Abstract
Background
Varicose veins (VV) are one of the common human diseases, but the role of genetics in its development is not fully understood.
Methods
We conducted an exome-wide association study of VV using whole-exome sequencing data from the UK Biobank, and focused on common and rare variants using single-variant association analysis and gene-level collapsing analysis.
Findings
A total of 13,823,269 autosomal genetic variants were obtained after quality control. We identified 36 VV-related independent common variants mapping to 34 genes by single-variant analysis and three rare variant genes (PIEZO1, ECE1, FBLN7) by collapsing analysis, and most associations between genes and VV were replicated in FinnGen. PIEZO1 was the closest gene associated with VV (P = 5.05 × 10−31), and it was found to reach exome-wide significance in both single-variant and collapsing analyses. Two novel rare variant genes (ECE1 and METTL21A) associated with VV were identified, of which METTL21A was associated only with females. The pleiotropic effects of VV-related genes suggested that body size, inflammation, and pulmonary function are strongly associated with the development of VV.
Conclusions
Our findings highlight the importance of causal genes for VV and provide new directions for treatment.
Author summary
In this study, whole-exome sequencing data from the UK Biobank were used to explore the effect of genetic variants on varicose veins (VV) and search for new VV-related genes. In contrast to traditional association studies, large-scale whole-exome sequencing analysis is more capable of identifying rare genetic variants (MAF < 1%) in diseases. The current study identified 34 VV-associated common variant genes by single-variant analysis and three rare variant genes (PIEZO1, ECE1, FBLN7) by collapsing analysis, and most associations were validated in FinnGen. In addition to replicating several genes reported in previous genome-wide association studies, we identified 17 novel genes that may be associated with VV. Through subsequent phenome-wide association analyses of identified genes, we found that these genes are also strongly associated with body size, inflammation, and pulmonary function. These findings contribute to understanding the underlying mechanisms of pathogenesis and developing novel therapeutic strategies for VV.
Introduction
Varicose veins (VV) are the most common chronic condition of the venous system, often affecting the lower extremities and manifesting as dilated, stretched, or tortuous superficial veins [1]. The majority of VV patients suffer from complications such as pain, swelling, hyperpigmentation, and ulcers. About 23% of adults aged 40–80 in the United States develop VV, including 22 million women and 11 million men [2]. Today, endovenous laser ablation is considered clinically the first-line treatment option for VV but has a post-operative recurrence rate of up to 20% [3,4]. Therefore, it is particularly important to define the etiology of VV. Previous epidemiological studies have shown that VV are associated with several risk factors, including advanced age, being female, pregnancy, obesity, height, and history of deep vein thrombosis [5–8]. A positive family history is one of the important risk factors for VV, which suggests that VV are likely to be modulated by genetic factors [9–12]. Most of the previous genetic studies on VV were genome-wide association studies (GWAS) [13–15]. Within the last decade, dozens of genomic loci associated with VV have been identified. However, GWAS are more limited in scope and mostly focused on common variants (minor allele frequency (MAF > 1%)), which usually have a small effect size and cannot directly identify the causal gene.
In contrast to traditional studies, large-scale whole-exome sequencing (WES) analysis is more capable of identifying rare genetic variants (MAF < 1%) in diseases [16]. Rare variants are genetic markers of high disease risk and help to identify novel genetic targets for drug interventions [17]. Apart from revealing the full spectrum of protein-coding variants, WES can facilitate the identification of novel loss-of-function (LOF) variants and enhance the ability to detect the associations between LOF variants and diseases. LOF variants can identify drivers of genetic risk, novel disease genes, and therapeutic targets [16]. Also, large-scale sequencing can evaluate the disease prevalence and carrier frequencies of rare genetic variants [18]. A recent study focused on the effect of PIEZO1 on altered VV risk and identified the association of VV with rare protein-truncating variants in PIEZO1 [19]. A multi-phenotype exome-wide association study (ExWAS) from the UK Biobank (UKB) involving 49,960 participants [16] identified novel LOF variants with significant disease impact, including PIEZO1 on VV (P = 3.2 × 10−8). In addition, a phenome-wide level ExWAS [20] exploring the contribution of rare variants to human disease also found the association between PIEZO1 and VV (P = 3.24 × 10−24). In contrast to their studies, we conducted a more comprehensive ExWAS of VV using the updated exome sequencing data of more than 350,000 UKB individuals, with additional validation and exploration of the findings and biological functions. We identified 19 known and 17 novel causal genes significantly associated with VV, with most of these associations validated in FinnGen [21]. In addition, our study explored the phenotypic associations between VV genes and multiple traits, showing that cardiovascular disease, height, biochemistry, and inflammatory indicators might contribute to the development of VV.
Results
Participant characteristics
In this study, we performed an ExWAS by utilizing phenotypic and genetic data from UKB, including exome sequencing data and VV diagnosis data (S1 Table). Following stringent quality control (QC) of genotypes and samples, we identified 350,770 unrelated Caucasian participants (21,643 VV cases and 329,127 controls) with a mean age of 56.94 years at enrollment, of which 53.76% were female (S2 Table). The case and control groups had similar age distribution (S1 Fig). The clustering of the VV case and control groups in the principal component analysis is shown in S2 Fig. A total of 13,823,269 autosomal genetic variants were obtained after QC, including 100,098 common variants (MAF > 1%) and 13,723,171 rare variants (MAF < 1%).
Exome-wide single-variant association analysis for VV
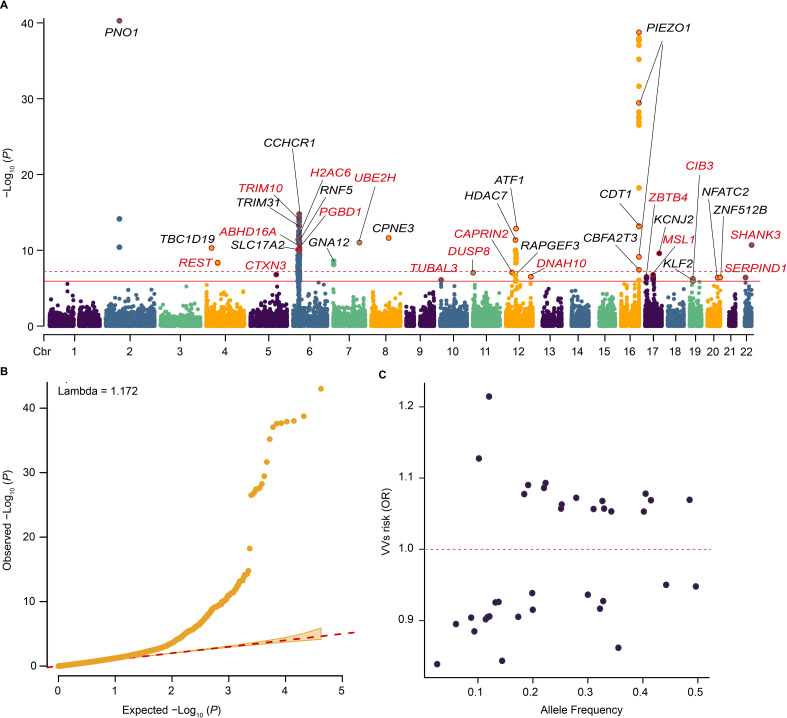
The mixed-effects logistic models adjusted for age, sex, and the top 10 principal components (PCs) were used to assess the association between VV and common coding variants. Firstly, 169 variants and 114 variants were found to reach the exome-wide significance (P < 1.19 × 10−6) and the genome-wide significance (P < 5 × 10−8) in the single-variant analysis (Fig 1A and S3 Table). After clumping 169 variants in strong linkage disequilibrium, 36 independent common variants associated with VV were identified (Fig 1A), of which 34 were successfully validated in FinnGen [21] (Table 1 and S4 Table). Among these independent common variants, 19 were significantly protective against VV, while the other 17 were significantly associated with an elevated risk of VV (Table 1). In addition, 36 independent common variants were mapped to 34 genes, of which 18 have been reported previously and the other 16 genes were novel (including TRIM10, UBE2H, TUBAL3, DUSP8, DNAH10, CAPRIN2, MSL1, ZBTB4, CIB3, SERPIND1, SHANK3, REST, CTXN3, H2AC6, PGBD1, ABHD16A). PNO1 and PIEZO1 had the most significant associations with VV, and the P-values were 9.81 × 10−44 and 1.75 × 10−39, respectively. TRIM10 showed the strongest association with VV among 16 novel common variant genes (P = 5.29 × 10−14). The quantile-quantile (Q-Q) plot of single common variants is shown in Fig 1B. It was noteworthy that these independent common variants were characterized by larger effect sizes at lower frequencies (Fig 1C). Furthermore, we conducted a conditional analysis of the identified common variants linked to VV using conditional and joint analysis (COJO). The results revealed that 27 independent common variants were retained, 25 of which were consistent with variants identified after clumping (S5 Table).
Fig 1. Summary of single-variant associations with VV among the unrelated Caucasians.
(A) Manhattan plot of single-variant associations for common variants and VV. The red solid line indicates the significance threshold (P = 1.19 × 10−6), and the red dashed line indicates the genome-wide significance threshold (P = 5.0 × 10−8). The gene name corresponds to the gene closest to the variant. Black fonts indicate previously identified genes, while red fonts indicate newly identified genes. (B) The quantile-quantile (Q-Q) plot for single-variant analysis. (C) The plot of effect size (odds ratio) versus effect allele frequency of identified independent common variants. Abbreviation: OR, odds ratio.
