Abstract
Expression of mouse mammary tumor virus (MMTV)-encoded superantigens in B lymphocytes is required for viral transmission and pathogenesis. We have previously established a critical role of an enhancer element within the long terminal repeat (LTR) for MMTV sag gene expression in B-lymphoid progenitor cells. We now demonstrate enhancer activity of this element in a promyelocytic progenitor cell line. We also map the position of the enhancer within the U3 region of the MMTV LTR and show that the progenitor cell enhancer shares functional elements with a previously described mammary gland-specific enhancer.
Mouse mammary tumor virus (MMTV), a murine B-type retrovirus, causes a high incidence of mammary gland carcinoma in infected females (15). MMTV-induced T-cell lymphomas are observed at a lower incidence. Infectious MMTV is produced by infected mammary gland epithelial cells and transmitted from mother to offspring via milk (3, 8). MMTV gene expression and virus production in mammary gland epithelial cells is constitutively activated through a mammary gland cell-specific enhancer element within the proviral long terminal repeats (LTRs) (14) and is further increased during pregnancy by receptor-mediated glucocorticoid hormone interaction with steroid-hormone-responsive elements near the major MMTV promoter. After passage through the gastrointestinal tract, the virus infects B lymphocytes, and these cells play an important role in virus spread in an infected animal (2, 9). MMTV encodes a superantigen (Sag) which, when it is expressed on the surfaces of B cells or other antigen-presenting cells, activates a large number of T cells by interaction with specific T-cell-receptor β-chains (1, 17). The resulting T-cell response in turn stimulates the infected B cells to proliferate (9) and thus amplifies the numbers of virus-infected cells (10) and potential target bystander cells. The subsequent clonal elimination of activated T cells results in a progressive depletion of a specific T-cell subset during the course of the infection (11). Infection of mammary gland epithelial cells completes the viral life cycle.
MMTV can also be genetically inherited in the form of endogenous proviruses (Mtv proviruses) in the host germ line. Sag expression from inherited proviruses usually leads to depletion of T-cell subsets that express reactive T-cell-receptor β-chains (6) but can also, as in the SJL mouse model of follicular B-cell lymphoma, induce T-cell-dependent, interleukin (IL)-mediated proliferation of B-lymphoma cells (22).
The viral sag gene encoding the MMTV superantigen is located within the 3′ viral LTR of a provirus (Fig. 1). The mechanism of MMTV sag gene expression has been controversial, and a total of four different promoters have been implicated in superantigen expression (19). The relative functional significance of putative MMTV promoters and enhancers for sag gene expression from an infectious, oncogenic MMTV provirus (21) has been tested by a major histocompatibility complex class II-independent superantigen reporter assay on the basis of a recombinant superantigen-luciferase gene that we developed earlier (19, 20). With this system we were able to detect sag gene expression in cells of the B-lymphocyte lineage and to demonstrate that promoters within the 5′ LTR are not required for sag gene expression. We have also described a transcription enhancer within the viral pol gene that is necessary for efficient expression of the sag gene in B-cell lines (19) (Fig. 1). In the B-cell progenitor line Ba/F3, sag gene expression and general gene expression from the MMTV LTR require a separate enhancer within the viral LTRs (20). Our recent data demonstrate expression of the MMTV sag gene in the myeloid progenitor cell line FDC-P1 (V. A. Tovar Sepulveda, B. Berdel, J. M. Coffin, and F. U. Reuss, submitted for publication). FDC-P1 cells were also found in electron micrographic experiments to produce MMTV-like virions in a dexamethasone-dependent fashion from an endogenous provirus. These data raise the question of whether MMTV gene expression in myeloid progenitor cells is also regulated by the enhancer in B-cell progenitor cells that we previously characterized. In the present study we demonstrate the activity of this enhancer element in the myeloid progenitor cell line FDC-P1 and show that the transcription enhancers for hematopoietic progenitor and mammary gland cells share functional elements.
