Abstract
EBNA2 is an Epstein-Barr virus (EBV)-encoded protein that regulates the expression of viral and cellular genes required for EBV-driven B-cell immortalization. Elucidating the mechanisms by which EBNA2 regulates viral and cellular gene expression is necessary to understand EBV-induced B-cell immortalization and viral latency in humans. EBNA2 targets to the latency C promoter (Cp) through an interaction with the cellular DNA binding protein CBF1 (RBPJk). The EBNA2 enhancer in Cp also binds another cellular factor, C promoter binding factor 2 (CBF2), whose protein product(s) has not yet been identified. Within the EBNA2 enhancer in Cp, we have previously identified the DNA sequence required for CBF2 binding and also determined that this element is required for efficient activation of Cp by EBNA2. In this study, the CBF2 activity was biochemically purified and microsequenced. The peptides sequenced were identical to the hnRNP protein AUF1. Antibodies against AUF1 but not antibodies to related hnRNP proteins reacted with CBF2 in gel mobility shift assays. In addition, stimulation of the cellular cyclic AMP (cAMP)/protein kinase A (PKA) signal transduction pathway results in an increase in detectable CBF2/AUF1 binding activity extracted from stimulated cells. Furthermore, the CBF2 binding site was able to confer EBNA2 responsiveness to a heterologous promoter when transfected cells were treated with compounds that activate PKA or by cotransfection of plasmids expressing a constitutively active catalytic subunit of PKA. EBNA2-mediated stimulation of the latency Cp is also increased in similar cotransfection assays. These results further support an important role for CBF2 in mediating EBNA2 transactivation; they identify the hnRNP protein AUF1 as a major component of CBF2 and are also the first evidence of a cis-acting sequence other than a CBF1 binding element that is able to confer responsiveness to EBNA2.
Epstein-Barr virus (EBV) is a human lymphocryptovirus associated with various human malignancies, including Burkitt's lymphoma (BL), Hodgkin's disease, nasopharyngeal carcinoma, and lymphoma in immunosuppressed individuals (53). EBV establishes a lifelong infection in humans, where B lymphocytes are the primary latent reservoir from which occasional virus reactivation occurs (53).
EBV also establishes latent infection in B lymphocytes in vitro and transforms them into continuously proliferating lymphoblastoid cell lines (LCLs). LCLs generally express 11 viral genes (referred to as latency III) that include EBNAs 1, 2, 3A, 3B, and 3C and EBNA-LP, latent membrane proteins LMP-1, -2A, and 2B, and two small noncoding RNAs (EBERs 1 and 2) (37). EBNA2 is essential for EBV-induced immortalization of B lymphocytes (9, 26). One of the major roles for EBNA2 is that it stimulates the expression of the major latency C promoter (Cp) as well as the LMP-1 and LMP-2A genes (18, 56, 63, 65, 69). EBNA2 also stimulates expression of several cellular proteins, including c-Myc, which is likely to be crucial for the immortalization process (11, 32, 38, 40, 47, 61, 62). Thus, EBNA2 is a key regulator of both viral and cellular gene expression during the latent stage of viral infection.
The regulation of EBNA2 expression in EBV-infected cells is likely to be important for viral persistence and the development of EBV-associated malignancies. With the exception of EBNA1, all viral latent proteins possess epitopes which induce a strong cytotoxic T-cell response (43, 53). Downregulation of EBNA2 expression and the concomitant downregulation of other latent proteins may allow a latently infected B cell to escape immune surveillance and also permit proliferation of malignant cells containing EBV. In healthy individuals, latent EBV infection appears to be primarily confined to resting B cells. The only EBV-expressed gene detected in these cells is LMP-2A (a pattern of gene expression termed latency 0) (49, 50, 58). In BL cells, only EBNA1 is expressed (latency I) (53, 58). In Hodgkin's disease, nasopharyngeal carcinoma, and T-cell lymphomas, EBNA1 and one or both LMPs are expressed (latency II) (53, 58). However, all EBNAs and LMPs are expressed when a competent immune response is not present, such as in primary infection, infectious mononucleosis, lymphoproliferative syndromes of immunocompromised individuals, and also LCLs (latency III) (53, 58). The association of these specific patterns of protein expression with various physiological and pathological states underscores the importance of viral gene regulation for persistence and oncogenesis. Cp and EBNA2 expression are likely to be major targets of this regulation. Elucidating the mechanisms that regulate transcription mediated by EBNA2 is therefore important for understanding EBV biology.
EBNA2 interacts with target promoters through contact with the ubiquitous cellular DNA binding protein C promoter binding factor 1 (CBF1 or RBPJk) (24, 28, 45, 69). CBF1 binding sites are found in all of the well-characterized EBNA2-responsive enhancers, and recombinant viruses expressing a mutant EBNA2 protein unable to interact with CBF1 fail to immortalize B cells (66). In addition to CBF1, another cellular factor(s) binds the Cp EBNA2-responsive enhancer, called C promoter binding factor 2 (CBF2) (33, 44). We have recently reported that the CBF2 binding site is also very important for EBNA2 stimulation of Cp in transient cotransfection assays (19). Moreover, this site is conserved in Cp from EBV-like lymphocryptoviruses of nonhuman primates and, like EBV Cp, is required for optimal stimulation by EBNA2 (20). In more biological assays, EBV recombinant viruses with a Cp containing a mutant CBF1 binding site had a modest average reduction in Cp activity relative to wild-type viruses (17). In another study, EBV recombinant viruses deleted for the Cp EBNA2 enhancer, which binds both CBF1 and CBF2, are strongly biased for Bam W promoter (Wp) usage and have little detectable Cp activity (67). These results suggest that in the context of the viral genome, Cp activity is dependent on factors other than CBF1, possibly including CBF2.
In this study, we sought to identify the cellular protein(s) that comprise CBF2. We have determined that a principal component of CBF2 is the hnRNP protein AUF1. Interestingly, we also found that CBF2 is regulated by the cyclic AMP-protein kinase A (cAMP/PKA) signaling pathway, and this pathway contributes to the regulation of Cp activity.
MATERIALS AND METHODS
Cell culture.
EBV-negative BL cell lines DG75 and CA46 were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and incubated in 5% CO2 at 37°C. In some experiments, PKA was induced by treatment of cells with 1 μM dibutyryl cAMP (Sigma).
Plasmids.
Reporter plasmids (pEFP40 and CpLUC) containing EBV sequences from nucleotides 10312 to 11336, which correspond to nucleotides −1021 to +3 relative to the C promoter (Cp) transcriptional initiation site, have been described previously (19). Derivatives of pEFP40 containing mutations in the CBF2 (pEFP56 or mut.CBF2CpLUC) and CBF1 (pEFP76 or mut.CBF1CpLUC) binding sites have also been described previously (19). Plasmid pEFP100 carries 16 copies of the wild-type Cp CBF2 binding site (16xCBF2LUC), and pEFP101 carries 16 copies of a mutant CBF2 binding site (GGTTCA→GGGGCA) cloned into the pGL3 promoter vector. Construction of multiple tandem copies of the CBF2 binding oligonucleotides was done as previously described (45). The oligonucleotides used for these constructions are as follows: CBF2 binding site: 5′ GATCTAAAAATTTATGGTTCAGTGCGTCGAGTGCTG 3′ (sense strand) and 5′ GATCCAGCACTCGACGCACTGAACCATAAATTTTTA 3′ (antisense strand); and mutant CBF2 binding site (transversion changes are underlined): 5′ GATCTAAAAATTTATGGGGCAGTGCGTCGAGTGCTG 3′ (sense strand) and 5′ GATCCAGCACTCGACGCACTGCCCCATAAATTTTTA 3′ (antisense strand).