Table 1. Independent common variants significantly associated with VV in the single-variant association analysis.
| rsID | CHR | POS | A0 | A1 | OR | 95% CI | P | Gene | Exonic function | FinnGen VV P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2044693 | 2 | 68157965 | G | A | 0.86 | 0.84–0.88 | 9.81×10−44 | PNO1 | Synonymous | 6.00×10−24 |
| rs2227901 | 4 | 56932023 | G | A | 1.08 | 1.05–1.11 | 4.23×10−9 | REST | Synonymous | 1.94×10−6 |
| 4 | 26584259 | C | T | 1.07 | 1.05–1.09 | 4.97×10−11 | TBC1D19 | Synonymous | / | |
| rs248709 | 5 | 127657571 | T | A | 1.06 | 1.03–1.08 | 1.59×10−7 | CTXN3 | Synonymous | 6.28×10−3 |
| rs34525648 | 6 | 25914625 | G | A | 0.91 | 0.88–0.93 | 4.87×10−10 | SLC17A2 | Nonsynonymous | 7.02×10−7 |
| 6 | 26124406 | C | T | 1.09 | 1.06–1.11 | 3.81×10−12 | H2AC6 | Synonymous | / | |
| rs33932084 | 6 | 28301047 | A | G | 0.90 | 0.88–0.93 | 6.28×10−10 | PGBD1 | Nonsynonymous | 1.08×10−7 |
| rs2239529 | 6 | 30110553 | C | T | 0.91 | 0.88–0.93 | 3.07×10−13 | TRIM31 | Synonymous | 5.61×10−4 |
| rs3094134 | 6 | 30154377 | C | T | 1.13 | 1.09–1.16 | 5.29×10−14 | TRIM10 | Synonymous | 4.20×10−4 |
| rs1576 | 6 | 31142614 | G | C | 0.92 | 0.90–0.94 | 1.65×10−15 | CCHCR1 | Nonsynonymous | 4.23×10−6 |
| rs1475865 | 6 | 31689636 | C | T | 1.07 | 1.05–1.10 | 2.73×10−10 | ABHD16A | Synonymous | 1.08×10−4 |
| rs3130349 | 6 | 32179919 | G | A | 0.92 | 0.89–0.94 | 7.02×10−12 | RNF5 | Synonymous | 1.47×10−4 |
| rs12539800 | 7 | 129839298 | T | C | 0.88 | 0.85–0.92 | 9.67×10−12 | UBE2H | Synonymous | 2.92×10−8 |
| rs798488 | 7 | 2762888 | T | C | 0.94 | 0.92–0.96 | 2.75×10−9 | GNA12 | Start loss | 6.84×10−3 |
| rs2304789 | 8 | 86554965 | C | T | 0.93 | 0.91–0.95 | 2.42×10−12 | CPNE3 | Nonsynonymous | 2.99×10−9 |
| rs7097775 | 10 | 5393955 | A | G | 1.05 | 1.03–1.08 | 8.09×10−7 | TUBAL3 | Synonymous | 3.46×10−4 |
| rs7129499 | 11 | 1556860 | G | A | 0.95 | 0.93–0.97 | 9.14×10−8 | DUSP8 | Synonymous | 9.48×10−7 |
| rs4930721 | 12 | 123933342 | C | T | 1.06 | 1.03–1.08 | 3.14×10−7 | DNAH10 | Synonymous | 7.74×10−5 |
| rs12146709 | 12 | 30735067 | T | C | 1.06 | 1.04–1.09 | 8.44×10−8 | CAPRIN2 | Nonsynonymous | 4.72×10−3 |
| rs12422983 | 12 | 47748853 | C | T | 0.93 | 0.90–0.95 | 2.32×10−7 | RAPGEF3 | Nonsynonymous | 4.99×10−4 |
| rs7306788 | 12 | 47784123 | C | T | 1.09 | 1.06–1.12 | 4.46×10−12 | HDAC7 | Synonymous | 8.05×10−15 |
| rs1129406 | 12 | 50809588 | C | T | 1.08 | 1.06–1.10 | 1.37×10−13 | ATF1 | Synonymous | 3.91×10−18 |
| rs8043924 | 16 | 88721296 | C | G | 1.21 | 1.18–1.25 | 1.75×10−39 | PIEZO1 | Synonymous | 4.56×10−37 |
| rs34908386 | 16 | 88734505 | C | T | 0.84 | 0.82–0.87 | 5.74×10−29 | PIEZO1 | Synonymous | 4.28×10−28 |
| rs6500493 | 16 | 88737574 | G | C | 1.07 | 1.05–1.09 | 7.48×10−10 | PIEZO1 | Synonymous | 2.52×10−6 |
| rs510862 | 16 | 88806103 | C | T | 1.09 | 1.07–1.12 | 5.79×10−14 | CDT1 | Synonymous | 4.93×10−14 |
| rs61734177 | 16 | 88898149 | C | G | 0.90 | 0.87–0.94 | 3.77×10−8 | CBFA2T3 | Nonsynonymous | 6.78×10−4 |
| rs143879261 | 17 | 40129440 | C | A | 0.84 | 0.78–0.90 | 1.71×10−7 | MSL1 | Synonymous | 0.03 |
| rs173135 | 17 | 70176185 | C | T | 0.90 | 0.87–0.93 | 2.58×10−10 | KCNJ2 | Synonymous | 1.18×10−5 |
| rs34914463 | 17 | 7463300 | T | C | 0.93 | 0.90–0.95 | 3.10×10−7 | ZBTB4 | Nonsynonymous | 2.19×10−3 |
| rs6512087 | 19 | 16164844 | T | C | 0.94 | 0.91–0.96 | 5.86×10−7 | CIB3 | Synonymous | 0.01 |
| rs3745318 | 19 | 16325451 | C | T | 1.06 | 1.03–1.08 | 1.18×10−6 | KLF2 | Synonymous | 4.16×10−10 |
| rs3746420 | 20 | 51524088 | G | C | 0.89 | 0.86–0.94 | 4.15×10−7 | NFATC2 | Synonymous | 1.13×10−8 |
| rs817329 | 20 | 63966341 | G | T | 1.05 | 1.03–1.07 | 3.92×10−7 | ZNF512B | Synonymous | 1.02×10−9 |
| rs4675 | 22 | 20787012 | T | C | 0.95 | 0.93–0.97 | 3.94×10−7 | SERPIND1 | Synonymous | 7.97×10−3 |
| rs9616915 | 22 | 50679152 | C | T | 1.07 | 1.05–1.09 | 2.07×10−11 | SHANK3 | Synonymous | 8.60×10−6 |
Genes highlighted in bold are not previously reported. rsID was obtained from FinnGen. The “/” represents the variants not tested in FinnGen.
Abbreviations: VV, varicose veins; CHR, chromosome; POS, position; OR, odds ratio; CI, confidence interval.
Three VV genes identified by rare variant collapsing analysis
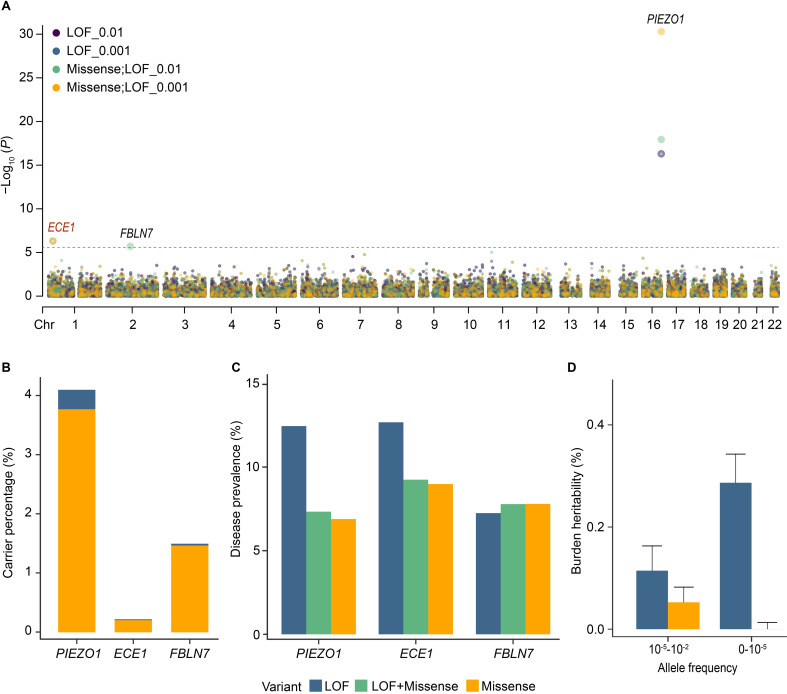
We performed the gene-level collapsing analysis to determine rare variant associations with VV. To elucidate the differences in genetic structures between genes, our analysis included 19,897 genes under 8 different models (Methods). Collapsing analysis showed 13 significant associations involving 3 genes (PIEZO1, ECE1, FBLN7) after Bonferroni correction (P < 2.51 × 10−6) (Fig 2A and S6 Table). The corresponding Q-Q plots are shown in S3 Fig. The significance level of the genetic variant associations between these significant genes and VV was enhanced by the inclusion of missense variants. In the LOF + missense model with MAF < 1%, these 3 genes were significantly associated with an elevated risk of VV (PIEZO1, odds ratio (OR) = 1.010, 95% confidence interval (CI) = 1.008–1.013, P = 1.15 × 10−18; ECE1, OR = 1.028, 95% CI = 1.016–1.039, P = 4.89 × 10−7; FBLN7, OR = 1.012, 95% CI = 1.007–1.017, P = 1.93 × 10−6) (S7 Table). The gene which had the strongest correlation with VV was PIEZO1 in the LOF model with MAF < 10−3 (PIEZO1, OR = 1.037, 95% CI = 1.029–1.046, P = 5.01 × 10−17). The associations of PIEZO1 and FBLN7 with VV have been reported in previous genetic studies [14,19]. The significant difference between the LOF + missense model (P = 4.89 × 10−7) and the LOF-only model (P = 0.02) for ECE1, a novel identified VV-related gene, suggested its association with VV was primarily driven by the missense mutation (Table 2). The associations of the three genes we identified in collapsing analysis with VV were further successfully validated in the VV genomic data from FinnGen (P ranged from 2.39 × 10−59 to 4.93 × 10−5, Table 2).
