Abstract
Coronavirus contains three envelope proteins, M, E and S, and a nucleocapsid, which consists of genomic RNA and N protein, within the viral envelope. We studied the macromolecular interactions involved in coronavirus assembly in cells infected with a murine coronavirus, mouse hepatitis virus (MHV). Coimmunoprecipitation analyses demonstrated an interaction between N protein and M protein in infected cells. Pulse-labeling experiments showed that newly synthesized, unglycosylated M protein interacted with N protein in a pre-Golgi compartment, which is part of the MHV budding site. Coimmunoprecipitation analyses further revealed that M protein interacted with only genomic-length MHV mRNA, mRNA 1, while N protein interacted with all MHV mRNAs. These data indicated that M protein interacted with the nucleocapsid, consisting of N protein and mRNA 1, in infected cells. The M protein-nucleocapsid interaction occurred in the absence of S and E proteins. Intracellular M protein-N protein interaction was maintained after removal of viral RNAs by RNase treatment. However, the M protein-N protein interaction did not occur in cells coexpressing M protein and N protein alone. These data indicated that while the M protein-N protein interaction, which is independent of viral RNA, occurred in the M protein-nucleocapsid complex, some MHV function(s) was necessary for the initiation of M protein-nucleocapsid interaction. The M protein-nucleocapsid interaction, which occurred near or at the MHV budding site, most probably represented the process of specific packaging of the MHV genome into MHV particles.
Assembly of virus particles is an essential step for a productive viral replication cycle. The intracellular sites of virus assembly vary among different viruses (35, 43). Assembly of enveloped viruses requires complex interactions between the lipid envelope, envelope proteins, and internal viral components. Budding of enveloped viruses, through cellular membranes, involves the process of envelopment of the viral nucleocapsid. The interaction of the viral nucleocapsid with envelope proteins is believed to drive the incorporation of the nucleocapsid in enveloped viruses (41). Indeed, interactions between viral envelope protein and nucleocapsid protein are required for the formation of alphaviruses (25, 45). In other enveloped viruses, such as rhabdovirus and paramyxovirus, a matrix protein mediates the interaction between the viral envelope, envelope proteins, and the nucleocapsid (6, 36). Studies of viral assembly mechanisms not only provide an excellent model system for understanding the macromolecular interactions in cells, but also offer valuable information for the development of preventive and therapeutic agents against viral infection.
Coronavirus is an enveloped virus containing a large, positive-stranded RNA genome. The prototypic coronavirus, mouse hepatitis virus (MHV), contains three envelope proteins, M, E, and S. S protein forms 180/90-kDa peplomers that bind to receptors (9) on coronavirus-susceptible cells and induce cell fusion (7, 12). M protein, the most abundant glycoprotein in the virus particle and in infected cells, is characterized as having three domains: a short N terminal ectodomain, a triple-spanning transmembrane domain, and a C-terminal endodomain (1). E protein is present only in minute amounts in infected cells and in the virus envelope (13, 23, 37, 47, 51), yet it is an essential protein for coronavirus envelope formation; coronavirus-like particles (VLPs) are assembled and released from cells that express both E and M proteins (4, 49). Furthermore, expression of E protein alone results in the production of membrane vesicles, which contain E protein (27). E protein also affects coronavirus morphogenesis, as it was shown that MHV mutants, encoding mutated E protein, are morphologically aberrant compared to wild-type MHV (10). Viral genomic RNA and N protein are found inside the viral envelope (44). A generally accepted model of coronavirus structure proposes that viral genomic RNA and N protein form a helical nucleocapsid (44).
In coronavirus-infected cells, genomic-size RNA, mRNA 1, and six to eight species of subgenomic mRNAs are produced. These virus-specific mRNAs comprise a nested set with common 3′ cotermini (20, 22) and a common leader sequence of approximately 60 to 80 nucleotides at the 5′ end (19, 42). Each of the coronavirus-specific proteins is translated from only one of these mRNAs. Among the mRNAs, only mRNA 1 is efficiently packaged into coronavirus particles, while subgenomic mRNAs either are not incorporated into virus particles (21, 30, 32) or are incorporated at a low efficiency (40); incorporation of MHV subgenomic mRNAs into MHV particles is usually undetectable (32). Studies of MHV defective interfering (DI) RNAs suggest that the specific packaging of mRNA 1 is mediated by a 69-nucleotide packaging signal, present only in mRNA 1 (11). The packaging signal is located 21 kb from the 5′ end of MHV genomic RNA (11, 48) and is necessary and sufficient for packaging RNA into MHV particles (50). The mechanism by which the packaging signal mediates specific packaging of MHV mRNA 1 into MHV particles is unknown.
MHV assembly takes place at the “budding compartment,” the smooth membranes of the intermediate compartment between the endoplasmic reticulum (ER) and the Golgi complex (18, 46). M protein itself does not determine the budding site; when M protein is expressed in the absence of other viral proteins, it migrates beyond the budding compartment and localizes in the late-Golgi complex (18). This indicates that an unidentified viral factor(s) restricts the migration of M protein to the budding compartment. One of the candidates that may restrict the migration of M protein is the viral nucleocapsid. It is reasonable to speculate that the binding of the nucleocapsid to M protein restricts the migration of M protein to the budding compartment and that this M protein-nucleocapsid interaction facilitates the envelopment of the nucleocapsid at the budding compartment. Although the envelopment of the nucleocapsid is an important step in coronavirus assembly, this process is poorly characterized. We know that S protein is dispensable for MHV nucleocapsid envelopment and production of MHV particles (15, 17, 39). A possible role of E protein in envelopment of the nucleocapsid is less obvious. Furthermore, interaction between M protein and the nucleocapsid, in infected cells, has not been experimentally demonstrated.
