Abstract
Purpose:
Erdafitinib is the only FDA-approved targeted therapy for FGFR2/3-altered metastatic urothelial cancer. We characterized the genetic landscape of FGFR-altered urothelial carcinoma and real-world clinical outcomes with erdafitinib, including on-treatment genomic evolution.
Methods:
Prospectively collected clinical data were integrated with institutional genomic data to define the landscape of FGFR2/3-altered urothelial carcinoma. To identify mechanisms of erdafitinib resistance, a subset of patients underwent prospective cell-free (cf)DNA assessment.
Results:
FGFR3 alterations predictive of erdafitinib sensitivity were identified in 39% (199/504) of patients with non-muscle invasive, 14% (75/526) with muscle-invasive, 43% (81/187) with localized upper tract, and 26% (59/228) with metastatic specimens. One patient had a potentially sensitizing FGFR2 fusion. Among 27 FGFR3-altered cases with a primary tumor and metachronous metastasis, 7 paired specimens (26%) displayed discordant FGFR3 status. Erdafitinib achieved a response rate of 40% but median progression-free and overall survival of only 2.8 and 6.6 months, respectively (n = 32). Dose reductions (38%, 12/32) and interruptions (50%, 16/32) were common. Putative resistance mutations detected in cfDNA involved TP53 (n=5), AKT1 (n=1), and second-site FGFR3 mutations (n=2).
Conclusion:
FGFR3 mutations are common in urothelial carcinoma, whereas FGFR2 alterations are rare. Discordance of FGFR3 mutational status between primary and metastatic tumors occurs frequently and raises concern over sequencing archival primary tumors to guide patient selection for erdafitinib therapy. Erdafitinib responses were typically brief and dosing was limited by toxicity. FGFR3, AKT1, and TP53 mutations detected in cfDNA represent putative mechanisms of acquired erdafitinib resistance.
Keywords: erdafitinib, FGFR, urothelial carcinoma, bladder cancer, cell-free DNA
INTRODUCTION
Oncogenic fibroblast growth factor receptor 3 (FGFR3) mutations and fusions are common in urothelial carcinoma, with significant variation in frequency reported as a function of tumor grade, stage, and primary tumor site (1–3). FGFR3 mutations and FGFR2/3 fusions are the only genomic alterations currently recognized as standard-of-care predictive biomarkers of response to an FDA-approved kinase inhibitor (erdafitinib) in metastatic urothelial carcinoma (RRID:SCR_012945). Accelerated FDA approval of erdafitinib was based on the results of a phase II single arm trial (BLC2001) which demonstrated an objective response rate (ORR) of 40% and progression-free survival (PFS) of 5.5 months in patients with metastatic urothelial carcinoma whose tumors harbored an oncogenic FGFR3 mutation or FGFR2/3 fusion (4). Retrospective studies also suggest an association between FGFR3 alterations and response to chemotherapy and immunotherapy for urothelial carcinoma (4–9).
Resistance mechanisms to erdafitinib remain largely undefined. Based on observations with other kinase inhibitors, both on-target mutations, such as FGFR3 gatekeeper mutations, and activation of parallel or downstream signaling pathways (oncogenic bypass) may mediate drug resistance (10). Analysis of cell-free (cf)DNA collected longitudinally in patients with metastatic urothelial carcinoma treated with the FGFR inhibitor infigratinib detected FGFR3 gatekeeper mutations in a minority of patients, but similar findings have yet to be reported with erdafitinib (11). Moreover, the degree of discordance of FGFR3 alterations between primary and metastatic tumors, a potential determinant of FGFR inhibitor sensitivity, warrants further exploration (12–14).
To define the landscape of FGFR2/3 alterations in urothelial carcinoma across disease states and treatment exposures, the mutational concordance of FGFR2/3 alterations between primary and metastatic disease sites, and the pattern of co-altered genes that could influence sensitivity to FGFR inhibition, we integrated tumor genomic data with detailed patient demographic and treatment response data generated as part of the largest prospective institutional sequencing initiative of urothelial carcinoma to date. We additionally characterized the clinical outcomes and toxicity profiles of patients with metastatic urothelial carcinoma treated with erdafitinib, with longitudinal profiling of cfDNA to characterize evolution of tumor genomes under the selective pressure of erdafitinib therapy.
MATERIALS AND METHODS
Patients
To characterize the frequency and landscape of FGFR2 and FGFR3 genomic alterations across urothelial carcinoma disease states, we identified all patients of both male and female sex at Memorial Sloan Kettering Cancer Center (MSK) with urothelial carcinoma-predominant histology (centrally reviewed by H.A.A.) who had undergone tumor genomic sequencing using the FDA-authorized MSK-IMPACT [Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets] targeted sequencing platform (15). In all patients with urothelial carcinoma who had received erdafitinib as standard care, baseline clinical characteristics, treatment outcomes, and adverse events were documented using CTCAE v5 (B.G.). Best overall response to erdafitinib was assessed by RECIST v1.1 (C.D.) (16). All patients who initiated erdafitinib at MSK and consented to genomic sequencing were eligible for and included in our prospective, longitudinal cfDNA sequencing analysis. Plasma samples were collected and banked at baseline, on-treatment every 4 weeks, and at progression. Samples collected at baseline, on-treatment at 8 weeks, and progression as well as individually selected additional timepoints were analyzed using MSK-ACCESS, a cfDNA assay designed to detect mutations and select copy number alterations in 129 cancer-associated genes (17, 18). MSK-ACCESS uses unique molecular indexes and >15,000x depth of coverage that allow for an allele frequency detection threshold of 0.1% (18). All patients included in the genomic sequencing portion of this study either signed written informed consent to a prospective institutional protocol approved by the Internal Review Board of Memorial Sloan Kettering Cancer Center (12–245, clinicaltrials.gov identifier NCT01775072), or were included under a limited waiver of authorization. The study was conducted in accordance with the Declaration of Helsinki, CIOMS, Belmont Report, and U.S. Common Rule.
All oncogenic FGFR3 mutations and all oncogenic FGFR2/3 fusions were classified as erdafitinib-sensitive. Oncogenic alterations were differentiated from variants of unknown significance using OncoKB (http://www.oncokb.org; RRID:SCR_014782) (19). Total and allele-specific copy number, tumor purity, and ploidy were estimated using the FACETS algorithm (version 0.5.6) (20). To model effects of FGFR3 gatekeeper mutations on erdafitinib binding, in silico analysis was performed using CHARMM-GUI and the crystal structure of FGFR1 complexed with erdafitinib (PDB ID: 5EW8). Final visualization was performed using VMD (21, 22).
Statistical Analysis
Patient characteristics were summarized across relevant clinical and biological variables, including sex, age, and race. Tests for associations of categorical data and continuous data were assessed using Fisher’s exact or Χ2 test and the Wilcoxon rank sum test, respectively. Survival endpoints were estimated using the Kaplan-Meier method, and differences in survival between groups were assessed by log-rank test. Overall survival (OS) was calculated as time from therapy initiation to death, while progression free survival (PFS) was calculated as time from therapy initiation to disease progression or death.
