Abstract
Nuclear export of the incompletely spliced mRNAs encoded by several complex retroviruses, including human immunodeficiency virus type 1 (HIV-1), is dependent on a virally encoded adapter protein, termed Rev in HIV-1, that directly binds both to a cis-acting viral RNA target site and to the cellular Crm1 export factor. Human endogenous retrovirus K, a family of ancient endogenous retroviruses that is not related to the exogenous retrovirus HIV-1, was recently shown to also encode a Crm1-dependent nuclear RNA export factor, termed K-Rev. Although HIV-1 Rev and K-Rev display little sequence identity, they share the ability not only to bind to Crm1 and to RNA but also to form homomultimers and shuttle between nucleus and cytoplasm. We have used mutational analysis to identify sequences in the 105-amino-acid K-Rev protein required for each of these distinct biological activities. While mutations in K-Rev that inactivate any one of these properties also blocked K-Rev-dependent nuclear RNA export, several K-Rev mutants were comparable to wild type when assayed for any of these individual activities yet nevertheless defective for RNA export. Although several nonfunctional K-Rev mutants acted as dominant negative inhibitors of K-Rev-, but not HIV-1 Rev-, dependent RNA export, these were not defined by their inability to bind to Crm1, as is seen with HIV-1 Rev. In total, this analysis suggests a functional architecture for K-Rev that is similar to, but distinct from, that described for HIV-1 Rev and raises the possibility that viral RNA export mediated by the ∼25 million-year-old K-Rev protein may require an additional cellular cofactor that is not required for HIV-1 Rev function.
Retroviral replication requires the coordinated expression of both unspliced and spliced forms of the initial, genome-length viral transcript. However, cells have evolved mechanisms to prevent the nuclear export of incompletely spliced cellular mRNAs, i.e., pre-mRNAs, via the canonical cellular mRNA export pathway. Because these same mechanisms can also lead to nuclear retention of incompletely spliced retroviral mRNAs, these viruses have had to evolve ways to target their incompletely spliced transcripts to the cytoplasm via export pathways that are not normally used by cellular mRNA molecules (reviewed by in references 8 and 34). One such mechanism, first identified in the pathogenic lentivirus human immunodeficiency virus type 1 (HIV-1), targets unspliced viral transcripts into a nuclear export pathway normally used for small nuclear RNAs and certain proteins that shuttle between nucleus and cytoplasm (10, 14, 29, 32, 42). Specifically, HIV-1 encodes an adapter protein, termed Rev, that can bind to, and multimerize on, a cis-acting viral RNA target site termed the HIV-1 Rev response element (H-RRE) (28, 29, 44). In the nucleus, Rev also binds, via a leucine-rich nuclear export signal (NES), to the cellular export factor Crm1 in an interaction that is dependent on the presence of the GTP-bound form of the cellular G-protein Ran (3, 11, 31, 38). Crm1 in turn interacts with components of the nuclear pore and thereby targets the resultant ribonucleoprotein complex to the cytoplasm (31). Subsequently, other lentiviruses were also shown to encode Rev proteins, while members of the distinct human T-cell leukemia virus type 1 (HTLV-1) family of oncoretroviruses encode a Rev homolog, termed Rex, that acts to induce the nuclear export of unspliced HTLV-1 transcripts by recruiting Crm1 to a cis-acting RNA target site, the Rex response element (3, 16, 21).
Until recently, only these two retroviral subgroups had been shown to encode a Rev-like nuclear RNA export activity. In contrast, other exogenous retroviruses either directly recruit a cellular nuclear export factor, distinct from Crm1, to an RNA target site termed a constitutive transport element or induce nuclear export of their incompletely spliced transcripts via an unknown mechanism (8, 34). Recently, however, a third type of Crm1-dependent retroviral RNA export factor, termed K-Rev, was identified in the human endogenous retrovirus K (HERV-K) and shown to induce the nuclear export of unspliced mRNAs bearing a copy of a HERV-K-derived RNA target site, the K-RRE (27, 43). The HERV-K family of endogenous retroviruses is present at between 50 and 100 copies per haploid human genome and entered the human germ line over a ∼25 million-year period starting ∼30 million years ago (1, 39). No exogenous retrovirus closely related to the HERV-K family is known to exist (17), and these endogenous viruses can therefore be viewed as fossil remnants of an exogenous virus family that is now probably extinct. However, the presence in the HERV-K genome of a nuclear RNA export factor and a cis-acting RNA target site that together are closely comparable in function to HIV-1 Rev (H-Rev) and the H-RRE does suggest that this RNA export activity arose fairly early in retroviral evolution (27, 43).
Previously, it has been demonstrated that the 116-amino-acid (aa) H-Rev protein and the 105-aa K-Rev protein share several biological properties, although they lack any evident sequence similarity (27, 43). Both H-Rev and K-Rev can specifically bind to their viral RNA response element, form homomultimers, and bind to Crm1 in a Ran · GTP-dependent manner (43). Curiously, although both H-Rev and K-Rev shuttle between nucleus and cytoplasm, H-Rev displays a predominantly nuclear and nucleolar localization at steady state whereas K-Rev is largely cytoplasmic (3, 43). However, K-Rev does localize to nuclei and nucleoli when Crm1 function is inhibited, thus demonstrating that the cytoplasmic localization of K-Rev is dependent on ongoing, Crm1-dependent nuclear export (27, 43).
The H-Rev protein has been subjected to intense mutational analysis, and the sequences involved in RNA binding, multimerization, and Crm1-dependent nuclear export are now well defined (8, 34). In contrast, no mutational analysis of K-Rev has been previously described. Here, we report the identification of sequences within K-Rev that are critical for K-RRE and Crm1 binding, multimerization, and nucleocytoplasmic shuttling and identify mutants of K-Rev that can function as specific dominant negative inhibitors of wild-type K-Rev function.
MATERIALS AND METHODS
Construction of molecular clones.
