Abstract
We present a novel mechanism by which viruses may inhibit the alpha/beta interferon (IFN-α/β) cascade. The double-stranded RNA (dsRNA) binding protein NS1 of influenza virus is shown to prevent the potent antiviral interferon response by inhibiting the activation of interferon regulatory factor 3 (IRF-3), a key regulator of IFN-α/β gene expression. IRF-3 activation and, as a consequence, IFN-β mRNA induction are inhibited in wild-type (PR8) influenza virus-infected cells but not in cells infected with an isogenic virus lacking the NS1 gene (delNS1 virus). Furthermore, NS1 is shown to be a general inhibitor of the interferon signaling pathway. Inhibition of IRF-3 activation can be achieved by the expression of wild-type NS1 in trans, not only in delNS1 virus-infected cells but also in cells infected with a heterologous RNA virus (Newcastle disease virus). We propose that inhibition of IRF-3 activation by a dsRNA binding protein significantly contributes to the virulence of influenza A viruses and possibly to that of other viruses.
Cells have the potential to respond quickly to virus infection by activation of the interferon (IFN) pathway. This rapid response is possible because of constitutively expressed transcriptional regulators, such as IFN regulatory factor 3 (IRF-3). The importance of IRF-3 in the initiation of the innate antiviral response has only recently been demonstrated (30, 31, 38, 41). IRF-3 acts as a sentinel of sorts: upon virus infection or exposure to double-stranded RNA (dsRNA), IRF-3 is rapidly activated by phosphorylation on several C-terminal serine and threonine residues (2, 22). Phosphorylation of IRF-3 results in its nuclear accumulation, where it assembles with other transcription factors and contributes to the induction of the transcription of specific defense genes, including beta interferon (IFN-β) (19, 30, 37). This in turn initiates the IFN cascade to induce an antiviral state in the cell.
Several viruses encode factors that target specific mediators of the antiviral state (reviewed in reference 34). Among the best-characterized viral targets are protein kinase R (PKR) and (2′-5′) oligoadenylate synthetase (12, 18). The IRF family has also recently been shown to be a strategic target for viruses such as human herpesvirus 8, which encodes viral IRF homologues (3, 4, 28). The E6 protein of human papillomavirus is known to bind directly to IRF-3, inhibiting the transcriptional activation of the IFN-β gene (29). The ability of viruses to inhibit the host IFN system is most likely a major determinant of their pathogenic properties.
We have demonstrated that an influenza A virus lacking the NS1 protein (delNS1 virus), but not wild-type influenza A virus, strongly induces the production of IFN in vivo and the stimulation of an IFN-regulated gene (13; M. Salvatore and A. Garcia-Sastre, unpublished data). Results from the present study indicate that differential activation of IRF-3 correlates with differences in IFN induction between these two influenza viruses. The NS1 protein of influenza virus has been associated with multiple activities, including the binding of dsRNA (reviewed in reference 20). We hypothesize that the influenza virus NS1 protein, by binding to the dsRNA generated during influenza virus infection, prevents the activation of this key mediator of the innate cellular immune response. The inhibition of IRF-3 activation by an influenza virus-encoded dsRNA binding protein represents a novel mechanism by which IFN synthesis can be inhibited. This mechanism may be relevant to the pathogenesis of other viruses.
MATERIALS AND METHODS
Viruses and cells.
DelNS1 virus was generated by ribonucleoprotein transfection of an altered viral influenza NS gene, using the temperature-sensitive helper virus 25A-1 as described previously (13). Influenza A viruses, Sendai virus, and Newcastle disease virus (NDV) were grown in embryonated chicken eggs (SPAFAS, Inc.) at 37°C for 48 h. Seven-day-old embryonated eggs were used to grow delNS1 virus stock, since delNS1 virus is unable to counteract the IFN response present in older eggs (25, 32). Ten-day-old embryonated eggs were used to grow wild-type PR8 virus, Sendai virus, or NDV. Approximately 100 PFU of virus was injected into each egg. Plaque assay of influenza A virus or NDV stocks was performed on MDCK cells in the presence of 2 μg of trypsin (Difco Laboratories)/ml at 37°C.
HEC-1b cells (ATCC) are endometrial carcinoma cells of human epithelial origin. These cells have been used by several groups for IRF-3 analyses since they are reportedly nonresponsive to interferon (5, 11). U3A cells have homozygous mutations in the STAT1 gene, a signal transducer required for alpha/beta IFN (IFN-α/β) signaling (24). Therefore, any effect on the IFN response pathway in either of these cell lines should be independent of IFN signaling. 293T cells are human epithelial cells which are known to express simian virus 40 large T antigen.
Expression plasmids.
pCAGGS-NS1(SAM) and pCAGGS-NS1-R38AK41A(SAM) contain the open reading frame of NS1 from wild-type influenza A (PR8) virus in an expression vector containing a chicken β-actin promotor (pCAGGS) (26). The Stratagene Quikchange site-directed mutagenesis kit was used to mutate the splice acceptor in the plasmid pT3NS using the oligonucleotides NS3ss (5′-ATTGCCTTCTCTTCCCGGACATACTGCTGAG-3′) and NS2ss2 (5′-CTCAGCAGTATGTCCGGGAAGAGAAGGCAAT-3′) as instructed by the manufacturer. The splice acceptor mutation was confirmed by sequencing. NS1 “SAM” was then amplified by PCR using the oligonucleotides NS1EcoRI5′ (5′-GCGCGAATTCAATAATGGATCCAAACACTG-3′) and NS1XhoI3′ (5′-GCGCCTCGAGTCAAACTTCTGACCTAATT-3′) and cloned between the EcoRI and XhoI sites of pCAGGS, creating pCAGGS-NS1(SAM). pCAGGS-NS1-R38AK41A(SAM) was constructed by PCR amplification of two fragments from pCAGGS-NS1(SAM) and encodes an NS1 protein with mutations at amino acid positions 38 (R→A) and 41 (K→A) and which contains a silent NheI site. PCRs were performed using primers NS1EcoRI5′ and R38AK41A.b (5′-GATCGGCTTCGCGCAGATCAGGCTAGCCTAAGAGGAAGA-3′) and R38AK41A.t (5′-TCTTCCTCTTAGGCTAGCCTGATCTGCGCGAAGCCGATC-3′) and NS1XhoI3′. The PCR products were digested with EcoRI and NheI and with NheI and XhoI, respectively, and cloned into pCAGGS between the EcoRI and XhoI sites. Mutations were confirmed by sequencing.
