Abstract
A number of studies have shown that replication-defective mutant strains of herpes simplex virus (HSV) can induce protective immunity in animal systems against wild-type HSV challenge. However, all of those studies used viruses with single mutations. Because multiple, stable mutations provide optimal levels of safety for live vaccines, we felt that additional mutations needed to be engineered into a candidate vaccine strain for HSV-2 and genital herpes. We therefore isolated an HSV-2 strain with deletion mutations in two viral DNA replication protein genes, UL5 and UL29. The resulting double deletion mutant virus strain, dl5-29, fails to form plaques or to give any detectable single cycle yields in normal monkey or human cells. Nevertheless, dl5-29 expresses nearly the same pattern of gene products as the wild-type virus or the single mutant viruses and induces antibody titers in mice that are equivalent to those induced by single deletion mutant viruses. Therefore, it is feasible to isolate a mutant HSV strain with two mutations in essential genes and with an increased level of safety but which is still highly immunogenic.
Herpes simplex virus 2 (HSV-2) is the major cause of genital herpes, a disease of significant morbidity in infected individuals (34). Primary infection in the genital tract may be asymptomatic or may cause a genital lesion, and the virus spreads to and establishes a latent infection in lumbosacral ganglia. At later times the virus can reactivate and may cause recurrent genital lesions or be shed without apparent symptoms. The last two decades have seen an alarming rise in the incidence of HSV-2 infections in the United States, with certain socioeconomic groups showing nearly 50% seropositivity (12). Immunodeficient individuals are very susceptible to disseminated herpes infection, and the individuals at highest risk of contracting a usually fatal disseminated disease are neonates whose mothers are undergoing a primary infection and actively shedding HSV-2 during childbirth. Individuals with active HSV-2 genital lesions have an increased risk of acquiring human immunodeficiency virus if exposed to the virus (2, 16, 18, 19). Asymptomatic shedding of HSV-2 from the genital tract has been shown to be more frequent than previously realized (33). Thus, an effective vaccine against genital herpes is needed to induce protective immunity that prevents or reduces primary infection and even, ideally, reduces recurrent disease and transmission.
Numerous approaches, including use of glycoprotein subunits, inactivated virus, attenuated mutant HSV strains, DNA, recombinant vectors expressing HSV antigens, and replication-defective mutant HSV strains, have been utilized for immunization against HSV infection in animal model systems (reviewed in references 23, 20, and 31). Clinical trials of HSV glycoprotein subunit vaccines have failed to show any significant protection against genital herpes (6, 32), presumably because insufficient levels of cellular immunity were induced. Clinical trials of attenuated HSV strains showed limited immunogenicity (M. Cadoz, M. Micoud, J. M. Seigneurin, M. R. Mallaret, C. Baccard, P. Morand, P. Chatel, B. Meignier, R. Whitely, and B. Roizman, Program Abstr. 32nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 341, 1992); thus, additional strategies are needed for an effective vaccine for genital herpes. HSV-1 replication-defective mutant virus strains have induced protective immunity in animal studies against (i) lethal intraperitoneal challenge with HSV-1 (27), (ii) ocular challenge with HSV-1 (25), (iii) HSV-1 inoculation by the ear pinna route (11), and (iv) HSV-2 genital challenge (22, 23). HSV-2 replication-defective mutant virus strains have induced protective immunity against HSV-2 genital challenge in guinea pigs (4, 8) and in mice (24). Phase I clinical trials have been performed with an HSV-2 glycoprotein H (gH) mutant virus, and this strain is reported to be immunogenic in humans (3).
Live microbial vaccines need more than one nonreverting mutation for adequate safety (7). Because all of the mutant viruses used in the studies cited above were only single mutants, we felt that a candidate HSV-2 replication-defective mutant vaccine for genital herpes should have at least two deletion mutations. We therefore deleted two genes encoding DNA replication genes, UL5 and UL29, to produce a mutant strain that was completely defective for DNA synthesis and viral replication. This report describes the genotypic and phenotypic characterization of the double deletion mutant and shows that the double deletion mutant is an immunogenic as single mutant viruses.
MATERIALS AND METHODS
Cells.
Vero (American Type Culture Collection [ATCC], Manassas, Va.) and Vero-derived cell lines were grown and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Diploid human lung fibroblast HEL299 and MRC-5 cell lines (ATCC) were grown in Vitacell medium (Eagle's minimum essential medium with Earle's balanced salt solution; ATCC) supplemented with 10% FBS. The S2, V827, and L2-5 cell lines were maintained in Dulbecco's modified Eagle's medium containing G418 (400 μg/ml) and used to construct single deletion mutant viruses. S2 cells contain the HSV-1 ICP8 gene (15), and V827 cells contain the HSV-1 ICP8 and ICP27 genes (X. Da Costa and D. M. Knipe, unpublished results). L2-5 cells, kindly provided by Sandra Weller, contain the HSV-1 UL5 gene. The new V529 cell line was isolated as described in Results by cotransfection of the neomycin resistance gene and HSV genes into Vero cells and selection in medium containing G418 (Mediatech, Herndon, Va.) as described elsewhere (15).
Viruses.
The parental wild-type (wt) HSV-2 strain was 186 syn+-1 (29). Marker transfer in the construction of HSV-2 replication-defective recombinant viruses was performed by calcium phosphate cotransfection and screening of viruses containing lacZ cassettes as blue plaques in medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) or viruses lacking lacZ cassettes as white plaques as described previously (8). Virus stocks were grown on Vero cells (wt HSV-2 only) or V529 cells in medium 199—1% calf serum, and titers were determined by plaque assay on V529 cells.
The HSV-2 mutant virus, 5BlacZ, contains an ICP8-lacZ gene fusion in the UL29 gene locus (8).
Plasmids.
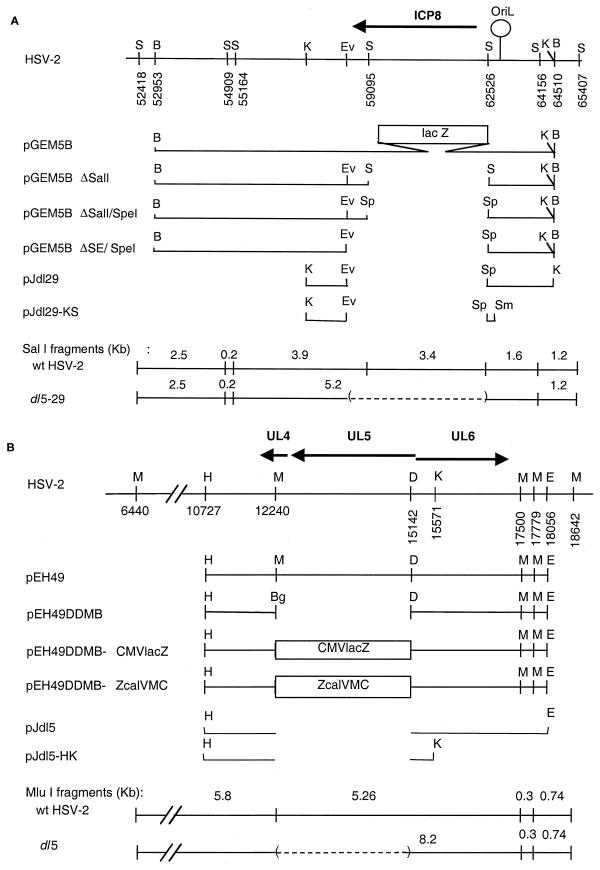
The HSV-2 UL29 and ICP8 gene plasmids (Fig. 1A) were constructed as follows. The 13.8-kbp BamHI fragment containing the ICP8-lacZ fusion gene and flanking sequences was isolated from 5BlacZ viral DNA and subcloned into pGEM7Zf(+) (Promega, Madison, Wis.) to generate plasmid pGEM5B. Deletion of the ICP8-lacZ gene fusion in pGEM5B by partial SalI digestion generated plasmid pGEM5BΔSalI. An SpeI linker (New England Biolabs, Beverly, Mass.) replaced the SalI site, resulting in plasmid pGEM5BΔSalI/SpeI. The deletion was extended approximately 300 bp by using an available EcoRV site, resulting in plasmid pGEM5BΔSE/SpeI.