The following mammalian expression plasmids have been previously described: the indicator plasmids pG6(−31)HIVLTRΔTARCAT (37), pDM128/CMV (23, 30), and pDM128/K-RRE (43); effector plasmids pcRev (29), pSG424/Crm1, pSG424/K-Rev, pBC12/CMV/K-Rev/VP16, pBC12/CMV/K-Rev (43), and pBC12/CMV/ΔCAN (3); and control plasmids pBC12/CMV and pBC12/CMV/β-gal (3). Derivatives of pBC12/CMV/K-Rev encoding K-Rev mutants K1 through K14 were constructed by PCR mutagenesis using primers that substituted a NotI site (i.e., 5′-GCG · GCC · GC-3′) for the sequences underlying the residues boxed in the K-Rev protein sequence shown in Fig. 1. This results in the substitution of alanines for the indicated residues. After PCR amplification, the resultant K-Rev DNA sequences were cleaved at flanking 5′ BspHI and 3′ XhoI sites and inserted into pBC12/CMV cleaved with NcoI and XhoI. Plasmids encoding fusions of the VP16 activation domain to K-Rev mutants K1 through K14 were prepared as previously described (43) for the wild-type pBC12/CMV/K-Rev/VP16 plasmid.
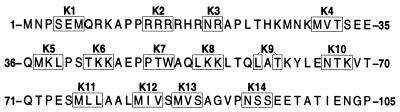
FIG. 1.
Predicted amino acid sequence of the K-Rev protein. The 14 introduced mutations resulted in the substitution of alanine for the indicated residues.
Yeast K-Rev expression plasmids were generated by PCR amplification of wild-type and mutant K-Rev genes (K1 through K14) using primers that introduced flanking EcoRI sites. After cleavage with EcoRI, the resultant DNA fragments were cloned in pPGK/VP16, in frame with the VP16 transcription activation domain and simian virus 40 T-antigen nuclear localization signal (NLS) present in pPGK/VP16 (5). The wild-type K-Rev EcoRI fragment was also cloned into pGBT9 (Clontech Inc.), to generate pGBT9/K-Rev, which encodes the GAL4 DNA binding domain fused to full-length K-Rev. The full-length K-RRE (HERV-K residues 8719 to 9152) was cloned into the RNA expression plasmid pMS2-2 (36) in both orientations to generate pMS2/K-RRE/s and pMS2/K-RRE/as for use in yeast three-hybrid assays.
Transfection of human cells.
For analysis of K-Rev function, 293T cells (35-mm-diameter plates) were transfected by the calcium phosphate method using 25 ng of the pDM128/K-RRE reporter plasmid, 25 ng of the pBC12/CMV/β-gal internal control, 50 ng of a pBC12/CMV/K-Rev effector plasmid (wild type or mutant) or pBC12/CMV as a negative control, and 1,000 ng of pBC12/CMV. To assay for dominant negative phenotypes of selected K-Rev mutants, 293T cells were transfected with 25 ng of pDM128/K-RRE, 25 ng of pBC12/CMV/β-gal, 50 ng of pBC12/CMV/K-Rev (wild type) or pBC12/CMV, and 1,000 ng of pBC12/CMV or pBC12/CMV/K-Rev encoding wild-type or mutant K-Rev. In a parallel experiment, the ability of K-Rev mutants to inhibit H-Rev function was assessed by cotransfection into 293T cells together with 25 ng of pDM128/CMV and 50 ng of pcRev. At ∼48 h after transfection, cells were harvested and lysed, and induced chloramphenicol acetyltransferase (CAT) and β-galactosidase (β-Gal) activities determined as previously described (3). In all experiments, the observed CAT enzyme activities were adjusted for minor differences in transfection efficiency or sample recovery, using the β-Gal enzyme activity as an internal control.
Two-hybrid assays in human 293T cells were performed essentially as previously described (3, 43). To assay for K-Rev multimerization, 293T cells were transfected with 250 ng of the pG6(−31)HIVLTRΔTARCAT reporter plasmid, 100 ng of the internal control plasmid pBC12/CMV/β-gal, 500 ng of pSG424/K-Rev, and 2,000 ng of pBC12/CMV/K-Rev/VP16 (wild type or mutant) or pBC12/CMV as a negative control. Similarly, to measure binding of K-Rev to Crm1 by mammalian two-hybrid assay, 293T cells were transfected with 50 ng of the pG6(−31)HIVLTRΔTARCAT reporter, 250 ng of the pBC12/CMV/β-gal internal control, 500 ng of pSG424/CRM1, and 2,000 ng of pBC12/CMV/K-Rev/VP16 (wild type or mutant) or pBC12/CMV. In each case, induced CAT and β-Gal activities were assayed at ∼48 h as previously described (3).
Immunofluorescence and Western blot assays.
The subcellular localization of K-Rev mutants in transfected 293T cells, in the presence or absence of the ΔCAN protein, was assayed as previously described (43). Briefly, 293T cells were transfected with 500 ng of pBC12/CMV/K-Rev (wild type or mutant) together with 1 μg of pBC12/CMV or pBC12/CMV/ΔCAN. At ∼50 h after transfection, cells were fixed and stained with a 1:500 dilution of a rabbit polyclonal anti-K-Rev antiserum followed by a 1:2,000 dilution of a fluorescein isothiocyanate-conjugated donkey anti-rabbit antiserum (The Jackson Laboratory). Images were collected using a Leica DMRB fluorescence microscope and converted to grayscale using Adobe Photoshop 4.0 software.
To determine the steady-state expression level of selected VP16/K-Rev fusion proteins, Western blot assays were performed essentially as previously described (2). Briefly, 293T cells (35-mm-diameter cultures) were transfected with 1 μg of the relevant VP16/K-Rev expression plasmid, and cell lysates were prepared at ∼48 h after transfection. After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose membranes and then probed with the anti-VP16 mouse monoclonal antibody SC-7545 (Santa Cruz Biotechnology) followed by a horseradish peroxidase-conjugated sheep anti-mouse antiserum (Amersham). Bound antibodies were visualized by enhanced chemiluminescence and autoradiography.
Yeast two- and three-hybrid assays.
