Abstract
Viral assembly is an ideal system in which to investigate the transient recognition and interplay between proteins. During morphogenesis, scaffolding proteins temporarily associate with structural proteins, stimulating conformational changes that promote assembly and inhibit off-pathway reactions. Microviridae morphogenesis is dependent on two scaffolding proteins, an internal and an external species. The external scaffolding protein is the most conserved protein within the Microviridae, whose canonical members are φX174, G4, and α3. However, despite 70% homology on the amino acid level, overexpression of a foreign Microviridae external scaffolding protein is a potent cross-species inhibitor of morphogenesis. Mutants that are resistant to the expression of a foreign scaffolding protein cannot be obtained via one mutational step. To define the requirements for and constraints on scaffolding protein interactions, chimeric external scaffolding proteins have been constructed and analyzed for effects on in vivo assembly. The results of these experiments suggest that at least two cross-species inhibitory domains exist within these proteins; one domain most likely blocks procapsid formation, and the other allows procapsid assembly but blocks DNA packaging. A mutation conferring resistance to the expression of a chimeric protein (chiDr) that inhibits DNA packaging was isolated. The mutation maps to gene A, which encodes a protein essential for packaging. The chiDr mutation confers resistance only to a chimeric D protein; the mutant is still inhibited by the expression of foreign D proteins. The results presented here demonstrate how closely related proteins could be developed into antiviral agents that specifically target virion morphogenesis.
The proper assembly of viral proteins and nucleic acids into a biologically active virion involves numerous and diverse macromolecular interactions. While structural proteins must correctly interact with other structural proteins, proper morphogenesis is equally dependent on interactions between structural and scaffolding proteins. Viral scaffolding proteins are transiently associated with morphogenetic intermediates but are not found in the mature viral particle (10, 12, 17, 22). These proteins promote the efficiency and fidelity of particle formation by ensuring proper interactions between viral proteins, promoting the nucleation of assembly, and aiding in the formation of a precisely sized capsid (13, 16, 19). In the Microviridae, assembly is dependent on two scaffolding proteins, an internal and an external species. The assembly pathway has been extensively characterized (8), and the atomic structures of both the φX174 virion and a morphogenetic intermediate containing a full complement of scaffolding proteins have been solved (2, 3, 14, 15). Therefore, the results of genetic and biochemical analyses can be interpreted within a structural context.
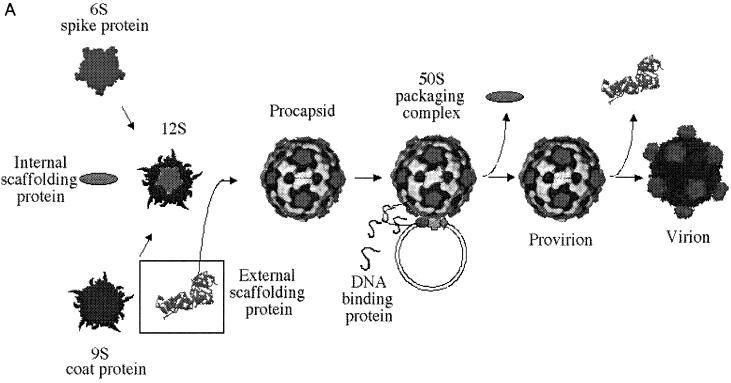
The Microviridae assembly pathway is illustrated in Fig. 1. The first detectable morphogenetic intermediates are the 9S and 6S particles, respective pentamers of the viral coat and spike proteins. In a reaction mediated by the internal scaffolding (or B) protein, these intermediates associate, forming the 12S particle. Twelve 12S particles are then organized into the procapsid by 240 copies of the external scaffolding (or D) protein. The preinitiation complex, consisting of one copy of viral proteins A and C, replicative-form (RF) DNA, and the host cell Rep protein, associates with the procapsid, probably along a twofold axis of symmetry (5), forming the 50S complex. Single-stranded genomic DNA is then concurrently synthesized and packaged. The internal scaffolding protein is extruded during this reaction, yielding the infectious provirion, which may represent the end of the intracellular assembly pathway. The external scaffolding protein may dissociate upon cell lysis. External scaffolding proteins are not unique to the Microviridae. The coliphage P4 Sid protein and the triplex proteins of the Herpesviridae perform analogous functions (13, 23). Although the Microviridae morphogenetic pathway has been well characterized, the mechanism in which viral scaffolding proteins enable nascent intermediates to reach a biologically active form remains somewhat obscure. Here we report that the expression of closely related external scaffolding proteins inhibits morphogenesis. Using chimeric proteins, we have defined the existence of two inhibitory domains. These results have implications for the use of closely related proteins in the development of antiviral analogs specific to viral morphogenesis.