pGFP-mIRF-3 expresses green fluorescent protein (GFP) fused to mouse IRF-3 (71% identical to human IRF-3) and was a gift of Tak Mak (University of Toronto, Toronto, Ontario, Canada). GFP-mIRF-3 was found to be concentrated in the nucleus in response to delNS1 virus or NDV infection in a variety of cell lines, including U3A, HEC-1b, and CV1 (data not shown).
pCAGGS-hIRF3 expresses the human IRF-3 open reading frame, which was amplified from HeLa cell mRNA by reverse transcription using an oligodeoxyribosylthymine primer, followed by PCR using the primers IRF-3/5′EcoRI (5′-CCGGAATTCATAATGGGAACCCCAAAG-3′) and IRF-3/3′XhoI (5′-CCGCTCGAGTCAGCTCTCCCCAGGGCC-3′). The IRF-3 PCR product was cloned into the pCAGGS vector between the EcoRI and XhoI sites.
Transfection and infection.
Transfections were performed using Lipofectamine 2000 (Gibco) as instructed by the manufacturer. Where indicated, an empty vector (pCAGGS) was added to maintain a constant amount of 0.5 μg of DNA for each transfection per 24-well coverslip. HEC-1b and U3A transfections were performed in duplicate and incubated at 37°C for 24 h before the addition of virus. Transfected cells were infected with a multiplicity of infection (MOI) of 1 with delNS1 or PR8 virus and with an MOI of 2 for NDV. For the Western blot of transfected IRF-3 expression, 293T cells were transfected in suspension and then plated onto 35-mm dishes. Twenty-four hours later, cells were infected with an MOI of approximately 3 for delNS1 virus or approximately 10 for PR8 virus or Sendai virus. Infections were performed in phosphate-buffered saline (PBS) for 1 h at 37°C. After 1 h, medium (Dulbecco modified Eagle medium and 10% fetal calf serum) was added (t = 0) and the mixture was incubated for the indicated time.
Fluorescence.
Cells were fixed and permeabilized for 20 min at room temperature with 2.5% formaldehyde–0.5% Triton X-100. Fixed cells were washed extensively with PBS and, where indicated, incubated with a monoclonal antibody to hIRF-3 (SL-12, kindly provided by Peter Howley) (dilution, 1:500) and a rabbit polyclonal antibody directed against the influenza virus NS1 protein (27) (dilution, 1:2,000) for 12 h. Washed cells were incubated with secondary antibodies directed against mouse (dilution, 1:750) or rabbit (dilution, 1:1,500) immunoglobulin G (IgG) (Texas Red or fluorescein isothiocyanate, respectively). All antibodies were diluted in PBS with 3% bovine serum albumin. Cells were affixed to slides, using mounting medium containing an antifade reagent (Molecular Probes). Approximately 100 to 500 cells for each experimental condition were counted using an Olympus IX-70 fluorescence microscope fitted with a Sony Catseye digital camera. Slide labels were covered during counting to minimize bias. Digital images were processed using Photoshop 4.01 software on a Macintosh Power PC.
Western analysis.
Approximately 106 infected 293T cells were pelleted and lysed in 100 μl of sodium dodecyl sulfate (SDS) buffer (0.5% SDS, 0.05 M Tris 7.5, 1 mM dithiothreitol). One percent of the cell extract was resolved by SDS–10% polyacrylamide gel electrophoresis (PAGE), and the gel was transferred to a nitrocellulose membrane. Transfected IRF-3 was detected by chemiluminescence using a monoclonal antibody against IRF-3 (SL-12; dilution, 1:1,000 in 5% milk), and an anti-mouse IgG peroxidase-labeled antibody (Amersham; dilution, 1:10,000 in 5% milk).
RT-PCR of IFN-β mRNA.
Total RNA was prepared from confluent 100-mm dishes of HEC-1b cells or HEC-1b cells infected with NDV, influenza A/PR8 virus, or delNS1 virus at an MOI of 1 for 14 h using Trizol reagent (Gibco). The RNA was digested with DNase 1 and subjected to reverse transcription (RT)-PCR analysis essentially as previously described (17).
RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase using random hexamer primers. A mock reaction was carried out with no reverse transcriptase added (−RT). One tenth of the resulting cDNA was used as a template for 25 cycles of PCR in the presence of [32P]dATP using specific primers for IFN-β or glyceraldehyde-3-phosphate-dehydrogenase (GAPDH). As a control for genomic DNA contamination, PCR was carried out with GAPDH primers using the mock (−RT) reactions as a template. Following polyacrylamide gel electrophoresis (PAGE), products were detected by autoradiography. The following primer sequences were used: GAPDH a, 5′-GTGAAGGTCGGAGTCAAC-3′; GAPDH b, 5′-TGGAATTTGCCATGGGTG-3′; IFN-β a, 5′-CACGACAGCTCTTTCCATGA-3′; and IFN-β b, 5′-AGCCAGTGCTCGATGAATCT-3′.