Data availability statement
All MSK-IMPACT and MSK-ACCESS results, including the gene sequencing panel version used to analyze each individual tumor, and associated clinical data are available via the cBioPortal for Cancer Genomics under study title “Bladder Cancer (MSK, Clin Cancer Research 2023)” at https://www.cbioportal.org/study/summary?id=bladder_msk_2023 (23).
RESULTS
Clinical and genomic characteristics of FGFR2/3-altered urothelial carcinoma
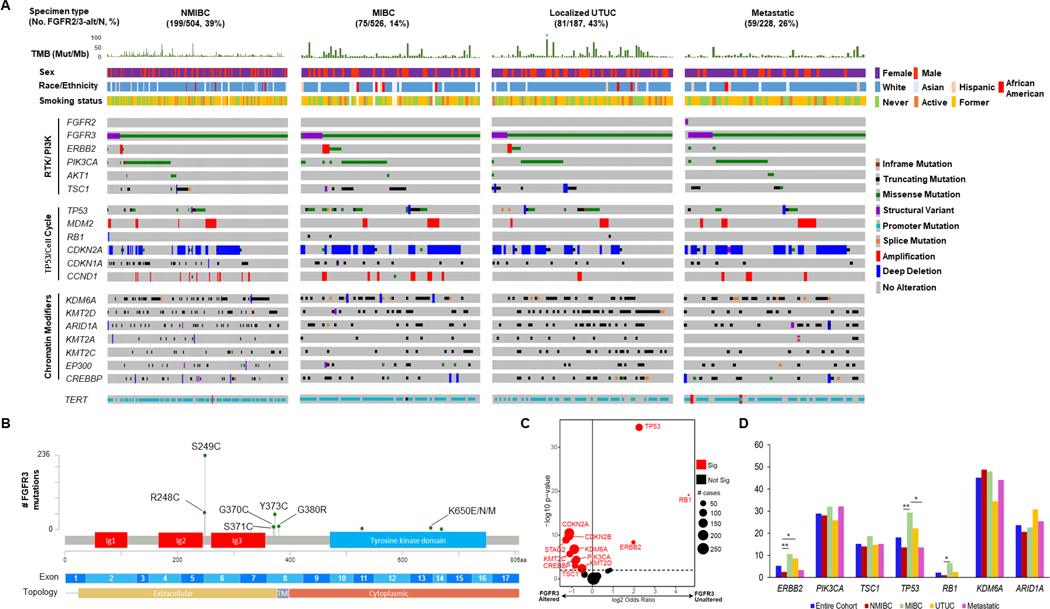
From a cohort of 1,421 patients with sequenced urothelial carcinoma tumors, FGFR2/3 alterations predictive of response to erdafitinib were identified in 414 of 1,507 (27.5%) urothelial carcinoma tumors (Figure 1A; Supplemental Figure 1). The 414 altered tumors were collected from 391 individual patients, with oncogenic FGFR3 alterations detected in at least one tumor in 27.5% of patients (391/1,421). Only one tumor had a potentially actionable FGFR2 alteration (an FGFR2:MARVELD3 gene fusion of unclear biologic and clinical significance). Oncogenic FGFR3 alterations were identified in 39% (199/504) of patients with non-muscle invasive bladder cancer (NMIBC) tumors, 14% (75/526) with muscle-invasive bladder cancer (MIBC) tumors, 43% (81/187) with clinically localized upper tract tumors, and 26% (59/228) with distant metastases (Supplemental Figure 2). The low prevalence of FGFR3 alterations in NMIBC tumors compared to prior series likely reflects the significant proportion of high-grade tumors in our dataset (88%) (24, 25). FGFR3 S249C was the most frequent FGFR3 alteration across clinical states (Figure 1B), while the R248C mutation was more common in upper tract (22%, 18/81) versus bladder tumors (11%, 37/333, p = .01, Χ2 test; Supplemental Figure 3).
Figure 1. The genomic landscape of FGFR3 mutant urothelial cancer.
(A) Oncoprint showing co-alterations of selected cancer-associated genes among patients with urothelial carcinoma whose tumor harbored FGFR2/3 alterations eligible for treatment with erdafitinib in the metastatic setting, stratified by disease state. (B) Lollipop plot depicting the spectrum of oncogenic FGFR3 mutations observed in 1,507 urothelial carcinoma tumors. (C) Volcano plot of selected genes co-altered with FGFR3, comparing alterations of tumors with versus without FGFR3 alteration. (D) Frequency of select co-altered genes among FGFR3-altered tumors as a function of primary site (bladder versus UTUC) and disease state.
Alt, altered; Mb, megabase; MIBC, muscle-invasive bladder cancer; Mut, mutations; NMIBC, non-muscle invasive bladder cancer; No., number; RTK, receptor tyrosine kinase; Sig, significant; TM, transmembrane; TMB, tumor mutational burden; UTUC, upper tract urothelial carcinoma.
†Hypermutated tumor with TMB of 409 mutations/megabase.
* p < .05
** p < .01
The clinical demographics of patients with oncogenic FGFR3 alterations are listed in Table 1. Representativeness of the study cohort is provided in Supplemental Table 1. Compared to patients without oncogenic FGFR3 alterations, a higher proportion of patients with oncogenic FGFR3 alterations were female (p = .0002). Patients with FGFR3 fusions were younger (p = .03) and less likely to have a history of tobacco use (p = .005) than those with mutations, consistent with a prior study (26). In contrast to that study, we observed no statistically significant association between FGFR3 fusions and Asian race, although the total number of Asian patients in our study was low limiting statistical power (n = 53 patients with Asian race, of whom 2 and 9 had oncogenic FGFR3 fusions and mutations, respectively). Moreover, tumors with oncogenic FGFR3 fusions had a lower tumor mutational burden (TMB) (median 5 versus 9 mutations/megabase, p = .0006) than tumors with FGFR3 mutations; however, there was no statistically significant difference in TMB between tumors with versus without an oncogenic FGFR3 alteration in general (p = .44).
Table 1.
Demographic characteristics of patients with urothelial carcinoma with oncogenic FGFR3 alterations.
