Abstract
Deubiquitinating enzymes (DUBs) are a large group of proteases that reverse ubiquitination process and maintain protein homeostasis. The DUBs have been classified into seven subfamilies according to their primary sequence and structural similarity. As a small subfamily of DUBs, the ubiquitin C-terminal hydrolases (UCHs) subfamily only contains four members including UCHL1, UCHL3, UCHL5, and BRCA1-associated protein-1 (BAP1). Despite sharing the deubiquitinase activity with a similar catalysis mechanism, the UCHs exhibit distinctive biological functions which are mainly determined by their specific subcellular localization and partner substrates. Besides, growing evidence indicates that the UCH enzymes are involved in human malignancies. In this review, the structural information and biological functions of the UCHs are briefly described. Meanwhile, the roles of these enzymes in tumorigenesis and the discovered inhibitors against them are also summarized to give an insight into the cancer therapy with the potential alternative strategy.
Keywords: Deubiquitinase, UCH family, cancer, inhibitors
Introduction
Post-translational modifications of proteins play a key role in regulating their cellular functions with high efficiency [1,2]. Ubiquitination, an essential protein post-translational modification, occurs through the sequential actions of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligase enzymes (E3s) [3-5]. It involves the formation of an isopeptide bond between the C-terminal glycine of ubiquitin and a lysine residue in the substrate. Deubiquitinating enzymes (DUBs) counteract the effects of ubiquitination by removing ubiquitin from target proteins and disassembling ubiquitin polymers [6,7]. Since protein ubiquitination and deubiquitination are involved in various biological processes, extensive studies have been conducted to understand their roles in cell cycle progression, DNA damage repair, cancer-related cellular signaling, tumorigenesis, etc. [6-10].
Human genome encodes approximately 100 unique DUBs, which can be generally divided into seven distinct subfamilies. Among these subfamilies, six of them belong to cysteine proteases, including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor-like proteases (OTUs), Machado-Joseph disease protein domain proteases (MJDs), MIU containing novel DUB family (MINDY) proteases, and zinc-binding metalloprotease (ZUFSP/ZUP1), while the remaining Jab1/Mov34/Mpr1 (JAMM) protease family members are metalloproteinases [7,11-14]. So far, four UCH family deubiquitinating enzymes have been identified in humans, and they are UCHL1, UCHL3, UCHL5, and BAP1 (BRCA1-associated protein-1). In yeast, there is only one UCH family member, YUH1 [15-17]. Except for BAP1, all of the remaining three UCHs have a molecular weight of 20-30 kDa. Meanwhile, the identified four UCHs share a highly conserved three-dimensional structure for their catalytic domains. The main function of UCHs is to recognize and hydrolyze the isopeptide bond at the C-terminal glycine of ubiquitin [15-17].
UCHL1, a unique deubiquitinating enzyme due to its dual ligase and hydrolase activity, is predominantly expressed in the brain. Dysfunction of its hydrolase activity would cause cellular accumulation of α-synuclein, and thus might contribute to the pathological process of neurodegenerative diseases such as Parkinson’s disease (PD) and dementia with Lewy bodies [18]. Moreover, a growing body of evidence indicates that UCHL1 has a debatable role in cancer, both in terms of its expression level and the signaling pathways it participates in. Therefore, it is crucial to fully elucidate the tumorigenesis-related pathways with UCHL1 involved through performing clinical and translational investigations. As the closest homolog of UCHL1 (53% sequence identity among human UCH members), UCHL3 is featured with its ability to hydrolyze the isopeptide bond involving the C-terminal glycine of either ubiquitin or ubiquitin-like molecule Nedd8 (neuronal precursor cell-expressed developmentally downregulated protein 8) [19]. When discussing the role of UCHL3 in tumorigenesis, one of the potential mechanisms could be attributed to its regulation of DNA repair [20]. In addition, UCHL3 also regulates cell cycle progression and epithelial-to-mesenchymal transition, indicative of its role as a tumor promoter [21,22]. UCHL5 is one of the deubiquitinases associated with the 19S regulatory subunit of the 26S proteasome. It interacts with 19S component RPN13 and trims the distal Ub moiety of Ub chains, which consequently rescues target proteins from proteolysis [23]. Besides, UCHL5 could interact with human INO80 chromatin-remodeling complex (hINO80) in nucleus and modulate nucleosome remodeling [23]. UCHL5 has been found to participate in regulating TGF-β signaling, the Hedgehog (Hh) signaling pathway, and the Wnt/β-catenin signaling pathway, indicating its role as a tumor promoter in cancer development [24-26]. BAP1, which is initially characterized as a tumor suppressor and predominantly distributes in nucleus, is known as the catalytic subunit of the polycomb repressive deubiquitinase (PR-DUB) complex. BAP1 has been demonstrated to remove mono-ubiquitin from histone 2A lysine 119 (H2AK119Ub) [27]. Besides, BAP1 participates in the formation of different multi-protein complexes by interacting with partners such as host cell factor 1 (HCF-1) and Ying Yang 1 (YY1). The multi-protein complexes with BAP1 as one of the key components are involved in cell cycle regulation, transcription, and cell death [28]. However, recent studies have revealed that BAP1 may have gain-of-function and carcinogenic features [29-31]. Therefore, uncovering its cellular functions associated with tumorigenesis in different types of human cancers is still in urgent need.
As aforementioned, UCH family members are involved in the key processes associated with the occurrence and development of malignant carcinomas, which include cell proliferation and epithelial-to-mesenchymal transition, etc. Therefore, interfering with UCHs’ activities utilizing small molecules might provide new strategies for cancer therapy. In this review, we summarize the structural information and biological functions of UCH family members and also their roles in cancer-related signaling pathways. Additionally, the developed inhibitors targeting UCH family members are also included for a better understanding of their potential working mechanisms in the treatment of human cancer.
Structure and function of UCH family members
UCHL1
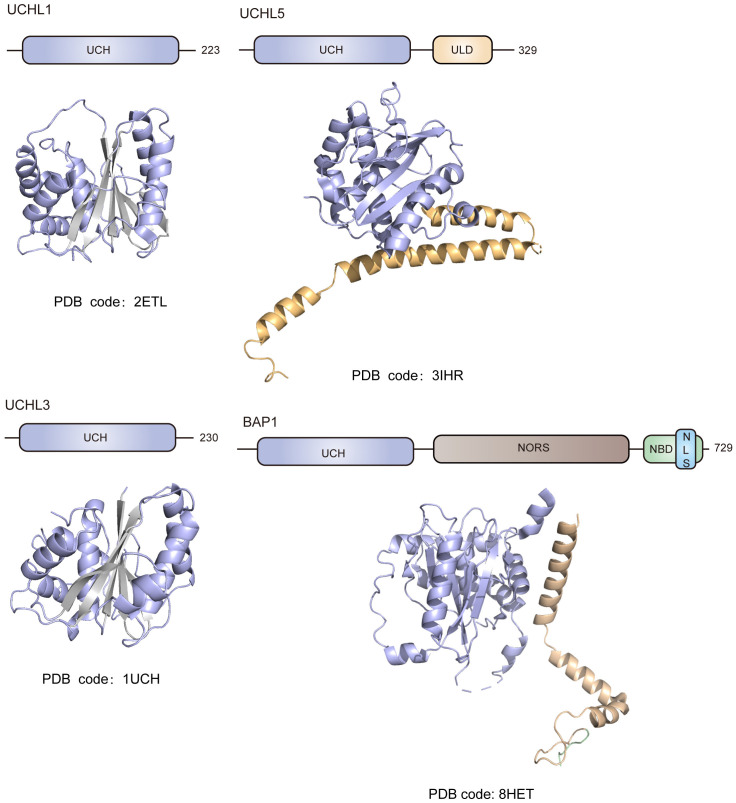
UCHL1 contains a conserved UCH catalytic domain with short N-terminal and C-terminal extensions. The structure of monomeric UCHL1 is composed of two ‘lobes’ of α-helices tightly packed against the hydrophobic core of six β-strands (Figure 1). The secondary structure elements of UCHL1 form an α-β-α sandwich fold that is the canonical structure for the catalytic domain of UCH family members. The catalytic cysteine (C90) of UCHL1 resides at the cleft formed between the N-terminal α-helix lobe and the hydrophobic core of β-strands [32]. In the apo state, the spatial alignment of UCHL1’s catalytic triad (C90, H161 & D176) is out of order, which thus represents the unproductive state of the enzyme [32] (Figure 2). Upon the binding of ubiquitin substrate, the catalytic triad rearranges to form an aligned conformation, which leads to the activation of UCHL1’s enzymatic activity [33]. Moreover, as revealed by the solved crystal structure of UCHL1, Ub polymers with larger space requirements could not be accommodated by the enzyme, which is in line with UCHL1’s primary recognition of monomeric ubiquitin and small adducts of ubiquitin [32]. The crossover loop positioned over the active site is a distinctive structure feature for all of the UCH family members, which prevents larger substrates from accessing the active site of UCHs. Among the UCH family members, UCHL1 has the shortest crossover loop spanning E149-N159 [17,32].
Figure 1.
Schematic demonstration of the structural domains and ribbon representation of the solved crystal structures for the UCH family members. All UCH family members including UCHL1, UCHL3, UCHL5 and BAP1 share a conserved UCH catalytic domain featured with a canonical αβα fold. Besides the UCH domain, UCHL5 contains an additional C-terminal ULD structural domain composed of four α-helices in its structure. BAP1 is composed of three domains including the UCH catalytic domain, the C-terminal domain, and a long non-organized regions (NORS) inserted in the middle of the enzyme. The NORS insertion unique to vertebrates contains multiple binding motifs which are responsible for BAP1’s interacting with chromosome-associated proteins, and the C-terminal domain of BAP1 contains a nuclear localization signal (NLS) at its tail. ULD, UCHL5-like domain; NBD, nucleosome binding domain; NLS, nuclear localization signal.
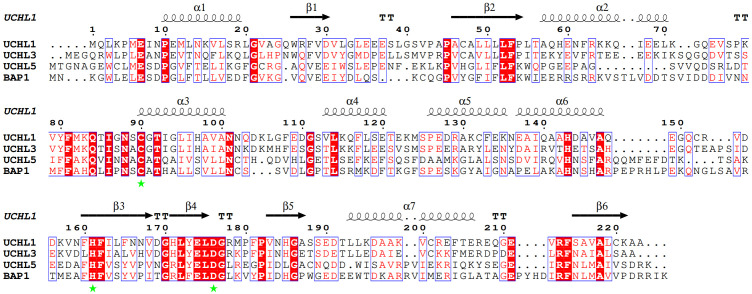
Figure 2.
Sequence alignment of UCH catalytic domain for UCH family members. The conserved residues through the UCH family are highlighted in red. The catalytic triad (C90, H161 & D176 for UCHL1) is marked with green star. The secondary structural elements observed in the crystal structure of UCHL1 are shown and labeled. The sequence alignment result was generated by using MEGA and ESPript 3.0.
UCHL1 could upregulate ubiquitin pool by hydrolyzing ubiquitinated peptides and stabilizing free ubiquitin [34]. In addition, although contradictory data has been reported, it looks like that the UCHL1 homodimer may exhibit ATP-independent ubiquitin ligase activity, which generates K63-linked ubiquitin chains on substrate protein and further prevents substrate from degradation [34]. As it has been known, a decent amount of UCHL1, which accounts for 1-2% of the total brain proteins, is expressed in neurons [35]. And α-synuclein is a neuronal protein and a key component of the pathogenic insoluble proteinaceous deposits known as Lewy bodies, which are commonly found in PD patients [36]. Studies have shown that dimeric UCHL1 could block α-synuclein’s proteasomal degradation and promote its accumulation by upregulating the K63-linked polyubiquitination of the protein. As an expected consequence, upregulated α-synuclein aggregates to form pathogenic protofibrils. Meanwhile, the UCHL1 S18Y variant exhibits reduced ubiquitin ligase activity which leads to the downregulation of α-synuclein and a possibly decreased risk of PD [18,37]. Besides potentially playing a role in the pathogenesis of typical Parkinson’s disease and dementia with Lewy bodies, UCHL1 has also been implicated as either an oncoprotein or a tumor suppressor. Not surprisingly, UCHL1 exhibits a high expression level in certain malignancies including small cell lung cancer, bone cancer, and renal cell carcinoma, while it demonstrates a deep deletion in head and neck cancer, and esophagogastric cancer. Up to date, the mechanisms underlying these observations remain obscure.
UCHL3
Same as that for UCHL1, UCHL3 forms a typical bilobal structure, which is featured with a central six-stranded antiparallel β-sheet and α-helices packing on both sides of the sheet (Figure 1). One of the two lobes is composed of the central β-sheet and two long α-helices, and the other structural lobe contains a helix bundle with one of them housing the active site nucleophile of C95 [38]. UCHL3 also possesses a disordered crossover loop spanning E154-D167 which positions over the active site cleft and finely tunes the substrate specificity of the enzyme [17]. When catalyzing the deubiquitination reaction, the flexible crossover loop stabilizes and exposes the active site of UCHL3 for accommodating ubiquitinated substrates [39]. As a typical deubiquitinating enzyme, UCHL3 is involved in the hydrolysis of ester, thioester, amide, peptide, and isopeptide bond, which is formed between the C-terminal glycine of ubiquitin and the modified molecules. Additionally, UCHL3 could interact with the ubiquitin-like protein Nedd8, which shares a 57% sequence similarity with ubiquitin, and removes Nedd8 from the modified proteins [19,40]. Thus, UCHL3 functions as a hydrolase for both ubiquitin and Nedd8. Meanwhile, due to the restriction of the crossover loop, UCHL3 is capable of handling ubiquitin adducts with substrates consisting of approximately 20 residues or less [41]. Interestingly, UCHL3 can interact with diubiquitin, but instead of hydrolyzing diubiquitin to monoubiquitin, the interaction between them inhibits the enzymatic activity of UCHL3 on other substrates [42]. The recently published crystal structure of UCHL3 complexed with K27-linked diubiquitin reveals that the active site residue C95 of the enzyme forms a thioester bond with K27-linked diubiquitin, which causes the activity inhibition of UCHL3 through a kinetic trapping mechanism [43].