RESULTS
Inhibition of hIRF3 activation by wild-type influenza virus but not by a virus lacking NS1.
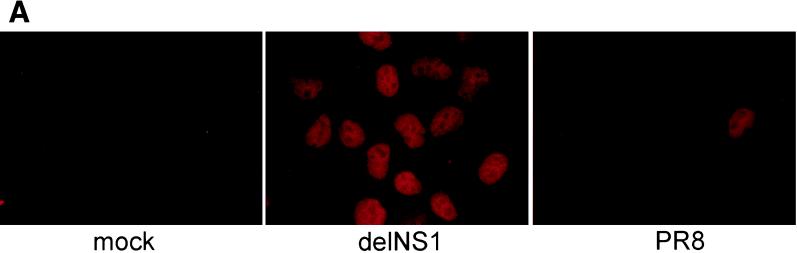
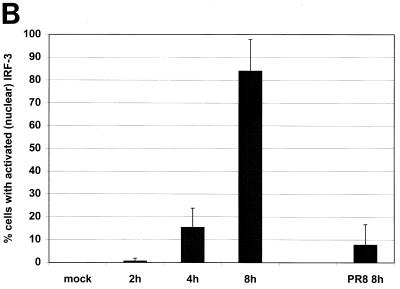
Several groups have demonstrated that a hallmark of IRF-3 activation in virus-infected cells is its nuclear accumulation (22, 38). We examined whether infection by wild-type influenza A PR/8/34 virus (PR8) or by an isogenic virus lacking the NS1 open reading frame (delNS1 virus) resulted in the nuclear accumulation of IRF-3. To address this question, HEC-1b cells were infected with either PR8 or delNS1 virus at an MOI of 1. As shown in Fig. 1A, delNS1 virus infection resulted in the nuclear accumulation of hIRF-3. At 8 h postinfection, more than 80% of delNS1 virus-infected cells showed a strong nuclear hIRF-3 signal (Fig. 1B). This nuclear IRF-3 immunofluorescence signal was clearly distinguishable from the staining pattern of IRF-3 in mock-infected cells, which showed a diffuse IRF-3 signal spread throughout the nucleus and the cytoplasm. Very few cells (<10%) with bright nuclear IRF-3 could be found among cells infected with wild-type PR8 influenza virus at 8 h postinfection. The IRF-3 staining pattern of the majority (>90%) of the cells infected with PR8 virus was indistinguishable from that of mock-infected cells (Fig. 1A and B). A time course of infection with delNS1 virus showed a gradual increase in the percentage of cells showing nuclear hIRF-3 over time, with a rapid increase between 4 and 8 h postinfection (Fig. 1B).
FIG. 1.
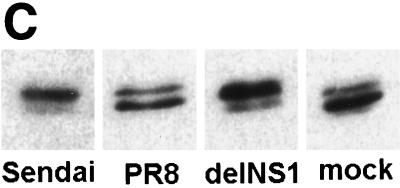
Nuclear accumulation of hIRF-3 in delNS1 virus-infected cells. (A) Coverslips coated with Hec-1b cells were mock infected or infected with delNS1 or wild-type influenza A virus (PR8) at an MOI of 1. At 8 h postinfection, fixed cells were stained with a monoclonal antibody directed against human IRF-3 (SL-12). (B) HEC-1b cells were infected with delNS1 or PR8 virus for 2, 4, or 8 h. At the given time points, fixed cells were stained with SL-12 and scored according to subcellular distribution of hIRF-3. (C) 293T cells were transfected with pCAGGS-hIRF-3 and infected 24 h later with either Sendai, PR8, or delNS1 virus or were mock infected. At 16 h postinfection, total cell lysates were probed with α-IRF-3 as indicated in Materials and Methods.
Furthermore, we wanted to confirm that the differences seen in IRF-3 localization in PR8 virus-infected cells versus delNS1 virus-infected cells were indeed due to the phosphorylation state of IRF-3. Others have shown that the phosphorylation of IRF-3 results in a more slowly migrating species when resolved by SDS-PAGE (22, 38). We therefore examined the migration patterns of IRF-3 in response to either PR8 or delNS1 virus infection. As shown by Western analysis in Fig. 1C, infection with either Sendai virus or delNS1 virus results in an increase in the amount of a more slowly migrating (activated) species of IRF-3, whereas levels of this form in PR8 virus-infected cells appear to be roughly equivalent to those found in mock-infected cells.
Inhibition of hIRF-3 activation by transient expression of wild-type NS1 but not by expression of a dsRNA binding mutant of NS1.