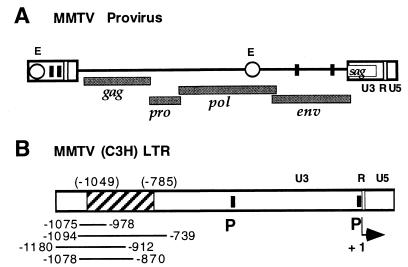
FIG. 1.
Schematic representation of an MMTV provirus and regulatory elements for MMTV gene expression. (A) MMTV provirus. The positions of viral genes (gag, pro, pol, env, and sag), promoters (filled squares), LTR and pol gene enhancers (E) (open circles), functional regions within the LTRs (U3, R, and U5), and the sag gene are indicated. (B) MMTV LTR. Shown is the LTR of MMTV strain C3H, with the positions of the mammary gland cell enhancer, as described by four different labs (7, 12, 14, 24), represented as solid lines below the LTR. The location of the newly characterized hematopoietic progenitor cell enhancer from positions −1025 to −761 of the Mtv-1 LTR, corresponding to the region from positions −1049 to −785 of the C3H LTR, is shown as a hatched box. The positions of promoters (P) within the LTR, functional regions (U3, R, and U5), and the transcription start site (+1, arrow) are indicated.
The MMTV progenitor cell enhancer is active in B-cell progenitor and promyelocytic cell lines.
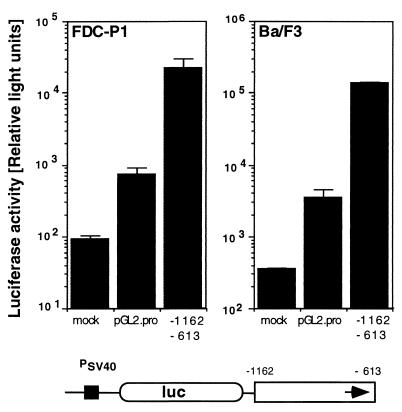
The murine, bone marrow-derived hematopoietic progenitor cell lines Ba/F3 and FDC-P1 are IL-3 dependent and nontransformed and represent pro-B and promyelocytic cell types, respectively. MMTV gene expression in the progenitor B-cell line Ba/F3 depends on the function of an enhancer element within a 548-bp region (nucleotides 9 to 556, positions −1162 to −613) of the MMTV(Mtv-1) LTR (4) present at the 5′ terminus of the Mtv-1/C3H provirus called hybrid MMTV (20, 21). The corresponding region within the MMTV(C3H) LTR (13) at the 3′ position of the reporter provirus has the same stimulatory effect. To address the question of whether this LTR region has enhancer activity in the murine FDC-P1 progenitor cell line, we have tested the ability of this enhancer to stimulate reporter gene expression from a heterologous promoter in these promyelocytic cells. We used the enhancer test plasmid pGL2-Promoter (Promega). Plasmid pGL2-Promoter carries an enhancerless simian virus 40 (SV40) promoter upstream of the luciferase reporter gene. DNA fragments containing putative enhancer elements can be inserted in either orientation upstream or downstream of the promoter-luciferase gene unit. Plasmid pGL2-Promoter carrying the Mtv-1 LTR fragment from nucleotides 6 to 556 (positions −1126 to −613 relative to the position of the transcription start site in Mtv-1) either 5′ or 3′ of the promoter-luciferase unit in either the plus or minus orientation has been described (20). For our enhancer test with FDC-P1 cells, we selected plasmid pGL2-E3+, which contains the LTR fragment 3′ of the promoter-luciferase unit in the positive orientation. The position of the enhancer 3′ of the luciferase transcription unit was chosen to exclude contributions from potential promoters within the fragment. Test plasmids were linearized within the plasmid backbone with a restriction enzyme and used for transfection of the mouse progenitor cell lines FDC-P1 (promyelocytic) and Ba/F3 (pro-B). Cells were harvested after 24 h and lysed. The luciferase activity and protein concentration were determined from the lysate. We found that the 548-bp enhancer fragment from positions −1162 to −613 of Mtv-1 stimulated gene expression in this assay system from an enhancerless SV40 promoter in both Ba/F3 and FDC-P1 cells by factors of 38 and 30, respectively (Fig. 2). We conclude that this enhancer is similarly active in both Ba/F3 B-cell progenitor and FDC-P1 promyelocytic progenitor cells. The effect of this enhancer element in primary hematopoietic progenitor cells remains to be tested.