UCHL5
UCHL5 primarily consists of two structural domains: the N-terminal UCH catalytic domain and the C-terminal ULD structural domain composed of four α-helices (Figure 1) [44]. The catalytic domain of UCHL5 adopts a classical αβα fold, with a central six-stranded β-sheet surrounded by α-helices [44]. UCHL5, along with the later-described BAP1, possesses relatively larger crossover loops compared to UCHL1 and UCHL3, indicating their ability to handle larger substrates [45]. In line with the aforementioned structural feature of UCHL5, it has been reported that UCHL5 could cleave diubiquitin [45]. As it has been demonstrated, UCHL5 functions as a component of the 26S proteasome and the INO80 chromatin remodeling complex. Upon the interaction of UCHL5 with either the partner in 26S proteasome or the partner in INO80 chromatin remodeling complex, the enzymatic activity of the protein would be enhanced and inhibited, respectively [23,46]. In the apo state, UCHL5 exhibits a relatively low catalytic activity against ubiquitin adducts, which is mainly caused by the modulation of its C-terminal ULD domain. When the RPN13 DEUBAD domain binds to the ULD domain of UCHL5, the enzyme switches to a conformation competent for ubiquitin binding, thus upregulating its catalytic activity and affinity for ubiquitin substrates [23,46]. On the other hand, when the INO80G DEUBAD domain interacts with UCHL5, the enzymatic activity of the enzyme is inhibited due to the blockage of the ubiquitin binding site [23,46]. As an exchangeable component of the 26S proteasome, UCHL5 can trim the distal polyubiquitin chains attached to protein substrates [47]. A recent study has demonstrated that proteasome-associated UCHL5 could selectively remove K48-branched ubiquitin chains from a mixture of ubiquitin conjugates [48]. For the specific role of UCHL5 in the INO80 chromatin remodeling complex, no detailed information has been uncovered yet. However, it has been demonstrated that UCHL5 and INO80G are key factors for the DNA double-strand break response [49].
BAP1
BAP1 contains 729 amino acid residues, which consist of three domains including the evolutionarily conserved UCH catalytic domain, the C-terminal nucleosome binding domain (NBD), and a long non-organized region (NORS) inserted in the middle of the enzyme (Figure 1). The NORS insertion predicted to be unstructured is unique to vertebrates. Meanwhile, it contains multiple binding motifs which are responsible for BAP1’s interaction with chromosome-associated proteins. Additionally, the C-terminal domain of BAP1 contains a nuclear localization signal (NLS) at its tail (Figure 1) [50,51]. It has been reported that the NLS region of BAP1 can be ubiquitinated upon the catalysis of the E2/E3 heterotrimeric enzyme UBE2O. While the attached ubiquitin could be removed through self-deubiquitination [52].
Unlike the other UCH family members, the complexity of BAP1’s role in cellular determines its dual function either as a tumor suppressor or a tumor promoter. The typical working model of BAP1 is dependent on its interaction with partner proteins. H2A is ubiquitinated by the PRC1 complex and deubiquitinated by the PR-DUB complexes, which mainly comprise BAP1, ASX-like proteins ASXL1/2/3, HCF-1, OGT, YY1, and so on [27,28,50,53-57]. Maintaining the balance between the PRC1 complex and the PR-DUB complex is crucial, as an upregulated level of PRC1 or mutations in BAP1 can lead to the accumulation of H2AK119Ub [58]. For the PR-DUB complexes, the initially determined molecular weight data suggested a binding stoichiometry of 2:1 for BAP1 to ASXL, while subsequently solved crystal structure of Calypso (Drosophila ortholog of human BAP1) complexed with ASXDEU revealed the formation of a complex with the corresponding binding stoichiometry at 2:2 [27,59]. In 2023, Weiran Ge et al. solved the specific nucleosome-binding mode of PR-DUB, which displaces the H2A C-terminal tail from the nucleosome surface and allows the access of H2AK119Ub to the enzymatic active site of BAP1 [60]. The cryo-electron microscopy structure of human PR-DUB, consisting of BAP1 and ASXL1, complexed with the chromatosome provides a strong basis for subsequent studies.
Potential roles of UCH family members in tumorigenesis
UCHL1 in tumorigenesis
According to the published literature, it looks like that UCHL1 could function as both tumor promoter and tumor suppressor. Meanwhile, the Human Protein Atlas data indicates that a significantly enhanced expression level of UCHL1 is observed in glioma, and the enzyme also serves as an unfavorable prognostic marker for both endometrial cancer (unfavorable) and urothelial cancer (https://www.proteinatlas.org/ENSG00000154277-UCHL1/pathology).
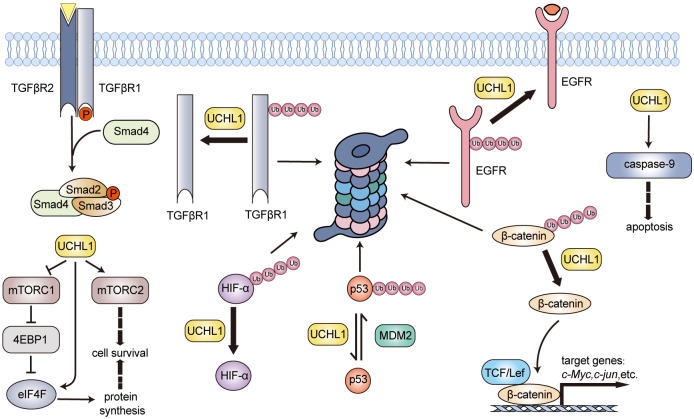
As an oncogene, it is evidenced that UCHL1’s mRNA level is linked to the recurrence in patients with invasive breast cancer [61]. In breast cancer cells, UCHL1 stabilizes EGFR through its deubiquitination activity, resulting in hyperactivated MAPK signal transduction (Figure 3), which consequently inhibits ERα expression by transcriptional repression [62]. The inhibition of UCHL1 might upregulate ERα by promoting the degradation of EGFR, thereby sensitizing ERα-positive breast cancer cells to selective ER modulator such as tamoxifen [62]. In triple-negative breast cancer (TNBC), UCHL1 also participates in TGF-β signaling pathways by deubiquitinating TGFR1 and SMAD2/3, thereby accelerating target genes’ transcription (Figure 3) [63]. Moreover, UCHL1 could stabilize hypoxia-inducible factor 1 (HIF-1) and enhance cancer metastasis (Figure 3) [64]. The UCHL1-HIF-1 axis reprograms carbohydrate metabolism and the pentose phosphate pathway to generate GSH, improving cancer cells’ antioxidant and radioresistance capabilities [65]. The β-catenin/TCF/Lef signaling pathway up-regulates several oncogenic cellular pathways through modulating genes such as c-Myc, Cyclin D, c-Jun, and others [66]. Studies have revealed that UCHL1 can prevent β-catenin degradation through its deubiquitinating activity, thus potentially promoting colorectal cancer progression (Figure 3) [67,68]. UCHL1 also plays a specific role in the mTOR signaling pathway in B-cell lymphoma [69]. It balances the mTOR complexes by disrupting mTORC1 and promoting mTORC2 assembly. UCHL1 interferes with the ubiquitination of the mTORC1 subunit raptor and impairs mTORC1 integrity (Figure 3). This action would result in the generation of free mTOR by enhancing the raptor dissociation, which then forms mTORC2 by interacting with the rictor. mTORC2 activates the survival and proliferative kinase Akt and promotes tumorigenesis [70]. However, the downregulation of mTORC1 activity may impair protein biosynthesis due to the decreased phosphorylation of its substrate 4EBP1 [71,72]. Recent research has found that UCHL1 promotes the assembly of eIF4F, which is the translation initiation complex and the target of 4EBP1, then stimulates protein synthesis through directly upregulating translation initiation in B-cell malignancy (Figure 3) [73]. Meanwhile, UCHL1 is also involved in the Akt signaling pathway. Poor prognosis of NSCLC is associated with UCHL1’s overexpression [74]. Abnormal activation of the Akt signaling pathway induced by UCHL1 could enhance the invasion of NSCLC cells [75]. Besides, overexpression of UCHL1 could upregulate thymidylate synthase, thus reducing pemetrexed-induced DNA degradation and cell cycle disruption in NSCLC cells [76]. As illustrated, UCHL1 could also lead to the proliferation and metastasis of osteosarcoma and gastric cancer through regulating Akt and MAPK/ERK signaling pathways [77,78]. Apart from its role in classical signaling pathways, UCHL1 promotes the development of high-grade serous ovarian cancer by maintaining protein homeostasis through the PSMA7-APEH-proteasome axis [79]. Additionally, UCHL1 could restore the H2O2-generating activity of NOX4 and facilitate the invasion of cancer cells [80,81].
Figure 3.
Schematic illustration of the roles for UCHL1 in tumorigenesis. As an oncogene, UCHL1 stabilizes EGFR, TGFβR1, HIF-α, and β-catenin through its DUB activity. Meanwhile, it balances the mTOR complexes by disrupting mTORC1 and promoting mTORC2 assembly. UCHL1 could also act as a tumor suppressor by regulating the p14ARF-MDM2-p53 tumor suppression pathway and promoting cell apoptosis through the caspase-dependent pathway. TGFβR, transforming growth factor-β receptor; EGFR, epidermal growth factor receptor; Smad, drosophila mothers against decapentaplegic protein; mTORC1, mechanistic target of rapamycin complex 1; mTORC2, mechanistic target of rapamycin complex 2; 4EBP1, eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1; eIF4F, eukaryotic translation initiation factor 4; HIF-α, hypoxia-inducible factor α; TCF/Lef, T-cell factor/lymphoid enhancer-binding factor.
Besides serving as an oncogene, evidence also indicates that UCHL1 could act as a tumor suppressor primarily by modulating cell cycle and cell apoptosis. UCHL1 could downregulate the degradation of p53 (a well-known tumor suppressor) and induce G0/G1 cell cycle arrest and apoptosis [82]. As it has been reported, p14ARF promotes the degradation of MDM2, known as a ubiquitin E3 ligase for p53, and thereby enhances the stability of p53 [83,84]. Ubiquitination analysis has revealed that UCHL1 could deubiquitinate p14ARF and p53, and finally result in p53 accumulation (Figure 3) [85]. Meanwhile, researchers have also discovered that UCHL1 may be involved in the p53 signaling pathway in LNCap cells by reducing Akt phosphorylation and upregulating the level of cell cycle inhibitor p27 kip1 [86]. Besides functioning through the p53 signaling pathway, UCHL1 is demonstrated to induce phosphorylation of protein phosphatase cdc25C at S216, leading to G2/M phase arrest and cell proliferation suppression of human colorectal carcinoma cells HCT116 [87]. UCHL1’s role in apoptosis has also been verified in hepatocellular carcinoma (HCC) cells. UCHL1 increases the cellular levels of cleaved caspase-9, cleaved caspase-3, and caspase-7, and enhances the cleavage of PARP, indicating that UCHL1 upregulates apoptosis through the intrinsic caspase-dependent mechanism [87]. Interestingly, UCHL1’s ligase activity seems also to play a role in tumor suppression. Cortactin, a member of the actin-binding protein family, is involved in tumor proliferation, migration, and invasion. In nasopharyngeal carcinoma, UCHL1 functions as a tumor suppressor by increasing K48-linked ubiquitination and degradation of Cortactin, which is achieved through its ligase activity [88].
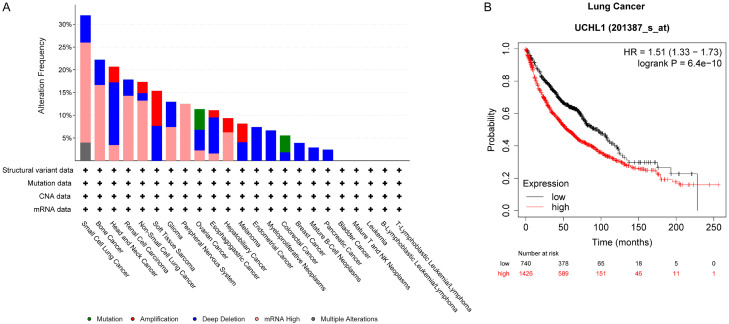
Analyzing the genetic alterations of UCHL1 across different cancer types by utilizing the Cancer Cell Line Encyclopedia (Broad, 2019) dataset can provide additional insights into UCHL1’s potential oncogenic or tumor-suppressive functions in various types of cancer (Figure 4A). For lung cancer, bone cancer, and renal cell carcinoma, high mRNA levels of UCHL1 suggest its potential role in promoting cancer progression. The amplification of gene encoding UCHL1 in non-small cell lung cancer and melanoma could also lead to UCHL1 overexpression, thus potentially contributing to oncogenic signal transductions that promote tumor growth and metastasis. Consistent with the aforementioned data, Kaplan-Meier survival analysis further reveals that lung cancer patients with high expression level of UCHL1 exhibit worse overall survival (OS) outcomes compared with those lung cancer patients with low expression level of UCHL1 (Figure 4B). Except for the upregulation of UCHL1 in specific types of cancer, loss of function alterations such as homozygous deletions have also been detected in certain malignancies such as head and neck cancer, and esophagogastric cancer. This observation might indicate a loss of tumor suppressor function of UCHL1, which would consequently promote tumorigenesis. The clinically relevant data confirms that UCHL1 plays a complicated role in controlling cell growth and cell death. Targeted analysis utilizing pan-cancer proteogenomics data and systematic target validation in certain types of cancer might be necessary to fully reveal UCHL1’s potential as the therapeutic target for a specific cancer.
