Abstract
Background
The clinical role of claudin 8 (CLDN8) in kidney renal clear cell carcinoma (KIRC) remains unclarified. Herein, the expression level and potential molecular mechanisms of CLDN8 underlying KIRC were determined.
Methods
High-throughput datasets of KIRC were collected from GEO, ArrayExpress, SRA, and TCGA databases to determine the mRNA expression level of the CLDN8. In-house tissue microarrays and immunochemistry were performed to examine CLDN8 protein expression. A summary receiver operating characteristic curve (SROC) and standardized mean difference (SMD) forest plot were generated using Stata v16.0. Single-cell analysis was conducted to further prove the expression level of CLDN8. A clustered regularly interspaced short palindromic repeats knockout screen analysis was executed to assess the growth impact of CLDN8. Functional enrichment analysis was conducted using the Metascape database. Additionally, single-sample gene set enrichment analysis was implied to explore immune cell infiltration in KIRC.
Results
A total of 17 mRNA datasets comprising 1,060 KIRC samples and 452 non-cancerous control samples were included in this study. Additionally, 105 KIRC and 16 non-KIRC tissues were analyzed using in-house immunohistochemistry. The combined SMD was -5.25 (95% confidence interval (CI): -6.13 to -4.37), and CLDN8 downregulation yielded an SROC area under the curve (AUC) close to 1.00 (95% CI: 0.99 - 1.00). CLDN8 downregulation was also confirmed at the single-cell level. Knocking out CLDN8 stimulated KIRC cell proliferation. Lower CLDN8 expression was correlated with worse overall survival of KIRC patients (hazard ratio of CLDN8 downregulation = 1.69, 95% CI: 1.2 - 2.4). Functional pathways associated with CLDN8 co-expressed genes were centered on carbon metabolism obstruction, with key hub genes ACADM, ACO2, NDUFS1, PDHB, SDHD, SUCLA2, SUCLG1, and SUCLG2.
Conclusions
CLDN8 is downregulated in KIRC and is considered a potential tumor suppressor. CLDN8 deficiency may promote the initiation and progression of KIRC, potentially in conjunction with metabolic dysfunction.
Keywords: Kidney renal clear cell carcinoma, CLDN8, Tissue microarray, Prognosis, Mechanism
Introduction
According to the 2020 global cancer statistics from the World Health Organization, renal cell carcinoma (RCC) accounts for 2.2% of all cancers, ranking as the 16th most common cancer. It is the third most prevalent malignant tumor of the urogenital system, following prostate (7.3%) and bladder cancers (3.0%) [1, 2]. RCC is classified into several subtypes, including kidney renal clear cell carcinoma (KIRC), kidney chromophobe, and kidney renal papillary cell carcinoma, with KIRC being the most common, comprising about 70% of cases [3, 4]. Nephrectomy, including radical and partial nephrectomy, remains the standard treatment for patients with localized RCC [5]. Although local nephrectomy has significantly reduced side effects, 20% of patients still experience complications, and 0.4% do not survive beyond 60 days post-surgery [5]. For advanced KIRC patients, targeted therapies and immunotherapies, such as nivolumab and bevacizumab, are the first-line treatment options. Despite these advancements, the prognosis for KIRC patients remains unsatisfactory. Therefore, there is a pressing need to explore novel therapeutic targets and develop new targeted drugs to alleviate the suffering of advanced KIRC patients and reduce the adverse effects associated with traditional treatments.
Claudins, also known as tight junction (TJ) proteins, form the basic framework of TJ, which are crucial for intercellular signaling. TJs coordinate gene expression through endocellular scaffolding proteins that support transcription factor regulation [6, 7]. Alterations in claudin levels can disrupt TJs and compromise barrier integrity [8]. Loss of TJ function in tumors leads to the loss of cellular polarity and impairs epithelial integrity during tumorigenesis [9]. Consequently, the expression of claudin proteins is considered to contribute to cancer progression and is associated with the loss of cell adhesion. CLDN8 is located on chromosome 21 and encodes the CLDN8 protein [10]. CLDN8 is primarily expressed at TJ structures located in distal aldosterone-sensitive nephrons and the posterior thin descending limb segments of long-looped nephrons in the mammalian kidney [11]. CLDN8 knockout significantly reduces Cl- permeability, highlighting its importance in the function of collecting ducts. In renal collecting ducts, the localization of CLDN4 to TJs is dependent on its interaction with CLDN8. In the absence of CLDN8, CLDN8 is the unique binding partner for CLDN4 [12]. Additionally, CLDN8 expression is induced by diabetes, leading to abnormal contraction of TJ proteins, which is associated with diabetic kidney injury [13].
In addition to its role in organ injury, CLDN8 expression varies in cancer. CLDN8 is downregulated in breast cancer tissues [14] and has been shown to support prostate cancer cell hyperplasia and metastasis [15]. However, few studies have clarified the role of CLDN8 in KIRC. In this study, we aimed to confirm the expression level of CLDN8 in KIRC, observe the effect of CLDN8 on the growth of KIRC cell line, and uncover potential molecular mechanism using public RNA-seq datasets and in-house tissue microarrays.
Materials and Methods
Datasets gathered from public databases
Microarray profiles utilized in this research were sourced from the gene expression omnibus (GEO) [16], ArrayExpress [17], sequence read archive, and the cancer genome atlas (TCGA) databases [18]. RNA-seq datasets for KIRC were retrieved from the GEO database using the search string (“kidney clear cell carcinoma” OR KIRC OR “clear cell renal cell carcinoma” OR ccRCC). The datasets were filtered according to the following criteria: 1) studies involving Homo sapiens; 2) sequencing of coding RNA; 3) datasets including both KIRC tissues (with no fewer than three cases) and non-KIRC tissues (with no fewer than three cases). Datasets were excluded if they met the following conditions: 1) involvement of genetic modification or drug treatment; 2) presence of substantial missing values. Additionally, transcripts per million (TPM) data for KIRC from the TCGA database were obtained using the TCGAbiolinks package (accessed July 12, 2021). Subsequently, all non-standardized datasets were normalized by log2(x + 1). To avoid batch effects, independent datasets from the same platform were combined to generate a unified platform dataset.
Immunohistochemical experiments
In this study, immunohistochemical (IHC) staining was performed to determine the CLDN8 protein levels in KIRC. The in-house tissue microarrays included 105 KIRC samples from different patients and 16 non-KIRC tissues, which were purchased from Fanpu Biotech, Inc. (Guilin, China). The anti-CLDN8 antibody (ab211439) was procured from Abcam, Inc. The IHC staining utilized the horseradish peroxidase system. The IHC experiment results were independently evaluated by two pathologists using the immunoreactive score (IRS) system (0 - 12 points) [19-21]. Staining intensity was graded on a scale of 0 - 3, indicating negative, weak, medium, and strong expression, respectively. The proportion of CLDN8-positive staining was categorized as follows: 0 (less than 10%), 1 (10-25%), 2 (26-50%), 3 (51-75%), and 4 (76-100%). The final score for each field was calculated by multiplying the positively stained CLDN8 ratio by the intensity score. Each independent sample was assessed in five different visual fields, with the arithmetic average representing the final score for each sample. The scores for all 121 samples were compiled into a dataset for subsequent analysis. Additionally, CLDN8 protein expression in KIRC tissues was also evaluated using the human protein atlas (HPA) database [22].