| Characteristic, no. (%) or median (range) | Patients with oncogenic FGFR3 alterations (n = 390) | Patients with oncogenic FGFR3 mutations (n = 354) | Patients with oncogenic FGFR3 fusions (n = 36) | Patients without oncogenic FGFR3 alterations (n = 1031) | With versus without FGFR3 oncogenic alterations | FGFR3 oncogenic mutation versus no oncogenic FGFR3 alteration | FGFR3 oncogenic fusions versus FGFR3 oncogenic mutations |
|---|---|---|---|---|---|---|---|
| Age | 67.8 (18.3–90.3) | 68.4 (29.6–89.5) | 61.8 (18.3–90.3) | 68.4 (32.2–99.6) | p = .030a | ||
| Male gender | 264 (67.5) | 239 (67.3) | 25 (69.4) | 798 (77.4) | p = .0002b | p = .0003b | |
| Race/Ethnicity | |||||||
| Caucasian, Non-Hispanic | 333 (90.0) | 304 (90.5) | 29 (85.3) | 820 (87.3) | |||
| Hispanic | 13 (3.5) | 10 (3.0) | 3 (8.8) | 36 (3.8) | |||
| African American | 12 (3.2) | 12 (3.6) | 0 (0) | 34 (3.6) | |||
| Asian | 11 (3.0) | 9 (2.7) | 2 (5.9) | 42 (4.5) | |||
| Native American | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | |||
| Other | 1 (0.3) | 1 (0.3) | 0 (0.0) | 6 (0.6) | |||
| Unknown | 20 | 18 | 2 | 92 | |||
| Smoking history | 256 (67.0) | 240 (69.4) | 16 (44.4) | 627 (63.4) | p = .005b |
Wilcoxon rank-sum test
Fisher’s exact test
We next examined the pattern of co-alterations in FGFR2/3-altered tumors (Figure 1A). Oncogenic alterations of the PI3K signaling pathway were common, most frequently involving PIK3CA (28%, n = 115/414), TSC1 (13%, n = 52/414), and AKT1 (2.7%, n = 11/414) (37% total, n = 154/414). Co-alterations of ERBB2 and FGFR3 were found in 5% of FGFR3-altered tumors (19/414), ranging from 2.5% (5/199) in NMIBC to 11% (8/75) of MIBC tumors. The spectrum of PIK3CA and HER2 alterations observed is shown in Supplemental Figure 4. Oncogenic alterations in the cell cycle regulatory genes CDKN2A and CDKN1A occurred in 42% (174/414) and 15% (62/414), respectively, whereas RB1 mutations and deletions were detected in 1.2% (5/414) of tumors. When comparing the genomic profile of FGFR3-altered versus wild-type tumors, an inverse association between oncogenic FGFR3 alterations and alterations of ERBB2, TP53, and RB1 was seen, while CDKN2A, CDKN2B, and KDM6A were frequently co-altered with FGFR3 (Figure 1C; Supplemental Figure 5). Co-alteration rates of select genes across disease states among FGFR3-altered tumors is shown in Figure 1D. The frequency of ERBB2 co-alteration was significantly lower among FGFR3-altered NMIBC tumors than FGFR3- altered MIBC and FGFR3-altered localized upper tract tumors, while the frequency of TP53 co-alteration was higher among FGFR3-altered MIBC tumors than FGFR3-altered NMIBC and FGFR3-altered metastatic tumors. RB1 co-alterations were more frequent in FGFR3-altered MIBC than FGFR3-altered NMIBC (Figure 1D).
Analysis of both a primary and metastatic tumor site was performed in 27 patients with FGFR3-altered tumors, with discordance noted in seven (26%). Among the discordant cases, FGFR3 alterations were found only in metastatic biopsies in four cases and were restricted to the archival primary tumor in two cases (Supplemental Figure 6). In one discordant case, the patient’s primary and metastatic tumors harbored different FGFR3 alterations.
Clinical outcomes of FGFR3-altered urothelial carcinoma treated with immune checkpoint blockade
Prior studies of association between FGFR3 alterations and immunotherapy efficacy in metastatic urothelial carcinoma have reported conflicting results (5, 6, 8). Of 1,421 patients with sequenced urothelial carcinoma tumors, 181 patients with metastatic urothelial carcinoma received immune checkpoint blockade and there was no statistically significant difference in PFS or OS comparing patients with an oncogenic FGFR3 alteration (n = 26) vs without (n = 155; p for PFS = 0.47; p for OS = 0.52, Supplemental Figure 7).
Erdafitinib in FGFR2/3 mutant metastatic urothelial carcinoma
Between 8/2019 and 7/2022, 32 patients with metastatic urothelial carcinoma were treated with erdafitinib. Baseline patient demographics are summarized in Table 2. Pre-treatment, FGFR3 S249C was the most frequent FGFR3 alteration detected (19/32, 59%), followed by FGFR3-TACC3 fusions (4/32, 13%), Y373C (3/32, 9%), and R248C (3/32, 9%). Twenty-seven of 32 patients had both tumor and cfDNA sequencing performed pre-treatment, with the oncogenic FGFR3 alteration detected in 70% (19/27) of cfDNA samples.
Table 2.
Baseline characteristics for erdafitinib-treated patients
| Characteristic | All erdafitinib-treated patients (N = 32) | Erdafitinib-treated patients with cfDNA (n = 27) |
|---|---|---|
| Median age, years (range) | 72 (57–86) | 72 (57–86) |
| Female, n (%) | 9 (28) | 7 (26) |
| Race, n (%) | ||
| African American | 2 (6) | 2 (7) |
| Caucasian | 29 (91) | 24 (89) |
| Other/Unknown | 1 (3) | 1 (4) |
| ECOG PS, n (%) | ||
| 0 | 6 (19) | 5 (19) |
| 1 | 20 (63) | 17 (63) |
| 2 | 5 (16) | 5 (19) |
| Unknown | 1 (3) | 0 (0) |
| Tumor histology, n (%) | ||
| Urothelial cancer, NOS | 25 (78) | 24 (89) |
| Urothelial cancer with squamous differentiation | 5 (16) | 1 (4) |
| Urothelial cancer with micropapillary features | 1 (3) | 1 (4) |
| Poorly differentiated urothelial carcinoma | 1 (3) | 1 (4) |
| Primary site of disease, n (%) | ||
| Bladder/urethral | 20 (63) | 16 (59) |
| Upper tract | 6 (19) | 5 (19) |
| Bladder/urethral & upper tract | 6 (19) | 6 (22) |
| Visceral metastases, n (%) | 29 (91) | 26 (96) |
| Liver metastases, n (%) | 15 (47) | 13 (48) |
| Prior anti-PD-1/L1, n (%) | 24 (89) | 21 (78) |
| Prior ADC, n (%) | 12 (44) | 11 (41) |
| Prior lines of therapy, n (%) | ||
| 0 | 0 (0) | 0 (0) |
| 1 | 8 (25) | 6 (22) |
| 2 | 13 (41) | 11 (41) |
| 3 | 4 (13) | 4 (15) |
| 3+ | 7 (22) | 6 (22) |
Abbreviations: ADC, antibody-drug conjugate; cfDNA, cell-free DNA; ECOG PS, Eastern Cooperative Oncology Group Performance Status; NOS, not otherwise specified
A recent analysis suggested low uptake of erdafitinib for the treatment of metastatic urothelial carcinoma in real-world practice (27). At our institution, of 38 patients with FGFR2/3-altered metastatic urothelial carcinoma who died after the accelerated FDA approval of erdafitinib, 24 (63%) received erdafitinib or an investigational FGFR inhibitor (Supplemental Figure 8). Eight of the 38 (21%) elected best supportive care in situations where erdafitinib treatment may have been feasible, and 14 of the 38 (16%) experienced rapid clinical decline, precluding erdafitinib use. Of the 8 patients who elected best supportive care, 7 had previously received chemotherapy, 7 an anti-PD-1/L1 agent, and 2 had received antibody-drug conjugates (ADCs). Five of 6 patients who did not receive ADC therapy died prior to accelerated approval of ADCs for metastatic urothelial carcinoma.