Figure 4.
Genetic alterations of UCHL1 in specific types of cancer. A. Data from Cancer Cell Line Encyclopedia (Broad, 2019), including mutations, structural variant, copy number alterations (CNA) and mRNA expression z-scores relative to diploid samples [RNA Seq RPKM; threshold 2.0], was analyzed using cBioportal software and visualized using the standard Oncoprint output. The Onco Query Language (OQL) used in the analysis was ‘UCHL1: MUT AMP GAIN EXP ≥ 2 HOMDEL HETLOSS EXP ≤ -2’. B. Lung cancer patients with high UCHL1 expression level suffered from shorter overall survival time as compared to those with low UCHL1 expression level.
UCHL3 in tumorigenesis
UCHL3 has been identified as an oncoprotein in various types of cancer. Elucidating the targets of UCHL3 will provide valuable insights into the working mechanisms of the enzyme in modulating the pathogenesis process of cancer.
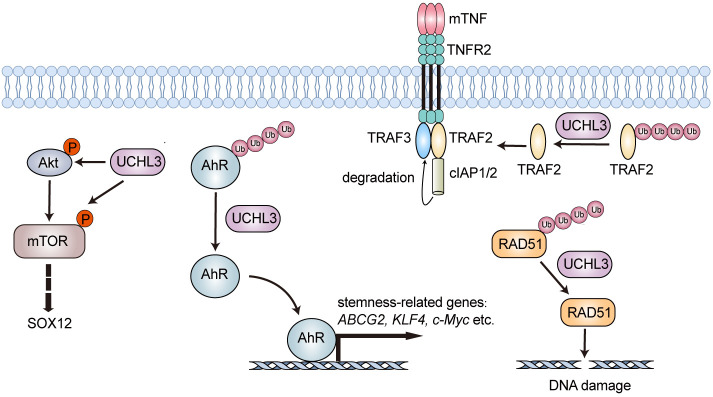
One of the well-known functions of UCHL3 is its involvement in the regulation of DNA repair. As revealed, the recruitment of RAD51 to double-strand breaks through its binding to BRCA2 would facilitate homologous recombination. And UCHL3 could deubiquitinate RAD51, which consequently favors the interaction between RAD51 and BRCA2 (Figure 5). UCHL3 targets three lysine sites (56, 57, and 63) near E59 of RAD51, which is critical for the interaction between RAD51 and BRCA2 [20]. Besides playing a role in the regulation of DNA repair, UCHL3 also affects tumorigenesis through its actions in multiple cellular pathways. UCHL3 promotes NF-κB signal transduction in ovarian cancer by stabilizing TNF receptor-associated factor 2 (TRAF2), which would lead to the acceleration of cell proliferation and migration (Figure 5) [89]. Consistently, UCHL3 is overexpressed in ovarian cancer and associated with a poor prognosis. In UCHL3-deficient ES-2 cells, the expressions of NF-κB target genes such as TNF-α, NFKB1, IL-6, and B94 are downregulated, which indicates that UCHL3 could influence ovarian cancer cells by regulating the NF-κB-mediated inflammatory response [89]. Moreover, UCHL3 is involved in the PI3K/Akt/mTOR signaling pathway, in which it upregulates the phosphorylation levels of Akt and mTOR and the expression level of SOX12 (Figure 5) [90,91]. In colorectal cancer, UCHL3 is found to promote the pathological process by modulating Akt-dependent SOX12 upregulation and facilitating EMT through upregulating Snail, Slug, and ZEB1 [90]. Researchers have also discovered that UCHL3 downregulates the level of p21 and upregulates the level of Cyclin D1 in non-small cell lung cancer [22]. Meanwhile, UCHL3 is up-regulated in both pancreatic cancer tissues and cell lines, contributing to the enhanced proliferation and aerobic glycolysis of pancreatic cancer cells. As demonstrated, UCHL3 could deubiquitinate and stabilize cell cycle modulator FOXM1 [92], which is involved in both the cell cycle modulation and the cellular metabolism. UCHL3 could interfere with the FOXM1-LDHA signaling pathway and influence aerobic glycolysis through enhancing the stability of FOXM1. Silence of LDHA (shLDHA) is capable of compromising the aforementioned effects of UCHL3 [93]. Besides the conventional working mechanisms, recent investigations have also established a correlation between UCHL3 and cancer stemness. UCHL3 has been found to play a role in enhancing the stability of the aryl hydrocarbon receptor (AhR), which is a transcription factor that controls the expression of stemness-related genes such as ABCG2, KLF4, and c-Myc (Figure 5) [94]. Meanwhile, AhR contributes to the overexpression of PD-L1, which promotes immune escape during the radiotherapy of cancer [95]. In line with the reported findings, LINC00665 could upregulate the level of UCHL3 and promote the immune escape of non-small cell lung cancer (NSCLC) by acting as a sponge of miR-582-5p and thus enhancing the stability of AhR protein. And miR-582-5p is a validated microRNA targeting UCHL3 [96]. In fact, the UCHL3/AhR/PD-L1 axis may serve as a potential therapeutic target since its downregulation could enhance the effectiveness of radiotherapy in patients with NSCLC [97].
Figure 5.
Schematic illustration of the roles for UCHL3 in tumorigenesis. UCHL3 stabilizes TRAF2, RAD51, and AhR through its DUB activity. Also, it upregulates the phosphorylation levels of Akt and mTOR. mTOR, mammalian target of rapamycin; AhR, aryl hydrocarbon receptor; TRAF, tumor necrosis factor receptor-associated factors; RAD51, RADiation sensitive 51.
Due to its role in tumorigenesis, studies exploring whether UCHL3 could serve as a target for cancer therapy are currently underway. Specially, an improved understanding of the signaling pathways involving UCHL3 is driving the development of UCHL3-targeted inhibitors which might serve as new therapeutic selections for cancer intervention.
UCHL5 in tumorigenesis
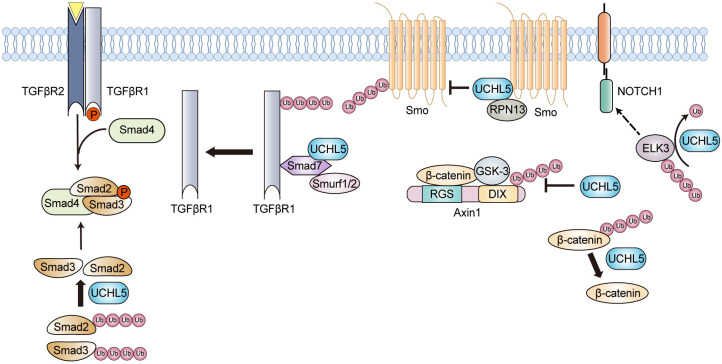
Overexpression of UCHL5 has been reported to be associated with a lower survival rate and an increased risk of cancer recurrence in cancer patients [98], and the enzyme modulates the pathogenesis process of cancer through multiple cellular pathways. UCHL5 plays a role in the TGF-β pathway. It interacts with Smad7 and downregulates the level of the Smurf1/2-Smad7 complex, thus rescuing TGFβRI from proteasomal degradation and enhancing TGF-β signaling activity (Figure 6) [24,25]. Furthermore, UCHL5 can deubiquitinate and stabilize Smad2/Smad3, thereby promoting TGF-β signaling and contributing to the pathogenesis of cancers associated with TP53-mutations (Figure 6). Inhibiting UCHL5 activity with bAP15 could suppress the survival of TP53-mutation containing ovarian cancer cells by regulating TGF-β signaling, which consequently inducing apoptosis. Furthermore, bAP15 exhibits a significant anti-tumor effect in nude mice with subcutaneous SKOV3 xenografts [99]. UCHL5 also acts as a positive regulator of the Hedgehog (Hh) signaling pathway by regulating the stability of Smoothened protein (Figure 6) [26]. UCHL5 forms a trimetric complex with Smo and RPN13, leading to a decrease in Smo ubiquitination and promoting its cell surface accumulation [26]. For the Wnt/β-catenin signaling pathway, it seems like UCHL5 plays a controversial role in it. In 2019, Zijian Li et al. reported that UCHL5 deubiquitinates β-catenin, resulting in its accumulation and the upregulation of downstream genes such as c-Myc and Cyclin D1 (Figure 6) [100]. Deletion of UCHL5 could downregulate the levels of c-Myc and Cyclin D1 mRNAs and inhibit the migration of HeLa cells [100]. In a recent study using endometrial cancer cells, it has been reported that UCHL5 contributes to cancer progression by enhancing the Wnt signaling pathway [101]. However, in 2022, Wonhee Han et al. discovered that UCHL5 could interact with the Axin1 DIX domain and stabilize the Axin1 protein, which acts as a negative regulator of the Wnt signaling pathway (Figure 6) [102]. Studies utilizing MCF7, SW480, and A549 cell lines also demonstrate a negative impact of UCHL5 on the Wnt signaling pathway [102]. These observations suggest that UCHL5’s modulation of the Wnt signaling pathway varies and might be dependent on the biological context. For the Akt/mTOR pathway, UCHL5 mainly exhibits an activating effect. UCHL5 enhances the migration and proliferation of bladder cancer via activating the Akt/mTOR pathway [103]. The downstream targets of this pathway, such as c-Myc, SLC25A19, and ICAM5, show reduced mRNA levels and protein expressions in response to UCHL5 silencing [103]. Studies also demonstrate that UCHL5 could deubiquitinate and stabilize ELK3, a protein related to Notch1 signaling (Figure 6). Targeting the UCHL5/ELK3/Notch1 axis may provide a possible therapeutic option for the treatment of pancreatic adenocarcinoma (PAAD) [104]. Besides the aforementioned modulation effects, UCHL5 regulates cell cycle and cell lifespan (apoptosis) through affecting specific pathways in the meantime. In lung adenocarcinoma (LUAD) cells, the expression level of UCHL5 is positively correlated with the mRNA and protein levels of Cyclin D1, CDK4, Cdc25C, p21, p27, and RB [105]. With the treatment of a dual UCHL5/USP14 inhibitor b-AP15, the expression level of COPS5, a regulator associated with p53 degradation, was downregulated, and the cellular levels of cell cycle-related proteins p27 and Cyclin E1 were upregulated [106]. In human pulmonary epithelial cell A549, UCHL5 silencing was capable of inducing apoptosis by activating Bax/Bcl-2, caspase-9, and caspase-3 [107]. Apart from the involvement of UCHL5 in the canonical signal transduction pathways, a study indicates that the CDK4/6-UCHL5-BRD4 axis may serve as a potential therapeutic target for the BETi-resistant MLL-r leukemia with a hallmark of BRD4 overexpression [108]. Moreover, UCHL5 is revealed to participate in the pathogenesis of hepatocellular carcinoma by deubiquitinating PRP19, an essential RNA splicing factor, and interacts with glucose-regulated protein 78 (GRP78) [109].
Figure 6.
Schematic illustration of the roles for UCHL5 in tumorigenesis. UCHL5 modulates the pathogenesis process of cancer mainly through stabilizing the key players of tumorigenesis-associated signaling pathways. TGFβR, transforming growth factor-β receptor; Smad, drosophila mothers against decapentaplegic protein; Smurf1/2, Smad ubiquitination regulatory factor 1/2; Smo, Smoothened; ELK3, ETS Transcription Factor ELK3; Axin1, axis inhibition protein 1.
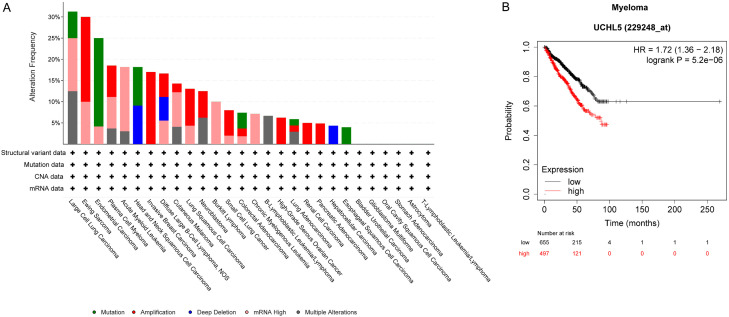
Consistent with the aforementioned findings, the data presented in the Cancer Cell Line Encyclopedia (Broad, 2019) shows alterations in UCHL5 across different cancer types, which supports its potential role in oncogenesis. UCHL5 mutations are observed in cancers such as large cell lung carcinoma and endometrial carcinoma. High mRNA level and DNA amplification for UCHL5 are noted in several cancer types, which include Ewing sarcoma, plasma cell myeloma (also known as multiple myeloma), acute myeloid leukemia, and invasive breast carcinoma (Figure 7A). The overexpression of UCHL5 could lead to the stabilization of oncogenic proteins and degradation of tumor suppressor proteins, therefore promoting the survival and proliferation of cancer cells. As known, VLX1570, a dual inhibitor of USP14 and UCHL5, had entered into clinical trials for patients with multiple myeloma. We thus performed Kaplan-Meier survival analysis to evaluate the correlation between UCHL5 and patients with myeloma. The obtained data indicates that patients with high UCHL5 expression level tend to have worse overall survival (OS) outcomes (Figure 7B).
Figure 7.
Genetic alterations in UCHL5 in specific types of cancer. A. Data from Cancer Cell Line Encyclopedia (Broad, 2019), including mutations, structural variant, copy number alterations (CNA) and mRNA expression z-scores relative to diploid samples [RNA Seq RPKM; threshold 2.0], was analyzed using cBioportal software and visualized using the standard Oncoprint output. The Onco Query Language (OQL) used in the analysis was ‘UCHL5: MUT AMP GAIN EXP ≥ 2 HOMDEL HETLOSS EXP ≤ -2’. B. Myeloma patients with high UCHL5 expression level suffered from shorter overall survival time as compared to those with low UCHL5 expression level.
