Abstract
Integrated into the sheep genome are 15 to 20 copies of type D endogenous loci that are highly related to two exogenous oncogenic viruses, jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus (ENTV). The exogenous viruses cause infectious neoplasms of the respiratory tract in small ruminants. In this study, we molecularly cloned three intact type D endogenous retroviruses of sheep (enJS56A1, enJS5F16, and enJS59A1; collectively called enJRSVs) and analyzed their genomic structures, their phylogenies with respect to their exogenous counterparts, their capacity to form viral particles, and the expression specificities of their long terminal repeats (LTRs). In addition, the pattern of expression of enJSRVs in vivo was studied by in situ hybridization. All of the three enJSRV proviruses had open reading frames for at least one of the structural genes. In particular, enJS56A1 had open reading frames for all structural genes, but it could not assemble viral particles when highly expressed in human 293T cells. We localized the defect for viral assembly in the first two-thirds of the gag gene by making a series of chimeras between enJS56A1 and the exogenous infectious molecular clone JSRV21. Phylogenetic analysis distinguished five ovine type D retroviruses: enJSRV groups A and B, ENTV, and two exogenous JSRV groups (African versus United Kingdom/North America isolates). Transient transfection assays indicated that the LTRs of the three enJSRVs were not preferentially active in differentiated lung epithelial cells. This suggests that the pulmonary tropic JSRV developed from a type D retrovirus that did not have lung specificity. Consistent with this, in situ hybridization of a panel of normal ovine tissues revealed high expression of enJSRV mRNA in the luminal epithelium and glandular epithelium of the uterus; lower expression was localized in the lamina propria of the gut and in the bronchiolar epithelium of the lungs.
The genomes of virtually all vertebrates, humans included, have been colonized during evolution by retroviruses via integration of their genomes into the germ line and subsequent fixation in the gene pool of the host population. These viruses, referred as endogenous retroviruses (ERV), are inherited by the host vertically in a Mendelian fashion. In contrast, exogenous retroviruses are horizontally transmitted and do not efficiently infect the germ line (6, 32, 50). Most ERVs are replication defective due to point mutations or deletions in their coding and/or regulatory regions; this presumably is necessary to avoid deleterious effects of replicating retroviruses in the hosts. The biological significance of the majority of ERVs is apparently minimal. On the other hand, some ERVs have maintained the capacity to express at least some of their genes and have either beneficial or detrimental effects. In mice, for example, expression during ontogeny of the superantigen by some endogenous mouse mammary tumor virus loci leads to clonal deletions of T cells required for successful infection by related exogenous and pathogenic mouse mammary tumor viruses (16, 24). The expression of some ERV proteins in mice and chickens can prevent infection by related exogenous viruses by receptor interference or postentry mechanisms (5, 29, 51, 56). In terms of deleterious effects, the generation of more pathogenic retroviruses can result from reactivation and/or recombination among different endogenous loci (19, 25, 60, 62, 65–67) or by recombination with related exogenous viruses (4, 17, 18, 20, 40, 55). This has been well documented in mice and selected lines of chickens. In humans, speculations on the pathogenic involvement of some endogenous loci in autoimmunity (39), testicular tumors (33, 64), or multiple sclerosis (52) have been reported, although there is no conclusive evidence to date.
Renewed interest in ERVs derives from the potential viral hazards associated with xenotransplantation or the use of retroviral vectors in gene therapy. There is concern about possible generation of pathogenic viruses originating (totally or partially) from ERVs (48, 49, 53, 61).
Sheep and goat genomes harbor 15 to 20 copies of endogenous type D retroviruses (22, 23, 70) highly related to two type D exogenous retroviruses that are oncogenic. The exogenous viruses, jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus (ENTV), cause, respectively, ovine pulmonary carcinoma (OPC) and enzootic nasal tumor in small ruminants (9, 12, 13, 44, 46, 70). At least some of the endogenous type D retroviruses (referred to as enJSRVs) are transcribed in a variety of sheep tissues (41, 59). Sequence analyses of the LTR, portions of env, and orf-x (2, 3, 41, 54) of some of the endogenous loci have been described and have been useful in distinguishing the exogenous and endogenous viruses. However, because all previous sequence data were obtained from PCR amplifications of portions of enJSRV proviruses, no information has been available on the total genomic structures of enJSRVs. Cloning and analysis of three intact enJSRV proviruses are described in this report. The results provided insight into the genomic structure of enJSRVs, their phylogeny, their relationship to exogenous JSRV and ENTV, the relative timing of insertion into the sheep genome, and the tissue specificity of exogenous versus enJSRV expression.
MATERIALS AND METHODS
Molecular cloning.
The construction of a lambda phage genomic DNA library from a sheep OPC lung tumor was described previously (46). The library was initially divided into 15 sublibraries, and each was independently amplified. Aliquots of the 15 sublibraries were screened for the presence of exogenous JSRV proviruses by using a JSRV U3-specific heminested PCR (45). The sublibraries negative for exogenous JSRV were further screened for the presence of enJSRVs. In particular, sublibraries 5 and 6 were plated onto bacterial agar plates and subjected to hybridization of plaque lifts with two 32P-labeled probes on replica filters: a JSRV gag-specific probe and an env-specific probe. Under the hybridization conditions used, these probes hybridized with both endogenous and exogenous JSRV sequences. Primary plaques positive for both probes were picked and further purified by dilution and plating for isolated plaques on bacterial lawns, followed by hybridization with both gag and env probes. The presence of exogenous JSRV was ruled out by exogenous LTR-specific PCR and by the lack of an exogenous JSRV-specific ScaI restriction site in gag. Three recombinant phages carrying distinct enJSRV loci were subcloned into pBlueScript (Stratagene) to give penJS56A1, penJS59A1, and penJS51F6. Both strands of the three clones were completely sequenced on an ABI Prism 310 genetic analyzer (Perkin-Elmer), using a BigDye Terminator DNA cycle sequencing kit (PE Applied Biosystems) as recommended by the manufacturer.
Computer analysis of sequence data.
Sequences were analyzed using the DNASTAR 1.59 software package (DNASTAR, Inc.) and DNA Strider 1.2 (37). Sequences alignments were performed using ClustalW 1.8 (63). Phylogenetic analysis calculating the genetic distances between sequence pairs was carried out by the DNADIST program in PHYLIP version 3.5 (15). Neighbor-joining trees were estimated by use of the NEIGHBOR program; bootstrap analyses used 1,000 bootstrap replications (14).
Plasmids.