To understand the macromolecular interactions that occur during coronavirus assembly, we studied the interaction of the MHV M protein and nucleocapsid in infected cells. Our study revealed an interaction between intracellular M protein and the viral nucleocapsid, containing N protein and mRNA 1, in infected cells. This interaction occurred in a pre-Golgi compartment and did not require the presence of S and E proteins. The specific interaction between M protein and the viral nucleocapsid most probably represented the process of specific packaging of mRNA 1 into MHV particles.
MATERIALS AND METHODS
Viruses and cells.
The plaque-cloned JHM strain of MHV (MHV-JHM) (28) was used as a helper virus. A 19-fold undiluted, serially passaged MHV-JHM preparation (16, 28) and an RNA− temperature-sensitive (ts) mutant of MHV-A59, LA16 (16), were used for the preparation of the DIssA/LA16 virus sample, which contained self-replicating MHV-JHM DI RNA, DIssA, as described previously (16). Mouse DBT cells were used for the propagation of viruses (14). BHK cells were used for the preparation of Sindbis virus pseudovirions. MHV was grown in 88% Eagle's minimum essential medium (pH 6.5), 10% tryptose phosphate broth, and 2% heat-inactivated fetal bovine serum.
Antibodies.
The anti-MHV M protein monoclonal antibodies J1.3 and J2.7 and the anti-MHV N protein monoclonal antibody J3.3 were kindly provided by J. O. Fleming of the University of Wisconsin at Madison. The anti-MHV M protein monoclonal antibody J2.7 was used for immunofluorescence studies. The non-MHV monoclonal antibody H2KkDk (H2K), which is against major histocompatibility complex class I antigen, was a kind gift from P. Gottlieb of The University of Texas at Austin.
Plasmid construction.
A Sindbis virus recombinant vector expressing MHV N protein (pSinN) was constructed by inserting the entire open reading frame of MHV-JHM N protein into the StuI site of a Sindbis virus expression vector, pSinRep5 (5) (Invitrogen, San Diego, Calif.).
Preparation of Sindbis virus pseudovirions.
Four Sindbis virus pseudovirions, SinM, expressing MHV-JHM M protein (27), SinE, expressing MHV-A59 E protein (27), SinN, expressing MHV-JHM N protein, and SinLacZ, expressing β-galactosidase protein, were produced as described previously (27). Briefly, recombinant Sindbis virus vectors were linearized by XhoI digestion and transcribed in vitro with SP6 RNA polymerase. BHK cells were cotransfected with the recombinant RNA transcripts and a Sindbis virus helper RNA, DH(26S), which expresses the Sindbis virus structural proteins (5, 26, 27), by electroporation. Culture fluid, containing the pseudovirions released from the transfected cells, was collected 30 h after transfection and used for the expression studies.
Labeling of intracellular proteins, immunoprecipitation, and SDS-PAGE.
DBT cells were infected with MHV-JHM at a multiplicity of infection (MOI) of 10. Infected cells were labeled with 50 to 100 μCi of Tran[35S] label for 30 min from 8 to 8.5 h postinfection (p.i.) or were pulse-labeled for 5 min at 8 h p.i. For the radiolabeling of expressed MHV proteins, DBT cells were infected with Sindbis virus pseudovirions and then metabolically labeled with 50 to 100 μCi of Tran[35S] label from 5 to 5.5 h p.i. DBT cells were infected with DIssA/LA16 at 39.5°C. At 3.5 h p.i., cells were superinfected with Sindbis virus pseudovirions and incubated at 39.5°C. The intracellular proteins were labeled with 50 to 100 μCi of Tran[35S] label from 8.5 to 9 h post-DIssA/LA16 infection. Cell lysates were prepared using lysis buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in phosphate-buffered saline [PBS]) (29), and the intracellular MHV-specific proteins were immunoprecipitated with monoclonal antibody J1.3, J3.3, or H2K as described previously (17). The immunoprecipitated proteins were incubated at 37°C for 30 min in sample buffer to prevent M protein aggregation (44) and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). The [3H]glucosamine-labeled proteins were resolved by SDS-PAGE and visualized by fluorography with Entensify (Dupont).
Preparation of virus-specific RNA.
The intracellular virus-specific RNAs were labeled with 32Pi and extracted from virus-infected cells as described previously (29). The nonradiolabeled intracellular virus-specific RNAs were extracted from DIssA/LA16-infected cells as described previously (29).
Immunoprecipitation of MHV-specific RNAs.
The cytoplasmic lysates that were prepared by using lysis buffer from MHV-infected cells were incubated with a monoclonal antibody at 4°C. The immune complexes were incubated with Pansorbin cells (Calbiochem) at 4°C and subsequently collected by centrifugation. The pellets were washed three times in PBS containing 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS. The final pellets were suspended in a buffer containing 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 12.5 mM EDTA, and 1% SDS. Proteinase K was added to the suspension at a final concentration of 0.1 mg/ml, and the sample was incubated at 37°C for 30 min. The RNA was extracted with phenol-chloroform as described previously (31).
