Abstract
To study the existence of GB virus C/hepatitis G virus (GBV-C/HGV) variants with different tropism, we have analyzed the heterogeneity and quasispecies composition of GBV-C/HGV isolated from in vitro-infected peripheral blood mononuclear cells (PBMC) and from sera, livers, and PBMC from two chronically infected patients. For this purpose, the GBV-C/HGV 5′ noncoding region (5′NCR) was amplified by reverse transcription-PCR and the amplified products were cloned and sequenced. These analyses showed that the master 5′NCR sequences isolated from the in vitro-infected PBMC and from the PBMC isolated from the patient whose serum was used as the inoculum were identical but different from that of the inoculum. Furthermore, phylogenetic analysis revealed that all PBMC sequences grouped together into a branch which was separate from those of the inoculum. For one of the two chronically infected patients, all the sequences from the PBMC and one from the liver clustered into a single branch while the sequences from the serum and all the other liver sequences grouped together in the other branch. For the other patient, the sequences from the serum and PBMC and three sequences from the liver grouped together into one branch, while the remaining five sequences from the liver were separated in a different cluster. In conclusion, our results support the existence of different GBV-C/HGV variants with different tissue tropism.
The GB virus C/hepatitis G virus (GBV-C/HGV) is a positive-sense, single-stranded RNA (9.4 kb in length) virus whose genetic structure resembles the hepatitis C virus (HCV) and which is considered belong to the Flaviviridae family of animal viruses (17, 18, 31, 32). Although GBV-C/HGV was discovered as a putative agent of non-A-E hepatitis (2, 7) and GBV-C/HGV RNA has been detected in the sera of patients with various liver diseases including fulminant hepatitis (39), chronic hepatitis C (1, 4, 34, 38), and cirrhosis with or without hepatocellular carcinoma (13, 14), recent works have shown that GBV-C/HGV may play a minor role in causing liver disease (2, 3, 10, 11, 22).
The replication of GBV-C/HGV presumably occurs via a negative-strand RNA intermediate. However, its replication site is still unknown, since no conclusive evidence regarding the GBV-C/HGV cell tropism has been reported. Thus, it remains unclear whether the liver is the main target for GBV-C/HGV infection and replication, because, although several authors have reported the detection of negative-polarity viral RNA in the liver (16, 19, 29, 30), others have been unable to detect this putative replicative intermediate (6, 15, 25, 28). Similarly, the results have been contradictory when peripheral blood mononuclear cells (PBMC) from GBV-C/HGV-infected patients have been examined for the presence of GBV-C/HGV-RNA of both positive and negative polarity (19, 21, 27, 29). On the other hand, in vitro studies have provided evidence that GBV-C/HGV is able to replicate in established cell lines of hematopoietic origin (MT-2C, a human T-cell leukemia virus type 1-infected human T-cell line) and in immortalized hepatocytes (PH5CH, a non-neoplastic human hepatocyte cell line immortalized with simian virus 40 large T antigen) (12). Furthermore, we have recently demonstrated that GBV-C/HGV can infect and replicate in PBMC from healthy donors after incubation of these cells with GBV-C/HGV-RNA-positive serum (8). In that work, in which a relatively small number of clones were analyzed, we also demonstrated that only a fraction of the GBV-C/HGV variants present in serum are able to infect and replicate in PBMC in vitro. This finding suggests the existence of lymphotropic variants of this virus. However there are no data on the existence of these lymphotropic variants in vivo. Furthermore, whether the GBV-C/HGV variants that infect PBMC in vivo are the same as those that infect the liver and circulate in serum is not known.
In the present study, we have analyzed the genomic heterogeneity and quasispecies composition of the GBV-C/HGV 5′ noncoding region (5′NCR) recovered from the PBMC of four healthy donors infected in vitro with a GBV-C/HGV RNA-positive serum. We have also studied the in vivo quasispecies composition in the 5′NCR from the GBV-C/HGV isolated from the sera, livers, and PBMC of two chronically infected patients, and our results clearly demonstrated that different GBV-C/HGV strains have a different tropism.
MATERIALS AND METHODS
In vitro infection of PBMC.
PBMC from four healthy blood donors (who were not infected by GBV-C/HGV, HCV, hepatitis B virus, human immunodeficiency virus, Epstein-Barr virus, or cytomegalovirus) were isolated as previously described (8). One million viable cells from each individual donor and a cell pool consisting of equal parts of PBMC from each donor at a final density of 106 cells were incubated with 10 μl of a GBV-C/HGV RNA-positive serum (inoculum). The culture method and the characteristics of the inoculum have been described previously (8). After the 30-day culture period, the cells were washed five times with phosphate-buffered saline (PBS), and the last wash was saved to be used as a negative control for GBV-C/HGV RNA detection. Total RNAs from each cell culture, from the serum used as the inoculum, and from the PBMC isolated from the patient whose serum was used as the inoculum were extracted by a modification of the guanidinium-phenol-chloroform method described by Chomczynski and Sacchi (5).
