Abstract
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that has been implicated in several disorders, including an association between HCMV reactivation and the overproliferation of arterial smooth muscle cells observed in restenosis. Although HCMV can mediate a growth-arrest phenotype in infected cells, the virus can also promote an environment conducive to proliferation. Here, we present evidence that the HCMV immediate-early (IE) proteins, IE1-72 and IE2-86, may be responsible for inducing this proliferative environment by altering cell cycle control. We find that expression of either of these IE proteins can alter the cell cycle distribution of randomly cycling cells towards S and G2/M phases. Additionally, we find that expression of IE2-86, but not IE1-72, induces quiescent cells into S phase and delays cell cycle exit. In the absence of p53, IE1-72 expression can induce S phase and delay cell cycle exit. We also demonstrate that p53 protein levels increase in fibroblasts following the expression of IE1-72. The observed accumulation of p53 protein in IE1-72-expressing cells may account for the inability of IE1-72 to induce S phase and delay cell cycle exit. Our data suggest that expression of HCMV IE1-72 and IE2-86 is sufficient to alter the cell cycle to generate an environment conducive to proliferation.
Human cytomegalovirus (HCMV) is a ubiquitous, species-specific beta-herpesvirus that, like other herpesviruses, can establish lifelong latency following primary infection (27). Reactivation of HCMV is observed in immunocompromised and immunosuppressed individuals. HCMV infection is endemic within the human population but rarely causes symptomatic disease in healthy, immunocompetent individuals (15). It has recently been demonstrated that prior HCMV infection may greatly enhance the risk of restenosis, a proliferative disorder characterized by the overproliferation of arterial smooth muscle cells (SMCs) along the vascular wall (46), following coronary angioplasty. It is not clear whether the accumulation of SMCs following coronary angioplasty is influenced by cellular or viral growth factors. The focus of attention has shifted towards manipulation of the intracellular pathways regulating proliferation, such as those regulated by p53 and the retinoblastoma (Rb) protein family, as a means to control restenosis. Mounting evidence supports a link between the reactivation of latent HCMV following coronary angioplasty, p53 inactivation, and restenosis (10, 36, 37), as HCMV was preferentially detected in a subset of restenotic lesions that exhibited high levels of p53 protein (37).
While it appears that HCMV may contribute to the development of restenosis, there is no definitive proof that the virus directly causes the overproliferation of SMCs. Rather, HCMV infection of human fibroblasts appears to trigger an opposite effect, namely, cell cycle arrest. In previous studies, HCMV infection was shown to induce arrest of cell growth either in late G1 or in G2/M (7, 8, 16, 24). These findings conflict with the conclusions from an earlier report that connected HCMV infection with the induction of cellular DNA synthesis (2). In support of the notion that HCMV modulates the host cell cycle machinery, HCMV infection also has a positive influence on factors that promote cell proliferation. Among the many factors that are upregulated during HCMV infection, the protooncogenes c-myc, c-fos, and c-jun are all rapidly activated in infected cells (4, 28). Additionally, viral infection appears to activate several cellular S-phase genes, including those for DNA polymerase α, dihydrofolate reductase (DHFR), and thymidine kinase (TK), as well as induce E2F transactivational activity and expression of cyclin E, cyclin A, and Cdk2 proteins (6, 14, 41). Importantly, HCMV-infected cells exhibit increased levels of hyperphosphorylated pRb, a key cell cycle regulator that governs the transition from G1 into S phase. The phosphorylation status of pRb, in particular, serves as the G1 restriction point by controlling the commitment to enter S phase and the subsequent continuation through the cell cycle (42). Taken together, these findings suggest that HCMV may influence the host cell cycle machinery to create an environment that is fully conducive to the replication of its viral DNA.
There is an existing precedent for viruses altering cell cycle control to their advantage. In particular, the small DNA tumor viruses, e.g., adenovirus, simian virus 40 (SV40), and human papillomavirus (HPV), can each perturb the cellular replication machinery to better facilitate the replication of their viral DNA. Their ability to overcome the normal regulation of cell proliferation control is dependent upon their oncogene products, which target p53 and members of the Rb family of proteins and inactivate their respective functions (reviewed in reference 30). In keeping with this concept, HCMV expresses two immediate-early gene products, IE1-72 and IE2-86, that may function in a similar manner. Both HCMV immediate-early (IE) proteins can bind to members of the Rb family of proteins. The IE1-72 protein interacts with the p107 protein, while the IE2-86 protein binds to pRb (11, 13, 17, 33). IE1-72 and IE2-86 expression can alleviate the repression of E2F transcriptional activity mediated by p107 and pRb, respectively (11, 17, 33). IE2-86 but not IE1-72 has been shown to interact with p53 in vitro and in vivo (5, 36, 40), and this interaction results in the downregulation of p53 transactivation function (36, 40).
A recent study suggested that IE2-86 expression leads to a cell cycle arrest at G1 (44). Though this finding is consistent with studies suggesting a G1 growth-arrest phenotype in HCMV-infected fibroblasts (7, 8, 16, 24), it contradicts the outcome one would expect given that IE2-86 targets and inhibits the functions of both pRb and p53. Moreover, IE2-86 can induce E2F activity as well as activate the cyclin E-Cdk2 and cyclin A-Cdk2 complexes (6), key rate-limiting steps in the progression from G1 into and through S phase. By inducing E2F activity, IE2-86 expression should lead to the induction of several genes involved in cellular DNA replication (DHFR and TK genes, etc.) since they are regulated by E2F functions. Analogous to IE2-86, the expression of the IE1-72 protein can induce several S-phase-associated genes, including those for DHFR and DNA polymerase α, through E2F induction or possibly by directly transactivating their promoters (14, 26, 41). The fact that the IE1-72 and IE2-86 proteins positively influence numerous factors related to S-phase progression suggests that expression of these proteins should promote progression through the cell cycle.
The objective of our study was to determine whether the HCMV IE proteins could effectively modulate the host cell cycle, presumably to promote an environment favorable for DNA replication. We focused our analysis on IE1-72 and IE2-86 and examined their ability to induce proliferation in rat embryo fibroblast and mouse embryo fibroblast (MEF) models of cell cycle control. We found that both IE1-72 and IE2-86 can perturb the normal cell cycle distribution of asynchronously cycling cells. We found that expression of IE2-86 is sufficient to induce cell cycle entry in quiescent, G0 cells. IE2-86 expression also delays the exiting of cells from the cell cycle. We also found that, in the absence of p53, expression of IE1-72 can induce growth-arrested cells to proliferate and delay cell cycle exit following serum depletion. Furthermore, we demonstrated that IE1-72 and IE2-86 expression can each induce the accumulation of p53 protein in cells. Taken together, our results support a model whereby expression of the HCMV IE proteins can promote progression through the cell cycle.
MATERIALS AND METHODS
Cell culture.
Cells from a rat embryo fibroblast cell line, REF52, were maintained in Dulbecco's modified Eagle medium (DMEM; GIBCO BRL) supplemented with 5% fetal bovine serum (HyClone, Inc.), 5% fetal calf serum (HyClone, Inc.), and 1% penicillin-streptomycin (GIBCO BRL) (19, 20). Early-passage wild-type (p53+/+) and genetically matched p53-null (p53−/−) MEFs (passages 2 to 7), a generous gift from Stephen Jones (University of Massachusetts Medical School, Worcester, Mass.), and human embryonic lung (HEL) fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (9, 21). To induce cells to undergo growth arrest, cells were washed twice with phosphate-buffered saline (PBS) and then cultured in reduced serum concentrations: REF52 cells and p53+/+ MEFs were cultured in medium containing 0.25% serum for a minimum of 36 and 48 h, respectively. The p53−/− MEFs were cultured in medium containing 0.1% serum for 60 h. The conditions selected were based on experiments done to optimize serum concentration and culturing times (unpublished observations). Rodent fibroblasts were seeded at a plating density of between 2 × 103 and 3 × 103 cells/cm2, prior to infection.