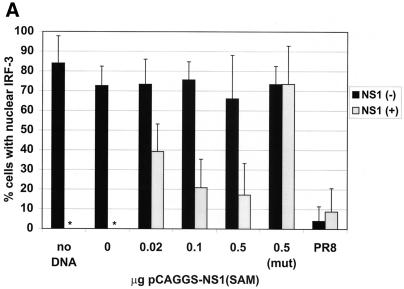
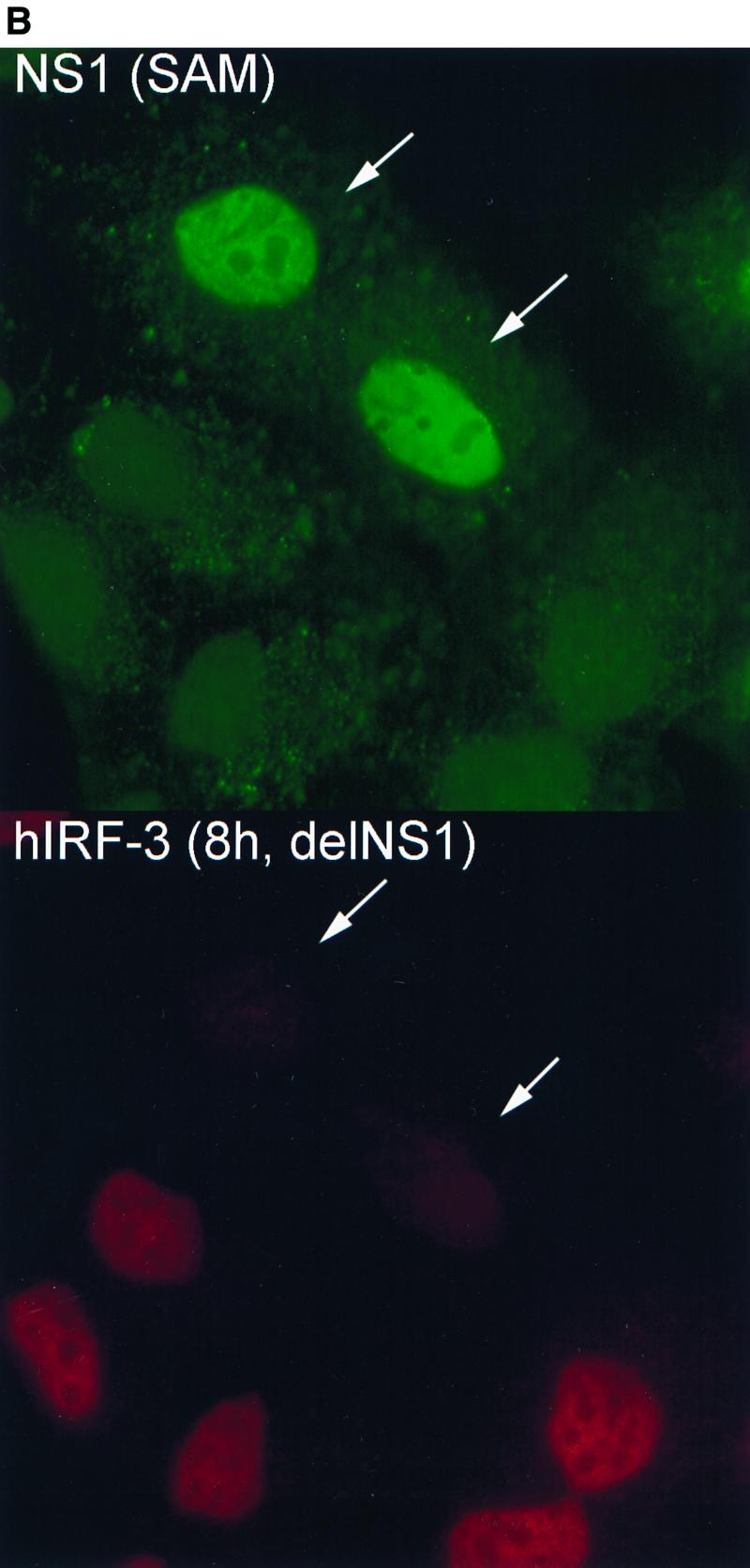
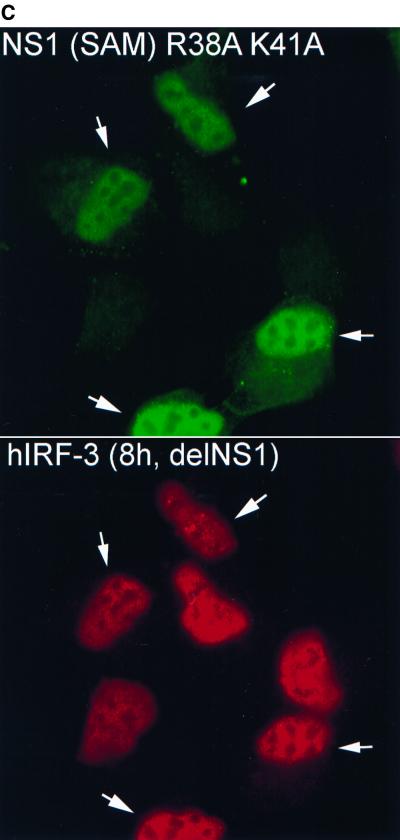
To examine whether a correlation exists between expression of wild-type NS1 and inhibition of hIRF-3 activation during virus infection, a plasmid encoding wild-type NS1 was transfected into HEC-1b cells. Transfected cells were then infected the next day with either delNS1 or PR8 virus (MOI = 1) for 8 h. Cells were scored according to NS1 expression and IRF-3 distribution. As shown in Fig. 2A, the majority of cells (between 70 and 80%) which did not show detectable levels of NS1 expression [NS1 (−)] showed nuclear accumulation of endogenous hIRF-3 in response to delNS1 infection. Transfection with 0.02 to 0.5 μg of pCAGGS-NS1(SAM), however, had a discernible effect on the nuclear accumulation of IRF-3 in response to delNS1 virus infection. Among cells expressing NS1 [NS1 (+)], a marked reduction in the percentage of cells showing activated (strong nuclear) IRF-3 compared to NS1 (−) cells was evident (Fig. 2A and B). Roughly 15 to 40% of NS1 (+) cells showed a nuclear accumulation of hIRF-3. The fraction of NS1 (+) cells with nuclear accumulation of IRF-3 did not appear to vary substantially with increasing amounts of pCAGGS-NS1(SAM) transfected per coverslip. However, a modest reduction in the fraction of NS1 (+) cells with activated hIRF-3 was observed between coverslips transfected with 0.02 μg and those transfected with 0.1 μg of the expression plasmid (from approximately 40 to 20%, respectively). We also tested whether the dsRNA binding activity of NS1 is required to inhibit IRF-3 activation. To assess this, a plasmid encoding an NS1 mutant [NS1-R38AK41A-(SAM)] was transfected into cells. The NS1-R38AK41A-(SAM) open reading frame encodes a protein with two mutations (R38 to A and K41 to A) which have been shown by others to abolish or severely diminish, respectively, the ability of NS1 to bind to RNA (36). As shown in Fig. 2A and C, expression of the mutant NS1 protein had no effect on the nuclear accumulation of IRF-3. Approximately 70% of both NS1-R38AK41A (+) and NS1-R38AK41A (−) cells showed nuclear accumulation of IRF-3.
