Abstract
Here we present the first complete genomic sequence, with analysis, of a very virulent strain of Marek's disease virus serotype 1 (MDV1), Md5. The genome is 177,874 bp and is predicted to encode 103 proteins. MDV1 is colinear with the prototypic alphaherpesvirus herpes simplex virus type 1 (HSV-1) within the unique long (UL) region, and it is most similar at the amino acid level to MDV2, herpesvirus of turkeys (HVT), and nonavian herpesviruses equine herpesviruses 1 and 4. MDV1 encodes 55 HSV-1 UL homologues together with 6 additional UL proteins that are absent in nonavian herpesviruses. The unique short (US) region is colinear with and has greater than 99% nucleotide identity to that of MDV1 strain GA; however, an extra nucleotide sequence at the Md5 US/short terminal repeat boundary results in a shorter US region and the presence of a second gene (encoding MDV097) similar to the SORF2 gene. MD5, like HVT, encodes an ICP4 homologue that contains a 900-amino-acid amino-terminal extension not found in other herpesviruses. Putative virulence and host range gene products include the oncoprotein MEQ, oncogenicity-associated phosphoproteins pp38 and pp24, a lipase homologue, a CxC chemokine, and unique proteins of unknown function MDV087 and MDV097 (SORF2 homologues) and MDV093 (SORF4). Consistent with its virulent phenotype, Md5 contains only two copies of the 132-bp repeat which has previously been associated with viral attenuation and loss of oncogenicity.
Marek's disease (MD) is a lymphoproliferative disease of chickens caused by the highly infectious cell-associated alphaherpesvirus MD virus serotype 1 (MDV1) (18). Yearly economic losses from MD total $1 billion worldwide (18). MDV1 infection results in a rapid onset of malignant T-cell lymphomas within several weeks of infection. Tumor infiltration results in a neural form of disease, which causes progressive paralysis, or a visceral form of disease, which is usually very acute and accompanied by high mortality. Productive virus replication in the skin and feather follicle epithelia with subsequent virus shedding is responsible for disease transmission (18).
MD is controlled by vaccination and good management practices (18). Naturally occurring nonpathogenic strains of MDV1, MDV2, and herpesvirus of turkey (HVT or MDV3) have been used individually or together in bivalent vaccines (18, 40). Recent increases in MD-related mortality and condemnations among vaccinated poultry have occurred in the United States. These increases in disease have occurred approximately 6 years after the introduction of new vaccines (99). In the late 1970s, following the introduction of HVT vaccines, and since 1992, after the introduction of bivalent MDV2-HVT-based vaccines, new MDV1 strains of greater virulence (very virulent [vv] and very virulent plus [vv+] MDV1) were isolated. These viruses are characterized by higher cytolytic activity, unusual tissue tropism, increased atrophy of lymphoid organs, immunosuppression, enhanced capacity to transform T cells, and earlier host death (7, 17, 99). It has been suggested that emergence of vv and vv+ MDV1 strains may be due to strong selective pressure generated by extensive vaccination and enhanced genetic resistance of commercial flocks (99).
To date, MDV1 genome characterization has involved partial sequencing of several different virus strains, accounting for approximately 40% of the complete genome (reviewed in reference 8). However, the genetic basis and molecular mechanisms underlying viral virulence and oncogenicity remain poorly understood. Genes encoding proteins involved in T-cell transformation (MEQ) and others with potential involvement in tumorigenicity, viral virulence, and host range (pp24, pp38, interleukin 8 [IL-8], SORF2) have been described (14, 24, 43, 57, 65, 66, 79, 89, 90, 102, 109). Additionally, virus attenuation has been associated with amplification of a 132-bp repeat within the long repeats (10–12, 34, 61, 78, 90). To improve understanding of MDV virulence and the mechanisms associated with enhanced viral virulence, more-complete information about the MDV genome and its gene complement is needed. Here we present the first complete genome sequence, with analysis, of a vv MDV1 isolate, Md5 (100).
MATERIALS AND METHODS
DNA isolation, cloning, and sequencing.
The Md5 strain of MDV was obtained from the American Type Culture Collection (Manassas, Va.) and passaged three times in primary chicken embryo fibroblast cell cultures. Viral DNA was extracted from the cytoplasm of infected cells as previously described (98). Random DNA fragments were obtained by incomplete enzymatic digestion with TaqI and AciI endonucleases (New England Biolabs, Beverly, Mass.). DNA fragments of 1.5 to 2.5 kbp were isolated after separation on agarose gels, cloned into the dephosphorylated AccI site of pUC19 plasmids, and grown in Escherichia coli DH10B cells (Gibco BRL, Gaithersburg, Md.). Plasmids were purified by alkaline lysis according to the manufacturer's instruction (Eppendorf 5 Prime, Boulder, Colo.). DNA templates were sequenced from both ends with M13 forward and reverse primers using dideoxy chain terminator sequencing chemistries (82) and the Applied Biosystem PRISM 377 automated DNA sequencer (PE Biosystems, Foster City, Calif.). ABI sequencing analysis software (version 3.3) was used for lane tracking and trace extraction. Bases were called from chromatogram traces with Phred (30), which also produced a quality file containing a predicted probability of error at each base position.
DNA sequence analysis.
DNA sequences were assembled with Phrap (29) using the quality files and default settings to produce a consensus sequence, which was manually edited with Consed (37). An identical sequence was assembled using the TIGR assembler with quality files and clone length constraints (95). Gap closure was achieved by primer walking of gap-spanning clones and sequencing of PCR products. The final DNA consensus sequence represented on average sixfold redundancy at each base position. The predicted restriction map for Md5 matched published data for the MDV1 GA strain (15). For descriptive purposes, we have presented Md5 in a linearized fashion as described by Dolan et al. (28). Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (2). Open reading frames (ORFs) encoding proteins of greater than or equal to 60 amino acids with a methionine start codon (92, 93) were evaluated for coding potential using the Hexamer (ftp.sanger.ac.uk/pub/rd) and Glimmer (81) computer programs. Other criteria included similarity to other herpesvirus or cellular proteins, published evidence for MDV proteins, and compact gene arrangement with little gene overlap (25, 96). Homology searches were conducted using Blast (3), PsiBlast (4), FASTA (70), and HMMER (16) programs with the following databases: PROSITE, Pfam, Prodom, Sbase, Blocks, Domo, and GenBank (16). GCG (26), MEMSAT (50), and SAPS (13) programs were used for gene analysis. Published mRNA and cDNA data were compared to the Md5 genomic sequence using the Est_genome (ftp.sanger.ac.uk/pub/EMBOSS) and Sim4 (33) alignment programs.
Nucleotide sequence accession number.
The MDV1 Md5 genome sequence has been deposited in GenBank under accession no. AF243438.
RESULTS AND DISCUSSION
Genome organization.
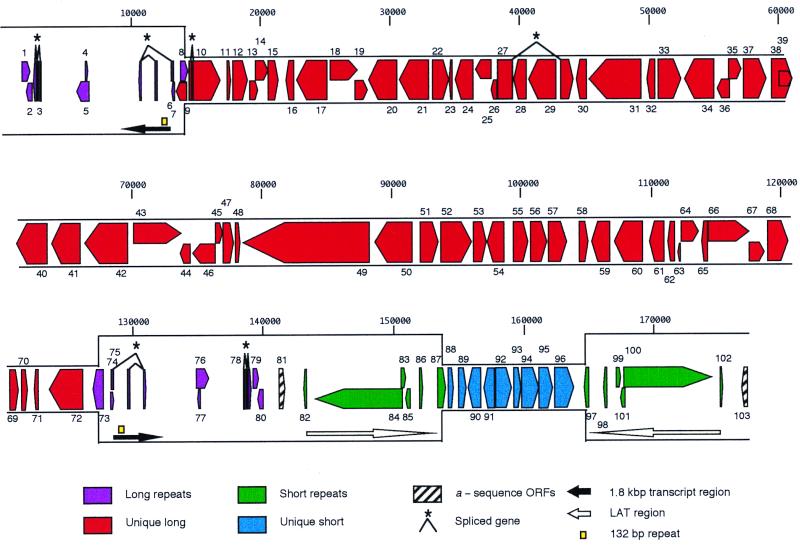
The Md5 genome is 177,874 bp long and contains a 44% G+C base composition. Md5 is organized in the same overall manner as other alphaherpesviruses (75). Long and short unique regions (UL and US regions, respectively) are 113,563 and 10,847 bp in length, respectively. Each unique region is bounded by identical inverted repeats. The terminal and internal UL repeats (TRL and IRL, respectively) are 13,065 bp, and the internal and terminal US repeats (IRS and TRS, respectively) are 12,264 bp. As with other herpesviruses, the G+C content in the repeat regions is higher than that in the unique regions (49 to 50% in repeats versus 41 to 42% in unique regions) (25, 36, 96). Md5 does not contain the retroviral long terminal repeat sequences previously reported at the US/short repeat boundary of cell culture-passaged MDV1 strains (44, 47, 48).