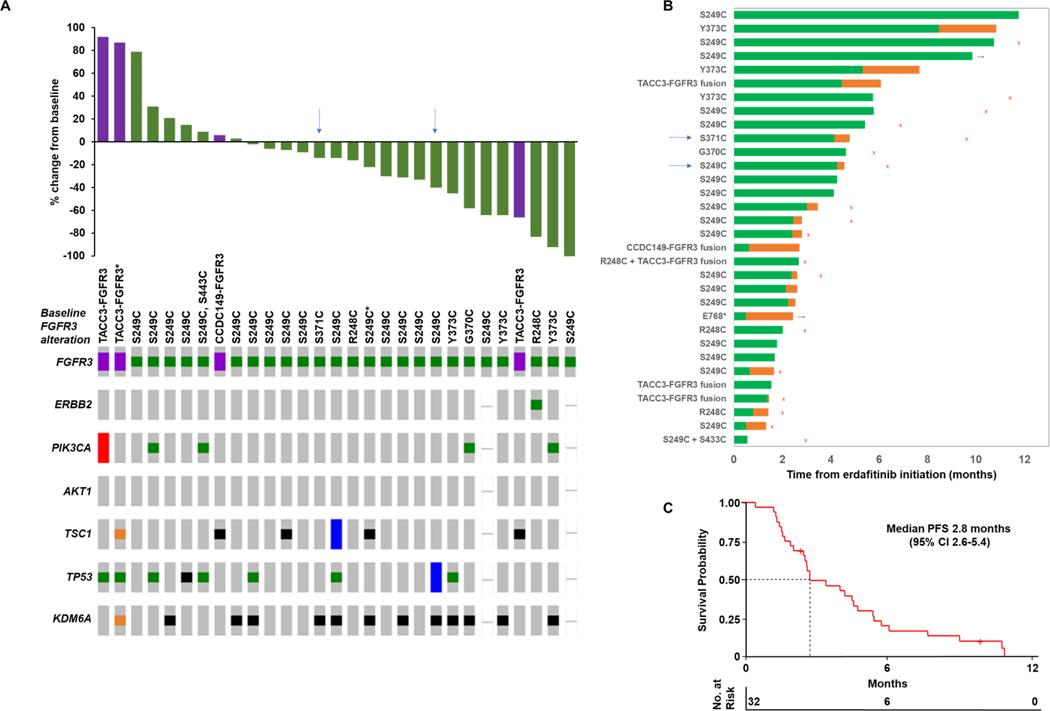
Among evaluable patients, the ORR to erdafitinib was 40% (12/30) and alterations in downstream signaling elements did not associate with response (Figure 2A). Although PIK3CA, TSC1, and ERBB2 oncogenic alterations could theoretically predict for resistance to receptor tyrosine kinase inhibition due to activation of signaling downstream of FGFR3, we did not observe an association between baseline alterations of these genes and response to erdafitinib. Among 17 evaluable patients who previously received immune checkpoint blockade, the ORR to erdafitinib was 35% (6/17). Notably in BLC2001, the ORR among 22 patients previously treated with immune checkpoint blockade was 59% (4). Patients with progression as best response had a five-fold lower FGFR3 variant allele frequency (VAF) in pre-treatment cfDNA compared to patients with partial response (median VAF 0.007 vs 0.035, p = 0.4; Supplemental Figure 9). All three patients with an FGFR3 Y373C mutation on pre-treatment tumor tissue experienced an objective response. As of last follow-up, only one patient remained on erdafitinib at 9.8 months, and one was on a treatment break but had not progressed at 2.4 months (Figure 2B). Median PFS and OS on erdafitinib were 2.8 months (95% CI 2.6–5.4) and 6.6 months (95% CI 4.6 to 17.3), respectively (Figure 2C, Supplemental Figure 10).
Figure 2. Real-world cohort of metastatic urothelial carcinoma patients treatment with erdafitinib.
(A) Waterfall plot depicting best overall response to erdafitinib therapy as well as pre-treatment FGFR3 alteration status and oncoprint depicting selected genomic co-alterations. (B) Swimmer’s plot displaying time on treatment with erdafitinib (Green bars). Orange bars indicate breaks between the last dose of erdafitinib and progression of disease or last follow-up—such breaks in erdafitinib were generally related to treatment-related toxicities. Black arrows denote patients who remain without progression of disease at last follow-up. Red “x”s denote patient deaths. Blue arrows denote patients who developed second site mutations of FGFR3 detected in cell-free DNA after initiating erdafitinib. (C) Kaplan-Meier analysis of progression-free survival of patients treated with erdafitinib therapy.
CI, confidence interval; PFS, progression-free survival
*FGFR3 alteration detected in pre-treatment cell-free DNA; all other baseline FGFR3 alterations displayed were detected by tumor sequencing.
The toxicity profile of erdafitinib was consistent with prior studies (Supplemental Table 2). Five of 32 patients (16%) underwent dose up-titration to 9mg daily based on phosphorus levels on day 14 as per dosing guidelines. Twelve of 32 patients (38%) required dose reductions and 16 of 32 patients (50%) required dose interruptions. Twenty-one of 32 patients (66%) had treatment discontinued for progression of disease, 7 for toxicity, and 2 for deaths unrelated to treatment-related toxicity or disease progression. The most common treatment-emergent adverse events (TEAE) of any grade were hyperphosphatemia (84%, 27/32), fatigue (59%, 19/32), mucositis (47%, 15/32), palmar-plantar erythrodysesthesia (38%, 12/32), and dysgeusia (31%, 10/32). The most common grade ≥3 TEAEs were mucositis (16%, 5/32) and palmar-plantar erythrodysesthesia (9%, 3/32).
