Abstract
Introduction
Norovirus is widely recognized as a leading cause of both sporadic cases and outbreaks of acute gastroenteritis (AGE) across all age groups. The GII.4 Sydney 2012 variant has consistently prevailed since 2012, distinguishing itself from other variants that typically circulate for a period of 2–4 years.
Objective
This review aims to systematically summarize the prevalence of norovirus gastroenteritis following emergence of the GII.4 Sydney 2012 variant.
Methods
Data were collected from PubMed, Embase, Web of Science, and Cochrane databases spanning the period between January 2012 and August 2022. A meta-analysis was conducted to investigate the global prevalence and distribution patterns of norovirus gastroenteritis from 2012 to 2022.
Results
The global pooled prevalence of norovirus gastroenteritis was determined to be 19.04% (16.66–21.42%) based on a comprehensive analysis of 70 studies, which included a total of 85,798 sporadic cases with acute gastroenteritis and identified 15,089 positive cases for norovirus. The prevalence rate is higher in winter than other seasons, and there are great differences among countries and age groups. The pooled attack rate of norovirus infection is estimated to be 36.89% (95% CI, 36.24–37.55%), based on a sample of 6,992 individuals who tested positive for norovirus out of a total population of 17,958 individuals exposed during outbreak events.
Conclusion
The global prevalence of norovirus gastroenteritis is always high, necessitating an increased emphasis on prevention and control strategies with vaccine development for this infectious disease, particularly among the children under 5 years old and the geriatric population (individuals over 60 years old).
Keywords: norovirus, gastroenteritis, prevalence, meta-analysis, genotype
1. Introduction
Norovirus (NoV) is a non-enveloped, single-stranded RNA virus belonging to the Caliciviridae family, with a genome length of approximately 7.5 kb and a diameter ranging from 26 to 40 nm (1). Genogroups are further classified into capsid genogroup (genotypes) or P-genogroup (genotypes), which are determined based on the divergence of the VP1 capsid (ORF2) amino acid sequence or nucleotide diversity in the RNA-dependent RNA polymerase (RdRp; ORF1) region, respectively. Based on the capsid genogroup, NoV have been classified into ten genogroups (GI ~ GXI). Among these genogroups, GI, GII, GIV, GVIII, and GIX have been identified in humans (2). GII genogroup has a significantly higher prevalence compared to others (3, 4). Presently, there are 48 distinct capsid genotypes and 60 unique P-genotypes. Additionally, a dual-nomenclature system has been proposed to integrate both the RdRp and VP1 sequences, in response to the possibility of recombination events occurring between ORF1 and ORF2 (2). NoV is widely recognized as a leading cause of both sporadic cases and outbreaks of acute gastroenteritis (GE) across all age groups. The populations most susceptible to norovirus gastroenteritis (NoVGE) may encompass infants, the older adult, and individuals with compromised immune systems (5). According to Nadim et al., the global prevalence of NoV in community cases of GE was reported as 24%, while the prevalence in outbreaks was reported as 38% (6). NoV caused more than 130,000 people globally in 2019, with over 43,000 deaths occurring among children under the age of five and an additional 54,000 deaths among individuals aged 70 or above. NoVGE resulted in substantial direct health system costs amounting to $4.2 billion (95% UI: $3.2–5.7 billion) annually, along with societal costs reaching $60.3 billion (95% UI: $44.4–83.4 billion) (7, 8).
NoV is highly contagious and can spread rapidly in closed settings such as hospitals, schools, and cruise ships (9). NoV outbreaks occur year-round, with a higher incidence during colder seasons. Furthermore, NoV is responsible for a significant proportion of foodborne illnesses worldwide, with contaminated food and water serving as the main routes of transmission (10). The majority of these outbreaks predominantly manifest in educational institutions such as schools and kindergartens in China, primarily through person-to-person transmission, foodborne transmission, and waterborne transmission, and multiple ways of co-transmission (11). Prior to 2012, there existed five distinct NoV GII.4 variants that caused a worldwide pandemic and underwent replacement every three to four years. However, since the emergence of the GII.4 Sydney 2012 variant, it has gained extensive global circulation. To enhance our understanding of the distribution of NoVGE during the GII.4 Sydney 2012 variant era, we conducted a comprehensive meta-analysis spanning from 2012 to 2022 with the aim of assessing the current global pooled prevalence of this disease.
2. Materials and methods
2.1. Study area and period
This study analyzed the global prevalence and distribution characters of NoVGE from 2012 to 2022. The distribution characteristics included: (1) the geographical distribution: the prevalence distribution in different countries, in the northern and southern hemispheres, in developed and developing countries, as well as in China and its neighboring countries; (2) temporal distribution: prevalence distribution in different years, in months, in seasons adjusted for local temperature variations, and in cold season and warm seasons; (3) population distribution: prevalence variation in human populations with regards to age groups and gender stratification; (4) examination of the composition of NoV genotypes among sporadic cases; (5) Based on the relevant literature data obtained through the strategy for retrieving literature, this study analyzed the size of outbreaks, their attack rate, as well as the composition of NoV genotypes. We excluded papers which did not have an English abstract, did not show the number of patients with acute GE or patients positive for NoV, or percentages that could be used for calculating prevalence.
2.2. Literature source
2.2.1. Search strategy
We searched the PubMed, Embase, Web of Science, and Cochrane databases between January 2012 and August 2022. The following search terms were used as a text word in each database:“Norovirus” or “Norwalk,” “caliciviruse” and “Morbidity,” “positive rate,” “detection rate,” “attack rate” or “prevalence”, limited GE cases or Diarrhea cases. The literature was screened by reading the title, and after eliminating irrelevant literature, study eligibility was further assessed by reading the abstract and full text.
2.2.2. Quality control, criteria for inclusion, and exclusion
The literature underwent independent screening, selection, and cross-validation by two researchers. In the event of any disagreements, a third researcher was consulted for resolution. Initially, two reviewers independently selected articles that fulfilled the study requirements based on their titles and abstracts. During the initial screening process, articles meeting the following conditions were excluded: (1) articles published in languages other than English; (2) The GE cases were not caused by NoV but by other caliciviruses; (3) The subjects were not humans; (4) data derived from specific patient groups, such as transplant recipients and immunocompromised patients; (5) The pathogens were detected by antigen assays such as ELISA and immune-assays, not by PCR-based diagnostics methods. (6) The subjects were infected with NoV by human intervention rather than natural infection, such as volunteer challenge studies. (7) The articles were opinion articles and editorial articles, such as review articles, case reports, posters, and conference abstracts.
We conducted a comprehensive analysis of the entire texts of the remaining articles to identify those that met our research criteria. During this phase, we excluded the articles with the following characteristics: (1) papers that did not report the number of patients with acute GE or patients testing positive for NoV, or provide percentages that could be used to calculate pooled prevalence; (2) papers with a study period less than 12 months for sporadic cases surveillance; (3) studies with a sample size of less than 30 participants; and (4) If multiple studies present the same data, priority was given to the study with the highest level of comprehensiveness. In addition, we have excluded studies that monitor sporadic cases conducted by sub-municipal units and those focusing on an isolated outbreak event.
2.2.3. Data extraction
The following information was extracted from each eligible article: the last name of the first author, year of publication, study location, specimen Research Topic time (year / month), disease type, number of cases, age ranges and gender of participants, number of NoV-positive cases, genogroup and genotype of NoV. The classification of a country as developed or developing was determined based on the World Bank’s economic development criteria outlined in this website (12). The extracted data were imported into a pre-designed Excel spreadsheet (Microsoft Corporation) (13).
2.3. Statistical analysis
The statistical analysis in this study was conducted using R (4.1.2) software, the shapiro.test function from the stats package was employed for assessing the normality of the pooled original prevalence of NoVGE. Use the original incidence rate or perform a logarithmic transformation to make it follow a normal distribution, and perform meta-analysis on it using the metaprop function included in the metapackage. The forested.meta function was utilized to generate the forest plot, while the funnel function was used for drawing the funnel plot. The Egger test and sensitivity analysis are performed by metabios function. Additionally, sensitivity analysis forest plots were created using the forest function. A significance test level of p < 0.05 was employed in the meta-analysis, while heterogeneity levels were categorized as high when I2 exceeded 50% and very high when I2 surpassed 75%. The comparison of rates and component ratios was conducted using the chi-square test, with statistical significance defined as p < 0.05.
3. Results
3.1. Study characteristics
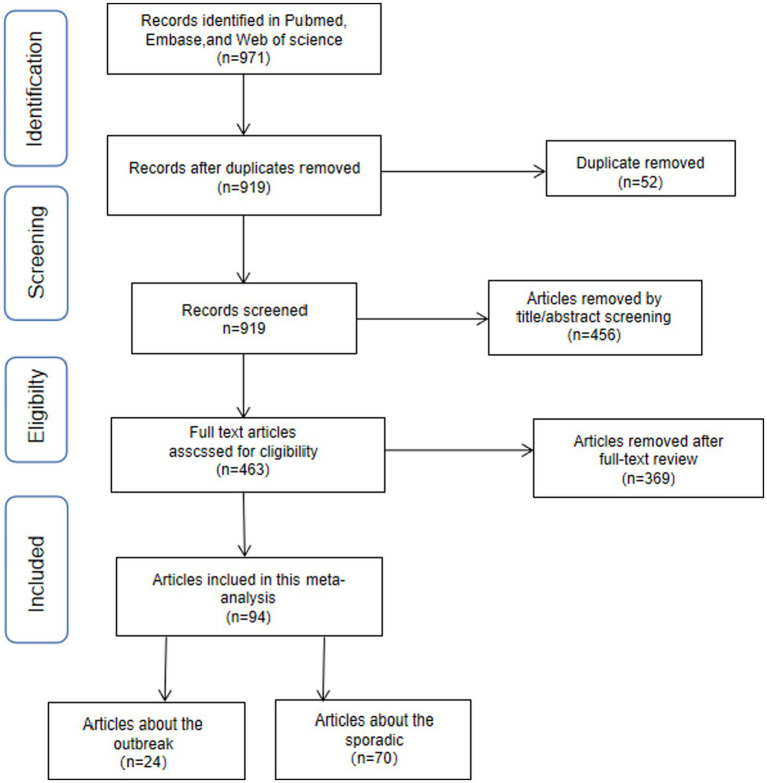
A total of 971 articles were initially identified in the search, with 883 retrieved from PubMed, 52 from Web of Science, 5 from Cochrane, and 31 from Embase. Fifty-two duplicate articles were excluded first, and 456 additional articles were excluded after review of titles and abstracts. The remaining 463 articles underwent a thorough assessment through full text reading. Among these, a total of 283 articles lacking data or figures or tables necessary for calculating the pooled prevalence of NoVGE were excluded. Furthermore, after careful evaluation, another set of 86 articles was excluded due to incomplete or unusable data. Ultimately, a total of 94 papers were considered to have good quality. A comprehensive overview illustrating the selection process is presented in Figure 1 and Table 1.
Figure 1.
Flowchart presenting the steps of literature search and selection Distribution of the sporadic NoV cases.
Table 1.