Alphaherpesvirus α-type sequences are located at the genomic termini and at the IRL/IRS junction of Md5. They consist of 7 tandem copies of a 60-bp repeat associated with the long direct repeat, a 43-bp unique spacer sequence, and 64 tandem copies of a 6-bp repeat associated with the short direct repeat (28, 52, 53, 96). As previously reported, the 6-bp repeat is identical to repetitive sequences in the direct repeats of human herpesvirus 6 and in eukaryotic telomeres (53).
Gene characterization.
Md5 contains 338 ORFs encoding proteins of 60 or more amino acids of which 103 are likely to be functional genes (Table 1). Seventy-three genes are present as single copies and initiate within unique regions. Thirty genes initiate and are partially or completely located within repeat regions, including two genes within α-sequence regions. MDV004, MDV007, MDV075, MDV077, MDV086, and MDV098 ORFs were annotated here because previous reports indicated that these ORFs were protein coding (41, 67, 72). Most Md5 genes are virtually identical (99% nucleotide and amino acid identity) to published genes from other MDV1 strains and less similar to those from MDV2 (40 to 86% amino acid identity) and HVT (38 to 81% amino acid identity). Among nonavian herpesviruses, EHV1 and EHV4 are generally the most similar to Md5 (25 to 61% amino acid identity).
TABLE 1.
Md5 ORFs
| ORF | Position (nt) (length [aa])a | MDV1
|
MDV2
|
HVT
|
Closest non-MDV species
|
HSV-1 homologue | Predicted structure and/or functionc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no.b | Reference | Accession no.b | Blast score | Length (aa) (% identity) | Accession no.b | Blast score | Length (aa) (% identity) | Named | Accession no.b | Blast score | Length (aa) (% identity) | ||||
| MDV001 | 1517–2173 (219) | ||||||||||||||
| MDV002 | 2478–1885 (198) | Arg-rich protein | |||||||||||||
| MDV003 | 3555–3492 (134) | AF065430 | 57 | Mus musculus | P10889 | 153 | 81 (41) | CxC chemokine | |||||||
| 3316–3181 | |||||||||||||||
| 3082–2881 | |||||||||||||||
| MDV004 | 5878–6285 (136) | U55025 | 72 | 23-kDa nuclear protein | |||||||||||
| MDV005 | 6753–5737 (339) | M89471 | 49 | MEQ protein | |||||||||||
| MDV006c | 10719–10508 | 14-kDa lytic phase protein, C-terminal exon | |||||||||||||
| MDV006b | 11821–11755 (93) | M77343 | 41 | 14-kDa lytic phase protein, alternate b N-term. exon | |||||||||||
| MDV006a | 13147–13105 (85) | L26394 | 41 | 14-kDa lytic phase protein, alternate a N-term. exon | |||||||||||
| MDV007 | 13392–13048 (115) | M82861 | 78 | ||||||||||||
| MDV008 | 13833–14297 (155) | D21060 | 109 | AB024414 | 169 | 91 (41) | 24-kDa phosphoprotein, pp24 | ||||||||
| MDV009 | 14338–13340 (333) | S72466 | 9 | ||||||||||||
| MDV010 | 14430–14535 (756) | S72466 | 9 | AB024414 | 1,985 | 757 (53) | FAdV | AF007578 | 271 | 205 (31) | Lipase | ||||
| 14701–16872 | |||||||||||||||
| MDV011 | 17431–17685 (85) | S72466 | 9 | ||||||||||||
| MDV012 | 17828–18979 (384) | AB024414 | 822 | 384 (48) | |||||||||||
| MDV013 | 19172–19756 (195) | U04994 | 105 | AB024414 | 532 | 180 (57) | SVHV | AF108378 | 123 | 130 (29) | UL1 | Virion surface glycoprotein L | |||
| MDV014 | 19641–20579 (313) | U04994 | 105 | AB024414 | 1,070 | 318 (64) | BHV-1 | AJ004801 | 683 | 228 (56) | UL2 | Uracil-DNA glycosylase | |||
| MDV015 | 20607–21290 (228) | U04994 | 105 | AB024414 | 754 | 215 (71) | EHV-4 | AF030027 | 551 | 202 (61) | UL3 | Nuclear phosphoprotein | |||
| MDV016 | 22615–21812 (268) | AB024414 | 768 | 268 (53) | EHV-1 | P28943 | 282 | 137 (43) | UL4 | Nuclear protein | |||||
| MDV017 | 25245–22672 (858) | AB024414 | 3,617 | 848 (80) | EHV-4 | AF030027 | 2,766 | 857 (60) | UL5 | DNA helicase-primase-associated protein | |||||
| MDV018 | 25313–27478 (722) | AB024414 | 2,544 | 724 (68) | EHV-1 | P28944 | 1,591 | 729 (46) | UL6 | Minor capsid protein, DNA packaging | |||||
| MDV019 | 27318–28232 (305) | AB024414 | 966 | 303 (59) | DEV | AF043730 | 634 | 279 (46) | UL7 | ||||||
| MDV020 | 30578–28272 (769) | AB024414 | 2,621 | 765 (63) | EHV-4 | AF030027 | 1,127 | 744 (35) | UL8 | DNA helicase-primase-associated protein | |||||
| MDV021 | 33117–30595 (841) | U28785 | 101 | AB024414 | 3,073 | 875 (69) | EHV-1 | P28947 | 2,089 | 838 (50) | UL9 | Ori binding protein | |||
| MDV022 | 33216–34487 (424) | AF118111 | AB021169 | 1,719 | 424 (77) | EHV-4 | AF030027 | 656 | 415 (34) | UL10 | Virion membrane glycoprotein M | ||||
| MDV023 | 34798–34547 (84) | AF118111 | AB024414 | 176 | 76 (52) | HSV-2 | P13294 | 81 | 28 (42) | UL11 | Myristylated tegument protein | ||||
| MDV024 | 36348–34777 (524) | AF118111 | AB024414 | 1,487 | 507 (56) | EHV-4 | AF030027 | 780 | 492 (38) | UL12 | DNase | ||||
| MDV025 | 37880–36342 (513) | AB024414 | 1,751 | 512 (65) | EHV-1 | P28966 | 851 | 461 (39) | UL13 | Serine/threonine protein kinase, tegument | |||||
| MDV026 | 38329–37601 (243) | AB024414 | 585 | 237 (54) | VZV | P09295 | 227 | 148 (35) | UL14 | Minor tegument protein | |||||
| MDV027 | 38360–39403 (737) | AB024414 | 3,086 | 736 (80) | EHV-1 | P28969 | 2,301 | 735 (61) | UL15 | DNA packaging protein (terminase) | |||||
| 42885–44051 | |||||||||||||||
| MDV028 | 40525–39446 (360) | AB024414 | 1,257 | 352 (66) | EHV-4 | AF030027 | 543 | 321 (37) | UL16 | Tegument protein | |||||
| MDV029 | 42741–40555 (729) | AB024414 | 2,093 | 716 (57) | EHV-4 | AF030027 | 877 | 713 (35) | UL17 | Tegument protein, DNA packaging | |||||
| MDV030 | 45132–44176 (319) | AB024414 | 1,412 | 320 (83) | EHV-1 | P28921 | 754 | 307 (46) | UL18 | Capsid protein | |||||
| MDV031 | 49439–45261 (1,393) | AB024414 | 6,508 | 1,393 (86) | Z54369 | 5,503 | 1,393 (75) | EHV-1 | P28920 | 4,209 | 1,360 (58) | UL19 | Major capsid protein | ||
| MDV032 | 50447–49746 (234) | AB018252 | 687 | 228 (56) | EHV-1 | P28971 | 260 | 200 (31) | UL20 | Membrane protein, virus egress | |||||
| MDV033 | 50708–52345 (546) | AB018252 | 1,506 | 544 (58) | EHV-1 | AF030027 | 525 | 544 (32) | UL21 | Tegument protein | |||||
| MDV034 | 54936–52498 (813) | P36336 | 85 | AB024414 | 2,451 | 808 (57) | S62554 | 2,360 | 806 (56) | CeHV-9 | U25806 | 587 | 642 (26) | UL22 | Envelope glycoprotein H |
| MDV035 | 56132–57043 (304) | A04086 | AB024414 | 727 | 303 (50) | BHV-1 | L39072 | 366 | 183 (44) | UL24 | |||||
| MDV036 | 56177–55122 (352) | P17653 | 84 | D85421 | 1,426 | 350 (74) | P25987 | 1,030 | 342 (58) | EHV-4 | AF030027 | 510 | 334 (36) | UL23 | Thymidine kinase |
| MDV037 | 57141–58889 (583) | A04086 | AB012137 | 2,274 | 581 (74) | EHV-1 | P28928 | 1,292 | 582 (48) | UL25 | DNA packaging | ||||
