Abstract
A number of mutations, including deletions, linker scan substitutions, and point mutations, were performed in the promoter of the late African swine fever virus (ASFV) gene coding for the capsid protein p72. The consequences of the mutations in terms of promoter activity were analyzed by luciferase assays using plasmids transfected into infected cells. The results showed that the promoter function is contained between nucleotides −36 and +5 relative to the transcription initiation site. Moreover, two major essential regions for promoter activity, centered at positions −13 and +3, were located along the 41-bp sequence, the latter mapping in the transcription start site. Sequence alignment with other ASFV late promoters showed homology in the region of transcriptional initiation, where the presence of the sequence TATA was observed in most of the promoters. Substitution of these four residues in three other late viral promoters strongly reduced their respective activities. These results show that cis-acting control elements of ASFV p72 gene transcription are restricted to a short sequence of about 40 bp and suggest that transcription of late genes is initiated around a TATA sequence that would function as an initiator element.
African swine fever virus (ASFV) is a large enveloped virus with icosahedral morphology whose genome is a double-stranded DNA molecule of about 170 kbp. Although the early events in the replication of the viral DNA occur in the nucleus of the host cells (17), ASFV is mainly a cytoplasmic virus. Virion assembly takes place in discrete cytoplasmic areas called viral factories, where the virus becomes engulfed by a two-membraned collapsed cisterna derived from the endoplasmic reticulum (6). Mature particles contain the enzymes required for early RNA synthesis and processing (19, 30, 31), most of which are known to be encoded by the virus genome (34). ASFV gene expression appears to follow a cascade mechanism similar to that described for poxviruses (22). Thus, four temporal classes of ASFV genes have been described. The expression of immediate-early and early genes begins immediately after infection, the latter being silenced at the time of maximal DNA replication (3). Intermediate gene expression is dependent on the synthesis of viral DNA, decreasing sharply when late mRNA synthesis becomes maximal (29).
At present, very little is known about the molecular mechanisms controlling ASFV gene transcription. Individual genes are tightly regulated independent transcription units whose mRNA start sites are located at a short distance from the corresponding translation initiation codon (3, 29). However, the structure of the promoters and the identity of transcription factors which regulate the expression of viral genes are unknown. In order to investigate the structural requirements for late promoter activity, we have obtained a number of mutants of the capsid protein p72 gene promoter and analyzed their transcriptional activities when directing the expression of the luciferase gene in transient-expression assays with ASFV-infected cells. The results obtained have allowed us to localize cis-acting elements required for late gene transcription and suggest the existence of sequence specificity in the region of transcriptional initiation.
Transient expression of luciferase gene regulated by the p72 promoter.
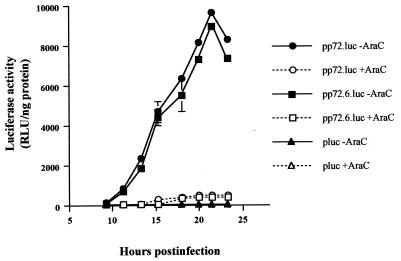
To study the promoter sequences required for transcription of ASFV late genes, we decided to analyze the 5′ flanking region of the gene encoding protein p72. This protein is the major component of the icosahedral capsid of the virus (10, 18) and constitutes about 35% of the virion mass (7). Therefore, we assumed that the promoter that controls its expression is one of the strongest late-functioning promoters and could thus be very useful for our purposes. Previously, we demonstrated that a DNA fragment including the first eight codons of the p72 gene and the 197-bp upstream sequence was capable of directing the expression of the lacZ gene at late times postinfection, either from a recombinant virus (v72-βgal) or from a plasmid transfected into ASFV-infected cells (27). We therefore cloned the firefly luciferase gene downstream of the 197-bp upstream sequence and the first A of the translation initiation codon of the p72 gene. A 218-bp PCR fragment was obtained using ASFV DNA as a template and oligonucleotides 5′-A and 3′-A (Table 1), which were cut with restriction enzymes BamHI and BglII and cloned into BamHI-linearized pUC118, generating plasmid pp72. The luciferase gene of Photinus pyralis was then inserted as a 1.4-kbp BamHI fragment from pKluc (16) into the pp72 plasmid that had been linearized with BamHI enzyme, thus obtaining the pp72.luc plasmid, in which the reporter is placed downstream of the 5′ flanking sequence of the p72 gene. Plasmid pp72.luc was capable of inducing the synthesis of high levels of luciferase at late times postinfection when transfected into ASFV-infected Vero cells (Fig. 1). A kinetic analysis of transient expression of luciferase showed that maximal levels of reporter activity were obtained at 21 h postinfection. Moreover, when the cells were infected in the presence of AraC, a specific inhibitor of the replication of ASFV DNA and therefore of late transcription, the luciferase activity was almost undetectable. Therefore, we conclude that the 198-bp DNA fragment cloned contains the promoter elements of the p72 gene.
