Abstract
It has been shown that cats can be protected against infection with the prototypic Petaluma strain of feline immunodeficiency virus (FIVPET) using vaccines based on either inactivated virus particles or replication-defective proviral DNA. However, the utility of such vaccines in the field is uncertain, given the absence of consistent protection against antigenically distinct strains and the concern that the Petaluma strain may be an unrepresentative, attenuated isolate. Since reduction of viral pathogenicity and dissemination may be useful outcomes of vaccination, even in the absence of complete protection, we tested whether either of these vaccine strategies ameliorates the early course of infection following challenge with heterologous and more virulent isolates. We now report that an inactivated virus vaccine, which generates high levels of virus neutralizing antibodies, confers reduced virus loads following challenge with two heterologous isolates, FIVAM6 and FIVGL8. This vaccine also prevented the marked early decline in CD4/CD8 ratio seen in FIVGL8-infected cats. In contrast, DNA vaccines based on either FIVPET or FIVGL8, which induce cell-mediated responses but no detectable antiviral antibodies, protected a fraction of cats against infection with FIVPET but had no measurable effect on virus load when the infecting virus was FIVGL8. These results indicate that the more virulent FIVGL8 is intrinsically more resistant to vaccinal immunity than the FIVPET strain and that a broad spectrum of responses which includes virus neutralizing antibodies is a desirable goal for lentivirus vaccine development.
Vaccines are required urgently to contain the current pandemic of human immunodeficiency virus (HIV). Unfortunately, the induction of immunity to lentiviruses by vaccination poses particular problems since, under natural conditions, these viruses establish persistent infections despite vigorous antiviral antibody and cell-mediated immune responses by the host. Hence, neither the nature of the viral immunogens nor the mode of vaccine delivery that might protect people from natural exposure is clear from the study of naturally occurring immune responses. This is particularly true for HIV, since the means to determine whether a vaccine might protect against infection have so far, by necessity, been indirect. Until recently (4, 22), HIV vaccine trials have been limited to observation of the immunological responses induced in human volunteers by candidate vaccines. While the chimpanzee is a realistic surrogate host for HIV vaccine testing, this endangered species is not available in sufficient numbers for statistically valid trials to be conducted; this has encouraged researchers to perform vaccine trials using macaques challenged with simian immunodeficiency virus (SIV)/HIV hybrids (SHIVs) expressing the HIV type 1 (HIV-1) env, tat, and rev genes in an SIV genomic background (12).
Despite this daunting challenge, direct evidence of successful vaccination has been obtained in comparative animal systems, particularly feline immunodeficiency virus (FIV) and SIV. For example, protection against FIV infection has been achieved by immunization with inactivated virus vaccines. In this way, cats immunized with inactivated FIV, derived from the FL4 cell line that is infected with the Petaluma isolate (FIVPET), were consistently protected from challenge with the homologous virus (37). However, protection did not necessarily extend to challenge with other strains of FIV. Thus, following vaccination with inactivated FIVPET, Johnson et al. observed no protection against challenge by the Shizuoka isolate (18), and we found no protection against the Glasgow-8 isolate of FIV (FIVGL8) (15). Clearly, for the development of effective vaccines for use in the field, it is important to know the extent to which a vaccine will protect against viruses other than those in the vaccine, and in particular those that are prevalent in the population to be immunized. Thus, for HIV it is very important to know if vaccines containing immunogens of a single clade will protect against natural infection with viruses of other clades. The FIV system may have useful predictive potential, since similar genetic variation occurs in FIV and HIV (31).
To examine the extent of heterologous protection, we tested the effect of vaccination with the inactivated FIVPET vaccine against the antigenically distinct isolates FIVAM6 and FIVGL8 (87 and 93% similarity, respectively, with FIVPET in the V3-V5 region of the envelope gene). Our previous observation that cats vaccinated with the inactivated FIVPET vaccine had much higher virus neutralizing antibody (VNA) levels to FIVPET than to FIVGL8 suggested that the difference in the extent of protection might be related to the extent of cross-neutralization by vaccine-induced antibodies (15). This explanation was supported by later work indicating that a threshold of VNA was required for protection in the period shortly after vaccination (16). Subsequently, as described in this report, we found that FIVGL8 was more virulent than FIVPET, establishing a higher virus load in cats than FIVPET and, unlike FIVPET, decreasing the CD4/CD8 ratio. Therefore, the FIVGL8 challenge provides a robust system to test the utility of the inactivated FIVPET vaccine in ameliorating the early course of infection, as determined by viral load or changes in CD4/CD8 ratio.
In the experiments reported here, cats were immunized with the inactivated FIVPET vaccine and then challenged with the homologous virus FIVPET or with either of two other FIV strains, FIVGL8 and FIVAM6. The third virus, FIVAM6, was chosen since it had been found to be more closely related to FIVPET than to FIVGL8, as assessed by cross-neutralization with a panel of cat sera. In the event, the FIVAM6 challenge stock was found to be intermediate between FIVPET and FIVGL8 in its behavior in neutralization tests. The degree of protection provided by vaccination against challenge with these three viruses was determined in terms of virus load and changes in CD4/CD8 ratio. Possible correlations between VNA titers and protection were examined.
In addition, heterologous protection was assessed in cats immunized with FIVPET or FIVGL8 DNA vaccine. Such DNA vaccinations have been shown previously to protect cats against challenge without inducing detectable VNA (17). This experiment also provided the opportunity to determine whether protection from FIVGL8 challenge, or indeed any decrease in viral load, might be achieved when the immunogen was precisely matched with the challenge virus.
MATERIALS AND METHODS
Cells and virus stocks.
FL4 cells were a generous gift of J. K. Yamamoto, University of Florida. These cells were maintained in RPMI 1640 medium (Gibco Biocult, Paisley, United Kingdom) containing 10% fetal bovine serum (Biological Industries Ltd., Cumbernauld, United Kingdom). 2 mM l-glutamine, 5 × 10−5 M 2-mercaptoethanol, penicillin (100 IU/ml), and streptomycin (100 mg/ml) (complete RPMI 1640 medium). CrFK cells were maintained in Dulbecco's modification of minimal Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, sodium pyruvate (0.11 mg/ml), penicillin (100 IU/ml), and streptomycin (100 μg/ml). MYA-1 cells and peripheral blood T cells were maintained in RPMI 1640 medium supplemented with recombinant human interleukin 2 (100 IU/ml; kind gift from T. Miyazawa, University of Tokyo, and M. Hattori, University of Kyoto).
Immunization of cats.
