Abstract
This article comments on:
Li Z, Zhang D, Liang X, Liang J. 2024. Receptor for Activated C Kinase 1 counteracts ABSCISIC ACID INSENSITIVE5-mediated inhibition of seed germination and post-germinative growth in Arabidopsis. Journal of Experimental Botany 75, 3932–3945.
Keywords: bZIP, degradation, development, interactor, scaffold, stress
Scaffold proteins are key elements in signaling pathways, helping other proteins to assemble in order to complete and enhance their function. General scaffold proteins promote the destabilization of transcription factors and/or connect them with repression complexes of gene expression, resulting in both cases in a negative regulation of their functions. Using a genetic and molecular approach, Li et al. (2024) reveal how the conserved scaffold protein Receptor for Activated C Kinase 1 (RACK1) negatively regulates abscisic acid (ABA) signaling through interaction with and inhibition of ABA-insensitive 5 (ABI5) during seed germination and early development.
As one of the most important phytohormones, ABA is crucial for plant development, including seed dormancy, seed germination, and seedling development (Chen et al., 2020). The ABI5 transcription factor is a key player in ABA-triggered processes (Lopez-Molina et al., 2001) and also acts as a molecular hub in the balance between early development and stress, integrating external and internal cues into cellular responses (Albertos et al., 2015). To coordinate these processes, several partners have been described to be involved in the control of the transcriptional activity and/or the protein stability of ABI5. This tight regulation encompasses different post-translational modifications (PTMs) that allow a specific and efficient control of ABI5, playing a significant role in the ABA signaling pathway. In this scenario, the identification of scaffold proteins is especially interesting. The functional role of scaffold proteins enables recruitment from a huge variety of substrate adaptors, which in turn recognize specific proteins and direct them to ubiquitination and subsequent proteasomal degradation. In their study, Li et al. (2024) reveal that RACK1 recruits two CUL4-based E3 ligases (DWA1 and DWA2) which function together to mediate the turnover of ABI5, thereby efficiently turning down ABA signaling during germination and early seedling establishment.
ABI5 interactors in ABA signaling
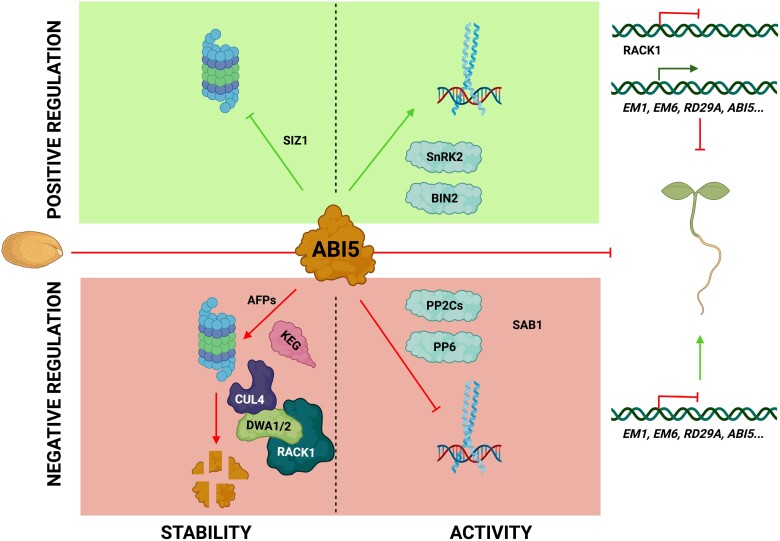
Several studies have demonstrated that the activity of ABI5 can be transcriptionally and post-translationally regulated by the interplay with different partners. Seedling establishment requires ABI5 degradation via the ubiquitin–26S proteolytic pathway (Fig. 1). Many proteins serve as negative interactors, promoting ABI5 destabilization, such as CULLIN4 (CUL4)-based and KEEP ON GOING (KEG) E3 ligases (Liu and Stone, 2010) and DWD HYPERSENSITIVE TO ABA1 (DWA1) and 2 (Lee et al., 2010), or involved in ABI5 stabilization but in an inactive form, like the SMALL UBIQUITIN RELATED MODIFIER E3 ligase (SIZ1) (Miura et al., 2009). Another important checkpoint is the modulation of transcriptional activity to switch the downstream gene expression network on and off (Fig. 1). ABA induces ABI5 phosphorylation, promoting its function as a transcriptional activator through the binding to ABA-responsive elements (ABREs) and controlling ABA-inducible gene expression (Lopez-Molina et al., 2001). Turning off ABA responses also requires the capacity to inactivate the binding of ABI5 to the promoters of the downstream targets. Different kinases such as SnRK2 (Nakashima et al., 2009) and BIN2 (Hu and Yu, 2014), and PROTEIN PHOSPHATASES type 2C (PP2Cs) (Lynch et al., 2012) and PROTEIN PHOSPHATASE6 (PP6) (Dai et al., 2013) have been described to regulate this step.