Fig 2. Summary of rare genetic associations with VV among the unrelated Caucasians.
(A) Manhattan plot of the gene-based collapsing analysis. The red dashed line indicates the significance threshold (P = 2.51 × 10−6). We only showed the results of the collapsing test with four models including two MAF cutoffs (0.01 and 0.001) and two variant annotation groups (LOF and LOF + missense), and the other results are shown in S6 Table. All models were adjusted for age, sex, and the top 10 PCs. (B) Bar chart showing carrier frequencies for rare LOF and missense variants for three identified rare variant genes. (C) Disease prevalence of LOF and missense variants in VV-related rare variant genes. (D) Heritability of the burden of genetic variants in VV. Abbreviation: LOF, loss-of-function.
Table 2. Significant associations with VV in the gene-level collapsing analysis.
| Gene | Group | MAF | Number of rare variants | OR | 95% CI | P | FinnGen VV P |
|---|---|---|---|---|---|---|---|
| PIEZO1 | LOF | 0.01 | 204 | 1.037 | 1.029–1.046 | 5.01 × 10−17 | 2.39 × 10−59 |
| LOF + Missense | 0.01 | 735 | 1.010 | 1.008–1.013 | 1.15 × 10−18 | ||
| LOF | 0.001 | 204 | 1.037 | 1.029–1.046 | 5.01 × 10−17 | ||
| LOF + Missense | 0.001 | 730 | 1.021 | 1.017–1.024 | 5.05 × 10−31 | ||
| LOF | 1 × 10−4 | 203 | 1.056 | 1.045–1.067 | 3.24 × 10−24 | ||
| LOF + Missense | 1 × 10−4 | 716 | 1.028 | 1.023–1.033 | 6.70 × 10−28 | ||
| LOF | 1 × 10−5 | 183 | 1.049 | 1.033–1.066 | 2.20 × 10−9 | ||
| LOF + Missense | 1 × 10−5 | 578 | 1.037 | 1.028–1.046 | 2.67 × 10−16 | ||
| ECE1 | LOF | 0.01 | 30 | 1.051 | 1.007–1.096 | 0.02 | 4.93 × 10−5 |
| LOF + Missense | 0.01 | 189 | 1.028 | 1.016–1.039 | 4.89 × 10−7 | ||
| LOF | 0.001 | 30 | 1.051 | 1.007–1.096 | 0.02 | ||
| LOF + Missense | 0.001 | 189 | 1.028 | 1.016–1.039 | 4.89 × 10−7 | ||
| LOF | 1 × 10−4 | 30 | 1.051 | 1.007–1.096 | 0.02 | ||
| LOF + Missense | 1 × 10−4 | 186 | 1.034 | 1.020–1.049 | 1.12 × 10−6 | ||
| LOF | 1 × 10−5 | 30 | 1.051 | 1.007–1.096 | 0.02 | ||
| LOF + Missense | 1 × 10−5 | 174 | 1.041 | 1.024–1.059 | 1.37 × 10−6 | ||
| FBLN7 | LOF | 0.01 | 29 | 1.007 | 0.978–1.036 | 0.60 | 7.25 × 10−10 |
| LOF + Missense | 0.01 | 136 | 1.012 | 1.007–1.017 | 1.93 × 10−6 | ||
| LOF | 0.001 | 29 | 1.007 | 0.978–1.036 | 0.60 | ||
| LOF + Missense | 0.001 | 135 | 1.011 | 1.003–1.019 | 0.01 | ||
| LOF | 1 × 10−4 | 29 | 1.007 | 0.978–1.036 | 0.60 | ||
| LOF + Missense | 1 × 10−4 | 128 | 1.010 | 0.998–1.022 | 0.20 | ||
| LOF | 1 × 10−5 | 25 | 0.986 | 0.941–1.034 | 0.56 | ||
| LOF + Missense | 1 × 10−5 | 102 | 1.004 | 0.982–1.027 | 0.70 |
Genes highlighted in bold are not previously reported. Number of rare variants: number of markers with MAF < 0.01.
Abbreviations: VV, varicose veins; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; LOF, loss of function.
Considering the high prevalence and incidence of VV in the general population, we calculated the carrier frequencies and disease prevalence rates of the putative pathogenic variants. PIEZO1 had the highest carrier frequency of missense variants (3.77%, the probability of being loss-of-function intolerant (pLI) = 0.54), followed by FBLN7 (1.47%, pLI = 0) and ECE1 (0.20%, pLI = 1) (Fig 2B and S8 and S9 Tables). The high pLI of ECE1 underscores its pronounced intolerance to LOF variants, consistent with our findings of lower carrier frequency (S4 Fig). These variant carriers showed generally modest disease prevalence rates (< 15%), and the disease prevalence rates of missense variants were lower than those of LOF variants in carriers (Fig 2C).
Burden heritability of VV
Then we calculated the gene-wise burden heritability of VV using the method developed by Weiner et al. [22]. In this analysis, ultra-rare variants were defined as MAF < 1 × 10−5 and rare variants were defined as having a MAF between 1 × 10−5 and 1 × 10−2. The burden heritability of rare variants focuses on variants that predict the most serious functional consequences. Among these, ultra-rare coding LOF variants explained 0.287% (se = 0.056%) of phenotypic variance, more than rare LOF variants explained. But rare missense variants accounted for more burden heritability than ultra-rare (rare, 0.053%; ultra-rare, -7.63 × 10−7) (Fig 2D and S10 Table).
Robustness of the association of rare variant genes with VV
We performed leave-one-variant-out (LOVO) analyses to assess the robustness of associations identified in gene-based collapsing analysis and to find the variants that affect each VV-related rare variant gene (S11 Table). After the removal of the variant chr1:21227980:A:G, the association between ECE1 and VV became non-significant (P = 3.81 × 10−6). Similarly, the highest LOVO P value attained for the association was P = 9.90 × 10−3 for FBLN7 and VV after removing chr2:112165024:C:G. Importantly, the association of VV and PIEZO1 remained robust in the LOVO analysis (S5 Fig). This suggested that the significant associations of VV with ECE1 and FBLN7 were strongly influenced by single rare variants, whereas the association with PIEZO1 were more influenced by a combined effect of multiple rare variants. We further assessed whether the significant rare variants were independent of nearby common variants by a conditional analysis. The results showed that the nearby common variants did not affect the effect size and significance of these three rare variant genes (S12 Table).
Sensitivity analysis of the effects of sex on VV
To explore whether the effects of genes on VV differ by sex, we re-performed single-variant analysis and collapsing analysis after stratifying participants by sex. These analyses included 162,210 males and 188,560 females. After clumping, we identified 5 and 21 independent common variants significantly related to VV in males and females, respectively, with four variants mapping to novel genes only in females (ARHGEF26, BTN1A1, H1-5, TRIM27, S13 Table). The PIEZO1 gene was strongly associated with VV, with different significance levels across sexes (male, P = 4.66 × 10−14; female, P = 6.81 × 10−21). The significant association between FBLN7 and VV was found only in females (P = 1.12 × 10−6). Notably, we additionally identified a significant association between a novel gene (METTL21A, P = 1.38 × 10−6) and VV in females in the model of LOF variants with MAF < 1 × 10−4 (S14 Table).
Enrichment analysis of VV genes and their expression
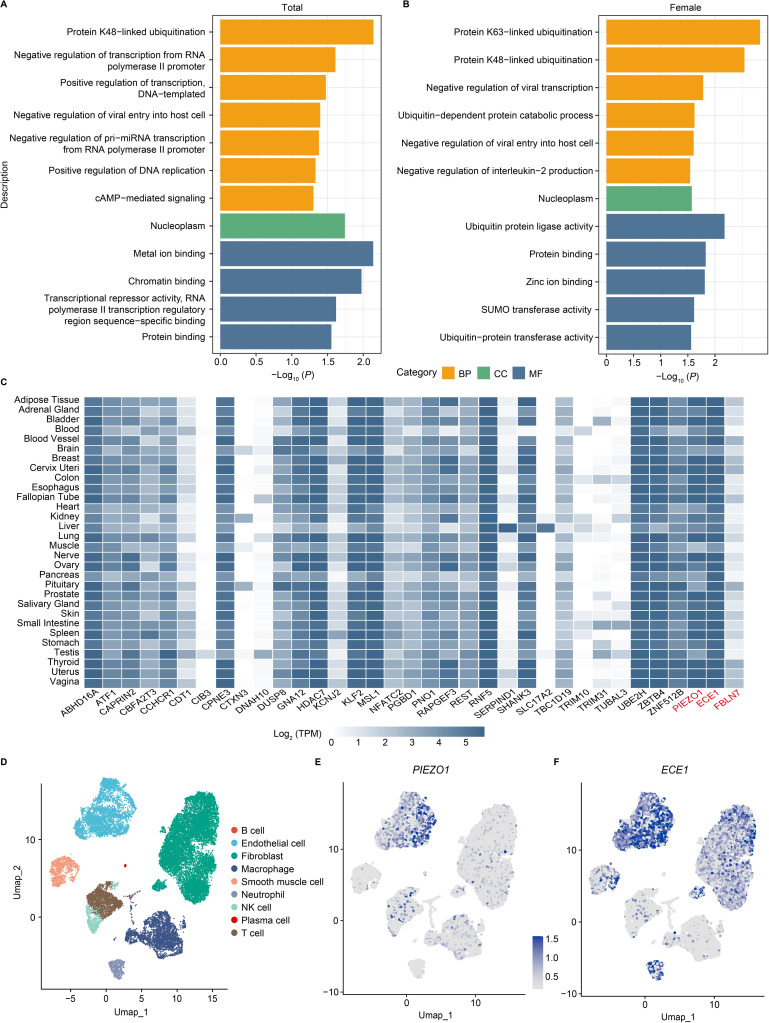
We conducted a pathway enrichment analysis using the Gene Ontology (GO) Consortium [23,24] for all 36 genes identified in the study as being significantly associated with VV. There were 12 GO terms significantly enriched for these genes (false discovery rate (FDR) correction, corrected α = 0.05). It was found that several VV genes (including UBE2H, TRIM31, and RNF5) were significantly enriched in protein K48-linked ubiquitination (P = 7.20 × 10−3) and 30 genes were enriched in protein binding (P = 2.79 × 10−2) (Fig 3A and S15 Table). In females, GO enrichment analysis showed that the identified VV-related genes were significantly enriched in protein ubiquitination (P = 1.47 × 10−3) and most genes were enriched in protein binding (P = 1.48 × 10−2) (Fig 3B and S16 Table). However, the VV genes identified in males were not successfully enriched for the same biological processes.
