Abstract
Candida auris is an emerging fungal pathogen responsible for healthcare-associated outbreaks that arise from persistent surface and skin colonization. We characterized the arsenal of adhesins used by C. auris and discovered an uncharacterized adhesin, Surface Colonization Factor (SCF1), and a conserved adhesin, IFF4109, that are essential for colonization of inert surfaces and mammalian hosts. SCF1 is apparently specific to C. auris and its expression mediates adhesion to inert and biological surfaces across isolates from all five clades. Unlike canonical fungal adhesins, which function through hydrophobic interactions, SCF1 relies on exposed cationic residues for surface association. SCF1 is required for C. auris biofilm formation, skin colonization, virulence in systemic infection, and colonization of inserted medical devices.
One-Sentence Summary:
Surface attachment, colonization, and disease caused by an emerging human pathogen is mediated by a lineage-specific adhesion factor.
Since initial reports of its discovery in 2009, the emerging fungal pathogen Candida auris has become an increasingly common source of life-threatening infection worldwide (1, 2). C. auris is frequently reported in association with nosocomial outbreaks, a characteristic rarely described with other Candida species, and is of urgent concern for public health authorities (3-7). C. auris outbreaks are characterized by persistent colonization of patient skin and abiotic surfaces, which can remain positive for extensive periods and serve as a source of contaminative transmission (8-14). C. auris also colonizes indwelling medical devices, which act as a risk factor for the development of invasive disease (15-21). Lapses in diagnostic screening and infection prevention measures are thought to contribute to the increasing rate of C. auris outbreaks (20). The ability of C. auris to robustly colonize a range of living and abiotic substrates is central to its emergence as a global health threat.
Colonization requires the initial physical association and attachment between fungal cells and substrate. For fungal pathogens, attachment is largely mediated by cell surface-exposed adhesin proteins (22). In Candida species, genetic expansion has resulted in the formation of adhesin families containing genes similar in sequence and domain architecture, with adhesive functions that are redundant or specific across family members (23, 24). C. auris encodes genes similar to members of the conserved ALS and IFF/HYR adhesin families found across the genus, although these genes may have expanded independently in C. auris and lack clear one-to-one homology with adhesins from well-characterized species. Moreover, their phenotypic importance in C. auris is not well understood (24-26).
To interrogate the role of individual C. auris adhesins in colonization phenotypes, we measured the adhesion between fungal cells and polymer substrates as a model for surface association. We found that C. auris does not primarily rely on conserved adhesins for surface attachment. Instead, we identified Surface Colonization Factor (SCF1), an adhesin specific to C. auris. SCF1 is necessary and sufficient for robust attachment of C. auris cells to polymer substrates. C. auris isolates from diverse and similar genetic lineages exhibit striking divergence in terms of substrate association, and this phenotypic plasticity is tightly correlated with strain-specific transcriptional control of SCF1. The nonspecific surface association driven by SCF1 does not occur through canonical hydrophobic interactions, but rather through cation-substrate interactions. To explore the clinical relevance of these findings, we investigated the importance of SCF1 in long-term colonization models. SCF1 is critical for biofilm formation in vitro, robust colonization of in vivo central venous catheters, colonization of both human and murine skin, and virulence in disseminated infection. These findings offer insight into the genetic and molecular mechanisms by which C. auris mediates surface association, a trait critical to the increasing disease burden of this emerging pathogen.
Results
Polymer surface attachment by the adhesin SCF1
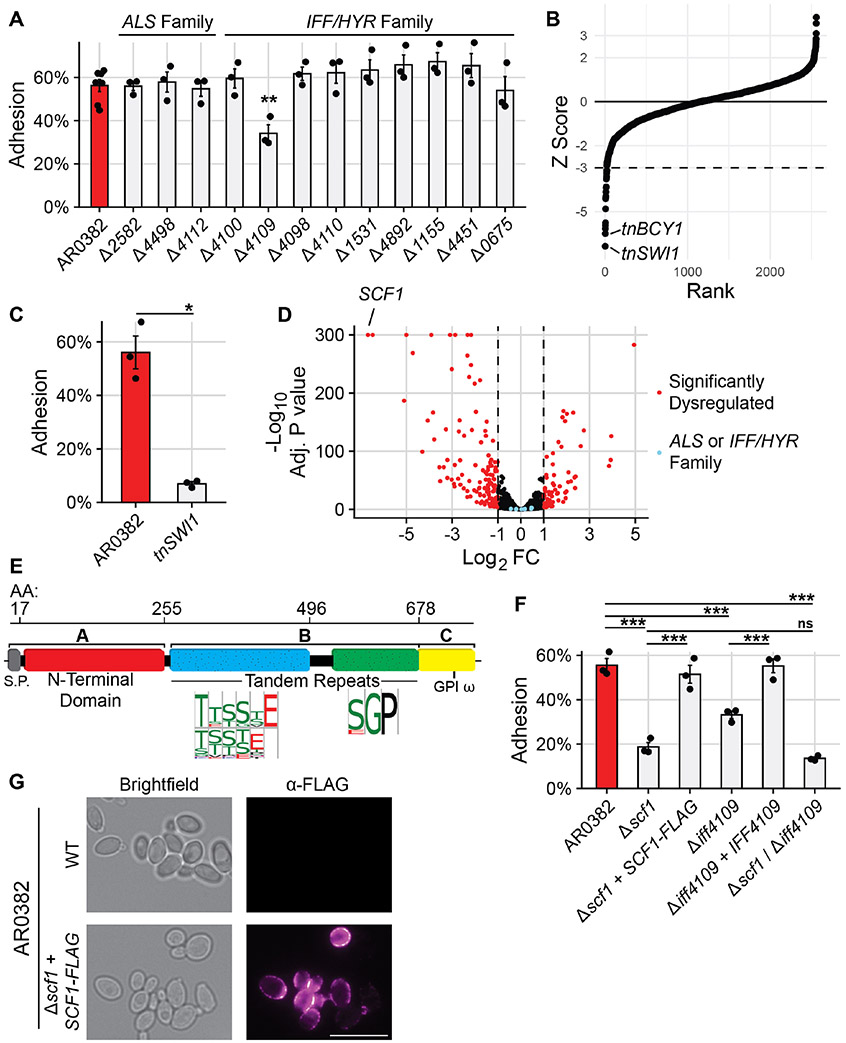
C. auris encodes twelve genes homologous to members of the characterized ALS and IFF/HYR adhesin families (24, 25, 27). We generated individual deletion mutants in the clade I AR0382 background for each adhesin gene to model their impact on surface association. We employed a flow cytometric adhesion assay that measures the ability of cells to attach to dispersed polystyrene microspheres in suspension (Fig. S1) (28). Of the twelve adhesin mutants, only deletion of IFF4109 (B9J08_004109) conferred an adhesive defect, while still failing to completely ablate attachment (Fig. 1A). To investigate the possibility that there were occult adhesive factors, we screened a library of 2,560 insertional mutants, prioritizing mutants exhibiting the most significant defects (Fig. 1B). The greatest loss of adhesive capacity was observed in tnSWI1 (B9J08_003460) and tnBCY1 (B9J08_002818) mutants (Fig. 1B, Fig. 1C, Fig. S2). Compared to the AR0382 parent, the tnSWI1 mutant exhibited no significant transcriptional dysregulation of the ALS or IFF/HYR adhesins, suggesting alternative mediators of adhesion (Fig. 1D). The strongest, most significantly dysregulated gene in tnSWI1 was an uncharacterized ORF (B9J08_001458), which had no significant primary sequence homology to characterized genes (Fig. 1D). This gene, however, exhibited a putative three-domain architecture consistent with canonical GPI-anchored fungal adhesins (Fig. 1E) (23). Notably, this same gene was also strongly downregulated in the tnBCY1 mutant while IFF4109 was not (Fig. S2). Deletion of the B9J08_001458 ORF in AR0382 conferred a substantial adhesive defect, thus we refer to the gene as Surface Colonization Factor (SCF1) (Fig. 1F). Complementation with an epitope-tagged SCF1 allele in the endogenous locus rescued the adhesive defect, and the epitope-tagged Scf1 protein localized to the cell surface, consistent with its role as an adhesin (Fig. 1F, Fig. 1G). Notably, deletion of IFF4109 in the Δscf1 background did not significantly reduce attachment beyond deletion of SCF1 alone, suggesting non-additive roles for these adhesins (Fig. 1F).