TABLE 1.
Names and sequences of the oligonucleotides used for the generation of promoter mutants
| Oligonucleotide namea | Sequenceb | Promoter(s) |
|---|---|---|
| 5′ and 3′ deletions | ||
| 5′-A | cgcgagatctGGGTCGCCGGAGAAAAGTCAAAAGG | p72, p72.1, p72.2 |
| 5′-B | cgcgagatCTAGCAAGAGCGTGTCAATAATT | p72.3, p72.5 |
| 5′-C | cgcgagaTCTTTGTTATTATCAAGATCC | p72.4 |
| 3′-A | gcgcggatccTATATAATGTTATAAAAATAATTT | p72, p72.3, p72.4 |
| 3′-B | gcgcggatccAATTATTGACACGCTCTTGCT | p72.1 |
| 3′-C | gcgcggatccGGATCTTGATAATAACAAAGA | p72.2, p72.5 |
| 6A | gatcTATTTAATAAAAACAATAAATTATTTTTATAACATTATATAg | |
| 6B | gatccTATATAATGTTATAAAAATAATTTATTGTTTTTATTAAATA | p72.6 |
| 7A | gatctTATTTTTATAACATTATATAg | |
| 7B | gatccTATATAATGTTATAAAAATAA | p72.7 |
| Linker scan substitutions | ||
| 5′-8 | gcggtaccgcgcgcATAAAAACAATAAATTATTTTTATAACATTATATAggatccccccccc | p72.8 |
| 5′-9 | gcggtaccTATTTAgcgcgAACAATAAATTATTTTTATAACATTATATAggatccccccccc | p72.9 |
| 5′-10 | gcggtaccTATTTAATAAAgcgcgTAAATTATTTTTATAACATTATATAggatccccccccc | p72.10 |
| 5′-11 | gcggtaccTATTTAATAAAAACAAgcgcgTATTTTTATAACATTATATAggatccccccccc | p72.11 |
| 5′-12 | gcggtaccTATTTAATAAAAACAATAAATgcgcgTTATAACATTATATAggatccccccccc | p72.12 |
| 5′-13 | gcggtaccTATTTAATAAAAACAATAAATTATTTgcgcgACATTATATAggatccccccccc | p72.13 |
| 5′-14 | gcggtaccTATTTAATAAAAACAATAAATTATTTTTATAgcgcgATATAggatccccccccc | p72.14 |
| 5′-15 | gcggtaccTATTTAATAAAAACAATAAATTATTTTTATAACATTgcgcgggatccccccccc | p72.15 |
| 3′-G | gggggggggatcc | From p72.8 to p72.15 |
| Single-nucleotide substitutionsc | ||
| 5′-D | cgcgggtaccTATTTAATAAAAACAATAAATTA | From p72.16A to p72.21T |
| 3′-16 | gcgcggatccTATATBATGTTATAAAAATAATTTATTGTTTTT | p72.16A, p72.16C, p72.16G |
| 3′-17 | gcgcggatccTATAVAATGTTATAAAAATAATTTATTGTTTTT | p72.17C, p72.17G, p72.17T |
| 3′-18 | gcgcggatccTATBTAATGTTATAAAAATAATTTATTGTTTTT | p72.18A, p72.18C, p72.18G |
| 3′-19 | gcgcggatccTAVATAATGTTATAAAAATAATTTATTGTTTTT | p72.19C, p72.19G, p72.19T |
| 3′-20 | gcgcggatccTBTATAATGTTATAAAAATAATTTATTGTTTTT | p72.20A, p72.20C, p72.20G |
| 3′-21 | gcgcggatccVATATAATGTTATAAAAATAATTTATTGTTTTT | p72.21C, p72.21G, p72.21T |
| TATA substitutions in p10, p11.5, and p54 promoters | ||
| 5′-p11.5 | cgcgggtaccTTTAAAATAAGCCATTTAAAGATTTAGAATTTA | p11.5, p11.5mut |
| 3′-p11.5 | gcgcggatccTATACATATAAATTCTAAATCTT | p11.5 |
| 3′-p11.5mut | gcgcggatccgcgcCATATAAATTCTAAATCTT | p11.5mut |
| 5′-p10 | cgcgggtaccATAATAAAAAATATTTTTTACTTTTTTTTCTTC | p10, p10mut |
| 3′-p10 | gcgcggatccTATATTATGAAGAAAAAAAAGTA | p10 |
| 3′-p10mut | gcgcggatccgcgcTTATGAAGAAAAAAAAGTA | p10mut |
| 5′-p54 | cgcgggtaccGTAATTTCATTGCGCCACAACATTTTTATATAT | p54, p54mut |
| 3′-p54 | gcgcggatccTATAAATAATATATAAAAATGTT | p54 |
| 3′-p54mut | gcgcggatccgcgcAATAATATATAAAAATGTT | p54mut |
Oligonucleotides are listed according to the type of mutagenesis for which each oligonucleotide was used.