Fig 3. Enrichment and expression analysis of significant effect associations in VV.
(A) Results of the functional enrichment in biological pathway databases (P < 0.05). (B) Results of the functional enrichment in biological pathway databases in females. Only the top 12 significant pathways (P < 0.05) are shown, and the complete results are shown in S16 Table. (C) The heat map shows the tissue enrichment results of each VV-related gene. The H2AC6 gene was not recognized in FUMA Ensembl ID. (D) Single-cell RNA sequencing analysis results of adipose tissue. (E, F) Cell-specific expression of PIEZO1 and ECE1 in adipose tissue. Abbreviation: BP, biological process; CC, cellular component; MF, molecular function; TPM, transcripts per million; Umap, Uniform manifold approximation and projection.
In addition, we explored the expression of VV-related independent common and rare variant genes in 30 general tissues in FUMA GTEx. We discovered that several genes were highly expressed in adipose tissue, including PIEZO1 and the novel identified ECE1 (Fig 3C and S17 Table). Next, single-cell expression data from human adipose tissue showed that PIEZO1 and ECE1 were most strongly expressed in the endothelial cell subpopulation, and higher expression of ECE1 was also detected in fibroblasts and neutrophils (Fig 3D, 3E and 3F).
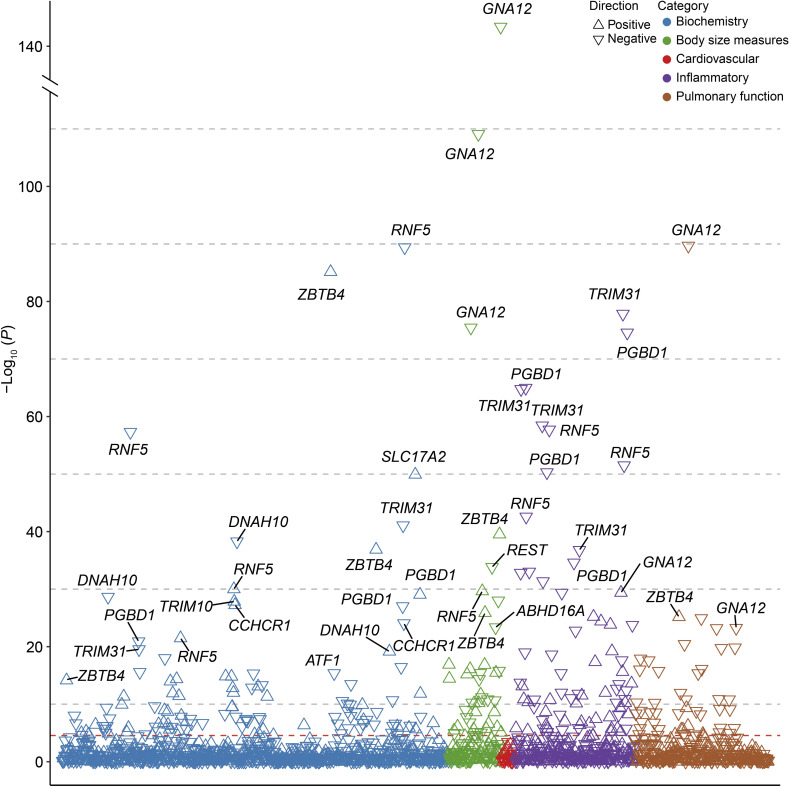
Phenotypic associations between identified VV genes and multiple traits
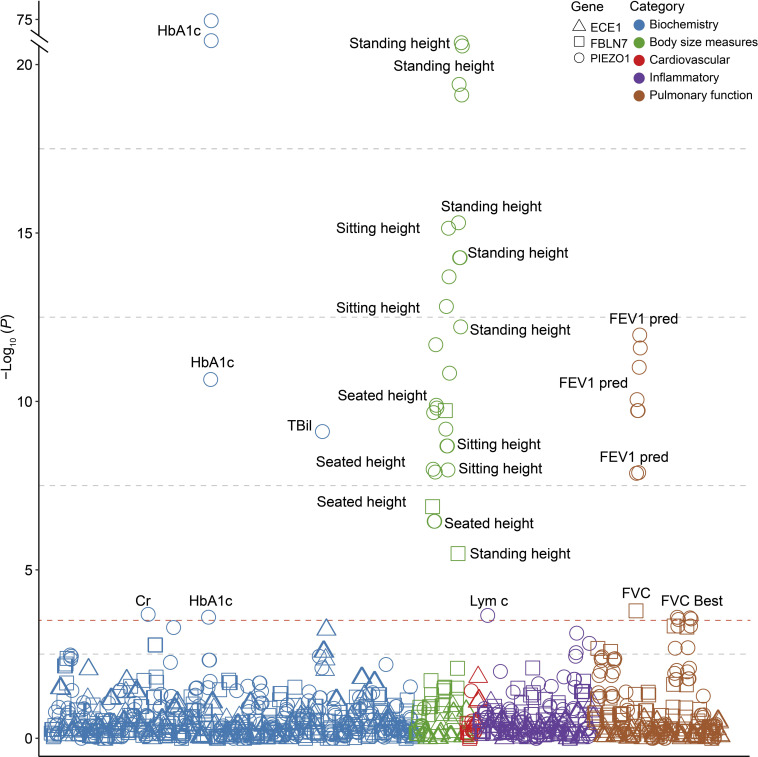
To further explore the association between VV-related genes and other phenotypes, we performed phenome-wide association analysis (Phe-WAS) for multiple quantitative traits and binary phenotypes, including biochemical indicators, cardiovascular disease, body size measures, inflammatory indicators, and pulmonary function (S18 Table). As expected, the majority of 53 traits showed significant associations with the identified VV-related genes (P < 2.77 × 10−5), especially common variant genes such as RNF5 (27/53), PGBD1 (24/53), TRIM31 (24/53), and GNA12 (14/53) (Fig 4). Furthermore, among these significant genotype-phenotype associations of independent common variant genes, 168 associations were negative and 113 were positive (S19 Table). Among three rare variant genes, PIEZO1 was significantly associated with a variety of curated phenotypes, such as HbA1c (P = 1.02 × 10−75), standing height (P = 5.41 × 10−23), sitting height (P = 7.23 × 10−16), and forced expiratory volume in 1 second (FEV1 pred, P = 1.07 × 10−12), and FBLN7 was mainly linked to height-related traits and pulmonary function indices (Fig 5 and S20 Table).
Fig 4. Phenotypic associations of the independent common variant genes associated with VV.
The P-values shown are two-sided. Multiple-trait Manhattan plot representing the association results between common variant genes and 53 phenotypes. The X-axis shows various phenotypes listed in the S19 Table. The dashed line represents the Bonferroni-corrected significance threshold for 53 traits and 34 genes (P = 2.77 × 10−5).
Fig 5. Phenotypic associations of the rare variant genes associated with VV.
The P-values shown are two-sided. Multiple-trait Manhattan plot representing the association results between 3 rare variant genes and 53 phenotypes. X-axis shows various phenotypes listed in the S20 Table. The dashed line represents Bonferroni-corrected significance for 53 traits and 3 genes (P = 3.14 × 10−4). Abbreviations: HbA1c, glycated hemoglobin; FEV1 pred, forced expiratory volume in 1-second, predicted; FVC, forced vital capacity; TBil, total bilirubin; Lym c, lymphocyte count; FVC Best, forced vital capacity, best measure.
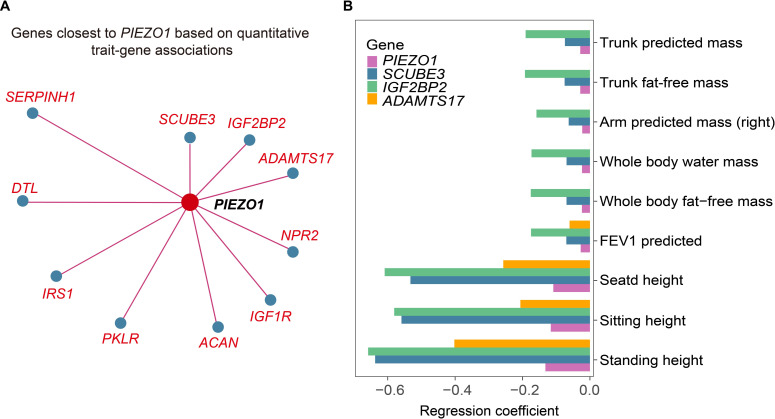
In addition, we tried to find genes with similar continuous trait fingerprints to these rare variant genes on the Gene-SCOUT website [25]. SCUBE3 and IGF2BP2 were found to be the most similar to the PIEZO1, respectively (Fig 6A). The module of gene signatures showed that both these similar genes and PIEZO1 were negatively associated with height related traits (Fig 6B). Recent evidence using machine learning methods [15] has identified that height is a risk factor for VV. Our findings further validated the effect of height on the risk of developing VV.
