Abstract
Early detection of cancer recurrence using specific biomarkers remains a clinically unmet need, although methodologies for monitoring tumor markers, cell-free DNA, and circulating tumor cells have been established for decades. Tumor recurrence develops in metastatic or dormant cancer cells under continuous immune surveillance. Alterations in the population and function of immune cells may contribute to cancer recurrence. Here, we utilized an animal model to imitate breast tumor recurrence after surgical resection and investigated the abundance and gene expression profiles of immune cells using NanoString analysis. Bioinformatic analysis of a published single-cell RNA sequencing database of myeloid-derived suppressor cells (MDSCs) was performed to identify common targets between the two studies. Identified biomarkers were validated using human peripheral blood mononuclear cell (PBMC) datasets. The inhibitory effect of MDSCs on T-cell proliferation was assessed in vitro. Our data demonstrated that the number of MDSCs significantly increased during recurrence. Comparison of our NanoString data with a single-cell RNA sequencing dataset of MDSCs in another spontaneous breast cancer model identified colony-stimulating factor 3 receptor (Csf3r)-positive MDSCs as a potential marker for predicting tumor relapse. We validated our findings using two previously published PBMC databases of patients with breast cancer with or without recurrence and confirmed the elevated MDSC gene signature and CSF3R expression in patients with tumor recurrence. 35 patients with breast cancer were also included in our study, that patients with higher levels of CSF3R had worse survival. In vitro experiments demonstrated that Csf3r+ MDSCs exhibited enhanced reactive oxygen species (ROS) levels and robust T-cell suppression ability. We conclude that an increase in CSF3R+ MDSCs is a potential biomarker for early detection of tumor recurrence in patients with breast cancer.
Keywords: Breast cancer, tumor recurrence, myeloid-derived suppressor cells, NanoString, CSF3R
Introduction
The development of novel therapeutic drugs or interventions, such as tyrosine kinase inhibitors, monoclonal antibodies, and immunotherapy, has greatly improved the survival of patients with breast cancer over the past decades. However, tumor recurrence at different metastatic sites is frequently observed after treatment and is a major cause of death. Early detection of tumor recurrence using specific blood biomarkers remains a clinically unmet need for breast cancer.
Different biomarkers have been tested for the early diagnosis and disease monitoring of patients with breast cancer [1], of which one is a protein marker. Recent studies have used two protein markers, the carcinoembryonic antigen and cancer antigen 15-3, to detect breast cancer recurrence [2,3]. Although the results suggest that these markers are useful, their sensitivity and specificity are unsatisfactory. Another method is liquid biopsy, which identifies circulating tumor cells (CTC) or cell-free tumor DNA (ctDNA). Ortolan et al. showed that ctDNA levels are positively associated with breast cancer relapse [4]. Cailleux et al. also demonstrated that the presence of ctDNA in the blood of patients with breast cancer who received neoadjuvant chemotherapy was correlated with an increased risk of recurrence [5]. The third method involves immune-cell profiling. The abundance of monocytes in the peripheral blood has been linked to good prognosis and reduced relapse in breast cancer patients [6]. A decrease in the absolute lymphocyte count is associated with shorter overall survival [7]. Additionally, a low neutrophil-to-lymphocyte ratio and high lymphocyte-to-monocyte ratio predict longer locoregional recurrence-free and distant metastasis-free survival [8].
Myeloid-derived suppressor cells (MDSC) are a heterogeneous group of immune cells derived from myeloid progenitors that originate from hematopoietic stem cells [9]. MDSCs comprise two distinct subgroups: granulocytic MDSCs (g-MDSCs) and monocytic MDSCs (m-MDSCs). MDSCs can inhibit T-cell function through several mechanisms, including (A) induction or expansion of regulatory T-cells (Tregs), (B) depletion of key amino acids that are essential for T-cell growth and differentiation, and (C) interference with T-cell migration and viability [10]. In patients with breast cancer, MDSC levels in the blood are approximately 10-fold higher than those in healthy individuals and correlate with advanced stage and lymph node invasion [11]. However, it remains unclear whether the abundance of MDSCs is altered during tumor relapse and whether a specific subset of MDSCs can predict recurrence.
In this study, we established an orthotopic animal model by inoculating 4T1 mouse breast cancer cells into the mammary fat pad of BALB/c mice and investigated the alteration in the gene expression profiles in different immune cell populations in the mouse peripheral blood at different experimental stages. Next, we analyzed a single-cell transcriptomic database of MDSCs generated from a spontaneous breast cancer model to confirm our findings. Functional assays were also performed to determine the suppressive effect of CSF3R+ MDSCs on T-cell proliferation. Finally, clinical validation was performed using two public peripheral blood mononuclear cells (PBMC) datasets from patients with breast cancer, with or without recurrence. Our findings suggest that an increase in blood CSF3R+ MDSCs is an important biomarker for predicting breast cancer recurrence.
Materials and methods
Mice
BALB/c mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan). The mice were housed in a specific pathogen-free environment with a 12/12 h light/dark cycle. All the experiments were approved by the Animal Care and Use Committee of the National Health Research Institute. In vivo experiments were conducted according to the AAALAC Verification Letter #001596 with protocol No. 109037-A.
PBMC isolation and RNA extraction
Mouse blood was collected in Microtainer® (BD) with ethylenediaminetetraacetic acid (EDTA) and incubated with 1 ml of ACK lysis buffer (Gibco, Grand Island, NY, USA) for 5 min at room temperature to lyse red blood cells. PBMCs were spun down by centrifugation at 1500 rpm for 10 min at 4°C and washed twice with phosphate-buffered saline (PBS). RNA was extracted using the Direct-Zol RNA Miniprep Kit (ZYMO RESEARCH) according to the manufacturer’s instructions.
NanoString panels and data preprocessing
The total RNA of PBMCs was subjected to NanoString nCounter analysis to examine the expressions of immune-related genes using the PanCancer Immune Profiling Panel and genes involved in myeloid innate immunity using the Myeloid Innate Immunity Panel. The raw data files in the RCC format were imported into the nSolver analysis software (version 4.0, NanoString Technologies) to generate gene expression profiles. Count data without nSolver software processing were exported and used for quality control (QC) and data normalization in the R language environment (https://www.r-project.org). Overlapping genes between these two panels showed high correlations (Supplementary Figure 1A). Principal component analysis (PCA) showed no significant batch effects between panels (Supplementary Figure 1B). These samples were merged into one dataset for subsequent data normalization. This dataset comprised 24 samples (12 mice × 2 immune/myeloid panels) across 1190 probes, including 1156 endogenous genes, 20 housekeeping genes, 8 negative control probes, and 6 positive control probes. Each count was transformed to a log2 scale and normalized using positive control probes and housekeeping genes to reduce variation.