Plasmid pRSP18 is a derivative of pPDL176A, which contains the wild-type version of EBNA2 cloned in the SG5 expression vector and an N-terminal FLAG sequence which was introduced in-frame with the EBNA2 initiation codon. The LMP-2A reporter plasmid contains the LMP-2A promoter region from −276 to +91 and was generously provided by Ursula Zimber-Strobl (LMP2ALUC) (69). Plasmid pPDL151 expressing the wild-type form of EBNA2 has been described previously (45). The plasmid expressing the catalytic subunit of αPKA was generously provided by Richard Maurer (46). AUF1 plasmids have been described previously (68).
Protein purification and microsequencing.
CA46 cells were used to produce nuclear extracts by a standard Dignam procedure. Nuclear extracts from 2 × 1010 cells were fractionated by application to a heparin-Sepharose column and eluted with a linear 0.1 to 1.0 M KCl gradient. After elution from the heparin-Sepharose column, the CBF2 activity of the fractions was determined by an electrophoretic mobility shift assay (EMSA) using the Cp CBF2 binding site as a probe. Fractions with binding activity were combined and further purified by DNA affinity chromatography using the Cp CBF2 oligonucleotide coupled to agarose, using procedures described previously (28). Fractions obtained from the DNA affinity column containing CBF2 binding activity were tested by EMSA. The proteins present in the fractions with CBF2 activity were visualized by silver staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel. UV cross-linking analysis was performed as previously described to reveal the protein which bound to the CBF2 binding oligonucleotide (19). To obtain the amino acid sequence from this protein, the gel fragment containing the activity was excised and digested in situ with Lys-C endoprotease. Single peptides were subjected to NH2-terminal microsequencing (Baylor College of Medicine protein sequencing core facility).
EMSA.
EMSAs were performed as described previously (19). Unless otherwise indicated, pooled fractions containing CBF2 activity derived from CA46 nuclear extracts that had been purified by heparin-Sepharose chromatography were used for EMSAs (44). For competition assays, unlabeled mutant oligonucleotides were annealed and added to the binding reaction at the indicated molar concentrations relative to the labeled wild-type oligonucleotide probe. For competition assays, unlabeled competitor oligonucleotides were incubated for 10 min with the nuclear extracts before addition of the radioactive probe. After EMSA, the amount of bound CBF2 activity was quantitated with a Molecular Dynamics PhosphorImager. Relative affinities of oligonucleotide binding to CBF2 were calculated as described previously (19). The following series of oligonucleotides were used in EMSAs (top strand only shown): wild-type Cp CBF2, 5′ GATCAATTTATGGTTCAGTGCGTCGAGTGCTAG 3′ (19); Mut Cp CBF2, 5′ GATCAATTTATGGGGCAGTGCGTCGAGTGCTAG 3′ (19); hnRNP A, 5′ GATCTATGATAGGGACTTAGGGTG 3′ (5); hnRNP C, 5′ GATCCTGGGAGGAGTTGGGGGAGGAGATTAGG 3′ (57); CarG, 5′ GATCTTTTTTACCTAATTAGGAAATG 3′ (55); and AUF1, 5′ GATCTAGATTACTTCAAAATAA 3′ (59).
Each top-strand oligonucleotide contains a GATC 5′ overhang when annealed to its complimentary oligonucleotide and enables the duplex oligonucleotides to be labeled by a Klenow fill-in procedure (44).
Antibodies.
Monoclonal antibodies 4B10 (anti-hnRNP A), 4F4 (anti-hnRNP C), and 5B9 (anti-AUF1/hnRNP D) were kindly provided by Gideon Dreyfuss (7, 52). Monoclonal antibodies to nucleolin, CREB/ATF, and Jun/Fos proteins were purchased from Santa Cruz Biotechnology. Polyclonal serum against AUF1 was prepared as described previously (68).
Immunoblotting.
DG75 cells were transfected with expression plasmids and 48 h later were lysed in radioimmunoprecipitation assay buffer containing protease inhibitors. Protein (30 μg) was resolved on an SDS–8% polyacrylamide gel and then transferred to nitrocellulose membranes (Schleicher and Schuell). The membrane was blocked overnight with 5% nonfat dried milk and 0.05% Tween 20 in phosphate-buffered saline (PBS). Anti-FLAG M5 monoclonal antibody (Sigma) was added at a 1:5,000 dilution and incubated for 1 h at room temperature. After several washes in PBS, a secondary antibody, horseradish peroxidase-conjugated anti-mouse immunoglobulin (Ig) (Santa Cruz Biotechnology) was added at a 1:3,000 dilution. The proteins recognized by the antibodies were detected by an enhanced chemiluminescence Western detection system (Pierce). Bands were quantified with a densitometer (Bio-Rad).
Transfections and luciferase assays.
Cells were transfected by the DEAE-dextran method as previously described (19). Transfections were harvested after 2 days of incubation, and the luciferase dual system (Promega) was used to determine the luciferase activity of the reporter genes and of the internal control (pRL-SV40; Promega) according to the manufacturer's instructions.
RESULTS
AUF1/hnRNP D binds the Cp CBF2 binding site.
In order to identify the protein component(s) that mediates CBF2 activity, nuclear extracts from CA46 cells were purified by heparin-Sepharose and DNA affinity chromatography (see Materials and Methods). Fractions obtained from the DNA affinity column containing the CBF2 binding activity were determined by EMSA using the CBF2 binding oligonucleotide as a probe. While several polypeptides of 15 to 45 kDa were observed, UV cross-linking showed that only a peptide of 40 kDa reacted with the CBF2 binding oligonucleotide probe (data not shown). The 40-kDa protein was excised from the polyacrylamide gel, and the amino acid sequence was determined. Three internal peptides derived from digestion of the protein with endonuclease Lys-C were sequenced. The sequences of the three peptides had 100% homology to the protein AUF1/hnRNP D (Fig. 1).
FIG. 1.
Amino acid sequences of three peptides derived from purified CBF2 polypeptide aligned with the AUF1/hnRNP D protein. The sequence of the AUF1/hnRNP D protein has been reported previously (68). The peptide sequences are shown below the region matching the AUF1/hnRNP D protein sequence (top line).
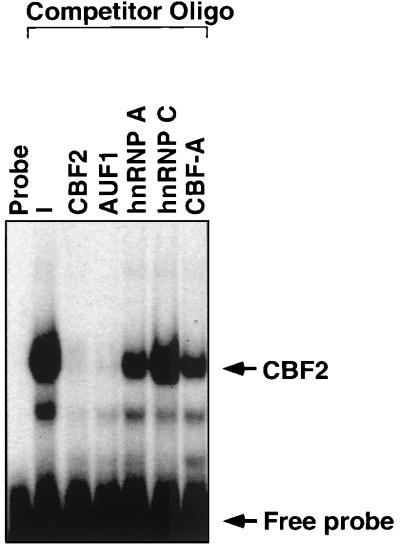
Because of the high homology between members of the hnRNP proteins, oligonucleotides containing the binding sites for some of the hnRNPs were synthesized, and the affinity of each hnRNP binding site for the CBF2 activity was measured by EMSA. hnRNPs are mostly single-stranded RNA binding proteins, although they have also been shown to bind single-stranded DNA with high affinity. Single-stranded DNA oligonucleotides containing the binding sites for AUF1, hnRNP A and C, and the hnRNP-related protein CarG binding factor (CBF-A) were used as competitors in an EMSA. CA46 nuclear extracts (see Materials and Methods) were used as a source of CBF2 for these studies. The affinity of each hnRNP binding site for the CBF2 activity was measured by its ability to compete for binding with the double-stranded Cp CBF2 binding site. Competitor oligonucleotides were used at a 40 M excess relative to the concentration of the probe. The competition assay showed that the oligonucleotide containing the binding site for AUF1 had the highest affinity for CBF2 relative to the other hnRNP binding oligonucleotides (Fig. 2). Competition with this binding site was similar to competition with the CBF2 binding site itself (Fig. 2). Using a 40 M excess of competitor, the hnRNP A oligonucleotide competes with 50% efficiency relative to the CBF2 binding oligonucleotide. This oligonucleotide contains the telomeric sequence TTAGGG that has also been reported to bind AUF1, which may explain the partial competition. At this concentration of competitor, there is also a partial competition with the CBF-A oligonucleotide, while there is no competition with the hnRNP C binding site. These results indicate that the activity that binds the Cp CBF2 binding site has a similar binding specificity to AUF1.