To assay for multimerization of K-Rev, Saccharomyces cerevisiae strain Y190 cells (19) were transformed with pGBT9/K-Rev together with pPGK/VP16/K-Rev (wild type or mutant) or pPGK/VP16 as a negative control. After selection on Leu− Trp− plates for 3 days, double transformants were harvested and grown overnight in Leu− Trp− medium. Similarly, for analysis of RNA binding by K-Rev, yeast strain L40 coat cells (Invitrogen) were transformed with an RNA expression plasmid (pMS2/K-RRE/s or pMS2/K-RRE/as as a negative control) and pPGK/VP16/K-Rev (wild type or mutant). After selection on Ura− Leu− plates for 3 days, double transformants were harvested and grown overnight in Ura− Leu− medium. In each case, cell equivalents were then lysed and assayed for β-Gal activity as previously described (5).
RESULTS
A set of 14 missense mutants in the context of the previously described K-Rev cDNA expression plasmid pBC12/CMV/K-Rev was constructed. In each case, the introduced mutation modified eight nucleotides to the sequence 5′-GCG · GCC · GC-3′ (with the reading frame indicated). This sequence encodes triple alanine, so that these mutations each introduced alanines in place of the boxed K-Rev residues shown in Fig. 1. These mutations were largely random in design, although K2 and K3 were targeted to an arginine-rich sequence that bears some similarity to the arginine-rich RNA binding domain and NLS seen in both H-Rev and Rex (16, 28, 33, 40). In addition, K7, K8, and K9 all affect a motif (50-WAQLKKLTQL-59) that has been suggested to be similar to the leucine-rich NES present in H-Rev (27).
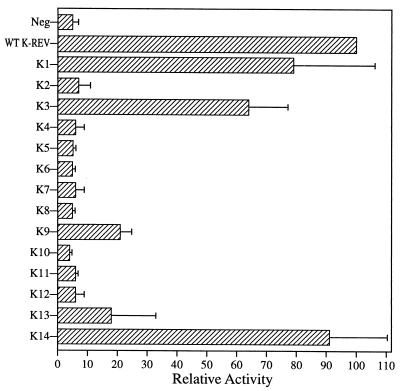
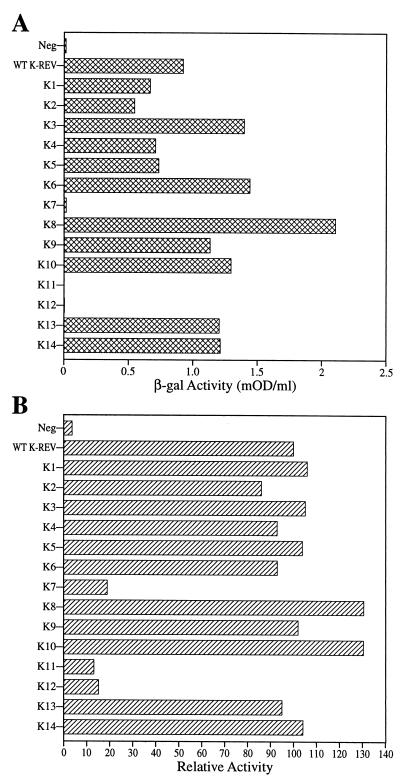
Figure 2 shows the ability of each K-Rev mutant to induce the nuclear export and expression of the unspliced cat mRNA encoded by the previously described indicator construct pDM128/K-RRE (43), which also contains the full-length K-RRE in cis. These data, which are normalized to values for the wild-type K-Rev expression plasmid, reveal that mutants K1 and K14 are fully active whereas K3 is almost wild type in activity. While K9 and possibly K13 displayed weak partial activity, all of the nine other K-Rev mutants were inactive.
FIG. 2.
Biological activities of K-Rev mutants. 293T cells were cotransfected with the pDM128/K-RRE indicator construct and expression plasmids encoding wild-type (WT) or mutant K-Rev. The parental pBC12/CMV expression plasmid served as a negative control (Neg). Induced CAT activities were assayed at ∼48 h after transfection as previously described (3). These data are normalized to the activity of wild-type K-Rev, which is arbitrarily set at 100. Results were also adjusted for nonspecific variability using the β-Gal internal control. Data shown represent the average of three independent experiments with standard deviation indicated.
Subcellular localization of K-Rev mutants.
Although K-Rev is normally largely cytoplasmic when expressed in human cells, inhibition of Crm1 function either by overexpression of ΔCAN, a dominant negative form of the nucleoporin Nup214/CAN that acts as a specific inhibitor of Crm1 function (3, 12, 24), or by addition of leptomycin B, a specific Crm1 inhibitor (11), results in the nuclear accumulation of K-Rev (27, 43). Therefore, we reasoned that K-Rev mutants that had lost the ability to bind to Crm1 should differ from wild-type K-Rev in being constitutively nuclear. Further, the localization of each defective mutant by immunofluorescence would also confirm that these proteins are indeed expressed.
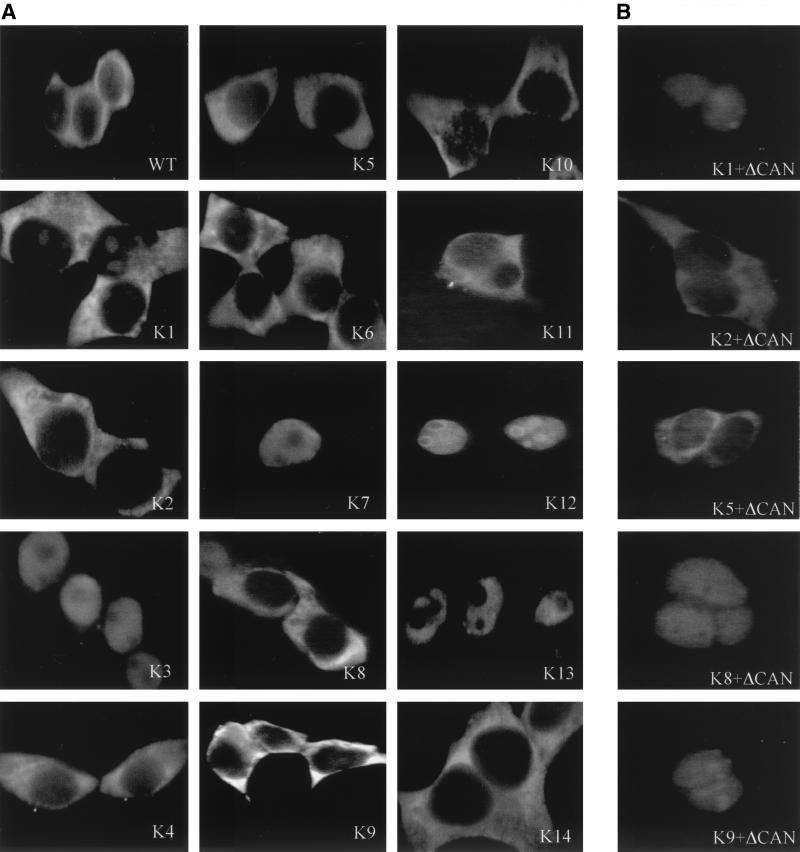
Figure 3A exemplifies the normal subcellular localizations of all 14 K-Rev mutants in human 293T cells. As may be readily observed, four of these mutants, K3, K7, K12, and K13, are nuclear, while the other 10 mutants share the cytoplasmic localization characteristic of wild-type K-Rev. The K12 mutant tended to concentrate in nucleoli, as previously also observed for wild-type K-Rev in cells lacking functional Crm1 (27, 43). In contrast, K3, K7, and, particularly, K13 were all excluded from the nucleoli (Fig. 3). Although K3 is nuclear at steady state, it is nevertheless active in inducing K-RRE-dependent RNA export (Fig. 2). Therefore, K3 must be able to exit the nucleus. As the K3 mutation is adjacent to the arginine motif of K-Rev, which, by analogy to H-Rev and Rex (33, 40), could function as an NLS, it appears possible that the K3 mutation may actually enhance the nuclear import of K-Rev rather than inhibit its export.