FIG. 1.
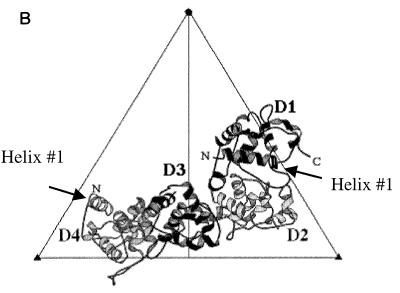
(A) The Microviridae morphogenetic pathway. In a reaction mediated by the internal scaffolding (or B) protein, 9S and 6S pentamers associate, forming the 12S particle. Twenty external scaffolding (or D) proteins add to this intermediate, which is then organized into the procapsid. The DNA binding (or J) protein enters the morphogenetic pathway during the packaging reaction, perhaps mediating the extrusion of the internal scaffold. Maturation is complete upon dissociation of the external scaffolding protein. (B) The four D proteins associated with each asymmetric unit (2). The subunits are designated D1 through D4; the amino and carboxyl termini are indicated where possible. The first α helices of D1 and D4 are depicted.
MATERIALS AND METHODS
Phage plating, media, burst experiments, stock preparation, generation of RF DNA, generation of radioactive lysates, and DNA isolation.
The reagents, media, buffers, and protocols for burst experiments and single-stranded DNA isolation have been previously described (6), as has the generation and isolation of RF DNA (1). The protocols for producing radioactive lysates and subsequent sucrose gradient sedimentation analyses are the same as those previously described exception that [35S]methionine and [35S]cysteine were used to label viral proteins (5).
Bacterial strains.
The Escherichia coli C strains C122 (supo), BAF5 (supE), and BAF30 recA have been previously described (1, 6). The C900 slyD host, used for the generation of radioactive lysates, was obtained from R. Young, Texas A&M University. The slyD mutation confers resistance to φX174 E-protein-mediated lysis (18).
Phage mutants.
The protocol used for oligonucleotide-mediated mutagenesis has been previously described (7). To generate the φX174 NheI/PvuI strain, φX174 am(D)Q22 DNA (7) was annealed to a mutagenic primer designed to concurrently eliminate the amber mutation and add the NheI restriction site. The mutant was selected for by the loss of the amber phenotype by transfection into C122 (supo). The NheI restriction site is located just after the region of the gene encoding the first α helix in the atomic structure of the D protein (2, 3). φX174 NheI DNA was mutagenized to introduce the previously described (9) am(J)S7 mutation. The PvuI site was then introduced with a mutagenic primer designed to concurrently eliminate the amber mutation and add the PvuI restriction site. Again, selection against the amber phenotype was used. The PvuI site served as a marker for the identification of the chimeric genes (see below). The am(E)W4 mutation was then introduced as previously described (4). Similar methods were used to generate the α3 NheI, α3 NheI/am(E)W4, α3am(J)S7, and α3am(D)Q20 mutations. The PvuI and NheI restriction sites do not alter the amino acid sequences of the φX174 or α3 D and J proteins.