Characteristics of studies included in the systematic review and meta-analysis.
| Author | Publication year | Location | Disease type | Number of Cases/exposed in outbreak | Number of positive cases |
|---|---|---|---|---|---|
| Saho Honjo (14) | 2022 | Japan | Sporadic | 457 | 182 |
| Jing Ai (15) | 2022 | Jiangsu, China | Outbreak | 396,105 | 10,306 |
| Yaoska Reyes (16) | 2022 | Nicaragua | Sporadic | 1,353 | 229 |
| Pattara Khamrin (17) | 2022 | Thailand | Sporadic | 889 | 154 |
| Noemi Navarro-Lleó (18) | 2022 | Spain | Sporadic | 4,950 | 471 |
| Gillian A. M. Tarr (19) | 2021 | Albert, Canada | Sporadic | 3,319 | 898 |
| Mahadeb Lo (20) | 2021 | East India | Sporadic | 2,812 | 170 |
| Gédéon Prince Manouana (21) | 2021 | Gabon | Sporadic | 177 | 26 |
| Takako Utsumi (22) | 2021 | East Java, Indonesia | Sporadic | 966 | 119 |
| Sylvia Kahwage Sarmento (23) | 2021 | Brazil | Sporadic | 1,546 | 496 |
| Alberto Ignacio (24) | 2021 | Amazon | Sporadic | 485 | 184 |
| Mahadeb Lo (20) | 2021 | East India | Sporadic | 2,812 | 170 |
| Ignacio Parrón (25) | 2021 | Catalonia, Spain | Outbreak | 4,631 | 1,201 |
| Chengxi Sun (26) | 2021 | Shandong, China | Outbreak | / | / |
| Scott Grytdal (27) | 2020 | U.S.A | Sporadic | 1,603 | 103 |
| Meghana P. Parikh (28) | 2020 | Tennessee, USA | Outbreak | 3,273 | 755 |
| Lei Ji (29) | 2020 | Huzhou, China | Sporadic | 551 | 100 |
| Daniel Hungerford (30) | 2020 | Blantyre, Malawi | Sporadic | 683 | 83 |
| Weiwei Shen (31) | 2020 | Taizhou, China | Sporadic | 1,464 | 139 |
| Baisong Li (32) | 2020 | Chongqing, China | Outbreak | / | 1,637 |
| Liping Chen (33) | 2020 | Huzhou, China | Outbreak | 450 | 199 |
| Miao Jin (34) | 2020 | China | Outbreak | / | / |
| Juan I. Degiuseppe (35) | 2020 | Argentina | Outbreak | / | 189 |
| Yi He (36) | 2020 | Shanghai, China | Outbreak | / | / |
| Ignacio Parrón (37) | 2020 | Catalonia, Spain | Outbreak | 451 | 175 |
| Marco André Loureiro Tonini (38) | 2020 | Brazil | Sporadic | 272 | 65 |
| Belinda L. Lartey (39) | 2020 | Ghana | Sporadic | 1,337 | 485 |
| Weiwei Shen (31) | 2019 | Taizhou, China | Sporadic | 1,464 | 139 |
| Mohammad Enayet Hossain (40) | 2019 | Bangladesh | Sporadic | 613 | 109 |
| A. Gelaw (41) | 2019 | Northwest Ethiopia | Sporadic | 450 | 60 |
| Hera Nirwati (42) | 2019 | Indonesia | Sporadic | 406 | 75 |
| Liang Xue (43) | 2019 | China | Sporadic | 217 | 43 |
| Evandro Leite Rodrigues Bitencurt (44) | 2019 | Brazil | Sporadic | 240 | 38 |
| Julianne R. Brown (45) | 2019 | London, UK | Outbreak | 182 | / |
| Hui-ying Li (46) | 2019 | Hohhot, China | Sporadic | 1863 | 450 |
| Lijuan Lu (47) | 2019 | Shanghai China | Sporadic | 1,433 | 220 |
| Takumi Motoya (48) | 2019 | Ibaraki, Japan | Outbreak | 4,588 | 2,681 |
| Shilu Mathew (49) | 2019 | Qatar | Sporadic | 600 | 177 |
| Zhiyong Gao (50) | 2019 | Beijing, China | Outbreak | 762 | 661 |
| Kanti Pabbaraju (51) | 2019 | Canada | Outbreak; Sporadic | /; 755 | /; 94 |
| Vivaldie Mikounou Louya (52) | 2019 | Congo | Sporadic | 545 | 148 |
| Betina Hebbelstrup Jensen (53) | 2019 | Denmark | Sporadic | 688 | 103 |
| Jianguang Fu (54) | 2019 | Jiangsu, China | Outbreak | / | 3,951 |
| Kgomotso Makhaola (55) | 2018 | Botswana | Sporadic | 484 | 45 |
| Jiankang Han (56) | 2018 | Huzhou, China | Sporadic | 1,001 | 204 |
| Mahsa Farsi (57) | 2018 | Tehran, Iran | Sporadic | 210 | 36 |
| E. Pagani (58) | 2018 | Northern Italy | Sporadic | 702 | 162 |
| Young Eun Kim (59) | 2018 | Seoul, South Korea | Sporadic; Outbreak | 1,659; 2,414 | 271; 518 |
| Massimiliano Bergalloa (60) | 2018 | Northern Italy | Sporadic | 192 | 78 |
| Victoria Kiseleva (61) | 2018 | Russia | Sporadic | 429 | 49 |
| Lanzheng Liu (62) | 2018 | Jinan, China | Outbreak | 414 | 238 |
| Caoyi Xue (63) | 2018 | Shanghai, China | Sporadic | 5,927 | 1,363 |
| Rosa Joosten (64) | 2017 | Netherlands | Outbreak | / | / |
| Leigh M. Howard (65) | 2017 | Lusaka, Zambia | Sporadic | 454 | 52 |
| Victor S. Santos Ricardo Q (66) | 2017 | Brazil | Sporadic | 1,432 | 280 |
| Mehme Özkan Timukan (67) | 2017 | Erzurum, Turkey | Sporadic | 427 | 86 |
| Giovanni Maurizio Giammanco (68) | 2017 | Italy | Sporadic | 2,603 | 316 |
| T. N. Hoa-Tran (69) | 2017 | Vietnam | Sporadic | 350 | 99 |
| S. Niendorf (70) | 2017 | Germany | Outbreak | 240 | 175 |
| Sonam Wangchuk (71) | 2017 | Bhutan | Sporadic | 623 | 147 |
| Jennifer L. Cannon (72) | 2017 | United States | Outbreak | / | / |
| Nafissatou Ouédraogo (73) | 2016 | Ouagadougou, Burkina Faso | Sporadic | 263 | 55 |
| Seyed Dawood Mousavi Nasab (74) | 2016 | Tehran, Iran | Sporadic | 170 | 15 |
| Zufan Sisay (75) | 2016 | Ethiopia | Sporadic | 213 | 54 |
| Peng Zhang (76) | 2016 | Huzhou, China | Sporadic | 746 | 196 |
| Leesa D (77). | 2016 | Australia | Outbreak | / | / |
| Casey L. (78) | 2016 | Bolivia | Sporadic | 201 | 69 |
| Yaqing He (79) | 2016 | Shenzhen, China | Outbreak | / | / |
| Makoto Kumazaki (80) | 2016 | Japan | Outbreak | 1,497 | 1,050 |
| Nada M Melhem (81) | 2016 | Lebanon | Sporadic | 739 | 83 |
| Hee Soo Koo (82) | 2016 | Busan, South Korea | Sporadic | 2,174 | 49 |
| C. F. Manso (83) | 2015 | Spain | Sporadic | 2,750 | 747 |
| Sonia Etenna Lekana-Douki (84) | 2015 | Gabon | Sporadic | 317 | 73 |
| Xiaofang Wu (85) | 2015 | Huzhou, China | Sporadic | 796 | 211 |
| Ben A. Lopman (86) | 2015 | Ecuador | Sporadic | 438 | 79 |
| Dongmei Tan (87) | 2015 | Nanning, China | Sporadic | 342 | 101 |
| Zhiyong Gao (88) | 2015 | Beijing, China | Sporadic | 3,832 | 263 |
| Heejin Ham, M. S (89). | 2015 | Seoul, South Korea | Sporadic | 1,685 | 302 |
| Jiankang Han (85) | 2015 | Huzhou, China | Sporadic | 809 | 193 |
| Jae-Seok Kim (90) | 2015 | Korea | Sporadic | 2,980 | 349 |
| Makoto Kumazak (91) | 2015 | Japan | Outbreak | 947 | |
| Ji-Hyuk Park (90) | 2015 | Korea | Outbreak | / | / |
| Masaki Yoneda (92) | 2014 | Nara, Japan | Sporadic | 274 | 32 |
| Ainara Arana (93) | 2014 | Chipsqua, Spain | Sporadic | 4,574 | 714 |
| James Ayukepi Ayukekbong (94) | 2014 | Cameroon Linbei | Sporadic | 2,458 | 100 |
| João Rodrigo Mesquita (95) | 2013 | Global | Sporadic | 373 | 83 |
| Meng-Bin Tang (96) | 2013 | Taiwan, China | Sporadic | 155 | 17 |
| Carmen F. Manso (97) | 2013 | Southern Spain | Sporadic | 2,643 | 747 |
| Amy A. Saupe (98) | 2013 | U.S.A | Sporadic | 1,060 | 127 |
| Maria E. Hasing (99) | 2013 | Canada | Outbreak | / | / |
| Hyun Soo Kim (100) | 2013 | Korea | Sporadic | 1718 | 254 |
| Pascale Huynena (101) | 2013 | Burkina Faso | Sporadic | 418 | 93 |
| Nguyen V. Trang (102) | 2012 | Vietnam | Sporadic | 501 | 180 |
| Mei Zeng (103) | 2012 | China | Sporadic | 4,440 | 1,148 |
3.2. Distribution of the sporadic norovirus cases
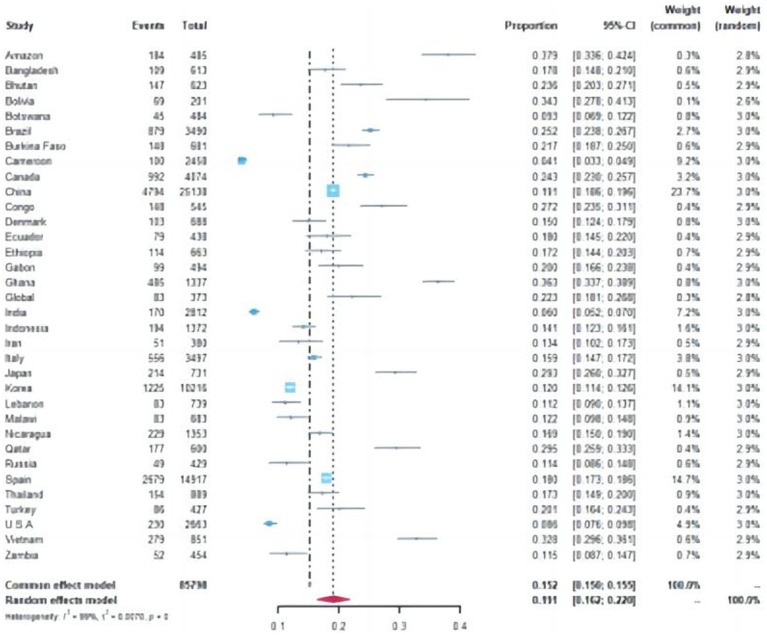
A total of 15,089 cases tested positive for NoV out of 85,798 sporadic cases with acute GE from 70 studies. The global average pooled prevalence of NoV in patients with acute GE was 19.04% (95%Cl:16.66–21.42%). Notably, the highest prevalence was observed in the Amazon region (37.94%), while Cameroon recorded the lowest prevalence at a mere 4.07%. Statistical analysis revealed an I2 value of 99% and a significance level of p < 0.001 (Figures 2, 3).