Plasmid pCMV2JS21 is a construct derived from the JSRV21 infectious molecular clone where the viral genes are under the control of the cytomegalovirus (CMV) immediate-early promoter (46). Plasmid pCMV2en56A1 was derived by replacing the 5′ LTR of penJS56A1 with the CMV promoter and JSRV R and U5 of pCMV2JS21 by standard molecular cloning techniques (57). Chimeric constructs between pCMV2JS21 and pCMV2en56A1 were obtained taking advantage of the common HpaI and BamHI restriction sites in gag (position 1274 of JSRV21) and at the end of pol (position 5265 of JSRV21, 135 bp before the end of the pol reading frame and 56 bp before the start codon of env). Plasmid pGPxEe has gag and pol of pCMV2JS21 and env from pCMV2en56A1, while plasmid pGPeEx has the gag and pol from pCMV2en56A1 and env from pCMV2JS21. Plasmid pGePEx has the majority of gag from pCMVen56A1 and pol and env from pCMV2JS21, while pGxPEe has the first two-thirds of gag from the exogenous pCMV2JS21 and the rest of the genome from pCMVen56A1.
The LTRs of penJS5F16, penJS56A1, and penJS59A1 were cloned into pGL3-basic (Promega) by standard PCR cloning techniques. The resulting plasmids (pen5F16-luc, pen56A1-luc, and pen59A1-luc) had the firefly luciferase gene under the control of the various endogenous LTRs. pJS21-luc contains the LTR of JSRV21 which drives the luciferase gene (42). pRL-null (Promega), a promoterless plasmid with the Renilla luciferase gene, was used to correct for transfection efficiency by cotransfection with the LTR reporter plasmids as described below.
Plasmids pCMV-HNF3α and pCMV-HNF3β, expressing hepatocyte nuclear factors 3α and -β (HNF-3α and -β; provided by R. H. Costa, University of Illinois, Chicago) were used in transactivation experiments.
Cell cultures.
MLE-15 (69), a mouse type II pneumocyte-derived cell line (provided by J. Whitsett), was grown in RPMI 1640 (Gibco BRL)–2% fetal bovine serum (FBS)–0.5% ITS (Sigma) modified with the addition of 5 mg of transferrin per liter, 10 mM HEPES, 10−8 M β-estradiol, and 10−8 M hydrocortisone. Human 293T cells (30), mtCC1-2 cells (34) (derived from mouse Clara cells and provided by F. DeMayo), and NIH 3T3 cells (ATCC [American Type Culture Collection] CCL-92) were grown in Dulbecco modified Eagle medium (DMEM; ATCC)–10% FBS. TCMK cells (derived from mouse kidney; ATCC CCL-139) were grown in DMEM (ATCC)–1× nonessential amino acids (Cellgro)–10% FBS. The ovine uterine endometrial LE cell line (27) was grown in F12-K (Gibco BRL)–10% FBS. All cell lines were grown in an incubator at 37°C with 5% CO2.
Transient transfections and luciferase assays.
Transient transfections were performed on 2 × 105 to 4 × 105 cells on six-well plates (Falcon) approximately 24 h before transfection. For each well, 500 ng of reporter plasmid and 50 ng of pRL-null were used with 6 μl of Fugene (Boehringer) as recommended by the manufacturers. Experiments were done in six replicates in at least two independent experiments. Cells were lysed 48 h after transfection and analyzed using the Promega dual luciferase reporter system protocol in a TD 20/20 luminometer (Turner Design) as recommended by the manufacturer. Values for the various endogenous LTR reporters were compared to the activity of pJS21-luc, which was taken as 100%.
For transactivation experiments, 200 ng of pJS21-luc, 1 to 200 ng of transactivating plasmid (or control plasmid containing the same promoter as the transactivating plasmid), and 50 ng of pRL-null were used in NIH 3T3 cells. The activation of JS21-luc by HNF-3 expression plasmids was calculated by comparing the relative activity of pJS21-luc cotransfected with either pCMVHNF-3α (or pCMVHNF-3β) or a plasmid with the CMV promoter alone. Transfection efficiencies were normalized as above, using pRL-null. For the production of viral particles, 293T cells were transfected with pCMV2JS21 or pCMV2en56A1 (or the various chimeras), and viral particles were collected from concentrated supernatants as previously described (46).
Western blotting.
Western blotting of concentrated 293T supernatants for the detection of JSRV major capsid (CA) protein was performed as already described (46).
Tissue samples.
Tissue samples used for in situ hybridizations were collected during the necroscopy of a healthy sheep. Tissues analyzed were lungs, liver, kidney, spleen, uterus, intestine/jejunal Peyer's patches, mediastinal lymph nodes, precrural lymph nodes, and jejunal lymph nodes. Samples were fixed in 10% neutral buffered formalin, processed routinely in an automatic tissue processor, embedded in paraffin wax, and sectioned (5 to 7 μm).
In situ hybridization.
In situ hybridization was performed essentially as previously described (58, 59). Deparaffinized, rehydrated, and deproteinated tissue sections were hybridized with radiolabeled antisense or sense cRNA probe generated from linearized plasmid template (DD54 [59]) by in vitro transcription with [α-35S]UTP (3,000 Ci/nmol; Amersham-Pharmacia). DD54 contains 436 bp from the env region of an enJSRV and is 96 to 98% identical to enJS56A1 and enJS5F16. Autoradiographs of slides were prepared using Kodak NTB-2 liquid photographic emulsion. Slides were kept at 4°C for 1 week, developed in Kodak D-19 developer, counterstained with Harris' modified hematoxylin in acetic acid (Fisher), dehydrated through a graded series of alcohol to xylene, and coverslipped. Photomicrographs were taken under bright-field and dark-field illumination using a Carl Zeiss Axioplan2 photomicroscope fitted with a Hamamatsu chilled 3CCD color camera.
Nucleotide sequences accession numbers.
Sequences of enJS56A1, enJS5F16, and enJS59A1 have been deposited in GenBank with accession numbers AF153615, AF136224, and AF136225.
RESULTS
enJSRV proviral structures and comparison with exogenous type D retroviruses.
We screened a lambda phage library from a sheep with JSRV-induced OPC and obtained three complete endogenous proviral loci termed enJS56A1, enJS5F16, and enJS59A1. The lengths of the proviruses were 6,915 bp for enJS5F16, 7,939 bp for enJS56A1, and 6,695 bp for enJS59A1; their genomic structures are schematically shown in Fig. 1.
FIG. 1.