Fig. 1. Surface Colonization Factor (SCF1) mediates C. auris adhesion to polymer surfaces.
(A) Adhesion of wild type AR0382 or mutants lacking one of twelve genes from ALS or IFF/HYR adhesin families. (B) 2,560 insertional mutants in the AR0382 strain background were screened for adhesion defects by measuring the proportion of cells able to remain attached to a cyclic olefin polymer surface after 3 washes with PBS. Strains are ordered by Z-score rank. Mutants with a Z-score more negative than −3 were considered to have a significant adhesive defect. (C) Adhesion of AR0382 and an insertional SWI1 mutant. (D) RNA-seq comparing the transcriptome of tnSWI1 to AR0382. SCF1 (B9J08_001458) is the strongest dysregulated gene. (E) Predicted domain architecture of Scf1, based on the clade I primary sequence, is consistent with canonical fungal adhesins. (F) Adhesion of adhesin mutants and complements compared to AR0382 (G) Immunofluorescence microscopy using an α-FLAG antibody. Representative images shown for WT AR0382 and AR0382 Δscf1 + SCF1-FLAG. Scale bar = 5 μm. Statistical differences were assessed using one-way ANOVA with Dunnett’s post-hoc test (A), student’s t-test (C), or one-way ANOVA with Tukey’s post-hoc test (G); *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: p > 0.05.
The specific reliance on SCF1 and IFF4109 for adhesion despite potential redundancy with other adhesins is reminiscent of other fungal pathogens. For instance, loss of ALS1 alone reduces Candida albicans adhesion, despite the presence of seven other ALS genes (Fig. S3A) (29). In C. auris, adhesins exhibit structural and transcriptional variation, which may explain their functional specificity (Fig. S3B) (25, 26). However, IFF4892 encodes the entire canonical adhesin architecture and shows similar expression to IFF4109, but of the two, only IFF4109 is required for adhesion, suggesting individual adhesins mediate specific adhesive mechanisms (Fig. 1A, Fig. S3B) (25). Such functional specificity is shown by increased flocculation and aggregation associated with overexpression of ALS4112, while these phenotypes are not associated with SCF1, despite its transcriptional expression being among the highest 2.5% of all genes in this strain background (Fig. S3B, Fig. S3C, Fig. S3D) (30, 31). These findings suggest functional specificity for surface association for SCF1 and IFF4109.
C. auris relies on SCF1 for adhesive plasticity
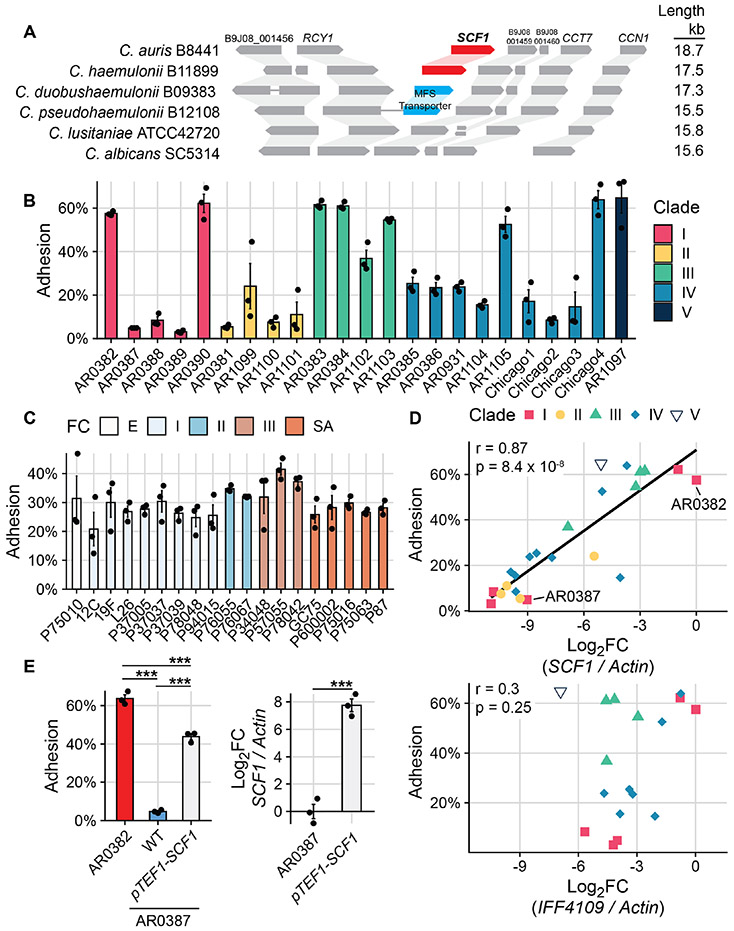
While many Candida and Saccharomyces adhesins belong to conserved gene families, we identified homologs of SCF1 only in C. auris and the closely related Candida haemulonii species and not in other members of the haemulonii complex (Fig. 2A). SCF1 is encoded in a genomic locus in C. auris and C. haemulonii that is syntenic, lacking an SCF1 homolog, even to distantly related species (Fig. 2A). Although the C. haemulonii SCF1 homolog functionally complements Δscf1 in C. auris, it is not essential for adhesion in C. haemulonii and shows poor expression across isolates, indicating reliance on SCF1 for adhesion is specific to C. auris (Fig. S4).