Fig 6. Quantitative traits and disease characteristics of PIEZO1.
(A) Genes with the most similar quantitative trait characteristics to PIEZO1 in UK Biobank from Gene-SCOUT. (B) Signatures of PIEZO1 and its similar genes (similarity to PIEZO1 sorted from top to bottom).
Discussion
Previous GWAS [14,15,26] have identified multiple risk loci and pathways involved in the pathophysiology of VV, improving the understanding of the polygenic architecture of the disease. The WES data from the UKB could provide a new direction for exploring the genetic mechanisms of VV [27]. VV-related genes and genetic variants identified in population-based WES studies were more abundant and reliable. It is perhaps that whole-exome data are usually obtained directly from sequencing rather than by imputation. WES typically focuses on coding regions of the genome and is more cost-effective and widely available than whole-genome sequencing [28]. Compared to non-coding variants, WES can directly assess variants that alter protein sequence, making it easier to explain their functional consequences. It also provides a clearer pathway to deeper mechanistic insights, and can potentially be useful for therapeutic target discovery [29–32] and precision medicine [33,34].
Recently, several studies of human gene-phenotype association for rare coding variants have identified the association between PIEZO1 and VV by using exome sequencing data from UKB participants [16,20]. Compared to the studies by Van Hout et al.[16] and Wang et al.[20], our study has a larger sample size and more validation of the findings (replication in the FinnGen cohort) and exploration of biological function (e.g. functional enrichment analysis, tissue expression analysis, and single-cell RNA sequencing analysis). In addition to the replication of 19 known VV-related genes from previous studies, our study further identified 17 novel genes. Most of these genes were successfully validated in FinnGen. Tissue analysis and single-cell gene expression analysis emphasized the importance of adipose tissue in the association between these identified genes and VV risk. Our Phe-WAS revealed the strong associations of these genes with body size measures, biological indicators, and inflammatory markers.
We identified a total of 36 VV-associated genes in our large-scale exome sequencing study. First, the exome-wide single-variant analysis identified 36 VV-related independent common variants mapping to 34 genes, half of which have been previously established [14,15,26,35,36]. The significant effects found in the single-variant analysis were the effect of PNO1 on chromosome 2 and that of PIEZO1 on chromosome 16. PNO1 plays an important role in both proteasome and ribosome biogenesis [37], and PIEZO1 is required for vascular development and function [38]. PNO1 is predominantly expressed in the liver and spleen, and slightly in the thymus, testis, and ovary, but not in the heart or brain [39]. In addition, it has been suggested that PNO1 may be involved in the expression of calcineurin phosphatase, a key signaling component in angiogenesis, and calcineurin phosphatase activity is essential for vascular development [15,40]. As the most significant novel gene identified in the single-variant analysis, TRIM10 was thought to be potentially involved in the body’s immune response [41]. Phenotypic association analysis revealed that TRIM10 had associations with several inflammatory markers, such as platelets, monocytes, and neutrophils. As a protein-coding gene, UBE2H encodes an E2 ubiquitin-conjugating enzyme family protein, which is involved not only in ubiquitination but also in cytoskeletal regulation and calcium signaling [42]. Our study also indicated that UBE2H is involved in protein binding and protein K48-linked ubiquitination by functional enrichment of the gene.
The other findings in this study were the associations of VV with three rare variant genes (PIEZO1, ECE1, and FBLN7) identified by the gene-based collapsing analysis. Both single-variant analysis and gene-based collapsing analysis revealed an association between VV and PIEZO1, supporting the previously reported hypothesis that common and rare variants may involve overlapping genes [43,44]. A study by Backman et al. [43] found that missense variants in PIEZO1 might have a gain-of-function effect and reduce VV risk, which offers the possibility of treating VV by pharmacological interventions. PIEZO1 is a mechanically activated ion channel that plays a decisive role in vascular architecture [38,45]. Its integral or endothelial-specific disruption can severely damage the growing vascular system and even lead to embryonic death.
As a newly discovered VV-related gene, ECE1 is an estrogen-regulatory gene in mesenteric arteries that could catalyze the conversion of pro-endothelin into endothelin-1, a potent vasoconstrictor [46,47]. A mouse model showed that estrogen receptor stimulation suppressed ECE1 expression [48]. In addition, previous studies have found an increase in ECE1 in pathological conditions, for instance, hypertension, coronary atherosclerosis, and vascular injury models [49–52]. Our expression analysis of ECE1 revealed its high expression in adipose tissue, especially in endothelial cells, and a positive association between obesity and VV risk has been demonstrated before [15,26]. It was well known that the development of VV was associated with a variety of vascular factors, such as changes in hemodynamics, endothelial cell activation, inflammation, and hypoxia [53–55]. As a cell adhesion molecule associated with diseases and developmental abnormalities, FBLN7 plays an essential role in specialized tissues such as the placenta, blood vessels, and cartilage [56–58]. FBLN7 protein fragments have been found to regulate endothelial cells and leukocyte function [59,60]. With anti-angiogenic function [57], they can be used to treat inflammatory diseases by modulating immune cell activity [61].
Moreover, although the carrier frequencies of LOF variants in the population were low, LOF mutations confer greater disease prevalence and genetic susceptibility to VV. As for the burden heritability, ultra-rare LOF variants had the highest heritability, followed by rare LOF variants and rare missense variants, which was in line with the previous finding by Weiner et al. [22] that LOF variants explained the majority of burden heritability.
We evaluated the robustness of the three genes identified in the collapsing analysis. The rare variant association results remained stable after adjusting for nearby common variants, probably due to our strict variant filter. Our sensitivity analysis revealed a novel gene in females (METTL21A), which prompted us to speculate whether the association between VV and sex might be influenced by genetic factors. Moreover, the current study revealed an overlap between VV-related genes identified by single-variant analyses and those identified by burden tests, suggesting the robustness of the association between overlapping genes and VV risk. A previous genetic study showed that vascular diseases were tightly linked to anthropometric indices [36]. The Phe-WAS of the identified genes showed VV genes were also associated with VV-related phenotypes, such as cardiovascular disease, body size measures, and pulmonary function indices. Phe-WAS can further explore genetic contributions to disease and identify the mechanisms of sharing across diseases [62].
There were some limitations in our study. First, the UKB was generally recruited from populations of European ancestry between the ages of 37 and 73, which makes our findings potentially inapplicable across all ethnic and age groups. Second, disease diagnoses are typically confirmed based on participant self-reports, ICD diagnosis codes, surgical records, and death registries. This may lead to the misclassification of disease phenotypes. Nevertheless, previous studies have been able to replicate genetic loci findings for common variants using the phenotype definitions in GWAS [14,15,26]. Third, we focused on the coding regions of the genome captured by WES, which may lead to the neglect of variants in non-coding regulatory regions. Due to the difficulty in obtaining exome sequencing data for VV from other databases, we validated our findings using summary statistics from FinnGen. The strength of our study is that it has the largest sample size of any VV-related whole-exome association study to date. Exome-wide analyses allow for more efficient targeting of variant genes that play an essential role in complex diseases compared to GWAS [63]. We defined the VV phenotype mainly based on the study of Ahmed et al.[14], which successfully replicated most of the VV-related risk loci identified in previous studies using this definition [15,64]. We additionally validated the results in FinnGen to determine the reproducibility of our findings.
In conclusion, large-scale exome-wide association studies are required for exploring the genetic associations of diseases. In this study, we identified many known genes and several novel effector genes significantly associated with VV. We found that LOF variants explained more burden heritability than missense variants. Furthermore, investigating genotype–phenotype associations would be critical for our insight into the underlying biological mechanisms of the disease, as well as for disease prevention and treatment. The study of genetic variants and diseases helps to develop strategies to target risk genes for prevention and treatment purposes.
Methods
Ethics statement
The UKB has received ethical approval from the National Health Service National Research Ethics Service and obtained written informed consent from all participants. This study was conducted under application number 19542.
Study population
The UKB is a large-scale prospective cohort of nearly 500,000 individuals aged 37–73 recruited between 2006 and 2010 at 22 centers in England, Scotland, and Wales [65]. Each participant provided biological samples, phenotypic endpoints, registry information on cancer and death, and health-related information, including biometric measures, lifestyle indicators, biomarkers in blood and urine, and imaging data [66].
In the discovery cohort, the VV cases were identified from the UKB using the following diagnostic, operative, and self-report codes (S1 Table): primary and/or secondary ICD-10 codes for varicose veins (I83), primary and/or secondary OPCS code for varicose vein surgery (L84-L88), self-reported operation code for varicose vein surgery (1479), self-reported non-cancer illness code for varicose veins (1494). Finally, 30,130 individuals who held at least one of these codes were classified as cases of VV.