| MDV038 | 58933–60921 (663) | AB012137 | 1,834 | 661 (58) | EHV-1 | P28936 | 695 | 365 (42) | UL26 | Capsid maturation protease, minor capsid scaffold protein | |||||
| MDV039 | 59887–60921 (345) | UL26.5 | Minor capsid scaffold protein | ||||||||||||
| MDV040 | 63652–61058 (865) | P18538 | 77 | AB024414 | 3,786 | 865 (82) | U01887 | 3,749 | 865 (81) | CHV | S72091 | 834 (54) | UL27 | Virion membrane glycoprotein B | |
| MDV041 | 66108–63730 (793) | AB024414 | 2,917 | 790 (71) | EHV-1 | P28973 | 1,795 | 756 (48) | UL28 | DNA packaging protein | |||||
| MDV042 | 69875–66303 (1,191) | AB024414 | 5,154 | 1,191 (80) | EHV-1 | P28932 | 2,943 | 1,209 (46) | UL29 | Single-stranded DNA binding protein | |||||
| MDV043 | 70144–73803 (1,220) | L40431 | 94 | AB024309 | 4,674 | 1,220 (74) | EHV-1 | P28858 | 3,346 | 1,211 (56) | UL30 | DNA polymerase catalytic subunit | |||
| MDV044 | 74632–73733 (300) | AB024309 | 1,254 | 304 (75) | EHV-4 | AF030027 | 847 | 258 (62) | UL31 | Nuclear phosphoprotein | |||||
| MDV045 | 76573–76974 (134) | AB024309 | 442 | 134 (64) | HSV-1 | P10217 | 240 | 132 (43) | UL33 | Role in DNA packaging | |||||
| MDV046 | 76574–74652 (641) | AB024309 | 2,235 | 641 (68) | EHV-1 | AF030027 | 1,223 | 626 (45) | UL32 | Role in DNA packaging | |||||
| MDV047 | 77070–77900 (277) | AB024309 | 885 | 269 (64) | HSV-1 | P10218 | 459 | 183 (50) | UL34 | Membrane phosphoprotein | |||||
| MDV048 | 77987–78379 (131) | AB024309 | 449 | 127 (70) | BHV-1 | Z78205 | 135 | 94 (39) | UL35 | Capsid protein | |||||
| MDV049 | 88471–78446 (3,342) | AB024309 | 7,666 | 3,339 (51) | EHV-1 | P28955 | 3,120 | 2,865 (31) | UL36 | Large tegument protein | |||||
| MDV050 | 91826–88689 (1,046) | AB024309 | 3,396 | 1,046 (62) | EHV-4 | AF030027 | 1,292 | 1,033 (31) | UL37 | Tegument protein | |||||
| MDV051 | 92197–93606 (470) | AB024309 | 1,578 | 470 (62) | EHV-1 | M86664 | 817 | 451 (40) | UL38 | Capsid protein | |||||
| MDV052 | 93831–96296 (822) | AB024309 | 3,162 | 792 (76) | PRV | X72087 | 2,002 | 729 (55) | UL39 | Ribonucleotide reductase, large subunit | |||||
| MDV053 | 96352–97380 (343) | AB024309 | 1,350 | 323 (79) | EHV-1 | P28847 | 1,026 | 320 (62) | UL40 | Ribonucleotide reductase, small subunit | |||||
| MDV054 | 98756–97434 (441) | AB012572 | 1,425 | 438 (65) | BHV-1 | Z54206 | 762 | 458 (39) | UL41 | Virion host shutoff protein, tegument | |||||
| MDV055 | 99413–100519 (369) | AB012572 | 1,451 | 365 (74) | A51521 | 1,407 | 369 (69) | HSV-1 | P10226 | 1,407 | 270 (31) | UL42 | DNA polymerase processivity subunit | ||
| MDV056 | 100682–101941 (420) | AB012572 | 1,116 | 420 (54) | A51521 | 1,040 | 422 (49) | BHV-1 | Z54206 | 144 | 270 (24) | UL43 | Probable membrane protein | ||
| MDV057 | 102164–103666 (501) | P22651 | 42 | AB012572 | 1,939 | 501 (72) | P18535 | 1,999 | 488 (75) | EHV-1 | P12889 | 468 | 444 (27) | UL44 | Virion membrane glycoprotein C |
| MDV058 | 104532–105164 (211) | P22653 | 42 | AB012572 | 788 | 211 (70) | P18536 | 861 | 212 (73) | EHV-4 | AF030027 | 112 | 154 (31) | UL45 | Envelope/membrane protein, cell fusion |
| MDV059 | 107006–105303 (568) | L10283 | 103 | AB012572 | 1,035 | 553 (42) | EHV-4 | AF030027 | 511 | 413 (32) | UL46 | Tegument phosphoprotein | |||
| MDV060 | 109574–107151 (808) | L10283 | 103 | AB012572 | 1,793 | 807 (50) | EHV-4 | AF030027 | 299 | 491 (25) | UL47 | Tegument phosphoprotein | |||
| MDV061 | 111096–109816 (427) | L10283 | 103 | AB012572 | 1,211 | 409 (60) | Z54368 | 1,195 | 422 (55) | EHV-4 | Q00028 | 743 | 406 (43) | UL48 | Immediate-early gene transactivator, tegument |
| MDV062 | 111953–111207 (249) | L10283 | 103 | AB012572 | 592 | 249 (52) | VZV | P09272 | 228 | 182 (32) | UL49 | Tegument phosphoprotein | |||
| MDV063 | 112366–113673 (436) | AB012572 | 1,383 | 389 (65) | EHV-1 | P28892 | 311 | 161 (42) | UL50 | dUTPase | |||||
| MDV064 | 112386–112102 (95) | L10283 | 103 | AB012572 | 342 | 95 (70) | ILT | Y14300 | 101 | 59 (32) | UL49.5 | Envelope/tegument protein | |||
| MDV065 | 114513–113767 (249) | AB012572 | 624 | 181 (69) | EHV-1 | P28961 | 337 | 184 (42) | UL51 | Virion phosphoprotein | |||||
| MDV066 | 114515–117736 (1,074) | AB021976 | 3,786 | 1,071 (66) | VZV | P09270 | 1,859 | 1,072 (39) | UL52 | DNA helicase-primase-associated protein | |||||
| MDV067 | 117718–118779 (354) | U10040 | 74 | AB016433 | 1,218 | 351 (64) | EHV-1 | P28933 | 456 | 345 (31) | UL53 | Glycoprotein K | |||
| MDV068 | 118929–120347 (473) | U10040 | 74 | AB016433 | 1,119 | 476 (54) | PRV | X87246 | 461 | 352 (34) | UL54 | Posttranslational gene regulation | |||
| MDV069 | 121289–120483 (269) | AB016433 | 619 | 192 (61) | A51541 | 742 | 259 (54) | ||||||||
| MDV070 | 121457–121954 (166) | AB016433 | 476 | 162 (56) | A51541 | 423 | 164 (49) | EHV-4 | AF030027 | 186 | 162 (29) | UL55 | |||
| MDV071 | 122897–122316 (194) | A51541 | 337 | 143 (46) | VZV | P09267 | 143 | 112 (34) | VZV orf2, EHV-4 gene 3 | ||||||
| MDV072 | 126241–123533 (903) | D13389 | 78 | AB016433 | 2,455 | 897 (54) | A51541 | 1,255 | 771 (40) | ||||||
| MDV073 | 127787–126918 (290) | P30023 | 21 | S70347 | 287 | 157 (40) | L37202 | 115 | 72 (38) | 38-kDa phosphoprotein, pp38 | |||||
| MDV074 | 128228–128572 (115) | M82861 | 78 | ||||||||||||
| MDV075a | 128473–128515 (85) | L26394 | 41 | 14-kDa lytic phase protein, alternate a N-term. exon | |||||||||||
| MDV075b | 129799–129865 (93) | M77343 | 41 | 14-kDa lytic phase protein, alternate b N-term. exon | |||||||||||
| MDV075c | 130901–131112 | 14-kDa lytic phase protein, C-terminal exon | |||||||||||||
| MDV076 | 134867–135883 (339) | M89471 | 49 | MEQ protein | |||||||||||
| MDV077 | 135742–135335 (136) | U55025 | 72 | 23-kDa nuclear protein | |||||||||||
| MDV078 | 138065–138128 (134) | AF065430 | 57 | Mus musculus | P10889 | 153 | 81 (41) | CxC chemokine | |||||||
| 138304–138439 | |||||||||||||||
| 138538–138739 | |||||||||||||||
| MDV079 | 139142–139735 (198) | Arg-rich protein | |||||||||||||
| MDV080 | 140103–139447 (219) | ||||||||||||||
| MDV081 | 141149–141634 (162) | a sequence | |||||||||||||
| MDV082 | 143447–143124 (108) | ||||||||||||||
| MDV083 | 150610–150945 (112) | L29643 | 56 | Antisense RNA protein | |||||||||||
| MDV084 | 150769–143807 (2,321) | Q02362 | 5 | EHV-1 | P17473 | 1,009 | 1,296 (30) | RS1 | Immediate-early gene transactivator, ICP4 | ||||||
| MDV085 | 151396–151001 (132) | ||||||||||||||
| MDV086 | 151988–152248 (87) | L13604 | 67 | Cytoplasmic protein | |||||||||||
| MDV087 | 153443–153979 (179) | L22174 | 14 | FPV | P14362 | 210 | 95 (42) | MDV1 SORF2 | |||||||