Isolation of mouse T-cells and MDSCs
Mice splenocytes were isolated with syringe rubber in a 70-μm cell strainer (Meltenyi Biotec). Ice-cold RPMI-1640 with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) were utilized to resuspend splenocytes. The cells were then centrifuged at 1500 rpm for 5 min at room temperature. ACK lysis buffer was added to lyse red blood cells, and mononuclear cells were collected. T-cells were isolated using a mouse CD4+ and CD8+ T-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany), and MDSCs were collected using a mouse MDSC isolation kit (Miltenyi Biotec).
In vitro T-cell suppression assay
Isolated T-cells were activated using 2 µg/ml anti-mouse CD28 (#102115, BioLegend, San Diego, CA, USA) and 0.5 µg/ml CD3ε antibody (#100339, BioLegend) in RPMI-1640 with 5% FBS, 1% PS, and 5 ng/ml recombinant mouse IL-2 (#575404, BioLegend). Activated T-cells were co-cultured with Csf3r- or Csf3+ MDSCs in a 1:1 ratio in 2 ml RPMI-1640 for 48 h. After co-culture, cells were harvested and incubated with TruStain FcX™ (anti-mouse CD16/32) antibody (Biolegend, #101319) in a 1:100 dilution on ice for 10 min. Cells were then stained with anti-CD3 antibody (#561388, BD) in a 1:100 dilution for 40 min at 4°C. The samples were washed twice with PBS and subjected to flow cytometry using an Attune NxT cytometer (Thermo Fisher Scientific, Waltham, MA, USA).
Reactive oxygen species (ROS) assay
Cells were incubated at 37°C for 30 min in Hanks’ balanced salt solution with 5 µmol/L dichlorofluorescin diacetate (DCF-DA) in the dark. After incubation, the cells were washed twice with PBS, and fluorescence was analyzed using an Attune NxT cytometer (Thermo Fisher Scientific).
Analysis of the single-cell RNA sequencing dataset
A single-cell RNA-sequencing dataset was downloaded from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/gds) under the accession number GSE139125 [12]. The dataset was generated from fluorescence-activated cell sorting-isolated Cd11b+/Gr1+ cells collected from the spleens of control wild-type (WT) and tumor-bearing mouse mammary tumor virus promoter-driven polyomavirus middle T antigen (MMTV-PyMT) mice using the 10× Genomics Chromium platform (10× Genomics). Two cell-gene UMI matrices were imported into the R package Seurat (version 4.0.1) [13], and the Seurat pipeline was used to integrate and identify clusters. Cells expressing fewer than 500 genes or cells with more than 8% of the total UMI counts in the mitochondrial genome were considered low-quality, dead, or dying and were excluded from subsequent analyses. Normalization was performed by scaling the total UMI counts to over 10000 to reduce cell-to-cell differences in the capture efficiency of the transcripts. Each expression value was transformed to a log2 scale after adding one count to avoid undefined values at zero counts. Canonical correlation analysis (CCA) was performed to integrate cells from WT and MMTV-PyMT mice based on the default parameters, and PCA was used to categorize the genes in each cell. Consequently, 20 clusters were identified from the first 20 principal components. The distribution of cells in each cluster was summarized using t-distributed stochastic neighbor embedding (t-SNE) plots (Supplementary Figure 2A and 2B), as implemented in the R package Rtsne. Cell type annotation was based on the canonical marker genes used by Alshetaiwi et al. [12]. The dotted plot represents the average and percentage expression levels of the marker genes in each cluster (Supplementary Figure 2C). m-MDSCs and g-MDSCs were annotated based on the MDSC signature, and 105 genes showing differential expression between WT and MMTV-PyMT mice in both m-MDSCs and g-MDSCs were identified [12]. The reanalysis of the data is available from the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) under the accession number SCP1868.
GEO datasets
For the analysis, we downloaded two public datasets, GSE65517 [14] and GSE201085 [6]. The GSE65517 dataset contained information on the Illumina HumanHT-12 V4 bead chip generated from monocytes of four patients with metastatic breast cancer and three healthy controls. The raw data were log2 (count+1) transformed and normalized using the Robust Spline Normalization algorithm implemented in the R package lumi. Gene expression was identified using probes showing the maximum average expression across seven samples. The GSE201085 dataset contained RNA-seq data generated from the whole blood of nine patients with triple-negative breast cancer with recurrence information. The quality control of these data was evaluated at different levels, including RNA quality, raw read data, alignment, and gene expression. Raw RNA-seq paired ends were mapped to the human reference genome hg19 using STAR 2.7.3. The raw read count matrix (GSE201085_featurecounts.csv) was downloaded from the GEO and normalized to the Trimmed Mean of M-values (TMM). A quasi-likelihood negative binomial generalized log-linear model was used to assess the significance of CSF3R expression between patients with and without recurrence using the R package edgeR [15]. Specifically, the “glmQLFit” function was used to fit the model, and the “estimateGLMTrendedDisp” function was used to estimate the dispersion.
Estimation of MDSC abundance
MDSC abundance in our NanoString dataset was estimated based on the average expression of 29 genes among the 105 genes defined as the MDSC signature [16]. We used the modified MDSC signature available in our study panels to estimate the abundance of MDSC in blood samples.
Gene set variation analysis (GSVA)
For each sample, GSVA was used to evaluate the enrichment level of a priori-defined gene set as implemented in the R package GSVA [17]. The hallmark gene sets and KEGG pathway databases in the Molecular Signatures Database (MSigDB; version 7.1 (https://www.gsea-msigdb.org/gsea/msigdb/)) were used as previously defined gene sets. The differentially enriched gene sets in MSigDB between the two conditions were identified using a t-test with a false discovery rate of less than 0.05 (q-value < 0.05).
Patient samples
The buffy coats of female patients with breast cancer were obtained from an ongoing clinical study. This study was approved by the Institutional Review Board (IRB) of the National Cheng Kung University Hospital (IRB No. A-ER-112-265). Buffy coat samples were collected from female patients with breast cancer who visited the outpatient clinic of the National Cheng Kung University Hospital, Tainan, Taiwan, between October 2017 and October 2019.
Human buffy coat sample preparation
Buffy coat samples collected from 35 breast cancer patients were stored at -80°C. After thawing on ice, the samples were mixed with 1 ml of ACK lysis buffer (Thermo Fisher, # A1049201) for 5 min at room temperature and centrifuged at 1500 rpm. Cell pellets of the buffy coat were stained with BD Pharmingen™ Human BD Fc Block according to the manufacturer’s instructions. The cells were stained for CD14 (BD Pharmingen, #555397), CD15 (BD Pharmingen, #560828), and CSF3R (BD Pharmingen, #554538) for 30 min at room temperature. After washing with 1X FACS buffer, the samples were fixed with 3% formaldehyde and analyzed by flow cytometry.