Fig. 2. C. auris uniquely relies on SCF1 for adhesive plasticity.
(A) Synteny schema depicting SCF1 and the conservation and orientation of adjacent ORFs. Genomic loci are shown in comparison to C. auris. Putative SCF1 homologs were only identified in C. auris and C. haemulonii. (B) Adhesion of 23 C. auris clinical isolates from all 5 clades. (C) Adhesion of 19 C. albicans clinical isolates from five clades. FC = Fingerprint Clade. (D) SCF1 transcript abundance (top panel) but not IFF4109 transcript abundance (bottom panel) is associated with adhesion to polystyrene in the same 23 C. auris isolates from (A). Log2FC are expressed relative to AR0382. Each point signifies the mean of three biological replicates. Pearson correlation coefficient and p-value indicated. Isolates that do not encode IFF4109 are not indicated in the bottom panel. (E) Comparison of adhesion between two Clade I isolates: AR0382 and AR0387. Overexpression of SCF1 using the strong TEF1 promoter (right panel) is sufficient to drive adhesion in the poorly adhesive AR0387 background (left panel). Statistical differences were assessed using one-way ANOVA (A) and (B), with Tukey’s post-hoc test (D) or student’s t-test (D); *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: p > 0.05.
To investigate the generalizability of the reliance on SCF1 and the variability between C. auris strains, we measured adhesion for 23 C. auris isolates representing all five clades and diverse geographic origins of C. auris. These strains exhibited substantial adhesive variation, regardless of clade (p=2x10−16, F=35.06, one-way ANOVA) (Fig. 2B). In contrast, a similar analysis of 19 genetically diverse C. albicans clinical isolates showed no significant adhesive variation (p=0.054, F=1.856, one-way ANOVA), indicating the surface association strategies of C. auris are more plastic than C. albicans (Fig. 2C). Interestingly, substantial variation in adhesion was observed even between genetically similar isolates of C. auris, e.g., AR0382 and AR0387, which differ by only 206 coding SNPs (Fig. 2B). In the poorly adhesive AR0387, SCF1 was the most down-regulated gene compared to the highly adhesive AR0382, reminiscent of the poorly adhesive tnSWI1 mutant (Fig. S5A, Fig. 1D). The transcriptome of AR0387 showed little overlap with that of the tnSWI1 strain, however, indicating dysregulation of SCF1 in AR0387 is not caused by a SWI/SNF complex defect (Fig. S5B). Furthermore, we observed no nucleotide variants in the SCF1 ORF or neighboring intragenic regions between AR0382 and AR0387.
Transcript abundance of SCF1 was tightly positively correlated with adhesion across isolates, regardless of clade (r=0.87, p=8.4 x 10−8) (Fig. 2D). In contrast, we observed no association between transcriptional control of IFF4109 and adhesion (r=0.3, p=0.25) (Fig. 2D). Experimentally, transcriptional overexpression of SCF1 was sufficient to elevate adhesion in the otherwise poorly adhesive isolate AR0387 (Fig. 2E). Importantly, the magnitude of overexpression using the TEF1 promoter (approximately 28-fold increase) was similar to and did not exceed the naturally varying magnitude of expression difference between the two wild type isolates AR0382 and AR0387 (approximately 29-fold change) (Fig. 2D, Fig. 2E). These data show that adhesive variation between C. auris isolates is associated with SCF1 expression variation.
In AR0382 and AR0387, the SCF1 locus is invariant, but other isolates exhibit allelic variation in, primarily concentrated in the low complexity tandem repeats (Table S1). We tested whether allelic variation also contributed to the adhesive variation among isolates. Overexpression of the native SCF1 allele in AR0381, a poorly adhesive clade II isolate, was sufficient to increase attachment (Fig. S6A). However, overexpression of the clade I SCF1 allele from AR0382 further elevated adhesion, despite similar levels of overexpression (Fig. S6A). Interestingly, in the clade I AR0382 background, which relies strongly on SCF1 for adhesion, complementation of the Δscf1 mutant with either the clade I or the clade II SCF1 allele resulted in similar levels of rescue of the adhesive phenotype (Fig. S6B). These findings show that sequence variation between these two SCF1 alleles does not intrinsically contribute to functional differences in adhesion, and that other factors may also influence adhesive capacity.
SCF1 and IFF4109 have distinct nonspecific mechanisms
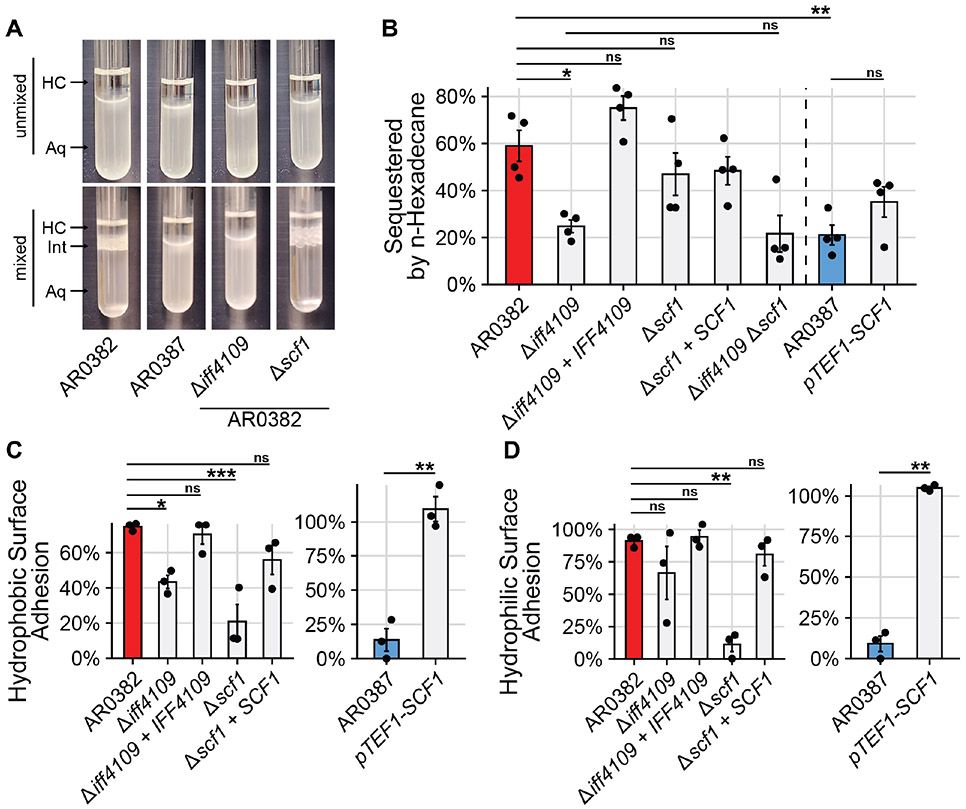
The reliance on SCF1 for surface association is complicated by the genetic interaction with IFF4109, where deletion of both does not result in a more severe adhesive defect than deletion of SCF1 alone (Fig. 1F). Loss of one adhesin did not result in dysregulation of the other, suggesting the interaction is not a regulatory one (Fig. S7). One possibility is that the two genes contribute to adhesion through distinct but complementary physical mechanisms. For other Candida species, adhesion to abiotic substrates is often nonspecific, with adhesins promoting affinity for hydrophobic substrates (32-34). The highly adhesive AR0382 strain exhibited elevated cell surface hydrophobicity compared to the poorly adhesive AR0387 (Fig. 3A, Fig. 3B). Deletion of the IFF4109 adhesin in AR0382 reduced cell surface hydrophobicity, which was rescued to wild type levels by complementation (Fig. 3A, Fig. 3B). In contrast, deletion or overexpression of SCF1 did not significantly impact cell surface hydrophobicity in either AR0382 or AR0387 (Fig. 3A, Fig. 3B).
