Abstract
Rotavirus is one of very few viruses that utilize the endoplasmic reticulum (ER) for assembly, and therefore it has been used as an attractive model to study ER-associated protein folding. In this study, we have examined the requirements for metabolic energy (ATP) for correct folding of the luminal and ER-associated VP7 of rotavirus. We found that VP7 rapidly misfolds in an energy-depleted milieu and is not degraded within 60 min. We also found that VP7 attained a stable minimum-energy state soon after translation in the ER. Most surprisingly, energy-misfolded VP7 could be recovered and establish correct disulfide bonds and antigenicity following a shift to an ATP-rich milieu. Using a Semliki Forest virus expression system, we observed that VP7 requires ATP and cellular, but not viral, factors for correct disulfide bond formation. Our results show for the first time that the disulfide bond formation of rotavirus VP7 is an ATP-dependent process. It has previously been shown that chaperones hydrolyze ATP during interaction with newly synthesized polypeptides and prevent nonproductive intra- and intermolecular interactions. The most reasonable explanation for the energy requirement of VP7 is thus a close interaction during folding with an ATP-dependent chaperone, such as BiP (Grp78), and possibly with protein disulfide isomerase. Taken together, our observations provide new information about folding of ER-associated proteins in general and rotavirus VP7 in particular.
Rotavirus undergoes a unique maturation process in the endoplasmic reticulum (ER). The assembly process, which includes translocation of subviral particles across the ER membrane and retention of mature virus in the ER, has provided a system in which posttranslational events, such as folding, targeting, and retention, can be studied (1, 13, 15, 19, 22, 23, 26). One of the rotavirus glycoproteins is the VP7 outer capsid protein, which is a luminal and ER-associated polypeptide with only N-linked high-mannose oligosaccharide residues (10). The VP7 protein of group A contains eight cysteine residues. These cysteines are highly conserved and important for the conformation of VP7 (10). Biochemical and morphological studies have established that calcium, an oxidizing milieu, and N-linked glycosylation are critical for the correct folding of VP7 (8, 15, 18, 20, 24). It has also been shown that VP7 becomes endo-β-N-acetylglucosaminidase H (endo-H)-resistant after brefeldin A treatment, which suggests modifications by Golgi-associated enzymes (16).
While it has previously been shown that protein folding in bacteria and mitochondria requires metabolic energy (2, 9), the role of ATP in the folding of proteins in the ER is still incompletely known. There are only a few studies of the effect of energy-depleting conditions on the posttranslational process of exporting proteins (3, 7), and there is no information available concerning the role of ATP in the folding of ER-associated proteins.
Previous studies have shown that major histocompatibility complex class I antigens require ATP for assembly in vitro (11), and it has also been shown that the hemagglutinin protein of influenza virus and the G protein of vesicular stomatis virus require ATP for posttranslational processing (3, 7). Furthermore, it has been demonstrated that ATP depletion blocks the export of proteins from the ER to the Golgi apparatus (25). More recently, it has also been shown that ATP is required for the assembly of herpesvirus and the release of retrovirus (6, 27).
In previous studies, we found that calnexin and protein disulfide isomerase (PDI) interact with NSP4 and VP7 during maturation (14, 15). In this study, we extend these observations and report that metabolic energy is required for correct disulfide bond formation and folding of VP7. We also found that VP7 reaches a stable minimum-energy state immediately after translation. These results provide new information about the role of ATP in the posttranslational processing of VP7.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
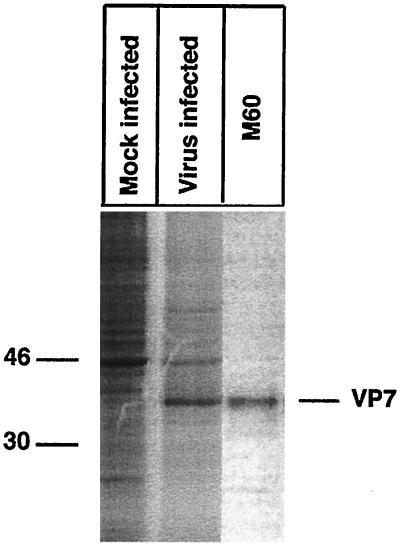
MA-104 cells were grown in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 10% fetal calf serum. Rhesus rotavirus (RRV) was obtained from infected MA-104 cells by freezing and thawing. The monoclonal antibody (MAb) used in this study, M60, recognizes a cross-reactive nonneutralizing epitope on VP7 (21) which is dependent on correct disulfide bond formation (15, 24).