Statistical analysis
Pearson’s correlation was used to assess correlations between the two NanoString panels. Student’s t-test was used to assess the differences in means between groups. To address multiple comparison issues, the q-value was calculated using the R package q-value. A gene was considered differentially expressed if the q-value was less than 0.05 and the fold change was greater than 1. All data were analyzed using the R statistical software (version 4.1.3).
Results
Accumulation of MDSCs and generation of an immunosuppressive microenvironment during breast tumor recurrence
To study the alterations in immune cell populations in the blood during tumor recurrence, we established an orthotopic model by inoculating luciferase-tagged mouse 4T1 (4T1-luci) breast cancer cells into the 4th mammary fat pad of BALB/c mice (Figure 1). Three weeks after inoculation, paired normal and tumor-bearing mice were sacrificed for blood collection and mononuclear cell isolation. Tumors of another three tumor-bearing mice were removed by surgical resection, and tumor recurrence was monitored by detecting bioluminescence using an IVIS imaging system. When a bioluminescent signal appeared, mice were sacrificed for blood collection. Tumor recurrence in mice was variable and was detected approximately 2-4 weeks after tumor dissection (5 to 7 weeks after cancer cell inoculation). Isolated mononuclear cells were subjected to NanoString analysis using the nCounter Mouse Myeloid Innate Immunity V2 panel (754 genes) and the Mouse PanCancer Immune Profiling Panel (770 genes). The count data were exported and used for quality control and data normalization using the R language. Pearson’s correlation and PCA showed no batch effects between the two panels (Supplementary Figure 1A and 1B).
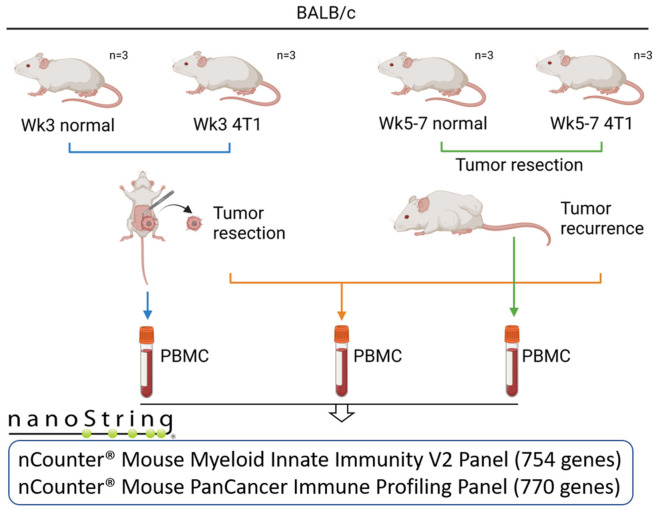
Figure 1.
Scheme of blood collection and NanoString analysis in tumor recurrence animal model. Twelve (four groups, n = 3/group) 6-week-old female BALB/c mice were included in this study. Six mice were subcutaneously transplanted with luciferase-tagged 4T1 (4T1-luc) mouse breast cancer cells (1×106 cells per mouse) into the 4th mammary fat pad. At week 3, one set of paired normal (n = 3) and tumor-bearing mice (n = 3) were euthanized, and PBMCs were collected. Another 3 tumor-bearing mice underwent tumor resection and suture surgery. The mice were monitored for tumor recurrence weekly using an IVIS imaging system. When a bioluminescent signal appeared (between weeks 5 and 7), tumor-recurrent paired mice were sacrificed for PBMC collection. PBMCs from 12 mice were subjected to NanoString analysis using nCounter® Mouse Myeloid Innate Immunity V2 panel (754 genes) and nCounter® Mouse PanCancer Immune Profiling Panels (770 genes).
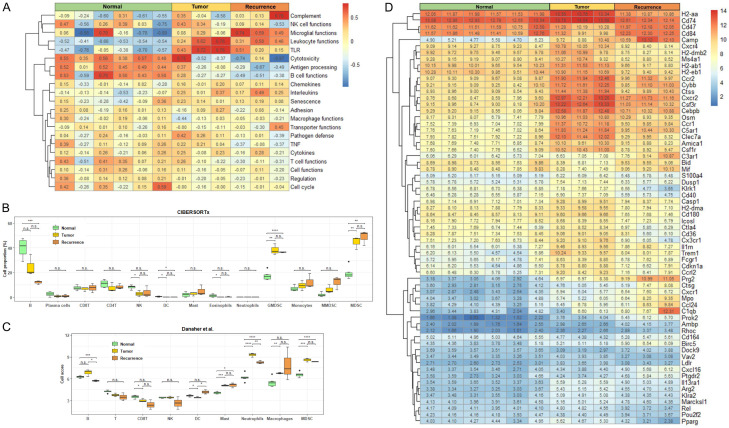
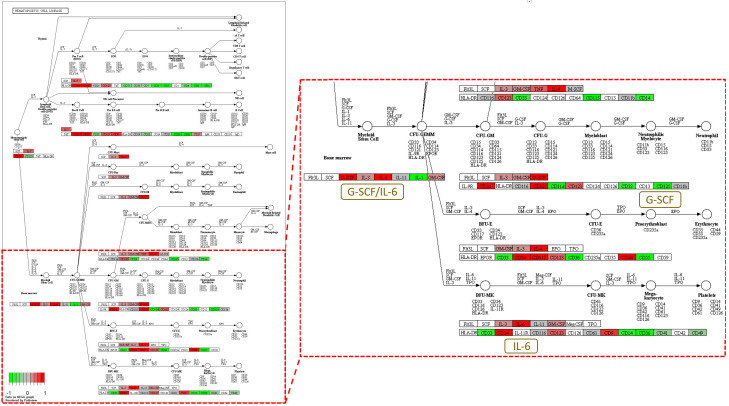
GSVA demonstrated elevated gene pathways related to leukocyte function, toll-like receptor (TLR) activation, and interleukin upregulation in tumor-recurrent mice, indicating a host defense against cancer cells by the immune system (Figure 2A). In contrast, decreased T-cell-mediated cytotoxicity, decreased B-cell function, and attenuation of antigen processing suggested immunosuppression. We further defined our cell clusters of the NanoString data by CIBERSORTx proportion using these markers and found that MDSCs, especially g-MDSCs, increased significantly in tumor-bearing (week 3) and tumor-recurrent (week 5-7) mice (Figure 2B). A decrease in the number of B-cells was also observed in mice with tumor recurrence. No significant alterations were observed in other immune cells. In cell score analysis by gene cluster averaging, both tumor-bearing and tumor-recurrent mice showed elevated neutrophil, macrophage, and MDSC signatures (Figure 2C). Moreover, a reduction in the abundance of CD8+ T and natural killer (NK) cells was observed. Sixty-four differentially expressed genes between normal, tumor-bearing, and tumor-recurrent mice were identified using NanoString analysis (Figure 2D). Among them, several genes, including Camp, Cxcr2, Csf3r, Cebpb, C5ar1, Osm, Ccr1, and Prg2, were consistently upregulated in the immune cells of tumor-bearing and tumor-recurrent mice. KEGG pathway analysis revealed a correlation between the hematopoietic cell lineage and the upregulation of G-scf, Il-6, Cd14, Cd33, Cd34, and Cd11b in tumor-recurrent mice compared to healthy controls (Figure 3). These data suggest that circulating MDSCs may build an immunosuppressive microenvironment to promote tumor relapse.