Expression level of CLDN8 in single cells
The expression level of CLDN8 in KIRC was investigated from a single-cell perspective using dataset GSE152938, retrieved from the GEO database, platform GPL20795 (HiSeq X Ten (Homo sapiens)) [23]. Quality control parameters included 200 < nFeature_RNA < 2,500 and mitochondrial proportion < 5%. Optimal clustering was achieved with resolution set to 0.8 and dims to 18, and the results were visualized using UMAP.
The role of CLDN8 in the propagation of KIRC cell lines
To elucidate the impact of CLDN8 on the growth of KIRC cells, we employed clustered regularly interspaced short palindromic repeats (CRISPR) knockout screen technology. The CERES algorithm was applied to compute dependency scores for CLDN8 across various KIRC cell lines. A negative score in a KIRC cell line indicated that CLDN8 inhibited the proliferation of that specific cell line, while a positive score suggested that CLDN8 promoted the growth of that cell line [24].
Statistical analysis and clinical value of CLDN8 in KIRC
The receiver operating characteristic (ROC) curve was drawn to calculate the true positive, true negative, false positive, and false negative rates using the pROC package. A summary receiver operating characteristic (SROC) curve was generated with Stata (version 16.0, StataCorp 2019, Stata Statistical Software: Release 16) based on both mRNA datasets and in-house IHC data to evaluate the diagnostic capacity of CLDN8 for KIRC. Additionally, standardized mean difference (SMD), sensitivity, specificity, and likelihood ratios were obtained. To detect publication bias, Egger’s test and Begg’s test were employed. CLDN8 expression was analyzed using a t-test, one-way analysis of variance (ANOVA) analysis, or a nonparametric test on unpaired independent samples with IBM SPSS Statistics 26 and R software (version 4.2.1), as propriate. Survival analysis for KIRC patient was conducted using the Kaplan-Meier plotter [25].
Pathway enrichment analysis
Differential expression genes (DEGs) of KIRC were identified by calculating SMDs (|SMD| > 0, P < 0.05). Co-expressed genes (CEGs) of CLDN8 were determined via Pearson correlation analysis (correlation coefficient > 0.30, P < 0.05 for positively CLDN8-correlated genes; correlation coefficient < -0.30, P < 0.05 for negatively CLDN8-correlated genes). Metascape was utilized to identify oncology terms for the input gene list using the hypergeometric test and Benjamini-Hochberg P-value correlation algorithm, noted for its frequent updates and robust pathway enrichment analysis including gene ontology (GO), the Kyoto encyclopedia of genes and genomes (KEGG), Reactome, MSigDB, and more. Downregulated CLDN8-related genes, indicating overlap between downregulated DEGs and positively correlated CEGs, were analyzed via Metascape for GO and KEGG enrichment. The results were visualized using the ggplot2 package on R (version 4.1.1). The STRING database [26] was employed to construct the protein-protein interaction (PPI) network, and closely connected hub genes were identified using the CytoHubba plugin in Cytoscape (version 3.8.0). The top 10 scoring genes were further analyzed using the GEPIA database [27] for survival analysis. Gene-gene interrelations for hub genes significantly associated with KIRC patient survival were explored using the GeneMANIA database [28].
Exploration of the immune microenvironment of KIRC
Gene expression values of the KIRC samples were extracted from TCGA-KIRC TPM data. Single-sample gene set enrichment analysis (ssGSEA), a popular algorithm for evaluating the patient’s immune microenvironment, was conducted using the GSVA package to predict immune cell concentrations in KIRC. The immune cells gene set collection was obtained from the TISIDB database, predicting a total of 28 types of immune cells. A heat map was generated to illustrate the infiltration of immune cells in each KIRC sample. Additionally, the correlation between CLDN8 expression and immune cell infiltration was analyzed using the TISIDB database [29]. Further analysis was performed on immune cells that showed statistically significant correlations (P < 0.05).
Ethics approval
The research was approved by ethics committees of Fanpu Biotech, Inc. (No. FANPU (2018) 23). The authors confirm that all methods in this study were carried out in accordance with relevant guidelines and regulations.
Results
mRNA expression datasets
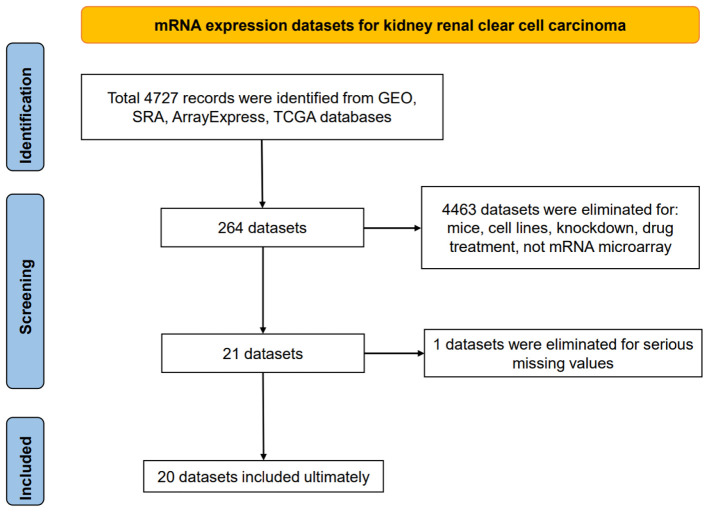
Twenty independent mRNA expression datasets with a total of 1,096 KIRC and 487 non-KIRC control tissue specimens were studied (Fig. 1, Supplementary Material 1, www.wjon.org). Eventually, 14 platform datasets were generated to calculate DEGs between KIRC tissue samples and non-cancerous kidney tissue samples. Since GPL97 and GSE16441 lacked CLDN8 expression data, the remaining 12 platform datasets, containing 1,060 KIRC and 452 non-KIRC from 17 independent mRNA expression matrices, were used to analyze CLDN8 differential expression and calculate CLDN8-related genes.
Figure 1.
Flow chart of mRNA expression datasets for kidney renal clear cell carcinoma tissues.
Downregulated CLDN8 in KIRC and the clinical value
CLDN8 was significantly downregulated in KIRC tissues at the mRNA level (Supplementary Material 2, www.wjon.org), demonstrating strong discriminative potential between KIRC and non-KIRC tissues, with an area under the curve (AUC) greater than 0.90 for all comparisons (Supplementary Material 3, www.wjon.org). CLDN8 protein localized in the cytoplasm and membrane of renal tubules. IHC images from in-house tissue microarrays and the HPA database showed significantly weaker staining intensity of CLDN8 protein in 105 KIRC tissues compared to 16 non-tumor tissues (unpaired Wilcoxon test, P < 0.001) (Figs. 2, 3, and Supplementary Material 4a, www.wjon.org). This diminished expression of CLDN8 protein also exhibited a strong discriminatory ability between KIRC and non-KIRC tissues (Supplementary Material 4b, www.wjon.org).
