Abstract
We cloned the sequence encoding murine interleukin-4 (mIL-4), including the secretory signal, into the genome of CVB3/0, an artificially attenuated strain of coxsackievirus B3, at the junction of the capsid protein 1D and the viral protease 2Apro. Two strains of chimeric CVB3 were constructed using, in one case, identical sequences to encode 2Apro cleavage sites (CVB3/0-mIL4/47) on either side of the inserted coding sequence and, in the other case, nonidentical sequences that varied at the nucleotide level without changing the amino acid sequences (CVB3-PL2-mIL4/46). Transfection of HeLa cells yielded progeny viruses that replicated with rates similar to that of the parental CVB3/0 strain, although yields of mIL-4-expressing strains were approximately 10-fold lower than those of the parental virus. Western blot analysis of viral proteins isolated from HeLa cells inoculated with either strain of chimeric virus demonstrated that the chimeric viruses synthesized capsid protein 1D at approximately twofold-higher levels than the parental virus. mIL-4 protein was detected by enzyme-linked immunosorbent assay (ELISA) in HeLa cells inoculated with either strain of chimeric virus. Lysates of HeLa cells inoculated with either chimeric virus induced the proliferation of the mIL-4-requiring murine MC-9 cell line, demonstrating biological activity of the CVB3-expressed mIL-4. Reverse transcription (RT)-PCR analysis of viral RNA derived from sequential passaging of CVB3/0-mIL4/47 in HeLa cells demonstrated deletion of the mIL-4 coding sequence occurring by the fourth passage, while similar analysis of CVB3-PL2-mIL4/46 RNA demonstrated detection of the mIL-4 coding sequence in the virus population through 10 generations in HeLa cells. mIL-4 protein levels determined by ELISA were consistent with the stability and loss data determined by RT-PCR analysis of the passaged viral genomes. Studies of insert stability of CVB3-PL2-mIL4/46 during replication in mice showed the presence of the viral mIL-4 insert in pancreas, heart, and liver at 14 days postinfection. Comparison of the murine antibody responses to CVB3-PL2-mIL4/46 and the parental CVB3/0 strain demonstrated an increased level of CVB3-binding serum immunoglobulin G1 in mice inoculated with CVB3-PL2-mIL4/46.
The six serotypes of the group B coxsackieviruses (CVB1 to CVB6) are human enteroviruses (23). The 7,400-nucleotide-long CVB genome encodes 11 proteins in a single open reading frame (ORF). The viral proteins are processed by the two viral proteases, termed 2Apro and 3Cpro (reviewed in references 53 and 61). The CVB induce both humoral (including mucosal) and cell-mediated antiviral immunities in humans as well as in mice (7–9, 27). CVB replicate in diverse human, primate, and murine cell cultures as well as in all mice and can induce myocarditis and pancreatitis in mice, both inflammatory diseases that closely resemble the human counterparts (24, 29, 56, 59, 69). Murine and human cells can express the receptor that is used by the CVB to bind to and enter cells (CAR, or coxsackievirus adenovirus receptor); the two proteins are similar in sequence (10, 11, 13, 14, 68).
Enteroviruses can be engineered to express foreign antigenic epitopes and larger polypeptides in a variety of ways. Shortly after elucidation of the capsid structures for poliovirus type 1 (PV1) (28) and human rhinovirus type 14 (60), short antigenic peptides were expressed in exterior capsid protein loop domains of a variety of human picornaviruses (1, 5, 18, 20, 38, 43, 47, 57, 71). Because expression of oligopeptides in external capsid protein loops was beset by instability problems as well as by a limitation on the length of peptides that could be expressed, expression from within the enteroviral RNA genome was studied. Two sites have since been used successfully, at the start of translation and at the junction of the viral capsid protein 1D and the viral protease 2Apro. Both sites permit the introduction of considerably larger foreign coding sequences. Expression at the start of enteroviral translation uses the viral protease 3Cpro to cleave at an artificially engineered recognition site to release the foreign polypeptide from the viral capsid protein 1A and nascent viral polyprotein (3, 42). However, expression at the start of translation suffered from instability of inserted sequence as well as slowed viral replication (41, 46, 66). In another approach, expression of foreign polypeptides between the enteroviral capsid protein 1D and the viral 2Apro relies on 2Apro to cleave in trans after it has performed its initial autocatalytic cis cleavage directly following translation (66). Similar to expression using 3Cpro, 2Apro recognition sites are engineered between the foreign polypeptide and the viral protein to permit separation of the foreign polypeptide from the viral protein. In this way, a variety of PV-based vectors have been engineered to express antigenic polypeptides from human (HIV) and simian (SIV) immunodeficiency viruses (3, 19, 66) as well as from rotavirus, hepatitis B virus, and herpes simplex virus (41, 42, 72). A PV vector expressing SIV Gag (p17) and Env (p41) induced both T-cell-mediated and humoral (including mucosal) immunities in primates (19). An ovalbumin cytotoxic T lymphocyte epitope was expressed in a PV vector (39) as well as the antigenic urease B protein from Helicobacter pylori (50) and human interleukin-2 (IL-2) (6). Two studies have used a CVB vector to express a foreign sequence. The exterior capsid protein 1D BC loop of CVB3 was used to express seven amino acids comprising the 1D BC loop of CVB4; this chimera induced neutralizing antibodies in mice against both parental viruses (57). More recently, Höfling and colleagues expressed the antigenic L1 loop of adenovirus type 2 (Ad2) hexon protein from within the ORF of an attenuated strain of CVB3 (27). This chimera induced protective neutralizing antibodies against both Ad2 and the CVB3 vector, even in the presence of existing host anti-CVB3 immunity, suggesting the possibility of a CVB vaccine against the primary viruses that cause human myocarditis (34).
To determine whether a CVB vector could stably express an intact biologically active protein that would also have a measurable biological effect in the mouse host, we used a CVB3 vector in which to clone the sequence encoding murine IL-4 (mIL-4), including the secretory signal. The mIL-4 coding sequence was placed at the P1-P2 junction, i.e., between the capsid protein 1D and the viral protease 2Apro, employing 2Apro to cleave the cytokine from the growing viral polyprotein. Two constructs were made to test whether foreign sequences were less stable in constructs with direct as opposed to degenerate repeats encoding the 2Apro cleavage sites. In this report, we demonstrate that mIL-4 expressed from a CVB vector is biologically active and that the mIL-4 coding sequence is detected in the viral genome through 10 passages in HeLa cells and through 14 days postinoculation (p.i.) in mice. Although coexpression of mIL-4 in the virus does not increase the murine host neutralizing antiviral antibody response with a single inoculation, it does increase the extent of CVB3-binding immunoglobulin G1 (IgG1), indicating that viral expression of this cytokine is biologically functional in the experimentally inoculated mouse host.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were propagated as monolayers in minimal essential medium (MEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 50 μg of gentamicin per ml at 37°C in a humidified 5% CO2–air atmosphere. The murine mast cell line MC/9 was obtained from the American Type Culture Collection (ATCC, Manassas, Va.) and maintained as a suspension culture in MEM as described above. CVB3/0, an artificially attenuated CVB3 strain (16, 70), was derived from the infectious cDNA copy of the viral genome (16). Virus stocks were prepared by transfection of HeLa cell monolayers as described (16), counted on HeLa cells, and stored in aliquots at −74°C.
Construction of murine IL-4-expressing CVB3 strains.
We generated chimeric genomes that expressed mIL-4 (GenBank accession no. M25892) between the last encoded viral capsid protein (P1-D; hereafter called 1D) and the P2-A viral protease (hereafter called 2Apro). Flanking the foreign insert on either side are short sequences that encode the cleavage site recognized by 2Apro, the viral protease used to excise the foreign protein from the nascent viral polyprotein. The first CVB3/0-based cDNA construct that resulted in the virus CVB3/0-mIL4/47 contains a perfect 72-bp repeat (nucleotides [nt] 3275 to 3346; numbering according to GenBank accession no. M88483) encoding 24 amino acids (aa) (Fig. 1B). This construct was obtained by PCR amplification of mIL-4 nt 88 to 507 (GenBank accession no. M25892) from the mIL-4 cDNA-containing plasmid p2A-E3 (ATCC) with primers IL41 (5′-AATATGAATTCCCGGGTAATGGGTCTCAACCCCCAG) and IL42 (5′-GGCGCCCGTATTTGTCATTGTAGTGATCGAGTAATCCATTTG). These primers added a SmaI site upstream and a NarI site downstream of the mIL-4 cDNA. The SmaI-NarI digest of this amplified DNA was ligated to two restriction fragments of pCVB3-0 (16), an EcoRI-SfcI fragment (nt 2766 to 3337; all CVB3 numbering is according to GenBank accession number M88483) filled in at the SfcI cut end and a HinP1I-SpeI fragment (nt 3298 to 3838). The blunted SfcI end was ligated to the SmaI end of the amplified mIL-4 cDNA, and the NarI end was ligated to the HinP1I cut end of the second CVB3 cDNA fragment using a common overlap. The chimeric DNA was subcloned in a plasmid vector, pGem-3 (Promega, Madison, Wis.). The added sequences were designed so that when ligated with these restriction sites to the CVB3 cDNA, the mIL-4-encoding sequence would be in frame with the CVB3 ORF. The chimeric cDNA of the subclone was ligated to restriction fragments of pCVB3-0 using the EcoRI site (nt 2765) and an ApaLI site (nt 3466) to generate pCVB3/0-mIL4-47, a plasmid encoding the full-length CVB3/0 genome with an mIL-4 cDNA insert in the pSVN vector (16).