Agarose gel electrophoresis of RNA and Northern (RNA) blotting.
Radiolabeled RNAs were denatured and electrophoresed through a 1% agarose gel containing formaldehyde as described previously (31). For Northern blot analysis, the nonradiolabeled RNAs were electrophoresed through a 1% agarose gel containing formaldehyde and then transferred onto nylon filters (29). Northern blot analysis was performed using a digoxigenin (DIG)-labeled random-primed probe (Boehringer), corresponding to 85 to 474 nucleotides (nt) from the 5′ end of MHV genomic RNA, and visualized with a DIG luminescent detection kit (Boehringer) according to the manufacturer's protocol.
RNase A treatment.
The cytoplasmic protein lysates were incubated with 10 μg of RNase A for 15 min at room temperature. The RNase-treated lysates were immunoprecipitated with anti-N protein monoclonal antibody J3.3 and analyzed by SDS-PAGE.
RESULTS
Presence of an interaction between M protein and N protein in infected cells.
We speculated that the MHV nucleocapsid interacts with M protein in infected cells and that this interaction facilitates the incorporation of the nucleocapsid into MHV particles. To examine the presence of the M protein-nucleocapsid interaction in infected cells, MHV-infected DBT cells were labeled with Tran[35S] label from 8 to 8.5 h p.i. and cell lysates were prepared. Radioimmunoprecipitation of MHV-specific proteins, using an anti-N protein monoclonal antibody, showed coimmunoprecipitation of M protein with N protein (Fig. 1A), demonstrating M protein-N protein interaction in infected cells. The anti-N protein antibody also coimmunoprecipitated MHV S protein (Fig. 1A); coimmunoprecipitation of S protein by the anti-N protein antibody may be due to the interaction between S protein and M protein in MHV-infected cells (34). Reciprocal immunoprecipitation analysis with an anti-M protein monoclonal antibody showed coimmunoprecipitation of N protein with M protein (Fig. 1A). The non-MHV-related control monoclonal antibody, anti-H2K, did not precipitate any proteins. These data demonstrated that M protein and N protein interacted in MHV-infected cells.
FIG. 1.
Interaction between N protein and M protein in MHV-infected cells. (A) DBT cells were infected with MHV-JHM, and intracellular proteins were labeled with Tran[35S] label from 8 to 8.5 h p.i. (lanes 2 to 4) or with [3H]glucosamine from 6.5 to 8.5 h p.i. (lane 5). The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2), an anti-M protein monoclonal antibody (lanes 3, 5) or an anti-H2K monoclonal antibody (lane 4), and viral proteins were analyzed by SDS–15% PAGE. Lane 1, 14C-labeled protein size marker. (B) MHV-JHM-infected DBT cells were pulse-labeled with Tran[35S] label for 5 min at 8 h p.i., and intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2). Lane 1, 14C-labeled protein size marker. Ab, antibody.
M protein is initially synthesized as an unglycosylated protein, M0, in the ER and then glycosylated to an intermediate form, M1, in the intermediate compartment (24). Further glycosylation of M protein, resulting in the mature forms, M3 to M5, takes place in the Golgi apparatus (24). The mobility of M protein in SDS-PAGE suggested that the majority of M protein that was coimmunoprecipitated by the anti-N protein antibody was in the M0 form. Indeed, analysis of [3H]glucosamine-labeled M protein confirmed that the anti-N protein antibody predominantly coprecipitated the unglycosylated M0 form (Fig. 1A, lane 5), implying that the N protein-M protein interaction occurred in pre-Golgi membranes. Furthermore, the anti-N protein antibody radioimmunoprecipitated the unglycosylated form of M protein, M0, from cell extracts prepared from MHV-infected cells, that were pulse-labeled with Tran[35S] label for 5 min (Fig. 1B). These data demonstrated that N protein interacted with newly synthesized M protein, very rapidly, in a pre-Golgi compartment.
Specific interaction between M protein and mRNA 1 in infected cells.
Next, we examined whether M protein interacted with a nucleocapsid, consisting of N protein and genomic-length MHV mRNA, mRNA 1. MHV-specific RNAs in infected cells were labeled with 32Pi in the presence of actinomycin D; under this condition MHV-specific mRNAs were preferentially radiolabeled. The radiolabeled cell lysates, prepared at 8 h p.i., were immunoprecipitated with an anti-M protein antibody or an anti-N protein antibody. MHV-specific RNAs were extracted from the immunoprecipitated samples and analyzed by agarose gel electrophoresis. Consistent with a previous report (2), the anti-N protein antibody immunoprecipitated all the MHV mRNAs, indicating the association of N protein with all MHV mRNAs. The result of immunoprecipitation using the anti-M protein antibody was striking; the anti-M protein antibody coimmunoprecipitated only the genomic-length mRNA, mRNA 1 (Fig. 2), clearly demonstrating that M protein specifically interacted with mRNA 1, but not with other subgenomic mRNAs. Since MHV particles preferentially package mRNA 1, among all intracellular MHV mRNAs, the specific interaction between M protein and mRNA 1 most probably represented the process of packaging of mRNA 1 into MHV particles.