FIG. 2.
Affinity of hnRNP binding sites for CBF2. Different hnRNP binding oligonucleotides were used to compete for CBF2 binding activity in an EMSA. CA46 nuclear extracts were mixed with a 40 M excess of the unlabeled hnRNP oligonucleotides. A 32P-labeled oligonucleotide containing the CBF2 binding site, Cp sequences −339 to −368, was then added to the shift reaction. The unlabeled competitor oligonucleotide (see Materials and Methods) used are indicated above the autoradiograph. Lanes with probe only (probe) and nuclear extract with no competitor added (−) are also shown.
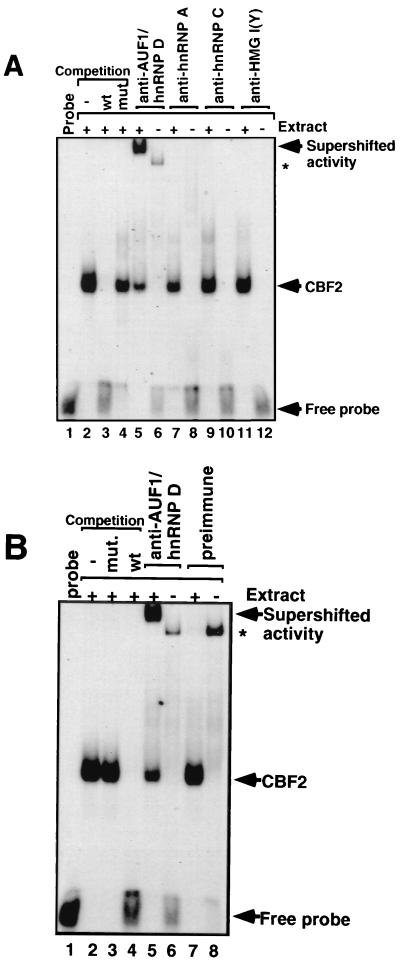
To further identify the component of the CBF2 binding activity, different antibodies against hnRNPs were tested for their ability to react with CBF2 complexes in an EMSA (Fig. 3A). Antibodies to AUF1/hnRNP D supershifted CBF2, while antibodies to hnRNP A or C or a negative control, HMG I(Y), were unable to supershift CBF2 (Fig. 3A). The anti-AUF1 serum was able to complex with approximately 70% of CBF2 binding activity as determined by quantitation of the EMSA bands using a PhosphorImager. A nonspecific binding activity is present when antibody is incubated without nuclear extract which is also present in the preimmune sera (Fig. 3B). Preimmune antiserum does not affect CBF2 binding activity or produce a supershifted complex (Fig. 3B). This result indicates that AUF1 is a major part of the cellular activity that binds the Cp CBF2 binding site.
FIG. 3.
Anti-AUF1/hnRNP D binds CBF2. Anti-hnRNP antibodies were incubated with CA46 nuclear extracts, and after 30 min of incubation, a 32P-labeled oligonucleotide probe containing the binding site for CBF2 was added to the reaction. (A) Autoradiography of a supershift analysis with antibodies against hnRNPs AUF1/hnRNP D (lane 5), A (lane 7), and C (lane 9) and control antibody against the HMG I(Y) protein (lane 11). Antibodies added to gel shift reactions without any added nuclear extract are shown in lanes 6 (AUF1/hnRNP D), 8 (hnRNP A), 10 (hnRNP C), and 12 [HMG I(Y)]. The CBF2-specific complex is confirmed by competitions with wild-type (wt) and mutant (mut) oligonucleotides shown in lanes 2 to 4. (B) Autoradiography of the supershift with preimmune and anti-AUF1/hnRNP D antisera. Lanes 1 to 6 are the same as in panel A. Preimmune serum was added to reactions in lanes 7 and 8 with and without nuclear extract, respectively. Both preimmune and immune sera form minor complexes with the labeled DNA probe and are indicated by asterisks.
CBF2/AUF1 activity binds both double- and single-stranded DNA.
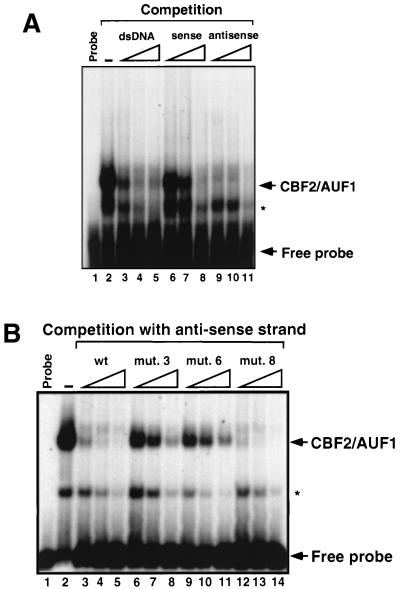
Like other RNA binding proteins, AUF1 has been reported to bind single-stranded DNA. We therefore estimated by EMSA the preference of AUF1 binding for double-stranded and the sense and antisense strands of the Cp CBF2 binding site. CA46 nuclear extracts were mixed with increasing concentrations of the unlabeled double-stranded or single-stranded sense and antisense CBF2 binding oligonucleotides, and CBF2/AUF1 binding activity was detected by EMSA. Single-stranded antisense DNA had the highest affinity for CBF2, while the double-stranded DNA and single-stranded sense DNA oligonucleotides had twofold and sixfold decreasing affinities, respectively (Fig. 4A). The ability of CBF2 to bind single-stranded DNA and to prefer binding to the antisense strand is similar to previous studies of AUF1 binding preferences (60).
FIG. 4.
CBF2/AUF1 is a sequence-specific DNA binding protein that binds single- and double-stranded DNA. (A) Autoradiograph of a competitive EMSA in which double- and single-stranded DNAs were used as competitors. A duplex oligonucleotide containing the Cp CBF2 binding site was used as a probe, and the same oligonucleotide in its double- or single-stranded form was used as the competitor. Competitor oligonucleotides were added at a 2.5-, 5-, and 25-fold molar excess relative to the probe, as indicated by the open triangles. Probe alone and probe bound with nuclear extract only are shown in lanes 1 and 2, respectively. (B) Autoradiograph of an EMSA in which different mutant CBF2 binding sites were used as competitors for CBF2 binding to the Cp CBF2 oligonucleotide probe. The indicated competitor oligonucleotide probes (see text) were added at a 2.5-, 5-, and 25-fold molar excess relative to the probe, as indicated by the open triangles. Probe alone and probe bound with nuclear extract only are shown in lanes 1 and 2, respectively. The asterisks denote a smaller shifted complex that is seen occasionally and is dependent on the batch of nuclear extract used for the experiments.
We then proceeded to test if the binding to single-stranded DNA had the same specificity as binding to double-stranded DNA. We have shown that the most critical residues for CBF2 binding in Cp are to the sequence GGTTCA (or TGAACC of the antisense strand) (19). A competitive EMSA was performed using several mutant CBF2 oligonucleotide probes that have been characterized previously. Mutants 3 (GGTTCA→ ttTTCA) and 4 (GGTTCA→GGggCA) have changes within the residues important for CBF2 binding. Mutant 8 has changes of two nucleotides eight bases downstream of the GGTTCA motif, and this mutant has an increased affinity for binding (19). The [32P]antisense strand was used as a probe, and mutant antisense oligonucleotides were added as unlabeled competitors to the shift reaction. Competition results show that mutant 8 has the higher affinity for binding, while mutants 3 and 4 have a greatly reduced affinity (Fig. 4B). These competitions indicate that the binding to single-stranded DNA is also highly specific and has the same requirement for the GGTTCA sequence binding to the double-stranded binding site.