Due to the high reversion frequencies of the am(D) mutations, double am(D) mutants and frameshift mutants were also generated. A second amber mutation was introduced into φX174 am(D)Q22 DNA at codon 18. Cells carrying a clone of the φX174 D gene were transfected. The double amber mutant was distinguished from the parental single amber mutant by its inability to propagate in strain BAF5 (supE), which is unable to support the growth of the double amber mutant. A frameshift mutation within the α3 D gene was generated by cutting α3 NheI RF DNA with NheI and filling in the overhangs, followed by blunt-end ligation using standard protocols (20). Cells harboring a clone of the α3 D gene were transfected, and the mutant was identified by complementation-dependent phenotype. All mutations were verified by a direct sequence analysis. φX174 chiDr mutants that had gained the ability to propagate in the presence of the chimeric φX/α3 D protein were selected by plating 108 wild-type phage on cells harboring a clone of the chimeric φX/α3 D gene (see below).
Cloning of the φX174 and α3 DJ, φX and α3 J, and both chimeric D genes.
Single-stranded DNA from both φX174 NheI/PvuI am(E)W4 and α3 NheI/PvuI am(E)W4 mutants served as the template for PCR amplification of the D and J genes. The DJ fragments were cloned directly into the TOPO TA vector (Invitrogen) and then subcloned into the pSE420 expression vector (Invitrogen). The cloned genes are under lactose induction. The α3 J gene was cloned into pSE420 by digesting α3 RF DNA with HinPI and Bsp120I and digesting plasmid DNA with BstBI and NotI. All clones were verified by complementation experiments and RF digests. The clone of the φX174 J gene has been previously described (9).
To construct the chimeric D genes, plasmid DNA from the α3 and φX174 DJ clones was digested with NheI and Bsp120I, yielding fragments encoding α helices 2 to 7 of gene D and the entire J gene. Following ligation and transformation, putative chimeric clones were identified by the ability to complement an am(J) mutant under low induction and verified by restriction enzyme digestion with EcoRV and PvuI. The chimeric D genes were also verified by direct DNA sequence analysis.
RESULTS
Cross-functional and cross-species inhibition analysis of the external scaffolding proteins.
To determine whether the φX174 and α3 external scaffolding proteins were capable of cross-complementation, nullD mutants were plated on cell lines harboring clones of the φX174 or α3 D and J genes (Table 1). Despite 70% identity in amino acid sequence of the proteins, no cross-complementation was observed. Both null D mutants plated with efficiencies at least 1 order of magnitude below the reversion frequencies of the nullD mutations, as assayed on the C122 (supo) host. The failure to recover revertants suggested that the expression of the foreign proteins might confer cross-species inhibition. To determine if the expression of the foreign external scaffolding protein or DNA binding protein was affecting wild-type morphogenesis, wild-type φX174 and α3 were plated on DJ plasmid-containing hosts and hosts harboring a clone of only the J genes. Expression of the J proteins failed to cross-inhibit either species of wild type, indicating that cross-inhibition was due to the overexpression of the scaffolding proteins.
TABLE 1.
EOPs of wild-type, nullD, and chiDr strains of φX174 and α3 in cells expressing foreign and chimeric external scaffolding proteinsa
| Virus | EOP for BAF30 (recA) with:
|
|||||
|---|---|---|---|---|---|---|
| pφXDJ | pα3DJ | pφXJ | pα3J | pφX/α3D | pα3/φXD | |
| φX174 nullDb | 1.0 | <10−6 | 10−6 | 10−6 | 10−6 | 10−6 |
| α32 nullDc | <10−6 | 1.0 | 10−6 | 10−6 | 10−6 | 10−6 |
| Wild-type φX174 | 1.0 | 1.0–10−6 | 1.0 | 1.0 | 10−3–10−5 | 1.0–10−2 |
| Wild-type α3 | 1.0–10−6 | 1.0 | 1.0 | 1.0 | 1.0–10−2 | 10−3–10−5 |
| φX174 chiDr | 0.5 | 1.0–10−2 | 0.5 | 0.5 | 1.0 | 1.0–10−2 |
The construction of chimeric genes is depicted in Fig. 2.