FIG. 2.
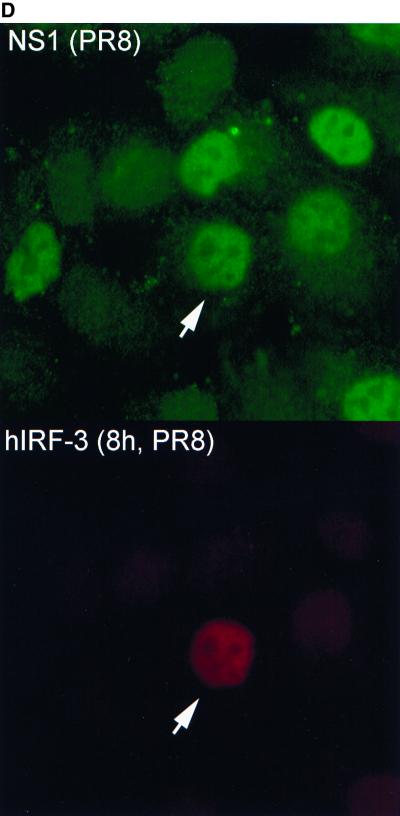
Effect on delNS1 virus-induced nuclear accumulation of hIRF-3 in HEC-1b cells by transient transfection of influenza virus NS1 proteins. (A) Coverslips coated with HEC-1b cells were transfected with a plasmid expressing the wild-type influenza virus NS1 protein [pCAGGS-NS1(SAM)] or an NS1 RNA binding mutant [pCAGGS-NS1-R38AK41A(SAM)] (mut) or with an empty vector [pCAGGS]. Both NS1 open reading frames contain a splice acceptor mutation (SAM) to ensure that spliced NEP mRNA is not produced. (The NEP is a second protein normally produced by alternative splicing from the influenza A virus NS gene.) Except with PR8-infected cells and with the no-DNA control, the amount of DNA in all transfected cells was standardized to 0.5 μg per coverslip using pCAGGS. Coverslips were transfected with either 0, 0.02, 0.1, or 0.5 μg of pCAGGS-NS1(SAM) or with 0.5 μg of pCAGGS-NS1-R38AK41A(SAM) and infected with delNS1 virus at an MOI of 1 for 8 h. Fixed cells were stained with antibodies directed against the NS1 protein and endogenous hIRF-3. Cells were scored according to NS1 expression and hIRF-3 distribution. Bars represent the mean cell number from between 5 and 13 random fields per experimental condition. Asterisks indicate that there was no NS1 expression. (B) HEC-1b cells were transfected with 0.1 μg of pCAGGS-NS1(SAM) and infected with delNS1 virus as indicated for panel A. Cells were stained with a rabbit polyclonal antibody against NS1 and a mouse monoclonal antibody against hIRF-3. Anti-rabbit IgG (fluorescein isothiocyanate) and anti-mouse IgG (Texas Red) were used as secondary antibodies. In the top panel, arrows indicate two cells expressing the NS1 protein (green). The surrounding cells do not express detectable levels of NS1. In the bottom panel, arrows indicate the same two cells, demonstrating a lack of nuclear accumulation of hIRF-3. Surrounding cells which did not express the NS1 protein show a bright, nuclear accumulation of endogenous IRF-3. (C) Cells were transfected with 0.1 μg of pCAGGS-NS1-R38AK41A(SAM) and infected with delNS1 for 8 h as described for panel A. Fixed cells were stained as described above. In the top panel, arrows indicate cells which show expression of the NS1-R38AK41A protein (green). The immunofluorescence staining pattern of NS1-R38AK41A was essentially the same as that of wild-type NS1, with bright nuclear staining. In addition to the nuclear signal, a diffuse cytoplasmic signal could also be detected in some cells expressing NS1-R38AK41A. The bottom panel shows NS1-R38AK41A-expressing cells with bright, nuclear accumulation of hIRF-3 (red). (D) HEC-1b cells were infected for 8 h with PR8 virus at an MOI of 1. Fixed cells were stained as described for panel B. The top panel shows virally expressed NS1 protein (green). The arrows indicate a cell which, when examined for IRF-3 localization (bottom panel, red), showed nuclear accumulation of IRF-3. Most cells are infected and are expressing viral NS1. The majority of PR8-infected cells do not, however, show nuclear accumulation of IRF-3. As indicated by the arrow (bottom panel), only one out of six infected cells in this field showed nuclear accumulation of IRF-3.