Fig. 1.
ABI5 interactome based on the post-translational protein modifications. Different post-translational modifications may positively (green) or negatively (red) affect ABI5 protein stability (left) or transcriptional activity (right). Among the positive effectors, sumoylation by SUMO E3 ligases (SIZ1) has been reported, while ubiquitination by the corresponding E3 ubiquitin ligases (KEG, CUL4) promotes ABI5 destabilization. In addition, phosphorylation and dephosphorylation of ABI5 exerted by specific protein kinases and phosphatases is a well-known mechanism impacting on ABI5 functionality. SnRK2s promote extensive changes in gene expression through the control of the phosphorylation level. Moreover, BIN2 interacts physically with ABI5 to modulate the crosstalk between ABA and brassinosteroids through phosphorylation. In contrast, ABI5 is dephosphorylated by PROTEIN PHOSPHATASES type 2C (PP2Cs) and PROTEIN PHOSPHATASE6 (PP6), which inhibits its activity. Also, Sensitive to ABA 1 (SAB1) interacts with ABI5 and is involved in the reduction of its phosphorylation level affecting its protein stability (Ji et al., 2019). Finally, members of the AFP-related Topless (TPL) co-repressors have the ability to physically associate with ABI5 and alter the transcriptional regulation of target genes. Created with BioRender.com
Many proteins involved in the transcriptional and post-translational regulation of ABI5 are related to the control of stability and/or activity, while other interactors that are becoming relevant have been described to modulate its function. These interacting partners serve as versatile scaffolds for the interactions between proteins and are promising targets for manipulation to improve agronomic traits related to the responses to changing environments or adjust the early seedling development under a variety of stress conditions. In this context, ABI Five binding Protein (AFP) facilitates ABI5 ubiquitin degradation, attenuating the ABA signal (Lopez-Molina et al., 2003). INDUCER OF CBF EXPRESSION1 (ICE1) negatively affects the transcriptional activity of ABI5 to maintain the correct ABA signal during germination (Hu et al., 2019). XPO1-interacting WD40 protein 1 (XIW1) was also described to maintain the ABI5 stability in the nucleus (Xu et al., 2019), acting as a positive switch in ABA responses. In the current work by Li et al. (2024), the scaffold protein RACK1 has emerged as a novel regulator of ABI5, repressing its transcriptional activity and protein stability. Interestingly, RACK1 also interacts with DWA1 and 2, functioning together to promote ABI5 degradation.
ABI5 post-translational regulation in plants: what for?
ABI5 serves as a nexus in the crosstalk between external conditions, hormone responses, and signaling events. Thus, it is tightly regulated both by a high number of interacting partners and by different PTMs that ensure the correct molecular coordination to maintain the balance between stress conditions and growth. Upon exposure to abiotic stresses (such as salinity or drought), ABI5 proteasome-dependent turnover decreases, and its accumulation induces the expression of ABA-responsive genes (Uno et al., 2000). This turnover is required for the inhibition of the growth and the activation of the stress tolerance response.