Contains two amber mutations in the gene.
Contains a frameshift mutation in codon 20.
Inhibition was very dependent on cell growth phase and induction conditions; therefore, ranges are reported. Low induction conditions were defined as those needed to obtain EOP (efficiency of plating) values of 1.0 in complementation experiments. For example, the cloned φX174 D gene will complement the φX174 nullD mutant at an isopropyl-β-d-thiogalactopyranoside (IPTG) concentration of 5.0 μM, using either log- or stationary-phase cells to seed the plate. This standard condition gave similar results with every cloned gene. High induction conditions were obtained by using an IPTG concentration of 50 μM and seeding plates with exponentially growing cells.
Characterization of inhibitory domains.
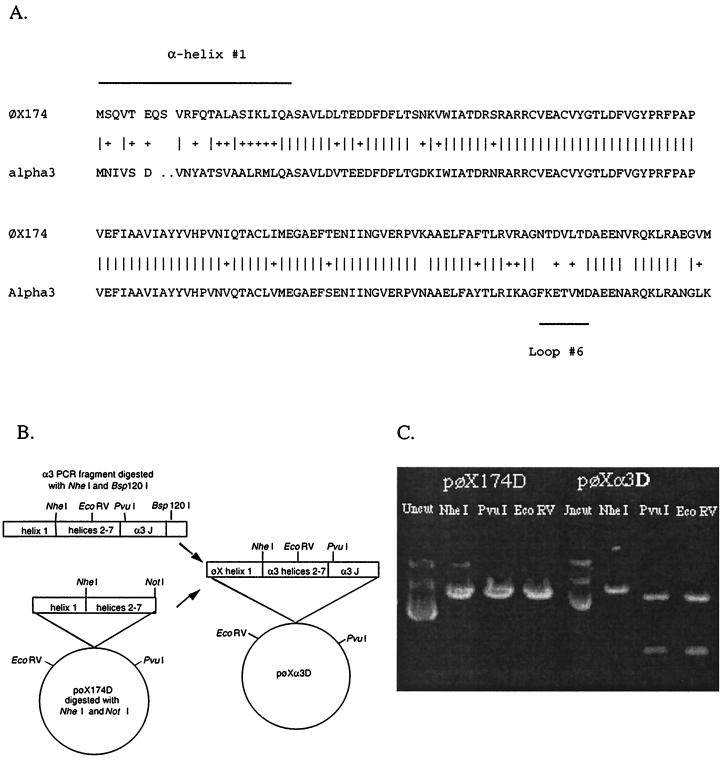
As shown in Fig. 2, divergent residues are localized to two distinct regions within the D proteins: amino acids comprising α helix 1 and the C terminus comprising loop 6 and the tip of α helix 7 in the atomic structure (2, 3, 11, 21). To determine whether one or both of these domains confer inhibitory effects, chimeric genes were constructed and the proteins were expressed in vivo. The φX/α3 protein contains α helix 1 from φX174; the remaining part of the protein is derived from α3. The opposite construct (α3/φX) contains α helix 1 from α3, with the remaining portion of the protein derived from φX174 (Fig. 2). Table 1 shows that neither chimeric gene can complement a nullD mutant. However, inhibition of the wild type was observed, suggesting that the chimeric proteins retain enough function to enter into the morphogenetic pathway. Expression of the chimeric protein possessing the φX174 α helix 1 inhibits φX174 morphogenesis (Table 1) even under low induction conditions and weakly inhibits α3 morphogenesis. Likewise, expression of the construct with α helix 1 from α3 strongly inhibits α3 morphogenesis but weakly inhibits φX174 morphogenesis. These results suggest that both domains confer some level of inhibition.