Nonviral sequences are in lowercase letters, and recognition sequences for restriction enzymes BglII, BamHI, and KpnI are underlined.
Positions indicated as B are C, G, or T residues, and positions indicated as V are A, C, or G residues.
FIG. 1.
Regulated expression of the luciferase gene by the p72 late promoter. Preconfluent monolayers of Vero cells (1.5 × 105) were transfected for 1 h with 16 μg of Lipofectamine reagent (Gibco BRL) and 1 μg of pluc, pp72.luc, or pp72.6.luc plasmids. Transfected cells were then infected with the BA71V strain of ASFV (13) at a multiplicity of infection of 5 PFU per cell in the absence or presence of AraC (40 μg/ml). Cells were washed and harvested at different times postinfection, and the luciferase activity was measured as previously described (16). The data are represented as the average ± the standard deviation of three assays. Luciferase values were normalized to the protein content.
Location and length of the p72 promoter.
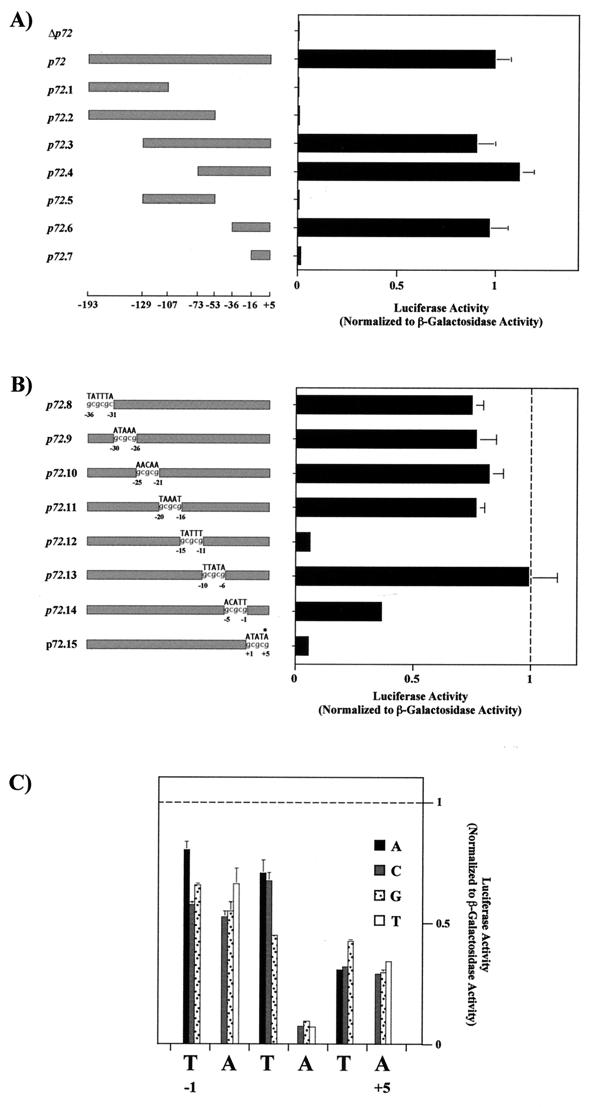
In order to determine the location of essential residues within the p72 promoter sequence, a deletion analysis was first undertaken. A series of luc plasmids containing 5′, 3′, or 5′ and 3′ promoter sequence deletions was constructed. Five mutants (p72.1 to p72.5) were obtained by PCR from pp72.luc by using the oligonucleotides 5′-A, 5′-B, 5′-C, 3′-A, 3′-B, and 3′-C (Table 1). The amplified fragments were cloned into pUC118 as described for plasmid pp72. Mutants p72.6 and p72.7 were obtained by hybridization of oligonucleotides 6A with 6B and 7A with 7B, respectively (Table 1). The resulting hybrids contained one end that was compatible with the BamHI enzyme in such a way that the cloning of one copy of the fragments into a BamHI site resulted in the retention of a single BamHI site in the derived plasmids. After annealing, the DNA fragments were incubated with T4 polynucleotide kinase and ATP and were inserted into the plasmid pUC118, which was linearized with BamHI endonuclease. The luc gene was cloned in the plasmids containing mutants p72.1 to p72.7 as in the case of pp72.luc. A control plasmid, p72GAL10T, in which the p72 promoter and nucleotides up to the eighth codon of the p72 gene are fused to the lacZ gene (16), was cotransfected with luc plasmids for 1 h into Vero cells just prior to infection with ASFV. Luciferase activities were normalized to that of β-galactosidase.