FIG. 1.
Plasmids used in this study. (A) UL29 gene plasmids. The top line shows the HSV-2 genome from bp 52000 to 66000. The arrow shows the location and orientation of the UL29 ORF encoding ICP8. Letters above the line indicate restriction endonuclease cleavage sites, and numbers below the line show base pair numbers for the cleavage sites. The next six lines diagram the viral DNA inserts from this region in various plasmids, and the bottom two lines show the predicted DNA fragments arising from this region in wt and deletion mutant viral DNAs upon cleavage with SalI. (B) UL5 gene plasmids. The top line shows the HSV-2 genome from bp 6400 to 18842. The arrows show the locations and orientations of the UL4, UL5, and UL6 ORFs. Letters above the line indicate the locations of restriction endonuclease cleavage sites, and numbers denote the base pair number for the cleavage site. The next six lines diagram the viral DNA inserts from this region in various plasmids, and the bottom two lines show the predicted DNA fragments from this region upon cleavage of wt or deletion mutant viral DNAs with MluI. Restriction sites: B, BamHI; S, SalI; K, KpnI; Ev, EcoRV; Sp, SpeI; Sm, SmaI; H, HindIII; M, MluI; Bg, BglII; D, DraI; E, EcoRI.
The HSV-2 UL5 gene plasmids (Fig. 1B) were made as follows. Plasmid pEH49 (29) contains an EcoRI-HindIII fragment from bp 10727 to 18056 of HSV-2 186 syn+-1 DNA. From this plasmid, a 3-kbp DraI-MluI fragment was excised, and a BglII linker (New England Biolabs) was ligated into the deletion site, creating the UL5 deletion plasmid, pEH49ΔDMB. A cytomegalovirus (CMV)-lacZ cassette, derived from plasmid pCL4 (C. G. Murphy and D. M. Knipe, unpublished results) in a BamHI-BglII fragment, was cloned into the BglII site of plasmid pEH49ΔDMB. The two plasmids obtained were pEH49ΔDMB-CMVlacZ and pEH49ΔDMB-ZcalVMC, which differed only in the orientation of the CMV-lacZ cassette.
Plasmid pZeoSV-2/UL5 containing an HSV-2 UL5 expression cassette was kindly provided by K. Metcalfe (K. Metcalfe, J. Metcalfe, D. McAllister, and M. Morin, unpublished results). This expression cassette was generated by PCR amplification of the HSV-2 strain 186 UL5 open reading frame (ORF; bp 12604 to 15249 (11) and insertion into the pZeoSV-2 expression vector (Invitrogen).
Plasmid pSV2neo (28) and HSV-1 UL29 or ICP8 gene plasmid p8B-S (14) have been described previously.
Southern hybridization.
Purified viral DNA was digested with restriction endonuclease(s), resolved by agarose gel electrophoresis, and transferred to Nytran membrane (Schleicher & Schuell, Keene, N.H.). Plasmid pEH49 (29) was linearized with HindIII, labeled, and used as the UL5 gene probe. Plasmid pGEM5B was linearized with BamHI, labeled, and used as the UL29 gene probe. Probes were labeled with [α-32P]dCTP (NEN, Boston, Mass.) using a random primer kit (Boehringer Mannheim, Indianapolis, Ind.). Following hybridization and washes, the blot was analyzed by autoradiography. To analyze the origin of replication, oriL, viral and plasmid DNA was digested with SalI and SphI (wt HSV-2, plasmid pEH51) or SalI, SphI, and SpeI (dl5-29). Plasmid pEH51, which contains the oriL region upstream of UL29 (29), was used as a control for analysis of oriL, and a 1.1-kb SalI/SphI fragment was purified and used as a probe. The probe was labeled with digoxigenin (Boehringer Mannheim). Following hybridization and washes, the blot was analyzed by chemiluminescence as specified by the manufacturer.
DNA sequencing.
Viral DNA fragments were isolated from purified HSV DNA and cloned into pGEM7Zf (Promega). Using standard M13/pUC sequencing primer, approximately 500 nucleotides of each subclone were sequenced in the Department of Microbiology and Molecular Genetics Core Sequencing Facility with an ABI Prism 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.).
Sequence analyses.
Viral genome sequence base pair numbers correspond to the published sequences of HSV-1 strain 17 (21) and HSV-2 strain HG52 (10). Sequence alignment and restriction enzyme fragment analysis were performed with the MacVector 6.5.3 program.
Single cycle growth assay.
Cells were grown to confluence in 25-cm2 culture flasks, infected with the indicated viruses at a multiplicity of infection (MOI) of 3, and incubated for 1 h at 37°C. To remove residual virus, we washed the cells with acidic buffer in a procedure modified from that of Highlander et al. (17). Briefly, virus inoculum was removed, cells were rinsed twice with phosphate-buffered saline (PBS), washed twice in succession for 90 s each time with 5 ml of acid buffer solution (40 mM citric acid, 10 mM KCl, 135 mM NaCl [pH 3.0]), and then rinsed twice with growth medium to neutralize the pH. The cells were overlaid with 5 ml of medium 199–1% calf serum and incubated at 37°C for 2 or 18 h. Uninfected cells treated with acid buffer were indistinguishable in appearance and viability from uninfected, untreated cells for at least 18 h. To harvest virus, we added 2.5 ml of sterile milk to the medium in each flask, and the infected cells were frozen at −80°C. After thawing, cell monolayers were scraped, transferred to 50-ml conical centrifuge tubes, sonicated for one 30-s pulse, and serially diluted for titration. Titers were determined on V529 cells and expressed as PFU per milliliter.
Analysis of viral proteins.