In vivo study of GBV-C/HGV quasispecies.
Genetic heterogeneity in the 5′NCR region of the GBV-C/HGV genome was analyzed in paired serum (stored at −20°C), liver, and PBMC (stored in liquid N2) specimens from two patients coinfected with HCV. The clinical features of these patients are summarized in Table 1. The patients presented anti-HCV antibodies as detected by enzyme-linked immunosorbent assay III (Ortho Diagnostic Systems, Raritan, N.J.) and confirmed by RIBA III (Ortho Diagnostic Systems). These two patients were selected according to the following criteria: (i) no previous antiviral or immunomodulatory treatment and (ii) presence of GBV-C/HGV RNA by reverse transcription-PCR (RT-PCR) in serum, liver, and PBMC samples obtained at the same time. Total RNA was extracted from 200 μl of serum, 100 mg of liver tissue, and 106 PBMC as mentioned above (5). Total RNAs from PBMC and liver tissue were quantitated, and 1.5 μg of cell-derived RNA or the whole-serum-derived RNA was used for GBV-C/HGV amplification. To prevent any possible contamination of the liver and PBMC samples by the serum GBV-C/HGV particles, the samples were washed five times with PBS before RNA extraction, and the last wash of each sample was stored for further analysis by RT-PCR as a negative control.
TABLE 1.
Clinical characteristics of the patients included in the study
| Parameter | Patient 1 | Patient 2 |
|---|---|---|
| Gender | Male | Male |
| Age (yr) | 31 | 48 |
| ALTa (IU/liter) | 214 | 100 |
| Known duration of disease (mo) | 72 | 180 |
| Knodell index value | 9 | 8 |
| Fibrosis score | 2 | 1 |
| Epidemiology | Intravenous drug abuse | Polytransfused |
ALT, alanine aminotransferase.
Amplification of the GBV-C/HGV 5′NCR.
GBV-C/HGV RNA from the inoculum, the PBMC corresponding to the inoculum, the 30-day-cultured cells, and the serum, liver, and PBMC specimens from the two patients studied were amplified by RT-nested PCR with primers derived from the 5′NCR of the GBV-C/HGV genome, as previously described (8). The amplified fragments covered nucleotides 36 to 356 of the 5′NCR (17).
Cloning and sequencing.
The amplified GBV-C/HGV RNA 5′NCR products were cloned into the pCR II-TOPO vector (TOPO TA Cloning kit; Invitrogen, San Diego, Calif.). An average of 20 clones (range, 11 to 50 clones) from each PCR product were automatically sequenced in both directions using the ALF Express DNA Sequencing System. In order to exclude the possibility that the changes observed in the sequences of the samples were not due to nucleotide misincorporation induced by the Taq polymerase, an in vitro-transcribed RNA from plasmid pCR-GNC, containing a cDNA corresponding to a GBV-C/HGV 5′NCR (nucleotides 10 to 387) whose nucleotide sequence was previously known, was included in each PCR assay. Subsequently, the amplification products of this synthetic RNA were cloned and sequenced at the same time as the in vitro and in vivo samples.
Analysis of the 5′NCR nucleotide sequences.
The sequences were aligned and edited using the Clustal X (35) and GeneDoc (version 2.5.000; K. B. Nicholas and H. B. Nicholas, Jr., 1997) programs, respectively. Phylogenetic analysis was performed using the DNADIST, NEIGHBOR, SEQBOOT, and CONSENSE programs from the Phylogeny Inference Package (PHYLIP, version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle, 1993). The evolutionary distances were estimated by the Kimura two-parameter method, and the unrooted phylogenetic trees were constructed by the neighbor-joining method. The final outputs of the trees were obtained with TreeView, version 1.5.2 (24). Bootstrap analyses were determined on 1,000 resamplings of the data sets. Bootstrap values of ≥70% were considered statistically significant for the observed grouping. In order to study the quasispecies composition of each sample type, the complexity coefficient (CC) was defined as the number of different clones from each sample divided by the total number of clones analyzed (23).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences presented in this paper are AF125468 to AF12505 and AF197346 to AF197447.
RESULTS
Specificity of GBV-C/HGV detection.