Adenovirus vectors.
Recombinant adenoviruses expressing HCMV immediate-early gene products IE1-72 (AdIE1-72) or IE2-86 (AdIE2-86) were kindly provided by Gary Hayward (John Hopkins University, Baltimore, Md.) (1). An empty vector virus, AdCon (19), was used as a control in all of the experiments. An E2F1-expressing adenovirus, AdE2F1 (19), has been described previously. Viruses were grown and titered in a human embryonic kidney cell line (293) and subsequently purified on cesium-chloride gradients as described previously (31). Virus titers were determined by immunohistochemical staining of the adenovirus type-2 hexon with an anti-adenovirus antibody (Biodesign International) and a 3,3′-diaminobenzidine (DAB) substrate kit from Vector Laboratories.
Virus infections.
Cells were infected with either AdIE1-72 or AdIE2-86 at various multiplicities of infection (MOIs). AdE2F1 was used to infect cells at an MOI of 50. Cells were washed with PBS and with serum-free DMEM prior to infection. DMEM containing adenovirus was added to the cells, and infections were carried out at 37°C in 5% carbon dioxide (CO2) for 1 h (31). Afterwards, the viral inoculum was removed and replaced with DMEM containing the appropriate serum concentration and cultured under the conditions described above. HCMV (Towne) infections of HEL cells (MOI = 5) were as described previously (21).
Western blot analysis.
Whole-cell extracts from REF52 cells infected with the adenovirus constructs and HEL cells infected with HCMV Towne were harvested at the indicated times. The harvested cells were washed twice with cold PBS and lysed in 100 μl of whole-cell extract buffer (50 HEPES [pH 7.9], 250 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.3 mM sodium orthovanadate, 2 mM sodium fluoride, 2 μg of apoprotinin per ml, 1 μg of pepstatin per ml, 2 μg of leupeptin per ml) by incubation for 30 min on ice. Soluble proteins were collected by centrifugation at 13,000 × g in a microcentrifuge, and this supernatant was stored at −70°C. Aliquots were analyzed by electrophoresis through denaturing sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis gels, and the resolved proteins were transferred to nitrocellulose membranes by electroblotting. HCMV IE1-72 and IE2-86 protein expression was detected with a mouse anti-cytomegalovirus monoclonal antibody (MAB810; Chemicon International, Inc.) using an enhanced chemiluminescence kit (Amersham) according to the manufacturer's recommendations.
Cell cycle analysis by flow cytometry.
Cells were infected with the appropriate recombinant adenoviruses and processed for flow cytometry as described previously (19). Briefly, cells were trypsinized, combined with any floating cells, pelleted, washed with PBS, repelleted, and resuspended in 400 μl of PBS. All centrifugations were at 500 × g for 5 min at 4°C. Subsequently, cells were fixed in cold ethanol (final concentration, 70%) and stored at 4°C. The fixed cells were washed twice with PBS and then incubated in 2 N HCl in water containing 0.2 mg of pepsin per ml at room temperature for 30 min. Afterwards, 0.1 M sodium tetraborate was added, and the cells were then washed with PBS and subsequently blocked with PBS containing 1% bovine serum albumin (BSA). Cells were resuspended in 0.5 ml of PBS containing propidium iodide (PI) and RNase A (0.5 mg/ml) and incubated for 30 min to overnight at 4°C. Flow cytometric analysis performed by the University of Massachusetts Medical School (UMMS) Flow Cytometry Core Facility.
Cell cycle analysis by BrdU incorporation.
At 12 h prior to harvesting, cells were incubated with 10 μM bromodeoxyuridine (BrdU). Cells were fixed in 90% ethanol at room temperature for 5 min. Fixed cells were washed twice with PBS and then incubated in 2 N HCl in water at room temperature for 30 min. Afterwards, 0.1 M sodium tetraborate was added; the cells were then washed twice with PBS and once with PBS–0.5% Tween 20 and subsequently blocked with PBS–0.5% Tween 20 containing 1% BSA. Immunohistochemical staining for BrdU incorporation was done by incubating the cells with a mouse anti-BrdU monoclonal antibody (Boehringer Mannheim) diluted in PBS–0.5% Tween 20–BSA for 1 h at room temperature in a humidified chamber. The cells were washed with PBS–0.5% Tween 20 and then incubated with a horseradish peroxidase-conjugated goat anti-mouse immunoglobulin secondary antibody (Vector Laboratories) diluted in PBS–0.5% Tween 20 for 1 h at room temperature in a humidified chamber. Following this incubation, cells were washed with PBS–0.5% Tween 20 and then incubated with a streptavidin substrate (Vector Laboratories) diluted in PBS for 30 min at room temperature in a humidified chamber. The cells were washed with PBS and then incubated with DAB substrate (Vectastain kit; Vector Laboratories) at room temperature for 20 min to visualize stained nuclei. Scoring for BrdU-positive cells was done by counting the number of cells stained positive for BrdU incorporation per cell population. A minimum of 10 fields and 300 total cells was scored for each cell population.
Immunohistochemical staining for p53 protein accumulation.
Semiconfluent cultures of REF52 cells were infected with the appropriate adenovirus constructs and immunohistochemically stained for p53 protein (20). At the time of harvest, the cells were washed three times with PBS and then fixed for 15 min each in formaldehyde (final concentration, 0.37%) followed by methanol. The cells were then washed twice in PBS–0.5% Tween 20. Cells were then incubated with either an anti-p53 monoclonal antibody (PAB421; Oncogene Science) or an anti-HCMV IE monoclonal antibody (MAB8130; Chemicon International, Inc.) in the presence of 1% BSA in PBS–0.5% Tween 20 for 45 min at room temperature. The cells were washed three times with PBS–0.5% Tween 20, and bound antibody was detected using a Vectastain DAB substrate kit as described by the manufacturer.
RESULTS
Using recombinant adenoviruses to express HCMV IE1-72 and IE2-86 in cells.
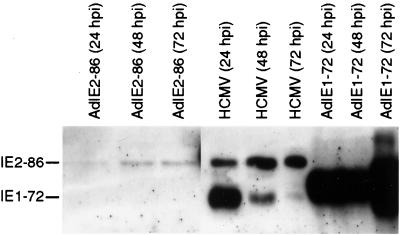
In order to identify the potential effects of the HCMV IE proteins on cell cycle control, we utilized recombinant adenovirus technology to express the individual IE proteins in cells. There are numerous advantages to using recombinant adenoviruses to express cDNAs of interest as compared to the conventional transfection techniques. Besides yielding higher transduction efficiencies and applying minimal selection pressure to the cells, recombinant adenoviruses can be used to express cDNAs in quiescent, G0 cells. We utilized two recombinant adenoviruses, AdIE1-72 and AdIE2-86, in our study (1). To confirm that both recombinant viruses expressed each IE product and to determine if their expression could alter cell growth control, we infected REF52 cells or MEFs with either AdIE1-72 or AdIE2-86. The REF52 cell line is an immortalized cell line that is permissive to infection with recombinant adenoviruses. REF52 cells can efficiently undergo growth arrest in response to serum withdrawal, and they express wild-type p53 and Rb family members, so they were used to examine the effects of the adenovirus E1A and E1B proteins on cell growth control (23). Extracts from REF52 cells infected with either AdIE1-72 or AdIE2-86 were analyzed for HCMV IE protein expression. As shown in Fig. 1, cells transduced with AdIE1-72 expressed the IE1-72 protein at levels exceeding those observed in HCMV-infected HELs. In contrast, cells transduced with AdIE2-86 expressed lower levels of IE2-86a protein as compared to the HCMV-infected cells.