Sequencing and quality control
The Regeneron Genetics Center (RGC) performed whole-exome sequencing on all participants from the UKB. The complete sequencing protocols were described in detail in the previous study [16]. Samples were subjected to double exponential 75 × 75bp paired-end reads using the IDT xGen Exome Research Panel v1.0 on the NovaSeq 6000 platform to capture exomes with over 20X coverage at 95.2% of sites. We performed similar additional QC on the OQFE WES pVCF files in the GRCh38 human reference genome construct [67] provided by the UKB (https://biobank.ctsu.ox.ac.uk/showcase/label.cgi?id=170), which was similar to that in a previous study [68]. Due to the limited quality control (QC) and filtering of the published dataset, we used a high-quality dataset generated from an extensive genotype, variant, and sample-level pipeline for further analysis (S1 Text). We utilized Hail to perform the genotype quality control and split multi-allelic sites into bi-allelic sites. All calls with low genotype quality or critically low/high genotype depth were set to no calls. And then we excluded variants based on the call rate < 90%, Hardy-Weinberg P-value < 1 × 10−15, and in Ensembl low-complexity regions. We selected a set of high-quality independent autosomal variants for the relationship inference, which was selected as: MAF > 0.1%, missingness < 1%, Hardy–Weinberg equilibrium (HWE) P-value > 1 × 10−6 and two rounds of pruning using—indep-pairwise 200 100 0.1 and—indep-pairwise 200 100 0.05. KING [69] software was applied to calculate pairwise heterozygote concordance rates for each pair of samples and the kinship coefficient using high-quality variants. Sample-level QC involved removing samples with Ti/Tv, Het/Hom, SNV/indel, number of singletons exceeding the mean ± 8SD, and discrepancies between self-reported and genetically inferred sex, as well as duplicates and withdrawn consent. The kinship coefficient threshold of 0.0884 (the 2nd relatedness) was used to define a related sample. For pairs with kinship coefficients higher than 0.0884, we removed samples associated with multiple other individuals iteratively until none remained to maximize the sample size. Then we randomly removed one from the remaining pairs. Finally, we restricted the sample to Caucasians (field 22006) and used high-quality independent autosomal variants to calculate the PCs within the descent.
Variant annotation
The preliminary analysis was performed by using SnpEff [70] to determine the most severe predicted consequences for specific variants in each gene transcript, with variants filtered to rare. We classified the variants annotated as stop gained, start/stop lost, splice donor/acceptor, or frameshift as LOF variants. A missense variant was considered damaging when it was predicted to be deleterious by SIFT (sorting intolerant from tolerant) [71], PolyPhen-2 HDIV, PolyPhen-2 HVAR [72], LRT (Likelihood Ratio Test)[73], and MutationTaster [74,75] consistently. The definitions of deleterious variants for each software are given in S1 Text.
Single-variant association analysis
Single-variant association analysis of common exonic single nucleotide polymorphisms (SNPs) in VV was performed using SAIGE [76] software. The analysis was adjusted for age, sex, and the top 10 PCs, and the significance threshold was set at 1.19 × 10−6 (Bonferroni correction for 42,123 exonic SNPs). Then, we clumped the results to remove possible associations due to strong linkage disequilibrium (LD) using the—clump function in PLINK v2. Significant SNPs within each risk locus were selected to identify the lead SNPs, which were defined as those with the lowest P value when a strong LD (r2 > 0.01) was observed for multiple SNPs within a 1000 kilobases (kb) window. Risk loci were identified as regions of ±1 Mb around lead SNPs.
Gene-based exome-wide association analysis
We conducted gene-based collapsing analysis to define rare variant genes associated with VV. Rare variants were defined as MAF < 1% [77]. In our preliminary analysis, carriers of rare LOF variants and missense variants were decomposed into individual variables for each gene. We performed two functional category masks: LOF and LOF + missense variants, and subdivided each functional mask into four frequency masks: < 0.01, < 0.001, < 0.0001, and < 0.00001. It means that we constructed eight gene-burden models for each gene. Following adjustment for age, sex, and the top 10 PCs, gene-based collapsing analysis was conducted in SAIGE-GENE+ software [78] using a logistic mixed model approach with a saddle-point approximation. This approach is effective for the analysis of large sample data, and provides accurate P-values even when the case-control ratio is highly unbalanced [76]. In our collapsing analysis, we mainly reported the P value of SKAT-O test [79], and the exome-wide significance threshold (P = 2.51 × 10−6) was determined by employing the Bonferroni correction (0.05/19,897). After identifying the VV-related genes, we further calculated the disease prevalence and carrier frequencies of rare variants for these genes.
Burden heritability estimation
In this study, we estimated the heritability explained by the rare coding variants using burden heritability regression (BHR) [22]. BHR estimates burden heritability from the regression slope of the gene burden statistic on the burden score. We focused on estimating burden heritability for two functional categories (LOF and missense variants) and two allele frequency bins (MAF < 1 × 10−5, and MAF between 1 × 10−5 and 1 × 10−2). Ultra-rare coding variants were defined as MAF < 1 × 10−5 in this study. A single-variant association test was performed on the VV to obtain summary statistics of variant-level association. The “aggregate” mode was used to calculate the total burden heritability of disease across multiple frequency-function strata. Single-variant association summary statistics were used as input utilizing the baseline-BHR file provided by Weiner et al.[22]. These analyses were performed based on the R package bhr (v0.1.0).
Replication of genes in the FinnGen cohort
We replicated the VV-related genes that we have identified using summary statistics from the FinnGen Consortium online results (version 8, 21). The FinnGen cohort recruited 342,499 individuals of Finnish descent with genotype and health register data, including 26,720 VV cases and 295,014 controls. The cohort collected and confirmed participants’ diagnostic information through national healthcare registries and used the International Classification of Diseases (ICD) for documentation. Genetic association tests were performed using logistic mixed models in SAIGE, and GWAS summary statistics for “Varicose veins” are publicly available online (see Data availability). To validate the findings of single-variant association analysis, we directly searched for our lead SNPs in summary statistics. For VV-related genes identified in gene-based collapsing analysis, we selected the variants with the strongest FinnGen VV correlation mapped to these genes. The significance threshold used for validation in the summary statistics was P < 0.05.
Gene set enrichment and expression analyses
To identify terms and metabolic pathways with functional enrichment in differentially expressed genes, we performed the GO enrichment analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [80]. The enrichment significance threshold was set at 0.05 (correction method = "FDR"). Normalized gene expression (transcripts per million, TPM) was obtained from the Genotype-Tissue Expression (GTEx) v8 for 30 general tissue types using the GENE2FUNC core program in Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) [81]. We used the average log2 (TPM) to compare expression levels between genes and tissue types. Data from each gene set was enriched for testing and multiple corrections (default for Benjamini-Hochberg). We leveraged the adipose tissue single-cell RNA sequencing data provided by the Tabula Sapiens Consortium to identify the expression levels of the genes in different cell types [82]. The Tabula Sapiens is a single-cell transcriptomic atlas of 24 human tissues and organs from 15 unique donors. The adipose tissues from two donors were obtained from surgery and immediately prepared from a fluorescence-activated cell sorting-sorted smart-seq2 platform or 10x Genomics 3’ V3.1 droplet-based sequencing platform. The quality control, data integration, clustering, and cell type annotation procedures of single-cell RNA sequencing data were conducted, as described in the previous study [82]. The R package Seurat was used for downstream data analysis (i.e., normalization and data scaling) and visualization [83].
Sensitivity and leave-one-variant-out analyses
As being female is one of the risk factors for VV, sensitivity analyses stratified by sex were performed to explore the sex heterogeneity of the genetic associations identified. There were 162,210 participants in the male group and 188,560 in the female group after stratification by sex. Enrichment analysis was then performed separately for the VV-related genes identified by sex. In addition, we assessed the robustness of the genetic results in the variant masks using LOVO analysis. After removing each variant from the significant models one at a time, we reran the association analysis for the variants included in the original mask. The variant that reached the maximum LOVO P value after removal was regarded as the most important variant in each gene [68].
Conditional analyses
We conducted multi-SNP-based conditional and joint association analyses (GCTA-COJO) using GCTA software [84,85] to identify conditionally independent variants from common variants associated with VV and to validate the results after clumping. The significance threshold was set at P = 1.19 × 10−6, and other parameters were left at their default settings.
We performed the conditional analysis to demonstrate that the significant rare variant signals were independent of nearby common variants. First, we ran association analyses for common variants (MAF > 0.5%) within the ±500 kb genomic region of the identified genes. Then we used the—clump function (—clump-p1 1 × 10−5,—clump-r2 0.01) in PLINK [86] to aggregate and threshold the results, identifying independent index common variants. Finally, gene-based collapsing analysis was rerun after adding the clumped common variant to the model as a covariate [68].
Phenome-wide association analyses
To better explain the mechanisms of associations between risk genes and VV, we performed Phe-WAS. This analysis mainly focused on 53 phenotypes selected from UKB that may be associated with VV, including cardiovascular disease, 29 biochemical traits, 4 body size traits, 9 inflammatory traits, and 10 spirometry traits. Associations of 53 phenotypes with rare variant genes associated with VV were conducted using gene-level collapsing analysis (P < 0.05/159), whereas linear or logistic models were applied to independent common variants (P < 0.05/1,802). All models were adjusted for age, sex, and the top 10 PCs. To correct for multiple testing, the Bonferroni test was utilized.
Gene-SCOUT
The Gene-SCOUT (Gene-Similarity from Continuous Traits, https://astrazeneca-cgr-publications.github.io/gene-scout/) [25] aims to find genes similar to specific genes and to construct a unique signature for each gene using associations derived from 450,000 exomes and 120,000 samples of metabolomic data sequenced from the UKB. This tool was used to identify the genes most similar to VV-related genes and to show significant associations of these genes with all quantitative traits.
Supporting information
Abbreviation: VV, varicose veins.
(TIF)
The x-axis and y-axis represent the values of the two components of PCA (PC1, PC2), and each point in the figure represents an individual. Abbreviation: VV, varicose veins; PC, principle components.
(TIF)
The y-axis represents the observed–log10 P values across all tests, while the x-axis represents the expected under the null-hypothesis. P values were obtained from the results of the gene-based collapse tests for varicose veins, using SAIGE-GENE+ software. In the gene-based collapse test, we applied two different maximum MAF cutoffs (0.01 and 0.001) and two different variant annotation groups (LOF and LOF + missense) to perform burden tests. All models were adjusted for age, sex, and top ten principal components. Abbreviation: VV, varicose veins; LOF, loss of function; λ, lambda.
(TIF)
P-values for differences between groups were calculated by t-test. MAF, minor allele frequency, NS., no significance, pLI, the probability of intolerance to loss of function.