BAP1 in tumorigenesis
BAP1 plays a key role in the modulation of chromatin’s epigenetic modifications, and it could modify the levels of H2AK119Ub and H3K27me3 through the formation of PR-DUB machinery (Figure 8). As it has been known, the PR-DUB complexes containing BAP1 and its partner proteins are capable of removing ubiquitin from histone H2A at its K119 residue, which is monoubiquitinated upon the catalysis of PRC1, thus consequently regulating DNA transcription processes and determining cell fate (Figure 8) [110,111]. Due to the counteracting functions of PRC1 and PR-DUB, the dynamic balance between them is required for the proper regulation of gene expression. Moreover, H2AK119Ub has been demonstrated to facilitate the recruitment of PRC2 complex (PRC2 complex methylates histone H3 at K27-H3K27me1, H3K27me2, and H3K27me3 [112]), and also the PR-DUB machinery is proved to play a role in the recruitment of MLL3 COMPASS complex containing the H3K27 demethylase UTX to gene enhancers (Figure 9) [113]. Consistent with the aforementioned findings, the depletion of BAP1 would result in an increase of H3K27me3 levels [114]. It is worth noting that the UTX subunit of MLL3 COMPASS removes H3K27me3 at the enhancers of tumor suppressors such as FRZB, GRHL2, and DACT2 [113].
Figure 8.
Schematic illustration of the roles for BAP1 in tumorigenesis. BAP1 plays a key role in the chromatin’s epigenetic modulation by modifying the levels of H2AK119Ub and H3K27me3 through the formation of multi-protein complexes composed of ASXLs, BAP1, HCF-1, YY1, FOXK1/2, OGT, MBD5/6 without or with COMPASS. Moreover, BAP1-containing complexes play an important role in cell cycle regulation through removing ubiquitin from non-histone substrates including its core component of HCF-1 and facilitating the recruitment of transcription factors of KLF5, YY1 and FOXK2. Besides cell cycle regulation, BAP1 modulates DNA repair through its interactions with BRCA1/BARD1 and INO80. Finally, BAP1 BAP1 could induce cytochrome C-dependent apoptosis by deubiquitinating the inositol-1,4,5-triphosphate receptor (IP3R3) and promote ferroptosis by downregulating the expression of SLC7A11. Me, H3K27me3; ASXLs, additional sex comb-like proteins; YY1, Ying Yang 1; HCF-1, Host cell factor 1; OGT, O-linked- N-acetylglucosamine transferase; FOXK1/2, factor forkhead box proteins K1/2; COMPASS, complex of proteins associated with Set1; E2F1, E2F transcription factor 1; KLF5, Kruppel-like factor 5; BRAD1, BRCA1 breast cancer type 1 susceptibility protein; SLC7A11, Solute Carrier Family 7 Member 11; IP3R, Inositol 1,4,5-triphosphate receptor.
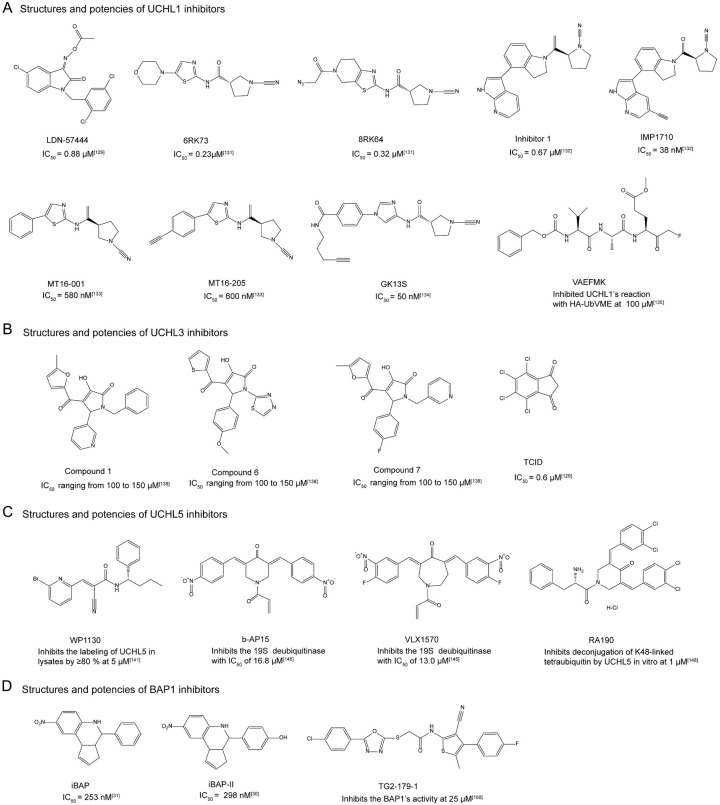
Figure 9.
Chemical structures and determined potencies of small molecule inhibitors against the UCH family members.
Apart from targeting histone substrate H2AK119Ub, BAP1 is involved in biological processes including cell cycle regulation, DNA repair modulation, and cell death by directly targeting non-histone substrates and indirectly transcription regulation of specific proteins, etc. (Figure 8). As known, BAP1 could form DUB complexes together with ASXL1/2/3, HCF-1, OGT, YY1, FOXK1/2, and so on, which play an important role in cell cycle regulation through removing ubiquitin from non-histone substrates including its core component of HCF-1 and facilitating the recruitment of transcription factors of KLF5, YY1, and FOXK2. It has been revealed that BAP1 interacts with the β-propeller domain of HCF-1 in nuclear and can remove K48-linked ubiquitin chains from HCF-1 [54]. HCF-1 modulates cell cycle by promoting the transition from G1 to S phase, which also involves E2F family transcription factors. In fact, HCF-1 interacts with E2F family transcription factors by recruiting the MLL family methyltransferases targeting H3K4 site of histone [115]. Meanwhile, BAP1 is found to bind to E2F1 targeted gene promoters mediated by the HCF-1:E2F1 complex and removes ubiquitin from H2A (Figure 8) [116]. Different from its interaction with E2F1, BAP1 directly recognizes transcription factor KLF5 and functions as a DUB enzyme for the protein. As reported, The KLF5/BAP1/HCF-1 protein complex promotes cell cycle progression by inhibiting p27 gene expression, which is the target gene of the transcription factor (Figure 8) [117]. Besides HCF-1, E2F1, and KLF5, the identified components of BAP1 DUB machinery YY1 and FOXK2 are also transcription factors targeting specific genes. Studies show that YY1 recruits BAP1 and HCF-1 to the promoter of the cox7c gene, which encodes a mitochondrial protein. BAP1 works as a coactivator for the expression of cox7c encoding protein [28]. And FOXK2 recruits BAP1 to its target genes through the forkhead-associated domain and then recruits HCF-1 through BAP1. Interestingly, BAP1 represses the transcription of FOXK2 target genes through its deubiquitination of H2A [55]. Furthermore, FOXK2 exhibits a correlation with cell cycle regulation as it undergoes phosphorylation catalyzed by cyclin-dependent kinases [118].
Besides cell cycle regulation, BAP1 modulates DNA repair through its interactions with BRCA1/BARD1 and INO80 (Figure 8). BAP1 is identified to interact with the RING finger domain of BRCA1 [55]. The binding of BAP1 would disrupt the formation of the RING heterodimer BRCA1-BARD1 and subsequently inhibit the E3 ligase activity of the complex. BAP1 silencing with shRNA would result in increased sensitivity of cells to ionizing radiation, which underscores the crucial role of the BAP1 and BRCA1-BARD1 interaction in DNA damage response [51]. Besides, experimental data suggests that BAP1 is phosphorylated by kinase ATM upon the occurrence of DNA damage, thus promoting its recruitment to the DNA double-strand breaks (DSB) site for H2A deubiquitination and allowing the efficient assembly of the homologous recombination factors BRCA1 and RAD51 (Figure 8) [119]. For INO80, which has a specific role in replication fork progression during normal DNA synthesis, recent studies indicated that BAP1 could deubiquitinate INO80 and recruit it to the replication fork through the interaction between BAP1 and H2AUb (Figure 8) [120-122].
BAP1 is also reported to modulate cell death through its functions in both cytoplasm and nucleus (Figure 8). When functioning in cytoplasm, BAP1 could induce cytochrome c-dependent apoptosis by deubiquitinating the inositol-1,4,5-triphosphate receptor (IP3R3), which promotes the release of Ca2+ into the cytoplasm and mitochondria, and interacts with the 14-3-3 protein to enhance the release of BAX protein (Figure 8) [123,124]. When functioning in nucleus, BAP1 could induce ferroptosis primarily by inhibiting the expression of SLC7A11, a glutamate/cystine reverse transporter protein [125]. BAP1 normally functions to repress the expression of SLC7A11 at least partly by deubiquitinating H2AUb, which consequently leads to the blockage of glutathione synthesis and the upregulation of lipid peroxidation, thereby inducing ferroptosis (Figure 8) [125].
Due to the involvement of BAP1 in numerous cellular pathways associated with tumorigenesis, BAP1 exhibits a dual role in pathological process of cancer. Initially achieved data suggested that BAP1 was a tumor suppressor since it suppressed the growth of MCF7 breast cancer cells through the BRCA1-related pathway [51,126]. Additionally, the germline mutations in BAP1 gene have been demonstrated to relate to an increased risk of developing various malignancies, such as mesothelioma, uveal melanoma, and other types of cancer. This condition has been named as “BAP1 cancer syndrome” [127,128]. In the meanwhile, BAP1’s oncogenic function could possibly be attributed to the direct and non-direct interactions of BAP1 with partner proteins such as HCF-1, KLF5, and ASXLs. It has been reported that the BAP1/HCF-1/KLF5 complex promotes the development and metastasis of breast cancer [117]. Additionally, truncated ASXL1, induced by nonsense and frameshift mutations of ASXL1, enhances BAP1’s recruitment to chromatin and stimulates the expression of pro-leukemic transcriptional signaling pathways and genes, such as HMGN5, STAT5A, HOXA11, TWIST1 and MBD2 [31,129]. Overall, elucidating the working mechanisms of BAP1 in different types of cancer remains a major focus.
Small molecule inhibitors against UCH family members
UCHL1 inhibitors
As aforementioned, UCHL1 may serve as a potential target for cancer therapy. Studies related to the development of UCHL1 inhibitors shed light on the discovery of novel therapeutic strategies for malignancies driven by UCHL1 dysfunctions.
Up to date, a few number of UCHL1 inhibitors have been reported, though they exhibit varying degrees of off-target effects. Isatin O-acyloxime LDN-57444 is an extensively studied competitive inhibitor against UCHL1, with the IC50 at 0.88 μM (Figure 9) [130]. Besides LDN-57444, the majority of the remaining inhibitors targeting the enzyme are featured with the cyanopyrrolidine moiety as the covalent warhead (Figure 9). As illustrated by recent pre-clinical studies, the administration of LDN-57444 could substantially postpone the growth of tumors in both cell line xenografts and PDX models of small cell lung cancer (SCLC) and neuroendocrine prostate cancer (NEPC). Besides, the intervention of LDN-57444 does not lead to any negative consequence on the body weight or cause histological damage to the liver and kidney tissues. LDN-57444 application could also antagonize the occurrence of liver and lymph node metastasis and reduce the size of liver and lymph node metastatic nodules in mice bearing SCLC. Importantly, the combination of LDN-57444 with cisplatin, a standard chemotherapy agent for specific types of cancer, has demonstrated a synergistic effect, which leads to an even more pronounced reduction in tumor growth and metastasis [131]. The positive result of the combination therapy highlights the potential of LDN-57444 to enhance the efficacy of existing treatment regimens, offering a new avenue for improving patient outcomes. Aaron D. Krabill et al. discovered that Compound 1, a cyanopyrrolidine-containing UCHL1 inhibitor, has an IC50 value of 0.67 μM towards UCHL1 [132] (Figure 9). NMR data unraveled that the binding of Compound 1 mainly affects those residues spatially close to the crossover loop and the active site (C90) of UCHL1. Additionally, Compound 1 is likely to interact with the oxyanion hole residues Q84 and N88, suggesting its binding to the active-site pocket of the enzyme [132]. Cellular assays derived data demonstrated that Compound 1 could inhibit the covalent attachment of HA-Ub-VME to UCHL1 in the cellular context and exhibit cytotoxicity against KMS11 and SW1271 cell lines [132]. Raymond Kooij et al. synthesized and characterized the compound of 6RK73, a UCHL1 inhibitor containing cyanopyrrolidine moiety (Figure 9) [133]. 6RK73 inhibits UCHL1 with an IC50 of 0.23 μM. Meanwhile, it suppresses TGFβ/SMAD-triggered transcription and markedly inhibits TGFβ-induced pSMAD2, pSMAD3, TβRI, and total SMAD levels in breast cancer cell MDA-MB-436. Additionally, 6RK73 could reduce breast cancer extravasation in zebrafish xenografts injected with breast cancer cell MDA-MB-436 [134]. Derived from compound 6RK73, the scientists further synthesized 8RK64 with azide group, which has an IC50 value of 0.32 μM for UCHL1 and could be potentially used as a two-step ABP (activity-based probe) due to its azide moiety (Figure 9) [133]. Furthermore, the fluorescent small-molecule activity-based DUB probe 8RK59, generated from 8RK64, is shown to effectively target UCHL1 both in vitro and in vivo [133]. Almost at the same time, Nattawadee Panyain et al. discovered IMP-1710, which was produced from the cyanamide-containing parent inhibitor 1 (Figure 9) [135]. IMP-1710 is the most potent covalent inhibitor against UCHL1 so far. The parent inhibitor 1 and its alkyne-tagged analogue IMP-1710 exhibit IC50 values of 90 nM and 38 nM for UCHL1, respectively. These two compounds have been demonstrated to target UCHL1 in breast cancer cells (Cal51) with IC50 values of 820 nM and 110 nM, respectively [135]. Proteome-wide competitive activity-based protein profiling (ABPP) data suggests that IMP-1710 selectively targets the active site C90 residue of UCHL1 and presents a minor off-target effect [135]. MT16-001 and ABP MT16-205 are also two cyanopyrrolidine inhibitors targeting UCHL1, and both of them contain a 5-phenylthiazole moiety linked to the cyanopyrrolidine warhead (Figure 9) [136]. MT16-205 is an alkyne-containing analog of MT16-001, while MT16-001 and MT16-205 show the IC50 values of 580 nM and 600 nM against UCHL1, respectively. Besides, both MT16-001 and MT16-205 exhibit high inhibition selectivity towards UCHL1 over the other UCH family members. However, gel-based and MS-based proteomic results indicate that they target UCHL1 and ALDH family members in HEK293 cells, suggesting a moderate off-target effect of UCHL1 inhibitors with this specific type of scaffold [136]. Recently, Christian Grethe’s team reported a novel UCHL1-targeted potent and nontoxic activity-based probe GK13S, which has an IC50 value at 50 nM (Figure 9) [137]. The co-crystal structure of UCHL1 complexed with the compound reveals that the 3-carboxy-N-cyanopyrrolidine probe GK13S binds to UCHL1 in a hybrid conformation of the apo and ubiquitin-bound states. GK13S covalently attaches to C90 of the enzyme through its cyanamide warhead and occupies the catalytic cleft. Its specificity is achieved by positioning the pyrrolidine moiety in a distinctive pocket of the apo conformation of UCHL1 [137]. Additionally, GK13S mimics the C-terminal LRGG peptide of ubiquitin and interacts with UCHL1 through hydrogen bonding and hydrophobic interactions. Cellular studies indicate that, despite primarily targeting UCHL1, GK13S also binds to PARK7 and another PARK7 family member C21orf33. Interestingly, although GK13S could inhibit UCHL1 activity in the cellular context, it does not impair the viability of HEK293 and U-87 MG cells [137].