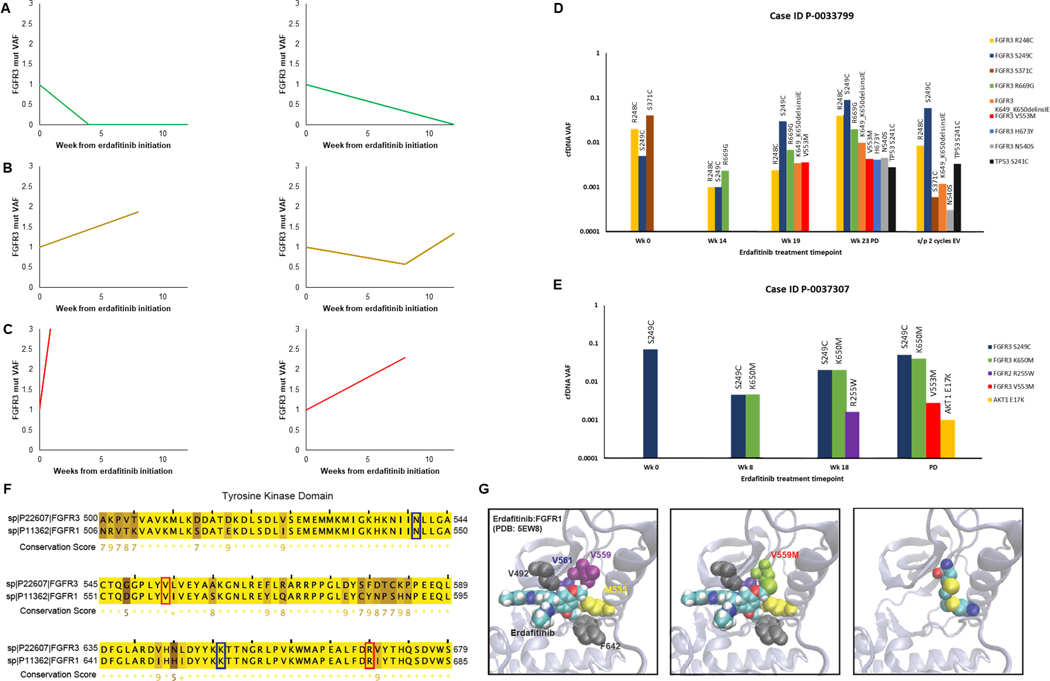
Serial cfDNA analyses were performed in 27 patients treated with erdafitinib to identify putative mechanisms of erdafitinib resistance. Changes in cfDNA VAF of FGFR3 hotspot mutations generally correlated with clinical benefit from erdafitinib, with a decrease in VAF corresponding to radiographic treatment response (Figures 3A-3C). While changes in cfDNA VAF and radiographic response were discordant in rare cases, such cases were characterized by extremely small VAF changes (on the order of 0.02%), that were not thought to reflect a meaningful change from baseline. During treatment, several patients acquired new somatic alterations in cfDNA prior to or at the time of radiographic disease progression, including oncogenic mutations of TP53 (n = 5/27), FGFR3 (n = 2/27), and AKT1 (n = 1/27) (Table 3; Figures 3D-E, Supplemental Figure 11). Of the ten patients who achieved partial response to erdafitinib and subsequently experienced disease progression, four displayed acquisition of new alterations in cfDNA related to FGFR3 signaling or TP53 loss prior to or at the time of disease progression. Patients demonstrating acquisition of these somatic alterations had a numerically longer median time on erdafitinib compared to patients without (p=0.16) (Supplemental Figure 12). Of 8 newly acquired FGFR3 alterations on treatment that were observed among a total of 2 individual patients, 2 of the FGFR3 alterations were previously reported hotspot mutations by OncoKB (19). Mutations of the kinase domain also occurred at residues that are evolutionarily conserved between FGFR3 and FGFR1, including FGFR3 N540S, V553M, K650M, and R669G, suggesting functional significance (Figure 3F). The N540S mutation interferes with erdafitinib binding in vitro (28), while in silico analysis predicted that V553M functions as a gatekeeper mutation that allosterically inhibits erdafitinib binding (Figure 3G). The latter mutation was acquired on treatment in two patients who initially experienced a partial response to erdafitinib followed by disease progression after FGFR3 V553M detection.
Figure 3. Cell-free DNA analysis of serial samples collected from metastatic urothelial cancer patients treated with erdafitinib.
(A-C) Changes in cfDNA FGFR3 VAF overtime on erdafitinib in patients who achieved a response (A), stable disease (B) or had primary disease progression (C). VAF was normalized to the pre-treatment cfDNA sample. (D-E) Bar graphs of cfDNA VAF for selected patients who acquired new alterations in FGFR3 signaling pathways or TP53. Plots include changes in the baseline cfDNA FGFR3 VAF as well as VAF of newly acquired alterations. (F) Paralogy analysis showing residues of the tyrosine kinase domain that are highly conserved between FGFR1 and FGFR3. Residues involved in FGFR3 alterations acquired on-treatment in the erdafitinib treated cohort are highlighted, with amino acid residues implicated in known drug resistant mutation highlighted in blue, and others highlighted in red. (G) In silico model of erdafitinib in binding pocket of FGFR1 based on previously reported crystal structure of FGFR1 (PDB:5EW8). Relevant amino acid residues that interact with erdafitinib are highlighted. FGFR1 V559M (paralogous to FGFR3 V553M, a novel cfDNA alteration acquired on erdafitinib in our cohort) could result in sulfur-sulfur bond formation that would sterically inhibit erdafitinib binding. cfDNA, cell-free DNA; EV, enfortumab vedotin; mut, mutation; PD, progression of disease; VAF, variant allele frequency; Wk, week
Table 3.
Cases of advanced/metastatic urothelial carcinoma treated with erdafitinib with subsequent acquisition of new notable cfDNA alterations prior to or at the time of disease progression.
| Case ID | Pre-treatment FGFR2/3 alterations | Alterations acquired on erdafitinib related to TP53 and FGFR signaling | Best response to erdafitinib by RECIST v1.1 |
|---|---|---|---|
| P-0004257 | FGFR3 Y373C | TP53 K132M; TP53 R158L | PR |
| P-0033799 | FGFR3 S249C; FGFR3 S371C; FGFR3 R399C; FGFR3 R248C; TACC3-FGFR3 fusion | FGFR3 R669Ga,b; FGFR3 V553Ma; FGFR3 N540Sa,b; FGFR3 H673Y; FGFR3 K649_K650delinsIEb,c; TP53 S241C; BRAF-CLIP2 fusion | SD |
| P-0044554 | FGFR3 S249C | TP53 E287Q | PD |
| P-0037307 | FGFR3 S249C | FGFR3 V553Ma; FGFR3 K650Mb,c; FGFR2 R255W; AKT1 E17K | PR |
| P-0044518 | FGFR3 S249C | TP53 I195T | PR |
| P-0042869 | FGFR3 Y373C | TP53 R248W; TP53 S241Y | PR |
Abbreviations: cfDNA, cell-free DNA; PD, progression of disease; PR, partial response; SD, stable disease
Putative FGFR3 gatekeeper mutation based on in silico analysis and/or preclinical data.
Likely pathogenic FGFR3 mutation based on OncoKB.
Hotspot FGFR3 mutation per OncoKB.
Based on the association between TP53 co-mutations and EGFR inhibitor resistance in EGFR mutant lung cancer (29, 30), we examined response as a function of TP53 co-alteration within our cohort. The objective response rate among patients with oncogenic TP53 alterations at baseline was 22% (2/9), lower than the overall 40% ORR (12/30). Oncogenic TP53 alterations were also found in 3/5 (60%) patients with primary disease progression on erdafitinib (Figure 2A). Within the urothelial TCGA, oncogenic alterations of FGFR3 and TP53 rarely co-occur, with a Log2 odds ratio of −1.8 (p < 0.001; Supplemental Figure 13) (1, 23, 31). Interrogation of our cohort of 1,507 urothelial carcinoma tumors also confirmed this inverse association between FGFR3 and TP53 alteration (Supplemental Figure 13).
DISCUSSION
With the goal of optimizing use of FGFR inhibitors in metastatic urothelial carcinoma, we performed an integrated analysis of clinical and genomic data from an institutional, prospective sequencing initiative. We confirmed that the frequency of FGFR3 alterations varied significantly across clinical states of urothelial carcinoma with enrichment in NMIBC and upper tract tumors compared to metastatic and MIBC tumors, consistent with prior reports (1, 2, 32). Notably, in a subset of patients who underwent analysis of both a primary and metastatic disease site, FGFR3 mutation was exclusive to either the primary tumor or the metastatic tumor biopsy. These results indicate that analysis of archival primary tumor tissue may not always accurately characterize the FGFR3 alteration status of a patient’s metastatic disease and potentially lead to erroneous patient selection for FGFR-inhibitor therapy. Finally, while the FDA label for erdafitinib includes oncogenic FGFR2 fusions, only a single FGFR2 fusion of unknown biologic and clinical significance was detected in our cohort.