(TIF)
The x-axis indicates variants removed from the gene-based collapsed test, and the y-axis indicates the -log10 P-value for associations without that variant P-values were derived from the gene-based collapsed test using the SAIGE-GENE+ software, adjusted for age, sex, and the top 10 principal components.
(TIF)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We gratefully thank all the participants and professionals for collecting and preparing data in UKB and FinnGen study.
Data Availability
The main data, including individual-level phenotype and sequencing data, used in this study were accessed from the UK Biobank under application number 19542 (https://www.ukbiobank.ac.uk/). Statistics for varicose veins in FinnGen are available on the website (http://r8.finngen.fi) and are indicated with the phenotype code I9_VARICVE. The scRNA-seq data used in the present study were acquired from GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the accession number: GSE201333. The following software and packages were used for data analysis: FUMA v.1.3.8 (https://fuma.ctglab.nl/), SnpEff (https://pcingola.github.io/SnpEff/), GCTA v.1.93 (https://yanglab.westlake.edu.cn/software/gcta/#COJO), SAIGE-GENE+ (https://github.com/saigegit/SAIGE), BHR (https://github.com/ajaynadig/bhr), PLINK (https://www.cog-genomics.org/plink/) and R v.4.2.0 (https://www.r-project.org/), DAVID, (https://david.ncifcrf.gov/), Gene-SCOUT (https://astrazeneca-cgr-publications.github.io/gene-scout/). Scripts used to perform the analyses are available at https://github.com/ddzhang877/vv_wes.
Funding Statement
JT, Yu was supported by grants from the Science and Technology Innovation 2030 Major Projects (2022ZD0211600), National Natural Science Foundation of China (82071201, 82071997), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), Research Start-up Fund of Huashan Hospital (2022QD002), Excellence 2025 Talent Cultivation Program at Fudan University (3030277001), and Shanghai Talent Development Funding for The Project (2019074). W Cheng was supported by grants from the Shanghai Rising-Star Program (21QA1408700). JF Feng was supported by grants from the 111 Project (B18015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Evans CJ, Fowkes FG, Ruckley CV, Lee AJ. Prevalence of varicose veins and chronic venous insufficiency in men and women in the general population: Edinburgh Vein Study. J Epidemiol Community Health. 1999;53(3):149–53. doi: 10.1136/jech.53.3.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamdan A. Management of varicose veins and venous insufficiency. Jama. 2012;308(24):2612–21. doi: 10.1001/jama.2012.111352 [DOI] [PubMed] [Google Scholar]
- 3.Marsden G, Perry M, Kelley K, Davies AH. Diagnosis and management of varicose veins in the legs: summary of NICE guidance. BMJ (Clinical research ed). 2013;347:f4279. doi: 10.1136/bmj.f4279 [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell TF, Balk EM, Dermody M, Tangney E, Iafrati MD. Recurrence of varicose veins after endovenous ablation of the great saphenous vein in randomized trials. Journal of vascular surgery Venous and lymphatic disorders. 2016;4(1):97–105. doi: 10.1016/j.jvsv.2014.11.004 [DOI] [PubMed] [Google Scholar]
- 5.Brand FN, Dannenberg AL, Abbott RD, Kannel WB. The epidemiology of varicose veins: the Framingham Study. American journal of preventive medicine. 1988;4(2):96–101. [PubMed] [Google Scholar]
- 6.Sisto T, Reunanen A, Laurikka J, Impivaara O, Heliövaara M, Knekt P, et al. Prevalence and risk factors of varicose veins in lower extremities: mini-Finland health survey. The European journal of surgery=Acta chirurgica. 1995;161(6):405–14. [PubMed] [Google Scholar]
- 7.Kurz X, Kahn SR, Abenhaim L, Clement D, Norgren L, Baccaglini U, et al. Chronic venous disorders of the leg: epidemiology, outcomes, diagnosis and management. Summary of an evidence-based report of the VEINES task force. Venous Insufficiency Epidemiologic and Economic Studies. International angiology: a journal of the International Union of Angiology. 1999;18(2):83–102. [PubMed] [Google Scholar]
- 8.Lee AJ, Evans CJ, Allan PL, Ruckley CV, Fowkes FG. Lifestyle factors and the risk of varicose veins: Edinburgh Vein Study. Journal of clinical epidemiology. 2003;56(2):171–9. doi: 10.1016/s0895-4356(02)00518-8 [DOI] [PubMed] [Google Scholar]
- 9.Zöller B, Ji J, Sundquist J, Sundquist K. Family history and risk of hospital treatment for varicose veins in Sweden. Br J Surg. 2012;99(7):948–53. doi: 10.1002/bjs.8779 [DOI] [PubMed] [Google Scholar]
- 10.Carpentier PH, Maricq HR, Biro C, Ponçot-Makinen CO, Franco A. Prevalence, risk factors, and clinical patterns of chronic venous disorders of lower limbs: a population-based study in France. J Vasc Surg. 2004;40(4):650–9. doi: 10.1016/j.jvs.2004.07.025 [DOI] [PubMed] [Google Scholar]
- 11.Scott TE, LaMorte WW, Gorin DR, Menzoian JO. Risk factors for chronic venous insufficiency: a dual case-control study. J Vasc Surg. 1995;22(5):622–8. doi: 10.1016/s0741-5214(95)70050-1 [DOI] [PubMed] [Google Scholar]
- 12.Lim CS, Davies AH. Pathogenesis of primary varicose veins. Br J Surg. 2009;96(11):1231–42. doi: 10.1002/bjs.6798 [DOI] [PubMed] [Google Scholar]
- 13.Ellinghaus E, Ellinghaus D, Krusche P, Greiner A, Schreiber C, Nikolaus S, et al. Genome-wide association analysis for chronic venous disease identifies EFEMP1 and KCNH8 as susceptibility loci. Sci Rep. 2017;7:45652. doi: 10.1038/srep45652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed WU, Kleeman S, Ng M, Wang W, Auton A, Lee R, et al. Genome-wide association analysis and replication in 810,625 individuals with varicose veins. Nature communications. 2022;13(1):3065. doi: 10.1038/s41467-022-30765-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukaya E, Flores AM, Lindholm D, Gustafsson S, Zanetti D, Ingelsson E, et al. Clinical and Genetic Determinants of Varicose Veins. Circulation. 2018;138(25):2869–80. doi: 10.1161/CIRCULATIONAHA.118.035584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Hout CV, Tachmazidou I, Backman JD, Hoffman JD, Liu D, Pandey AK, et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586(7831):749–56. doi: 10.1038/s41586-020-2853-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momozawa Y, Mizukami K. Unique roles of rare variants in the genetics of complex diseases in humans. Journal of human genetics. 2021;66(1):11–23. doi: 10.1038/s10038-020-00845-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi SH, Jurgens SJ, Weng LC, Pirruccello JP, Roselli C, Chaffin M, et al. Monogenic and Polygenic Contributions to Atrial Fibrillation Risk: Results From a National Biobank. Circ Res. 2020;126(2):200–9. doi: 10.1161/CIRCRESAHA.119.315686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smelser DT, Haley JS, Ryer EJ, Elmore JR, Cook AM, Carey DJ. Association of varicose veins with rare protein-truncating variants in PIEZO1 identified by exome sequencing of a large clinical population. Journal of vascular surgery Venous and lymphatic disorders. 2022;10(2):382–9.e2. doi: 10.1016/j.jvsv.2021.07.007 [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Dhindsa RS, Carss K, Harper AR, Nag A, Tachmazidou I, et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature. 2021;597(7877):527–32. doi: 10.1038/s41586-021-03855-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–18. doi: 10.1038/s41586-022-05473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiner DJ, Nadig A, Jagadeesh KA, Dey KK, Neale BM, Robinson EB, et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature. 2023;614(7948):492–9. doi: 10.1038/s41586-022-05684-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330-d8. [DOI] [PMC free article] [PubMed]
- 25.Middleton L, Harper AR, Nag A, Wang Q, Reznichenko A, Vitsios D, et al. Gene-SCOUT: identifying genes with similar continuous trait fingerprints from phenome-wide association analyses. Nucleic Acids Res. 2022;50(8):4289–301. doi: 10.1093/nar/gkac274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shadrina AS, Sharapov SZ, Shashkova TI, Tsepilov YA. Varicose veins of lower extremities: Insights from the first large-scale genetic study. PLoS genetics. 2019;15(4):e1008110. doi: 10.1371/journal.pgen.1008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiabor Barrett KM, Bolze A, Ni Y, White S, Isaksson M, Sharma L, et al. Positive predictive value highlights four novel candidates for actionable genetic screening from analysis of 220,000 clinicogenomic records. Genet Med. 2021;23(12):2300–8. doi: 10.1038/s41436-021-01293-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro DM, Delaneau O. Non-coding rare variant associations with blood traits on 166 740 UK Biobank genomes. 2023:2023.12.01.569422. [Google Scholar]
- 29.Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. The New England journal of medicine. 2018;378(12):1096–106. doi: 10.1056/NEJMoa1712191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. The New England journal of medicine. 2017;377(3):211–21. doi: 10.1056/NEJMoa1612790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen JC, Boerwinkle E, Mosley TH Jr., Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. The New England journal of medicine. 2006;354(12):1264–72. doi: 10.1056/NEJMoa054013 [DOI] [PubMed] [Google Scholar]
- 32.Scott RA, Freitag DF, Li L, Chu AY, Surendran P, Young R, et al. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci Transl Med. 2016;8(341):341ra76. doi: 10.1126/scitranslmed.aad3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science (New York, NY). 2016;354(6319). doi: 10.1126/science.aaf7000 [DOI] [PubMed] [Google Scholar]