Besides the UCHL1 inhibitors described above, peptidomimetics have also been developed to chemically modulate the function of UCHL1. VAEFMK, a peptide-based fluoromethylketone molecule, was originally discovered serendipitously as a hit in a screening of halo-methylketone tripeptides against the herpes simplex virus cysteine protease UL36 (Figure 9) [138]. However, during the counter-screening against a panel of human DUBs, VAEFMK was found to present an inhibition activity towards UCHL1. The co-crystal structure of VAEFMK complexed with UCHL1 shows that its fluoromethylketone moiety covalently attaches to the active-site cysteine (C90) of the enzyme, and the remaining part of the compound resides near the active-site cleft from the other side of the crossover loop. According to the solved co-crystal structure, Aaron D. Krabill et al. carried out structure-activity relationship (SAR) studies aiming to optimize the irreversible covalent UCHL1 inhibitor VAEFMK. They finally found that the 4-alkynylbenzyloxy compound 34 exhibits an IC50 of 7.7 µM against UCHL1, which is about 3-fold lower than the parent compound of VAEFMK. In-gel fluorescence data of SW1271 cells reveals that this molecule shows a weak off-target effect [139].
In brief summary, quite an amount of efforts have been made to the inhibitor development targeting UCHL1. However, the therapeutic effects of the developed inhibitors against UCHL1 are still questionable due to their varying degrees of off-target effects. UCHL1 inhibitors with both satisfying potency and high selectivity in vivo are needed.
UCHL3 inhibitors
As an emerging target for antitumor drug discovery, the development of UCHL3 inhibitors is on the rise. However, only a few inhibitors have been reported so far.
As previously reported, UCHL3 could deubiquitinate RAD51, and thus favor its recruitment to DNA damage sites for DNA repair purpose. Due to the critical role of UCHL3 in tumorigenesis, Zhiwang Song et al. conducted a repurposing study and found that perifosine, a previously reported Akt inhibitor, could inhibit UCHL3 and UCHL3-mediated deubiquitination of RAD51 in MDA-MB-231 cells, thereby disrupting DNA repair via homologous recombination [140]. Hirayama et al. conducted a three-step virtual screening (DOCK, high-speed GOLD, and low-speed GOLD) against human UCHL3 (PDB code: 1XD3) by using the ChemBridge CNS-Set virtual library and identified 10 candidate inhibitors. Biochemical data reveals that compounds 1, 6, and 7 are competitive inhibitors of UCHL3, with the IC50 values ranging from 100 to 150 μM (Figure 9) [141]. All three compounds share the 1,5-dihydro-2H-pyrrol-2-one group, indicating its importance in UCHL3 inhibition. Besides, compounds 1, 6, and 7 form a hydrogen bond with A11 of UCHL3, while the false hit compound 10 does not, suggesting that the hydrogen bond is critical for targeting UCHL3. Besides the aforementioned hit compounds, TCID was also discovered as a UCHL3 inhibitor, and its IC50 value against the enzyme is 0.6 μM (Figure 9) [130,142]. According to the data reported by Lianlian Ouyang et al., the application of TCID would downregulate the level of AhR and weaken the stem cell properties of NSCLC cells [94].
UCHL3’s actions in tumorigenesis have been extensively studied, and the development of inhibitors against the enzyme is becoming a new focus. Meanwhile, due to the high sequence similarity between UCHL3 and UCHL1, it is challenging to discover inhibitors selectively targeting UCHL3.
UCHL5 inhibitors
UCHL5 primarily functions as an oncoprotein. Specially, due to the involvement of UCHL5 in 26S proteasome machinery, the inhibition of its activity is expected to significantly affect the cellular function which may lead to cell death. Therefore, UCHL5 could serve as a target for anti-cancer drug development. Currently, a limited number of small molecule inhibitors against UCHL5 have been developed, and they all act as either pan-DUB inhibitors or dual-targeted inhibitors for UCHL5 and USP14. Moreover, although no complex structure has been solved yet, experimental results suggest that most UCHL5-targeted inhibitors function through covalently binding to the enzyme. These inhibitors all contain covalent warheads in their structures.
Most of UCHL5 inhibitors such as WP1130, b-AP15, VLX1570, and RA190 contain α,β-unsaturated ketone group in their chemical structures that can covalently modify the active site cysteine of the enzyme through Michael addition (Figure 9). The well-known pan-DUB inhibitor, WP1130, has been reported to inhibit the activity of USP9X, USP5, USP14, and UCHL5 [143]. WP1130 is found to be more effective in reducing leukemic colony formation, and it could strongly inhibit the colony formation of cells derived from chronic myeloid leukemia patients featured with Bcr/Abl T315I mutant expression. Additionally, WP1130 is also capable of suppressing the growth of K562 heterotransplanted tumors, as well as both wild-type Bcr/Abl and Bcr/Abl T315I mutant-expressing BaF/3 cells transplanted into nude mice [143]. Upon the screening of a library containing candidate anti-cancer drugs that promote lysosomal membrane permeabilization-mediated apoptosis, b-AP15 was identified as a selective inhibitor of UCHL5 and USP14. It exhibits an IC50 of 16.8 ± 2.8 μM against the 19S regulatory particle of proteasome when using Ub-AMC as the hydrolysis substrate (Figure 9) [98]. It was found that b-AP15 may inhibit cell growth and induce G2/M cell cycle arrest. Meanwhile, the administration of b-AP15 could induce c-Myc-regulated, Noxa-dependent apoptosis [144]. According to the reported literature, b-AP15 is also capable of enhancing ER stress/unfolded protein response (UPR) and suppressing Wnt/Notch1 signaling pathways, thereby promoting cytotoxicity in hepatocellular carcinoma cells [145]. The administration of b-AP15 to mice with FaDu squamous carcinoma xenografts and HCT116 colon carcinoma xenografts could significantly reduce tumor burden. The same team has also administered b-AP15 to C57BL/6J mice with Lewis lung carcinomas (LLCs) using a 2-days-on, 2-days-off schedule, and to BALB/c mice with orthotopic breast carcinoma (4T1) using a 1-day-on, 3-days-off schedule. b-AP15 could significantly inhibit tumor growth in both models without affecting the body weight of experimental mice. Meanwhile, a decrease in the number of pulmonary metastases was observed for the group of mice with 4T1 breast carcinomas treated with b-AP15 [146]. Moreover, b-AP15 inhibits tumor cell invasion in multiple myeloma cells and overcomes bortezomib resistance by downregulating CDC25C, CDC2, and cyclin B1, and inducing caspase-dependent cell death [147]. According to the results of in vivo studies, b-AP15 also exhibits potent activity against multiple myeloma (MM) in MM.1S and KMS-11 disseminated xenograft mouse models, and the compound could significantly inhibit tumor growth and prolong survival of experimental mice without showing notable toxicity. Furthermore, b-AP15 shows synergistic anti-MM activity when being jointly applied with other conventional and novel anti-MM agents, such as the HDAC inhibitor SAHA, dexamethasone, and lenalidomide. This suggests that combining b-AP15 with other therapeutic agents could enhance its antitumor efficacy and potentially overcome the acquired drug resistance in MM treatment [147]. Although it seems like b-AP15 has a satisfying efficacy against specific types of cancer, the water solubility and stability of this compound has limited its potential clinical application. To optimize the drug-like properties of b-AP15, VLX1570, a dual inhibitor of USP14 and UCHL5 was generated [148,149]. VLX1570 exhibits promising drug-like characteristics such as increased potency and enhanced solubility in aqueous solutions. When using Ub-AMC as the substrate of hydrolysis assay, VLX1570 shows slightly higher potency in comparison to b-AP15 (VLX1570: IC50 = 13.0 ± 2.7 μM; b-AP15: IC50 = 16.8 ± 2.8 μM) towards 19S deubiquitinase (Figure 9) [148,149]. Targeting USP14 and UCHL5 with VLX1570 could trigger apoptosis in Waldenstrom macroglobulinemia (WM) cells, which is accompanied by the accumulation of polyubiquitinated protein conjugates with high molecular weight. Following a 21-day VLX1570 treatment, a reduction in tumor burden was detected in WM-xenografted mice, as evidenced by both bioluminescent radiance and direct measurements using calipers, and extended survival was observed for mice treated with the compound in comparison to mice treated with the vehicle (median survival, 62 vs 44 days, respectively, P = 0.0008) [150]. In vivo studies demonstrate that VLX1570 could significantly inhibit the DUB activity of proteasome machinery and induce apoptosis in multiple myeloma cells [152,153]. VLX1570 is the first DUB inhibitor that goes into clinical trial. However, the phase 1/2 trial (NCT02372240) for assessing the safety and efficacy of VLX1570 intravenous (IV) infusion administered with low dose dexamethasone on days 1, 2, 8, 9, 15, 16 of a 28-day cycle in patients with confirmed diagnosis of multiple myeloma with relapsed or relapsed and refractory disease was terminated due to the reason associated with the dose-limiting toxicity. In a recent study, anti-myeloma effects of VLX1570 were noted at doses at or above 0.6 mg/kg, however, two patients treated at the 1.2 mg/kg dose level experienced severe, abrupt, and progressive respiratory insufficiency [151]. RA190 is another reported UCHL5 covalent inhibitor. It has a bis-benzylidine piperidone scaffold and was identified as a dual inhibitor of RPN13 and UCHL5, which could cause the accumulation of K48-linked polyubiquitinated proteins (Figure 9) [152-154]. NMR analysis reveals that RA190 covalently binds to C88 of RPN13 in the 19S proteasome, while mass spectrometry analysis shows its interaction with several cysteine residues of UCHL5 [154]. Meanwhile, cellular studies have demonstrated that RA190 could induce endoplasmic reticulum stress and upregulate p53 and p53-regulated genes in cervical cancer cells [153]. Multiple myeloma (MM) cell lines, including those that are resistant to the treatment of bortezomib, show sensitivity to RA190. RA190 also could stabilize targets of the human papillomavirus (HPV) E6 oncoprotein, leading to a higher efficacy of the compound in killing cells transformed with HPV. When being administered, RA190 could significantly inhibit the growth of multiple myeloma NCI-H929 and ovarian cancer cell line ES2 xenografts. Additionally, oral administration of RA190 could slow down the tumor growth of HPV16+ syngeneic mouse model [153].
Up to now, most of the identified UCHL5 inhibitors show low specificity against the enzyme, which limits the working mechanism elucidation coupled to their inhibition on UCHL5. Same as the other UCH family members, UCHL5 also has a catalytic pocket with a small size. This structural feature and also the lack of high-resolution structure information for UCHL5 complexed with compound make it very challenging to discover high selectivity inhibitors against the enzyme. Therefore, making more efforts in studies aiming to fully reveal the structure characteristics of UCHL5 may accelerate the anti-cancer drug design process.
BAP1 inhibitors
Although BAP1 is primarily demonstrated as a tumor suppressor, evidence suggests that it might also act as an oncoprotein under certain circumstances. Therefore, the development of BAP1-targeted inhibitors has been carried out by scientists.