Analysis of the mutants showed that 3′ deletions completely abolished luciferase activity (p72.1, p72.2, or p72.5) (Fig. 2). By contrast, deletion of 5′ sequences in mutants p72.3, p72.4, and p72.6 did not affect promoter activity. The results indicate that the promoter elements are located within the 41 bp at the 3′ end of the sequence. More importantly, this region (p72.6 promoter) contains all the essential residues which drive late expression of a reporter gene, as shown by time course tran sient-expression assays and the inhibitory effect of AraC in infected Vero cells (Fig. 1). Moreover, the sites for transcription initiation of the luc gene under the control of this promoter in transfection assays are the same as those of the p72 gene in the viral genome. Thus, as can be seen in Fig. 3, the 5′ ends of the reporter gene mRNA map at residues −5 to −2 relative to the translation initiation codon, as did those described for the p72 gene mRNA (26). We will designate in this work the second residue (−4) used for initiation as the +1 position.
FIG. 2.
(A) Mapping of the p72 late promoter. Luciferase activity was measured in ASFV-infected Vero cells previously transfected with plasmids containing deletion fragments of the p72 promoter and was normalized to β-galactosidase activity produced from cotransfected control plasmid p72GAL10T. Transfections were carried out as described in the legend to Fig. 1 by using 12 ng of luciferase plasmids and 1 μg of p72GAL10T plasmid per 1.5 × 105 cells. Cells were infected with the BA71V strain of ASFV at a multiplicity of infection of 5 PFU per cell, and they were washed and harvested at 18 h postinfection. Luciferase and β-galactosidase activities were determined using the Dual-Light kit from Tropix Inc. The data are represented as the average plus the standard deviation of three assays and are normalized to p72 activity. The positions of the deletion fragment ends with respect to the transcriptional start point (+1) are indicated below the diagram. Δp72 corresponds to the luciferase gene without the p72 promoter. (B) Linker scanning analysis of the p72 promoter. The substituted sequence is indicated above each promoter, and the positions of mutated residues are indicated. Activities were determined as in panel A. All values were normalized to p72.6 activity, indicated with a vertical dashed line. The asterisk indicates the position of the first nucleotide of the p72 gene translation start codon. (C) Effect of single-nucleotide substitutions at or near the transcriptional start site. Activities were determined as for panel A. All values were normalized to p72.6 activity, indicated with a horizontal dashed line. Symbols for base substitutions are shown.
FIG. 3.
Analysis of RNA synthesized from p72 promoter mutants. Preconfluent monolayers of Vero cells (1.5 × 105) were transfected with 16 μg of Lipofectamine reagent and 1 μg of plasmids containing different mutations of the p72 promoter regulating the luciferase gene. The primer for the luciferase gene was complementary to nucleotides between bp 35 and 64 of the noncoding strand of the gene. The primer for the D117R gene has been previously described (32). Total RNA was obtained at 18 h postinfection, hybridized with end-labeled primer, and extended with avian myeloblastosis virus reverse transcriptase. The sizes (in nucleotides) of the major bands, calculated by using products of an irrelevant DNA sequencing reaction for markers, are indicated. The lower panel shows the quantitated values of the transcripts produced by each mutant normalized first to the internal D117R transcript value and then to the p72.6 value. A diagram showing the relative promoter lengths and sequences of the promoters used is shown on the left. Mutant p72.19C has a point mutation in which the A residue at position +3 was replaced by C.
On the other hand, complete loss of reporter activity occurred when the length of the promoter fragment in the transfected plasmid was reduced from 41 to 21 bp (p72.7) (Fig. 2A), suggesting that ASFV sequences between −36 and −16 are critical for activity. However, the possibility that transcription of the reporter gene was inhibited by bringing vector sequences closer to the mRNA start site cannot be ruled out at present.