FIG. 3.
Subcellular localization of K-Rev. The subcellular localization of K-Rev in transfected 293T cells, in the absence (A) or presence (B) of the ΔCAN inhibitor of Crm1 function, was determined as previously described (43), using a rabbit polyclonal anti-K-Rev antiserum. WT, wild type.
While the nuclear export of K-Rev is dependent on Crm1 (27, 43), the mechanism mediating K-Rev nuclear import is not known. However, if a K-Rev mutant that is normally cytoplasmic (Fig. 3) can relocalize to the nucleus in the absence of Crm1 function, then this would imply that it retains a functional NLS. Conversely, the continued cytoplasmic localization of a K-Rev mutant, in the absence of Crm1 function, would imply that the introduced mutation had inhibited K-Rev nuclear import. We therefore examined the subcellular localization of normally cytoplasmic K-Rev mutants in the presence of the Crm1 inhibitor ΔCAN (Fig. 3B). As may be seen, mutants K2 and K5 (as well as mutants K4 and K11 [data not shown]) fail to accumulate in the nucleus in the presence of ΔCAN. Conversely, mutants K1, K8, and K9 (as well as K6, K10, and K14 [data not shown]) are similar to wild-type K-Rev (43) in that ΔCAN induces their relocalization from the cytoplasm to the nucleus. These data therefore suggest that mutations K2, K4, K5, and K11 all block the effective nuclear import of K-Rev. While this result was expected for K2, which has lost three arginine residues from the K-Rev basic motif (Fig. 1), it is unexpected in the case of the other three K-Rev mutants.
Crm1 binding by K-Rev mutants.
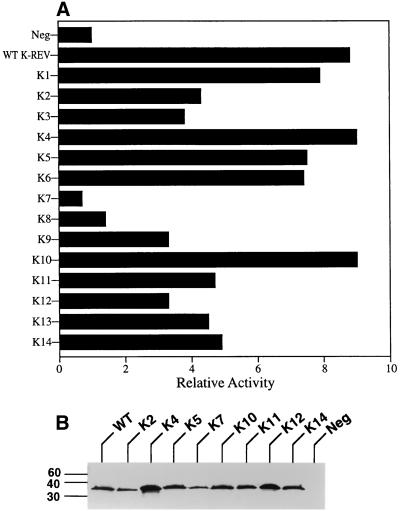
We have previously reported that K-Rev can interact specifically with the Crm1 nuclear export factor both in a mammalian two-hybrid assay and in vitro, in the latter case only in the presence of Ran · GTP (43). Figure 4A demonstrates the abilities of the K-Rev mutants to interact with Crm1 in vivo. In this assay, 293T cells were transfected with the indicator construct pG6(−31)HIVLTRΔTARCAT, which contains six GAL4 DNA binding sites flanking a minimal HIV-1 promoter element linked to the cat indicator gene (37). These same cells were also transfected with a plasmid expressing Crm1 fused to the GAL4 DNA binding domain and a third plasmid expressing the VP16 transcription activation domain fused to wild-type or mutant K-Rev. The parental pBC12/CMV plasmid served as a negative control. Binding of K-Rev to Crm1 would result in recruitment of the VP16 activation domain to the indicator construct and, hence, activation of CAT expression.
FIG. 4.
Crm1 binding by K-Rev mutants. (A) Abilities of K-Rev mutants to bind to the Crm1 nuclear export factor were determined by two-hybrid assay in transfected 293T cells, as described previously (3) and in the text. These data were adjusted using the β-Gal internal control and then expressed as a multiple of the level of CAT enzyme activity seen in 293T cells transfected with the indicator construct, the GAL4-Crm1 fusion protein expression plasmid, and the negative (Neg) control plasmid pBC12/CMV. These are representative data reflective of four independent transfection experiments. (B) Western analysis of K-Rev/VP16 fusion protein expression levels in transfected 293T cells. Blots were probed with a mouse monoclonal antibody directed against the VP16 activation domain. Sizes are indicated in kilodaltons.
These data demonstrate that mutants K1, K4, K5, K6, and K10 are essentially identical to wild-type K-Rev in the ability to interact with Crm1, while K2, K3, K9, K11, K12, K13, and K14 all appear to be ∼2-fold less active. Finally, mutant K7 does not detectably interact with Crm1, while K8 is at most slightly active. While the inability of mutant K7 to bind to Crm1 is clearly consistent with the nuclear localization of K7 observed by immunofluorescence (Fig. 3), K8 proved fully able to shuttle between nucleus and cytoplasm (Fig. 3) despite its apparently very weak interaction with Crm1. Conversely, K12 and K13, which also localize to the nucleus when Crm1 is functional (Fig. 3), nevertheless bind to Crm1 as effectively as mutants that clearly do retain the ability to shuttle (Fig. 4A).