A PCR fragment of 320 bp, corresponding to the GBV-C/HGV 5′NCR amplification product, was detected in the inoculum, the PBMC corresponding to the inoculum, the 30-day-cultured PBMC of the healthy donors, and the cell pool after experimental infection, as well as in the serum, liver, and PBMC samples of the two infected patients. In contrast, the last PBMC and liver PBS washes and all the negative controls included in each PCR assay were always GBV-C/HGV negative. On the other hand, sequencing of the amplification products from the RNA transcribed in vitro from plasmid pCR-GNC always gave the expected nucleotide sequence previously known, confirming that no nucleotide misincorporation due to Taq polymerase activity occurred during the PCR procedures.
Analysis of the GBV-C/HGV 5′NCR sequence of in vitro-infected PBMC.
To study the sequence heterogeneity of PBMC after the experimental infection, the PCR products from the inoculum and the different cell cultures were cloned. A total of 50 clones from the inoculum and 29 clones from the PBMC corresponding to the inoculum were sequenced. With respect to the in vitro-infected PBMC and cell pool, an average of 22 clones (range, 19 to 27) were sequenced.
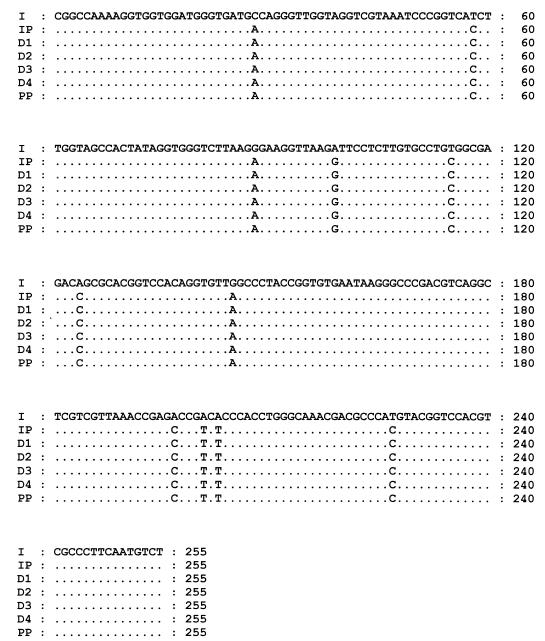
As shown in Table 2, CCs of 0.50 and 0.34 were obtained for the sequences isolated from the inoculum and the PBMC corresponding to the inoculum, respectively. On the other hand, CCs ranged from 0.33 to 0.55 for the 5′NCR sequences of the PBMC infected in vitro, and the CC for the sequences of the GBV-C/HGV isolates recovered from the cell pool was 0.29. When the master sequences (the master sequence for each sample type is the sequence which occurs with the highest frequency) were aligned and compared, we found that those recovered from the PBMC corresponding to the inoculum and the in vitro-infected PBMC and cell pool were all identical, but different from the master sequence recovered from the inoculum (Fig. 1). None of the GBV-C/HGV 5′NCR sequences derived from the serum used as the inoculum were identical to the master sequences of the PBMC corresponding to the inoculum or the in vitro-infected PBMC and cell pool, and the mean genetic distance between the sequences was 0.0508 ± 0.0045 (range, 0.0406 to 0.0619).
TABLE 2.
Heterogeneity of the GBV-C/HGV 5′NCR nucleotide sequences observed in the PBMC infected in vitro
| Sample typea | No. of clones sequenced | No. of different sequences found | CC | Frequency of sequencesb |
|---|---|---|---|---|
| I | 50 | 25 | 0.50 | 1 (24), 1 (3), 23 (1) |
| IP | 29 | 10 | 0.34 | 1 (19), 1 (2), 8 (1) |
| D1 | 20 | 11 | 0.55 | 1 (10), 10 (1) |
| D2 | 19 | 10 | 0.53 | 1 (9), 1 (2), 8 (1) |
| D3 | 21 | 7 | 0.33 | 1 (15), 6 (1) |
| D4 | 27 | 9 | 0.33 | 1 (19), 8 (1) |
| PP | 21 | 6 | 0.29 | 1 (16), 5 (1) |
I, inoculum; IP, PBMC corresponding to the inoculum; D1 through D4, PBMC of the four healthy subjects infected in vitro with the inoculum; PP, cell pool made by the mixture of the PBMC of the four healthy donors infected in vitro.
The number preceding the parentheses represents a sequence, or sequences; the number within the parentheses is the number of clones in which that sequence or those sequences appear. For instance, for I, 1 sequence appears in 24 clones, 1 appears in 3 clones, and 23 appear in 1 clone each.
FIG. 1.
Nucleotide alignment of the master sequences recovered from the inoculum (I), the PBMC of the patient whose serum was used as the inoculum (IP), the PBMC from the four healthy donors infected in vitro with GBV-C/HGV (D1 through D4), and the pool of cells from the four donors infected in vitro with GBV-C/HGV (PP).
