Abstract
The transmission of H9N2 influenza viruses to humans and the realization that the A/Hong Kong/156/97-like (H5N1) (abbreviated HK/156/97) genome complex may be present in H9N2 viruses in southeastern China necessitated a study of the distribution and characterization of H9N2 viruses in poultry in the Hong Kong SAR in 1999. Serological studies indicated that H9N2 influenza viruses had infected a high proportion of chickens and other land-based birds (pigeon, pheasant, quail, guinea fowl, and chukka) from southeastern China. Two lineages of H9N2 influenza viruses present in the live-poultry markets were represented by A/Quail/Hong Kong/G1/97 (Qa/HK/G1/97)-like and A/Duck/Hong Kong/Y280/97 (Dk/HK/Y280/97)-like viruses. Up to 16% of cages of quail in the poultry markets contained Qa/HK/G1/97-like viruses, while about 5% of cages of other land-based birds were infected with Dk/HK/Y280/97-like viruses. No reassortant between the two H9N2 virus lineages was detected despite their cocirculation in the poultry markets. Reassortant viruses represented by A/Chicken/Hong Kong/G9/97 (H9N2) were the major H9N2 influenza viruses circulating in the Hong Kong markets in 1997 but have not been detected since the chicken slaughter in 1997. The Qa/HK/G1/97-like viruses were frequently isolated from quail, while Dk/HK/Y280/97-like viruses were predominately associated with chickens. The Qa/HK/G1/97-like viruses were evolving relatively rapidly, especially in their PB2, HA, NP, and NA genes, suggesting that they are in the process of adapting to a new host. Experimental studies showed that both H9N2 lineages were primarily spread by the aerosol route and that neither quail nor chickens showed evidence of disease. The high prevalence of quail infected with Qa/HK/G1/97-like virus that contains six gene segments genetically highly related to HK/156/97 (H5N1) virus emphasizes the need for surveillance of mammals including humans.
The key to influenza pandemic preparedness is good surveillance for influenza viruses in humans and lower animals, as was demonstrated in the H5N1 “bird flu” incident in Hong Kong SAR (16, 18). In that incident, an avian H5N1 influenza virus infected humans with a high fatality rate (2, 21, 24). Surveillance of live poultry markets in December 1997 revealed that H9N2 influenza viruses were the second most commonly isolated virus, making up about 4% of infected poultry, mainly chickens (19). Prior to 1990, H9N2 influenza viruses were mainly reported from avian species in North America (reviewed in reference 4) and were reported only in ducks in southeastern China (15). By 1997, the H9N2 viruses had been isolated from northern China, Korea, Pakistan, India, Saudi Arabia, Germany, Italy, Ireland, and South Africa (1, 5, 9). Three lineages of H9N2 influenza viruses were circulating in southeastern China at that time (3). One, represented by an isolate from a quail, Qa/HK/G1/97 (for virus nomenclature, see Table 3, footnote a), is of particular concern to public health authorities because it contains six internal genes that are genetically closely related to H5N1 (HK/156/97).
TABLE 3.
Antigenic analysis of H9N2 influenza viruses
| Virusa | HI titers with chicken antiserab to Dk/HK/Y280/97 | HI titers with ferret antisera to:
|
||
|---|---|---|---|---|
| Qa/HK/G1/97 | Ck/HK/G9/97 | HK/1073/99 | ||
| Qa/HK/G1/97 | 80 | 320 | <c | 80 |
| Qa/HK/A17/99 | 80 | 160 | < | 1,280 |
| Pg/HK/FY6/99 | 80 | 40 | < | 640 |
| Ck/HK/NT16/99 | 160 | 160 | < | 1,280 |
| Qa/HK/SSP10/99 | 80 | 160 | < | 1,280 |
| Ph/HK/SSP11/99 | 80 | 40 | < | 640 |
| Qa/HK/AP26/99 | 80 | 160 | < | 1,280 |
| Qa/HK/SF105/99 | 40 | 40 | < | 320 |
| Qa/HK/SF107/99 | 40 | 80 | < | 640 |
| Qa/HK/AP22/99 | 80 | 320 | < | 640 |
| HK/1073/99 | 160 | 160 | < | 1,280 |
| Dk/HK/Y280/97 | 640 | < | 320 | < |
| Ck/HK/SF2/99 | 2,560 | 40 | 1,280 | 80 |
| Ck/HK/FY20/99 | 2,560 | < | 1,280 | 80 |
| Qa/HK/NT28/99 | 640 | < | 160 | < |
| Ck/HK/KC12/99 | 1,280 | < | 640 | 80 |
| SCk/HK/SF44/99 | 2,560 | < | 1,280 | 80 |
| Ck/HK/SF3/99 | 2,560 | 40 | 1,280 | 40 |
| Ck/HK/SF4/99 | 2,560 | 40 | 1,280 | 40 |
| Ck/HK/SF5/99 | 2,560 | < | 1,280 | 80 |
| Ck/HK/SF6/99 | 1,280 | < | 320 | < |
| Ck/HK/SF9/99 | 1,280 | < | 640 | < |
| Ck/HK/SF10/99 | 1,280 | < | 320 | 40 |
| Ck/HK/SF11/99 | 2,560 | 40 | 2,560 | 40 |
| Ck/HK/SF12/99 | 1,280 | < | 640 | 40 |
| SCk/HK/SF15/99 | 2,560 | 40 | 2,560 | 160 |
| SCk/HK/SF43/99 | 2,560 | < | 1,280 | 80 |
| Ck/HK/FY16/99 | 2,560 | < | 640 | 40 |
| Ck/HK/G9/97 | 5,120 | 40 | 2,560 | 40 |
| Ck/HK/G23/97 | 2,560 | < | 640 | 40 |
| Dk/HK/Y439/97 | 80 | < | < | < |
| Ck/Kor/006/96 | 160 | < | < | < |
The poultry markets in Hong Kong were depopulated in late December 1997, and land-based poultry was reintroduced in February 1998. H5N1 influenza viruses similar to the pathogenic 1997 H5N1 strain have not been detected since then despite extensive routine serological and virological surveillance. On the other hand, H9N2 viruses have been isolated from market chickens and other poultry to the present (this report) as well as from chickens in southern and northern China (4). Recently, H9N2 viruses were also isolated from patients with influenza-like illnesses, both in southeastern China and in Hong Kong SAR (5, 13). The H9N2 influenza viruses isolated from two children in Hong Kong were highly related in all gene segments with Qa/HK/G1/97 (Yi Pu Lin, personal communication), but the H9N2 isolates from China have not been genotyped. The transmission to humans of influenza viruses containing H5N1-like internal genes emphasizes the need for better understanding the occurrence and molecular epidemiology of these viruses in the live-poultry markets of Hong Kong and on their stability and host range.
Whereas H9N2 viruses were isolated from over 4% of chickens in the live-bird markets of Hong Kong in 1997 (19), only one of these isolates belonged to the Qa/HK/G1/97-like genotype that possess internal genes similar to the pathogenic H5N1 viruses of 1997. However, on that occasion, surveillance was focused predominantly on chickens and other types of poultry, but only 15 quail were sampled. Influenza surveillance of live-poultry markets resumed in 1999 with the objective of determining the prevalence of viruses with the Qa/HK/G1/97 genotype and their distribution among different types of poultry.