Lu Wang et al. discovered a small molecule BAP1 inhibitor iBAP, which exhibits high selectivity for BAP1 over other DUBs, and the IC50 value of this compound against BAP1 is 253 nM (Figure 9) [31]. Meanwhile, Lu Wang et al. observed that leukemia cells carrying truncated ASXL1 gain-of-function mutations, such as K562 cells (ASXL1-Y591*) and THP1 cells with ASXL1-Y591 reading frameshift (fs) mutations, are more sensitive to the treatment of iBAP. RNA-seq analysis of THP1-ASXL1-WT and THP1-ASXL1-Y591fs cells reveals that iBAP administration would lead to the downregulation of a few of leukemia-associated genes such as HMGN5, STAT5A, HOXA11, TWIST1, and MBD2. These genes are known targets of ASXL1 [31]. In vivo study demonstrates that iBAP (50 mg/kg/day; i.p.; 4 weeks) could delay the progression of ASXL1-mutant bearing leukemia and improve the survival of K562 (ASXL1-WT/Y591*) xenograft NSGS mice model and patient-derived tumor cells (ASXL1-WT/Q588*) NSGS mice model [31]. The same research group designed and synthesized a series of analogs of iBAP, and discovered that iBAP analogs with -NO2 or -OH modifications at the -R1 position exhibit higher inhibition efficacy than the parent compound (Figure 9) [30]. Among the iBAP analogs, iBAP-II is the most potent one, and it shows an inhibition activity towards BAP1 with the IC50 at 298 nM. Besides, iBAP-II exhibits an inhibition selectivity against BAP1 over its evolutionarily closest UCH family member UCHL5. The IC50 value of iBAP-II towards UCHL5 is around 664 nM. According to the generated docking models of iBAP and iBAP-II complexed with Calypso (Drosophila homolog of human BAP1), the compounds bind to the catalytic cavity of Calypso. The cellular inhibition effect of iBAP-II is stronger than iBAP against BAP1-WT SCLC cell lines (NCI-H1963, NCI-H748, NCI-H1882, and KP3 cells) [30]. Additionally, the treatment of iBAP-II against NCI-H1963 cells leads to a decrease in the protein level of histone H2AK119Ub, confirming the inhibition of BAP1 by the compound. Moreover, the BAP1 inhibition by iBAP-II could significantly delay tumor progression in the SCLC xenograft model [30]. Besides the discovery of aforementioned BAP1 inhibitors, a recent high-throughput screening identified TG2-179-1 as a BAP1 inhibitor (Figure 9). According to the in silico docking model, TG2-179-1 is likely to be covalently bound to the active site of BAP1. Notably, TG2-179-1 inhibits BAP1 activity not only towards the Ub-AMC substrate but also towards the physiological substrate H2AK119Ub [155]. Cellular assay data has demonstrated that TG2-179-1 exhibits toxicity against multiple colon cancer cell lines, with the IC50 value smaller than 10 μM. TG2-179-1 also could inhibit the growth of colon tumors in the HCT116 xenograft mice without showing significant adverse events, including the loss of body weight. However, it is worth noting that TG2-179-1 also presents toxicity against normal colon cells with low BAP1 expression level, indicating a possible off-target effect [155].
Overall, since BAP1 exhibits both tumor-suppressing and tumor-promoting effects, it is crucial to elucidate the specific roles of BAP1 in different types of cancer. BAP1 inhibitors may serve as a potential therapeutic choice for those cancers driven by the BAP1 upregulation.
The translational study for deubiquitinase inhibitors is still in the early stage, with only two DUB inhibitors VLX1570 and KSQ-4279 progressing into clinical trials. As mentioned above, in 2015, VLX1570 was approved for a phase I/II clinical trial in patients with multiple myeloma (NCT02372240). However, VLX1570 clinical trials were terminated in 2017 owing to severe dose-limiting toxicity that resulted in patient death. KSQ-4279 is a highly potent and selective first-in-class USP1 inhibitor, and now it is undergoing phase I trials for the treatment of patients with advanced solid tumors (NCT05240898). KSQ-4279 was primarily developed for the treatment of ovarian and triple-negative breast cancer. The previous research established KSQ-4279’s potential for monotherapy and its synergistic effects with PARP inhibitors (PARPi) in PARPi-naive BRCA-mutant cancer. Combining KSQ-4279 with PARPi (Olaparib) could result in significant anti-tumor effects in PARPi-resistant, BRCA mutant, ovarian orthotopic PDX models and PARPi-resistant, BRCA-mutant, triple-negative breast cancer (TNBC) PDX models [156,157].
DUBs are involved in multiple biological processes, and some of them exhibit low substrate specificities [158]. The complicated biological network involving DUBs and also the function redundancy of the enzymes pose a significant challenge in understanding how DUB inhibitors might exert therapeutic effects against cancer. Despite the well-defined catalytic pockets of DUBs that are amenable to drug design, identifying compounds that exhibit both satisfying potency and high selectivity remains a major challenge.
Conclusions and future perspectives
The UCH family deubiquitinases play crucial regulatory roles in multiple biological processes such as protein degradation, cell cycle control, DNA repair, and signaling transduction, and so on. Currently, studies on UCH family members mainly focus on the following aspects: (1) Biological function studies aiming to elucidate the working mechanisms of UCHs in various biological processes, as well as their roles in the pathogenesis of human diseases. (2) Biochemical and biophysical studies focusing on the fully understanding of the catalytic mechanism and substrate specificity of the UCH family members. (3) Drug candidate development aiming to develop inhibitors that target UCH family members for the treatment of related diseases including neurodegenerative diseases and cancers.
Due to the pivotal role of the UCH family members in tumorigenesis, the development of inhibitors targeting these enzymes has emerged as a promising therapeutic strategy against cancer. Up to date, a number of small molecule inhibitors towards UCHs have been developed, and the anti-cancer effects of the compounds have also been demonstrated in preclinical studies. However, UCH-targeted inhibitor development still faces quite a few of challenges including achieving selective inhibition of specific UCH family members, discovering UCHs inhibitors with high potency, and developing candidate compounds with acceptable off-target effects and toxicity. Despite these challenges, scientists are actively engaged in developing UCHs-targeted inhibitors through rational drug design and high-throughput screening. With the extensive input and the advancements in technology, a broader range of UCH inhibitors with satisfying drug-like properties might be obtained, and they will offer novel options for cancer therapy. Besides, the developed small molecule inhibitors with high potency and specificity could also facilitate the function elucidation of UCHs by serving as chemical probes.
Acknowledgements
We thank the National Natural Science Foundation of China (Grant Nos. 32000890, 32171220 and 22107111) for supporting our research.
Disclosure of conflict of interest
None.
References
- 1.Lee JM, Hammaren HM, Savitski MM, Baek SH. Control of protein stability by post-translational modifications. Nat Commun. 2023;14:201. doi: 10.1038/s41467-023-35795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farley AR, Link AJ. Identification and quantification of protein posttranslational modifications. Methods Enzymol. 2009;463:725–763. doi: 10.1016/S0076-6879(09)63040-8. [DOI] [PubMed] [Google Scholar]
- 3.Callis J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book. 2014;12:e0174. doi: 10.1199/tab.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montagut AM, Armengol M, de Pablo GG, Estrada-Tejedor R, Borrell JI, Roue G. Recent advances in the pharmacological targeting of ubiquitin-regulating enzymes in cancer. Semin Cell Dev Biol. 2022;132:213–229. doi: 10.1016/j.semcdb.2022.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 7.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 8.Daviet L, Colland F. Targeting ubiquitin specific proteases for drug discovery. Biochimie. 2008;90:270–283. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen M, Schmitt S, Buac D, Dou QP. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin Ther Targets. 2013;17:1091–1108. doi: 10.1517/14728222.2013.815728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermanns T, Pichlo C, Woiwode I, Klopffleisch K, Witting KF, Ovaa H, Baumann U, Hofmann K. A family of unconventional deubiquitinases with modular chain specificity determinants. Nat Commun. 2018;9:799. doi: 10.1038/s41467-018-03148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwasna D, Abdul Rehman SA, Natarajan J, Matthews S, Madden R, De Cesare V, Weidlich S, Virdee S, Ahel I, Gibbs-Seymour I, Kulathu Y. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol Cell. 2018;70:150–164. e6. doi: 10.1016/j.molcel.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Y, Fu D, Shen XZ. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Bett JS, Ritorto MS, Ewan R, Jaffray EG, Virdee S, Chin JW, Knebel A, Kurz T, Trost M, Tatham MH, Hay RT. Ubiquitin C-terminal hydrolases cleave isopeptide- and peptide-linked ubiquitin from structured proteins but do not edit ubiquitin homopolymers. Biochem J. 2015;466:489–498. doi: 10.1042/BJ20141349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronau JA, Beckmann JF, Hochstrasser M. Substrate specificity of the ubiquitin and Ubl proteases. Cell Res. 2016;26:441–456. doi: 10.1038/cr.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmine Belin A, Westerlund M, Bergman O, Nissbrandt H, Lind C, Sydow O, Galter D. S18Y in ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) associated with decreased risk of Parkinson’s disease in Sweden. Parkinsonism Relat Disord. 2007;13:295–298. doi: 10.1016/j.parkreldis.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- 20.Luo K, Li L, Li Y, Wu C, Yin Y, Chen Y, Deng M, Nowsheen S, Yuan J, Lou Z. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 2016;30:2581–2595. doi: 10.1101/gad.289439.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song HM, Lee JE, Kim JH. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochem Biophys Res Commun. 2014;452:722–727. doi: 10.1016/j.bbrc.2014.08.144. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Jin X, Zheng J, Jiang N, Shi W. UCH-L3 promotes non-small cell lung cancer proliferation via accelerating cell cycle and inhibiting cell apoptosis. Biotechnol Appl Biochem. 2021;68:165–172. doi: 10.1002/bab.1909. [DOI] [PubMed] [Google Scholar]
- 23.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cutts AJ, Soond SM, Powell S, Chantry A. Early phase TGF beta receptor signalling dynamics stabilised by the deubiquitinase UCH37 promotes cell migratory responses. Int J Biochem Cell Biol. 2011;43:604–612. doi: 10.1016/j.biocel.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 25.Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, Yao X, Pang S, Chen P, Jiang W, Shan Z, Zhang Q. The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J Mol Cell Biol. 2018;10:243–257. doi: 10.1093/jmcb/mjx036. [DOI] [PubMed] [Google Scholar]
- 27.Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun. 2016;7:10292. doi: 10.1038/ncomms10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ 3rd, Drobetsky E, Milot E, Shi Y, Affar el B. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szczepanski AP, Zhao Z, Sosnowski T, Goo YA, Bartom ET, Wang L. ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med. 2020;12:63. doi: 10.1186/s13073-020-00760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuboyama N, Wang R, Szczepanski AP, Chen H, Zhao Z, Shi L, Wang L. Therapeutic targeting of BAP1/ASXL3 sub-complex in ASCL1-dependent small cell lung cancer. Oncogene. 2022;41:2152–2162. doi: 10.1038/s41388-022-02240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Birch NW, Zhao Z, Nestler CM, Kazmer A, Shilati A, Blake A, Ozark PA, Rendleman EJ, Zha D, Ryan CA, Morgan MAJ, Shilatifard A. Epigenetic targeted therapy of stabilized BAP1 in ASXL1 gain-of-function mutated leukemia. Nat Cancer. 2021;2:515–526. doi: 10.1038/s43018-021-00199-4. [DOI] [PubMed] [Google Scholar]
- 32.Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci U S A. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci U S A. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 35.Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40:1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Seo JH, Suh YH. Alpha-synuclein, Parkinson’s disease, and Alzheimer’s disease. Parkinsonism Relat Disord. 2004;10(Suppl 1):S9–13. doi: 10.1016/j.parkreldis.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Liu YC, Fallon L, Lashuel HA, Liu ZH, Lansbury PT. The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 38.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 40.Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, Kolli N, Gan-Erdene T, Wilkinson KD, Gill G, Lima CD, Ploegh HL, Ovaa H. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 42.Setsuie R, Sakurai M, Sakaguchi Y, Wada K. Ubiquitin dimers control the hydrolase activity of UCH-L3. Neurochem Int. 2009;54:314–321. doi: 10.1016/j.neuint.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 43.van Tilburg GBA, Murachelli AG, Fish A, van der Heden van Noort GJ, Ovaa H, Sixma TK. K27-Linked diubiquitin inhibits UCHL3 via an unusual kinetic trap. Cell Chem Biol. 2021;28:191–201. e8. doi: 10.1016/j.chembiol.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Maiti TK, Permaul M, Boudreaux DA, Mahanic C, Mauney S, Das C. Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme. FEBS J. 2011;278:4917–4926. doi: 10.1111/j.1742-4658.2011.08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou ZR, Zhang YH, Liu S, Song AX, Hu HY. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