While targeted therapies have been approved in over 15 cancer types, only a minority of patients experience durable clinical responses due to intrinsic and acquired drug resistance. One mechanism of resistance is alteration in the drug target, such as EGFR T790M mutations following exposure to first-generation EGFR inhibitors in non-small cell lung cancer, and alternative BRAF splicing, a common mechanism of resistance to BRAF inhibition in melanoma (33, 34). In our cohort of erdafitinib-treated patients, longitudinal analysis of cfDNA identified on-treatment acquisition of potential gatekeeper mutations in FGFR3, including V553M and N540S, activation of downstream signaling (AKT1 activating mutation), and oncogenic alterations of TP53 as potential mechanisms of acquired drug resistance. Limited preclinical data suggest that acquired FGFR3 mutations can interfere with drug binding, leading to erdafitinib resistance (28). On-treatment acquisition of second-site mutations that might alter the binding properties of erdafitinib occurred rarely in our cohort and corresponded to longer duration on treatment which presumably allowed for development of drug-resistant kinase mutations over time. The relatively low rate of acquired resistance mutations may in part stem from the inability for most patients to remain on long-term continuous drug dosing as a result of erdafitinib-related toxicities. However, our data suggesting that FGFR3 V553M and N540S can act as mediators of acquired resistance to erdafitinib hold implications for the ongoing development of novel FGFR inhibitors that may have the ability to overcome therapeutic resistance mediated by these putative gatekeeper mutations.
The on-treatment acquisition of TP53 alterations observed in our erdafitinib-treated patient cohort is intriguing given the strong inverse association between FGFR3 and TP53 alterations in both the urothelial TCGA and MSK-IMPACT cohorts (1). TP53 alterations are also associated with poor outcomes with several targeted therapies, including EGFR inhibitors in non-small cell lung cancer (35) and neratinib in HER2-positive breast cancer (36). In our cohort, baseline co-alterations of TP53 were also numerically higher in patients with primary progression on erdafitinib. While the mechanism underlying the potential association between TP53 alterations and targeted therapy resistance cannot be determined by the current study, prior studies have established loss of TP53 function as a progenitor of cellular plasticity and epithelial-mesenchymal transition (EMT) (37), which itself is associated with FGFR-inhibitor resistance in vitro (38). If validated by future studies, an association between TP53 alteration and erdafitinib resistance could suggest implications for clinical practice. For example, TP53 co-alteration may provide rationale for selection of alternative therapies to erdafitinib, when available. Moreover, the relatively low rate of TP53 co-alterations in NMIBC compared to MIBC may suggest a higher likelihood of response to erdafitinib in non-muscle invasive disease. Indeed, a phase 2 study for FGFR2/3-altered recurrent intermediate-risk NMIBC recently demonstrated a complete response rate to erdafitinib in 6 of 8 evaluable patients (39).
In our cohort, intrinsic resistance to erdafitinib was common and may reflect the presence of co-alterations within bypass pathways or pre-existing heterogeneity of FGFR3 alterations, both within a tumor and across lesions. The significant discordance of FGFR3 alteration status between primary and metastatic tumors and between tumor tissue and cfDNA observed in our cohort underscores the high degree of pre-existing genomic heterogeneity in bladder cancer, which is greater than that seen with BRAF or EGFR driven melanoma and lung cancer, respectively. These findings also highlight the utility of liquid biopsies to capture subclonal tumor cell populations resistant to FGFR-directed therapy and to detect emerging resistance on treatment.
While some studies suggest a correlation between FGFR3 alterations, the presence of a “cold” immune microenvironment (40), and resistance to immune checkpoint blockade in FGFR3-altered tumors (41, 42), other analyses (6), including ours, have shown that benefit from immune checkpoint blockade does not significantly differ as a function of FGFR3 alteration status. Therefore, patients with FGFR3-altered tumors should be offered checkpoint blockade as standard of care (4, 6).
Erdafitinib is currently the only FDA-approved targeted therapy for patients with metastatic urothelial carcinoma, yet a recent analysis of 761 patients with metastatic disease progressing on platinum chemotherapy found that only 45% underwent genetic testing for FGFR2/3 alterations and of these, only 42% with an FGFR2/3 alteration received erdafitinib (27). Our single-institution experience similarly indicates that erdafitinib is frequently reserved as a 3rd line treatment or later and was not offered to every patient with a potentially eligible genetic alteration, in striking contrast to the near-universal use of EGFR inhibition in EGFR mutant NSCLC (43). While insufficient genomic testing may play a role, the significant toxicities of erdafitinib therapy likely contribute to this low uptake. Moreover, the median PFS of only 2.8 months in our real-world patient population indicates a significant unmet need for FGFR inhibitors with enhanced anti-tumor activity and a more tolerable toxicity profile. The relatively poor clinical outcomes experienced by our erdafitinib-treated cohort in comparison to published trial data from BLC2001 (4) likely reflect the relatively adverse prognostic factors present in real-world patient cohorts such as our own. Such adverse factors included more frequent visceral metastases, more prior lines of therapy, poor performance status, and older age, which can unfortunately limit the generalizability of clinical trial data to real-world practice. Notably, of four patients with FGFR3 fusions in our erdafitinib-treated cohort, three experienced progression of disease as best response to erdafitinib. This is consistent with trial data reporting a lower objective response rate among patients with FGFR3 fusion compared to FGFR3 mutations (4).
Our study had several limitations. First, the retrospective nature of our genomic analysis is subject to sampling bias and occult confounders. However, our cohort of 1,507 tumors represents the largest single institution urothelial carcinoma cohort sequenced to date, and our current practice of offering next-generation sequencing to all patients with metastatic urothelial carcinoma treated at our institution reduces the likelihood of significant patient selection bias. Second, not all patients on erdafitinib had collection of cfDNA at the same timepoints, due primarily to fewer in-person visits during the COVID19 pandemic. Finally, while the MSK-IMPACT gene panel includes all genes commonly mutated in urothelial carcinoma, additional mechanisms of resistance involving genes not included in the panel or stemming from transcriptomic changes would not be detected. For example, changes in expression of FGFR3 or other mitogenic tyrosine kinases would not be captured by the DNA sequencing panels employed in the current study.