This study enhances the epidemiological understanding of the occurrence of H9N2 influenza viruses. They are widely distributed in poultry in southeastern China, being isolated from most types of land-based poultry tested. The genetic characterization of these H9N2 influenza viruses from poultry in Hong Kong markets indicates that the lineage represented by Qa/HK/G1/97 is now frequently isolated from quail and is evolving more rapidly than that represented by Dk/HK/Y280/97. The study also characterizes the stability of H9N2 viruses in the environment, assesses their replication and pathogenicity in quail, and demonstrates that Qa/HK/G1/97-like viruses are in the process of adapting to land-based poultry.
MATERIALS AND METHODS
Poultry marketing in Hong Kong.
Prior to the H5N1 incident in Hong Kong SAR in 1997, all types of live poultry for sale to the public were housed in about 1,000 retail markets across the region. Thus, aquatic birds, e.g., domestic ducks, geese, wild ducks, and occasional wild waterfowl, were kept in separate cages in the immediate vicinity of land-based birds, e.g., chickens, silkie chickens, pigeons, quail, pheasant, chukka, and guinea fowl. The most abundant bird in the market was chicken, the mix of other birds varying with the market and locality. After the slaughter of poultry across the SAR at the end of 1997, the markets were cleaned and the policy for holding birds was changed. Aquatic birds are slaughtered in one central facility, and live aquatic birds are not available from retail outlets. The land-based birds are imported by truck after being screened for evidence of antibodies to H5N1 viruses to a separate wholesale market; approximately 120,000 chickens and more than 20,000 other land-based birds are imported each day. Live land-based birds continue to be sold through the approximately 1,000 retail markets. While the majority of chicken at retail outlets are sold each day, there are always a number that remain for more than 1 day. The more expensive birds (pheasant, silkie chicken, and guinea fowl) may remain in the markets for several days. The turnover rate for the different birds is not available and differs from market to market. The retail poultry markets have a carryover population of birds that are topped up each day from the wholesale market.
Serological studies in poultry.
Thirteen birds from each consignment (approximately 1,000 birds per consignment) going to the wholesale market were bled daily. The number of samples tested for H5 antibody per consignment would detect a serologically positive bird with 99% confidence if the seroprevalence of the disease was 30% or with 95% confidence at a seroprevalence of 22%. The sampling rate also had to consider the logistics of testing every consignment of birds every day of the year and providing results before the birds were sold at the wholesale poultry markets each evening from midnight. All sera were tested for antibodies to H5 influenza viruses; once weekly, all sera were also tested for antibodies to H9 influenza viruses (Ck/HK/G9/97 and Ck/HK/G1/97) in hemagglutination inhibition (HI) assays (12). The majority of the sera were tested without receptor-destroying enzyme (RDE) treatment. A subgroup was also tested after RDE treatment, and this did not affect the results obtained; preimmune chicken sera contained no detectable inhibitors to the H9 viruses in HI tests.
Virus sampling.
From 13 April through 25 November 1999, a total of 19 live-poultry markets, eight in Hong Kong, five in Kowloon, and six in the New Territories, were sampled for influenza viruses as described elsewhere (17). On average, 50 individual samples were collected from the fecal trays under the poultry in each market. Only land-based birds were studied, as aquatic birds are no longer present in these markets (see Table 2). The number of samples collected from each type of poultry was approximately in proportion to the poultry in the market. Thus, chickens were most common, constituting 90% of total. Fecal samples were collected into tissue culture medium 199 containing penicillin G (2 × 106 U/liter), polymyxin B (2 × 106 U/liter), gentamicin (250 mg/liter), nystatin (0.5 × 106 U/liter), ofloxacin HCl (60 mg/liter), and sulfamethoxazole (0.2 g/liter).
TABLE 2.
Surveillance of poultry markets for H9N2 influenza viruses, 1999a
| Species | No. of dropping trays
|
% Positive | |
|---|---|---|---|
| Tested | Containing H9N2 virus | ||
| Chicken | 1,180 | 55 | 4.7 |
| Pigeon | 137 | 4 | 2.9 |
| Silkie chicken | 101 | 4 | 3.6 |
| Quail | 101 | 16 | 16.0 |
| Pheasant | 59 | 3 | 5.0 |
| Guinea fowl | 53 | 2 | 3.8 |
| Chukka | 49 | 2 | 4.1 |
| Mixedb | 50 | 6c | 12.0 |
| Partridge | 3 | 0 | |
| Environment | 6 | 0 | |
Thirty-five poultry markets were surveyed for influenza virus as described in Materials and Methods from 10 April to 25 November 1999; 27 markets contained H9N2 in one or more kinds of poultry. The viruses were identified serologically with monospecific antisera (14), and representative viruses were genotyped by sequence analysis (Fig. 1).
Mixtures of two or three species per cage (e.g., silkie chicken plus guinea fowl; pigeon plus quail plus pheasant).
Two of the six positive cages contained quail.
Virus was isolated in embryonated chicken eggs and identified serologically with monospecific antisera to each known influenza virus subtype (14). The poultry in the markets were generally healthy. The only evidence of respiratory infection was a very mild “snick” in some that was observed only after careful examination and was not readily detectable.
Replication and transmission of H9N2 virus in quail.
To study replication, groups of quail (Coturnix coturnix; young adult) were infected orally, intranasally, and orbitally with 103 50% egg infectious doses (EID50) in 0.5 ml of different H9N2 viruses. The dose of virus was distributed between three routes (see Table 6). The birds were swabbed on days 3, 5, and 7 from the trachea and cloaca, and virus was titrated for infectivity in chicken embryos (12). The birds were observed daily for disease signs. To study transmission, two quail were infected orally, intranasally, and orbitally with 103 EID50 of HK/1073/99 influenza virus in 0.5 ml. Infected birds were placed in a cage with two susceptible contact birds of the same age. A group of four quail was placed in a cage directly below the infected birds. The fecal dropping tray was removed from the upper cage, allowing fecal contact with the uninfected birds. Other groups of birds were housed in cages directly adjacent to the infected birds, with a distance of 1 ft between cages. The airflow was from the bottom of the isolation cubicle and out through a top filter. Tracheal and cloacal samples were collected daily and titrated for infectivity of viruses in embryonated chicken eggs.
TABLE 6.
Replication of H9N2 influenza viruses in quaila
| Virus | No. with virus shedding on indicated day (infectivity titer [log10/0.1 ml])
|
|||||
|---|---|---|---|---|---|---|
| Trachea
|
Cloaca
|
|||||
| 3 | 5 | 7 | 3 | 5 | 7 | |
| Qa/HK/G1/97 | 4 (>5.0) | 4 (>5.0) | 3 (NT) | 4 (<1) | 2 (<1) | 0 |
| Ck/HK/G9/97 | 4 (5.0) | 4 (3.8) | 2 (2.5) | 2 (<1) | 0 | 0 |
| Qa/HK/A17/99 | 4 (4.8) | 4 (5.0) | 4 (3.2) | 2 (<1) | 2 (<1) | 1 |
Each group contained four animals, none of which showed signs of disease from days 1 to 11. NT, not tested.
Infection of pigeons.
Four pigeons (young adult white) were inoculated orally, intranasally, and orbitally with 106 EID50 of either Pg/HK/FY6/99, Qa/HK/G1/97, or Ck/HK/G9/97. The dose of virus was distributed between the three routes. The infected birds were caged with four uninfected pigeons to test for transmission, and the birds were swabbed daily tracheally and cloacally for evidence of virus infection.
Stability of the H9N2 virus in the environment.