- 34.Manickam K, Buchanan AH, Schwartz MLB, Hallquist MLG, Williams JL, Rahm AK, et al. Exome Sequencing-Based Screening for BRCA1/2 Expected Pathogenic Variants Among Adult Biobank Participants. JAMA network open. 2018;1(5):e182140. doi: 10.1001/jamanetworkopen.2018.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shadrina A, Tsepilov Y, Sokolova E, Smetanina M, Voronina E, Pakhomov E, et al. Genome-wide association study in ethnic Russians suggests an association of the MHC class III genomic region with the risk of primary varicose veins. Gene. 2018;659:93–9. doi: 10.1016/j.gene.2018.03.039 [DOI] [PubMed] [Google Scholar]
- 36.Levin MG, Huffman JE, Verma A, Sullivan KA, Rodriguez AA, Kainer D, et al. Genetics of varicose veins reveals polygenic architecture and genetic overlap with arterial and venous disease. Nature Cardiovascular Research. 2023;2(1):44–57. [Google Scholar]
- 37.Wang X, Wu T, Hu Y, Marcinkiewicz M, Qi S, Valderrama-Carvajal H, et al. Pno1 tissue-specific expression and its functions related to the immune responses and proteasome activities. PloS one. 2012;7(9):e46093. doi: 10.1371/journal.pone.0046093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515(7526):279–82. doi: 10.1038/nature13701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou GJ, Zhang Y, Wang J, Guo JH, Ni J, Zhong ZM, et al. Cloning and characterization of a novel human RNA binding protein gene PNO1. DNA sequence: the journal of DNA sequencing and mapping. 2004;15(3):219–24. doi: 10.1080/10425170410001702159 [DOI] [PubMed] [Google Scholar]
- 40.Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105(7):863–75. doi: 10.1016/s0092-8674(01)00396-8 [DOI] [PubMed] [Google Scholar]
- 41.Jia X, Zhao C, Zhao W. Emerging Roles of MHC Class I Region-Encoded E3 Ubiquitin Ligases in Innate Immunity. Frontiers in immunology. 2021;12:687102. doi: 10.3389/fimmu.2021.687102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lausen J, Pless O, Leonard F, Kuvardina ON, Koch B, Leutz A. Targets of the Tal1 transcription factor in erythrocytes: E2 ubiquitin conjugase regulation by Tal1. The Journal of biological chemistry. 2010;285(8):5338–46. doi: 10.1074/jbc.M109.030296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backman JD, Li AH, Marcketta A, Sun D, Mbatchou J, Kessler MD, et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599(7886):628–34. doi: 10.1038/s41586-021-04103-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604(7906):509–16. doi: 10.1038/s41586-022-04556-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy SE, Cahalan SM, et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(28):10347–52. doi: 10.1073/pnas.1409233111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell FD, Skepper JN, Davenport AP. Endothelin peptide and converting enzymes in human endothelium. Journal of cardiovascular pharmacology. 1998;31 Suppl 1:S19-21. doi: 10.1097/00005344-199800001-00008 [DOI] [PubMed] [Google Scholar]
- 47.Kido T, Sawamura T, Masaki T. The processing pathway of endothelin-1 production. Journal of cardiovascular pharmacology. 1998;31 Suppl 1:S13-5. doi: 10.1097/00005344-199800001-00006 [DOI] [PubMed] [Google Scholar]
- 48.Rodrigo MC, Martin DS, Eyster KM. Vascular ECE-1 mRNA expression decreases in response to estrogens. Life sciences. 2003;73(23):2973–83. doi: 10.1016/j.lfs.2003.05.001 [DOI] [PubMed] [Google Scholar]
- 49.Disashi T, Nonoguchi H, Iwaoka T, Naomi S, Nakayama Y, Shimada K, et al. Endothelin converting enzyme-1 gene expression in the kidney of spontaneously hypertensive rats. Hypertension (Dallas, Tex: 1979). 1997;30(6):1591–7. doi: 10.1161/01.hyp.30.6.1591 [DOI] [PubMed] [Google Scholar]
- 50.Grantham JA, Schirger JA, Williamson EE, Heublein DM, Wennberg PW, Kirchengast M, et al. Enhanced endothelin-converting enzyme immunoreactivity in early atherosclerosis. Journal of cardiovascular pharmacology. 1998;31 Suppl 1:S22-6. doi: 10.1097/00005344-199800001-00009 [DOI] [PubMed] [Google Scholar]
- 51.Maguire JJ, Davenport AP. Increased response to big endothelin-1 in atherosclerotic human coronary artery: functional evidence for up-regulation of endothelin-converting enzyme activity in disease. British journal of pharmacology. 1998;125(2):238–40. doi: 10.1038/sj.bjp.0702102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.d’Uscio LV, Barton M, Shaw S, Lüscher TF. Endothelin in atherosclerosis: importance of risk factors and therapeutic implications. Journal of cardiovascular pharmacology. 2000;35(4 Suppl 2):S55-9. doi: 10.1097/00005344-200000002-00013 [DOI] [PubMed] [Google Scholar]
- 53.Raffetto JD. Pathophysiology of Chronic Venous Disease and Venous Ulcers. The Surgical clinics of North America. 2018;98(2):337–47. doi: 10.1016/j.suc.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 54.Pfisterer L, König G, Hecker M, Korff T. Pathogenesis of varicose veins ‐ lessons from biomechanics. VASA Zeitschrift fur Gefasskrankheiten. 2014;43(2):88–99. doi: 10.1024/0301-1526/a000335 [DOI] [PubMed] [Google Scholar]
- 55.Lim CS, Kiriakidis S, Sandison A, Paleolog EM, Davies AH. Hypoxia-inducible factor pathway and diseases of the vascular wall. J Vasc Surg. 2013;58(1):219–30. doi: 10.1016/j.jvs.2013.02.240 [DOI] [PubMed] [Google Scholar]
- 56.de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, Fisher LW, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. The Journal of biological chemistry. 2007;282(42):30878–88. doi: 10.1074/jbc.M705847200 [DOI] [PubMed] [Google Scholar]
- 57.Ikeuchi T, de Vega S, Forcinito P, Doyle AD, Amaral J, Rodriguez IR, et al. Extracellular Protein Fibulin-7 and Its C-Terminal Fragment Have In Vivo Antiangiogenic Activity. Sci Rep. 2018;8(1):17654. doi: 10.1038/s41598-018-36182-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell MW, Raeker MO, Geisler SB, Thomas PE, Simmons TA, Bernat JA, et al. Functional analysis of candidate genes in 2q13 deletion syndrome implicates FBLN7 and TMEM87B deficiency in congenital heart defects and FBLN7 in craniofacial malformations. Human molecular genetics. 2014;23(16):4272–84. doi: 10.1093/hmg/ddu144 [DOI] [PubMed] [Google Scholar]
- 59.de Vega S, Suzuki N, Nonaka R, Sasaki T, Forcinito P, Arikawa-Hirasawa E, et al. A C-terminal fragment of fibulin-7 interacts with endothelial cells and inhibits their tube formation in culture. Archives of biochemistry and biophysics. 2014;545:148–53. doi: 10.1016/j.abb.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Vega S, Hozumi K, Suzuki N, Nonaka R, Seo E, Takeda A, et al. Identification of peptides derived from the C-terminal domain of fibulin-7 active for endothelial cell adhesion and tube formation disruption. Biopolymers. 2016;106(2):184–95. doi: 10.1002/bip.22754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarangi PP, Chakraborty P, Dash SP, Ikeuchi T, de Vega S, Ambatipudi K, et al. Cell adhesion protein fibulin-7 and its C-terminal fragment negatively regulate monocyte and macrophage migration and functions in vitro and in vivo. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2018;32(9):4889–98. doi: 10.1096/fj.201700686RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bastarache L, Denny JC, Roden DM. Phenome-Wide Association Studies. Jama. 2022;327(1):75–6. doi: 10.1001/jama.2021.20356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flannick J, Mercader JM, Fuchsberger C, Udler MS, Mahajan A, Wessel J, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570(7759):71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helkkula P, Hassan S, Saarentaus E, Vartiainen E, Ruotsalainen S, Leinonen JT, et al. Genome-wide association study of varicose veins identifies a protective missense variant in GJD3 enriched in the Finnish population. Communications biology. 2023;6(1):71. doi: 10.1038/s42003-022-04285-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen NE, Sudlow C, Peakman T, Collins R. UK biobank data: come and get it. Sci Transl Med. 2014;6(224):224ed4. doi: 10.1126/scitranslmed.3008601 [DOI] [PubMed] [Google Scholar]
- 66.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szustakowski JD, Balasubramanian S, Kvikstad E, Khalid S, Bronson PG, Sasson A, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet. 2021;53(7):942–8. doi: 10.1038/s41588-021-00885-0 [DOI] [PubMed] [Google Scholar]
- 68.Jurgens SJ, Choi SH, Morrill VN, Chaffin M, Pirruccello JP, Halford JL, et al. Analysis of rare genetic variation underlying cardiometabolic diseases and traits among 200,000 individuals in the UK Biobank. Nat Genet. 2022;54(3):240–50. doi: 10.1038/s41588-021-01011-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics (Oxford, England). 2010;26(22):2867–73. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nature protocols. 2016;11(1):1–9. doi: 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- 72.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome research. 2009;19(9):1553–61. doi: 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7(8):575–6. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 75.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11(4):361–2. doi: 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 76.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335–41. doi: 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou W, Bi W, Zhao Z, Dey KK, Jagadeesh KA, Karczewski KJ, et al. SAIGE-GENE+ improves the efficiency and accuracy of set-based rare variant association tests. Nat Genet. 2022;54(10):1466–9. doi: 10.1038/s41588-022-01178-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics (Oxford, England). 2012;13(4):762–75. doi: 10.1093/biostatistics/kxs014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–w21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature communications. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science (New York, NY). 2022;376(6594):eabl4896. doi: 10.1126/science.abl4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American journal of human genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–75, s1-3. doi: 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]