The CBF2 oligonucleotide contains an A/T-rich region upstream of the GGTTCA sequence that resembles binding sites for AUF1/hnRNP D, hnRNP A, and CarG. Mutagenesis of the A/T-rich region did not significantly affect the binding of the CBF2/AUF1 activity, indicating that the GGTTCA sequence is the only binding site for CBF2/AUF1 present in the Cp CBF2 oligonucleotide (data not shown).
AUF1/hnRNP D activity is induced by stimulation of the cAMP/PKA signaling pathway.
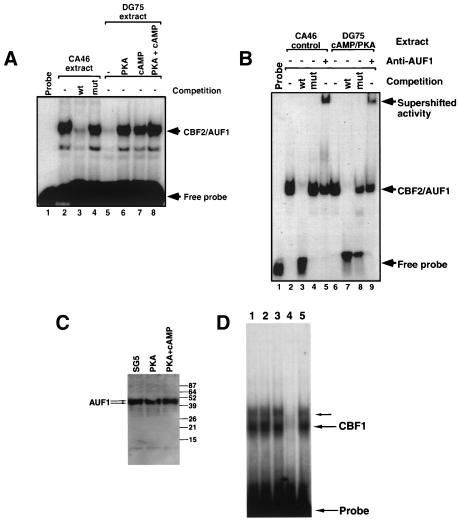
In vitro binding of AUF1 to the cellular CD21 promoter is enhanced in cellular extracts derived from cells treated with cAMP analogs (59). Activation of the cAMP/PKA signaling pathway also results in upregulation of the CD21 promoter (59). Since the cAMP/PKA signaling pathway appeared to influence AUF1-mediated activation of the CD21 promoter, we wanted to determine if induction of the cAMP/PKA signaling pathway would also result in increased CBF2/AUF1 binding activity. DG75 cells were treated with different conditions to stimulate the cAMP/PKA pathway. DG75 cells were used for these studies because they are more amenable to transfection than CA46 cells (unpublished observations) (41). CA46 cells were used in the earlier biochemical studies because they are more easily adapted for growth in spinner cultures that were used to generate large numbers of cells for preparation of nuclear extracts for biochemical purification. Previous studies have indicated that EBNA2 activity is indistinguishable in the two cell types (41). DG75 cells were either transfected with a plasmid that expresses the catalytic subunit of αPKA, incubated with the cAMP analog dibutyryl cAMP, or given both stimuli. Cellular extracts were obtained 48 h posttreatment, and equal concentrations of protein were used in an EMSA to test the CBF2/AUF1 binding activity after induction. Unstimulated or untransfected cell extracts were used as a negative control. Highly purified and concentrated cellular extracts from CA46 cells were used as a positive control. CBF2/AUF1 binding activity was induced ninefold when the cells were transfected with a PKA expression plasmid and treatment with dibutyryl cAMP, while individual stimulation with plasmid transfection or the cAMP analog caused a six- and fivefold induction, respectively (Fig. 5A). Both the CA46 CBF2/AUF1 activity and the cAMP/PKA-stimulated activity of DG75 extracts had the same specificity for binding to the Cp CBF2/AUF1 site, as demonstrated by competition with wild-type and mutant binding sites (Fig. 5B). Furthermore, the cAMP/PKA-stimulated activity is also recognized by an anti-AUF1 antibody (Fig. 5B). These results strongly suggest that the activity from DG75 cells that is upregulated by stimulation of the cAMP/PKA pathway is the same CBF2/AUF1 activity present in the heparin-purified nuclear extracts from CA46 cells. Thus, the CBF2/AUF1 activity that binds the EBNA2 enhancer in the Cp is targeted for regulation by signals transduced by the cAMP/PKA cellular pathway. To address whether stimulation through the cAMP/PKA pathway modifies existing CBF2/AUF1 protein to bind DNA or induces transcriptional or translational mechanisms that result in greater synthesis of CBF2/AUF1, we also performed immunoblot analysis on the cellular extracts used in Fig. 5A. Using antibodies to the AUF1 protein (68), the total amounts of AUF1 detected by immunoblot did not change when the cAMP/PKA pathway was induced (Fig. 5C). Thus, induction of cAMP/PKA is likely to modify existing pools of AUF1 that result in induction of AUF1 binding. To test whether other transcription factors were affected by PKA/cAMP treatment, we also performed an EMSA using a CBF1-specific DNA probe. DG75 cells treated cotransfected with a PKA expression plasmid or treated with dibutyryl cAMP did not have any demonstrable increase in CBF1 binding. Thus, PKA/cAMP induction does not appear to have global effects on DNA binding activity of other transcription factors.
FIG. 5.
CBF2/AUF1 binding activity is increased by induction of the cAMP/PKA signal transduction pathway. (A) Autoradiograph of an EMSA that shows CBF2/AUF1 binding from heparin-Sepharose-purified CA46 control extracts (lanes 2 to 4) and cell extracts from DG75 cells that were untreated (lane 5), transfected with a plasmid constitutively expressing PKA (lane 6), treated with a cAMP analog dibutyryl cAMP (lane 7), or both (lane 8). Heparin-Sepharose-purified CA46 extracts were used as a control, and competitions with wild-type (wt) and mutant (mut) CBF2 binding oligonucleotides were used as a control for specificity of binding (lanes 2 to 4). (B) Autoradiograph of an EMSA that shows CBF2 binding from partially purified CA46 control extracts (lanes 2 to 4) and cell extracts from DG75 cells transfected with a plasmid constitutively expressing PKA and treated with the cAMP analog dibutyryl cAMP. Wild-type competitor oligonucleotide inhibits CBF2 (lanes 3 and 7), while the mutant competitor does not (lanes 4 and 8). Antibody to AUF1 was added to the gel shift reaction mixtures in lanes 5 and 9. (C) Immunoblot of AUF1 protein from SG5- or PKA-transfected DG75 cells and DG75 cells transfected with PKA and treated with dibutyryl cAMP. Blots were probed with AUF1 antiserum as described previously (66). Molecular size standards are shown on the right (in kilodaltons). AUF1 appears as a doublet of p40 and p42 proteins. (D) Autoradiograph of an EMSA that shows CBF1 binding from DG75 cells transfected with pSG5 expression vector alone (lane 1), a PKA expression vector (lane 2), or a PKA expression vector and treatment with dibutyryl cAMP (lane 3). Lanes 4 and 5 show CBF1 binding in the presence of an unlabeled wild-type CBF1 binding competitor oligonucleotide and a mutant oligonucleotide unable to bind CBF1, respectively. Probe- and CBF1-specific complexes are indicated to the right. In some cases CBF1 binding activity is detected as two bands, with the slower migrating form in less abundance (43; unpublished observations).
cAMP/PKA signaling enhances EBNA2 stimulation of Cp and confers EBNA2 responsiveness to CBF2 binding sites.
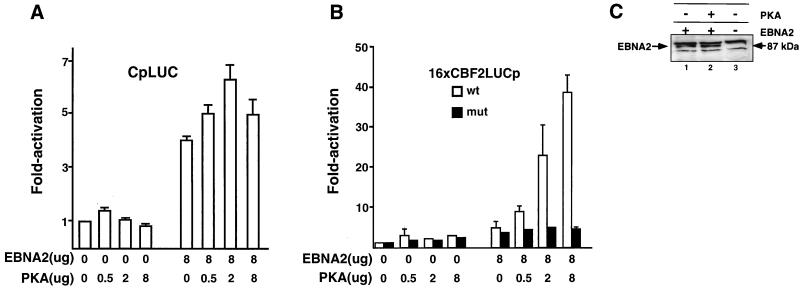
Because the binding of CBF2/AUF1 to the response element in Cp is increased severalfold by cAMP/PKA signaling, we wanted to know if the cAMP/PKA pathway regulates expression of Cp. DG75 cells were cotransfected with a Cp reporter construct (CpLUC) and plasmids constitutively expressing EBNA2 (pPDL151) and the catalytic subunit of αPKA (pPKA). Cp-driven reporter activity was then measured. Interestingly, we found that while activity of Cp is not induced by PKA expression alone, PKA together with EBNA2 expression resulted in higher levels of Cp stimulation (6.3-fold) than observed with EBNA2 expression alone (4-fold) (Fig. 6A). Cotransfection of pPDL151 and pPKA with the control reporter vector without Cp sequences failed to stimulate gene activity (data not shown).