Levels of gene expression in virus- and mock-infected cells were examined as described previously (8). Briefly, cells were infected with the indicated virus or were mock infected and then incubated in medium 199 with 1% calf serum. At the indicated times, cells were overlaid with 2 ml of labeling medium (Eagle's minimum essential medium, without methionine or cysteine [Sigma, St. Louis, Mo.], containing 100 μCi of EXPRE35S35S™ protein labeling mix with [35S]methionine and [35S]cysteine [NEN] and supplemented with 10% dialyzed FBS [GIBCO/BRL, Gaithersburg, Md.]) for 30 min. Cells were solubilized with 1.0 ml of gel sample buffer (0.5 M Tris-HCl [pH 6.8], 20% glycerol, 20% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 0.5% bromphenol blue) containing 50 μg of N-p-tosyl-l-lysine chloromethyl ketone (TLCK) per ml. Proteins were resolved by electrophoresis in a 9.25% DATD-cross-linked polyacrylamide gel. The gels were dried under vacuum and exposed to Biomax MR single-sided emulsion film.
Immunoblotting for glycoproteins was performed as described previously (8). Briefly, 2 × 106 Vero cells infected with dl5, dl29, or dl5-29 virus at an MOI of 20 were harvested in sample buffer containing TLCK at 16 h postinfection (hpi). Equivalent volumes of each infected cell lysate were resolved by electrophoresis in a 9.25% DATD-cross-linked SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Schleicher & Schuell) by electroelution. Rabbit polyclonal serum R1161 (kindly provided by R. Courtney, Pennsylvania State University) and R-45 (kindly provided by G. Cohen and R. Eisenberg, University of Pennsylvania) were used as primary antibodies to detect gB and gD, respectively. Immune complexes were detected using goat anti-rabbit immunoglobulin G (IgG) alkaline phosphatase-conjugated secondary antibody (Sigma). Western blots were developed using nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) substrate reagents as directed by the supplier (Sigma).
Animals and animal protocols.
Female BALB/c mice were obtained from Taconic Farms (Germantown, N.Y.) and housed in accordance with institutional and National Institutes of Health guidelines on the care and use of animals in research. Immunization of mice was performed as described previously (26). Six-week-old female BALB/c mice were randomized into four groups of six mice each. All animals were injected twice, 4 weeks apart, by the subcutaneous route with 2 × 106 PFU of dl5, dl29, or dl5-29 or an equivalent dose of control cell extract. Mice were bled from the tail vein 21 days after the primary inoculation and a second time 9 days after the boost inoculation. Serum was isolated using Becton Dickinson Microtainer serum separator tubes (VWR, Boston, Mass.) and stored at −20°C.
ELISA.
Ninety six-well plates were coated with HSV-2 antigen (Advanced Biotechnologies Inc., Columbia, Md.) at 50 ng per well in 0.05 M carbonate-bicarbonate buffer (pH 9.6) (Sigma) overnight at 4°C. Plates were blocked with PBS containing 5% milk at 37°C for 1 h, washed three times with PBS containing 5% milk and 0.05% Tween 20, and incubated with serial twofold dilutions of mouse sera for 2 h at 37°C. Plates were then washed three times with PBS containing 0.05% Tween 20 and incubated with goat anti-mouse IgG antibody conjugated with alkaline phosphatase (1:1,000; Sigma) for 1 h at 37°C. Finally, plates were washed three times with PBS containing 0.1% Tween 20, developed using SigmaFast p-nitrophenyl phosphate (Sigma), and read at 405 nm. Antibody titers indicate the final reciprocal dilution resulting in optical density readings greater than 0.2 unit above background and are expressed as the geometric mean (log2) ± standard deviation, where log2 6 = 1:64 and log2 12 = 1:4,096. Enzyme-linked immunosorbent assay (ELISA) results were analyzed for statistical significance by Student's t test.
RESULTS
Construction of HSV-2 deletion mutant strains.
Replication-defective mutant strains of HSV-2 had been shown to induce protective immunity against wt viral challenge (4, 8). Because these studies had used strains with only one mutation, we felt that additional mutations needed to be engineered into a candidate HSV-2 vaccine strain for optimal safety. We therefore attempted to isolated an HSV-2 strain with two independent deletion mutations in viral DNA replication protein genes, i.e., UL5 and UL29 (Fig. 1).
We first isolated a cell line, V529, that contained the UL5 and UL29 genes and complemented HSV-1 UL5 and UL29 single mutant viruses. We cotransfected Vero cells with plasmid pSV2neo, plasmid pZeoSV-2/UL5 expressing UL5 (Fig. 1B), and the HSV-1 plasmid p8B-S expressing HSV-1 ICP8 (Fig. 1A). Plasmid pZeoSV-2/UL5 contains the HSV-2 UL5 ORF (bp 12604 to 15249) (10) expressed from the pZeoSV-2 expression vector. Plasmid p8B-S contains HSV-1 sequences from bp 62655 to 56772, which extends upstream and downstream from the HSV-1 UL29 ORF (sequence coordinates 62053 to 58465) and contains the promoter and polyadenylation signal of the UL29 gene. The cells were subjected to a brief glycerol shock, grown to confluency, and then replated in medium containing G418. The cells were refed every 3 to 4 days with medium containing G418 and incubated until colonies were visible on each plate. Individual colonies were picked, expanded, and screened for the ability to support replication of the HSV-1 hr99 UL5 mutant virus (35) and of the HSV-2 5BlacZ UL29 mutant virus (8). The cell clone that was best able to complement the two mutant viruses was named V529 and used for further study.
The UL29 deletion mutant virus was derived from the HSV-2 5BlacZ mutant virus. The HSV-2 5BlacZ virus containing the lacZ coding sequences fused at the 5′ end of the UL29 ORF was described previously (8). The UL29 deletion mutant virus was constructed by cotransfection of 5BlacZ viral DNA with linearized pGEM5BΔSE/SpeI (Fig. 2A) into S2 cells. The progeny viruses were harvested, and white plaques isolated in an X-Gal agarose overlay were purified three times and screened for the ability to grow in S2 but not Vero cells. The deletion was verified by Southern hybridization (see below). One virus clone was chosen for further study and named dl29.
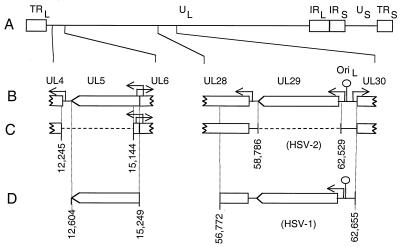
FIG. 2.
Genome structure of wt and mutant HSV-2 strains. (A) Genome structure of the wt virus. Boxes represent repeated sequences in the viral genome, and lines represent the unique sequences. (B) Expanded regions showing sequence features in the vicinity of the UL5 (left) and UL29 (right) viral genes. Boxes indicate the locations and orientations of ORFs. Arrows indicate the start sites and direction of transcription. (C) Genomic locations of the dl5 (left) and dl29 (right) deletion mutations. Numbers correspond to base pairs in the HSV-2 strain HG-52 sequence (10). (D) Sequence coordinates of the HSV-2 UL5 gene (left) and HSV-1 UL29 gene (right) transformed into Vero cells to make V529 cells.