FIG. 2.
Specific M protein-mRNA 1 interaction in MHV-infected cells. MHV-JHM-infected DBT cells were labeled with 32Pi from 6 to 8 h p.i. in the presence of actinomycin D, and cytoplasmic protein lysates were prepared. The intracellular (i.c.) proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2), an anti-M protein monoclonal antibody (lane 3), or an anti-H2K monoclonal antibody (lane 4). MHV-specific RNAs were extracted from the immunoprecipitated samples and analyzed by agarose-formaldehyde gel electrophoresis. Lane 1, virus-specific RNAs extracted from 32P-labeled MHV-infected cells at 8 h p.i.
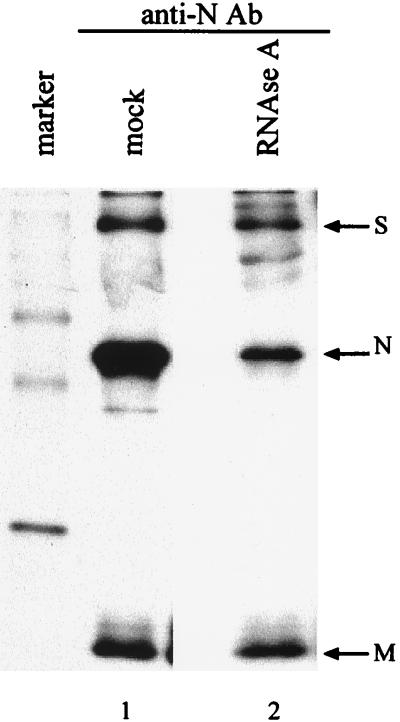
N protein-M protein interaction retained after removal of mRNA 1 by RNase A treatment.
Next, we tested the possibility that M protein interacts only with mRNA 1 in the nucleocapsid. For this analysis MHV-infected cell lysates were treated with RNase A or mock treated. RNase A treatment of cell extracts would result in degradation of all RNAs, including mRNA 1. If M protein interacts only with mRNA 1 in the nucleocapsid, degradation of mRNA 1 by RNase A treatment would result in the dissociation of M protein from the N protein-mRNA 1 complex. 35S-labeled and nonradiolabeled MHV-infected cell lysates were treated with RNase A or mock treated. After treatment, intracellular RNAs were extracted from nonradiolabeled cell lysates to determine the effect of RNase A on the integrity of intracellular viral RNAs. Northern blot analysis of MHV-specific RNAs, using a random-primed MHV-specific cDNA probe, which hybridizes with all MHV mRNAs, showed extensive degradation of MHV mRNAs in the RNase A-treated sample; no MHV-specific RNAs were detected after RNase A treatment, while all MHV mRNA species were detected in mock-treated cells (data not shown). Radioimmunoprecipitation analysis of RNase-treated, 35S-labeled cell lysates showed the coimmunoprecipitation of M protein by an anti-N protein antibody (Fig. 3), demonstrating that the M protein-N protein interaction was maintained even after the removal of viral mRNA 1. These data suggested that there was an RNA-independent interaction between M protein and N protein in the nucleocapsid.
FIG. 3.
Interaction between N protein and M protein after RNase A treatment. MHV-JHM-infected DBT cells were labeled with Tran[35S] label from 8 to 8.5 h p.i., and cytoplasmic lysates were prepared. Equal volumes of the lysates were either treated with RNase A (lane 2) or mock treated (lane 1) for 15 min at room temperature. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody and analyzed by SDS–15% PAGE.
The anti-N protein antibody immunoprecipitated a smaller amount of N protein in the RNase-treated sample than in the mock-treated sample, while this antibody coimmunoprecipitated similar amounts of M protein in both samples (Fig. 3). Although the reason for the less efficient immunoprecipitation of N protein by the anti-N protein antibody in the RNase A-treated sample is unknown, removal of mRNAs from the complexes of N protein-MHV mRNAs by RNase treatment may alter the conformation of N protein. The anti-N protein antibody may bind less efficiently to the conformationally altered N protein. However, this putative structural alteration of N protein did not drastically affect the interaction of N protein with M protein, because the amounts of M protein that coimmunoprecipitated with N protein in the RNase-treated sample and the mock-treated sample were similar.
Analysis of interaction between expressed M protein and N protein.