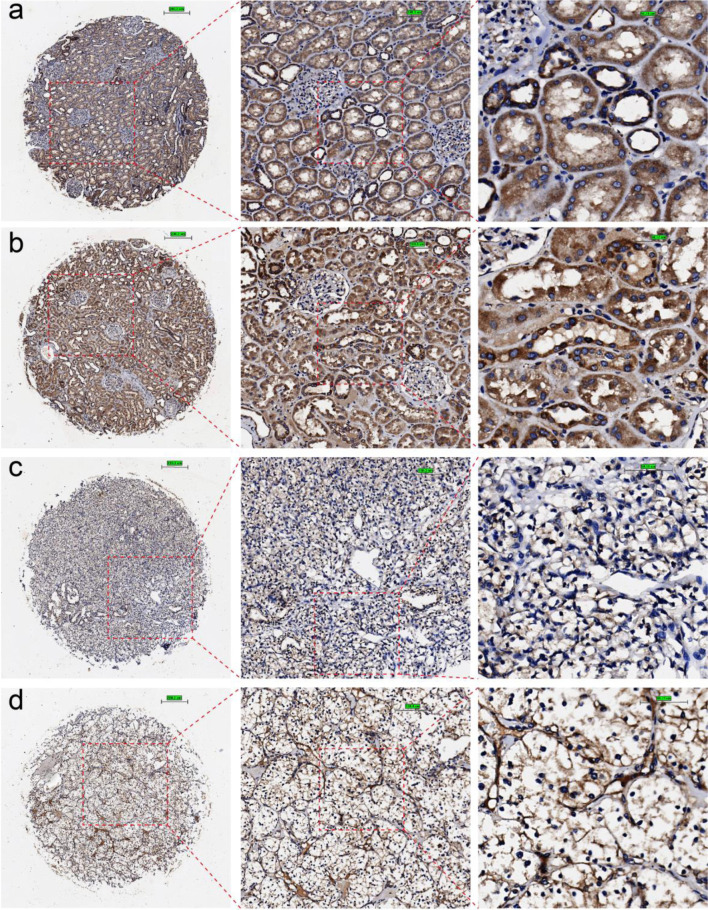
Figure 2.
Immunohistochemical staining of claudin 8 (CLDN8) protein in non-KIRC and KIRC tissues. (a, b) Non-KIRC tissues. (c, d) KIRC tissues. KIRC: kidney renal clear cell carcinoma.
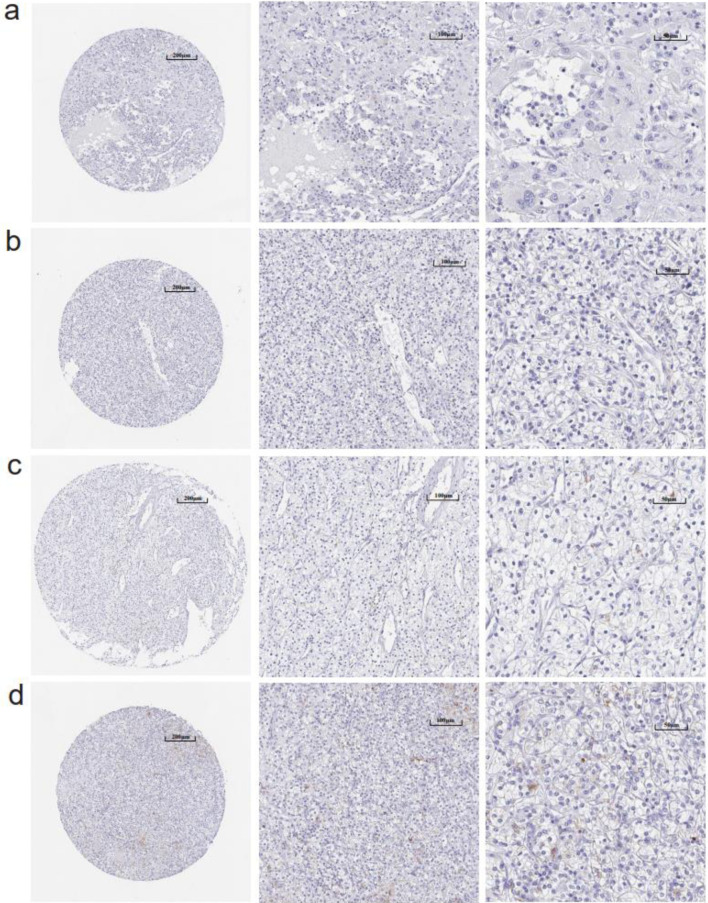
Figure 3.
(a-d) Immunohistochemistry images of claudin 8 (CLDN8) protein in kidney renal clear cell carcinoma tissues derived from the human protein atlas (HPA) database.
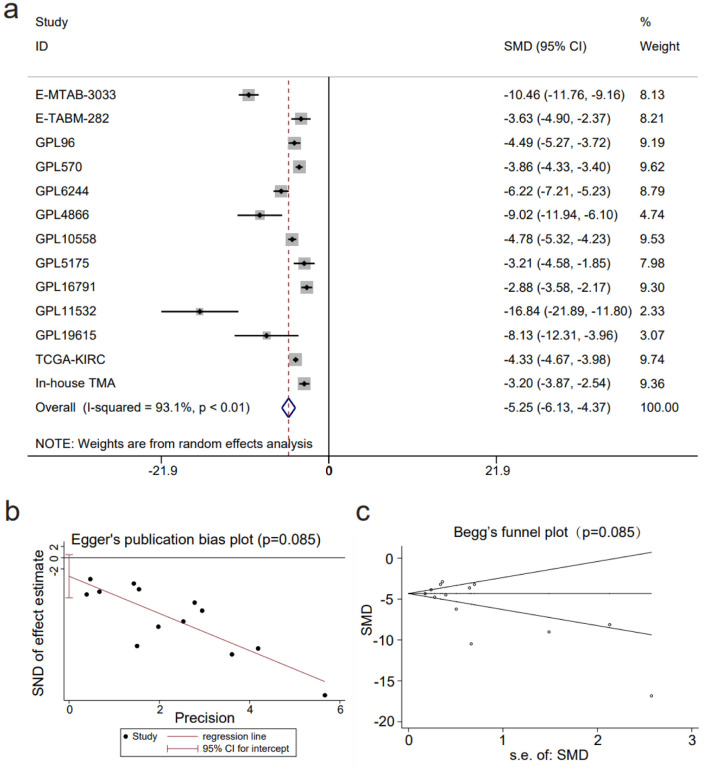
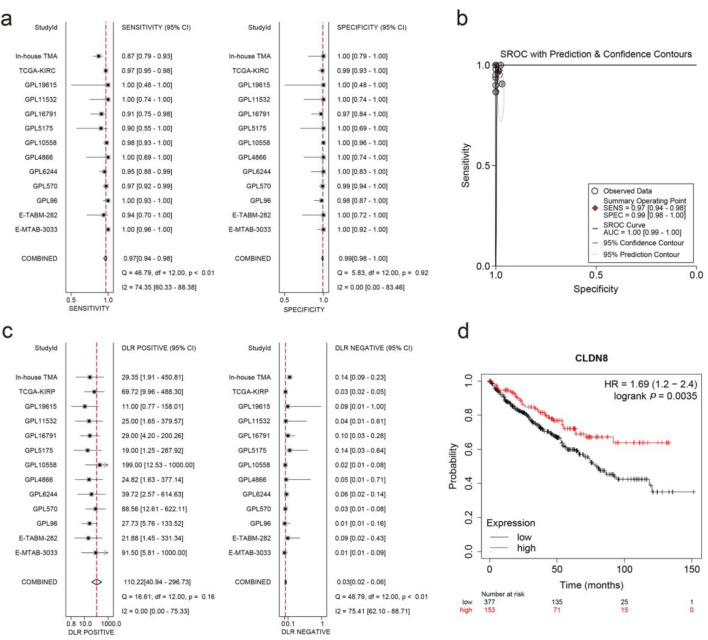
An integrated SMD analysis of mRNA datasets and in-house tissue microarrays revealed significantly lower CLDN8 expression in 1,165 KIRC tissues compared to 468 non-KIRC tissues (SMD = -5.25; 95% confidence interval (CI): -6.13 to -4.37; I2 = 93.1%; P < 0.01) (Fig. 4a). No publication bias was detected (P = 0.09) (Fig. 4b, c). The SROC curve and forest plot confirmed the robust potential of CLDN8 in distinguishing KIRC from non-KIRC tissues (AUC = 1.00, 95% CI: 0.99 - 1.00; sensitivity = 0.97, 95% CI: 0.94 - 0.96; specificity = 0.99, 95% CI: 0.98 - 1.00; positive likelihood ratio = 110.22, 95% CI: 40.94 - 296.73; negative likelihood ratio = 0.03, 95% CI: 0.02 - 0.06) (Fig. 5a-c). Furthermore, survival analysis indicated that lower CLDN8 expression in KIRC was associated with shorter survival time (hazard ratio of CLDN8 downregulation = 1.69, 95% CI: 1.2 - 2.4) (Fig. 5d). Apart from ethnicity, CLDN8 expression showed no significant association with clinicopathological features such as gender, age, tumor grade, and tumor stage (Supplementary Material 5, www.wjon.org).
Figure 4.