PR8 virus-infected cells were also examined for NS1 expression and IRF-3 localization. A very small percentage of PR8 virus-infected cells demonstrated a nuclear accumulation of IRF-3, and this fraction did not appear to differ significantly between NS1 (+) and NS1 (−) cells. The NS1(−) cells in the PR8-infected coverslips are most likely uninfected cells, which would explain their low level of IRF-3 activation (<5%). Expression of either wild-type or mutant (R38AK41A) NS1 from an expression plasmid did not appear to effect the percentage of PR8-infected cells showing nuclear accumulation of IRF-3 (data not shown).
Influenza virus NS1-mediated inhibition of IRF-3 activation induced by a heterologous virus.
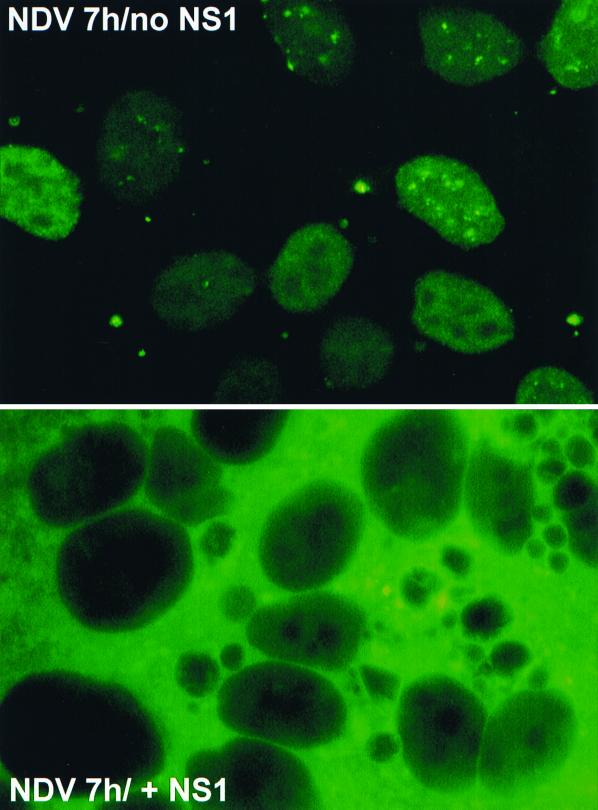
NDV is a known activator of IFN-α/β in several cell types and has been demonstrated to be a potent inducer of IRF-3 activation (41). We asked whether the influenza virus NS1 protein could prevent or delay the activation of IRF-3 by NDV or whether this inhibitory effect of NS1 was specific to influenza virus infection. For this experiment we used a plasmid expressing GFP fused to mouse IRF-3 (pGFP-mIRF-3) and human U3A cells, which have a homozygous disruption in the STAT1 gene. U3A cells have been shown to be nonresponsive to IFN (24). When transfected with pGFP-mIRF-3, U3A cells show a bright nuclear accumulation of GFP–mIRF-3 in response to delNS1 infection, confirming results obtained with endogenous hIRF-3 (data not shown). U3A cells were transfected with pGFP-mIRF3 and cotransfected with either pCAGGSNS1(SAM) or an empty vector (pCAGGS). At 24 h posttransfection, cells were infected with NDV as indicated. At 7 h postinfection, cells transfected with an empty pCAGGS vector showed distinct nuclear localization of GFP–mIRF-3 in more than 90% of the fields examined (Fig. 3, top). However, several fields of cells in coverslips which had received the NS1 expression plasmid showed striking cytoplasmic GFP-mIRF3 localization (Fig. 3, bottom). The apparent increase in the intensity of GFP expression in the NS1-transfected cells over that in control plasmid-transfected cells is likely due to the ability of NS1 to enhance translation (9, 10). Cotransfection of an NS1 expression plasmid resulted in a marked reduction (from 94.8 to 38.2%) in the mean percentage of NDV-infected cell fields showing nuclear GFP–mIRF-3 accumulation.
FIG. 3.
Inhibition of NDV-induced nuclear accumulation of GFP-mIRF3 in U3A cells by transient transfection of the influenza virus NS1 protein. Coverslips coated with U3A fibroblasts were transfected with pGFP-mIRF3 plus pCAGGS-NS1(SAM) or an empty vector (pCAGGS). At 24 h posttransfection, cells were infected with NDV at an MOI of 2 for 7 h. Washed cells were then fixed and examined for GFP–mIRF-3 distribution. Fluorescence of GFP–mIRF-3 in NDV infected cells ± NS1 is shown.
Differential induction of IFN-β mRNA in influenza A PR8 virus-infected cells versus that in delNS1 virus-infected cells.
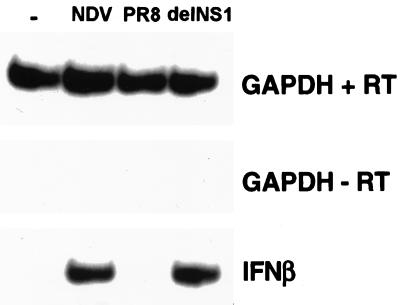
The correlation between the nuclear accumulation of IRF-3 and its activation as a positive regulator of the transcription of a subset of IFN-related genes, including IFN-β, is well documented (22, 30, 38, 41). Given the pronounced inhibition of IRF-3 nuclear accumulation in PR8-infected cells compared to that in cells infected with delNS1 virus, we examined whether a difference in the induction of IFN-β mRNA by these two viruses could be detected. Confluent 100-mm dishes of HEC-1b cells were infected at an MOI of 1 for 14 h as indicated. RT-PCR of infected cell extracts using primers specific for human IFN-β mRNA resulted in a prominent band in NDV- and delNS1-infected cells. RT-PCR of cell extracts from cells infected with the same MOI of wild-type PR8 influenza virus, however, did not produce a detectable band corresponding to IFN-β mRNA. Amplification of a housekeeping gene, GAPDH, was consistent for all viruses tested as well as for mock-infected cells.