FIG. 1.
Expression of HCMV proteins IE1-72 and IE2-86 in REF52 cells infected with AdIE1-72 or AdIE2-86. Western blot analysis of protein from whole-cell lysates from REF52 cells infected with either AdIE1-72 or AdIE2-86 (MOI = 250) (100 μg/lane) or HEL cells infected with HCMV (Towne) (MOI = 5) (25 μg/lane). Recombinant adenovirus infections were performed at an MOI of 250. Cell lysates were harvested at the times postinfection indicated. HCMV IE proteins were identified by probing with an anti-cytomegalovirus monoclonal antibody specific for a common determinant.
Expression of HCMV IE1-72 and IE2-86 disrupts cell cycle distribution in asynchronously cycling cells.
Previous experiments have shown that infections with HCMV exhibit a negative influence on cell cycle progression. Several groups have reported that HCMV-infected fibroblasts undergo an arrest at G1 and G2/M phases following infection (7, 8, 16, 24, 34). However, the small DNA tumor viruses appear to have a different effect on proliferation. These viruses (i.e., adenovirus, SV40, and HPV), through the ability of their IE proteins to bind to the Rb family members and induce E2F transactivational functions, alter the distribution of cells towards S phase (reviewed in reference 30). Given that both HCMV IE1-72 and IE2-86 bind Rb family members (11, 13, 17, 33) and induce E2F activity (11, 17, 33) and that HCMV infection results in the induction of not only E2F activity (reviewed in reference 15), but also cyclin-dependent kinase (Cdk) activities associated with S-phase induction (16), we wanted to determine if HCMV IE1-72 or IE2-86 expression could alter the cell cycle distribution of randomly cycling cells. To address this issue, we infected REF52 cells with either AdIE1-72 or AdIE2-86 and examined them for alterations in cell cycle distribution over time.
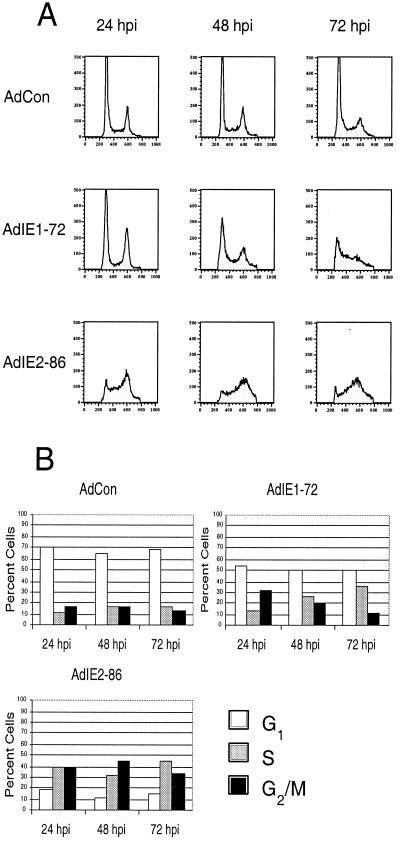
With the expression of either IE1-72 or IE2-86, the distribution of randomly cycling REF52 cells was altered relative to the distribution observed with control-infected cells (Fig. 2) or mock-infected cells (data not shown). Infection with AdIE1-72 caused an increase in the percentage of REF52 cells in S phase at 48 and 72 h postinfection (Fig. 2). Cells expressing IE1-72 exhibited almost a twofold increase in the percentage of S-phase cells compared to control-infected cells at both the 48 h postinfection (p.i.) (26.4 versus 16.3%) and 72 h p.i. (35.4 versus 17.5%) time points. Concomitant with this increase in S-phase cells, there was a decrease in the percentage of cells in the G1 phase of the cell cycle following infection with AdIE1-72 compared to following infection with AdCon at both the 48-h-p.i. (49.8 versus 65.6%) and 72-h-p.i. (51.1 versus 69.5%) time points.
FIG. 2.
Expression of HCMV IE1-72 and IE2-86 disrupts cell cycle progression in asynchronously cycling cells. Cells were infected with AdIE1-72, AdIE2-86, or AdCon at an MOI of 500 as described in Materials and Methods. Following infection, cells were fixed at the times indicated, and stained with PI. Flow cytometric analysis for cellular DNA content was performed by assessing the levels of PI incorporated by the cells. (A) Fluorescence-activated cell sorter (FACS) analysis from infected REF52 cells stained with PI; (B) histograms summarizing FACS data from infected REF52 cells.
Infection with AdIE2-86 had a dramatic effect on the cell cycle distribution of REF52 cells (Fig. 2). As was observed for cells expressing IE1-72, the distribution of the cells was altered so that there was an increase in the percentage of the cell population in S phase and a concomitant decrease in the percentage of the cell population in G1. Specifically, the expression of IE2-86 caused an increase in the percentage of cells in S phase (two- to threefold higher than the percentage observed with control samples (Fig. 2B). At 24 h p.i. over one-third of the IE2-86-expressing cells were in S phase whereas a smaller percentage of AdCon-infected cells were in S phase (38.8 versus 12.3%). IE2-86 expression also caused an increase in the percentage of G2/M-phase cells compared to the percentage observed for control-infected cells. At 24 h p.i. close to half of the IE2-86-expressing cells (44.1%) were in G2/M phase, whereas a smaller proportion of the control-infected cells (17.1%) were in G2/M phase.
Taken together, our data indicate that HCMV IE1-72 and IE2-86 can, like many other factors, influence the cell cycle of randomly cycling fibroblasts by biasing the distribution towards S phase and G2/M phase.
HCMV IE2-86 expression induces quiescent cells to proliferate and delays cell cycle exit.
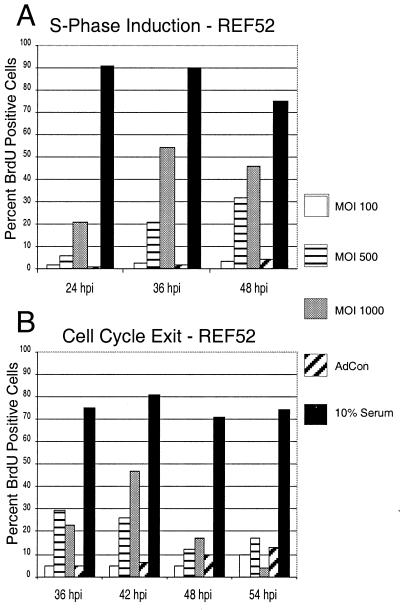
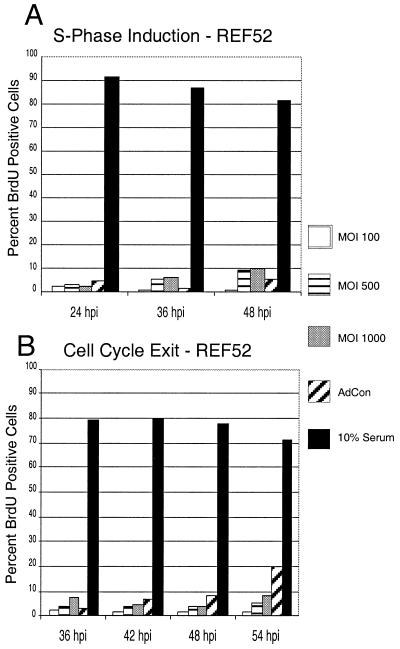
Since expression of HCMV IE2-86 disrupted the normal distribution of randomly cycling cells following infection with AdIE2-86, we wanted to examine whether IE2-86 can alter the more stringent measures of growth control by inducing quiescent cells into S phase and inhibiting cells from exiting the cell cycle. To address this question, we infected quiescent, serum-depleted cultures of REF52 cells with increasing MOIs of AdIE2-86 and examined the cells for S-phase induction by determining the percentage of cells that incorporated BrdU. As data in Fig. 3A indicate, more than 90% of control-infected cells underwent growth arrest following serum withdrawal. Most of the cells reentered the cell cycle upon the addition of serum to the culture medium. Expression of IE2-86 in growth-arrested fibroblasts resulted in up to a 10-fold increase in the population of cells incorporating BrdU relative to the control population (MOI at 36 h p.i., 500; 21.1 versus 2.0%, respectively). This ability of IE2-86 to induce S phase appeared to be dose dependent, since higher doses of AdIE2-86 induced a stronger proliferative response than lower doses did.