The decline in claudin 8 (CLDN8) expression in kidney renal clear cell carcinoma. (a) Forest plot depicting significant downregulation of CLDN8 expression in kidney renal clear cell carcinoma tissues. (b) Egger’s plot and (c) Begg’s funnel plot indicate the absence of publication bias (P = 0.09).
Figure 5.
Integrated analysis of effect size forest plots, summary receiver operating characteristics curve, and survival analysis. (a) Sensitivity and specificity assessment. (b) Summary receiver operating characteristics curve. (c) Positive and negative likelihood ratios. (d) Kaplan-Meier survival curve.
Downregulation of CLDN8 in KIRC cells at the single-cell level
After quality control and clustering optimization, a total of 3,306 KIRC cells and 204 normal cells were obtained (Fig. 6a). CLDN8 mRNA was significantly downregulated in KIRC single cells (P < 0.0001) (Fig. 6b).
Figure 6.
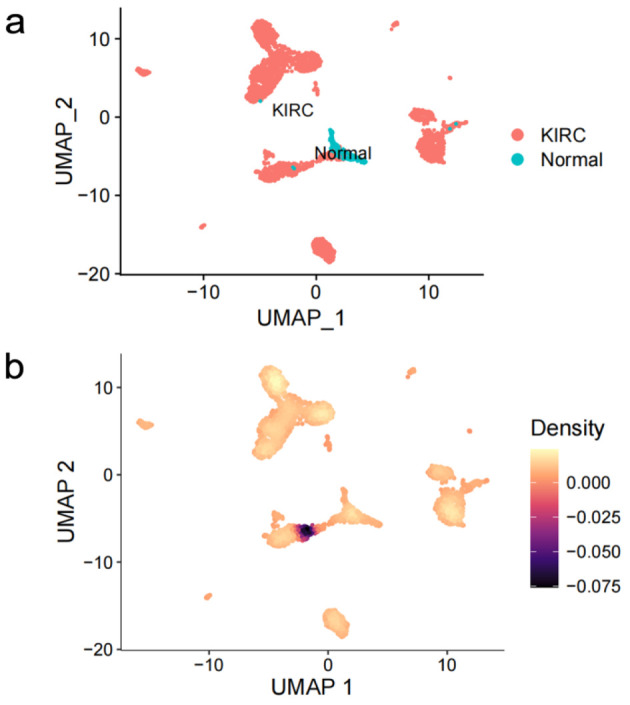
Expression profiling of claudin 8 (CLDN8) in single cells of kidney renal clear cell carcinoma. (a) Cell distribution in kidney renal clear cell carcinoma (KIRC). (b) Comparative analysis of CLDN8 mRNA expression levels in KIRC cells and normal cells.
CLDN8 inhibited KIRC cell propagation
We assessed the dependency scores of CLDN8 using CRISPR knockout screen technology to evaluate its effect on the growth of KIRC cell lines. As was shown in Figure 7, knocking out the CLDN8 gene resulted in faster growth in 20 KIRC cell lines. This finding suggests that CLDN8 may play an inhibitory role in the proliferation of KIRC cells.
Figure 7.
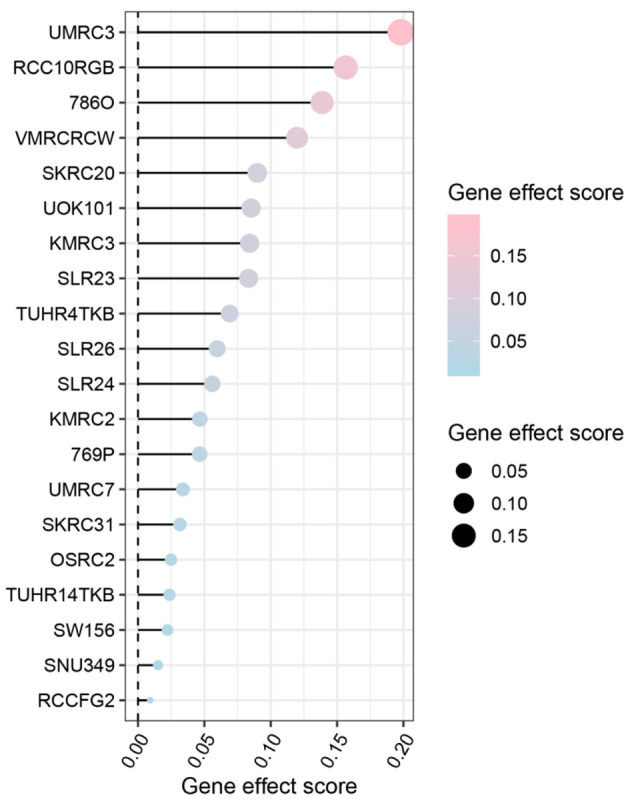
Gene effect score analysis of claudin 8 (CLDN8) in 20 kidney renal clear cell carcinoma cell lines. The findings suggest a potential inhibitory role of CLDN8 in the growth of kidney renal clear cell carcinoma cells.