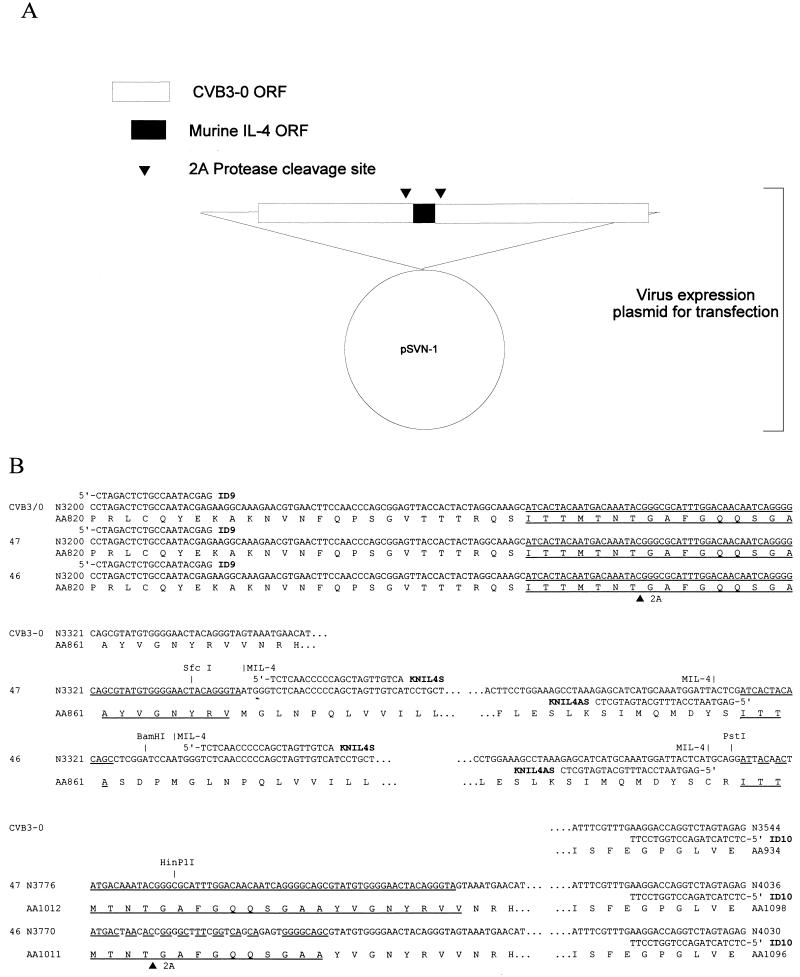
FIG. 1.
Structure of CVB3-IL-4 plasmid and cDNA sequence. (A) Outline of the plasmid construct that gives rise to the mIL-4-expressing CVB3 strains. Both plasmids encode the full ORF of mIL-4 (GenBank accession no. M25892) between the capsid coding region (P1) and the 2Apro coding region, with a repeated 2Apro cleavage site (▾) bordering the insert. (B) pCVB3/0-mIL/47 (termed 47 in the figure) has a perfect repeat of 72 bp, 24 aa; pCVB3-PL2-mIL4/46 (termed 46 in the figure) has 70% identity between the 50-nt sequence encoding a perfect 17-aa repeat 2Apro cleavage site. Due to the length of the sequence, it is presented in three contiguous segments. The CVB3 sequences of both recombinant cDNA genomes are derived from pCVB3/0 and cloned in the plasmid vector pSVN (16). The repeated sequences encoding 2Apro cleavage sites are underlined, and the restriction sites used in the construction of the plasmids are indicated. The sequence and annealing site of primers (ID9, ID10, KNIL4S, and KNIL4AS) used in the detection of chimeric and deleted genomes are indicated. N, nucleotide; AA, amino acid.
We then generated a CVB3/0-based subclone with an inserted polylinker (containing BamHI, AvrII, EcoRV, and PstI sites) flanked by a nucleotide sequence encoding 2Apro cleavage sites: the downstream sequence encoding the 2Apro cleavage site has the altered nucleotide sequence. The duplication of the encoded 2Apro cleavage site was generated by PCR amplification of pCVB3/0 cDNA with primers containing mutated nucleotide sequence with restriction sites to generate two fragments: with ID5 (5′-CGACCTGGAGCTGACGTTTGT) and ID8 (5′-CTAGGATCCGAGGCTGCCCCTGATTGTTGTCC) to generate nt 2788 to 3323 with an added BamHI site downstream, and with 2APLY (5′-CTCGGATCCTAGGATATCCTGCAGGATTACAACTATGACTAACACCGGGGCTTTCGGTCAGCAGAGTGGGGCAGCGTATGTGGGGAACTACAGG) and DI-5 (5′-GGCAGTCACAGTGATCAGG) to generate a polylinker (BamHI, AvrII, EcoRV, and PstI) and a 50-nt sequence (70% identity with the wild-type 1D/2Apro) encoding 17 aa with the 2Apro cleavage site upstream of nt 3307 to 3949. The ID5/ID8 amplified cDNA was digested with BamHI and ScaI and ligated to a pCVB3/0 AvrII-ScaI restriction fragment (nt 2035 to 2818) in the XbaI (overlap with AvrII) and BamHI sites of the pBluescript II plasmid vector (Stratagene) to generate subclone 1. The 2APOLY/DI-5 amplified cDNA was digested with BamHI and SpeI and ligated with an SpeI-ScaI restriction fragment (nt 3838 to 5138) of pCVB3/0 into a BamHI-EcoRV restriction fragment to generate pBSPL1, a subclone of pCVB3/0 from nt 2035 to 5138 containing an in-frame insert with a polylinker and the downstream-encoded 2Apro cleavage site (with nucleotide sequence altered from the wild type). Because the ORF of the insert was in frame with that of the lacZ gene encoded by pBluescript II, the plasmid was digested with NotI to linearize the DNA, and the 3′-terminal overhangs were filled in with the Klenow fragment of DNA polymerase I and then religated to generate pBSPL2.
Enzymatic amplification of mIL-4 cDNA (nt 88 to 507) from plasmid p2A-E3 (ATCC) with MIL4C (5′-GGATCCAATGGGTCTCAACCCCCAGCTAGTTGTCATCCTGCTCTTCTTTCTC) and MIL4D (5′-GAGCATGCATGAGTAATCCATTTGCATGATGCTCTTTAGGCTTTCCAG) generated DNA which was digested with BamHI and NsiI and then ligated into the BamHI and PstI sites of the pBSPL2 polylinker. A BglII-XbaI fragment of the pBSPL2-IL4 subclone was ligated into the BglII (CVB3 nt 2043) and XbaI (CVB3 nt 4948) sites of the pCVB3-0 plasmid to generate the infectious CVB3-IL4 chimeric genome pCVB3-PL2-mIL4-46.
Generation of virus stocks from plasmid-encoded cDNA.
Both pCVB3/0-mIL4/47 and pCVB3-PL2-mIL4/46 were electroporated into cells in culture using 10 μg of plasmid, 500 μg of herring sperm DNA, and 106 trypsinized HeLa cells in 1 ml of OPTI-MEM medium (Life Technologies, Gaithersburg, Md.) using a Cell-Porator apparatus (Life Technologies) set to 300 V and 330 μF. Electroporated cells were plated in MEM containing 10% FBS and 50 μg of gentamicin per ml and incubated at 37°C in a humidified 5% CO2–air atmosphere for 72 h. This culture, termed pass 1, was harvested by two cycles of freeze-thaw lysis and cleared of cellular debris by centrifugation, and 0.25 ml was used to infect 5 × 105 HeLa cells in monolayer culture to generate CVB3/0-mIL4/47 and CVB3-PL2-mIL4/46 stocks. Passages of virus were similarly performed, and the cells were then incubated at 37°C in 5% CO2 for 72 h or until greater than 95% of cells showed cytopathic effect (CPE), followed by freeze-thaw lysis and centrifugation as described above. All stocks were stored at −74°C until used.
Single-step growth curves.
The replication and yields of the chimeric viruses were compared to those of the parental strain CVB3/0 in single-step growth curves as described elsewhere (70). Briefly, 105 HeLa cells were plated per well in 24-well clusters. On the following day, cell cultures were rinsed and the medium was replaced with 0.25 ml per well of medium containing virus at a multiplicity of infection (MOI) of 10 50% tissue culture-infective doses (TCID50) per cell. After incubation at 37°C for 30 min, the cells were rinsed twice with 0.1 M NaCl and then refed with 0.25 ml of MEM. Cells were harvested by freeze-thaw lysis at various times p.i. and cleared as described above. Growth curves were determined in triplicate.
ELISA and biological activity assays for mIL-4.