FIG. 2.
(A) Sequence alignment of the φX174 and α3 external scaffolding proteins. (B) Generation of the φX/α3 chimeric gene. (C) Verification of the φX/α3 chimeric gene.
Isolation of a φX174 mutant resistant to the expression of the φX/α3 chimeric external scaffolding protein.
To isolate a φX174 mutant that could efficiently propagate in the presence of the chimeric φX/α3 D protein, 108 wild-type phage were plated on the cells expressing the chimeric protein at high induction conditions. The chiDr mutant was identified by its ability to consistently plate with EOPs near 1.0 in the presence of the chimeric protein. Initially 80% of the chiDr genome was sequenced. Only one base change was found. The mutation confers a valine→alanine change at amino acid 286 in protein A. This region of gene A does not overlap gene B. To confirm that this base change was solely responsible for the ChiDr phenotype, the mutant A gene was moved into another wild-type background. The phenotype bred true.
The A protein initiates both stage II and stage III DNA synthesis. During stage II DNA synthesis, protein A mediates the semiconservative replication of double-stranded RF DNA. This stage of DNA replication occurs independently of procapsid morphogenesis. In stage III DNA replication, single-stranded genomic DNA is concurrently synthesized and packaged. This stage of DNA synthesis cannot occur in the absence of procapsids (8). The packaging preinitiation complex, consisting of viral proteins A and C and the host cell Rep protein, must dock to a fully formed procapsid. The location of the chiDr substitution suggests that chimeric φX/α3 D protein, more specifically α helices 2 to 6, inhibits DNA packaging, not procapsid formation (see below).
The φX174 chiDr mutant was assayed for the ability to grow in cells overexpressing the wild-type α3 and chimeric α3/φX external scaffolding proteins. Although inhibition was observed in cells expressing the α3 D protein (Table 1), it was considerably less than that obtained for wild-type φX174, comparable to that observed for wild-type φX174 in cells expressing the α3/φX chimera. In addition, the chiDr mutation appears to have little or no effect on growth in the presence of the α3/φX protein. These data suggest that two cross-species inhibitory domains exist in the external scaffolding proteins.
A φX174 chiDr/amD/amD mutant was constructed to determine whether chiDr mutation allowed for the utilization of the chimeric φX/α3 protein in plating assays. The chimeric gene was unable to complement the amD mutation in the chiDr background (data not shown). The low level of inhibition (10−2) of wild-type φX174 growth by the α3/φX chimeric protein proved too refractory for the isolation of φX174 resistant mutants and experiments described below.
In vivo analysis of wild-type inhibition by foreign and chimeric scaffolds.
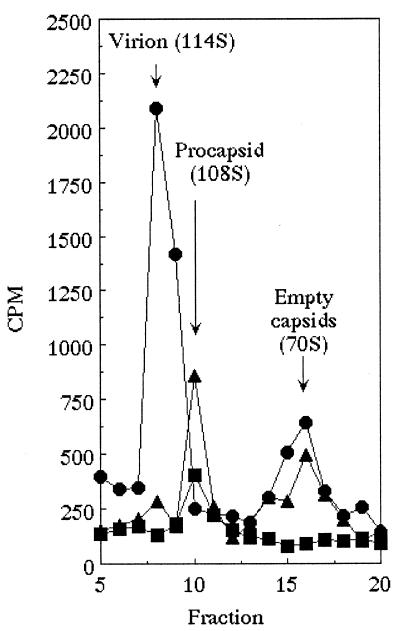
The location of the chiDr mutation in gene A suggests that the φX/α3 protein does not block procapsid formation but blocks DNA packaging. To test this hypothesis, the assembly intermediates synthesized in wild-type φX174-infected cells were analyzed by sucrose gradient sedimentation as described in Materials and Methods. The sedimentation profiles of 35S-labeled viral products are presented in Fig. 3. In cells expressing the φX174 D protein, infectious virion (114S) and degraded procapsid structures (70S) were detected. However, in the presence of the chimeric φX/α3 protein, only procapsids (108S) and degraded procapsids were present. This result is consistent with a block in DNA packaging and the location of the chiDr mutation in gene A, since the A protein must interact with the procapsid during DNA packaging. In cells expressing the α3 external scaffolding protein, only low levels of procapsids and/or degraded procapsids were detected, suggesting that morphogenesis is inhibited before procapsid formation.
