Abstract
We have tested the effect of metabolic inhibitors, membrane cholesterol depletion, and detergent extraction of cell surface molecules on the susceptibility of MA104 cells to infection by rotaviruses. Treatment of cells with tunicamycin, an inhibitor of protein N glycosylation, blocked the infectivity of the SA-dependent rotavirus RRV and its SA-independent variant nar3 by about 50%, while the inhibition of O glycosylation had no effect. The inhibitor of glycolipid biosynthesis d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) blocked the infectivity of RRV, nar3, and the human rotavirus strain Wa by about 70%. Sequestration of cholesterol from the cell membrane with β-cyclodextrin reduced the infectivity of the three viruses by more than 90%. The involvement of N-glycoproteins, glycolipids, and cholesterol in rotavirus infection suggests that the virus receptor(s) might be forming part of lipid microdomains in the cell membrane. MA104 cells incubated with the nonionic detergent octyl-β-glucoside (OG) showed a ca. 60% reduction in their ability to bind rotaviruses, the same degree to which they became refractory to infection, suggesting that OG extracts the potential virus receptor(s) from the cell surface. Accordingly, when preincubated with the viruses, the OG extract inhibited the virus infectivity by more than 95%. This inhibition was abolished when the extract was treated with either proteases or heat but not when it was treated with neuraminidase, indicating the protein nature of the inhibitor. Two protein fractions of around 57 and 75 kDa were isolated from the extract, and these fractions were shown to have rotavirus-blocking activity. Also, antibodies to these fractions efficiently inhibited the infectivity of the viruses in untreated as well as in neuraminidase-treated cells. Five individual protein bands of 30, 45, 57, 75, and 110 kDa, which exhibited virus-blocking activity, were finally isolated from the OG extract. These proteins are good candidates to function as rotavirus receptors.
Rotaviruses, the leading cause of severe dehydrating diarrhea in infants and young children worldwide, are nonenveloped viruses that possess a genome of 11 segments of double-stranded RNA contained in a triple-layered protein capsid. The outermost layer is composed of two proteins, VP4 and VP7. VP4 forms spikes that extend from the surface of the virus and has been associated with a variety of functions, including the initial attachment of the virus to the cell membrane and the penetration of the cell by the virion (14).
Rotaviruses have a very specific cell tropism, infecting only enterocytes on the tips of intestinal villi (26), which suggests that specific host receptors must exist. In vitro, they also display a strict tropism, binding to a variety of cell lines but infecting efficiently only those of renal or intestinal epithelium origin (15). Despite the advances in the molecular and structural biology of the virus, little is known about the rotavirus cell receptors. Some animal rotavirus strains interact with sialic acid (SA) on the cell surface, and this interaction is a requirement for the efficient attachment and infection of the virus to susceptible cells (9, 17, 27, 34, 39, 57). Accordingly, a number of glycoconjugates bind to and block the infectivity of animal rotaviruses in vitro and in vivo (3, 4, 6, 17, 32, 46, 52–54, 56, 57). Some of these glycoconjugates may play a role as possible receptors, like GM3 gangliosides in newborn piglet intestine (47), GM1 in LLC-MK2 cells (52), and 300- to 330-kDa glycoproteins in murine enterocytes (3). More recently, it has also been suggested that α2β1, αxβ2, and α4β1 integrins may be involved in rotavirus cell entry (11, 24).
The binding of animal rotaviruses RRV and SA11 to an SA-containing cell receptor is nonessential since variants whose infectivity is no longer dependent on the binding to these acid sugars have been isolated (35, 39). The secondary importance of SA as an attachment site for rotaviruses, at least under laboratory conditions of infection, is also reflected by the fact that the infectivity of most, if not all, human rotavirus (HRV) strains is not affected by neuraminidase treatment of cells (9, 17, 19, 41). Recently, through competition infection assays using the SA-dependent RRV, its SA-independent variant nar3, and the naturally neuraminidase-resistant HRV strain Wa, the existence of at least three cell surface sites involved in the interaction of rotaviruses with MA104 cells during the early steps of infection was determined (41).
In this study we used two approaches to characterize the cell surface structures that could serve as rotavirus receptors. In the first approach, MA104 cells were treated with metabolic inhibitors of glycosylation as well as of glycolipid synthesis to determine the effects on the infectivity of rotaviruses RRV, nar3, and Wa. In the second approach, the putative receptors for rotaviruses were extracted with the nonionic detergent octyl-β-glucoside (OG) under noncytolytic conditions. The molecules present in the extract, which were shown to inhibit rotavirus infectivity when incubated with the viruses in solution, were biochemically characterized and partially purified.
MATERIALS AND METHODS
Cells and viruses.
The human rotavirus strain Wa and the rhesus strain RRV were obtained from Harry B. Greenberg, Stanford University, Stanford, Calif. The SA-independent rotavirus RRV variant nar3 has been previously described (39, 40). All rotavirus strains were propagated in MA104 cells as described previously (13). The rhesus monkey epithelial cell line MA104 was grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and was used for all experiments carried out in this work. BHK-21, CHO, and L929 cells were grown in MEM containing 10% FBS. Reovirus type 1 and poliovirus type 3, Leon strain, were kindly provided by C. Ramos (CISEI, National Institute of Public Health, Cuernavaca, Mexico) and R. M. del Angel (CINVESTAV-IPN, Mexico City, Mexico).
Infectivity assay.
MA104 cells in 96-well plates were washed twice with phosphate-buffered saline (PBS) and then about 1,000 focus-forming units (FFUs) of a trypsin-activated cell lysate containing rotaviruses RRV, nar3, Wa, or control viruses, reovirus and poliovirus, was adsorbed to the cells for 45 min at 4°C. After the adsorption period, the virus inoculum was removed, the cells were washed once with PBS, MEM was added, and the infection was left to proceed for 14 h at 37°C. The infected cells were detected by an immunoperoxidase focus detection assay, using as the detecting antibody a rabbit polyclonal hyperimmune serum to porcine rotavirus YM, as described previously (33). The FFUs were counted with the help of a Visiolab 1000 station (Biocom). This station, which was used for both image acquisition and analysis, is configured with a Matrox Meteor RGB frame grabber and a 8295 Cohu RGB CCD color TV camera. Motorized stages (Marzhauser) were adapted to an inverted Nikon Diaphot 300 microscope. The stage control unit was a Marzhauser Multicontrol MC2000, piloted by Explo (Biocom). Macro command files for Explo were developed to perform a semiautomated counting of the infected cells. In this manner, an accurate positioning in the center of each well was achieved automatically for later predefined scanning and visual counting of infected cells within a selected well area.
Treatment of MA104 cells with metabolic inhibitors.
Monolayers of MA104 cells in 96-well plates were grown to confluence; either 2 μg of tunicamycin (Boehringer) per ml or 2 mM benzyl N-acetyl-α-d-galactosamide (benzylGalNAc) (Oxford Glyco Systems) in MEM was added, and the cells were further incubated for either 24 h (tunicamycin) or 3 days (benzylGalNAc). To inhibit the synthesis of glycolipids, 60% confluent MA104 cells were treated with 25 μM d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) (Matreya, Inc.) in MEM–10% FBS for 3 days, replacing the medium with fresh drug daily. After treatment with the respective drug, the cells were washed twice with PBS and then infected with rotaviruses, reovirus, or poliovirus, as described above. To determine the reconstitution of the susceptibility of the cells to virus infection after drug treatment, the cells were washed twice with PBS at time zero, MEM was added, and the cell monolayers were kept at 37°C. At the indicated times, the cells were washed once with PBS and infected as described above. Cell viability was determined by exclusion of trypan blue (22).
The effect of tunicamycin and benzylGalNAc on the cellular synthesis of N- and O-glycans, respectively, was evaluated by detection of sugars on immunoblots, using a digoxigenin glycan detection kit (Boehringer Mannheim; no. 1500-783). Under the treatment conditions described above, glycoconjugates were reduced by about 50% in both tunicamycin- and benzylGalNAc-treated cells compared to untreated cells (data not shown).
Extraction and immunochemical analyses of lipids from MA104 cells.
PDMP-treated or untreated MA104 cells were harvested by centrifugation and washed twice with PBS. Total lipids were extracted essentially as described by Guo et al. (19). Thin-layer chromatography was carried out in a solvent system of chloroform-methanol-water (5:4:1) containing 12 mM MgCl2. The plastic plate was dried for 2 h at 37°C and then soaked by capillarity in n-hexane containing 10% poly(isobutyl methacrylate) (Aldrich). The glycolipids were then detected immunochemically on the thin-layer chromatograms, as reported previously (30), employing the same carbohydrate detection kit described above. After treatment of MA104 cells with PDMP, as described above, the content of mono- and disialogangliosides was about 30 to 40% of that found in untreated cells (data not shown).