- 46.Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell. 2015;61:901–911. doi: 10.1016/j.molcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 48.Deol KK, Crowe SO, Du J, Bisbee HA, Guenette RG, Strieter ER. Proteasome-bound UCH37/UCHL5 debranches ubiquitin chains to promote degradation. Mol Cell. 2020;80:796–809. e9. doi: 10.1016/j.molcel.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct modes of regulation of the UCH37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ 3rd. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 52.Mashtalir N, Daou S, Barbour H, Sen NN, Gagnon J, Hammond-Martel I, Dar HH, Therrien M, Affar el B. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell. 2014;54:392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O’Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okino Y, Machida Y, Frankland-Searby S, Machida YJ. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem. 2015;290:1580–1591. doi: 10.1074/jbc.M114.609834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daou S, Barbour H, Ahmed O, Masclef L, Baril C, Sen Nkwe N, Tchelougou D, Uriarte M, Bonneil E, Ceccarelli D, Mashtalir N, Tanji M, Masson JY, Thibault P, Sicheri F, Yang H, Carbone M, Therrien M, Affar E. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat Commun. 2018;9:4385. doi: 10.1038/s41467-018-06854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR 3rd, Yang XY. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbour H, Daou S, Hendzel M, Affar EB. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat Commun. 2020;11:5947. doi: 10.1038/s41467-020-19722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foglizzo M, Middleton AJ, Burgess AE, Crowther JM, Dobson RCJ, Murphy JM, Day CL, Mace PD. A bidentate Polycomb repressive-deubiquitinase complex is required for efficient activity on nucleosomes. Nat Commun. 2018;9:3932. doi: 10.1038/s41467-018-06186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge W, Yu C, Li J, Yu Z, Li X, Zhang Y, Liu CP, Li Y, Tian C, Zhang X, Li G, Zhu B, Xu RM. Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase. Nature. 2023;616:176–182. doi: 10.1038/s41586-023-05841-y. [DOI] [PubMed] [Google Scholar]
- 61.Miyoshi Y, Nakayama S, Torikoshi Y, Tanaka S, Ishihara H, Taguchi T, Tamaki Y, Noguchi S. High expression of and mRNA predicts early recurrence in patients with invasive breast cancer. Cancer Sci. 2006;97:523–529. doi: 10.1111/j.1349-7006.2006.00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen XS, Wang KS, Guo W, Li LY, Yu P, Sun XY, Wang HY, Guan YD, Tao YG, Ding BN, Yin MZ, Ren XC, Zhang Y, Chen CS, Ye YC, Yang JM, Cheng Y. UCH-L1-mediated down-regulation of estrogen receptor alpha contributes to insensitivity to endocrine therapy for breast cancer. Theranostics. 2020;10:1833–1848. doi: 10.7150/thno.39814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S, Gonzalez-Prieto R, Zhang M, Geurink PP, Kooij R, Iyengar PV, van Dinther M, Bos E, Zhang X, Le Devedec SE, van de Water B, Koning RI, Zhu HJ, Mesker WE, Vertegaal ACO, Ovaa H, Zhang L, Martens JWM, Ten Dijke P. Deubiquitinase activity profiling identifies UCHL1 as a candidate oncoprotein that promotes TGF beta-induced breast cancer metastasis. Clin Cancer Res. 2020;26:1460–1473. doi: 10.1158/1078-0432.CCR-19-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, Tanimoto K, Torii M, Sowa T, Menju T, Sonobe M, Kakeya H, Toi M, Date H, Hammond EM, Hiraoka M, Harada H. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1 alpha. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakashima R, Goto Y, Koyasu S, Kobayashi M, Morinibu A, Yoshimura M, Hiraoka M, Hammond EM, Harada H. UCHL1-HIF-1 axis-mediated antioxidant property of cancer cells as a therapeutic target for radiosensitization. Sci Rep. 2017;7:6879. doi: 10.1038/s41598-017-06605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 67.Bheda A, Yue W, Gullapalli A, Whitehurst C, Liu R, Pagano JS, Shackelford J. Positive reciprocal regulation of Ubiquitin C-terminal hydrolase L1 and beta-Catenin/TCF signaling. PLoS One. 2009;4:e5955. doi: 10.1371/journal.pone.0005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong J, Zhao M, Ma Y, Luo Q, Liu J, Wang J, Yuan X, Sang J, Huang C. UCHL1 acts as a colorectal cancer oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity. Int J Mol Med. 2012;30:430–436. doi: 10.3892/ijmm.2012.1012. [DOI] [PubMed] [Google Scholar]
- 69.Hussain S, Ziesmer SC, Feldman AL, Ansell SM, Galardy PJ. UCHL1 Is a novel regulator of mTOR signaling that may predict a poor response to mTOR inhibition in patients with B-Cell lymphoma. Blood. 2011;118:1577–1577. [Google Scholar]
- 70.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hussain S, Feldman AL, Das C, Ziesmer SC, Ansell SM, Galardy PJ. Ubiquitin hydrolase UCH-L1 destabilizes mTOR complex 1 by antagonizing DDB1-CUL4-mediated ubiquitination of raptor. Mol Cell Biol. 2013;33:1188–1197. doi: 10.1128/MCB.01389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hussain S, Bedekovics T, Liu Q, Hu W, Jeon H, Johnson SH, Vasmatzis G, May DG, Roux KJ, Galardy PJ. UCH-L1 bypasses mTOR to promote protein biosynthesis and is required for MYC-driven lymphomagenesis in mice. Blood. 2018;132:2564–2574. doi: 10.1182/blood-2018-05-848515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu J, Yu S, Jia M, Sun PL, Gao H. Ubiquitin C-terminal hydrolase-L1 expression in non-small-cell lung cancer and its association with clinicopathological features and prognosis. Virchows Arch. 2022;480:577–585. doi: 10.1007/s00428-021-03199-y. [DOI] [PubMed] [Google Scholar]
- 75.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 76.Ding X, Gu Y, Jin M, Guo X, Xue S, Tan C, Huang J, Yang W, Xue M, Zhou Q, Wang W, Zhang Y. The deubiquitinating enzyme UCHL1 promotes resistance to pemetrexed in non-small cell lung cancer by upregulating thymidylate synthase. Theranostics. 2020;10:6048–6060. doi: 10.7150/thno.42096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu YY, Yang M, Zhao M, Luo Q, Yang L, Peng H, Wang J, Huang SK, Zheng ZX, Yuan XH, Liu P, Huang CZ. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumour Biol. 2015;36:8379–8387. doi: 10.1007/s13277-015-3566-0. [DOI] [PubMed] [Google Scholar]
- 78.Zheng S, Qiao G, Min D, Zhang Z, Lin F, Yang Q, Feng T, Tang L, Sun Y, Zhao H, Li H, Yu W, Yang Y, Shen Z, Yao Y. Heterogeneous expression and biological function of ubiquitin carboxy-terminal hydrolase-L1 in osteosarcoma. Cancer Lett. 2015;359:36–46. doi: 10.1016/j.canlet.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 79.Tangri A, Lighty K, Loganathan J, Mesmar F, Podicheti R, Zhang C, Iwanicki M, Drapkin R, Nakshatri H, Mitra S. Deubiquitinase UCHL1 maintains protein homeostasis through the PSMA7-APEH-proteasome axis in high-grade serous ovarian carcinoma. Mol Cancer Res. 2021;19:1168–1181. doi: 10.1158/1541-7786.MCR-20-0883. [DOI] [PubMed] [Google Scholar]
- 80.Song IK, Kim HJ, Magesh V, Lee KJ. Ubiquitin C-terminal hydrolase-L1 plays a key role in angiogenesis by regulating hydrogen peroxide generated by NADPH oxidase 4. Biochem Biophys Res Commun. 2018;495:1567–1572. doi: 10.1016/j.bbrc.2017.11.051. [DOI] [PubMed] [Google Scholar]
- 81.Kim HJ, Magesh V, Lee JJ, Kim S, Knaus UG, Lee KJ. Ubiquitin C-terminal hydrolase-L1 increases cancer cell invasion by modulating hydrogen peroxide generated via NADPH oxidase 4. Oncotarget. 2015;6:16287–16303. doi: 10.18632/oncotarget.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang T, Li L, Yin X, Yuan C, Tan C, Su X, Xiong L, Putti TC, Oberst M, Kelly K, Ren G, Tao Q. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS One. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 84.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 85.Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, Zeng MS, Jia WH, Zeng YX, Chan AT, Cao Y. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949–2958. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- 86.Ummanni R, Jost E, Braig M, Lohmann F, Mundt F, Barett C, Schlomm T, Sauter G, Senff T, Bokemeyer C, Sultmann H, Meyer-Schwesinger C, Brummendorf TH, Balabanov S. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer. 2011;10:129. doi: 10.1186/1476-4598-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, Li H, Cheng YY, Rocken C, Ebert MP, Chan AT, Sung JJ. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
- 88.Zhao Y, Lei Y, He SW, Li YQ, Wang YQ, Hong XH, Liang YL, Li JY, Chen Y, Luo WJ, Zhang PP, Yang XJ, He QM, Ma J, Liu N, Tang LL. Hypermethylation of UCHL1 promotes metastasis of nasopharyngeal carcinoma by suppressing degradation of Cortactin (CTTN) Cells. 2020;9:559. doi: 10.3390/cells9030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang MH, Zhang HH, Du XH, Gao J, Li C, Shi HR, Li SZ. UCHL3 promotes ovarian cancer progression by stabilizing TRAF2 to activate the NF-kappa B pathway. Oncogene. 2020;39:322–333. doi: 10.1038/s41388-019-0987-z. [DOI] [PubMed] [Google Scholar]
- 90.Li J, Zheng Y, Li X, Dong X, Chen W, Guan Z, Zhang C. UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via Akt/mTOR signaling pathway. Am J Transl Res. 2020;12:6445–6454. [PMC free article] [PubMed] [Google Scholar]
- 91.Castillo SD, Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin Ther Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 92.Song Z, Li J, Zhang L, Deng J, Fang Z, Xiang X, Xiong J. UCHL3 promotes pancreatic cancer progression and chemo-resistance through FOXM1 stabilization. Am J Cancer Res. 2019;9:1970–1981. [PMC free article] [PubMed] [Google Scholar]
- 93.Fan Y, Hu D, Li D, Ma C, Tang Y, Tao Q, Deng L, Tang D. UCHL3 promotes aerobic glycolysis of pancreatic cancer through upregulating LDHA expression. Clin Transl Oncol. 2021;23:1637–1645. doi: 10.1007/s12094-021-02565-1. [DOI] [PubMed] [Google Scholar]
- 94.Ouyang L, Yan B, Liu Y, Mao C, Wang M, Liu N, Wang Z, Liu S, Shi Y, Chen L, Wang X, Cheng Y, Cao Y, Xiao D, Zhang L, Liu S, Tao Y. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct Target Ther. 2020;5:78. doi: 10.1038/s41392-020-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang GZ, Zhang L, Zhao XC, Gao SH, Qu LW, Yu H, Fang WF, Zhou YC, Liang F, Zhang C, Huang YC, Liu Z, Fu YX, Zhou GB. The Aryl hydrocarbon receptor mediates tobacco-induced PD-L1 expression and is associated with response to immunotherapy. Nat Commun. 2019;10:1125. doi: 10.1038/s41467-019-08887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu LM, Yuan YJ, Yu H, Wang S, Wang P. LINC00665 knockdown confers sensitivity in irradiated non-small cell lung cancer cells through the miR-582-5p/UCHL3/AhR axis. J Transl Med. 2022;20:350. doi: 10.1186/s12967-022-03516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han SC, Wang GZ, Yang YN, Fang WF, Sun BB, Zhang JD, Zhou HQ, Zhang L, Wang Y, Zhou GB. Nuclear AhR and membranous PD-L1 in predicting response of non-small cell lung cancer to PD-1 blockade. Signal Transduct Target Ther. 2023;8:191. doi: 10.1038/s41392-023-01416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X, Stafford W, Mazurkiewicz M, Fryknas M, Brjnic S, Zhang X, Gullbo J, Larsson R, Arner ES, D’Arcy P, Linder S. The 19S deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Mol Pharmacol. 2014;85:932–945. doi: 10.1124/mol.113.091322. [DOI] [PubMed] [Google Scholar]
- 99.Fukui S, Nagasaka K, Miyagawa Y, Kikuchi-Koike R, Kawata Y, Kanda R, Ichinose T, Sugihara T, Hiraike H, Wada-Hiraike O, Sasajima Y, Ayabe T. The proteasome deubiquitinase inhibitor bAP15 downregulates TGF-beta/Smad signaling and induces apoptosis via UCHL5 inhibition in ovarian cancer. Oncotarget. 2019;10:5932–5948. doi: 10.18632/oncotarget.27219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Zhou L, Jiang T, Fan L, Liu X, Qiu X. Proteasomal deubiquitinase UCH37 inhibits degradation of beta-catenin and promotes cell proliferation and motility. Acta Biochim Biophys Sin (Shanghai) 2019;51:277–284. doi: 10.1093/abbs/gmy176. [DOI] [PubMed] [Google Scholar]
- 101.Liu D, Song Z, Wang X, Ouyang L. Ubiquitin C-terminal hydrolase L5 (UCHL5) accelerates the growth of endometrial cancer via activating the Wnt/β-catenin signaling pathway. Front Oncol. 2020;10:865. doi: 10.3389/fonc.2020.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han W, Koo Y, Chaieb L, Keum BR, Han JK. UCHL5 controls beta-catenin destruction complex function through Axin1 regulation. Sci Rep. 2022;12:3687. doi: 10.1038/s41598-022-07642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao Y, Yan X, Bai X, Tang F, Si P, Bai C, Tuoheti K, Guo L, Yisha Z, Liu T, Liu T. UCHL5 promotes proliferation and migration of bladder cancer cells by activating c-Myc via Akt/mTOR signaling. Cancers (Basel) 2022;14:5538. doi: 10.3390/cancers14225538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Y, Cao L, Guo Z, Gu H, Zhang K, Qiu Z. Deubiquitinase UCHL5 stabilizes ELK3 to potentiate cancer stemness and tumor progression in pancreatic adenocarcinoma (PAAD) Exp Cell Res. 2022;421:113402. doi: 10.1016/j.yexcr.2022.113402. [DOI] [PubMed] [Google Scholar]
- 105.Zhang J, Xu H, Yang X, Zhao Y, Xu X, Zhang L, Xuan X, Ma C, Qian W, Li D. Deubiquitinase UCHL5 is elevated and associated with a poor clinical outcome in lung adenocarcinoma (LUAD) J Cancer. 2020;11:6675–6685. doi: 10.7150/jca.46146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang ZY, Hong J, Zhang JH, Wang XF, Ma YS, Xiong ZX, Sun HR, Cheng C, Xie BZ, Liu JB, Ouyang YG, Fu D. Treatment with b-AP15 to inhibit UCHL5 and USP14 deubiquitinating activity and enhance p27 and Cyclin E1 for tumors with p53 deficiency. Technol Cancer Res Treat. 2022;21:15330338221119745. doi: 10.1177/15330338221119745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen Z, Niu X, Li Z, Yu Y, Ye X, Lu S, Chen Z. Effect of ubiquitin carboxy-terminal hydrolase 37 on apoptotic in A549 cells. Cell Biochem Funct. 2011;29:142–148. doi: 10.1002/cbf.1734. [DOI] [PubMed] [Google Scholar]