FIG. 3.
Sedimentation analysis of particles produced in the presence of foreign and chimeric scaffolding proteins. Symbols: ●, wild-type α3 grown in cells overexpressing the α3 D protein (control); ■, wild-type φX174 grown in cells overexpressing the α3 D protein; ▴, wild-type φX174 grown in cells overexpressing the φXα3 chimeric D protein.
Isolation of a φX174 multiple mutant resistant to the expression of the α3 chimeric external scaffolding protein.
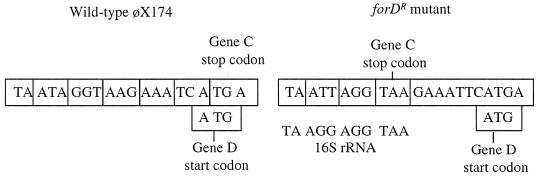
Although it was not possible to isolate a φX174 mutant resistant to expression of the α3 external scaffolding protein via one mutational step (data not shown), a mutant was obtained in a multistep selection. Cells expressing the α3 external scaffolding protein were infected at a multiplicity of 0.01 in liquid culture. Induction conditions were kept at levels that produced an EOP of 10−2 in plating assays. The ForDr (foreign D resistance) phenotype also confers resistance to both chimeric scaffolding proteins. The entire forDr genome has been sequenced. Two single-base insertions have been found at the end of gene C, which is adjacent to gene D. One insertion creates a premature stop codon in gene C, three codons upstream from the natural stop codon (Fig. 4). The natural stop codon overlaps with the gene D start codon. The premature stop codon overlaps with the ribosome binding site (RBS) of gene D. C protein termination and D protein initiation may now be more efficiently coupled than in the wild type. In addition, the insertion may create an RBS that can more easily anneal to the 16S rRNA. The second insertion is found between the D gene RBS and start codon. These considerations suggest that part of the ForDr phenotype may involve increasing wild-type D protein, as opposed to a truncated C protein, synthesis.
FIG. 4.
Nucleotide sequence of the upstream D regulatory region in the forD mutant.
DISCUSSION
Foreign external scaffolding proteins are potent cross-species inhibitors of viral morphogenesis. The Microviridae scaffolding proteins have diverged only 26% on the amino acid level, and this divergence is localized to two distinct regions: α helix 1 and loop 6/α helix 7 in the atomic structure. Chimeric proteins were designed to separate these two domains in order to determine their individual inhibitory effects. The results of this analysis suggest that both domains are responsible for some level of inhibition and demonstrate the feasibility of using closely related proteins as antiviral agents.
The efficiency of inhibition ascribed to each domain differs and appears to be concentration dependent. Low levels of induction are sufficient when the inhibited virus and α helix 1 of the chimeric protein are of the same origin. This suggests that α helix 1 may play a critical role in the self-association of D proteins into dimers or dimer recognition of other structural proteins. Within the atomic structure of the procapsid, four D proteins associate with the viral coat protein. The D proteins are arranged as asymmetric dimers of dimers, D1D2 and D3D4, and are not related by quasi-equivalence (2, 3). Residues between α helices 2 and 6 mediate the vast majority of the intradimer and interdimer contacts. This region of the φX174 and α3 D proteins exhibits 96% conservation on the amino acid level. This high level of conservation most likely allows interspecies oligomerization, effectively diluting the amount of indigenous protein capable of progressing through assembly as homodimers. However, α helix 1 mediates some intradimer contacts, but only in the D1D2 species. Without these contacts, dimers with a foreign α helix 1 may not be efficiently placed into the D1D2 position. However, once placed into the D1D2 position, that helix makes no contacts with other scaffolding or structural proteins.