Genomic structures of endogenous and exogenous type D retroviruses of sheep. Premature stop codons are indicated by a vertical bar underlined by an asterisk. For convenience, the gag open reading frame has been fixed in the same reading frame of all sequences shown. The numbered bar at the bottom indicates distances in kilobases. The exogenous JSRV and ENTV show the canonical retroviral gag, pro, pol, and env with pro in a different open reading frame from pol, the same for all type D and B retroviruses. An additional open reading frame (orf-x) overlapping pol is present in JSRV but is interrupted by two stop codons in ENTV (8). enJS56A1 is the only one of the three endogenous proviruses cloned in this study to maintain full (or nearly full) open reading frames in all structural genes. enJS59A1 has premature stop codons in gag and pol and a major deletion in env. enJS5F16 has a deletion in pol. Different peptide sequences at the 3′ end of the pol gene in enJS56A1 due to a frameshift are indicated by cross-hatching. The LTRs are indicated by solid boxes.
(i) LTRs.
All three endogenous proviral loci had an upstream and a downstream LTR, the hallmark of complete proviruses. In the U3 region of the LTR there were major differences with respect to the exogenous JSRV, as previously reported (3, 41). The U3 regions of the endogenous loci were longer than those of the exogenous JSRV and ENTV. The U3 of the endogenous loci varied between 301 (enJS59A1) and 319 (enJS56A1 and enJS5F16) bp, while that of the exogenous JSRV21 is 266 bp (46, 70) or, for ENTV, only 250 bp (9). The U3 regions of enJS5F16 and enJS56A1 were 98% identical, and they had 85% sequence identity with respect to enJS59A1. The endogenous loci showed approximately 74% sequence identity with respect to the JSRV21 U3, while R and U5 were highly homologous among the endogenous loci and with respect to JSRV21 (92 to 96% identity). The upstream and downstream LTRs of enJS5F16 were identical, while those of enJS56A1 and enJS59A1 displayed two- and four-base changes respectively (see below).
(ii) gag.
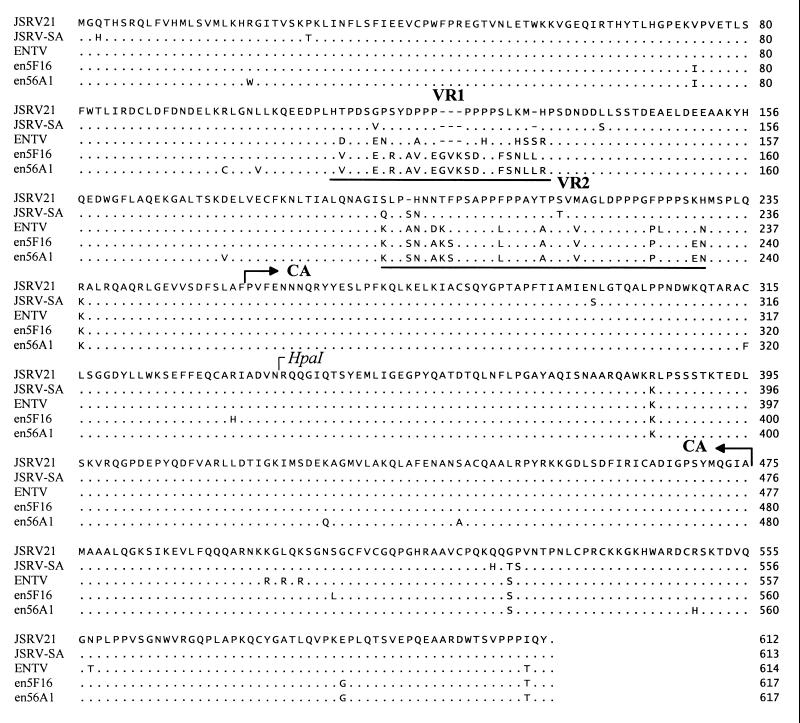
All three endogenous loci had a conserved tRNA1,2Lys primer binding site, the same used by exogenous JSRV and ENTV (9, 46, 70). The gag gene had an intact open reading frame in enJS56A1 and enJS5F16, while a 1-bp insertion created a frameshift with a termination codon at position 820 in enJS59A1 provirus. The whole Gag predicted polyprotein was 98.2% identical in the endogenous clones and was highly conserved between endogenous and exogenous viruses (94 to 95% identity), with the exception of a short region corresponding to the predicted matrix of JSRV21 (nucleotides 624 to 661). This region showed a proline-rich motif in the exogenous JSRV and ENTV, but there was no meaningful protein sequence similarity with the endogenous clones (Fig. 2). Interestingly, this region also showed polymorphism between JSRV and ENTV in that there was only 50% identity at the amino acid level compared to the 95.8% identity for the entire Gag polyprotein. We termed this region VR1 (variable region 1) for the type D retroviruses of sheep. Downstream of VR1 (50 amino acid residues) there was another region of polymorphism between endogenous and exogenous viruses; we termed this region VR2. This region was also relatively proline rich for both endogenous and exogenous viruses. It was interesting that ENTV showed a closer relationship to the endogenous loci in VR2 than to the exogenous JSRV.
FIG. 2.
Alignment of deduced gag amino acid sequences of sheep type D retroviruses: exogenous JSRV21 (AF105220), JSRV-SA (M80216), and ENTV (Y16627) and the endogenous enJS5F16 and enJS56A1 that maintain full-length gag open reading frames. Dots refer to identical sequences, while dashes indicate lack of sequence. VR1 and VR2 are underlined. Note that the prolines in VR1 of the exogenous JSRVs and ENTV are absent in the endogenous proviruses. In VR2, ENTV is more similar to enJSRVs than JSRV. The putative CA region and the HpaI site used to generate exogenous-endogenous chimeras (Fig. 4) are indicated.
(iii) pro.
The pro region showed an uninterrupted open reading frame for all three endogenous clones and was highly homologous for all of them. The endogenous clones and the exogenous JSRV21 showed very high homology in this region (95 to 99.7% amino acid identity). The dUTPase motifs found in the 5′ half of the pro gene (70) of JSRV are conserved in the endogenous loci.
(iv) pol.
pol showed major defects in enJS5F16 and enJS59A1: in enJS5F16 there were two large deletions of 154 and 872 bp, while a point mutation in enJS59A1 created a stop codon (position 4071 of the provirus sequence) in the reverse transcriptase domain. In enJS56A1 there was a 2-bp deletion with respect to exogenous JSRV at the 3′ end of the pol gene (corresponding to the end of the integrase [IN] domain) that would yield a polypeptide 14 aa shorter respect the native JSRV IN, with the last 33 aa having no similarity due to the frameshift. Besides this difference, the Pol polyproteins of enJS56A1 and JSRV21 were 97.8% identical.