FIG. 3.
IE2-86 induces quiescent cells to proliferate and delays cell cycle exit in cells. (A) REF52 cells were rendered quiescent by culturing in the presence of 0.25% serum and then infected with AdIE2-86 (at the MOI indicated) or AdCon (MOI = 500). Cells were maintained in media containing 0.25% serum and pulsed with 10 μM BrdU for 12 h prior to harvest at the indicated times postinfection. (B) Asynchronous cultures of REF52 cells were infected with AdIE2-86 (at the MOI indicated) or AdCon (MOI = 500) and then subjected to culture in reduced serum (0.25%). Cells were pulsed with BrdU for 12 h prior to harvest at the indicated times postinfection. For both panels, cells were immunohistochemically stained for BrdU incorporation with an anti-BrdU monoclonal antibody, and the number of BrdU-positive cells was scored against the total number of cells counted per well.
To determine if IE2-86 expression would influence exit from the cell cycle, we infected a population of random cycling REF52 cells with AdIE2-86 or control virus and monitored for changes in cell cycle exit kinetics following culture in reduced serum. Optimization experiments demonstrated that culturing REF52 cells for 36 h in reduced serum (0.25%) was sufficient to induce the majority of cells to exit the cell cycle as measured by BrdU incorporation (data not shown). Fibroblasts were cultured in reduced serum for a minimum of 36 h after infection with the recombinant adenoviruses and pulsed with BrdU for 12 h prior to harvest to assess the impact of IE2-86 expression on cell cycle exit. As the data in Fig. 3B demonstrate, approximately 90% of the REF52 cells became quiescent by 36 h after serum withdrawal. As expected, cells cultured under normal conditions (10% serum in the medium) continued to proliferate. Cell cycle exit in fibroblasts infected with AdIE2-86 (expressing IE2-86) was delayed compared with that in control virus-infected cells. We found that at the highest MOI, almost half of the IE2-86-expressing cells (46.7%) continued to incorporate BrdU through 42 h following serum withdrawal, whereas the proportion was much lower for control-infected cells (6.4%). To confirm that cell cycle alterations were due to IE2-86 expression and were not a consequence of the recombinant adenovirus approach we employed, we transfected two different plasmids containing IE2-86 cDNAs (from J. Nelson, Oregon Health Sciences Center, Portland, Oreg., and T. Stamminger, University of Erlangen-Nurnberg, Erlangen, Germany) into REF52 cells prior to serum withdrawal. Using this approach, we obtained BrdU incorporation results that were similar to those obtained with the AdIE2-86-infected cells (40% of IE2-86-transfected cells were BrdU positive) (data not shown). These results suggest that IE2-86 expression can delay cell cycle exit for extended periods following serum withdrawal. Together, these experiments indicate that IE2-86 can influence cellular proliferation control by spurring growth-arrested cells to proliferate and by delaying exit from the cell cycle.
Expression of HCMV IE2-86 induces proliferation and delays cell cycle exit in p53+/+ and p53−/− MEFs.
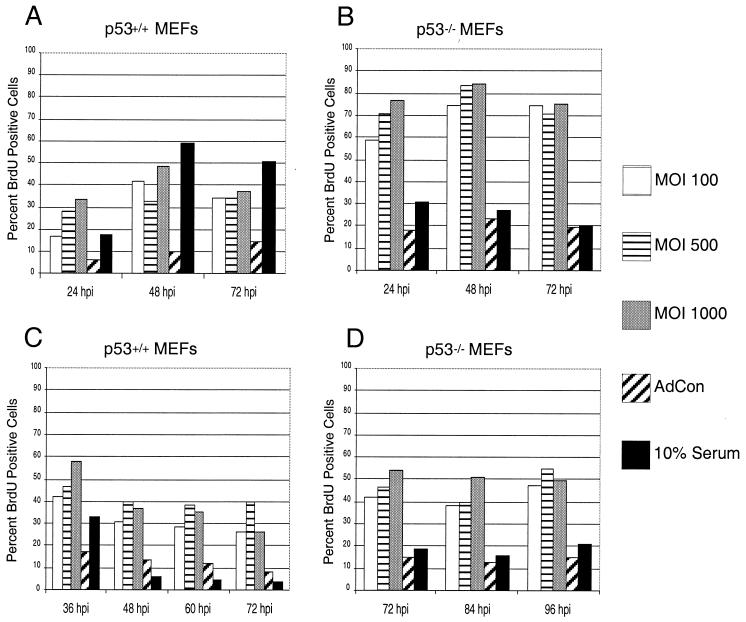
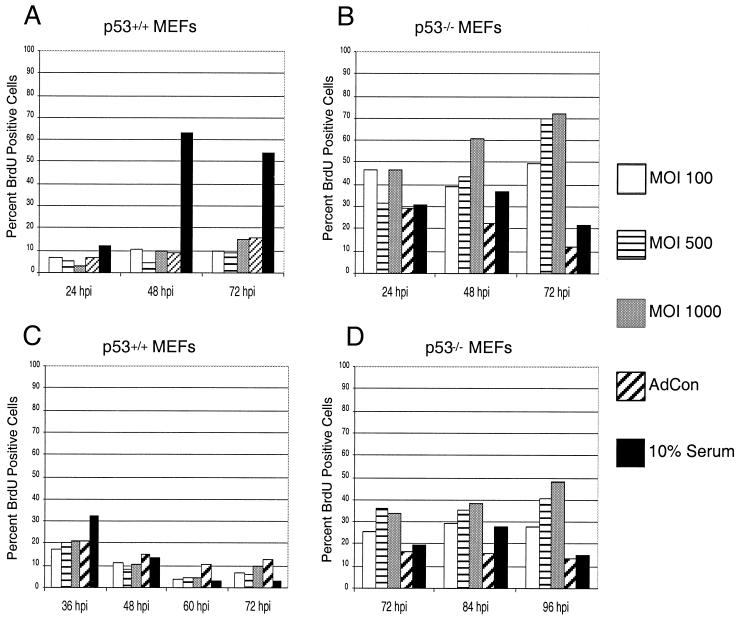
Viral oncoproteins from several DNA tumor viruses such as adenovirus, SV40, and HPV have the ability to bind to p53 and inhibit its activity. Several groups have demonstrated that HCMV IE2-86, in addition to interacting with pRb, can bind p53 (5, 40) and can block apoptosis (48). Additionally, p53 protein levels are elevated in HCMV infections (12, 29) and this may be coincident with the HCMV-mediated growth arrest observed in these viral infections (7, 8, 16, 24). Since IE2-86 can bind to p53, we wanted to determine whether targeting of p53 by IE2-86 was required for its ability to induce proliferation as well as delay growth arrest. To address this issue, we examined the effects of IE2-86 expression in early-passage MEFs lacking p53 (p53−/−) and their wild-type counterparts (p53+/+). Although both cell types were derived from the same strain of mice (9), the p53+/+ and p53−/− MEFs required different culturing conditions to induce an optimal level of growth arrest. The p53+/+ MEFs required culturing in 0.25% serum for 48 h to induce quiescence in these cells, and the p53−/− MEFs required 0.1% serum for 60 h (data not shown). Growth-arrested MEFs were infected with increasing MOIs of AdIE2-86 or control adenovirus (MOI = 500) and then pulsed with BrdU to assess DNA replication levels. As shown in Fig. 4A, quiescent p53+/+ MEFs were induced to incorporate BrdU following infection with AdIE2-86. At 48 h p.i. we found that over 40% of the IE2-86-expressing p53+/+ MEFs incorporated BrdU, compared to 10% of the control-infected cells. IE2-86 exhibited a similar effect when expressed in quiescent p53−/− MEFs. While the culture conditions were sufficient to arrest growth for approximately 80% of the p53−/− MEFs, AdIE2-86 transduction induced almost a fourfold increase in the percentage of BrdU-positive p53−/− MEFs (MOI = 500; 84%) compared to control-infected cells (MOI = 500; 23%) at the 48 h-p.i. point. The fold induction in BrdU positive cells is similar in p53+/+ cells and p53−/− cells. Therefore, the ability of IE2-86 to induce S phase from quiescent cells is apparently independent of p53 protein.