CLDN8 participating in KIRC via significant signaling pathways
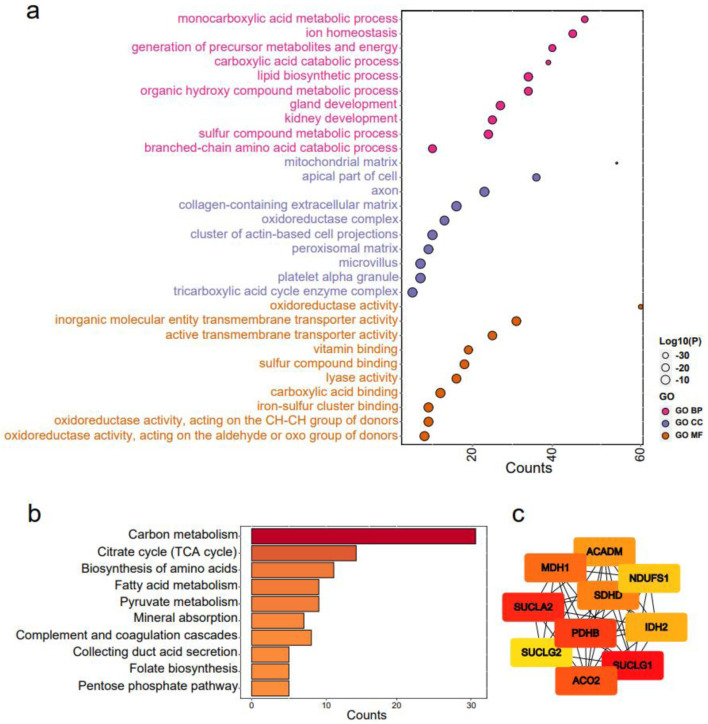
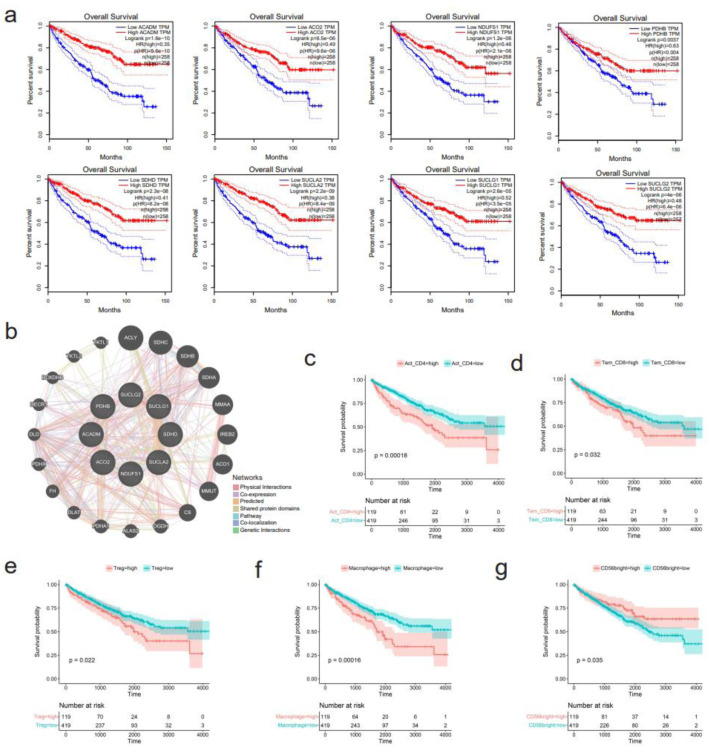
For GO enrichment analysis, the terms “carboxylic acid catabolic process” in biological processes (BP), “mitochondrial matrix” in cellular components (CC), and “oxidoreductase activity” in molecular functions (MF) were identified as highly enriched (Fig. 8a). In KEGG pathways analysis, “carbon metabolism” and “citrate cycle (TCA cycle)” were emphasized (Fig. 8b). Ten hub genes, including ACADM, ACO2, NDUFS1, PDHB, SDHD, SUCLA2, SUCLG1, SUCLG2, MDH1, and IDH2, were identified (Fig. 8c). Notably, eight of these hub genes, namely ACADM, ACO2, NDUFS1, PDHB, SDHD, SUCLA2, SUCLG1, and SUCLG2, exhibited significant correlations with overall survival in KIRC patients (Fig. 9a). A functional interaction network was constructed using these eight hub genes, with FH being identified as prominent (Fig. 9b).
Figure 8.
Functional enrichment analysis of claudin 8 (CLDN8) and hub genes. (a) Gene ontology terms encompassing biological processes, cell components, and molecular functions. (b) Kyoto encyclopedia of genes and genomes pathways. (c) Identification of top 10 hub genes via protein-protein interaction network analysis using Cytohubba.
Figure 9.
Survival analysis of hub genes and several immune cells. (a) Downregulation of eight hub genes, including ACADM, ACO2, NDUFS1, PDHB, SDHD, SUCLA2, SUCLG1, and SUCLG2, correlates with shorter overall survival of kidney renal clear cell carcinoma (KIRC) patients. (b) Functional interaction network of the eight hub genes using GeneMANIA algorithm. (c-g) Correlation between infiltration levels of various immune cells and survival of KIRC patients. (c) Activated CD4+ T cells. (d) Effector memory CD8+ T cells. (e) Regulatory T cells. (f) Macrophage. (g) CD56bright natural killer cells.
Correlation between CLDN8 downregulation and the immune infiltration levels of KIRC tissues
The infiltration levels of 28 immune cells were predicted in the immune microenvironment of KIRC tissues (Supplementary Material 6a, www.wjon.org). Among them, seven kinds of immune cells, including activated CD4+ T cells, effector memory CD4+ T cells, effector memory CD8+ T cells, regulatory T cells, natural killer cells, memory B cells, and macrophages showed potentially negative correlation with CLDN8 expression (Supplementary Material 6b, www.wjon.org). Conversely, the abundance of CD56bright natural killer cells had a potentially positive correlation with CLDN8 expression. High infiltration of activated CD4+ T cells, effector memory CD8+ T cells, regulatory T cells, and macrophages significantly shortened the survival time of KIRC patients, as did lower levels of CD56bright natural killer cells (Fig. 9c-g).
Discussion
Combining public high-throughput datasets with in-house tissue microarrays and IHC, we demonstrate that CLDN8 expression is consistently diminished at both mRNA and protein levels. Moreover, our single-cell analysis reveals downregulation of CLDN8 in KIRC, which correlates with poorer prognosis. Furthermore, through CRISPR knockout screen analysis, we suggest a potential tumor-suppressive role for CLDN8 in vitro. The downregulation of CLDN8 may accelerate KIRC progression, possibly in association with oncometabolites. Additionally, increased macrophages levels in KIRC are identified as an adverse factor in disease prognosis.