Enzyme-linked immunosorbent assays (ELISAs) were performed upon cell lysates from single-step growth curves and upon lysates of 106 HeLa cells inoculated at an MOI of 20 TCID50 per cell with stocks from virus passages that were then harvested at 6 h p.i. The presence of mIL-4 protein was detected by ELISA using the Mouse IL-4 Cytoscreen Immunoassay Kit (BioSource International, Camarillo, Calif.). Briefly, virus-infected HeLa cell lysates and standards were bound to wells coated with mIL-4-specific antibody. A second biotinylated mIL-4-specific antibody was applied, which in turn was detected by streptavidin-peroxidase. Action of the peroxidase upon the substrate tetramethylbenzidine produced a chromogen measured by an EAR400AT ELISA plate reader (Labinstruments GMBH, Salzburg, Austria) at 450 nm. The reading from duplicate lysates was compared to a curve of readings of duplicate wells of standards from 3.9 to 250 pg/ml. Lysates were thawed only once prior to ELISA.
The biological activity of the mIL-4 was determined using proliferation of MC/9 murine mast cells (37) measured using a modified MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium] assay (25). Briefly, duplicate serial dilutions of lysates from cells infected with recombinant mIL-4 and uninfected (control) cells were plated with 5 × 103 MC/9 cells in Dulbecco's MEM with 116 mg of l-arginine per liter, 36 mg of l-asparagine per liter, 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids, supplemented with 6 mg of folic acid per liter, 0.05 mM 2-mercaptoethanol, 2 mM glutamine, and 10% FBS. A standard curve was derived using recombinant mIL-4 (BioSource International). Cells were incubated at 37°C for 40 h, after which MTT was added to 250 μg/ml for 2 h at 37°C. An equal volume of freshly made 50% (vol/vol) dimethyl formamide–20% (vol/vol) sodium dodecyl sulfate (SDS; pH 4.7) solution was added and incubated at 37°C overnight to lyse and dissolve the chromagen. Absorbance at 570 nm was measured, and concentration of mIL-4 was determined by comparison to the recombinant mIL-4 standard.
Western blot analysis of viral proteins in infected cells.
HeLa cells were inoculated with virus at an MOI of 50 and incubated for specific times, after which the medium was removed and the cell monolayer was lysed in reducing Laemmli buffer (36). Lysates were stored at −74°C until use. The lysates were electrophoresed in SDS-containing 14% polyacrylamide gels (Novex, San Diego, Calif.) followed by electroblotting to Immobilon-P membranes (Millipore, Bedford, Mass.). Blots were blocked for 2 h in phosphate-buffered saline (PBS) containing 5% (wt/vol) dry milk and 0.5% (vol/vol) Tween 80 (PBS-Tween) and then washed four times with PBS-Tween. Blots were then probed for 1 h with a 1:1,500 dilution of the primary polyclonal horse anti-CVB3 neutralizing antibody (ATCC), which detects CVB3 capsid protein 1D (27). The blots were washed four times as described above, then the primary antibody was detected using peroxidase-conjugated rabbit anti-horse IgG (Jackson ImmunoResearch Laboratories, West Grove, Pa.) at a dilution of 1:125,000. Detection of the capsid protein 1D was performed with an ECL+ kit and Hyperfilm (Amersham, Arlington Heights, Ill.) as directed by the supplier. A Nucleo Vision Gel Documentation System (Nucleo Tech, San Mateo, Calif.) and software (GelExpert version 3.5) were used for the analysis of the visualized-detected capsid protein on the film.
RT-PCR analysis of viral RNA.
Viral genomic RNA from chimeric virus stocks from each passage was isolated using Trizol LS reagent (Life Technology) according to manufacturer's instructions. Synthesis of cDNA was carried out in 20-μl reactions containing 20 U of SuperscriptII reverse transcriptase (Life Technologies) in 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphates (dNTPs), 1 U of RNasin (Promega) per ml, and 1 μl of specific primer (ID10 or KNIL4AS, each at 2.5 optical density units at 260 nm [ODU260]/ml) for 1 h at 42°C. Following ethanol precipitation, the cDNA was used as template in 50-μl PCRs containing 200 U of Taq polymerase (Promega) per ml and 1 μl of each oligonucleotide primer (at 2.5 ODU260/ml) with 0.2 mM dNTPs, 10 mM Tris (pH 9), 40 mM MgCl2, and 50 mM KCl with the following cycling conditions: 1 min at 95°C, 1 min at 57°C, 1 min at 72°C; 35 cycles of 30 s at 95°C, 30 s at 57°C, and 35 s at 72°C; finishing with 10 min at 72°C and chilling. The CVB3 primers ID9 (5′-CTAGACTCTGCCAATACGAG; CVB3 numbering, nt 3201 to 3220) and ID10 (5′-CTCTACTAGACCTGGTCCTT, reverse complement of CVB3 nt 3525 to 3544), which anneal to the CVB3 cDNA flanking the encoded 2Apro cleavage sites and the inserted mIL-4 cDNA, were used in reactions to amplify cDNA from viral genomes containing mIL-4 insert and deleted genomes. The DNA amplified with ID9 and ID10 from CVB3/0-mIL4/47 is 836 bp, that from CVB3-PL2-mIL4/46 is 830 bp, and that from the parental CVB3/0 is 344 bp. The mIL-4-specific primers KNIL4S (5′-TCTCAACCCCCAGCTAGTTGTCA; mIL-4 nt 93 to 115) and KNIL4AS (5′-GAGTAATCCATTTGCATGATGCTC; reverse complement of mIL-4 nt 483 to 506) were used to amplify DNA from within the CVB3-encoded mIL-4 sequences. The amplified DNA is 414 bp. Amplimers were analyzed on 1.5% agarose gels, and DNA migrating at the appropriate size was purified for sequencing verification.
Inoculation of mice and serological assessment of host response.
C3H/HeJ male mice 3 to 4 weeks of age (Jackson Laboratories, Bar Harbor, Maine) were obtained, rested for 3 days, and then inoculated intraperitoneally with 5 × 105 TCID50 of either CVB3-PL2-mIL4/46 or CVB3/0 in 0.1 ml of unsupplemented medium as the diluent. Mice were sacrificed on days 1, 2, 4, and 14 p.i. Sera were isolated from clotted blood, aliquoted, and stored at −74°C. Neutralizing antibody titers against CVB3 were measured in a standed CPE reduction assay (27). Virus-binding antibody was measured by a modification of a protocol described elsewhere (27). Briefly, the ELISA was set up using 96-well flat-bottomed plates coated with a 1:1,000 dilution of hyperimmune horse anti-CVB3 serum (ATCC). CVB3 was then bound to the coated plates, followed by experimental mouse sera. IgG1 and IgG2a subtypes were assayed using reagents of a commercially available ELISA (mouse hybridoma subtyping kit; Roche Molecular Biochemicals, Indianapolis, Ind.). Relative amounts of CVB3-binding IgG1 or IgG2a in equal amounts of mouse serum were determined by the action of bound peroxidase-conjugated goat anti-mouse IgG1 or anti-mouse IgG2a upon 2,2′-azino-di-(3-ethylbenzthiazoline sulfonate) (ABTS) measured by absorbance at 405 nm.
RT-PCR evaluation of CVB3-PL2-mIL4/46 insert stability during replication in mice.
Heart, pancreas, and liver were taken on days 1, 2, 4, and 14 p.i. from C3H/HeJ mice inoculated as described above with CVB3-PL2-mIL4/46. Frozen tissues were homogenized in Trizol LS (Life Technologies) followed by purification of total RNA per the manufacturer's recommendations. Complementary DNA was synthesized from RNA as described above, and PCR was performed using the CVB3-specific primers ID9 and ID10 on 5% of the total cDNA yield as described above. These CVB3-based primers do not amplify cDNA from uninfected mouse tissues.
RESULTS
Construction of CVB3/0-based strains that express mIL-4.
Two CVB3 cDNA genomes encoding mIL-4 between the capsid coding (P1) region and the 2Apro coding region were constructed (Fig. 1A). In the first construct, pCVB3/0-mIL4/47, the mIL-4 insert was flanked by a perfect repeat of 72 nt encoding 24 aa (P7 to P17′, notations as per Schecter and Berger [62]) (Fig. 1B). To test whether the rate of loss of insert, presumably by recombination due to the perfect duplications, could be minimized, the vector genome was redesigned (based on the attenuated pCVB3-0 cDNA) with a polylinker followed by a sequence encoding 16 aa of the 2Apro cleavage site (P7 to P10′) (62) downstream of the wild-type sequence encoding the 1D/2Apro cleavage site (Fig. 1B). The repeated 2Apro cleavage site is encoded by 50 nt with 70% identity to the upstream nucleotide sequence. The polylinker restriction sites were then used to insert mIL-4 cDNA that had been enzymatically amplified to contain the appropriate restriction sites in frame with the CVB3 ORF.
Characterization of mIL-4-expressing CVB3 strains in cell culture.