The inactivated FIVPET vaccine was prepared from the culture fluid of the FL4 feline lymphoblastoid cell line that is persistently infected with FIVPET (36). The vaccine was prepared by a method similar to that previously described (15) in which culture fluid was inactivated with 0.5% (vol/vol) paraformaldehyde prior to partial purification by two cycles of sucrose gradient centrifugation. Thirty-five 11-week-old specific-pathogen-free kittens were randomized into seven groups of five kittens. Four groups of five kittens were immunized subcutaneously at 0, 3, and 6 weeks with 250 μg of inactivated virus in MF59.0 citrate adjuvant (kindly provided by Chiron Corporation), and three groups received adjuvant alone. Three weeks following the final inoculation, five cats inoculated with the inactivated virus vaccine and five cats inoculated with adjuvant alone were challenged intraperitoneally with 10 50% infectious doses (ID50) of either the homologous FIVPET, FIVGL8, or FIVAM6 as shown in Table 1. The final group of five vaccinates was left unchallenged, allowing comparisons to be made between vaccinates following challenge.
TABLE 1.
Immunization with inactivated FIV vaccine
| Group | Vaccine | Challenge virus |
|---|---|---|
| 1 | Inactivated virus | FIVPET |
| 2 | Adjuvant | FIVPET |
| 3 | Inactivated virus | FIVGL8 |
| 4 | Adjuvant | FIVGL8 |
| 5 | Inactivated virus | FIVAM6 |
| 6 | Adjuvant | FIVAM6 |
| 7 | Inactivated virus | None |
The DNA vaccines were prepared as in a previous study (17) from reverse transcriptase (RT) deletion mutants generated from the F14 molecular clone of FIVPET (25) or from the 414 molecular clone of FIVGL8 (N. Spibey and J. Macdonald, unpublished data). The resulting DNA vaccines were designated PETΔRT and GL8ΔRT, respectively. Thirty-six 14-week-old kittens were randomized into six groups of six kittens. The kittens were immunized intramuscularly at 0, 4, and 8 weeks with either PETΔRT plus gamma interferon (IFN-γ) DNA, GL8ΔRT plus IFN-γ DNA, or IFN-γ DNA alone as shown in Table 2. Each kitten received 100 μg of each DNA in a total volume of 200 μl of phosphate-buffered saline at four sites in the gastrocnemius and quadriceps muscles. On week 12, the cats were challenged intraperitoneally with 10 50% cat ID50 of FIVPET or FIVGL8 derived from the relevant molecular clone.
TABLE 2.
Immunization with DNA vaccine
| Group | DNA vaccine | Challenge virus |
|---|---|---|
| 1 | PETΔRT + IFN-γ | FIVPET |
| 2 | GL8ΔRT + IFN-γ | FIVPET |
| 3 | IFN-γ | FIVPET |
| 4 | PETΔRT + IFN-γ | FIVGL8 |
| 5 | GL8ΔRT + IFN-γ | FIVGL8 |
| 6 | IFN-γ | FIVGL8 |
Serological tests.
Plasma samples were tested for the presence of VNA using a focus reduction assay in CrFK cells (6) that has been described previously (26). Titers of antibodies recognizing FIV p17 or FIV p24 were determined by enzyme-linked immunosorbent assay (ELISA). Microtiter plates (high binding; Greiner Laboritechnik, Dursley, Gloucestershire, United Kingdom) were coated overnight either with the synthetic peptide RAISSWKQRNRWEWRPD, representing an immunodominant linear neutralization site in the third variable region of FIV gp120 (8, 23), or with an immunodominant epitope in the transmembrane glycoprotein (TM) (1, 27, 30) represented by the synthetic peptide CNQNQFFCK. Antibodies recognizing these FIV peptides were detected as described previously (1, 10). These peptide sequences are conserved between the FIVPET and FIVGL8 isolates.
Flow cytometry.
Samples of whole blood were collected into EDTA and processed for flow cytometry as described previously (33). CD4+ lymphocytes were detected using a 1:1:1 mixture of monoclonal antibodies vpg31, vpg33, and vpg34; CD8+ lymphocytes were detected with monoclonal antibody vpg9. Primary antibodies were detected using fluorescein isothiocyanate-conjugated F(ab′)2 fragment of sheep anti-mouse immunoglobulin G whole molecule. Samples were analyzed on an EPICS Elite flow cytometer, 5,000 events being collected in listmode for each sample.
Isolation of FIV.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized venous peripheral blood by centrifugation over Ficoll-Hypaque (Pharmacia LKB, Biotechnology Inc., Piscataway, N.J.). Then 106 PMBC were cocultivated with 106 MYA-1 cells, which are highly sensitive for FIV replication (24). The cultures were maintained in complete RPMI 1640 medium supplemented with 100 IU of interleukin 2 per ml. Samples of culture supernatant were tested at intervals for the presence of FIV p24 by ELISA (IDEXX Laboratories, Portland, Maine), and cultures were maintained for 21 days before being scored as negative.
Semiquantitative virus isolation.
The initial number of infected cells per 2 × 106 PBMC for each cat was measured as described previously (17). Briefly, decreasing numbers of PBMC (2 × 106, 2 × 105, 2 × 104, 2 × 103, 2 × 102, 20, and 2) were cocultivated, in duplicate, in 48-well plates with 106 MYA-1 cells in a total volume of 1.5 ml of complete RPMI 1640 medium, and samples of culture supernatant were tested on day 14 for the presence of FIV p24 by ELISA.
Quantification of proviral load.
The FIV proviral load in PBMC was quantified using real-time PCR measuring PCR product accumulation through a dual-labeled fluorogenic TaqMan probe (13). The primers used were FIV0771f (5′-AGA ACC TGG TGA TAT ACC AGA GAC-3′) and FIV1081r (5′-TTG GGT CAA GTG CTA CAT ATT G-3′). The probe used in this system was FIV1010p (5′-FAM-TAT GCC TGT GGA GGG CCT TCC T-TAMRA-3′). The oligonucleotides were designed to detect a variety of FIV A-subtype isolates and have been previously shown to detect FIVPET, FIVGL8, and FIVAM6 with only minor differences in the PCR efficiency (19, 21). The 50-μl PCR mixtures contained 10 mM Tris (pH 8.3), 50 mM KCl, 3 mM MgCl2, 200 nM dATP, dCTP, dGTP, 400 nM dUTP, 300 nM each primer, 200 nM fluorogenic probe, and 2.5 U of Taq DNA polymerase. After the initial denaturation (2 min at 95°C), amplification was performed with 45 cycles of 15 s at 95°C and 60 s at 60°C. The PCR and the online measurement of the emitted fluorescence were performed on an ABI 7700 sequence detector system (Perkin-Elmer, Foster City, Calif.). The copy number per PCR was calculated by the Sequence Detection software version 1.6 (Perkin-Elmer), using a 10-fold dilution series (ranging from 5 to 5 × 105 copies) of FIV Zurich 2 containing plasmid pBSCompZ2 (kind gift from H. Lutz, University of Zurich), which served as standard in each PCR run. The DNA content per PCR was estimated by optical density measurement at 260 nm and comparison of sample aliquots on agarose gel electrophoresis after ethidium bromide staining. To calculate the percentage of infected PBMC, a mean DNA content of 6 pg per cell and one proviral copy per cell was assumed.