Figure 2.
Forest plot of the pooled norovirus prevalence in the sporadic gastroenteritis cases from difference countries. CI, confidence interval.
Figure 3.
Funnel plot of the pooled norovirus prevalence in the sporadic gastroenteritis cases from difference countries. CI, confidence interval.
Specifically, the pooled prevalence in the Southern Hemisphere reached 20.02% (95% Cl: 19.32–20.72%), whereas it stood at 17.15% (95% Cl: 16.88–17.42%) in the Northern Hemisphere (Chi-square value = 41.6, p < 0–05) (Supplementary Figures S4.1, S4.2). Furthermore, developed countries demonstrated a lower positivity rate of 16.25% (95% Cl:15–88%-16–63%), whereas developing countries displayed a relatively higher positivity rate of 18–58% (95%C1:18–23%-18–93%) (Chi-square value = 55.34, p < 0.05).
Out of the 70 articles reviewed, 43 originated from China and its neighboring countries. The pooled prevalence of NoVGE patients, based on the 43 articles, was found to be 17.63% (95% Cl: 13.35–21.91%) (Supplementary Figures S1.1, S1.2). This prevalence was lower than that of China alone (19.07, 95% Cl: 18.59–19.56%) Additionally, Bangladesh and Thailand exhibited similar prevalence rates to China (Chi-square value = 0.44, p < 0.05; Chi-square value = 1.18, p < 0.05) (Supplementary Figures S2.1, S2.2). Japan and Vietnam demonstrated higher prevalence rates at 29.27% (95% Cl: 25.98–32.57%) and 32.78% (95% Cl: 29.63–35.94%), respectively. Conversely, Korea, Indonesia, and Russia showed relatively lower prevalence rates, with India reporting the lowest prevalence at 6.05% (95% Cl: 5–7). Furthermore, the prevalence of NoV in Zhejiang (19.12, 95% Cl: 18.63–19.61%) was significantly higher than that in Taiwan Province (10.97, 95% Cl: 6.05–15.89%) (Chi-square value = 4.84, p < 0.05) (Figure 4).
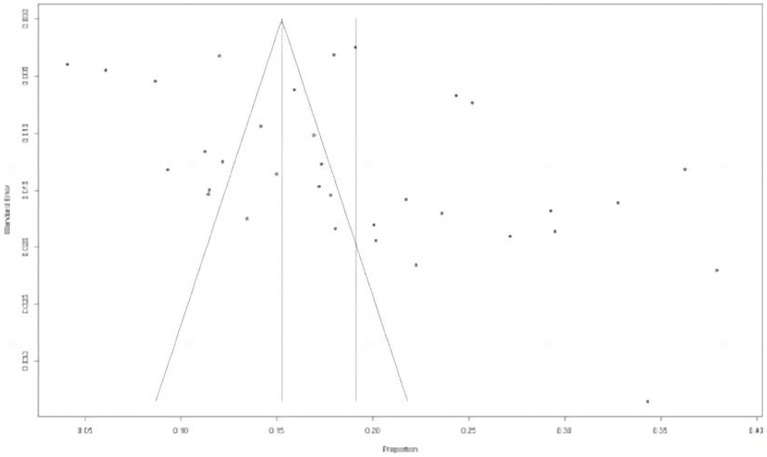
Figure 4.
Funnel plot for assessment of publication bias.
A total of 20 studies provided information on sample Research Topic, with contributions from China (5), Korea (3), Indonesia (2), the U.S.A. (2), and one each from Japan, the Amazon, Botswana, Brazil, India, Italy, and Thailand. Additionally, a separate study conducted simultaneous monitoring in multiple countries. The value of I2 indicated a high level of heterogeneity at 98%, with p < 0.05 (Supplementary Figure S11.1). The average pooled prevalence based on these papers was approximately 16.01% (95% Cl: 15.59–16.44%). Notably, in 2015 the highest positive rate observed was 23.21% (95% Cl: 21.79–24.64%), followed by 21.07% (95% Cl:19 0.55%-22 0.59%) in 2017.
A total of nine articles, including two from China, four from Korea, and one each from Amazon, Indonesia, Italy, and the USA were analyzed to provide data on the prevalence of NoV in patients with GE on a monthly basis. The analysis revealed a significant level of heterogeneity (I2 = 98%, p < 0.05) (Supplementary Figures S5.1, S5.2). The overall prevalence of NoV was determined to be 16.52% (95% Cl: 12.49–20.54%). To account for seasonal differences between the northern and southern hemispheres, adjustments were made for their respective seasons as described above. Following these adjustments, it was found that December had the highest prevalence of NoV at 26.31% (95% Cl: 24.15–28.46%), followed by January at 23.44% (95% Cl: 21.46%-25–42%) and November at 23.32% (95% Cl: 21–18%-25–47%). In contrast, June exhibited the lowest prevalence of NoV at only 6–83% (95% Cl: 5 0.43%-8 0.23%).
Based on the aforementioned nine articles, an additional two papers were included in this analysis, revealing a prevalence of 16.25% (95% Cl: 8.84–23.66%), with a high level of heterogeneity (I2 = 100%, p < 0.05) (Supplementary Figures S6.1, S6.2). It is worth noting that during the winter months (December–February in the Northern Hemisphere and June–August in the Southern Hemisphere), a significantly higher prevalence of NoV was observed at 22.98% (95% Cl: 21.82–24.14%). In contrast, the summer months (June–August in the Northern Hemisphere and December–February in the Southern Hemisphere) exhibited a relatively lower prevalence of NoV at 7.05% (95% Cl: 6.23–7.79%).
A comprehensive analysis was conducted using data from the 11 aforementioned studies and an additional publication to examine the prevalence of NoV during warm seasons (April to September in the Northern Hemisphere, October to March in the Southern Hemisphere) and cold seasons (October to March in the Northern Hemisphere, April to September in the Southern Hemisphere). The statistical analysis revealed a high level of heterogeneity (I2 = 100%, p < 0.05) (Supplementary Figures S7.1, S7.2). The overall prevalence of NoV was determined to be 15.21% (95% Cl: 5.02–25.39%). Specifically, during warm seasons, the prevalence rate was 10.01% (95% Cl: 9.42–10.61%); however, this significantly increased to 20.41% (95% Cl: 19.66–21.15%) during cold seasons (p < 0.01).
3.3. Distribution of the norovirus outbreak
A total of 7,054 outbreak events were pooled from 24 studies across 10 countries: China (10), the United States (2), Japan (2), Canada (2), Spain (2), Argentina, Australia, Germany, South Korea, the Netherlands, and the United Kingdom. However, 15 articles were excluded due to insufficient information on the number of outbreaks or lack of information on cases or exposed individuals. Of the remaining nine articles, one reported a total of 10,563 individuals testing positive for NoV out of 396,797 exposed individuals with an attack rate of 2.66% (95%Cl: 2.55%–2.65%). Upon review of their raw data, it shows that the number of exposed people in a large collective unit is the same as the total number of people in the unit, particularly in settings such as universities where not all individuals may have been exposed. Therefore, exclusion is necessary. In the remaining 8 articles, a total of 6,992 individuals tested positive for NoV out of a population of 17,958 who were exposed to outbreaks, allowing for an estimated prevalence of NoV infection at 36.89% (95% CI: 36.24–37.55%). From 2012 to 2019, a total of 10 studies in these papers reported the time information of the NoV outbreak from [China (3), the United States (2), Spain (1), Canada (1), Netherlands (1), Australia (1), Japan (1)]. These data showed that there are 334, 1,389, 1,028, 776, 148, 183, 259, and 26 outbreaks events in each respective year from 2012 to 2019.
There were 6 studies [China (2), Spain (1), Canada (1), Netherlands (1), South Korea (1)] provide the months distribution of the outbreaks, showing that December had the highest number of outbreak events (155) followed by November (80), July had the lowest count of outbreak events (4). Analyzing seasonal distribution revealed that spring accounted for 621 norovirus outbreak events while summer recorded 122 events, Autumn and winter experienced relatively higher incidences with counts reaching 282 and 401 events, respectively, based on the data obtained from 12 studies [China (6), Spain (1), the United States (2), Canada (1), Italy (1), South Africa (1), and the Netherlands (1)].
3.4. Genotype of norovirus
A total of 34 articles from 17 countries were used to genotype analysis in sporadic cases, including Brazil (2 articles), Botswana (2 articles), Bhutan (1 articles), China (8 articles), Ethiopia (1 article), Ghana (1 article), Germany (1 article), Italy (2 articles), Indonesia (2 articles), Japan (2 articles), Korea (4 articles), Malawi (1 article), Nicaragua (1 article), Qatar (1 article), Spain (3 articles), Thailand (1 article) and Vietnam (1 article). These studies reported 4,944 patients infected with various strains including GI.2 to GI.19, GI. untyped, GII.1 to GII.17, and GII.20 to GII.22. Among them, the proportion of GII.4 was the highest, 1722 cases of GII.4 genotype were reported in 16 countries, accounting for 34.83% of the total. The second was GII.3, with 1,245 cases (25.18%) reported from 15 countries. The detection rate of GII.2 genotype was 8.54% in 15 countries, while GII.17, GII.6, GII.14, GII.5, and GII.1 was 4.25, 2.89, 2.60, 2.74, and 2.53%, respectively.
In 8 articles reporting norovirus genotype from1292 cases in China, GI accounted for 6.89% and GII accounted for 93.11%. The most genotype was GII.4 with 481 cases (37.23%). The second was GII.3, with 308 cases (23.84%). Then there were 124 cases of GII.17, accounting for 9.60%, and 117 cases of GII.2, accounting for 9.06%.
A total of 14 studies [China (5), Spain (2), Canada (1), Germany (1), Japan (1)] reported the genotype of norovirus in 5166 outbreak events, with GInorovirus accounting for 12.25% and GII norovirus accounting for 87.75%, with GII.4 being the most prevalent (41.54%%), followed by GII.2 (22.11%), GII.6 (5.50%), and GII.3 (5.34%).
3.5. Sensitivity analysis
Sensitivity analysis revealed no statistically significant differences, except for a few outlier studies that deviate from the overall estimate. Since all the studies fall within the 95% confidence interval, the pooled prevalence remains unaffected by individual study findings.
4. Discussion
Norovirus is a major human pathogen caused severe GE affecting people from vulnerable populations of all age (104). Over the past three decades, a multitude of genotypes, including antigenically distinct variants of GII.4 noroviruses, have emerged and circulated in the world. GII.4 result more than 50% of all norovirus infections globally (13). The last variant to emerge, Sydney_2012, has been circulating at high incidence worldwide for over a decade caused the most severe disease burden (105). In this study, the pooled prevalence and genetic diversity of NoV were analyzed from studies conducted on patients with GE and published between January 2011 and April 2012.
In this study, it showed that the significant positive rate of norovirus in patients with acute GE worldwide. The positive rate of norovirus in GE patients is 19.04% (95%Cl:16.66–21.42%), which is higher than others reported. Ahmed et al. estimated the pooled prevalences of NoV infection was 18% in all age (106) while Mohammad Farahmand estimated the pooled prevalence of NoV infection was 17.7%% (95Cl: 16.3–19.2%) among children with GE from 45 countries across the world from 2015 to 2020 (13). Manish M Patel estimated that the pooled positive rate is 12% in patients with severe GE cases among children <5 years of age and 12% (95% Cl:9–15%) of mild and moderate diarrhea cases among persons of all ages (107) (Supplementary Figures S9.1, S9.2). The positive rate of the norovirus in patients is influenced by multiple factors, including viral variations, the number of susceptible individuals, and the surveillance and monitoring capabilities of participating institutions. Consequently, inconsistencies in pooled positive rates may arise due to variations in literature sources selected based on different inclusion and exclusion criteria. The high positive rate showed that we need more considering targeted intervention.