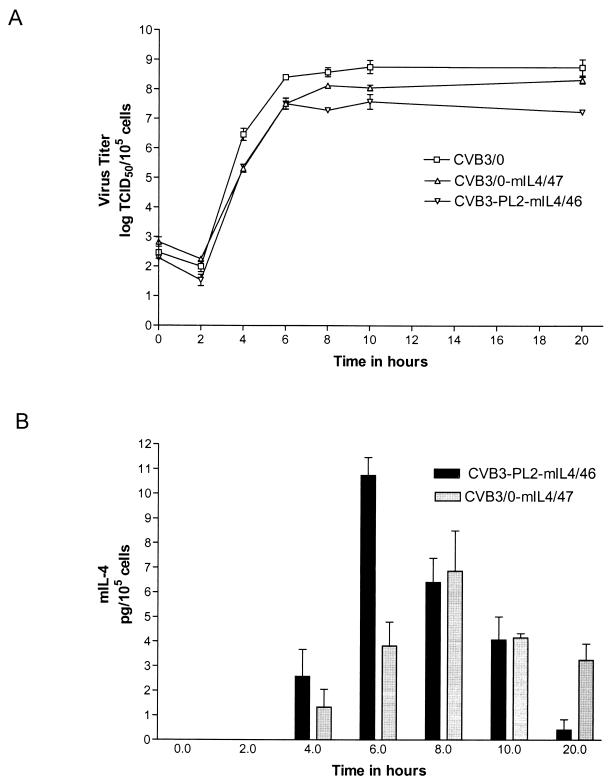
The strains CVB3/0-mIL4/47 and CVB3-PL2-mIL4/46 were assayed for replication rate and yield in HeLa cells using one-step growth curves, using the parental strain CVB3/0 as the control (Fig. 2A). Both chimeric CVB3 strains replicated at rates similar to or slightly slower than that of the parental virus strain. CVB3-PL2-mIL4/46 yielded approximately 10-fold fewer virus than did CVB3/0-mIL4/47 when assayed between 8 and 20 h p.i. By 8 to 9 h p.i. under these conditions, HeLa cell monolayers show nearly complete CPE. To determine whether either chimeric CVB3 strain were expressing mIL-4 protein by ELISA, we assayed cleared lysates of HeLa cells that had previously been inoculated with CVB3/0-mIL4/47 or CVB3-PL2-mIL4/46. No mIL-4 protein was detected in uninoculated or CVB3/0-inoculated HeLa cells; according to the manufacturer, the ELISA used in this assay has no cross-reactivity with human IL-4 at up to 5,000 pg/ml. mIL-4 was detected in both cultures, with a peak of mIL-4 production occurring at approximately 6 h p.i. for CVB3-PL2-mIL4/46 and 8 h p.i. for CVB3/0-mIL4/47 (Fig. 2B). The chimeric virus CVB3-PL2-mIL4/46 expressed 1.5-fold-more mIL-4 (11 pg/105 cells) at its peak of production at 6 h p.i. than the did the CVB3/0-mIL4/47 chimeric strain (7 pg/105 cells) at 8 h p.i. (P < 0.05).
FIG. 2.
Characterization of mIL-4-expressing CVB3 strains in cell culture. (A) Single-step growth curves of the viruses on HeLa cells. (B) Concentration of mIL-4 in lysates as measured by ELISA. HeLa cells were inoculated with the parental virus CVB3/0 or the chimeric strains CVB3/0-mIL4-47 and CVB3-PL2-mIL4-46. Inoculated cell cultures were harvested by freeze-thaw lysis at the stated times. Error bars indicate standard error of the mean.
As MC/9 cells proliferate as a function of the concentration of mIL-4, the biological activity of the CVB3-expressed mIL-4 in HeLa cell lysates harvested at 6 h p.i. was determined on MC/9 cell cultures. Supernatants from uninoculated HeLa cultures or control CVB3/0-inoculated HeLa cultures did not stimulate MC/9 growth, consistent with the absence of detectable mIL-4 in these cultures by ELISA. Specific activities of the CVB3-expressed mIL-4 were derived from the ELISA protein concentration data and the biological activity data measured on MC/9 cells. The CVB3-expressed mIL-4 specific activities were between 0.13 and 0.61 U/pg compared to a standard curve using purified recombinant mIL-4 as determined by ELISA and did not vary significantly as a function of the recombinant CVB3 strain used; however, the specific activity of purified recombinant IL-4 is approximately 100-fold greater (55) than that expressed by either CVB3 strain. These data demonstrated that both chimeric CVB3 strains efficiently produced biologically active mIL-4. Although CVB3/0-mIL4/47 replicated to higher titers than CVB3-PL2-mIL4/46, ELISA quantitation of mIL-4 production in the growth curve demonstrated that CVB3-PL2-mIL4/46 produced mIL-4 earlier and at higher yields than CVB3/0-mIL4/47.
Western blot analysis of CVB3 capsid protein 1D processing.
Efficient replication of either chimeric virus requires efficient cleavage by 2Apro of the mIL-4 protein from the nascent CVB3 polyprotein. Although the processing of the mIL-4 protein was inferred to be efficient from the similar growth curves of the chimeric and parental viruses, we used Western blots to assay the relative amounts of capsid protein 1D in HeLa cells inoculated with either of the chimeric strains or with the parental virus. Analysis of inoculated cells isolated between 5 and 9 h p.i. demonstrated that CVB3/0-mIL4/47- and CVB3-PL2-mIL4/46-inoculated cells yielded slightly more capsid protein 1D than cells inoculated with the parental CVB3/0 virus (Fig. 3). Comparison of the 1D yields in CVB3/0-infected HeLa cells showed approximately twofold less 1D protein than in cells inoculated with either of the chimeric virus strains. The difference in viral protein production was not inferred from the single-step growth curves (Fig. 2A), in which chimeric strains generated lower yields of virus than the control. We did not observe evidence of a larger capsid protein 1D band, such as might be expected from a fusion protein consisting of protein 1D and mIL-4 had the 2Apro been significantly delayed in cleaving at that site and as has been observed for another, similarly constructed CVB3 chimeric strain (27). We could not detect mIL-4 on Western blots despite repeated attempts with a commercially available anti-mIL-4 antibody (R&D Systems, Minneapolis, Minn.).
FIG. 3.
Western blot analysis of viral proteins in inoculated HeLa cells. Proteins were harvested from HeLa cells inoculated with the parental CVB3/0 or either of the chimeric strains by lysing the cell monolayers in Laemmli loading buffer. Lysates were prepared at the times shown (in hours p.i.) and electrophoresed in SDS–14% PAGE followed by electroblotting. Capsid protein 1D was detected with a polyclonal horse anti-CVB3 neutralizing antibody followed by peroxidase-conjugated rabbit anti-horse IgG. C, purified CVB3/0; TC, uninfected HeLa cell lysate. 1D, location of the detected capsid protein at 34 kDa is shown. Lane M, size markers (in kilodaltons).
Stability of the mIL-4 coding sequence in the chimeric CVB3 strains.
Although foreign polypeptides have been successfully expressed at the site between capsid protein 1D and the viral protease 2Apro in PV vectors, partial or complete deletions of the foreign insert have been reported. Andino and colleagues ascribed a degree of insert instability in a PV vector to the presence of identical repeat sequences encoding the 2Apro cleavage sites, an instability that could be largely overcome by the use of nonidentical coding sequences that presumably slowed the rate of homologous recombination (66). We examined the stability of the mIL-4 coding sequence in the presence of direct repeats encoding the 2Apro cleavage sites (in strain CVB3/0-mIL4/47) versus 2Apro cleavage sites that had been engineered to be 30% nonidentical without changing the amino acid sequence (strain CVB3-PL2-mIL4/46).
Beginning with transfection stocks, both chimeric viruses were passaged on HeLa cells. Virus stocks made at each passage were used to prepare viral RNA for RT-PCR of the region containing the insert. Using PCR primers located inside the mIL-4 coding sequence, RT-PCR analysis of RNA from CVB3-PL2-mIL4/46 demonstrated maintenance of the mIL-4 coding sequence through 10 passages in HeLa cells (Fig. 4C). In contrast, we observed rapid loss of the mIL-4 coding sequence in CVB3/0-mIL4/47 (Fig. 4D) beginning with the second pass and an apparently complete loss by pass 4 to 5. Examining all viral genomes in the population using PCR primers located outside the insert site, we observed that a smaller amplimer, consistent with a deleted genome, began to appear in CVB3-PL2-mIL4/46 passages 5 and 6 (Fig. 4A). Sequence analysis of this band verified it as from a CVB3 genome in which the insert had been lost through recombination in the sequences encoding the repeated 2Apro, encoding a protein sequence identical to CVB3/0; the deletion returned the CVB3 genome to the wild-type sequence and maintained the ORF of the virus, suggesting that the deleted genome was viable (data not shown). Although this deleted genome was present in the population, it did not become the primary population in the passage of CVB3-PL2-mIL4/46 until pass 8 or 9. These data concur with those (Fig. 4C) showing that the mIL-4-specific amplimer is present through pass 10. However, in a similar analysis of the passaged CVB3/0-mIL4/47, the deleted genome appears and becomes the major population by the fourth passage of the virus (Fig. 4B). This rapid loss of the CVB3/0-mIL4/47 mIL-4 coding sequence and the rapid dominance of the deleted viral genome is consistent with the loss of mIL-4-specific signal at pass 4 or 5 in Fig. 4D.
FIG. 4.