The UL5 deletion virus was constructed in two stages. First, a mutant virus was generated by insertion of a lacZ expression cassette in place of the UL5 ORF. Infectious wt HSV-2 genomic DNA was cotransfected with either pEH49ΔDMB-CMVlacZ or pEH49ΔDMB-ZcalVMC (Fig. 1B) into L2-5 cells. Blue plaques isolated in an X-Gal agarose overlay were purified three times and screened for the ability to grow on L2-5 but not Vero cells. The replacement of sequences at the UL5 locus was confirmed by Southern hybridization. One virus clone, designated UL5-lacZ, was used for subsequent constructions. To delete the inserted sequences, we cotransfected UL5-lacZ viral DNA with linearized plasmid pEH49ΔDMB (Fig. 1B) into L2-5 cells. White plaques isolated in X-Gal agarose were purified and screened for the ability to grow on L2-5 but not Vero cells. The UL5 deletion was verified by Southern hybridization analysis. One virus clone was named dl5 and used for further study.
The UL5/UL29 double deletion mutant virus was generated by crossing the two single deletion mutant viruses, dl5 and dl29. V529 cells were coinfected with dl5 and dl29, each at an MOI of 3. Cells were incubated until cytopathic effect was observed and then harvested as described previously (8). Progeny virus titers were determined on V529 and Vero cells to determine the frequency of recombination, based on the percentage of wt recombinants. Fifty-four single plaques picked at random were plated on L2-5, V827, and V529 cells. Three isolates that grew on V529 cells but not on L2-5 or V827 cells were purified through three more rounds on V529 cells. The double mutations in two isolates were confirmed by Southern hybridization (see below), and one of these was chosen for further study and named dl5-29.
Southern blot analysis of the deletion mutant genomes.
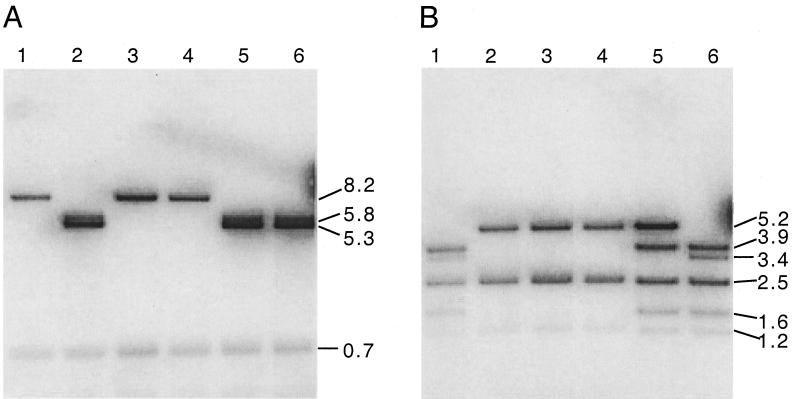
We confirmed the deletions in the UL5 and UL29 genes of dl5-29 virus (Fig. 2) using Southern blot hybridization. To analyze the UL5 gene, wt and recombinant viral DNAs were cleaved with MluI and hybridized with labeled plasmid pEH49 (Fig. 1B). With HSV-2 wt viral DNA, bands of 5.8 and 5.3 kbp were observed (Fig. 3A, lane 6), as expected (Fig. 1B). Similarly, hybridization to restricted dl29 and 5BlacZ DNAs yielded the wt bands of 5.8 and 5.3 kbp (Fig. 3A, lanes 2 and 5, respectively). Hybridization to MluI-cleaved dl5 DNA showed only a single 8.2-kbp DNA band (Fig. 3A, lane 1), as a result of the 2.8-kbp deletion (Fig. 1B). Two isolates of dl5-29 showed only the 8.2-kbp band (Fig. 3A, lanes 3 and 4), showing that they contained the same deletion mutation.
FIG. 3.
Southern blot analysis of deletion mutations in dl5-29. (A) UL5; viral DNAs digested with MluI and probed with plasmid pEH49. Lane 1, dl5; lane 2, dl29; lanes 3 and 4, dl5-29 (two isolates); lane 5, 5BlacZ; lane 6, wt HSV-2. (B) UL29; viral DNA digested with SalI and probed with plasmid pGEM5B. Lane 1, dl5; lane 2, dl29; lanes 3 and 4, dl5-29 (two isolates); lane 5, 5BlacZ; lane 6, wt HSV-2.
To analyze the UL29 gene, wt and recombinant viral DNAs were cleaved with SalI endonuclease and hybridized with labeled pGEM5B (Fig. 1A). With wt HSV-2 DNA, bands of 3.9, 3.4, 2.5, 1.6, and 1.2 kbp were observed (Fig. 3B, lane 6), as expected (Fig. 1A). Similarly, the dl5 mutant DNA contained the wt bands of 3.9, 3.4, 2.5, 1.6, and 1.2 kbp (Fig. 3B, lane 1). In contrast, the 5BlacZ DNA showed bands of 5.2, 3.9, 2.5, 1.6, and 1.2 kbp (Fig. 3B, lane 5), consistent with the insertion of lacZ sequences in the wt 3.4-kbp band (Fig. 1A). The dl29 genomic DNA showed bands of 5.2, 2.5, and 1.2 kbp (Fig. 3B, lane 2), consistent with deletion of the 3.4-kbp band (Fig. 1A). The two dl5-29 isolates showed the same bands (Fig. 3B, lanes 3 and 4), indicating that they contain the same deletion. Thus, dl5-29 virus contained deletions of the expected sizes in UL5 and UL29.
To ensure the integrity of oriL, located 333 bp upstream of the dl29 deletion, we performed Southern hybridization analysis of wt and recombinant genomes using an oriL probe (Fig. 4). Wild-type viral DNA showed a 1.2-kbp band that hybridized with the oriL probe (Fig. 4, lane 1), and dl5-29 DNA showed the same size band, consistent with an intact oriL in dl5-29 (lane 2). Deletions in the oriL region could be detected on this gel, as plasmid pEH51, which sustains deletions in the oriL sequences, showed several faster-migrating bands (lane 3). These results demonstrate that the origin of replication in dl5-29 mutant virus remained intact.
FIG. 4.
Southern blot analysis of the oriL region in dl5-29. Viral or plasmid DNA was digested with specific restriction endonucleases, separated electrophoretically, transferred, and probed with a purified 1.1-kb SalI/SphI fragment of plasmid pEH51 containing the HSV-2 oriL region. Lane 1, wt HSV-2 DNA digested with SalI and SphI; lane 2, dl5-29 DNA digested with SalI, SpeI, and SphI; lane 3, pEH51 digested with SalI and SphI.
Sequence analysis of the deletions.