Our analysis reveals significant disparities in positive rates across regions and countries, with the highest rate observed in the Amazon region reaching up to 37.9%(95%Cl:33.6–42.4%), while the lowest rate was recorded in Cameroon at only 4.1% (95%Cl:3.3–4.9%). These highest and lowest positive rate countries is same as other meta-analysis (108). Yingyin Liao et al. showed that there was no statistical difference in terms of norovirus prevalence among different national income level (109). But our data indicates that developing countries exhibit higher rates of positivity compared to developed nations. Furthermore, our combined positive rate surpasses the findings from the meta-analysis conducted by Gia Thanh Nguyen et al., which revealed a decline in prevalence from 18% (95% CI: 16–20%) for upper middle-income countries to 15% (95%Cl:13–18%) and 6% (95%Cl:3–10%) for lower middle- and low-income countries, respectively, in both developed and developing regions (110) (Supplementary Figures S3.1, S3.2). Similarly, there is a greater prevalence of norovirus in the Southern Hemisphere than in the Northern Hemisphere. These findings emphasize substantial regional variations in norovirus occurrence. Insufficient or limited data on sporadic cases and outbreaks have been collected from numerous countries; however, it is worth noting that unreported sporadic cases are prevalent in these regions, particularly Africa. Consequently, the global incidence of norovirus may be underestimated due to these factors. All above highlight the necessity for further investigation into underlying factors contributing to these differences in positivity rates such as implementation of pandemic control measures, availability of healthcare resources, and testing capabilities.
According to our analysis of population distribution characteristics, we found that infants below the age of two had a higher positivity rate of norovirus infection, with a recorded rate of 23.1% (95%Cl: 21.7–24.5%). Notably, adolescents aged between five and fourteen demonstrated peak infection rates at 27.8% (95%Cl: 23.8–32.1%), while individuals within the age range of five to twenty also displayed high levels at 27.6% (95%Cl: 24.2–31.3%). In contrast, older adult individuals over sixty exhibited significantly lower infection rates at 12.3% (95%Cl:10.9–13 0.8%). In previous studies, Gia Thanh Nguyen reported prevalence rates of 16% (95%Cl:14–18%) for children under five years and 17% (95%Cl:13–21%) for adults. Ahmed et al. highlighted the global prevalence of norovirus in different age groups, with rates of 18%(95% CI: 15–20%)for children under five years, 18% (95% CI: 13–24%) for individuals over the age of five, and 19% (95% CI: 17–21%)for mixed age groups. The highest positive rate was shown in adolescents (14.74%) and adults (14.74%), followed by children (14.17%) and older adults (14.05%) (110). The pooled attack rate of norovirus was 6.73% based on the data of 899 outbreak events, with a higher attack rate in North than in South China, while the highest attack rate was found among older adults (11.85%), followed by children (9.48%), adolescents (5.53%) and adults (4.55%) (111). Yingyin Liao’s study revealed a comparable prevalence of norovirus among children under 5 years old, individuals over 5 years old, and across all age groups (109). The variations in prevalence observed among the age groups in these studies can be attributed to disparities in data sources, which focus on different populations under surveillance across various regions, as well as the distinct categorization of age groups employed within the studies. Furthermore, a higher prevalence of males (19.3% [18–20%]) compared to females (18.% [17–19%]) was observed, which is consistent with the findings reported by Mohammad Farahmand et al., indicating that boys are more susceptible than girls to NoVGE (13). This gender disparity may be attributed to occupational or lifestyle factors, as males tend to have a higher likelihood of dining outside the home or having increased contact with larger social networks.
The years 2015–2017 witnessed a notable increase in norovirus positive rates, with values of 23.2 (21.8–24.6), 20.8 (19.6–22.0), and 21.1 (19.6–22.6) respectively. The positive rate during the cold season, at 20.4 (19.7–21.2), significantly exceeded that of the warm season, particularly during winter where it reached a notably higher rate of 23.0% (21.8–24.2%). Among all months, December and January exhibited the highest rates with values of 26.3 (24.2–28.5) and 23.4 (21.5–25.5), respectively. Changes in temperature had the greatest attributable risk for norovirus incidence in a long-term study of England and Wales (112). Our results also suggested that there is a positive correlation between rainfall and seasonality as previous study (113). Although our findings align with previous studies (113), it is imperative to acknowledge the inherent limitations in our data sources. To gain a comprehensive understanding of the global seasonality of norovirus disease and its influencing factors, further research is needed in monitoring regions with limited capacity over an extended period of time (Table 2).
Table 2.
Analysis of the prevalence of norovirus in gastroenteritis.
| Characteristics | Categories | Cases of studies | Pooled prevalence (%) (95% CI) | Heterogeneity test I2%, p-value | Differences between subgroups; χ2 test (p-value) |
|---|---|---|---|---|---|
| Age groups (1) | <2 year | 827 | 23.1 (21.7–24.5) | 97%, p < 0.01 | 148.9 (p < 0.01) |
| 2–5 years | 94 | 13.5 (11.0–16.2) | |||
| 5–14 years | 130 | 27.8 (23.8–32.1) | |||
| 14-60 years | 782 | 19.0 (17.8–20.3) | |||
| >60 years | 249 | 12.3 (10.9–13.8) | |||
| Total | 2082 | 19.0 (14.3–23.7) | |||
| Age groups (2) | <5 years | 1,047 | 22.1 (21.0–23.3) | 98%, p < 0.01 | 136.4 (p < 0.01) |
| 5-20 years | 175 | 27.6 (24.2–31.3) | |||
| 20-60 years | 532 | 17.3 (16.0–18.7) | |||
| >60 years | 249 | 12.3 (10.9–13.8) | |||
| Total | 2003 | 19.7 (14.5–24.9) | |||
| Sex | Male | 1,337 | 19.3 (18.3–20.2) | 0, p = 0.48 | 0.5 (p = 0.48) |
| Female | 787 | 18.7 (17.6–19.9) | |||
| Total | 2,124 | 19.1 (18.3–19.8) | |||
| Years | 2012 | 518 | 16.4 (15.1–17.7) | 98%, p < 0.01 | 374.8 (p < 0.01) |
| 2013 | 927 | 14.9 (14.0–15.8) | |||
| 2014 | 492 | 15.2 (14.0–16.4) | |||
| 2015 | 783 | 23.2 (21.8–24.6) | |||
| 2016 | 929 | 20.8 (19.6–22.0) | |||
| 2017 | 582 | 21.1 (19.6–22.6) | |||
| 2018 | 222 | 9.8 (8.6–11.0) | |||
| 2019 | 155 | 10.2 (8.7–11.7) | |||
| Total | 4,608 | 16.2 (15.7–16.6) | |||
| Month | N January, S July | 413 | 23.4 (21.5–25.5) | 98%, p < 0.01 | 668.5 (p < 0.01) |
| N February, S August | 270 | 18.8 (16.8–20.9) | |||
| N March, S September | 326 | 22.0 (19.9–24.2) | |||
| N April, S October | 215 | 17.1 (15.1–19.3) | |||
| N May, S November | 159 | 12.2 (10.5–14.1) | |||
| N June, S December | 85 | 6.8 (5.5–8.4) | |||
| N July, S January | 96 | 6.9 (5.5–8.3) | |||
| N August, S February | 133 | 9.1 (7.7–10.7) | |||
| N September, S March | 166 | 12.2 (10.5–14.1) | |||
| N October, S April | 296 | 20.3 (18.3–22.5) | |||
| N November, S May | 348 | 23.3 (21.2–25.6) | |||
| N December, S June | 422 | 26.3 (24.2–28.5) | |||
| Total | 2,929 | 16.5 (12.5–20.5) | |||
| Season | Spring | 734 | 16.9 (15.8–18.0) | 99%, p < 0.01 | 650.2 (p < 0.01) |
| Summer | 327 | 7.1 (6.3–7.8) | |||
| Autumn | 847 | 18.1 (17.0–19.3) | |||
| Winter | 1,160 | 23.0 (21.8–24.2) | |||
| Total | 3,068 | 16.3 (8.8–23.7) | |||
| Temperature | Warm season | 981 | 10.0 (9.4–10.6) | 100%, p < 0.01 | 459.4 (p < 0.01) |
| Cold season | 2,315 | 20.4 (19.7–21.2) | |||
| Total | 3,296 | 15.2 (5.0–25.4) | |||
| Counties | Amazon | 184 | 37.9 (33.6–42.4) | 99%, p < 0.01 | 2947.1 (p < 0.01) |
| Bangladesh | 109 | 17.8 (14.8–21.0) | |||
| Bhutan | 147 | 23.6 (20.3–27.1) | |||
| Bolivia | 69 | 34.3 (27.8–41.3) | |||
| Botswana | 45 | 9.3 (6.9–12.2) | |||
| Brazil | 879 | 25.2 (23.8–26.7) | |||
| Burkina Faso | 148 | 21.7 (18.7–25.0) | |||
| Cameroon | 100 | 4.1 (3.3–4.9) | |||
| Canada | 992 | 24.3 (23.0–25.7) | |||
| China | 4,794 | 19.1 (18.6–19.6) | |||
| Congo | 148 | 27.2 (23.5–31.1) | |||
| Denmark | 103 | 15.0 (12.4–17.9) | |||
| Ecuador | 79 | 18.0 (14.5–22.0) | |||
| Ethiopia | 114 | 17.2 (14.4–20.3) | |||
| Gabon | 99 | 20.0 (16.6–23.8) | |||
| Ghana | 485 | 36.3 (33.7–38.9) | |||
| Global | 83 | 22.3 (18.1–26.8) | |||
| India | 170 | 6.0 (5.2–7.0) | |||
| Indonesia | 194 | 14.1 (12.3–16.1) | |||
| Iran | 51 | 13.4 (10.2–17.3) | |||
| Italy | 556 | 15.9 (14.7–17.2) | |||
| Japan | 214 | 29.3 (26.0–32.7) | |||
| Korea | 1,225 | 12.0 (11.4–12.6) | |||
| Lebanon | 83 | 11.2 (9.0–13.7) | |||
| Malawi | 83 | 12.2 (9.8–14.8) | |||
| Nicaragua | 229 | 16.9 (15.0–19.0) | |||
| Qatar | 177 | 29.5 (25.9–33.3) | |||
| Russia | 49 | 11.4 (8.6–14.8) | |||
| Spain | 2,679 | 18.0 (17.3–18.6) | |||
| Thailand | 154 | 17.3 (14.9–20.0) | |||
| Turkey | 86 | 20.1 (16.4–24.3) | |||
| U.S.A | 230 | 8.6 (7.6–9.8) | |||
| Vietnam | 279 | 32.8 (29.6–36.1) | |||
| Zambia | 52 | 11.5 (8.7–14.7) | |||
| Total | 15,089 | 19.0 (16.7–21.4) | |||
| Regions | Developed country | 6,048 | 16.3 (15.9–16.6) | 99%, p < 0.01 | 79.9 (p < 0.01) |
| Developing Countries | 8,958 | 18.6 (18.2–18.9) | |||
| Total | 15,006 | 17.4 (15.1–19.7) | |||
| The Southern Hemisphere | 2,485 | 20.0 (19.3–20.7) | 98%, p < 0.01 | 55.5 (p < 0.01) | |
| The Northern Hemisphere | 12,521 | 17.1 (16.9–17.4) | |||
| Total | 15,006 | 18.6 (15.6–21.3) | |||
| Chinese Mainland | 4,777 | 19.1 (18.6–19.6) | 83%, p = 0.02 | 10.5 (p < 0.01) | |
| Taiwan | 17 | 11.0 (6.5–17.0) | |||
| Total | 4,794 | 15.4 (7.5–23.4) |
N: The Northern Hemisphere; S: The Southern Hemisphere. Spring: In the Northern Hemisphere, the period from March to May is considered spring, while in the Southern Hemisphere it occurs from September to November. Summer: From June to August, the Northern Hemisphere experiences summer, whereas in the Southern Hemisphere it takes place between December and February. Autumn: The months of September to November mark autumn in the Northern Hemisphere, while in the Southern Hemisphere it falls within March to May. Winter is defined as the period from December to February in the Northern Hemisphere and from June to August in the Southern Hemisphere, according to seasonal divisions. The warm and cold seasons were determined based on the mean monthly temperature of each city included in this study. In the Northern Hemisphere, the warm season spanned from April to September, while in the Southern Hemisphere it extended from October to March; all other months were designated as the cold season. The pooled prevalence of each subgroup was calculated using the random effects model in R software.