In this context, PTMs have been revealed to be key regulatory switches to modulate transcriptional activity and/or stability, that tailor the ABA response through modulating the status of ABI5 (Table 1). These PTMs include phosphorylation, carried out by kinases and reversed by the action of phosphatases, that finally coordinate the binding of ABI5 to specific promoters. Regarding the control of ABI5 stability, ubiquitination promotes its degradation, while sumoylation prohibits it. In addition, the specific S-nitrosylation of Cys153 also regulates ABI5, leading to its interaction with the E3 ubiquitin ligases KEG and CUL4. Through the regulation of the stability and activity of ABI5, PTMs turn the signaling events off and on to respond efficiently to internal and external challenges.
Table 1.
Compilation of ABI5 post-translational regulation
| PTM | Interactor | Effects | Phenotype | References |
|---|---|---|---|---|
| Phosphorylation | SnRK2s | Transcriptional activation and stabilization | Mutants are sensitive to desiccation, showed severe growth defects during seed development, a loss of dormancy level, and elevated seed ABA content. | Nakashima et al. (2009) |
| BIN2 | Transcriptional activation and stabilization | BIN2-overexpressing plants have an ABA-hypersensitive phenotype. | Hu and Yu (2014) | |
| Dephosphorylation | PP2Cs | Transcriptional inactivation | Triple mutants manifest pleiotropic effects during the life cycle, related to the great reduced phosphorylation level of the bZIP transcription factor. | Lynch et al. (2012) |
| PP6 | Transcriptional inactivation and destabilization | Mutants exhibit an ABA0hypersensitive phenotype. | Dai et al. (2013) | |
| SAB1 | Inhibition of phosphorylation. Transcriptional inactivation and destabilization | Mutants are ABA hypersensitive at both the seed germination and post-germination stages. | Ji et al. (2019) | |
| Ubiquitination | KEG | Degradation | Mutants show growth arrest and hypersensitivity to ABA and other ABA-related abiotic stresses. | Liu and Stone (2010) |
| CUL4, DWA1, DWA2 | Degradation | Mutants manifest delayed germination and ABA hypersensitive phenotypes. | Lee et al. (2010) | |
| AFP | Degradation | afp mutants are hypersensitive to ABA | Lopez-Molina et al. (2003) | |
| Sumoylation | SIZ1 | Inhibition of degradation | Mutants present ABA hypersensitivity for seed germination arrest and seedling primary root growth inhibition. | Miura et al. (2009) |
| S-nitrosylation | GSNO | Degradation | Germination promotion. | Albertos et al. (2015) |
RACK1 scaffold protein in ABA signaling
Scaffold proteins of mitogen-activated protein kinase (MAPK) cascades, such as Ste5 in yeast and KSR (Kinase Suppressor of Ras 1) in mammals, have been studied in depth. Similarly, RACK1 proteins were identified as MAPK scaffold proteins in Arabidopsis thaliana. These proteins present seven WD-repeat domains that confer the scaffolding properties to interact with other signaling proteins (Ullah et al., 2008; Adams et al., 2011). Different RACK1 functions have been described in plants, such as responses to abiotic and biotic stresses and protein synthesis (Guo et al., 2011). However, one of the more relevant roles of RACK1 in plants is its involvement in hormone responses and, more specifically, in the ABA signaling pathway (Chen et al., 2006; Guo and Chen, 2008).
There are three RACK1 proteins in A. thaliana, RACK1A, B, and C, and the three of them show unequal genetic redundancy, with RACK1A being indispensable (Guo and Chen, 2008). The loss-of-function rack1a single mutant as well as rack1a/b and rack1a/c double mutants are hypersensitive to ABA during seed germination, cotyledon greening, and root growth (Guo et al., 2009, 2011). ABA-responsive marker genes (i.e. RD29B and RAB18) are up-regulated in rack1a mutants, correlating with the hypersensitive phenotype to this hormone. Contrarily, overexpressing RACK1A resulted in reduced ABA sensitivity. Together, these data support the role of RACK1 as a negative regulator of ABA signaling.