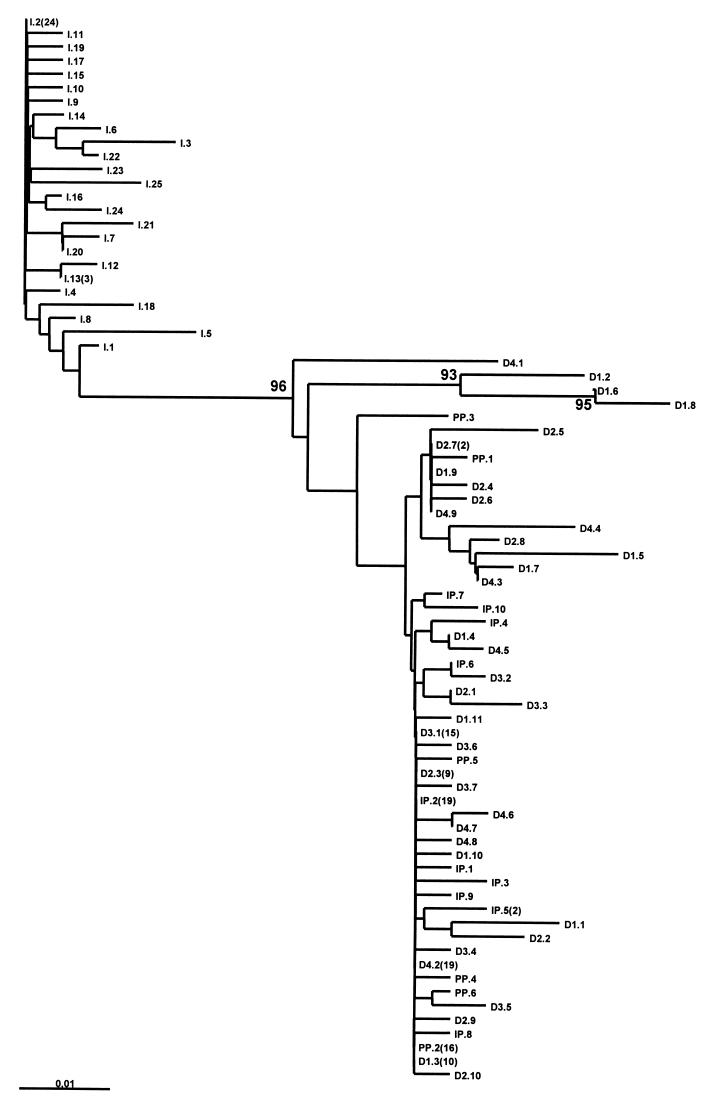
The unrooted phylogenetic tree constructed with all these GBV-C/HGV sequences revealed that the sequences derived from the serum used as the inoculum were grouped together in a cluster, while the sequences from the PBMC corresponding to the inoculum and the in vitro-infected PBMC and cell pool clustered together into a separate branch, with a bootstrap value of 96% (Fig. 2).
FIG. 2.
Unrooted neighbor-joining tree constructed with the 5′NCR nucleotide sequences from the in vitro-infected PBMC. The isolates are designated I (inoculum), IP (PBMC of the patient whose serum was used as the inoculum), D1 through D4 (PBMC from the four healthy donors infected in vitro with GBV-C/HGV), or PP (pool of cells from the four donors infected in vitro), followed by the clone number and, in parentheses, the number of clones bearing the same sequence. Bootstrap values are shown in the nodes of the tree.
Analysis of the 5′NCR sequence from serum, liver, and PBMC samples of the GBV-C/HGV-infected patients.
To study the complexity of the GBV-C/HGV quasispecies in the 5′NCR infecting the serum, liver, and PBMC in vivo, the PCR products from each compartment corresponding to two patients with chronic GBV-C/HGV infection were cloned and sequenced. A total of 41 clones from patient 1 (13 from the serum, 11 from the liver, and 17 from PBMC) and 45 clones from patient 2 (16 from the serum, 15 from the liver, and 14 from PBMC) were analyzed.
In patient 1, the CCs in the serum and liver were similar (0.61 and 0.64, respectively) and higher than that found in the isolates recovered from the PBMC (0.35). In contrast, in patient 2, the CCs in the three compartments studied were similar (0.81 in the serum, 0.80 in the liver, and 0.86 in PBMC).