Genogroup GII is the leading NoV genogroup in the world. The GII.4 NoV genotype have been dominating in the past thirty years, in our review, a great diversity of NoV genotypes were reported ranging from GII.1 to GII.20, except GII.18, with the predominance of GII.4 (31.19%) from all identified genotypes. This report is also in agreement with previous studies where an increased prevalence of non-GII.4 NoV had been reported in different parts of China and Africa as a single variant or as a recombinant type (108, 114). In our study, the prevailing genotypes of norovirus in sporadic cases were found to be GII.4 (31.19%), GII.3 (24.73%), GII.2 (8.58%), GII.17 (4.31%), and GII.6 (2.94%); whereas the predominant genotypes in outbreak cases were observed as follows: GII.441.54%%, followed by GII.2 (22.11%), GII.6 (5.50%), and GII.3 (5.34%). The period from 2012 to 2014 accounted for approximately 66.40%(2751/4143) of outbreaks events maybe resulted by the endemic of norovirus GII.4 which caused about 41.54% outbreak events. In the United States, a surveillance study conducted from 2012 to 2016 also identified disparities in the prevalence of dominant norovirus genotypes between sporadic cases and outbreaks of GE (28). In the winter of 2014/15 GII.P17-GII.17 norovirus became a major cause of GE outbreaks in China and Japan and have replaced the previously dominant GII.4 genotype Sydney 2012 variant in some areas in Asia, which had been predicted as a strain to end the GII.4 era (115). However, the detection rate of GII.17 appears to be relatively lower, consistent with the findings reported by Yingyin Liao (109). This observation may potentially be attributed to the limited duration and geographical scope of the epidemic caused by GII.17 (105).
Previous research has indicated that males may have a higher susceptibility to NoVGE compared to females (male-to-female odds ratio: 1.1; 95%Cl: 1.03–1.3; I2 = 45.3%). This suggests both sex hormones and notable endocrine and genetic differences between males and females early in life may contribute to an increased vulnerability to NoV infection (13). In our analysis, we did not find a statistically significant gender-based distribution, although there was a slightly higher overall prevalence in men compared to women. This observation could be attributed to the inclusion of data from the adult population in our dataset (Table 3).
Table 3.
The genotype composition of the sporadic NoVGE.
| Country | Total | GI | GII | GII0.2 | GII0.3 | GII0.4 | GII0.6 | GII0.17 | Other GII Genotypes |
|---|---|---|---|---|---|---|---|---|---|
| Bhutan | 88 | 0 | 88 (100%) | 2 (2.27%) | 43 (48.86%) | 26 (29.55%) | 3 (3.41%) | 2 (2.27%) | 12 (13.63%) |
| Botswana | 61 | 0 | 61 (100%) | 3 (4.92%) | 23 (37.70%) | 23 (37.70%) | 0 | 0 | 12 (19.67%) |
| Brazil | 227 | 14 (6.17%) | 213 (93.83%) | 8 (3.52%) | 36 (15.86%) | 149 (65.64%) | 4 (1.76%) | 0 | 30 (13.22%) |
| China | 1,292 | 89 (6.89%) | 1,203 (93.11%) | 117 (9.06%) | 308 (23.84%) | 481 (37.23%) | 44 (3.41%) | 123 (9.52%) | 219 (16.95%) |
| Ethiopia | 60 | 1 (1.67%) | 59 (98.33%) | 4 (6.67%) | 0 | 21 (35.00%) | 12 (20.00%) | 8 (13.03%) | 15 (25.00%) |
| Germany | 134 | 13 (9.70%) | 121 (90.30%) | 61 (45.52%) | 3 (2.23%) | 40 (29.85%) | 5 (3.73%) | 6 (4.48%) | 19 (14.18%) |
| Ghana | 159 | 18 (11.32%) | 141 (88.68%) | 13 (8.18%) | 86 (54.09%) | 7 (4.40%) | 0 | 0 | 53 (33.3%) |
| Indonesia | 144 | 7 (4.86%) | 137 (95.14%) | 12 (8.33%) | 21 (14.58%) | 59 (40.97%) | 11 (7.64%) | 8 (5.56%) | 33 (22.92%) |
| Italy | 362 | 6 (1.66%) | 356 (98.34%) | 25 (6.91%) | 11 (3.03%) | 248 (68.51%) | 21 (5.80%) | 34 (9.39%) | 23 (6.35%) |
| Japan | 154 | 0 | 154 (100%) | 53 (34.42%) | 10 (6.49%) | 70 (45.45%) | 5 (3.25%) | 12 (7.79%) | 4 (2.60%) |
| Korea | 1,225 | 100 (8.16%) | 1,125 (91.84%) | 78 (6.37%) | 500 (40.82%) | 201 (16.41%) | 19 (1.55%) | 1 (0.08%) | 426 (34.78%) |
| Malawi | 42 | 0 | 42 (100%) | 2 (4.76%) | 6 (14.29%) | 20 (47.62%) | 2 (4.76%) | 1 (2.38%) | 11 (26.19%) |
| Nicaragua | 114 | 41 (35.96%) | 73 (64.04%) | 0 | 0 | 44 (38.60%) | 1 (0.88%) | 8 (7.02%) | 61 (53.51%) |
| Qatar | 177 | 2 (1.13%) | 175 (98.87%) | 24 (13.56%) | 28 (15.82%) | 110 (62.15%) | 3 (1.69%) | 5 (2.82%) | 7 (3.95%) |
| Spain | 498 | 27 (5.42%) | 471 (94.58%) | 13 (2.61%) | 85 (17.07%) | 173 (34.74%) | 8 (1.61%) | 1 (0.20%) | 218 (43.78%) |
| Thailand | 131 | 1 (0.76%) | 130 (99.24%) | 7 (5.34%) | 66 (50.38%) | 0 | 1 (0.76%) | 0 | 57 (43.51%) |
| Vietnam | 76 | 0 | 76 (100%) | 0 | 19 (25.00%) | 50 (65.79%) | 4 (5.26%) | 1 (1.32%) | 2 (2.63%) |
| Total | 4,944 | 319 (6.45%) | 4,625 (93.55%) | 422 (8.54%) | 1,245 (25.18%) | 1722 (34.83%) | 143 (2.89%) | 210 (4.25%) | 1,202 (24.31%) |
Efforts are warranted to advance the development of effective vaccines and control programs aimed at mitigating the burden of this substantial disease. Due to the considerable and rapid variability of norovirus, the coexistence of multiple dominant genotypes in the same time and space, the low cross-protection between genotypes, and the high incidence and severity outcomes among infants, older adult individuals, and patients with underlying diseases, we suggest that polyvalent vaccines containing multiple genotype-specific antigen epitopes as proposed by other researchers should be adopted and recommended for the above-mentioned population and tailored for the aforementioned high-risk populations. Several norovirus vaccine candidates have shown limited progress in clinical trials. However, Vaxart Pharmaceutical Inc. has developed an oral norovirus vaccine candidate utilizing recombinant adenovirus-based vectors carrying genes encoding norovirus VP1s to express antigens locally in the epithelial cells within the intestine of vaccine recipients, thereby inducing mucosal immunity (116, 117). This candidate vaccine has recently demonstrated a statistically significant reduction in infection rate, a non-statistically significant reduction in NoVGE, and a substantial reduction in viral shedding during challenge studies (118). These findings suggest that oral administration holds promise for the successful development of an effective norovirus vaccine.
5. Limitations
There are several limitations associated with this study. Firstly, there are publication bias which influence the results in our research findings. The majority of these publications have predominantly originated from developed regions and areas where English serves as the primary language, which may introduce a potential source of bias. Secondly, we collected as much published data as possible to mitigate the potential impact of inadequate sample size on our findings. Notably, the actual subgroup sample size significantly exceeded the calculated sample size, thereby enhancing the robustness and reliability of our results (S12). However, it is important to acknowledge that due to the diverse range of data sources utilized and inherent biases in available literature and data sources, there are certain limitations in achieving consistency across variables between studies, which may have some influence on our outcomes. In summary, the presence of heterogeneity in the literatures is apparent and should be taken into account when interpreting the findings of this review. Although our systematic approach and stringent inclusion criteria likely mitigated heterogeneity, biases inherent in the original studies, variations in study design and population, as well as publication bias cannot be completely eliminated.
6. Conclusion
Our findings reveal a high prevalence of NoVGE, yet significant data gaps exist in various regions, particularly in developing countries, indicating the absence of systematic surveillance for NoVGE in these areas. To reduce the incidence and diseases burden of NoVGE, it is crucial to enhance global standardized monitoring and collaboration, as well as develop cost-effective vaccines.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
PZ: Writing – original draft, Writing – review & editing. CH: Writing – original draft, Writing – review & editing. XD: Writing – original draft. XC: Methodology, Formal analysis, Writing – original draft. LJ: Data curation, Methodology, Supervision, Writing – review & editing. ZG: Supervision, Writing – review & editing. LH: Supervision, Writing – review & editing. DZ: Project administration, Supervision, Writing – review & editing.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by funding from Beijing Natural Science Foundation (No. L232011), Epidemiological characteristics and economic burden of gastroenteritis caused by gene variation of norovirus in China and National Key Research, and Development Program of China (No. 2021YFC2301000), Research on targeted culture technology of difficult culture and trace pathogens.