Cholesterol depletion of MA104 cell monolayers.
Confluent MA104 cell monolayers in 96-well plates were washed twice with PBS and then incubated with 10 mM methyl-β-cyclodextrin (Aldrich) in PBS for 1 h at 37°C with occasional shaking. After this time the cells were washed twice with PBS and infected as described above.
To replenish the cells with cholesterol after methyl-β-cyclodextrin treatment, the drug was removed and the cells in 96-well plates were washed twice with PBS and then underwent essentially the same treatment as that described by Falconer et al. (16). Briefly, 200 μl of MEM–7%FBS with or without 0.1 mM cholesterol (5-cholesten-3β-ol-3β-hydroxy-5-cholestene) (Sigma), which was freshly made in 100% ethanol, was added per well and left for the indicated periods. At the end of the incubation time the cells were washed twice with MEM and infected as described above.
To determine the cholesterol content of untreated or cyclodextrin-treated MA104 cells, the cells in suspension were pelleted, the pellet was suspended in 0.8% OG by extensive vortexing, and the suspension was cleared by centrifugation for 5 min at 2,000 × g in an Eppendorf centrifuge. The cholesterol present in the supernatant or in the cyclodextrin extract of cells was assayed spectrophotometrically using the Boehringer Mannheim diagnostic kit (no. 139050). All cholesterol determinations were made in the presence of 0.2% OG.
OG treatment of MA104 cell monolayers.
Confluent MA104 cell monolayers in 96-well plates were washed twice with PBS. The cells were then incubated with 0.2% OG (Pierce) in MEM for 90 min at room temperature with occasional shaking. After this time, the cells were washed twice with PBS and infected as described above. To determine the cell viability and the degree of cell membrane permeabilization that may have been caused by the detergent, we evaluated the ability of the cells to exclude the vital dye trypan blue and the level of the cytoplasmic enzyme lactate dehydrogenase in the OG extract (25). Treatment of cells with 0.2% OG was shown to release less than 5% of the total lactate dehydrogenase activity. A 100% lysis was determined by homogenization of the cells in 0.2% OG.
Binding assay.
Rotavirus binding was determined by a nonradioactive assay, essentially as described by Zárate et al. (59). Briefly, a suspension of 5 × 104 MA104 cells either untreated, previously treated with PDMP or tunicamycin, or extracted with OG or cyclodextrin, as described above, was incubated for 1 h at 4°C with 300 ng of trypsin-activated purified viruses in MEM–1% bovine serum albumin. The cell-virus complexes were washed three times with ice-cold PBS containing 0.5% bovine serum albumin. During the last wash, the cells were transferred to a fresh Eppendorf tube and then lysed in 50 mM Tris (pH 7.5)–150 mM NaCl–0.1% Triton X-100. The viruses present in the lysates were quantified by an enzyme-linked immunosorbent assay (59). In all assays, a binding control with no cells was performed.
To assay the binding-blocking activity of the OG extract, 300 ng of purified virus particles was incubated with 20 μg of OG-extracted proteins per ml for 90 min at 37°C. The virus-OG extract mixture was then added to MA104 cells in suspension, and the assay was performed as described above. The blocking activity of the hyperimmune sera to the 57- and 75-kDa protein fractions (see below) was assayed by preincubating the MA104 cells with a 1:5 dilution of the corresponding preimmune or hyperimmune sera for 1 h at 4°C. After the cells were washed with PBS, the viruses were added and the assay was carried out as described above.
Effect of the OG extract on rotavirus infectivity.
Confluent MA104 cell monolayers in T-flasks were washed twice with PBS–0.5 mM EDTA and left to detach in this buffer for 30 min at 37°C. The cells were counted, pelleted at 85 × g for 5 min at 4°C, resuspended at a concentration of 2.2 × 107 to 2.5 × 107 cells/ml in MEM–0.2% OG, and incubated with gentle shaking for 90 min at room temperature. After this time the cells were pelleted, and the concentration of extracted proteins in the supernatant was determined by the method of Lowry (Bio-Rad); a typical concentration was approximately 5 μg of protein/106 cells. The inhibitory activity of this extract on the infectivity of rotaviruses was measured by incubating dilutions of the extract in MEM with the virus for 90 min at 37°C. As a control, the viruses were incubated with 0.2% OG in MEM. To test for the specificity of inhibition, reovirus and poliovirus were assayed in the same manner as were rotaviruses. The biochemical nature of the inhibitory factor present in the OG extract from untreated cells was determined by boiling (95°C) for 15 min or by incubation of the extract (50 μg of protein/ml) with 2 mg of tosyl phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma) per ml for 1 h at 37°C or with 36 mU of neuraminidase per ml for 2 h at 37°C.
Preparative gel electrophoresis.
The proteins extracted from about 5 × 107 cells (in 2 ml of 0.2% OG) were adjusted with nonreducing Laemmli sample buffer to give the following final concentrations: 62.5 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol, and 0.025% bromophenol blue. These proteins were immediately loaded, without heating, in a single lane of a 3-mm-thick, 14-cm-wide preparative SDS–11% polyacrylamide gel. The gel was run at 8 mA until the bromophenol blue ran out of the gel. After electrophoresis, the gel was stained in 1% Coomassie blue R-250 in water (22) and slices about 3 mm wide were cut and minced in PBS; the proteins in the gel pieces were eluted into PBS by mild shaking for 48 h at room temperature. The eluted proteins were split into several aliquots, precipitated with 5 volumes of acetone, washed twice with 80% cold acetone, and dried for 1 min in a Savant evaporator. To analyze the precipitated proteins, one protein aliquot was resuspended in reducing (5% β-mercaptoethanol) Laemmli sample buffer, boiled for 3 min, and run in an SDS–11% polyacrylamide gel. The ability of the eluted proteins to block rotavirus infectivity was tested, as described above, using a second protein aliquot resuspended in MEM with 1 mM β-mercaptoethanol. After the first round of gel purification, the protein fractions with inhibitory activity were run in a second preparative 7% polyacrylamide gel and all the Coomassie blue-stained bands were cut out again, eluted, and assayed for inhibitory activity. After three rounds of preparative gel electrophoresis, five protein bands, all of which blocked rotavirus infectivity, were isolated (see Fig. 8). In each case, after the bands had been cut out, the proteins were eluted, acetone precipitated, and resuspended in MEM–1 mM β-mercaptoethanol, as described above. Starting from the second preparative gel, the proteins were recovered by electroelution: the gel slices were immersed in sample buffer (2% SDS, 19.2 mM glycine, 2.5 mM Tris base) and electroeluted in an ISCO chamber for 45 min (3 W) using 0.1% SDS–192 mM glycine–25 mM Tris base as a running buffer.
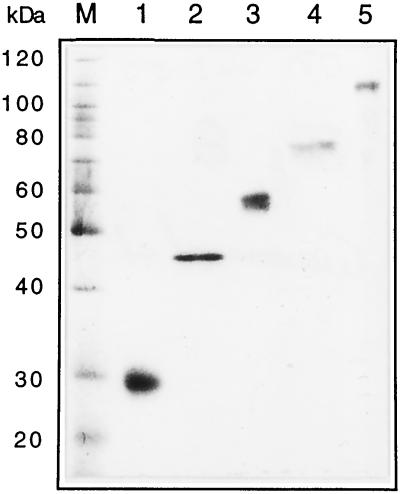
FIG. 8.
Isolated proteins with inhibitory activity for rotavirus infectivity. The protein bands that were shown to block rotavirus infectivity after three rounds of purification by preparative gel electrophoresis (see Table 4) were analyzed in an 11% polyacrylamide gel under reducing conditions. The protein bands were detected by silver staining.
Preparation of polyclonal antibodies.
The proteins eluted from fractions 6 (∼57 kDa) and 10 (∼75 kDa) (see Fig. 5A), which were shown to have the maximal inhibitory activity for rotavirus infection, were used to immunize rabbits, as described previously (22). Briefly, New Zealand White rabbits (3 to 4 kg) were immunized subcutaneously with 500 μg of protein in Freund's complete adjuvant. Two booster injections were given subcutaneously at 2-week intervals with the same amount of protein emulsified in Freund's incomplete adjuvant. The rabbits were bled after the third immunization. A sample of serum was obtained from each animal before immunization.