FIG. 6.
Cotransfection of EBNA2 and constitutively active PKA activates Cp and confers EBNA2 responsiveness to a minimal promoter containing CBF2 binding sites. (A) Target plasmids were cotransfected with or without EBNA2- and PKA-expressing plasmids. The amounts of effector plasmids are indicated below the bar graph, and fold activation is indicated to the left. The CpLUC reporter plasmid pEFP40 was used as the target reporter plasmid. Results are averages from three independent experiments. Bars show standard errors of the means. (B) Cotransfection assays using either 16xCBF2LUC (open bars) or 16xmut.CBF2LUC (solid bars) as target plasmids and EBNA2 and/or PKA expression plasmids as effector plasmids. Concentrations of effector plasmids are indicated below the bar graph, and fold activation is indicated to the left. Target plasmids were generally used at 2.0 μg unless otherwise specified. Results are averages from three independent experiments ± standard errors of the mean. (C) Western blot to estimate the abundance of EBNA2 in transfected DG75 cells in the presence and absence of PKA induction. Extracts derived from cells transfected with FLAG-EBNA2 (lane 1), EBNA2 and PKA (lane 2), or vector alone (lane 3) were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted using the M5 anti-FLAG antibody. The EBNA2-specific band is indicated by the arrow on the left. Nonspecific bands were detected by the FLAG monoclonal antibody in all cellular extracts.
Since the CBF2/AUF1 binding activity is increased when the cAMP/PKA signaling pathway is stimulated, we tested whether Cp CBF2/AUF1 binding sites alone could confer EBNA2 responsiveness when the cAMP/PKA pathway was activated. Reporter plasmids containing multimerized wild-type (pEFP100 or 16xCBF2LUC) and mutant CBF2 (pEFP101 or 16xmut.CBF2LUC) binding sites were cotransfected with EBNA2 or PKA expression plasmids. In the absence of PKA cotransfection, EBNA2 did not stimulate either reporter plasmid containing wild-type or mutant binding sites to any appreciable level (Fig. 6B). However, cotransfection of both EBNA2 and PKA expression plasmids resulted in stimulation of reporter gene activity up to 38-fold for wild-type reporter plasmids, but stimulated reporter plasmids containing mutant CBF2 binding sites to only 2- or 3-fold above background levels (Fig. 6B). While these data demonstrate a powerful ability of PKA to confer EBNA2 responsiveness on CBF2 reporter plasmids, it is likely that in the context of natural promoters containing only a single CBF2 binding site, the role of CBF2 will be somewhat reduced. This is in agreement with data in Fig. 6A that shows an increase in EBNA2-mediated Cp activation by PKA stimulation that, while statistically significant, is somewhat more modest. Levels of endogenous PKA modulation may also mask some of CBF2's effect on Cp activation and are addressed in the experiments shown in Fig. 8. As a control, cotransfection of a plasmid expressing the α isoform of PKC was unable to stimulate the expression of 16xCBF2LUC, indicating a specific requirement for the cAMP/PKA pathway (data not shown). A different level of specificity was demonstrated in experiments in which plasmids expressing the active cytoplasmic form of human Notch 1 and Notch 2, which also activate promoters through interaction with CBF1, were used instead of EBNA2. Neither form of Notch was able to activate the 16xCBF2LUC reporter plasmid either alone or combined with PKA (data not shown).
FIG. 8.
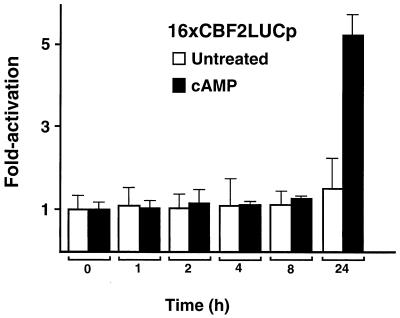
Kinetics of PKA enhancement of EBNA2-mediated transactivation after treatment with dibutyryl cAMP. DG75 cells were cotransfected with the 16XCBF2LUC reporter and the EBNA2 expression plasmid (8.0 μg of each) and 2.0 μg of the 16xCBF2LUC reporter plasmid. At 24 h after transfection, cells were stimulated by adding 1 μM dibutyryl cAMP to the culture medium (solid bars). Transfected but untreated cells were used as a control (open bars). Cells were harvested at different points after induction, and the luciferase activity of the reporter gene was measured. Time points are shown below the graph in hours, and fold activation is shown on the left. Results are representative of three independent experiments ± standard errors of the mean.
To rule out the possibility that PKA expression affects EBNA2 expression in the transient cotransfection assays, we performed an immunoblot analysis to detect EBNA2 protein levels. An N-terminally FLAG-tagged EBNA2 (pRSP18) was transfected into DG75 cells with and without pPKA. Western blot analysis using an anti-FLAG antibody shows that the abundance of EBNA2 in the transfected cells is not significantly induced by cotransfection with PKA (Fig. 6C). Dose-response experiments using pRSP18 did not show any difference in its ability to stimulate the 16xCBF2LUC reporter compared to stimulation by nontagged EBNA2 (data not shown).
Enhancement of EBNA2-mediated stimulation of Cp by PKA is dependent on a functional CBF2/AUF1 binding site.
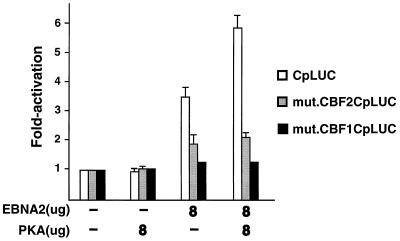
Because EBNA2 stimulation of both Cp and CBF2/AUF1 reporter plasmids is enhanced by PKA signaling, we hypothesized that the activation of the full-length Cp would be in part dependent on the CBF2/AUF1 binding site. A Cp reporter plasmid containing a mutation in the CBF1 binding site was also examined to determine if there was a dependence on CBF1 for this activity. As expected, there was an almost fourfold activation of the CpLUC with EBNA2 alone and a sixfold activation with EBNA2 and PKA coexpression (Fig. 7). The mut.CBFCp1LUC reporter plasmid was not stimulated under any condition, while the mut.CBF2CpLUC reporter was slightly activated. We have previously reported that mut.CBF2CpLUC can be activated to low levels when cotransfected with high levels of EBNA2 expression plasmid (19). These results indicate that the PKA-dependent enhancement of EBNA2 stimulation of Cp requires both CBF2/AUF1 and CBF1 elements. Alternatively, the presence of only a single CBF2 binding site in the context of Cp may not be sufficient to confer EBNA2 responsiveness under these conditions.
FIG. 7.
Enhancement of EBNA2-mediated stimulation of Cp is dependent on CBF1 and CBF2 binding sites. DG75 cells were transfected with target promoters CpLUC (open bars), CBF2mut.CpLUC (shaded bars), and CBF1mut.CpLUC (solid bars). Cells were harvested 48 h after transfection, and the luciferase activity was calculated. Target plasmids were cotransfected with EBNA2- or PKA-expressing plasmids alone or together. Target plasmids were generally used at 2.0 μg unless otherwise specified. Concentrations of effector plasmids used are indicated below the bar graph, and fold activation is indicated to the left. Results represent an average of three independent experiments ± standard errors of the mean.