Conflict of interest
XD was employed by Chengdu Kanghua Biological Products Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1373322/full#supplementary-material
References
- 1.Hardy ME. Norovirus protein structure and function. FEMS Microbiol Lett. (2005) 253:1–8. doi: 10.1016/j.femsle.2005.08.031 [DOI] [PubMed] [Google Scholar]
- 2.Chhabra P, de Graaf M, Parra GI, Chan MC, Green K, Martella V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. (2019) 100:1393–406. doi: 10.1099/jgv.0.001318, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu SC, Hu SC, Liao LM, Chen YH, Lin JH. Norovirus Genogroup II Epidemics and the Potential Effect of Climate Change on Norovirus Transmission in Taiwan. Viruses. (2022) 14:641. doi: 10.3390/v14030641, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tohma K, Lepore CJ, Martinez M, Degiuseppe JI, Khamrin P, Saito M, et al. Genome-wide analyses of human noroviruses provide insights on evolutionary dynamics and evidence of coexisting viral populations evolving under recombination constraints. PLoS Pathog. (2021) 17:e1009744. doi: 10.1371/journal.ppat.1009744, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karst SM. Pathogenesis of noroviruses, emerging RNA viruses. Viruses. (2010) 2:748–81. doi: 10.3390/v2030748, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng D-P, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. (2006) 346:312–23. doi: 10.1016/j.virol.2005.11.015, PMID: [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Chen C, du Y, Yan D, Jiang D, Liu X, et al. Global Burden and Trends of Norovirus-Associated Diseases From 1990 to 2019: An Observational Trend Study. Front Public Health. (2022) 10:905172. doi: 10.3389/fpubh.2022.905172, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global Economic Burden of Norovirus Gastroenteritis. PLoS One. (2016) 11:e0151219. doi: 10.1371/journal.pone.0151219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang LJ, Zhou M, Deng J, Zuo KJ, Shi JB, Lai YY. Study on the relationship between 11β-hydroxysteroid dehydrogenase and glucocorticoid response in nasal polyps. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. (2019) 54:198–202. doi: 10.3760/cma.j.issn.1673-0860.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 10.Niziolek PJ, Bullock W, Warman ML, Robling AG. Missense Mutations in LRP5 Associated with High Bone Mass Protect the Mouse Skeleton from Disuse- and Ovariectomy-Induced Osteopenia. PLoS One. (2015) 10:e0140775. doi: 10.1371/journal.pone.0140775, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu F, Jiang B, Guo X, Hou L, Tian Y, Zhang J, et al. Norovirus outbreaks in China, 2000-2018: A systematic review. Rev Med Virol. (2022) 32:e2382. doi: 10.1002/rmv.2382, PMID: [DOI] [PubMed] [Google Scholar]
- 12.World Bank Classification . (n.d.) Developed and Developing Countries: World Bank Classification | Economics. Available at: https://www.economicsdiscussion.net/economic-development/developed-and-developing-countries-world-bank-classification-economics/30010
- 13.Farahmand M, Moghoofei M, Dorost A, Shoja Z, Ghorbani S, Kiani SJ, et al. Global prevalence and genotype distribution of norovirus infection in children with gastroenteritis: A meta-analysis on 6 years of research from 2015 to 2020. Rev Med Virol. (2022) 32:e2237. doi: 10.1002/rmv.2237, PMID: [DOI] [PubMed] [Google Scholar]
- 14.Honjo S, Kuronuma K, Fujiya Y, Nakae M, Ukae S, Nihira H, et al. Genotypes and transmission routes of noroviruses causing sporadic acute gastroenteritis among adults and children, Japan, 2015-2019. Infect Genet Evol. (2022) 104:105348. doi: 10.1016/j.meegid.2022.105348, PMID: [DOI] [PubMed] [Google Scholar]
- 15.Ai J, Zhu Y, Fu J, Cheng X, Zhang X, Ji H, et al. Study of risk factors for total attack rate and transmission dynamics of norovirus outbreaks, Jiangsu Province, China, From 2012 to 2018. Front Med (Lausanne). (2021) 8:786096. doi: 10.3389/fmed.2021.786096, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes Y, González F, Gutiérrez L, Blandón P, Centeno E, Zepeda O, et al. Secretor status strongly influences the incidence of symptomatic norovirus infection in a genotype-dependent manner in a Nicaraguan Birth Cohort. J Infect Dis. (2022) 225:105–15. doi: 10.1093/infdis/jiab316, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khamrin P, Kumthip K, Yodmeeklin A, Jampanil N, Phengma P, Yamsakul P, et al. Changing Predominance of Norovirus Recombinant Strains GII.2[P16] to GII.4[P16] and GII.4[P31] in Thailand, 2017 to 2018. Microbiol Spectr. (2022) 10:e0044822. doi: 10.1128/spectrum.00448-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro-Lleó N, Santiso-Bellón C, Vila-Vicent S, Carmona-Vicente N, Gozalbo-Rovira R, Cárcamo-Calvo R, et al. Recombinant Noroviruses Circulating in Spain from 2016 to 2020 and Proposal of Two Novel Genotypes within Genogroup I. Microbiol Spectr. (2022) 10:e0250521. doi: 10.1128/spectrum.02505-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarr GAM, Pang XL, Zhuo R, Lee BE, Chui L, Ali S, et al. Attribution of Pediatric Acute Gastroenteritis Episodes and Emergency Department Visits to Norovirus Genogroups I and II. J Infect Dis. (2021) 223:452–61. doi: 10.1093/infdis/jiaa391, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo M, Mitra S, de P, Banerjee A, Deb AK, Miyoshi SI, et al. Genetic characterization and evolutionary analysis of norovirus genotypes circulating among children in eastern India during 2018-2019. Arch Virol. (2021) 166:2989–98. doi: 10.1007/s00705-021-05197-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manouana GP, Nguema-Moure PA, Mbong Ngwese M, Bock CT, Kremsner PG, Borrmann S, et al. Genetic Diversity of Enteric Viruses in Children under Five Years Old in Gabon. Viruses. (2021) 13. doi: 10.3390/v13040545, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utsumi T, Lusida MI, Dinana Z, Wahyuni RM, Soegijanto S, Athiyyah AF, et al. Molecular epidemiology and genetic diversity of norovirus infection in children hospitalized with acute gastroenteritis in East Java, Indonesia in 2015-2019. Infect Genet Evol. (2021) 88:104703. doi: 10.1016/j.meegid.2020.104703, PMID: [DOI] [PubMed] [Google Scholar]
- 23.Sarmento SK, de Andrade JDSR, Miagostovich MP, Fumian TM. Virological and Epidemiological Features of Norovirus Infections in Brazil, 2017-2018. Viruses. (2021) 13:1724. doi: 10.3390/v13091724, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivares AIO, Leitão GAA, Pimenta YC, Cantelli CP, Fumian TM, Fialho AM, et al. Epidemiology of enteric virus infections in children living in the Amazon region. Int J Infect Dis. (2021) 108:494–502. doi: 10.1016/j.ijid.2021.05.060, PMID: [DOI] [PubMed] [Google Scholar]
- 25.Parrón I, Barrabeig I, Alseda M, Rius C, Cornejo-Sánchez T, Jané M, et al. Norovirus outbreaks in long-term care facilities in Catalonia from 2017 to 2018. Sci Rep. (2021) 11:23218. doi: 10.1038/s41598-021-02348-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun C, Zhao Y, Wang G, Huang D, He H, Sai L. Molecular epidemiology of GII noroviruses in outpatients with acute gastroenteritis in Shandong Province, China. Arch Virol. (2021) 166:375–87. doi: 10.1007/s00705-020-04883-1, PMID: [DOI] [PubMed] [Google Scholar]
- 27.Grytdal S, Browne H, Collins N, Vargas B, Rodriguez-Barradas MC, Rimland D, et al. Trends in Incidence of Norovirus-associated Acute Gastroenteritis in 4 Veterans Affairs Medical Center Populations in the United States, 2011-2015. Clin Infect Dis. (2020) 70:40–8. doi: 10.1093/cid/ciz165, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikh MP, Vandekar S, Moore C, Thomas L, Britt N, Piya B, et al. Temporal and Genotypic Associations of Sporadic Norovirus Gastroenteritis and Reported Norovirus Outbreaks in Middle Tennessee, 2012-2016. Clin Infect Dis. (2020) 71:2398–404. doi: 10.1093/cid/ciz1106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji L, Hu G, Xu D, Wu X, Fu Y, Chen L. Molecular epidemiology and changes in genotype diversity of norovirus infections in acute gastroenteritis patients in Huzhou, China, 2018. J Med Virol. (2020) 92:3173–8. doi: 10.1002/jmv.26247, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hungerford D, Jere KC, Bar-Zeev N, Harris JP, Cunliffe NA, Iturriza-Gómara M. Epidemiology and genotype diversity of norovirus infections among children aged <5 years following rotavirus vaccine introduction in Blantyre, Malawi. J Clin Virol. (2020) 123:104248. doi: 10.1016/j.jcv.2019.104248, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Shen W, Sheng Y, Weng J, Li G, Wang D, Qiu D, et al. Molecular epidemiology of norovirus associated with acute gastroenteritis in Taizhou, China: A retrospective study. J Infect Public Health. (2020) 13:34–9. doi: 10.1016/j.jiph.2019.06.006, PMID: [DOI] [PubMed] [Google Scholar]
- 32.Li B, Xiao D, Li Y, Wu X, Qi L, Tang W, et al. Epidemiological analysis of norovirus infectious diarrhea outbreaks in Chongqing, China, from 2011 to 2016. J Infect Public Health. (2020) 13:46–50. doi: 10.1016/j.jiph.2019.06.019, PMID: [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Xu D, Wu X, Liu G, Ji L. An increasing prevalence of non-GII.4 norovirus genotypes in acute gastroenteritis outbreaks in Huzhou, China, 2014-2018. Arch Virol. (2020) 165:1121–8. doi: 10.1007/s00705-020-04599-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin M, Wu S, Kong X, Xie H, Fu J, He Y, et al. Norovirus Outbreak Surveillance, China, 2016-2018. Emerg Infect Dis. (2020) 26:437–45. doi: 10.3201/eid2603.191183, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degiuseppe JI, Barclay L, Gomes KA, Costantini V, Vinjé J, Stupka JA. Molecular epidemiology of norovirus outbreaks in Argentina, 2013-2018. J Med Virol. (2020) 92:1330–3. doi: 10.1002/jmv.25684, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y, Lu Y, Xue C, Li E, Zhang Q, Xu F, et al. Surveillance of the 'bud event of norovirus-associated gastroenteritis' in schools: does it work in the prevention of norovirus infection outbreaks in Shanghai? Epidemiol Infect. (2020) 148:e104. doi: 10.1017/S0950268820000965, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parrón I, Barrabeig I, Alseda M, Cornejo-Sánchez T, Guix S, Jané M, et al. Involvement of Workers in Closed and Semiclosed Institutions in Outbreaks of Acute Gastroenteritis Due to Norovirus. Viruses. (2020) 12. doi: 10.3390/v12121392, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loureiro Tonini MA, Pires Gonçalves Barreira DM, Bueno de Freitas Santolin L, Bondi Volpini LP, Gagliardi Leite JP, le Moullac-Vaidye B, et al. FUT2, Secretor Status and FUT3 Polymorphisms of Children with Acute Diarrhea Infected with Rotavirus and Norovirus in Brazil. Viruses. (2020) 12. doi: 10.3390/v12101084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lartey BL, Quaye O, Damanka SA, Agbemabiese CA, Armachie J, Dennis FE, et al. Understanding Pediatric Norovirus Epidemiology: A Decade of Study among Ghanaian Children. Viruses. (2020) 12. doi: 10.3390/v12111321, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hossain ME, Rahman R, Ali SI, Islam MM, Rahman MZ, Ahmed S, et al. Epidemiologic and Genotypic Distribution of Noroviruses Among Children With Acute Diarrhea and Healthy Controls in a Low-income Rural Setting. Clin Infect Dis. (2019) 69:505–13. doi: 10.1093/cid/ciy915, PMID: [DOI] [PubMed] [Google Scholar]