Rotavirus infection.
RRV was activated with 10 μg of trypsin (Sigma, St. Louis, Mo.) per ml for 30 min at 37°C before inoculation of MA-104 cells. After 1 h of infection, the inoculum was replaced with serum-free Eagle's MEM. Virus titers were determined by peroxidase staining, as previously described (15, 16).
DNA constructs.
cDNAs encoding the open reading frame of VP7 (SA11) were synthesized by reverse transcription followed by PCR. The primers used (5′-3′) were 5′-ACTAGTCCTAGGTAGCTCCTTTTAATG-3′ and 5′-ACTAGTCCTAGGCTAACCTAAGTTATA-3′. The underlined sequences are restriction sites for SpeI and AvrIII. The PCR product was cloned in a PCR-script plasmid (Stratagene, La Jolla, Calif.). After transfection into Escherichia coli (Stratagene), positive clones were purified and plasmids were digested with SpeI. The gene was then ligated into the SpeI site of a Semliki Forest virus (SFV) expression vector plasmid. The recombinant plasmid (pSFV-VP7) was then transfected into E. coli as described previously (12).
Generation of recombinant SFV.
Recombinant plasmids were purified using plasmid minipreps (Qiagen, Valencia, Calif.), linearized, and used as templates for in vitro transcription with SP6 RNA polymerase as described previously (12, 17), with the exception that recombinant plasmids were linearized with NruI instead of SpeI.
In vitro transcripts made from a recombinant pSFV-VP7 plasmid were electroporated into BHK-21 cells together with an equal amount of mRNA transcript from SpeI-linearized pSFV-Helper 1 (12, 17). Electroporated BHK-21 cells were then diluted in medium, seeded onto tissue culture dishes, and incubated for 24 h at 37°C. The media, including recombinant SFV, were collected and frozen at −70°C until use.
Treatment of cells with DTT and ATP-depleting medium.
To deplete cells of intracellular ATP, monolayers were incubated with glucose-free DMEM containing 20 mM 2-deoxy-d-glucose and 10 mM sodium azide as described previously (3, 6, 27).
To produce metabolically labeled proteins synthesized under reducing conditions, 2 mM dithiothreitol (DTT) was added to the medium 30 min before a pulse and maintained during the chase periods (15).
Metabolic labeling of viral proteins.
To produce metabolically labeled cell lysates, MA-104 cells were infected with trypsin-activated RRV at a multiplicity of infection (MOI) of 10 as described previously (14, 16). At 7 h postinfection (p.i.), infected cells were starved for 1 h in methionine- and cysteine-free medium before being labeled for 5 min with 250 μCi of [35S]methionine-cysteine (Trans-label; Dupont). For chase experiments, cells were washed and incubated with Eagle's MEM or ATP-depleting medium containing an excess of methionine (10 mM) and 1 mM cycloheximide. At the end of each radioactive pulse or after a chase period, the cells were incubated with ice-cold phosphate-buffered saline containing 40 mM N-ethylmaleimide (NEM) (Sigma) for 2 min to prevent disulfide bond rearrangement (15, 24). The cells were then lysed in ice-cold lysis buffer {2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 150 mM NaCl, and 50 mM HEPES}, and the lysate was clarified of cell debris by centrifugation at 13,000 × g for 2 min in a microcentrifuge before use.
Endo-H digestion.
Digestion with Endo-H (Boehringer, Mannheim, Germany) was performed essentially as described previously (16). Briefly, cell lysates were diluted 1/10 with 50 mM sodium acetate (pH 5.5) and digested with 5 mU of Endo-H at 37°C overnight.