Linker scan substitutions along the p72 promoter.
To localize the critical residues within the 41-bp promoter, linker scan substitutions were performed. A set of eight mutants (p72.8 to p72.15) containing GCGCGC or GCGCG substitutions along the 41-bp sequence was constructed as follows. The nontemplate strands of the promoters were synthesized as oligonucleotides which contained KpnI and BamHI sites at their 5′ and 3′ ends, respectively (Table 1, oligonucleotides 5′-8 to 5′-15). In addition, a tail of seven C residues was included at their 3′ ends in such a way that the BamHI site and the C tail together would create a 13-bp sequence common to all of them. The single-stranded oligonucleotides were annealed to a 13-bp primer complementary to this 3′ end common sequence (Table 1, oligonucleotide 3′-G). Extension of the second strand was performed with DNA polymerase I (Klenow fragment) and all four deoxynucleotides. The double-stranded products were then digested with KpnI and BamHI and cloned into pUC118 linearized with the same endonucleases, and the luciferase gene was inserted downstream of the promoters. The plasmids thus obtained were cotransfected with control p72GAL10T into infected cells, and the normalized luciferase activity was determined. The results are summarized in Fig. 2B. Two regions, from −15 to −11 (p72.12) and from +1 to +5 (p72.15), in which substitution of the original sequence for a GCGCG sequence greatly decreased luciferase levels were found. The activity of these mutants was reduced to <6% of the wild-type value. On the other hand, substitutions from −5 to −1 (p72.4) significantly inhibited reporter activity (by approximately 60%), while mutations in the region from −36 to −16 reduced this activity to a much lesser extent. The deletion of this latter region, however, considerably diminished luciferase levels (Fig. 2A, p72.7 construct), suggesting that sequences with a length of more than five or six bases are involved in the promoter function of this region.
Single-nucleotide substitutions at the mRNA initiation region of the p72 promoter.
Mutants p72.14 and p72.15 have substitutions in residues which are sites for mRNA initiation. To better analyze the effect of changes in these and surrounding nucleotides, we obtained a complete set of all three possible single-nucleotide substitutions in positions from −1 to +5. Mutant promoters (p72.16A to p72.21T) were generated using mutually priming long oligonucleotides (8). Oligonucleotide 5′-D, which contains the 23-bp 5′ sequence of the promoter nontemplate strand, was independently mixed with oligonucleotides 3′-16 to 3′-21, which contain the 33-bp 5′ sequence of the promoter template strand mutated at different residues (Table 1). These pairs of oligonucleotides annealed at a 15-bp segment at their 3′ ends and acted as both template and primer. The extension of the oligonucleotides was performed with Sequenase (Pharmacia Biotech) in the presence of all four deoxynucleotides. The double-stranded DNA mutant promoters obtained in this way had a structure similar to that of linker scan mutants, with KpnI and BamHI sites at their ends, allowing their cloning into pUC118 plasmid. Plasmids containing point mutations were transfected together with control plasmid p72GAL10T, and the luciferase activity was measured and normalized to β-galactosidase activity. The results show that position +3 is notable for its intolerance to any deviation from the wild-type sequence (Fig. 2C). Changes at nucleotides +4 and +5 reduced the activity of the promoter about threefold, while only a small effect was observed when positions −1 to +2 were replaced.
Compilation of the linker scanning and point mutation data defined two critical regions within the p72 late promoter: an upstream region located around positions −15 to −11 and a downstream region encompassing the transcription initiation site.
Effects of p72 promoter mutations on transcription.
As described above, the relative strengths of the p72 promoter and promoter mutants were examined by measuring luciferase activity, which provided a convenient and precise method of assessing the promoter activity. The enzymatic activity is assumed to be an accurate reflection of the amount of mRNA synthesized. This was confirmed by direct analysis of the RNA isolated from infected cells that had been transfected with the wild type and representative mutations of the p72 promoter. As an internal control, transcripts from the D117R gene of the viral genome, which encodes the major structural protein p17 (32), were analyzed in parallel. As probes for this analysis, we used oligonucleotides that contained sequences of the coding strands of the luciferase and D117R genes close to their 5′ ends. These primers were end labeled, hybridized with total RNA, and elongated with avian myeloblastosis virus reverse transcriptase as described elsewhere (29). The elongated products were electrophoresed on a polyacrylamide gel. Quantitation of the luciferase transcripts (Fig. 3) demonstrated a good correlation between the RNA synthesized and the activity measured (Fig. 2).
A TATA sequence is required for efficient late viral transcription.