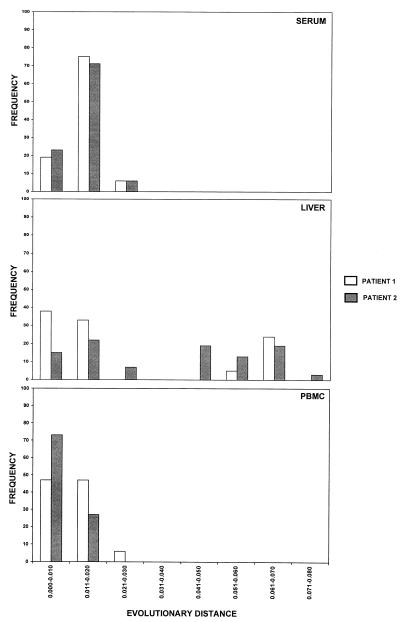
When the frequency with which the evolutionary distances occurred in the GBV-C/HGV isolates from the three compartments was analyzed, the distances in serum and PBMC samples were distributed around a single peak in both patients (Fig. 3). In contrast, the distribution of the distances among the GBV-C/HGV isolates from the liver showed two independent peaks in both cases (Fig. 3).
FIG. 3.
Histograms of the frequency of each evolutionary distance in the GBV-C/HGV 5′NCR sequences from serum, liver, and PBMC specimens of the two patients with GBV-C/HGV infection.
With respect to the master sequences in the three compartments studied, in patient 1, the master sequences from the serum and the liver were identical and represented 30.8 and 36.4% of the total sequences from the serum and liver, respectively; however, this master sequence was different from that of the PBMC, which represented 70.% of total sequences from this compartment. In contrast, in patient 2, the master sequences from the serum and PBMC, which represented 18.7 and 21%, respectively, of total sequences, were identical, but different from the predominant sequence from the liver, which represented 20% of all liver sequences.
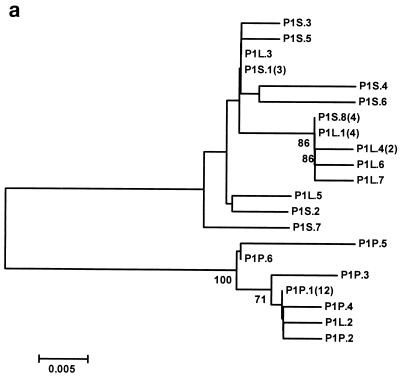
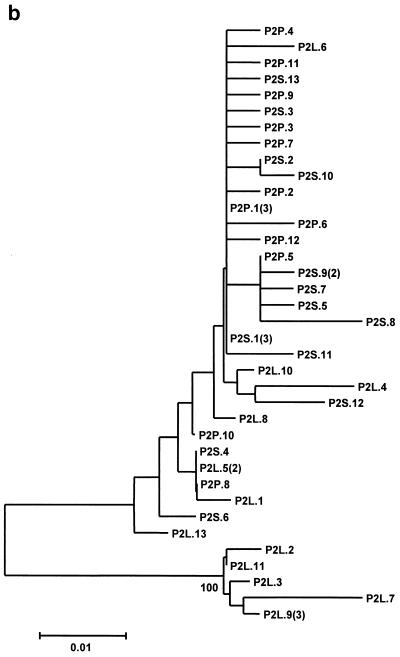
The neighbor-joining phylogenetic trees constructed with the GBV-C/HGV 5′NCR nucleotide sequence of the isolates from both patients are shown in Fig. 4. In patient 1, all the isolates from the PBMC samples and one isolate from the liver are separated into a single branch, while the remaining sequences from the serum and liver are clustered in another branch (Fig. 4a). In the other patient, all the sequences from the serum and PBMC samples and seven sequences from the liver are grouped into a single branch, while the remaining five different sequences from the liver biopsy specimen are separated into a different branch (Fig. 4b).
FIG. 4.
Unrooted neighbor-joining trees constructed with the 5′NCR nucleotide sequences of GBV-C/HGV isolated from serum, liver, and PBMC samples of patients 1 (a) and 2 (b). The clones are designated P1 or P2 (indicating those isolated from patient 1 or 2, respectively), plus S, L, or P (indicating the source as serum, liver, or PBMC, respectively), followed by the clone number and, in parentheses, the number of clones with identical sequences in each compartments. The bootstrap values are given in the nodes of the trees.
DISCUSSION
The tissue tropism of the recently described GBV-C/HGV is unknown. In vitro studies have provided evidence which indicates that this virus is able to infect and replicate in lymphoid cells and in immortalized hepatocytes (8, 30). In contrast, contradictory results have been reported with respect to the detection of GBV-C/HGV RNA of negative polarity (the putative replicative intermediate) in liver and PBMC samples from chronically infected patients (6, 15, 25). These discrepancies could be partially explained if there are GBV-C/HGV variants with different cell tropism.
