2). Some samples were run undiluted to account for low parasite DNA amount. The BamHI restriction enzyme was chosen specifically due to a predicted restriction site located between the tandem pfpmp2 and pfpmp3 genes, while generating similar lengths for all target-containing DNA fragments including pfhsp70 and pfmdr1 (10–60 kb).
Fresh primer/probe master mixes were prepared for each assay at 10x final concentrations (Tables 1 and 2). Each 23 µL ddPCR reaction contained 5 µL of restriction-digested DNA (diluted as mentioned above), 1:10 dilution of each 10 x primer/probe mix (four sets), and 1:2 dilution of ddPCR Supermix for probes (Bio-Rad Laboratories). Reactions underwent droplet generation using a QX200 Droplet Generator with droplet generation oil for probes (Bio-Rad Laboratories) and subsequent PCR using a C1000 Thermal Cycler (Bio-Rad Laboratories). Thermal cycling conditions are as follows: 10 min at 95°C initial denaturation step, 1 min at 95°C second denaturation step, and 2 min at 58°C annealing and extension step (ramp rate of 1°C per second), the second denaturation step and the annealing/extension step repeated 60 times, and then 10 min at 98°C to halt the reaction. This longer cycling program (60 cycles versus the standard 40 cycles) increased ddPCR assay success for lower-quality clinical samples (Fig. S3). Samples were kept at 4°C until use for analysis (no longer than 24 hours).
After amplification, probe fluorescence was read using a QX200 Droplet Reader (Bio-Rad Laboratories) and analyzed using QuantaSoft Software version 1.7.4 (Bio-Rad Laboratories). Populations were manually defined as instructed by the manufacturer using QuantaSoft software. Samples were only considered valid if a minimum of 10,000 total droplets were observed. Poisson confidence intervals provided by the software were used to assess sample quality.
Samples were screened for quality using the lambda (λ) value in accordance with the Minimum Information for Publication of Quantitative Digital PCR Experiments (22, 45, 46), where λ = ln(#accepted droplets/#negative droplets). Samples passed quality screening if λ was between 0.005 and 1.1. This threshold was chosen as it represented high-quality clinical samples, where the difference between the maximal 95% CI value and the determined copy number was less than 30% of the copy number value ([(MaxCI – CopyNumber) / CopyNumber] <30%, Fig. S11, blue box). Furthermore, during this calculation, if the accuracy (defined as: %100 * |1 – CopyNumber|) deviates from the expected value of “1”, it is either a product of insufficient quantification or potential CNV signal (Fig. S11, green box).
CNVs were identified with QuantaSoft Software. Each droplet was automatically designated as positive or negative for template DNA through the detection of the fluorescence signal after amplification (Fig. 1 ). The relative abundance of a single-copy reference (pfhsp-70, PF3D7_0818900) compared to a target amplicon was used to detect CNVs (pfmdr1, PF3D7_0523000; pfpmp2, PF3D7_1408000; pfpmp3, PF3D7_1408100, e.g., a target/reference abundance of 2:1 indicates a duplication of the target). Samples that passed quality screening (see above) and possessed a ratio ≥1.5 target/reference were considered positive for CNVs. Samples with copy numbers between 1.2 and 1.5 were manually inspected and considered potentially positive if samples were high quality and droplet populations were well-resolved.