To confirm that these VP16/K-Rev fusion proteins are indeed expressed at equivalent levels in vivo, we also performed a Western blot analysis on transfected 293T cells. As shown in Fig. 4B, all tested VP16 fusion proteins were readily detectable in cellular extracts, although K4 appears to be slightly overexpressed, and K7 slightly underexpressed, compared to their peers.
Multimerization of K-Rev in vivo.
The ability of H-Rev and HTLV-1 Rex to multimerize is required for the nuclear export of their target viral RNA molecules (6, 18, 28, 44). Previously, we have shown by two-hybrid assay in human 293T cells that K-Rev also has the ability to multimerize (43), although it has not been demonstrated formally that this multimerization extends beyond dimer formation. To address this issue for the K-Rev mutants listed in Fig. 1, we used two-hybrid analysis in yeast cells (9) (Fig. 5A) or in human cells (Fig. 5B) to measure, in each case, the ability of a fusion protein consisting of the GAL4 DNA binding domain linked to wild-type K-Rev to bind to wild-type or mutant K-Rev fused to the VP16 activation domain and to an active NLS. Gratifyingly, these two assays yielded very similar data (Fig. 5). Specifically, K7, K11, and K12 were all profoundly defective for multimerization, while all other K-Rev mutants were comparable in multimerization activity to wild-type K-Rev.
FIG. 5.
Multimerization of the K-Rev protein. (A) Multimerization of K-Rev was detected in yeast cells using the two-hybrid assay (9). Cells were transformed with expression plasmids encoding (i) the GAL4 DNA binding domain fused to wild-type (WT) K-Rev and (ii) wild-type or mutant K-Rev fused to the VP16 transcription activation domain. Induced β-Gal activities were measured as previously described (5) and are given in milli-optical density units (mOD) per milliliter of yeast culture. Data are representative of three independent experiments. (B) Similar to panel A except that the two-hybrid assay was performed in transfected human 293T cells, as described in the text. Induced CAT enzyme activities were measured ∼48 h after transfection, adjusted using the β-Gal internal control plasmid, and then normalized to the activity seen with wild-type K-Rev, which was arbitrarily set at 100. Data are representative of three independent experiments.
K-RRE binding by K-Rev.
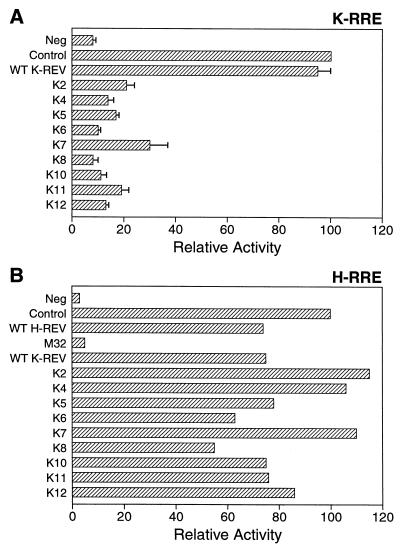
Previously, we demonstrated that K-Rev can bind specifically to the K-RRE in vitro but not to irrelevant RNA targets, such as the H-RRE (43). Elsewhere, we have recently also documented that K-Rev can specifically bind to the K-RRE, but not to the antisense K-RRE or to irrelevant RNA targets, in the yeast three-hybrid assay (36, 42a). We used this three-hybrid assay to examine the ability of K-Rev mutants to bind the K-RRE (Fig. 6). In this in vivo assay, the sense or antisense K-RRE was expressed in yeast L40 coat cells as a fusion to the MS2 bacteriophage operator RNA, while wild-type or mutant K-Rev was expressed fused to the VP16 activation domain, as in Fig. 5A. As may be observed in Fig. 6, wild-type K-Rev bound the sense orientation K-RRE effectively but interacted only very poorly with the antisense K-RRE. Only one mutant, K2, was entirely negative for K-RRE binding, although K3, K4, K7, K11, and K12 were all clearly less active than wild-type K-Rev. K7, K11, and K12 are defective for multimerization (Fig. 5); it is therefore possible that their lower K-RRE binding activity reflects an inability to multimerize effectively on the K-RRE rather than a reduced affinity for the putative primary K-Rev RNA binding site on the K-RRE.
FIG. 6.
Binding of K-Rev to the K-RRE. Abilities of K-Rev mutants to bind to the K-RRE in vivo were determined by three-hybrid assay in yeast (36). L40 coat cells were transformed with an expression plasmid encoding the full-length sense or antisense K-RRE fused to the MS2 operator RNA and with a second plasmid expressing a fusion protein consisting of the VP16 activation domain linked to wild-type (WT) or mutant K-Rev, as also described for Fig. 5A. The negative control (Neg) consists of yeast cells expressing the MS2/K-RRE/as (antisense) RNA and the wild-type VP16/K-Rev fusion protein. Induced β-Gal activities were determined as previously described (5).
Inactive K-Rev mutants exert a dominant negative phenotype.
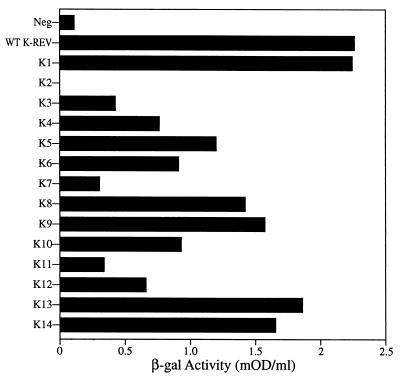
The mutational analysis presented in this report identified nine K-Rev mutants that have lost the ability to mediate the nuclear export of unspliced mRNAs bearing the K-RRE. Previously, it has been demonstrated that certain inactive mutants of H-Rev or HTLV-1 Rex can exert a potent dominant negative phenotype when coexpressed with the wild-type form of the same protein, particularly if expressed in excess (7, 30, 35). To test whether this phenotype would also be detected for any of the nine inactive K-Rev mutants identified in this study, we transfected 293T cells with the pDM128/K-RRE indicator construct and 50 ng of the wild-type K-Rev expression plasmid pBC12/CMV/K-Rev together with 1,000 ng of pBC12/CMV/K-Rev expressing either wild-type or defective mutant K-Rev. As shown in Fig. 7A, each of the nine inactive K-Rev mutants, but not wild-type K-Rev, proved able to potently inhibit the expression of the unspliced cat mRNA containing the K-RRE when expressed at ∼20-fold excess over the wild-type protein. This inhibition was specific, as none of these K-Rev mutants had any significant effect on the level of expression of the β-Gal enzyme encoded by the cotransfected pBC12/CMV/β-gal internal control plasmid (data not shown).