DISCUSSION
Activation of IRF-3 in delNS1-infected cells versus that in PR8 virus-infected cells.
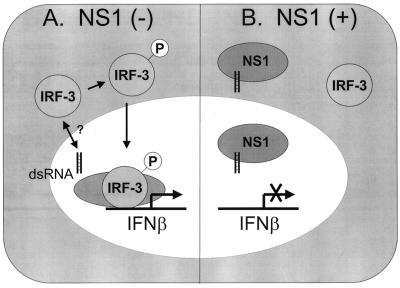
Two influenza A viruses, which differ genetically by the presence of the NS1 open reading frame, were compared for their ability to induce the activation of IRF-3. Infection with delNS1 virus dramatically induced the activation and nuclear accumulation of IRF-3 (Fig. 1). Infection with PR8 virus, on the other hand, resulted in IRF-3 nuclear accumulation in a very limited number of cells. dsRNA generated during virus infection is thought to be one of the key signals in the induction of the IFN cascade, initiating the response by activation of IRF-3. The NS1 protein is produced early in wild-type influenza virus infection (33) and is one of the most abundant proteins in the infected cell. In the context of viral infection, NS1 can be found in both the nucleus and the cytoplasm (21, 39). One of the pleiotropic activities associated with NS1 is the ability to bind to dsRNA in vitro (14, 16, 23). We therefore hypothesize that the NS1 protein present in PR8-infected cells is directly responsible for the differences seen in IRF-3 activation (see Fig. 5).
FIG. 5.
Model for the mechanism of inhibition of IFN-α/β induction by the influenza virus NS1 protein. (A) In the absence of the NS1 protein, dsRNA generated during the course of virus infection promotes the phosphorylation and nuclear accumulation of IRF-3 by a mechanism which has not yet been precisely defined. Once inside the nucleus, IRF-3 associates with other transcription factors and initiates the induction of the antiviral state by upregulating the transcription of cellular defense genes, including IFN-β. (B) In the presence of NS1, dsRNA generated during viral infection is sequestered (in the nucleus and/or in the cytoplasm) and is unable to induce the nuclear accumulation of IRF-3. The transcription of IFN-β mRNA is prevented, and the virus is able to avert the antiviral state induced by the IFN cascade.
Induction of the IFN-β promotor by dsRNA or viral infection appears to be mediated by the coordinated activation of at least three transcription factors: IRF-3, NF-κB, and ATF/c-Jun (19, 30, 37, 38). The role of the dsRNA protein kinase PKR in the induction of the IFN cascade is not well understood. We and others have demonstrated that the influenza virus NS1 protein is able to inhibit the activation of this cellular kinase (1, 15, 23, 35). While it is becoming clear that PKR plays a critical role in NF-κB activation by dsRNA or viral infection in cell culture (6), there is no evidence suggesting a role of PKR in IRF-3 activation. Moreover, IFN-α/β mRNA induction in response to virus infection has been reported to be unimpaired in mice lacking functional PKR (40). Nevertheless, if PKR is directly involved in IRF-3 activation, NS1-mediated inhibition of PKR may partially explain the low levels of IRF-3 phosphorylation and IFN-β mRNA induction seen in PR8 virus-infected cells. The specific role of PKR in IRF-3 activation, therefore, awaits further clarification. In addition, the effect of NS1 expression on the activation of other dsRNA-responsive transcription factors, such as NF-κB and ATF/c-Jun, is currently under investigation.
NS1 is a general inhibitor of IRF-3 activation which requires an intact dsRNA binding domain.
We next asked whether the exogenous expression of the NS1 protein from a plasmid would have an effect on IRF-3 localization in delNS1 virus-infected cells. Transfected cells were infected with delNS1 virus for 8 h and examined for NS1 expression and for the subcellular distribution of IRF-3 (Fig. 2). Even when expressed in trans, NS1 clearly had an effect on IRF-3 localization in delNS1-infected cells. Furthermore, this effect could be observed in a different system using a heterologous virus (NDV) and a different readout (pGFP-mIRF-3 expression) (Fig. 3). Since the influenza virus NS1 protein is able to bind both to model influenza virus RNA and nonspecifically to dsRNA in vitro (14, 16, 23), it is conceivable that the influenza virus NS1 protein may act in a general sense to bind and sequester any viral and/or double-stranded forms of RNA which could potentially activate IRF-3. This hypothesis is strengthened by the fact that cells expressing a mutant NS1 which lacks a functional RNA binding domain (R38AK41A-NS1) (36) show the same level of IRF-3 activation as control cells (Fig. 2).
IFN-β mRNA induction.
Activation of IRF-3 results in its nuclear accumulation, where it assembles with CREB-binding protein (CBP/p300) as part of the transcriptional complex DRAF1 (dsRNA-activated factor 1) (38). dsRNA-activated factor 1 binds to IFN-β- and IFN-α4 promoters, contributing to their transcriptional activation (7, 8). We therefore examined whether the differences we saw in IRF-3 activation between delNS1 virus- and PR8 virus-infected cells correlated with differences in IFN-β mRNA induction. We found that delNS1 virus infection induced the transcription of detectable levels of IFN-β mRNA, whereas PR8 virus infection did not (Fig. 4). Infection with PR8 virus, which encodes the NS1 protein, is therefore much less likely to induce the downstream antiviral effects of IFN-α/β than the isogenic virus lacking this IFN antagonist. Which specific antiviral pathways are affected, and to what degree, await further analysis.