An analysis of the 5′-flanking sequences of ASFV late genes whose transcription initiation site has been mapped showed that late transcription starts very frequently at A or T residues. A comparison of these sequences showed a conservation around the mRNA start site (Fig. 4). Thus, the sequence TATA of the coding strand is located at or near the start point in 14 out of 17 late genes. No clear regions of similarity were observed in upstream sequences, although tracts of T or A residues were present in most cases.
FIG. 4.
Alignment of 5′ flanking sequences of ASFV late genes. Sequences were aligned by maximizing the identities around the transcriptional start site. Initiation sites (underlined) mapped within or close to a TATA sequence. The translation initiation codons are indicated in bold. References are as follows: p72 (26), p10 (J. M. Rodríguez, unpublished data), EP153R (14), IAP-like (11), p12 (4), B438L (15), S273R (A. Alejo, unpublished data), L83L (1), p17 (32), p11.5 (data not shown), CD2-like (28), p54 (25), I226R and I243L (29), Geranylgeranyl pyrophosphate synthase (GGPPS) (2), J154R (5), and dUTPase (24). The consensus sequence shows the most common nucleotide(s) at each position upstream of residue +5. Also listed is the next most common nucleotide that differs by only one from the most common residue. Alternative TATA sequences are indicated over a gray background.
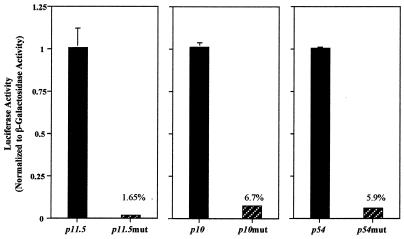
As determined above, the corresponding TATA sequence in the p72 promoter is a critical region. To assess if the TATA sequence acts as a general essential element for initiation of late transcription, we cloned the putative promoter sequences of three other late genes. The 41-bp DNA fragments containing the putative TATA elements and upstream sequences of the late genes encoding the p11.5, p10, and p54 major structural proteins were cloned. A mutant version of each promoter in which the TATA sequence was replaced by GCGC was also cloned. The cloning strategy was similar to that described for single-nucleotide mutagenesis of the p72 promoter, using the oligonucleotides shown in Table 1. Plasmids containing the promoters and the luciferase gene were cotransfected with p72GAL10T, and the normalized luciferase levels were determined. The activity of the wild type versus the TATA mutant promoter for each case is shown in Fig. 5. As can be seen, the percentage of mutant promoter activities ranged from less than 2% (p11.5 promoter) to about 6% (p10 and p54 promoters). These results show that a TATA sequence located at the transcription start point is essential for the activity of four late promoters and suggest that ASFV late transcription is dependent on the presence of this sequence at or near the region of transcriptional initiation.
FIG. 5.
Requirement of the TATA sequence in the transcription initiation region for activity of other viral late promoters. Plasmids containing wild-type and mutant versions of late promoters directing the expression of the luciferase gene were cotransfected with p72GAL10T as described in the legend of Fig. 2, and luciferase activity was normalized to that of β-galactosidase. All values were normalized to that of wild-type activity in each promoter. The percentages of mutant activities with respect to the wild-type levels are indicated.
A model for regulation of ASFV late gene expression.
Expression of ASFV late genes takes place after the onset of viral DNA replication and is dependent on it. It has been proposed that viral trans-acting factors regulating late transcription are expressed from newly synthesized viral genomes (29). These factors may be involved in the transactivation of the luciferase gene under the control of the late viral promoters, which was observed in the infected cells. The luciferase activities found under these conditions are correlative to mRNA levels, as demonstrated by primer extension assays of the accumulated messengers during ASFV infection (Fig. 2 and 3). The results show that the system is very useful for analyzing the structure of cis-acting late control elements using mutated promoters.
Two regions essential for p72 promoter activity have been detected. One region is located at positions −15 to −11, as replacement of these nucleotides by a GCGCG sequence abolished the promoter activity. Whether this effect is a consequence of the deletion of some essential sequence motif is unknown. No evident nucleotide conservation in that region of other late promoters is observed. On the other hand, A and T tracts are frequently found along ASFV late promoters, but the significance of this characteristic in transcription activation remains to be determined. In relation to this, it has been suggested that T-rich sequences upstream from a number of eukaryotic genes activate RNA polymerase II transcription (21). Similarly, tracts of A or T residues activate late promoter function in vaccinia virus (12).