(v) orf-x.
The orf-x region (an alternate reading frame in pol) was uninterrupted in enJS59A1. In enJS5F16 there was a major orf-x truncation as a consequence of the deletion in pol, while in enJS56A1 there was a stop codon 39 bp before the usual stop codon in JSRV21 orf-x.
(vi) env.
The env gene was deleted in enJS59A1 but was a fully open reading frame in enJS5F16 and enJS56A1. This region was 98% identical at the amino acid level between the two endogenous loci and ca. 92% identical between endogenous and exogenous JSRV21 sequences. In the last 67 aa of Env (in the transmembrane region) there was another region of high divergence between endogenous and exogenous sequences (57 to 59% amino acid identity). This region also was shown to be highly variable between JSRV type 1 (composed of isolates from the African continent) and JSRV type 2 (from the United Kingdom and United States) sequences (2). We termed this region VR3. VR3 was also highly variable between exogenous JSRV and ENTV sequences (9) (Fig. 3).
FIG. 3.
Alignment of deduced env amino acid sequences of sheep type D endogenous retroviruses: exogenous JSRV21, JSRV-SA, and ENTV and the endogenous proviruses (enJS5F16 and enJS56A1) that maintain an open reading frame along the entirety of env. The boundary between the surface (SU) and transmembrane (TM) regions is indicated. VR3 is underlined; note the polymorphism between all sequences in VR3.
enJS56A1 does not produce viral particles.
Of the three endogenous loci cloned in this study, enJS56A1 had uninterrupted open reading frames in all of the structural genes. The only coding defects of enJS56A1 were a premature stop codon in orf-x and a frameshift leading to the last 33 aa of IN with no homology with the exogenous amino acid sequence. The orf-x is not necessary for viral particle formation and infectivity in vitro (M. Palmarini and H. Fan, unpublished results). It therefore seemed possible that this provirus could encode virus particles.
To test whether enJS56A1 had the potential to express viral particles, we generated a construct where transcription is driven by the CMV immediate-early promoter (termed pCMV2en56A1) (Fig. 4). In pCMV2en56A1, the upstream LTR of penJS56A1 was replaced with the CMV promoter and the JSRV21 R and U5 regions (from pCMV2JS21) (46). pCMV2JS21, a derivative of pJSRV21 where the CMV immediate-early promoter drives JSRV transcription, has been a useful tool to produce JSRV infectious virus in vitro by transiently transfecting 293T cells and collecting viral particles in the resulting supernatant (46, 47). In this study we transfected 293T cells in parallel with pCMV2JS21 and with pCMV2en56A1 and harvested the supernatant at 24, 48, and 72 h posttransfection. Viral particles harvested from the resultant pools were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using a rabbit antiserum toward the CA protein of JSRV (43). We observed a CA-specific band of 26 kDa in the concentrated supernatants of pCMV2JS21-transfected cells, as expected, but no band was detected in the pCMV2en56A1 supernatants (Fig. 4B). This indicated that enJS56A1 was unable to form virus particles. To localize the region(s) responsible for this defect, we made chimeric constructs between pCMV2JS21 and pCMV2en56A1 (Fig. 4A). pGPxEe had gag and the majority of pol from exogenous JSRV21 and the 3′ 180 bp of pol and the entire env from enJS56A1; pGPeEx was the opposite chimera, with gag and the majority of pol from the endogenous locus and env from JSRV21. pGePEx had the first two-thirds of gag from the endogenous enJS56A1 and the rest of the genome from pCMV2JS21; pGxPEe was the opposite chimera, with exogenous gag and endogenous pol and env. pGPxEe was able to produce viral particles (Fig. 4B); the defect for viral production was therefore not due to the frameshift of the 3′ portion of the pol open reading frame that is after the BamHI site used to make this chimera. Conversely, neither pGPeEx or pGePEx was able to produce viral particles, while pGxPEe did produce viral particles. The defect for particle formation is therefore localized in the first two-thirds of gag, upstream of the HpaI site (position 1274 in JSRV21); interestingly, this region contains the gag VR1 and VR2. However, single amino acid changes outside VR1 and VR2 or polymorphism in the untranslated gag (Fig. 5) might also determine the assembly defect of enJS56A1.
FIG. 4.
Virus production by endogenous-exogenous chimeras. (A) Schematic structure of the parental (JSRV21 and enJS56A1) and chimeric plasmids. The restriction enzyme sites used for the cloning are indicated. In pCMV2JS21 and in various chimeric constructs, expression is driven by the CMV immediate-early promoter (indicated by the arrow). (B) Western blot of 300-fold-concentrated supernatant from equal numbers of 293T cells transiently transfected with the constructs shown in panel A. Lung secretions collected from an OPC-affected animal were used as a positive control (LF), while mock-transfected 293T supernatants were used as negative controls. The filters were incubated with a rabbit antiserum against the capsid (CA) protein of JSRV (43). The 26-kDa CA protein is indicated.
FIG. 5.
Nucleotide sequence alignment of the 5′ untranslated regions of sheep type D retroviruses: exogenous JSRV21, JSRV-SA, and ENTV and the endogenous enJS5F16, enJS59A1, and enJS56A1 proviruses. The primer binding site (PBS) is underlined.
Phylogenetic analysis and evolution.
We generated unrooted neighbor-joining phylogenetic trees to assess the phylogenetic relationships between the three endogenous loci cloned in this study and with other known sequences of endogenous and exogenous type D retroviruses of sheep (2, 3, 9, 41, 46). We generated a tree for the U3 region, one for env and one for gag and pol (Fig. 6). In each tree it was possible to distinguish three major branches: one for the endogenous loci, one for the exogenous ENTV sequence, and one for the exogenous JSRV sequence, confirming previous analyses with limited gag sequences (10). The exogenous JSRVs could be further divided into two branches corresponding to sequences derived from Africa or from the United States and the United Kingdom as previously described (2, 3). In all of the generated trees, the enJS59A1 loci branched apart from the other two endogenous loci cloned in this study as well as from most of the previous endogenous sequences isolated by PCR cloning.
FIG. 6.