In conclusion, our real-world experience with erdafitinib in patients with FGFR3-altered tumors indicates that this treatment approach has modest clinical activity and the potential for significant toxicity which limit its use in routine clinical practice. This result underscores the need for FGFR3 isoform-specific inhibitors to ameliorate the on-target, off-tumor toxicities associated with disruption of pan-FGFR signaling, a significant contributor to the toxicity profile observed with erdafitinib and similar agents. Two isoform-specific agents, TYRA300 and LOXO435, are currently accruing patients with FGFR3 alterations in early-phase trials, and tolerability and efficacy data from these studies are highly anticipated. Finally, the high degree of discordance in FGFR3 alterations between primary and metastatic tumors suggests that sequencing of metastatic tumor specimens or cfDNA analysis following diagnosis of metastatic disease may improve identification of patients likely to benefit from FGFR inhibition or detect second site mutations in FGFR3 that are early harbingers of evolving resistance to therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Erdafitinib is the only FDA-approved targeted therapy for FGFR2/3-altered metastatic urothelial carcinoma. From an institutional repository of 1,507 sequenced urothelial carcinoma tumors, we examined the largest clinically annotated cohort of 414 FGFR2/3-altered tumors to date. Discordance of FGFR3 mutational status between matched primary and metachronous metastatic tumors occurred frequently, raising concern over reliance upon sequencing of archival tumor tissue to guide patient selection for erdafitinib therapy. We also presented the first real-world outcomes data for erdafitinib therapy, and found that progression-free survival was shorter than expected compared to trial data; dosing was frequently limited by toxicity, highlighting the need for more tolerable FGFR inhibitors. Prospective sequencing of cfDNA from patients treated with erdafitinib revealed on-treatment acquisition of FGFR3, AKT1, and TP53 mutations, suggesting putative mechanisms of acquired resistance. A novel putative FGFR3-gatekeeper mutation was identified, potentially informing development of future FGFR inhibitors.
Acknowledgements
The authors thank members of the Memorial Sloan Kettering Kravis Center for Molecular Oncology, and Diagnostic Molecular Pathology for their work establishing and maintaining the MSK-IMPACT datasets. This work and all authors were supported by the Marie-Josee and Henry R. Kravis Center for Molecular Oncology, Cycle for Survival, NIH/NCI P01-CA221757and NIH/NCI SPORE in Bladder Cancer P50-CA221745, and the NIH/NCI Cancer Center Support Grant P30-CA008748. B. Guercio was supported by NIH/NCI T32-CA009207.
Abbreviations:
- ADC
antibody-drug conjugate
- Alt
altered
- cfDNA
cell-free DNA
- CI
confidence interval
- ECOG PS
Eastern Cooperative Oncology Group Performance Status
- EMT
epithelial-mesenchymal transition
- EV
enfortumab vedotin
- mut
mutation
- FGFR3
fibroblast growth factor receptor 3
- Mb
megabase
- MIBC
muscle-invasive bladder cancer
- MSK
Memorial Sloan Kettering Cancer Center
- MSK-IMPACT
Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets
- Mut
mutations
- NMIBC
non-muscle invasive bladder cancer
- No
number
- NOS
not otherwise specified
- ORR
objective response rate
- OS
overall survival
- PD
progression of disease
- PFS
progression-free survival
- PR
partial response
- RTK
receptor tyrosine kinase
- SD
stable disease
- Sig
significant
- TEAE
treatment-emergent adverse events
- TM
transmembrane
- TMB
tumor mutational burden
- UTUC
upper tract urothelial carcinoma
- VAF
variant allele frequency
- Wk
week
Footnotes
Conflicts of Interest: B. Guercio reports honoraria from Medscape; institutional research funding from Oncocyte and Genentech; as well as expenses from Seagen/Astellas, EMD Serono, Natera, Gilead, Aveo Oncology, and Exelixis. M. Sarfaty reports honoraria from Pfizer, Bristol Myers Squibb/Pfizer, Janssen, Astellas Pharma, Astellas Pharma, MSD Oncology, MSD Oncology; consulting or advisory role with Astellas Pharma and MSD Oncology; as well as institutional research funding from AstraZeneca. M.Y. Teo reports consulting/advisory role for Janssen Oncology and institutional research funding from Bristol-Myers Squibb, Clovis Oncology, and Pharmacyclics. S.A. Funt reports employment with ByHeart; stock and other ownership interests in Kite, a Gilead company, UroGen pharma, Allogene Therapeutics, Neogene Therapeutics, Kronos Bio, Vida Ventures, IconOVir Bio, and Doximity, 76Bio; consulting or advisory roles with Merck, Immunai, and BioNTech; institutional research funding from Genentech/Roche, AstraZeneca, Decibel Therapeutics; as well as travel, accommodations, or expenses from Bristol Myers Squibb, and AstraZeneca/MedImmune. C.H. Lee reports honoraria from Intellisphere, Research to Practice, and American Institute of Continuing Medical Education; consulting or advisory role for Bristol Myers Squibb, Eisai, EMD Serono, Exelixis, Merck, and Pfizer; as well as institutional research funding from Bristol Myers Squibb, Calithera Biosciences, Eisai, Eli Lilly, Exelixis, Merck, and Pfizer. D.H. Aggen reports research funds from Seattle Genetics and Astellas. M. Lattanzi reports consulting with Janssen, Aveo, Merck, Aadi; and speaking fees from Seagen, Janssen, Astrazeneca, EMD Serono. H.A. Al-Ahmadie has provided consultation to AstraZeneca, Janssen Biotech, Bristol-Myers-Squibb, and Paige.ai. E. Pietzak reports honoraria from UpToDate; consulting or advisory roles with Merck, Chugai Pharma, QED Therapeutics, Janssen, and Urogen pharma; as well as research funding from Janssen. B.H. Bochner reports consulting or advisory role for Olympus. M.F. Berger reports a patent pending on “Systems and Methods for Detecting Cancer Via cfDNA Screening;” research funding from Grail; consulting fees from Eli Lilly and PetDx. L.D.E.: and is an employee of and shareholder in Puma Biotechnology, Inc. D.B. Solit has served as a consultant for/received honorarium from Pfizer, Loxo/Lilly Oncology, Vividion Therapeutics, Scorpion Therapeutics, Fore Therapeutics, FOG Pharma, Rain Therapeutics, and BridgeBio. J.E. Rosenberg has served as a consultant for Astellas, Seagen, Merck, Roche, Genentech, AstraZeneca, Janssen Biotech, Gilead, Pfizer, EMD-Serono, Mirati, Boehringer Ingelheim, Pharmacyclis, GSK, Infinity, Tyra BioSciences, Bayer, and QED Therapeutics and received honoraria from EMD-Serono. D.F. Bajorin reports honoraria from Bristol Myers Squibb/Medarex; consulting or advisory roles with Merck, Bristol Myers Squibb Foundation; as well as institutional research funding from Novartis, Merck, Bristol Myers Squibb, AstraZeneca, Astellas Pharma, and Seattle Genetics/Astellas. G. Iyer reports consulting or advisory roles with Bayer, Janssen, Mirati Therapeutics, Basilea, Flare Therapeutics, and Loxo/Lilly; speakers’ bureau activities with Gilead Sciences and Lynx Group; as well as institutional research funding from Mirati Therapeutics, Novartis, Debiopharm Group, Bayer, Janssen, and Seattle Genetics. N. Ratna, C. Duzgol, A.M. Regazzi, Z. Chen, A.R. Brannon, R. Shah, C. Chu, A.T. Lenis declare no potential conflicts of interest.