| MDV088 | 154149–154685 (179) | M80595 | 80 | AB016432 | 417 | 171 (50) | X68653 | 237 | 120 (40) | FHV-1 | D42113 | 195 | 140 (34) | US1 | Immediate-early phosphoprotein, ICP22 |
| MDV089 | 154978–155616 (213) | M80595 | 80 | AB016432 | 612 | 214 (57) | A18267 | 430 | 185 (43) | FHV-1 | D42113 | 229 | 178 (32) | US10 | Virion protein |
| MDV090 | 156785–155733 (351) | L22174 | 80 | AB016432 | 948 | 345 (52) | X68653 | 757 | 345 (46) | MDV1 SORF3 | |||||
| MDV091 | 157824–157015 (270) | PRF1710264A | 76 | AB016432 | 1,061 | 265 (72) | X68653 | 908 | 259 (66) | EHV-1 | P28964 | 317 | 172 (40) | US2 | |
| MDV092 | 157936–159141 (402) | PRF1710264C | 76 | AB016432 | 1,168 | 388 (59) | A18267 | 1,201 | 386 (59) | CHV | U84223 | 595 | 351 (37) | US3 | Serine/threonine protein kinase |
| MDV093 | 159254–159694 (147) | PRF1710264D | 76 | MDV1 SORF4 | |||||||||||
| MDV094 | 159865–161073 (403) | U60532 | 14 | S83367 | 937 | 339 (56) | X68653 | 831 | 347 (48) | BHV-1 | A25176 | 237 | 210 (30) | US6 | Membrane glycoprotein D |
| MDV095 | 161183–162247 (355) | L22174 | 14 | D85420 | 829 | 346 (49) | X68653 | 610 | 334 (40) | FHV-1 | S72415 | 208 | 255 (28) | US7 | Membrane glycoprotein I |
| MDV096 | 162389–163879 (497) | L22174 | 14 | D86926 | 1,034 | 442 (47) | X68653 | 1,121 | 493 (44) | EHV-4 | AF030027 | 305 | 410 (26) | US8 | Membrane glycoprotein E |
| MDV097 | 165001–164555 (149) | M80595 | 80 | FPV | P14362 | 90 | 47 (40) | N terminus MDV1 SORF2 | |||||||
| MDV098 | 166456–166196 (87) | L13604 | 67 | Cytoplasmic protein | |||||||||||
| MDV099 | 167048–167443 (132) | ||||||||||||||
| MDV100 | 167675–174637 (2,321) | Q02362 | 5 | EHV-1 | P17473 | 1,009 | 1,296 (30) | RS1 | Immediate-early gene transactivator, ICP4 | ||||||
| MDV101 | 167834–167499 (112) | L29643 | 56 | Antisense RNA protein | |||||||||||
| MDV102 | 174997–175320 (108) | ||||||||||||||
| MDV103 | 177295–176810 (162) | a sequence | |||||||||||||
aa, amino acids; nt, nucleotides.
Accession numbers are from GenBank, SwissProt, or PRF databases.
Function was deduced either from the degree of amino acid similarity to products of known genes or by the presence of Prosite signatures. term., terminal.
FAdV, fowl adenovirus; SVHV, simian varicella herpesvirus; BHV-1, bovine herpesvirus 1, DEV, duck enteritis virus; CeHV-9, cercopithecine herpesvirus 9; CHV, canine herpesvirus; ILT, infectious laryngotracheitis virus; FPV, fowlpox virus; FHV-1, feline herpesvirus 1.
UL region.
The UL region, extending from nucleotide positions 14029 to 127591, contains 64 genes of which 38 have not been previously described (Fig. 1, Table 1). MDV013 to MDV070 genes, which represent 57% of the Md5 genome, are colinear with UL1 to UL55 genes of herpes simplex virus type 1 (HSV-1). Proteins encoded by these Md5 genes are 22 to 61% identical to HSV-1 homologues and 42 to 86% identical to MDV2 homologues. Capsid proteins MDV030, MDV031, and MDV048, DNA replication proteins MDV017, MDV021, MDV042, MDV043, MDV055, and MDV066, nuclear proteins MDV015 and MDV044, and glycoproteins MDV022, MDV040, and MDV057 encoded in the UL are highly conserved with homologues in MDV2 (66 to 86% amino acid identity). Tegument proteins (MDV023, MDV033, MDV049, MDV059, MDV060, and MDV062) and membrane proteins MDV032 and MDV056 are less conserved (42 to 65% amino acid identity). Viral enzymes MDV025, MDV052, MDV055, and MDV063 contain notable insertions and deletions compared to MDV2 homologues. MDV009, MDV010, MDV011, MDV012, MDV069, and MDV072 genes, located at the ends of the UL region, are absent in nonavian herpesviruses such as HSV and equine herpesvirus (EHV), suggesting a possible role for these genes in avian host range.
FIG. 1.
Linear map of the Md5 genome. Genes (colored arrows) are numbered from left to right based on position of methionine initiation codons. Genes and RNA transcript regions (black-and-white arrows) are transcribed in the directions indicated. Genomic regions are defined in the color key. Yellow boxes, regions of 132-bp repeats. Nucleotide positions are indicated above the map.
The MDV010 gene encodes a 684-amino-acid protein that is similar to other viral proteins and to known eukaryotic lipases (Fig. 2). The gene for the MDV010 homologue in MDV1 strain GA (MDV1 GA) has previously been shown to be spliced (9). The predicted protein contains the serine active site within the lipase signature motif (Prosite PS00120) (Fig. 2) and conserved cysteines involved in disulfide bond formation (amino acid positions 416 and 438). The region between amino acids 216 and 374 is similar to those in eukaryotic lipases such as phospholipase A1 and triacylglycerol lipase. Similarity to predicted proteins from MDV2 and fowl adenovirus extends beyond this region, suggesting the presence of virus-specific domains. The presence of a signal peptide in the amino-terminal domain and a transmembrane domain at amino acid positions 540 to 553 suggest that MDV010 may be membrane localized.
FIG. 2.
Multiple amino acid sequence alignment of MDV010 with lipases. Asterisks, conserved sites at lipase Prosite signature (PS00120); boldface, serine active sites; shaded residues, amino acid identity to MDV010. Amino acid positions are indicated on the right. Avianadeno, avian adenovirus 8, accession no. AF021254; squirrel, Spermophilus tridecemlineatus, accession no. AF027293; pig, Sus scrofa domestica, accession no. P00591; wasp, Vespula vulgaris, accession no. L43561; MDV2, accession no. AB024414; MDV1, Md5 isolate, MDV010.
Type A1 phospholipases have been identified in many mammalian tissues (platelets, liver, and heart) as membrane-bound or cytosolic enzymes which catalyze transacylation reactions (39, 64, 83). Phospholipase A1 activity on phosphatidic acid substrates may modify intracellular second-messenger pathways (39). Modification of host cell second-messenger pathways has been observed with other virus infections (1, 27, 87). Human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) encodes a G protein-coupled receptor which activates phospholipase C and which stimulates cell proliferation and transformation (35). Both cytomegalovirus and adenovirus affect arachidonic acid metabolism through pathways involving phospholipase A2 (1, 27). Arachidonic acid is a precursor to prostaglandins, leukotrienes, and lipoxins, molecules which modify inflammatory responses. Altered lipid metabolism has been observed both in vivo and in cell cultures during MDV infection (32, 38). MDV010 may perform host range functions involving alteration of host lipid metabolism and/or modification of second-messenger signaling pathways.