Phylogenetic analysis of sheep type D retroviruses of sheep. Unrooted phylogenetic trees for U3 (A), env (B), and gag and pol (C) were derived by neighbor joining. To show consistency, all bootstrap values obtained with 1,000 replications of bootstrap sampling are shown. Sequences used for the analysis are termed as in their original references with the exception of loci 1 to 6, which are indicated as L1 to L6 in panel A. GenBank accession numbers: AF105220 (JSRV21); M80216 (JSRV-SA); X95445-X95452 (endogenous loci 1 to 6 and exogenous type I and II LTRs); Y16627 (ENTV); Y18301 to Y18305 (JS7, 809T, 83RS28, and 92K3); Z66531 to Z66533 (enJSRV1 to -3); Z71304 (LTR-UK); (AF136224) enJS5F16; (AF136225) enJS59A1; AF153615 (enJS56A1). In all trees there are five distinct phylogenetic groups: enJSRV-A and -B for the endogenous loci; and the ENTV group and two groups for exogenous JSRV, JSRV-I (African isolates), and JSRV-II (isolates from the United States and United Kingdom).
The endogenous type D retrovirus loci seemed to be quite young from the evolutionary point of view. An estimate of the time of integration in the sheep germ line of these elements could be made by taking into account the variability between 5′ and 3′ LTRs of a single locus. The intragenomic differences would reflect these changes that have accumulated since the integration into the sheep germ line, as these LTRs presumably were identical at the time of integration. The LTRs would presumably mutate at the general rate of noncoding sequences and pseudogenes. Thus, intragenomic variability of the LTRs can be used as a molecular clock to estimate time of integration (11, 35, 38). By this analysis, the integration events of enJS56A1 and enJS59A1 happened ca. 0.9 to 1.8 million years ago, calculated from an average value of 4.85 × 10−9 substitutions per nucleotide site per year relative to pseudogenes (31). enJS5F16 might have integrated less than 500,000 years ago, based on sequence identity of the upstream and downstream LTRs. These numbers are subject to a wide margin of error (1) and do not take into account the possibility of gene conversion (28).
From the constructed trees (Fig. 6), we could divide the endogenous loci into at least two phylogenetic groups, enJSRV-A and -B. Another one or two groups might arise (e.g., loci 5 and 6 might form a group separate from enJSRV-A), but complete proviral sequences need to be obtained in order to fully classify these elements.
Expression of enJSRVs in vivo.
To evaluate the expression of enJSRVs in vivo, we performed in situ hybridization to a panel of tissues collected from healthy sheep. We used the DD54 probe, which contains 436 bp of the env gene that is 96 to 98% identical to enJS56A1 and enJS5F16 env. DD54 was previously identified in a study designed to isolate mRNAs differentially expressed in the endometrial lumena (LE) and glandular epithelia (GE) of the ovine uterus (59). Indeed, we detected a very strong hybridization signal in the endometrial LE and GE of the ovine uterus (Fig. 7A to C). Cells in the lamina propria of the gut also showed some specific mRNA expression (Fig. 7D to F), and low levels of mRNA expression was detected in the bronchiolar epithelium of the lungs (Fig. 7G to I). The alveolar epithelium did not show signal above background. Very weak or no signals above background were detected in the liver (Fig. 7J to L), kidney, spleen, tonsils, and peripheral lymph nodes (data not shown).
FIG. 7.
Expression of enJSRVs in vivo. In situ localization of enJSRV env mRNA in selected sheep tissues. Cross-sections of sheep tissues were hybridized with an α-35S-labeled antisense or sense DD54 cRNA probes. Hybridized sections were digested with RNase, and protected transcripts were visualized by liquid emulsion autoradiography. Developed slides were counterstained lightly with hematoxylin, and photomicrographs taken under bright-field or dark-field illumination. (A to C) Day 11 cyclic ovine uterus; (D to F) intestine; (G to I) lung; (J to L) liver. Shown are bright-field exposures (A, D, G, and J); hybridization with antisense probes, dark-field exposures (B, E, H, and K); and hybridizations with sense probes, dark-field exposures (C, F, I, and L). All photomicrographs are shown at a magnification of ×600.
enJSRV LTRs do not show pulmonary tropism and are not transactivated by HNF-3.
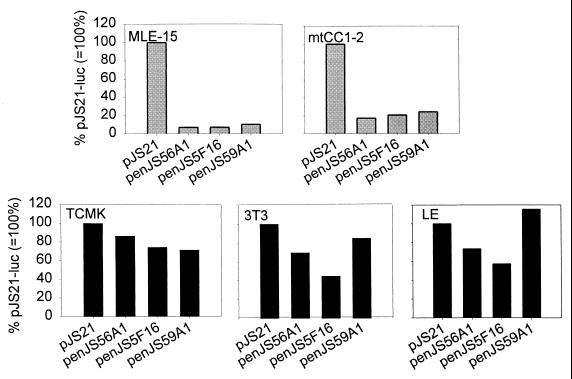
Recently we have shown that the LTR of JSRV is preferentially transcriptionally active in mouse cell lines derived from differentiated epithelial cells of the lungs (type II pneumocytes and Clara cells) (42). To assess whether the pulmonary tropism was common to exogenous and endogenous viruses, we performed reporter assays with luciferase-expressing constructs driven by the exogenous JSRV21 LTR (pJS21-luc) or by the LTR of each of the three endogenous loci (enJS56A1-luc, enJS5F16-luc, and enJS59A1-luc). Results were expressed as percentage of the luciferase activity of pJS21-luc after adjustment for transfection efficiency (measured by Renilla luciferase values induced by a cotransfected reporter plasmid [pRL-null]).
We performed the experiments in five different cell lines: MLE-15 (a mouse cell line derived from type II pneumocytes), mtCC1-2 (derived from mouse Clara cells), TCMK (derived from mouse kidney), NIH 3T3 (mouse embryo fibroblasts), and LE (a sheep cell line derived from the uterine endometrial epithelium). In a previous study (42) we showed that JS21-luc had the highest relative luciferase activity in MLE-15 and mtCC1-2; NIH 3T3 had an intermediate level of JS21-luc expression, while TCMK had low expression. The LE cells were chosen because of a previous report that enJSRV transcripts were found in endometrial epithelium (59). As shown in Fig. 8, the endogenous LTRs had a much lower luciferase activity compared to pJS21-luc in the lung-derived cell lines MLE-15 and mtCC1-2, ranging from 7 to 11% in MLE-15 cells and from 17 to 24% in mtCC1-2 cells. In contrast, in TCMK, 3T3, and LE cells, the activities of the endogenous LTR clones were much more comparable (45 to 115%) to that of pJS21-luc. These results suggested that the JSRV-like exogenous virus that infected the sheep germ line to give the enJSRV elements did not have preferential tropism for differentiated epithelial cells of the lungs and that the current lung tropism of the exogenous JSRV arose relatively recently.