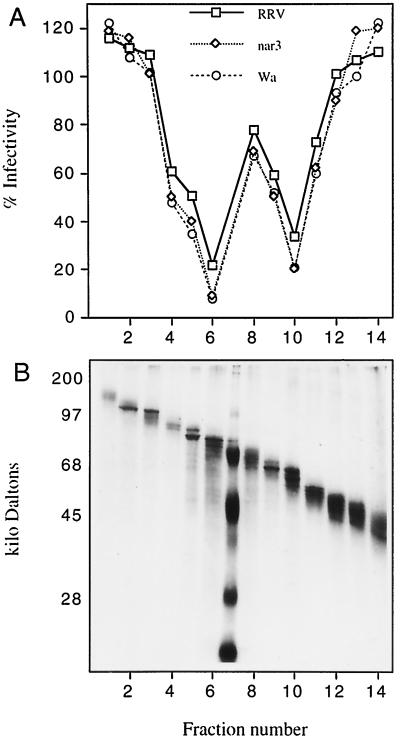
FIG. 5.
Inhibition of rotavirus infectivity by OG-extracted proteins fractionated by gel electrophoresis. About 250 μg of proteins extracted with 0.2% OG from MA104 cells was separated by preparative SDS-polyacrylamide gel electrophoresis under nonreducing conditions. After electrophoresis, the gel was stained with Coomassie blue in water, gel slices were cut out, and the proteins were eluted. (A) Inhibitory activity of the eluted proteins present in the fractions shown in panel B. (B) Gel electrophoresis of the eluted protein fractions. Only the portion of the gel where inhibitory activity was found is shown; the remaining higher- and lower-molecular-mass protein fractions had no inhibitory activity.
The ability of the sera to block rotavirus infectivity was assayed by incubating dilutions of the sera with monolayers of MA104 cells in 96-well plates for 90 min at 37°C. The cells were washed twice with PBS and then infected as described above. The preimmune sera were used as negative controls. The hyperimmune sera were tested for their ability to recognize viruses RRV, nar3, and Wa by an enzyme-linked immunosorbent assay, as described by Menchaca et al. (37); at the lowest dilution tested (1:100), no reactivity was found (data not shown). These antisera did not inhibit the hemagglutination of RRV and nar3 (data not shown).
Western immunoblotting.
The proteins present in the 0.2% OG extract were separated in an 11% polyacrylamide gel and transferred to nitrocellulose. The transferred proteins were incubated with the sera to the 57- and 75-kDa fractions, diluted 1:1,000 in PBS. The bound antibodies were developed by incubation with protein A-peroxidase, and 3-amino-9-ethyl-carbazole (Sigma) was added as a substrate, as previously described by Arias et al. (1).
Immunofluorescence.
MA104 cells grown on glass coverslips to approximately 80% confluence were fixed with 4% paraformaldehyde in PBS for 20 min at 37°C. After this time the cells were washed twice with PBS, either permeabilized or not by incubation with PBS–0.5% Triton X-100 for 5 min at room temperature, and then washed twice with PBS with gentle swirling. The fixed cells were blocked with 1 M glycine for 1 h at 37°C, washed twice with PBS, and then incubated with a 1:1,000 (anti-57-kDa fraction) or 1:1,500 (anti-75 kDa fraction) dilution of the sera for 90 min at 37°C. The cells were washed four times with PBS and then incubated in the dark for 1 h at 4°C with a goat anti-rabbit immunoglobulin G coupled to fluorescein isothiocyanate (Dako Co.), diluted 1:100 in PBS. The cells were washed four times with PBS and mounted on glass slides on 10% glycerol in PBS. The slides were analyzed using a Bio-Rad MRC-600 microscope. The preimmune sera were used as negative controls.
RESULTS
Inhibitors of N glycosylation and glycolipid synthesis block rotavirus infection.
To assess the biochemical nature of the cellular receptor for rotaviruses, MA104 cells were treated with specific inhibitors of glycosylation prior to infection. Two inhibitors were used: tunicamycin, which blocks an early step in the N-glycosylation pathway involving transfer between UDP-GlcNAc and dolichol-1-phosphate (12), and benzylGalNAc, which is a competitive inhibitor of the transferase (N-acetyl-α-d-galactosaminyltransferase) involved in the first step of the biosynthesis of most types of O-linked carbohydrates (5). In addition, we used the synthetic analog of ceramide, PDMP, to inhibit the biosynthesis of the glycosphingolipid precursor glucosylceramide (45). The cells pretreated with the inhibitors were then infected with either wild-type RRV, the neuraminidase-resistant RRV variant nar3, or the HRV strain Wa.
Treatment of cells with 2 μg of tunicamycin per ml for 24 h before infection inhibited the infectivity of rotaviruses RRV and nar3 by about 50%, while preincubation of the cells for 3 days with PDMP, the inhibitor of glycolipids synthesis, blocked the infectivity of the viruses by about 80% (RRV and Wa) or 60% (nar3) (Table 1). On the other hand, inhibition of O glycosylation by benzylGalNAc had no effect on the infectivity of RRV but increased the infectivity of nar3 and Wa by about 50%, indicating that under conditions where the levels of cell surface O-linked carbohydrates are decreased, these viruses infect the cell more efficiently. The total cell content of N- and O-glycoproteins was reduced by at least 50% by the corresponding inhibitory drug (data not shown). The infectivity of reovirus and poliovirus, which were used as controls, was not inhibited by any of these three drugs, with poliovirus actually showing a twofold increase in infectivity in the cells treated with tunicamycin (Table 1), as has been reported for other viruses like human immunodeficiency virus type 2 and B-lymphotropic papovavirus (28, 43).
TABLE 1.
Effect of metabolic inhibitors, cell membrane cholesterol depletion, and OG on the infectivity of rotaviruses in MA104 cells
| Inhibitora | % Infectivity (SE)b of:
|
||||
|---|---|---|---|---|---|
| RRV | nar3 | Wa | Reovirus | Poliovirus | |
| None | 100 | 100 | 100 | 100 | 100 |
| PDMP (25 μg/ml) | 20 (2) | 40 (9.4) | 23 (3.8) | 95 (3) | 114 (0) |
| Tunicamycin (2 μg/ml) | 56 (2.5) | 48 (2.8) | —c | 91 (5.5) | 192 (15) |
| BenzylGalNAc (2 mM) | 101 (0.5) | 150 (4.8) | 147 (7.2) | 110 (4.5) | 108 (4.5) |
| OG (0.2%) | 41 (5.4) | 41 (2.4) | 39 (4.8) | 89 (2) | 199 (29) |
| β-Cyclodextrin (1 mM) | 9 (1.8) | 6 (2.3) | 5 (1.8) | 96 (0) | 95 (3) |
MA104 cell monolayers were incubated with the indicated concentration of inhibitor for 1 h (β-cyclodextrin), 24 h (tunicamycin), or 72 h (PDMP and benzylGalNAc) at 37°C or for 90 min (OG) at room temperature before virus infection.
SE, one standard error of the mean of at least three independent experiments carried out in duplicate.
The infectivity of Wa was inhibited by about 50% regardless of whether tunicamycin was added to the cells 24 h before or immediately after the virus adsorption; thus, this inhibition was considered nonspecific.
Under the conditions employed, the inhibitors did not have a significant effect on cell protein synthesis, as judged by electrophoresis of 35S-labeled proteins, or on the viability of cells, as judged by trypan blue exclusion (data not shown). To control for a possible nonspecific, toxic effect of the drugs on the replication of rotaviruses, in a separate experiment we added the inhibitors immediately after the virus had been adsorbed for 45 min at 4°C. Under these conditions the drugs did not affect rotavirus infectivity, with the exception of rotavirus Wa, whose infectivity was decreased about 50% by tunicamycin; for this reason, this inhibition was considered to be nonspecific. The effect of the inhibitors was reversible since the cells became fully susceptible to rotavirus infection by about 20 and 24 h after removing tunicamycin and PDMP, respectively (data not shown). Taken together these results suggest that glycolipids and N-glycosylated but not O-glycosylated proteins are important for rotavirus infection. To determine if the treatment of cells with tunicamycin and PDMP inhibited the attachment of the virus to the cell surface or if the inhibition of infectivity occurred at a postattachment step, we performed binding assays using cells treated with the different drugs. We found that treatment of cells with tunicamycin did not affect the binding of either of the three viruses tested while treatment of cells with PDMP did not affect the attachment of RRV and Wa but decreased the binding of nar3 by 54% (Table 2). This level of inhibition in the attachment of the virus to cells is very similar to the 60% inhibition in the infectivity of nar3 caused by PDMP (Table 1), which suggests that most if not all of the blockage in the infectivity of nar3 in PDMP-treated cells is due to an inhibition of the binding of this variant to the cell surface.