US region.
The US region, extending from positions 153799 to 164645 (10,847 bp), contains nine genes (encoding MDV088 to MDV097), which include homologues of the HSV-1 US1, US2, US3, US6, US7, US8, and US10 genes. The MDV1 GA US region has previously been completely sequenced (11,160 bp), and the MDV1 RB1B US region has been partially sequenced (14, 76). Md5 ORFs are colinear with and virtually identical (>99% nucleotide identity) to ORFs from these two MDV1 strains. Md5 contains sequences at the US/TRS boundary (nucleotide positions 164033 to 164518 and 164636 to 165464) that are absent in MDV1 GA. Due to the expansion of the TRS, the Md5 US region is 313 nucleotides shorter than the US region of MDV1 GA. As has been previously reported, the arrangement of genes in the MDV1 US region differs from those of other alphaherpesviruses. The MDV089 gene (US10 gene homologue) is inverted and translocated compared to the US10 gene in HSV-1, and no homologues of HSV-1 US genes are located in the short repeat regions as they are in pseudorabies virus (PRV), EHV, and varicella-zoster virus (VZV) (14, 25, 62, 76, 96, 108).
The arrangement of genes in the Md5 US region is similar to that in the US regions of MDV2 and HVT (46, 106). Proteins encoded in the Md5 US region are 47 to 72% and 40 to 66% identical to their homologues in MDV2 and HVT, respectively. MDV090, previously described as SORF3, is unique to the avian herpesviruses MDV1, MDV2, and HVT (106). MDV093 (SORF4) is unique to MDV1 (14, 46, 106). Absence of the MDV093 gene in nonpathogenic MDV2 and HVT suggests a possible role in viral virulence.
Long repeats.
The long repeat regions are 13,065 bp located at nucleotide positions 964 to 14028 and 127592 to 140656. These repeat regions contain 16 genes, all of which are unique to MDV1. Proteins from three of these genes associated with cellular transformation include the Marek's EcoRI Q fragment protein (MEQ), which is present in two copies (MDV005 and MDV076), pp38 (MDV073), and pp24 (MDV008) (24, 79, 90, 102, 109).
MDV005 and MDV076 genes encode MEQ, a 339-amino-acid basic region leucine zipper protein (49). MEQ is a transcriptional transactivator and potential oncoprotein detected in MDV-induced tumors and cell lines and has been shown to induce and maintain transformed cell phenotypes, protect transformed cells from apoptosis, and colocalize with cyclin-dependent kinase 2 in a cell cycle-dependent manner (49, 58, 59, 73, 102). Compared to MEQ from MDV1 GA, MDV005 and MDV076 contain amino acid substitutions at positions Ala 217 within the second full proline-rich repeat, Val 283, and Thr 320. The MDV004 and MDV077 genes are antisense to the MEQ gene and homologues of an ORF previously shown to encode a 23-kDa nuclear protein expressed in MDV-transformed lymphoblastoid cells (72).
The MDV008 and MDV073 genes encode oncogenicity-related phosphoproteins pp24 and pp38, respectively (21). pp24 and pp38 are among the first MDV proteins expressed in MDV-induced tumors and are part of a phosphorylated protein complex present in MDV-induced lymphoblastoid cell lines (43, 65, 66, 89). These genes span the long repeat/UL boundary (60, 109). The two proteins share 65 amino acids at their amino termini, which are encoded in the long repeats, while their carboxyl termini are encoded at either end of the UL region (60, 109). Interestingly, these two proteins have carboxyl-terminal amino acid similarity that has not been previously described. Conserved amino acids include a DLLVEAE motif (amino acid positions 85 to 91 in MDV008 and 163 to 169 in MDV073) and a region of 30% amino acid identity (amino acid positions 93 to 155 of MDV008 and 213 to 275 of MDV073). MDV2 pp24 and pp38 homologues and an HVT pp38 homologue do not contain the amino-terminal domains present in MDV1 proteins (68, 91). Given that MDV2 and HVT are nononcogenic, the novel amino-terminal regions present in MDV1 homologues may play some role in viral virulence and/or oncogenicity.
The MDV003 and MDV078 genes are spliced genes with homology to genes encoding mammalian CxC chemokines (Table 1). This gene was previously described as encoding an IL-8 homologue; however, IL-8 activity was not demonstrated (57). The MDV003 and MDV078 genes each comprise three exons, and the proteins share 41% amino acid identity with murine macrophage inhibitory protein 2 (MIP-2) and contain the four cysteine residues necessary for disulfide bonding. The amino-terminal region, which defines receptor-binding specificity, is less similar to those of MIP-2 and other CxC chemokines than is the carboxyl-terminal region (23). Chemokines mediate immune cell activation and migration during inflammation (6). Chemokine homologues encoded by other herpesviruses have been shown to function as either agonists or antagonists (54). An MDV-encoded chemokine may function in immune evasion by affecting host inflammatory responses. Chemokines have also been associated with vasculopathologies such as atherosclerosis and transplant vascular sclerosis (97). Atherosclerotic lesions including proliferative changes in arteries have been observed in MDV-infected chickens (31). Conceivably, the MDV-encoded chemokine may be involved in MDV-associated atherosclerosis.
A family of 1.8-kb RNAs mapping to the long repeat region has been associated with oncogenicity (11, 12, 51, 78) (Fig. 1). Transcripts originate from the same promoter/enhancer regions as the pp24 and pp38 genes but are transcribed in the opposite orientation (11, 22, 24, 88). Loss of oncogenicity and altered transcription have been associated with an expanded number of 132-bp repeats (4 to >35 units) within this 1.8-kb RNA region (10–12, 34, 61, 78, 90). Complex patterns of bidirectional transcription, which both initiate and terminate within the 132-bp repeats, have been previously reported (22). Md5 contains a single pair of 132-bp repeats located at nucleotide positions 12282 to 12546 and 129074 to 129338 (Fig. 1). This finding is consistent with the number of repeats (one to three copies) present in other pathogenic MDV strains (11, 61, 78). Although there are no readily identifiable genes in this region based on our criteria, there are four ORFs of greater than 60 codons present. Three of these ORFs have been previously found in cDNA clones (41, 45, 71). We have annotated the alternatively spliced MDV006 and MDV075 genes based on the work of Hong and Coussens (41), who identified a 14-kDa protein from this region. Given the complex transcriptional patterns and alternate splicing which occur within this region, additional protein-coding sequences which have not been annotated here may be present (12, 22, 41, 71).
Short repeats.
The short repeat regions are 12,264 bp at nucleotide positions 141535 to 153798 and 164646 to 176909 and contain 12 genes (Fig. 1). The MDV084 and MDV100 genes encode homologues of the HSV-1 major immediate-early transactivating protein ICP4. These proteins, which contain 2,321 amino acids and comprise over 57% of the short repeat regions, contain a 900-amino-acid amino-terminal extension compared with ICP4 homologues of other herpesviruses. A similarly sized ICP4 homologue is present in HVT and MDV1 GA (5, 107). Md5 encodes homologues of two additional immediate-early proteins found in HSV-1: ICP27 (MDV068), which is essential for HSV-1 replication, and ICP22 (MDV088). Md5 lacks homologues of immediate-early proteins ICP0, which is nonessential for the replication of HSV-1 in cell culture, and ICP47, a host range protein which blocks HSV-1 antigen presentation (104).
The region containing the US/short repeat junction is variable in MDV1, MDV2, and HVT (14, 46, 106). Expansion of the Md5 short repeat compared to those of the MDV1 Md11 strain and less-virulent JM and Cu-2 strains has been previously noted by restriction enzyme analysis (48). The MDV087 and MDV097 genes span the US/short repeat boundary (Fig. 1). The products of these two genes have 119 identical amino-terminal amino acids that are encoded within the short repeat region. MDV087 was previously described as SORF2 in MDV1 GA (14). Unlike MDV087, SORF2 is encoded entirely within the US region of MDV1 GA (14). A homologue of MDV097 is absent in MDV1 GA. SORF2 has been shown to be nonessential for replication of MDV1 in cell culture, and it is absent in the nonpathogenic MDV2 (46, 69). MDV087 and MDV097 are similar to putative proteins from fowlpox virus and fowl adenovirus, suggesting an avian host range function for these proteins (2, 14, 69, 86). Thus, differences at the US/short repeat junction, including the presence of a second gene (encoding MDV097) similar to the SORF2 gene, may affect viral virulence and contribute to Md5's enhanced virulence.