Fig. 3. IFF4109, but not SCF1, mediates adhesion through cell surface hydrophobicity.
(A) Representative images from Microbial Attachment to Hydrocarbons (MATH) assay. Hydrophobic cells are sequestered from the aqueous phase (Aq) to the aqueous-hydrocarbon interface (Int) after mixing with the hydrocarbon phase (HC). (B) Proportion of cells sequestered out of the aqueous phase during MATH assay. (C), (D) Cells were allowed to attach to a hydrophobic, untreated polystyrene surface (C) or a hydrophilic, vacuum plasma treated polystyrene surface (D) for 1 hour. The surface was washed and the proportion of cells that remained attached after washing was measured. Statistical differences were assessed using one-way ANOVA with Tukey’s post-hoc test (B), (C), and (D) or student’s t-test (C) and (D); *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: p > 0.05.
Elevated cell surface hydrophobicity likewise promotes affinity for hydrophobic substrates (33). We measured the adhesion of C. auris isolates to both an untreated hydrophobic polystyrene surface and a polystyrene surface modified using a vacuum plasma treatment to become strongly hydrophilic. Both IFF4109 and SCF1 mediated adhesion to the hydrophobic substrate (Fig. 3C). However, only SCF1 mediated adhesion to the hydrophilic substrate, showing that SCF1 is not dependent on hydrophobicity (Fig. 3D). Notably, AR0382 and AR0387 still exhibited differential adhesion to the hydrophilic surface, indicating hydrophobic interactions are not primarily responsible for the differential strain phenotypes (Fig. 3D).
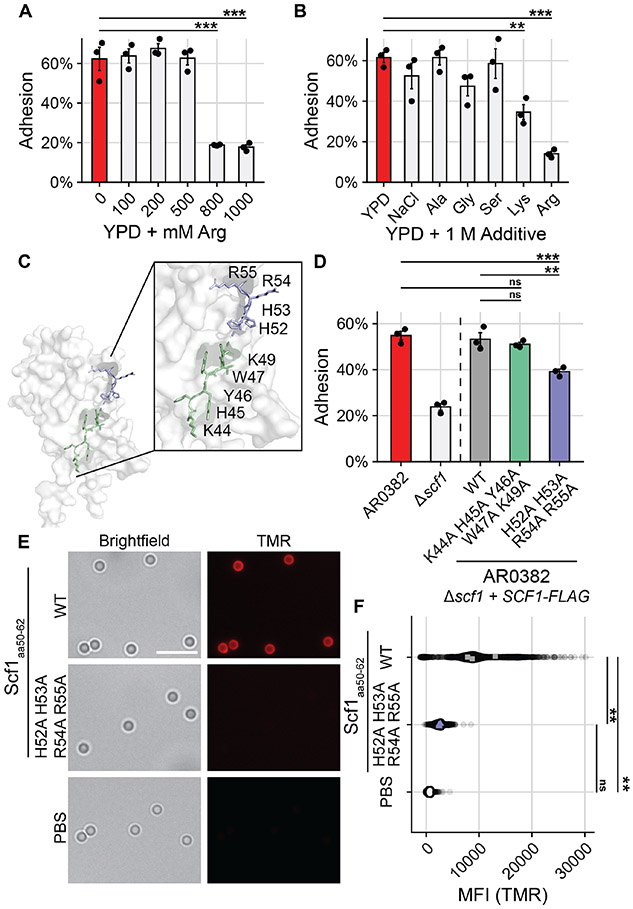
To investigate the mechanism of Scf1 adhesion, we examined the apical N-terminal domain using AlphaFold2, which suggested this domain contains a core Fibronectin-type III fold similar to the FLO11 family of adhesins characterized in Saccharomyces cerevisiae and conserved throughout Ascomycota (Fig. S8A) (32, 35). However, SCF1 does not exhibit significant primary sequence homology to S. cerevisiae FLO11 and lacks conservation of the canonical aromatic bands responsible for adhesive functions in true FLO11 homologs (Fig. S8A, Fig. S8B) (32, 35). Furthermore, model confidence dwindles outside the Fibronectin fold, suggesting substantial variation from Flo11 adhesins (Fig. S9). In its primary sequence, the SCF1 N-terminal domain exhibits an enrichment of arginine and lysine residues compared to other yeast adhesins (Table S2). Adhesive systems in many marine organisms rely on similarly cation-rich proteins, which act through displacement of hydrated ions at the surface-liquid interface or direct cation-π interactions with substrates (36-40). We reasoned that if Scf1 relied on such interactions, adhesion could be inhibited by a saturating concentration of cations at the substrate interface that could not be competitively displaced by SCF1. Consistent with this hypothesis, high concentrations of arginine in solution were sufficient to ablate AR0382 adhesion (Fig. 4A). Similar concentrations of NaCl or other non-cationic amino acids did not produce the same effect, while exogenous lysine produced a more modest inhibition of attachment, consistent with lysine’s weaker ability to form electrostatic interactions (Fig. 4B) (41).
Fig. 4. Specific cationic residues are critical for Scf1-mediated surface association.
(A), (B) WT AR0382 adhesion in the presence of increasing concentrations of arginine (A) or 1 M additives (B). (C) Predictive model of the Scf1 N-terminal domain with two neighboring cationic-aromatic clusters highlighted. (D) Adhesion of wild type AR0382, a mutant lacking SCF1, or AR0382 Δscf1 + SCF1-FLAG mutants encoding the wild type SCF1 allele or alleles containing the indicated mutations. (E) TMR-labelled 13-amino acid peptides corresponding to the wild type Scf1 sequence (residues 50-62) or the same sequence with the indicated mutations incubated with the same polystyrene microspheres used to measure adhesion in (D). Scale Bar = 5 μm. (F) Quantification (MFI) of peptide binding to individual polystyrene microspheres as in (E) measured by TMR epifluorescence, corrected for background fluorescence. Each point represents an individual microsphere. Colored points represent averages of individual experiments, used for statistical analysis. Statistical differences were assessed using one-way ANOVA with Dunnett’s post-hoc test (A), (B) or one-way ANOVA with Tukey’s post-hoc test (D) and (F); *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: p > 0.05.