RESULTS
Whole inactivated FIVPET virus vaccine protects against FIVPET but not FIVGL8 or FIVAM6 challenge.
We had shown previously that the inactivated virus vaccine containing FIVPET grown in FL4 cells protected against infection with FIVPET but not against FIVGL8. In the present experiment, cats were vaccinated as before and then challenged with either of these two viruses or with FIVAM6, which was found to be antigenically more closely related to FIVPET than to FIVGL8 (26). It was expected that if the degree of antigenic difference between the viruses influenced the outcome of challenge, the vaccinates might also be protected against FIVAM6.
Following challenge, virus could not be isolated from any of the inactivated virus vaccinates challenged with FIVPET, whereas virus was isolated from three of five controls by 6 weeks after infection (Table 3). At 24 weeks after challenge, virus was not isolated from two of these three infected cats, but a fourth control cat did yield virus at that time; at 30 weeks after challenge, virus was isolated from all four of the cats from which virus had been isolated previously. Therefore, significant protection was achieved against the homologous FIVPET (P = 0.048, Fisher's exact test). Following challenge with FIVGL8, virus was isolated consistently from three of five vaccinates and all of the five controls from 6 weeks postchallenge onward; however, this difference was not statistically significant, indicating that vaccination did not confer significant protection against FIVGL8 challenge (P = 0.4). Of the cats challenged with FIVAM6, virus was isolated from two of five vaccinates and four of five controls from 9 weeks after challenge, but again this difference was not statistically significant (P = 0.2).
TABLE 3.
Results of virus isolation at intervals following challenge
| Challenge | Inoculum | Cat | Virus isolation at indicated wk after challengea
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 6 | 9 | 12 | 18 | 24 | 30 | |||
| FIVPET | Inactivated virus | 1 | − | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | − | ||
| 3 | − | − | − | − | − | − | − | ||
| 4 | − | − | − | − | − | − | − | ||
| 5 | − | − | − | − | − | − | − | ||
| Adjuvant | 1 | − | − | − | − | − | − | − | |
| 2 | + | + | + | + | + | + | + | ||
| 3 | − | + | + | + | + | − | + | ||
| 4 | − | + | + | − | + | − | + | ||
| 5 | − | − | − | − | − | + | + | ||
| FIVAM6 | Inactivated virus | 1 | − | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | − | ||
| 3 | − | − | + | + | + | − | + | ||
| 4 | + | − | + | + | + | + | + | ||
| 5 | − | − | − | − | − | − | − | ||
| Adjuvant | 1 | + | + | + | + | + | + | + | |
| 2 | + | + | + | + | + | + | + | ||
| 3 | + | + | + | + | + | + | + | ||
| 4 | + | + | + | + | − | + | + | ||
| 5 | − | − | − | − | − | − | − | ||
| FIVGL8 | Inactivated virus | 1 | − | − | − | − | − | − | − |
| 2 | + | + | + | + | + | + | + | ||
| 3 | + | + | + | + | + | + | + | ||
| 4 | − | − | − | − | − | − | − | ||
| 5 | + | + | + | + | + | + | + | ||
| Adjuvant | 1 | + | + | + | + | + | + | + | |
| 2 | − | + | + | + | + | + | + | ||
| 3 | + | + | + | + | + | + | + | ||
| 4 | + | + | + | + | + | + | + | ||
| 5 | + | + | + | + | + | + | + | ||
No virus was isolated from any cat at the time of challenge.
FIVPET vaccine significantly suppresses virus load and CD4+ T-cell loss in cats challenged with FIVGL8.
Even though the vaccine failed to protect a significant number of cats against challenge with the heterologous viruses, we found that it did have a notable ameliorating effect, particularly on the challenge with FIVGL8, as indicated by greatly reduced virus load and maintenance of CD4/CD8 T-cell ratios compared with the values in unvaccinated cats.
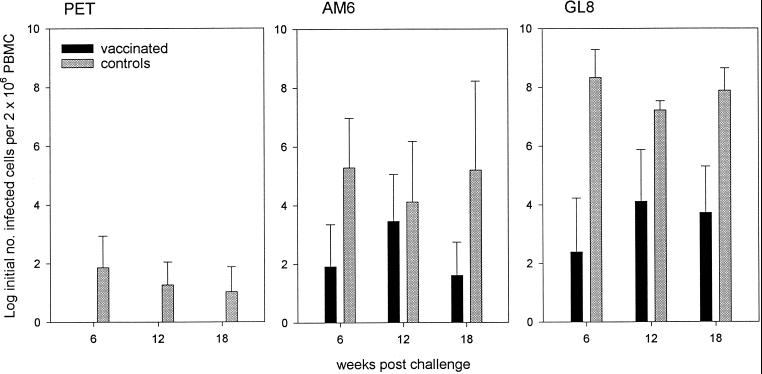
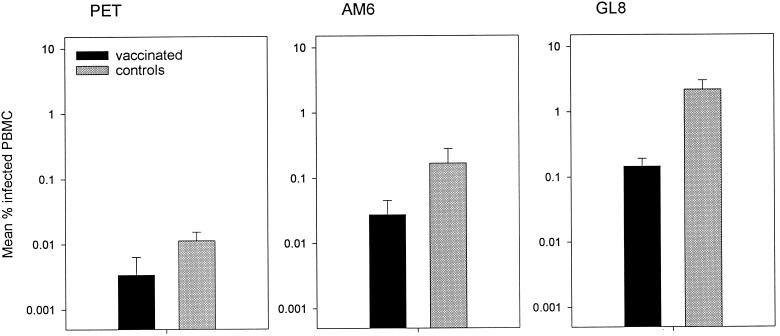
The infectious viral burden in PBMC was measured by semiquantitative virus isolation at 6, 12, and 18 weeks postchallenge. As shown in Fig. 1, the loads were different for each strain. Notably, for each strain the loads were lower in the vaccinated cats than in the controls. The viral burden of the FIVPET-infected control cats was consistently greater than in the vaccinated cats, in which no virus was detected at any time point, but the difference just failed to reach statistical significance. Likewise, the differences in viral burden between the vaccinated cats and controls challenged with FIVGL8 were clear and consistent but not statistically significant. Nevertheless, the trend suggested that with more data, interesting differences might be revealed. Consequently, we assessed the proviral loads in PBMC 18 weeks after challenge using real-time PCR and compared the proviral loads between vaccinates and controls for each challenge virus. As shown in Fig. 2, the proviral loads of the FIVGL8-challenged vaccinates were significantly lower than those of the controls (means ± standard errors of the means [SEM]), 0.15 ± 0.04 and 2.18 ± 0.8, respectively; P = 0.050, Student's t test). Therefore the GL8-challenged vaccinated cats developed lower proviral loads than the controls.