RIPA.
Immunoprecipitation was performed essentially as described previously (24). Briefly, radiolabeled lysates (100 μl) were incubated with 10 μl of the desired antibody and 400 μl of radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 50 mM HEPES, and 0.5% CHAPS) overnight at 4°C. Fifty microliters of Staphylococcus aureus protein A–Sepharose CL-4B (Pharmacia, Uppsala, Sweden) was subsequently added to the mixture, which was then incubated for 2 h at 4°C. The immune complexes were washed three times with RIPA buffer, suspended in nonreducing (10 mM Tris-HCl [pH 6.8], 0.5% sodium dodecyl sulfate [SDS], and 10% glycerol) or reducing (nonreducing sample buffer with 2% β-mercaptoethanol) sample buffer and boiled for 5 min before separation by SDS-polyacrylamide gel electrophoresis (PAGE).
SDS-PAGE.
Polypeptide separation was performed by SDS-PAGE with a 4.5% stacking gel and a 10% separation gel as previously described (24). Electrophoresis was carried out at a constant voltage of 50 V at room temperature, followed by fixation with 10% glacial acetic acid and 35% methanol for 1 h at room temperature. Autoradiography was performed as previously described (24). Molecular mass standards (Amersham, Little Chalfont, United Kingdom) included myosin (200 kDa), phosphorylase B (97 kDa), bovine serum albumin (69 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), and lysozyme (14 kDa).
RESULTS AND DISCUSSION
Metabolic energy is required to support correct folding and disulfide bond formation of VP7.
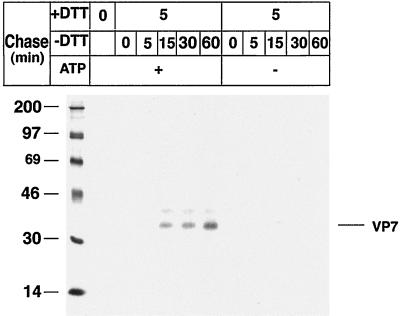
To study the role of ATP in the posttranslational folding of rotavirus VP7, RRV-infected cells were treated with 2 mM DTT in Eagle's MEM for 30 min before and during a metabolic pulse. To study posttranslational processing, the cells were chased in ATP or ATP-depleted medium (3, 6, 27) for up to 60 min (Fig. 1). The cells were then harvested in lysis buffer and immunoprecipitated by MAb M60, a disulfide bond-dependent MAb (15, 24).
FIG. 1.
Correct disulfide bond formation of rotavirus VP7 requires metabolic energy during posttranslational processing. MA-104 cells were infected with RRV (MOI, 10) and, at 7 h p.i., starved for 1 h in methionine-cysteine-free medium and subsequently labeled with [35S]methionine-cysteine (250 μCi) for 5 min. DTT (2 mM) was added to the medium at 7.5 h p.i. and maintained during the 5-min metabolic pulse and a 5-min chase. To examine posttranslational processing, labeled proteins were chased in ATP medium (Eagle's MEM supplemented with 1 mM cycloheximide and 10 mM methionine) or ATP-depleted medium (20 mM 2-deoxy-d-glucose, 10 mM sodium azide, 1 mM cycloheximide, and 10 mM methionine in glucose-free DMEM) for the indicated times. At the end of the pulse and each chase, the monolayers were incubated with ice-cold phosphate-buffered saline containing 40 mM NEM for 2 min, followed by cell lysis, immunoprecipitation by MAb M60, and analysis by reducing SDS-PAGE. Molecular mass markers (in kilodaltons) are shown on the left side of the panel.
As expected (24), shifting the conditions from a reducing (with DTT) to an oxidizing (without DTT) milieu (Fig. 1) rapidly restored the disulfide-dependent VP7 M60 epitope. However, in contrast to the ATP-rich milieu, VP7 folding was not restored within 60 min in the ATP-depleted milieu (Fig. 1), suggesting that VP7 misfolds in the absence of metabolic energy during posttranslational processing. We have previously (24) shown that misfolded VP7 can be restored and can establish correct disulfide bonds upon incubation in oxidizing medium. Here we extend this information and show for the first time that the folding process not only requires an oxidizing milieu but also metabolic energy.