A family of latency-associated transcripts (LATs) antisense to the ICP4 homologue has been described in MDV1 (19, 20, 55, 56, 63). Although the role of LATs in viral latency and cell transformation is poorly understood, a gene homologue of the MDV086 and MDV098 genes has been shown to encode a protein from this region (67). The MDV083 and MDV101 genes also have been annotated here as potential genes. Given the complex splicing of LAT transcripts within this region, additional protein-coding sequences may be present.
Conclusions.
MDV1 genome analysis confirms the structural and functional relatedness of MDV1 to other alphaherpesviruses in gene complement and organization, particularly with regard to genes involved in basic replicative functions. Novel DNA sequences in direct repeat regions and near unique/repeat junctions contain genes likely involved in virulence and host range. The complete Md5 genome provides a basis from which comparisons with MDV strains of lesser or greater virulence may be made, thus contributing to our overall understanding of pathogen-host interactions and the evolution of MDV virulence. Additionally, this information will permit the engineering of novel MDV1 vaccine viruses and expression vectors with enhanced efficiency and greater versatility.
ACKNOWLEDGMENTS
We thank A. Ciupryk and G. Smoliga for excellent technical assistance and W. H. Martinez, F. P. Horn, and R. G. Breeze for their interest and encouragement.
REFERENCES
- 1.AbuBakar S, Boldogh I, Albrecht T. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholipase A2 and protein kinase C. Biochem Biophys Res Commun. 1990;166:953–959. doi: 10.1016/0006-291x(90)90903-z. [DOI] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson A S, Francesconi A, Morgan R W. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology. 1992;189:657–667. doi: 10.1016/0042-6822(92)90589-h. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 7.Barrow A, Venugopal K. Molecular characteristics of very virulent European MDV isolates. Acta Virol. 1999;43:90–93. [PubMed] [Google Scholar]
- 8.Becker Y, Asher Y, Bujanover S, Darai G. The dynamic herpesvirus DNA genome: the case of MDV-1 and HSV-1. Acta Virol. 1999;43:81–89. [PubMed] [Google Scholar]
- 9.Becker Y, Asher Y, Tabor E, Davidson I, Malkinson M. Open reading frames in a 4556 nucleotide sequence within MDV-1 BamHI-D DNA fragment: evidence for splicing of mRNA from a new viral glycoprotein gene. Virus Genes. 1994;8:55–69. doi: 10.1007/BF01703602. [DOI] [PubMed] [Google Scholar]
- 10.Becker Y, Tabor E, Asher Y, Davidson I, Malkinson M, Witter R L. PCR detection of amplified 132 bp repeats in Marek's disease virus type 1 (MDV-1) DNA can serve as an indicator for critical genomic rearrangement leading to the attenuation of virus virulence. Virus Genes. 1993;7:277–287. doi: 10.1007/BF01702588. [DOI] [PubMed] [Google Scholar]
- 11.Bradley G, Hayashi M, Lancz G, Tanaka A, Nonoyama M. Structure of the Marek's disease virus BamHI-H gene family: genes of putative importance for tumor induction. J Virol. 1989;63:2534–2542. doi: 10.1128/jvi.63.6.2534-2542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley G, Lancz G, Tanaka A, Nonoyama M. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed with BamHI-H. J Virol. 1989;63:4129–4135. doi: 10.1128/jvi.63.10.4129-4135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunovskis P, Velicer L F. The Marek's disease virus (MDV) unique short region: alphaherpesvirus-homologous, fowlpox virus-homologous, and MDV-specific genes. Virology. 1995;206:324–338. doi: 10.1016/s0042-6822(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 15.Buckmaster A E, Scott S D, Sanderson M J, Boursnell M E G, Ross N L J, Binns M M. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J Gen Virol. 1988;69:2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- 16.Burks C. Molecular biology database list. Nucleic Acids Res. 2000;27:1–9. doi: 10.1093/nar/27.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calnek B W, Harris R W, Buscaglia C, Schat K A, Lucio B. Relationship between the immunosuppressive potential and the pathotype of Marek's disease virus isolates. Avian Dis. 1998;42:124–132. [PubMed] [Google Scholar]
- 18.Calnek B W, Witter R L. Marek's disease. In: Calnek B W, editor. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1991. pp. 369–413. [Google Scholar]
- 19.Cantello J L, Anderson A S, Morgan R W. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek's disease virus. J Virol. 1994;68:6280–6290. doi: 10.1128/jvi.68.10.6280-6290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantello J L, Parcells M S, Anderson A S, Morgan R W. Marek's disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J Virol. 1997;71:1353–1361. doi: 10.1128/jvi.71.2.1353-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Sondermeijer P J A, Velicer L F. Identification of a unique Marek's disease virus gene which encodes a 38-kilodalton phosphoprotein and is expressed in both lytically infected cells and latently infected lymphoblastoid tumor cells. J Virol. 1992;66:85–94. doi: 10.1128/jvi.66.1.85-94.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Velicer L F. Multiple bidirectional initiations and terminations of transcription in the Marek's disease virus long repeat regions. J Virol. 1991;65:2445–2451. doi: 10.1128/jvi.65.5.2445-2451.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark-Lewis I, Kim K-S, Rajarathnam K, Gong J-H, Dewald B, Moser B, Baggiolini M, Sykes B D. Structure-activity relationships of chemokines. J Leukoc Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 24.Cui Z, Lee L F, Liu J-L, Kung H-J. Structural analysis and transcriptional mapping of the Marek's disease virus gene encoding pp38, an antigen associated with transformed cells. J Virol. 1991;65:6509–6515. doi: 10.1128/jvi.65.12.6509-6515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 26.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitrov T, Krajcsi P, Hermiston T W, Tollefson A E, Hannink M, Wold W S M. Adenovirus E3-10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J Virol. 1997;71:2830–2837. doi: 10.1128/jvi.71.4.2830-2837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 30.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 31.Fabricant C G, Fabricant J, Minick C R, Litrenta M M. Herpesvirus-induced atherosclerosis in chickens. Fed Proc. 1983;42:2476–2479. [PubMed] [Google Scholar]
- 32.Fabricant C G, Hajjar D P, Minick C R, Fabricant J. Herpesvirus infection enhances cholesterol and cholesteryl ester accumulation in cultured arterial smooth muscle cells. Am J Pathol. 1981;105:176–184. [PMC free article] [PubMed] [Google Scholar]
- 33.Florea L, Hartzell G, Zhang Z, Rubin G, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuchi K, Tanaka A, Schierman L W, Witter R L, Nonoyama M. The structure of Marek's disease virus DNA: the presence of unique expansion in nonpathogenic viral DNA. Proc Natl Acad Sci USA. 1985;82:751–754. doi: 10.1073/pnas.82.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geras-Raaka E, Arvanitakis L, Bais C, Cesarman E, Mesri E A, Gershengorn M C. Inhibition of constitutive signaling of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor by protein kinases in mammalian cells in culture. J Exp Med. 1998;187:801–806. doi: 10.1084/jem.187.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 37.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:192–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 38.Hajjar D P, Fabricant C G, Minick C R, Fabricant J. Virus-induced atherosclerosis. Herpesvirus infection alters aortic cholesterol metabolism and accumulation. Am J Pathol. 1986;122:62–70. [PMC free article] [PubMed] [Google Scholar]
- 39.Higgs H N, Glomset J A. Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. J Biol Chem. 1996;271:10874–10883. doi: 10.1074/jbc.271.18.10874. [DOI] [PubMed] [Google Scholar]
- 40.Hirai K, Nakajima K, Ikuta K, Kirisawa R, Kawakami Y, Mikami T, Kato S. Similarities and dissimilarities in the structure and expression of viral genomes of various virus strains immunologically related to Marek's disease virus. Arch Virol. 1986;89:113–130. doi: 10.1007/BF01309883. [DOI] [PubMed] [Google Scholar]