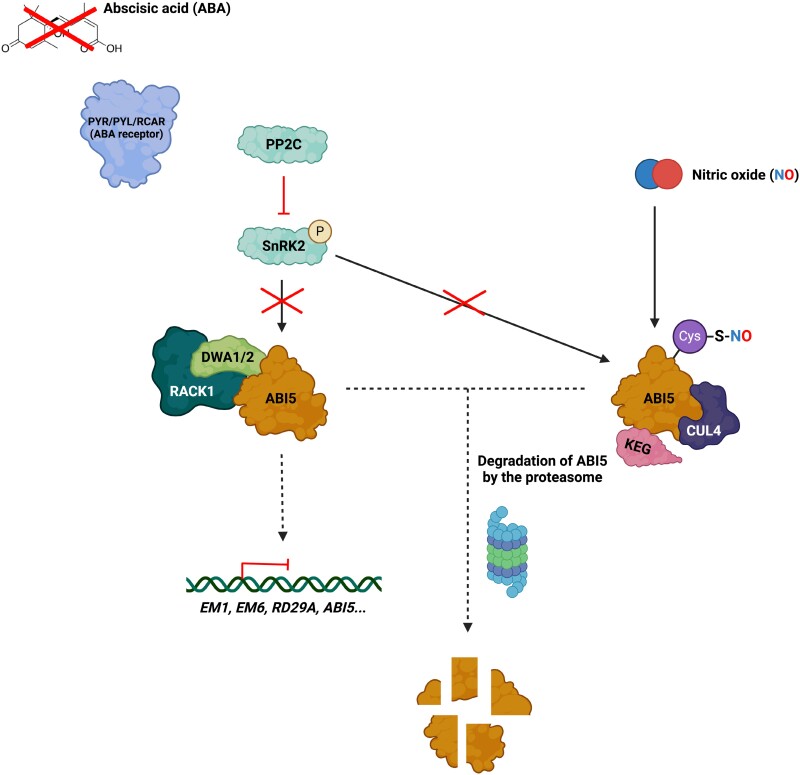
In their study, Li et al. (2024) demonstrated that RACK1 acts upstream of ABI5, a central hub of growth repression, and they observed the interaction between both proteins. Phosphorylation is key to ABI5 function, and Thr201 in ABI5 was identified as an important residue mediating the interaction between RACK1 and ABI5, as identified by bimolecular fluorescene complementation (BiFC) and co-immunoprecipitation (CoIP) assays using phosphomimic and phosphodead versions of ABI5. RACK1 was also observed to interact with DWA1 and DWA2, both having a role in ABI5 destabilization by the proteasome, confirming the key role of RACK1 in ABI5 degradation. However, this regulation occurs not only at the protein level but also at the transcriptional level, where a feedback regulatory loop includes RACK1 interaction with ABI5 to inhibit ABI5 expression and, on the other hand, ABI5 also has a role in the repression of RACK1 by direct binding to its promoter (Fig. 1). Indeed, transcription of ABI5 and RACK1 has an opposite pattern during seed germination, and in the presence of ABA RACK1 expression is highly reduced. Li et al. (2024) were able to identify a region of the RACK1 promoter containing G-box-like motifs that could be recognized by ABI5 in order to regulate its expression. This work, together with previous knowledge on the role of this scaffold protein in the ABA signaling pathway and, specifically, in how the complex of RACK1 and DWA1/2 regulates ABI5, provides a deeper understanding of one of the mechanisms through which this hormone regulates Arabidopsis seed germination (Fig. 2).
Fig. 2.
Scaffold proteins and their postulated role in ABA signaling. RACK1 recruits two substrate receptors for CUL4-based E3 ligases (DWA1 and DWA2) which function together to mediate the turnover of ABI5, thereby efficiently turning down ABA signaling downstream of the module of PYR/PYL/RCAR receptors, the PP2C-negative regulators- and the SnRK2-positive partners. Similarly, alterations modulating the ABI5 redox status include S-nitrosylation of specific Cys residues that enables the interaction to the E3 ubiquitin ligases KEG and CUL4 to promote ABI5 destabilization. Created with BioRender.com
Stepping forward
Scaffold proteins can bind multiple signaling molecules and are able to recruit molecular hubs within specific subcellular domains or modulate the efficiency of signal transduction. In doing so, scaffolds organize plant signaling pathways and ensure an efficient and specific flow of information, serving as regulatory targets. This modus operandi provides specific checkpoints to tailor the transcriptional control of plant development and stress conditions that could be translated into crop improvement in the agricultural field. Understanding how and when scaffolds control all these pathways could be used to achieve increases in crop stress tolerance and consequently crop production. The molecular changes of the interaction dynamics under stress would result in the identification of useful targets that could be manipulated to increase plant resistance to different stressors.