Kinetics of CBF2LUCIp activation after cAMP induction.
To further characterize the PKA enhancement of EBNA2 stimulation of the 16xCBF2LUC reporter plasmid, the kinetics of promoter activation were determined after treatment with dibutyryl cAMP. DG75 cells were cotransfected with the 16xCBF2LUC- and EBNA2-expressing plasmids. At 24 h after transfection, fresh culture medium containing dibutyryl cAMP was added to the cells. Cells were harvested at different times postinduction, and luciferase activity was measured. No promoter activity was detected at early times postinduction. However, at 24 h there was a fivefold stimulation (Fig. 8). Similar transfections with the CpLUC reporter also indicated that enhancement by PKA induction required at least 24 h (data not shown). The levels of 16xCBF2LUC induction are lower than those observed when constitutively active PKA is expressed by cotransfection. The difference is most likely due to the fact that cAMP only activates existing PKA in the cell, while the PKA expression plasmid provides higher levels of constitutively active PKA expression. These data suggest that direct modification of CBF2/AUF1 by PKA is not responsible for PKA-mediated enhancement and that events further downstream are likely mediating transcriptional enhancement through EBNA2.
Kinase activity of PKA required for enhancement of EBNA2 stimulation of Cp.
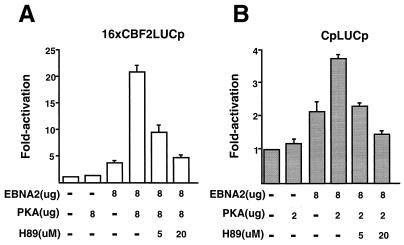
To determine whether PKA kinase activity is required for its ability to enhance EBNA2-mediated transactivation, a kinase inhibitor was tested. H89 is a Ser/Thr kinase inhibitor which has a 10-fold-higher specificity for PKA than for other cellular Ser/Thr kinases. DG75 cells were transfected with the 16xCBF2LUCp or CpLUC reporter plasmid and expression plasmids for EBNA2 and PKA. In addition, in some of the transfections, the H89 inhibitor was also added. H89 treatment was able to block EBNA2/PKA-mediated stimulation of both 16xCBF2LUC and CpLUC to levels of activation observed with EBNA2 alone (Fig. 9A and B). The results obtained suggest that the kinase activity of PKA is required to enhance EBNA2-mediated stimulation of Cp and to reporters containing CBF2 binding sites alone.
FIG. 9.
Effect of Ser/Thr kinase inhibitor H89 on the enhancement of EBNA2-mediated stimulation of Cp by PKA. (A) DG75 cells were cotransfected with the 16xCBF2LUC reporter and EBNA2 and/or PKA expression plasmids. Target reporter plasmids were used at 2.0 μg, and the effector plasmid concentrations are indicated below the graph. The concentrations of H89 are also indicated below the graph. Cells were harvested 48 h after transfection, and the luciferase activity was calculated. Fold activation levels are shown on the left. Results represent an average of three independent experiments ± standard errors of the mean. (B) Same as panel A except the CpLUC reporter plasmid was used.
Enhancement of EBNA2 transactivation by PKA-mediated signaling is not a general characteristic of all EBNA2-responsive promoters.
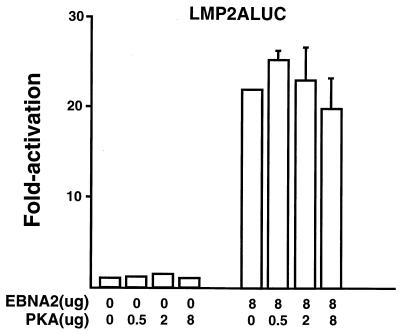
Among the EBV latency promoters, EBNA2 stimulates the Cp, LMP-1, and LMP-2A promoters. Because the cAMP/PKA pathway targets both the Cp and LMP-1 promoters, it is possible that this type of regulation is a general characteristic of all latent EBNA2-responsive promoters. In order to test this question, the EBNA2 enhancer present in the LMP-2A promoter was tested for its ability to respond to PKA in transient cotransfection experiments (Fig. 10). The LMP-2Ap is activated 22-fold when cotransfected with EBNA2. However, when cotransfected with a PKA-expressing plasmid, no further stimulation occurs (Fig. 10). This indicates that signaling through PKA does not target all EBNA2-responsive promoters. An alternative explanation is that at the concentrations of EBNA2 effector plasmid used, the LMP-2A promoter is already saturated and remains unresponsive to further stimulation. Similar transfection experiments with the LMP-2A promoter were performed using different concentrations of the EBNA2-expressing plasmid, with the same results (data not shown). In agreement with these transfection results, our previous results showed that CBF2/AUF1 does not bind to the EBNA2 enhancer located in LMP-2A promoter sequences (19).
FIG. 10.
Analysis of the ability of EBNA2-responsive elements to be activated by PKA. DG75 cells were transfected with the LMP-2A promoter. Some cells were also transfected with EBNA2-expressing plasmids or the PKA expression plasmid. Cells were harvested 48 h after transfection, and the luciferase activity was calculated. Results represent an average of three independent experiments ± standard errors of the mean.
Expression of AUF1 cDNA results in a dose-dependent increase in EBNA2 stimulation of Cp.
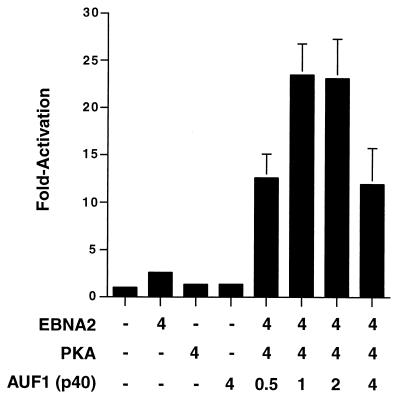
To further confirm that AUF1 was conferring EBNA2 responsivness on CBF2-containing promoters, we performed cotransfection experiments with AUF1 cDNA expression plasmids. Cotransfection of EBNA2 with PKA or AUF1 alone did not result in any activation of the 16xCBF2LUC reporter (Fig. 11). However, cotransfection of all three plasmids resulted in a 10-fold activation of 16xCBF2LUC that was further increased by cotransfection with increasing amounts of AUF1 expression plasmid (Fig. 11). Since AUF1 exists as four different isoforms (p37, p40, p42, and p45), we used the p40 isoform first, since this isoform was identical in size to the purified protein identified previously (Fig. 1) and also identified in immunoblot analysis (Fig. 5C). However, subsequent experiments using the other AUF1 isoforms also have shown identical results (data not shown).
FIG. 11.
Overexpression of an AUF1 cDNA results in stimulation of a minimal promoter containing CBF2 binding sites in a dose-dependent manner. Target plasmids were cotransfected with plasmids expressing EBNA2, PKA, or AUF1 or not transfected. The amounts of effector plasmids are indicated below the bar graph, and fold activation is indicated to the left. The 16xCBF2LUC reporter plasmid was used as the target reporter plasmid. Results are averages from three independent experiments ± standard errors of the mean.
DISCUSSION
In this study, we demonstrated that AUF1/hnRNP D binds the CBF2 binding site from the EBV latency Cp. AUF1/hnRNP D binds to both single- and double-stranded DNA containing the CBF2 element in a sequence-specific manner. Stimulation of the cAMP/PKA signaling pathway results in an increase in detectable AUF1/hnRNP D binding to the CBF2 element from Cp. We also found that Cp CBF2 binding sites alone were able to confer EBNA2 responsiveness to a heterologous promoter in the presence of stimulatory signals from the cAMP/PKA pathway. Finally, the ability of EBNA2 to stimulate Cp is enhanced by cAMP/PKA signaling, and this regulation requires functional CBF2 and CBF1 binding elements.