Claudins, initially defined as dominant integral membrane proteins of TJs, function as fences, signaling molecules, and barriers. Recently research has revealed their involvement in carcinogenesis through signal transduction, inflammatory response, hyperplasia, epithelial-mesenchymal transition (EMT), migration, and survival [30]. As a member of the claudin family, CLDN8 has been increasingly associated with malignant tumors. In this study, we analyzed RNA-seq datasets from major public databases, performed in-house tissue microarrays, and conducted IHC staining to examine CLDN8 mRNA and protein levels in KIRC. Our results showed significantly lower CLDN8 expression in KIRC, detected in 1,060 KIRC samples versus 452 non-KIRC samples in RNA-sequencing datasets and in 105 KIRC tissues versus 16 non-tumor tissues in in-house tissue microarrays. IHC images also displayed weak CLDN8 protein expression in KIRC. Additionally, single-cell analysis indicated downregulation of CLDN8 in 3,306 KIRC cells. Previous studies have demonstrated CLDN8 dysfunction in prostate cancer [15], breast cancer [14], osteosarcoma [31], and retinoblastoma [32]. Specifically, low CLDN8 expression in KIRC was found to inhibit hyperplasia, metastasis, and invasion of cancer cells through EMT and AKT (AKT/PKB, protein kinase B) pathways. Our CRISPR knockout screen analysis further revealed that CLDN8 may hinder the proliferation of 20 KIRC cell lines. Moreover, decreased CLDN8 expression exhibited a strong discriminatory potential between KIRC and non-KIRC tissues, correlating with worse prognosis in KIRC patients, indicating its potential as a biomarker for KIRC. Overall, our findings provide a comprehensive understanding of CLDN8 expression in KIRC tissues, highlighting its role in carcinogenesis.
To further explore the role of CLDN8 in KIRC, we analyzed its co-expressed network. The results highlighted the “carboxylic acid catabolic process” in BP, “mitochondrial matrix” in CC, and “oxidoreductase activity” in MF. The most enriched KEGG pathways included “carbon metabolism,” and “TCA cycle”. Intriguingly, the eight hub genes - ACADM, ACO2, NDUFS1, PDHB, SDHD, SUCLA2, SUCLG1, and SUCLG2 - exhibited tumor-suppressive function, with ACO2, SDHD, SUCLA2, SUCLG1, and SUCLG2 being crucial in the TCA cycle [33, 34]. PDHB is involved in converting pyruvate to acetyl coenzyme A [33], ACADM catalyzes mitochondrial fatty acid oxidation [35], and NDUFS1 is a component of mitochondrial complex I [36]. The disruption of aerobic metabolism, particularly the TCA cycle, is a notable characteristic of KIRC. Recent studies have revealed that metabolic changes support cancer invasion by degrading the extracellular matrix, reducing cell-cell/matrix contact, increasing invadopodia formation, and activating EMT pathways [37]. Lipid accumulation favors cancer cell motility and exacerbates tumor progression in ACADM-deficient HCC [35]. Increased pyruvate due to PDHB loss aids malignant cell invasion [38], and NDUFS1 knockout enhances invasion and migration in human lung adenocarcinoma cell lines [39]. Consequently, blocking the TCA cycle or decreasing critical enzymes leads to the accumulation of metabolites that contribute to tumor initiation and progression [33, 40]. For instance, succinate accumulation resulting from SDH deletion stabilizes hypoxia-inducible factor 1 (HIF-1), promoting malignant cell survival, proliferation, and angiogenesis [40]. Succinate-CoA accumulation from the inactivation of SDH, SUCLA2, SUCLG1, and SUCLG2, promotes tumor growth through global succinylation and provides a bioenergetic source when oxidative phosphorylation or glycolysis is hindered [33]. Elevated citrate flux due to ACO2 blockade enhances colorectal cancer growth by upregulating stearoyl-CoA desaturase, boosting lipid desaturation [41]. Additionally, gene-gene interaction analysis using the GeneMANIA database suggested that FH, together with hub genes, may play a role in KIRC. FH deficiency leads to fumarate overload, contributing to oncogenic transformation by stabilizing HIF-1α [40]. Notably, the loss of function of key enzymes or accumulation of oncometabolites in the TCA cycle is associated with disrupted cell-cell adherence in cancer cells. According to Wang et al [42], fumarate can drive EMT by inhibiting miR-200ba429 demethylation, a metastasis repressor. Additionally, SDHB knockout promotes EMT in colorectal cancer cells through TGF-β signaling or by enhancing the TJ transcriptional suppression complex SNAIL1-SMAD3/SMAD4 [43]. Furthermore, SDH downregulation, whether by exogenous succinate treatment or SDH-subunit (B, D) knockout, enhances KIRC cell invasion [44]. In this setting, CLDN8 and essential genes in the TCA cycle are markedly decreased in KIRC, and oncometabolites from the blocked TCA cycle likely accelerate KIRC progression by degrading intercellular TJs or through the EMT pathway. Supporting this, Zhu et al have conducted preliminary gene interference and cell function experiments to determine how CLDN8 influences KIRC cell migration and invasion by regulating the EMT process.
The KIRC microenvironment exhibits a high infiltration of immune cells. Our findings indicate a negative correlation between CLDN8 expression and activated CD4+ T cells, effector memory CD8+ T cells, regulatory T cells, and macrophages, while a positive correlation is observed with CD56bright natural killer cells. Notably, these five cell types are closely linked to KIRC patient prognosis. Building upon the research by Wu et al, it is suggested that succinate secretion by malignant cells could activate succinate receptor signaling, leading to the polarization of macrophages into tumor-associated macrophages, thereby promoting tumor progression [45].
However, this study is limited by the lack of functional experiment verification. Additional evidence is needed to determine whether insufficient CLDN8 accelerates KIRC progression through EMT or oncometabolite pathways. Furthermore, the sample size of non-KIRC control tissues for IHC was relatively small. While a comparative arm with a similar number of cases would strengthen our analysis, constraints such as limited sample availability and ethical considerations prevented us from obtaining a larger number of normal kidney samples for IHC. Nevertheless, we assure that the non-KIRC control samples used are representative and that rigorous experimental methods and techniques were employed to ensure the accuracy and reliability of our results. The unpaired Wilcoxon test clearly demonstrates significantly lower expression of CLDN8 protein in KIRC tissues. This finding, along with results from mRNA platform datasets, supports the marked downregulation of CLDN8 in KIRC. Future research should include clinical verification with larger sample sizes to further substantiate these findings.
Conclusion
Through our immunochemistry experiments, we observed a significant downregulation of CLDN8 in KIRC, which appears to be associated with a less favorable survival outcome for KIRC patients. In addition, lower CLDN8 expression may facilitate KIRC occurrence and progression by the EMT pathway.
Supplementary Material
mRNA datasets of kidney renal clear cell carcinoma obtained from GEO, ArrayExpress, and TCGA databases.
Comparative analysis of CLND8 mRNA expression levels in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues.
The discriminative potential of CLND8 mRNA downregulation in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues.