FIG. 2.
Activity of the MMTV LTR enhancer in hematopoietic progenitor cell lines. FDC-P1 and Ba/F3 cells were transfected either in the absence of a reporter gene plasmid (mock) or with the enhancer test plasmid pGL2-Promoter (pGL2-Pro) or pGL2-Pro containing the Mtv-1 LTR fragment from positions −1162 to −613 cloned into the unique BamHI site 3′ of the promoter-reporter gene cassette (−1162 to −613). The schematic structure of the reporter plasmid is shown below the graph. An enhancerless SV40 promoter (PSV40) controls the expression of the luciferase reporter gene (luc). The MMTV enhancer element is represented by an open box, with an arrow indicating the positive orientation of the fragment. The IL-3-dependent mouse progenitor cell lines Ba/F3 (16) and FDC-P1 (5), gifts from U. Klingmüller, were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 10% WEHI 3B-conditioned medium, penicillin (100 IU/ml), and streptomycin (100 μg/ml). DNA was introduced by electroporation with a Bio-Rad Genepulser. Equimolar amounts of test plasmids (2 pmol) were linearized outside of the cloned provirus with a restriction enzyme. Carrier plasmid pGEM-2 and TE (10 mM Tris, 1 mM EDTA [pH 7.0]) were added to a constant DNA amount and volume. Cells were harvested after 24 h, washed twice in phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4), and lysed in reporter lysis buffer (Promega). The luciferase assay system (Promega) was used for detection of firefly luciferase (duplicate assays). Light emission was determined for 10 s in a model ILA911 (Tropix) or a model Lumat LB 9501 (Berthold, Wildbad, Germany) luminometer. Protein concentration in cellular extracts was measured by the Bio-Rad protein assay. Results are expressed as relative light units and represent the arithmetic means ± standard deviations from at least three parallel experiments with two different DNA preparations.
The MMTV enhancer for B-cell progenitors is located within a 265-bp region of the LTR and overlaps with a previously described mammary gland cell enhancer.
Also within the MMTV LTR, mammary gland-specific enhancer elements have been variously described at positions −1075 to −978 (24), positions −1094 to −739 (14), positions −1180 to −902 (7), and positions −1078 to −870 (12) of MMTV (strain C3H) (Fig. 1B). The different limits of this enhancer may derive from differences in the levels of resolution of the individual analyses and the use of different cell lines or assay systems by these investigators. This enhancer promotes general MMTV gene expression in mammary epithelial cells. The region of the MMTV (Mtv-1) LTR that we found to be active as the transcription enhancer in the hematopoietic progenitor cell lines Ba/F3 and FDC-P1 corresponds to positions −1186 to −638 of MMTV(C3H) and includes all of the previously characterized mammary gland enhancer elements. To determine the position of the newly identified progenitor cell enhancer within the MMTV LTR and to answer the question of whether the mammary gland and pro-B-cell enhancers are separate entities, we selected the luciferase reporter plasmid pGL2-Pro with the MMTV(Mtv-1) enhancer region (positions −1162 to −613) integrated 5′ of the SV40 promoter luciferase reporter gene in either the plus (pGL2-E5+) or minus (pGL2-E5−) orientation (20). Relative enhancer strength in Ba/F3 cells was tested after analysis of progressive exonuclease deletion from the 5′-end-flanking plasmid region (Fig. 3). We found that truncation from the 5′ terminus up to position −1025 did not affect reporter gene expression. Deletion to position −972 resulted in a partial loss (to 22%) and deletion to position −840 and beyond resulted in a complete loss (≤1%) of enhancer activity. Removal of sequences from the 3′ end to position −761 left enhancer activity unchanged. A moderate reduction in enhancer activity to 46% was noticed when the enhancer was shortened to position −838. Truncation of the enhancer to position −893 and beyond resulted in a complete loss of activity (≤2%). These data suggest that a functional pro-B-cell enhancer is present within a region from positions −1025 to −761 of the MMTV(Mtv-1) U3 region. The 5′ limits of the B-cell progenitor enhancer are found between positions −1025 and −972, and the 3′ border is located between positions −761 and −838.