Figure 2.
Immune profiling of the PBMCs collected from tumor-bearing and tumor-recurred mice using NaonoString analysis. A. Cellular functions of PBMCs of individual groups analyzed by GSVA. B. Cell type proportions among normal mice (light green box), tumor-bearing mice (yellow box), and tumor-resected mice with recurrence (orange box) estimated using CIBERSORTx with LM22 and custom MDSC signature matrix. The results were summarized to the 13 cell types, including B-cells, plasma cells, CD8+ T-cells, CD4+ T-cells, NK cells, DC cells, mast cells, eosinophils, G-MDSCs, monocytes, m-MDSCs, and total MDSCs. C. Cell type abundances were estimated based on the average expression of the cell type-specific gene sets [12,16]. D. The expression of 64 differentially expressed genes in PBMCs of normal, tumor-bearing, and tumor-recurred mice in the NanoString analysis. n.s.: not significant; *: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.001; ****: p-value < 0.0001.
Figure 3.
Enrichment of pathway genes in hematopoietic cell lineage in tumor-recurred mice. The NanoString results reported in Figure 2 were subjected to KEGG pathway analysis. The gene expression profile of the PBMCs of tumor-recurred mice showed enrichment of pathway genes in hematopoietic cell lineage when compared to that of normal mice. Genes, including G-CSF and IL-6, are strongly expressed in tumor-recurred mice.
Csf3r+ MDSCs are significantly increased during tumor recurrence and are capable of inhibiting T-cell proliferation
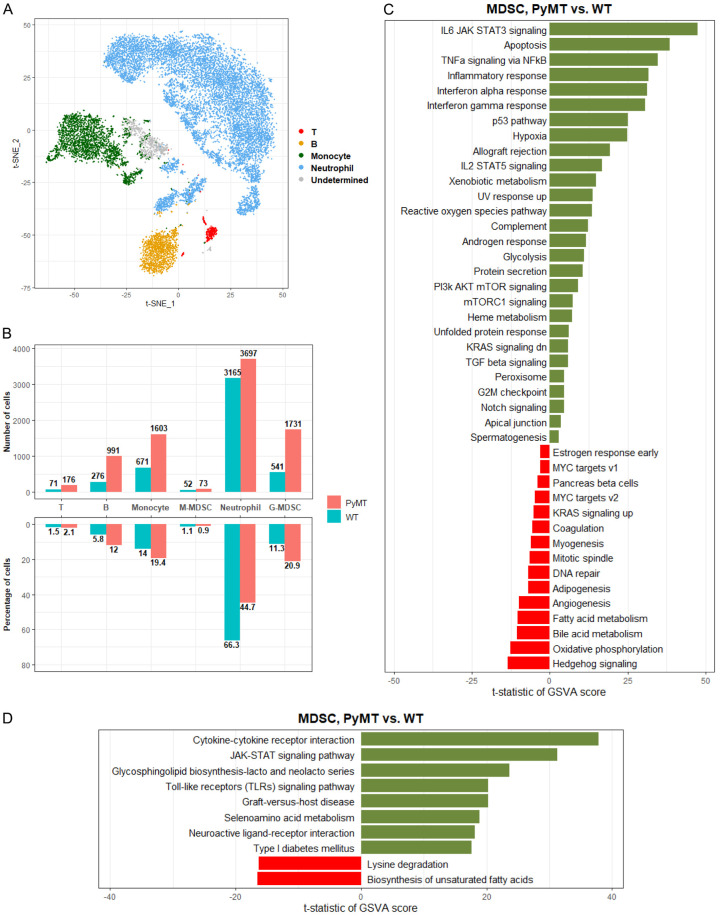
Constitutive overexpression of the polyomavirus middle T antigen (PyMT) using the mouse mammary tumor virus (MMTV) promoter results in the spontaneous development of breast tumors in mice that closely recapitulate human breast tumorigenesis [18]. To verify our findings, we analyzed single-cell RNA sequencing data of MDSCs collected from an MMTV-PyMT breast cancer mouse model [12]. Using classical immunophenotyping markers, we identified Cd3g (T-cells), Cd79a and Cd19 (B-cells), Csf1r and Ccr2 (monocytes), Ly6g and Cxcr2 (neutrophils), and a subset of undetermined cells (Figure 4A). The cell numbers and proportions of various cell types were compared between the WT and MMTV-PyMT mice, and an obvious increase in MDSCs was observed (Figure 4B). In hallmark pathway analysis, several signaling pathways were upregulated in tumor MDSCs, including interleukin-6 (Il-6), Janus kinase-signal transducer and activator of transcription (Jak-Stat), tumor necrosis factor-alpha (Tnf-α) signaling via nuclear factor kappa B (Nf-κb), and interferon (Ifn) family response (Figure 4C). Conversely, metabolic pathways, including oxidative phosphorylation, bile acid metabolism, and fatty acid metabolism, were downregulated. KEGG pathway analysis revealed cytokine-cytokine receptor interaction and JAK-STAT signaling as the top two enriched pathways, suggesting that stimulation of type I and II cytokine receptors by cytokines and interferons leads to JAK phosphorylation and STAT activation (Figure 4D). Interestingly, the pathways involved in the regulation of lysine degradation and the biosynthesis of unsaturated fatty acids were dramatically suppressed in tumor MDSCs.
Figure 4.
Identification and annotation of MDSCs in scRNA-seq data. (A) In the t-SNE plot, the cells were colored based on the cell types identified from the canonical marker genes. The cell type was annotated as “undetermined” because all marker genes identifying monocytes or neutrophils were either expressed or non-expressed simultaneously in this cell type. (B) The upper bar chart shows the numbers of T-cells, B-cells, monocytes, m-MDSC, neutrophils, and g-MDSC in MMTV-PyMT and WT mice; the bottom bar chart shows the proportions of these cell types in MMTV-PyMT and WT mice. The differentially enriched (C) hallmark and (D) KEGG gene sets analysis between MMTV-PyMT and WT mice within MDSCs.