Downregulation of CLDN8 protein in kidney renal clear cell carcinoma tissues. (a) Comparison of immunohistochemical staining scores of CLDN8 protein in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues (unpaired Wilcoxon test, P < 0.001). (b) The robust discriminatory ability of CLDN8 protein between KIRC and non-KIRC tissues.
Correlations between CLDN8 mRNA expression and clinicopathological parameters of kidney renal clear cell carcinoma patients (*P < 0.05).
Infiltration analysis of immune cells in kidney renal clear cell carcinoma tissue. (a) Heatmap presenting 28 immune cells enriched in kidney renal clear cell carcinoma tissues. (b) Spearman correlation analysis between CLDN8 mRNA expression and immune cells in TISIDB database.
Acknowledgments
The authors thank the technical assistance of Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis.
Funding Statement
The study was supported by the Guangxi Higher Education Undergraduate Teaching Reform Project (2022JGA146), Guangxi Educational Science Planning Key Project (2022ZJY2791), Guangxi Medical University Undergraduate Education and Teaching Reform Project (2023Z10), and Guangxi Natural Science Foundation (2021GXNSFAA075033).
Conflict of Interest
The authors declare no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
HCJ, ZGH, GC, JWC, SHL, and BQ designed the study. HCJ, JDL, GLZ, ZGH, and CYZ collected public datasets and participated in data analysis. ZGH, YXT, KQ, and GC performed the in-house experiments with tissue microarrays. HCJ, GLZ, and CYZ wrote the manuscript. GC, JWC, SHL, and BQ contributed to guiding and polishing the draft. HCJ, JDL, GC, and YLM revised the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets generated and/or analyzed during the current study are available in gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), and the cancer genome atlas (https://portal.gdc.cancer.gov/). The accession number of datasets were documented in Supplemental Material 1 (www.wjon.org). Data related to this manuscript can be made available from the corresponding author Bin Qin upon reasonable request.
Abbreviations
- ANOVA
analysis of variance
- AUC
area under the curve
- BP
biological process
- CC
cellular component
- CEGs
co-expressed genes
- CI
confidence interval
- CLDN4
claudin 4
- CLDN8
claudin 8
- DEGs
differential expression genes
- EMT
epithelial-mesenchymal transition
- GEO
gene expression omnibus
- GO
gene ontology
- HPA
human protein atlas
- IHC
immunohistochemical
- IRS
immunoreactive score
- KEGG
Kyoto encyclopedia of genes and genomes
- KIRC
kidney renal clear cell carcinoma
- MF
molecular function
- PPI
protein-protein interaction
- RCC
renal cell carcinoma
- ROC
receiver operating characteristic
- SMD
standardized mean difference
- ssGSEA
single-sample gene set enrichment analysis
- SROC
summary receiver operating characteristic curve
- TCA
tricarboxylic acid
- TCGA
the cancer genome atlas
- TJ
tight junction
- TPM
transcripts per million
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Gu YY, Chen G, Lin P, Cheng JW, Huang ZG, Luo J, Zhai GQ. et al. Development and validation of an immune prognostic classifier for clear cell renal cell carcinoma. Cancer Biomark. 2020;27(2):265–275. doi: 10.3233/CBM-191017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao Z, Zheng Z, Zheng X, Wu H, Zhao W, Mu X, Sun F. et al. Comprehensive characterization of metabolism-associated subtypes of renal cell carcinoma to aid clinical therapy. Oxid Med Cell Longev. 2022;2022:9039732. doi: 10.1155/2022/9039732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao L, He RQ, Huang ZG, Dang YW, Gu YY, Yan HB, Li SH. et al. Genome-wide analysis of the alternative splicing profiles revealed novel prognostic index for kidney renal cell clear cell carcinoma. J Cancer. 2020;11(6):1542–1554. doi: 10.7150/jca.36998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trevisani F, Floris M, Minnei R, Cinque A. Renal oncocytoma: the diagnostic challenge to unmask the double of renal cancer. Int J Mol Sci. 2022;23(5):2603. doi: 10.3390/ijms23052603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei M, Zhang Y, Yang X, Ma P, Li Y, Wu Y, Chen X. et al. Claudin-2 promotes colorectal cancer growth and metastasis by suppressing NDRG1 transcription. Clin Transl Med. 2021;11(12):e667. doi: 10.1002/ctm2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JL, Ekambaram P, Carleton NM, Hu D, Klei LR, Cai Z, Myers MI. et al. MALT1 is a targetable driver of epithelial-to-mesenchymal transition in claudin-low, triple-negative breast cancer. Mol Cancer Res. 2022;20(3):373–386. doi: 10.1158/1541-7786.MCR-21-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marincola Smith P, Choksi YA, Markham NO, Hanna DN, Zi J, Weaver CJ, Hamaamen JA. et al. Colon epithelial cell TGFbeta signaling modulates the expression of tight junction proteins and barrier function in mice. Am J Physiol Gastrointest Liver Physiol. 2021;320(6):G936–G957. doi: 10.1152/ajpgi.00053.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Zhao J, Cao M, Liu R, Chen G, Li S, Xie Y. et al. Mast cells-derived MiR-223 destroys intestinal barrier function by inhibition of CLDN8 expression in intestinal epithelial cells. Biol Res. 2020;53(1):12. doi: 10.1186/s40659-020-00279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuang X, Chen B, Huang S, Han J, Zhou G, Xu S, Chen M. et al. Hypermethylation of miR-145 promoter-mediated SOX9-CLDN8 pathway regulates intestinal mucosal barrier in Crohn's disease. EBioMedicine. 2022;76:103846. doi: 10.1016/j.ebiom.2022.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sassi A, Wang Y, Chassot A, Roth I, Ramakrishnan S, Olivier V, Staub O. et al. Expression of claudin-8 is induced by aldosterone in renal collecting duct principal cells. Am J Physiol Renal Physiol. 2021;321(5):F645–F655. doi: 10.1152/ajprenal.00207.2021. [DOI] [PubMed] [Google Scholar]
- 12.Hou J, Renigunta A, Yang J, Waldegger S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci U S A. 2010;107(42):18010–18015. doi: 10.1073/pnas.1009399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina-Jijon E, Rodriguez-Munoz R, Gonzalez-Ramirez R, Namorado-Tonix C, Pedraza-Chaverri J, Reyes JL. Aldosterone signaling regulates the over-expression of claudin-4 and -8 at the distal nephron from type 1 diabetic rats. PLoS One. 2017;12(5):e0177362. doi: 10.1371/journal.pone.0177362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Zheng A, Lu H, Jin Z, Peng Z, Jin F. The expression and prognostic significance of claudin-8 and androgen receptor in breast cancer. Onco Targets Ther. 2020;13:3437–3448. doi: 10.2147/OTT.S242406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashikari D, Takayama KI, Obinata D, Takahashi S, Inoue S. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci. 2017;108(7):1386–1393. doi: 10.1111/cas.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. https://www.ncbi.nlm.nih.gov/geo/
- 17. https://www.ebi.ac.uk/arrayexpress/
- 18. https://portal.gdc.cancer.gov/