Although the M protein-N protein interaction was retained after the removal of MHV mRNA 1, this finding does not mean that MHV mRNA 1 is dispensable for the initiation of the M protein-nucleocapsid interaction; mRNA 1 may play an important role in the establishment of the M protein-nucleocapsid interaction. We used Sindbis virus pseudovirions expressing M protein (SinM pseudovirion) (27) and N protein (SinN pseudovirion) to examine whether the interaction between M protein and N protein could be established in the absence of mRNA 1 or other MHV functions; we examined the interaction between expressed M protein and N protein. Sindbis virus pseudovirions expressing β-galactosidase (SinLacZ) (27) were used as a negative control. Immunofluorescence analysis showed that approximately 90% of cells expressed N protein and M protein after 5 h p.i. with SinN pseudovirions and SinM pseudovirions, respectively (data not shown). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining showed that approximately 90% of SinLacZ pseudovirion-infected cells expressed β-galactosidase (data not shown). These data demonstrated that most of the cells were infected with these pseudovirions. Radioimmunoprecipitation analysis using an anti-M protein antibody or an anti-N protein antibody showed excellent expression levels of both M protein and N protein in Sindbis virus pseudovirion-infected DBT cells (Fig. 4A). We confirmed the specificities of the anti-N protein antibody and the anti-M protein antibody by radioimmunoprecipitation analysis of a mixture of two cell lysates, each of which was independently infected with SinM pseudovirions and SinN pseudovirions; the anti-M protein antibody and anti-N protein antibody specifically immunoprecipitated M protein and N protein, respectively (Fig. 4B). We noticed that the anti-M protein antibody frequently immunoprecipitated a faint band that migrated very close to N protein (Fig. 4, asterisks). This minor band was not an MHV-specific protein, as it did not comigrate with N protein and was easily separated from N protein in gels with different concentrations (see Fig. 4B). Some Sindbis virus-derived proteins were also immunoprecipitated by the anti-N protein antibody and the anti-M protein antibody (Fig. 4, arrows); these bands were not detected in uninfected cells (data not shown). In an experimental group, in which cells were coinfected with SinN pseudovirions and SinM pseudovirions, the anti-N protein antibody immunoprecipitated N protein but not M protein (Fig. 4C). Similarly, the anti-M protein antibody immunoprecipitated only M protein, but not N protein (Fig. 4C). These data indicated that coexpressed M and N proteins did not interact with each other. Some MHV function(s) appeared to be necessary to establish the M protein-N protein interaction in infected cells.
FIG. 4.
Analysis of interaction between expressed N protein and M protein. (A) DBT cells that were infected with SinN pseudovirions (lanes 1 and 2) or SinM pseudovirions (lanes 3 and 4) were labeled with Tran[35S] label from 5 to 5.5 h p.i. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lanes 1 and 4) or an anti-M protein monoclonal antibody (lanes 2 and 3), and viral proteins were analyzed by SDS–15% PAGE. (B) Equal volumes of 35S-labeled intracellular protein lysates from DBT cells, infected with SinM pseudovirions alone and SinN pseudovirions alone, were mixed, and intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 1), an anti-M protein monoclonal antibody (lane 2), or an anti-H2K monoclonal antibody (lane 3). The viral proteins were analyzed by SDS–12% PAGE. (C) DBT cells were coinfected with SinM pseudovirions and SinN pseudovirions, and intracellular proteins were labeled with Tran[35S] label from 5 to 5.5 h p.i. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 1), an anti-M protein monoclonal antibody (lane 2), or an anti-H2K monoclonal antibody (lane 3). Viral proteins were analyzed by SDS–15% PAGE. The marked protein bands (indicated by arrows and an asterisk) are non-MHV proteins.
The M protein-nucleocapsid interaction occurred in the absence of S and E proteins.
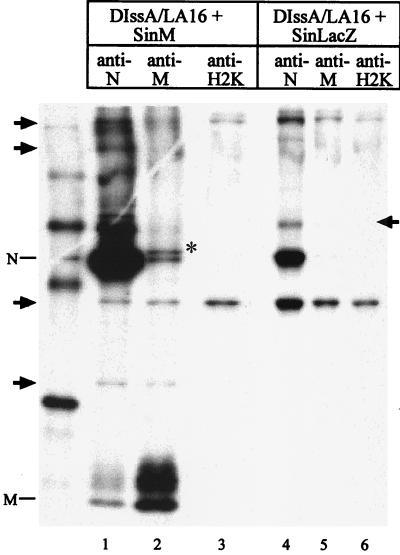
The coimmunoprecipitation studies of MHV-infected cells shown above demonstrated that both the anti-N protein antibody and the anti-M protein antibody coimmunoprecipitated S protein (Fig. 1 and 3). Our interpretation was that the buffer used for radioimmunoprecipitation did not disrupt the M protein-S protein interaction (34) and that S protein was coimmunoprecipitated due to this interaction. It is highly unlikely that S protein is necessary for the M protein-nucleocapsid interaction, because MHV particles containing the nucleocapsid are produced in the absence of S protein (15, 17, 39). In contrast, the role of another MHV envelope protein, E protein, in the M protein-nucleocapsid interaction is unknown. We further examined the roles of S protein and E protein in the M protein-nucleocapsid interaction by using a unique self-replicating MHV DI RNA, DIssA (16). DIssA is a naturally occurring MHV DI RNA that carries gene 1, encoding the RNA polymerase function, and gene 7, encoding N protein. Importantly, DIssA has a deletion of the entire S, E, and M genes; MHV gene 1 proteins and N protein are produced in DIssA-replicating cells, whereas S, M, and E envelope proteins are not produced in these cells (16). For the preparation of DIssA DI particles, an RNA− ts mutant of MHV-A59, LA16, was used as a helper virus as described previously (16). Briefly, DBT cells were coinfected with LA16 and the MHV-JHM sample, obtained after 19 undiluted passages of MHV-JHM that contained DIssA DI particles, at the permissive temperature (32.5°C) for LA16. The samples were passaged three times at 32.5°C to replace the helper virus of DIssA DI particles from MHV-JHM to LA16 (16). Infection of this LA16 sample containing DIssA DI particles, DIssA/LA16, at 39.5°C, the nonpermissive temperature for LA16, results in synthesis of only DIssA and N protein-encoding mRNA 7, but not LA16 RNAs; S protein and E protein are not produced in DIssA/LA16-infected cells (16).