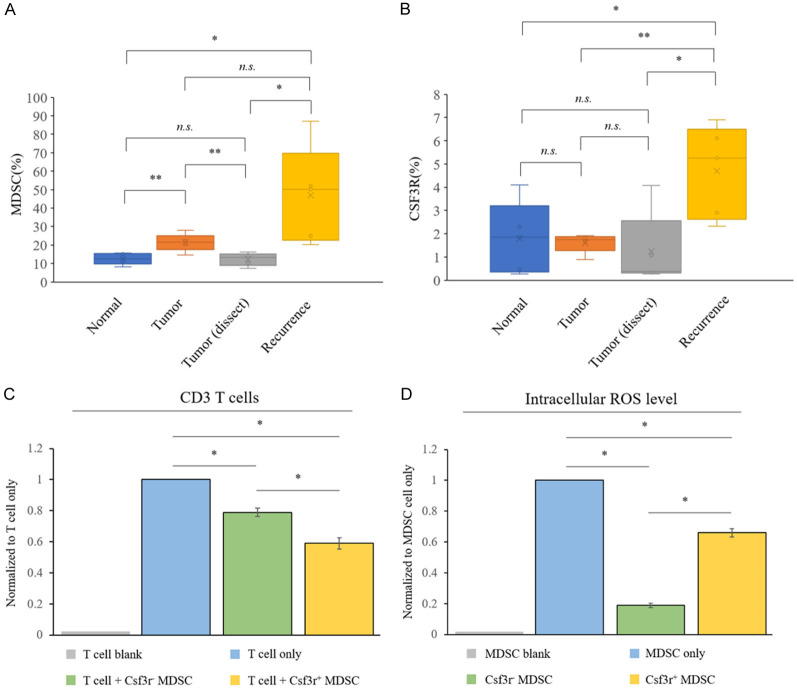
A comparison of the 64 differentially expressed genes in our NanoString analysis and the 105 differentially expressed genes altered in tumor MDSCs in the single-cell RNA sequencing data identified ten common genes (Cd84, Cxcr2, Csf3r, Osm, Ccr1, C5ar1, Amica1, Dusp1, Prok2, and Arg2; Figure 2D). Among them, Cxcr2, Csf3r, Osm, Ccr1, and C5ar1 were also upregulated in tumor-recurrent mice, indicating the possibility of using these markers to identify a subset of MDSCs to detect tumor relapse. We focused on Cxcr2, Csf3r, and Ccr1 because these chemokine receptors are associated with the accumulation and trafficking of MDSCs to primary or metastatic tumors. We repeated our mouse experiments and collected blood samples for MDSC analysis. As shown in Figure 5A, the percentage of total MDSCs increased in the blood of tumor-bearing mice. After tumor dissection, the percentage of total MDSCs decreased to normal control levels. When the tumor recurred, the total MDSC abundance increased again (P = 0.033 and 0.04, compared with the levels in the normal control and tumor dissection groups, respectively). Csf3r+ MDSCs represented a small proportion (approximately 2%) of the total MDSCs in normal mice (Figure 5B). Interestingly, the percentage of Csf3r+ MDSCs was not altered in tumor-bearing and tumor-dissected mice. However, a robust increase in Csf3r+ MDSCs in tumor-recurrent mice (P = 0.02 and 0.017, compared to the levels in the normal control and tumor dissection groups, respectively) was found, suggesting that this subset of MDSCs could be a more reliable biomarker for predicting tumor relapse. Next, we investigated the inhibitory effects of Csf3r+ MDSCs on T-cell proliferation. Because the abundance of CSF3R+ MDSCs was very low in the peripheral blood, we isolated Csf3r- and Csf3r+ MDSCs from the spleens of tumor-bearing mice. As shown in Figure 5C, Csf3r+ MDSCs suppressed T-cell proliferation more effectively than Csf3r- MDSCs. In addition, we detected ROS production, an indicator of MDSC function, using DCF-DA staining and demonstrated a 3.7-fold increase in ROS in Csf3r+ MDSCs compared to that in Csf3r- MDSCs (Figure 5D). These results suggest that blood Csf3r+ MDSCs are potential biomarkers for the prediction of tumor relapse and that these MDSCs exhibit strong T-cell suppression activity.
Figure 5.
CSF3R+ MDSCs are elevated in the PBMC of tumor-recurred mice and exhibit higher immunosuppressive activity. The PBMC samples of various groups of mice were collected and subjected to marker staining. The populations of (A) CD11b+/Gr1+ and (B) CD11b+/Gr1+/Csf3r+ MDSCs studied by flow cytometry. (C) CD3+ T-cell populations assessed by flow cytometry after co-culture with Csf3r- or Csf3r+ MDSCs isolated from the spleen of tumor-bearing mice, with the results normalized to the T-cell group, defined as 1 (gray bar). (D) Intracellular ROS levels of various sub-populations of MDSCs measured by DCF-DA staining, with the fluorescent intensities normalized to the total MDSC group, defined as 1 (gray bar). n.s.: no significance, *: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.001; ****: p-value < 0.0001.
MDSC signature and CSF3R expression are elevated in the blood of patients with breast cancer with tumor recurrence
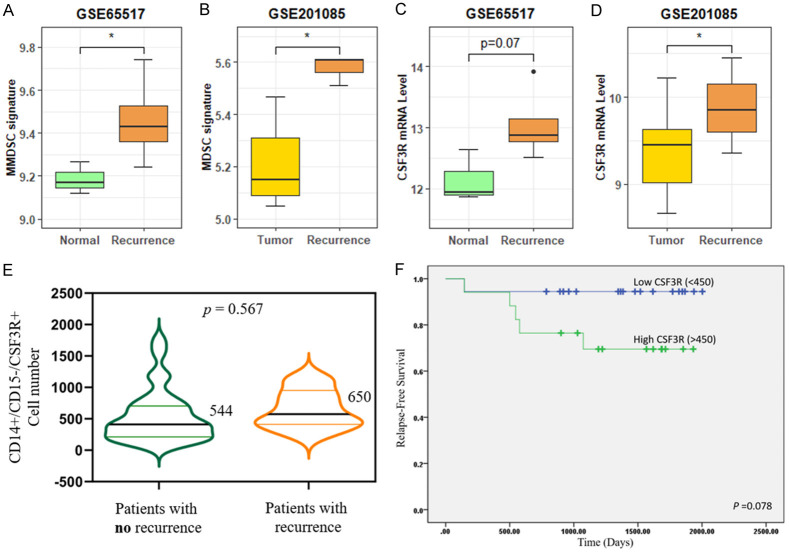
Most public databases contain genetic information on tumors but not on PBMCs in patients with cancer. Two public datasets (GSE65517 and GSE201085), which included microarray and RNA sequencing data of PBMCs collected from patients with breast cancer, with or without locoregional relapse or metastasis, were used for clinical validation. We first compared the expression of the MDSC signature reported in a previous single-cell RNA sequencing study [12] using these two datasets. The MDSC signature scores significantly increased in patients with breast cancer with tumor recurrence in both the datasets (P = 0.045 and 0.002 in GSE65517 and GSE201085, respectively; Figure 6A and 6B). Next, we investigated the CSF3R mRNA levels in these two datasets and found higher CSF3R expression in patients with metastatic or recurrent tumors (P = 0.071 and 0.019 in GSE65517 and GSE201085, respectively; Figure 6C and 6D). To obtain more reliable results, human buffy coat specimens (n = 35) were collected for further analysis. A total of 35 patients with invasive ductal carcinoma were included, with six patients showing relapse. Characteristics of the 35 patients enrolled in this study are listed in Table 1. The age range of the patients was 30-78 years, with a median age of 58 years. All patients with breast cancer were clinically classified as having stages I-III. 5 patients with primary breast cancer received neoadjuvant chemotherapy, 24 received adjuvant chemotherapy, and 6 did not receive chemotherapy (nil). Flow cytometric analyses revealed the presence of CD14+/CD15-/CSF3R+ cells in the PBMCs of patients with breast cancer. The median number was 544/10000 cells (n = 29) in patients without recurrence and 650/10000 cells (n = 6) in those with recurrence (Figure 6E). However, there was no significant difference in CSF3R expression between patients with and without recurrence (P = 0.567). We next performed a Kaplan-Meier plot by utilizing a log-rank test and found higher relapse-free survival in patients with low CSF3R expression (cut-off value < 450) than in patients with high CSF3R expression (P = 0.078; Figure 6F). These data suggest that the MDSC population and CSF3R expression were both increased in the PBMCs of patients with breast cancer with metastatic or recurrent tumors, in agreement with the findings in our orthotopic and MMTV-PyMT models.