Four chickens were infected with the Qa/HK/G1/97 virus orally, intranasally, and orbitally with 1.0 ml containing approximately 100 EID50, the dose of virus being distributed equally between the three routes. Tracheal swabs were collected daily from days 3 to 7. Fecal samples were collected from pans under the cages and made into 10% suspensions with phosphate-buffered saline or dried at room temperature (25°C). Aliquots were stored at different temperatures and titrated in embryonated eggs for residual infectious virus. Dried samples were rehydrated and titrated similarly.
Chicken and ferret antisera.
Three-week-old specific-pathogen-free chickens were inoculated intranasally and orally with 1.0 ml of infectious allantoic fluid (EID50 = 106.5). The chickens were bled 3 weeks postinfection and boosted intravenously with 1.0 ml of infectious virus allantoic fluid. Ferret antiserum was kindly provided by Yi Pu Lin, World Influenza Center, Mill Hill, London, England.
Gene sequencing and analysis.
Viruses used in this study are characterized in Table 3 and Fig. 1. Viral gene sequencing and analysis were carried out as described by Guan et al. (3). In brief, viral RNA was extracted from infective allantoic fluid using an RNEasy Mini kit (Qiagen, Inc., Valencia, Calif.). Reverse transcription followed by PCR was performed using specific primers for each gene segment (primer sequences are available on request). PCR products were purified with a QIAQuick PCR purification kit (Qiagen) and sequenced by using synthetic oligonucleotides produced by the Center for Biotechnology at St. Jude Children's Research Hospital. Reactions were performed with Rhodamine Dye-Terminator Cycle Sequencing Ready Reaction kits used with AmpliTaq DNA polymerase FS (Perkin-Elmer/Applied Biosystems, Inc.). Samples were electrophoresed and analyzed on Perkin-Elmer model 377 DNA sequencers (Perkin-Elmer/Applied Biosystems).
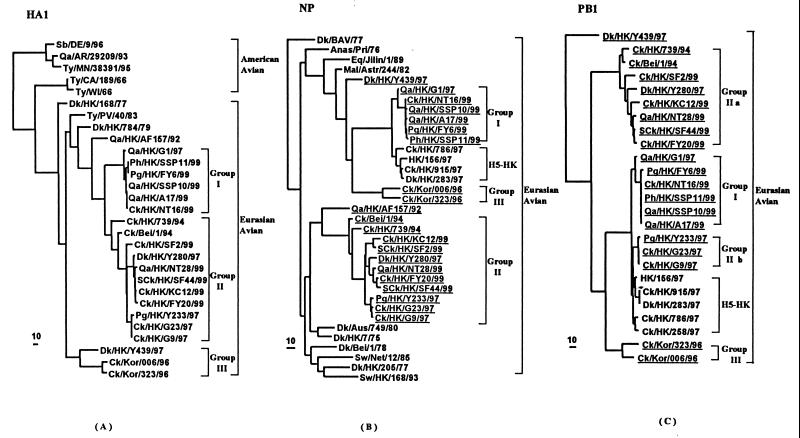
FIG. 1.
Phylogenetic trees for the H9 HA1 (A), NP (B), and PB1 (C) genes of influenza A viruses. The nucleotide sequences of the HA1, NP, and PB1 genes were analyzed with PAUP by using a maximum-parsimony algorithm. Nucleotides 55 to 1014 (960 bp) of H9 the HA1 gene, nucleotides 48 to 1398 (1,351 bp) of NP gene, and nucleotides 67 to 1428 (1,362 bp) of the PB1 gene were used for the phylogenetic analysis. The HA1 phylogenetic tree is rooted to A/Dk/Alberta/60/76 (H12N5). The NP phylogenetic tree is rooted to A/Equine/Prague/1/56 (H7N7), and the PB1 tree is rooted to B/Lee/40. Lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Viruses are characterized in Tables 3 and 4; information can also be found in reference 3 or GenBank (accession numbers D00050, M63775, M63776, M63782, M36812, M63786, M30764, M63783, M22573, and M30749). All viruses underlined in the NP and PB1 gene trees are H9N2 influenza viruses. Group I, II, and III designations are based on phylogenetic relationships of the HA1 gene.
Sequence data were edited and analyzed using the Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.). Phylogenetic analyses were carried out using PAUP (Phylogenetic Analysis Using Parsimony), version 4.0 (developed by David Swofford, Illinois Natural History Survey, Champaign).
Analysis of the rate of amino acid changes in different H9N2 virus groups.
A defined region (given in Table 5) of the eight gene segments of six influenza viruses isolated in 1997 or 1999 was analyzed. Based on the topology of phylogenetic trees and the date of virus isolation, five viruses were determined as the reference strains for each group tested. A matrix analysis was performed with GeneDoc, version 2.3 (developed by K. B. Nicholas) to quantitate the nucleotide and amino acid changes in each gene of each isolate. The nucleotide and amino acid changes per site were determined by dividing them by the length of the sequence analyzed. The rate of amino acid change was calculated as indicated in Table 5. The average values in each group were compared between virus groups. The P values were determined by the Student t test.
TABLE 5.
Rates of change in different sublineages of H9N2 and H5N1 influenza viruses from Hong Kong
| Gene | Nucleotide range analyzed | Nucleotide change/sitea
|
Amino acid change/sitee
|
Rate of amino acid change/nucleotide changef (mean ± SE)
|
P valueg
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qa/HK/G1/97-likeb (group I) | Dk/HK/Y280/97-likec (group II) | H5N1d | Qa/HK/G1/97-like (group I) | Dk/HK/Y280/97-like (group II) | H5N1 | Qa/HK/G1/97-like (group I) | Dk/HK/Y280/97-like (group II) | H5N1 | I:II | I:H5 | II:H5 | ||
| PB2 | 988–2289 | 0.0117 | 0.0148 | 0.0081 | 0.0046 | 0.0005 | 0.0069 | 0.39 ± 0.01 | 0.05 ± 0.11 | 0.74 ± 0.42 | <0.05* | NS | <0.05* |
| PB1 | 67–1428 | 0.0081 | 0.0216 | 0.0128 | 0.0000 | 0.0013 | 0.0022 | 0.00 | 0.057 ± 0.08 | 0.19 ± 0.08 | NS | <0.05* | <0.05 |
| PA | 19–1677 | 0.0129 | 0.0170 | 0.0097 | 0.0043 | 0.0065 | 0.0036 | 0.34 ± 0.07 | 0.42 ± 0.29 | 0.39 ± 0.09 | NS | NS | NS |
| HA | 88–1077 | 0.0126 | 0.0148 | 0.018 | 0.0066 | 1.43 ± 0.07 | 0.48 ± 0.14 | <0.05* | |||||
| NP | 34–1398 | 0.0085 | 0.0174 | 0.0066 | 0.0035 | 0.0022 | 0.0044 | 0.41 ± 0.21 | 0.14 ± 0.12 | 0.64 ± 0.37 | <0.05 | NS | <0.05 |
| NA | 20–1421 | 0.0082 | 0.012 | 0.0068 | 0.0055 | 0.83 ± 0.10 | 0.46 ± 0.14 | <0.05* | |||||
| M1 | 50–982 | 0.0103 | 0.0073 | 0.0071 | 0.0016 | 0.0008 | 0.0016 | 0.12 ± 0.27 | 0.15 ± 0.34 | 0.24 ± 0.54 | NS | NS | <0.05 |
| NS1 | 41–879 | 0.0006 | 0.023 | 0.0137 | 0.0009 | 0.0161 | 0.0141 | 0.59 ± 1.32 | 0.68 ± 0.17 | 1.02 ± 0.04 | NS | NS | <0.05* |
Rate of nucleotide change at each nucleotide site compared to the reference strain (number of nucleotide changes/length of sequence analyzed).