While it is known that protein folding in mitochondria and the cytosol of bacteria is energy dependent (2, 9), the situation is less clear for the ER of mammalian cells. The fact that ATP is present in the ER lumen became evident recently with the discovery of an ATP translocator in the ER membrane (5), but there is only a very limited amount of information about the role of ATP in protein folding in the ER (3, 7). However, it has previously been shown that molecular chaperones require ATP in order to prevent misfolding of newly synthesized polypeptides (3). As we and others have previously shown that PDI and BiP (Grp78) interact with VP7 during maturation (15, 28), we believe that the most reasonable explanation for the energy requirement for correct folding of VP7 is that ATP is required for chaperone-assisted maturation of VP7.
Misfolded VP7 is not degraded in energy-depleted cells.
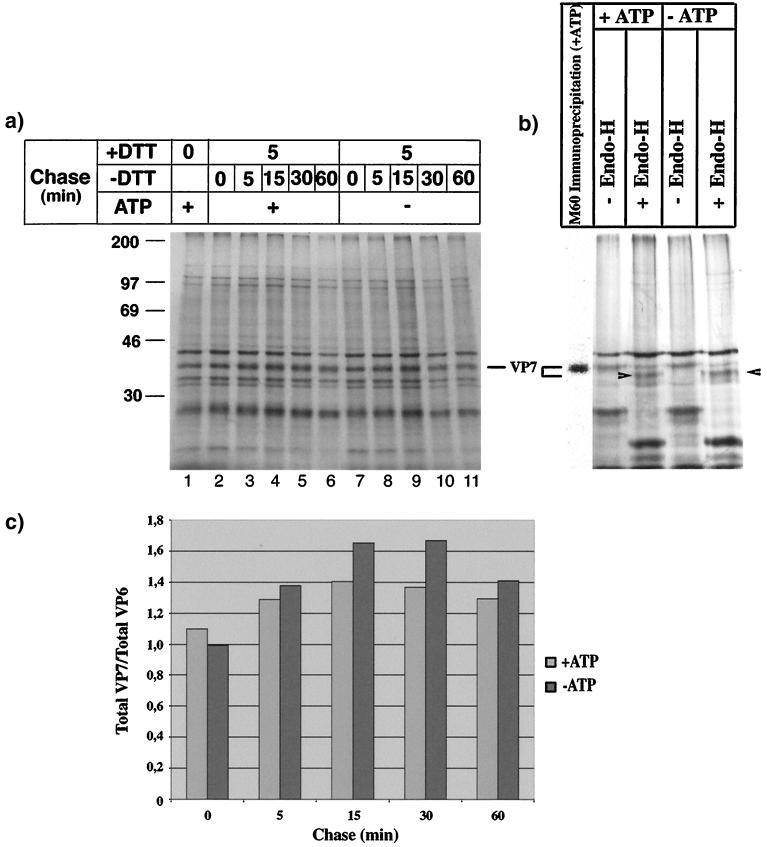
To examine if VP7 in ATP-depleted cells was degraded during chase, we analyzed the cell lysates shown in Fig. 1 under reducing conditions. To identify VP7, an Endo-H experiment was performed (Fig. 2b). Quantification by densitometry revealed that VP7 was not degraded within 60 min (Fig. 2a and c). This observation leads to the conclusion that the lack of MAb M60 reactivity to VP7 in the ATP-depleted cells (Fig. 1) could not be explained as degradation of VP7. As no VP7 aggregates could be detected in nonreducing gels (not shown), we propose two possible explanations for the disappearance of VP7: VP7 remains in a reduced form during ATP depletion, or VP7 oxidizes and obtains aberrant intradisulfide bonds during ATP depletion. It has previously been shown that an ATP-depleted milieu does not diminish disulfide bond formation (3). Furthermore, we show below that VP7 establishes disulfide bonds during ATP deficiency, which suggests that VP7 does not remain in a reduced form after a shift from reducing to oxidizing conditions during ATP depletion.