Although AUF1/hnRNP D was initially characterized as a factor that regulated mRNA stability, recent studies have also shown that it functions as a transcription factor. The LR1 factor, which is composed of AUF1 complexed with nucleolin, regulates the c-myc and EBV Fp promoters (3, 4, 15, 27). The CD21 promoter is also regulated by AUF1 (59). Like CBF2, both of these activities appear to be B-cell specific, and the binding sites have some similarity to the CBF2 element (Fig. 12) (3, 59, 60). None of the DNA response elements for these factors appears to resemble the previously reported RNA binding sites (14, 23, 30, 31, 35, 36, 48). It is worth noting that both c-Myc and CD21 are direct targets for EBNA2 activation and that AUF1/hnRNP D might be a general cellular cofactor required for EBNA2-mediated activation of some viral and cellular promoters (11, 34).
FIG. 12.
Comparison of the CBF2 binding sequence to AUF1 binding sequences from cellular promoters. Nucleotides shown in bold represent shared sequences from c-myc and CD21 promoters with the core CBF2 binding site. Underlined bases indicate nucleotide positions that, when mutated, affected AUF1 binding to the sequence shown.
The AUF1/hnRNP D activity that binds Cp may also comprise additional cellular proteins. Two observations support this possibility. First, binding of AUF1/hnRNP D to Cp sequences is partially inhibited by oligonucleotides containing the hnRNP A and CarG binding sites. Second, anti-hnRNP A antibody partially depletes the AUF1/hnRNP D binding activity in gel shifts, although a supershifted complex was not observed. Further characterization of the activity binding the Cp AUF1/hnRNP D site is required in order to conclude if AUF1/hnRNP D is working alone or associated with other proteins. Supershift analysis using an antinucleolin antibody (Santa Cruz Biotechnology) was negative, suggesting that this protein does not participate in this activity as it does in LR1 (data not shown). Additionally, we also found that antibodies to CREB and ATF proteins failed to react with CBF2/AUF1 in gel mobility shift assays.
HnRNP proteins are also regulated by phosphorylation (8, 21, 29). Phosphorylation of LR1 is required for binding to DNA target sequences, but the kinase that mediates LR1 phosphorylation is currently unknown. AUF1/hnRNP D has been shown to be phosphorylated in vivo, and such a modification could affect its RNA binding and protein-protein interaction capacity (68). Although the specific signal(s) that results in AUF1/hnRNP D phosphorylation has not been elucidated, there is some evidence that AUF1/hnRNP D expression and/or function may be regulated by the G protein/adenylyl cyclase signal transduction pathway (10, 39, 51, 64). Stimulation of the β-adrenergic receptor with norepinephrine or the agonist isoproterenol results in activation of adenylyl cyclase and an increased abundance of AUF1/hnRNP D (10, 39). Furthermore, treatment of cells with analogs of cAMP results in increased binding of AUF1/hnRNP D to the CD21 promoter that also correlates with its induction (59).
The AUF1/hnRNP D activity that binds Cp is also increased in the presence of inducers of the cAMP/PKA signaling pathway. This result illustrates the possible targeting of cellular cAMP/PKA signaling pathways for control of C promoter expression. Similarly, the LMP-1 promoter is also regulated by cAMP and PKA but, unlike Cp, is mediated through a cAMP response element (CRE) (54). The LMP-1 promoter is also responsive to signaling through the cAMP/PKA pathway and EBNA2, but analysis of whether both are needed for activation was not done (54).
The mechanism by which PKA is conferring EBNA2 responsiveness to the Cp AUF1/hnRNP D binding site is uncertain at this point, and there are different possible explanations. First, PKA may directly modify EBNA2, AUF1/hnRNP D, or CBF1 (e.g., by phosphorylation), and this modification promotes interactions between these proteins. Phosphorylation of EBNA2 at this point seems unlikely, as the bulk of EBNA2 detected in cells cotransfected with pPKA migrates with a mobility similar to that of EBNA2 from untreated cells (Fig. 6C). Second, PKA could modify a different protein, which then promotes or is part of the complex that activates promoter expression. Finally, a more indirect effect would be that PKA is upregulating cellular gene expression through CREB/ATF sites and one of the products of this upregulation is stimulating AUF1/hnRNP D activation effects.
A kinetic analysis of the induction after cAMP treatment shows no detectable activity of the promoter at early times posttreatment. Induction is detected after 16 h of treatment (data not shown), and a four- to sixfold activation is reached after 24 h (Fig. 8). Dibutyryl cAMP is a liposoluble analog of cAMP which is able to diffuse through and penetrate the cellular membrane. It is known that the levels of intracytoplasmic dibutyryl cAMP rise in the first 4 h after cells are placed in the presence of this drug (1, 22, 25, 41). Once cAMP levels have risen, activation of PKA and translocation to the nuclear compartment occur in about 30 min. Therefore, gene expression of promoters with binding sites for transcription factors that are directly modified by PKA have a peak of expression 4 to 8 h after treatment with cAMP analogs (1, 22, 25, 41). Because Cp activation occurs between 16 and 24 h post-cAMP induction, our results seem to be in agreement with a secondary or indirect effect of PKA.
The role of signaling through the cAMP/PKA pathway in the establishment and maintenance of latency in the EBV-infected B lymphocyte has not been studied. However, it is known that activation of this pathway blocks viral reactivation in the presence of stimuli that otherwise would result in activation of the lytic cycle and expression of the EBV immediate-early genes (12). A variety of treatments have been reported to induce viral reactivation of EBV latent infection. The most extensively studied and most consistently effective are treatment with phorbol esters and immune cross-linking of IgG receptors (13, 42, 70). Activation of the EBV lytic cycle by either phorbol esters or anti-IgG receptors is blocked in the presence of an active cAMP/PKA pathway (12). These results suggest a positive role for PKA in the maintenance of latency. Taken together, both positive roles of signaling through the cAMP/PKA pathway in latency and B-cell proliferation may explain the significance of the regulation of the expression of Cp and LMP-1 promoters by this pathway.
Both phorbol esters and cross-linking of the B-cell receptor result in activation of the PKC and downstream activation of mitogen-activated protein kinase (MAPK). In several cell lines, activations of cAMP/PKA and MAPK pathways have antagonistic functions (6, 16). Therefore, the contribution of the cAMP/PKA pathway to EBV latency may be given at different levels: (i) by counteracting PKC and MAPK function; (ii) by stimulating latent gene expression; and (iii) by providing the appropriate cellular milieu where latency is maintained. It is interesting to note that upregulation of genes coding G protein-coupled receptors by EBV infection has been reported, and cAMP/PKA signaling through these receptors may be an important latency function (2).
In summary, we have demonstrated that AUF1 is a component of CBF2. In addition, stimulation of the cAMP/PKA signaling pathway, which is known to regulate AUF1 activity, has positive effects on EBNA2-mediated stimulation of Cp. Future studies can now be directed towards identification of the mechanisms that mediate this phenomenon.
ACKNOWLEDGMENTS
This work was supported by NIH grants R29 CA69437 (P.D.L.) and RO1 CA52443 (G.B.) and by an award from the William Stamps Farish Foundation (P.D.L.).