- 108.Amari K, Sasagawa S, Imayoshi N, Toda Y, Hosogi S, Imamura T, Ashihara E. The CDK4/6-UCHL5-BRD4 axis confers resistance to BET inhibitors in MLL-rearranged leukemia cells by suppressing BRD4 protein degradation. Biochem Biophys Res Commun. 2022;588:147–153. doi: 10.1016/j.bbrc.2021.12.063. [DOI] [PubMed] [Google Scholar]
- 109.Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H, Xue R, Liu T, Huang X, Dong L, Wu H, Shen X. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559–572. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 110.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 112.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 113.Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, Cozzolino KA, Louis N, He X, Morgan MA, Takahashi YH, Collings CK, Smith ER, Ntziachristos P, Savas JN, Zou L, Hashizume R, Meeks JJ, Shilatifard A. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med. 2018;24:758–769. doi: 10.1038/s41591-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, Micol JB, Durham B, Knutson SK, Campbell JE, Blum G, Shi X, Doud EH, Krivtsov AV, Chung YR, Khodos I, de Stanchina E, Ouerfelli O, Adusumilli PS, Thomas PM, Kelleher NL, Luo M, Keilhack H, Abdel-Wahab O, Melnick A, Armstrong SA, Levine RL. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 116.Pan H, Jia R, Zhang L, Xu S, Wu Q, Song X, Zhang H, Ge S, Xu XL, Fan XQ. BAP1 regulates cell cycle progression through E2F1 target genes and mediates transcriptional silencing via H2A monoubiquitination in uveal melanoma cells. Int J Biochem Cell Biol. 2015;60:176–184. doi: 10.1016/j.biocel.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 117.Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, Shao M, You D, Fan Z, Xia H, Liu R, Chen C. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi: 10.1038/ncomms9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marais A, Ji Z, Child ES, Krause E, Mann DJ, Sharrocks AD. Cell Cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK·cyclin complexes. J Biol Chem. 2010;285:35728–35739. doi: 10.1074/jbc.M110.154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, Barbour H, Corbeil L, Hébert J, Drobetsky E, Masson JY, Di Noia JM, Affar el B. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A. 2014;111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- 121.Lee HS, Seo HR, Lee SA, Choi S, Kang D, Kwon J. BAP1 promotes stalled fork restart and cell survival via INO80 in response to replication stress. Biochem J. 2019;476:3053–3066. doi: 10.1042/BCJ20190622. [DOI] [PubMed] [Google Scholar]
- 122.Poli J, Gasser SM, Papamichos-Chronakis M. The INO80 remodeller in transcription, replication and repair. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160290. doi: 10.1098/rstb.2016.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, Pellegrini L, Signorato V, Olivetto F, Pastorino S, Nasu M, Napolitano A, Gaudino G, Morris P, Sakamoto G, Ferris LK, Danese A, Raimondi A, Tacchetti C, Kuchay S, Pass HI, Affar EB, Yang H, Pinton P, Carbone M. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature. 2017;546:549–553. doi: 10.1038/nature22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sime W, Niu Q, Abassi Y, Masoumi KC, Zarrizi R, Kohler JB, Kjellstrom S, Lasorsa VA, Capasso M, Fu H, Massoumi R. BAP1 induces cell death via interaction with 14-3-3 in neuroblastoma. Cell Death Dis. 2018;9:458. doi: 10.1038/s41419-018-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, Sirohi K, Li X, Wei Y, Lee H, Zhuang L, Chen G, Xiao ZD, Hung MC, Chen J, Huang P, Li W, Gan B. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20:1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carbone M, Ferris LK, Baumann F, Napolitano A, Lum CA, Flores EG, Gaudino G, Powers A, Bryant-Greenwood P, Krausz T, Hyjek E, Tate R, Friedberg J, Weigel T, Pass HI, Yang H. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med. 2012;10:179. doi: 10.1186/1479-5876-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ohar JA, Cheung M, Talarchek J, Howard SE, Howard TD, Hesdorffer M, Peng H, Rauscher FJ, Testa JR. Germline BAP1 mutational landscape of asbestos-exposed malignant mesothelioma patients with family history of cancer. Cancer Res. 2016;76:206–215. doi: 10.1158/0008-5472.CAN-15-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, Yonezawa T, Fujino T, Hayashi Y, Kawabata KC, Fukuyama T, Tanaka Y, Yokoyama A, Yamazaki S, Kozuka-Hata H, Oyama M, Kojima S, Kawazu M, Mano H, Kitamura T. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun. 2018;9:2733. doi: 10.1038/s41467-018-05085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT Jr. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–846. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 131.Liu S, Chai T, Garcia-Marques F, Yin Q, Hsu EC, Shen M, Shaw Toland AM, Bermudez A, Hartono AB, Massey CF, Lee CS, Zheng L, Baron M, Denning CJ, Aslan M, Nguyen HM, Nolley R, Zoubeidi A, Das M, Kunder CA, Howitt BE, Soh HT, Weissman IL, Liss MA, Chin AI, Brooks JD, Corey E, Pitteri SJ, Huang J, Stoyanova T. UCHL1 is a potential molecular indicator and therapeutic target for neuroendocrine carcinomas. Cell Rep Med. 2024;5:101381. doi: 10.1016/j.xcrm.2023.101381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Krabill AD, Chen H, Hussain S, Feng C, Abdullah A, Das C, Aryal UK, Post CB, Wendt MK, Galardy PJ, Flaherty DP. Ubiquitin C-terminal hydrolase L1: biochemical and cellular characterization of a covalent cyanopyrrolidine-based inhibitor. Chembiochem. 2020;21:712–722. doi: 10.1002/cbic.201900434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kooij R, Liu S, Sapmaz A, Xin BT, Janssen GMC, van Veelen PA, Ovaa H, Dijke PT, Geurink PP. Small-molecule activity-based probe for monitoring ubiquitin C-terminal hydrolase L1 (UCHL1) activity in live cells and zebrafish embryos. J Am Chem Soc. 2020;142:16825–16841. doi: 10.1021/jacs.0c07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu S, González-Prieto R, Zhang M, Geurink PP, Kooij R, Iyengar PV, van Dinther M, Bos E, Zhang X, Le Dévédec SE, van de Water B, Koning RI, Zhu HJ, Mesker WE, Vertegaal ACO, Ovaa H, Zhang L, Martens JWM, Ten Dijke P. Deubiquitinase activity profiling identifies UCHL1 as a candidate oncoprotein that promotes TGFβ-induced breast cancer metastasis. Clin Cancer Res. 2020;26:1460–1473. doi: 10.1158/1078-0432.CCR-19-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Panyain N, Godinat A, Lanyon-Hogg T, Lachiondo-Ortega S, Will EJ, Soudy C, Mondal M, Mason K, Elkhalifa S, Smith LM, Harrigan JA, Tate EW. Discovery of a potent and selective covalent inhibitor and activity-based probe for the deubiquitylating enzyme UCHL1, with antifibrotic activity. J Am Chem Soc. 2020;142:12020–12026. doi: 10.1021/jacs.0c04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Panyain N, Godinat A, Thawani AR, Lachiondo-Ortega S, Mason K, Elkhalifa S, Smith LM, Harrigan JA, Tate EW. Activity-based protein profiling reveals deubiquitinase and aldehyde dehydrogenase targets of a cyanopyrrolidine probe. RSC Med Chem. 2021;12:1935–1943. doi: 10.1039/d1md00218j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grethe C, Schmidt M, Kipka GM, O’Dea R, Gallant K, Janning P, Gersch M. Structural basis for specific inhibition of the deubiquitinase UCHL1. Nat Commun. 2022;13:5950. doi: 10.1038/s41467-022-33559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 139.Krabill AD, Chen H, Hussain S, Hewitt CS, Imhoff RD, Muli CS, Das C, Galardy PJ, Wendt MK, Flaherty DP. Optimization and anti-cancer properties of fluoromethylketones as covalent inhibitors for ubiquitin C-terminal hydrolase L1. Molecules. 2021;26:1227. doi: 10.3390/molecules26051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Song Z, Tu X, Zhou Q, Huang J, Chen Y, Liu J, Lee S, Kim W, Nowsheen S, Luo K, Yuan J, Lou Z. A novel UCHL3 inhibitor, perifosine, enhances PARP inhibitor cytotoxicity through inhibition of homologous recombination-mediated DNA double strand break repair. Cell Death Dis. 2019;10:398. doi: 10.1038/s41419-019-1628-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hirayama K, Aoki S, Nishikawa K, Matsumoto T, Wada K. Identification of novel chemical inhibitors for ubiquitin C-terminal hydrolase-L3 by virtual screening. Bioorg Med Chem. 2007;15:6810–6818. doi: 10.1016/j.bmc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 142.de Juan-Sanz J, Nunez E, Lopez-Corcuera B, Aragon C. Constitutive endocytosis and turnover of the neuronal glycine transporter GlyT2 is dependent on ubiquitination of a C-terminal lysine cluster. PLoS One. 2013;8:e58863. doi: 10.1371/journal.pone.0058863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bartholomeusz GA, Talpaz M, Kapuria V, Kong LY, Wang S, Estrov Z, Priebe W, Wu J, Donato NJ. Activation of a novel Bcr/Abl destruction pathway by WP1130 induces apoptosis of chronic myelogenous leukemia cells. Blood. 2007;109:3470–3478. doi: 10.1182/blood-2006-02-005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sha B, Chen X, Wu H, Li M, Shi J, Wang L, Liu X, Chen P, Hu T, Li P. Deubiquitylatinase inhibitor b-AP15 induces c-Myc-Noxa-mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis. 2019;24:826–836. doi: 10.1007/s10495-019-01561-9. [DOI] [PubMed] [Google Scholar]
- 145.Ding Y, Chen X, Wang B, Yu B, Ge J. Deubiquitinase inhibitor b-AP15 activates endoplasmic reticulum (ER) stress and inhibits Wnt/Notch1 signaling pathway leading to the reduction of cell survival in hepatocellular carcinoma cells. Eur J Pharmacol. 2018;825:10–18. doi: 10.1016/j.ejphar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 146.D’Arcy P, Brnjic S, Olofsson MH, Fryknäs M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 147.Tian Z, D’Arcy P, Wang X, Ray A, Tai YT, Hu Y, Carrasco RD, Richardson P, Linder S, Chauhan D, Anderson KC. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–716. doi: 10.1182/blood-2013-05-500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang X, D’Arcy P, Caulfield TR, Paulus A, Chitta K, Mohanty C, Gullbo J, Chanan-Khan A, Linder S. Synthesis and evaluation of derivatives of the proteasome deubiquitinase inhibitor b-AP15. Chem Biol Drug Des. 2015;86:1036–1048. doi: 10.1111/cbdd.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X, Mazurkiewicz M, Hillert EK, Olofsson MH, Pierrou S, Hillertz P, Gullbo J, Selvaraju K, Paulus A, Akhtar S, Bossler F, Khan AC, Linder S, D’Arcy P. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci Rep. 2016;6:26979. doi: 10.1038/srep26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Paulus A, Akhtar S, Caulfield TR, Samuel K, Yousaf H, Bashir Y, Paulus SM, Tran D, Hudec R, Cogen D, Jiang J, Edenfield B, Novak A, Ansell SM, Witzig T, Martin P, Coleman M, Roy V, Ailawadhi S, Chitta K, Linder S, Chanan-Khan A. Coinhibition of the deubiquitinating enzymes, USP14 and UCHL5, with VLX1570 is lethal to ibrutinib- or bortezomib-resistant Waldenstrom macroglobulinemia tumor cells. Blood Cancer J. 2016;6:e492. doi: 10.1038/bcj.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rowinsky EK, Paner A, Berdeja JG, Paba-Prada C, Venugopal P, Porkka K, Gullbo J, Linder S, Loskog A, Richardson PG, Landgren O. Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Invest New Drugs. 2020;38:1448–1453. doi: 10.1007/s10637-020-00915-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Osei-Amponsa V, Sridharan V, Tandon M, Evans CN, Klarmann K, Cheng KT, Lack J, Chari R, Walters KJ. Impact of losing hRpn13 Pru or UCHL5 on proteasome clearance of ubiquitinated proteins and RA190 cytotoxicity. Mol Cell Biol. 2020;40:e00122-20. doi: 10.1128/MCB.00122-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, Hung CF, Chen X, Walters KJ, Roden RB. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell. 2013;24:791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR Jr, Walters KJ. Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and UCH37 as anticancer targets. Nat Commun. 2017;8:15540. doi: 10.1038/ncomms15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kang M, Park SG, Lee SA, Kim S, Lee D, Shirbhate ME, Youn SY, Kim KM, Cha SS, Kwon J. Targeting BAP1 with small compound inhibitor for colon cancer treatment. Sci Rep. 2023;13:2264. doi: 10.1038/s41598-023-29017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cadzow L, Brenneman J, Sullivan P, Liu H, Shenker S, McGuire M, Grasberger P, Sinkevicius K, Hafeez N, Histen G, Chipumuro E, Hixon J, Krall E, Cogan S, Wilt J, Schlabach M, Stegmeier F, Olaharski A, Wylie A. Development of KSQ-4279 as a first-in-class USP1 inhibitor for the treatment of BRCA-deficient cancers. Eur J Cancer. 2020;138:S52. [Google Scholar]
- 157.Cadzow L, Gokhale PC, Ganapathy S, Sullivan P, Nayak S, Shenker S, Schlabach M, Tobin E, Matulonis UA, Liu JF, Stegmeier F, Wylie A. KSQ-4279, a first-in-class USP1 inhibitor shows strong combination activity in BRCA mutant cancers with intrinsic or acquired resistance to PARP inhibitors. Eur J Cancer. 2022;174:S37–S38. [Google Scholar]
- 158.Rossio V, Paulo JA, Chick J, Brasher B, Gygi SP, King RW. Proteomics of broad deubiquitylase inhibition unmasks redundant enzyme function to reveal substrates and assess enzyme specificity. Cell Chem Biol. 2021;28:487–502. e5. doi: 10.1016/j.chembiol.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]