- 41.Gelaw A, Pietsch C, Mann P, Liebert UG. Molecular detection and characterisation of sapoviruses and noroviruses in outpatient children with diarrhoea in Northwest Ethiopia. Epidemiol Infect. (2019) 147:e218. doi: 10.1017/S0950268819001031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nirwati H, Donato CM, Mawarti Y, Mulyani NS, Ikram A, Aman AT, et al. Norovirus and rotavirus infections in children less than five years of age hospitalized with acute gastroenteritis in Indonesia. Arch Virol. (2019) 164:1515–25. doi: 10.1007/s00705-019-04215-y, PMID: [DOI] [PubMed] [Google Scholar]
- 43.Xue L, Cai W, Gao J, Zhang L, Dong R, Li Y, et al. The resurgence of the norovirus GII.4 variant associated with sporadic gastroenteritis in the post-GII.17 period in South China, 2015 to 2017. BMC Infect Dis. (2019) 19:696. doi: 10.1186/s12879-019-4331-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bitencurt ELR, Siqueira JAM, Medeiros TB, Bandeira RDS, de Souza Oliveira D, de Paula Souza e Guimarães RJ, et al. Epidemiological and molecular investigation of norovirus and astrovirus infections in Rio Branco, Acre, Northern Brazil: A retrospective study. J Med Virol. (2019) 91:997–1007. doi: 10.1002/jmv.25395, PMID: [DOI] [PubMed] [Google Scholar]
- 45.Brown JR, Roy S, Shah D, Williams CA, Williams R, Dunn H, et al. Norovirus Transmission Dynamics in a Pediatric Hospital Using Full Genome Sequences. Clin Infect Dis. (2019) 68:222–8. doi: 10.1093/cid/ciy438, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li HY, Zhang YG, Lei X, Song J, Duan ZJ. Prevalence of noroviruses in children hospitalized for acute gastroenteritis in Hohhot, China, 2012-2017. BMC Infect Dis. (2019) 19:595. doi: 10.1186/s12879-019-4230-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu L, Zhong H, Xu M, Su L, Cao L, Jia R, et al. Genetic diversity and epidemiology of Genogroup II noroviruses in children with acute sporadic gastroenteritis in Shanghai, China, 2012-2017. BMC Infect Dis. (2019) 19:736. doi: 10.1186/s12879-019-4360-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motoya T, Umezawa M, Saito A, Goto K, Doi I, Fukaya S, et al. Variation of human norovirus GII genotypes detected in Ibaraki, Japan, during 2012-2018. Gut Pathog. (2019) 11:26. doi: 10.1186/s13099-019-0303-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathew S, Alansari K, Smatti MK, Zaraket H, Al Thani AA, Yassine HM. Epidemiological, Molecular, and Clinical Features of Norovirus Infections among Pediatric Patients in Qatar. Viruses. (2019) 11. doi: 10.3390/v11050400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Z, Liu B, Yan H, Li W, Jia L, Tian Y, et al. Norovirus outbreaks in Beijing, China, from 2014 to 2017. J Infect. (2019) 79:159–66. doi: 10.1016/j.jinf.2019.05.019, PMID: [DOI] [PubMed] [Google Scholar]
- 51.Pabbaraju K, Wong AA, Tipples GA, Pang XL. Emergence of a Novel Recombinant Norovirus GII.P16-GII.12 Strain Causing Gastroenteritis, Alberta, Canada. Emerg Infect Dis. (2019) 25:1556–9. doi: 10.3201/eid2508.190059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikounou Louya V, Vouvoungui C, Koukouikila-Koussounda F, Veas F, Kobawila SC, Ntoumi F. Molecular characterization of norovirus infection responsible for acute diarrhea in Congolese hospitalized children under five years old in Brazzaville, Republic of Congo. Int J Infect Dis. (2019) 88:41–8. doi: 10.1016/j.ijid.2019.07.034, PMID: [DOI] [PubMed] [Google Scholar]
- 53.Hebbelstrup Jensen B, Jokelainen P, Nielsen ACY, Franck KT, Rejkjær Holm D, Schønning K, et al. Children Attending Day Care Centers are a Year-round Reservoir of Gastrointestinal Viruses. Sci Rep. (2019) 9:3286. doi: 10.1038/s41598-019-40077-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu J, Bao C, Huo X, Hu J, Shi C, Lin Q, et al. Increasing Recombinant Strains Emerged in Norovirus Outbreaks in Jiangsu, China: 2015-2018. Sci Rep. (2019) 9:20012. doi: 10.1038/s41598-019-56544-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makhaola K, Moyo S, Lechiile K, Goldfarb DM, Kebaabetswe LP. Genetic and epidemiological analysis of norovirus from children with gastroenteritis in Botswana, 2013-2015. BMC Infect Dis. (2018) 18:246. doi: 10.1186/s12879-018-3157-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han J, Wu X, Chen L, Fu Y, Xu D, Zhang P, et al. Emergence of norovirus GII.P16-GII.2 strains in patients with acute gastroenteritis in Huzhou, China, 2016-2017. BMC Infect Dis. (2018) 18:342. doi: 10.1186/s12879-018-3259-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farsi M, Roodbari F, Nejati B, Arashkia A, Jalilvand S, Nateghian A, et al. Prevalence and genetic diversity of norovirus genogroup II in children less than 5 years of age with acute gastroenteritis in Tehran, Iran. MedMicrobiolImmunol. (2018) 207:201–10. doi: 10.1007/s00430-018-0541-6, PMID: [DOI] [PubMed] [Google Scholar]
- 58.Pagani E, Folli F, Tofani S, Ruggeri FM, Ostanello F, di Bartolo I. Pilot survey of norovirus in Northern Italy: an example of surveillance of norovirus gastroenteritis. Epidemiol Infect. (2018) 146:291–6. doi: 10.1017/S0950268817002989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim YE, Song M, Lee J, Seung HJ, Kwon EY, Yu J, et al. Phylogenetic characterization of norovirus strains detected from sporadic gastroenteritis in Seoul during 2014-2016. Gut Pathog. (2018) 10:36. doi: 10.1186/s13099-018-0263-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergallo M, Daprà V, Rassu M, Bonamin S, Cuccu R, Calvi C, et al. Prevalence and Clinical Profile of Human Salivirus in Children with Acute Gastroenteritis in Northern Italy, 2014-2015. Intervirology. (2018) 61:49–52. doi: 10.1159/000490568, PMID: [DOI] [PubMed] [Google Scholar]
- 61.Kiseleva V, Faizuloev E, Meskina E, Marova A, Oksanich A, Samartseva T, et al. Molecular-Genetic Characterization of Human Rotavirus A Strains Circulating in Moscow, Russia (2009–2014). Virol Sin. (2018) 33:304–13. doi: 10.1007/s12250-018-0043-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L, Guan H, Zhang Y, Wang C, Yang G, Ruan S, et al. The prevalence of non-GII.4 norovirus genotypes in acute gastroenteritis outbreaks in Jinan, China. PLoS One. (2018) 13:e0209245. doi: 10.1371/journal.pone.0209245, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue C, Pan L, Zhu W, Wang Y, Fu H, Cui C, et al. Molecular epidemiology of genogroup II norovirus infections in acute gastroenteritis patients during 2014-2016 in Pudong New Area, Shanghai, China. Gut Pathog. (2018) 10:7. doi: 10.1186/s13099-018-0233-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joosten R, Sonder G, Parkkali S, Brandwagt D, Fanoy E, Mughini-Gras L, et al. Risk factors for gastroenteritis associated with canal swimming in two cities in the Netherlands during the summer of 2015: A prospective study. PLoS One. (2017) 12:e0174732. doi: 10.1371/journal.pone.0174732, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howard LM, Mwape I, Siwingwa M, Simuyandi M, Guffey MB, Stringer JSA, et al. Norovirus infections in young children in Lusaka Province, Zambia: clinical characteristics and molecular epidemiology. BMC Infect Dis. (2017) 17:92. doi: 10.1186/s12879-017-2206-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santos VS, Gurgel RQ, Cavalcante SM, Kirby A, Café LP, Souto MJ, et al. Acute norovirus gastroenteritis in children in a highly rotavirus-vaccinated population in Northeast Brazil. J Clin Virol. (2017) 88:33–8. doi: 10.1016/j.jcv.2016.10.015, PMID: [DOI] [PubMed] [Google Scholar]
- 67.Timurkan MÖ, Aydin H, Aktaş O. Frequency and molecular characterization of human norovirus in Erzurum, Turkey. Turk J Med Sci. (2017) 47:960–6. doi: 10.3906/sag-1509-87, PMID: [DOI] [PubMed] [Google Scholar]
- 68.Giammanco GM, de Grazia S, Bonura F, Cappa V, Muli SL, Pepe A, et al. Norovirus GII.17 as Major Epidemic Strain in Italy, Winter 2015-16. Emerg Infect Dis. (2017) 23:1206–8. doi: 10.3201/eid2307.161255, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoa-Tran TN, Nakagomi O, Dao ATH, Nguyen AT, Agbemabiese CA, Vu HM, et al. Molecular epidemiology of noroviruses detected in Vietnamese children with acute gastroenteritis from 2012 to 2015. J Med Microbiol. (2017) 66:34–45. doi: 10.1099/jmm.0.000417, PMID: [DOI] [PubMed] [Google Scholar]
- 70.Niendorf S, Jacobsen S, Faber M, Eis-Hübinger AM, Hofmann J, Zimmermann O, et al. Steep rise in norovirus cases and emergence of a new recombinant strain GII.P16-GII.2, Germany, winter 2016. Euro Surveill. (2017) 22:30447. doi: 10.2807/1560-7917.ES.2017.22.4.30447, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wangchuk S, Matsumoto T, Iha H, Ahmed K. Surveillance of norovirus among children with diarrhea in four major hospitals in Bhutan: Replacement of GII.21 by GII.3 as a dominant genotype. PLoS One. (2017) 12:e0184826. doi: 10.1371/journal.pone.0184826, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magaña LC, et al. Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses. J Clin Microbiol. (2017) 55:2208–21. doi: 10.1128/JCM.00455-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ouédraogo N, Kaplon J, Bonkoungou IJ, Traoré AS, Pothier P, Barro N, et al. Prevalence and Genetic Diversity of Enteric Viruses in Children with Diarrhea in Ouagadougou, Burkina Faso. PLoS One. (2016) 11:e0153652. doi: 10.1371/journal.pone.0153652, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mousavi Nasab SD, Sabahi F, Makvandi M, Mirab Samiee S, Nadji SA, Ravanshad M. Epidemiology of Rotavirus-Norovirus Co-Infection and Determination of Norovirus Genogrouping among Children with Acute Gastroenteritis in Tehran, Iran. IBJ. (2016) 20:280–6. doi: 10.22045/ibj.2016.05, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sisay Z, Djikeng A, Berhe N, Belay G, Gebreyes W, Abegaz WE, et al. Prevalence and molecular characterization of human noroviruses and sapoviruses in Ethiopia. Arch Virol. (2016) 161:2169–82. doi: 10.1007/s00705-016-2887-7, PMID: [DOI] [PubMed] [Google Scholar]
- 76.Zhang P, Chen L, Fu Y, Ji L, Wu X, Xu D, et al. Clinical and molecular analyses of norovirus-associated sporadic acute gastroenteritis: the emergence of GII.17 over GII.4, Huzhou, China, 2015. BMC Infect Dis. (2016) 16:717. doi: 10.1186/s12879-016-2033-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruggink LD, Dunbar NL, Marshall JA. Emergence of GII.Pg norovirus in gastroenteritis outbreaks in Victoria, Australia. J Med Virol. (2016) 88:1521–8. doi: 10.1002/jmv.24511, PMID: [DOI] [PubMed] [Google Scholar]