RT-PCR analysis of mIL-4 insert stability following passage of chimeric viruses in HeLa cells. Amplimers were obtained and analyzed on 1.5% agarose gels following RT-PCR of viral RNA isolated from sequentially passaged CVB3-PL2-mIL4/46 (A and C) and CVB3/0-mIL4/47 (B and D). ELISA detection of mIL-4 protein is also shown for comparison (C and D). Viruses were passaged 10 times in HeLa cells as described in the text. Amplification of the region containing the insert in the viral RNA populations was accomplished with the primers ID9 and ID10 (A and B); the intact mIL-4 insert was detected as an 836-bp (or 830-bp) product and as a 344-bp product when the insert was deleted. Specific detection of the mIL-4-containing insert was accomplished using primers KNIL4S and KNIL4AS (C and D); the presence intact insert was detected as a 414-bp product, whereas deletion of the product resulted in no amplimer produced. mIL-4 ELISA (C and D), concentration of mIL-4 protein as detected by ELISA for each pass (5 × 105 HeLa cells infected at an MOI of ≥10 with designated viral passage stock, harvested at 6 h p.i.). Specific activities of mIL-4 ranged between 0.13 and 0.61 U/pg. Lanes P2 to P10, results from passages 2 to 10, respectively; lane M, 100-bp ladder (Life Technologies); lane PC, positive control amplification using pCVB3-PL2-mIL4/46 DNA; lane −, negative control (no DNA added) amplification; lane 0, PCR of CVB3/0 virus stock. Gel was stained with Cyber Green (FMC, Philadelphia, Pa.); images were captured using a Nucleo Vision gel documentation system (Nucleo Tech Corp.).
To some degree, the discrepancy between the two RT-PCR assays of the viral genomes (the greater signal with the mIL-4-specific RT-PCR than of the undeleted genome with ID9 and ID10) can be explained by competition for amplification of the multiple products generated by priming with ID9 and ID10. Amplification of mixes of pCVB3-0 and pCVB3/-mIL4/47 at different ratios have demonstrated that the shorter product is preferentially amplified (data not shown), an observation that has also been made by others (46). Furthermore, expression of mIL-4 protein in lysates from cells infected with serially passaged virus stocks is consistent with the loss of the mIL-4 sequences and the dominance of the deleted genome.
The mIL-4 protein concentration falls sharply in lysates of cells infected with passage 2 to 3 of CVB3/0-mIL4/47 and is not detectable in those infected with passage 5, in which mIL-4 cDNA is also undetectable by RT-PCR (Fig. 4D). The mIL-4 concentration remains constant in lysates of cells infected with passages 2 to 7 of CVB3-PL2-mIL4/46 but begins to decrease with passages 8 and 9, virus populations in which a significant proportion of the deleted genome is detected (Fig. 4C). Although reversion to the wild-type genome through loss of the mIL-4 coding sequence suggests that there is a cost to the increase in the length of the polyprotein and to the requirement for processing by 2Apro in trans, the slower loss of the mIL-4-encoding genomes in the passage of CVB3-PL2-mIL4/46 demonstrated that alteration of the sequence encoding one of the two 2Apro cleavage sites correlates with a reduced rate at which the reversion occurs.
Stability of the mIL-4 coding sequence in CVB3-PL2-mIL4/46 as a function of replication in the murine host.
To determine the stability of the mIL-4 coding insert in the more stable chimeric virus CVB3-PL2-mIL4/46 during replication in the murine host, C3H/HeJ mice were inoculated with the virus. Mice were sacrificed at 1, 2, 4, and 14 days p.i., and liver, heart, and pancreas were assayed for the presence of CVB3 RNA containing the mIL-4 insert by RT-PCR (Fig. 5). In this assay using RNA from mouse tissues, as opposed to the assay in HeLa cells, which do not transcribe a IL-4 mRNA, we used primers in the CVB3 genome on either side of the insert site (ID9 and ID10) for one PCR. Assays of heart, pancreas, liver, serum, and fecal contents from five individual mice at days 1 to 4 p.i. showed that mixed populations of full-length and deleted genomes were present (data not shown). Although deleted genomes were present in these tissues within 24 h of infection, the mIL-4-encoding genomes remained detectable in pancreas, liver, and heart as long as 14 days p.i. (Fig. 5).
FIG. 5.
Detection of CVB3-associated mIL-4 insert at 14 days p.i. in mice. Amplimers were obtained and analyzed on 1.5% agarose gels following RT-PCR of viral RNA in total tissue RNA isolated from hearts (A), pancreas (B), and liver (C) of mice inoculated 14 days previously with 5 × 105 TCID50 of CVB3-PL2-mIL4/46. cDNA was made from RNA of tissues from five individual mice. Amplification of the region containing the insert in the viral RNA populations was accomplished using the CVB3-based primers ID9 and ID10. The intact mIL-4 insert was detected as an 830-bp product and as a 344-bp product when the insert was deleted. Lanes 1 to 5, PCR from cDNA of individual mouse tissues; lane 46, PCR from CVB3-PL2-mIL4/46 virus stock; lane 0, PCR from CVB3/0 virus stock; lane M, 100-bp ladder (Life Technologies). Gel was stained with Cyber Green (FMC) and images were captured using a Nucleo Vision gel documentation system (Nucleo Tech Corp.).
Murine antibody response to mIL-4 expressed in CVB3-PL2-mIL4/46.
In order to determine whether mIL-4 expressed by the recombinant virus CVB3-PL2-mIL4/46 induced a measurable biological effect in mice, we assayed both titers of antiviral antibodies and levels of IgG subtypes as a function of inoculation with the chimeric versus the parental virus strain. Although mice inoculated 14 days previously with either CVB3/0 or CVB3-PL2-mIL4/46 demonstrated detectable levels of serum anti-CVB3 neutralizing antibodies, mice inoculated with CVB3/0 neutralized CVB3 at dilutions of between 1:64 and 1:512, while anti-CVB3 antibody titers in sera from mice inoculated with the CVB3-PL2-mIL4/46 chimeric strain were lower at 1:32 to 1:128.
Sera were also assayed by ELISA for IgG1 and IgG2a isotypes of CVB3-binding antibody, comparing them to sera from control mice not inoculated with CVB3 (Fig. 6). Sera from mice inoculated with CVB3/0 had an average 5-fold increase in CVB3-binding IgG1 and 37-fold increase in CVB3-binding IgG2a compared to sera from uninoculated control mice (Fig. 6A). Mice inoculated with CVB3-PL2-mIL4/46 averaged a 15-fold increase in CVB3-binding IgG1 (three times more than that induced by CVB/0) and a 30-fold increase in CVB3-binding IgG2a. Of the five sera from mice inoculated with the mIL-4-expressing CVB3-PL2-mIL4/46 strain, two showed no significant increase in CVB3-binding IgG1 compared to CVB3-binding IgG1 from CVB3/0-inoculated mice. However, sera from the other three mice contained a 21- to 24-fold increase in the concentration of IgG1 over control mice sera, compared to the average 5-fold increase in the sera from CVB3/0-inoculated mice, a significant difference (P < 0.05). No significant difference was observed in the levels of CVB3-binding IgG2a from either group of CVB3-inoculated mice (Fig. 6B).
FIG. 6.
Increase in CVB3-binding IgG1 in mice inoculated with CVB3-PL2-mIL4/46 compared to CVB3/0. Quantity of CVB3-binding IgG1 (A) or IgG2a (B) in individual mouse serum samples from mice inoculated 14 days previously with CVB3-PL2-mIL4/46 (46) or CVB3-0 (0) or cell culture medium (control). CVB3-binding isotype was detected by ELISA using peroxidase activity of bound goat anti-mouse IgG1 or bound goat anti-mouse IgG2a and quantitated by absorbance at 405 nm.
DISCUSSION
We describe here the first reported cloning and expression of a biologically active cytokine from within the genome of an artificially attenuated strain of CVB3. The CVB3 protease 2Apro cleaves mIL-4 from the nascent CVB3 polyprotein efficiently, as judged by similarity of growth curve kinetics and virus yields between chimeric strains and the parental virus, as well as by Western blot analysis. The mIL-4 that is synthesized by the chimeric CVB3 strains is biologically active, promoting the proliferation of the mIL-4-requiring cell line MC/9. Using two variations of chimeric virus that differ in the sequences encoding the 2Apro recognition sites, we demonstrate that significantly greater stability of the mIL-4 coding sequence in the CVB3 genome is associated with nonidentical sequences encoding the 2Apro cleavage recognition sites. Indeed, inoculation of mice with a single dose of CVB3-PL2-mIL4/46 generates viral populations in heart, liver, and pancreas which still contain the mIL-4 coding sequence at 14 days p.i. Mice inoculated with the more stable of the two chimeric strains have higher levels of IgG1 than mice inoculated with the parental CVB3 strain or uninoculated mice, demonstrating that expression of mIL-4 by an enterovirus has a measurable impact on the host immune system.