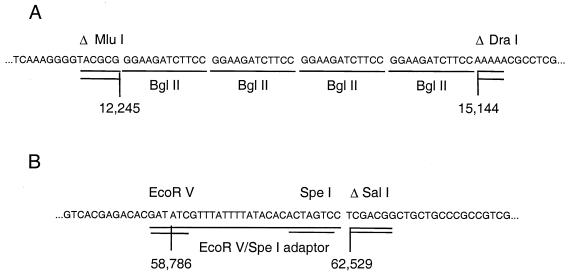
To precisely define the engineered mutations in the dl5-29 genome, we isolated DNA fragments that spanned the UL5 and UL29 deletions from dl5-29 viral DNA and then cloned and sequenced them. The 4.4-kbp HindIII-EcoRI fragment bearing the deletion at the UL5 locus and a 2.7-kbp KpnI fragment bearing the deletion at the UL29 locus were isolated (Fig. 1). These were cloned into pGEM7Zf vector (Promega) to generate two plasmids, designated pJdl5 and pJdl29, respectively (Fig. 1). Two subclones, pJdl5-HK and pJdl29-KS, were constructed for direct sequencing by subcloning the 2.5-kbp HindIII-KpnI viral DNA fragment from pJdl5 and deleting a 1.5-kbp SmaI fragment from pJdl29, respectively. The nucleotide sequence of pJdl5-HK revealed that the 2.9-kbp MluI/DraI fragment (bp 12246 to 15143 of HSV-2 strain HG-52) had been deleted and replaced with four BglII linkers (Fig. 5A). The nucleotide sequence of pJdl29-KS revealed that the 3.7-kbp EcoRV/SalI fragment (bp 58787 to 62528) was deleted in the dl5-29 genome and replaced with the synthetic EcoRV/SpeI adapter, bearing an ICP8 poly(A) signal flanked by EcoRV and SpeI restriction endonuclease cleavage sites (Fig. 5B). Sequence comparisons with the published HSV-2 sequence (10) revealed that the viral sequences flanking the deletion sites were intact (100% identity with published sequence). Thus, dl5-29 contained the expected gene deletions.
FIG. 5.
(A) Sequence of the dl5 deletion site in dl5-29 viral DNA; (B) sequence of the dl29 deletion site in dl5-29 viral DNA.
The sequences bounding the deletion sites were compared with the viral DNA sequences contained in V529 cells (Fig. 1C and D). The 3′ end of the UL5 gene contains no homologous sequences in the cell line, and so homologous recombination between the virus and cell should not be possible. This deletion also removed the 5′ end of the nonessential UL4 gene. The 5′ end of the UL5 gene is retained in dl5-29 because it contains the 5′ end of the essential UL6 gene. The UL29 deletion was designed to remove the UL29 ORF but retain its promoter, oriL, the UL30 promoter, and the UL28 promoter. The HSV-1-derived sequences encoding ICP8 in V529 cells and the HSV-2 sequences spanning the UL29 deletion site were compared by a sequence alignment program to determine the similarity between the UL29 flanking regions. Upstream, 608 nucleotides of HSV-1 corresponded to 573 nucleotides (plus gaps) of HSV-2, with 383 identical nucleotides, yielding 63% identity. Downstream, 1,625 nucleotides (plus gaps) of HSV-1 corresponded to 1,719 nucleotides of HSV-2, with 1,344 identical nucleotides, yielding 78% identity. The limited homology and the sequence gaps greatly reduced homologous recombination in this region because no wt UL29 recombinant viruses have been derived from dl5-29 propagated in these cells (Table 1; Da Costa and Knipe, unpublished).
TABLE 1.
Plaque formation by wt and mutant viruses on Vero and Vero-derived cell lines
| Virus | Titer (PFU/ml)a on cell line:
|
|||
|---|---|---|---|---|
| Vero | L2-5 | S2 | V529 | |
| wt HSV-2 | 3.2 × 108 | NDb | ND | ND |
| dl5 | <102 | 1.7 × 108 | ND | 1.6 × 108 |
| dl29 | <102 | ND | 2.5 × 108 | 2.9 × 108 |
| dl5-29 | <102 | <102 | <102 | 1.3 × 108 |
Plaque assays were performed on Vero, L2-5, S2, and V529 cells for each of the viruses indicated. L2-5 cells express HSV-1 UL5; S2 cells express HSV-1 ICP8; V529 expresses HSV-2 UL5 and HSV-1 ICP8.
ND, not determined.
Growth phenotype of the deletion mutants.
HSV-1 UL5 and UL29 mutants grow only on complementing cell lines and do not replicate viral DNA in normal cells (15, 35). The HSV-2 mutants isolated in this study were also examined for their growth phenotypes. The single deletion mutants dl5 and dl29 formed plaques only on the appropriate complementing cell lines derived from Vero cells (Table 1). At low dilutions of virus, the cytopathic effect of the deletion mutant viruses on normal cells prevented determination of virus titers. However, efficiencies of plating on noncomplementing versus complementing cell lines were less than 10−6, as observed previously with 5BlacZ (8). To determine if human cells could complement the growth of either dl5 or dl29, we examined the single cycle yields of dl5, dl29, dl5-29, and wt HSV-2 infections in two human cell lines, HEL299 and MRC-5, as well as Vero cells (Table 2). Cells were infected at an MOI of 3 and harvested at 2 hpi, a time corresponding to the viral eclipse phase, or at 18 hpi, a time corresponding to the end of a single growth cycle. Titers were subsequently determined by plaque assay on V529 cells. The wt HSV-2 titer increased by several orders of magnitude between 2 to 18 hpi in both human and Vero cells. Conversely, in the same time interval, the titers of the dl5, dl29, and dl5-29 mutant viruses remained constant or declined. There was no detectable yield of deletion mutant virus, even in a single cycle experiment. Thus, these mutant viruses are absolutely replication defective, and it is unlikely that the deletion mutants will grow in any mammalian host.
TABLE 2.
Single cycle yields of wt and mutant virus strains in human and Vero cell lines
| Virus | Titera (PFU/ml) on cell line:
|
|||||
|---|---|---|---|---|---|---|
| MRC-5
|
HEL299
|
Vero
|
||||
| 2 hpi | 18 hpi | 2 hpi | 18 hpi | 2 hpi | 18 hpi | |
| wt HSV-2 | 8.9 × 101 | 6.6 × 105 | 1.2 × 103 | 1.3 × 107 | 8.0 × 102 | 2.7 × 106 |
| dl5 | 6.6 × 101 | 6.8 × 101 | 1.2 × 103 | 2.1 × 102 | NDb | ND |
| dl29 | 1.2 × 102 | 5.5 × 101 | 1.2 × 103 | 3.0 × 102 | ND | ND |
| dl5-29 | 6.6 × 101 | 2.8 × 101 | 2.9 × 103 | 2.2 × 102 | 9.0 × 103 | 4.0 × 103 |
At the times indicated, total infectious virus in each culture was determined by plaque assay on V529 cells.
ND, not determined.
Protein expression by the deletion mutant viruses in noncomplementing cells.
We wished to determine if the double mutant virus showed any impairment for gene expression compared to the single mutant viruses. We therefore examined the patterns of protein synthesis in Vero cells infected with the dl5, dl29, dl5-29, or 5BlacZ mutant virus, or with wt HSV-2, at early and late times postinfection. The double mutant, dl5-29, showed a pattern of protein expression very similar to those of the single deletion mutants, except that dl5-29 and dl29 did not express ICP8 (Fig. 6). All of the deletion mutants showed a pattern of protein expression similar to that of wt virus, except that late proteins ICP5, gB, and ICP25 were expressed at lower levels (Fig. 6). To look more specifically at viral glycoproteins, we performed Western blot analysis of gB and gD. dl5-29 expressed gB-2, although at lower levels than dl5, dl29, or wt virus (Fig. 7A). In contrast, all of the mutant viruses expressed gD at levels similar to the wt virus level (Fig. 7B). Thus, these replication-defective mutant viruses generated nearly wt levels of viral proteins in infected cells and therefore may be expected to generate relevant antigens in the mammalian host. The combined effect of UL5 and UL29 in stimulating gB-2 expression remains to be explained, however.