Figure 6.
Validation of MDSC abundance and Csf3r mRNA expression in clinical PBMC datasets. Boxplots of MDSC signature in (A) monocytes from patients with metastatic breast cancer compared with healthy donors and (B) whole blood from patients with TNBC with recurrence compared with those without recurrence. Boxplot of Csf3r mRNA expression in (C) monocytes and (D) whole blood from patients with metastatic breast cancer and patients with TNBC with recurrence, respectively. (E) CD14+/CD15-/CSF3R+ cell population of 35 patients with breast cancer, 29 with no recurrence (median = 544, 544/10000 cells) and 6 with recurrence (median = 650, 650/10000 cells). (F) Kaplan-Meier survival curve of 35 patients.
Table 1.
Clinicopathologic characteristics of the invasive ductal carcinoma cohort
| Characteristics | No. (%) |
|---|---|
| Age (years):median (range) | 58 (30-78) |
| Gender | |
| Male | 35 (100) |
| Female | 0 (0) |
| Cancer type | |
| Breast invasive carcinoma | 35 (100) |
| c T stage | |
| 1B | 2 (5) |
| 1C | 10 (29) |
| 2 | 21 (60) |
| 3 | 1 (3) |
| 4 | 1 (3) |
| c N stage | |
| 0 | 23 (66) |
| 1 | 10 (29) |
| 2 | 2 (5) |
| c Stage | |
| IA | 11 (31) |
| IIA | 12 (34) |
| IIIA | 2 (5) |
| IIB | 9 (26) |
| IIIB | 1 (3) |
| Chemotherapy | |
| Adjuvant | 24 (69) |
| Neoadjuvant | 5 (15) |
| Nil | 6 (16) |
| Surgery | |
| Breast conserving surgery | 16 (46) |
| Total mastectomy | 19 (54) |
| Recurrence | |
| Yes | 6 (16) |
| No | 29 (84) |
| Death | |
| Yes | 5 (15) |
| No | 30 (85) |
Discussion
Immune surveillance is a critical process for preventing the relapse of metastatic or dormant cancer cells. When immunity is suppressed in patients with cancer, tumor recurrence may develop. In this study, we found that an increase in MDSCs in the peripheral blood is a potential biomarker of tumor relapse. MDSCs are immunosuppressive cells that effectively inhibit the function and proliferation of cytotoxic T-cells. Accumulating evidence suggests that MDSCs play a pivotal role in the progression of breast cancer. Bergenfelz et al. reported that m-MDSC levels are elevated in patients with metastatic breast cancer and are potential biomarkers for monitoring disease progression [14]. In addition, breast cancer patients with high levels of m-MDSCs had worse survival rates [19]. Using a similar tumor formation and resection model as that shown in this study, Monteran et al. investigated the effect of adjuvant chemotherapy on the modulation of immunosuppression and metastatic relapse [20]. They found that doxorubicin treatment stimulated the production of complement factors in lung fibroblasts, which enhanced the recruitment of MDSCs to lung tissues, leading to T-cell dysfunction and increased lung metastasis. Inhibition of MDSC recruitment by blocking chemotherapy-triggered complement signaling reduces metastatic relapse in the lungs. This study clearly demonstrated the important role of MDSCs in the establishment of an immunosuppressive microenvironment in the premetastatic niche. Recently, Liu et al. demonstrated that upregulation of aldehyde dehydrogenase 1A1 (ALDH1A1) in breast cancer cells enhances the production of granulocyte-macrophage colony-stimulating factor (GM-CSF), resulting in the expansion of MDSCs and the generation of an immunosuppressive microenvironment that accelerate tumor progression [21]. The increase in MDSCs also correlates with malnutrition, bone destruction, and worse clinical outcomes in patients with breast cancer [22,23]. Furthermore, a meta-analysis of 40 studies revealed that breast cancer patients showed the most significant association between high MDSC levels and poor OS (pooled hazard ratio = 3.053) [24]. Although these studies clearly demonstrated the association between MDSC levels and clinicopathological features, whether MDSC abundance in the peripheral blood could be a predictor of tumor recurrence is not known. Using animal studies and bioinformatics analysis, we provide the first evidence that an increase in CSF3R+ MDSCs is a potential biomarker for predicting tumor relapse in patients with breast cancer.