FIG. 1.
Infectious viral burdens in cats vaccinated with inactivated virus and adjuvant controls measured at 6, 12, and 18 weeks postchallenge by quantitative virus isolation. The results shown represent means ± SEM.
FIG. 2.
Proviral loads in cats vaccinated with inactivated virus and adjuvant controls measured 18 weeks postchallenge by real-time PCR. The results shown represent means ± SEM.
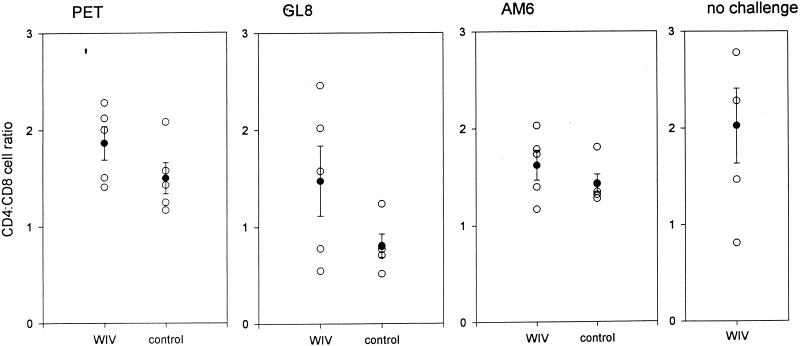
A second indicator of a positive effect of the vaccine in cats infected with FIVGL8 was that the reduction in the CD4/CD8 ratio that accompanies infection with FIVGL8 (33) was not observed in the vaccinated cats. CD4+ and CD8+ lymphocyte numbers were measured as an indicator of the effect of infection 37 weeks following challenge. Since total lymphocyte numbers may be highly variable, the CD4/CD8 ratios were compared between the groups (Fig. 3). The mean CD4/CD8 ratio was significantly lower for the unvaccinated controls infected with FIVGL8 than for the unvaccinated controls infected with either FIVPET (0.81 ± 0.12 and 1.50 ± 0.16, respectively; P = 0.008) or FIVAM6 (0.81 ± 0.12 and 1.63 ± 0.15, respectively; P = 0.003), indicating that FIVGL8 represented the most virulent challenge. Consistent with these findings, the ratios in the FIVGL8-infected control cats were significantly lower than those of the unchallenged inactivated virus vaccinates (0.81 ± 0.12 and 2.02 ± 0.39, respectively, P = 0.017), while there was no significant reduction in the ratios of the FIVPET- and FIVAM6-infected control cats. It was notable that the CD4/CD8 ratios of the inactivated virus vaccinates challenged with FIVGL8 were not significantly lower than those of the unchallenged vaccinates. This result indicated that although these cats were not protected from the FIVGL8 challenge, the decreased CD4/CD8 ratios noted in the unvaccinated, FIVGL8-infected cats were abolished in the vaccinated, FIVGL8-infected cats.
FIG. 3.
Scatter plot of CD4/CD8 lymphocyte ratios in cats vaccinated with inactivated virus (WIV) and adjuvant control cats measured 37 weeks after challenge with FIVPET, FIVGL8, FIVAM6, or no challenge, as indicated. The mean ratios ± SEM are superimposed.
FIVPET vaccine induces significantly higher titers of VNA against FIVPET than against FIVGL8 or FIVAM6.
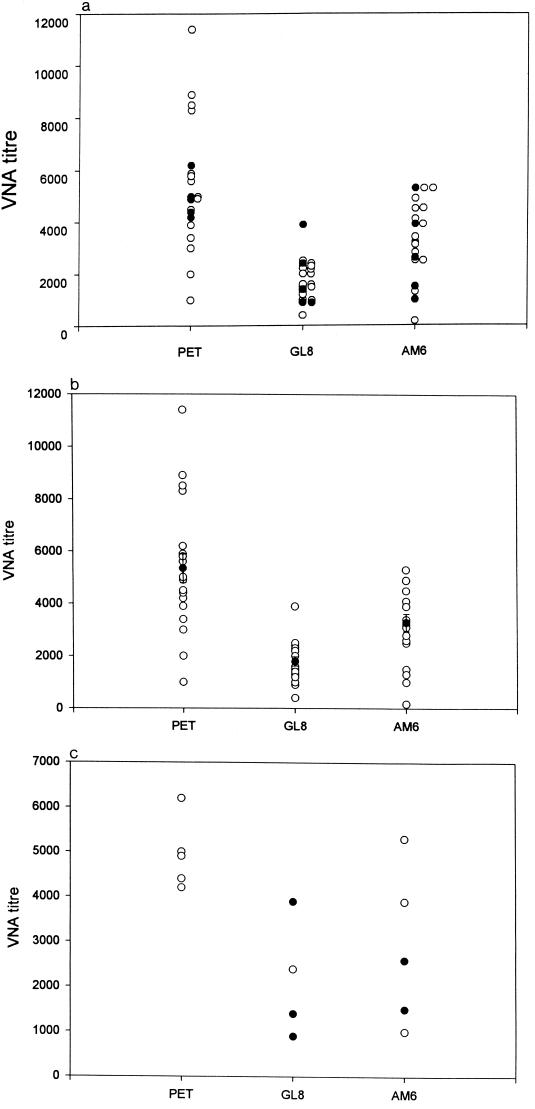
The VNA titers induced by the vaccine against the three challenge strains of FIV used in the experiment were measured and compared. As shown in Fig. 4a, there was considerable variation in the VNA response against each isolate, especially against FIVPET, with a range of <1,000 to 11,400. It was shown retrospectively that the vaccinates challenged with FIVPET all had VNA titers on the day of challenge which were close to the mean titer, not these extreme values. Also, the VNA titers of the vaccinates that were challenged with FIVGL8 or FIVAM6 were representative of the titers induced against the corresponding challenge strains. Figure 4b demonstrates that the VNA titers induced against the FIVGL8 and FIVAM6 strains were significantly lower than those induced against FIVPET. Since vaccine protection did not extend to the FIVGL8 or FIVAM6 challenge, these results were consistent with the hypothesis that high VNA titers are associated with resistance to infection with FIV (15, 16) and that the failure of the vaccine to protect against challenge with heterologous viruses might be due to antigenic differences between the viruses. However, within the FIVGL8 and FIVAM6 groups there was no absolute correlation between VNA titer to a virus and protection from challenge with that virus (Fig. 4c).