- 41.Hong Y, Coussens P M. Identification of an immediate-early gene in the Marek's disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide. J Virol. 1994;68:3593–3603. doi: 10.1128/jvi.68.6.3593-3603.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ihara T, Kato A, Ueda S, Ishihama A, Hiari K. Comparison of the sequence of the secretory glycoprotein A (gA) gene in Md5 and BC-1 strains of Marek's disease virus type 1. Virus Genes. 1989;3:127–140. doi: 10.1007/BF00125125. [DOI] [PubMed] [Google Scholar]
- 43.Ikuta K, Nakajima K, Naito M, Ann S H, Ueda S, Kato S, Hirai K. Identification of Marek's disease virus-specific antigens in Marek's disease lymphoblastoid cell lines using monoclonal antibody against virus-specific phosphorylated polypeptides. Int J Cancer. 1985;35:257–264. doi: 10.1002/ijc.2910350219. [DOI] [PubMed] [Google Scholar]
- 44.Isfort R J, Qian Z, Jones D, Silva R F, Witter R, Kung H-J. Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology. 1994;203:125–133. doi: 10.1006/viro.1994.1462. [DOI] [PubMed] [Google Scholar]
- 45.Iwata A, Ueda S, Ishihama A, Hirai K. Sequence determination of cDNA clones of transcripts from the tumor-associated region of the Marek's disease virus genome. Virology. 1992;187:805–808. doi: 10.1016/0042-6822(92)90483-6. [DOI] [PubMed] [Google Scholar]
- 46.Jang H-K, Ono M, Kim T-J, Izumiya Y, Damiani A M, Matsumura T, Niikura M, Kai C, Mikami T. The genetic organization and transcriptional analysis of the short unique region in the genome of nononcogenic Marek's disease virus serotype 2. Virus Res. 1998;58:137–147. doi: 10.1016/s0168-1702(98)00110-5. [DOI] [PubMed] [Google Scholar]
- 47.Jones D, Brunovskis P, Witter R, Kung H-J. Retroviral insertional activation in a herpesvirus: transcriptional activation of Us genes by an integrated long terminal repeat in a Marek's disease virus clone. J Virol. 1996;70:2460–2467. doi: 10.1128/jvi.70.4.2460-2467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones D, Isfort R, Witter R, Kost R, Kung H-J. Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences. Proc Natl Acad Sci USA. 1993;90:3855–3859. doi: 10.1073/pnas.90.9.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones D, Lee L, Liu J-L, Kung H-J, Tillotson J K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 51.Kawamura M, Hayashi M, Furuichi T, Nonoyama M, Isogai E, Namioka S. The inhibitory effects of oligonucleotides, complementary to Marek's disease virus mRNA transcribed from the BamHI-H region, on the proliferation of transformed lymphoblastoid cells, MDCC-MSB1. J Gen Virol. 1991;72:1105–1111. doi: 10.1099/0022-1317-72-5-1105. [DOI] [PubMed] [Google Scholar]
- 52.Kishi M, Bradley G, Jessip J, Tanaka A, Nonoyama M. Inverted repeat regions of Marek's disease virus DNA possess a structure similar to that of the α sequence of herpes simplex virus DNA and contain host cell telomere sequences. J Virol. 1991;65:2791–2797. doi: 10.1128/jvi.65.6.2791-2797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishi M, Harada H, Takahashi M, Tanaka A, Hayashi M, Nonoyama M, Josephs S F, Buchbinder A, Schachter F, Ablashi D V, Wong-Staal F, Salahuddin S Z, Gallo R C. A repeat sequence, GGGTTA, is shared by DNA of human herpesvirus 6 and Marek's disease virus. J Virol. 1988;62:4824–4827. doi: 10.1128/jvi.62.12.4824-4827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lalani A S, Barrett J W, McFadden G. Modulating chemokines: more lessons from viruses. Immunol Today. 2000;21:100–106. doi: 10.1016/s0167-5699(99)01556-x. [DOI] [PubMed] [Google Scholar]
- 55.Li D, O'Sullivan G, Greenall L, Smith G, Jiang C, Ross N. Further characterization of the latency-associated transcription unit of Marek's disease virus. Arch Virol. 1998;143:295–311. doi: 10.1007/s007050050287. [DOI] [PubMed] [Google Scholar]
- 56.Li D-S, Pastorek J, Zelnik V, Smith G D, Ross L J N. Identification of novel transcripts complementary to the Marek's disease virus homologue of the ICP4 gene of herpes simplex virus. J Gen Virol. 1994;75:1713–1722. doi: 10.1099/0022-1317-75-7-1713. [DOI] [PubMed] [Google Scholar]
- 57.Liu J-L, Lin S-F, Xia L, Brunovskis P, Li D, Davidson I, Lee L F, Kung H-J. MEQ and V-IL8: cellular genes in disguise? Acta Virol. 1999;43:94–101. [PubMed] [Google Scholar]
- 58.Liu J-L, Ye Y, Lee L F, Kung H-J. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J Virol. 1998;72:388–395. doi: 10.1128/jvi.72.1.388-395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J-L, Ye Y, Qian Z, Qian Y, Templeton D J, Lee L F, Kung H-J. Functional interactions between herpesvirus oncoprotein MEQ and cell cycle regulator CDK2. J Virol. 1999;73:4208–4219. doi: 10.1128/jvi.73.5.4208-4219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makimura K, Peng F-Y, Tsuji M, Hasegawa S, Kawai Y, Nonoyama M, Tanaka A. Mapping of Marek's disease virus genome: identification of junction sequences between unique and inverted repeat regions. Virus Genes. 1994;8:15–24. doi: 10.1007/BF01703598. [DOI] [PubMed] [Google Scholar]
- 61.Maotani K, Kanamori A, Ikuta K, Ueda S, Kato S, Hirai K. Amplification of a tandem direct repeat within inverted repeats of Marek's disease virus DNA during serial in vitro passage. J Virol. 1986;58:657–660. doi: 10.1128/jvi.58.2.657-660.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 63.McKie E A, Ubukata E, Hasegawa S, Zhang S, Nonoyama M, Tanaka A. The transcripts from the sequences flanking the short component of Marek's disease virus during latent infection from a unique family of 3′-coterminal RNAs. J Virol. 1995;69:1310–1314. doi: 10.1128/jvi.69.2.1310-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagai Y, Aoki J, Sato T, Amano K, Matsuda Y, Arai H, Inoue K. An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J Biol Chem. 1999;274:11053–11059. doi: 10.1074/jbc.274.16.11053. [DOI] [PubMed] [Google Scholar]
- 65.Naito M, Nakajima K, Iwa N, Ono K, Yoshida I, Konobe T, Ikuta K, Ueda S, Kato S, Hirai K. Demonstration of a Marek's disease virus-specific antigen in tumour lesions of chickens with Marek's disease using monoclonal antibody against a virus phosphorylated protein. Avian Pathol. 1986;15:503–510. doi: 10.1080/03079458608436311. [DOI] [PubMed] [Google Scholar]
- 66.Nakajima K, Ikuta K, Naito M, Ueda S, Kato S, Hirai K. Analysis of Marek's disease virus serotype 1-specific phosphorylated polypeptides in virus-infected cells and Marek's disease lymphoblastoid cells. J Gen Virol. 1987;68:1379–1389. doi: 10.1099/0022-1317-68-5-1379. [DOI] [PubMed] [Google Scholar]
- 67.Ohashi K, O'Connell P H, Schat K A. Characterization of Marek's disease virus BamHI-A-specific cDNA clones obtained from a Marek's disease lymphoblastoid cell line. Virology. 1994;199:275–283. doi: 10.1006/viro.1994.1125. [DOI] [PubMed] [Google Scholar]
- 68.Ono M, Kawaguchi Y, Maeda K, Kamiya N, Tohya Y, Kai C, Niikura M, Mikami T. Nucleotide sequence analysis of Marek's disease virus (MDV) serotype 2 homolog of MDV serotype 1 pp38, an antigen associated with transformed cells. Virology. 1994;201:142–146. doi: 10.1006/viro.1994.1275. [DOI] [PubMed] [Google Scholar]
- 69.Parcells M S, Anderson A S, Cantello J L, Morgan R W. Characterization of Marek's disease virus insertion and deletion mutants that lack US1 (ICP22 homolog), US10, and/or US2 and neighboring short-component open reading frames. J Virol. 1994;68:8239–8253. doi: 10.1128/jvi.68.12.8239-8253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 71.Peng F, Bradley G, Tanaka A, Lancz G, Nonoyama M. Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek's disease virus. J Virol. 1992;66:7389–7396. doi: 10.1128/jvi.66.12.7389-7396.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Q, Shirazi Y. Isolation and characterization of Marek's disease virus (MDV) cDNAs from a MDV-transformed lymphoblastoid cell line: identification of an open reading frame antisense to the MDV Eco-Q protein (Meq) Virology. 1996;221:368–374. doi: 10.1006/viro.1996.0388. [DOI] [PubMed] [Google Scholar]