FIG. 8.
enJSRV LTR transcriptional activity. Plasmids penJS56A1-luc, penJS5F16-luc, and penJS59A1-luc were transfected into various cell lines as described in Materials and Methods. Cell lines were derived from mouse differentiated lung epithelial cells (MLE-15 and mtCC1-2) and extrapulmonary tissues such as mouse fibroblasts (NIH 3T3), mouse kidney (TCMK), and sheep endometrium (LE). Luciferase activities of the various endogenous locus LTRs as percentages of the activity of pJS21-luc, a reporter plasmid driven by the JSRV21 LTR, are shown (average of 6 to 12 replicates).
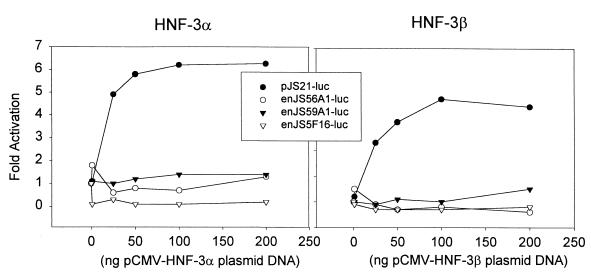
In addition, we tested whether the LTRs of the endogenous clones could be transactivated by HNF-3. HNF-3 is a transcription factor that has been shown to play a major role in lung-specific transcription (7, 21, 36, 68). We have shown that the JSRV LTR has two potential HNF-3-responsive element and that this LTR can be transactivated in 3T3 cells by coexpression of HNF-3α or -β (42). In the transactivation experiments shown in Fig. 9, none of the endogenous LTR clones responded to cotransfection with HNF-3α or HNF-3β expression plasmids, while the exogenous JSRV was activated by both HNF-3α and -β as expected. The lack of response to HNF-3 strengthened the hypothesis that the enJSRV elements do not show the pulmonary tropism exhibited by JSRV.
FIG. 9.
Effects of HNF-3α and HNF-3β on enJSRV LTRs. pJS21-luc, penJS56A1-luc, penJS5F16-luc, and penJS59A1-luc were cotransfected into NIH 3T3 cells (that do not efficiently support JSRV enhancer activity) along with an expression plasmid for either HNF-3α or HNF-3β. Different amounts of the transcription factor expression plasmids were cotransfected with a set amount (200 ng) of the reporter plasmid DNA. The amounts of luciferase activity for the different cotransfections are shown as fold activation over the activity of reporter plasmid cotransfected with a plasmid having the CMV promoter but not HNF-3 insert.
DISCUSSION
In this study we molecularly cloned three enJSRVs and investigated their proviral structure, phylogeny, and pattern of expression. All three proviruses contained open reading frames for one or more structural genes. In particular, enJS56A1 was a virtually full-length provirus, with open reading frames for gag, most of pol, and env. However enJS56A1 was unable to make viral particles even when it was highly expressed. By construction of viral chimeras between exogenous JSRV21 and enJS56A1, we identified the first two-thirds of gag of enJS56A1 as the region where the main defect for particle formation lies. Also, we identified two short regions in gag (VR1 and VR2) containing major differences between ovine endogenous and exogenous type D retroviral proteins. In particular, VR1 contains a proline-rich region in both JSRV and ENTV that is absent in the endogenous proviruses. A third region that is divergent between exogenous and endogenous sequences was located in the carboxy-terminal portion of the transmembrane protein (that we termed VR3) and was previously reported (2, 9). Interestingly, in these variable regions there is also polymorphism between JSRV and ENTV. In the future, it will be interesting to investigate the influence of VR1, VR2, and VR3 on the replication or pathogenicity of the oncogenic exogenous viruses.
With the exception of these three variable regions, the endogenous proviruses were remarkably similar in protein-coding sequences to their exogenous counterparts (except for deletions). However, a strong polymorphism was localized in the U3 region of the LTR (3, 41) where the retroviral promoter and enhancers are located. We have recently shown that the exogenous JSRV LTR is a main determinant of viral tropism for the differentiated epithelial cells of the lungs (42). By in situ hybridization, we have shown that the strongest expression of enJSRVs seems to be in the LE and GE of the uterus. Weaker expression was detected in the lamina propria of the gut and in the bronchiolar epithelium of the lung, but no signal above background was detected in the alveolar epithelium. In a previous study, however, low levels of enJSRV expression were detected by sensitive reverse transcription-PCR assays in several sheep tissues of different origins (41). The tissue-specific pattern of enJSRV expression could reflect tissue specificity of the enJSRV LTRs, at least for those enJSRVs that are expressed. However, it is also possible that expression (or nonexpression) of different enJSRVs is determined by the host cell sequences surrounding the integrated enJSRV proviruses. This is a general question for all endogenous retroviruses. We used transient transfections and reporter assays to determine that the LTRs of the three cloned endogenous proviruses do not exhibit the same lung specificity as exogenous JSRV. The most likely explanation is that JSRV became lung tropic during its evolution as an exogenous virus, but that its progenitor (more likely to resemble enJSRVs) did not originally have strong pulmonary tropism. Consistent with this, the LTRs of the various enJSRVs were not transactivated by HNF-3, a transcription factor involved in lung-specific gene expression and capable of transactivating the JSRV21 LTR in 3T3 cells (7, 21, 36, 68). However, the presence of a specific enJSRV that gave rise to the exogenous JSRV and that is expressed primarily in differentiated lung epithelial cells cannot be excluded.
Based on analysis of the variability between 5′ and 3′ LTRs of the same locus, we estimated that enJS56A1 and enJS59A1 were integrated into the sheep genome between 0.9 million and 1.8 million years ago, while enJS5F16 might have integrated less than 500,000 years ago. This estimation, though subject to high variability, is in general agreement with the conclusions of a previous study based on different observations (23). Hecht et al. showed that sheep (and wild members of the genus Ovis) and goats (and wild members of the genus Capra) both have approximately 20 copies of endogenous type D-related retroviruses. Moreover, the restriction endonuclease profiles of these elements are different between the two genera but are similar among members of the same genus. Thus, most of the enJSRV loci were acquired after the divergence between sheep and goats (4 million to 10 million years ago) (26). Thus, the enJSRVs are rather young from the evolutionary point of view and can be considered “modern” endogenous retroviruses (8); the existence of the closely related exogenous JSRV and ENTV is also consistent with these elements being evolutionarily young (6).
ACKNOWLEDGMENTS
Massimo Palmarini and Claus Hallwirth contributed equally to this work.