Some coat-scaffolding interactions may not be apparent in the current atomic structure. During crystallization, the φX174 procapsid matured at the threefold axes of symmetry, producing a closed structure. The native, or open, structure has pores at the threefold axes. Genetic data (4, 7) suggest that an interaction may occur between D4 α helix 1 and α helix 4 of the viral coat protein. Both helices are found at the threefold axis of symmetry in the closed structure. In an open structure, the coat protein helix could be shifted upward and might contact α helix 1 of the D4 subunit, which is the most closely associated with underlying coat protein. Both helices are amphipathic and could interact via hydrophobic interfaces. If this interaction is indeed present in the native structure, this may exclude dimers with a foreign α helix 1 from the D3D4 position.
Finally, contacts made by α helix 1 may be important in forming an assembly naive dimer, from which the D1D2 and D3D4 are formed. All three models are consistent with the high-level induction conditions needed for proteins with a foreign α helix 1 and the absence of large particles. Further analyses will be required to distinguish between these models. The isolation of the resistant mutants described here is limited. The selection procedures would not have generated mutations within the cloned foreign or chimeric genes. Considering the low level of inhibition conferred by chimeric scaffolding proteins with foreign α helices 1, it should be possible to construct the chimeric gene directly within the φX174 genome and select for mutants which can propagate without concurrent expression of a cloned wild-type gene.
The mechanism of inhibition conferred by foreign COOH termini, on the other hand, is more apparent. The φX174 chiDr mutant alters viral protein A, a component of the genome biosynthesis/packaging machinery, which binds the procapsid along the twofold axis of symmetry. The location of the chiDr mutation and the isolation of procapsids from cells expressing the chimeric protein suggest that chimeric and wild-type scaffolding proteins form procapsids that cannot interact with the genome biosynthetic/packaging machinery. Because the chiDr phenotype does not circumvent the need for the wild-type protein, chimeric and wild-type proteins can achieve conformations needed for chiDr A protein recognition that chimeras alone cannot achieve. Nor does the mutation confer resistance to the expression of the wild-type α3 D protein or the α3/φX chimera, consistent with a model in which foreign scaffolding proteins confer multiple blocks in morphogenesis.
In the COOH termini, only the regions comprising loop 6 and α helix 7 have diverged between the φX174 and α3 proteins. α helix 7 is ordered only in the D4 subunit, in which it makes multiple contacts with the underlying coat. While the D protein amino acids participating in these contacts have diverged, albeit conservatively, the target residues in the φX174 and α3 coat proteins are identical. These observations suggest that the highly divergent loop 6 may be responsible for recognizing the genome biosynthetic/packaging machinery or forming part of its docking interface. With the identification of the chiDr mutations, the chimeric D gene can be placed directly into a chiDr background. The chimeric virus can be propagated in cells expressing the wild-type D protein. A direct and simple selection, the loss of complementation-dependent growth, can be used to isolate mutants that can utilize only the chimeric protein or a mutated form.
ACKNOWLEDGMENT
This study was supported by a grant from the National Science Foundation to B.A.F.