FIG. 4.
(a) Scatter plot of day-of-challenge VNA titers of the inactivated virus vaccinates against the three challenge isolates of FIV (open symbols), indicating the cats that were subsequently challenged with each of the isolates (closed symbols). (b) Scatter plot of day-of-challenge VNA titers of the three groups of inactivated virus vaccinates measured against the homologous challenge virus isolate, with the mean titers ± SEM superimposed (closed symbols). (c) Scatter plot of day-of-challenge VNA titers of the inactivated virus vaccinates measured against the homologous challenge virus isolate. The vaccinates that became infected following challenge are indicated by the closed symbols.
FIVGL8 is more virulent than FIVPET or FIVAM6.
An alternative reason that we considered for the clear lack of vaccinal protection against FIVGL8 and FIVAM6 was that these viruses were more virulent than the FIVPET used for challenge, from which the vaccinates were protected. The first indication that FIVGL8 was more virulent than either of the other two viruses was that in unvaccinated control cats, only the FIVGL8 challenge lowered the CD4/CD8 T-cell ratio. Furthermore, FIVGL8 established much higher viral loads than the other two viruses, as assessed by both quantitative virus isolation (Fig. 1) and real-time PCR (Fig. 2). Proviral loads in the PBMC of control cats challenged with 10 ID50 of FIVPET, FIVGL8, or FIVAM6 were compared pairwise 18 weeks after challenge. These comparisons revealed that the mean viral loads of the FIVGL8-infected cats were significantly greater than those of the FIVPET-infected (P = 0.035) and FIVAM6-infected (P = 0.048) cats. In contrast, there was no significant difference between the mean proviral loads measured in the FIVPET-infected cats compared to the FIVAM6-infected cats (P = 0.201).
DNA vaccination does not protect against challenge with GL8.
Vaccination with FIV DNA provided an opportunity to determine whether protection might be achieved against FIVGL8, using a different system in which the vaccine immunogen and challenge viruses could be matched. We demonstrated previously that a replication-defective FIVPET DNA vaccine with a deletion in the RT region of the pol gene (PETΔRT) protected cats against challenge with the homologous FIVPET isolate and that no antiviral antibodies were detected in the sera of vaccinated cats (16). Therefore, we extended our previous studies by testing whether the protective immune response induced by DNA vaccination, which should not include the induction of VNA, might confer protection against heterologous challenge. In this experiment we were able to carry out a cross-protection study, since we had available the original PETΔRT vaccine and a newly developed analogous construct of FIVGL8, GL8ΔRT.
In line with our previous studies (17), cats were inoculated with the FIV DNA construct plus a plasmid containing the feline IFN-γ gene as an adjuvant or with IFN-γ plasmid alone. Following challenge, vaccinated and control cats were monitored for serological responses indicative of infection postchallenge and for evidence of infection by virus isolation and real-time PCR. After FIVPET challenge, virus could be isolated from all six control cats inoculated with IFN-γ DNA, from five of six cats inoculated with PETΔRT plus IFN-γ DNA, and from three of six cats inoculated with GL8ΔRT plus IFN-γ DNA (Table 4). In contrast, following FIVGL8 challenge, virus was isolated from five of six control cats inoculated with IFN-γ DNA, from six of six cats inoculated with PETΔRT plus IFN-γ DNA, and from five of six cats inoculated with GL8ΔRT plus IFN-γ DNA (Table 4). Serological responses were detected in all cats from which virus was isolated. Hence while four of the vaccinates were protected from challenge with FIVPET, none was protected from FIVGL8 challenge.
TABLE 4.
Titers of antibody to FIV TM peptide, reactivity of plasma samples by immunoblotting, and results of virus isolation at intervals following PET challenge
| Inoculum | Finding at indicated wk postchallengea
|
||||||
|---|---|---|---|---|---|---|---|
| Cat | 6
|
9
|
13
|
||||
| VI | IB | TM | VI | IB | VI | ||
| Pet challenge | |||||||
| PETΔRT + IFNγ | 1 | + | + | 25 | + | + | − |
| 2 | + | + | 25 | + | + | + | |
| 3 | + | + | 25 | + | + | − | |
| 4 | + | + | 25 | + | + | + | |
| 5 | + | + | 5 | + | + | − | |
| 6 | − | − | 0 | − | − | − | |
| GL8ΔRT + IFNγ | 1 | − | − | 0 | − | − | − |
| 2 | − | − | 0 | + | + | − | |
| 3 | + | + | 25 | + | + | + | |
| 4 | − | − | 0 | − | − | − | |
| 5 | + | + | 25 | + | + | + | |
| 6 | + | + | 5 | + | + | + | |
| IFN-γ | 1 | + | + | 25 | + | + | + |
| 2 | + | + | 5 | + | + | − | |
| 3 | + | + | 5 | + | + | + | |
| 4 | + | + | 5 | + | + | + | |
| 5 | + | + | 25 | + | + | + | |
| 6 | + | − | 25 | + | + | − | |
| GL8 challenge | |||||||
| PETΔRT + IFNγ | 1 | + | + | 625 | + | + | + |
| 2 | + | + | 125 | + | + | + | |
| 3 | + | + | 125 | + | + | + | |
| 4 | + | ND | ND | ND | ND | ND | |
| 5 | + | + | 125 | + | + | + | |
| 6 | + | + | 625 | + | + | + | |
| GL8ΔRT + IFNγ | 1 | + | + | 625 | + | + | + |
| 2 | + | + | 625 | + | + | + | |
| 3 | + | + | 625 | + | + | + | |
| 4 | + | + | 25 | + | + | + | |
| 5 | + | + | 625 | + | + | + | |
| IFN-γ | 1 | + | + | 625 | + | + | + |
| 2 | + | + | 625 | + | + | + | |
| 3 | + | + | 625 | + | + | + | |
| 4 | − | − | 0 | − | − | − | |
| 5 | + | + | 625 | + | + | + | |
| 6 | + | + | 625 | + | + | + | |
At the time of challenge, all cats were negative with respect to immunoblotting (IB), presence (titer) of antibody to TM peptide, and virus isolation (VI). ND, not determined.