Reference virus is Qa/HK/G1/97; compared to the first second set of five viruses in Table 3 which were isolated in 1999.
Reference virus is Dk/HK/Y280/97; compared to the five boldfaced viruses in Table 3 which were isolated in 1999.
Reference virus is Ck/HK/258/97; compared to Dk/HK/Y283/97, Dk/HK/P46/97, Goose/HK/W355/97, Ck/HK/786/97, and Ck/HK/915/97.
Rate of amino acid change for each amino acid site compared to the reference strain (number of amino acid changes/length of amino acid sequence analyzed).
Calculated as amino acid change per site/nucleotide change per site.
Calculated for comparing the rates of amino acid change per nucleotide change between different groups: I:II (group I compared to group II), I:H5 (group I compared to H5N1 group); II:H5 (group II compared to H5N1 group). If P value is <0.05, there is significant difference between the two groups, unadjusted for multiple tests; if P value is <0.05*, the difference is still significant even adjusted for multiple pairwise testing by Bonferroni adjustment. If P value is >0.05, there is no significant (NS) difference between the two groups compared.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained from this study are available from GenBank under accession numbers AF222606 through AF222681.
RESULTS
Serological surveillance at the port of entry.
Serological surveillance was done at the port of entry to Hong Kong SAR to ascertain what types of poultry had antibodies to H9N2. Poultry imported into Hong Kong SAR are screened daily for serological evidence of H5N1 influenza virus infection; the samples collected at monthly intervals from April 1999 until March 2000 were examined in HI tests for antibodies to the hemagglutinin (HA) of H9N2 viruses. The majority of species of poultry tested had antibodies that reacted with both Qa/HK/G1/97 and Ck/HK/G9/97; this included most of the land-based as well as aquatic birds (Table 1). The largest number of consignments tested were chickens, where a higher percentage had antibodies to Ck/HK/G9/97 (68.7) than to Qa/HK/G1/97 (58.7); similarly, ducks had a higher percentage of antibodies to Ck/HK/G9/97 than to Qa/HK/G1/97, and geese consignments had antibodies only to Ck/HK/G9/97. The number of consignments of other species tested was insufficient to make comparisons. Over the 1 year of testing, there was minor fluctuation in the number of positive consignments of chickens for H9N2; Ck/HK/G9/97 was always higher than Qa/HK/G1/97, and there was no major change in prevalence (results not shown). The numbers of other poultry tested were too low to detect trends.
TABLE 1.
Serological study of poultry at ports of entry to Hong Kong SAR for HI antibodies to the H9 subtype
| Species tested | % of poultry consignments positive for HI antibodies toa:
|
|
|---|---|---|
| Qa/HK/G1/97 | Ck/HK/G9/97 | |
| Chicken | 58.7 (983) | 68.7 (908) |
| Chukka | 88.9 (9) | 66.7 (6) |
| Quail | 57.9 (19) | 54.5 (11) |
| Pheasant | 50.0 (9) | 40.0 (5) |
| Silkie chicken | 35.4 (48) | 41.7 (24) |
| Guinea fowl | 33.3 (9) | 0 (6) |
| Pigeon | 22.9 (35) | 31.6 (19) |
| Duck | 14.9 (134) | 37.5 (136) |
| Goose | 0 (125) | 38.2 (89) |
A consignment was considered positive if two or more birds had an HI titer of 1/32 or greater; the range was from 1/32 to 1/640. Figures in parentheses give the number of consignments tested; 13 sera from each consignment (ca. 1,000 birds per consignment) were tested in HI tests over the period from April 1999 to March 2000. On average, up to 50% of sera in the consignment were positive.
Selected chicken sera with HI antibody to H9N2 viruses were tested in serum neutralization tests, and the presence of antibody was confirmed. In both HI and neutralization tests, chicken sera gave higher antibody titers with the Ck/HK/G9/97 virus than with the Qa/HK/G1/97 virus, indicating that the H9N2 virus infecting these birds was more closely related to Ck/HK/G9/97.
Serological analyses of chicken sera collected between 1968 and 1979 from poultry markets and stored frozen until 1999 gave equivocal results. In HI tests, 29 of 111 reacted with HK/1073/99 (H9N2), with titers ranging from 1/40 to 1/160. However, after treatment with RDE (to remove nonspecific inhibitors of hemagglutination), none of the sera reacted with any of the viruses tested. Serum neutralization tests also failed to demonstrate convincing neutralization activity.
Isolation of H9N2 influenza viruses from poultry in 1999 from Hong Kong SAR.
Since the majority of poultry imported into Hong Kong SAR had evidence of past infection with H9N2 viruses, we next determined if poultry in the markets were infected with H9N2 and were the potential source of virus for humans. Of the 35 markets tested from April to November 1999, 27 contained H9N2 influenza viruses and overall 5.2% of fecal samples from cages of domestic poultry contained H9N2 viruses. H9N2 viruses were found in the feces of all species of birds tested except partridge, which was present in few markets (Table 2). The majority of H9N2 influenza virus isolates were from chickens (62%), quail (17%), and silkie chickens (7%), with the remainder from pheasant, guinea fowl, and pigeons. H9N2 influenza viruses were isolated during each of the months when tests were done and were present in markets throughout Hong Kong, Kowloon, and the New Territories of Hong Kong SAR.
The percentages of dropping trays containing influenza viruses were generally similar for the majority of land-based birds, ranging from 2.9% for pigeons to 5.0% for pheasant (Table 2). Two groups fell outside this range, with 16% of quail and 12.0% of trays from more than one species yielding H9N2 viruses. The isolation rate from quail in the first half of 1999 (11 isolates from 49; 22%) was higher than in the second half of the year (5 from 52; 10%).
Characterization of H9N2 viruses isolated in the poultry markets in Hong Kong in 1999. (i) Antigenic analysis.
At least three distinguishable lineages of H9N2 influenza viruses were detected in domestic poultry in Asia in 1997 (3). To determine which lineages of H9N2 influenza viruses are present in the poultry in the poultry markets in the SAR in 1999, the viruses were characterized antigenically and molecularly. Ferret or chicken antisera to the reference strain of the lineages found previously in the SAR were included in the analysis (Table 3). Ferret antisera discriminated between viruses belonging to the Qa/HK/G1/97-like group of viruses from the Dk/HK/Y280/97-like and Ck/Korea/006/96 (H9N2)-like groups of viruses. Similarly, ferret sera to Ck/HK/G9/97 and chicken sera to Dk/HK/Y280/97 clearly separated the viruses in this group from the Qa/HK/G1/97 and Ck/Korea/006/96 groups of viruses. The H9N2 influenza viruses isolated from 1999 all belong to the Qa/HK/G1/97 or the Dk/HK/Y280/97 group. It is noteworthy that most of the H9N2 viruses similar to Qa/HK/G1/97 were from quail, with one isolate each from a pheasant, a pigeon, and a chicken: similarly, the majority of the isolates in the group represented by Dk/HK/Y280/97 were from chickens, with fewer isolates from silkie chickens and one isolate from a quail. No virus belonging to the Ck/Korea/006/96 group was isolated in 1999 from poultry in Hong Kong. The human isolate A/Hong Kong/1073/97 (H9N2) (Y. P. Lin, personal communication) belongs to the group that is isolated mainly from quail. The Ck/HK/G9/97 and Dk/HK/Y280/97 viruses grouped together with chicken and ferret antisera, showing that antigenically they are similar.