High-resolution melting for evaluating Plasmodium species
High-resolution melt (HRM) assays were used to validate the Plasmodium species of samples used to test the specificity of the quadruplex assay (47). Plasmid controls containing partial species-specific 18S rRNA gene sequences were obtained through BEI Resources (Manassas, VA, USA) (P. falciparum (MRA-177, lot: 70042650), P. vivax (MRA-178, lot: 70041752), P. malariae (MRA-179, lot: 70051095), and P. ovale wallikeri (MRA-180, lot: 70043212). Each 25 µL reaction contained 2 x HRM PCR Master Mix (Type-it HRM PCR Kit, Qiagen), 700 nM of primers targeting the multicopy 18S rRNA gene: 5′-GTT CCT CTA AGA AGC TTT-3′ and 5′-TAA CGA ACG AGA TCT TAA-3′ (48) (IDT, USA), ~10 ng of the template DNA, and RNase-free water. The PCR was performed with the following cycling conditions: 95°C for 5 min, 45 cycles of 95°C for 10 s, 55°C for 20 s, followed by an HRM ramp from 65°C to 95°C, with an increase of 0.1°C every 2 s on the Rotor-Gene Q real-time PCR instrument with a 72-well rotor (Qiagen). Rotor-Gene Q software (version 2.3.5, build 1; Qiagen) was used to plot the change in fluorescence versus temperature (dF/T), which allowed us to compare the HRM peaks of the clinical samples and plasmid controls. Multiple peaks are diagnostic of genomic DNA samples (from clinical or laboratory sources) from specific Plasmodium species (47). These peaks are due to amplification of distinct 18S rRNA gene copies present in the genomes (# of total 18 s rRNA genes in genome/# of 18 s rRNA genes with primer homology: P. falciparum 5/5; P. vivax 3/2; and P. ovale wallikeri 2/2).
ACKNOWLEDGMENTS
The authors would like to acknowledge Fundação para a Ciência e a Tecnologia for funds to GHTM (UID/04413/2020) and LA-REAL (LA/P/0117/2020) and Instituto CAMÕES (https://www.instituto-camoes.pt/) BOLSAS CAMÕES, FUNDAÇÃO MILLENNIUM BCP (https://www.fundacaomillenniumbcp.pt/en/), Paróquia de São Nicolau – Lisboa (to C.D.S.), R01AI150856 (to J.L.G.), the University of Virginia Global Infectious Disease Institute (to C.W. and N.B.), and Center for Global Health Equity (to C.W.).
We thank Michelle Warthan (University of Virginia) for laboratory parasite culturing and Dr. Michael Klemba (Virginia Tech) for genomic DNA from the PM2GT clone F4 parasite line.
Contributor Information
Fatima Nogueira, Email: FNogueira@ihmt.unl.pt.
Jennifer L. Guler, Email: jlg5fw@virginia.edu.
Audrey Odom John, The Children's Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
ETHICS APPROVAL
The study protocol obtained ethical clearance from the National Bioethics Committee for Health of Mozambique (CNBS—IRB00002657, Ref: 131/CNBS/2021, dated: March 2021). The University of Virginia Institutional Review Board for Health Sciences Research provided ethical approval for samples used as species controls in this study from patients admitted into the University of Virginia hospital (IRB-HSR protocol #21081). All samples were handled in accordance with approved protocols and in agreement with the ethical standards of the Declaration of Helsinki. A waiver for informed consent was provided through the University of Virginia health facilities because our study design met the following criteria: (i) the research involved minimal risk to subjects, (ii) the waiver does not adversely affect the rights and welfare of subjects, and (iii) the research could not practicably be carried out without the waiver.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00346-24.
Figures S1 to S11.
Clinical sample ddPCR assessment including dilutions and repeats.