TABLE 2.
Effect of metabolic inhibitors, cell membrane cholesterol depletion, and OG on the binding of rotaviruses to MA104 cells
| Inhibitora | % Binding (SE)b of virus strain:
|
||
|---|---|---|---|
| RRV | nar3 | Wa | |
| None | 100 | 100 | 100 |
| PDMP (25 μg/ml) | 110 (19) | 46 (20) | 104 (12.5) |
| Tunicamycin (2 μg/ml) | 111 (14) | 101 (12.5) | 94 (21) |
| Octyl-β-glucoside (0.2%) | 32 (4.5) | 40 (7.5) | 33 (0.5) |
| β-cyclodextrin (1 mM) | 112 (6.5) | 109 (16) | 116 (4.5) |
MA104 cell monolayers were incubated with the indicated concentration of inhibitor for 1 h (β-cyclodextrin), 24 h (tunicamycin), or 72 h (PDMP) at 37°C or for 90 min (OG) at room temperature before the assay.
SE, one standard error of the mean of at least three independent experiments carried out in duplicate.
Infection of octyl-β-glucoside-extracted cells.
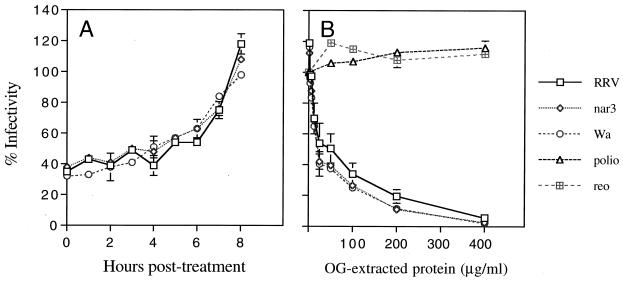
As a different approach to characterize the rotavirus receptor, we used the nonionic detergent OG to extract the receptor from the cell membrane under noncytolytic conditions, as has been described for other virus receptors (2, 10, 23, 36, 49, 55). MA104 cells were incubated with 0.2% OG for 90 min at room temperature; under these conditions the cells maintained their viability and integrity, as judged by trypan blue exclusion and the low levels of lactate dehydrogenase activity, a cytosolic marker detected in the OG extract (less than 5% of total enzyme activity). The cells extracted with the detergent were found to be about 60% refractory to infection by the three viruses tested (Table 1). As described above for the metabolic inhibitors, this effect was also found to be reversible; if the detergent was washed away after the treatment period, the cells fully regained their susceptibility for infection at about 8 h posttreatment (Fig. 1A), which most probably accounts for the time of synthesis, transport, and accumulation of the receptor in the cell membrane at the levels needed for the virus to efficiently infect the cell. Of interest, the attachment of all three viruses to OG-extracted cells was inhibited by 60 to 70% (Table 2), indicating that the reduced infectivity of the viruses in OG-treated cells might be due to a decreased ability of the virus particles to bind to the cell surface.
FIG. 1.
(A) Recovery of the susceptibility of MA104 cells to rotavirus infection after extraction with OG. Cell monolayers in 96-well plates were extracted with 0.2% OG and allowed to recover in MEM at 37°C. At the indicated times, the monolayers were washed with PBS and infected with rotaviruses. (B) Inhibition of rotavirus infectivity by the OG extract from MA104 cells. The indicated concentrations of OG-extracted protein were incubated with the viruses for 90 min at 37°C. The virus-protein mixtures were used to infect MA104 cell monolayers in 96-well plates. In both panels, the percent infectivity is relative to the infectivity of the viruses incubated in 0.2% OG. Error bars represent 1 standard error of the mean of three or more experiments carried out in duplicate.
The OG extract inhibits rotavirus infection.
Since treatment of MA104 cells with OG diminished the ability of the viruses to attach to and thus to infect cells, it is likely that the detergent was extracting cell surface molecules involved in the initial interaction of rotaviruses with the cell, possibly the rotavirus receptor(s). If this were the case, the molecule(s) present in the OG extract could interact with the virus in solution, preventing the binding of the virus to the cell membrane and thus blocking its infectivity. We found that incubation of the OG extract with either of the three rotavirus strains did block their infectivity in a concentration-dependent manner (Fig. 1B). At the maximum concentration tested, 400 μg of protein per ml, the infectivity of the viruses was inhibited by about 95%; 50% inhibition was achieved at about 40 μg of protein per ml. In contrast, the infectivity of poliovirus and reovirus was not affected by the extract (Fig. 1B). These results strongly suggest a specific interaction of the viruses with the OG-solubilized cell surface molecules. Preincubation of the viruses with a solution of 0.2% OG did not affect their infectivity. The infectivity in the presence of OG was taken as the 100% value for each virus.
The inhibition of rotavirus infectivity caused by the OG extract seems to be due to a blockage in cell attachment since preincubation of the viruses with 20 μg of the OG-extracted protein per ml decreased RRV binding to the cell by 40%, nar3 binding by 41%, and Wa binding by 43% (Table 3). These percentages are in close agreement with the degree of inhibition of infectivity achieved with this concentration of extract (Fig. 1B).
TABLE 3.
Effect of the OG extract, and antibodies to 75 kDa OG protein fraction, on the binding of rotaviruses to MA104 cellsa
| Inhibitor | % Binding (SE)b of virus strain:
|
||
|---|---|---|---|
| RRV | nar3 | Wa | |
| 0.2% OG (control) | 100 | 100 | 100 |
| OG extract (20 μg/ml) | 60 (2.5) | 59 (4) | 57 (1.5) |
| No serum (control) | 100 | 100 | 100 |
| Polyclonal antibodies to the 75-kDa OG fraction | |||
| Preimmune serum | 102 (10) | 97 (8) | 105 (5.5) |
| Hyperimmune serum | 92 (6.5) | 68 (2.5) | 28 (3.5) |
Rotaviruses were incubated with the indicated concentration of OG-extracted proteins for 90 min at 37°C. The virus-OG extract mixture was then added to MA104 cells in suspension, and the assay was performed as described in Materials and Methods. The blocking activity of the hyperimmune sera to the 75-kDa protein fractions was assayed by preincubating the MA104 cells with a 1:5 dilution of the preimmune or hyperimmune sera for 1 h at 4°C. After the cells were washed, the viruses were added and the assay was carried out as described in Materials and Methods.
SE, one standard error of the mean of at least three independent experiments carried out in duplicate.
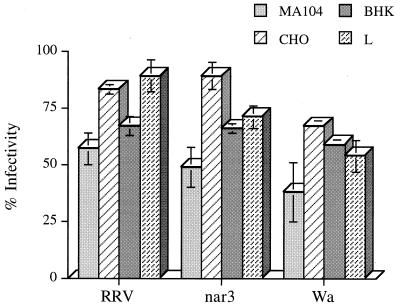
The inhibitory capacity of OG extracts obtained from BHK, CHO, and L cells, which are about 1,000-fold less susceptible to rotavirus infection than MA104 cells, was determined. As can be seen in Fig. 2, the OG extracts from the three poorly permissive cells showed some inhibitory activity, although in all cases this activity was less pronounced than that observed with the extract from MA104 cells.
FIG. 2.
Inhibition of rotavirus infectivity by OG extracts from cells poorly permissive to rotavirus infection. OG-extracted proteins (20 μg/ml) from CHO, BHK, L, or MA104 cells (as indicated) were incubated with the viruses for 90 min at 37°C. The virus-protein mixtures were used to infect MA104 cell monolayers in 96-well plates. The percent infectivity is relative to the infectivity of the viruses incubated in 0.2% OG. Error bars represent 1 standard error of the mean of three experiments carried out in duplicate.
Biochemical nature of the inhibitory component present in the MA104 OG cell extract.
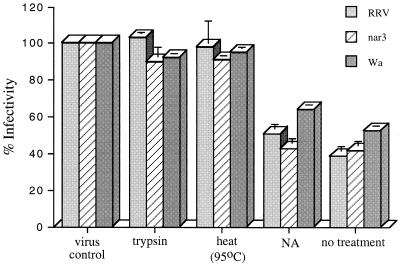
To determine the biochemical nature of the inhibitory component present in the OG cell extract, we tested the effect of heat inactivation, neuraminidase, and proteolytic treatment on the inhibitory activity of the extract. We found that either boiling for 15 min or treatment with trypsin completely abolished the inhibitory activity while treatment with neuraminidase had no effect on the blocking capacity of the extract (Fig. 3). These results indicate that the inhibitory component of the extract is a protein.