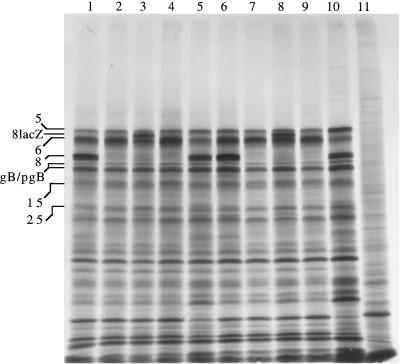
FIG. 6.
Protein expression by wt and mutant viruses in infected Vero cells. Vero cells were infected with the indicated viruses and labeled from 6 to 6.5 or 9.5 to 10 hpi. Lysates were prepared, and the proteins were resolved by SDS-PAGE. An autoradiogram is shown. Lanes: 1, dl5, 6 to 6.5 hpi; 2, dl29, 6 to 6.5 hpi; 3, 5BlacZ, 6 to 6.5 hpi; 4, dl5-29, 6 to 6.5 hpi; 5, wt, 6 to 6.5 hpi; 6, dl5, 9.5 to 10 hpi; 7, dl29, 9.5 to 10 hpi; 8, 5BlacZ, 9.5 to 10 hpi; 9, dl5-29, 9.5 to 10 hpi; 10, wt, 9.5 to 10 hpi; 11, mock-infected cells, 9.5 to 10 hpi. The positions of certain viral proteins are indicated at the left.
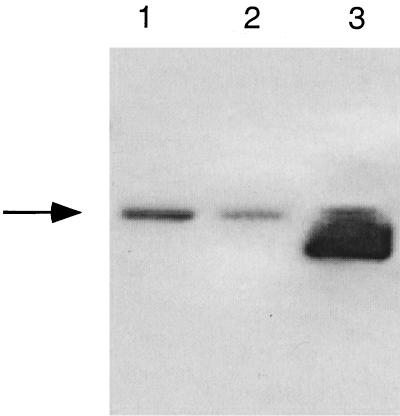
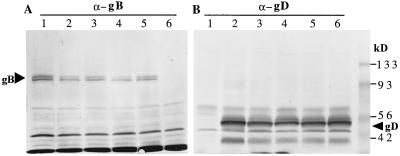
FIG. 7.
Western blot analysis of gB and gD expression. Proteins from infected cell lysates were electroblotted from SDS-polyacrylamide gels onto nitrocellulose membranes and probed with either anti-gB (A) or anti-gD (B) antibody. Arrowheads indicate the specific glycoprotein bands. (A) Lanes: 1, dl5; 2, dl29; 3, 5BlacZ; 4, dl5-29; lane 5, wt HSV-2; lane 6, mock-infected cells. (B) Lanes: 1, mock-infected cells; 2, wt HSV-2; 3, dl5-29; 4, 5BlacZ; lane 5, dl29; lane 6, dl5. Positions of molecular weight markers are indicated on the right.
The phenotype of certain HSV mutants has been different in human cells than in monkey cells (1). We therefore examined viral protein expression in human cells to confirm that the phenotype of dl5-29 was similar in human cells (Fig. 8). In HEL299 cells, dl5-29 expressed a similar pattern of proteins as wt virus, except that ICP8 was not expressed and expression of certain late proteins such as ICP5 and gB was reduced (Fig. 8, lanes 2 and 3). In MRC-5 cells, dl5-29 showed a pattern of viral protein expression similar to wt virus, except that ICP8 was absent and expression of all proteins was reduced somewhat. Therefore, dl5-29 shows similar gene expression properties in human and monkey cells.
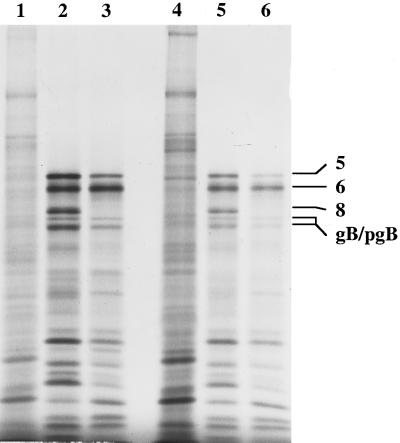
FIG. 8.
Protein expression by wt and dl5-29 viruses in human cell lines. The indicated cells were infected with wt or dl5-29 virus at an MOI of 10 for 9.5 h and pulse-labeled with from 9.5 to 10 hpi. Lysates were prepared, and the proteins were resolved by SDS-PAGE. An autoradiogram is shown. Lane 1, HEL299 cells mock infected; lane 2, HEL299 cells infected with wt virus; lane 3, HEL299 cells infected with dl5-29; lane 4, MRC-5 cells mock infected; lane 5, MRC-5 cells infected with wt virus; lane 6, MRC-5 cells infected with dl5-29.
Anti-HSV-2 antibody responses in immunized mice.
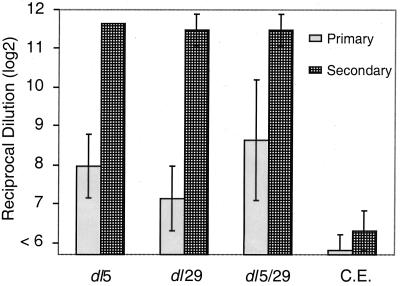
Previous studies had shown that dl5-29 induces a protective immune response (9), but we wished to obtain more quantitative information about the immune responses to the single and double mutant viruses. We therefore measured antibody responses to viral antigens in mice immunized with the mutant viruses. BALB/c mice were inoculated subcutaneously with 2 × 106 PFU of dl5, dl29, or dl5-29 virus or with control cell extract. Sera were collected at 21 days after primary immunization (primary response) and at 9 days after the boost immunization (secondary response), and anti-HSV-2 antibody titers were determined by ELISA. The primary and the secondary mean antibody titers generated following dl5-29 immunization were statistically indistinguishable from the antibody titers generated following immunization with either single mutant (Fig. 9). Mock immunization with cell extract alone gave low background values, demonstrating the specificity of this assay. These results indicate that the addition of a second mutation to enhance the safety of a single mutation replication-defective HSV-2 vaccine virus does not compromise the ability of the virus to elicit host immune responses to viral antigens.
FIG. 9.
Antibody responses to deletion mutant viruses. BALB/c mice were inoculated with 2 × 106 PFU of the indicated virus or control cell extract at days 0 and 28. Sera were obtained at day 21 (Primary) and, after a boost, at day 37 (Secondary). Individual samples were analyzed for anti HSV-2 antibody titer by ELISA as described in Materials and Methods. Values shown indicate the geometric mean (log2) ± standard deviation (n = 6).