Previously there had only been one isolate like Qa/HK/G1/97 characterized from the SAR in 1997 (3); the isolation of multiple isolates of this lineage in 1999 may reflect the larger number of quail tested; alternatively, these viruses may now be more widely represented than in 1997.
(ii) Molecular analysis.
To provide complete genotyping of representative H9N2 influenza viruses isolated in 1999, five isolates of the Qa/HK/G1/97-like and Dk/HK/Y280/97-like virus groups shown in boldface in Table 3 were analyzed (Table 4). Each of the gene segments was partially sequenced, and their homologies were compared with influenza virus strains from 1997. A/Qa/HK/A17/99 (H9N2) is representative of all viruses in its group (Table 3) and shows highest homology with the eight gene segments of Qa/HK/G1/97. These viruses show high homology with six gene segments of HK/156/97 and with the PB2 and PB1 genes of Ck/HK/G9/97 (Table 4). The A/Qa/HK/A17/99(H9N2) was distinguishable in all gene segments from Dk/HK/Y280/97. Conversely, A/Ck/HK/FY20/99 (H9N2) shows highest homology with the eight gene segments of Dk/HK/Y280/97 and with six gene segments of Ck/HK/G9/97. It is noteworthy that no virus containing the gene complement of Ck/HK/G9/97 have been found since 1997, nor have viruses similar to Ck/Kor/006/96 been detected since 1997.
TABLE 4.
Homology of the gene segments of 1999 H9N2 influenza viruses with reference strain
| Representative virus | Gene segment | % Homologya with:
|
|||
|---|---|---|---|---|---|
| Qa/HK/G1/97 (H9N2) | Dk/HK/Y280/97 (H9N2) | Ck/HK/G9/97 (H9N2) | HK/156/97 (H5N1) | ||
| Qa/HK/A17/99 | PB2 | 98.8 (98.9) | 85.6 (85.7) | 97.4 (97.5) | 97.8 (97.9) |
| PB1 | 99.2 (99.2) | 91.0 (90.8) | 98.2 (98.1) | 98.3 (98.3) | |
| PA | 98.7 (98.7) | 88.9 (88.9) | 88.4 (88.3) | 97.4 (97.4) | |
| HA | 98.9 (98.7) | 90.1 (90.1) | 90.7 (90.7) | —b | |
| NP | 99.1 (99.2) | 88.7 (88.7) | 89.0 (89.1) | 97.7 (97.8) | |
| NA | 99.2 (99.2) | 93.5 (93.4) | 93.1 (93.0) | — | |
| M | 99.3 (99.2) | 96.7 (96.6) | 96.5 (96.4) | 99.2 (99.1) | |
| NS | 99.9 (99.9) | 93.7 (93.7) | 93.3 (93.3) | 98.6 (98.6) | |
| Ck/HK/FY20/99 | PB2 | 86.4 (86.0) | 99.0 (98.5) | 86.3 (85.8) | 86.3 (86.0) |
| PB1 | 90.6 (90.7) | 97.9 (97.8) | 90.8 (90.8) | 90.4 (90.4) | |
| PA | 89.1 (89.1) | 98.9 (98.3) | 96.5 (96.7) | 88.7 (88.8) | |
| HA | 89.4 (90.0) | 98.5 (98.5) | 96.5 (97.0) | — | |
| NP | 89.2 (88.9) | 98.5 (98.3) | 98.2 (98.1) | 89.4 (89.1) | |
| NA | 93.5 (93.2) | 99.6 (98.8) | 94.5 (94.2) | — | |
| M | 95.9 (96.2) | 99.0 (99.1) | 98.5 (98.6) | 96.0 (96.3) | |
| NS | 93.0 (93.1) | 98.1 (97.8) | 97.9 (97.6) | 93.5 (93.7) | |
Calculated based on the nucleotide sequences of PB1 (nucleotides 988 to 2289), PB2 (67 to 1428), PA (19 to 1677), HA (88 to 1077), NP (34 to 1398), NA (20 to 1421), M (50 to 982), and NS (41 to 879). The highest homologies are in boldface. Values in parentheses are means for the five viruses shown in boldface in each group in Table 3 which were isolated in 1999.
—, different HA or NA subtypes.
(iii) Phylogenetic analysis.
In earlier characterization of H9N2 influenza viruses from Hong Kong poultry in 1997, only a single isolate containing the HK/156/97-like internal gene constellation, namely, Qa/HK/G1/97, had been characterized (3). This virus was assigned to a separate branch on the HA phylogenetic tree. Phylogenetic analyses of the HA1, nucleoprotein (NP), and PB1 of representative H9N2 influenza viruses isolated from 1999 are given in Fig. 1. There are three distinguishable lineages of viruses of the Eurasian clade in the HA1 phylogenetic tree. The contemporary quail, pigeon, and pheasant isolates represented by Qa/HK/A17/99 are antigenically (Table 3) and phylogenetically (Fig. 1A) similar to Qa/HK/G1/97 (group 1). Recent chicken and silkie chicken isolates and one quail isolate represented by Ck/HK/FY20/99 are antigenically and phylogenetically similar to Dk/HK/Y280/97 (group II). The Ck/HK/G9/97-like viruses form a distinguishable sublineage of group II viruses, of which there are no contemporary isolates. The group containing Ck/Kor/006/96 formed a separate lineage (group III) that contained a duck isolate from Hong Kong in 1997 (A/Dk/HK/Y439/97). Recent isolates of this lineage were not detected. This may reflect the lack of samples from aquatic birds because these species are no longer sold in the live-bird retail markets of Hong Kong.
In addition to the 10 strains of H9N2 isolates from 1999 shown in Fig. 1A, 34 other H9N2 isolates were genotyped by partial sequencing of the HA1 gene. Overall these 44 tested strains were from chickens (n = 18), silkie chickens (n = 4), chukka (n = 1), quail (n = 12), pigeons (n = 2), pheasant (n = 3), guinea fowl (n = 1), and cages with mixed birds (n = 3). Seventeen chicken isolates (94%) and one quail, one pigeon, one pheasant, one chukka, and all four silkie chicken isolates were of the Dk/HK/Y280/97 lineage. In contrast, 11 (92%) of 12 quail, 1 chicken, 1 pigeon, 2 pheasant, and 1 guinea fowl isolates belonged to the Qa/HK/G1/97 lineage. The apparent specificity of the two lineages for chicken and quail is maintained even within the same market on a given occasion.
Analysis of the NP phylogenetic tree confirmed that recent isolates represented by Qa/HK/A17/99 are phylogenetically related to Qa/HK/G1/97 (group 1)- and HK/156/97 (H5N1)-like viruses (Fig. 1B). The NP genes represented by the Ck/Kor/006/96 are in a separate sublineage. The NP genes of the group represented by Ck/HK/FY20/99 are phylogenetically closely related to Dk/HK/Y280/97 (group II). The NP of Ck/HK/G9/97 (H9N2) is part of this evolutionary pathway and forms a distinguishable sublineage. No isolates of the latter sublineage were isolated in 1999.