FIG. 4.
IE2-86 induces proliferation and delays cell cycle exit in p53+/+ and p53−/− MEFs. Early-passage p53+/+ (A) or p53−/− (B) MEFs were cultured under low serum conditions appropriate to induce growth arrest. Cells were then infected with AdIE2-86 at the indicated MOIs or with AdCon (MOI = 500) and maintained under low serum conditions. Cells were pulsed with BrdU for 12 h prior to harvesting at the indicated times postinfection. Early-passage p53+/+ (C) or p53−/− (D) MEFs were infected with AdIE2-86 (at the MOIs indicated) or with AdCon (MOI = 500) and then subjected to serum starvation. Cells were pulsed with BrdU for 12 h prior to harvesting at the indicated times postinfection. In each experiment shown, BrdU positive cells were identified by immunohistochemical staining and the number of BrdU-positive cells was scored against the total number of cells counted per well.
We next determined if p53 targeting was required for IE2-86 to delay growth arrest following serum withdrawal. Asynchronously cycling p53+/+ and p53−/− MEFs were first infected with AdIE2-86 and then cultured in reduced serum to induce growth arrest. Similar to the effects seen in the REF52 cells, expression of IE2-86 delayed the ability of the p53+/+ MEFs to exit the cell cycle following culture in 0.25% serum (Fig. 4C). The percentage of cells still BrdU positive following AdIE2-86 infection (MOI = 500) and serum withdrawal was almost threetimes higher than that observed for AdCon-infected cells (MOI = 500) at 48 h p.i. (39.8 versus 13.6%, respectively). Delayed growth arrest by IE2-86 was also evident in cells lacking p53. Infection with AdIE2-86 at an MOI of 500 resulted in a threefold-greater percentage of BrdU positive cells than was observed for AdCon-infected cells at 72 h p.i. (46.1 versus 15.2%) (Fig. 4D). Taken together, these findings suggest that p53 targeting is not required by IE2-86 to induce proliferation or to delay cell cycle exit.
HCMV IE1-72 expression does not induce quiescent cells to proliferate and fails to delay cell cycle exit.
Since expression of HCMV IE1-72 also disrupted the normal distribution of randomly cycling cells following infection with AdIE1-72, we wanted to determine whether IE1-72 could influence cell proliferation control in a manner similar to IE2-86. To address whether IE1-72 could induce growth-arrested cells to reenter the cell cycle, we infected REF52 cells with AdIE1-72 and measured S-phase induction by BrdU incorporation. As the data in Fig. 5A indicate, IE1-72 expression had no apparent effect on quiescence following infection with AdIE1-72 compared to quiescence following infection with the control virus (MOI at 48 h p.i. = 500; percentage of cells quiescent, 8.8 versus 5.4%). To address whether IE1-72 could retard cell cycle exit, we infected REF52 cells with AdIE1-72 and subjected them to reduced serum conditions. Unlike IE2-86 expression, IE1-72 expression did not delay cell cycle exit in fibroblasts infected with AdIE1-72 (Fig. 5B). Analysis at earlier times postinfection or postserum withdrawal did not show any delay in the kinetics of cell cycle exit following AdIE1-72 infection (data not shown).
FIG. 5.
IE1-72 fails to induce quiescent cells to proliferate and does not delay cell cycle exit in REF52 cells. (A) REF52 cells were rendered quiescent by culturing in the presence of 0.25% serum and infected with AdIE1-72 (at the MOIs indicated) or AdCon (MOI = 500). Cells were maintained in media containing 0.25% serum and pulsed with 10 μM BrdU for 12 h prior to harvest. (B) Asynchronous cultures of REF52 cells were infected with AdIE1-72 (at the MOIs indicated) or AdCon (MOI = 500) and then subjected to serum withdrawal. Cells were pulsed with BrdU 12 h prior to harvesting. For both panels, cells were immunohistochemically stained for BrdU incorporation with an anti-BrdU monoclonal antibody and the number of BrdU-positive cells was scored against the total number of cells counted per well.
Taken together, these results and those shown in Fig. 2 suggest that IE1-72 can influence cell cycle progression in asynchronously cycling cell populations but is ineffective in altering cell cycle parameters under the more stringent conditions such as inducing quiescent cells to proliferate or in delaying cell cycle exit following serum withdrawal.
Expression of HCMV IE1-72 induces proliferation and delays cell cycle exit in the absence of p53.
Although expression of IE1-72 did cause a modest change in the cell cycle distribution in randomly cycling cells, IE1-72 expression failed to induce proliferation and delay cell cycle arrest in rat fibroblasts. We found this outcome surprising given the fact that IE1-72 has been shown to bind to p107 and induce E2F activity (17, 33). Since p53 can induce growth arrest under certain conditions, we asked if the presence of p53 prevents IE1-72 from inducing proliferation. To address this issue, p53+/+ and p53−/− MEFs were infected with AdIE1-72 following a growth arrest induced by serum withdrawal. As shown in Fig. 6A, serum-starved p53+/+ MEFs failed to reenter the cell cycle and S phase following infection with AdIE1-72. Contrary to the effects observed for the p53+/+ MEFs, expression of IE1-72 was able to induce growth-arrested p53−/− MEFs to reenter the cell cycle (Fig. 6B). For example, at 48 h p.i. there was an increase of over 2.5-fold in BrdU-positive cells observed in the population of cells infected with the highest MOI of AdIE1-72 compared to that observed in control-infected cells (60.6 versus 22.3%) as measured by BrdU incorporation. This ability of IE1-72 to induce cell cycle reentry was apparent even at the 72-h-p.i. point where over six times as many IE1-72-expressing cells incorporated BrdU compared to control-infected cells (MOI = 500 for each infection).