We are grateful to Reza Omid for tissue culture work, to Lorenzo Gonzales for providing pathological material, and to Marcelo de las Heras and Mike Sharp for useful discussions.
M.P. was a recipient of an American Cancer Society Ray and Estelle Spehar fellowship. This work was supported in part by NIH grant RO1CA82564 and by funds from the South African National Research Foundation and the Ondersterpoort Veterinary Institute. Support from the UCI Cancer Research Institute and the DNA sequencing core of the Chao Family Comprehensive Cancer Center is acknowledged.
REFERENCES
- 1.Avise J. Molecular markers, natural history and evolution. New York, N.Y: Chapman & Hall; 1994. [Google Scholar]
- 2.Bai J, Bishop J V, Carlson J O, DeMartini J C. Sequence comparison of JSRV with endogenous proviruses: envelope genotypes and a novel ORF with similarity to a G-protein-coupled receptor. Virology. 1999;258:333–343. doi: 10.1006/viro.1999.9728. [DOI] [PubMed] [Google Scholar]
- 3.Bai J, Zhu R Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson S J, Ruis B L, Fadly A M, Conklin K F. The unique envelope gene of the subgroup J avian leukosis virus derives from ev/J proviruses, a novel family of avian endogenous viruses. J Virol. 1998;72:10157–10164. doi: 10.1128/jvi.72.12.10157-10164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 7.Bruno M D, Bohinski R J, Huelsman K M, Whitsett J A, Korfhagen T R. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- 8.Coffin J M. Retroviridae and their replication. In: Fields B N, Knipe D M, editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 1437–1500. [Google Scholar]
- 9.Cousens C, Minguijon E, Dalziel R G, Ortin A, Garcia M, Park J, Gonzalez L, Sharp J M, de las Heras M. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J Virol. 1999;73:3986–3993. doi: 10.1128/jvi.73.5.3986-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cousens C, Minguijon E, Garcia M, Ferrer L M, Dalziel R G, Palmarini M, de las Heras M, Sharp J M. PCR-based detection and partial characterization of a retrovirus associated with contagious intranasal tumors of sheep and goats. J Virol. 1996;70:7580–7583. doi: 10.1128/jvi.70.11.7580-7583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dangel A W, Baker B J, Mendoza A R, Yu C Y. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics. 1995;42:41–52. doi: 10.1007/BF00164986. [DOI] [PubMed] [Google Scholar]
- 12.de las Heras M, Sharp J M, Ferrer L M, Garcia de Jalon J A, Cebrian L M. Evidence for a type D-like retrovirus in enzootic nasal tumour of sheep. Vet Rec. 1993;132:441. doi: 10.1136/vr.132.17.441. [DOI] [PubMed] [Google Scholar]
- 13.DeMartini J C, York D F. Retrovirus-associated neoplasms of the respiratory system of sheep and goats. Ovine pulmonary carcinoma and enzootic nasal tumor. Vet Clin North Am Food Anim Pract. 1997;13:55–70. doi: 10.1016/s0749-0720(15)30364-9. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP: phylogeny inference package, 3.5 ed. Seattle: University of Washington; 1995. [Google Scholar]
- 16.Golovkina T V, Chervonsky A, Dudley J P, Ross S R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 17.Golovkina T V, Jaffe A B, Ross S R. Coexpression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J Virol. 1994;68:5019–5026. doi: 10.1128/jvi.68.8.5019-5026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovkina T V, Piazzon I, Nepomnaschy I, Buggiano V, de Olano Vela M, Ross S R. Generation of a tumorigenic milk-borne mouse mammary tumor virus by recombination between endogenous and exogenous viruses. J Virol. 1997;71:3895–3903. doi: 10.1128/jvi.71.5.3895-3903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golovkina T V, Prakash O, Ross S R. Endogenous mouse mammary tumor virus Mtv-17 is involved in Mtv-2-induced tumorigenesis in GR mice. Virology. 1996;218:14–22. doi: 10.1006/viro.1996.0161. [DOI] [PubMed] [Google Scholar]
- 20.Hartley J W, Wolford N K, Old L J, Rowe W P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay J G, Crystal R G. Lung-specific gene expression. In: Crystal R G, West J B, Weibel E R, Barnes P J, editors. The lung: scientific foundations. 2nd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 277–304. [Google Scholar]
- 22.Hecht S J, Carlson J O, DeMartini J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and unaffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 23.Hecht S J, Stedman K E, Carlson J O, DeMartini J C. Distribution of endogenous type B and type D sheep retrovirus sequences in ungulates and other mammals. Proc Natl Acad Sci USA. 1996;93:3297–3302. doi: 10.1073/pnas.93.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Held W, Waanders G A, Shakhov A N, Scarpellino L, Acha-Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 25.Imai S, Tsubura Y, Hilgers J, Michalides R. A new locus (Mtv-4) for endogenous mammary tumor virus expression and early mammary tumor development in the SHN mouse strain. J Natl Cancer Inst. 1983;71:517–521. [PubMed] [Google Scholar]
- 26.Irwin D M, Kocher T D, Wilson A C. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- 27.Johnson G A, Burghardt R C, Newton G R, Bazer F W, Spencer T E. Development and characterization of immortalized ovine endometrial cell lines. Biol Reprod. 1999;61:1324–1330. doi: 10.1095/biolreprod61.5.1324. [DOI] [PubMed] [Google Scholar]
- 28.Johnson W E, Coffin J M. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak C A, Gromet N J, Ikeda H, Buckler C E. A unique sequence related to the ecotropic murine leukemia virus is associated with the Fv-4 resistance gene. Proc Natl Acad Sci USA. 1984;81:834–837. doi: 10.1073/pnas.81.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebkowsky J S, Clancy S, Calos M P. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature. 1985;317:169–171. doi: 10.1038/317169a0. [DOI] [PubMed] [Google Scholar]
- 31.Li W H, Wu C I, Luo C C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 32.Lower R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350–356. doi: 10.1016/s0966-842x(99)01565-6. [DOI] [PubMed] [Google Scholar]
- 33.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magdaleno S M, Wang G, Jackson K J, Ray M K, Welty S, Costa R H, DeMayo F J. Interferon-gamma regulation of Clara cell gene expression: in vivo and in vitro. Am J Physiol. 1997;272:L1142–L1151. doi: 10.1152/ajplung.1997.272.6.L1142. [DOI] [PubMed] [Google Scholar]