The PB1 genes of contemporary H9N2 influenza viruses belong to two phylogenetic groups (Fig. 1C). Those represented by Qa/HK/A17/99 are closely related to Qa/HK/G1/97 (group I) and cluster with PB1 genes of HK/156/97-like H5N1 viruses and Ck/HK/G9/97 (group IIb). Recent isolates represented by Ck/HK/FY20/99 are closely related to Dk/HK/Y280/97 (group IIa) and are clearly distinct from the group IIb and H5-HK-related viruses. No additional examples of Ck/Kor/006/96 were isolated in Hong Kong markets recently.
Thus, the phylogenetic analysis confirms that two lineages of H9N2 viruses, one related to Qa/HK/G1/97 and the other related to Dk/HK/Y280/97, continue to circulate in southeastern China in 1999.
Evolutionary analysis of the two H9N2 virus lineages in Hong Kong.
It has been proposed that when an influenza virus transfers to a new host, there is a period of rapid evolution with an increased accumulation of amino acid changes (8, 22). To determine if there are differences between the rate of evolution of the two H9N2 virus lineages that continue to circulate in poultry in southeast China in 1999, we analyzed the rates of amino acid substitutions in each gene segment (Table 5). The number of nucleotide changes per site was higher in all gene segments for Dk/HK/Y280/97-like viruses in 1999 than in the Qa/HK/G1/98-like viruses, but few of these changes resulted in amino acid changes (Table 5). In fact, when the amino acid changes per nucleotide change were compared, the opposite was found; the PB2, HA, NP, and neuraminidase (NA) genes in the Qa/HK/G1/97-like viruses were evolving significantly faster than the same genes in the Dk/HK/Y280/97-like viruses. In contrast, compared with the H5N1 influenza viruses from 1997, it was found that the Qa/HK/G1/97-like viruses show fewer amino acid substitutions per nucleotide change than the H5N1 virus group, even though such differences do not reach statistical significance. Thus, among the three virus lineages tested, H5N1 viruses and Qa/HK/G1/97-like viruses were evolving relatively rapidly, suggesting that both of these viruses are recent introductions to their respective hosts.
Replication of H9N2 influenza viruses in quail.
Since the majority of H9N2 influenza viruses in the group represented by Qa/HK/G1/97 (Tables 3 and 4), containing internal genes similar to those of HK/156/97 (H5N1), have been isolated from quail, the possibility exists that this H5N1 isolate is primarily a quail influenza virus. We therefore wished to determine the ability of these viruses to replicate in quail and whether they caused disease. Groups of quail were inoculated with Qa/HK/G1/97, Ck/HK/G9/97, and Qa/HK/A17/99 H9N2 influenza viruses. Each of these viruses replicated in the respiratory tract with an initial titer of >5.0 log10/0.1 ml and was shed for at least 7 days (Table 6) but produced no disease signs. Replication of each virus in the intestinal tract was detected, but the number of birds shedding virus in feces was limited and low titers of virus were detected (<1.0 log10/0.1 ml) (Table 6). It is noteworthy that H9N2 influenza virus isolated from chickens (Ck/HK/G9/97) also replicated in quail for at least 7 days and produced no disease signs, and a minority of birds shed viruses in feces. In quail, these H9N2 viruses are primarily shed in the respiratory tract; none of the viruses caused disease signs.
Transmission of the H9N2 influenza viruses in quail.
Previous studies established that the H5N1 influenza virus that caused the bird flu incident in 1997 was spread mainly by the fecal route (17). Since the H9N2 virus that was isolated from two children in Hong Kong in 1999 is highly related in all genes to Qa/HK/G1/97 (Lin, personal communication), it was important to establish replication of the human virus in quail. HK/1073/99 (H9N2) was inoculated into the respiratory tract and orally into two quail; transmissibility was studied in quail in physical contact in the same cage, in quail housed in cages below the infected birds (fecal and water contact), and in birds housed in cages adjacent to the infected birds (aerosol contact) (Table 7). The HK/1073/99 virus was transmitted to all contact quails; the virus was shed primarily via the respiratory tract, being isolated from the trachea of contact birds for up to 9 days, with peak levels of virus shedding on day 6. Minimal numbers of quail shed virus in feces, and the titers were low. However, birds in the fecal and water contact group were infected before the aerosol group, possibly due to transmission through contaminated water from above. None of the infected quail showed disease signs. Thus, the human H9N2 influenza virus is nonpathogenic in quail and shed for an extended period of at least 9 days, primarily by the aerosol route. In contrast to what was found in quail, where virus titers of Qa/HK/G1/97 from fecal samples were low and intermittently detected, in chickens this virus was shed in both feces and tracheal samples (4).
TABLE 7.
Transmission of A/HK/1073/99 (H9N2) among quail
| Day after contact | No. infected (virus titer [log10/ml])
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Infected quaila
|
Physical contactb
|
Fecal contactc
|
Aerosol contactd
|
|||||
| Trachea | Cloaca | Trachea | Cloaca | Trachea | Cloaca | Trachea | Cloaca | |
| 2 | 2 (2.8) | 0 (<e) | 2 (4.8) | 0 | 4 (4.8) | 0 | 0 | 0 |
| 4 | 2 (2.8) | 0 (<) | 2 (NTf) | 2 (<1.0) | 4 (NT) | 1 (<1.0) | 2 (4.5) | 0 |
| 6 | 2 (4.5) | 0 (<) | 2 (4.5) | 0 (<) | 4 (4.5) | 1 (<1.0) | 4 (2.0) | 0 |
| 9 | 2 (<1.0) | 0 (<) | 2 (2.0) | 2 (<1.0) | 4 (2.5) | 0 | 4 (2.0) | 3 (<1.0) |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Two quail were infected with A/HK/1073/99 (H9N2).
The two infected quail were put into a cage with two contact quail.
Four quail were put into a cage below the infected birds with the litter tray removed.
Four quail were put into a cage at the same level as the infected birds but separated by 1 ft. Airflow was from the bottom and out through top filters.
<, below the level of detection.
NT, not tested.
Stability of Qa/HK/G1/97 (H9N2) influenza at ambient temperatures.
Since the probable source of the H9N2 influenza virus that infected the two humans in Hong Kong SAR was poultry (13), the stability of this virus was determined under environmental conditions. Tracheal and fecal samples of Qa/HK/G1/97 were collected from infected chickens at the peak of virus shedding (days 3 through 7) and held at different temperatures. In air-dried samples at 22°C, infectious virus was still detectable after 8 h but fell to undetectable levels within 1 day (results not shown). The infectivity titers in wet samples declined in titer over 2 to 3 days, but even at 35°C, infectious virus was still detectable (−2.5 log10 EID50) in fecal samples at 3 days. Tracheal samples at 22 or 35°C, either wet or dry, were more stable at 22°C than at 35°C: at 22°C, virus was detectable for 3 days, while at 35°C, infectivity was detectable for 4 h, but not for 8 h. At 4°C, infectious virus (>2.0 log10 units) was detectable for at least 4 days. Thus, air drying facilitates destruction of H9N2 influenza viruses, but virus was detectable for up to 8 h in feces. In wet conditions, fecal samples can maintain virus for at least 3 days at ambient temperatures; at lower temperatures, the virus was maintained at moderate levels of infectivity for at least 4 days.
Studies on H9N2 influenza viruses in pigeons.