In a previous study on in vitro infection of PBMC, we found that only a minor proportion of the GBV-C/HGV variants present in serum are able to infect and replicate in PBMC, which suggests the existence of lymphotropic variants (8). However, there are no data on the existence of these variants in vivo. Furthermore, it is not known whether there are any variants with tropism for the liver cells and whether all these variants are present in serum. Consequently we have studied the genomic heterogeneity and quasispecies composition of the 5′NCR of viral genomes recovered from in vitro-infected PBMC and from serum, liver, and PBMC samples from two chronically infected patients.
Regarding the in vitro-infected PBMC, we found that the predominant GBV-C/HGV 5′NCR sequence was the same in all in vitro-infected PBMC and in the PBMC isolated from the patient whose serum was used as the inoculum. This sequence was different from the predominant sequence found in the inoculum. In fact, the predominant sequence of the PBMC was not detected among the 50 clones sequenced from the inoculum, showing that this variant represents a minority of the circulating GBV-C/HGV variants. Furthermore, the phylogenetic analysis of these sequences shows that the sequences derived from the inoculum grouped together in a cluster, while those of PBMC grouped in a separate branch (bootstrap value, 96%).
Considered as a whole, all these findings show that there are GBV-C/HGV lymphotropic variants and that in vitro infection of PBMC depends on the presence of these variants in the inoculum but not on host factors, as the PBMC of the four unrelated donors were infected with similar efficiencies.
In the naturally infected patients, we have found that GBV-C/HGV in the liver and PBMC exists as a complex distribution of nonidentical but closely related genomes (quasispecies), as has been reported previously for serum (26, 36). This situation is similar to that of the hepatitis C virus (20, 23). When the frequency at which the evolutionary distances occurred in the GBV-C/HGV isolated from the three compartments was analyzed, it was seen that while sequences isolated from the serum and PBMC of both patients were distributed around a single peak, the sequences isolated from the livers of the two patients were distributed into two independent peaks. This finding shows that the GBV-C/HGV quasispecies composition is more complex in the liver than in the serum or PBMC.
When phylogenetic analysis of these sequences was performed, it was found that the sequences isolated from the serum and liver in patient 1 grouped into a single branch, while the sequences from PBMC (all except one) were grouped into a different branch. In contrast, for patient 2, all the sequences from the serum and PBMC and some of those from the liver grouped within in one branch, while the remaining liver sequences grouped into a different branch. Furthermore, for patient 1, the predominant sequence in the serum (representing 30.8% of the total serum sequences) and the predominant sequence in the liver (36.4% of all liver sequences) were identical, and this sequence was different from the predominant sequence found in PBMC (70.5% of the total PBMC sequences). A different situation occurs in patient 2, for whom the predominant sequence in the serum and the predominant sequence in PBMC (representing 18.7 and 21% of all sequences from the serum and PBMC, respectively) were identical to each other and different from the predominant sequence from the liver (20% of all liver sequences). All these results strongly support the existence of GBV-C/HGV variants with different tropism for the liver and lymphoid cells.
Finally, the fact that for patient 1 the sequences from serum and liver grouped together and that the predominant sequences were identical could indicate that the liver was the main contributor to the quasispecies composition, while in patient 2 the clustering of PBMC and serum sequences and some of the liver sequences in a single branch suggests that in this case both the liver and PBMC contribute to the quasispecies composition in the serum. However, it should be noted that both patients for whom serum, liver and PBMC samples were analyzed were also coinfected by HCV. Since HCV infects and replicates in both the liver and PBMC (9, 37), the possibility that HCV may influence the tropism of the GBV-C/HGV variants by facilitating the replication of some strains of these lines cannot be excluded. So, to definitively conclude that there are GBV-C/HGV variants with different tropism, further studies analyzing patients infected only by GBV-C/HGV should be performed. Furthermore, the samples analyzed were obtained at a single time point from each patient; therefore, changes in GBV-C/HGV quasispecies composition in the serum, liver, and PBMC over time cannot be excluded. However, the fact that GBV-C/HGV does not seem to have a high mutation rate (33) argues against this hypothesis.
In conclusion, our results have demonstrated tissue compartmentalization of GBV-C/HGV 5′NCR sequences from PBMC, liver, and serum samples obtained at the same time from chronic GBV-C/HGV carriers coinfected by HCV, suggesting the existence of GBV-C/HGV variants with different tropism. The question of the factors involved in this different tropism, as well as its pathological implications, deserves future research.