FIG. 2.
VP7 is not degraded during ATP depletion. (a) Analysis of the cell lysates shown in Fig. 1 under reducing SDS-PAGE conditions. Molecular mass markers (in kilodaltons) are shown on the left side of the panel. (b) Aliquots of cell lysates (panel a, lanes 6 and 11) were diluted 1/10 with sodium acetate and mock or Endo-H treated (5 mU) at 37°C for 4 h. After digestion, the cell lysates were separated by SDS-PAGE under reducing conditions. The cell lysate in panel a, lane 6, was immunoprecipitated with MAb M60. Arrowheads, digested VP7. (c) The amount of VP7 was measured by densitometry from the fluorograph shown in panel a, lanes 2 to 11, and the results are presented relative to the total amount of VP6.
It is possible that ATP-depleted VP7 forms large aggregates, which may not enter the gel. However, as we did not detect any intermediate forms of aggregates during our chase protocol under nonreducing conditions, we believe that VP7 does not aggregate in ATP-depleted media. As no VP7 aggregates could be detected in nonreducing gels and no degradation was observed within 60 min, we favor the idea that VP7 obtains aberrant intradisulfide bonds during energy deficiency. This is further supported by the lack of reactivity with MAb M60 during ATP depletion (Fig. 1).
Energy-misfolded VP7 is rescued and establishes correct disulfide bonds following a shift to an oxidizing and ATP-rich milieu.
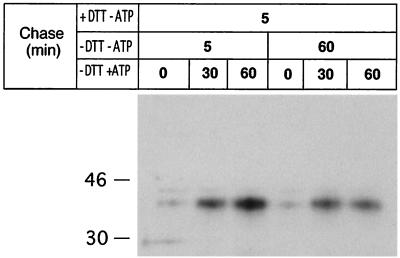
Next, we asked if energy-misfolded VP7 could be recovered if ATP was restored. To address this question, RRV-infected cells were treated with 2 mM DTT for 30 min before and during a 5-min metabolic pulse followed by chase as described in the legend to Fig. 3. The figure shows that the disulfide bond-dependent epitope was absent before ATP was restored. After about 30 min in ATP medium, correct disulfide bonds were catalyzed and a significant amount of VP7 was recognized by MAb M60. Careful examination of Fig. 3 also reveals that slightly more VP7 was immunoprecipitated after 5 min than after 60 min of chase (−DTT −ATP), and as we did not detect any measurable degradation of VP7 during ATP deficiency, we suggest that a prolonged ATP deficiency may lead to permanent misfolding of VP7. We and others have previously shown that reduced and misfolded proteins can be rescued by incubation in oxidizing medium (4, 15, 24). We now extend these observations and show that not only is an oxidizing milieu required to rescue misfolded proteins, metabolic energy is also required, an observation not previously reported for ER-associated proteins.
FIG. 3.
Energy-misfolded VP7 is refolded and establishes correct disulfide bonds following a shift to an oxidizing and ATP-rich milieu. At 7 h p.i., RRV-infected cells were starved for 1 h in methionine- and cysteine-free medium and subsequently metabolically labeled (250 μCi) for 5 min. DTT (2 mM) was added to the medium at 7.5 h p.i. and maintained during pulse and chase for 5 min. The reducing medium (+DTT −ATP) was then replaced with oxidizing medium (−DTT −ATP) for 5 or 60 min followed by incubation in oxidizing ATP medium (−DTT +ATP). At the end of each chase, the monolayers were incubated with ice-cold PBS-NEM, followed by cell lysis, immunoprecipitation by M60, and analysis under reducing SDS-PAGE conditions. Molecular mass markers (in kilodaltons) are shown on the left side of the panel.
Rotavirus VP7 reaches a stable minimum-energy state during or soon after translation.