Furthermore, knowledge of the key protein residues involved in the interaction of scaffolds with different targets and the hierarchy that governs the different PTMs, could result in the fine-tuning of protein regulation by new genomic techniques, undergoing an innovation that can drive more sustainable, competitive, and resilient agriculture. There is no doubt that this study is just a stepping stone towards more exciting research to come.
Funding
The research of OL is supported by the projects PID2020-119731RB-I00 from the Spanish Ministry of Science, Innovation and Universities (MICIU/AEI), SA142P23 from the Regional Government of Castile and Leon, and ‘Escalera de Excelencia’ CLU-2018-04 co-funded by the FEDER Operative Program of Castile and Leon 2014–2020 Spain.
Contributor Information
Fátima Pollo-Rodríguez, Departamento de Botánica y Fisiología Vegetal, Instituto de Investigación en Agrobiotecnología (CIALE), Facultad de Biología, Universidad de Salamanca, C/ Río Duero 12, 37185 Salamanca, Spain.
Inmaculada Sánchez-Vicente, Departamento de Botánica y Fisiología Vegetal, Instituto de Investigación en Agrobiotecnología (CIALE), Facultad de Biología, Universidad de Salamanca, C/ Río Duero 12, 37185 Salamanca, Spain.
Oscar Lorenzo, Departamento de Botánica y Fisiología Vegetal, Instituto de Investigación en Agrobiotecnología (CIALE), Facultad de Biología, Universidad de Salamanca, C/ Río Duero 12, 37185 Salamanca, Spain.
Conflict of interest
The authors have no conflicts to declare.
References
- Adams DR, Ron D, Kiely PA.. 2011. RACK1, a multifaceted scaffolding protein: structure and function. Cell Communication and Signaling 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertos P, Romero-Puertas MC, Tatematsu K, Mateos I, Sánchez-Vicente I, Nambara E, Lorenzo O.. 2015. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nature Communications 6, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, JonesA M.. 2006. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. Journal of Experimental Botany 57, 2697–2708. [DOI] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y.. 2020. Abscisic acid dynamics, signaling, and functions in plants. Journal of Integrative Plant Biology 62, 25–54. [DOI] [PubMed] [Google Scholar]
- Dai M, Xue Q, Mccray T, et al. 2013. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. The Plant Cell 25, 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chen JG.. 2008. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biology 8, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG.. 2009. RACK1 is a negative regulator of ABA responses in Arabidopsis. Journal of Experimental Botany 60, 3819–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG.. 2011. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiology 155, 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Han X, Yang M, Zhang M, Pan J, Yu D.. 2019. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. The Plant Cell 31, 1520–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yu D.. 2014. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. The Plant Cell 26, 4394–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang S, Cheng C, Li R, Wang Z, Jenkins GI, Kong F, Li X.. 2019. The RCC1 family protein SAB1 negatively regulates ABI5 through multidimensional mechanisms during postgermination in Arabidopsis. New Phytologist 222, 907–922. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW.. 2010. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. The Plant Cell 22, 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang D, Liang X, Liang J.. 2024. Receptor for Activated C Kinase 1 counteracts ABSCISIC ACID INSENSITIVE5-mediated inhibition of seed germination and post-germinative growth in Arabidopsis. Journal of Experimental Botany 75, 3932–3945. [DOI] [PubMed] [Google Scholar]
- Liu H, Stone SL.. 2010. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. The Plant Cell 22, 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH.. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH.. 2003. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes & Development 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T, Erickson BJ, Finkelstein RR.. 2012. Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase (PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Molecular Biology 80, 647–658. [DOI] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM.. 2009. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA 106, 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, et al. 2009. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology 50, 1345–1363. [DOI] [PubMed] [Google Scholar]
- Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC.. 2008. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Science 17, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K.. 2000. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences, USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wan W, Jiang G, et al. 2019. Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Molecular Plant 12, 1598–1611. [DOI] [PubMed] [Google Scholar]