REFERENCES
- 1.Aikawa T, Sugay Y, Okamoto H. Hepatitis G infection in drug abusers with chronic hepatitis C. N Engl J Med. 1996;334:195. doi: 10.1056/NEJM199601183340316. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J, Nakatsuyi Y, Melpolder J, Wages J, Wesley R, Shih J W K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 3.Alter M J, Gallagher M, Morris T T, Moyer L A, Meeks E L, Krawczynski K, Kim J P, Margolis H S. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. doi: 10.1056/NEJM199703133361101. [DOI] [PubMed] [Google Scholar]
- 4.Berenguer M, Terrault N A, Piatak M, Yun A, Kim J P, Lau J Y N, Lake J R, Roberts J R, Ascher N, Ferrel L, Wright T. Hepatitis G virus infection in patients with hepatitis C virus infection undergoing liver transplantation. Gastroenterology. 1996;111:1569–1575. doi: 10.1016/s0016-5085(96)70019-7. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Fan X, Xu Y, Solomon H, Ramrakhiani S, Neuschwander-Tetri B A, Di Bisceglie A M. Detection of hepatitis G/GB virus-C viral RNA in liver and serum. J Med Virol. 1999;58:160–164. [PubMed] [Google Scholar]
- 7.Fiordalisi G, Zanella I, Mantero G, Bettinardi A, Stellini R, Paraninfo G, Cadeo G, Primi D. High prevalence of GB virus C infection in a group of Italian patients with hepatitis of unknown etiology. J Infect Dis. 1996;174:181–183. doi: 10.1093/infdis/174.1.181. [DOI] [PubMed] [Google Scholar]
- 8.Fogeda M, Navas S, Martín J, Casqueiro M, Rodríguez E, Arocena C, Carreño V. In vitro infection of human peripheral blood mononuclear cells by GB virus C/hepatitis G virus. J Virol. 1999;73:4052–4061. doi: 10.1128/jvi.73.5.4052-4061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong T S, Shindo M, Feinstone S M, Hoofnagle J H, Di Bisceglie A M. Detection of the replicative intermediates of hepatitis C viral RNA in liver and serum samples of patients with chronic hepatitis C. J Clin Investig. 1991;88:1058–1060. doi: 10.1172/JCI115368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried M W, Khudyakov Y E, Smallwood G A, Cong M-E, Nichols B, Diaz E, Siefert P, Gutekunst K, Gordon R D, Boyer T D, Fields H A. Hepatitis G virus co-infection in liver transplant recipients with chronic hepatitis C and non-viral chronic liver disease. Hepatology. 1997;25:1271–1275. doi: 10.1002/hep.510250536. [DOI] [PubMed] [Google Scholar]
- 11.Hadziyannis S J. The “hepatitis” G virus: biology, epidemiology and search for disease. Prog Liver Dis. 1997;14:219–245. [Google Scholar]
- 12.Ikeda M, Sugiyama K, Mizutani T, Tanaka T, Shimotohno K, Kato N. Hepatitis G virus replication in human cultured cells displaying susceptibility to hepatitis C virus infection. Biochem Biophys Res Commun. 1997;235:505–408. doi: 10.1006/bbrc.1997.6818. [DOI] [PubMed] [Google Scholar]
- 13.Kanda T, Yokosuka O, Imazeki F, Tagawa M, Ehata T, Saisho H. GB virus-C RNA in Japanese patients with hepatocellular carcinoma and cirrhosis. J Hepatol. 1997;27:464–469. doi: 10.1016/s0168-8278(97)80349-2. [DOI] [PubMed] [Google Scholar]
- 14.Kao J H, Chen P J, Lai M Y, Chen W, Liu D P, Wang J T, Shen M C, Chen D S. GB virus-C/hepatitis G virus infection in an area endemic for viral hepatitis, chronic liver disease, and liver cancer. Gastroenterology. 1997;112:1265–1270. doi: 10.1016/s0016-5085(97)70139-2. [DOI] [PubMed] [Google Scholar]
- 15.Laskus T, Radkowski M, Wang L-F, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71:7804–7806. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskus T, Radkowski M, Wang L-F, Vargas H, Rakela J. Detection of hepatitis G virus replication sites by using highly strand-specific Tth-based reverse transcriptase PCR. J Virol. 1998;72:3072–3075. doi: 10.1128/jvi.72.4.3072-3075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Linnen J, Wages J, Zhank-Kek Z-Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji I, Shih J W K, Young L, Piatak M, Jr, Hoover C, Fernandez J, Cheng S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 19.Madejón A, Fogeda M, Bartolomé J, Pardo M, González C, Cotonat T, Carreño V. GB virus C RNA in serum, liver, and peripheral blood mononuclear cells from patients with chronic hepatitis B, C, and D. Gastroenterology. 1997;113:573–578. doi: 10.1053/gast.1997.v113.pm9247478. [DOI] [PubMed] [Google Scholar]