REFERENCES
- 1.Burch A D, Ta H J, Fane B A. Cross-functional analysis of the Microviridae internal scaffolding protein. J Mol Biol. 1999;286:95–104. doi: 10.1006/jmbi.1998.2450. [DOI] [PubMed] [Google Scholar]
- 2.Dokland T, McKenna R, Ilag L L, Bowen B R, Incardona N L, Fane B A, Rossmann M G. Structure of a viral assembly intermediate with molecular scaffolding. Nature. 1997;389:308–313. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- 3.Dokland T, Bernal R A, Burch A D, Pletnev S, Fane B A, Rossmann M G. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus φX174. J Mol Biol. 1999;288:595–608. doi: 10.1006/jmbi.1999.2699. [DOI] [PubMed] [Google Scholar]
- 4.Ekechukwu M C, Fane B A. Characterization of the morphogenetic defects conferred by cold-sensitive prohead accessory and scaffolding proteins of φX174. J Bacteriol. 1995;177:829–830. doi: 10.1128/jb.177.3.829-830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekechukwu M C, Oberste D J, Fane B A. Host and φX174 mutations which affect the morphogenesis or stabilization of the 50S complex, a single stranded DNA synthesizing intermediate. Genetics. 1995;140:1167–1174. doi: 10.1093/genetics/140.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fane B A, Hayashi M. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage φX174. Genetics. 1991;128:663–671. doi: 10.1093/genetics/128.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fane B A, Shien S, Hayashi M. Second-site suppressors of a cold sensitive external scaffolding protein of bacteriophage φX174. Genetics. 1993;134:1003–1011. doi: 10.1093/genetics/134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi M, Aoyama A, Richardson D L, Hayashi M N. Biology of the bacteriophage φX174. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Publishing Corporation; 1988. pp. 1–71. [Google Scholar]
- 9.Jennings B, Fane B A. Genetic analysis of the φX174 DNA binding protein. Virology. 1997;227:370–377. doi: 10.1006/viro.1996.8351. [DOI] [PubMed] [Google Scholar]
- 10.King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 11.Kodaira K, Nakano K, Okada S, Taketo A. Nucleotide sequence of the genome of bacteriophage α3: interrelationship of the genome structure and the gene products with those of the phages φX174, G4 and øK. Biochem Biophys Acta. 1992;113:277–288. doi: 10.1016/0167-4781(92)90440-b. [DOI] [PubMed] [Google Scholar]
- 12.Liebowitz J, Horowitz M S. Synthesis and assembly of adenovirus polypeptides. III. Reversible inhibition of hexon assembly in adenovirus type 5 temperature-sensitive mutants. Virology. 1975;66:10–24. doi: 10.1016/0042-6822(75)90175-0. [DOI] [PubMed] [Google Scholar]
- 13.Marvik O J, Dokland T, Nokling R H, Jacobsen E, Larsen T, Lindqvist B J. The capsid size-determining protein sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 14.McKenna R, Ilag L L, Rossmann M G. Analysis of the single-stranded DNA bacteriophage φX174 at a resolution of 3.0 Å. J Mol Biol. 1994;237:517–543. doi: 10.1006/jmbi.1994.1253. [DOI] [PubMed] [Google Scholar]
- 15.McKenna R, Xia D, Willingmann P, Ilag L L, Krishnaswamy S, Rossmann M G, Olson N H, Baker T S, Incardonna N L. Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature. 1992;355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prevelige P E, Thomas D, King J. Nucleation and growth phage in the polymerization of coat scaffolding subunits into icosadhedral procapsid shells. J Biophys. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rixon F J. Structure and assembly of herpes viruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 18.Roof W D, Horne S M, Young K D, Young R. SlyD, a host gene required for φX174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 19.Rossmann M G. Constraints on the assembly of spherical virus particles. Virology. 1984;134:1–13. doi: 10.1016/0042-6822(84)90267-8. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Sanger F, Coulson A R, Friedmann C T, Air G M, Barrell B G, Brown N L, Fiddes J C, Hutchison III C A, Slocombe P M, Smith M. The nucleotide sequence of bacteriophage φX174. J Mol Biol. 1978;125:225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- 22.Siden E J, Hayashi M. Role of gene B product in bacteriophage φX174 development. J Mol Biol. 1974;89:1–16. doi: 10.1016/0022-2836(74)90159-4. [DOI] [PubMed] [Google Scholar]
- 23.Spencer J V, Newcomb W W, Thomsen D R, Homa F L, Brown J C. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]