Pigeons are the second most common type of poultry in the live-poultry markets in the Hong Kong SAR. To date, only a limited number of influenza viruses have been obtained from pigeons (Table 2). In this study, Pg/HK/FY6/99 (H9N2), genetically closely related to Qa/HK/G1/97, was inoculated into pigeons. To determine if this isolate as well as Qa/HK/G1/97 and Ck/HK/G9/97 can replicate and be transmitted between pigeons, four birds were infected and put in contact with four pigeons in the same cage. Virus was detected on the first day after infection in two of four birds inoculated with Ck/HK/G9/97 and in one of the four birds inoculated with Pg/HK/FY6/99, but virus was not detected in Qa/HK/G1/97-inoculated birds; also, no viruses spread to contact birds (results not shown). Thus, two of the viruses tested replicated poorly if at all in pigeons, one failed to replicate, and none of the H9N2 viruses tested were transmitted to contact pigeons.
DISCUSSION
This study establishes that two phylogenetic lineages of H9N2 influenza viruses continue to circulate in the poultry of southeast China. One lineage is circulating predominantly in quail and is represented by Qa/HK/G1/97; members of this lineage carry six internal gene segments genetically closely related to those found in HK/156/97 that spread directly to humans from chickens, causing lethal infection in 6 of 18 persons infected (20, 21, 24). Viruses identical in all gene segments with Qa/HK/G1/97 spread to two children in Hong Kong in 1999 (13; Lin, personal communication).
Serological surveillance at ports of entry into Hong Kong SAR provided evidence of H9N2 infection in most species of poultry examined, including both land-based and aquatic birds; over 50% of consignments of chickens and quail had been infected with H9N2 viruses. In chickens and ducks, where the most extensive studies were done, a higher percentage had antibodies to Ck/HK/G9/97 than to Qa/HK/G1/97 (Table 1). Since postinfection chicken sera do not completely discriminate between Qa/HK/G1/97 and Ck/HK/G9/97 (Table 3), we are not sure which of these H9N2 viruses infected the poultry. The results indicate a widespread distribution of these viruses in different types of poultry and were fairly constant over 1 year. It is interesting that geese showed no detectable antibodies to Qa/HK/G1/97, suggesting that this species may not be susceptible to this H9N2 virus.
Studies in the poultry markets in 1999 indicated that the two H9N2 lineages might have different host preferences. The viruses similar to Qa/HK/G1/97 were predominantly from quail, and the percentage of fecal samples in quail cages yielding H9N2 virus was as high as 16%. This contrasts with the percentage of chickens and other land-based poultry, where the percentage of cages containing H9N2 viruses was between 3 and 5%. The high percentage of positive fecal samples from quail in the markets (Table 2) contrasts with the finding that the H9N2 viruses are transmitted primarily by the aerosol route in experimental studies (Table 7). A possible explanation is that virus from respiratory secretions contaminates the drinking water which is splashed into the dropping trays. The other cages that yielded above the 3 to 5% range were those containing mixed species, and this may be due to the presence of quail in the cages. The reason for the high incidence of H9N2 virus in quail in the poultry markets was not resolved in this study. One possibility is that a relatively high percentage of quail entering the markets are already infected. Alternatively, quail entering the market were not carrying virus and were not immune, and the virus was acquired in the market, where it is maintained by regular introduction of naive birds. While the Qa/HK/G1/97-like virus was also isolated from cages containing pigeons and pheasant and one cage of chickens, we cannot be certain that these isolates actually originated from those species, from fecal contamination, or from aerosol contamination of fomites. While chickens could be experimentally infected with Qa/HK/G1/97 virus, we failed to infect pigeons experimentally with this virus.
The high proportion of quail shedding Qa/HK/G1/97-like viruses (16%) is close to the proportion of chickens shedding H5N1 in 1997 (21.4%) (19). The presence of such a high percentage of birds with Qa/HK/G1/97 that is known to have infected two children (13; Lin, personal communication) is cause for concern. While it cannot be said for certain that the incidence of Qa/HK/G1/97-like virus was higher than in 1997 due to the small sample size in 1997 (15 quail), it is apparent that in 1999 the percentage was high. This situation may smolder for an extended period, but the high rate of evolution in the quail virus indicates that further interspecies transmission might occur.
Antigenic and genetic analyses confirmed that the two lineages of H9N2 viruses circulating in southeastern China in 1999 are distinguishable and are clearly separated phylogenetically. This applies to each of the eight gene segments of the viruses in each lineage. It is noteworthy that we failed to detect reassortants between the Qa/HK/G1/97-like virus and the Dk/HK/Y280/97-like viruses even though both viruses were isolated from the same market on the same day. Earlier studies demonstrated that reassortants between these lineages were detected in 1997 (3). The Ck/HK/G9/97 is a reassortant that possesses the PB1 and PB2 genes, similar to Qa/HK/G1/97, with the remainder of the genome most homologous with Dk/HK/Y280/97-like virus (Table 4). The Ck/HK/G9/97-like viruses were widespread in the poultry markets in 1997 with H5N1 strains, and neither has been detected since the depopulation and cleaning of the markets in December 1997. The possibility exists that both the HK/156/97-like and the Ck/HK/G9/97-like viruses are reassortants (3, 23) that arose in the poultry markets in Hong Kong SAR. The failure to detect reassortants between the two cocirculating H9N2 lineages in 1999 may reflect the relatively small number of viruses that have been completely genotyped, but it is apparent that no reassortants between the H9N2 lineages have become dominant in the poultry markets in 1999.
This study establishes that viruses with internal genes closely related to H5N1/97 and Qa/HK/G1/97 are still circulating in poultry in southeastern China. The transmission of H9N2 influenza viruses to humans was reported in Guangdong Province adjacent to Hong Kong SAR (5) and was confirmed in two children in Hong Kong (13; Lin, personal communication). The virus that transmitted to humans in Hong Kong belonged to the Qa/HK/G1/97-like lineage and contained six gene segments of the HK/156/97 human genotype. In the H9N2 envelope, the virus seemingly has less propensity to spread to humans than in the H5N1 envelope. Whether this reflects a lower intensity of exposure (quail are less widespread in markets than chickens) or an intrinsic property of the virus is unclear. The absence of basic amino acids at the cleavage site in the H9 HA (4) may modulate the pathogenicity of the H9N2 viruses. The fact that H9N2 viruses of the Qa/HK/G1/97-like genotype can transmit to and cause respiratory disease in humans (13) confirms that the surface glycoproteins can fulfill their primary functions in mammals. The presence of the H5N1 internal genome complex probably contributes significantly to the ability of these viruses to transfer and replicate in humans. Investigation of the internal genome complex of HK/156/97-like viruses implicate the PB2, PB1, PA, and NP genes (7, 11, 25) in host range transmission, but the actual residues involved are not known. However, the available evidence indicates that the H5N1 genome has a special propensity for interspecies transmission to mammals. The continued circulation of Qa/HK/G1/97-like viruses in poultry, especially in quail, alerts us to the continuing need for active surveillance for these viruses in humans and pigs in this region.
Transmission studies of the Qa/HK/G1/97-like viruses in quail indicate that this virus is transmitted primarily by the aerosol route (Tables 6 and 7). In comparison, the HK/156/97-like viruses (Ck/HK/220/97) are transmitted primarily through the feces (17). Since the internal genomes of Qa/HK/G1/97 (H9N2) and HK/156/97 (H5N1) are largely homologous, the differences in mode of transmission are probably linked to the HA and NA genes. It was demonstrated that mutations in the HA gene were associated with ability to multiply in the intestinal tract of ducks (10). The receptor binding site on the HA of Qa/HK/G1/97 has a leucine residue 226, which is typically found in human influenza viruses and may in fact explain the ability of this virus to transmit to humans.