- 20.Maggi F, Fornai C, Vatteroni M L, Giorgi M, Morrica A, Pistello M, Cammarota G, Marchi S, Ciccorossi P, Bionda A, Bendelli M. Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol. 1997;78:1521–1525. doi: 10.1099/0022-1317-78-7-1521. [DOI] [PubMed] [Google Scholar]
- 21.Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol. 1998;79:705–714. doi: 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- 22.Mushahwar I K, Zuckerman J N. Clinical implications of GB virus C. J Med Virol. 1998;56:1–3. doi: 10.1002/(sici)1096-9071(199809)56:1<1::aid-jmv1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Navas S, Martín J, Quiroga J A, Castillo I, Carreño V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page R D M. TreeView. An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Pessoa M G, Terrault N A, Detmer J, Kolberg J, Collins M, Hassoba H M, Wright T L. Quantitation of hepatitis G and C viruses in the liver: evidence that hepatitis G virus is not hepatotropic. Hepatology. 1998;27:877–880. doi: 10.1002/hep.510270335. [DOI] [PubMed] [Google Scholar]
- 26.Pickering J M, Thomas H C, Karayiannis P. Genetic diversity between hepatitis G virus isolates: analysis of nucleotide variation in the NS-3 and putative ‘core’ peptide genes. J Gen Virol. 1997;78:53–60. doi: 10.1099/0022-1317-78-1-53. [DOI] [PubMed] [Google Scholar]
- 27.Radkowski M, Wang L-F, Vargas H, Rakela J, Laskus T. Lack of evidence for GB virus C/hepatitis G virus replication in peripheral mononuclear cells. J Hepatol. 1998;28:179–183. doi: 10.1016/0168-8278(88)80002-3. [DOI] [PubMed] [Google Scholar]
- 28.Radkowski M, Wang L-F, Cianciara J, Rakela J, Laskus T. Analysis of hepatitis G virus/GB virus C quasispecies and replication sites in human subjects. Biochem Biophys Res Commun. 1999;258:296–299. doi: 10.1006/bbrc.1999.0632. [DOI] [PubMed] [Google Scholar]
- 29.Saito S, Tanaka K, Kondo M, Morita K, Kitamura T, Kiba T, Numata K, Sekihara H. Plus- and minus-stranded hepatitis G virus RNA in liver tissue and in peripheral blood mononuclear cells. Biochem Biophys Res Commun. 1997;237:288–291. doi: 10.1006/bbrc.1997.7103. [DOI] [PubMed] [Google Scholar]
- 30.Seipp S, Scheidel M, Hofmann W J, Töx U, Theilmann L, Goeser T, Kallinowski B. Hepatotropism of GB virus C (GBV-C): GBV-C replication in human hepatocytes and cells of human hepatoma cell lines. J Hepatol. 1999;30:570–579. doi: 10.1016/s0168-8278(99)80186-x. [DOI] [PubMed] [Google Scholar]
- 31.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 32.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desay S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, Van Sant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki Y, Katayama K, Fukushi S, Kageyama T, Oya A, Okamura H, Tanaka Y, Mizokami M, Gojobori T. Slow evolutionary rate of GB virus C/hepatitis G virus. J Mol Evol. 1999;48:383–389. doi: 10.1007/pl00006482. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka E, Alter J, Nakatsuji Y, Shih W-K, Kim J P, Matsumoto A, Kobayashi M, Kiyosawa K. Effect of hepatitis G virus infection on chronic hepatitis C. Ann Intern Med. 1996;125:772–773. doi: 10.7326/0003-4819-125-9-199611010-00007. [DOI] [PubMed] [Google Scholar]
- 35.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viazov S, Riffelmann M, Khoudyakov Y, Fields H, Varenholz C, Roggendorf M. Genetic heterogeneity of hepatitis G virus isolates from different parts of the world. J Gen Virol. 1997;78:577–581. doi: 10.1099/0022-1317-78-3-577. [DOI] [PubMed] [Google Scholar]
- 37.Wang J T, Sheu J C, Lin J T, Wang T H, Chen D S. Detection of replicative form of hepatitis C virus RNA in peripheral blood mononuclear cells. J Infect Dis. 1992;166:1167–1169. doi: 10.1093/infdis/166.5.1167. [DOI] [PubMed] [Google Scholar]
- 38.Wang J T, Tsai F C, Lee C-Z, Chen P-J, Sheu J C, Wang T H, Chen D S. A prospective study of transfusion-associated GB virus C infection: similar frequency but different clinical presentation compared with hepatitis C virus. Blood. 1996;88:1881–1886. [PubMed] [Google Scholar]
- 39.Yoshiba M, Okamoto H, Mishiro S. Detection of the GBV-C hepatitis virus genome in serum from patients with fulminant hepatitis of unknown aetiology. Lancet. 1995;346:1131–1132. doi: 10.1016/s0140-6736(95)91802-7. [DOI] [PubMed] [Google Scholar]