Expression of foreign protein sequences in enteroviruses has evolved from the expression of oligopeptide sequences in external capsid protein loops (4, 5, 18, 20, 22, 31, 35, 38, 40, 43, 47, 57, 58, 74) to the expression of polypeptides and proteins from within the enteroviral ORF. Chimeric PV have been made with inserted sequences encoding foreign antigens placed at the 5′ end of the PV ORF (3, 42) and between the P1 and P2 region (41, 66). Constructs with inserts at the start of the viral ORF produced temperature-sensitive virus (41, 66), which rapidly generated deleted viral genomes in which most or part of the foreign insert was lost; the virus population in passage rapidly became dominated by these viruses (46), which entailed a concomitant loss of potential for generating an immune response to the foreign antigen (66). Inserted sequences in PV-based chimerae at the P1-P2 junction utilizing 2Apro to process the foreign protein from the PV polyprotein were more viable and not temperature sensitive, although inserts were deleted rapidly by recombination at the direct repeat of sequence encoding the 2Apro cleavage site (66). When one of these repeat sequences encoding the 2Apro recognition site was mutated to decrease the percent nucleotide identity, the stability of the recombinant viral genome in passage was substantially increased (66). Work presented here with two CVB3/mIL-4 chimeric strains confirms and extends these stability data to CVB, showing that enhanced foreign insert stability is associated with the use of dissimilar sequences to encode the 2Apro cleavage sites. PV-based chimerae with inserts of up to 700 nt (resulting in genomes 110% of the wild type in size) have been retained in the viral population for at least three passages (66), but work with diverse antigens demonstrates that stability is to some extent dependent upon the sequence inserted in addition to the increase in genome size. By comparison, the mIL-4 coding sequences used in the current study added 420 nt to these genomes (106% of wild-type size) and were stably maintained through 10 generations in cell culture and were still detected at 14 days p.i. in mice. Creation of a vaccinia virus-based helper packaging system permitted the production of larger chimeric PV genomes in which the capsid coding region could be replaced with entire HIV-1 proteins or with IL-2 (6, 54); similar work using CVB3 capsid proteins donated in trans from a plasmid vector have been reported (32). Such enteroviruses lacking the ability to produce infectious virus are limited to a single round of replication, potentially limiting their use as a vector for immunomodulatory proteins such as ILs.
We noted that while both chimeric CVB3 strains in this study replicated to 5- to 10-fold lower levels than the parental CVB3/0 virus, these strains also demonstrated an approximately 2-fold increase in production of viral proteins, as judged from detection of the viral capsid protein 1D on Western blots. We hypothesize that this effect may be due to somewhat slowed processing of the viral polyprotein at the 1D carboxy terminus because of the change from a cis to a trans cleavage by 2Apro in the chimeric constructs. In turn, slower capsid protein processing could result in slowed encapsidation of the viral mRNA. Slowed encapsidation might then be reflected as lowered production of infectious virus and would increase the amount of viral mRNA potentially available for translation, as only newly replicated RNA is encapsidated (51). We have observed a similar but more pronounced effect of delayed processing at the 1D carboxy terminus in another chimeric CVB3 genome that encodes an Ad antigenic polypeptide (27). In this CVB3-Ad chimera, the impaired 2Apro processing resulted not only in greater amounts of viral protein translated earlier in the replication cycle than in the control parental virus-infected cells, but in nearly equimolar levels of a fusion protein consisting of the capsid protein 1D and the Ad polypeptide as well. We did not detect this type of fusion product in the CVB3/mIL-4 strains characterized in this report. The overproduction of mIL-4 from CVB3-PL2-mIL4/46 in comparison to CVB3/0-mIL4/47 correlated with a slight decrease in virus yield from CVB3-PL2-mIL4/46-infected cells compared to CVB3/0-mIL4/47-infected cells. Although these two viruses are very similar, there is a decrease in the size of the repeated amino acid sequence containing the 2Apro cleavage site from 24 aa in CVB3/0-mIL4/47 to 17 aa in CVB3-PL2-mIL4/46. Studies of enterovirus and rhinovirus 2Apro cleavage specificities have indicated that the amino acids from P8 to P1′ have the most importance for proteolytic processing (26, 64; reviewed in reference 61). However, it is possible that the in vitro experiments which defined the importance of P8 to P1′ for cleavage would not detect slight effects upon cleavage from neighboring sequence. Indeed, we did not detect fusion proteins consisting of CVB3 capsid protein 1D and mIL-4 in cell lysates. Slight effects on protein processing can be amplified by the interdependence of translation, RNA replication, and encapsidation of the picornavirus replication cycle, as only newly synthesized viral RNA is packaged (51) and only translatable RNAs are replicated (49). This suggests that a decrease in packaging due to a slightly slowed processing at the carboxy terminus of 1D could result in enhanced viral protein production and decreased virus yield, even when decreased processing at the 1D carboxy terminus is too small to detect in Western blots. We are investigating the connection between delayed processing and overproduction of viral proteins using inserts for which more sensitive detection methods exist.
The delayed processing and overexpression effect that we have observed would also attenuate the chimeric virus for replication to some extent and may also help to explain, at least in part, the increase in the viral population of deleted genomes as the virus strain is passaged in cell culture. Minimized identity of the sequence encoding the 2Apro cleavage site in CVB3-PL2-mIL4/46 correlated with increased stability of the mIL-4 coding sequence, and we speculate that the decrease in identity of the repeated sequence decreases the overall rate of homologous recombination, similar to results reported in PV systems (66). Although by passages 8 and 9, the CVB3-PL2-mIL4/46 virus population contained deleted genomes, the chimeric genomes were still present and expressing mIL-4 upon infection of cells.
Although we demonstrated that CVB3-PL2-mIL4/46 retained the mIL-4 coding sequence for days following inoculation and during replication in mice, we observed no potentiation of the murine humoral response against the CVB3 vector in terms of an increase in antiviral neutralizing antibody titers. Although the chimeric strain induced a robust host immune response against CVB3, the lower level of anti-CVB3 antibodies observed may be due to lower titers of virus replicating in the mice. The responses of the T helper cells to antigens have been characterized as Th1, Th2, or a predifferentiation state, Th0 (45; reviewed in reference 44). Characteristic Th1 immune responses are cell-mediated immune response include the production of IL-2 and gamma interferon, and promote the production of cytotoxic T lymphocytes and switching of antibody isotypes to IgG2a. Th2 responses promote humoral or allergic immunity, generate a cytokine profile which includes IL-4, IL-6, IL-9, and IL-10, and promote switching to IgG1 and IgE. Expression of IL-4 can promote the establishment of Th2-type immune responses (65). As pathogenic Th1 immune responses have been noted both in myocarditis (30, 33, 63) and in insulin-dependent diabetes myelitis (reviewed in reference 67), treatment with cytokines has been used to help alter immune responses: treatment with IL-4 and IL-10 has been used experimentally to decrease myocarditis and diabetes (12, 48) and can be used to enhance recruitment of T helper cells to an appropriate response or to suppress the effects of a pathogenic response (reviewed in reference 52). Our observation of an increase in CVB3-binding IgG1 in CVB3-PL2-mIL4/46-infected mice relative to mice inoculated with the parental CVB3 strain indicates that mIL-4 encoded in the viral genome is capable of impacting the murine immune response. As the cytokine is also produced in cells infected with the virus, the effect on the immune response to the virus is that most likely to be changed by the cytokine. This occurs despite low levels of neutralizing antibody in these mice. The CVB3-PL2-mIL4/46 virus replicates to 10-fold lower titers than the parental virus strain in cell culture, and it is also attenuated for replication relative to the parental strain in mice (data not shown). That it generated a significant immune response and increased Th2-type isotype switching despite this attenuation following a single inoculation of virus demonstrates the ability of this vector to deliver a biologically active molecule. Increased biological effect may require increases in dosage or repeated inoculations of such viruses which are attenuated for replication. We are pursuing these questions as well as the extent of expression of CVB3-expressed mIL-4 in tissues using mIL-4 knockout mice.
We cloned and expressed the entire ORF encoding mIL-4 (73), a sequence that also encodes the secretory signal. We did not observe secretion of mIL-4 into the medium in HeLa cells infected with CVB3-PL2-mIL4/46 that could not also be accounted for by cell lysis due to the infection; biologically active mIL-4 was retained within infected cells. The lack of secretion of mIL-4 may be due to effects of this enterovirus upon the secretory pathway in HeLa cells; the closely related PV have been shown to rapidly shut off this process (17, 21). Although signal sequences in the context of dicistronic PV have been shown to have a lethal effect on chimeras (38, 39), other PV chimeras with signal sequences inserted at the P1-P2 junction have shown secretion, as measured by glycosylation of encoded foreign proteins expressed in cell culture (66). The ability of PV replicons to also express glycosylated proteins from downstream in the ORF (2) suggests that a secretory sequence at the start of an ORF may be more attenuating than those imbedded within a polyprotein. Although it may be possible to have the encoded foreign protein enter the endoplasmic reticulum, the activity of the CVB3-expressed mIL-4 is probably not dependent upon secretion. Biologic effect of mIL-4 in mice alone is not sufficient to infer secretion; the encoded IL-4 (fused with P1 proteins or processed by 2Apro) would be released upon cell lysis, and fusion proteins incorporating IL-4 have been shown to promote the immunogenicity of proteins (44). In this work we have shown an effect on the immune response of the murine host to the vector (CVB3) of the encoded mIL-4. Further work will demonstrate the extent to which the encoded ILs affect other immune responses in infected hosts.