In the present study, the SinM pseudovirion was used to express M protein in DIssA/LA16-infected cells. The SinLacZ pseudovirion was used as a negative control. DIssA/LA16-infected DBT cells were superinfected with SinM pseudovirions or SinLacZ pseudovirions at 3.5 h p.i. Virus-infected cells were incubated at 39.5°C throughout the infection, and intracellular proteins were radiolabeled with Tran[35S] label from 8.5 h to 9 h, post-DIssA/LA16 infection. Coimmunoprecipitation analysis showed that the anti-N protein antibody coimmunoprecipitated M protein with N protein from the lysates of cells infected with DIssA/LA16 and SinM pseudovirions (Fig. 5, lane 1). The anti-M protein antibody also coimmunoprecipitated N protein with M protein from the same lysate (Fig. 5, lane 2). Several other bands were also detected in the immunoprecipitation analysis of the cell lysates from cells infected with DIssA/LA16 and SinM pseudovirions; some were derived from Sindbis virus, while the origins of others were unclear. None of these bands comigrated with MHV S protein. The anti-M protein antibody and anti-N protein antibody did not immunoprecipitate M protein from cells infected with DIssA/LA16 and SinLacZ pseudovirions, indicating that M protein expression was undetectable in cells infected with DIssA/LA16. It is unlikely that a very small amount of E protein, which may be expressed from revertant LA16 in the DIssA/LA16 virus preparation, facilitated the M protein-N protein interaction, because the M protein-N protein interaction did not occur in cells coinfected with SinM pseudovirions, SinN pseudovirions, and SinE pseudovirions (data not shown). These data demonstrated that M protein interacted with N protein in the absence of S and E proteins.
FIG. 5.
Interaction between the nucleocapsid and M protein in the absence of S and E proteins. DBT cells were infected with DIssA/LA16 at 39.5°C. At 3.5 h post-DIssA/LA16 infection, cells were superinfected with either SinM pseudovirions (lanes 1 to 3) or SinLacZ pseudovirions (lanes 4 to 6) and incubated at 39.5°C. The intracellular proteins were labeled with Tran[35S] label from 8.5 to 9 h post-DIssA/LA16 infection, and cytoplasmic lysates were prepared. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lanes 1 and 4), an anti-M protein monoclonal antibody (lanes 2 and 5), or an anti-H2K monoclonal antibody (lanes 3 and 6). The viral proteins were analyzed by SDS–12% PAGE. The 14C-labeled protein size marker is shown on the left of the gel. The marked protein bands (indicated by arrows and an asterisk) are non-MHV proteins.
To further confirm that M protein interacted with the nucleocapsid, consisting of N protein and DIssA RNA, cell lysates from cells infected with DIssA/LA16 and SinM pseudovirions were immunoprecipitated with an anti-M protein antibody. MHV-specific RNAs that were coprecipitated by the anti-M protein antibody were extracted from the immunoprecipitated samples. Northern blot analysis, using a cDNA probe that binds to DIssA RNA, showed that the anti-M protein antibody coimmunoprecipitated DIssA RNA from cells infected with DIssA/LA16 and SinM pseudovirions, while the same antibody failed to coimmunoprecipitate DIssA RNA from cells infected with DIssA/LA16 and SinLacZ pseudovirions (Fig. 6). These data demonstrated that M protein interacted with the nucleocapsid, consisting of N protein and DIssA RNA, in cells expressing DIssA and M protein. The studies, using DIssA/LA16 and Sin M pseudovirions, further confirmed that the observed M protein-nucleocapsid interaction indeed occurred within cells and not in intracellular virus particles, since no MHV particles are produced in cells infected with DIssA/LA16 and expressing M protein (17). We concluded that S and E proteins were dispensable for the intracellular M protein-nucleocapsid interaction.
FIG. 6.
Specific interaction of M protein with DIssA RNA in the absence of S and E proteins. DBT cells were infected with DIssA/LA16 at 39.5°C. At 3.5 h post-DIssA/LA16 infection, cells were superinfected with either SinM pseudovirions (lanes 1 and 2) or SinLacZ pseudovirions (lanes 3 and 4) and incubated at 39.5°C. At 9 h post-DIssA/LA16 infection, cytoplasmic lysates were prepared and separated into two groups. Intracellular RNAs (lanes 1 and 3) were extracted from one group of lysates. An anti-M protein monoclonal antibody was added to another group, and immunoprecipitation was performed. RNAs were extracted from the immunoprecipitated samples (lanes 2 and 4). Extracted RNAs were separated by 1% agarose gel electrophoresis, and DIssA RNA was detected by Northern blot analysis. The part of the autoradiogram that contains DIssA RNA is indicated.