FIG. 6.
IE1-72 induces proliferation and delays cell cycle exit in the absence of p53. Early passage p53+/+ (A) and p53−/− (B) MEFs were growth arrested as described earlier and then infected with AdIE1-72 (at the MOI indicated) or AdCon (MOI = 500). Cells were pulsed with BrdU for 12 h prior to harvesting at the indicated times postinfection. Early passage p53+/+ (C) and p53−/− (D) MEFs were infected with AdIE1-72 (at the MOI indicated) or AdCon (MOI = 500) and then subjected to serum withdrawal. Cells were pulsed with BrdU for 12 h prior to harvesting at the indicated times postinfection. In each experiment, cells were immunohistochemically stained with an anti-BrdU monoclonal antibody (MAb) and the number of BrdU-positive cells scored against the total number of cells counted per well.
To address whether p53 hinders IE1-72 from delaying cell cycle exit, p53+/+ and p53−/− MEFs were infected with AdIE1-72 prior to serum withdrawal. As shown in Fig. 6C, IE1-72 expression did not perturb the ability of the p53+/+ MEFs to undergo growth arrest following serum withdrawal. These results are consistent with the results obtained by expressing IE1-72 in REF52 cells (Fig. 5B). However, a different pattern was observed in the p53−/− MEFs transduced with AdIE1-72. Expression of IE1-72 delayed cell cycle exit in the p53−/− MEFs following serum withdrawal (Fig. 6D). After 72 h of culturing AdIE1-72-infected cells in serum-depleted medium, over one-third of these cells (MOI = 500; 36.0%) remained cycling compared to a smaller percentage of the control-infected cells (MOI = 500; 16.5%). This ability of IE1-72 to delay cell cycle exit was apparent even at 96 h p.i. Taken together, these findings suggest that p53 can mask the proliferative capacity of IE1-72.
Expression of either IE1-72 or IE2-86 results in accumulation of p53 protein.
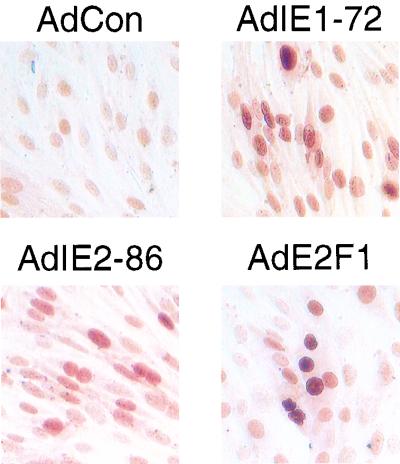
We have shown that both IE1-72 and IE2-86 can modulate the cell cycle. The IE2-86 protein has a more prominent effect on proliferation control in that it induces S phase and delays growth arrest under all of the conditions tested. We found that IE1-72 has the capacity to exhibit a similar phenotype but only in cells devoid of p53 expression. In response to various types of stress including inactivation of Rb family members by DNA tumor virus oncoproteins, there is an increase in p53 protein levels (reviewed in reference 18). This change in p53 protein levels has been shown to correlate with an increase in p53 activity leading to growth arrest and/or apoptosis (22). Given that expression of the IE proteins of the small DNA tumor viruses can lead to accumulation of p53 protein, and given that under certain circumstances p53 accumulation leads to growth arrest at G1, we determined whether expression of the HCMV IE proteins affected p53 protein levels. To test this possibility, we infected REF52 cells with AdIE1-72 or AdIE2-86 and then immunohistochemically stained for p53 protein. As a positive control, we utilized AdE2F1, a recombinant adenovirus that expresses E2F1, a factor that members of our group have previously shown to cause an accumulation of p53 protein in rodent fibroblasts (20). The results of these experiments are shown in Fig. 7. As expected, infection with the E2F1-expressing adenovirus caused p53 protein to accumulate in almost all of the cells. Infection with AdCon did not have an effect on p53 levels in cells. For IE2-86, most of the cells infected with AdIE2-86 stained positive for p53. Likewise, p53 protein accumulation was also observed in REF52 cells transfected with cDNAs encoding IE2-86 (data not shown). Unlike the few AdCon-infected cells that stained positive for p53, localization of p53 protein in the IE2-86-expressing cells was in the nucleus. This finding was expected since IE2-86 has been shown to interact with p53 and inhibit its function (5, 37, 40). p53 accumulation in conjunction with HCMV IE protein expression has been observed in SMCs grown from restenotic lesions (10, 37).
FIG. 7.
IE1-72 and IE2-86 expression promotes p53 accumulation in REF52 cells. Near-confluent cultures of REF52 cells were infected with AdIE1-72, AdIE2-86, or AdCon at an MOI of 500 and cultured under normal conditions. Cells infected with AdE2F1 served as a positive control for the experiment. At 24 h p.i., cells were harvested, fixed with formaldehyde, and immunohistochemically stained for p53 using a commercial anti-p53 MAb.
Accumulation of p53 protein was also observed in cells expressing IE1-72. Analogous to the effects seen in the AdIE2-86-infected cells, the increased staining for p53 protein was localized to the nucleus. This result was unexpected since IE1-72 has not previously been shown to interact with p53. Our observation that IE1-72 expression promotes p53 accumulation suggests a possible link between this particular HCMV IE protein and its inability to induce proliferation in p53-expressing cells. In infections with the small DNA tumor viruses, increased levels of p53 protein usually result in growth arrest or apoptosis unless inactivated by a viral protein such as adenovirus E1B protein or the SV40 large T antigen (3, 45). Given that IE1-72 has not been shown to interact with p53, the increased levels of p53 protein may, in essence, mask the growth promoting effects of IE1-72 by blocking cells in G1.
DISCUSSION
The goal of our study was to characterize the effects of HCMV IE1-72 and IE2-86 on the cell cycle. Here, we present evidence that expression of the HCMV IE proteins can modulate the cell cycle, presumably to promote an environment favorable for viral replication. Specifically, we found that expression of IE2-86 can drive cells out of quiescence and into S phase as well as delay cells from exiting the cell cycle into G0. We also found that IE1-72 can mediate effects similar to those observed with IE2-86, but only in cells deficient for p53. Finally, we observed an accumulation of p53 protein upon the expression of either IE1-72 or IE2-86 in rat fibroblasts.
We modeled our analysis of the HCMV IE proteins on that of the studies performed on the human adenovirus E1A and E1B proteins since these proteins can perturb the cell cycle by targeting members of the Rb family of proteins and p53 (35, 43, 45). Analogous to those studies, we utilized a rat fibroblast cell line (REF52) which is not permissive to adenovirus or HCMV replication in this study. These cells express wild-type p53 and Rb family members and respond very well to serum withdrawal by undergoing growth arrest. Using a recombinant adenovirus expressing the cDNA encoding green fluorescent protein, workers in our laboratory have found that >95% of the REF52 cells express the fluorescent protein when infected at MOIs similar to the ones used in the present study (data not shown). Furthermore, using immunofluorescence staining for IE1-72 or IE2-86 protein expression following infection with either AdIE1-72 or AdIE2-86, we also found that essentially all of the cells express HCMV IE proteins at the MOIs utilized in our experiments (data not shown). Use of the REF52 cells in our preliminary experiments also allowed us to establish the conditions necessary for our subsequent studies in wild-type and p53-null mouse fibroblasts. There is a clear advantage to using the wild-type and the p53-knockout MEFs in our study of the relationship between the HCMV IE proteins and p53 compared to using cancer cell lines that lack p53. By using fibroblasts from wild-type and p53-null mouse embryos, we could effectively assess the effect of IE1-72 and IE2-86 on the cell cycle in the presence or absence of p53 in cells that are essentially genetically identical without having to worry about complications, such as genetic variability, arising from the use of cancer cell lines.