ACKNOWLEDGMENTS
We thank the Barrick family, the Jurgensmier family, E. Barnett, and M. Guthrie for generous donations in support of this work in memory of Mary Barrick, Jason Jurgensmier, Sharon Ann Watson, and Richie, respectively.
This work was also supported in part by grants from the USPHS (N.M.C.), the American Heart Association (N.M.C. and S.T.), and the Technology Advancement Group at UNMC (N.M.C. and S.T.).
REFERENCES
- 1.Altmeyer R, Murdin A D, Harber J J, Wimmer E. Construction and characterization of a poliovirus/rhinovirus antigenic hybrid. Virology. 1991;184:636–644. doi: 10.1016/0042-6822(91)90433-c. [DOI] [PubMed] [Google Scholar]
- 2.Anderson M, Porter D, Fultz P, Morrow C. Poliovirus replicons that express the gag or the envelope surface protein of simian immunodeficiency virus SIV (smm) PBj14. Virology. 1996;219:140–149. doi: 10.1006/viro.1996.0231. [DOI] [PubMed] [Google Scholar]
- 3.Andino R, Silvera D, Suggett S D, Achaso P L, Miller C J, Baltimore D, Feinberg M. Engineering poliovirus as a vaccine vector for the expression of diverse antigens. Science. 1994;265:1448–1451. doi: 10.1126/science.8073288. [DOI] [PubMed] [Google Scholar]
- 4.Arnold G F, Resnick D, Li Y, Zhang A, Smith A D, Geisler S C, Jacobo-Molina A, Lee W, Webster R G, Arnold E. Design and construction of rhinovirus chimeras incorporating immunogens from polio, influenza, and human immunodeficiency viruses. Virology. 1994;198:703–708. doi: 10.1006/viro.1994.1082. [DOI] [PubMed] [Google Scholar]
- 5.Arnold G F, Resnick D A, Smith A D, Geisler S C, Holmes A K, Arnold E. Chimeric rhinoviruses as tools for vaccine development and characterization of protein epitopes. Intervirology. 1996;39:72–78. doi: 10.1159/000150477. [DOI] [PubMed] [Google Scholar]
- 6.Basak S, McPherson S, Kang S, Collawn J F, Morrow C D. Construction and characterization of encapsidated poliovirus replicons that express biologically active murine interleukin 2. J Interferon Cytokine Res. 1998;18:305–313. doi: 10.1089/jir.1998.18.305. [DOI] [PubMed] [Google Scholar]
- 7.Beck M, Chapman N, McManus B, Mullican J, Tracy S. Secondary enterovirus infection in the murine model of myocarditis: pathologic and immunologic aspects. Am J Pathol. 1990;136:669–681. [PMC free article] [PubMed] [Google Scholar]
- 8.Beck M, Tracy S. Murine cell-mediated immune response recognizes an enterovirus group-specific antigen(s) J Virol. 1989;63:4148–4156. doi: 10.1128/jvi.63.10.4148-4156.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck M, Tracy S. Evidence for a group-specific enteroviral antigen(s) recognized by human T cells. J Clin Microbiol. 1990;28:1822–1827. doi: 10.1128/jcm.28.8.1822-1827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 11.Bergelson J M, Krithivas A, Celi L, Droguett G, Horwitz M S, Wickham T, Crowell R L, Finberg R W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron M J, Arreaza G A, Zucker P, Chensue S W, Strieter R M, Chakrabati S, Delovitch T L. IL4 prevents insulitis and insulin dependent diabetes mellitus in nonobese diabetic mice by potentiation of regulatory T helper 2 cell function. J Immunol. 1997;159:4686–4692. [PubMed] [Google Scholar]
- 13.Carson S, Chapman N, Tracy S. Purification of the putative coxsackievirus B receptor from HeLa cells. Biochem Biophys Res Commun. 1997;233:325–328. doi: 10.1006/bbrc.1997.6449. [DOI] [PubMed] [Google Scholar]
- 14.Carson S, Hobbs J T, Tracy S, Chapman N. Expression of the coxsackievirus and adenovirus receptor in cultured human umbilical vein endothelial cells: regulation in response to cell density. J Virol. 1999;73:7077–7079. doi: 10.1128/jvi.73.8.7077-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman N, Romero J, Pallansch M, Tracy S. Sites other than nucleotide 234 determine cardiovirulence in natural isolates of coxsackievirus B3. J Med Virol. 1997;52:258–261. [PubMed] [Google Scholar]
- 16.Chapman N, Tu Z, Tracy S, Gauntt C. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3 strain: its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch Virol. 1994;135:115–130. doi: 10.1007/BF01309769. [DOI] [PubMed] [Google Scholar]
- 17.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 18.Colbere-Garapin F, Christodoulou C, Crainic R, Garapin A C, Candrea A. Addition of a foreign oligopeptide to the major capsid protein of poliovirus. Proc Natl Acad Sci USA. 1988;85:8668–8672. doi: 10.1073/pnas.85.22.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crotty S, Lohman B L, Lu F S, Tang S, Miller C J, Andino R. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: stimulation of humoral, mucosal, and cellular immunity. J Virol. 1999;73:9485–9495. doi: 10.1128/jvi.73.11.9485-9495.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedieu J F, Ronco J, van der Werf S, Hogle J M, Henin Y, Girard M. Poliovirus chimeras expressing sequences from the principal neutralization domain of human immunodeficiency virus type 1. J Virol. 1992;66:316–327. doi: 10.1128/jvi.66.5.3161-3167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans D, McKeating J, Meredith J M, Burke K L, Katrack K, John A, Ferguson M, Minor P D, Weiss R, Almond J W. An engineered poliovirus chimaera elicits broadly reactive HIV1 neutralizing antibodies. Nature. 1989;339:385–388. doi: 10.1038/339385a0. [DOI] [PubMed] [Google Scholar]
- 23.Francki R, Fauquet C, Knudson D, Brown F. Classification and nomenclature of viruses. Vienna, Austria: Springer-Verlag; 1991. [Google Scholar]
- 24.Godeny E, Arizpe H M, Gauntt C J. Characterization of the antibody response in vaccinated mice protected against coxsackievirus B3 induced myocarditis. Viral Immunol. 1987;1:305–314. doi: 10.1089/vim.1987.1.305. [DOI] [PubMed] [Google Scholar]
- 25.Hansen M B, Nielsen S E, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 26.Hellen C U, Lee C K, Wimmer E. Determinants of substrate recognition by poliovirus 2A proteinase. J Virol. 1992;66:3330–3338. doi: 10.1128/jvi.66.6.3330-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofling K, Tracy S, Chapman N, Leser S L. Expression of the antigenic adenovirus type 2 hexon protein L1 loop region in a group B coxsackievirus. J Virol. 2000;74:4570–4578. doi: 10.1128/jvi.74.10.4570-4578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 angstrom resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 29.Huber S A, Job L P. Cellular immune mechanisms in coxsackievirus group B, type 3 induced myocarditis in Balb/C mice. Adv Exp Med Biol. 1983;161:491–508. doi: 10.1007/978-1-4684-4472-8_29. [DOI] [PubMed] [Google Scholar]
- 30.Huber S A, Pfaeffle B. Differential Th1 and Th2 cell responses in male and female BALB/c mice infected with coxsackievirus group B type 3. J Virol. 1994;68:5126–5132. doi: 10.1128/jvi.68.8.5126-5132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins O, Cason J, Burke K, Lunney D, Gillen A, Patel D, McCance D J, Almond J W. An antigen chimera of poliovirus induces antibodies against human papillomavirus type 16. J Virol. 1990;64:1201–1206. doi: 10.1128/jvi.64.3.1201-1206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia X, Van Eden M, Busch M, Eherenfeld E, Summers D E. trans-Encapsidation of a poliovirus replicon by different picornavirus capsid proteins. J Virol. 1998;72:7922–7977. doi: 10.1128/jvi.72.10.7972-7977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelleher-Doyle M, Telford S R 3, Criscione L, Lin S R, Spielman A, Gravallese E M. Cytokines in murine lyme carditis: Th1 cytokine expression follows expression of proinflammatory cytokines in a susceptible mouse strain. J Infect Dis. 1998;177:242–246. doi: 10.1086/517364. [DOI] [PubMed] [Google Scholar]
- 34.Kim, K.-S., K. Hofling, S. D. Carson, N. M. Chapman, and S. Tracy. The primary viruses of myocarditis, in press. In L. T. Cooper and K. Knowlton (ed.), Myocarditis. Mayo Academic Press, Chicago, Ill.