- 19.Wen JY, Qin LT, Chen G, Huang HQ, Shen MJ, Pang JS, Tang YX. et al. Downregulation of MicroRNA-1 and its potential molecular mechanism in nasopharyngeal cancer: an investigation combined with in silico and in-house immunohistochemistry validation. Dis Markers. 2022;2022:7962220. doi: 10.1155/2022/7962220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang WY, Chen G, Chen SW, Dang YW, Deng Y, Zhou HF, Kong JL. et al. The indication of poor prognosis by high expression of ENO1 in squamous cell carcinoma of the lung. J Oncol. 2021;2021:9910962. doi: 10.1155/2021/9910962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He RQ, Li JD, Du XF, Dang YW, Yang LJ, Huang ZG, Liu LM. et al. LPCAT1 overexpression promotes the progression of hepatocellular carcinoma. Cancer Cell Int. 2021;21(1):442. doi: 10.1186/s12935-021-02130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. https://www.proteinatlas.org/
- 23.Su C, Lv Y, Lu W, Yu Z, Ye Y, Guo B, Liu D. et al. Single-cell RNA sequencing in multiple pathologic types of renal cell carcinoma revealed novel potential tumor-specific markers. Front Oncol. 2021;11:719564. doi: 10.3389/fonc.2021.719564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho KH, Huang TW, Liu AJ, Shih CM, Chen KC. Cancer Essential Genes Stratified Lung Adenocarcinoma Patients with Distinct Survival Outcomes and Identified a Subgroup from the Terminal Respiratory Unit Type with Different Proliferative Signatures in Multiple Cohorts. Cancers (Basel) 2021;13(9):2128. doi: 10.3390/cancers13092128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. http://kmplot.com/analysis/
- 26. https://string-db.org/
- 27. http://gepia.cancer-pku.cn/
- 28. http://genemania.org/
- 29. http://cis.hku.hk/TISIDB/
- 30.Li J. Targeting claudins in cancer: diagnosis, prognosis and therapy. Am J Cancer Res. 2021;11(7):3406–3424. [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Yang Y, Hao P, Ding X. Claudin 8 Contributes to Malignant Proliferation in Human Osteosarcoma U2OS Cells. Cancer Biother Radiopharm. 2015;30(9):400–404. doi: 10.1089/cbr.2015.1815. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Lu B, Wang X, Jiang H, Kuang W. MiR-361-5p inhibits cell proliferation and induces cell apoptosis in retinoblastoma by negatively regulating CLDN8. Childs Nerv Syst. 2019;35(8):1303–1311. doi: 10.1007/s00381-019-04199-9. [DOI] [PubMed] [Google Scholar]
- 33.Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene. 2021;40(19):3351–3363. doi: 10.1038/s41388-020-01639-8. [DOI] [PubMed] [Google Scholar]
- 34.Gut P, Matilainen S, Meyer JG, Pallijeff P, Richard J, Carroll CJ, Euro L. et al. SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease. Nat Commun. 2020;11(1):5927. doi: 10.1038/s41467-020-19743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma APY, Yeung CLS, Tey SK, Mao X, Wong SWK, Ng TH, Ko FCF. et al. Suppression of ACADM-mediated fatty acid oxidation promotes hepatocellular carcinoma via aberrant CAV1/SREBP1 signaling. Cancer Res. 2021;81(13):3679–3692. doi: 10.1158/0008-5472.CAN-20-3944. [DOI] [PubMed] [Google Scholar]
- 36.Ellinger J, Poss M, Bruggemann M, Gromes A, Schmidt D, Ellinger N, Tolkach Y. et al. Systematic expression analysis of mitochondrial complex I identifies NDUFS1 as a biomarker in clear-cell renal-cell carcinoma. Clin Genitourin Cancer. 2017;15(4):e551–e562. doi: 10.1016/j.clgc.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Elia I, Doglioni G, Fendt SM. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018;28(8):673–684. doi: 10.1016/j.tcb.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Jobard E, Pontoizeau C, Blaise BJ, Bachelot T, Elena-Herrmann B, Tredan O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014;343(1):33–41. doi: 10.1016/j.canlet.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Zhan J, Sun S, Chen Y, Xu C, Chen Q, Li M, Pei Y. et al. MiR-3130-5p is an intermediate modulator of 2q33 and influences the invasiveness of lung adenocarcinoma by targeting NDUFS1. Cancer Med. 2021;10(11):3700–3714. doi: 10.1002/cam4.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scagliola A, Mainini F, Cardaci S. The tricarboxylic acid cycle at the crossroad between cancer and immunity. Antioxid Redox Signal. 2020;32(12):834–852. doi: 10.1089/ars.2019.7974. [DOI] [PubMed] [Google Scholar]
- 41.You X, Tian J, Zhang H, Guo Y, Yang J, Zhu C, Song M. et al. Loss of mitochondrial aconitase promotes colorectal cancer progression via SCD1-mediated lipid remodeling. Mol Metab. 2021;48:101203. doi: 10.1016/j.molmet.2021.101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YP, Li JT, Qu J, Yin M, Lei QY. Metabolite sensing and signaling in cancer. J Biol Chem. 2020;295(33):11938–11946. doi: 10.1074/jbc.REV119.007624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Chen Y, Wu G. SDHB deficiency promotes TGFbeta-mediated invasion and metastasis of colorectal cancer through transcriptional repression complex SNAIL1-SMAD3/4. Transl Oncol. 2016;9(6):512–520. doi: 10.1016/j.tranon.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aggarwal RK, Luchtel RA, Machha V, Tischer A, Zou Y, Pradhan K, Ashai N. et al. Functional succinate dehydrogenase deficiency is a common adverse feature of clear cell renal cancer. Proc Natl Acad Sci U S A. 2021;118(39):e2106947118. doi: 10.1073/pnas.2106947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, Yeh CC. et al. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol Cell. 2020;77(2):213–227.e215. doi: 10.1016/j.molcel.2019.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA datasets of kidney renal clear cell carcinoma obtained from GEO, ArrayExpress, and TCGA databases.
Comparative analysis of CLND8 mRNA expression levels in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues.
The discriminative potential of CLND8 mRNA downregulation in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues.
Downregulation of CLDN8 protein in kidney renal clear cell carcinoma tissues. (a) Comparison of immunohistochemical staining scores of CLDN8 protein in kidney renal clear cell carcinoma (KIRC) and non-KIRC tissues (unpaired Wilcoxon test, P < 0.001). (b) The robust discriminatory ability of CLDN8 protein between KIRC and non-KIRC tissues.
Correlations between CLDN8 mRNA expression and clinicopathological parameters of kidney renal clear cell carcinoma patients (*P < 0.05).
Infiltration analysis of immune cells in kidney renal clear cell carcinoma tissue. (a) Heatmap presenting 28 immune cells enriched in kidney renal clear cell carcinoma tissues. (b) Spearman correlation analysis between CLDN8 mRNA expression and immune cells in TISIDB database.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in gene expression omnibus (https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), and the cancer genome atlas (https://portal.gdc.cancer.gov/). The accession number of datasets were documented in Supplemental Material 1 (www.wjon.org). Data related to this manuscript can be made available from the corresponding author Bin Qin upon reasonable request.