CSF3R is a cognate receptor for granulocyte colony-stimulating factor (G-CSF), an important cytokine that participates in the regulation of granulocyte proliferation, differentiation, and survival [25]. CSF3R activation by G-CSF stimulates downstream signaling, including the JAK/STAT, AKT, and MAPK pathways. Currently, G-CSF is used to prevent chemotherapy-induced neutropenia in patients with breast cancer [26]. However, recent studies have highlighted possible detrimental effects of G-CSF on breast cancer. Waight et al. showed that tumor-released G-CSF acts via MDSCs to promote breast tumorigenesis in multiple mouse models [27]. Ravindranathan et al. demonstrated that G-CSF derived from breast cancer cells decreased the efficacy of autologous tumor cell vaccination and that inhibition of G-CSF signaling enhanced protective immunity in experimental animals [28]. In addition, patients with breast cancer with high serum levels of G-CSF have a shorter disease-free survival after chemotherapy with bisphosphonate zoledronic acid [29]. Because G-CSF plays a critical role in the regulation of the development and differentiation of neutrophils, this cytokine may also affect the neutrophil phenotype and heterogeneity to accelerate breast cancer progression. Geng et al. also demonstrated that an elevated neutrophil-to-lymphocyte ratio correlated with worsened disease-free survival and a higher risk of recurrence in patients with breast cancer who underwent curative resection [30]. Similarly, an increase in neutrophil-to-lymphocyte ratio also predicts late relapse in patients with breast cancer [31]. In patients with neoadjuvant chemotherapy-treated breast cancer, higher levels of tumor-infiltrating neutrophils are significantly associated with shorter disease-free survival and are a predictor of a high incidence of recurrence [32]. These studies support the notion that tumor-derived G-CSF may expedite the recruitment of tumor-promoting immune cells, such as MDSCs, N2 neutrophils, and M2 macrophages, to create a favorable microenvironment for tumor relapse.
The two critical findings of this study are noteworthy. First, the increase in CSF3R+ MDSCs in the peripheral blood predicts tumor recurrence more precisely than the increase in total MDSCs, although they only account for 2% of the total MDSCs. In a clinical setting, the detection of this specific subset of MDSCs is important. Second, CSF3R+ MDSCs may play an important role in promoting tumor relapse. We hypothesized that minimal residual metastatic breast cancer cells release G-CSF to stimulate the expansion of CSF3R+ MDSCs, as detected in the peripheral blood. These cells are then recruited to metastatic cancer cells to suppress immune surveillance and promote cancer cell outgrowth. The fact that the number of CD14+/CD15-/CSF3R+ cells in patients with recurrence was not significantly different from that in patients without recurrence may be due to the limited number of patients analyzed. Indeed, we found that patients with higher levels of CSF3R had worse survival and that CSF3R+ MDSCs were more effective in inhibiting T-cell proliferation, suggesting a stronger immunosuppressive activity of these MDSCs. However, our hypothesis warrants further experimental and clinical validation.
Collectively, our results identified CSF3R+ MDSCs as biomarkers for predicting tumor recurrence in patients with breast cancer and suggested the possibility of preventing metastatic relapse by targeting the G-CSF-CSF3R axis.
Conclusion
In this study, we provide evidence that MDSCs in the peripheral blood are significantly increased during tumor relapse in experimental animal models and patients with breast cancer. More importantly, we report for the first time that a subset of CSF3R+ MDSC may predict breast cancer recurrence more precisely. In vitro functional assays also demonstrated that Csf3r+ MDSCs exhibited stronger T-cell suppression activity, which may help cancer cells escape immune surveillance and promote tumor recurrence. A large population study is required to validate the clinical significance of CSF3R+ MDSCs for the early detection of metastatic relapse in patients with breast cancer after surgical resection or chemotherapy.
Acknowledgements
We acknowledge the use of BioRender software to generate Figure 1. This study was supported by grants 111-2320-B-400-024 to WCH from the Ministry of Science and Technology, Taiwan, and CA-111-PP-17 to WCH from the Ministry of Health and Welfare, Taiwan.
Disclosure of conflict of interest
None.
Abbreviations
- CCA
canonical correlation analysis
- CSF3R
colony-stimulating factor 3 receptor
- CTC
circulating tumor cells
- ctDNA
cell-free tumor DNA
- DCF-DA
dichlorofluorescin diacetate
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- g-CSF
granulocyte colony-stimulating factor
- m-MDSC
monocytic MDSCs
- GSVA
gene set variation analysis
- IFN
interferon
- IL-6
interleukin-6
- JAK
Janus kinase
- MDSC
myeloid-derived-suppressor cells
- g-MDSC
granulocytic MDSC
- m-MDSC
monocytic MDSC
- MMTV
mouse mammary tumor virus
- NF-κB
nuclear factor kappa B
- PBMC
peripheral blood mononuclear cells
- PS
penicillin-streptomycin
- PyMT
polyomavirus middle T antigen
- ROS
reactive oxygen species
- STAT
signal transducer and activator of transcription
- TLR
Toll-like receptor
- TMM
trimmed mean of M-values
- TNF-α
tumor necrosis factor-alpha
- Treg
regulatory T cells
Supporting Information
References
- 1.Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M, Esener S, Fitzgerald RC, Gambhir SS, Kuhn P, Rebbeck TR, Balasubramanian S. Early detection of cancer. Science. 2022;375:eaay9040. doi: 10.1126/science.aay9040. [DOI] [PubMed] [Google Scholar]
- 2.Hing JX, Mok CW, Tan PT, Sudhakar SS, Seah CM, Lee WP, Tan SM. Clinical utility of tumour marker velocity of cancer antigen 15-3 (CA15-3) and carcinoembryonic antigen (CEA) in breast cancer surveillance. Breast. 2020;52:95–101. doi: 10.1016/j.breast.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uygur MM, Gümüş M. The utility of serum tumor markers CEA and CA 15-3 for breast cancer prognosis and their association with clinicopathological parameters. Cancer Treat Res Commun. 2021;28:100402. doi: 10.1016/j.ctarc.2021.100402. [DOI] [PubMed] [Google Scholar]
- 4.Ortolan E, Appierto V, Silvestri M, Miceli R, Veneroni S, Folli S, Pruneri G, Vingiani A, Belfiore A, Cappelletti V, Vismara M, Dell’Angelo F, De Cecco L, Bianchi GV, de Braud FG, Daidone MG, Di Cosimo S. Blood-based genomics of triple-negative breast cancer progression in patients treated with neoadjuvant chemotherapy. ESMO Open. 2021;6:100086. doi: 10.1016/j.esmoop.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cailleux F, Agostinetto E, Lambertini M, Rothé F, Wu HT, Balcioglu M, Kalashnikova E, Vincent D, Viglietti G, Gombos A, Papagiannis A, Veys I, Awada A, Sethi H, Aleshin A, Larsimont D, Sotiriou C, Venet D, Ignatiadis M. Circulating tumor DNA after neoadjuvant chemotherapy in breast cancer is associated with disease relapse. JCO Precis Oncol. 2022;6:e2200148. doi: 10.1200/PO.22.00148. [DOI] [PubMed] [Google Scholar]