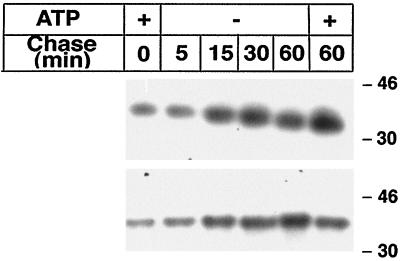
To determine if VP7 reaches a stable minimum-energy conformation during or soon after translation, infected cells were labeled for 5 min in oxidizing ATP medium and chased in ATP-deficient medium for up to 60 min. As illustrated in Fig. 4, and in contrast to results with reducing ATP medium (with DTT) (Fig. 1), VP7 was recognized by MAb M60 after the pulse, suggesting that the disulfide bond-dependent epitope critical for recognition by MAb M60 was catalyzed during translation. Most surprisingly, the epitope was intact during ATP depletion for up to 60 min of chase, suggesting that VP7 had reached a conformation during translation which did not depend on metabolic energy to remain intact.
FIG. 4.
Rotavirus VP7 reaches an ATP-resistant conformation during or soon after translation. Infected cells were labeled for 5 min with [35S]methionine-cysteine (250 μCi) in oxidizing and ATP-containing medium followed by chase in ATP-deficient medium (20 mM 2-deoxy-d-glucose, 10 mM sodium azide, 1 mM cycloheximide, and 10 mM methionine in glucose-free DMEM). At the end of the pulse and each chase, monolayers were incubated with ice-cold PBS-NEM for 2 min, followed by cell lysis, immunoprecipitation by M60, and analysis under nonreducing (upper panel) or reducing (lower panel) SDS-PAGE conditions. Molecular mass markers (in kilodaltons) are shown on the right side of the panel.
Critical examination of Fig. 4 reveals that migration of VP7 increased during the chase (Fig. 4, upper panel), suggesting additional processing of VP7 in the absence of metabolic energy. Analysis of the same lysates under reducing conditions (Fig. 4, lower panel) revealed that the migration differences disappeared (Fig. 4), indicating that the increased mobility of VP7 under nonreducing conditions was not a result of oligosaccharide trimming of VP7 but rather an effect of the catalysis of intramolecular disulfide bonds.
Based on the results presented in Fig. 4, we propose that VP7 reaches a minimum-energy state during or soon after translation (detected by MAb M60) which does not depend on ATP to remain intact. This is in contrast to influenza hemagglutinin, whose monomers depend on energy to remain oxidized and correctly folded and which do not reach a stable minimum-energy state soon after translation (3). The most reasonable explanation for the two different folding pathways would be that influenza hemagglutinin is a secretory protein that undergoes a series of trimming and folding events during its transport through the secretory pathway, whereas rotavirus VP7 remains in the ER and only contains a few N-linked carbohydrates.
Careful examination of Fig. 3 also reveals that more VP7 precipitated in the chase than in the pulse (Fig. 4); we therefore believe that additional or shifted disulfide bonds (posttranslationally) make the M60 epitope more available for this antibody during the chase.
VP7 establishes correct disulfide bonds without the presence of additional rotavirus proteins.
The results presented so far were obtained with infectious rotavirus under various experimental conditions. To examine the ability of VP7 to assemble and establish correct disulfide bonds in the absence of additional rotavirus proteins, VP7 was cloned and expressed by an SFV expression system. To analyze the expression efficiency of VP7, cells were infected with recombinant SFV VP7. Figure 5 shows that VP7 was efficiently expressed concomitant with a significant reduction in host cell protein synthesis. To ensure that VP7 established correct disulfide bonds, it was immunoprecipitated with the disulfide bond-dependent MAb M60. Figure 5 shows that M60 recognized VP7, indicating that VP7 expressed by SFV established correct disulfide bonds and that this conformation was not dependent on additional rotavirus proteins.
FIG. 5.