FIG. 3.
Biochemical nature of the inhibitory factor present in the OG extract. A 0.2% OG extract was obtained from cells in suspension. Just prior to the incubation with the virus, the extract was either boiled (95°C) for 15 min (heat), incubated with 2 mg of trypsin per ml of extract for 1 h at 37°C (trypsin), or incubated with 36 mU of neuraminidase per ml (NA). The untreated extract (no treatment) was used as a positive control. Viruses and extract (100 μg of protein extract per ml of virus) were mixed and incubated for 90 min at 37°C, and then MA104 cells in 96-well plates were infected with the virus-protein mixtures. The percent infectivity is relative to the infectivity of viruses incubated with a solution of 0.2% OG in MEM (virus control). Error bars represent 1 standard error of the mean of three or more experiments carried out in duplicate.
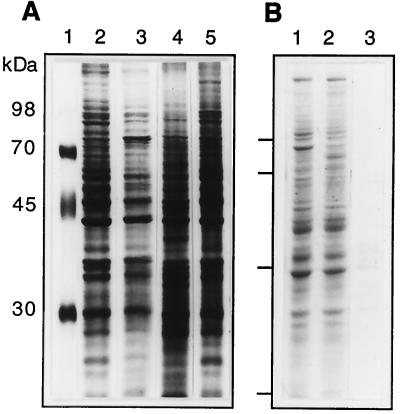
The profile of proteins extracted with OG is shown in Fig. 4A, lane 5. Treatment with tunicamycin, PDMP, or neuraminidase modifies this profile (lanes 2 to 4), reflecting the modification in the carbohydrate content of glycoproteins caused by tunicamycin and neuraminidase. In the case of PDMP, this result suggests that the impaired synthesis of glycolipids alters either the transport of proteins to the plasma membrane or their extractability from the cell surface by OG. In this regard, it is of interest that the OG extract from PDMP-treated cells failed to block rotavirus infectivity (data not shown), suggesting that the inhibitory proteins could not be extracted from these cells.
FIG. 4.
Analysis of the proteins extracted from MA104 cells. Cell monolayers were treated with either neuraminidase, tunicamycin, or PDMP, as described in Materials and Methods, and the cells were then extracted with 0.2% OG for 90 min at room temperature. (A) The extracted proteins were separated by electrophoresis under reducing conditions in an SDS–11% polyacrylamide gel and silver stained. OG-extracted proteins from MA104 cells treated with neuraminidase (lane 2), tunicamycin (lane 3), or PDMP (lane 4) or left untreated (lane 5) are shown. Lane 1 contains molecular mass markers. (B) Cells in suspension were extracted with 10 mM β-cyclodextrin for 1 h at 37°C, as described in Materials and Methods. Proteins in untreated cells (lane 1), extracted cells (lane 2), and the cyclodextrin extract (lane 3) were analyzed by gel electrophoresis.
Cholesterol depletion of MA104 cells inhibits rotavirus infectivity.
It has been proposed that glycosphingolipids, cholesterol, and proteins can interact specifically in cell membranes to form microdomains termed rafts (46). Given the involvement of glycolipids and N-glycosylated proteins on rotavirus infectivity, we tested if depletion of the cell cholesterol would have any effect on virus infectivity. To do this, we incubated the cells with 10 mM β-cyclodextrin for 1 h at 37°C; this treatment has been shown to selectively extract cholesterol from the plasma membrane in preference to other membrane lipids (28). Under these conditions, about two-thirds (65%) of the cell cholesterol was removed (see Materials and Methods). The treatment of cells with β-cyclodextrin inhibited the infectivity of RRV, nar3, and Wa rotavirus strains by more than 90% but had no effect on the infectivity of reovirus and poliovirus (Table 1). It is noteworthy that the binding of the three rotavirus strains was not affected (Table 2), indicating that the decrease in the cholesterol content of the cell affects virus infectivity at a postattachment step.
The protein profile of cells treated with β-cyclodextrin was not very different from that of untreated cells (Fig. 4B, lanes 1 and 2), even though this antibiotic extracted a small amount of protein from the cells (lane 3).
To demonstrate that the depletion of cholesterol was the cause of the reduction of virus infectivity after the β-cyclodextrin treatment, cells in 96-well plates were washed twice with MEM, and then either MEM alone, MEM–7% FBS, or MEM–7% FBS containing 0.1 mM cholesterol was added for different times. At the end of the incubation period, the cells were washed twice and infected with rotaviruses. At 8 h posttreatment, the cells incubated in the presence of cholesterol had fully recovered their susceptibility to rotaviruses while the cells incubated with MEM alone or MEM–7% FBS were still about 80 and 50% refractory to rotavirus infection, respectively (data not shown).
Fractionation of the inhibitory components present in the OG extract from MA104 cells.
To characterize the proteins that block rotavirus infection, we fractionated the OG extract obtained from MA104 cells by preparative SDS-polyacrylamide gel electrophoresis. After the gel electrophoresis, slices of a single-lane gel were cutout, and the proteins were eluted in PBS, concentrated by precipitation with acetone, and resuspended in PBS with 1 mM β-mercaptoethanol. This method has been successful for recovering proteins with enzymatic activity (20, 48). The proteins obtained from the different fractions (Fig. 5B) were tested for their ability to block rotavirus infection. Proteins eluted from two well-defined regions of the gel, around 57- and 75 kDa, had the ability to efficiently inhibit the infectivity of all three rotaviruses tested (Fig. 5A). The pattern of inhibition observed in Fig. 5A was found to be consistent in independent gel fractionation experiments.
Antibodies to the OG extract protein fractions inhibit rotavirus infection.
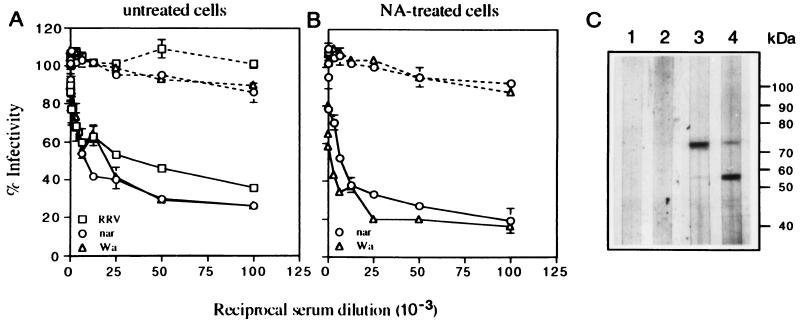
Protein fractions 6 and 10 in Fig. 5A, which represent the peak of inhibitory activity, were used to immunize rabbits. The hyperimmune sera obtained against these two fractions were found to block the infectivity of all three strains of rotavirus when preincubated with the cells for 90 min at 37°C prior to addition of the virus, while the preimmune sera had no effect (shown for the serum to the 75-kDa protein fraction in Fig. 6A). The inhibitory effect of the two antisera was not additive since a mixture of the two inhibited rotavirus infectivity by about 70% at a dilution of 1/100 (data not shown). Of interest, both sera blocked the infectivity of HRV Wa and that of the SA-independent variant nar3 in cells treated with neuraminidase (shown for the serum to the 75-kDa fraction in Fig. 6B), suggesting that they contain antibodies to an SA-independent rotavirus receptor. In a binding inhibition assay, the serum to the 75-kDa fraction did not inhibit the attachment of rotavirus RRV to MA104 cells but inhibited 32% of the binding of nar3 and 72% of that of Wa (Table 3). The blocking specificity of these antisera was confirmed by the following assays: they did not recognize any of the three viruses by enzyme-linked immunosorbent assay, and they did not inhibit the hemagglutination activity of RRV and nar3. Furthermore, the sera were shown not to inhibit the infectivity of poliovirus or that of reovirus in an FFU reduction assay as described in Materials and Methods for rotavirus (data not shown).
FIG. 6.
Inhibitory activity of hyperimmune sera to OG-extracted proteins. (A and B) OG protein fractions 6 and 10 shown in Fig. 5B, containing polypeptides of around 57 and 75 kDa, respectively, were used to raise antibodies in rabbits. Serial dilutions of the preimmune (dashed lines) and hyperimmune (continuous lines) sera to the 75-kDa protein fraction were incubated with untreated (A) or neuraminidase (NA)-treated (B) MA104 cells for 90 min at 37°C before addition of the virus. Similar inhibition results were obtained with the serum to the 57-kDa protein fraction (data not shown). Error bars represent 1 standard error of the mean of three or more experiments carried out in duplicate. (C) Immunoblot analysis of the OG-extracted proteins. The proteins extracted from MA104 cells with 0.2% OG were separated in an SDS–11% polyacrylamide gel under reducing conditions and transferred to nitrocellulose. The transferred proteins were incubated with a 1,000-fold dilution of the preimmune (lanes 1 and 2) or hyperimmune (lanes 3 and 4) sera to the 57-kDa (lanes 2 and 4) or 75-kDa (lanes 1 and 3) protein fractions. The bound antibodies were developed by incubation with protein A-peroxidase and a chromogenic substrate.