The consequence of aerosol transmission is obvious for interspecies transmission; thus, we might ask, with up to 16% of cages of quails shedding the Qa/HK/G1/97-like virus, why are there not more interspecies transmissions? It seems likely that the Qa/HK/G1/97-like viruses lack other genetic characteristics required for interspecies transmission and that these properties are probably associated with the HA and NA genes. As these viruses continue to circulate, it is possible that they will acquire the genetic changes needed for more successful interspecies transmission.
To obtain information on the evolutionary rates of the two H9N2 virus lineages circulating in 1999, Qa/HK/G1/97-like and Dk/HK/Y280/97-like viruses from 1999 were compared with the H5N1 viruses from 1997. The internal gene complex of HK/156/97-like viruses has been shown to be in relatively rapid evolution. This is consistent with the results of a previous report (25). It is postulated that the H5N1 virus in chickens is a reassortant between Goose/Guangdong/1/96 (H5N1) and either a Qa/HK/G1/97-like or an A/Teal/HK/W312/97-like (H6N1) virus (3, 6, 23). Comparison of the amino acid substitutions per nucleotide change of each gene segment shows that the PB2, HA, NP, and NA genes of Qa/HK/G1/97 are evolving significantly faster than the corresponding gene segments of Dk/HK/Y280/97-like viruses (Table 5). The high amino acid substitution in the PB2, HA, NP, and NA genes of Qa/HK/G1/97-like viruses indicates that these genes may be important for adaptation to quail; like the HK/156/97-like viruses, the Qa/HK/G1/97-like viruses may be also relatively recent introductions into quail. What is apparent is that the Qa/HK/G1/97-like viruses in 1999 are in rapid evolution in those genes associated with host range transmission to mammals.
It is to be noted that the amino acid substitution rates in the genes of Qa/HK/G1/97-like and Dk/HK/Y280/97-like viruses are calculated from strains isolated 2 years apart, i.e., in 1997 to 1999, while those for H5N1 were calculated from viruses isolated in a single year, 1997, since no subsequent isolates are available. Thus, the rate of amino acid substitution of H5N1 viruses, underestimates, if anything, the change compared with the H9N2 viruses. Even so, H5N1 viruses still appear to have a higher amino acid substitution rate than Qa/HK/G1-like viruses, though these differences do not reach statistical significance except for the PB1 gene (Table 5). Overall, these results suggest that Dk/HK/Y280/97-like and Qa/HK/G1/97-like viruses were introduced into their land-based hosts earlier than the H5N1 viruses.
This study on H9N2 influenza viruses from poultry markets confirms that viruses belonging to two lineages represented by Qa/HK/G1/97 and Dk/HK/Y280/97 viruses are widespread in poultry in southeastern China and that the Qa/HK/G1/97-like viruses are prevalent in quail in the poultry markets. The studies provide preliminary characterization of the biological properties of these viruses in their hosts and emphasize the need for continued surveillance of viruses containing the H5N1 replicative complex in poultry and mammals.
ACKNOWLEDGMENTS
This study was supported by Public Health Service research grants AI29680 and AI95357 from the National Institute of Allergy and Infectious Diseases, Wellcome Trust grant 057476/Z/99/Z, Cancer Center Support CORE grant CA-21765, and the American Lebanese Syrian Associated Charities.
The continued support of the officers of the Food and Environmental Hygiene Department is gratefully acknowledged, and we kindly acknowledge Xiong Xiaoping, Biostatistics and Epidemiology, St. Jude Children's Research Hospital, for statistical analysis. We thank L. J. Zhang, P. Ghose, C. Y. Cheung, C. H. Lee, and D. Walker for excellent technical support and S. Gray and A. Goh for preparation of the manuscript.
REFERENCES
- 1.Alexander D J. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. doi: 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 2.Claas E C J, Osterhaus A D M E, Van Beck R, de Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A (H5N1) virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 3.Guan Y, Shortridge K F, Krauss S, Webster R G. Molecular characterization of H9N2 influenza viruses; were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y J, Krauss S, Senne D A, Mo I P, Lo K S, Xiong X P, Norwood M, Shortridge K F, Webster R G, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 5.Guo Y J, Li J W, Cheng I. Discovery of humans infected by avian influenza A (H9N2) virus. Chin J Exp Clin Virol. 1999;15:105–108. [PubMed] [Google Scholar]
- 6.Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin P S, Peiris M, Shortridge K F, Webster R G. Characterization of the influenza A gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J Virol. 2000;74:6309–6315. doi: 10.1128/jvi.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horimoto Y, Yamazaki Y, Fukushima T, Saito T, Lindstrom S E, Omoe K, Nerome R, Lim W, Sugita S, Nerome K. Evolutionary characterization of the six internal genes of H5N1 human influenza A virus. J Gen Virol. 2000;81:1293–1303. doi: 10.1099/0022-1317-81-5-1293. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig S, Stitz L, Planz O, Van H, Fitch W M, Scholtissek C. European swine virus as a possible source for the next influenza pandemic? Virology. 1995;212:555–561. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- 9.Naeem K, Ullah A, Manvell R J, Alexander D J. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet Rec. 1999;145:560. doi: 10.1136/vr.145.19.560. [DOI] [PubMed] [Google Scholar]
- 10.Naeve C W, Hinshaw V S, Webster R G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984;51:567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. Genetic analysis of the compatibility between polymerase proteins from humans and avian strains of influenza A viruses. J Gen Virol. 2000;81:1283–1291. doi: 10.1099/0022-1317-81-5-1283. [DOI] [PubMed] [Google Scholar]
- 12.Palmer D F, Coleman M, Dowdle W R, Schild G G. Advanced laboratory techniques for influenza diagnosis. Immunology Series no. 6. Washington, D.C.: U.S. Department of Health, Education, and Welfare; 1975. pp. 51–52. [Google Scholar]
- 13.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L, Lai R W, Orr W K, Shortridge K F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 14.Shortridge K F, Butterfield W K, Webster R G, Campbell C H. Isolation and characterization of influenza A viruses from avian species in Hong Kong. Bull WHO. 1977;55:15–19. [PMC free article] [PubMed] [Google Scholar]
- 15.Shortridge K F. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- 16.Shortridge K F. The next pandemic influenza virus? Lancet. 1995;346:1210–1212. doi: 10.1016/s0140-6736(95)92906-1. [DOI] [PubMed] [Google Scholar]
- 17.Shortridge K F, Zhou N N, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti K G, Norwood M, Senne D, Sims L, Takada A, Webster R G. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- 18.Shortridge K F. Influenza—a continuing detective story. Lancet. 1999;354(Suppl.):SIV29. doi: 10.1016/s0140-6736(99)90372-0. [DOI] [PubMed] [Google Scholar]
- 19.Shortridge K F. Poultry and the influenza H5N1 outbreak in Hong Kong, 1997: abridged chronology and virus isolation. Vaccine. 1999;17:S26–S29. doi: 10.1016/s0264-410x(99)00102-4. [DOI] [PubMed] [Google Scholar]
- 20.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subbarao K, Kilmov A, Katz J, Regenery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 22.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Subbarao K, Cox N J, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 24.Yuen K Y, Chan P K S, Peiris M, Tsang D N C, Que T L, Shortridge K F, Cheung P T, To W K, Ho E T F, Sung R, Cheng A F B Members of the H5N1 Study Group. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhou N N, Shortridge K F, Claas E J, Krauss S L, Webster R G. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]