DISCUSSION
To elucidate the macromolecular interactions that occur during coronavirus assembly, we examined interactions between M protein, MHV RNA, and N protein in MHV-infected cells. Coimmunoprecipitation analyses demonstrated that both N protein and mRNA 1 specifically interacted with M protein in MHV-infected cells. RNase treatment of cell extracts and subsequent immunoprecipitation analysis revealed that the M protein-N protein interaction could be maintained in the absence of viral RNAs in MHV-infected cells. These M protein-N protein and M protein-mRNA 1 interactions in infected cells have not been described previously. These data indicate that M protein interacts with the MHV nucleocapsid, consisting of N protein and mRNA 1.
M protein is initially synthesized as an unglycosylated protein in the ER and is then glycosylated in the intermediate compartment (24). Further glycosylation of M protein takes place in the Golgi apparatus (24). Pulse-labeling experiments demonstrated that newly synthesized, unglycosylated M protein interacted with N protein at the ER membrane (Fig. 1B), suggesting that M protein interacted with the nucleocapsid in a pre-Golgi compartment. Thus, the present study suggested that the site of M protein-nucleocapsid interaction overlaps with MHV budding sites, the ER membrane, and the intermediate compartment (18, 46). We believe that the M protein-nucleocapsid interaction, which appeared to occur near or at the MHV budding sites, represents the process of specific packaging of mRNA 1 into MHV particles.
A recent model of coronavirus structure proposed that coronavirus contains an internal proteinaceous spherical core shell that surrounds the helical nucleocapsid (38). In this model the core shell consisted of mostly M protein and a lesser amount of N protein (38). However, we found that MHV M protein existed exclusively on the viral envelope and not inside the virus particle (K. Narayanan and S. Makino, unpublished data). The presence of M protein on the spherical core shell may be due to the interaction of envelope M protein with the N protein-genomic RNA complex. Hence, the nucleocapsid interacted with M protein that was present exclusively on intracellular membranes.
Understanding the mechanism of initiation of the M protein-nucleocapsid interaction requires further studies. One possible mechanism is that direct M protein-mRNA 1 association initiates the interaction between the nucleocapsid and M protein. Sturman et al. showed that virion M protein and MHV genomic RNA cosediment in sucrose gradients (44). Their data suggested a direct interaction between mRNA 1 and M protein, which is consistent with this model. We have previously demonstrated that MHV mRNA 1 and DIssA RNA contain a 69-nt packaging signal, located about 21 kb from the 5′ end of the genome; the packaging signal is not present in other subgenomic mRNAs (11). The secondary structure of the packaging signal is important for its biological function (11), and the presence of the packaging signal in non-MHV RNA transcripts allows the packaging of these RNA transcripts into MHV particles (50). Recent studies of the MHV packaging signal and bovine coronavirus packaging signal confirmed the previous studies on the MHV packaging signal (3, 8, 33). M protein may directly interact with mRNA 1, through the packaging signal, to initiate the M protein-nucleocapsid interaction. RNase digestion of MHV RNAs in the infected cell extracts did not disrupt the M protein-N protein interaction, suggesting that there was an interaction between M protein and N protein in the nucleocapsid (Fig. 3). However, the possibility that a short RNA, which may remain even after extensive digestion with the nuclease, may be sufficient to mediate this interaction cannot be excluded. Nevertheless, these data imply that the process of RNA packaging is initiated by the mRNA 1-M protein interaction, which is further stabilized by an interaction between M protein and N protein in the nucleocapsid.
The present data, however, do not exclude the possibility that the binding of M protein to N protein initiates the M protein-nucleocapsid interaction. We demonstrated that expressed M protein and N protein did not interact, indicating that some unidentified MHV function(s) was necessary for the establishment of the M protein-N protein interaction. The viral genomic RNA, mRNA 1, may be a factor necessary for the initial interaction between M protein and N protein in the nucleocapsid. For example, it is possible that N protein binds to mRNA 1 to form a nucleocapsid. Binding of N protein to mRNA 1 may alter the conformation of N protein, and this altered conformation may allow N protein to bind to M protein. RNA-mediated alteration of N protein conformation was indeed suggested by the finding that only a relatively small amount of N protein was immunoprecipitated by the anti-N protein antibody in the RNase-treated sample (Fig. 3). If the binding of M protein to N protein initiates the M protein-nucleocapsid interaction, then how does M protein specifically bind only to the N protein-mRNA 1 complex and not to N protein interacting with other MHV subgenomic mRNAs? Coronavirus genomic RNA forms a helical nucleocapsid structure, whereas the status of N protein binding to subgenomic mRNAs in infected cells is not known. Binding of N protein to subgenomic RNAs may not form the helical nucleocapsid structure, and M protein may preferentially interact with N protein in the helical nucleocapsid structure.
We expected that S protein would not play a role in the M protein-nucleocapsid interaction, because MHV particles containing the nucleocapsid are produced in the absence of S protein (15, 17, 39). We confirmed this through the analysis of DIssA; an anti-M protein antibody coimmunoprecipitated N protein and DIssA RNA in cells expressing DIssA and M protein (Fig. 5 and 6); production of S protein in these cells was undetectable. The present study also showed that E protein was dispensable for the M protein-nucleocapsid interaction in MHV-infected cells. We previously demonstrated that membrane vesicles containing E protein, which are released from MHV-infected cells, do not contain a nucleocapsid (27), suggesting that E protein probably does not interact with the nucleocapsid in infected cells. The biological function of E protein in coronavirus assembly appears to be specific for viral envelope formation and budding but not nucleocapsid envelopment (27, 37).