FIG. 4.
Differential induction of IFN-β mRNA in response to either PR8 or delNS1 virus infection. Confluent 100-mm dishes of HEC-1b cells were infected with either NDV, PR8 virus, or delNS1 virus at an MOI of 1 for 14 h. One dish of mock (PBS)-infected cells was included as a negative control (−). At 14 h postinfection, cells were washed twice with cold PBS, and total RNA was subjected to RT-PCR analysis using specific primers for IFN-β or GAPDH. As a control for genomic DNA contamination, PCR was carried out with GAPDH primers using the mock (−RT) reactions as a template. Following PAGE, products were detected by autoradiography.
The ability of a virus to inhibit the expression of defense genes such as IFN-β is likely to be inextricably linked to its pathogenicity. This model is exemplified by the influenza virus system. We show that the presence of the NS1 protein in wild-type PR8 virus-infected cells is associated with an inhibition in the activation of IRF-3 and of the subsequent transcription of IFN-β mRNA (Fig. 5B). Combined with our previous results showing the extreme differences in pathogenicity between the PR8 and delNS1 viruses (13), we propose that the ability of the NS1 protein to inhibit the activation of IRF-3 is an important virulence factor for influenza virus. It should be noted that the NS1 of PR8 virus could not completely prevent the activation of IRF-3 in infected cells. We therefore propose that variability within the NS1 sequence among different strains of influenza virus may be responsible for differences in ability to inhibit this arm of the immune system. These differences may play a role in virus pathogenicity.
We also propose that other viral dsRNA binding proteins, such as the E3L protein of poxviruses (42), may have an analogous mechanism for preventing IFN-α/β synthesis. Finally, the design of compounds to specifically target the inhibitory activity on the IFN system could represent an alternative for the generation of novel antiviral agents. In the case of influenza viruses, compounds would be selected for their ability to prevent NS1-mediated abrogation of IRF-3 activation.
ACKNOWLEDGMENTS
We thank Peter Howley (Harvard University, Boston, Mass.) for kindly providing the SL-12 mouse monoclonal antibody directed against human IRF-3. We also thank Tak Mak (University of Toronto, Toronto, Ontario, Canada) for the pGFP-mIRF-3 expression plasmid.
This work was supported in part by grants to A.G.-S. and P.P. from the National Institutes of Health and by Austrian Science Fund Project 12548-MOB (T.M.). T.M. was supported by the Austrian Academy of Sciences. C.F.B. was supported by an NIH NRSA fellowship.
REFERENCES
- 1.Bergman M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braganca J, Civas A. Type I interferon gene expression: differential expression of IFN-A genes induced by viruses and double-stranded RNA. Biochimie. 1998;80:673–687. doi: 10.1016/s0300-9084(99)80021-2. [DOI] [PubMed] [Google Scholar]
- 3.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 5.Chen H Y, Sato T, Fuse A, Kuwata T, Content J. Resistance to interferon of a human adenocarcinoma cell line, HEC-1, and its sensitivity to natural killer cell action. J Gen Virol. 1981;52:177–181. doi: 10.1099/0022-1317-52-1-177. [DOI] [PubMed] [Google Scholar]
- 6.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 7.Daly C, Reich N C. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem. 1995;270:23739–23746. doi: 10.1074/jbc.270.40.23739. [DOI] [PubMed] [Google Scholar]
- 8.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuse A, Ashino-Fuse H, Kuwata T. Binding of 125I-labeled human interferon to cell lines with low sensitivity to interferon. Gann. 1984;75:379–384. [PubMed] [Google Scholar]
- 12.Gale M, Jr, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 14.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 15.Hatada E, Saito S, Fukuda R. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J Gen Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 17.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs B L, Langland J O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 19.Juang Y, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krug R M. Unique functions of the NS1 protein. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. Oxford, England: Blackwell Science Ltd.; 1998. pp. 82–92. [Google Scholar]
- 21.Li Y, Yamakita Y, Krug R M. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc Natl Acad Sci USA. 1998;95:4864–4869. doi: 10.1073/pnas.95.9.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 24.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morahan P S, Grossberg S E. Age-related cellular resistance of the chicken embryo to viral infections. I. Interferon and natural resistance to myxoviruses and vesicular stomatitis virus. J Infect Dis. 1970;121:615–623. doi: 10.1093/infdis/121.6.615. [DOI] [PubMed] [Google Scholar]
- 26.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 27.Norton G P, Tanaka T, Tobita K, Nakada S, Buonagurio D A, Greenspan D, Krystal M, Palese P. Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology. 1987;156:204–213. doi: 10.1016/0042-6822(87)90399-0. [DOI] [PubMed] [Google Scholar]
- 28.Pitha P M, Au W C, Lowther W, Juang Y T, Schafer S L, Burysek L, Hiscott J, Moore P A. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie. 1998;80:651–658. doi: 10.1016/s0300-9084(99)80018-2. [DOI] [PubMed] [Google Scholar]
- 29.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 31.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 32.Sekellick M J, Biggers W J, Marcus P I. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell Dev Biol. 1990;26:997–1003. doi: 10.1007/BF02624475. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro G I, Gurney T, Jr, Krug R M. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J Virol. 1987;61:764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 35.Tan S L, Katze M G. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Riedel K, Lynch P, Chien C Y, Montelione G T, Krug R M. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. . (Erratum, 3:813.) [DOI] [PubMed] [Google Scholar]
- 38.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff T, O'Neill R E, Palese P. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuwen H, Cox J H, Yewdell J W, Bennink J R, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]