DISCUSSION
The goal of this research was to isolate a mutant of HSV-2 containing deletions in two essential genes that could serve as a potential genital herpes vaccine strain. Viruses with single mutations in the ICP8 gene (8, 25, 27), the ICP27 gene (27), or the gH gene (4, 11) had been used to induce protective immunity in experimental animal systems, but standard practice in vaccine design is to include two or more nonreverting mutations in vaccine strains to increase safety of the vaccine (7). We therefore attempted to isolate a double deletion mutant strain. It was not immediately obvious that this would be straightforward because the first double mutant virus we isolated, an HSV-1 ICP8/ICP27 double mutant virus strain, expressed viral proteins at levels much lower either of the parental mutant viruses (Da Costa and Knipe, unpublished). The low level of viral gene expression appeared to be due to a synergism between the two mutations. We turned then to isolating a mutant virus with dual mutations in DNA replication protein genes for two reasons. First, we had observed that UL29 mutant and UL5 mutant virus strains showed similar phenotypes in expressing immediate-early, early, and even certain late genes at rates comparable to wt virus infection (8; M. Chen and D. Knipe, unpublished results). Second, a mutant virus with dual mutations in DNA replication protein genes should be absolutely defective for replication in normal cells. This would provide a high level of safety and, if the double mutant virus was immunogenic, argue against the idea that limited replication of the single mutants in vivo might be responsible for their ability to induce immune responses.
dl5-29 is absolutely defective for replication in normal cells.
The dl5 and dl29 mutant viruses are absolutely defective for plaque formation or growth in single cycle growth assays in normal monkey or human cells (this work) or mouse cells (9). dl5-29 is also completely defective for growth in these same assays; furthermore, the independent defects of the single mutants argued that dl5-29 has a double block for growth in normal cells. We believe that the single cycle growth assay in human cells is the most rigorous and appropriate assay for testing the growth phenotype of mutant viruses. Although the gH mutant viruses have been referred to as single cycle mutant viruses (11), the only information published on the growth properties of the gH mutants is the inability of the HSV-1 gH mutant virus to produce infectious virus in Vero cells (13). No information has been published on the properties of the HSV-2 gH mutant virus currently in clinical trials. The phenotype of a mutant virus in single cycle growth assays in human cells needs to be determined before a virus is considered replication defective or a single cycle mutant for vaccine trials. Indeed, the HSV-1 gH mutant virus is capable of establishing latency in mice (30), possibly indicating that it can replicate and spread at the site of inoculation.
Similarity of immune responses to double and single mutants.
Our previously published study had shown that dl5-29 induced protective immunity against genital challenge in mice with HSV-2 (9), but we measured the antibody responses to the single and double deletion mutant viruses to obtain a more quantitative measure of the level of the immune responses. The primary and secondary antibody responses to dl5-29 were equivalent to those elicited by the single deletion mutants, dl5 and dl29. Thus, the available evidence indicates that the immune responses to the double deletion mutant virus are similar to the responses to the single deletion mutant viruses. This would argue that under the conditions studied, replication and spread of the immunizing virus are not necessary for efficient immunization. Further studies with varied immunizing doses and examination of cellular and other immune responses are necessary to ensure that the immune responses to the different mutant viruses are equivalent.
Stability of the dl5-29 virus upon propagation in complementing cell lines.
dl5-29 has proven to be stable upon propagation in the V529 cell line because no viruses capable of growth on normal, ICP8-expressing, or UL5 protein-expressing cells have been detected in the dl5-29 stocks after three passages (Table 1; Da Costa and Knipe, unpublished). Homologous recombination at the 3′ end of the UL5 gene is precluded by the lack of homologous sequences in the virus and in the cell line (Fig. 2). Approximately 100 bp of homologous sequences remain at the 5′ end of the UL5 gene because of the overlap with the essential UL6 gene (Fig. 2). However, lack of homologous sequences at one end of a deletion is sufficient to prevent recombination. The flanking sequences of the ICP8 gene in the V5-29 cell line are from HSV-1, while the viral sequences in dl5-29 virus are from HSV-2. Thus, although these sequences are colinear within the two viruses, they are on average only 63% identical upstream of the UL29 ORF and 78% identical downstream of the UL29 ORF, which results in little or no recombination. An additional cell line capable of complementing the growth of dl5-29 virus has been isolated by transformation of expression vectors containing only the UL5 and UL29 ORFs (Metcalfe et al., unpublished). This cell line contains no sequences homologous with the UL29 gene region in dl5-29 and should provide even further safety during growth of the virus. Further studies of dl5-29 during repeated passages in this cell line are needed to define the stability of the vaccine stocks of this virus.
Vector potential of dl5-29 virus.
In addition to its potential for serving as a genital herpes vaccine, dl5-29 has the properties desired in a vaccine vector. It is replication defective in all types of normal mammalian cells (this work and reference 9), and so it should not spread and cause disease in any host. It also shows a defect in latent infection (9), and so its genome should not persist in the host, providing a further measure of safety. Furthermore, the two mutated genes, UL5 and UL29, retain their promoter and polyadenylation sites, and a heterologous ORF(s) could be inserted into these transcriptional units. HSV-1 and HSV-2 induce a strong Th1 T-cell response to heterologous antigens expressed from the viral genome (5), and so dl5-29 could serve as a vector to induce a strong CD8+ T-cell response against a heterologous antigen, providing the basis for a combination vaccine.
ACKNOWLEDGMENTS
This research was supported by Avant Immunotherapeutics (formerly Virus Research Institute) and grant CA26345 from the National Cancer Institute, NIH.
We thank Karen Metcalfe for generously supplying plasmid pZeoSV2/UL5.