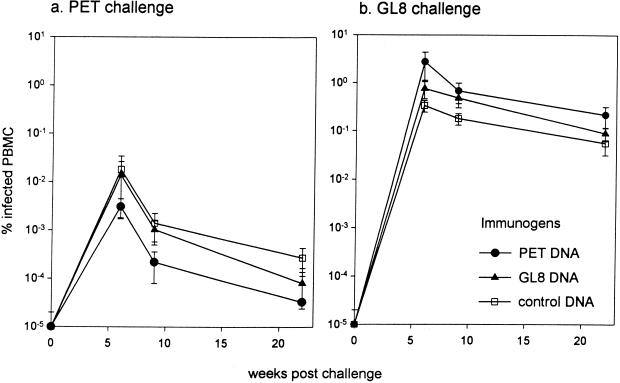
Comparison of proviral loads by real-time PCR revealed no significant reductions in loads between the vaccinated cats and the controls. The mean viral load in the infected PETΔRT DNA vaccinates was lower than in the control cats following FIVPET challenge, but in contrast to our earlier studies (17), this difference failed to reach statistical significance (Fig. 5a). Following FIVGL8 challenge, no reduction in viral load was evident in either group of vaccinates compared to the controls (Fig. 5b). Strikingly, examination of the proviral loads at the peak of viremia, 6 weeks after challenge, demonstrated that the mean proviral load of the FIVGL8-infected control group was approximately 20-fold greater than the mean load of the FIVPET-infected control group (0.28% versus 0.015% infected PBMC, respectively; P = 0.02). These findings with the challenge stocks produced from the FIVPET and FIVGL8 molecular clones were consistent with the higher mean viral load of cats infected with the biological isolate of FIVGL8 than of cats infected with the biological isolate of FIVPET.
FIG. 5.
Proviral loads in DNA vaccinates and controls measured at intervals postchallenge with FIVPET (a) or FIVGL8 (b) by real-time PCR. The results shown represent means ± SEM.
DISCUSSION
This study recapitulates previous findings on the strong protective effect of an inactivated virus vaccine against challenge with the homologous FIVPET strain and its relative ineffectiveness against infection with the heterologous FIVGL8 isolate (15). In the present study, another heterologous isolate, FIVAM6, was also found to establish infection in the majority of vaccinated cats. However, examination of the early course of infection in cats challenged with the heterologous viruses showed that those which became viremic despite receiving inactivated virus vaccine displayed no T-cell subset changes (for FIVGL8) and had lower virus loads as measured by real-time PCR (for FIVGL8 and FIVAM6), a trend which was statistically significant for the more virulent FIVGL8 isolate.
DNA vaccines based on a replication-defective mutant of FIVPET have also been shown to protect a fraction of cats against the homologous isolate (17). This study shows that analogous vaccines based on FIVPET and FIVGL8 proviruses have a limited protective effect against FIVPET. However, the DNA vaccines neither prevented infection with FIVGL8 nor led to reduced virus loads, even when the vaccine and challenge viruses were derived from the same molecularly cloned provirus. Together, these findings indicate that the virulent FIVGL8 isolate is intrinsically more resistant to vaccine-induced immune responses. The relative virulence of FIV strains has been tested systematically for only a few cases (9). However, it seems highly likely that in the field, animals will encounter primary isolate strains typified by FIVGL8, and it follows that candidate vaccines should be tested for their efficacy against such isolates.
The role of humoral immunity in inactivated virus vaccine protection has been the subject of previous studies. A critical role was indicated by the passive transfer of immunity to FIVPET using serum from vaccinated animals (14) and the observation that a threshold titer of VNA was associated with protection in experiments involving suboptimal immunization with inactivated virus (16). It is therefore tempting to ascribe the relative inefficiency of heterologous protection against FIVGL8 and FIVAM6 to the lower effective titer of cross-reactive neutralizing antibodies. While all three of the challenge viruses were of FIV clade A, comparison of their potentials to cross-neutralize showed that FIVPET and FIVGL8 were clearly distinct, while FIVAM6 was more closely related, if not identical, to FIVPET (26). The FIVAM6 stock used as the challenge virus in the present experiment was a modification of that used in the study of antigenicity, since it had been passaged once in cats and then grown in Q201 feline T cells before being used for challenge in order to raise its titer in vivo. Subsequently, in the present study, it was found to be intermediate between the other two viruses in cross-neutralization studies, and thus the challenge stock had undergone some antigenic changes relative to the strain used in the earlier neutralization studies.
However, the apparently greater effect of inactivated virus vaccination on FIVGL8 compared to FIVAM6 does not follow precisely the pattern of in vitro neutralization. This is not particularly surprising, as the efficiency of virus neutralization is strongly affected by cell substrate and may be markedly different in vivo (2). Although virus-specific effector cytotoxic T-lymphocyte (CTL) responses were not assessed in this study, it has been shown previously that inactivated virus vaccines elicit CTL responses that correlate with long-lived protection (16), and cross-reactive T-cell epitopes cannot be expected to have the same distribution among strains as neutralizing determinants. We conclude that serological responses, while clearly important, are not the sole determinant of protective immunity.
As in our previous studies, we found that DNA vaccines based on engineered defective proviruses carrying an in-frame deletion in pol (ΔRT) produced no detectable antiviral antibodies in the recipients. Therefore, the mechanism by which these vaccines protect against FIVPET may be quite different from that used by the inactivated virus vaccine. Strong CTL responses were induced to Env and Gag determinants by FIVΔRT, but previous studies showed no correlation between the magnitude of these responses and the outcome of infection (11), leading to the conclusion that cell-mediated responses to nonstructural genes or vaccine-induced innate immune responses are responsible for this phenomenon. However, in this study we found no evidence of efficacy against FIVGL8 with ΔRT vaccines. By constructing the analogous vaccine for FIVGL8, it was possible to conduct a reciprocal experiment in which FIVPET and FIVGL8 vaccines and challenge strains were interchanged. This experiment excludes antigenic polymorphism as the basis of the resistance of FIVGL8 to DNA vaccine protection. Rather, vaccine resistance appears to be due to the intrinsic virulence of the FIVGL8 isolate, which is manifested in a molecularly cloned virus that has not been repassaged in vivo. It should, therefore, be possible to dissect the determinants of FIV virulence by creating molecular chimeras between these two prototypic strains.