The second essential region of the p72 promoter is localized at positions +1 to +5 and includes sites for mRNA initiation. When the 5′ sequences of 17 late genes were aligned by maximizing the identities around the transcription start region, we found some similarity. Thus, TATA sequences are present in 14 out of 17 promoters. As for the p72 promoter, replacement of this sequence in three other late gene promoters was also deleterious for activity, suggesting that the TATA sequence could be a motif for late promoter function. Sequence motifs which map around the site for transcription initiation and are necessary for transcription have been described for other systems. Thus, initiator elements of eukaryotes are common for many promoters of protein-encoding genes and have been grouped into families (33). Similar elements are present in poxvirus and baculovirus promoters. Intermediate and late vaccinia virus transcriptions start in the essential sequences TAAA and TAAAT (23), respectively. In the case of baculovirus, transcription of late and very late genes initiates at a TAAG sequence which is an essential element for both classes of promoters (20). Transcription factors which associate with these initiator elements have been described for eukaryotes (33) and vaccinia virus late promoters (9). It is likely that ASFV late transcription will depend on late transcription factors with affinity for the TATA sequence, an affinity which could be weaker or stronger, depending on the surrounding sequences. On the other hand, the essentiality of the sequence could be related to the mechanism of transcriptional initiation. Thus, an essential step in this mechanism is the melting at the site of initiation, which requires the unwinding of the double helix, a process which is energy dependent. It is possible that the A/T-rich sequence at the transcription initiation site could play a role in facilitating the unwinding of the double-stranded DNA and, consequently, the initiation of transcription. The finding that replacement of the A/T-rich region by G/C residues strongly reduces transcription is in keeping with this.
This is the first mutational analysis of an ASFV promoter. Future work will focus on elucidating the function of the TATA element in ASFV late transcription as well as on identifying the possible transcriptional factors which could interact with it.
Acknowledgments
We thank J. F. Rodriguez for help in setting up the analysis of promoters in ASFV, M. L. Salas and J. Salas for helpful discussions and critical reading of the manuscript, and M. Mencía and A. Alejo for comments about the manuscript.
This work was supported by Dirección General de Investigación Científica y Técnica grant PB96-0902-C02-01, European Community grant FAIR5-CT97-3441, Comunidad Autónoma de Madrid grant 07B/0032/1997, Ministerio de Educación y Cultura grant AGF98-1352-CE, and an institutional grant from Fundación Ramón Areces.
REFERENCES
- 1.Alejo A. Ph.D. thesis. Madrid, Spain: Universidad Autónoma de Madrid; 1999. [Google Scholar]
- 2.Alejo A, Yáñez R J, Rodríguez J M, Viñuela E, Salas M L. African swine fever virus trans-prenyltransferase. J Biol Chem. 1997;272:9417–9423. doi: 10.1074/jbc.272.14.9417. [DOI] [PubMed] [Google Scholar]
- 3.Almazán F, Rodríguez J M, Andrés G, Pérez R, Viñuela E, Rodríguez J F. Transcriptional analysis of multigene family 110 of African swine fever virus. J Virol. 1992;66:6655–6667. doi: 10.1128/jvi.66.11.6655-6667.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almazán F, Rodríguez J M, Angulo A, Viñuela E, Rodríguez J F. Transcriptional mapping of a late gene coding for the p12 attachment protein of African swine fever virus. J Virol. 1993;67:553–556. doi: 10.1128/jvi.67.1.553-556.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almazán F, Murguía J R, Rodríguez J M, de la Vega I, Viñuela E. A set of African swine fever virus tandem repeats shares similarities with SAR-like sequences. J Gen Virol. 1995;76:729–740. doi: 10.1099/0022-1317-76-4-729. [DOI] [PubMed] [Google Scholar]
- 6.Andrés G, García-Escudero R, Simón-Mateo C, Viñuela E. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol. 1998;72:8988–9001. doi: 10.1128/jvi.72.11.8988-9001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrés G, Simón-Mateo C, Viñuela E. Assembly of African swine fever virus: role of polyprotein pp220. J Virol. 1997;71:2331–2341. doi: 10.1128/jvi.71.3.2331-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1994. [Google Scholar]
- 9.Broyles S S, Liu X, Zhu M, Kremer M. Transcription factor YY1 is a vaccinia virus late promoter activator. J Biol Chem. 1999;274:35662–35667. doi: 10.1074/jbc.274.50.35662. [DOI] [PubMed] [Google Scholar]