Though our findings do not appear to be consistent with the block in cell cycle progression observed in HCMV-infected human fibroblasts (7, 8, 16, 24), they are consistent with earlier reports that HCMV infection influences the host cell cycle by affecting the expression of various genes involved in DNA synthesis. HCMV-infected cells display many characteristics associated with cellular DNA replication, including the activation of many cellular S-phase genes, Rb hyperphosphorylation, induction of E2F transactivational activity, and expression of cyclin E and Cdk2 proteins (6, 14, 26, 32, 41). Moreover, HCMV infections also lead to the activation of both cyclin E- and cyclin A-associated kinase activity, important rate limiting events associated with cellular DNA replication (16). The apparent ability of the virus to influence the aforementioned proliferation-promoting activities reflects the capacity for HCMV to induce an S-phase environment in infected host cells. Despite the observations that imply a link between HCMV and the stimulation of cellular DNA synthesis, numerous groups have reported that HCMV-infected human fibroblasts arrest in either late G1 or G2/M in both quiescent and cycling cells (7, 8, 16, 24). However, the described growth arrest phenotype does not appear to be relevant to the permissiveness of cells to HCMV infection, since we have recently observed that HCMV infection of human umbilical vein endothelial cells induces these cells to proliferate (Yurochko et al., unpublished data).
Recently, Wiebusch and Hagemeier (44) reported that IE2-86 inhibits cell cycle progression by inducing a G1 arrest. This was observed following transient transfections of IE2-86 cDNA into cells from U-373, an astrocytoma cell line that is permissive to HCMV infection. They demonstrate that expression of the IE2-86 in asynchronously cycling U-373 cells causes an arrest in G1. Additionally, they show that IE2-86 blocked S-phase entry after growth stimulation of serum-deprived U-373 cells. Taken together, their data suggests that the IE2-86 protein is at least one of the HCMV factors responsible for the G1 arrest phenotype observed in HCMV infected cells. In contrast, our data insinuate a different role for IE2-86 in affecting the cell cycle. We employed a strategy where we tested the ability of IE2-86 to influence cellular proliferation under varying degrees of growth stringency. Specifically, we looked at the effect of IE2-86 expression on cycling cells (low stringency), cells undergoing growth arrest (medium stringency), and quiescent cells (high stringency). Using a recombinant adenovirus to express the IE2-86 cDNA in our cells of interest, we found that expression of IE2-86 has a growth-promoting effect on the cell cycle under all conditions tested. While we cannot account for the discrepancy between our data and the results reported by Wiesbusch and Hagemeier (44), we have made attempts to resolve the apparent differences between our studies. Using the same IE2-86 expression vector (pHM121) employed by Wiebusch and Hagemeier (44), we repeated our analysis using transient transfection rather than recombinant adenovirus infection to introduce the IE2-86 cDNA into cells. We found that IE2-86 expression in transiently transfected rat fibroblasts continue to incorporate BrdU when serum is withdrawn (data not shown). When another expression vector (pcDNA3-IE2-86) containing an independently isolated IE2-86 cDNA (from J. Nelson) was tested, the same outcome was observed, further supporting our claim that IE2-86 can modulate the host cell cycle to generate an S-phase-like environment.
Based on the results presented here, we define a phenotype for IE2-86 where cellular DNA replication is induced following IE2-86 expression. Although the data presented in our study appear to contradict the results published by Wiebusch and Hagemeier (44), our findings are consistent with the notion that IE2-86 promotes an environment conducive to proliferation. An earlier examination conducted by Fortunato et al. on the effects of IE2-86 and the cell cycle showed that transient expression of IE2-86 could affect proliferation (11). Specifically, they demonstrated that transient transfection of IE2-86 cDNA into SAOS-2 cells caused an increase in the population of cells in S/G2/M phase with a concomitant decrease in G1 cells (11). We observed a similar outcome when IE2-86 was expressed in asynchronously cycling rat and mouse fibroblasts. Furthermore, IE2-86 binding to pRb is sufficient to bypass pRb-mediated repression of E2F transcriptional activity (13). These findings, coupled with the fact that IE2-86 can induce the expression of cyclin E as well as positively influence factors associated with S-phase entry including E2F1, TK, and DNA polymerase α, strongly suggest that IE2-86 has the capacity to drive cells out of G0/G1 and into S phase.
Although our findings are consistent with the notion that IE2-86 promotes an environment conducive to proliferation, there is a discrepancy between our phenotype and the G1 arrest observed following HCMV infection of human fibroblasts (7, 8, 16, 24). In those studies, HCMV infection blocked cells in G1 as measured by the absence of cellular DNA replication. A recent report from the laboratory of Lu and Shenk has shown that a component of the HCMV tegument, the UL69 protein, could mediate a G1 arrest in cells expressing the viral protein (25). Since both HCMV infection and IE2-86 expression have been previously shown to positively influence factors associated with S phase, it is possible that the G1 arrest phenotype observed following HCMV infection is due to the presence of the UL69 protein or possibly other factors encoded by HCMV. Therefore, it is conceivable that upon infection with HCMV, expression of IE2-86 activates cellular factors associated with the transition from G1 to S. However, in the presence of the UL69 protein, G1 arrest occurs by disrupting the proliferative signals generated by the IE2-86 protein, thereby masking the growth-promoting effects of IE2-86 and inhibiting cellular DNA replication.
In addition to IE2-86, we also examined the IE1-72 protein to determine if it could influence cell cycle control. Like IE2-86, there are several lines of evidence that imply a link between IE1-72 and overcoming cell cycle control. First, we have previously shown that binding of the IE1-72 protein to p107 can overcome the p107-mediated repression of an E2F-responsive promoter (17, 33). Another observation supporting the relationship between IE1-72 and cell cycle control is that IE1-72 can phosphorylate the pocket proteins, p107 and p130, as well as several of members of the E2F family of transcription factors (32). Finally, the IE1-72 protein has been shown to activate the dihydrofolate reductase and DNA polymerase α promoters (14, 26, 41), both of which are induced during the transition from G1 to S phase and are factors necessary for cellular DNA synthesis. Based on these lines of evidence, we assumed that IE1-72 expression could mediate a proliferative phenotype similar to the one observed for IE2-86-expressing cells. Contrary to our observations for IE2-86-expressing cells, we find that IE1-72 expression does not dramatically affect cell proliferation control in normal rat or mouse fibroblasts. However, in the absence of p53, IE1-72 expression was sufficient to induce S-phase reentry and delay cell cycle exit in MEFs. The ability of IE1-72 to modulate the cell cycle in the absence of p53 implies a role for p53 in negatively influencing the proliferative capacity of IE1-72. This notion is reinforced by our observations of nuclear p53 protein accumulation in the presence of IE1-72 expression by recombinant adenovirus infection or by transient transfection (data not shown). Increased nuclear p53 protein levels following IE1-72 expression in the absence of a p53-inactivating function ascribed to IE1-72 (5) may account for the inability of IE1-72 to induce proliferation in our S-phase induction and cell cycle exit experiments. Therefore, it is feasible that the presence of increased p53 protein levels negates IE1-72 by inhibiting its proliferative effects through an undefined mechanism.