It appears that the immune response elicited by the present form of DNA vaccination is qualitatively and/or quantitatively inadequate to restrict the growth of the virulent FIVGL8, possibly because this virus can establish a significant level of replication before an anamnestic immune response is triggered. Nevertheless, it is conceivable that these vaccines may still be of benefit when combined with the partially efficacious inactivated virus vaccines, as the strategies induce markedly different immune effector mechanisms. One study has already indicated superior cross-protection against heterologous challenge by a prime-boost strategy using canarypox virus vectors and virus-infected cells (32). However, it is unclear whether the heterologous isolate in that case was as virulent as FIVGL8.
Like the FIV system, the SIV/macaque model has revealed hurdles to achieving vaccine protection against virulent isolates (12). DNA vaccines used alone have induced protection only against challenge strains with low replicative capacity, whether the challenge was SIV (3) or SHIVs expressing HIV-1 env, tat, and rev in an SIV genomic backbone (20). In addition, SIV envelope glycoprotein vaccines elicited only limited protection against heterologous isolates (28). However, it is encouraging that DNA priming followed by boosting with envelope glycoprotein induced protective responses superior to those obtained with either DNA or protein alone (29). Using a DNA prime and envelope glycoprotein boost protocol, it was shown recently that macaques could be protected against a pathogenic SHIV challenge, with a proportion of immunized macaques maintaining their CD4 cell counts (12). In addition, protection against wild-type, disease-inducing strains of SIV has been demonstrated following infection with live, attenuated deletion mutants lacking accessory genes such as nef, vpr, or vpx (5, 7, 35). However, even a live attenuated SIV vaccine based on the SIVmac239 isolate was only partially protective against challenge with the heterologous, uncloned pathogenic SIVsm660 isolate (34). These results reveal that we have much to learn with respect to the factors governing lentivirus vaccine efficacy and that no single virus/challenge system can be relied on to predict the behavior of HIV vaccines in human beings. While FIV vaccines suitable for use in the field may not yet be an immediate prospect, our present results encourage further studies aimed at optimizing immune responses and testing the longer-term effects of vaccines on the endpoints of disease progression and viral transmission.
ACKNOWLEDGMENTS
This work was supported by the UK Medical Research Council, Wellcome Trust, Intervet International BV, and EC Concerted Action, FAVEUR.
We are grateful to J. Norrie, Robertson Centre for Biostatistics, University of Glasgow, for statistical analyses, to J. K. Yamamoto, University of Florida, for providing the FL4 cell line, to Chiron Corporation for providing the adjuvant, and to D. Graham, R. Irvine, and the late J. Cole for technical assistance.
REFERENCES
- 1.Avrameas A, Strosberg A D, Moraillon A, Sonigo P, Pancino G. Serological diagnosis of feline immunodeficiency virus (FIV) infection based on synthetic peptides from Env glycoproteins. Res Virol. 1993;144:209–218. doi: 10.1016/s0923-2516(06)80031-2. [DOI] [PubMed] [Google Scholar]
- 2.Baldinotti F, Matteucci D, Mazzetti P, Giannelli C, Bandecchi P, Tozzini F, Bendinelli M. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J Virol. 1994;68:4572–4579. doi: 10.1128/jvi.68.7.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer J D, Ugen K E, Wang B, Agadjanyan M, Gilbert L, Bagarazzi M L, Chattergoon M, Frost P, Javadian A, Williams W, Refaeli Y, Ciccarelli R B, McCallus D, Coney L, Weiner D B. Protection of chimpanzees from high dose heterologous HIV-1 challenge by DNA vaccination. Nat Med. 1997;3:526–532. doi: 10.1038/nm0597-526. [DOI] [PubMed] [Google Scholar]
- 4.Connor R I, Korber B, Graham B S, Hahn B H, Ho D D, Walker B D, Neumann A U, Vermund S H, Mestecky J, Jackson S, Fenamore E, Cao Y, Gao F, Kalams S, Kunstman K J, McDonald D, McWilliams N, Trkola A, Moore J P, Wolinsky S M. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor R I, Montefiori D C, Binley J M, Moore J P, Bonhoeffer A, Gettie A, Fenamore K E, Sheridan K E, Ho D D, Dailey P J, Marx P A. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crandell R A, Fabricant C G, Nelson-Rees W A. Development, characterisation and virus susceptibility of a feline (Felis catus) renal cell line (CRFK) In Vitro. 1973;9:176–185. doi: 10.1007/BF02618435. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt H D, Kanki P, Essex M, Desrosiers R C. Protective effects of a live-attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 8.de Ronde A, Stam J G, Boers P, Langedijk H, Meloen R, Hesselink W, Keldermans L C E J M, van Vliet A, Verschoor E J, Horzinek M C, Egberink H F. Antibody response in cats to the envelope proteins of feline immunodeficiency virus: identification of an immunodominant neutralization domain. Virology. 1994;198:257–264. doi: 10.1006/viro.1994.1028. [DOI] [PubMed] [Google Scholar]
- 9.Diehl L J, Mathiason-DuBard C K, O'Neil L L, Obert L, Hoover E A. Induction of accelerated feline immunodeficiency virus disease by acute-phase virus passage. J Virol. 1995;69:6149–6157. doi: 10.1128/jvi.69.10.6149-6157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn J N, Cannon C A, Beatty J, Mackett M, Rigby M, Neil J C, Jarrett O. Induction of feline immunodeficiency virus-specific cytotoxic T cells in vivo with carrier-free synthetic peptide. J Virol. 1994;68:5835–5844. doi: 10.1128/jvi.68.9.5835-5844.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn J N, Hosie M J, Rigby M A, Mackay N, Dunsford T H, Cannon C A, Neil J C, Jarrett O. Factors influencing cellular immune responses to feline immunodeficiency virus induced by DNA vaccination. Vaccine. 2000;18:1118–1132. doi: 10.1016/s0264-410x(99)00375-8. [DOI] [PubMed] [Google Scholar]
- 12.Girard M, Habel A, Chanel C. New prospects for the development of a vaccine against human immunodeficiency virus type 1. An overview. C R Acad Sci. 1999;322:959–966. doi: 10.1016/s0764-4469(00)87193-0. [DOI] [PubMed] [Google Scholar]