By Western blotting, the sera to the 75-kDa fraction recognized a protein of about 73 kDa and, to a lesser extent, a protein of about 57 kDa in the 0.2% OG cell extract (Fig. 6C, lane 3). Of interest, the serum to the 57-kDa protein fraction also recognized proteins of 73 and 57 kDa, although the latter protein was recognized more efficiently by this serum (lane 4). The preimmune sera did not recognize any of these proteins (lanes 1 and 2).
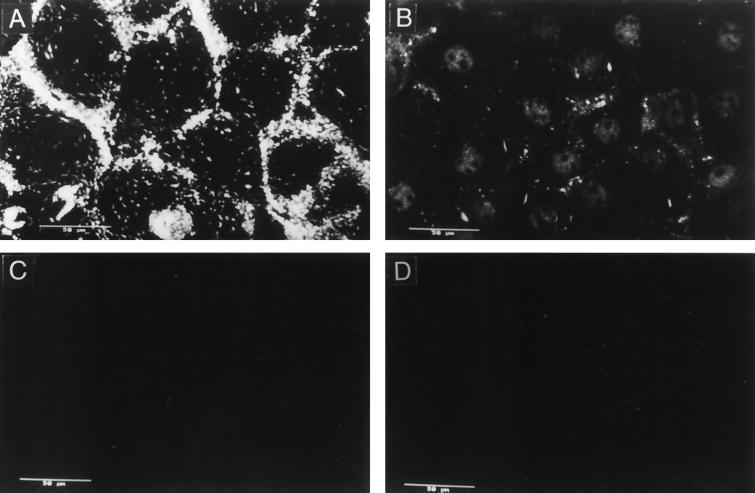
The hyperimmune sera were shown to recognize proteins on the surface of the MA104 cells, as judged by their reactivity with nonpermeabilized cells by flow cytometry (data not shown) and by indirect immunofluorescence (shown for the anti-75-kDa serum in Fig. 7). The pattern of immunofluorescence (for both anti-57- and anti-75-kDa sera) was patchy over the surface of the cell, but there was a higher concentration of the fluorescent signal on the intercellular junctions (Fig. 7A). In permeabilized cells, a weak signal associated mainly with the nuclei was found (Fig. 7B). No fluorescent signal was detected when the preimmune sera were used to stain either permeabilized or nonpermeabilized cells (Fig. 7C and D).
FIG. 7.
Immunofluorescence of cells incubated with the serum to the 75-kDa OG protein fraction. MA104 cells were fixed with paraformaldehyde and permeabilized with Triton X-100 (B and D) or not permeabilized (A and C). The cells were incubated with a 1:1,500 dilution of the preimmune (C and D) or hyperimmune (A and B) sera to the 75-kDa protein fraction for 90 min at 37°C and stained with a goat anti-rabbit immunoglobulin G coupled to fluorescein isothiocyanate.
Purification of the cellular proteins which block rotavirus infectivity.
The proteins with rotavirus blocking activity were purified by SDS-polyacrylamide gel electrophoresis from an OG extract obtained from MA104 cells. After three rounds of purification by gel electrophoresis, using the inhibitory activity of the proteins as marker, we were able to isolate five bands with molecular masses of approximately 110, 75, 57, 45, and 30 kDa (Fig. 8) which were able to inhibit the infectivity of all three rotavirus strains tested. Although these proteins consistently inhibited rotavirus infectivity through the rounds of purification carried out, the final amount of protein recovered was small, which prevented us from determining the precise specific inhibitory activity for each protein and testing if they were recognized by the hyperimmune sera. Table 4 shows the results of a blocking infectivity assay with the purified proteins; in this blocking assay, the same amount of protein shown in the gel in Fig. 8 was used. The relative inhibitory activity of each protein for all three viruses was found to be 75 kDa > 110 kDa > 45 kDa = 30 kDa > 57 kDa. Given the fractionation method employed, it is quite possible that each of these bands may represent more than one protein species.
TABLE 4.
Blocking of rotavirus infectivity by purified OG-extracted proteins from MA104 cells
| Protein (kDa) | Relative amta of protein | % Infectivityb of virus strain:
|
||
|---|---|---|---|---|
| RRV | nar3 | Wa | ||
| 110 | 2 | 50 | 51 | 38 |
| 75 | 1 | 41 | 17 | 29 |
| 57 | 5 | 62 | 50 | 68 |
| 45 | 10 | 55 | 23 | 31 |
| 30 | 15 | 44 | 25 | 23 |
The amount of each protein incubated with the rotavirus strains is the same as that shown in the gel of Fig. 8. The 75-kDa band was the least abundant and was set at 1 (about 10 ng) in relative terms. The concentration of this protein during the infectivity assay was about 100 ng/ml.
The infectivity-blocking assay was carried out only once due to the small amount of material available.
DISCUSSION
The entry of rotaviruses into MA104 cells seems to be a multistep process involving interactions of the virus surface protein VP4 and maybe of VP7 with more than one cell surface site present in either the same or a different cellular structure(s) (11, 41).
In the present study we employed two approaches to characterize the biochemical nature of the rotavirus receptor(s). In the first approach we used metabolic inhibitors of glycosylation and synthesis of glycolipids to study their effect on the infectivity of three different rotavirus strains. We found that tunicamycin, an inhibitor of protein N glycosylation, diminished the infectivity of rotaviruses RRV and nar3 despite their differential dependence on SA for infectivity, implying that these viruses interact with N-linked glycoproteins at some point during cell entry. The fact that the treatment of cells with this drug did not affect the binding of the viruses suggests that the blockage occurs after the initial attachment of the virion to the cell surface. Tunicamycin has been successfully used to specifically analyze the role of N-glycans as receptors for several viruses (7, 28, 43, 44).
Treatment of MA104 cells with PDMP, an inhibitor of glycolipid biosynthesis, resulted in the more pronounced inhibition of infectivity observed for all three rotavirus strains (Table 1). Interaction of rotaviruses with gangliosides GM1 and GM3 has been reported (47, 52), and this interaction has been shown to be SA dependent. In this case, however, the inhibition caused by PDMP seems not to be the result of a deficient attachment of the SA-dependent rotavirus RRV to the cell surface since it was not significantly affected (Table 2). This observation suggests that RRV does not interact, or at least does not interact exclusively, with the SA present on PDMP-sensitive gangliosides. On the other hand, the binding of the SA-independent variant nar3 was decreased in PDMP-treated cells, suggesting that either glycosphingolipids or, more probably, a protein whose correct transport or conformation depends on their presence might be used by nar3 to attach to the cell. Of interest, the binding of the HRV strain Wa was not affected by PDMP, in agreement with the suggestion that nar3 and Wa, despite having an infectivity resistant to neuraminidase treatment of cells, bind to different cell surface sites (41). Finally, the fact that PDMP, but not tunicamycin, affected the attachment of nar3 suggests that the inhibition caused by the N-glycosylation inhibitor is not due to its reported ability to inhibit ganglioside biosynthesis (50, 58).
In addition to the involvement of N-glycosylated proteins and glycolipids in rotavirus entry, we found that cholesterol depletion inhibited the infectivity of rotaviruses by more than 90% (Table 1). These findings are of interest with regard to the recent description of functional lipid microdomains, or rafts, in the cell membrane (51). These rafts have been proposed to be composed of cholesterol, glycosphingolipids (gangliosides among others), and a specific set of associated proteins. They are thought to function as specialized platforms for apical cell sorting of proteins and signal transduction. For some proteins which form part of these lipid microdomains (oxytocin receptor, placental alkaline phosphatase, gD1 decay-accelerating factor), the disassembly of the rafts by cholesterol depletion disrupts or modifies the receptor activity, even though the receptor might be present in the same abundance on the cell membrane (21, 29). In this regard, the finding that the attachment of RRV, nar3, and Wa to cholesterol-depleted cells is not affected while their infectivity is severely impaired is consistent with the possibility that the rotavirus receptor(s) might be forming part of some of these lipid microdomains. It is tempting to hypothesize that in cholesterol-depleted cells, the receptor(s) retains its ability to bind rotavirus particles but in order to fully promote virus entry it must be organized in a lipid microdomain. In addition, the fact that the OG extract from PDMP-treated cells failed to show inhibitory activity suggests that PDMP treatment may have disrupted the lipid raft organization such than one or more of the active proteins in Fig. 8 never became associated with or localized within these membrane microdomains and as result are not extracted with OG. Experiments are under way to test this hypothesis.