FIG. 3.
Localization of the MMTV pro-B-cell enhancer within the Mtv-1 LTR by deletion analysis in Ba/F3 cells. The enhancer test plasmid pGL2-Promoter with the Mtv-1 LTR fragment from positions −1162 to −613 cloned into the unique XhoI site 5′ of the promoter-reporter gene cassette in either the plus (pGL2-E5+, filled squares) or minus (pGL2-E5−, open triangles) orientation and mutant plasmids with truncations to positions −1025, −972, −840, −750, and −737 for pGL2-E5+ and to positions −761, −838, −893, −969, −1096, and −1154 for pGL2-E5− were linearized with BamHI and transfected into Ba/F3 cells by electroporation. For each plasmid, a total of four transfections with two different plasmid preparations was done. After 30 h, cells were harvested and lysed and luciferase activity was determined as described in the legend to Fig. 2. Results are expressed as relative light units and represent the arithmetic means ± standard deviations from at least three parallel experiments with two different DNA preparations. Shown below the graph are a diagram of the MMTV (Mtv-1) enhancer fragment from positions −1162 to −613 as described previously (20) and a representation of the MMTV LTR with the functional regions (U3, R, and U5), transcription start site (+1) (labeled by a bent arrow), and restriction sites indicated. A hatched box indicates the region required for full activity.
The MAF and NFI binding regions contribute to MMTV pro-B-cell enhancer activity.
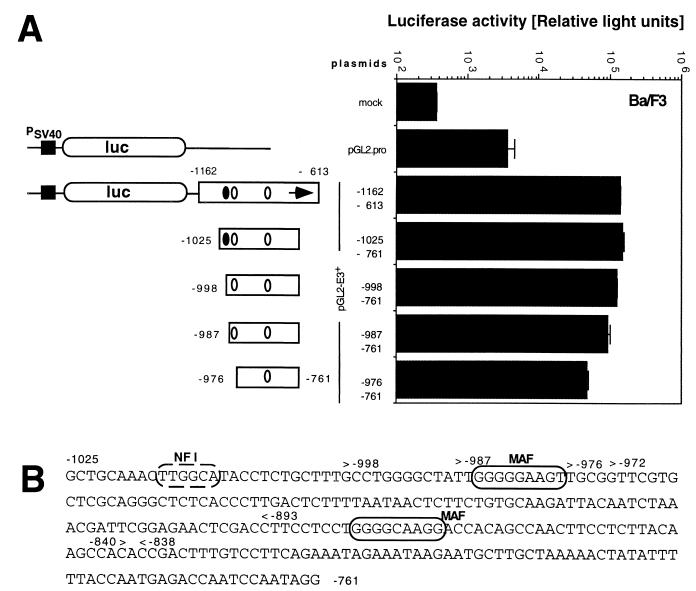
To test the hypothesis that the MMTV(Mtv-1) LTR region from positions −1025 to −761 retains the full enhancer activity, we isolated the corresponding region as a PCR-generated fragment and cloned this element between the BamHI and SalI sites of the luciferase reporter plasmid pGL2-Promoter. The resulting plasmid contained the potential enhancer in a position 3′ of the SV40 promoter luciferase gene reporter unit (pGL2-E3+[−1025/−761]). When the enhancer activity of this plasmid was compared to that of the pGL2-E3+ plasmid, which includes the original enhancer (positions −1162 to −613), in Ba/F3 cells, no significant difference was observed (Fig. 4A). We conclude from this experiment, that a fully functional pro-B-cell enhancer is located within a region from positions −1025 to −761 of the MMTV(Mtv-1) LTR. This region corresponds to positions −1049 to −785 of MMTV(C3H) and is largely identical to the mammary gland-specific enhancer previously characterized independently by four different laboratories (7, 12, 14, 24) (Fig. 1B). The B-cell progenitor enhancer region identified here is separate from the glucocorticoid response elements at positions −202 to −52. It contains the previously identified binding sites for mammary cell-activating factor (MAF) and CTF (also called NFI) (14) (Fig. 4B), which are involved in enhancer activity in mammary gland cells. MAF is an Ets-related transcription factor (23) and binds at two sites within the enhancer (Fig. 4B).