REFERENCES
- 1.Armstrong R, Wen W, Meinkoth J, Taylor S, Montminy M. A refractory phase in cyclic AMP-responsive transcription requires downregulation of protein kinase A. Mol Cell Biol. 1995;15:1826–1832. doi: 10.1128/mcb.15.3.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brys A, Maizels N. LR1 regulates c-myc transcription in B-cell lymphomas. Proc Natl Acad Sci USA. 1994;91:4915–4919. doi: 10.1073/pnas.91.11.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulfone-Paus S, Dempsey L A, Maizels N. Host factors LR1 and Sp1 regulate the Fp promoter of Epstein-Barr virus. Proc Natl Acad Sci USA. 1995;92:8293–8297. doi: 10.1073/pnas.92.18.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Iyengar R. Interactions between the Gs/protein kinase A and the Ras/MAP-kinase signalling pathways. Biochem Soc Trans. 1995;23:129–133. doi: 10.1042/bst0230129. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y D, Dreyfuss G. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci USA. 1984;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobianchi F, Calvio C, Stoppini M, Buvoli M, Riva S. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res. 1993;21:949–955. doi: 10.1093/nar/21.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohn J N, Levine T B, Olivari M T, Garberg V, Lura D, Francis G S, Simon A B, Rector T. Plasma norepinephrine as a guide to prognosis in patients with chronic congestive heart failure. N Engl J Med. 1984;311:819–823. doi: 10.1056/NEJM198409273111303. [DOI] [PubMed] [Google Scholar]
- 11.Cordier M, Calender A, Billaud M, Zimber U, Rousselet G, Pavlish O, Banchereau J, Tursz T, Bornkamm G, Lenoir G M. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J Virol. 1990;64:1002–1013. doi: 10.1128/jvi.64.3.1002-1013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daibata M, Humphreys R E, Takada K, Sairenji T. Activation of latent EBV via anti-IgG-triggered, second messenger pathways in the Burkitt's lymphoma cell line Akata. J Immunol. 1990;144:4788–4793. [PubMed] [Google Scholar]
- 13.Davies A H, Grand R J, Evans F J, Rickinson A B. Induction of Epstein-Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J Virol. 1991;65:6838–6844. doi: 10.1128/jvi.65.12.6838-6844.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 15.Dempsey L A, Hanakahi L A, Maizels N. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J Biol Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 16.Dumont J E, Jauniaux J C, Roger P P. The cyclic AMP-mediated stimulation of cell proliferation. Trends Biochem Sci. 1989;14:67–71. doi: 10.1016/0968-0004(89)90046-7. [DOI] [PubMed] [Google Scholar]
- 17.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahraeus R, Jansson A, Ricksten A, Sjoblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuentes-Pananá E M, Ling P D. Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation. J Virol. 1998;72:693–700. doi: 10.1128/jvi.72.1.693-700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuentes-Pananá E M, Swaminathan S, Ling P D. Transcriptional activation signals found in the Epstein-Barr virus (EBV) latency C promoter are conserved in the latency C promoter sequences from baboon and rhesus monkey EBV-like lymphocryptoviruses (cercopithicine herpesviruses 12 and 15) J Virol. 1999;73:826–833. doi: 10.1128/jvi.73.1.826-833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung P A, Labrecque R, Pederson T. RNA-dependent phosphorylation of a nuclear RNA binding protein. Proc Natl Acad Sci USA. 1997;94:1064–1068. doi: 10.1073/pnas.94.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 23.Gorlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 24.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara M, Alberts A, Brindle P, Meinkoth J, Feramisco J, Deng T, Karin M, Shenolikar S, Montminy M. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 26.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 27.Hanakahi L A, Dempsey L A, Li M J, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 29.Idriss H, Kumar A, Casas-Finet J R, Guo H, Damuni Z, Wilson S H. Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry. 1994;33:11382–11390. doi: 10.1021/bi00203a037. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarzembowski J A, Malter J S. Cytoplasmic fate of eukaryotic mRNA: identification and characterization of AU-binding proteins. Prog Mol Subcell Biol. 1997;18:141–172. doi: 10.1007/978-3-642-60471-3_7. [DOI] [PubMed] [Google Scholar]
- 32.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Jin X W, Speck S H. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm G W, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0: common modular structure and binding properties of the 2xRBD-Gly family. J Biol Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 36.Kamada S, Miwa T. A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene. 1992;119:229–236. doi: 10.1016/0378-1119(92)90276-u. [DOI] [PubMed] [Google Scholar]
- 37.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippencott-Raven Publishers; 1996. pp. 107–172. [Google Scholar]
- 38.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knutson J C. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64:2530–2536. doi: 10.1128/jvi.64.6.2530-2536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koh W S, Jeon Y J, Herring A C, Kaminski N E. Transient CRE- and kappa B site-binding is cross-regulated by cAMP-dependent protein kinase and a protein phosphatase in mouse splenocytes. Life Sci. 1997;60:425–432. doi: 10.1016/s0024-3205(96)00667-4. [DOI] [PubMed] [Google Scholar]
- 42.Lazdins J, Zompetta C, Grimaldi S, Barile G, Venanzoni M, Frati L, Faggioni A. TPA induction of Epstein-Barr virus early antigens in Raji cells is blocked by selective protein kinase-C inhibitors. Int J Cancer. 1987;40:846–849. doi: 10.1002/ijc.2910400624. [DOI] [PubMed] [Google Scholar]
- 43.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–658. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 44.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer R A. Both isoforms of the cAMP-dependent protein kinase catalytic subunit can activate transcription of the prolactin gene. J Biol Chem. 1989;264:6870–6873. [PubMed] [Google Scholar]
- 47.Meitinger C, Strobl L J, Marschall G, Bornkamm G W, Zimber S U. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J Virol. 1994;68:7497–7506. doi: 10.1128/jvi.68.11.7497-7506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miwa T, Kedes L. Duplicated CArG box domains have positive and mutually dependent regulatory roles in expression of the human alpha-cardiac actin gene. Mol Cell Biol. 1987;7:2803–2813. doi: 10.1128/mcb.7.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. . (Erratum, 72:9419, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 51.Pende A, Tremmel K D, DeMaria C T, Blaxall B C, Minobe W A, Sherman J A, Bisognano J D, Bristow M R, Brewer G, Port J. Regulation of the mRNA-binding protein AUF1 by activation of the beta-adrenergic receptor signal transduction pathway. J Biol Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- 52.Pinol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. . (Erratum, 2:490.) [DOI] [PubMed] [Google Scholar]
- 53.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 54.Sjoblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smidt M P, Russchen B, Snippe L, Wijnholds J, Ab G. Cloning and characterisation of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res. 1995;23:2389–2395. doi: 10.1093/nar/23.13.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung N S, Kenney S, Gutsch D, Pagano J S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991;65:2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tay N, Chan S H, Ren E C. Identification and cloning of a novel heterogeneous nuclear ribonucleoprotein C-like protein that functions as a transcriptional activator of the hepatitis B virus enhancer II. J Virol. 1992;66:6841–6848. doi: 10.1128/jvi.66.12.6841-6848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorley-Lawson D A, Miyashita E M, Khan G. Epstein-Barr virus and the B cell: that's all it takes. Trends Microbiol. 1996;4:204–208. doi: 10.1016/s0966-842x(96)90020-7. [DOI] [PubMed] [Google Scholar]
- 59.Tolnay M, Lambris J D, Tsokos G C. Transcriptional regulation of the complement receptor 2 gene: role of a heterogeneous nuclear ribonucleoprotein. J Immunol. 1997;159:5492–5501. [PubMed] [Google Scholar]
- 60.Tolnay M, Vereschagina L A, Tsokos G C. Heterogeneous nuclear ribonucleoprotein D0B is a sequence-specific DNA-binding protein. Biochem J. 1999;338:417–425. [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, Gregory C D, Rowe M, Rickinson A B, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci USA. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson G M, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acids Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 65.Woisetschlaeger M, Jin X W, Yandava C N, Furmanski L A, Strominger J L, Speck S H. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci USA. 1991;88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 67.Yoo L I, Mooney M, Puglielli M T, Speck S H. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J Virol. 1997;71:9134–9142. doi: 10.1128/jvi.71.12.9134-9142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimber-Strobl U, Kremmer E, Grasser F, Marschall G, Laux G, Bornkamm G W. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12:167–175. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.zur Hausen H, O'Neill F J, Freese U K, Hecker E. Persisting oncogenic herpesvirus induced by the tumour promoter TPA. Nature. 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]