We next investigated whether specific cationic regions or residues were critical for Scf1 activity. We generated point mutations in cationic residues in different areas of the N-terminal domain, focusing on residues that clustered with aromatic groups, as this pattern potentiates electrostatic adhesion (Fig. S10A) (42). Several mutations had no adhesive impact, but an R54A R55A mutant exhibited a modest adhesive defect, while showing no discernable effect on SCF1 transcription, protein expression, or localization (Fig. S10). Mutating the entire cation-aromatic cluster, H52 H53 R54 R55, resulted in a similar adhesive defect (Fig. 4C, Fig. 4D, Fig. S11). Notably, a nearby cation-aromatic cluster, K44-K49, which was modeled to be less surface-exposed, was not required for adhesion (Fig. 4C, Fig. 4D, Fig. S11). To determine whether surface exposure of the HHRR cluster (res. 52-55) would be sufficient to promote adhesion, we synthesized peptides corresponding to Scf1 residues 50-62 with the intact wild-type cluster or the HHRR residues mutated. The wild-type peptide adhered to polystyrene microspheres, but mutation of the HHRR cluster completely ablated this ability (Fig. 4E, Fig. 4F). Interestingly, similar patterns of cation-aromatic clusters are also abundant in some lipid-binding proteins (40, 43). The wild type Scf1 peptide was similarly able to adhere to phosphotidylcholine microparticles, and SCF1 expression potentiated lipid particle binding by C. auris cells, suggesting SCF1 may also contribute to association with biotic substrates (Fig. S12).
SCF1 promotes long-term colonization and virulence
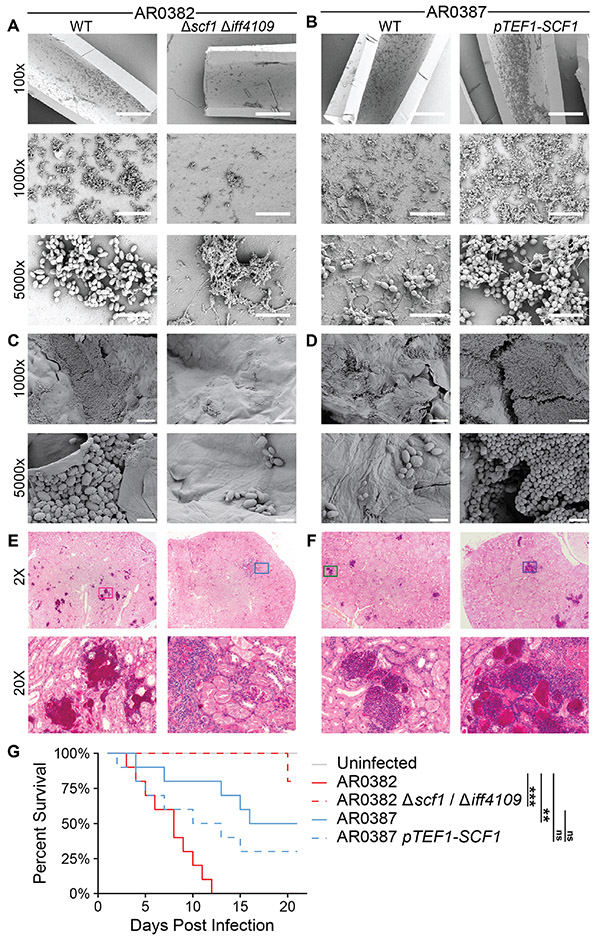
We next investigated the impact of SCF1 and IFF4109 on other aspects of surface colonization. We measured the importance of these adhesins for biofilm growth, which can promote prolonged environmental persistence (11, 44-46). The two adhesins were functionally redundant, and deletion of both was required to ablate biofilm formation in AR0382, suggesting the partial adhesive contributions of each is sufficient to establish colonization (Fig. S13A). Expression of SCF1 alone in otherwise biofilm-incompetent isolates was sufficient to establish biofilm colonization (Fig. S13B, Fig. S13C, Fig. S13D, Fig. S13E). This pattern continued for in vivo biofilms, where loss of SCF1 and IFF4109 ablated the ability of AR0382 to colonize the luminal surface of a polyethylene rat central venous catheter, and overexpression of SCF1 was sufficient to potentiate AR0387 colonization (Fig. 5A, Fig. 5B).
Fig. 5. SCF1 mediates host colonization and infection phenotypes.
(A), (B) Polyethylene central venous catheters were set in rat jugular veins and inoculated intraluminally with C. auris. Representative scanning electron microscopy images of the luminal catheter surface are shown from catheters collected 24 hrs after infection. Scale bars: 400 μm (100x), 40 μm (1000x), 5 μm (5000x). (C), (D) Full thickness human skin explants were colonized with C. auris for 24 hrs before washing to remove unassociated cells. Representative scanning electron microscopy images of the skin surface following washing are shown. Scale bars: 20 μm (1000x), 4 μm (5000x). (E), (F), (G) Immunosuppressed mice were infected intravenously (via tail vein injection) with 5 x 107 C. auris cells. Histopathology sections of the kidneys 7 days post infection (E), (F) were stained with PAS. Magenta color indicates lesion areas. Ten infected mice for each strain were monitored for survival for 21 days (G). Statistical comparisons of overall survival were assessed using the Mantel-Haenszel log-rank test with Benjamini-Hochberg correction. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns p > 0.05.
We then investigated whether biological surface association followed the same reliance on these adhesins. Again, we observed that loss of SCF1 and IFF4109 diminished the ability of AR0382 to colonize ex vivo human skin explants and in vivo murine skin, while overexpression of SCF1 potentiated skin colonization by AR0387 (Fig. 5C, Fig. 5D, Fig. S14). Given this potential for interaction with host tissues, we also investigated the importance of these adhesins in disseminated infection. Histopathological examination of tissues collected from mice 7 days after intravenous C. auris infection revealed that loss of SCF1 and IFF4109 reduced AR0382 dissemination to the kidneys and heart, while overexpression of SCF1 in AR0387 was sufficient to increase fungal lesions (Fig. 5E, Fig. 5F, Fig. S15). Loss of SCF1 and IFF4109 substantially attenuated the virulence of AR0382, with the wild type causing 100% mortality within 12 days of infection and the mutant causing 20% mortality after 21 days (Fig. 5G). Similarly, overexpression of SCF1 reduced the median survival of mice infected with AR0387 from 18.5 days to 11.5 days and ablated the difference in overall survival between the less virulent AR0387 and the more virulent AR0382 (Fig. 5G).