FIG. 4.
A 265-bp region from the Mtv-1 LTR is sufficient as an enhancer in Ba/F3 cells. (A) Ba/F3 cells were transfected without added reporter plasmid DNA (mock) or with the enhancer test plasmid pGL2-Pro or pGL2-Pro carrying the Mtv-1 regions indicated cloned into the unique BamHI and SalI sites 3′ of the promoter-reporter cassette. Cells were harvested and lysed, and luciferase activity was determined. Results are expressed as relative light units and represent the arithmetic means ± standard deviations from at least three parallel experiments with two different DNA preparations. PSV40, SV40 promoter (solid square). The NFI (solid oval) and MAF (open oval) sites are shown. (B) The DNA sequence of the Mtv-1 LTR region (8) from positions −1025 to −761 is presented below the graph. Established NFI and MAF binding sites (14) are shown by dashed and solid ovals, respectively.
The truncation analysis results shown in Fig. 3 suggest an important role for the 3′ MAF binding site: loss of the region between positions −838 and −893, including this MAF binding site, drastically reduced enhancer activity from 46% to less than 1% of the intact enhancer. The NFI and MAF sites at the 5′ end of the enhancer are not sufficient to maintain detectable enhancer activity. To specifically test the contribution of the MAF and NFI transcription factor binding sites at the 5′ terminus of the element to enhancer activity in pro-B cells, we successively removed these two sites (Fig. 4). We found that enhancer elements truncated in steps to position −976 showed gradually reduced enhancer activity (Fig. 4). In this analysis the loss of the region from −1025 to −998 that includes the NFI interaction site had only a minor effect. Further truncation to position −987 near the MAF binding site or removal of this site after deletion to position −976 reduced activity from 88 to 68 or 35%, respectively. The region from positions −976 to −761 that included only the 3′ MAF site still retained 35% of the maximal activity. This is in good agreement with our previous results shown in Fig. 3, where truncation from the 5′ terminus to position −972 was found to reduce enhancer activity to 22%.
We conclude that the MMTV transcription enhancer for B-cell progenitor and possibly also myeloid progenitor cells largely colocalizes with a previously characterized MMTV mammary gland enhancer (Fig. 1B). Both enhancers share specific functional regions that include the previously described NFI and MAF binding sites. Regions including the NFI and MAF binding sites at the 5′ end of the enhancer had a weak-to-moderate effect, while the region surrounding the 3′ MAF binding site had a strong effect on enhancer activity.