FIG. 7.
Dominant negative phenotypes of K-Rev mutants. (A) 293T cells were transfected with 25 ng of the indicator construct pDM128/K-RRE, 50 ng of pBC12/CMV (Neg), or 50 ng of the wild-type (WT) K-Rev expression plasmid pBC12/CMV/K-Rev. In addition, the cells were cotransfected with 1 μg of the parental pBC12/CMV plasmid (Neg and Control) or with 1 μg of a pBC12/CMV-based plasmid encoding wild-type or mutant K-Rev. Induced CAT activities were determined at ∼48 h posttransfection, adjusted using the β-Gal internal standard, and then normalized to the control activity, which was arbitrarily set at 100. These data represent the average of three independent experiments with the standard deviation indicated. (B) Similar to panel A except that 293T cells were transfected with 25 ng of the H-RRE-based indicator plasmid pDM128/CMV and with 50 ng of the H-Rev expression plasmid pcRev. M32 is a previously described (30) dominant negative mutant of H-Rev.
To further confirm the specificity of this inhibition, we transfected 293T cells as described for Fig. 7A except that pDM128/K-RRE was replaced by the similar indicator plasmid pDM128/CMV, which contains the H-RRE in place of the K-RRE (30), while pBC12/CMV/K-Rev was substituted with an expression plasmid, pcRev, encoding wild-type H-Rev. As shown in Fig. 7B, coexpression of H-Rev activated expression of the cat gene encoded by the pDM128/CMV indicator plasmid; this has previously been shown to result from the H-Rev-induced nuclear export of the encoded unspliced cat mRNA (23, 30). As a control, we cotransfected a previously described (30) dominant negative mutant of H-Rev, termed M32, that lacks a functional NES. As expected, this resulted in >20-fold inhibition of CAT expression (Fig. 7B). In contrast, the K-Rev mutants that potently inhibited wild-type K-Rev function (Fig. 7A) had no significant inhibitory effect on H-Rev (Fig. 7B); the inhibition of the K-Rev-induced cat mRNA expression reported in Fig. 7A therefore clearly reflects a specific, dominant negative phenotype.
DISCUSSION
The critical importance of H-Rev in the HIV-1 replication cycle and the fact that H-Rev was the first sequence-specific nuclear mRNA export factor to be identified have led to considerable interest in the mechanism of action of H-Rev (8, 34). As a result, H-Rev has been subjected to intense mutational analysis, which has led to the fairly precise definition of the sequences within H-Rev required for RNA binding, nuclear localization, homomultimerization, and Crm1 binding. Mutation of the leucine-rich Crm1 binding motif on H-Rev, which also blocks Rev nuclear export, gives rise to a potent dominant negative phenotype (10, 11, 30, 31). This inhibition most probably reflects the formation of inactive multimers, containing both mutant and wild-type H-Rev, on the K-RRE. On the other hand, H-Rev mutants that are unable to bind the K-RRE or to multimerize are essentially recessive negative (28, 30). Importantly, mutations that block H-Rev function can be shown to inactivate one or more of the biological properties of H-Rev enumerated above (28, 30, 34).
Although the HTLV-1 Rev protein has not yet been subjected to the level of experimental analysis applied to H-Rev, Rex has been shown to share a similar set of functional domains, albeit not in the same relative position within the linear protein sequence (6, 18, 22, 33, 41). Again, mutations that block Rex function have been shown to affect RNA binding, Crm1 binding, multimerization, or NLS function. Of interest, while certain Rex mutants are also dominant negative, these appear to be defective for multimerization rather than for Crm1 recruitment, and they have therefore been proposed to act by a squelching mechanism (20).
Previously, we had demonstrated that K-Rev, like H-Rev and Rex, is able to bind to Crm1 and to its cognate RNA target, the K-RRE, as well as to multimerize and to shuttle between nucleus and cytoplasm (43). We therefore sought to use mutational analysis to identify the functional domains important for these biological activities and to assess their role in K-Rev-mediated nuclear RNA export. It was anticipated that these experiments would permit a comparison of the functional architecture of K-Rev with that of H-Rev and Rex and hence perhaps shed new light on the evolution of these unusual retroviral regulatory proteins.
This report described 14 missense mutants of the 105-aa K-Rev protein and reports an extensive analysis of the biological properties of these mutants, 9 of which have lost the ability to induce K-RRE-dependent RNA export activity (summarized in Table 1). Importantly, all of these K-Rev mutants were expressed at readily detectable levels, as determined by immunofluorescence (Fig. 3) or Western blot analysis (Fig. 4B). Further, all K-Rev mutants retained one or more specific biological functions (Table 1), and therefore are not acting by simply inducing a global misfolding of the K-Rev protein.
TABLE 1.
Biological properties of K-Rev mutantsa
| Virus | Function | Wild-type subcellular localizationb | Crm1 binding | Multimerization | RNA binding | Dominant negative |
|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | + | NA |
| K1 | + | + | + | + | + | NA |
| K2 | − | − | + | + | − | + |
| K3 | + | − | + | + | +/− | NA |
| K4 | − | − | + | + | + | + |
| K5 | − | − | + | + | + | + |
| K6 | − | + | + | + | + | + |
| K7 | − | − | − | − | +/− | + |
| K8 | − | + | +/− | + | + | + |
| K9 | +/− | + | + | + | + | NA |
| K10 | − | + | + | + | + | + |
| K11 | − | − | + | − | +/− | + |
| K12 | − | − | + | − | +/− | + |
| K13 | +/− | − | + | + | + | NA |
| K14 | + | + | + | + | + | NA |
RNA binding and nuclear import.