- 73.Qian Z, Brunovskis P, Rauscher F I, Lee L, Kung H-J. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J Virol. 1995;69:4037–4044. doi: 10.1128/jvi.69.7.4037-4044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren D, Lee L F, Coussens P M. Identification and characterization of Marek's disease virus genes homologous to ICP27 and glycoprotein K of herpes simples virus-1. Virology. 1994;204:242–250. doi: 10.1006/viro.1994.1528. [DOI] [PubMed] [Google Scholar]
- 75.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 76.Ross L J N, Binns M M, Pastorek J. DNA sequence and organization of genes in a 5.5 kbp EcoRI fragment mapping in the short unique segment of Marek's disease virus (strain RB1B) J Gen Virol. 1991;72:949–954. doi: 10.1099/0022-1317-72-4-949. [DOI] [PubMed] [Google Scholar]
- 77.Ross L J N, Sanderson M, Scott S D, Binns M M, Doel T, Milne B. Nucleotide sequence and characterization of the Marek's disease virus homologue of glycoprotein B of herpes simplex virus. J Gen Virol. 1989;70:1789–1804. doi: 10.1099/0022-1317-70-7-1789. [DOI] [PubMed] [Google Scholar]
- 78.Ross N, Binns M M, Sanderson M, Schat K A. Alterations in DNA sequence and RNA transcription of the BamHI-H fragment accompany attenuation of oncogenic Marek's disease herpesvirus. Virus Genes. 1993;7:33–51. doi: 10.1007/BF01702347. [DOI] [PubMed] [Google Scholar]
- 79.Ross N, O'Sullivan G, Rothwell C, Smith G, Burgess S C, Rennie M, Lee L F, Davison T F. Marek's disease virus EcoRI-Q gene (meq) and a small RNA antisense to ICP4 are abundantly expressed in CD4+ cells and cells carrying a novel lymphoid marker, AV37, in Marek's disease lymphomas. J Gen Virol. 1997;78:2191–2198. doi: 10.1099/0022-1317-78-9-2191. [DOI] [PubMed] [Google Scholar]
- 80.Sakaguchi M, Urakawa T, Hirayama Y, Miki N, Yamamoto M, Hirai K. Sequence determination and genetic content of an 8.9-kb restriction fragment in the short unique region and the internal inverted repeat of Marek's disease virus type 1 DNA. Virus Genes. 1992;6:365–378. doi: 10.1007/BF01703085. [DOI] [PubMed] [Google Scholar]
- 81.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sato T, Aoki J, Nagai Y, Dohmae N, Takio K, Doi T, Arai H, Inoue K. Serine phospholipid-specific phospholipase A that is secreted from activated platelets. J Biol Chem. 1997;272:2192–2198. doi: 10.1074/jbc.272.4.2192. [DOI] [PubMed] [Google Scholar]
- 84.Scott S D, Ross N L J, Binns M M. Nucleotide and predicted amino acid sequences of the Marek's disease virus and turkey herpesvirus thymidine kinase genes; comparisons with thymidine kinase genes of other herpesviruses. J Gen Virol. 1989;70:3055–3065. doi: 10.1099/0022-1317-70-11-3055. [DOI] [PubMed] [Google Scholar]
- 85.Scott S D, Smith G D, Ross N L J, Binns M M. Identification and sequence analysis of the homologues of the herpes simplex virus type 1 glycoprotein H in Marek's disease virus and the herpesvirus of turkeys. J Gen Virol. 1993;74:1185–1190. doi: 10.1099/0022-1317-74-6-1185. [DOI] [PubMed] [Google Scholar]
- 86.Sheppard M, Werner W, Tsatas E, McCoy R, Prowse S, Johnson M. Fowl adenovirus recombinant expressing VP2 of infectious bursal disease virus induces protective immunity against bursal disease. Arch Virol. 1998;143:915–930. doi: 10.1007/s007050050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shibutani T, Johnson T M, Yu Z-X, Ferrans V J, Moss J, Epstein S E. Pertussis toxin-sensitive G proteins as mediators of the signal transduction pathways activated by cytomegalovirus infection of smooth muscle cells. J Clin Investig. 1997;100:2054–2061. doi: 10.1172/JCI119738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shigekane H, Kawaguchi Y, Shirakata M, Sakaguchi M, Hirai K. The bi-directional transcriptional promoters for the latency-relating transcripts of the pp38/pp24 mRNAs and the 1.8 kb-mRNA in the long inverted repeats of Marek's disease virus serotype 1 DNA are regulated by common promoter-specific enhancers. Arch Virol. 1999;144:1893–1907. doi: 10.1007/s007050050713. [DOI] [PubMed] [Google Scholar]
- 89.Silva R F, Lee L F. Monoclonal antibody-mediated immunoprecipitation of proteins from cells infected with Marek's disease virus or turkey herpesvirus. Virology. 1984;136:307–320. doi: 10.1016/0042-6822(84)90167-3. [DOI] [PubMed] [Google Scholar]
- 90.Silva R F, Witter R L. Genomic expansion of Marek's disease virus DNA is associated with serial in vitro passage. J Virol. 1985;54:690–696. doi: 10.1128/jvi.54.3.690-696.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith G D, Zelnik V, Ross L J N. Gene organization in herpesvirus of turkeys: identification of a novel open reading frame in the long unique region and a truncated homologue of pp38 in the internal repeat. Virology. 1995;207:205–216. doi: 10.1006/viro.1995.1067. [DOI] [PubMed] [Google Scholar]
- 92.Staden A, McLachlan A D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sui D, Wu P, Kung H J, Lee L F. Identification and characterization of a Marek's disease virus gene encoding DNA polymerase. Virus Res. 1995;36:269–278. doi: 10.1016/0168-1702(94)00114-r. [DOI] [PubMed] [Google Scholar]
- 95.Sutton G G, White O, Adams M D, Kerlavage A R. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 96.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 97.Terkeltaub R, Boisvert W A, Curtiss L K. Chemokines and atherosclerosis. Curr Opin Lipidol. 1998;8:397–405. doi: 10.1097/00041433-199810000-00003. [DOI] [PubMed] [Google Scholar]
- 98.Wesley R D, Tuthill A E. Genome relatedness among African swine fever virus field isolates by restriction endonuclease analysis. Prev Vet Med. 1984;2:53–62. [Google Scholar]
- 99.Witter R L. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 100.Witter R L, Sharma J M, Fadly A M. Pathogenicity of variant Marek's disease virus isolants in vaccinated and unvaccinated chickens. Avian Dis. 1980;24:210–232. [Google Scholar]
- 101.Wu T-F, Sun W, Boussaha M, Southwick R, Coussens P M. Cloning and sequence analysis of Marek's disease virus origin binding protein (OBP) reveals strict conservation of structural motifs among OBPs of divergent alphaherpesviruses. Virus Genes. 1996;13:143–157. doi: 10.1007/BF00568907. [DOI] [PubMed] [Google Scholar]
- 102.Xie Q, Anderson A S, Morgan R W. Marek's disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J Virol. 1996;70:1125–1131. doi: 10.1128/jvi.70.2.1125-1131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yanagida N, Yoshida S, Nazerian K, Lee L F. Nucleotide and predicted amino acid sequences of Marek's disease virus homologues of herpes simplex virus major tegument proteins. J Gen Virol. 1993;74:1837–1845. doi: 10.1099/0022-1317-74-9-1837. [DOI] [PubMed] [Google Scholar]
- 104.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 105.Yoshida S, Lee L F, Yanagida N, Nazerian K. Identification and characterization of a Marek's disease virus gene homologous to glycoprotein L of herpes simplex virus. Virology. 1994;204:414–419. doi: 10.1006/viro.1994.1546. [DOI] [PubMed] [Google Scholar]
- 106.Zelnik V, Darteil R, Audonnet J C, Smith G D, Riviere M, Pastorek J, Ross L J N. The complete sequence and gene organization of the short unique region of herpesvirus of turkeys. J Gen Virol. 1993;74:2151–2162. doi: 10.1099/0022-1317-74-10-2151. [DOI] [PubMed] [Google Scholar]
- 107.Zelnik V, Kopacek J, Rejholcova O, Kabat P, Pastorek J. ICP4 homologues of both Marek's disease virus and herpesvirus of turkeys are larger than their alphaherpesvirus counterparts. In: Silva R F, Cheng H H, Coussens P M, Lee L F, Velicer L F, editors. Current research on Marek's disease. Kennett Square, Pa: American Association of Avian Pathologists; 1996. pp. 164–169. [Google Scholar]
- 108.Zhang G, Leader D P. The structure of the pseudorabies virus genome at the end of the inverted repeat sequences proximal to the junction with the short unique region. J Gen Virol. 1990;71:2433–2441. doi: 10.1099/0022-1317-71-10-2433. [DOI] [PubMed] [Google Scholar]
- 109.Zhu G-S, Iwata A, Gong M, Ueda S, Hirai K. Marek's disease virus type 1-specific phosphorylated proteins pp38 and pp24 with common amino acid termini are encoded from the opposite junction regions between the long unique and inverted repeat sequences of viral genome. Virology. 1994;200:816–820. doi: 10.1006/viro.1994.1249. [DOI] [PubMed] [Google Scholar]