The MMTV sag gene within the LTR encodes isolate-specific superantigens that activate specific T-cell populations when they are expressed on the surfaces of antigen-presenting cells (1). Our previous screen for functional elements within the MMTV provirus that mediate sag gene expression demonstrated, surprisingly, that promoters within the 5′ LTR are not required for sag gene expression in B-lymphoid cell lines (19, 20). By contrast, expression of the MMTV sag gene in the mouse pro-B cell line Ba/F3 depends on an enhancer element within the viral U3 region (20). We demonstrate now that this enhancer is similarly active in a nonlymphoid hematopoietic progenitor cell line, FDC-P1, which is committed in its differentiation capacity to the myeloid cell lineage that includes monocytes/macrophages, granulocytes, platelets, and erythrocytes (5). These data suggest that the observed expression of the MMTV sag gene and the production of MMTV virions in FDC-P1 cells (Tovar Sepulveda et al., submitted) is regulated by the effect of the MMTV LTR enhancer in the LTR on individual promoters. For sag expression, our data indicate that a non-LTR promoter is used (Tovar Sepulveda et al., submitted), but the effect of the LTR enhancer on sag expression in these cells has not directly been tested yet. It is interesting that monocytes and macrophages are antigen-presenting cells and express the major histocompatibility complex class II antigens required for MMTV superantigen activity. MMTV transmission requires the presence of B cells, but MMTV superantigen presentation, T-cell stimulation, and T-cell deletion are possible in the absence of B cells (2). The non-B antigen-presenting cells that are able to mediate the superantigen effect have not yet been identified, but the involvement of myeloid cell types appears likely.
The MMTV transcription enhancer for pro-B cells maps to the region from positions −1025 to −761 of the MMTV(Mtv-1) LTR (4). This region corresponds to positions −1049 to −785 of MMTV(C3H) (13). The DNA sequences of the endogenous provirus Mtv-1 and the exogenous C3H virus isolate in this region differ in eight positions scattered throughout the region. As previously shown, there are no strain-specific differences with respect to enhancer activity between the Mtv-1 and C3H- LTRs in Ba/F3 B-cell progenitor cells (20). A region from the MMTV(C3H) LTR that contains this enhancer element has the same stimulatory effect on sag gene expression in the context of our hybrid MMTV reporter provirus.
The B-cell progenitor enhancer described here largely colocalizes with the mammary gland-specific enhancer previously characterized by four independent laboratories (7, 12, 14, 24) of the MMTV (C3H) LTR. Specifically, the MMTV enhancer for gene expression in pro-B cells contains the previously identified binding sites for MAF and NFI (14) (Fig. 4B). The regions surrounding the NFI, 5′ MAF, and 3′ MAF binding sites have only a weak, a moderate, and a strong effect on enhancer activity in Ba/F3 cells, respectively. At the 3′ border of the enhancer, still uncharacterized functional elements downstream of the MAF site have a moderate effect on enhancer function. These data suggest that functional elements are shared between the mammary gland cell and B-cell progenitor enhancers. The involvement of additional transcription factors and binding sites for enhancer activity in hematopoietic progenitors is possible. Additionally, the specificity of MMTV gene expression may be influenced by regulatory elements outside the previously characterized mammary gland and progenitor cell enhancers (18).
Acknowledgments
We are grateful to H. zur Hausen for support, to U. Klingmüller and H. Varmus for generous gifts of cell lines or reagents, and to S. Fenner for technical assistance.
J.M.C. is an American Cancer Society professor of molecular biology and microbiology. This work was supported by National Cancer Institute award R35CA44385 to J.M.C. and a Leukemia Society of America fellowship to F.U.R.