Clinical sample CNV calls and quality metrics.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Sato S. 2021. Correction to: Plasmodium—a brief introduction to the parasites causing human malaria and their basic biology. J Physiol Anthropol 40:3. doi: 10.1186/s40101-021-00254-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO . 2022. World malaria report 2022. Geneva, Switzerland: World Health Organization [Google Scholar]
- 3. World Health Organization . 2018. Status report on artemisinin resistance and ACT efficacy (August 2018). Vol. 10. World Health Organization [Google Scholar]
- 4. Plowe CV, Roper C, Barnwell JW, Happi CT, Joshi HH, Mbacham W, Meshnick SR, Mugittu K, Naidoo I, Price RN, Shafer RW, Sibley CH, Sutherland CJ, Zimmerman PA, Rosenthal PJ. 2007. World antimalarial resistance network (WARN) III: molecular markers for drug resistant malaria. Malar J 6:1–10. doi: 10.1186/1475-2875-6-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MISAU . 2017. Normas de tratamento DA Malária em Moçambique, p 82. 3rd ed. MISAU, editor. Maputo. [Google Scholar]
- 6. Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. 2020. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania. Sci Rep 10:3500. doi: 10.1038/s41598-020-60549-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leroy D, Macintyre F, Adoke Y, Ouoba S, Barry A, Mombo-Ngoma G, Ndong Ngomo JM, Varo R, Dossou Y, Tshefu AK, Duong TT, Phuc BQ, Laurijssens B, Klopper R, Khim N, Legrand E, Ménard D. 2019. African isolates show a high proportion of multiple copies of the Plasmodium falciparum plasmepsin-2 gene, a piperaquine resistance marker. Malar J 18:126. doi: 10.1186/s12936-019-2756-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uwimana A, Legrand E, Stokes BH, Ndikumana J-L, Warsame M, Umulisa N, Ngamije D, Munyaneza T, Mazarati J-B, Munguti K, Campagne P, Criscuolo A, Ariey F, Murindahabi M, Ringwald P, Fidock DA, Mbituyumuremyi A, Menard D. 2020. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 26:1602–1608. doi: 10.1038/s41591-020-1005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanboonkunupakarn B, White NJ. 2022. Advances and roadblocks in the treatment of malaria. Br J Clin Pharmacol 88:374–382. doi: 10.1111/bcp.14474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin RE, Shafik SH, Richards SN. 2018. Mechanisms of resistance to the partner drugs of artemisinin in the malaria parasite. Curr Opin Pharmacol 42:71–80. doi: 10.1016/j.coph.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 12. Boonyalai N, Thamnurak C, Sai-Ngam P, Ta-Aksorn W, Arsanok M, Uthaimongkol N, Sundrakes S, Chattrakarn S, Chaisatit C, Praditpol C, et al. 2021. Plasmodium falciparum phenotypic and genotypic resistance profile during the emergence of Piperaquine resistance in Northeastern Thailand. Sci Rep 11:13419. doi: 10.1038/s41598-021-92735-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sidhu ABS, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 194:528–535. doi: 10.1086/507115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, Hallett RL. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 51:991–997. doi: 10.1128/AAC.00875-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ngalah BS, Ingasia LA, Cheruiyot AC, Chebon LJ, Juma DW, Muiruri P, Onyango I, Ogony J, Yeda RA, Cheruiyot J, Mbuba E, Mwangoka G, Achieng AO, Ng’ang’a Z, Andagalu B, Akala HM, Kamau E. 2015. Analysis of major genome loci underlying artemisinin resistance and pfmdr1 copy number in pre- and post-acts in western Kenya. Sci Rep 5:8308. doi: 10.1038/srep08308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, et al. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis 17:174–183. doi: 10.1016/S1473-3099(16)30415-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim P, Alker AP, Khim N, Shah NK, Incardona S, Doung S, Yi P, Bouth DM, Bouchier C, Puijalon OM, Meshnick SR, Wongsrichanalai C, Fandeur T, Le Bras J, Ringwald P, Ariey F. 2009. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J 8:1–9. doi: 10.1186/1475-2875-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta H, Macete E, Bulo H, Salvador C, Warsame M, Carvalho E, Ménard D, Ringwald P, Bassat Q, Enosse S, Mayor A. 2018. Drug-resistant polymorphisms and copy numbers in Plasmodium falciparum, Mozambique, 2015. Emerg Infect Dis 24:40–48. doi: 10.3201/eid2401.170864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83:8604–8610. doi: 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z, Sun K, Jing C, Cao H, Ma R, Wu J. 2019. Comparison of droplet digital PCR and direct Sanger sequencing for the detection of the BRAF V600E mutation in papillary thyroid carcinoma. J Clin Lab Anal 33:e22902. doi: 10.1002/jcla.22902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor SC, Laperriere G, Germain H. 2017. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep 7:2409. doi: 10.1038/s41598-017-02217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Srisutham S, Suwannasin K, Sugaram R, Dondorp AM, Imwong M. 2021. Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR. Malar J 20:120. doi: 10.1186/s12936-021-03659-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz-Villalba A, Ruijter JM, van den Hoff MJB. 2021. Use and misuse of Cq in qPCR data analysis and reporting. Life 11:496. doi: 10.3390/life11060496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tong Y, Shen S, Jiang H, Chen Z. 2017. Application of digital PCR in detecting human diseases associated gene mutation. Cell Physiol Biochem 43:1718–1730. doi: 10.1159/000484035 [DOI] [PubMed] [Google Scholar]
- 25. Cheeseman IH, Gomez-Escobar N, Carret CK, Ivens A, Stewart LB, Tetteh KKA, Conway DJ. 2009. Gene copy number variation throughout the Plasmodium falciparum genome. BMC Genomics 10:353. doi: 10.1186/1471-2164-10-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chugh M, Scheurer C, Sax S, Bilsland E, van Schalkwyk DA, Wicht KJ, Hofmann N, Sharma A, Bashyam S, Singh S, Oliver SG, Egan TJ, Malhotra P, Sutherland CJ, Beck H-P, Wittlin S, Spangenberg T, Ding XC. 2015. Identification and deconvolution of cross-resistance signals from antimalarial compounds using multidrug-resistant Plasmodium falciparum strains. Antimicrob Agents Chemother 59:1110–1118. doi: 10.1128/AAC.03265-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sijwali PS, Kato K, Seydel KB, Gut J, Lehman J, Klemba M, Goldberg DE, Miller LH, Rosenthal PJ. 2004. Plasmodium falciparum cysteine protease falcipain-1 is not essential in erythrocytic stage malaria parasites. Proc Natl Acad Sci U S A 101:8721–8726. doi: 10.1073/pnas.0402738101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ansbro MR, Jacob CG, Amato R, Kekre M, Amaratunga C, Sreng S, Suon S, Miotto O, Fairhurst RM, Wellems TE, Kwiatkowski DP. 2020. Development of copy number assays for detection and surveillance of piperaquine resistance associated plasmepsin 2/3 copy number variation in Plasmodium falciparum. Malar J 19:181. doi: 10.1186/s12936-020-03249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galatas B, Saúte F, Martí-Soler H, Guinovart C, Nhamussua L, Simone W, Munguambe H, Hamido C, Montañà J, Muguande O, Maartens F, Luis F, Paaijmans K, Mayor A, Bassat Q, Menéndez C, Macete E, Rabinovich R, Alonso PL, Candrinho B, Aide P. 2020. A multiphase program for malaria elimination in southern Mozambique (the Magude project): a before-after study. PLoS Med 17:e1003227. doi: 10.1371/journal.pmed.1003227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cuinhane CE, Galatas B, Lopez JM, Djive H, Nhantumbo H, Murato I, Saúte F, Aide P, Munguambe K, Torres N. 2023. Acceptability and perceived barriers to reactive focal mass drug administration in the context of a malaria elimination program in Magude district, Southern Mozambique: a qualitative study. PLoS One 18:e0283160. doi: 10.1371/journal.pone.0283160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. da Silva C, Matias D, Dias B, Cancio B, Silva M, Viegas R, Chivale N, Luis S, Salvador C, Duarte D, Arnaldo P, Enosse S, Nogueira F. 2023. Anti-malarial resistance in Mozambique: absence of Plasmodium falciparum Kelch 13 (K13) propeller domain polymorphisms associated with resistance to artemisinins. Malar J 22:160. doi: 10.1186/s12936-023-04589-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phuc BQ, Rasmussen C, Duong TT, Dong LT, Loi MA, Ménard D, Tarning J, Bustos D, Ringwald P, Galappaththy GL, Thieu NQ. 2017. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg Infect Dis 23:715–717. doi: 10.3201/eid2304.161872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacob CG, Thuy-nhien N, Mayxay M, Maude RJ, Quang HH, Hongvanthong B, et al. 2021. Genetic surveillance in the greater mekong subregion and south Asia to support malaria control and elimination, p 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nsanzabana C. 2021. Time to scale up molecular surveillance for anti-malarial drug resistance in sub-saharan Africa. Malar J 20:401. doi: 10.1186/s12936-021-03942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nsanzabana C, Djalle D, Guérin PJ, Ménard D, González IJ. 2018. Tools for surveillance of anti-malarial drug resistance: an assessment of the current landscape. Malar J 17:75. doi: 10.1186/s12936-018-2185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leonard CM, Hwang J, Assefa A, Zulliger R, Candrinho B, Dimbu PR, Saifodine A, Plucinski M, Rogier E. 2022. Missed Plasmodium ovale infections among symptomatic persons in Angola, Mozambique, and Ethiopia. Open Forum Infect Dis 9:ofac261. doi: 10.1093/ofid/ofac261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller I, Zimmerman PA, Reeder JC. 2007. Plasmodium malariae and Plasmodium ovale - the ‘bashful’ malaria parasites. Trends Parasitol 23:278–283. doi: 10.1016/j.pt.2007.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDaniels JM, Huckaby AC, Carter SA, Lingeman S, Francis A, Congdon M, Santos W, Rathod PK, Guler JL. 2021. Extrachromosomal DNA amplicons in antimalarial-resistant Plasmodium falciparum. Mol Microbiol 115:574–590. doi: 10.1111/mmi.14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haynes JD, Diggs CL, Hines FA, Desjardins RE. 1976. Culture of human malaria parasites Plasmodium falciparum. Nature 263:767–769. doi: 10.1038/263767a0 [DOI] [PubMed] [Google Scholar]
- 40. Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. doi: 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 41. Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, DeRisi J, Rathod PK. 2013. Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog 9:e1003375. doi: 10.1371/journal.ppat.1003375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bereczky S, Mårtensson A, Gil JP, Färnert A. 2005. Short report: rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am J Trop Med Hyg 72:249–251. [PubMed] [Google Scholar]
- 43. Whale AS, Huggett JF, Tzonev S. 2016. Fundamentals of multiplexing with digital PCR. Biomol Detect Quantif 10:15–23. doi: 10.1016/j.bdq.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu S, Huckaby AC, Brown AC, Moore CC, Burbulis I, McConnell MJ, Güler JL. 2021. Single-cell sequencing of the small and AT-skewed genome of malaria parasites. Genome Med 13:75. doi: 10.1186/s13073-021-00889-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhat S, Herrmann J, Armishaw P, Corbisier P, Emslie KR. 2009. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem 394:457–467. doi: 10.1007/s00216-009-2729-5 [DOI] [PubMed] [Google Scholar]
- 46. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, Bustin SA. 2013. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem 59:892–902. doi: 10.1373/clinchem.2013.206375 [DOI] [PubMed] [Google Scholar]
- 47. Kassaza K, Operario DJ, Nyehangane D, Coffey KC, Namugosa M, Turkheimer L, Ojuka P, Orikiriza P, Mwanga-Amumpaire J, Byarugaba F, Bazira J, Guler JL, Moore CC, Boum Y. 2018. Detection of Plasmodium species by high-resolution melt analysis of DNA from blood smears acquired in southwestern Uganda. J Clin Microbiol 56:e01060-17. doi: 10.1128/JCM.01060-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mangold KA, Manson RU, Koay ESC, Stephens L, Regner M, Thomson RB, Peterson LR, Kaul KL. 2005. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol 43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S11.
Clinical sample ddPCR assessment including dilutions and repeats.
Clinical sample CNV calls and quality metrics.