- 6.Axelrod ML, Wang Y, Xu Y, Sun X, Bejan CA, Gonzalez-Ericsson PI, Nunnery S, Bergman RE, Donaldson J, Guerrero-Zotano AL, Massa C, Seliger B, Sanders M, Mayer IA, Balko JM. Peripheral blood monocyte abundance predicts outcomes in patients with breast cancer. Cancer Res Commun. 2022;2:286–292. doi: 10.1158/2767-9764.CRC-22-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyama Y, Kawai S, Uenaka N, Okazaki M, Asaoka M, Teraoka S, Ueda AI, Miyahara K, Kawate T, Kaise H, Yamada K, Ishikawa T. Absolute lymphocyte count, platelet-to-lymphocyte ratio, and overall survival in eribulin-treated HER2-negative metastatic breast cancer patients. Cancer Diagn Progn. 2021;1:435–441. doi: 10.21873/cdp.10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia W, Wu J, Jia H, Yang Y, Zhang X, Chen K, Su F. The peripheral blood neutrophil-to-lymphocyte ratio is superior to the lymphocyte-to-monocyte ratio for predicting the long-term survival of triple-negative breast cancer patients. PLoS One. 2015;10:e0143061. doi: 10.1371/journal.pone.0143061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21:485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safarzadeh E, Hashemzadeh S, Duijf PHG, Mansoori B, Khaze V, Mohammadi A, Kazemi T, Yousefi M, Asadi M, Mohammadi H, Babaie F, Baradaran B. Circulating myeloid-derived suppressor cells: an independent prognostic factor in patients with breast cancer. J Cell Physiol. 2019;234:3515–3525. doi: 10.1002/jcp.26896. [DOI] [PubMed] [Google Scholar]
- 12.Alshetaiwi H, Pervolarakis N, McIntyre LL, Ma D, Nguyen Q, Rath JA, Nee K, Hernandez G, Evans K, Torosian L, Silva A, Walsh C, Kessenbrock K. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci Immunol. 2020;5:eaay6017. doi: 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergenfelz C, Larsson AM, von Stedingk K, Gruvberger-Saal S, Aaltonen K, Jansson S, Jernström H, Janols H, Wullt M, Bredberg A, Rydén L, Leandersson K. Systemic monocytic-MDSCs are generated from monocytes and correlate with disease progression in breast cancer patients. PLoS One. 2015;10:e0127028. doi: 10.1371/journal.pone.0127028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danaher P, Kim Y, Nelson B, Griswold M, Yang Z, Piazza E, Beechem JM. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. 2022;13:385. doi: 10.1038/s41467-022-28020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergenfelz C, Roxå A, Mehmeti M, Leandersson K, Larsson AM. Clinical relevance of systemic monocytic-MDSCs in patients with metastatic breast cancer. Cancer Immunol Immunother. 2020;69:435–448. doi: 10.1007/s00262-019-02472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monteran L, Ershaid N, Doron H, Zait Y, Scharff Y, Ben-Yosef S, Avivi C, Barshack I, Sonnenblick A, Erez N. Chemotherapy-induced complement signaling modulates immunosuppression and metastatic relapse in breast cancer. Nat Commun. 2022;13:5797. doi: 10.1038/s41467-022-33598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Qiang J, Deng Q, Xia J, Deng L, Zhou L, Wang D, He X, Liu Y, Zhao B, Lv J, Yu Z, Lei QY, Shao ZM, Zhang XY, Zhang L, Liu S. ALDH1A1 activity in tumor-initiating cells remodels myeloid-derived suppressor cells to promote breast cancer progression. Cancer Res. 2021;81:5919–5934. doi: 10.1158/0008-5472.CAN-21-1337. [DOI] [PubMed] [Google Scholar]
- 22.Danilin S, Merkel AR, Johnson JR, Johnson RW, Edwards JR, Sterling JA. Myeloid-derived suppressor cells expand during breast cancer progression and promote tumor-induced bone destruction. Oncoimmunology. 2012;1:1484–1494. doi: 10.4161/onci.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonda K, Shibata M, Ohtake T, Matsumoto Y, Tachibana K, Abe N, Ohto H, Sakurai K, Takenoshita S. Myeloid-derived suppressor cells are increased and correlated with type 2 immune responses, malnutrition, inflammation, and poor prognosis in patients with breast cancer. Oncol Lett. 2017;14:1766–1774. doi: 10.3892/ol.2017.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang PF, Song SY, Wang TJ, Ji WJ, Li SW, Liu N, Yan CX. Prognostic role of pretreatment circulating MDSCs in patients with solid malignancies: a meta-analysis of 40 studies. Oncoimmunology. 2018;7:e1494113. doi: 10.1080/2162402X.2018.1494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta HM, Corey SJ. G-CSF, the guardian of granulopoiesis. Semin Immunol. 2021;54:101515. doi: 10.1016/j.smim.2021.101515. [DOI] [PubMed] [Google Scholar]
- 26.Freyer G, Jovenin N, Yazbek G, Villanueva C, Hussain A, Berthune A, Rotarski M, Simon H, Boulanger V, Hummelsberger M, Falandry C. Granocyte-colony stimulating factor (G-CSF) has significant efficacy as secondary prophylaxis of chemotherapy-induced neutropenia in patients with solid tumors: results of a prospective study. Anticancer Res. 2013;33:301–307. [PubMed] [Google Scholar]
- 27.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PLoS One. 2011;6:e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravindranathan S, Nguyen KG, Kurtz SL, Frazier HN, Smith SG, Koppolu BP, Rajaram N, Zaharoff DA. Tumor-derived granulocyte colony-stimulating factor diminishes efficacy of breast tumor cell vaccines. Breast Cancer Res. 2018;20:126. doi: 10.1186/s13058-018-1054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubellacker JM, Baryawno N, Severe N, DeCristo MJ, Sceneay J, Hutchinson JN, Haider MT, Rhee CS, Qin Y, Gregory WM, Garrido-Castro AC, Holen I, Brown JE, Coleman RE, Scadden DT, McAllister SS. Modulating bone marrow hematopoietic lineage potential to prevent bone metastasis in breast cancer. Cancer Res. 2018;78:5300–5314. doi: 10.1158/0008-5472.CAN-18-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geng SK, Fu SM, Fu YP, Zhang HW. Neutrophil to lymphocyte ratio is a prognostic factor for disease free survival in patients with breast cancer underwent curative resection. Medicine (Baltimore) 2018;97:e11898. doi: 10.1097/MD.0000000000011898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon G, Noh H, Cho IJ, Lee JI, Han A. Prediction of late recurrence in patients with breast cancer: elevated neutrophil to lymphocyte ratio (NLR) at 5 years after diagnosis and late recurrence. Breast Cancer. 2020;27:54–61. doi: 10.1007/s12282-019-00994-z. [DOI] [PubMed] [Google Scholar]
- 32.Geng SK, Fu SM, Ma SH, Fu YP, Zhang HW. Tumor infiltrating neutrophil might play a major role in predicting the clinical outcome of breast cancer patients treated with neoadjuvant chemotherapy. BMC Cancer. 2021;21:68. doi: 10.1186/s12885-021-07789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.