- 10.Carrascosa J L, Gonzalez P, Carrascosa A L, Garcia-Barreno B, Enjuanes L, Viñuela E. Localization of structural proteins in African swine fever virus particles by immunoelectron microscopy. J Virol. 1986;58:377–384. doi: 10.1128/jvi.58.2.377-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacón M R, Almazán F, Nogal M L, Viñuela E, Rodriguez J F. The African swine fever virus IAP homolog is a late structural polypeptide. Virology. 1995;214:670–674. doi: 10.1006/viro.1995.0083. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J, Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 13.Enjuanes L, Carrascosa A L, Moreno M A, Viñuela E. Titration of African swine fever virus. J Gen Virol. 1976;32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- 14.Galindo I, Almazán F, Bustos M J, Viñuela E, Carrascosa A L. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology. 2000;266:340–351. doi: 10.1006/viro.1999.0080. [DOI] [PubMed] [Google Scholar]
- 15.Galindo I, Viñuela E, Carrascosa A L. Characterization of the African swine fever virus protein p49: a new late structural polypeptide. J Gen Virol. 2000;81:59–65. doi: 10.1099/0022-1317-81-1-59. [DOI] [PubMed] [Google Scholar]
- 16.García R, Almazán F, Rodríguez J M, Alonso M, Viñuela E, Rodriguez J F. Vectors for the genetic manipulation of African swine fever virus. J Biotechnol. 1995;40:121–131. doi: 10.1016/0168-1656(95)00037-q. [DOI] [PubMed] [Google Scholar]
- 17.García-Beato R, Salas M L, Viñuela E, Salas J. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology. 1992;188:637–649. doi: 10.1016/0042-6822(92)90518-t. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Escudero R, Andrés G, Almazán F, Viñuela E. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J Virol. 1998;72:3185–3195. doi: 10.1128/jvi.72.4.3185-3195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznar J, Salas M L, Vinuela E. DNA-dependent RNA polymerase in African swine fever virus. Virology. 1980;101:169–175. doi: 10.1016/0042-6822(80)90493-6. [DOI] [PubMed] [Google Scholar]
- 20.Lu A, Miller L K. Regulation of baculovirus late and very late gene expression. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum; 1997. pp. 193–216. [Google Scholar]
- 21.Lue N F, Buchman A R, Kornberg R D. Activation of yeast RNA polymerase transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci USA. 1989;86:486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss B, Ahn B-Y, Amegadzie B, Gershon P D, Keck J G. Cytoplasmic transcription system encoded by vaccinia virus. J Biol Chem. 1991;266:1355–1358. [PubMed] [Google Scholar]
- 23.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 24.Oliveros M, García-Escudero R, Alejo A, Viñuela E, Salas M L, Salas J. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J Virol. 1999;73:8934–8943. doi: 10.1128/jvi.73.11.8934-8943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez F, Ley V, Gómez-Puertas P, García R, Rodriguez J F, Escribano J M. The structural protein p54 is essential for African swine fever virus viability. Virus Res. 1996;40:161–167. doi: 10.1016/0168-1702(95)01268-0. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez J M. Ph.D. thesis. Madrid, Spain: Universidad Autónoma de Madrid; 1993. [Google Scholar]
- 27.Rodríguez J M, Almazán F, Viñuela E, Rodriguez J F. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the β-galactosidase gene. Virology. 1992;188:67–76. doi: 10.1016/0042-6822(92)90735-8. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez J M, Yáñez R J, Almazán F, Viñuela E, Rodriguez J F. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J Virol. 1993;67:5312–5320. doi: 10.1128/jvi.67.9.5312-5320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez J M, Salas M L, Viñuela E. Intermediate class of mRNAs in African swine fever virus. J Virol. 1996;70:8584–8589. doi: 10.1128/jvi.70.12.8584-8589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salas M L, Kuznar J, Viñuela E. Polyadenylation, methylation, and capping of the RNA synthesized in vitro by African swine fever virus. Virology. 1981;113:484–491. doi: 10.1016/0042-6822(81)90176-8. [DOI] [PubMed] [Google Scholar]
- 31.Salas M L, Rey-Campos J, Almendral J M, Talavera A, Vinuela E. Transcription and translation maps of African swine fever virus. Virology. 1986;152:228–240. doi: 10.1016/0042-6822(86)90387-9. [DOI] [PubMed] [Google Scholar]
- 32.Simón-Mateo C, Freije J M P, Andrés G, López-Otín C, Viñuela E. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology. 1995;206:1140–1144. doi: 10.1006/viro.1995.1039. [DOI] [PubMed] [Google Scholar]
- 33.Weis L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 34.Yáñez R J, Rodríguez J M, Nogal M L, Yuste L, Enríquez C, Rodriguez J F, Viñuela E. Analysis of the complete sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]