References
- 1.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171(3):540–56 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer Reveals Potential Biomarkers and Rational Therapeutic Targets. Eur Urol. 2017;72(6):952–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med. 2019;381(4):338–48. [DOI] [PubMed] [Google Scholar]
- 5.Rose TL, Weir WH, Mayhew GM, Shibata Y, Eulitt P, Uronis JM, et al. Fibroblast growth factor receptor 3 alterations and response to immune checkpoint inhibition in metastatic urothelial cancer: a real world experience. Br J Cancer. 2021;125(9):1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Gong Y, Saci A, Szabo PM, Martini A, Necchi A, et al. Fibroblast Growth Factor Receptor 3 Alterations and Response to PD-1/PD-L1 Blockade in Patients with Metastatic Urothelial Cancer. Eur Urol. 2019;76(5):599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teo MY, Mota JM, Whiting KA, Li HA, Funt SA, Lee CH, et al. Fibroblast Growth Factor Receptor 3 Alteration Status is Associated with Differential Sensitivity to Platinum-based Chemotherapy in Locally Advanced and Metastatic Urothelial Carcinoma. European urology. 2020;78(6):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powles T, Sridhar SS, Loriot Y, Bellmunt J, Mu XJ, Ching KA, et al. Avelumab maintenance in advanced urothelial carcinoma: biomarker analysis of the phase 3 JAVELIN Bladder 100 trial. Nat Med. 2021;27(12):2200–11. [DOI] [PubMed] [Google Scholar]
- 9.Guercio BJ, Iyer G, Rosenberg JE. Developing Precision Medicine for Bladder Cancer. Hematol Oncol Clin North Am. 2021;35(3):633–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau DK, Jenkins L, Weickhardt A. Mechanisms of acquired resistance to fibroblast growth factor receptor targeted therapy. Cancer Drug Resist. 2019;2(3):568–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal SK, Rosenberg JE, Hoffman-Censits JH, Berger R, Quinn DI, Galsky MD, et al. Efficacy of BGJ398, a Fibroblast Growth Factor Receptor 1–3 Inhibitor, in Patients with Previously Treated Advanced Urothelial Carcinoma with FGFR3 Alterations. Cancer Discov. 2018;8(7):812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nature genetics. 2016;48(12):1490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meeks JJ, Al-Ahmadie H, Faltas BM, Taylor JA 3rd, Flaig TW, DeGraff DJ, et al. Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat Rev Urol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinton TN, Chen Z, Wise H, Lenis AT, Chavan S, Donoghue MTA, et al. Genomic heterogeneity as a barrier to precision oncology in urothelial cancer. Cell Rep. 2022;41(12):111859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–47. [DOI] [PubMed] [Google Scholar]
- 17.Tsui DWY, Cheng ML, Shady M, Yang JL, Stephens D, Won H, et al. Tumor fraction-guided cell-free DNA profiling in metastatic solid tumor patients. Genome medicine. 2021;13(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose Brannon A, Jayakumaran G, Diosdado M, Patel J, Razumova A, Hu Y, et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat Commun. 2021;12(1):3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–65. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–8, 27–8. [DOI] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Amoros A, Tardon A, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24(22):3664–71. [DOI] [PubMed] [Google Scholar]
- 25.Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer Reveals Potential Biomarkers and Rational Therapeutic Targets. European urology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nassar AH, Lundgren K, Pomerantz M, Van Allen E, Harshman L, Choudhury AD, et al. Enrichment of FGFR3-TACC3 Fusions in Patients With Bladder Cancer Who Are Young, Asian, or Have Never Smoked. JCO Precis Oncol. 2018;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nimgaonkar V, Hubbard RA, Carpenter EL, Mamtani R. Biomarker Testing, Treatment Uptake, and Survival Among Patients With Urothelial Cancer Receiving Gene-Targeted Therapy. JAMA Oncol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patani H, Bunney TD, Thiyagarajan N, Norman RA, Ogg D, Breed J, et al. Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use. Oncotarget. 2016;7(17):24252–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non-Small-Cell Lung Cancer. JCO Precis Oncol. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canale M, Andrikou K, Priano I, Cravero P, Pasini L, Urbini M, et al. The Role of TP53 Mutations in EGFR-Mutated Non-Small-Cell Lung Cancer: Clinical Significance and Implications for Therapy. Cancers (Basel). 2022;14(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassler MR, Bray F, Catto JWF, Grollman AP, Hartmann A, Margulis V, et al. Molecular Characterization of Upper Tract Urothelial Carcinoma in the Era of Next-generation Sequencing: A Systematic Review of the Current Literature. Eur Urol. 2020;78(2):209–20. [DOI] [PubMed] [Google Scholar]
- 33.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480(7377):387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res. 2017;23(9):2195–202. [DOI] [PubMed] [Google Scholar]
- 36.Smyth LM, Piha-Paul SA, Won HH, Schram AM, Saura C, Loi S, et al. Efficacy and Determinants of Response to HER Kinase Inhibition in HER2-Mutant Metastatic Breast Cancer. Cancer Discov. 2020;10(2):198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi ZD, Pang K, Wu ZX, Dong Y, Hao L, Qin JX, et al. Tumor cell plasticity in targeted therapy-induced resistance: mechanisms and new strategies. Signal Transduct Target Ther. 2023;8(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grygielewicz P, Dymek B, Bujak A, Gunerka P, Stanczak A, Lamparska-Przybysz M, et al. Epithelial-mesenchymal transition confers resistance to selective FGFR inhibitors in SNU-16 gastric cancer cells. Gastric Cancer. 2016;19(1):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daneshmand S, Zaucha R, Gartrell BA, Lotan Y, Hussain SA, Lee EK, et al. Phase 2 study of the efficacy and safety of erdafitinib in patients (pts) with intermediate-risk non–muscle-invasive bladder cancer (IR-NMIBC) with FGFR3/2 alterations (alt) in THOR-2: Cohort 3 interim analysis. Journal of Clinical Oncology. 2023;41(6_suppl):504-. [Google Scholar]
- 40.Sweis RF, Spranger S, Bao R, Paner GP, Stadler WM, Steinberg G, et al. Molecular Drivers of the Non-T-cell-Inflamed Tumor Microenvironment in Urothelial Bladder Cancer. Cancer Immunol Res. 2016;4(7):563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siefker-Radtke AO, Necchi A, Rosenbaum E, Culine S, Burgess EF, O’Donnell PH, et al. Efficacy of programmed death 1 (PD-1) and programmed death 1 ligand (PD-L1) inhibitors in patients with FGFR mutations and gene fusions: Results from a data analysis of an ongoing phase 2 study of erdafitinib (JNJ-42756493) in patients (pts) with advanced urothelial cancer (UC). J Clin Oncol. 2018;36(6_suppl):450-. [Google Scholar]
- 42.Joerger M, Cassier PA, Penel N, Cathomas R, Richly H, Schostak M, et al. Rogaratinib in patients with advanced urothelial carcinomas prescreened for tumor FGFR mRNA expression and effects of mutations in the FGFR signaling pathway. J Clin Oncol. 2018;36(15_suppl):4513-. [Google Scholar]
- 43.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017;7(6):596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MSK-IMPACT and MSK-ACCESS results, including the gene sequencing panel version used to analyze each individual tumor, and associated clinical data are available via the cBioPortal for Cancer Genomics under study title “Bladder Cancer (MSK, Clin Cancer Research 2023)” at https://www.cbioportal.org/study/summary?id=bladder_msk_2023 (23).