The present study and previous studies illustrate a possible mechanism for the envelopment of the MHV nucleocapsid. The nucleocapsid, consisting of viral genomic-size mRNA 1 and N protein, interacts with M protein in a pre-Golgi compartment, probably at the ER membrane. The interaction between the nucleocapsid and M protein may be initiated either by the binding of M protein to the viral genomic RNA, through the packaging signal, or by direct interaction between N protein and M protein. In the former case, the M protein-packaging signal interaction could lead to the association of M protein with N protein, thereby stabilizing the complex between M protein and the nucleocapsid. In the latter case, the association of mRNA 1 with N protein may alter the conformation of N protein; the altered form of N protein may specifically bind to M protein. E protein does not play a role in the interaction between M protein and the nucleocapsid, yet E protein facilitates the budding of virus particles, containing the nucleocapsid, at the budding compartment. An E protein-M protein interaction probably occurs during or after the establishment of the M protein-nucleocapsid interaction; the direct interaction between E protein and M protein remains to be demonstrated. S protein is incorporated into the virus particle through its interaction with M protein (34). Finally, E protein and M protein mediate the budding of MHV particles from the budding compartment.
ACKNOWLEDGMENTS
We thank John Fleming for anti-M protein and anti-N protein monoclonal antibodies. We also thank Paul Gottlieb for anti-H2K monoclonal antibody.
This work was supported by Public Health Service grant AI29984 from the National Institutes of Health.
REFERENCES
- 1.Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric R S, Nelson G W, Fleming J O, Deans R J, Keck J G, Casteel N, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos E C, Dobbe J C, Luytjes W, Spaan W J. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J Virol. 1997;71:5684–5687. doi: 10.1128/jvi.71.7.5684-5687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos E C, Luytjes W, van der Meulen H V, Koerten H K, Spaan W J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capone J, Ghosh H P. Association of the nucleocapsid protein N of vesicular stomatitis virus with phospholipid vesicles containing the matrix protein M. Can J Biochem Cell Biol. 1984;62:1174–1180. doi: 10.1139/o84-151. [DOI] [PubMed] [Google Scholar]
- 7.Collins A R, Knobler R L, Powell H, Buchmeier M J. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology. 1982;119:358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cologna R, Hogue B G. Identification of a bovine coronavirus packaging signal. J Virol. 2000;74:580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frana M F, Behnke J N, Sturman L S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godet M, L'Haridon R, Vautherot J F, Laude H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano N, Fujiwara K, Hino S, Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 15.Holmes K V, Doller E W, Sturman L S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K H, Makino S. Two murine coronavirus genes suffice for viral RNA synthesis. J Virol. 1995;69:2313–2321. doi: 10.1128/jvi.69.4.2313-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K H, Narayanan K, Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J Virol. 1997;71:3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumperman J, Locker J K, Meijer A, Horzinek M C, Geuze H J, Rottier P J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M, Baric R S, Brayton P R, Stohlman S A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M M, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai M M, Stohlman S A. RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibowitz J L, Wilhelmsen K C, Bond C W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locker J K, Griffiths G, Horzinek M C, Rottier P J. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N- and O-linked glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez S, Yao J S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lustig S, Jackson A C, Hahn C S, Griffin D E, Strauss E G, Strauss J H. Molecular basis of Sindbis virus neurovirulence in mice. J Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda J, Maeda A, Makino S. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology. 1999;263:265–272. doi: 10.1006/viro.1999.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino S, Shieh C K, Keck J G, Lai M M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makino S, Taguchi F, Hirano N, Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984;139:138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makino S, Yokomori K, Lai M M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molenkamp R, Spaan W J. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opstelten D J, Raamsman M J, Wolfs K, Horzinek M C, Rottier P J. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 36.Portner A, Murti K G. Localization of P, NP, and M proteins on Sendai virus nucleocapsid using immunogold labeling. Virology. 1986;150:469–478. doi: 10.1016/0042-6822(86)90311-9. [DOI] [PubMed] [Google Scholar]
- 37.Raamsman M J, Locker J K, de Hooge A, de Vries A A, Griffiths G, Vennema H, Rottier P J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risco C, Anton I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rottier P J, Horzinek M C, van der Zeijst B A. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons K, Garoff H. The budding mechanisms of enveloped animal viruses. J Gen Virol. 1980;50:1–21. doi: 10.1099/0022-1317-50-1-1. [DOI] [PubMed] [Google Scholar]
- 42.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens E B, Compans R W. Assembly of animal viruses at cellular membranes. Annu Rev Microbiol. 1988;42:489–516. doi: 10.1146/annurev.mi.42.100188.002421. [DOI] [PubMed] [Google Scholar]
- 44.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suomalainen M, Liljestrom P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in Sac− cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 47.Tung F Y, Abraham S, Sethna M, Hung S L, Sethna P, Hogue B G, Brian D A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Most R G, Bredenbeek P J, Spaan W J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vennema H, Godeke G J, Rossen J W, Voorhout W F, Horzinek M C, Opstelten D J, Rottier P J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo K, Joo M, Narayanan K, Kim K H, Makino S. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol. 1997;71:824–827. doi: 10.1128/jvi.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]