Discussion
C. auris encodes genes similar to the conserved ALS and IFF/HYR adhesin families, and proposed models suggest differential utilization of these adhesins may contribute to epidemiological differences among isolates (25, 27). Our findings suggest the C. auris-specific adhesin SCF1 and the conserved adhesin IFF4109 are the principal mediators of association with abiotic surfaces, and additionally contribute substantially to infection and long-term colonization of both biological and abiotic surfaces. Interestingly, the other conserved adhesin genes appeared to not mediate surface association. Whether this is the product of functional or regulatory divergence remains to be explored. Notably, we observed widespread differential regulation of SCF1 among C. auris isolates regardless of clade, suggesting transcriptional control of this adhesin has adapted more recently than clade separation. The widespread plasticity around a single genetic element responsible for diverse clinically-relevant phenotypes could be problematic in outbreak settings. While SCF1 and IFF4109 contribute to host infection and colonization, the mechanisms of their interaction with host systems remain unclear. Understanding how variable adhesion allows C. auris to mediate infection is likely to offer therapeutic insights. Prior work suggests vaccination or monoclonal antibody therapy targeting Als or Iff/Hyr adhesins may offer protection against lethal C. auris infection (27, 47). Furthermore, the complementary function of SCF1 and IFF4109 with divergent mechanisms suggests C. auris has evolved the capacity for promiscuous surface association and colonization. Mediation of hydrophobic interactions is largely conserved among fungal adhesins, consistent with the adhesive mechanism of the conserved IFF4109 (32-34). The cation rich SCF1, however, appears to functionally resemble proteins from bivalve, barnacle, and Vibrio adhesion systems. For these organisms, cation-dependent surface interactions promote adhesion in aqueous and highly ionic environments (36-39). C. auris has been isolated from the coastal wetlands of the Andaman Islands and from a Colombian estuary, suggesting a possible marine natural habitat, and this ecological niche may have conferred similar selective pressures on adhesion mechanisms (48, 49). Development of unique adhesion biology may in part explain the tenacity of this organism on medically-relevant substrates. Still, differential utilization of SCF1 by different isolates suggests an unknown selective pressure may govern its expression. Understanding this adaptation and its clinical consequences more fully may offer important insights into the outbreak potential of this pathogen.
Overall, our work characterizes of the adhesin machinery used by C. auris for surface association and colonization. The identification of SCF1 and the characterization of the genetic determinants of adhesion add to the growing understanding of the pathobiology this emerging organism.
Supplementary Material
Acknowledgments:
We thank Jonathan Sexton (University of Michigan) for consultation on the development of a label-free high throughput imaging-based adhesion assay. We thank Mohammad Siddiq (University of Michigan) for consultation in investigating SCF1 homology and variants. We also thank Adam Abraham (University of Michigan) for consultation on surface modification and assistance with vacuum plasma treatment experiments.
Funding:
National Institutes of Health grant R21AI169186 (DJS, TRO)
National Institutes of Health grant T32AI007528 (DJS)
National Institutes of Health grant F31AI169823 (DJS)
National Institutes of Health grant T32AI007413 (GZ)
National Institutes of Health grant R01AI073289 (DA)
National Institutes of Health grant R01AI145939 (JEN)
National Institutes of Health grant R21AI159583 (JEN)
American Heart Association grant 938451 (SS)
National Institutes of Health UCLA CTSI grant KL2TR001882 (SS)
National Institute of Health grant R01AI141202 (ASI)
WACCBIP-World Bank ACE Masters Fellowship WACCBIP-NCDs:Awandare (JAEA)
Footnotes
Competing interests: TRO and DJS are inventors on U.S. Provisional Patent 63/502,704 filed 05/17/23 and U.S. Provisional Patent 63/514,470 filed 07/19/23 related to this work.
Data and materials availability: Data from Illumina sequences are available in the NCBI SRA under BioProject accession number PRJNA904261 (RNA-Seq data) or PRJNA904262 (AtMT mutant whole genome sequencing). Strains and constructs generated in this study will be provided for research purposes upon request. All data are available in the main text or the supplementary materials.
References
- 1.Chakrabarti A, Sood P, On the emergence, spread and resistance of Candida auris: host, pathogen and environmental tipping points. J. Med. Microbiol 70 (2021), doi: 10.1099/jmm.0.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akinbobola AB, Kean R, Hanifi SMA, Quilliam RS, Environmental reservoirs of the drug-resistant pathogenic yeast Candida auris. PLoS Pathog. 19, e1011268 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkenazi-Hoffnung L, Rosenberg Danziger C, Navigating the New Reality: A Review of the Epidemiological, Clinical, and Microbiological Characteristics of Candida auris, with a Focus on Children. J Fungi (Basel). 9 (2023), doi: 10.3390/jof9020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP, US Candida auris Investigation Team, Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect. Dis 18, 1377–1384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts SC, Zembower TR, Ozer EA, Qi C, Genetic Evaluation of Nosocomial Candida auris Transmission. J. Clin. Microbiol 59 (2021), doi: 10.1128/JCM.02252-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, MSD, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM, Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus - United States, May 2013-August 2016. MMWR Morb. Mortal. Wkly. Rep 65, 1234–1237 (2016). [DOI] [PubMed] [Google Scholar]
- 7.WHO fungal priority pathogens list to guide research, development and public health action. Geneva: World Health Organization; (2022) (available from https://www.who.int/publications/i/item/9789240060241). [Google Scholar]
- 8.Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, Noble-Wang J, Pacilli M, McPherson TD, Black S, Kemble SK, Herzegh O, Ahmad A, Forsberg K, Jackson B, Litvintseva AP, Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin. Infect. Dis 73, 1142–1148 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, NISC Comparative Sequencing Program, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK, Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat. Med 27, 1401–1409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H, Candida auris Investigation Workgroup, Candida auris in Healthcare Facilities, New York, USA, 2013–2017. Emerg. Infect. Dis 24, 1816–1824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dire O, Ahmad A, Duze S, Patel M, Survival of Candida auris on environmental surface materials and low-level resistance to disinfectant. J. Hosp. Infect 137, 17–23 (2023). [DOI] [PubMed] [Google Scholar]