- 13.Heid C A, Stevens J A, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Hohdatsu T, Pu R, Torres B A, Trujillo S, Gardner M B, Yamamoto J K. Passive antibody protection of cats against feline immunodeficiency virus infection. J Virol. 1993;67:2344–2348. doi: 10.1128/jvi.67.4.2344-2348.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosie M, Osborne R, Yamamoto J K, Neil J C, Jarrett O. Protection against homologous but not heterologous challenge induced by inactivated feline immunodeficiency virus vaccines. J Virol. 1995;69:1253–1255. doi: 10.1128/jvi.69.2.1253-1255.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosie M J, Flynn J N. Feline immunodeficiency virus vaccination: characterization of the immune correlates of protection. J Virol. 1996;70:7561–7568. doi: 10.1128/jvi.70.11.7561-7568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosie M J, Flynn N, Rigby M A, Cannon C, Dunsford T, Mackay N, Argyle D, Willett B J, Miyazawa T, Onions D E, Jarrett O, Neil J C. DNA vaccination affords significant protection against feline immunodeficiency virus infection without inducing detectable antiviral antibodies. J Virol. 1998;72:7310–7319. doi: 10.1128/jvi.72.9.7310-7319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C M, Torres B A, Koyama H, Yamamoto J K. FIV as a model for AIDS vaccination. AIDS Res Hum Retroviruses. 1994;10:225–228. doi: 10.1089/aid.1994.10.225. [DOI] [PubMed] [Google Scholar]
- 19.Klein D, Janda P, Steinborn R, Muller M, Salmons B, Gunzburg W H. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis. 1999;20:291–299. doi: 10.1002/(SICI)1522-2683(19990201)20:2<291::AID-ELPS291>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Letvin N L, Montefiori D C, Yasutomi Y, Perry H C, Davies M E, Lekutis C, Alroy M, Freed D C, Lord C I, Handt L K, Liu M A, Shiver J W. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc Natl Acad Sci USA. 1997;94:9378–9383. doi: 10.1073/pnas.94.17.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leutenegger C, Klein D, Hofmann-Lehmann R, Mislin C, Hummel U, Boni J, Boretti F, Guenzburg W H, Lutz H. Rapid FIV provirus quantitation by PCR using the TaqMan fluorigenic real time detection system. J Virol Methods. 1999;78:105–116. doi: 10.1016/s0166-0934(98)00166-9. [DOI] [PubMed] [Google Scholar]
- 22.Locher C P, Grant R M, Collisson E A, Reyes-Teran G, Elbeik T, Kahn J O, Levy J A. Antibody and cellular immune responses in breakthrough infection subjects after HIV type 1 glycoprotein 120 vaccination. AIDS Res Hum Retroviruses. 1999;15:1685–1689. doi: 10.1089/088922299309720. [DOI] [PubMed] [Google Scholar]
- 23.Lombardi S, Garzelli C, La Rosa C, Zaccaro L, Specter S, Malvaldi G, Tozzini F, Esposito F, Bendinelli M. Identification of a linear neutralization site within the third variable region of the feline immunodeficiency virus envelope. J Virol. 1993;67:4742–4749. doi: 10.1128/jvi.67.8.4742-4749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa T M, Furuya T, Itagaki S, Tohya Y, Takahashi E, Mikami T. Establishment of a feline T-lymphoblastoid cell line highly sensitive for replication of feline immunodeficiency virus. Arch Virol. 1989;108:131–135. doi: 10.1007/BF01313750. [DOI] [PubMed] [Google Scholar]
- 25.Olmsted R A, Hirsch V M, Purcell R H, Johnson P R. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne R, Rigby M, Siebelink K, Neil J C, Jarrett O. Virus neutralization reveals antigenic variation among feline immunodeficiency virus isolates. J Gen Virol. 1994;75:3641–3645. doi: 10.1099/0022-1317-75-12-3641. [DOI] [PubMed] [Google Scholar]
- 27.Pancino G, Chappey C, Saurin W, Sonigo P. B epitopes and selection pressures in feline immunodeficiency virus envelope glycoproteins. J Virol. 1993;67:664–672. doi: 10.1128/jvi.67.2.664-672.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polacino P, Stallard V, Klaniecki J E, Montefiori D C, Langlois A J, Richardson B A, Overbaugh J, Morton W R, Benveniste R E, Hu S-L. Limited breadth of the protective immunity elicited by simian immunodeficiency virus SIVmne gp160 vaccines in a combination immunization regimen. J Virol. 1999;73:618–630. doi: 10.1128/jvi.73.1.618-630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson H L, Montefiori D C, Johnson R P, Manson K H, Kalish M L, Lifson J D, Rizvi T A, Lu S, Hu S L, Mazzara G P, Panicali D L, Herndon J G, Glickman R, Candido M A, Lydy S L, Wyand M S, McClure H M. Neutralizing antibody-independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat Med. 1999;5:526–534. doi: 10.1038/8406. [DOI] [PubMed] [Google Scholar]
- 30.Sibille P, Avrameas A, Moraillon A, Richardson J, Sonigo P, Pancino G, Strosberg A D. Comparison of serological tests for the diagnosis of feline immunodeficiency virus infection of cats. Vet Microbiol. 1995;45:259–267. doi: 10.1016/0378-1135(94)00128-j. [DOI] [PubMed] [Google Scholar]
- 31.Sodora D L, Shpaer E G, Kitchell B E, Dow S W, Hoover E A, Mullins J I. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J Virol. 1994;68:2230–2238. doi: 10.1128/jvi.68.4.2230-2238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tellier M C, Pu R, Pollock D, Vitsky A, Tartaglia J, Paoletti E, Yamamoto J K. Efficacy evaluation of prime-boost protocol: canarypoxvirus-based feline immunodeficiency virus (FIV) vaccine and inactivated FIV-infected cell vaccine against heterologous FIV challenge in cats. AIDS. 1998;12:11–18. doi: 10.1097/00002030-199801000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Willett B J, Hosie M J, Neil J C, Jarrett O. Infection with feline immunodeficiency virus is followed by the rapid expansion of a CD8+ lymphocyte subset. Immunology. 1993;78:1–6. [PMC free article] [PubMed] [Google Scholar]
- 34.Wyand M S, Manson K, Montefiori D C, Lifson J D, Johnson R P, Desrosiers R C. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J Virol. 1999;73:8356–8363. doi: 10.1128/jvi.73.10.8356-8363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto J K, Ackley C D, Zochlinski H, Louie H, Pembroke E, Torten M, Hansen H, Munn R, Okuda T. Development of IL-2-independent feline lymphoid cell lines chronically infected with feline immunodeficiency viruses: importance for diagnostic reagents and vaccines. Intervirology. 1991;32:361–375. doi: 10.1159/000150220. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto J K, Okuda T, Ackley C D, Louie H, Pembroke E, Zochlinski H, Munn R J, Gardner M B. Experimental vaccine protection against feline immunodeficiency virus. AIDS Res Hum Retroviruses. 1991;7:911–921. doi: 10.1089/aid.1991.7.911. [DOI] [PubMed] [Google Scholar]