REFERENCES
- 1.Aubert M, Blaho J A. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J Virol. 1999;73:2803–1813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augenbraun M H, McCormack W M. Sexually transmitted diseases in HIV-infected persons. Infect Dis Clin North Am. 1994;8:439–448. [PubMed] [Google Scholar]
- 3.Bernstein D I, Stanberry L R. Herpes simplex virus vaccines. Vaccine. 1999;17:1681–1689. doi: 10.1016/s0264-410x(98)00434-4. [DOI] [PubMed] [Google Scholar]
- 4.Boursnell M E G, Entwisle C, Blakeley D, Roberts C, Duncan I A, Chisholm S E, Martin G M, Jennings R, Ni Challanain D, Sobek I, Inglis S C, McLean C S. A genetically inactivated herpes simplex virus type 2 (HSV-2) vaccine provides effective protection against primary and recurrent HSV-2 disease. J Infect Dis. 1997;175:16–25. doi: 10.1093/infdis/175.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brubaker J O, Thompson C M, Morrison L A, Knipe D M, Siber G B, Finberg R W. Th1-associated immune responses to beta-galactosidase expressed by replication-defective herpes simplex virus. J Immunol. 1996;157:1598–1604. [PubMed] [Google Scholar]
- 6.Corey L, Langenberg A G M, Ashley R, Sekulovich R E, Izu A E, Douglas J M, Jr, Handsfield H H, Warren T, Marr L, Tyring S, DiCarlo R, Adimora A A, Leone P, Dekker C L, Burke R L, Leong W P, Straus S E. Recombinant glycoprotein vaccine for the prevention of genital HSV-2 infection: two randomized controlled trials. JAMA. 1999;282:331–340. doi: 10.1001/jama.282.4.331. [DOI] [PubMed] [Google Scholar]
- 7.Curtiss R, III, Kelly S M, Tinge S A, Tacket C O, Levine M M, Srinivasan J, Koopman M. Recombinant salmonella vectors in vaccine development. Dev Biol Stand. 1994;82:23–33. [PubMed] [Google Scholar]
- 8.Da Costa X J, Bourne N, Stanberry L R, Knipe D M. Construction and characterization of a replication-defective herpes simplex virus 2 ICP8 mutant strain and its use in immunization studies in a guinea pig model of genital herpes. Virology. 1997;232:1–12. doi: 10.1006/viro.1997.8564. [DOI] [PubMed] [Google Scholar]
- 9.Da Costa X J, Jones C A, Knipe D M. Immunization against genital herpes with a vaccine virus that has defects in productive and latent infection. Proc Natl Acad Sci USA. 1999;96:6994–6998. doi: 10.1073/pnas.96.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeogh D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell H E, McLean C S, Harley C, Efstathiou S, Inglis S, McLean A C. Vaccine potential of a herpes simplex virus type 1 mutant with an essential glycoprotein deleted. J Virol. 1994;68:927–932. doi: 10.1128/jvi.68.2.927-932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming D T, McQuillan G M, Johnson R E, Nahmias A J, Aral S O, Lee F K, St. Louis M E. Herpes simplex virus type 2 in the United States, 1976 to 1994. N Engl J Med. 1997;337:1105–1111. doi: 10.1056/NEJM199710163371601. [DOI] [PubMed] [Google Scholar]
- 13.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao M, Bouchey J, Curtin K, Knipe D M. Genetic identification of a portion of the herpes simplex virus ICP8 protein required for DNA-binding. Virology. 1988;163:319–329. doi: 10.1016/0042-6822(88)90272-3. [DOI] [PubMed] [Google Scholar]
- 15.Gao M, Knipe D M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989;63:5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenblatt R M, Lukehart S A, Plummer F A, Quinn T C, Critchlow C W, Ashley R L, D'Costa L J, Ndinya-Achola J O, Corey L, Ronald A R, et al. Genital ulceration as a risk factor for human immunodeficiency virus infection. AIDS. 1988;2:47–50. doi: 10.1097/00002030-198802000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Highlander S L, Cai W H, Person S, Levine M, Glorioso J C. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol. 1988;62:1881–1888. doi: 10.1128/jvi.62.6.1881-1888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmberg S D, Stewart J A, Gerber A R, Byers R H, Lee F K, O'Malley P M, Nahmias A J. Prior herpes simplex virus type 2 infection as a risk factor for HIV infection. JAMA. 1988;259:1048–1051. [PubMed] [Google Scholar]
- 19.Hook E W, III, Cannon R O, Nahmias A J, Lee F F, Campbell C H, Jr, Glasser D, Quinn T C. Herpes simplex virus infection as a risk factor for human immunodeficiency virus infection in heterosexuals. J Infect Dis. 1992;165:251–255. doi: 10.1093/infdis/165.2.251. [DOI] [PubMed] [Google Scholar]
- 20.Krause P R, Straus S E. Herpesvirus vaccines: development, controversies, and applications. Infect Dis Clin North Am. 1999;13:61–81. doi: 10.1016/s0891-5520(05)70043-x. [DOI] [PubMed] [Google Scholar]
- 21.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 22.McLean C S, Challanain N, Duncan D I, Boursnell M E, Jennings R, Inglis S C. Induction of a protective immune response by mucosal vaccination with a DISC HSV-1 vaccine. Vaccine. 1996;14:987–992. doi: 10.1016/0264-410x(95)00259-4. [DOI] [PubMed] [Google Scholar]
- 23.McLean C S, Erturk M, Jennings R, Nichallanain D, Minson A C, Duncan I, Boursnell M E G, Inglis S C. Protective vaccination against primary and recurrent disease caused by herpes simplex virus (HSV) type 2 using a genetically disabled HSV-1. J Infect Dis. 1994;170:1100–1109. doi: 10.1093/infdis/170.5.1100. [DOI] [PubMed] [Google Scholar]
- 24.Morrison L A, Da Costa X J, Knipe D M. Influence of mucosal and parenteral immunization with a replication-defective mutant of HSV-2 on immune responses and protection from genital challenge. Virology. 1998;243:178–187. doi: 10.1006/viro.1998.9047. [DOI] [PubMed] [Google Scholar]
- 25.Morrison L A, Knipe D M. Immunization with replication-defective mutants of herpes simplex virus type 1: sites of immune intervention in pathogenesis of challenge virus infection. J Virol. 1994;68:689–696. doi: 10.1128/jvi.68.2.689-696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison L A, Knipe D M. Mechanisms of immunization with a replication-defective mutant of herpes simplex virus 1. Virology. 1996;220:402–413. doi: 10.1006/viro.1996.0328. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen L H, Knipe D M, Finberg R W. Replication-defective mutants of herpes simplex virus (HSV) induce cellular immunity and protect against lethal HSV infection. J Virol. 1992;66:7067–7072. doi: 10.1128/jvi.66.12.7067-7072.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 29.Spang A E, Godowski P J, Knipe D M. Characterization of herpes simplex virus 2 temperature-sensitive mutants whose lesions map in or near the coding sequences for the major DNA-binding protein. J Virol. 1983;45:332–342. doi: 10.1128/jvi.45.1.332-342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speck P G, Efstathiou S, Minson A C. In vivo complementation studies of a glycoprotein H-deleted herpes simplex virus-based vector. J Gen Virol. 1996;77:2563–2568. doi: 10.1099/0022-1317-77-10-2563. [DOI] [PubMed] [Google Scholar]
- 31.Stanberry L R, Cunningham A L, Mindel A, Scott L L, Spruance S L, Aoki F Y, Lacey C J. Prospects for control of herpes simplex virus disease through immunization. Clin Infect Dis. 2000;30:549–566. doi: 10.1086/313687. [DOI] [PubMed] [Google Scholar]
- 32.Straus S E, Wald A, Kost R G, McKenzie R, Langenberg A G, Hohman P, Lekstrom J, Cox E, Nakamura M, Sekulovich R, Izu A, Dekker C, Corey L. Immunotherapy of recurrent genital herpes with recombinant herpes simplex virus type 2 glycoproteins D and B: results of a placebo-controlled vaccine trial. J Infect Dis. 1997;176:1129–1134. doi: 10.1086/514103. [DOI] [PubMed] [Google Scholar]
- 33.Wald A, Zeh J, Selke S, Warren T, Ryncarz A J, Ashley R, Kriegger J N, Corey L. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N Engl J Med. 2000;342:844–850. doi: 10.1056/NEJM200003233421203. [DOI] [PubMed] [Google Scholar]
- 34.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 35.Zhu L A, Weller S K. The UL5 gene of herpes simplex virus type 1: isolation of a lacZ insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J Virol. 1992;66:458–468. doi: 10.1128/jvi.66.1.458-468.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]