- 12.de Almeida JN Jr, Brandão IB, Francisco EC, Luis S de Almeida R, de Oliveira Dias P, Pereira FM, Santos Ferreira F, Souza de Andrade T, de Miranda Costa MM, de Souza Jordão RT, Meis JF, Colombo AL, Favarello LM, Lima SL, Lima R, Miranda IO, de Jesus Lopes TL, Soares da Silva DC, Nobre de Moura L, Ribeiro LC, Carlos de Albuquerque Bandeira A, Moreira Urpia T, Rubia Gonçalves M, de Groot T, The Candida auris Brazilian Study Group, Axillary Digital Thermometers uplifted a multidrug-susceptible Candida auris outbreak among COVID-19 patients in Brazil. Mycoses 64, 1062–1072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM, A Candida auris Outbreak and Its Control in an Intensive Care Setting. N. Engl. J. Med 379, 1322–1331 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Patterson CA, Wyncoll D, Patel A, Ceesay Y, Newsholme W, Chand M, Mitchell H, Tan M, Edgeworth JD, Cloth Lanyards as a Source of Intermittent Transmission of Candida auris on an ICU. Crit. Care Med 49, 697–701 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Vila T, Montelongo-Jauregui D, Ahmed H, Puthran T, Sultan AS, Jabra-Rizk MA, Comparative Evaluations of the Pathogenesis of Candida auris Phenotypes and Candida albicans Using Clinically Relevant Murine Models of Infections. mSphere. 5 (2020), doi: 10.1128/mSphere.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinayagamoorthy K, Pentapati KC, Prakash H, Prevalence, risk factors, treatment and outcome of multidrug resistance Candida auris infections in Coronavirus disease (COVID-19) patients: A systematic review. Mycoses. 65, 613–624 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajni E, Jain A, Gupta S, Jangid Y, Vohra R, Risk Factors for Candidemia in Intensive Care Unit: A Matched Case Control Study from North-Western India. Acta Medica . 65, 83–88 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Mulet Bayona JV, Tormo Palop N, Salvador García C, Guna Serrano MDR, Gimeno Cardona C, Candida auris from colonisation to candidemia: A four-year study. Mycoses 00, 1–9 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Allaw F, Haddad SF, Habib N, Moukarzel P, Naji NS, Kanafani ZA, Ibrahim A, Zahreddine NK, Spernovasilis N, Poulakou G, Kanj SS, COVID-19 and C. auris: A Case-Control Study from a Tertiary Care Center in Lebanon. Microorganisms. 10 (2022), doi: 10.3390/microorganisms10051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyman M, Forsberg K, Sexton DJ, Chow NA, Lockhart SR, Jackson BR, Chiller T, Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann. Intern. Med 176, 489–495 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benedict K, Forsberg K, Gold JAW, Baggs J, Lyman M, Candida auris–Associated Hospitalizations, United States, 2017-2022. Emerg. Infect. Dis 29, 1485–1487 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Groot PWJ, Bader O, de Boer AD, Weig M, Chauhan N, Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot. Cell 12, 470–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Essen L-O, Vogt MS, Mösch H-U, Diversity of GPI-anchored fungal adhesins. Biol. Chem 401, 1389–1405 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Smoak RA, Snyder LF, Fassler JS, He BZ, Parallel Expansion and Divergence of an Adhesin Family in Pathogenic Yeasts. Genetics 223 (2023), doi: 10.1093/genetics/iyad024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muñoz JF, Welsh RM, Shea T, Batra D, Gade L, Howard D, Rowe LA, Meis JF, Litvintseva AP, Cuomo CA, Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics 218 (2021), doi: 10.1093/genetics/iyab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh S-H, Schliep K, Isenhower A, Rodriguez-Bobadilla R, Vuong VM, Fields CJ, Hernandez AG, Hoyer LL, Using Genomics to Shape the Definition of the Agglutinin-Like Sequence (ALS) Family in the Saccharomycetales. Front. Cell. Infect. Microbiol 11, 794529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, French S, Lockhart SR, Chiller T, Edwards JE Jr, Ibrahim AS, The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 15, e1007460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva-Dias A, Miranda IM, Rocha R, Monteiro-Soares M, Salvador A, Rodrigues AG, Pina-Vaz C, A novel flow cytometric protocol for assessment of yeast cell adhesion. Cytometry A. 81, 265–270 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP, Portrait of Candida albicans adherence regulators. PLoS Pathog. 8, e1002525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier C, Brown AJP, Lorenz A, Candida auris undergoes adhesin-dependent and - independent cellular aggregation. bioRxiv p. 2023.04.21.537817 (2023) doi: 10.1101/2023.04.21.537817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bing J, Guan Z, Zheng T, Zhang Z, Fan S, Ennis CL, Nobile CJ, Huang G, Clinical isolates of Candida auris with enhanced adherence and biofilm formation due to genomic amplification of ALS4. PLoS Pathog. 19, e1011239 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraushaar T, Brückner S, Veelders M, Rhinow D, Schreiner F, Birke R, Pagenstecher A, Mösch H-U, Essen L-O, Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure. 23, 1005–1017 (2015). [DOI] [PubMed] [Google Scholar]
- 33.El-Kirat-Chatel S, Beaussart A, Derclaye S, Alsteens D, Kucharíková S, Van Dijck P, Dufrêne YF, Force Nanoscopy of Hydrophobic Interactions in the Fungal Pathogen Candida glabrata. ACS Nano. 9 1648–1655 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Valotteau C, Prystopiuk V, Cormack BP, Dufrêne YF, Atomic Force Microscopy Demonstrates that Candida glabrata Uses Three Epa Proteins To Mediate Adhesion to Abiotic Surfaces. mSphere. 4 (2019), doi: 10.1128/mSphere.00277-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brückner S, Schubert R, Kraushaar T, Hartmann R, Hoffmann D, Jelli E, Drescher K, Müller DJ, Oliver Essen L, Mösch H-U, Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family. Elife. 9 (2020), doi: 10.7554/eLife.55587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier GP, Rapp MV, Waite JH, Israelachvili JN, Butler A, BIOLOGICAL ADHESIVES. Adaptive synergy between catechol and lysine promotes wet adhesion by surface salt displacement. Science. 349, 628–632 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Liang C, Gao L, Li S, Zhang Y, Zhang J, Cao Y, Hidden complexity of synergistic roles of Dopa and lysine for strong wet adhesion. Mater. Chem. Front 1, 2664–2668 (2017). [Google Scholar]
- 38.Liang C, Strickland J, Ye Z, Wu W, Hu B, Rittschof D, Biochemistry of Barnacle Adhesion: An Updated Review. Frontiers in Marine Science. 6 (2019), doi: 10.3389/fmars.2019.00565. [DOI] [Google Scholar]