- 35.Mager D L, Freeman J D. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology. 1995;213:395–404. doi: 10.1006/viro.1995.0012. [DOI] [PubMed] [Google Scholar]
- 36.Margana R K, Boggaram V. Functional analysis of surfactant protein B (SP-B) promoter. Sp1, Sp3, TTF-1, and HNF-3alpha transcription factors are necessary for lung cell-specific activation of SP-B gene transcription. J Biol Chem. 1997;272:3083–3090. doi: 10.1074/jbc.272.5.3083. [DOI] [PubMed] [Google Scholar]
- 37.Mark C. DNA Strider: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acid Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medstrand P, Mager D L. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa K, Harrison L C. The potential roles of endogenous retroviruses in autoimmunity. Immunol Rev. 1996;152:193–236. doi: 10.1111/j.1600-065x.1996.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 40.Neil J C, Fulton R, Rigby M, Stewart M. Feline leukaemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol. 1991;171:67–93. doi: 10.1007/978-3-642-76524-7_4. [DOI] [PubMed] [Google Scholar]
- 41.Palmarini M, Cousens C, Dalziel R G, Bai J, Stedman K, DeMartini J C, Sharp J M. The exogenous form of Jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmarini M, Datta S, Omid R, Murgia C, Fan H. The long terminal repeat of Jaagsiekte sheep retrovirus is preferentially active in differentiated epithelial cells of the lungs. J Virol. 2000;74:5776–5787. doi: 10.1128/jvi.74.13.5776-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmarini M, Dewar P, De las Heras M, Inglis N F, Dalziel R G, Sharp J M. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 44.Palmarini M, Fan H, Sharp J M. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 45.Palmarini M, Holland M J, Cousens C, Dalziel R G, Sharp J M. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J Gen Virol. 1996;77:2991–2998. doi: 10.1099/0022-1317-77-12-2991. [DOI] [PubMed] [Google Scholar]
- 46.Palmarini M, Sharp J M, De las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmarini M, Sharp J M, Lee C, Fan C. In vitro infection of ovine cell lines by jaagsiekte sheep retrovirus (JSRV) J Virol. 1999;73:10070–10078. doi: 10.1128/jvi.73.12.10070-10078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patience C, Takeuchi Y, Cosset F L, Weiss R A. Packaging of endogenous retroviral sequences in retroviral vectors produced by murine and human packaging cells. J Virol. 1998;72:2671–2676. doi: 10.1128/jvi.72.4.2671-2676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 50.Patience C, Wilkinson D A, Weiss R A. Our retroviral heritage. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 51.Payne L N, Pani P K. A dominant epistatic gene which inhibits cellular susceptibility to RSV (RAV-0) J Gen Virol. 1971;13:455–462. doi: 10.1099/0022-1317-13-3-455. [DOI] [PubMed] [Google Scholar]
- 52.Perron H, Garson J A, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke P W, Voisset C, Blond J L, Lalande B, Seigneurin J M, Mandrand B. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc Natl Acad Sci USA. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell D F, Broscius C M, Vanin E F, Buckler C E, Nienhuis A W, Martin M A. An array of murine leukemia virus-related elements is transmitted and expressed in a primate recipient of retroviral gene transfer. J Virol. 1996;70:887–897. doi: 10.1128/jvi.70.2.887-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosati S, Pittau M, Alberti A, Pozzi S, York D F, Sharp J M, Palmarini M. An accessory open reading frame (orf-x) of jaagsiekte sheep retrovirus is conserved between different virus isolates. Virus Res. 2000;66:109–116. doi: 10.1016/s0168-1702(99)00118-5. [DOI] [PubMed] [Google Scholar]
- 55.Ruis B L, Benson S J, Conklin K F. Genome structure and expression of the ev/J family of avian endogenous viruses. J Virol. 1999;73:5345–5355. doi: 10.1128/jvi.73.7.5345-5355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruscetti S, Davis L, Feild J, Oliff A. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J Exp Med. 1981;154:907–920. doi: 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Spencer T E, Ing N H, Ott T L, Mayes J S, Becker W C, Watson G H, Mirando M A, Brazer F W. Intrauterine injection of ovine interferon-tau alters oestrogen receptor and oxytocin receptor expression in the endometrium of cyclic ewes. J Mol Endocrinol. 1995;15:203–220. doi: 10.1677/jme.0.0150203. [DOI] [PubMed] [Google Scholar]
- 59.Spencer T E, Stagg A G, Joyce M M, Jenster G, Wood C G, Bazer F W, Wiley A A, Bartol F F. Discovery and characterization of endometrial epithelial messenger ribonucleic acids using the ovine uterine gland knockout model. Endocrinology. 1999;140:4070–4080. doi: 10.1210/endo.140.9.6981. [DOI] [PubMed] [Google Scholar]
- 60.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoye J P, Le Tissier P, Takeuchi Y, Patience C, Weiss R A. Endogenous retroviruses: a potential problem for xenotransplantation? Ann N Y Acad Sci. 1998;862:67–74. doi: 10.1111/j.1749-6632.1998.tb09118.x. [DOI] [PubMed] [Google Scholar]
- 62.Stoye J P, Moroni C, Coffin J M. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tonjes R R, Lower R, Boller K, Denner J, Hasenmaier B, Kirsch H, Konig H, Korbmacher C, Limbach C, Lugert R, Phelps R C, Scherer J, Thelen K, Lower J, Kurth R. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:S261–S267. doi: 10.1097/00042560-199600001-00039. [DOI] [PubMed] [Google Scholar]
- 65.van Nie R, Verstraeten A A. Studies of genetic transmission of mammary tumour virus by C3Hf mice. Int J Cancer. 1975;16:922–931. doi: 10.1002/ijc.2910160606. [DOI] [PubMed] [Google Scholar]
- 66.van Nie R, Verstraeten A A, De Moes J. Genetic transmission of mammary tumour virus by GR mice. Int J Cancer. 1977;19:383–390. doi: 10.1002/ijc.2910190316. [DOI] [PubMed] [Google Scholar]
- 67.Verstraeten A A, van Nie R. Genetic transmission of mammary tumour virus in the DBAf mouse strain. Int J Cancer. 1978;21:473–475. doi: 10.1002/ijc.2910210412. [DOI] [PubMed] [Google Scholar]
- 68.Whitsett J A, Glasser S W. Regulation of surfactant protein gene transcription. Biochim Biophys Acta. 1998;1408:303–311. doi: 10.1016/s0925-4439(98)00076-3. [DOI] [PubMed] [Google Scholar]
- 69.Wikenheiser K A, Vorbroker D K, Rice W R, Clark J C, Bachurski C J, Oie H K, Whitsett J A. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90:11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the Jaaksiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]