The RNA binding and NLS sequences in H-Rev and Rex both coincide with an arginine-rich motif (16, 28, 33, 40); we therefore anticipated that the similar arginine-rich sequence present in K-Rev (Fig. 1) might fulfill a similar function. Of the 14 mutations introduced into K-Rev, 2 (K2 and K3) were targeted to this basic motif. The K2 mutant was found to be inactive, while K3 retained almost full activity (Fig. 2), thus implying that the latter mutation was not particularly deleterious.
Analysis of K-RRE binding by the K-Rev mutants showed that K2 lacked any binding ability whereas the ability of K3 was significantly attenuated (Fig. 6). Therefore, it appears that the K-Rev arginine motif is indeed critical for binding to the K-RRE. All other K-Rev mutants except K7, K11, and K12 bound the K-RRE effectively. However, these three exceptions are all defective for multimerization (Fig. 5), and we believe their lower K-RRE binding activity likely reflects their inability to effectively multimerize on the K-RRE.
While the wild-type K-Rev protein is normally cytoplasmic, specific inactivation of Crm1 function results in localization of K-Rev to the nucleus (27, 43). This result demonstrates that K-Rev is a nucleocytoplasmic shuttle protein whose cytoplasmic localization requires ongoing nuclear export mediated by Crm1 and implies the existence of an NLS whose function is revealed only when K-Rev nuclear export is inhibited. However, a fusion protein consisting of the amino-terminal 29 aa of K-Rev, including the entire basic domain, linked to the heterologous β-Gal protein only weakly localized to the nucleus (H. Wiegand, unpublished observation), thus implying that the K-Rev NLS is not active in this fusion protein context or that this NLS extends significantly beyond the K-Rev arginine motif (Fig. 1). We therefore instead attempted to define the putative K-Rev NLS by subcellular localization of mutants of full-length K-Rev using immunofluorescence analysis (Fig. 3).
The phenotype expected for a K-Rev protein lacking a functional NLS is continued cytoplasmic localization even in the absence of Crm1 function. This is in fact exactly what is seen with K2 (Fig. 3), thus supporting the hypothesis that the K-Rev arginine motif is important for K-Rev nuclear import. Surprisingly, however, mutants K4, K5, and K11 were also all cytoplasmic when Crm1 was inhibited. Therefore, these mutants appear to have lost the ability to import into the nucleus. This finding suggests the surprising possibility that nuclear import of K-Rev is mediated by the combined action of several discontinuous K-Rev sequences. If true, this result would clearly explain why fusion of only the amino-terminal 29 aa of K-Rev to β-Gal did not result in effective nuclear import (see above).
Analysis of the subcellular localization of K-Rev mutants (Fig. 3) revealed that K3 was nuclear at steady state. Because K3 is, however, biologically active (Fig. 2), it seems improbable that this reflects an inability to export from the nucleus. We therefore hypothesize that the K3 mutation may actually enhance the function of the otherwise relatively ineffective NLS present in K-Rev, so that import becomes more efficient than Crm1-dependent export, i.e., the situation normally seen with H-Rev and Rex (33, 40). However, this hypothesis can be substantiated only by a more complete analysis of K-Rev nucleocytoplasmic transport.
Crm1 binding and nuclear export.
Analysis of Crm1 binding to K-Rev in 293T cells (Fig. 4A) showed that K7 was defective for Crm1 binding whereas the activity of K8 was highly attenuated. K9 and K12 were also somewhat attenuated for Crm1 binding, while all other mutants showed ≥50% of the wild-type K-Rev activity. The K7 mutant was found to be nuclear at steady state (Fig. 3), consistent with a block in Crm1-dependent nuclear export. Surprisingly, K8 retained cytoplasmic localization in the presence of functional Crm1 and became nuclear in the absence of Crm1 function (Fig. 3), thus implying that the weak interaction observed between K8 and Crm1 (Fig. 4A) was sufficient to maintain nucleocytoplasmic shuttling. Not only K7 but also K3, K12, and K13 were nuclear at steady state. Of interest, K13 was specifically excluded from nucleoli whereas K12 tended to concentrate in the nucleoli (Fig. 3), as also seen for wild-type K-Rev in the absence of Crm1 function (27, 43). The significance of this difference is not, however, clear. We have argued above that the active K3 mutant is likely nuclear due to enhanced NLS function. Both K12 and K13 retain substantial Crm1 binding activity, and it is therefore possible that they also exhibit enhanced nuclear import. Alternately, they may be retained in the nucleus for unknown reasons distinct from any inability to recruit Crm1.
H-Rev and Rex, as well as most other proteins that depend on Crm1 to mediate their nuclear export, have been shown to contain a short, leucine-rich sequence that serves as a specific Crm1 binding site and hence as an NES (3, 4, 10, 11, 25, 31, 38, 42). The function of this NES is dependent on the appropriate spacing of four hydrophobic, most commonly leucine, residues interspersed by smaller, hydrophilic residues. A consensus sequence for leucine-rich NESs that envisages a spacing of two or three residues between leucines 1 and 2 and leucines 2 and 3 and a single residue between leucine residues 3 and 4 has been proposed (4, 25). This latter spacing appears invariant in functional leucine-rich NESs (25). While the K-Rev sequence 50-WAQLKKLTQL-59 bears some similarity to known leucine-rich NESs (8, 27, 34), it clearly does not have a single-residue spacer between the two most carboxy-terminal leucines and therefore does not conform to this consensus. Nevertheless, mutation K7, which changes tryptophan 50 to alanine, blocks Crm1 binding, while mutations K8, which changes leucine 53 to alanine, and K9, which changes leucine 59 to alanine, reduce Crm1 binding (Fig. 4A). However, the latter two mutants differ from K7 in that they retain the ability to shuttle between nucleus and cytoplasm (Fig. 3). Although this K-Rev motif could represent an unusual leucine-rich NES, substitution of this sequence for the authentic leucine-rich NES present in H-Rev did not rescue H-Rev function (Bogerd, unpublished), even though leucine-rich NESs from several proteins have previously been shown to support H-Rev-induced nuclear export when switched for the H-Rev NES (13–15, 22, 41). To test whether repair of the spacing of the leucine residues in the putative K-Rev NES would enhance activity, we exchanged the last two residues to give 50-WAQLKKLTLQ-59, a sequence that closely matches the NES consensus (4, 25). However, this did not result in enhanced K-Rev function (Bogerd, unpublished).