- 78.Challappa R, Saito M, Mejia C, Bern C, Webman R, Pajuelo M, et al. Burden of Norovirus and Rotavirus in Children After Rotavirus Vaccine Introduction, Cochabamba, Bolivia. Am J Trop Med Hyg. (2016) 94:212–7. doi: 10.4269/ajtmh.15-0203, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He Y, Jin M, Chen K, Zhang H, Yang H, Zhuo F, et al. Gastroenteritis Outbreaks Associated with the Emergence of the New GII.4 Sydney Norovirus Variant during the Epidemic of 2012/13 in Shenzhen City, China. PLoS One. (2016) 11:e0165880. doi: 10.1371/journal.pone.0165880, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumazaki M, Usuku S. Norovirus genotype distribution in outbreaks of acute gastroenteritis among children and older people: an 8-year study. BMC Infect Dis. (2016) 16:643. doi: 10.1186/s12879-016-1999-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melhem NM, Zaraket H, Kreidieh K, Ali Z, Hammadi M, Ghanem S, et al. Clinical and epidemiological characteristics of norovirus gastroenteritis among hospitalized children in Lebanon. World J Gastroenterol. (2016) 22:10557–65. doi: 10.3748/wjg.v22.i48.10557, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koo HS, Lee MO, Ku PT, Hwang SJ, Park DJ, Baik HS. Molecular epidemiology of norovirus in asymptomatic food handlers in Busan, Korea, and emergence of genotype GII.17. J Microbiol. (2016) 54:686–94. doi: 10.1007/s12275-016-6312-4, PMID: [DOI] [PubMed] [Google Scholar]
- 83.Manso CF, Romalde JL. Molecular epidemiology of norovirus from patients with acute gastroenteritis in northwestern Spain. Epidemiol Infect. (2015) 143:316–24. doi: 10.1017/S0950268814000740, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lekana-Douki SE, Kombila-Koumavor C, Nkoghe D, Drosten C, Drexler JF, Leroy EM. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int J Infect Dis. (2015) 34:90–5. doi: 10.1016/j.ijid.2015.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- 85.Wu X, Han J, Chen L, Xu D, Shen Y, Zha Y, et al. Prevalence and genetic diversity of noroviruses in adults with acute gastroenteritis in Huzhou, China, 2013-2014. Arch Virol. (2015) 160:1705–13. doi: 10.1007/s00705-015-2440-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopman BA, Trivedi T, Vicuña Y, Costantini V, Collins N, Gregoricus N, et al. Norovirus Infection and Disease in an Ecuadorian Birth Cohort: Association of Certain Norovirus Genotypes With Host FUT2 Secretor Status. J Infect Dis. (2015) 211:1813–21. doi: 10.1093/infdis/jiu672, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan D, Deng L, Wang M, Li X, Ma Y, Liu W. High prevalence and genetic diversity of noroviruses among children with sporadic acute gastroenteritis in Nanning City, China, 2010-2011. J Med Virol. (2015) 87:498–503. doi: 10.1002/jmv.24103, PMID: [DOI] [PubMed] [Google Scholar]
- 88.Gao Z, Li X, Yan H, Li W, Jia L, Hu L, et al. Human calicivirus occurrence among outpatients with diarrhea in Beijing, China, between April 2011 and March 2013. J Med Virol. (2015) 87:2040–7. doi: 10.1002/jmv.24265, PMID: [DOI] [PubMed] [Google Scholar]
- 89.Ham H, Oh S, Seung H, Jo S. Molecular characteristics of noroviruses genogroup I and genogroup II detected in patients with acute gastroenteritis. Ann Lab Med. (2015) 35:242–5. doi: 10.3343/alm.2015.35.2.242, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim JS, Kim HS, Hyun J, Kim HS, Song W. Molecular Epidemiology of Human Norovirus in Korea in 2013. Biomed Res Int. (2015) 2015:468304: 1–8. doi: 10.1155/2015/468304, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumazaki M, Usuku S. Genetic Analysis of Norovirus GII.4 Variant Strains Detected in Outbreaks of Gastroenteritis in Yokohama, Japan, from the 2006-2007 to the 2013-2014 Seasons. PLoS One. (2015) 10:e0142568. doi: 10.1371/journal.pone.0142568, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoneda M, Okayama A, Kitahori Y. Epidemiological Characteristics of Norovirus Associated with Sporadic Gastroenteritis among Children from the 2006/2007 to 2011/2012 Season in Nara Prefecture, Japan. Intervirology. (2014) 57:31–5. doi: 10.1159/000353852, PMID: [DOI] [PubMed] [Google Scholar]
- 93.Arana A, Cilla G, Montes M, Gomariz M, Pérez-Trallero E. Genotypes, recombinant forms, and variants of norovirus GII.4 in Gipuzkoa (Basque Country, Spain), 2009-2012. PLoS One. (2014) 9:e98875. doi: 10.1371/journal.pone.0098875, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ayukekbong JA, Fobisong C, Tah F, Lindh M, Nkuo-Akenji T, Bergström T. Pattern of circulation of norovirus GII strains during natural infection. J Clin Microbiol. (2014) 52:4253–9. doi: 10.1128/JCM.01896-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mesquita JR, Costantini VP, Cannon JL, Lin SC, Nascimento MS, Vinjé J. Presence of antibodies against genogroup VI norovirus in humans. Virol J. (2013) 10:176. doi: 10.1186/1743-422X-10-176, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang MB, Chen CH, Chen SC, Chou YC, Yu CP. Epidemiological and molecular analysis of human norovirus infections in Taiwan during 2011 and 2012. BMC Infect Dis. (2013) 13:338. doi: 10.1186/1471-2334-13-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manso CF, Torres E, Bou G, Romalde JL. Role of norovirus in acute gastroenteritis in the Northwest of Spain during 2010-2011. J Med Virol. (2013) 85:2009–15. doi: 10.1002/jmv.23680, PMID: [DOI] [PubMed] [Google Scholar]
- 98.Saupe AA, Kaehler D, Cebelinski EA, Nefzger B, Hall AJ, Smith KE. Norovirus Surveillance among Callers to Foodborne Illness Complaint Hotline, Minnesota, USA, 2011–2013. Emerg Infect Dis. (2013) 19:1293–6. doi: 10.3201/eid1908.130462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hasing ME, Lee BE, Preiksaitis JK, Tellier R, Honish L, Senthilselvan A, et al. Emergence of a new norovirus GII.4 variant and changes in the historical biennial pattern of norovirus outbreak activity in Alberta, Canada, from 2008 to 2013. J Clin Microbiol. (2013) 51:2204–11. doi: 10.1128/JCM.00663-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim HS, Hyun J, Kim HS, Kim JS, Song W, Lee KM. Emergence of GII.4 Sydney norovirus in South Korea during the winter of 2012-2013. J Microbiol Biotechnol. (2013) 23:1641–3. doi: 10.4014/jmb.1308.08053, PMID: [DOI] [PubMed] [Google Scholar]
- 101.Huynen P, Mauroy A, Martin C, Savadogo LG, Boreux R, Thiry E, et al. Molecular epidemiology of norovirus infections in symptomatic and asymptomatic children from Bobo Dioulasso, Burkina Faso. J Clin Virol. (2013) 58:515–21. doi: 10.1016/j.jcv.2013.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- 102.Trang NV, Luan LT, Kim-Anh LT, Hau VT, Nhung LTH, Phasuk P, et al. Detection and molecular characterization of noroviruses and sapoviruses in children admitted to hospital with acute gastroenteritis in Vietnam. J Med Virol. (2012) 84:290–7. doi: 10.1002/jmv.23185, PMID: [DOI] [PubMed] [Google Scholar]
- 103.Zeng M, Xu X, Zhu C, Chen J, Zhu Q, Lin S, et al. Clinical and molecular epidemiology of norovirus infection in childhood diarrhea in China. J Med Virol. (2012) 84:145–51. doi: 10.1002/jmv.22248, PMID: [DOI] [PubMed] [Google Scholar]
- 104.Parra GI, Tohma K, Ford-Siltz LA, Eguino P, Kendra JA, Pilewski KA, et al. Minimal Antigenic Evolution after a Decade of Norovirus GII.4 Sydney_2012 Circulation in Humans. J Virol. (2023) 97:e0171622. doi: 10.1128/jvi.01716-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kendra JA, Tohma K, Parra GI. Global and regional circulation trends of norovirus genotypes and recombinants, 1995-2019: A comprehensive review of sequences from public databases. Rev Med Virol. (2022) 32:e2354. doi: 10.1002/rmv.2354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. (2014) 14:725–30. doi: 10.1016/S1473-3099(14)70767-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. (2008) 14:1224–31. doi: 10.3201/eid1408.071114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Afework DT, Shumie MK, Endalew GF, Adugna AG, Tarekegn BG. Pooled prevalence and genetic diversity of norovirus in Africa: a systematic review and meta-analysis. Virol J. (2022) 19:115. doi: 10.1186/s12985-022-01835-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liao Y, Hong X, Wu A, Jiang Y, Liang Y, Gao J, et al. Global prevalence of norovirus in cases of acute gastroenteritis from 1997 to 2021: An updated systematic review and meta-analysis. Microb Pathog. (2021) 161:105259. doi: 10.1016/j.micpath.2021.105259, PMID: [DOI] [PubMed] [Google Scholar]
- 110.Nguyen GT, Phan K, Teng I, Pu J, Watanabe T. A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries. Medicine (Baltimore). (2017) 96:e8139. doi: 10.1097/MD.0000000000008139, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li TT, Xu Q, Liu MC, Wang T, Che TL, Teng AY, et al. Prevalence and Etiological Characteristics of Norovirus Infection in China: A Systematic Review and meta-analysis. Viruses. (2023) 15. doi: 10.3390/v15061336, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lopman B, Armstrong B, Atchison C, Gray JJ. Host, weather and virological factors drive norovirus epidemiology: time-series analysis of laboratory surveillance data in England and Wales. PLoS One. (2009) 4:e6671. doi: 10.1371/journal.pone.0006671, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ahmed SM, Lopman BA, Levy K. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS One. (2013) 8:e75922. doi: 10.1371/journal.pone.0075922, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu J, Fang L, Sun L, Zeng H, Li Y, Zheng H, et al. Association of GII.P16-GII.2 Recombinant Norovirus Strain with Increased Norovirus Outbreaks, Guangdong, China, 2016. Emerg Infect Dis. (2017) 23:1188–90. doi: 10.3201/eid2307.170333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Graaf M, van Beek J, Vennema H, Podkolzin AT, Hewitt J, Bucardo F, et al. Emergence of a novel GII.17 norovirus – End of the GII.4 era? Euro Surveill. (2015) 20. doi: 10.2807/1560-7917.es2015.20.26.21178, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim L, Liebowitz D, Lin K, Kasparek K, Pasetti MF, Garg SJ, et al. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight. (2018) 3:e121077. doi: 10.1172/jci.insight.121077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scallan CD, Tingley DW, Lindbloom JD, Toomey JS, Tucker SN. An adenovirus-based vaccine with a double-stranded RNA adjuvant protects mice and ferrets against H5N1 avian influenza in oral delivery models. Clin Vaccine Immunol. (2013) 20:85–94. doi: 10.1128/CVI.00552-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Precision Vaccinations . (2023). Oral Norovirus Vaccine Candidate Passes Challenge Study. Available at: https://www.precisionvaccinations.com/2023/09/09/oral-norovirus-vaccine-candidate-passes-challenge-study (Accessed September 9, 2023)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.