Besides IE1-72, we observed nuclear p53 protein accumulation in fibroblasts expressing IE2-86 following infection with AdIE2-86 or transfection with plasmids encoding the IE2-86 protein. However, IE2-86 interacts with p53 and can block its transactivation function (36, 39, 40).
We present evidence that the HCMV proteins IE1-72 and IE2-86 are capable of altering cell cycle control. Additional evidence supporting the proliferative capacity of IE1-72 and IE2-86 comes from a study conducted by Zhou et al. (47). Using a rat SMC model, Zhou et al. demonstrated that expression of the HCMV IE proteins significantly increased SMC proliferation following infection with HCMV (Towne). Although they incorporated cells that are nonpermissive for HCMV infection, they were still able to demonstrate that IE1-72 and IE2-86 are expressed under the conditions described in their analysis (47). Therefore, the data presented by Zhou et al. are entirely consistent with the notion that HCMV IE1-72 and IE2-86 proteins have a positive influence on cell proliferation. The perturbation of cell growth control by the HCMV IE proteins may be one of the ways the virus promotes the development of restenosis. It is widely believed that the accumulation of SMCs in the vessel intima is due to complex mechanisms that include factors influencing SMC migration and proliferation. It has been suggested that HCMV may indirectly enhance SMC migration by increasing the expression of the PDGF β-receptor in infected SMCs (47). The virus may also directly promote SMC migration through expression of the US28 gene product, and HCMV-encoded chemokine receptor (38). The expression of this chemokine receptor, coupled with the release of specific chemokines that activate this US28 protein, may induce the infected SMCs to migrate and localize to the site of vessel injury. These findings offer additional mechanisms by which HCMV can contribute to accumulation of SMCs along the vessel wall.
Based on our observations, we envision two related mechanisms by which expression of the HCMV IE proteins may promote the overproliferation of SMCs that is characteristic of restenotic plaque formation. In the absence of HCMV, damage resulting from the angioplasty-induced injury to the arterial vessel wall induces the migration of SMCs to the vessel intima and the subsequent proliferation of these cells to heal the wound. Under normal conditions, the SMCs terminate proliferation and undergo growth arrest that most likely involves a p53-dependent mechanism. However, in the presence of HCMV, the SMCs continue to proliferate because the expressed HCMV IE proteins prevent their exit from the cell cycle. In our second model, injury to the arterial vessel wall leads to the migration of SMCs to the wound site. The HCMV IE proteins then induce a greater than normal number of SMCs to proliferate. In both examples, expression of the HCMV IE proteins would lead to the overrepresentation of SMCs along the vessel wall.
ACKNOWLEDGMENTS
We thank the UMMS Flow Cytometry Core Facility and Rachel Gerstein for assistance in flow cytometric analyses. We thank Michelle Debatis for technical assistance and members of the Kowalik laboratory for technical advice during the course of this study. We also thank Trudy Morrison, Madelyn Schmidt, and Raymond Welsh for critically reviewing the manuscript.
J.P.C. was supported by an NIH grant for training in immunology (5T32 AI07349-09) and a National Research Service Award (1F31HL10334-01) from the National Heart, Lung, and Blood Institute. A.D.Y. was supported by a New Investigator Award from the state of Louisiana (YIA99). This research was supported in part by Center Grant DK3520 and by grants from the American Heart Association (9830085N) and the National Cancer Institute (R01CA86038-01) to T.F.K.
REFERENCES
- 1.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht T, Nachtigal M, St. Jeor S C, Rapp F. Induction of cellular DNA synthesis and increased mitotic activity in Syrian hamster embryo cells abortively infected with human cytomegalovirus. J Gen Virol. 1976;30:167–177. doi: 10.1099/0022-1317-30-2-167. [DOI] [PubMed] [Google Scholar]
- 3.Bargonetti J, Reynisdottir I, Friedman P N, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 4.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 5.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 8.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 10.Epstein S E, Speir E, Zhou Y F, Guetta E, Leon M, Finkel T. The role of infection in restenosis and atherosclerosis: focus on cytomegalovirus. Lancet. 1996;348(Suppl. 1):S13–S17. doi: 10.1016/s0140-6736(96)98005-8. [DOI] [PubMed] [Google Scholar]
- 11.Fortunato E A, Sommer M H, Yoder K, Spector D H. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortunato E A, Spector D H. p53 and Rpa are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J Virol. 1998;72:2033–2039. doi: 10.1128/jvi.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang E-S, Kowalik T. The pathogenicity of human cytomegalovirus: an overview. In: Becker G D Y, Huang E-S, editors. Molecular aspects of human cytomegalovirus diseases. New York, N.Y: Springer-Verlag; 1993. pp. 3–45. [Google Scholar]
- 16.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R A, Yurochko A D, Poma E E, Zhu L, Huang E S. Domain mapping of the human cytomegalovirus IE1-72 and cellular p107 protein-protein interaction and the possible functional consequences. J Gen Virol. 1999;80:1293–1303. doi: 10.1099/0022-1317-80-5-1293. [DOI] [PubMed] [Google Scholar]
- 18.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 19.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 21.Kowalik T F, Yurochko A D, Rinehart C A, Lee C Y, Huang E S. Productive infection of human endometrial stromal cells by human cytomegalovirus. Virology. 1994;202:247–257. doi: 10.1006/viro.1994.1340. [DOI] [PubMed] [Google Scholar]
- 22.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 23.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 24.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski E. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2447–2479. [Google Scholar]
- 28.Monick M M, Geist L J, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol. 1992;7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 29.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevins J R. Disruption of cell-cycle control by viral oncoproteins. Biochem Soc Trans. 1993;21:935–938. doi: 10.1042/bst0210935. [DOI] [PubMed] [Google Scholar]
- 31.Nevins J R, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F transcription factor. Methods Enzymol. 1997;283:205–219. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- 32.Pajovic S, Wong E L, Black A R, Azizkhan J C. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol Cell Biol. 1997;17:6459–6464. doi: 10.1128/mcb.17.11.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 36.Speir E, Huang E S, Modali R, Leon M B, Shawl F, Finkel T, Epstein S E. Interaction of human cytomegalovirus with p53: possible role in coronary restenosis. Scand J Infect Dis Suppl. 1995;99:78–81. [PubMed] [Google Scholar]
- 37.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 38.Steblow D N, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson J A. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka K, Zou J P, Takeda K, Ferrans V J, Sandford G R, Johnson T M, Finkel T, Epstein S E. Effects of human cytomegalovirus immediate-early proteins on p53-mediated apoptosis in coronary artery smooth muscle cells. Circulation. 1999;99:1656–1659. doi: 10.1161/01.cir.99.13.1656. [DOI] [PubMed] [Google Scholar]
- 40.Tsai H L, Kou G H, Chen S C, Wu C W, Lin Y S. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271:3534–3540. [PubMed] [Google Scholar]
- 41.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weingerg R A. The retinoblastoma gene and cell growth control. Trends Biochem Sci. 1990;15:199–202. doi: 10.1016/0968-0004(90)90162-5. [DOI] [PubMed] [Google Scholar]
- 43.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 44.Wiebusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G(1) J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y F, Leon M B, Waclawiw M A, Popma J J, Yu Z X, Finkel T, Epstein S E. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med. 1996;335:624–630. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y F, Yu Z X, Wanishsawad C, Shou M, Epstein S E. The immediate early gene products of human cytomegalovirus increase vascular smooth muscle cell migration, proliferation, and expression of PDGF beta-receptor. Biochem Biophys Res Commun. 1999;256:608–613. doi: 10.1006/bbrc.1999.0387. [DOI] [PubMed] [Google Scholar]
- 48.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]