K-Rev multimerization.
The ability of K-Rev to multimerize or, more accurately, to at least dimerize was assayed in both human and yeast cells (Fig. 5). In both systems, K7, K11, and K12 were defective for multimerization whereas all other mutants were essentially wild type. It has previously been demonstrated that Crm1 substantially enhances both H-Rev and Rex multimerization in vivo (6, 18, 26), and these latter two proteins, at least, are known to bind to Crm1 in not only human but also yeast cells (31). It therefore seems possible that the inability of the K7 mutant to bind to Crm1 (Fig. 4A) may underly the apparent defect in multimerization. If this is the case, then the true multimerization domain in K-Rev would actually coincide with the sequence defined by mutants K11 and K12, both of which lack K-RRE nuclear export activity.
Biological activities of K-Rev mutants.
The data discussed above suggest that RNA binding by K-Rev involves an obvious arginine-rich motif (residues 8 to 20) that bears a clear similarity to the arginine-rich RNA binding domains present in H-Rev and Rex (8, 16, 28, 34). This motif also appears important for nuclear import of K-Rev. Binding to Crm1 requires the integrity of a K-Rev sequence (residues 50 to 59) that features several large hydrophobic residues, although this motif appears distinct from the NESs previously defined in H-Rev and Rex (4, 8, 25, 34). Finally, K-Rev multimerization appears to require residues localized around 76 to 84.
Unfortunately, these data do not allow us to fully explain the RNA export activities observed for some K-Rev mutants. Particularly puzzling is why certain K-Rev mutants are unable to support K-RRE-dependent nuclear export when they appear wild type for all properties examined. Specifically, mutants K6, K8, and K10 appear fully active for RNA binding, Crm1 binding, multimerization, and nucleocytoplasmic shuttling yet are inactive for RNA export (Fig. 2; Table 1). No equivalent mutants have so far been described for H-Rev or Rex. A further mystery is that all nine inactive K-Rev mutants turn out to be potent dominant negative inhibitors of K-Rev (Fig. 7). This is clearly unlike the situation reported for H-Rev and Rex, where dominant negative mutants selectively block Crm1 binding and multimerization, respectively (6, 11, 18, 28, 30, 31, 38). In contrast, these nine K-Rev mutants are defective in several different ways (Table 1). However, it is unlikely that these K-Rev mutants act by sequestering a limiting supply of Crm1, both because K7, which does not bind to Crm1, is a dominant negative mutant (Fig. 4A and 7A) and because none of these K-Rev mutants proved able to inhibit H-Rev function (Fig. 7B).
In conclusion, we have defined sequences in K-Rev that function in Crm1 and K-RRE binding, multimerization, and K-Rev nucleocytoplasmic shuttling. While each of these activities appears critical for K-Rev function, their loss does not fully suffice to explain the phenotypes observed for certain K-Rev mutants in mediating K-RRE-dependent nuclear RNA export. Our data may therefore imply the existence of an additional, unknown interaction that is required to mediate K-Rev function but that may play no role in the nuclear mRNA export induced by the perhaps more highly evolved H-Rev and HTLV-1 Rex proteins.
ACKNOWLEDGMENT
The first two authors contributed equally to this report.
REFERENCES
- 1.Barbulescu M, Turner G, Seaman M I, Deinard A S, Kidd K K, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd H P, Fridell R A, Blair W S, Cullen B R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd H, Greene W C. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böhnlein S, Pirker F P, Hofer L, Zimmermann K, Bachmayer H, Böhnlein E, Hauber J. Transdominant repressors for human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev function. J Virol. 1991;65:81–88. doi: 10.1128/jvi.65.1.81-88.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 11.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod M, van Deursen J, Van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. A nuclear role for the fragile X mental retardation. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 14.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridell R A, Fischer U, Lührmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grassmann R, Berchtold S, Aepinus C, Ballaun C, Boehnlein E, Fleckenstein B. In vitro binding of human T-cell leukemia virus Rex proteins to the Rex-response element of viral transcripts. J Virol. 1991;65:3721–3727. doi: 10.1128/jvi.65.7.3721-3727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths D J, Venables P J W, Weiss R A, Boyd M T. A novel exogenous retrovirus sequence identified in humans. J Virol. 1997;71:2866–2872. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakata Y, Umemoto T, Matsushita S, Shida H. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type I. J Virol. 1998;72:6602–6607. doi: 10.1128/jvi.72.8.6602-6607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;19:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 20.Heger P, Rosorius O, Hauber J, Stauber R H. Titration of cellular export factors, but not heteromultimerization, is the molecular mechanism of trans-dominant HTLV-I Rex mutants. Oncogene. 1999;18:4080–4090. doi: 10.1038/sj.onc.1202762. [DOI] [PubMed] [Google Scholar]
- 21.Hidaka M, Inoue J, Yoshida M, Seiki M. Post-transcriptional regulator (Rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 27.Magin C, Löwer R, Löwer J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 1999;73:9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 29.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 30.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 32.Ohno M, Segref A, Bachi A, Wilm M, Mattaj I W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 33.Palmeri D, Malim M H. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 35.Rimsky L, Duc Dodon M, Dixon E P, Greene W C. Trans-dominant inactivation of HTLV-I and HIV-1 gene expression by mutation of the HTLV-I Rex transactivator. Nature. 1989;341:453–456. doi: 10.1038/341453a0. [DOI] [PubMed] [Google Scholar]
- 36.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southgate C D, Green M R. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 38.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 39.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weichselbraun I, Farrington G K, Rusche J R, Böhnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 42a.Yang, J., H. P. Bogerd, S.-Y. Le, and B. R. Cullen. The human endogenous retrovirus K Rev response element coincides with a predicted RNA folding region. RNA, in press. [DOI] [PMC free article] [PubMed]
- 43.Yang J, Bogerd H P, Peng S, Wiegand H, Truant R, Cullen B R. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci USA. 1999;96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zapp M L, Hope T J, Parslow T G, Green M R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]