- 35.Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. A recombinant virus between the Sabin 1 and Sabin 3 vaccine strains of poliovirus as a possible candidate for a new type 3 poliovirus live vaccine strain. J Virol. 1988;62:2828–2835. doi: 10.1128/jvi.62.8.2828-2835.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Lee F, Yokota T, Otsuka T, Meyerson P, Villaret D, Coffman R, Mosmann T, Rennick D, Roehm N, Smith D. Isolation and characterization of a mouse interleukin cDNA clone that expresses B-cell stimulatory factor 1 activities and T-cell- and mast cell-stimulating activities. Proc Natl Acad Sci USA. 1986;83:2061–2065. doi: 10.1073/pnas.83.7.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemon S M, Barcla W, Ferguson M, Murphy P, Jing L, Burke K, Wood D, Katrak K, Sangar D, Minor P D. Immunogenicity and antigenicity of chimeric picornaviruses which express hepatitis A virus (HAV) peptide sequences: evidence for a neutralization domain near the amino terminus of VP1 of HAV. Virology. 1992;188:285–295. doi: 10.1016/0042-6822(92)90758-h. [DOI] [PubMed] [Google Scholar]
- 39.Mandl S, Sigal L, Rock K, Andino R. Poliovirus vaccine vectors elicit antigenic specific cytotoxic T cells and protect mice against lethal challenge with malignant melanoma cells expressing a model antigen. Proc Natl Acad Sci USA. 1998;95:8216–8221. doi: 10.1073/pnas.95.14.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin A, Wychowski C, Couderc C, Crainic R, Hogle J, Girard M. Engineering a poliovirus type 2 antigenic site on a type 1 capsid results in a chimaeric virus which is neurovirulent for mice. EMBO J. 1988;7:2839–2847. doi: 10.1002/j.1460-2075.1988.tb03140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattion N M, Reilly P A, Camposano E, Wu S L, DiMichele S J, Ishizaka S T, Fantini S E, Crowley J C, Weeks-Levy C. Characterization of recombinant polioviruses expressing regions of rotavirus VP4, hepatitis B surface antigen, and herpes simplex virus type 2 glycoprotein D. J Virol. 1995;69:513–527. doi: 10.1128/jvi.69.8.5132-5137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattion N M, Reilly P A, DiMichele S J, Crowley J, Weeks-Levy C. Attenuated poliovirus strain as a live vector: expression of regions of rotavirus outer capsid protein VP7 by using recombinant Sabin 3 viruses. J Virol. 1994;68:3925–3933. doi: 10.1128/jvi.68.6.3925-3933.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minor P D, Ferguson G, Katrack K, Wood D, John A, Howlett J, Dunn G, Burke K, Almond J. Antigenic structure of chimeras of type 1 and type 3 polioviruses involving antigenic sites 2, 3 and 4. J Gen Virol. 1991;72:2475–2481. doi: 10.1099/0022-1317-72-10-2475. [DOI] [PubMed] [Google Scholar]
- 44.Mosmann T, Sad S. The expanding universe of T cell subsets: Th1, Th2 and more. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 45.Mosmann T R, Cherwinski H, Bond M W, Giedlin M A, Coffman R L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 46.Mueller S, Wimmer E. Expression of foreign proteins by poliovirus polyprotein fusion: analysis of genetic stability reveals rapid deletions and formation of cardioviruslike open reading frames. J Virol. 1998;72:20–31. doi: 10.1128/jvi.72.1.20-31.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murdin A D, Lu H H, Murray M G, Wimmer E. Poliovirus antigenic hybrids simultaneously expressing antigenic determinants from all three serotypes. J Gen Virol. 1992;73:607–611. doi: 10.1099/0022-1317-73-3-607. [DOI] [PubMed] [Google Scholar]
- 48.Nishio R, Matsumori A, Shioi T, Ishida H, Sasayama S. Treatment of experimental viral myocarditis with interleukin 10. Circulation. 1999;100:11021–11028. doi: 10.1161/01.cir.100.10.1102. [DOI] [PubMed] [Google Scholar]
- 49.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 50.Novak M J, Smythies L E, McPherson S A, Smith P D, Morrow C D. Poliovirus replicons encoding the B subunit of Helicobacter pylori urease elicit a Th1 associated immune response. Vaccine. 1999;17:2384–2391. doi: 10.1016/s0264-410x(99)00035-3. [DOI] [PubMed] [Google Scholar]
- 51.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. Virology. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Garra A, Murphy K. Role of cytokines in determining T lymphocyte function. Curr Opin Immunol. 1994;6:458–466. doi: 10.1016/0952-7915(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 53.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 54.Porter D, Ansardi D, Choi W, Morrow C. Encapsidation of genetically engineered poliovirus minireplicons which express human immunodeficiency virus type 1 Gag and Pol proteins upon infection. Vaccine. 1995;13:101–322. doi: 10.1128/jvi.67.7.3712-3719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramanathan L, Le H V, Labdon J E, Mays-Ichinco C A, Syto R, Arai N, Nagabhushan T L, Trotta P P. Multiple forms of recombinant murine interleukin-4 expressed in COS-7 monkey kidney cells. Biochim Biophys Acta. 1989;1007:283–288. doi: 10.1016/0167-4781(89)90149-8. [DOI] [PubMed] [Google Scholar]
- 56.Ramsingh A I. Coxsackievirus and pancreatitis. Front Biosci. 1997;2:53–62. [Google Scholar]
- 57.Reinmann B, Zell R, Kandolf R. Mapping of a neutralizing antigenic site of coxsackievirus B4 by construction of an antigen chimera. J Virol. 1991;65:3475–3480. doi: 10.1128/jvi.65.7.3475-3480.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rose C, Andrews W, Ferguson M, McKeating J, Almond J, Evans D. The construction and characterization of poliovirus antigen chimeras presenting defined regions of the human T lymphocyte marker CD4. J Gen Virol. 1994;75:969–977. doi: 10.1099/0022-1317-75-5-969. [DOI] [PubMed] [Google Scholar]
- 59.Rose N R, Neumann D A, Herskowitz A. Genetics of susceptibility to viral myocarditis in mice. Pathol Immunopathol Res. 1988;7:266–278. doi: 10.1159/000157122. [DOI] [PubMed] [Google Scholar]
- 60.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–156. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 61.Ryan M D, Flint M. Virus encoded proteinases of the picornavirus supergroup. J Gen Virol. 1997;78:699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 62.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157–162. [DOI] [PubMed]
- 63.Seko Y, Takahashi N, Yagita H, Okumura K, Yazaki Y. Expression of cytokine mRNAs in murine hearts with acute myocarditis caused by coxsackievirus B3. J Pathol. 1997;183:105–108. doi: 10.1002/(SICI)1096-9896(199709)183:1<105::AID-PATH1094>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 64.Skern T, Sommergruber W, Auer H, Volkmann P, Zorn M, Liebig H D, Fessl F, Blaas D, Kuechler E. Substrate requirements of a human rhinoviral 2A proteinase. Virology. 1991;181:46–54. doi: 10.1016/0042-6822(91)90468-q. [DOI] [PubMed] [Google Scholar]
- 65.Swain S L, Weinberg A D, English M, Huston G. IL4 directs the development of Th2 like helper effectors. J Immunol. 1990;145:3796–3806. [PubMed] [Google Scholar]
- 66.Tang S, van Rij R, Silvera D, Andino R. Toward a poliovirus-based simian immunodeficiency virus vaccine: correlation between genetic stability and immunogenicity. J Virol. 1997;71:7841–7850. doi: 10.1128/jvi.71.10.7841-7850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian J, Olcott A P, Hanssen L R, Zekzer D, Middleton B, Kaufman D L. Infectious Th1 and Th2 autoimmunity in diabetes prone mice. Immunol Rev. 1998;164:119–127. doi: 10.1111/j.1600-065x.1998.tb01214.x. [DOI] [PubMed] [Google Scholar]
- 68.Tomko R, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tracy, S., K. Hofling, S. Pirruccello, P. H. Lane, S. M. Reyna, and C. Gauntt. 1999. Group B coxsackievirus myocarditis and pancreatitis in mice: connection between viral virulence phenotypes. J. Med. Virol., in press. [DOI] [PubMed]
- 70.Tu Z, Chapman N, Hufnagel G, Tracy S, Romero J R, Barry W H, Currey K, Shapiro B. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J Virol. 1995;69:4607–4618. doi: 10.1128/jvi.69.8.4607-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vella C, Minor P D, Weller I V, Jenkins O, Evans D, Almond J. Recognition of poliovirus/HIV chimaeras by antisera from individuals with HIV infection. AIDS. 1991;5:425–430. doi: 10.1097/00002030-199104000-00011. [DOI] [PubMed] [Google Scholar]
- 72.Yim T, Tang S, Andino R. Poliovirus recombinants expressing hepatitis B virus antigens elicited a humoral immune response in susceptible mice. Virology. 1996;218:61–70. doi: 10.1006/viro.1996.0166. [DOI] [PubMed] [Google Scholar]
- 73.Yokota T, Otsuka T, Mosmann T, Banchereau J, DeFrance T, Blanchard D, De Vries J E, Lee F, Arai K. Isolation and characterization of a human interleukin cDNA clone, homologous to mouse B-cell stimulatory factor 1, that expresses B-cell- and T-cell-stimulating activities. Proc Natl Acad Sci USA. 1986;83:5894–5898. doi: 10.1073/pnas.83.16.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang A, Geisler S C, Smith A D, Resnick D A, Li M L, Wang C Y, Looney D J, Wong-Staal F, Arnold E, Arnold G F. A disulfide-bound HIV-1 V3 loop sequence on the surface of human rhinovirus 14 induces neutralizing responses against HIV-1. J Biol Chem. 1999;380:365–374. doi: 10.1515/BC.1999.048. [DOI] [PubMed] [Google Scholar]