The infectivity of the two nonenveloped viruses that were used as controls, poliovirus and reovirus, was not inhibited by the described drugs, showing that the effect observed on the infectivity of rotaviruses was specific. The human poliovirus receptor is an integral membrane protein with the conserved amino acids and domain structure characteristic of members of the immunoglobulin superfamily (31, 38). The nature of the reovirus receptor is less well defined; most of the available evidence suggest that reovirus binds to multiple sialoglycoproteins rather to a single homogeneous species on the cell surface (8, 18, 42).
In a second approach to characterize the rotavirus receptor, MA104 cells were incubated with a solution of 0.2% OG. It has been shown that at low concentrations, like the one used in this work, OG is able to extract proteins from the cell surface without impairing the viability of the cells (see Results) (23, 36). This nonionic detergent has been useful in experiments to obtain the receptors for Semliki Forest virus, parvovirus, vesicular stomatitis virus, polyomavirus, simian virus 40, and rabies virus from intact cell monolayers (2, 10, 23, 36, 49, 55). MA104 cells extracted with OG lost their ability to bind rotaviruses by about the same extent (60%) to which they became refractory to infection, suggesting that OG extracts from the cell surface the receptor molecules needed by all three strains of rotavirus to attach to and thus infect the cell. In agreement with this finding is the fact that the OG extract, when preincubated with these viruses, inhibited both their binding to and infection of MA104 cells. This suggests that the putative OG-solubilized cell receptors are able to interact with the viruses in solution. The inhibitory activity of the OG extract was lost by treatment with proteases and heat but not by treatment with neuraminidase, indicating that the active component is a protein.
To test for a correlation between the susceptibility of the cell line and the ability of the OG extract to inhibit rotavirus infection, we obtained OG extracts from BHK, CHO, and L cells, which are about 1,000-fold less susceptible to rotavirus infection than are MA104 cells. The extracts from these three cell lines inhibited the infectivity of rotaviruses to different degrees but in general to a lesser extent than that achieved with the MA104 cell extract (Fig. 2). As suggested in this work and by others (11, 41), these results might be explained if more than one cell surface molecule were implicated in rotavirus infection, which would make possible the absence of one of the receptor molecules in the less susceptible cell lines while other surface components, which could be extracted with OG and block rotavirus infectivity, would still be present.
Two protein fractions with blocking activity for rotavirus infectivity were obtained by gel fractionation of the OG extract of the MA104 cells. The hyperimmune sera prepared against these two fractions were shown to react primarily with two polypeptides of 73 and 57 kDa. Although it is not possible to be certain if the more immunogenic proteins are the active inhibitory components of the extract, it seems at least that the inhibitory antibodies present in both hyperimmune sera recognize the same cell surface molecule or different molecules in a protein complex since the blocking efficiency of the individual sera was not additive and since the cell surface recognition patterns obtained with the two antisera were strikingly similar.
Five individual protein bands with inhibitory activity for rotavirus infectivity were isolated from the OG extract. These proteins need to be assayed to test the specificity of their inhibitory activity and to investigate if they are somehow related to each other. However, the fact that all of these proteins block the infectivity of RRV, nar3, and Wa rotaviruses suggest that at least one of them, or a complex formed by more than one, could be a common cellular receptor for rotaviruses. The determination of the identity of these proteins should help to define the cell surface molecules involved in the interactions that seem to occur between rotaviruses and the cell surface during infection.
As a working hypothesis, we propose that the rotavirus receptor is likely to be a complex of several cell components including gangliosides, N-linked glycoproteins, and probably other proteins which might all associate in lipid rafts and need the lipid microdomain organization to function efficiently in the binding and internalization of rotavirus particles. The protein components of this proposed complex could include the integrin molecules that have been reported recently (11, 24).
ACKNOWLEDGMENTS
We thank Rafaela Espinosa for the immunofluorescence experiments, Pavel Isa for the flow cytometric analysis, and Leticia Vega Alvarado for her contribution to the development of the command files to semiautomatically count infected cells.
This work was partially supported by grants 75197-527106 from the Howard Hughes Medical Institute, G0012-N9607 from the National Council for Science and Technology—Mexico, and IN207496/IN201399 from DGAPA-UNAM.
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, López S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak S, Turner H, Parr S. Identification of a 40- to 42-kDa attachment polypeptide for canine parvovirus in A72 cells. Virology. 1994;205:7–16. doi: 10.1006/viro.1994.1614. [DOI] [PubMed] [Google Scholar]
- 3.Bass D M, Mackow E R, Greenberg H B. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology. 1991;183:602–610. doi: 10.1016/0042-6822(91)90989-o. [DOI] [PubMed] [Google Scholar]
- 4.Beisner B, Kool D, Marich A, Holmes I H. Characterisation of G serotype dependent non-antibody inhibitors of rotavirus in normal mouse serum. Arch Virol. 1998;143:1277–1294. doi: 10.1007/s007050050375. [DOI] [PubMed] [Google Scholar]
- 5.Brockhausen I, Moller G, Pollex-Kruger A, Rutz V, Paulsen H, Matta K L. Control of O-glycan synthesis: specificity and inhibition of O-glycan core 1 UDP-galactose:N-acetylgalactosamine-alpha-R-beta-3-galactosyltransferase from rat liver. Biochem Cell Biol. 1992;70:99–108. doi: 10.1139/o92-015. [DOI] [PubMed] [Google Scholar]
- 6.Chen C C, Baylor M, Bass D M. Murine intestinal mucins inhibit rotavirus infection. Gastroenterology. 1993;105:84–92. doi: 10.1016/0016-5085(93)90013-3. [DOI] [PubMed] [Google Scholar]
- 7.Chen M H, Benjamin T. Roles of N-glycans with alpha 2,6 as well as alpha 2,3 linked sialic acid in infection by polyoma virus. Virology. 1997;233:440–442. doi: 10.1006/viro.1997.8596. [DOI] [PubMed] [Google Scholar]
- 8.Choi A H, Paul R W, Lee P W. Reovirus binds to multiple plasma membrane proteins of mouse L fibroblasts. Virology. 1990;178:316–320. doi: 10.1016/0042-6822(90)90412-k. [DOI] [PubMed] [Google Scholar]
- 9.Ciarlet M, Estes M K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80:943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 10.Clayson E T, Compans R W. Characterization of simian virus 40 receptor moieties on the surfaces of Vero C1008 cells. J Virol. 1989;63:1095–1100. doi: 10.1128/jvi.63.3.1095-1100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulson B S, Londrigan S H, Lee D J. Rotavirus contains intergrin ligand sequences and a disintegrin-like domain implicated in virus entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbein A. Inhibitors of glycoprotein synthesis. Methods Enzymol. 1983;98:135–155. doi: 10.1016/0076-6879(83)98144-2. [DOI] [PubMed] [Google Scholar]
- 13.Espejo R, Martínez E, López S, Muñoz O. Different polypeptide composition of two human rotavirus types. Infect Immun. 1980;28:230–237. doi: 10.1128/iai.28.1.230-237.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. 3rd ed. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 1625–1655. [Google Scholar]
- 15.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukudome K, Yoshie O, Konno T. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell absorption. Virology. 1989;172:196–205. doi: 10.1016/0042-6822(89)90121-9. [DOI] [PubMed] [Google Scholar]
- 18.Gentsch J R, Pacitti A F. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology. 1987;161:245–248. doi: 10.1016/0042-6822(87)90192-9. [DOI] [PubMed] [Google Scholar]
- 19.Guo C T, Nakagomi O, Mochizuki M, Ishida H, Kiso M, Ohta Y, Suzuki T, Miyamoto D, Hidar K I, Suzuki Y. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J Biochem. 1999;126:683–688. doi: 10.1093/oxfordjournals.jbchem.a022503. [DOI] [PubMed] [Google Scholar]