SFV-expressed VP7 establishes correct disulfide bonds. Cells were mock or SFV VP7 infected (MOI, 10) and, at 18 h p.i., starved for 1 h in methionine- and cysteine-free medium and subsequently labeled with [35S]methionine-cysteine (250 μCi) for 45 min. At the end of the pulse, the monolayers were incubated with ice-cold PBS-NEM, harvested in lysis buffer, immunoprecipitated with M60, and analyzed under reducing SDS-PAGE conditions. Molecular mass markers (in kilodaltons) are shown on the left side of the panel.
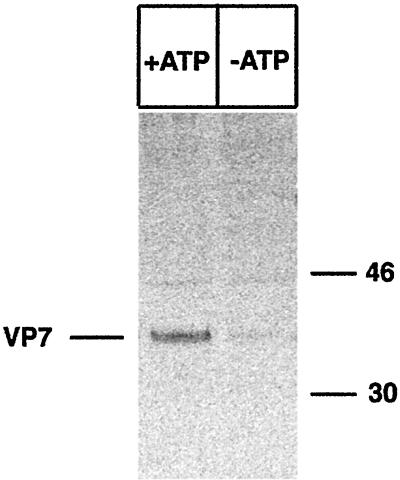
To analyze the role of ATP in VP7 folding, MA-104 cells were infected with recombinant SFV and treated with 2 mM DTT before a metabolic pulse. DTT was maintained during pulse and chase but removed 60 min before lysis. As shown in Fig. 6, energy depletion caused misfolding of VP7, most likely due to aberrant disulfide bond formation.
FIG. 6.
SFV-expressed VP7 requires ATP for correct folding. MA-104 cells were infected with recombinant SFV VP7 (MOI, 10). At 18 h p.i., the cells were starved for 1 h in methionine- and cysteine-free medium and subsequently labeled with [35S]methionine (250 μCi) for 15 min. DTT (2 mM) was added to the medium at 18.5 h p.i. and maintained during the pulse. The cells were chased in oxidizing ATP (+ATP) or ATP-deficient (−ATP) medium for 60 min. The monolayers were subsequently incubated with ice-cold PBS-NEM, harvested in lysis buffer, immunoprecipitated with M60, and analyzed by reducing SDS-PAGE. Molecular mass markers (in kilodaltons) are shown on the right side of the panel.
In summary, we report for the first time that metabolic energy is required to ensure the correct folding and disulfide bond formation of ER-associated proteins, such as rotavirus VP7. We have also shown that misfolded VP7 can be correctly refolded in the presence but not in the absence of ATP. Furthermore, we found that VP7 reaches a stable minimum-energy state during or immediately after translation. We and others (14, 28) have previously shown that chaperons and foldase enzymes, such as BiP and PDI, participate in VP7 folding. It is most reasonable to assume that the energy-dependent misfolding of VP7 was influenced by the ATP-dependent chaperone BiP (3) and possibly also by PDI, which participates in the disulfide bond formation of VP7 (15).
ACKNOWLEDGMENTS
This project received financial support from the Swedish Medical Council (grant K98-06X-10392-06B), the Karolinska Research Foundation, and the Swedish Society for Medical Research.
We are grateful to Harry Greenberg for MAb M60.