REFERENCES
- 1.Acha Orbea H, MacDonald H R. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 2.Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber B T. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J Exp Med. 1994;179:1457–1466. doi: 10.1084/jem.179.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bittner J J. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162–169. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 4.Crouse C A, Pauley R J. Molecular cloning and sequencing of the MTV-1 LTR: evidence for a LTR sequence alteration. Virus Res. 1989;12:123–137. doi: 10.1016/0168-1702(89)90059-2. [DOI] [PubMed] [Google Scholar]
- 5.Dexter T M, Garland J, Scott D, Scolnick E, Metcalf D. Growth of factor-independent hemopoietic precursor cell lines. J Exp Med. 1980;152:1036–1047. doi: 10.1084/jem.152.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frankel W N, Rudy C, Coffin J M, Huber B T. Linkage of Mls genes to endogenous mammary tumor viruses of inbred mice. Nature. 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 7.Gouilleux F, Sola B, Couette B, Richard Foy H. Cooperation between structural elements in hormono-regulated transcription from the mouse mammary tumor virus promoter. Nucleic Acids Res. 1991;19:1563–1569. doi: 10.1093/nar/19.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hainaut P, Vaira D, Francois C, Calberg B C M, Osterrieth P M. Natural infection of Swiss mice with mouse mammary tumor virus (MMTV): viral expression in milk and transmission of infection. Arch Virol. 1985;83:195–206. doi: 10.1007/BF01309916. [DOI] [PubMed] [Google Scholar]
- 9.Held W, Shakhov A N, Izui S, Waanders G A, Scarpellino L, MacDonald H R, Acha Orbea H. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J Exp Med. 1993;177:359–366. doi: 10.1084/jem.177.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Held W, Waanders G A, Shakhov A N, Scarpellino L, Acha Orbea H, MacDonald H R. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 11.Kappler J W, Staerz U, White J, Marrack P C. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre P, Berard D S, Cordingley M G, Hager G L. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol Cell Biol. 1991;11:2529–2537. doi: 10.1128/mcb.11.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majors J E, Varmus H E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983;47:495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mink S, Hartig E, Jennewein P, Doppler W, Cato A C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992;12:4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandi S, McGrath C M. Mammary neoplasia in mice. Adv Cancer Res. 1973;17:353–414. [Google Scholar]
- 16.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 17.Pullen A M, Wade T, Marrack P, Kappler J W. Identification of the region of T cell receptor beta chain that interacts with the self-superantigen MIs-1a. Cell. 1990;61:1365–1374. doi: 10.1016/0092-8674(90)90700-o. [DOI] [PubMed] [Google Scholar]
- 18.Qin W, Golovkina T V, Peng T, Nepomnaschy I, Buggiano V, Piazzon I, Ross S R. Mammary gland expression of mouse mammary tumor virus is regulated by a novel element in the long terminal repeat. J Virol. 1999;73:368–376. doi: 10.1128/jvi.73.1.368-376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuss F U, Coffin J M. Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene. J Virol. 1998;72:6073–6082. doi: 10.1128/jvi.72.7.6073-6082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuss F U, Coffin J M. Stimulation of mouse mammary tumor virus superantigen expression by an intragenic enhancer. Proc Natl Acad Sci USA. 1995;92:9293–9297. doi: 10.1073/pnas.92.20.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shackleford G M, Varmus H E. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc Natl Acad Sci USA. 1988;85:9655–9659. doi: 10.1073/pnas.85.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsiagbe V K, Yoshimoto T, Asakawa J, Cho S Y, Meruelo D, Thorbecke G J. Linkage of superantigen-like stimulation of syngeneic T cells in a mouse model of follicular center B cell lymphoma to transcription of endogenous mammary tumor virus. EMBO J. 1993;12:2313–2320. doi: 10.1002/j.1460-2075.1993.tb05885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welte T, Garimorth K, Philipp S, Jennewein P, Huck C, Cato A C, Doppler W. Involvement of Ets-related proteins in hormone-independent mammary cell-specific gene expression. Eur J Biochem. 1994;223:997–1006. doi: 10.1111/j.1432-1033.1994.tb19078.x. [DOI] [PubMed] [Google Scholar]
- 24.Yanagawa S-I, Tanaka H, Ishimoto A. Identification of a novel mammary cell line-specific enhancer element in the long terminal repeat of mouse mammary tumor virus, which interacts with its hormone-responsive element. J Virol. 1991;65:526–531. doi: 10.1128/jvi.65.1.526-531.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]