- 39.Kim S, Yoo HY, Huang J, Lee Y, Park S, Park Y, Jin S, Jung YM, Zeng H, Hwang DS, Jho Y, Salt Triggers the Simple Coacervation of an Underwater Adhesive When Cations Meet Aromatic π Electrons in Seawater. ACS Nano. 11, 6764–6772 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Huang X, Nero T, Weerasekera R, Matej KH, Hinbest A, Jiang Z, Lee RF, Wu L, Chak C, Nijjer J, Gibaldi I, Yang H, Gamble N, Ng W-L, Malaker SA, Sumigray K, Olson R, Yan J, Vibrio cholerae biofilms use modular adhesins with glycan-targeting and nonspecific surface binding domains for colonization. Nat. Commun 14, 2104 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokalingam S, Raghunathan G, Soundrarajan N, Lee S-G, A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS One. 7, e40410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan H, Wang J, Tao Z, Huang J, Rao P, Kurokawa T, Gong JP, Adjacent cationic-aromatic sequences yield strong electrostatic adhesion of hydrogels in seawater. Nat. Commun 10, 5127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin S, Wang J, Gambhir A, Murray D, PIP2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct 31, 151–175 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Kean R, McKloud E, Townsend EM, Sherry L, Delaney C, Jones BL, Williams C, Ramage G, The comparative efficacy of antiseptics against Candida auris biofilms. Int. J. Antimicrob. Agents 52, 673–677 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Kean R, Delaney C, Sherry L, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R, Williams C, Ramage G, Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere. 3 (2018), doi: 10.1128/mSphere.00334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Short B, Brown J, Delaney C, Sherry L, Williams C, Ramage G, Kean R, Candida auris exhibits resilient biofilm characteristics in vitro: implications for environmental persistence. J. Hosp. Infect 103, 92–96 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Singh S, Barbarino A, Youssef EG, Coleman D, Gebremariam T, Ibrahim AS, Protective Efficacy of Anti-Hyr1p Monoclonal Antibody against Systemic Candidiasis Due to Multi-Drug-Resistant Candida auris. J Fungi (Basel). 9 (2023), doi: 10.3390/jof9010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arora P, Singh P, Wang Y, Yadav A, Pawar K, Singh A, Padmavati G, Xu J, Chowdhary A, Environmental Isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India. MBio. 12 (2021), doi: 10.1128/mBio.03181-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escandón P, Novel Environmental Niches for Candida auris: Isolation from a Coastal Habitat in Colombia. J Fungi (Basel). 8 (2022), doi: 10.3390/jof8070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lutgring JD, Machado M-J, Benahmed FH, Conville P, Shawar RM, Patel J, Brown AC, FDA-CDC Antimicrobial Resistance Isolate Bank: a Publicly Available Resource To Support Research, Development, and Regulatory Requirements. J. Clin. Microbiol 56, e01415–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veri AO, Miao Z, Shapiro RS, Tebbji F, O’Meara TR, Kim SH, Colazo J, Tan K, Vyas VK, Whiteway M, Robbins N, Wong KH, Cowen LE, Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet. 14, e1007270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santana DJ, O’Meara TR, Forward and reverse genetic dissection of morphogenesis identifies filament-competent Candida auris strains. Nat. Commun 12, 7197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SO, Frazer C, Bennett RJ, An Adjuvant-Based Approach Enables the Use of Dominant HYG and KAN Selectable Markers in Candida albicans. mSphere. 7, e0034722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D, The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25 (2009), pp. 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee DW, Hong CP, Kang HA, An effective and rapid method for RNA preparation from non-conventional yeast species. Anal. Biochem 586, 113408 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL, Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13, 134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 60.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamprecht MR, Sabatini DM, Carpenter AE, CellProfiler™: free, versatile software for automated biological image analysis. Biotechniques. 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Bray M-A, Carpenter A, Imaging Platform, Broad Institute of MIT and Harvard, "Advanced Assay Development Guidelines for Image-Based High Content Screening and Analysis" in Assay Guidance Manual, Markossian S, Sittampalam GS, Grossman A, Brimacombe K, Arkin M, Auld D, Austin CP, Baell J, Caaveiro JMM, Chung TDY, Coussens NP, Dahlin JL, Devanaryan V, Foley TL, Glicksman M, Hall MD, Haas JV, Hoare SRJ, Inglese J, Iversen PW, Kahl SD, Kales SC, Kirshner S, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Saradjian P, Trask OJ Jr, Weidner JR, Wildey MJ, Xia M, Xu X, Eds. (Eli Lilly & Company and the National Center for AdvancingTranslational Sciences, Bethesda (MD), 2017; https://www.ncbi.nlm.nih.gov/pubmed/23469374). [Google Scholar]
- 66.Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, Warrenfeltz S, Aurrecoechea C, Stoeckert CJ Jr, Kissinger JC, Roos DS, Hertz-Fowler C, FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J Fungi (Basel). 4, e4010039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maguire SL, ÓhÉigeartaigh SS, Byrne KP, Schröder MS, O’Gaora P, Wolfe KH, Butler G, Comparative genome analysis and gene finding in Candida species using CGOB. Mol. Biol. Evol 30, 1281–1291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M, ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, Söding J, Steinegger M, Fast and accurate protein structure search with Foldseek. Nat. Biotechnol (2023), doi: 10.1038/s41587-023-01773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenberg M, Gutnick D, Rosenberg E, Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett 9, 29–33 (1980). [Google Scholar]
- 71.Li Y, Pham JQ, Johnston KP, Green PF, Contact angle of water on polystyrene thin films: effects of CO(2) environment and film thickness. Langmuir. 23, 9785–9793 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Gulati M, Lohse MB, Ennis CL, Gonzalez RE, Perry AM, Bapat P, Arevalo AV, Rodriguez DL, Nobile CJ, In Vitro Culturing and Screening of Candida albicans Biofilms. Curr. Protoc. Microbiol 50, e60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A, Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun 72, 6023–6031 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eix EF, Johnson CJ, Wartman KM, Kernien JF, Meudt JJ, Shanmuganayagam D, Gibson ALF, Nett JE, Ex vivo human and porcine skin effectively model C. auris colonization, differentiating robust and poor fungal colonizers. J. Infect. Dis 225, 1791–1795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horton MV, Johnson CJ, Kernien JF, Patel TD, Lam BC, Cheong JZA, Meudt JJ, Shanmuganayagam D, Kalan LR, Nett JE, Candida auris Forms High-Burden Biofilms in Skin Niche Conditions and on Porcine Skin. mSphere. 5 (2020), doi: 10.1128/mSphere.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GYC, McGavin MJ, Travers JB, Otto M, Inohara N, Núñez G, Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature. 503, 397–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pacilli M, Kerins JL, Clegg WJ, Walblay KA, Adil H, Kemble SK, Xydis S, McPherson TD, Lin MY, Hayden MK, Froilan MC, Soda E, Tang AS, Valley A, Forsberg K, Gable P, Moulton-Meissner H, Sexton DJ, Jacobs Slifka KM, Vallabhaneni S, Walters MS, Black SR, Regional Emergence of Candida auris in Chicago and Lessons Learned From Intensive Follow-up at 1 Ventilator-Capable Skilled Nursing Facility. Clin. Infect. Dis 71, e718–e725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.