REFERENCES
- 1.Bergmann C C, Maass D, Poruchynsky M S, Atkinson P H, Bellamy A R. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 1989;8:1695–1703. doi: 10.1002/j.1460-2075.1989.tb03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkareva E S, Lissin N M, Girshovich A S. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature. 1988;336:254–257. doi: 10.1038/336254a0. [DOI] [PubMed] [Google Scholar]
- 3.Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 4.Braakman I, Hoover-Litty H, Wagner K R, Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clairmont C A, De Maio A, Hirschberg C B. Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. J Biol Chem. 1992;267:3983–3990. [PubMed] [Google Scholar]
- 6.Dasgupta A, Wilson D W. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J Virol. 1999;73:2006–2015. doi: 10.1128/jvi.73.3.2006-2015.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doms R W, Keller D S, Helenius A, Balch W E. Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. J Cell Biol. 1987;105:1957–1969. doi: 10.1083/jcb.105.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormitzer P R, Greenberg H B. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology. 1992;189:828–832. doi: 10.1016/0042-6822(92)90616-w. [DOI] [PubMed] [Google Scholar]
- 9.Ellis R J, van der Vies S M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 10.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy F, Gabathuler R, Larsson R, Kvist S. ATP is required for in vitro assembly of MHC class I antigens but not for transfer of peptides across the ER membrane. Cell. 1991;67:265–274. doi: 10.1016/0092-8674(91)90178-2. [DOI] [PubMed] [Google Scholar]
- 12.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 13.Meyer J C, Bergmann C C, Bellamy A R. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology. 1989;171:98–107. doi: 10.1016/0042-6822(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 14.Mirazimi A, Nilsson M, Svensson L. The molecular chaperone calnexin interacts with enterotoxin NSP4 of rotavirus in vitro and in vivo. J Virol. 1998;72:8705–8709. doi: 10.1128/jvi.72.11.8705-8709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirazimi A, Svensson L. Carbohydrates facilitate correct disulfide bond formation and folding of rotavirus VP7. J Virol. 1998;72:3887–3892. doi: 10.1128/jvi.72.5.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirazimi A, von Bonsdorff C H, Svensson L. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology. 1996;217:554–563. doi: 10.1006/viro.1996.0150. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson M, von Bonsdorff C H, Weclewicz K, Cohen J, Svensson L. Assembly of viroplasm and virus-like particles of rotavirus by a Semliki Forest virus replicon. Virology. 1998;242:255–265. doi: 10.1006/viro.1997.8987. [DOI] [PubMed] [Google Scholar]
- 18.Poruchynsky M S, Maass D R, Atkinson P H. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J Cell Biol. 1991;114:651–656. doi: 10.1083/jcb.114.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poruchynsky M S, Tyndall C, Both G W, Sato F, Bellamy A R, Atkinson P H. Deletions into an NH2-terminal hydrophobic domain result in secretion of rotavirus VP7, a resident endoplasmic reticulum membrane glycoprotein. J Cell Biol. 1985;101:2199–2209. doi: 10.1083/jcb.101.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahrabadi M S, Lee P W. Calcium requirement for syncytium formation in HEp-2 cells by respiratory syncytial virus. J Clin Microbiol. 1988;26:139–141. doi: 10.1128/jcm.26.1.139-141.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 22.Stirzaker S C, Both G W. The signal peptide of the rotavirus glycoprotein VP7 is essential for its retention in the ER as an integral membrane protein. Cell. 1989;56:741–747. doi: 10.1016/0092-8674(89)90677-6. [DOI] [PubMed] [Google Scholar]
- 23.Stirzaker S C, Whitfeld P L, Christie D L, Bellamy A R, Both G W. Processing of rotavirus glycoprotein VP7: implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987;105:2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svensson L, Dormitzer P R, von Bonsdorff C H, Maunula L, Greenberg H B. Intracellular manipulation of disulfide bond formation in rotavirus proteins during assembly. J Virol. 1994;68:5204–5215. doi: 10.1128/jvi.68.8.5204-5215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verde C, Pascale M C, Martire G, Lotti L V, Torrisi M R, Helenius A, Bonatti S. Effect of ATP depletion and DTT on the transport of membrane proteins from the endoplasmic reticulum and the intermediate compartment to the Golgi complex. Eur J Cell Biol. 1995;67:267–274. [PubMed] [Google Scholar]
- 26.Weclewicz K, Kristensson K, Greenberg H B, Svensson L. The endoplasmic reticulum-associated VP7 of rotavirus is targeted to axons and dendrites in polarized neurons. J Neurocytol. 1993;22:616–626. doi: 10.1007/BF01181488. [DOI] [PubMed] [Google Scholar]
- 27.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu A, Bellamy A R, Taylor J A. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum-localized virion component. J Virol. 1998;72:9865–9872. doi: 10.1128/jvi.72.12.9865-9872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]