- 20.Hager D A, Burgess R R. Elution proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 21.Hannan L A, Edidin M. Traffic, polarity, and detergent solubility of a glycosylphosphatidylinositol-anchored protein after LDL deprivation of MDCK cells. J Cell Biol. 1996;133:1265–1276. doi: 10.1083/jcb.133.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Helenius A, Morien B, Fries E, Simons K, Robinson P, Schirrmacher V, Terhorst C, Strominger J. Human (HLA-A and HLA-B) and murine (H-2d) histocompatibility antigens are cell surface receptors for Semliki forest virus. Proc Natl Acad Sci USA. 1978;75:3846–3850. doi: 10.1073/pnas.75.8.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hewish M J, Takada Y, Coulson B S. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J Virol. 2000;74:228–236. doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson M K, Whittaker V P. Lactate dehydrogenase as a cytoplasmic marker in brain. Biochem J. 1963;88:404–409. doi: 10.1042/bj0880404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D N, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. 3rd ed. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 1657–1708. [Google Scholar]
- 27.Keljo D J, Smith A K. Characterization of binding of simian rotavirus SA-11 to cultured epithelial cells. J Pediatr Gastroenterol Nutr. 1988;7:249–256. doi: 10.1097/00005176-198803000-00015. [DOI] [PubMed] [Google Scholar]
- 28.Keppler O T, Herrmann M, Oppenlander M, Meschede W, Pawlita M. Regulation of susceptibility and cell surface receptor for the B-lymphotropic papovavirus by N glycosylation. J Virol. 1994;68:6933–6939. doi: 10.1128/jvi.68.11.6933-6939.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with β-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 30.Kniep B, Mühlradt P F. Immunochemical detection of glycosphingolipids on thin-layer chromatograms. Anal Biochem. 1990;188:5–8. doi: 10.1016/0003-2697(90)90519-f. [DOI] [PubMed] [Google Scholar]
- 31.Koike S, Horie H, Ise I, Okitsu A, Yoshida M, Iizuka N, Takeuchi K, Takegami T, Nomoto A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9:3217–3224. doi: 10.1002/j.1460-2075.1990.tb07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koketsu M, Nitoda T, Juneja L R, Kim M, Kashimura N, Yamamoto T. Sialyloligosaccharides from egg yolk as inhibitor of rotaviral infection. J Agric Food Chem. 1995;43:858–861. [Google Scholar]
- 33.Lizano M, López S, Arias C F. The amino-terminal half of rotavirus SA114fM VP4 protein contains a hemagglutination domain and primes for neutralizing antibodies to the virus. J Virol. 1991;65:1383–1391. doi: 10.1128/jvi.65.3.1383-1391.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludert J E, Feng N, Yu J H, Broome R L, Hoshino Y, Greenberg H B. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J Virol. 1996;70:487–493. doi: 10.1128/jvi.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludert J E, Mason B B, Angel J, Tang B Z, Hoshino Y, Feng N G, Vo P T, Mackow E M, Ruggeri F M, Greenberg H B. Identification of mutations in the rotavirus protein VP4 that alter sialic-acid-dependent infection. J Gen Virol. 1998;79:725–729. doi: 10.1099/0022-1317-79-4-725. [DOI] [PubMed] [Google Scholar]
- 36.Marriott S J, Griffith G R, Consigli R A. Octyl-β-d-glucopyranoside extracts polyomavirus receptor moieties from the surfaces of mouse kidney cells. J Virol. 1987;61:375–382. doi: 10.1128/jvi.61.2.375-382.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menchaca G, Padilla L, Méndez-Toss M, Contreras J F, Puerto F I, Guiscafré H, Mota F, Herrera I, Cedillo R, Muñoz O, Ward R, Hoshino Y, López S, Arias C F. Serotype specificity of the neutralizing-antibody response induced by the individual surface proteins of rotavirus in natural infections of young children. Clin Diagn Lab Immunol. 1998;5:328–334. doi: 10.1128/cdli.5.3.328-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 39.Méndez E, Arias C F, López S. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J Virol. 1993;67:5253–5259. doi: 10.1128/jvi.67.9.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Méndez E, Arias C F, López S. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J Virol. 1996;70:1218–1222. doi: 10.1128/jvi.70.2.1218-1222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Méndez E, López S, Cuadras M A, Romero P, Arias C F. Entry of rotaviruses is a multistep process. Virology. 1999;263:450–459. doi: 10.1006/viro.1999.9976. [DOI] [PubMed] [Google Scholar]
- 42.Paul R W, Choi A H, Lee P W. The alpha-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology. 1989;172:382–385. doi: 10.1016/0042-6822(89)90146-3. [DOI] [PubMed] [Google Scholar]
- 43.Potempa S, Picard L, Reeves J D, Wilkinson D, Weiss R A, Talbot S J. CD4-independent infection by human immunodeficiency virus type 2 strain ROD/B: the role of the N-terminal domain of CXCR-4 in fusion and entry. J Virol. 1997;71:4419–4424. doi: 10.1128/jvi.71.6.4419-4424.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y M, Peng S W, Hengst K, Evander M, Park D S, Zhou J, Frazer I H. Epithelial cells display separate receptors for papillomavirus VLPs and for soluble L1 capsid protein. Virology. 1996;216:35–45. doi: 10.1006/viro.1996.0032. [DOI] [PubMed] [Google Scholar]
- 45.Radin N S, Shayman J A, Inokuchi J I. Metabolic effects of inhibiting glucosylceramide synthesis with PDMP and other substances. Adv Lipid Res. 1993;26:183–213. [PubMed] [Google Scholar]
- 46.Rolsma M D, Gelberg H B, Kuhlenschmidt M S. Assay for evaluation of rotavirus-cell interactions: identification of an enterocyte ganglioside fraction that mediates group A porcine rotavirus recognition. J Virol. 1994;68:258–268. doi: 10.1128/jvi.68.1.258-268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rolsma M D, Kuhlenschmidt T B, Gelberg H B, Kuhlenschmidt M S. Structure and function of a ganglioside receptor for porcine rotavirus. J Virol. 1998;72:9079–9091. doi: 10.1128/jvi.72.11.9079-9091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheele G, Pash J, Bieger W. Identification of proteins according to biological activity following separation by two-dimensional isoelectric focusing/sodium dodecyl sulfate gel electrophoresis: analysis of human exocrine pancreatic proteins. Anal Biochem. 1981;112:304–313. doi: 10.1016/0003-2697(81)90298-0. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel R, Tralks T S, Willingham M C, Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell. 1983;32:639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- 50.Semmes O J, Sztein M S, Bailey J M, Merritt W D. Tunicamycin inhibits function and expression of the high-affinity IL-2 receptor in a murine IL-2-dependent cell line. Int J Immunopharmacol. 1992;14:583–593. doi: 10.1016/0192-0561(92)90119-6. [DOI] [PubMed] [Google Scholar]
- 51.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 52.Superti F, Donelli G. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J Gen Virol. 1991;72:2467–2474. doi: 10.1099/0022-1317-72-10-2467. [DOI] [PubMed] [Google Scholar]
- 53.Willoughby R E. Rotaviruses preferentially bind O-linked sialylglycoconjugates and sialomucins. Glycobiology. 1993;3:437–445. doi: 10.1093/glycob/3.5.437. [DOI] [PubMed] [Google Scholar]
- 54.Willoughby R E, Yolken R H. SA11 rotavirus is specifically inhibited by an acetylated sialic acid. J Infect Dis. 1990;161:116–119. doi: 10.1093/infdis/161.1.116. [DOI] [PubMed] [Google Scholar]
- 55.Wunner W H, Reagan K J. Nature of the rabies virus cellular receptor. In: Crowell R L, Lonberg-Holm K, editors. Virus attachment and entry into cells. Washington, D.C.: American Society for Microbiology; 1985. pp. 152–159. [Google Scholar]
- 56.Yolken R H, Peterson J A, Vonderfecht S L, Fouts E T, Midthun K, Newburg D S. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J Clin Investig. 1992;90:1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yolken R H, Willoughby R, Wee S B, Miskuff R, Vonderfecht S. Sialic acid glycoproteins inhibit in vitro and in vivo replication of rotaviruses. J Clin Investig. 1987;79:148–154. doi: 10.1172/JCI112775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yusuf H K, Pohlentz G, Sandhoff K. Ganglioside biosynthesis in Golgi apparatus: new perspectives on its mechanism. J Neurosci Res. 1984;12:161–178. doi: 10.1002/jnr.490120205. [DOI] [PubMed] [Google Scholar]
- 59.Zárate S, Espinosa R, Romero P, Méndez E, Arias C F, López S. The VP5 domain of VP4 can mediate the attachment of rotaviruses to cells. J Virol. 2000;74:593–599. doi: 10.1128/jvi.74.2.593-599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]